Abstract
Despite its important biological role, the evolution of recombination rates remains relatively poorly characterized. This owes, in part, to the lack of high-quality genomic resources to address this question across diverse species. Humans and our closest evolutionary relatives, anthropoid apes, have remained a major focus of large-scale sequencing efforts, and thus recombination rate variation has been comparatively well studied in this group—with earlier work revealing a conservation at the broad- but not the fine-scale. However, in order to better understand the nature of this variation, and the time scales on which substantial modifications occur, it is necessary to take a broader phylogenetic perspective. I here present the first fine-scale genetic map for vervet monkeys based on whole-genome population genetic data from ten individuals and perform a series of comparative analyses with the great apes. The results reveal a number of striking features. First, owing to strong positive correlations with diversity and weak negative correlations with divergence, analyses suggest a dominant role for purifying and background selection in shaping patterns of variation in this species. Second, results support a generally reduced broad-scale recombination rate compared with the great apes, as well as a narrower fraction of the genome in which the majority of recombination events are observed to occur. Taken together, this data set highlights the great necessity of future research to identify genomic features and quantify evolutionary processes that are driving these rate changes across primates.
Keywords: vervet monkey, African green monkey, genetic map, recombination
Introduction
Homologous meiotic recombination ensures proper pairing and segregation of chromosomes in most sexually reproducing organisms. In the absence of an alternative mechanism, as is the case in most Eutherian mammals (Zwick et al. 1999), errors in the recombination process can be highly deleterious, causing chromosomal nondisjunction, often leading to aneuploidy, as well as other chromosomal abnormalities, many of which underlie human disease and developmental disabilities (see review by Alves et al. [2017]). In addition to playing a critical role in the formation of viable gametes, recombination is of fundamental importance to the process of evolution. By breaking down linkage between loci and shuffling parental alleles into novel combinations, recombination facilitates the speed and efficacy of natural selection (Felsenstein and Yokoyama 1976; Otto and Barton 2001; Otto and Lenormand 2002). Recombination aids positive selection by mitigating interference between sites (Hill and Robertson 1966), either bringing advantageous alleles at different loci onto the same genetic background or decoupling advantageous alleles from deleterious ones (Fisher 1930; Muller 1932). By bringing deleterious mutations at different loci onto a common genetic background, purifying selection can purge them more efficiently from a population (Felsenstein 1974), thus slowing down “Muller’s ratchet” (Muller 1964). Moreover, by modulating the effects of both selective sweeps and background selection, recombination directly shapes the local genomic landscape and levels of genetic diversity observed within populations (Maynard Smith and Haigh 1974; Begun and Aquadro 1992; Charlesworth et al. 1993; Wiehe and Stephan 1993; Hudson and Kaplan 1995; Charlesworth 2012).
Given its crucial importance as a biological and evolutionary process, recombination rates might be expected to be similar across closely related species. Yet, tremendous heterogeneity in rates and patterns of recombination persists at every scale examined—from variation across the genome to differences between individuals, sexes, populations, and species (see review by Stapley et al. [2017]). Perhaps for anthropocentric reasons, a focal point of many primate studies has been the characterization of the causes and consequences of recombination rate variation in humans and their closest living evolutionary relatives, anthropoid apes, with comparative studies revealing a conservation at the broad- but not the fine-scale (Wall et al. 2003; Ptak et al. 2004, 2005; Winckler et al. 2005; Auton et al. 2012; Stevison et al. 2016), thus suggesting that diverse mechanisms may control genome-wide recombination rates at different scales among species. Although considerable efforts have been made to gain a better understanding of recombination rate evolution in great apes, less effort has been put toward deeper comparative genomic analyses by studying non-ape primates. Population genomic comparisons of the recombination landscape in monkeys with those of great apes offer a unique opportunity to provide much added clarity on how much variation in recombination rates exist in natural primate populations and over which time scales rates evolve. To date, coarse-scale genetic linkage maps exist for three biomedically relevant Old World monkey species—baboons (Papio hamadryas), rhesus macaques (Macaca mulatta), and vervet monkeys (Chlorocebus aethiops sabaeus)—constructed by mapping human orthologous genotype markers in large-scale pedigrees (Rogers et al. 2000, 2006; Cox et al. 2006; Jasinska et al. 2007). In contrast to the broad-scale correlation in recombination rate observed between the great apes, these maps demonstrate considerable rate variation over longer evolutionary time scales. Specifically, using information at homologous loci to extrapolate from the length ratio of the three Old World monkey maps to the human map, the authors reported total map lengths that are about 20–30% shorter than the human map, despite large overall synteny and similar karyotypes (Rogers et al. 2000, 2006; Cox et al. 2006; Jasinska et al. 2007), suggesting that humans might experience higher recombination rates than Old World monkeys. Due to the limited number of meioses that can practically be observed in pedigrees and the low density of markers, it had previously been hypothesized that a higher resolution in humans compared with the studied Old World monkeys could potentially have led to an overestimation of the observed differences (Rogers et al. 2000; Coop and Przeworski 2007)—though a recently published fine-scale recombination map for rhesus macaques confirmed the initial observation of a significantly lower broad-scale rate in rhesus compared with humans (Xue et al. 2016). Despite these initial insights, studies of fine-scale recombination rates in non-ape primates remain sparse and additional maps are required to aid the quantification of interspecies variation with regards to both broad- and fine-scale rates in order to shed light on the evolutionary forces and genetic determinants affecting recombination.
Vervet monkeys are of particular interest to the scientific community due to their importance as a model system for biomedical and biodevelopmental research—with genetic maps directly aiding the study of quantitative trait loci (QTL) that influence phenotypic variation with regards to human diseases or complex traits such as behavior (Cox et al. 2006). The currently available linkage map is based on 360 human orthologous short tandem repeats genotyped in >400 members from a characterized pedigree of a breeding colony, resulting in an average resolution of 9.8 cM (Jasinska et al. 2007). Although this map has been successfully utilized for biomedical research (see Jasinska et al. 2013 and references therein), its low resolution limits statistical power to identify QTL using genetic linkage analysis, which is sensitive to both sample size and marker density (Almasy and Blangero 1998). Moreover, the map retained several large gaps and no unique marker order could be determined for three of the autosomes, thus no genetic linkage map yet exists for chromosomes 18, 19, and 28 (Jasinska et al. 2007). Here, I have taken advantage of previously developed and validated statistical methodology (McVean et al. 2002, 2004; Auton and McVean 2007) to indirectly estimate a fine-scale recombination map for vervet monkeys from patterns of allelic association observed in whole-genome population genetic data, resulting in a map that contains orders of magnitude more markers than the coarse-scale map currently available for the species. This map provides a valuable resource for the scientific community, aiding a wide range of population genetic and evolutionary analysis, from fine-grained evolutionary comparative analysis investigating how quickly recombination rates evolve within the primate clade to genome-wide association studies related to human health and disease—ultimately improving our ability to study heritable genetic disorders in this widely used system.
Results and Discussion
Genome-Wide Polymorphism Data
Whole-genome sequencing data for ten unrelated captive vervet monkeys (C. aethiops sabaeus) (supplementary fig. 1, Supplementary Material online) with a genome-wide average coverage of 34× per individual (supplementary table 1, Supplementary Material online) allowed for the identification of genetic variants. As spurious variants can impact estimates of the population recombination rate from patterns of allelic association, variant calls were subjected to stringent filter criteria (see Materials and Methods for details). The final data set contains 10.7 million high-quality biallelic single-nucleotide polymorphisms (SNPs) distributed across 29 autosomes (supplementary table 2, Supplementary Material online) in the accessible parts of the genome (supplementary table 3, Supplementary Material online). The number of variants is highly consistent across samples and chromosomes (supplementary fig. 2, Supplementary Material online) and the transition–transversion ratio (Ts/Tv) of 2.37 (supplementary table 2, Supplementary Material online) compares well with the previously published Ts/Tv of 2.32 for the species (Huang et al. 2015). The SNP density of the ten individuals (5.1/kb) is higher than those previously reported for ten Western chimpanzees (1.9/kb) and nine humans from the 1000 Genomes Project (3.7/kb) (Auton et al. 2012) but lower than those observed in nine rhesus macaques (7.2 and 11.1/kb for Indian and Chinese rhesus macaques, respectively) (Xue et al. 2016) and baboons (19.1/kb) (Robinson et al. 2019).
To evaluate the power of this study to detect SNPs, the data set was compared with the previously generated Association Mapping Panel released by the Vervet Genetic Mapping Project (VGMP) (Huang et al. 2015). The Association Mapping Panel contains ∼500k SNPs genotyped in 721 Caribbean-origin vervet monkeys (including the ten individuals of this study) with a minor allele frequency of at least 25% (Huang et al. 2015). Based on comparisons to sites of the Association Mapping Panel (Huang et al. 2015), the estimated power to detect SNPs present at least once in the samples is >89% (supplementary table 2 and supplementary fig. 3, Supplementary Material online).
Fine-Scale Recombination Landscape
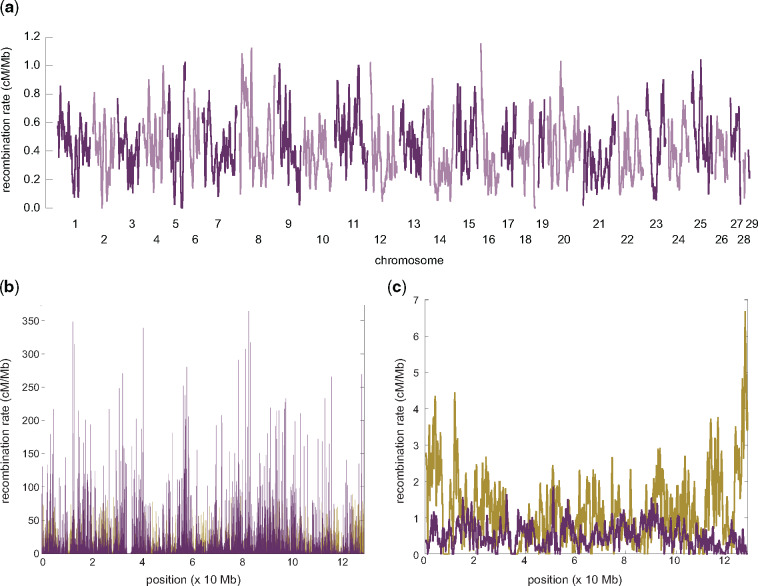
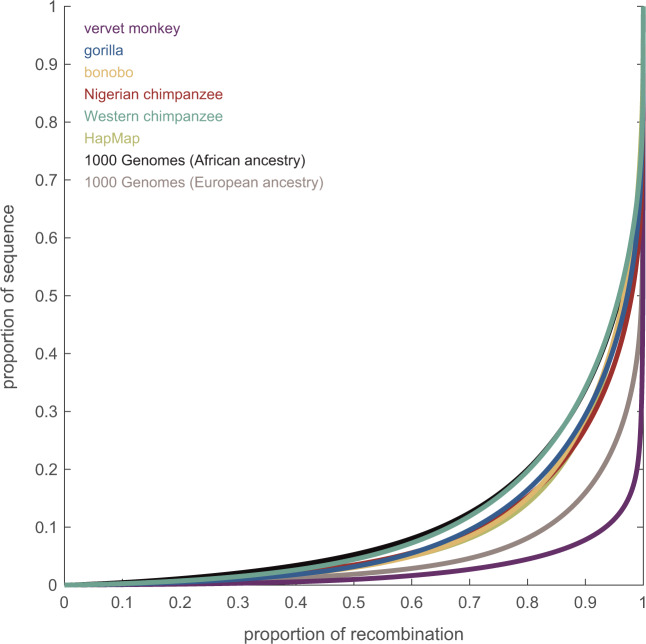
In order to compare recombination rates between vervets and humans, regions of broad-scale synteny were utilized to construct a fine-scale genetic map (fig. 1), following a similar approach than those previously employed for estimating fine-scale maps for the ten Western chimpanzees (Pan troglodytes verus) of the PanMap project (Auton et al. 2012), as well as the ten Nigerian chimpanzees (Pan troglodytes ellioti), 13 bonobos (Pan paniscus), and 15 Western gorillas (Gorilla gorilla gorilla) of the Great Ape Recombination Maps project (Stevison et al. 2016). Consistent with observations in great apes (Auton et al. 2012; Stevison et al. 2016) as well as other species (Jensen-Seaman et al. 2004), recombination rates in vervet are elevated in the subtelomeric regions of the chromosomes and lower near the centromeres (fig. 1a). The genome-wide average recombination rate is 0.434 ± 0.442 cM/Mb (100-kb windows), similar to rates reported in Indian rhesus macaques (0.433 ± 0.333 cM/Mb; Xue et al. 2016). Both Old World monkey species exhibit a notable overall reduction across chromosomes compared with humans (1.322 ± 1.399 cM/Mb; International HapMap Consortium 2007) as depicted in figure 1b and c (for illustration, vervet chromosome 11 and its primary homologous chromosome in human, chromosome 12, is shown) and anthropoid apes (∼1.193 cM/Mb for bonobos, Nigerian chimpanzee, and gorillas; see fig. 1 in Stevison et al. 2016). Relatedly, vervet recombination occurs in a much narrower fraction of the genome compared with the great apes studied to date (fig. 2). Specifically, ∼80% of recombination is concentrated in only 4% of the vervet genome compared with 20% in human populations of African ancestry (1000 Genomes Project Consortium 2010) as well as Western chimpanzees (Auton et al. 2012), 15% in Nigerian chimpanzees, bonobos, as well as gorillas (Stevison et al. 2016), and 8% in human populations of European ancestry (1000 Genomes Project Consortium 2010)—suggesting that recombination is more concentrated in hotspots in vervets than in humans and anthropoid apes. Thus, although differences on the fine-scale have been well documented even between closely related primate species (Wall et al. 2003; Ptak et al. 2004, 2005; Winckler et al. 2005; Auton et al. 2012; Stevison et al. 2016), this study adds to the growing literature documenting changes in both broad- and fine-scale recombination rate between great apes and Old World monkeys (Rogers et al. 2000, 2006; Cox et al. 2006; Jasinska et al. 2007; Xue et al. 2016; Robinson et al. 2019). The observed substantial differences raise the question of whether there are universal genomic features that shape the recombination landscapes among primates.
Fig. 1.
Genome-wide recombination rates in vervet monkeys. (a) Broad-scale recombination rates (averaged over 1-Mb regions). Autosomes are represented in alternating colors. (b) Fine-scale recombination rates for vervet (purple) along chromosome 11 and, for comparison, its primary homologous chromosome in human (i.e., chromosome 12; green). Human rates were taken from the population-averaged genetic map generated by the HapMap Project (International HapMap Consortium 2007). (c) As for (b) but on the broad-scale (averaged over 1-Mb regions).
Fig. 2.
Genome-wide distribution of fine-scale recombination rates. The concentration of recombination in genetic maps estimated from vervet monkeys (this study), humans (from the HapMap Project [International HapMap Consortium 2007] as well as from populations of European and African ancestry from the 1000 Genomes Project [1000 Genomes Project Consortium 2010]), Western chimpanzees (Auton et al. 2012), Nigerian chimpanzees, bonobos, and gorillas (Stevison et al. 2016). The fine-scale genetic map for rhesus macaques (Xue et al. 2016) was not made publicly available upon publication, unfortunately limiting the ability for comparative studies such as the work presented here.
Genomic Factors Influencing Recombination Rate
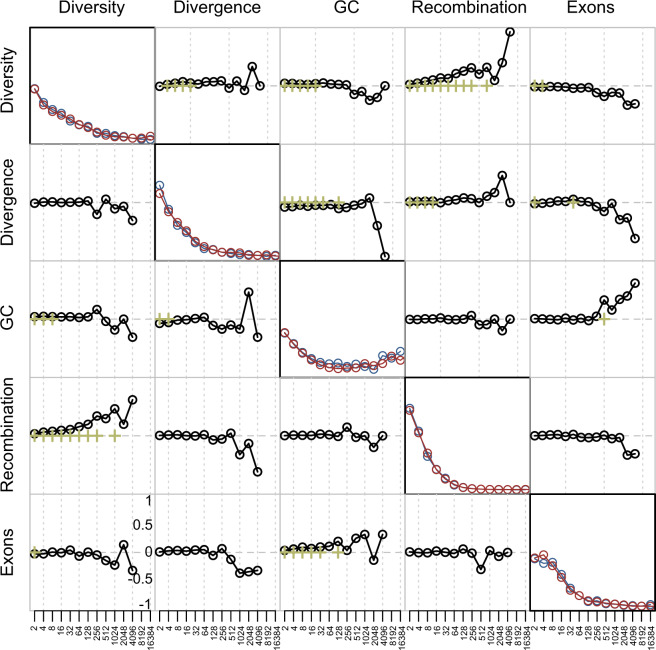
The influence of particular genomic features on rates and patterns of recombination may well be different between the fine- and the broad-scales (Stevison and Noor 2009). To characterize scale-specific relationships between recombination rate and potential correlates, summary statistics of genomic features, namely genetic diversity (average pairwise differences), divergence (based on the alignment of the vervet reads to the human reference genome assembly), recombination, GC-content, and evolutionary constraint (utilizing exon content as a surrogate), were calculated in 1-kb regions along the genome and a discrete (Haar) wavelet transformation applied to provide information on heterogeneity in each signal at successively broader scales. Specifically, a wavelet transform decomposes a series of observed features into a series of detail coefficients that quantify variation between neighboring observations captured at a range of (2n) scales as well as smooth coefficients that approximate the original signal by smoothing over these scales (Spencer et al. 2006). Figure 3 and supplementary figure 4, Supplementary Material online, summarize the wavelet transformations for each annotated genomic feature, including the pairwise correlations (calculated using Kendall’s rank correlation) between the detail coefficients at each scale. Once the data have been transformed, a linear model analysis can be carried out to investigate whether changes in these genomic features can predict changes in recombination rates at varying spatial scales. In contrast to a linear model analysis of the smoothed approximation which essentially characterizes relationships between correlates by averaging over successively broader scales (fig. 4a and supplementary fig. 5(top), Supplementary Material online), a linear model analysis of the detail coefficients characterizes the relationships between correlates at a specific scale (i.e., it provides information about how a change in one particular genomic feature, e.g., recombination rate, predicts a change in other genomic features, e.g., diversity and divergence, at the same scale; Spencer et al. 2006) (fig. 4b and supplementary fig. 5(bottom), Supplementary Material online). Thus, although the former generally has more power, the latter allows for the identification of (potentially different) scale-specific correlations between genomic features while, at the same time, controlling for autocorrelations and varying background rates (Spencer et al. 2006).
Fig. 3.
Power spectra and pairwise correlations of detail coefficients. The power spectrum of the wavelet decomposition (i.e., a summary of the total variance in the signal due to heterogeneity at distinct scales) of each genomic feature on the short (blue) and long (red) arms of chromosome 8 is depicted along the diagonal. Pairwise correlations between detail coefficients (calculated using Kendall's rank correlation) are depicted in the off-diagonal plots (bottom left: short arm; top right: long arm; green crosses highlight correlations significant at the 1% level in a two-tailed test). Signals are provided at increasingly broader kilobase scales.
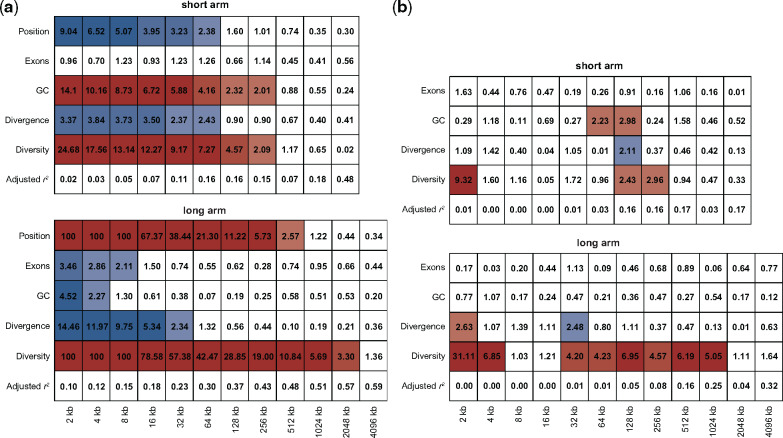
Fig. 4.
Genomic features associated with recombination rate. Marginal significance for the linear model analysis of the (a) smoothed and (b) detail coefficients using diversity, divergence, GC-content, and exon content as predictors of recombination. Colors indicate significant linear relationships (red: positive; blue: negative; with color intensity being proportional to the level of significance using a two-sided t-test to determine −log10P-values). Adjusted r2 indicate the proportion of variance in the signal that can be explained by the model.
The power spectrum of the wavelet decomposition (i.e., a summary of the total variance in the signal due to heterogeneity at distinct scales) of each genomic feature in the longest chromosome (i.e., chromosome 8) is depicted along the diagonal in figure 3 (other autosomes are depicted in supplementary fig. 4, Supplementary Material online). Mimicking previous observations in humans (Spencer et al. 2006), diversity and divergence exhibit the highest heterogeneity at the smallest scale (2 kb), whereas exon content shows the greatest heterogeneity at the 2–8-kb scale. GC-content follows a bimodal distribution, with the highest heterogeneity observed at both the fine (2–8 kb) as well as broad (8–16 Mb) scales—with the latter likely reflecting isochore structure. However, contrasting previous results in humans where the largest heterogeneity for recombination was observed at the intermediate scale (8 kb) (Spencer et al. 2006), the largest contribution to heterogeneity for recombination in vervets is at the finest scale (2 kb).
The pairwise correlations between detail coefficients are depicted on the off-diagonal plots in figure 3 and supplementary figure 4, Supplementary Material online. As expected from mutation rate variation across the genome (see Walsh and Lynch 2019 for a detailed discussion), nucleotide diversity is significantly positively correlated with divergence at fine to intermediate scales (4–32 kb). At the same scales, a significant positive correlation with base composition can be observed. A significant negative correlation between nucleotide diversity and exon content is present at the finest scales (2–4 kb), likely due to the pervasiveness of purifying selection acting on the genome. As expected from previous observations in great apes (Spencer et al. 2006; Auton et al. 2012; Pfeifer and Jensen 2016; Stevison et al. 2016), diversity shows a significant positive correlation with recombination rate at the fine-scale (2–4 kb). However, in vervets, this strong positive correlation extends to much broader scales (up to 2 Mb), an association often attributed to genetic hitchhiking effects associated with linkage to deleterious (i.e., background selection) and/or beneficial mutations (i.e., genetic hitchhiking) (Maynard Smith and Haigh 1974; Begun and Aquadro 1992; Charlesworth et al. 1993).
In order to predict changes in recombination, linear modeling of both the smoothed and detailed coefficients was performed. Linear model analysis of the smoothed coefficients (fig. 4a and supplementary fig. 5(top), Supplementary Material online) highlights the recombination-suppressing effect of the centromere, in concordance with previous observations in great apes (Spencer et al. 2006; Auton et al. 2012; Pfeifer and Jensen 2016; Stevison et al. 2016). Diversity is significantly positively correlated at scales up to 2 Mb, whereas divergence is negatively correlated at the fine and intermediate scales (2–64 kb), likely due to genetic hitchhiking effects. The effects of exon content differ between the short and long arms of chromosome 8. The long arm shows a weak negative correlation on the small (2–8 kb) scale, likely due to a preference for crossovers to occur outside of genes in primates (e.g., International HapMap Consortium 2007). The short arm shows no significant association, potentially due to lack of power (exon density is ∼40% lower compared with the long arm). Similar differences are observed between several other chromosome arms (supplementary fig. 5(top), Supplementary Material online). Biased gene conversion is thought to cause the correlation between recombination and GC-content observed in many organisms (Pessia et al. 2012), and indeed such a correlation is also observed in vervets. Confirming the results obtained from the smoothed coefficients, linear model analysis of the detailed coefficients (fig. 4b and supplementary fig. 5(bottom), Supplementary Material online) reveals a strong positive, but highly localized (2–4 kb), effect of recombination on diversity (i.e., on the scale of recombination hotspots), as well as a much weaker effect on the intermediate and broad scales (32 kb up to 2 Mb). Apart from diversity, other genomic features—including divergence—only show weak correlations on the small to intermediate scales (though it is important to note that the analysis of detailed coefficients has reduced power at the broad-scales owing to fewer observations). One potential reason for the observation of a strong negative correlation of recombination and divergence at the fine-scale in the linear model analysis of the smoothed, but not the detailed, coefficients might be the ephemeral nature of recombination hotspots, resulting in differences in the fine-scale recombination landscape between primates which may obscure associations with divergence (see discussion in Spencer et al. 2006).
Conclusions
Large-scale genome sequencing projects provide an opportunity to study fundamental evolutionary processes at a much broader phylogenetic scale. This is of particular importance for studying the evolution of recombination rates, as initial studies among the great apes have suggested substantial fine-scale variation, both with regards to the rates themselves but also with respect to the brief evolutionary periods necessary to observe such variation. I have here extended comparative analyses to vervet monkeys, an Old World monkey species which shares a most recent common ancestor with humans around 25 Ma (Kumar and Hedges 1998). By constructing the first fine-scale recombination rate map for the species, valuable insights in to recombination rate variation and evolution across this deeper primate time scale have emerged. Results demonstrate an overall reduction in recombination rates in vervets relative to the great apes, as well as a considerably reduced proportion of the genome in which recombination events have been observed to occur. The underlying reasons for this intensity of recombination in such a limited proportion of the genome—be they related to PRDM9 diversity, population history, or other—is a question in need of further investigation.
Additionally, the primary correlate of recombination rate is nucleotide diversity—a widely observed pattern (see Cutter and Payseur 2013) that has long been associated with both positive and negative selection effects on linked sites, as well as to the potential mutagenic effects of recombination itself (Halldorsson et al. 2019). However, contrasting with both the positive selection- and mutagenic-based explanations is the observation of a weak negative correlation between recombination rate and divergence—a pattern that is also found to be related to exon content. This, combined with the general expectation of a far greater input of deleterious compared with beneficial mutations in the genome (see Comeron 2014, 2017; Jensen et al. 2019) would suggest a dominant role for background selection in shaping variation across the vervet genome. Specifically, as lower recombination rate regions experience a decreased efficacy of natural selection as well as suppressed local effective population sizes compared with higher recombination rate regions, weakly deleterious mutations may periodically be fixed by genetic drift (Charlesworth et al. 1993; Charlesworth 2012; Campos and Charlesworth 2019).
In summary, this study provides the first fine-scale recombination map in vervets, a resource that is expected to greatly aid future QTL studies in this important and highly utilized biomedical species. Furthermore, results support earlier suggestions of reduced recombination rates outside the great apes—an emerging pattern which will necessitate a detailed study of underlying genomic features and evolutionary processes which may be driving these major, broad-scale differences between species.
Materials and Methods
Data
Publicly available whole-genome high-throughput sequencing data for 15 captive vervet monkeys (C. aethiops sabaeus) (four females and 11 males with a genome-wide average coverage of 32.6× per individual), housed at the Wake Forest University Primate Center Vervet Research Colony, was downloaded from SRA (i.e., females: SRS578075, SRS578771; SRS578938, SRS578090; males: SRS578089, SRS578723, SRS578082, SRS579180, SRS579162, SRS579182, SRS579165, SRS579166, SRS579161, SRS579164, and SRS579163) (supplementary table 1, Supplementary Material online).
Read Mapping
Following Pfeifer (2017a, 2017c), reads from each read group were aligned to the repeat-masked C. sabaeus reference assembly v.1.1 (chlSab2) (Warren et al. 2015), as downloaded from the NCBI GenBank website (accession number GCA_000409795.2), using BWA-MEM v.0.7.13 with default parameters (Li and Durbin 2009). To remove potential contamination, the Epstein-Barr virus genome (NCBI Reference Sequence NC_007605.1) was included in the reference assembly as a decoy. After mapping, duplicates were marked using Picard v.2.1.1 (http://broadinstitute.github.io/picard, last accessed April 7, 2020). Multiple sequence realignments were performed around indels using the Genome Analysis Toolkit (GATK) v.3.5 RealignerTargetCreator and IndelRealigner (McKenna et al. 2010; DePristo et al. 2011; Van der Auwera et al. 2013) and Base Alignment Qualities (Li 2011) were adjusted to downweight base qualities in regions that showed high ambiguity in the local alignment following Auton et al. (2012). Subsequently, base quality scores were recalibrated using GATK’s BaseRecalibrator, together with a training set of ∼500k known variants from the genome-wide SNP panel of the VGMP (Huang et al. 2015), downloaded from the European Variant Archive (study number PRJEB7923), to define characteristics underlying high-quality calls. After preprocessing each read group individually, reads originating from the same sample were merged and per-sample duplicates marked using Picard in order to eliminate polymerase chain reaction duplicates introduced during library construction.
Variant Calling, Genotyping, and Filtering
For each individual, variant calls were made using GATK’s v.3.5 HaplotypeCaller with default parameters (McKenna et al. 2010; DePristo et al. 2011; Van der Auwera et al. 2013). After the initial calls, variants were jointly genotyped using GATK’s GenotypeGVCFs.
As in many other primates, the assembly of the vervet sex chromosomes is of a lower quality than that of the autosomes. As a consequence, the data set was limited to autosomal, biallelic SNPs and subsequently filtered to decrease the number of artifactual variant calls. In the absence of an experimentally validated high-quality variant call set for the species, the initial data set was filtered following GATK’s Best Practice recommendations. Specifically, GATK’s VariantFiltration was run with the following default hard filter criteria (with acronyms as defined by the GATK package):
The nonreference allele was fixed in the studied population (AF = 1).
The root mean square mapping quality of all reads was <40 (MQ < 40).
There was a qualitative difference between the mapping qualities of the reads supporting the reference allele compared with those supporting the nonreference allele (MQRankSum < −12.5).
There was a bias in the position of alleles within the reads that support the reference and nonreference alleles (ReadPosRankSum < −8.0).
The variant quality score divided by the sum of depth across all samples with nonreference genotypes was <2.0 (QD < 2.0).
There was evidence of strand bias (using either Fisher’s exact test FS > 60.0 or the Symmetric Odds Ratio SOR > 4.0).
Extreme depth of read coverage is a frequent sign of false positive variant calls in regions, where read alignment is poor (such as regions of unresolved collapsed copy number variants or repeats; see review by Pfeifer [2017b]). Therefore, the total depth of coverage at a position was limited to remove the 2.5% and 97.5% percentiles of the distribution (i.e., removing sites with read depth DP < 190 or DP > 579).
After initial filtering, family relationships between individuals were inferred using the software KING (Manichaikul et al. 2010). Five individuals (i.e., SRS578075, SRS579162, SRS579163, SRS579164, and SRS579180) were excluded from further analyses as they were closely related to other individuals in the data set (supplementary fig. 1, Supplementary Material online). The remaining ten individuals (three females and seven males) exhibited a genome-wide average coverage of 34× per individual.
As an emerging model organism, there is still a shortage of large-scale high-quality genomic data for vervet monkeys that can be utilized for reliable genotype imputation (e.g., the majority of samples included in one of the largest genomic studies of the species to date [Huang et al. 2015] were sequenced to merely >4× coverage [407 individuals] and >1× coverage [302 individuals]). As the majority of variants discovered in this study (∼98%) contained genotype information for all individuals, sites with missing genotypes were excluded from further analysis to ensure the stringency of the data set by avoiding potential biases resulting from computational imputation of genotypes.
As spurious variants might lead to disruptions of linkage disequilibrium patterns, additional more stringent filtering criteria were applied to the data set:
False positive SNPs often arise in close proximity to other SNPs, thus if more than two SNPs fell within a 10-bp window, they were removed using GATK's VariantFiltration (clusterSize = 3; clusterWindowSize = 10).
SNPs showing an excess of heterozygosity were removed. Specifically, a P-value for Hardy–Weinberg Equilibrium was calculated using the “—hardy” option in VCFtools v.0.1.13 (Danecek et al. 2011), and SNPs with P < 0.01 removed.
Although the vervet reference assembly shows the second highest degree of sequence continuity among primates (Huang et al. 2015, Warren et al. 2015), the human reference assembly remains the highest quality primate genome assembly to date. Thus, SNPs were reciprocally lifted over between the original C. sabaeus (chlSab2) genome and the human (hg38) genome using the UCSC “liftOver” tool (Casper et al. 2018). Only sites that mapped back to their original position were retained for further analyses.
Using the hg38 annotations, the data set was limited to SNPs that fell within regions of the human genome that were uniquely mappable by a k36-mer (as defined by the UCSC Genome Browser [Casper et al. 2018] Hoffman Lab “unique mappability” track).
In addition, SNPs within regions blacklisted by ENCODE (ENCODE Project Consortium 2012), which often exhibit anomalous, high read counts in next generation sequencing experiments, were excluded.
The resulting SNP data set contained 10,795,556 biallelic autosomal SNPs, with a Ts/Tv of 2.37 (supplementary table 2, Supplementary Material online), in the accessible part of the genome (supplementary table 3, Supplementary Material online).
SNP Discovery Power
To evaluate the power to detect SNPs, the data set was compared with previously generated genotype data that contained the ten individuals from this study. Specifically, the data set was compared with 497,163 sites of the Association Mapping Panel released by the VGMP, genotyped in 721 Caribbean-origin vervet monkeys (minor allele frequency >= 25%; r2 = 0.9) (Huang et al. 2015). Overall, 95.9% of SNPs were novel compared with the SNPs of the Association Mapping Panel that were polymorphic in the ten individuals from this study, whereas 89.2% of SNPs were rediscovered (supplementary table 2, Supplementary Material online). Taken together, this study has 89.4% power to detect SNPs present at least once in the samples (supplementary figure 3, Supplementary Material online).
Divergence
In order to obtain a data set of sites divergent between vervets and humans, reads were additionally aligned to the repeat-masked human (hg38) reference assembly (downloaded from the UCSC Genome Browser [Casper et al. 2018]) and variants called, genotyped, and filtered using a similar methodology than descripted above (see Read Mapping section and Variant Calling, Genotyping, and Filtering section), with two exceptions. First, as the reads originated from vervets (rather than humans), no base quality score recalibration was performed. Second, as GATK’s HaplotypeCaller discovers variants by performing a local de novo haplotype assembly, multiple sequence realignments are now obsolete.
In total, 69,920,804 fixed differences between the ten vervet individuals of this study and the hg38 reference assembly were discovered. One hundred and six of these were polymorphic in the 721 vervets of the VGMP (Huang et al. 2015) and thus excluded from further analysis. In addition, 4,896,664 sites were excluded as they were polymorphic in the 1000 Genomes Phase 3 data set containing 84.4 million variants detected in 2,504 human individuals from 26 populations (1000 Genomes Project Consortium 2015), resulting in a total of 65,024,034 divergent sites.
Synteny
Vervet and human genomes display large-scale synteny (Finelli et al. 1999; Jasinska et al. 2007). In order to compare recombination rates, regions of broad-scale synteny between the two primates were identified following the guidelines outlined in the PanMap (Auton et al. 2012) and Great Ape Recombination Maps (Stevison et al. 2016) projects. Specifically, syntenic regions were defined as continuous parts of a chromosome whereby consecutive pairs of sites exhibit the same orientation (i.e., increasing or decreasing coordinates relative to the reference assembly) and are separated by no more than 50-kb sequence on both the vervet and its primary homologous human chromosome in order to avoid large gaps. Following Stevison et al. (2016), sites between syntenic regions were removed and consecutive regions with matching orientations were merged into syntenic blocks if 1) they were separated by fewer than 300 intervening sites and 2) the distance between adjacent sites remained smaller than 50 kb.
The resulting data set contained 457 synteny blocks between the vervet and human genomes, comprising a total of 9,632,875 SNPs (corresponding to 89.4% of the SNP data set) and ranging from 25.8 kb to 73.4 Mb in size, with an average size of 5.0 Mb (supplementary fig. 6, Supplementary Material online; and see supplementary fig. 7, Supplementary Material online, for the size distribution of the syntenic blocks). SNPs within these syntenic blocks were subsequently used to estimate recombination rates.
Phasing
Haplotypes were reconstructed from the population genomic data using the software PHASE, a Bayesian statistical method well suited for small sample sizes (Stephens et al. 2001; Stephens and Donnelly 2003; Stephens and Scheet 2005). Following Auton et al. (2012), syntenic blocks were split into regions containing 401 SNPs with a 100 SNP overlap between adjacent regions. For each region, PHASE v.2.1 was run for 200 iterations with a 300-iteration burn-in period using the following options “-MR -F.05 -l10 -x5 -X5.” Phased haplotypes were joined back together by selecting the phase with the minimum Hamming distance between haplotypes across the 100-SNP overlapping region. Phasing fixed 168,471 sites which were subsequently removed from the data set.
Generation of a Fine-Scale Genetic Map
Following the PanMap (Auton et al. 2012) and Great Ape Recombination Maps (Stevison et al. 2016) projects, LDhat v.2.2 (McVean et al. 2002, 2004; Auton and McVean 2007) was used to estimate the population recombination rate ρ. Specifically, the haplotype data were divided into regions containing 4,000 SNPs, with a 200-SNP overlap between regions. For each region, LDhat “interval” was run for 60 million iterations (-its 60000000) with a block penalty of 5 (-bpen 5), and samples were taken every 40,000 iterations (-samp 40000). LDhat “stat” was used to discard the initial 20 million iterations (-burn 500) of the Monte Carlo Markov Chain as burn-in. Following Auton et al. (2012), recombination rate estimates between adjacent SNPs were obtained by taking the mean across samples, and region-based estimates were combined at the midpoint of the overlap.
To circumvent incorrectly inferred or genotyped variants erroneously interrupting true patterns of linkage disequilibrium, a regional filtering strategy developed and validated by Auton et al. (2012) was applied to remove problematic regions that might lead to localized breakdowns of linkage disequilibrium. Specifically, recombination rate estimates in a region were filtered out if 1) the population recombination rate ρ between two adjacent SNPs was >100 or 2) there was a gap larger than 50 kb in the reference assembly (as determined by the chlSab2 “gap” track obtained from the UCSC TableBrowser [Karolchik et al. 2004]). The recombination rate of these filtered regions as well as their surrounding 100 SNPs (i.e., 50 SNPs upstream and downstream) was set to 0.
One hundred and twenty-two distinct regions with ρ > 100 were identified, leading to the exclusion of 12,928 SNPs. In addition, 19 gaps larger than 50 kb were annotated in the chlSab2 reference assembly, leading to the exclusion of a further 1,936 SNPs. Overall, the recombination rate of 14,664 SNPs was set to 0 (200 SNPs failed both filter criteria). The genetic map before and after filtering is depicted in supplementary figure 8, Supplementary Material online.
Estimation of the Effective Population Size
LDhat (McVean et al. 2002, 2004; Auton and McVean 2007) estimates the population recombination rate ρ = 4Ner, where Ne is the effective population size and r is the recombination rate per site per generation, making it thus necessary to estimate the effective population size in order to obtain per-generation estimates. Utilizing nucleotide diversity, π, measured in intergenic regions of the genome (i.e., excluding the 28,078 genes annotated by Ensembl [Zerbino et al. 2018]) and assuming a mutation rate μ of 0.94 × 10−8 per base pair per generation (Pfeifer 2017a), the effective population size was estimated to be 17,085—similar to previously obtained estimates for the species (Huang et al. 2015).
Genomic Factors Influencing Recombination Rate
A wavelet analysis was utilized to identify the correlation of recombination rate with several genomic features—namely diversity, divergence, GC-content, and evolutionary constraint utilizing exon content as a surrogate. Genomic features were calculated in 1-kb regions along a chromosome, excluding centromeric regions. Thereby, population recombination rates were calculated as the slope of a regression of genetic distances of markers. Diversity and divergence were estimated from the vervet and human alignments, respectively, taking into account the number of sites accessible to the variant discovery in this study. GC-content was obtained from the annotated C. sabaeus reference assembly v.1.1 (chlSab2) (Warren et al. 2015), as downloaded from the NCBI GenBank website (accession number GCA_000409795.2). Exon locations were taken from Ensembl build 98 (Zerbino et al. 2018). Following Spencer et al. (2006), a discrete (Haar) wavelet transformation was applied using the R packages “Rwave” and “wavethresh” to provide information on heterogeneity in each signal at different scales. Importantly, as the discrete wavelet transform is only defined for regularly sampled subsets, it cannot be applied to series containing gaps. Unfortunately, in genomic data, gaps remain an inevitable issue, even at high-coverage (e.g., several large gaps of >50 kb are present in the current chlSab2 reference assembly). As a consequence, each chromosome was split into the largest possible regularly sampled subsets in powers of 2.
Following Spencer et al. (2006), a linear model analysis was carried out with the intercept forced through the origin to identify scale-specific correlations between features. Prior to the linear model analysis, recombination, diversity, and divergence were log-transformed to account for the nonnormality of the residuals.
Results for chromosome 8 (i.e., the longest chromosome offering the most power) are presented using the largest possible subsets of the short (16 Mb) and long (65 Mb) arms (figs. 3 and 4); the statistics for the largest subsets of all other autosomes are depicted in supplementary figures 4 and 5, Supplementary Material online.
Data Availability
The fine-scale genetic map for vervet monkeys generated in this study is available at http://spfeiferlab.org/data.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
I am grateful to Adam Auton for sharing his code from previous publications and Jeffrey Jensen for helpful comments and discussion. I would also like to thank two anonymous reviewers whose suggestions helped to improve an earlier version of the manuscript. Computations were partially performed at Arizona State University’s High Performance Computing facility.
This study was based on whole-genome high-throughput sequencing data that are publicly available at NCBI’s SRA (accessions: SRS578075, SRS578771; SRS578938, SRS578090; SRS578089, SRS578723, SRS578082, SRS579180, SRS579162, SRS579182, SRS579165, SRS579166, SRS579161, SRS579164, and SRS579163).
References
- 1000 Genomes Project Consortium. 2010. A map of human genome variation from population-scale sequencing. Nature 467(7319):1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium. 2015. A global reference for human genetic variation. Nature 526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L, Blangero J.. 1998. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 62(5):1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves I, Houle AA, Hussin JG, Awadalla P.. 2017. The impact of recombination on human mutation load and disease. Philos Trans R Soc B 372(1736):20160465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, Fledel-Alon A, Pfeifer S, Venn O, Ségurel L, Street T, Leffler E, Bowden R, Aneas I, Broxholme J, et al. 2012. A fine-scale chimpanzee genetic map from population sequencing. Science 336(6078):193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, McVean G.. 2007. Recombination rate estimation in the presence of hotspots. Genome Res. 17(8):1219–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, Aquadro CF.. 1992. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature 356(6369):519–520. [DOI] [PubMed] [Google Scholar]
- Campos JL, Charlesworth B.. 2019. The effects on neutral variability of recurrent selective sweeps and background selection. Genetics 212(1):287–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper J, Zweig AS, Villarreal C, Tyner C, Speir ML, Rosenbloom KR, Raney BJ, Lee CM, Lee BT, Karolchik D, et al. 2018. The UCSC Genome Browser database: 2018 update. Nucleic Acids Res. 46(D1):D762–D769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. 2012. The effects of deleterious mutations on evolution at linked sites. Genetics 190(1):5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Morgan MT, Charlesworth D.. 1993. The effect of deleterious mutations on neutral molecular variation. Genetics 134(4):1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron JM. 2014. Background selection as baseline for nucleotide variation across the Drosophila genome. PLoS Genet. 10(6):e1004434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron JM. 2017. Background selection as null hypothesis in population genomics: insights and challenges from Drosophila studies. Philos Trans R Soc B 372(1736):20160471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G, Przeworski M.. 2007. An evolutionary view of human recombination. Nat Rev Genet. 8(1):23–34. [DOI] [PubMed] [Google Scholar]
- Cox LA, Mahaney MC, Vandeberg JL, Rogers J.. 2006. A second-generation genetic linkage map of the baboon (Papio hamadryas) genome. Genomics. 88(3):274–281. [DOI] [PubMed] [Google Scholar]
- Cutter AD, Payseur BA.. 2013. Genomic signatures of selection at linked sites: unifying the disparity among species. Nat Rev Genet. 14(4):262–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 27(15):2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489(7414):57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 1974. The evolutionary advantage of recombination. Genetics 78(2):737–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J, Yokoyama S.. 1976. The evolutionary advantage of recombination. II. Individual selection for recombination. Genetics 83(4):845–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finelli P, Stanyon R, Plesker R, Ferguson-Smith MA, O’Brien PC, Wienberg J.. 1999. Reciprocal chromosome painting shows that the great difference in diploid number between human and African green monkey is mostly due to non-Robertsonian fissions. Mamm Genome 10(7):713–718. [DOI] [PubMed] [Google Scholar]
- Fisher RA. 1930. The genetical theory of natural selection. Oxford: The Clarendon Press. [Google Scholar]
- Halldorsson BV, Palsson G, Stefansson OA, Jonsson H, Hardarson MT, Eggertsson HP, Gunnarsson B, Oddsson A, Halldorsson GH, Zink F, et al. 2019. Characterizing mutagenic effects of recombination through a sequence-level genetic map. Science 363(6425):eaau1043. [DOI] [PubMed] [Google Scholar]
- Hill WG, Robertson A.. 1966. The effect of linkage on limits to artificial selection. Genet Res. 8(3):269–294. [PubMed] [Google Scholar]
- Huang YS, Ramensky V, Service SK, Jasinska AJ, Jung Y, Choi OW, Cantor RM, Juretic N, Wasserscheid J, Kaplan JR, et al. 2015. Sequencing strategies and characterization of 721 vervet monkey genomes for future genetic analyses of medically relevant traits. BMC Biol. 13(1):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kaplan NL.. 1995. Deleterious background selection with recombination. Genetics 141(4):1605–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium. 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449(7164):851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinska AJ, Schmitt CA, Service SK, Cantor RM, Dewar K, Jentsch JD, Kaplan JR, Turner TR, Warren WC, Weinstock GM, et al. 2013. Systems biology of the vervet monkey. ILAR J. 54(2):122–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinska AJ, Service S, Levinson M, Slaten E, Lee O, Sobel E, Fairbanks LA, Bailey JN, Jorgensen MJ, Breidenthal SE, et al. 2007. A genetic linkage map of the vervet monkey (Chlorocebus aethiops sabaeus). Mamm Genome 18(5):347–360. [DOI] [PubMed] [Google Scholar]
- Jensen JD, Payseur BA, Stephan W, Aquadro CF, Lynch M, Charlesworth D, Charlesworth B.. 2019. The importance of the Neutral Theory in 1968 and 50 years on: a response to Kern and Hahn 2018. Evolution 73(1):111–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen-Seaman MI, Furey TS, Payseur BA, Lu Y, Roskin KM, Chen CF, Thomas MA, Haussler D, Jacob HJ.. 2004. Comparative recombination rates in the rat, mouse, and human genomes. Genome Res. 14(4):528–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ.. 2004. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32(Database issue):D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Hedges SB.. 1998. A molecular timescale for vertebrate evolution. Nature 392(6679):917–920. [DOI] [PubMed] [Google Scholar]
- Li H. 2011. Improving SNP discovery by base alignment quality. Bioinformatics 27(8):1157–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM.. 2010. Robust relationship inference in genome-wide association studies. Bioinformatics 26(22):2867–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J, Haigh J.. 1974. The hitch-hiking effect of a favourable gene. Genet Res. 23(1):23–35. [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next generation DNA sequencing data. Genome Res. 20(9):1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G, Awadalla P, Fearnhead P.. 2002. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics 160(3):1231–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean GA, Myers SR, Hunt S, Deloukas P, Bentley DR, Donnelly P.. 2004. The fine-scale structure of recombination rate variation in the human genome. Science 304(5670):581–584. [DOI] [PubMed] [Google Scholar]
- Muller HJ. 1932. Some genetic aspects of sex. Am Nat. 66(703):118–138. [Google Scholar]
- Muller HJ. 1964. The relation of recombination to mutational advance. Mutat Res. 1(1):2–9. [DOI] [PubMed] [Google Scholar]
- Otto SP, Barton NH.. 2001. Selection for recombination in small populations. Evolution 55(10):1921–1931. [DOI] [PubMed] [Google Scholar]
- Otto SP, Lenormand T.. 2002. Resolving the paradox of sex and recombination. Nat Rev Genet. 3(4):252–261. [DOI] [PubMed] [Google Scholar]
- Pessia E, Popa A, Mousset S, Rezvoy C, Duret L, Marais GA.. 2012. Evidence for widespread GC-biased gene conversion in eukaryotes. Genome Biol. Evol. 4(7):675–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer SP. 2017. a. Direct estimate of the spontaneous germ line mutation rate in African green monkeys. Evolution 71(12):2858–2870. [DOI] [PubMed] [Google Scholar]
- Pfeifer SP. 2017. b. From next-generation resequencing reads to a high-quality variant data set. Heredity 118(2):111–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer SP. 2017. c. The demographic and adaptive history of the African green monkey. Mol Biol Evol. 34(5):1055–1065. [DOI] [PubMed] [Google Scholar]
- Pfeifer SP, Jensen JD.. 2016. The impact of linked selection in chimpanzees: a comparative study. Genome Biol Evol. 8(10):3202–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptak SE, Hinds DA, Koehler K, Nickel B, Patil N, Ballinger DG, Przeworski M, Frazer KA, Pääbo S.. 2005. Fine-scale recombination patterns differ between chimpanzees and humans. Nat Genet. 37(4):429–434. [DOI] [PubMed] [Google Scholar]
- Ptak SE, Roeder AD, Stephens M, Gilad Y, Pääbo S, Przeworski M.. 2004. Absence of the TAP2 human recombination hotspot in chimpanzees. PLoS Biol. 2(6):e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JA, Belsare S, Birnbaum S, Newman DE, Chan J, Glenn JP, Ferguson B, Cox LA, Wall JD.. 2019. Analysis of 100 high-coverage genomes from a pedigreed captive baboon colony. Genome Res. 29(5):848–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J, Garcia R, Shelledy W, Kaplan J, Arya A, Johnson Z, Bergstrom M, Novakowski L, Nair P, Vinson A, et al. 2006. An initial genetic linkage map of the rhesus macaque (Macaca mulatta) genome using human microsatellite loci. Genomics. 87(1):30–38. [DOI] [PubMed] [Google Scholar]
- Rogers J, Mahaney MC, Witte SM, Nair S, Newman D, Wedel S, Rodriguez LA, Rice KS, Slifer SH, Perelygin A, et al. 2000. A genetic linkage map of the baboon (Papio hamadryas) genome based on human microsatellite polymorphisms. Genomics. 67(3):237–247. [DOI] [PubMed] [Google Scholar]
- Spencer CC, Deloukas P, Hunt S, Mullikin J, Myers S, Silverman B, Donnelly P, Bentley D, McVean G.. 2006. The influence of recombination on human genetic diversity. PLoS Genet. 2(9):e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapley J, Feulner PGD, Johnston SE, Santure AW, Smadja CM.. 2017. Variation in recombination frequency and distribution across eukaryotes: patterns and processes. Philos Trans R Soc Lond B Biol Sci. 372(1736):20160455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Donnelly P.. 2003. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 73(5):1162–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Scheet P.. 2005. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 76(3):449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P.. 2001. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 68(4):978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevison LS, Noor M.. 2009. Recombination rates in Drosophila In: Encyclopedia of Life Sciences (eLS). Chichester (United Kingdom: ): John Wiley and Sons, Ltd. [Google Scholar]
- Stevison LS, Woerner AE, Kidd JM, Kelley JL, Veeramah KR, McManus KFGreat Ape Genome ProjectBustamante CD, Hammer MF, Wall JD.. 2016. The time scale of recombination rate evolution in great apes. Mol Biol Evol. 33(4):928–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. 2013. From FastQ data to high-confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43:11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD, Frisse LA, Hudson RR, Di Rienzo A.. 2003. Comparative linkage-disequilibrium analysis of the beta-globin hotspot in primates. Am J Hum Genet. 73(6):1330–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh B, Lynch M.. 2019. Evolution and selection of quantitative traits Oxford: Oxford University Press. [Google Scholar]
- Warren WC, Jasinska AJ, García-Pérez R, Svardal H, Tomlinson C, Rocchi M, Archidiacono N, Capozzi O, Minx P, Montague MJ, et al. 2015. The genome of the vervet (Chlorocebus aethiops sabaeus). Genome Res. 25(12):1921–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiehe TH, Stephan W.. 1993. Analysis of a genetic hitchhiking model, and its application to DNA polymorphism data from Drosophila melanogaster. Mol Biol Evol. 10(4):842–854. [DOI] [PubMed] [Google Scholar]
- Winckler W, Myers SR, Richter DJ, Onofrio RC, McDonald GJ, Bontrop RE, McVean GA, Gabriel SB, Reich D, Donnelly P, et al. 2005. Comparison of fine-scale recombination rates in humans and chimpanzees. Science 308(5718):107–111. [DOI] [PubMed] [Google Scholar]
- Xue C, Raveendran M, Harris RA, Fawcett GL, Liu X, White S, Dahdouli M, Rio Deiros D, Below JE, Salerno W, et al. 2016. The population genomics of rhesus macaques (Macaca mulatta) based on whole-genome sequences. Genome Res. 26(12):1651–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Giron CG, et al. 2018. Ensembl 2018. Nucleic Acids Res. 46(D1):D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick ME, Cutler DJ, Langley CH.. 1999. Classic Weinstein: tetrad analysis, genetic variation and achiasmate segregation in Drosophila and humans. Genetics 152(4):1615–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The fine-scale genetic map for vervet monkeys generated in this study is available at http://spfeiferlab.org/data.