Abstract
Nodaviruses are small bisegmented RNA viruses belonging to the family Nodaviridae. Nodaviruses have been identified in different hosts, including insects, fishes, shrimps, prawns, dogs, and bats. A novel porcine nodavirus was first identified in the United States by applying next-generation sequencing on brain tissues of pigs with neurological signs, including uncontrollable shaking. RNA1 of the porcine nodavirus had the highest nucleotide identity (51.1%) to the Flock House virus, whereas its RNA2 shared the highest nucleotide identity (48%) with the RNA2 segment of caninovirus (Canine nodavirus). Genetic characterization classified porcine nodavirus as a new species under the genus Alphanodavirus. Further studies are needed to understand the pathogenicity and clinical impacts of this virus.
Keywords: porcine nodavirus, genome, detection, characterization, United States
1. Introduction
Nodaviruses are small, nonenveloped isometric bisegmented RNA viruses [1]. Nodavirus was first identified in Culex tritaeniorhynchus mosquitoes in the Japanese village of Nodamura [2]. Subsequently, other members of the Nodaviridae family were identified in a variety of insects, including the Black beetle virus, Flock House virus, Boolara virus, and Pariacoto virus. In addition to insects, nodavirus was also identified as the cause of neurological disease in farmed striped jack fish in 1992, and in many other fish species. Based on the genetic diversity of the RNA2 segment, nodaviruses are taxonomically classified into two genera, Alphanodavirus and Betanodavirus, which infect insects and fish, respectively. Recently, novel nodaviruses that infect shrimps and prawns had distinct genomic sequences and thus have been tentatively proposed as the genus Gammanodavirus [3,4,5].
The genome of nodavirus consists of two linear, single-stranded, and positive-sense RNA molecules. The two genome segments are capped at the 5′ ends but not polyadenylated at the 3′ ends. The larger segment RNA1 is approximately 3.1 kb in length, encoding the protein A, an RNA-dependent RNA polymerase (RdRp). A subgenomic transcript RNA3 is synthesized from RNA1 and encodes two small nonstructural proteins: B1, the function of which is unknown; and B2, which functions as an RNA silencing inhibitor during replication [6,7]. Not all nodaviruses encode B1. The smaller segment RNA2 is approximately 1.4 kb long, encoding the protein α, a viral capsid protein precursor. During the viral assembly, protein α is autocleaved into a 38 kDa protein β and a 5 kDa protein γ near its C-terminus at the conserved Asn/Ala site [1].
The presence of nodavirus in mammalian hosts is not common. A bat nodavirus detected in the brain of Eptesicus serotinus was reported in 2014 [8], and nodaviruses have also been detected in otter and feral dog feces [9,10]. Although originally isolated from insects, nodaviruses of the genus Alphanodavirus were able to infect mammals in experiments. Nodamura virus (NoV), the prototype species of the genus Alphanodavirus, was able to lethally infect both insects and mammals, including suckling mice and suckling hamsters [2,11,12,13]. Therefore, NoV is the only known arthropod-borne virus that can infect insects and vertebrate animals. Serological evidence also showed that NoV might be able to infect pigs. It was reported that 13 of 16 pigs (81%) had positive neutralizing antibodies against NoV in 1956, and 14 of 27 pigs (51%) in 1957, near Tokyo, Japan [14]. However, no nodavirus has been identified and genetically characterized from naturally infected pigs. In the present study, we report identification of porcine nodavirus for the first time in the world.
2. Materials and Methods
2.1. Pig Samples
Different types of pig samples including brain, spinal cord, heart, liver, kidney, spleen, intestine, lymphoid tissues, blood, and feed were submitted by a client to the Iowa State University Veterinary Diagnostic Laboratory (ISU-VDL).
2.2. Testing
Real-time PCR testing was performed on Pestivirus, Sapelovirus, Teschovirus, Astrovirus, Glaesserella parasuis, porcine circovirus 2, porcine reproductive and respiratory syndrome virus (PRRSV), and Streptococcus suis. The sequences of the primers and probes used are shown in Table 1. All the real-time PCR assays were performed in a reaction mixture of 20 μL containing 5 μL TaqMan Fast Virus 1-Step Master Mix (Applied Biosystems, Waltham, MA, USA), 0.8 μL (final concentration 0.4 µM) of each of the primers, 0.4 μL (final concentration 0.2 µM) of probe, 8 μL nuclease-free water, and 5 μL extracted RNA. The amplification was performed at 50 °C for 5 min, then at 95 °C for 20 s, followed by 40 cycles at 95 °C for 3 s and 60 °C for 30 s. The PRRSV real-time RT PCR was performed with TaqMan NA and EU PRRSV Reagents (Applied Biosystems) following the manufacturer’s instructions.
Table 1.
Primers and probes used in this study.
| Target Pathogen | Name | Sequence | Reference |
|---|---|---|---|
| Porcine nodavirus | Noda_F Noda_P Noda_R |
TTGTCCCAACCTACAGGA FAM-CCGAATCATGCGACCACACG-BHQ TTGGCCAGTGGACTTGAA |
|
| Porcine pestivirus | APPV_RTF APPV_RTR APPV_RTP |
TGCCTGGTATTCGTGGC TCATCCCATGTTCCAGAGT FAM-CCTCCGTCTCCGCGGCTTCTTTGG-BHQ |
[15] |
| Porcine sapelovirus | PSV_RTF PSV_RTR PSV_RTP |
GGCAGTAGCGTGGCGAGC CTACTCTCCTGTAACCAGT FAM-CGATAGCCATGTTAGTG-MGB |
[16] |
| Porcine teschovirus | PTV_RTF PTV_RTR PTV_RTP |
CACCAGCGTGGAGTTCCTGTA AGCCGCGACCCTGTCA FAM-TGCAGGACTGGACTTG-MGB |
[17] |
| Porcine astrovirus 3 | PoAstV3_RTF PoAstV3_RTR PoAstV3_RTP |
ATGACYCTCTATGGGAAACTCCTT GTGCCTRGCAACAACCTCCAA FAM-TTGGCCAYAACCTCCCTGA-MGB |
[18] |
| Porcine circovirus 2 | PCV2_RTF PCV2_RTR PCV2_RTP |
GACTGTWGAGACTAAAGGTGGAACTGTA GCTTCTACACCTGGGACAGCA FAM-CCCGTTGGAATGGT-MGB |
|
| Glaesserella parasuis | GPS_RTF GPS_RTR GPS_RTP |
CCACTTACTTGAAGCCATTCTTCTT CCGCTTGCCATACCCTCTT FAM-ATCGGAAGTATTAGAATTAAGKGC-MGB |
[19] |
| Streptococcus suis | Ssuis_RTF Ssuis_RTR Ssuis_RTP |
CTTTTGGACAGTTTCGGAGAAGA TTTTCGTTTTCAAGAACTCGTTTG FAM-AAGACCGTTATCAGACAAC-MGB |
2.3. Next-Generation Sequencing (NGS) and Sequence Analysis
Nucleic acids extracted from brain tissue were subjected to a sequence-independent, single-primer amplification, library preparation using an Illumina Nextera XT kit, and sequenced using an Illiumina MiSeq v2 sequencing kit (500 cycles) on MiSeq (Illumina, San Diego, CA, USA) as previously described [11]. Raw FastQ files were analyzed using Kraken software and assembled using a SPAdes assembler. A local BLAST (blastx) was performed on the assembler contigs. MEGA 7.0.26 was used for sequence alignment and phylogenetic tree analysis, and a BioEdit sequence alignment editor was used to calculate the sequence identity.
2.4. Real-Time RT-PCR for Porcine Nodavirus
The real-time RT-PCR was performed in a reaction mixture of 25 μL containing 12.5 μL 2× AgPath-ID RT-PCR Buffer (Applied Biosystems), 1 μL 25× RT-PCR Enzyme Mix, 1 μL (final concentration 0.4 µM) of each of the Noda_F and Noda_R primers (Table 1), 0.5 μL (final concentration 0.2 µM) of probe Noda_P, 4 μL nuclease-free water, and 5 μL extracted RNA. The amplification was performed at 48 °C for 10 min, then at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 45 s. For each amplification plot, a cycle threshold (CT) value was calculated to represent the cycle number at which the reporter signal was above threshold.
3. Results and Discussion
In November of 2017, an acute onset of neurologic signs was observed in one barn of 20-week-old pigs on a six-barn site in North Carolina. The affected pigs had abrupt onset of full-body tremors and uncontrollable shaking, followed by prostration and death within 24 h. Affected pigs were often bullied by unaffected pigs, which likely contributed to their rapid decline and death. Twenty-two pigs died over a two-day period, and a similar disease was reported in an adjacent three barn site. Samples including brain, heart, lung, liver, kidney, spleen, tonsil, lymph node, intestine, colon, blood, and feed were submitted to ISU-VDL. Neither gross lesions nor histopathological lesions were observed in brain, spinal cord, heart, liver, kidney, spleen, intestine, and lymphoid tissues. The polymerase chain reaction (PCR) was negative for common porcine pathogens, including Pestivirus, Sapelovirus, Teschovirus, Astrovirus, Glaesserella parasuis, and porcine circovirus 2, using routine assays at the ISU-VDL. Although both PRRSV and S. suis were detected in lung tissue, findings based on lesions and clinical signs did not favor them as causes of the observed clinical signs or deaths. The differential diagnoses included various metabolic diseases and intoxications, which were then excluded.
The brain-tissue sample was subsequently analyzed using NGS technology on a MiSeq platform. Raw FastQ files was analyzed using Kraken software and assembled using a SPAdes assembler [20,21]. MEGA 7.0.26 was used for sequence alignment and phylogenetic tree analysis, and a BioEdit sequence alignment editor was used to calculate the sequence identity [22]. Kraken taxonomical analysis of raw FastQ data and local nucleotide blast of assembled contigs revealed no hit to viral reads. However, the protein BLAST (blastx) of assembled contigs revealed a low level of amino acid sequence similarity to a RdRp encoded by the RNA1 genome segment in Nodaviridae family. Further analysis showed that two separated segments, RNA1 (3065 nt) and RNA2 (1370 nt) (Accession numbers: MK014575 and MK014576), with similar genome structures to those of nodavirus had been assembled, and the virus was tentatively named as porcine nodavirus. The nucleotide sequence alignment revealed that the RNA1 segment of pig nodavirus showed low identities (23.4% to 51.1%) to other nodaviruses (Table 2). Surprisingly, the pig nodavirus was least closely related to the RNA1 of feral dog and bat nodaviruses (23.4% and 32.1% nt identities, respectively); instead, it had the highest nucleotide identity to the Flock House virus (51.1%). The protein A encoded by the genome segment RNA1 was 1014 amino acids in length, and contained the core motifs defined for RdRp. The protein A of porcine nodavirus shared 14.2% to 44.6% of identities with other nodaviruses at the amino acid level (Table 2).
Table 2.
Identity (%) of porcine nodavirus to other nodaviruses.
| Strain | RNA1 | RNA2 | GenBank Accession # | ||
|---|---|---|---|---|---|
| nt | aa | nt | aa | ||
| Caninovirus_sp._isolate_South_Douro | 23.4 | 14.2 | 48 | 47.2 | KY214439, KY214441 |
| Pariacato_virus | 33.5 | 19 | 46.2 | 41.5 | NC_003691, NC_003692 |
| Flock_House_virus | 51.1 | 40.2 | 39.8 | 32.7 | NC_004146, NC_004144 |
| Drosophila_melanogaster_American_nodavirus | 50.3 | 39.9 | 39.7 | 30.3 | GQ342965, GQ342966 |
| Black_beetle_virus | 51 | 39.8 | 39.2 | 31.8 | NC_001411, NC_002037 |
| Nodamura_virus | 48 | 44.5 | 37.7 | 33.2 | NC_002690, NC_002691 |
| Tiger_puffer_nervous_necrosis_virus | 34.5 | 17.5 | 25.1 | 5.9 | NC_013460, NC_013461 |
| Wuhan_nodavirus | 31.3 | 16.3 | 25.1 | 6.5 | AY962576, DQ233638 |
| Striped_Jack_nervous_necrosis_virus | 33.8 | 17.5 | 25 | 6.4 | NC_003448, NC_003449 |
| Redspotted_grouper_nervous_necrosis_virus | 34.7 | 17 | 24.6 | 6.9 | FJ789783, FJ789784 |
| Redspotted_grouper_nervous_necrosis_virus_SGWak97 | 34.4 | 16.9 | 24.4 | 6.6 | NC_008040, NC_008041 |
| Barfin_flounder_nervous_necrosis_virus | 34.3 | 17.4 | 24.3 | 5.9 | NC_013458, NC_013459 |
| Mosinovirus_C36-CI-2004 | 32.9 | 18.6 | 21.7 | 6.4 | KJ632942, KJ632943 |
| Macrobrachium_rosenbergii_nodavirus | 46.4 | 42.9 | 29 | 8.6 | NC_005094, NC_005095 |
| Redspotted_grouper_nervous_necrosis_virus_MENNV1310 | 34.3 | 17.1 | 26 | 6.6 | KY315688, KY315689 |
| Penaeus_vannamei_nodavirus | 48 | 42.6 | 19.4 | 7 | NC_014978, NC_014977 |
| Lutzomyia_nodavirus_piaui | 45.3 | 34.9 | 18.4 | 4.4 | KR003799, KR003800 |
| Santeuil_nodavirus_JU1264 | 26.2 | 10.7 | 14.8 | 8.2 | HM030972, HM030973 |
| Orsay_virus_JU1580 | 27.4 | 12.3 | 14.2 | 4.2 | HM030970, HM030971 |
| Le_Blanc_nodavirus_JU1498 | 25.6 | 11.3 | 13.5 | 12.1 | JQ943579, JQ943580 |
| Bat_guano_associated_nodavirus | 32.1 | 19.2 | - | - | HM228873, |
| Shuangao_noda-like_virus_1_insectZJ94204 | 45.9 | 44.6 | - | - | KX883120 |
| Shuangao_insect_virus_11_insectZJ65889 | 45 | 45 | - | - | NC_033265 |
nt: nucelotide; aa: amino acid; -: not available.
The smaller genome segment RNA2 of the identified porcine nodavirus contained 1370 nt. Interestingly, unlike RNA1, the RNA2 of porcine nodavirus shared the highest identity (48% and 47.2%, at both nucleotide and amino acid levels, respectively) with the RNA2 segment of caninovirus (Canine nodavirus) (Table 2) [9], suggesting that porcine nodavirus could be a potential recombinant between nodaviruses of insects and mammals. The RNA2 of porcine nodavirus shared relatively higher identities (37.7–46.2%) with alphanodaviruses isolated from insects than Betanodavirus isolated from fishes (24.3–25.1%) and Gammanodavirus isolated from shrimps and prawns (19.4–29%) (Table 2). Similarly, the capsid precursor protein of porcine nodavirus encoded by RNA2 shared significantly higher similarities to those of other Alphanodavirus (33.2–41.5%) than Betanodavirus (5.9–6.9%) and Gammanodavirus (7–8.6%) at the amino acid level.
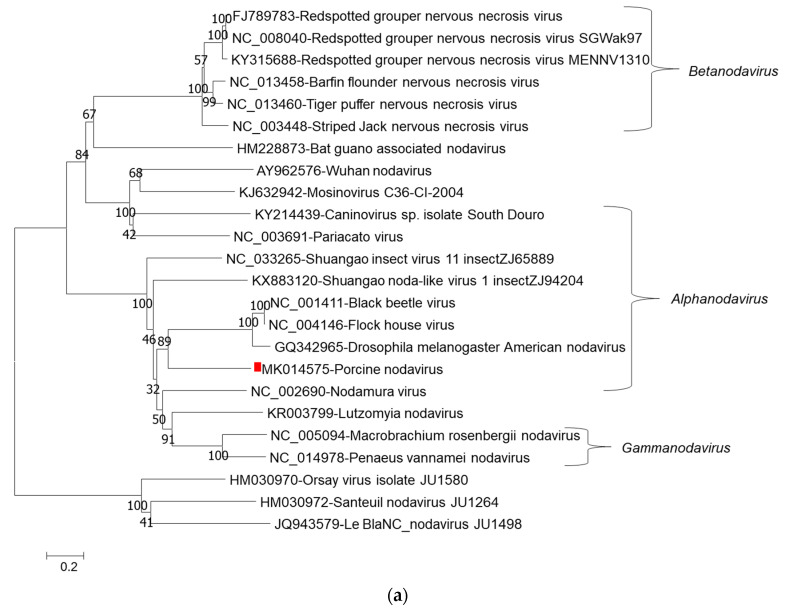
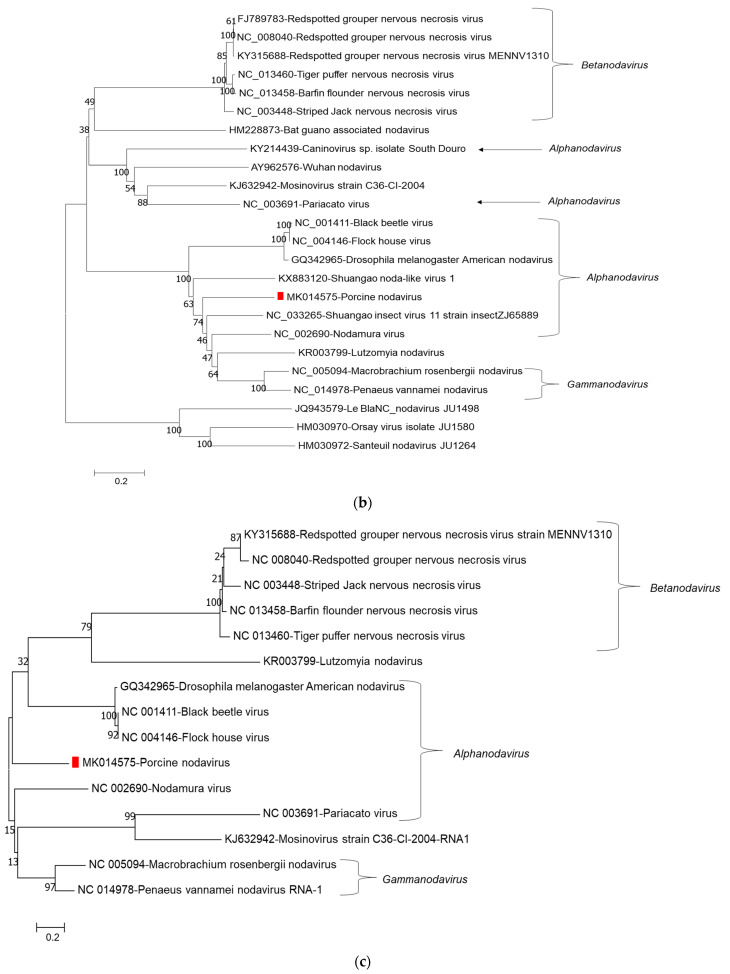
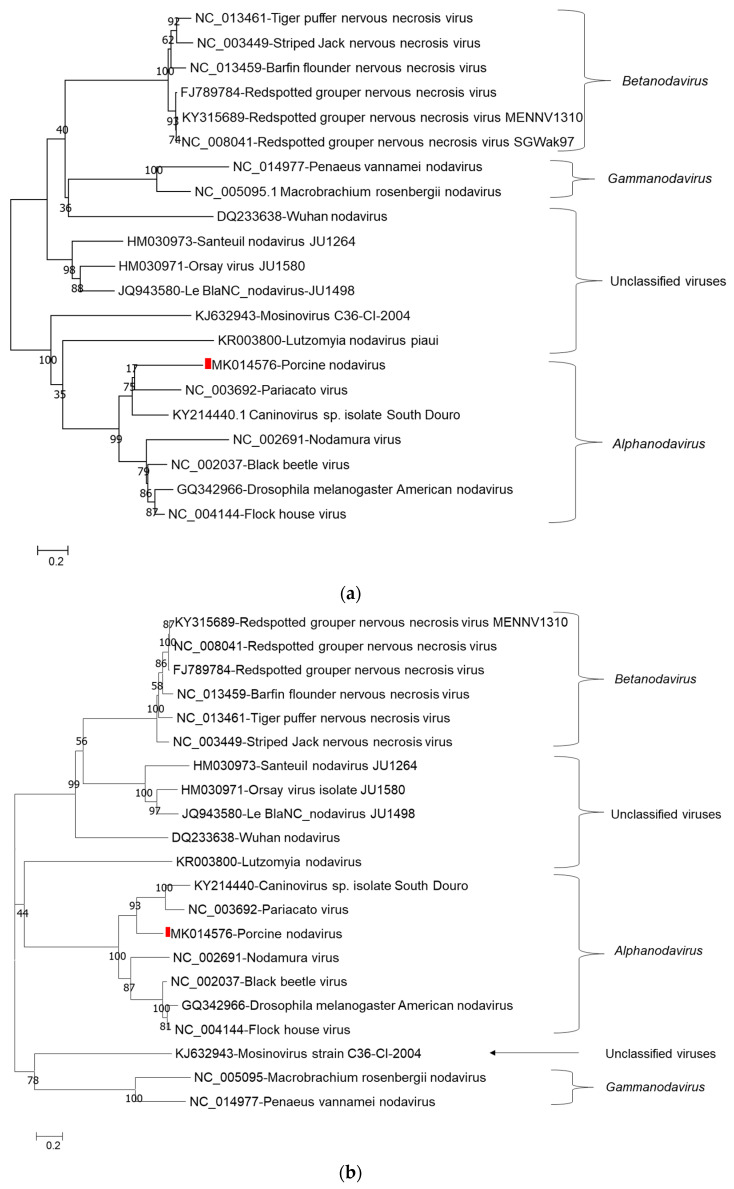
Phylogenetic tree analysis of nucleotide sequences of two segments confirmed that porcine nodavirus cluster together with isolates from Alphanodavirus. Specifically, porcine nodavirus clustered together with three viruses (Black beetle virus, Flock House virus, and Drosophila melanogaster American nodavirus) and two viruses (Shuangao insect virus 11 and Nodamura virus) in the RNA 1 nucleotide and amino acid trees, respectively (Figure 1a–c); and clustered together with two viruses (caninovirus and Pariacato virus) at both the nucleotide and amino acid levels of the RNA2 segment (Figure 2a,b).
Figure 1.
Phylogenetic relationship of porcine nodavirus (indicated by red square) with other representative nodaviruses. The tree was constructed by using the neighbor-joining method with the maximum composite likelihood model in MEGA version 7.0.26 (http://www.megasoftware.net) with 1000 bootstrap replicates, based on the nucleotide sequence of RNA1 (a), amino acid sequences of RNA dependent RNA polymerase (b), and amino acid sequences of B2 (c).
Figure 2.
Phylogenetic relationship of porcine nodavirus (indicated by red square) with other representative nodaviruses. The tree was constructed by using the neighbor-joining method with the maximum composite likelihood model in MEGA version 7.0.26 (http://www.megasoftware.net) with 1000 bootstrap replicates, based on nucleotide sequence of RNA2 (a) and amino acid sequence of capsid protein precursor alpha (b).
The classification of nodaviruses is based on the genetic diversity of the RNA2 segment, and a novel species can be classified if the identity of the RNA2 to the most closely related species is less than 80% and 87% at the nucleotide and amino acid levels, respectively. Porcine nodavirus shared the highest identities (<50%) of RNA2 to the feral dog isolate at nucleotide and amino acid levels (Table 2). Therefore, porcine nodavirus was classified as a novel species in the genus Alphanodavirus.
The fish nodavirus known as nervous necrosis virus (Betanodavirus genus) mainly targets the central nervous system, including the brain, spinal cord, and retina. The infected fish suffer neurological disorders characterized by viral encephalopathy and retinopathy [23,24]. In addition, the NoV, which belongs to the genus Alphanodavirus, could experimentally infect suckling mice leading to hind-limb paralysis [2]. The novel porcine nodavirus identified in our study was detected in the brain tissue of pigs showing neurological signs and uncontrollable shaking. The real-time RT-PCR testing detected the nodavirus in pig brain and spinal cord samples, with Ct values of 26.2 and 26.5. Future studies are required to determine the significance and potential pathogenicity of this porcine nodavirus, and epidemiological studies are needed to monitor its evolution and clinical impact.
Author Contributions
Conceptualization, formal analysis, and writing—original draft preparation, C.Y., L.W., G.L.; writing—review, editing, and data curation, C.Y., L.W., Y.Z., G.L.; methodology, C.Y., K.S., E.B., J.G.-T., Q.C., Y.Z., H.S., L.W. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Iowa State University Veterinary Diagnostic Laboratory.
Data Availability Statement
The data presented in this study are available in the present article. Genome sequences were deposited in GenBank under the accession numbers MK014575–MK014576.
Conflicts of Interest
All authors have declared no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sahul Hameed A.S., Ninawe A.S., Nakai T., Chi S.C., Johnson K.L. ICTV Virus Taxonomy Profile: Nodaviridae. J. Gen. Virol. 2018;100:3–4. doi: 10.1099/jgv.0.001170. [DOI] [PubMed] [Google Scholar]
- 2.Scherer W.F., Hurlbut H.S. Nodamura virus from Japan: A new and unusual arbovirus resistant to diethyl ether and chloroform. Am. J. Epidemiol. 1967;86:271–285. doi: 10.1093/oxfordjournals.aje.a120737. [DOI] [PubMed] [Google Scholar]
- 3.Naveen Kumar S., Karunasagar I., Karunasagar I. Protection of Macrobrachium rosenbergii against white tail disease by oral administration of bacterial expressed and encapsulated double-stranded RNA. Fish Shellfish Immunol. 2013;35:833–839. doi: 10.1016/j.fsi.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 4.Low C.F., Md Yusoff M.R., Kuppusamy G., Ahmad Nadzri N.F. Molecular biology of Macrobrachium rosenbergii nodavirus infection in giant freshwater prawn. J. Fish Dis. 2018;41:1771–1781. doi: 10.1111/jfd.12895. [DOI] [PubMed] [Google Scholar]
- 5.NaveenKumar S., Shekar M., Karunasagar I., Karunasagar I. Genetic analysis of RNA1 and RNA2 of Macrobrachium rosenbergii nodavirus (MrNV) isolated from India. Virus Res. 2013;173:377–385. doi: 10.1016/j.virusres.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Guo X., Lu R. Characterization of virus-encoded RNA interference suppressors in Caenorhabditis elegans. J. Virol. 2013;87:5414–5423. doi: 10.1128/JVI.00148-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H., Li W.X., Ding S.W. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- 8.Dacheux L., Cervantes-Gonzalez M., Guigon G., Thiberge J.M., Vandenbogaert M., Maufrais C., Caro V., Bourhy H. A preliminary study of viral metagenomics of French bat species in contact with humans: Identification of new mammalian viruses. PLoS ONE. 2014;9:e87194. doi: 10.1371/journal.pone.0087194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conceicao-Neto N., Godinho R., Alvares F., Yinda C.K., Deboutte W., Zeller M., Laenen L., Heylen E., Roque S., Petrucci-Fonseca F., et al. Viral gut metagenomics of sympatric wild and domestic canids, and monitoring of viruses: Insights from an endangered wolf population. Ecol. Evol. 2017;7:4135–4146. doi: 10.1002/ece3.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conceicao-Neto N., Zeller M., Heylen E., Lefrere H., Mesquita J.R., Matthijnssens J. Fecal virome analysis of three carnivores reveals a novel nodavirus and multiple gemycircularviruses. Virol. J. 2015;12:79. doi: 10.1186/s12985-015-0305-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bailey L., Scott H.A. The pathogenicity of Nodamura virus for insects. Nature. 1973;241:545. doi: 10.1038/241545a0. [DOI] [PubMed] [Google Scholar]
- 12.Scherer W.F. Variable results of sodium deoxycholate tests of Nodamura virus, an ether and chloroform resistant arbovirus. Proc. Soc. Exp. Biol. Med. 1968;129:194–199. doi: 10.3181/00379727-129-33282. [DOI] [PubMed] [Google Scholar]
- 13.Tesh R.B. Infectivity and pathogenicity of Nodamura virus for mosquitoes. J. Gen. Virol. 1980;48:177–182. doi: 10.1099/0022-1317-48-1-177. [DOI] [Google Scholar]
- 14.Scherer W.F., Verna J.E., Richter W. Nodamura virus, an ether- and chloroform-resistant arbovirus from Japan: Physical and biological properties, with ecologic observations. Am. J. Trop. Med. Hyg. 1968;17:120–128. doi: 10.4269/ajtmh.1968.17.120. [DOI] [PubMed] [Google Scholar]
- 15.Arruda B.L., Arruda P.H., Magstadt D.R., Schwartz K.J., Dohlman T., Schleining J.A., Patterson A.R., Visek C.A., Victoria J.G. Identification of a divergent lineage porcine pestivirus in nursing piglets with congenital tremors and reproduction of disease following experimental inoculation. PLoS ONE. 2016;11:e0150104. doi: 10.1371/journal.pone.0150104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matias Ferreyra F.S., Bradner L.K., Burrough E.R., Cooper V.L., Derscheid R.J., Gauger P.C., Harmon K.M., Madson D., Pineyro P.E., Schwartz K.J., et al. Polioencephalomyelitis in domestic swine associated with porcine astrovirus type 3. Vet. Pathol. 2020;57:82–89. doi: 10.1177/0300985819875741. [DOI] [PubMed] [Google Scholar]
- 17.Jimenez-Clavero M.A., Fernandez C., Ortiz J.A., Pro J., Carbonell G., Tarazona J.V., Roblas N., Ley V. Teschoviruses as indicators of porcine fecal contamination of surface water. Appl. Environ. Microbiol. 2003;69:6311–6315. doi: 10.1128/AEM.69.10.6311-6315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rawal G., Matias Ferreyra F., R. Macedo N., K. Bradner L., M. Harmon K., Mueller A., Allison G., C. L. Linhares D., Arruda B. Detection and cellular tropism of porcine astrovirus type 3 on breeding farms. Viruses. 2019;11:1051. doi: 10.3390/v11111051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frandoloso R., Martinez-Martinez S., Rodriguez-Ferri E.F., Gutierrez-Martin C.B. Comparison of real-time PCR and culture isolation in colostrum-deprived pigs immunized and challenged with haemophilus parasuis. Lett. Appl. Microbiol. 2012;54:149–152. doi: 10.1111/j.1472-765X.2011.03187.x. [DOI] [PubMed] [Google Scholar]
- 20.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wood D.E., Salzberg S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munday B.L., Nakai T. Nodaviruses as pathogens in larval and juvenile marine finfish. World J. Microbiol. Biotechnol. 1997;13:375–381. doi: 10.1023/A:1018516014782. [DOI] [Google Scholar]
- 24.Ghiasi M., Binaii M., Ghasemi M., Fazli H., Zorriehzahra M.J. Haemato-biochemical disorders associated with nodavirus like-agent in adult leaping mullet Liza saliens (Risso, 1810) in the Caspian Sea. VirusDisease. 2016;27:12–18. doi: 10.1007/s13337-015-0289-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available in the present article. Genome sequences were deposited in GenBank under the accession numbers MK014575–MK014576.