Abstract
Porcine deltacoronavirus (PDCoV) is one of the emerged coronaviruses posing a significant threat to the swine industry. Previous work showed the presence of a viral accessory protein NS6 in PDCoV-infected cells. In this study, we detected the expression of the NS6 protein in small intestinal tissues of PDCoV-infected piglets. In addition, SDS-PAGE and Western blot analysis of sucrose gradient-purified virions showed the presence of a 13-kDa NS6 protein. Further evidences of the presence of NS6 in the PDCoV virions were obtained by immunogold staining of purified virions with anti-NS6 antiserum, and by immunoprecipitation of NS6 from purified virions. Finally, the anti-NS6 antibody was not able to neutralize PDCoV in cultured cells. These data establish for the first time that the accessory protein NS6 is expressed during infection in vivo and incorporated into PDCoV virions.
Keywords: Coronavirus, Porcine deltacoronavirus (PDCoV), NS6, Virions
1. Introduction
In the past two decades, several newly emerged coronaviruses (CoVs), including the severe acute respiratory syndrome coronavirus (SARS-CoV) (Zhong et al., 2003), Middle East respiratory syndrome coronavirus (MERS-CoV) (Zaki et al., 2012), SARS-CoV-2 (Zhou et al., 2020), porcine deltacoronavirus (PDCoV) (Woo et al., 2012), and swine enteric alphacoronavirus (SeACoV) (Yang et al., 2019a, 2019b, 2020b), have caused respiratory or gastrointestinal diseases in humans or animals. They have posed significant threats to public health or animal health. Among these, PDCoV (genus Deltacoronavirus; subfamily Orthocoronavirinae; family Coronaviridae; order Nidovirales) mainly causes acute diarrhea, vomiting, dehydration, and mortality in nursing pigs (Jung et al., 2015), and leads to a significant economic losses for the swine industry. Since being first reported in Hong Kong in 2012 (Woo et al., 2012), PDCoV has been detected in the United States (Ma et al., 2015; Wang et al., 2014) and many Asian countries including China (Dong et al., 2015; Wang et al., 2015). Most recently, in vitro study shown that PDCoV can infect cell lines derived from multiple species, such as humans, pigs, and chickens, indicating its potential cross-species transmissibility (Li et al., 2018).
Like other CoVs, PDCoV is an enveloped virus with a relatively large (25.4 kb) single-stranded, positive-sense RNA genome, which has a 5′ cap structure and a 3′ poly (A) tail, allowing it to act as an mRNA for translation of the replicase polyprotein (Woo et al., 2012). The first two-thirds of the genome encode polyprotein (pp) 1a and pp1ab, which are proteolytically cleaved into 15 mature nonstructural proteins related to viral replication and transcription (Wang et al., 2015). The remaining third of the genome contains ORFs encoding viral structural proteins, including the spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins. For all CoVs, M and S make up the majority of protein incorporated into the viral envelope. Trimers of S, a type I membrane glycoprotein, form the unique spike structure found on the surface of virions that mediates attachment to the host receptor as well as subsequent membrane fusion (Wang et al., 2018; Yang et al., 2020a). The S protein is incorporated into virions through noncovalent interactions with the M protein (Godeke et al., 2000). The N protein alone makes up the CoV nucleocapsid, and promotes completion of virion assembly via direct interaction with the M protein (Fehr and Perlman, 2015). The E protein is a small, 9-kDa integral membrane protein that plays a part in viral assembly and morphogenesis, with small quantities found within the virion (Liu et al., 2007).
In addition to these common structural proteins shared by all CoVs, there are a number of species-specific ORFs that encode accessory proteins, some of which appear to be incorporated in virions at low levels (Liu et al., 2014). The PDCoV accessory NS6 gene, which does not show significant homology to accessory proteins of the other CoVs, is located between the M and N genes, and encodes a 94-amino-acid (aa) protein with a predicted molecular mass of 11 kDa (Wang et al., 2015; Woo et al., 2012). A recent report found that the leader-body fusion site upstream of the NS6 subgenomic RNA (sgRNA) start codon is at nt −148, rather than the predicted nt −46, and confirmed the existence of a separate NS6 protein in vitro (Fang et al., 2016). NS6 was found to antagonize IFN-β production by interacting with RIG-I and MDA5 to impede their association with double-stranded RNA (Fang et al., 2018). Recently, a recombinant virus (rPDCoV-ΔNS6-GFP), in which the NS6 gene was replaced by the green fluorescent protein (GFP) gene, has been constructed by reverse genetics (Zhang et al., 2020).
Nevertheless, the additional function of NS6 in the PDCoV life cycle remains poorly understood, and further analysis is needed. The initial aim of the present study was to establish that the accessory NS6 protein is indeed expressed in target tissues of pigs during infection in vivo. Furthermore, we provided the first evidence that NS6 protein is incorporated into purified PDCoV virions. These data suggest that NS6 protein is not only a PDCoV accessory protein but also a component associated with PDCoV virions.
2. Results
2.1. Prokaryotic expression and purification of NS6 and generation of two specific antibodies
The expression of the full-length recombinant NS6 protein fused with a polyhistidine tag in E. coli was subjected to sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot (WB) analysis. The apparent molecular mass of the NS6 protein was approximately 13 kDa, which is slightly larger than the predicted molecular weight (11 kDa; Supplementary Figs. S1A and S1B). Subsequently, the purified protein was used to immunize ten, 6-week-old female BALB/c mice to produce anti-NS6 polyclonal antibodies. Immunofluorescence assay (IFA) revealed specific fluorescence in PDCoV-infected cells at 16 hpi (Supplementary Fig. S2A), whereas no signal was detected in mock-infected cells (data not shown). However, the antibody could not recognize NS6 protein in WB assay (data not shown). Thus, we prepared anti-NS6 peptide antibodies (anti-NS6-pt) by immunization of rabbits with two synthetic peptides derived from the putative NS6 aa 15–28 and aa 81–94 sequences (Table 1 ). IFA revealed specific fluorescence in PDCoV-infected cells, while no staining was observed with pre-immune rabbit serum (Supplementary Fig. S2A). Furthermore, WB analysis confirmed that the anti-NS6-pt reacted strongly with recombinant full-length NS6 protein expressed in E. coli or PDCoV-infected cell lysates (Supplementary Fig. S2B).
Table 1.
Summary of PDCoV antibodies used in the study.
| Name of Ab | Type | Antigen | Source animal | Applicationa |
|---|---|---|---|---|
| Anti-NS6 | pAb | Recombinant NS6 protein | Mouse | IFA, IHC, IE, VN |
| Anti-NS6-pt | pAb | Synthetic NS6 peptide | Rabbit | IFA, WB, IP |
| Hyperimmune serum | pAb | Purified PDCoV virions | Rabbit | WB, IEM, VN |
| Anti-S1 | pAb | Recombinant S1 protein | Rabbit | IEM |
| Anti-M | pAb | Synthetic M peptide | Rabbit | IHC, WB |
| Anti-Nb | mAb | Recombinant N protein | Mouse | IFA, IP |
IFA: Immunofluorescence assay; IHC: Immunohistochemistry; IEM: Immunoelectron microscopy; WB: Western blot; IP: Immunoprecipitation; VN: Virus neutralization.
Purchased from Medgene Labs, USA.
2.2. Evidence of NS6 protein expression in vivo
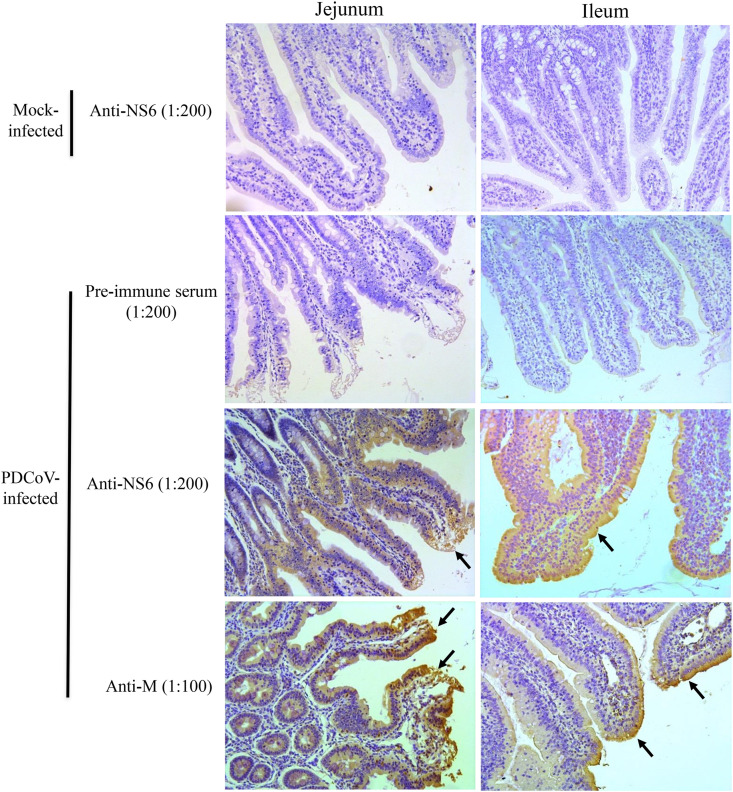
Though previous research had confirmed NS6 expression in PDCoV-infected cells, there has been no experimental evidence to confirm its expression during viral infection in vivo. Since the small intestine is the primary target of PDCoV infection (Jung et al., 2016; Ma et al., 2015), we searched for NS6 expression in the jejunum and ileum of infected piglets by immunohistochemical (IHC) staining. NS6 was detected mainly in the villous epithelium of the jejunum and ileum of experimentally infected pigs, consistent with the tissue distribution of the control PDCoV antigen M protein (Fig. 1 ; the lower four panels). No positive signals were detected in the corresponding tissues stained with anti-NS6 from mock-infected piglets (Fig. 1; the top two panels) and in the corresponding tissues stained with pre-immune serum from PDCoV-infected pigs (Fig. 1; the second top two panels).
Fig. 1.
Detection of NS6 protein expression in the PDCoV target tissues in vivo. Representative immunohistochemical (IHC) staining in the jejunum and ileum from PDCoV-infected or mock-infected piglets. Samples were stained with pre-immune mouse serum (1:200), mouse anti-NS6 antiserum (1:200) or anti-M (1:100) antibodies. Black arrows indicate positive antigens; magnification = 300 × .
2.3. PDCoV NS6 is incorporated into virions
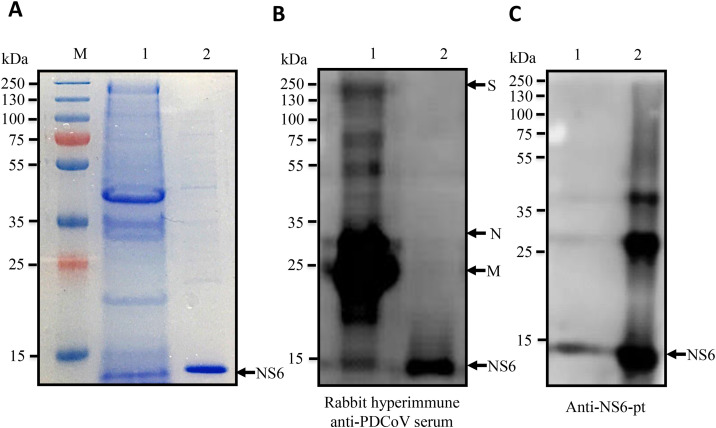
We next examined whether NS6 is a component of purified PDCoV virions by SDS–PAGE, WB, immunoelectron microscopy (IEM) or immunoprecipitation (IP) analysis. First, SDS-PAGE showed the presence of a protein of approximately 13 kDa, similar to that of the full-length recombinant protein expressed in E. coli (Fig. 2 A). A WB analysis using a hyperimmune rabbit anti-PDCoV serum (1:1000) was performed, showing all of the major viral structural proteins (S, N and M) with expected sizes along with a small amount of the 13 kDa NS6 band (Fig. 2B; lane 1). As a control, the purified NS6 protein expressed in E. coli was also recognized by the hyperimmune anti-PDCoV serum (Fig. 2B; lane 2). Finally, the identity of the band was confirmed by WB analysis using an anti-NS6-pt (Fig. 2C; lane 1). Although some non-specific signals were detected after expression in E. coli, only one band (with a molecular weight of approximately 13 kDa) was detected in purified virions (Fig. 2C). The molecular weight of the NS6 protein detected in virions was slightly larger than the recombinant NS6 protein expressed in E. coli, which is likely due to some post-translational modifications (PTMs) such as phosphorylation, glycosylation, or sumoylation of NS6 within the virions (as shown in Supplementary Fig. S3). To exclude cross-contamination of cellular and viral non-structural proteins from PDCoV-infected cells, the virions were purified by two steps of ultracentrifugation. The absence of contamination with cellular proteins was confirmed by WB using an anti-beta actin antibody (data not shown).
Fig. 2.
Identification of the NS6 protein in purified PDCoV virions. (A) SDS-PAGE, (B) Western blot (WB) using hyperimmune anti-PDCoV rabbit serum, and (C) WB using an anti-NS6 peptide antibody, showing PDCoV virions purified on a sucrose gradient (lane 1) and recombinant full-length NS6 protein expressed in E. coli (lane 2); M: protein marker.
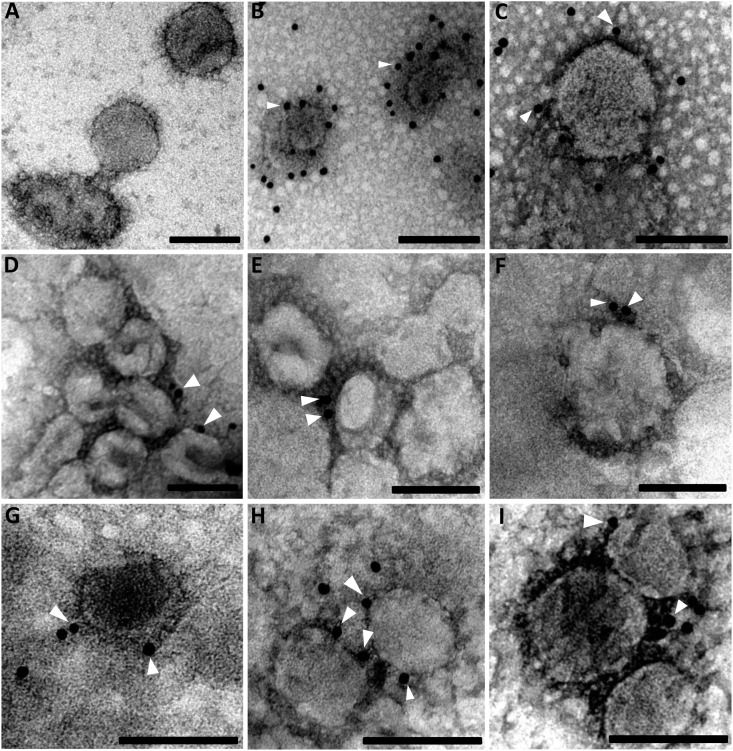
IEM was conducted to further confirm incorporation of NS6 into PDCoV particles by immunogold labeling. No staining was observed when purified virions were incubated with pre-immune rabbit serum (Fig. 3 A), whereas strong reactivity was observed with hyperimmune rabbit serum (Fig. 3B and C) and anti-S1 antibodies (Fig. 3D–F), shown by the number of gold particles surrounding the virus envelope. Gold particles were clearly associated with virions when incubated with the mouse anti-NS6 antiserum (Fig. 3G–I).
Fig. 3.
Identification of NS6 incorporated in PDCoV virions by IEM. Purified PDCoV particles were immunogold-labeled using different primary antibodies: (A) pre-immune rabbit serum (1:100); (B, C) hyper-immune rabbit anti-PDCoV (1:500); (D–F) rabbit anti-S1 polyclonal (1:100); and (G–I) mouse anti-NS6 polyclonal (1:50). Scale bars correspond to 100 nm. White arrowheads indicate the gold particles.
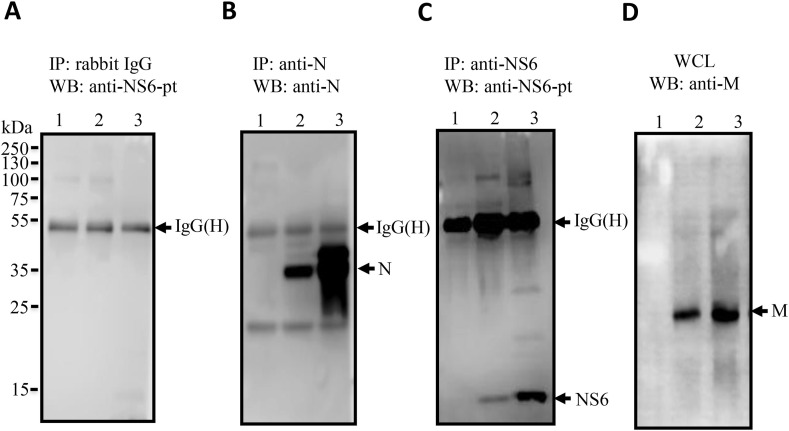
Moreover, we attempted to determine whether the anti-NS6 antibody could immunoprecipitate the NS6 protein from purified virions. IP was performed with rabbit IgG (as the negative control; Fig. 4 A), anti-N mAb (as the positive control) or anti-NS6-pt to capture protein complexes, respectively, in mock-infected, PDCoV-infected LLC-PK1 cells, or purified PDCoV particles (Fig. 4A–D). WB analysis demonstrated that, while the structural N protein could be pulled down by the anti-N mAb (Fig. 4B), the NS6 protein was also efficiently precipitated from PDCoV-infected cells or purified virions but not from the mock-infected cells by the anti-NS6 antibody (Fig. 4C). Therefore, these data suggest that the NS6 protein is attached to the PDCoV virion.
Fig. 4.
Immunoprecipitation of the NS6 protein from PDCoV-infected cells or purified virions by anti-NS6-pt. Mock-infected cells (lane 1), PDCoV-infected cells (lane 2), or purified virions (lane 3) were subjected to immunoprecipitation analysis using (A) rabbit IgG (as the negative control), (B) anti-N (as the positive control), and (C) anti-NS6 antibody, respectively. The immunoprecipitated proteins were probed with anti-NS6-pt or anti-N antibody. (D) Whole cell lysates were analyzed by Western blot analysis using the anti-M antibody. IP: immunoprecipitation; WB: Western blot; WCL: Whole cell lysate; IgG (H): IgG heavy chain.
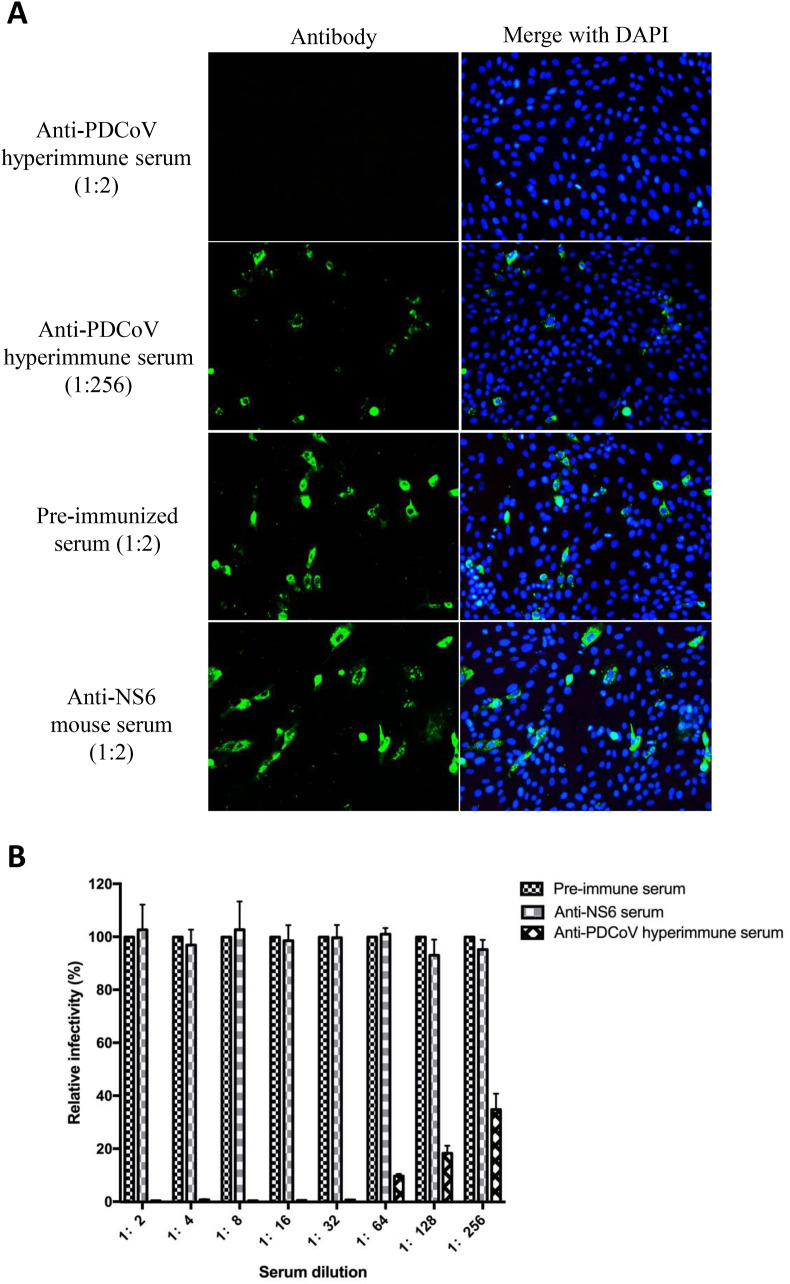
2.4. The anti-NS6 antibody does not have PDCoV-neutralizing activity
In order to test whether immunization with NS6 could induce neutralizing antibodies against PDCoV, we analyzed the neutralizing activity of the mouse anti-NS6 antiserum in LLC-PK1 cells by the virus neutralizing assay. As determined by IFA, the hyperimmune anti-PDCoV serum used as the positive control suppressed 100% of PDCoV infection at a 1:2 dilution and about 50% at a 1:256 dilution (Fig. 5 A; the top four panels). In contrast, neither the pre-immunized mouse serum used as the negative control nor the anti-NS6 antiserum at a 1:2 dilution showed the neutralizing activity against PDCoV (Fig. 5A; the lower four panels). The IFA results were further supported by PDCoV RNA quantification (Fig. 5B). The anti-NS6 antiserum did not block PDCoV infection between 1:2 to 1:256 dilutions, whereas the hyperimmune anti-PDCoV serum exhibited effective neutralizing activity in a dose-dependent manner (Fig. 5B).
Fig. 5.
Assessment of virus neutralizing activity of the anti-NS6 antibody. Two-fold serial dilutions (from 1:2 to 1:256) of the pre-immune serum, the hyperimmune anti-PDCoV serum and the anti-NS6 mouse serum were used to react with 100 TCID50 of PDCoV. The mixtures were inoculated in LLC-PK1 cells for 48 h. (A) Numbers of PDCoV-infected cells were counted by IFA; (B) PDCoV RNA titers were measured by qRT-PCR, which were then compared between different treatments and represented by the relative infectivity (viral RNA copies of tested sera/viral RNA copies of pre-immune sera × 100%). Data represent mean values plus standard deviation (SD) of at least 3 replicates.
3. Discussion
DeltaCoVs, which mainly infect birds and mammals, have the smallest genomes (25.4–26.7 kb) among all CoVs. Complete genome sequencing and comparative genome analysis has shown that deltaCoVs have similar genome characteristics and structures, including as many as four putative genes for accessory proteins (Woo et al., 2012). CoVs accessory proteins are species specific and some (such as SARS-CoV ORF6, 7a, 7b and 9b proteins) have been shown as minor components of virions (Huang et al., 2006; Schaecher et al., 2007; Xu et al., 2009). Interestingly, the SARS-CoV accessory ORF6, 7a, 7b and 9b proteins are not required for virus replication (Liu et al., 2014). Despite a recent increase in attention, few studies have looked at the accessory proteins of PDCoV (Fang et al., 2016, 2017). Previous work has shown that PDCoV NS6 was expressed and distributed in the cytoplasm during early virus infection and antagonizes IFN-β production in vitro (Fang et al., 2018). In addition, Zhang et al. generated a NS6-null recombinant PDCoV mutant by reverse genetics and showed that NS6 play a key role in promoting viral replication and pathogenesis (Zhang et al., 2020). However, whether the NS6 is expressed in target tissues of infected pigs was not assessed yet. This study fills this gap, confirming detection of NS6 protein in intestinal tissue of infected newborn piglets.
We further demonstrate that NS6 is incorporated into virus particles. First, we generated and validated the specificity of anti-NS6 antibodies and compared their reactivity with hyperimmune rabbit anti-PDCoV serum in WB analysis. The observed differences in band intensity indicate that NS6 is an integral, though minor component of PDCoV virions. This lower quantity may explain why detection of NS6 in purified virions required a higher concentration (1:200) of anti-NS6 peptide antibody. Both the IEM and IP results offered more visible evidence of NS6-specific antibodies reacting with PDCoV particles. However, as no putative transmembrane domain was predicted by bioinformatics software, the pattern or mechanism of NS6 protein assembly into virions remains for further study.
According to the previous reports, accessory proteins have been broadly implicated in cell proliferation, programmed cell death, inducing ER stress as well as antagonizing IFN-β production (Dedeurwaerder et al., 2014; Liu et al., 2014; Niemeyer et al., 2013; Yang et al., 2013). Although recent research has confirmed that ectopic expression of NS6 antagonizes IFN-β production (Fang et al., 2018), a more thorough and accurate functional analysis of the NS6 protein at the level of viral infection is needed. In this study, we also demonstrated that the anti-NS6 antibody does not have neutralizing activity against PDCoV, likely due to the NS6 proteins presenting in minor amounts within virions. Previously, we found that PDCoV infection induced significant changes in cell morphology, such as the presence of many cytoplasmic vesicles, including dilated rough ER autophagosome-like vesicles (Qin et al., 2019). Whether or not NS6 is involved in these cellular processes, and how it plays a role in PDCoV life cycle, are currently under investigation.
4. Materials and methods
4.1. Cells and virus
A porcine kidney epithelial cell line, LLC-PK1 (ATCC CL-101), was grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum and 1% (wt/vol) antibiotics (penicillin and streptomycin). The PDCoV Chinese “Hunan” strain (GenBank accession no. KY513724) was used (Pan et al., 2017; Wang et al., 2018; Yang et al., 2020a), which was propagated in LLC-PK1 cells with 5 μg/ml of trypsin at 37 °C with 5% CO2.
4.2. Virus purification
PDCoV was purified on sucrose gradients using a modification of the procedure described by Hofmann and Wyler for the purification of PEDV (Hofmann and Wyler, 1990). Briefly, PDCoV-infected LLC-PK1 cell cultures were harvested at 48 h post-infection (hpi), and intracellular virus was released by subjecting the cultures to three freeze-thaw cycles, followed by centrifugation at 4000×g for 30 min to remove coarse cell debris. Virions in the supernatant were then pelleted by high-speed centrifugation (105,000×g) at 4 °C for 2 h, and resuspended in TNE (20 mM Tris-HCl, pH 7.2, 100 mM NaCl, 2 mM EDTA) to 5% of the original volume. Virus preparations were layered onto two sucrose cushions (12 ml of 20% and 8 ml of 45%, prepared in TNE) and centrifuged at 146,000×g at 4 °C for 16 h. The resulting band between the two sucrose concentrations was collected with a syringe, and then used for analysis of virion proteins.
4.3. Expression of recombinant PDCoV NS6 protein in bacteria and generation of a mouse anti-NS6 polyclonal antibodies (pAb)
The putative NS6 gene was amplified from total RNAs extracted from PDCoV-infected LLC-PK1 cells by one-step RT-PCR and subsequently cloned into the pET-28a (+) vector (Novagen) with a polyhistidine tag at the carboxyl terminus between EcoRI and XhoI restriction sites. The recombinant plasmid was verified by Sanger sequencing and transformed into Rosetta (DE3) competent cells (Transgen biotech, China). After induction with 1 mM IPTG for 6 h, bacteria were collected by centrifugation (8000×g, 15 min) and lysed using the supersonic schizolysis method. The lysates were then centrifuged at 12,000×g for 15 min at 4 °C to collect supernatants and precipitates (inclusion bodies). The supernatants and precipitates were analyzed by SDS-PAGE. The resulting protein was subsequently purified using Ni-NTA His•Bind® Resin (Transgen biotech, China). Six-week-old female BALB/c mice were immunized with 20 μg of the purified NS6 protein in the complete Freund's adjuvant. The subsequent three immunizations were carried out at 3-week intervals with 20 μg of the same antigens mixed with the incomplete Freund's adjuvant. One week after the last immunization, anti-NS6 polyclonal antibodies were collected in the serum. Indirect enzyme-linked immunosorbent assay (ELISA), IFA and WB were used to determine the titer and specificity of the antibodies as described previously (Qin et al., 2017; Wang et al., 2018).
4.4. Generation of three rabbit pAbs specific to NS6, M or S subunit 1 (S1) of PDCoV
Two distinct short peptides predicted to suitable for immunization were selected from the deduced primary structure of PDCoV NS6 (acetyl-CNKRHIRREDVPEL-amide corresponding to aa 15–28, and acetyl-CIRPSLQVILEDELN-amide corresponding to aa 81–94). Two New Zealand white rabbits were immunized initially with 0.1 mg mixed peptides-keyhole limpet haemocyanin (KLH) conjugate in complete Freund's adjuvant. Booster injections with the same amount of the conjugate emulsified in incomplete Freund's adjuvant were given at three-week intervals, and sera were collected one week after the third booster injection. IFA and WB analysis were used to test the specificity of the anti-NS6-peptide pAb (anti-NS6-pt).
The PDCoV M protein antigenic peptide (acetyl-DTFHYTFKKPVESNNDPE-amide) that has been described previously (Chen et al., 2015) was used to produce anti-peptide pAb specific to M as mentioned above. The PDCoV S1 (aa 1–574) with a six-histidine tag was expressed by baculovirus system in SF9 insect cells as described previously (Wang et al., 2017). The purified proteins were used to immunize rabbits, and anti-S1 pAb were harvested at 55 days postimmunization.
4.5. Preparation of PDCoV hyperimmune serum
One New Zealand white rabbit was immunized four times with purified PDCoV particles, emulsified with Freund's complete adjuvant for the first injection and with Freund's incomplete adjuvant for the rest. Serum samples were collected periodically to measure the specific IgG by ELISA, and pre-immunization sera were collected for use as negative controls.
All of the generated pAbs or antisera described above are summarized in Table 1.
4.6. Immunofluorescence assay
PDCoV-infected cells were washed twice with phosphate-buffered saline (PBS) and fixed with acetone. Anti-N monoclonal antibody (Medgene Labs, USA; 1:1000 dilution), mouse anti-NS6 pAb (1:1000 dilution), rabbit anti-NS6-pt pAb (1:500 dilution), or pre-immune rabbit serum (1:1000 dilution), was added over the cells and incubated for 1 h at 37 °C. Cells were then washed thrice with PBS and Alexa Fluor 488- or 594-conjugated anti-rabbit or anti-mice IgG (Thermo Fisher Scientific) at a 1:1000 dilution was then added. After 30 min of incubation at 37 °C, the cells were washed thrice with PBS followed by 4′,6-diamidino-2-phenylindole (DAPI) staining, and were visualized under a fluorescence microscope.
4.7. SDS-PAGE and Western blot analysis
The purified PDCoV particles were resuspended in 1 × PBS and 5 × Protein Loading Buffer (Thermo Fisher Scientific, USA). Samples were separated by 12% SDS-PAGE. The proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane that was subsequently blocked with Tris-buffered saline (TBS) containing 3% bovine serum albumin (BSA) at room temperature for 2 h and then incubated overnight at 4 °C with a hyperimmune rabbit anti-PDCoV serum or an anti-NS6-pt antibody. The blots were then incubated with Goat horseradish peroxidase (HRP) conjugated anti-rabbit IgG (Thermo Fisher Scientific, USA).
For Western blot analysis, PDCoV-infected cells were lysed in lysis buffer (25 mM Tris-HCl, 200 mM NaCl, 10 mM NaF, 1 mM Na3VO4, 25 mM β-glycerophosphate, 1% NP40, and protease cocktail (Biotool, Houston, TX). Samples were resolved on SDS-PAGE and transferred onto polyvinylidene difluoride (PVDF) membrane that was subsequently blocked with Tris-buffered saline (TBS) containing 3% bovine serum albumin (BSA) overnight at 4 °C. Proteins were detected using the anti-N mAb or anti-M pAb at 1:1000 dilution followed by incubation with horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (1:5000 dilution; Thermo Fisher Scientific).
4.8. Immunoelectron microscopy (IEM)
Suspensions of purified PDCoV virions were placed onto nickel formvar-carbon coated grids (three grids per experiment) for approximately 10 min, followed by 1% bovine serum albumin (BSA) in 0.01 M PBS (pH 7.4) for 30 min. After, the samples were incubated with a mouse anti-NS6 antiserum for 30 min, or a pre-immune mouse serum as a negative control, with a rabbit anti-PDCoV hyperimmune serum and a rabbit anti-S1 antiserum as positive controls. All antibodies were diluted in 0.1% BSA and 0.05% Tween 20 in 0.01 M PBS (pH 7.4). The samples were then washed three times for 5 min per time with PBS and incubated with goat anti-mouse IgG conjugated to 10-nm gold particles (#G7652, Sigma) or goat anti-rabbit IgG conjugated to 10-nm gold particles (#G7402, Sigma). Grids were washed five times with PBS for 2 min per time, and then washed five times with Milli-Q water for 2 min per time, respectively. After the washing, samples were negatively stained with 3% uranyl acetate for 5 min and with lead citrate for 3 min and observed under electron microscopy.
4.9. Animal experiments and immunohistochemical staining
The animal experiment conducted in neonatal pigs was approved by the Experimental Animal Ethics Committee of Zhejiang University (no. ZJU20170026). Six, 3-day-old conventional piglets, shown to be RNA-negative for PEDV, PDCoV and TGEV in fecal samples, were divided into two groups (n = 3). Piglets in group An each received 2 ml of DMEM orally as negative controls, whereas piglets in groups B were each challenged orally with 2 ml of PDCoV at 1 × 106 plaque-forming units (PFU)/ml. At 3 days post-infection (dpi), all piglets were euthanized and jejunum and ileum were collected and fixed in 10% (v/v) phosphate-buffered formalin for IHC staining with mouse anti-NS6 antiserum. Rabbit anti-M antibodies prepared by our lab were used as a positive control.
4.10. Immunoprecipitation
Purified PDCoV particles, mock-infected, or PDCoV-infected LLC-PK1 cell were lysed in lysis buffer (25 mM Tris-HCl, 200 mM NaCl, 10 mM NaF, 1 mM Na3VO4, 25 mM β-glycerophosphate, 1% NP40) containing a protease inhibitor cocktail (Bimake, USA) and incubated on a shaker for 30 min at 4 °C. The lysates were then cleared by centrifugation at 14,000×g for 10 min. The supernatants were precipitated with anti-NS6-pt, rabbit IgG and anti-N mAb, respectively, and incubated with gentle rocking overnight at 4 °C. Protein A beads (Beyotime, China) washed with lysis buffer were added to supernatant fractions and incubated with gentle rocking for 2 h at 4 °C. After washing four times with lysis buffer, isolated immunoprecipitated proteins were boiled 10 min with PAGE sample loading buffer and then subjected to WB analysis with specific antibodies.
4.11. Virus neutralizing assay
The pre-immune mouse serum, the anti-NS6 mouse serum, and the hyperimmune anti-PDCoV serum were heat-inactivated for 30 min at 65 °C followed by 2-fold serial dilutions from 1:2 to 1:256 with DMEM, respectively. The diluted samples were subsequently mixed with 100 TCID50 (50% tissue culture infectious dose) of PDCoV and incubated for 60 min at 37 °C followed by inoculation and culture of LLC-PK1 cell monolayers per dilution for 48 h. Neutralization activity was evaluated by IFA using the anti-N antibody, and by comparison of viral RNA titers between different treatments using one-step quantitative reverse-transcription polymerase chain reaction (qRT-PCR) targeting the PDCoV M gene as described previously.
4.12. Predictions of post-translational modifications (PTMs) sites in NS6 protein
The serine, threonine, and tyrosine phosphorylation sites in NS6 were predicted by the NetPhos 2.0 server (http://www.cbs.dtu.dk/services/NetPhos/). The sites sumoylation were checked with the SUMOsp 2.0 program (http://sumosp.biocuckoo.org/). The potential glycosylation sites, including O-GalNAc (mucin-type) glycosylation (NetOGlyc), N-linked glycosylation (NetNGlyc), and C-mannosylation (NetCGlyc), were searched by the CBS (Center for Biological Sequence Analysis) server at the Technical University of Denmark (https://services.healthtech.dtu.dk/).
CRediT authorship contribution statement
Pan Qin: Methodology, Data curation, Funding acquisition, Writing - original draft. Wen-Ting Luo: Methodology, Investigation, Data curation. Quan Su: Investigation, Validation. Pengwei Zhao: Investigation, Validation. Yuqi Zhang: Investigation, Validation. Bin Wang: Investigation, Validation. Yong-Le Yang: Investigation, Validation. Yao-Wei Huang: Conceptualization, Funding acquisition, Supervision, Writing - review & editing.
Declaration of competing interest
There are no known conflicts of interest.
Acknowledgements
This work was funded by the National Key Research and Development Program of China (2016YFD0500102) and the National Natural Science Foundation for Young Scientists of China (31802205).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virol.2021.01.011.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Chen Q., Gauger P., Stafne M., Thomas J., Arruda P., Burrough E., Madson D., Brodie J., Magstadt D., Derscheid R., Welch M., Zhang J. Pathogenicity and pathogenesis of a United States porcine deltacoronavirus cell culture isolate in 5-day-old neonatal piglets. Virology. 2015;482:51–59. doi: 10.1016/j.virol.2015.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedeurwaerder A., Olyslaegers D.A., Desmarets L.M., Roukaerts I.D., Theuns S., Nauwynck H.J. ORF7-encoded accessory protein 7a of feline infectious peritonitis virus as a counteragent against IFN-alpha-induced antiviral response. J. Gen. Virol. 2014;95:393–402. doi: 10.1099/vir.0.058743-0. [DOI] [PubMed] [Google Scholar]
- Dong N., Fang L.R., Zeng S.L., Sun Q.Q., Chen H.C., Xiao S.B. Porcine deltacoronavirus in Mainland China. Emerg. Infect. Dis. 2015;21:2254–2255. doi: 10.3201/eid2112.150283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P., Fang L., Hong Y., Liu X., Dong N., Ma P., Bi J., Wang D., Xiao S. Discovery of a novel accessory protein NS7a encoded by porcine deltacoronavirus. J. Gen. Virol. 2017;98:173–178. doi: 10.1099/jgv.0.000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P., Fang L., Liu X., Hong Y., Wang Y., Dong N., Ma P., Bi J., Wang D., Xiao S. Identification and subcellular localization of porcine deltacoronavirus accessory protein NS6. Virology. 2016;499:170–177. doi: 10.1016/j.virol.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P., Fang L., Ren J., Hong Y., Liu X., Zhao Y., Wang D., Peng G., Xiao S. Porcine deltacoronavirus accessory protein NS6 antagonizes interferon beta production by interfering with the binding of RIG-I/MDA5 to double-stranded RNA. J. Virol. 2018;92 doi: 10.1128/JVI.00712-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godeke G.J., de Haan C.A., Rossen J.W., Vennema H., Rottier P.J. Assembly of spikes into coronavirus particles is mediated by the carboxy-terminal domain of the spike protein. J. Virol. 2000;74:1566–1571. doi: 10.1128/jvi.74.3.1566-1571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M., Wyler R. Enzyme-linked immunosorbent assay for the detection of porcine epidemic diarrhea coronavirus antibodies in swine sera. Vet. Microbiol. 1990;21:263–273. doi: 10.1016/0378-1135(90)90037-V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Ito N., Tseng C.T., Makino S. Severe acute respiratory syndrome coronavirus 7a accessory protein is a viral structural protein. J. Virol. 2006;80:7287–7294. doi: 10.1128/JVI.00414-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Hu H., Eyerly B., Lu Z.Y., Chepngeno J., Saif L.J. Pathogenicity of 2 porcine deltacoronavirus strains in gnotobiotic pigs. Emerg. Infect. Dis. 2015;21:650–654. doi: 10.3201/eid2104.141859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Hu H., Saif L.J. Porcine deltacoronavirus infection: etiology, cell culture for virus isolation and propagation, molecular epidemiology and pathogenesis. Virus Res. 2016;226:50–59. doi: 10.1016/j.virusres.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Hulswit R.J.G., Kenney S.P., Widjaja I., Jung K., Alhamo M.A., van Dieren B., van Kuppeveld F.J.M., Saif L.J., Bosch B.J. Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E5135–e5143. doi: 10.1073/pnas.1802879115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Fung T.S., Chong K.K., Shukla A., Hilgenfeld R. Accessory proteins of SARS-CoV and other coronaviruses. Antivir. Res. 2014;109:97–109. doi: 10.1016/j.antiviral.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Yuan Q., Liao Y. Coronavirus envelope protein: a small membrane protein with multiple functions. Cell. Mol. Life Sci.: CMLS. 2007;64:2043–2048. doi: 10.1007/s00018-007-7103-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Zhang Y., Liang X., Lou F., Oglesbee M., Krakowka S., Li J. Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. mBio. 2015;6 doi: 10.1128/mBio.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemeyer D., Zillinger T., Muth D., Zielecki F., Horvath G., Suliman T., Barchet W., Weber F., Drosten C., Muller M.A. Middle East respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J. Virol. 2013;87:12489–12495. doi: 10.1128/JVI.01845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Tian X., Qin P., Wang B., Zhao P., Yang Y.L., Wang L., Wang D., Song Y., Zhang X., Huang Y.W. Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China. Vet. Microbiol. 2017;211:15–21. doi: 10.1016/j.vetmic.2017.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin P., Du E.Z., Luo W.T., Yang Y.L., Zhang Y.Q., Wang B., Huang Y.W. Characteristics of the life cycle of porcine deltacoronavirus (PDCoV) in vitro: replication kinetics, cellular ultrastructure and virion morphology, and evidence of inducing autophagy. Viruses. 2019;11:455. doi: 10.3390/v11050455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin P., Li H., Wang J.W., Wang B., Xie R.H., Xu H., Zhao L.Y., Li L., Pan Y., Song Y., Huang Y.W. Genetic and pathogenic characterization of a novel reassortant mammalian orthoreovirus 3 (MRV3) from a diarrheic piglet and seroepidemiological survey of MRV3 in diarrheic pigs from east China. Vet. Microbiol. 2017;208:126–136. doi: 10.1016/j.vetmic.2017.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaecher S.R., Mackenzie J.M., Pekosz A. The ORF7b protein of severe acute respiratory syndrome coronavirus (SARS-CoV) is expressed in virus-infected cells and incorporated into SARS-CoV particles. J. Virol. 2007;81:718–731. doi: 10.1128/JVI.01691-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Liu Y., Ji C.M., Yang Y.L., Liang Q.Z., Zhao P., Xu L.D., Lei X.M., Luo W.T., Qin P., Zhou J., Huang Y.W. Porcine deltacoronavirus engages the transmissible gastroenteritis virus functional receptor porcine aminopeptidase N for infectious cellular entry. J. Virol. 2018;92 doi: 10.1128/JVI.00318-18. e00318-00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Lei X., Qin P., Zhao P., Wang B., Wang Y., Li Y., Jin H., Li L., Huang Y.W. [Development and application of real-time RT-PCR and S1 protein-based indirect ELISA for porcine deltacoronavirus] Sheng Wu Gong Cheng Xue Bao. 2017;33:1265–1275. doi: 10.13345/j.cjb.170119. [DOI] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA. Emerg. Infect. Dis. 2014;20:1227–1230. doi: 10.3201/eid2007.140296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.W., Yue H., Fang W., Huang Y.W. Complete genome sequence of porcine deltacoronavirus strain CH/Sichuan/S27/2012 from Mainland China. Genome Announc. 2015;3 doi: 10.1128/genomeA.00945-15. e00945–00915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Zheng B.J., Zeng R., Lu W., Lin Y.P., Xue L., Li L., Yang L.L., Xu C., Dai J., Wang F., Li Q., Dong Q.X., Yang R.F., Wu J.R., Sun B. Severe acute respiratory syndrome coronavirus accessory protein 9b is a virion-associated protein. Virology. 2009;388:279–285. doi: 10.1016/j.virol.2009.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Zhang L., Geng H., Deng Y., Huang B., Guo Y., Zhao Z., Tan W. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell. 2013;4:951–961. doi: 10.1007/s13238-013-3096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.L., Liang Q.Z., Xu S.Y., Mazing E., Xu G.H., Peng L., Qin P., Wang B., Huang Y.W. Characterization of a novel bat-HKU2-like swine enteric alphacoronavirus (SeACoV) infection in cultured cells and development of a SeACoV infectious clone. Virology. 2019;536:110–118. doi: 10.1016/j.virol.2019.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.L., Meng F., Qin P., Herrler G., Huang Y.W., Tang Y.D. Trypsin promotes porcine deltacoronavirus mediating cell-to-cell fusion in a cell type-dependent manner. Emerg. Microb. Infect. 2020;9:457–468. doi: 10.1080/22221751.2020.1730245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.L., Qin P., Wang B., Liu Y., Xu G.H., Peng L., Zhou J., Zhu S.J., Huang Y.W. Broad cross-species infection of cultured cells by bat HKU2-related swine acute diarrhea syndrome coronavirus and identification of its replication in murine dendritic cells in vivo highlight its potential for diverse interspecies transmission. J. Virol. 2019;93 doi: 10.1128/JVI.01448-19. e01448-01419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.L., Yu J.Q., Huang Y.W. Swine enteric alphacoronavirus (swine acute diarrhea syndrome coronavirus): an update three years after its discovery. Virus Res. 2020;285:198024. doi: 10.1016/j.virusres.2020.198024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhang M., Li W., Zhou P., Liu D., Luo R., Jongkaewwattana A., He Q. Genetic manipulation of porcine deltacoronavirus reveals insights into NS6 and NS7 functions: a novel strategy for vaccine design. Emerg. Microb. Infect. 2020;9:20–31. doi: 10.1080/22221751.2019.1701391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong N.S., Zheng B.J., Li Y.M., Poon, Xie Z.H., Chan K.H., Li P.H., Tan S.Y., Chang Q., Xie J.P., Liu X.Q., Xu J., Li D.X., Yuen K.Y., Peiris, Guan Y. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China. Lancet (Lond., Engl.) 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. February, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.