Abstract
Ethnopharmacological relevance
Reduning injection (RDNI) is a patented Traditional Chinese medicine that contains three Chinese herbal medicines, respectively are the dry aboveground part of Artemisia annua L., the flower of Lonicera japonica Thunb., and the fruit Gardenia jasminoides J.Ellis. RDNI has been recommended for treating Coronavirus Disease 2019 (COVID-19) in the “New Coronavirus Pneumonia Diagnosis and Treatment Plan".
Aim of the study
To elucidate and verify the underlying mechanisms of RDNI for the treatment of COVID-19.
Methods
This study firstly performed anti-SARS-CoV-2 experiments in Vero E6 cells. Then, network pharmacology combined with molecular docking was adopted to explore the potential mechanisms of RDNI in the treatment for COVID-19. After that, western blot and a cytokine chip were used to validate the predictive results.
Results
We concluded that half toxic concentration of drug CC50 (dilution ratio) = 1:1280, CC50 = 2.031 mg crude drugs/mL (0.047 mg solid content/mL) and half effective concentration of drug (EC50) (diluted multiples) = 1:25140.3, EC50 = 103.420 μg crude drugs/mL (2.405 μg solid content/mL). We found that RDNI can mainly regulate targets like carbonic anhydrases (CAs), matrix metallopeptidases (MMPs) and pathways like PI3K/AKT, MAPK, Forkhead box O s and T cell receptor signaling pathways to reduce lung damage. We verified that RDNI could effectively inhibit the overexpression of MAPKs, PKC and p65 nuclear factor-κB. The injection could also affect cytokine levels, reduce inflammation and display antipyretic activity.
Conclusion
RDNI can regulate ACE2, Mpro and PLP in COVID-19. The underlying mechanisms of RDNI in the treatment for COVID-19 may be related to the modulation of the cytokine levels and inflammation and its antipyretic activity by regulating the expression of MAPKs, PKC and p65 nuclear factor NF-κB.
Keywords: Reduning injection, COVID-19, Network pharmacology, SARS-CoV-2, Molecular docking, Western blot
Graphical abstract
1. Introduction
In December 2019, a coronavirus that is capable of human-to-human transmission was discovered and spread rapidly around the globe, thereby leading to a pandemic. Globally, as of 9:28am CET on 21 November 2020, the pathogenic SARS-coronavirus 2 (SARS-CoV-2) has caused at least 56,982,476 confirmed cases, including 1,361,847 deaths (Word Health Organization, 2020). Compared with severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS), Coronavirus Disease 2019 (COVID-19) is a beta coronavirus that affects the lower respiratory tract and manifests as pneumonia in humans (Sohrabi et al., 2020). The complications of this disease include acute respiratory distress syndrome (ARDS), RNA anemia, acute cardiac injury and secondary infection. Around 30% of the patients were admitted to an intensive care unit (ICU) and 15% were died (Huang et al., 2020). ARDS is the leading cause of death from COVID-19, and may cause similar immune-pathogenic characteristics compared to SARS-CoV and MERS-CoV infection (Huang et al., 2020). Moreover, cytokines were found to play a key role in the emergence of ARDS and are central to the development of inflammation (Coperchini et al., 2020). Genome studies found that coronavirus is evolutionarily related (80% identity) to the beta-coronavirus implicated in the SARS (Chen et al., 2020). Three proteins that can be built on homologous templates are considered targets for drug design, including papain-like protease (PLP), 3-chymotrypsin-like protease (3CLpro, aka main protease, Mpro) and angiotensin-converting enzyme-2 (ACE2) (Dong et al., 2020.). These first two proteins play a key role in human infection. Replication of the virus initiates the binding of the spike (S) glycoprotein to a host cell receptor and the release of the positive-sense RNA into the cytoplasm of the cells. The large viral genomic RNA is translated to produce two long polyproteins, pp1a and pp1ab, which are processed by viral proteases, including PLPs (PLP1 and/or PLP2) and Mpro into nonstructural proteins (Deng et al., 2019). The ACE2 gene encodes the ACE2 protein, which has been known to be a receptor for SARS-CoV (Zhou et al., 2020; Cao et al., 2020). A study found that 2019-nCoV S protein binds to ACE2 with higher affinity than SARS-CoV S protein (Wrapp et al., 2020).
COVID-19 has caused huge losses and led to great health and economic burdens to the world. However, no vaccines and therapeutics have been proved clinically effective for the treatment of severe COVID-19 (Dai et al., 2020). The combination of α-interferon and the anti-HIV drugs Lopinavir/Ritonavir (Kaletra®) has been used, but the effect is limited and it may induce side effects (Cao et al., 2020). Although recent studies have reported the significant efficacy of Remdesivir, until now, no drug has been shown to reduce mortality or length of hospital stay (Pan et al., 2020; Sanders et al., 2020; Ferner and Aronson, 2020). Thus, effective and safe drugs against COVID-19 are urgently needed.
Thousand-of-year's experiences have been accumulated for the use of Traditional Chinese medicine (TCM) in the treatment of pandemic and endemic diseases (Yang et al., 2020). Since 2003, TCM has been utilized to fight against SARS, H1N1, H7N9, MERS and other viral diseases (Luo et al., 2019). TCM has also been recommended for the treatment of COVID-19 in the “New Coronavirus Pneumonia Diagnosis and Treatment Plan” (Trial Version One to Seven) (General office of the national health commission, 2020), and over 91.5% of infected patients in China were receiving TCM treatment. Meanwhile, observation results showed that the total effective rate was more than 90% (Wan et al. (2020); Zhang et al. (2020)). Reduning injection (RDNI) is one of the patented TCM recommendation in the “New Coronavirus Pneumonia Diagnosis and Treatment Plan” (Trial Version Six and Seven), and it is especially recommended for severe cases. It contains three Chinese herbal medicines: the dry aboveground part of Artemisia annua L., the flower of Lonicera japonica Thunb., and the fruit Gardenia jasminoides J.Ellis (Geng et al., 2015). The ratio of the three drugs is 25:15:12. In the prescription of RDNI, Artemisia annua L. is the sovereign drug with the functions of clearing heat and cooling blood, relaxing surface tissue; Lonicera japonica Thunb. is good at clearing heat, removing toxicity and dispersing exterior evils. As the minister drug, Lonicera japonica Thunb. assists Artemisiae annuae enhancing the effect of clearing heat and dispersing; Gardenia jasminoides J.Ellis plays the part of assistant drug, and it has the effects of detoxification, heat-clearing, cooling blood and relieving restlessness by lowering the triple energizer's fire of heart, lung, stomach, helping minister drug Lonicera japonica Thunb. to clear heat and detoxify. The combination of all these medicines can not only disperse the wind-heat evil through the surface, but also clear heat-toxicity from the inside, so as to play the function of clearing heat and detoxifying, expelling wind, and relieving the exterior together. RDNI possesses multiple functions such as dispelling wind, clearing heat, and detoxification and has been widely used in China for the treatment of fever, viral infection, allergy and inflammation (Wang et al., 2016; Gao et al., 2020). For the syndrome of COVID-19, RDNI may have an antipyretic effect and reduce the symptoms.
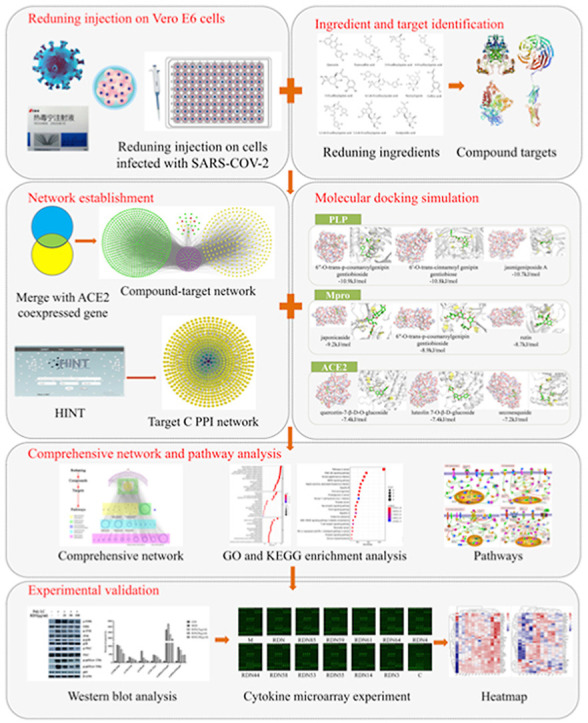
To elucidate the underlying mechanisms of RDNI for the treatment of COVID-19, this study performed anti-SARS-CoV-2 experiments in Vero E6 cells., Moreover, network pharmacology combined with molecular docking was adopted to explore the pharmacological mechanisms of RDNI against COVID-19 and to pick out the potential active compounds that showed a strong binding affinity with PLP, Mpro and ACE2. In order to verify the results from network pharmacology, we also used western blot and a cytokine chip to detect the effects of RDNI on Poly:IC-induced cytokine secretion in BEAS-2B cells.
2. Methods
2.1. Anti- SARS-CoV-2 in Vero E6 cells
2.1.1. Materials
RDNI (size:10 mL/tubule, Batch No. 191104) was obtained from Jiangsu Kanion Pharmaceutical Co., Ltd., China, the concentration used was 2.6 g crude drug/mL (60.45 mg solid/mL). Vero E6 cells were used in this experiment, which were preserved by the pathogen Center of Institute of Laboratory Animal Science (ILAS), Chinese Academy of Medical Sciences. The virus used was SARS-CoV-2, the titer of the stocks was 105. Fifty percent tissue culture infectious doses (TCID 50/mL), and was stored at −80 °C at the Pathogen Center of Institute of Laboratory Animal Science (ILAS), Chinese Academy of Medical Sciences. A virus titer of 100 TCID50/mL was used. All the above procedures were completed at the biosafety level-3 laboratory (BSL-3) laboratory.
2.1.2. Cytotoxicity test
Using a sterile 96-well culture plate, 100 μL of VeroE6 cells at a concentration of 5 × 104 cells/mL were seeded onto each empty well and cultured for 24 h at 37 °C with 5% CO2.Then the drug was diluted in DMEM medium (containing 2% dioxin and 16 μg/ml trypsin) with 1:20, 1:40, 1:80, …and 1:2560 total 8 dilutions. The cell culture medium in the 96-well culture plate was discarded, diluted drugs were added (100μL/well), replicates of wells were used, then each well was added with 100 μL blank DMEM medium (containing 2% double-antibody and 16 μg/mL trypsin). The control group was treated with 200 μL DMEM medium (containing 2% double-antibody and 16 μg/mL trypsin) The cells were incubated at 37 °C in a 5% CO2 incubator for 4–5 days. The cytopathic effect (CPE) was observed under the light microscope. The change of CPE in the cells was recorded as “+“, and no change of CPE was recorded as “-“.
2.1.3. Antiviral test of drugs
Using a sterile 96-well culture plate, 100 μL of Vero E6 cells at a concentration of 5 × 104 cells/mL was seeded onto each empty well and cultured for 24 h at 37 °C with 5% CO2. The test drugs were diluted for 8 concentrations (100 μL/well), 4 replicates of wells were used for each concentration, and then an equal volume (100 μL/well) of 100 TCID50 virus was added to each well, followed by incubation in a 5% CO2 incubator at 37 °C for 1 h. Meanwhile, the control group was treated with 200 μL of DMEM medium (containing 2% antibody and 16 μL/mL trypsin) per well, and the model group was treated with 100 μL virus solution containing 100 TCID50 per well at 37 °C for 1 h. The supernatant was aspirated after 1 h. 200 μL/well DMEM medium containing 2% antibody and 16 μL/mL trypsin was added to the cell wells. The cells were then incubated at 37 °C in a 5% CO2 incubator for 4–5 days. The CPE was observed under the light microscope. The change of CPE in the cells was recorded as "+", and no change of CPE was recorded as “-“.
2.1.4. Efficacy evaluation indicators
Selection index (SI) is the ratio of 50% toxic concentration of the drug to the 50% inhibitory concentration of the virus (CC50/EC50), or the ratio of the maximum non-toxic concentration of the drug to the minimum effective concentration of the virus (CC0/EC0).
2.2. Network pharmacology and molecular docking
2.2.1. Data collection
After combining the injection ingredients found from the literatures (Li et al., 2015; Yang et al., 2014), Chemoffice (www.cambridgesoft.com) software was used to obtain simplified molecular input line entry specification (SMILES) or three-dimensional chemical structure files of the chemical composition in RDNI. The details of the ingredients were list in Supplementary file 1. they were then imported into SuperPred (Nickel et al., 2014) (http://prediction.charite.de), SwissTargetPrediction (Gfeller et al., 2014) (http://swisstargetprediction.ch/) and STITCH (Szklarczyk et al., 2016) (http://stitch.embl.de/) to collect known or predicted targets of the compounds. To ensure the accuracy and comprehensiveness of target screening, we set the probability filter of SwissTargetPrediction to above zero, while “minimum required interaction score” was set at higher confidency (0.700) and “max number of interactors to show” was set at no more than 50 interactors. In the attachment of the literature report (Wang et al., 2020), single-cell sequencing of colonic epithelial cells identified 5556 genes that are co-expressed with ACE2. Cytoscape software (Shannon et al., 2003) (version 3.7.1, http://cytoscape.org/) was used to take the intersection to acquire the genes that are co-expressed with ACE2 for the targets of RDNI.
2.2.2. Targets network establishment and analysis
The collected targets were sorted and imported into Cytoscape to create a network to visualize the pharmacological mechanisms. We downloaded all the human high-quality binary protein-protein interactions from the high-quality proteomics database HINT (High-quality INTeractomes, last updated in April 2019) (Das and Yu (2012)), which was applied to construct the human genome-wide protein-protein interaction (PPI) network. All the targets of RDNI were mapped onto the HINT network. Compound targets and the edges between them were extracted, and only the largest connected branch was retained to build the target network.
2.2.3. Biological functional and pathway enrichment analysis
The DAVID (Kanehisa and Sato (2020)) platform (https://david.ncifcrf.gov/, Version 6.8) was employed for disease cluster analysis (functional annotation clustering), the species and background were set to " homo sapiens ". Subsequently, gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyze of important targets were conducted by the clusterProfiler package (Burley et al., 2017) in R (version 3.6.1, https://www.r-project.org/). The pathways were then classified through the KEGG BRITE database (https://www.genome.jp/kegg/brite.html, last updated: April 1, 2020). After that, the Pathway Builder Tool (http://www.proteinlounge.com/PathwayBuilder.aspx) was used to draw the path diagram.
2.2.4. Molecular docking
Firstly, we constructed the 3D structure of the compounds and saved them in * mol2 format with the energy minimized through ChemOffice software. Next, 3D structures’ * PDB format of ACE2 (PDB ID: 2AJF), PLP (PDB ID: 4OVZ), Mpro (PDB ID: 6LU7) were download from the PDB data (Yu et al., 2012) (https://www.rcsb.org/), and PyMOL (http://www.PyMOL.org) software was selected to perform dehydration and hydrogenation of proteins. Moreover, the AutoDock (http://autodock.scripps.edu/) software was used to convert the compounds and target proteins format to * pdbqt format. We set the active pocket site at the original inhibitor of the protein. Finally, Vina was used for molecular docking. If the binding energy is less than 0, it indicates that the ligand and the receptor can spontaneously bind. The compound with the lowest binding free energy can provide new drug design models for further experiments. The heatmap package in R was also used to draw the heatmap of molecular docking.
2.3. In vitro experimental verification
2.3.1. Western blot analysis
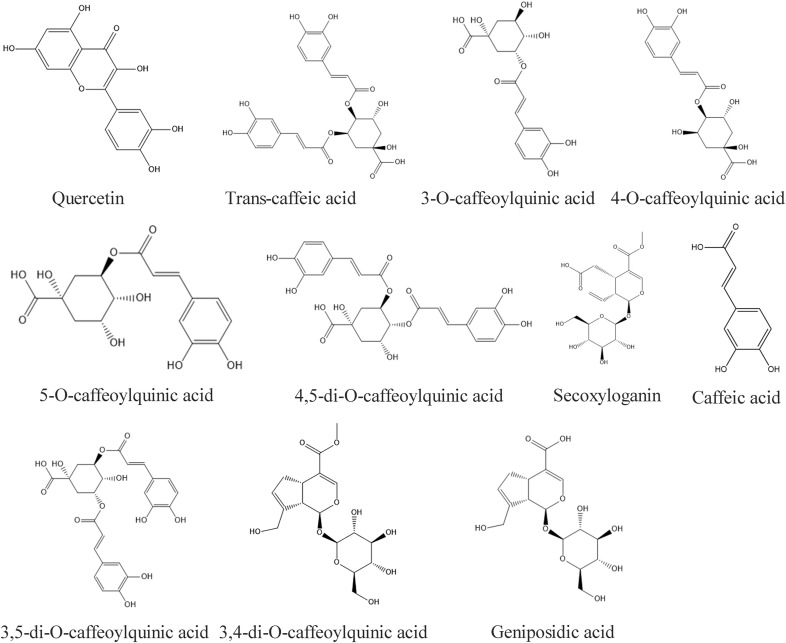
RDNI (Batch No: 130331, 10 mL/branch), 3-O-caffeoylquinic acid, 4-O-caffeoylquinic acid, 3,4-di-O-caffeoylquinic acid, trans-caffeic acid, geniposide, quercetin, 5-O-caffeoylquinic acid, 3,5-di-O-caffeoylquinic acid, 4,5-di-O-caffeoylquinic acid, geniposidic acid, secoxyloganin and secoxyloganin were obtained from Kanion Pharmaceutical Co., Ltd (Jiangsu, China). Each monomer was added at a concentration of 30, 60 and 120 μM, while the concentrations of RDNI were 25, 50 and 100 μg/mL. Antibodies including anti-β-actin (IPW0007), anti-extracellular signal-regulated kinase (ERK) 1/2 (IPW1497), anti-p-ERK1/2 (IPW1634), anti-c-Jun-NH2-terminal kinase (JNK) (IPW0737), anti-p-JNK (IPW0736), anti-protein kinase C (PKC) (IPW0725), anti-p-PKC (IPW0724), anti-nuclear factor-κB (NF-κB)-p65 (IPW1629), anti-p–NF–κB-p65 (ser276) (IPW0781), anti-p–NF–κB-p65 (ser536) (IPW1630) were purchased from Nanjing Pathway Biological Technology Co., Ltd (Nanjing, China). Anti-p38 (KGYT3514) and anti-p-p38 (KG11253) were obtained from Nanjing KeyGen Biotech Co., Ltd (Nanjing, China). While the secondary antibodies were purchased from Jackson Co., Ltd (Lancaster, Pennsylvania).
BEAS-2B cells were purchased from Shanghai Bioleaf Biotech Co., Ltd (Shanghai, China). PolyI:C was purchased from Sigma. The cells were cultured in tissue culture flasks, at 37 °C with 5% CO2. The culture medium was discarded when 90% of them were fused. These cells were washed twice with PBS and were digested with 0.25% trypsin-EDTA solution. After centrifugation at 800 rpm for 4 min, the supernatant was discarded. 3 mL of complete medium was added to resuspend the cell pellet, and 1 mL was inoculated in a tissue culture flask at a density of 1 × 106 cells/mL. 5 mL complete culture medium was added, and the cells were cultured at 37 °C with 5% CO2. After 60%–70% of them were fused, the culture medium was discarded and the cells were washed twice with PBS.
The cells were divided into 3 groups (blank, model and sample groups). The original medium in the culture flask was discarded. 2 mL serum-free medium was added to the cells for the blank group, and the cells were then incubated for 24 h. 50 μg/mL PolyI:C serum-free medium was added to the model and sample groups. After incubating for 60 min, the samples were collected for western blot analysis.
Total proteins were extracted using Tissue or Cell Total Protein Extraction Kit (KGP2100, Nanjing KeyGen Biotech, Nanjing, China). The protein concentration was determined using Lowry Protein Assay Kit (KGP4200, Nanjing KeyGen Biotech, Nanjing, China). Then the proteins were separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), with PageRuler™ Prestained Protein Ladder (26616, Thermo Fisher Scientific, Shanghai, China) as the pre-stained marker and 50 μg per well as the sample volume. Next, the proteins were transferred to polyvinylidene difluoride (PVDF) membranes after being blocking in blocking solution (105, Nanjing Lufei Biological Technology, Nanjing, China). Then, the membranes were incubated with primary antibodies β-actin (1:1000 dilution), ERK1/2 (1:1000 dilution), p-ERK1/2 (1:1000 dilution), JNK (1:1000 dilution), p-JNK (1:2000 dilution), PKC (1:1000 dilution), p-PKC (1:1000 dilution), NF-κB-p65 (1:1000 dilution), p–NF–κB-p65 (ser276) (1:1000 dilution), and p–NF–κB-p65 (ser536) (1:1000 dilution) at 4 °C overnight. Afterwards, they were incubated with the HRP-conjugated goat anti-mouse or anti-rabbit secondary antibodies. Finally, the protein bands were visualized by ECL reagent (Nanjing Lufei Biological Technology, Nanjing, China) with ChemiDoc™ Imaging System (Bio-Rad). The densitometric analyses of the immunoreactive bands were quantified by Image J software.
2.3.2. Cytokine microarray experiment
The QAH–INF–3 chip purchased from Raybiotech (Georgia, America) was used to detect the effect of RDNI on PolyI:C-induced cytokine secretion in BEAS-2B cells. In this study, blank, model and sample groups were used, each group had 3 replicate wells. Before the formal experiment, the medium was discarded and serum-free medium was added to starve the cells for 1 h. According to the needs of the experiment, 200 μL of the test drug solution was prepared and administered with serum-free medium (the monomer compound concentration was 60 μM and the compound concentration was 50 μg/mL). At the same time, 200 μL serum-free blank medium was added to the blank and model groups. After 1 h of drug treatment, a certain amount of PolyI:C mother liquor was added to the model and sample groups to make the final concentration of PolyI:C to be 50 μg/mL, and the incubation time was 24 h. After treatment, the supernatant was collected and centrifuged at 4 °C to take 100 μL for chip detection. The Raybiotech kit containing Blocking Buffer, Cytokine Standard Mix, Wash Buffer, Detection Antibody (Biotin-conjugated Anti-Cytokines), Sample Diluent and Cy3 equivalent dye-conjugated streptavidin was used for experimental samples, which was followed by Raybiotech's standard operating procedures including sample preparation, chip hybridization, washing and detection. The Axon Genepix chip scanner (GenePix 4000B, Axon Instruments, USA) was used to scan the cytokine chip.
3. Result
3.1. Anti- SARS-CoV-2 in vitro experiment
3.1.1. Cytotoxicity test of the subject drugs
The drug was diluted with eight different concentrations, and the results were shown in Table 1 . The results showed that dilution of 1:640 (4.063 crude drugs mg/mL, 0.094 mg solid content/mL) and above dilutions were toxic to the cells. When the dilution was 1:1280 (2.031 mg crude drugs/mL, 0.047 mg solid content/mL), 2 of the 4 compound pores were toxic to the cells. According to the calculation formula of CC50, CC50 (dilution ratio) = 1:1280, CC50 = 2.031 mg crude drugs/mL (0.047 mg solid content/mL).
Table 1.
The measurement of toxicity of the drugs.
| Subject's drugs | Dilution ratio | Crude drug concentration (μg/mL) | Solid content concentration (mg/mL) | Results(4 compound pores) | |||
|---|---|---|---|---|---|---|---|
| RDNI | 20 | 130.000 | 3.023 | + | + | + | + |
| 40 | 65.000 | 1.511 | + | + | + | + | |
| 80 | 32.500 | 0.756 | + | + | + | + | |
| 160 | 16.250 | 0.378 | + | + | + | + | |
| 320 | 8.125 | 0.189 | + | + | + | + | |
| 640 | 4.063 | 0.094 | + | + | + | + | |
| 1280 | 2.031 | 0.047 | + | + | – | – | |
| 2560 | 1.016 | 0.024 | – | – | – | – | |
| Control | 200 μL DMEM | – | – | – | – | ||
“+" means that the cells have CPE changes, "-" means that the cells have no CPE changes or was in normal cell morphology.
3.1.2. Antiviral experiment of the subject drugs
The drug was diluted with eight different concentrations, and the results showed that the drugs that were in 1:10000 dilution (260.000 μg crude drugs/mL, 6.045 μg solid content/mL) or higher concentration had inhibition effects on the virus. When the dilution ratio was 1:20,000 (130.000 μg crude drugs/mL, 3.023 μg solid content/mL), 3 of the 4 compound pores had inhibition effects on the virus, as shown in Table 2 . According to the formula of drug toxicity EC50, we concluded that EC50 (diluted multiples) = 1:25140.3, EC50 = 103.420 μg crude drugs/mL (2.405 μg solid content/mL).
Table 2.
Effects of drug against SARS-COV-2.
| Subjects drugs | Dilution ratio | Crude drug concentration (μg/mL) | Solid content concentration (μg/mL) | Results (4 compound pores) | |||
|---|---|---|---|---|---|---|---|
| RDNI | 2500 | 1040.000 | 24.180 | – | – | – | – |
| 5000 | 520.000 | 12.090 | – | – | – | – | |
| 10000 | 260.000 | 6.045 | – | – | – | – | |
| 20000 | 130.000 | 3.023 | – | – | – | + | |
| 40000 | 65.000 | 1.511 | + | + | + | + | |
| 80000 | 32.500 | 0.756 | + | + | + | + | |
| 160000 | 16.250 | 0.378 | + | + | + | + | |
| 320000 | 8.125 | 0.189 | + | + | + | + | |
| Control | 200 μL DMEM | – | – | – | – | ||
| Model | 100TCID50 | + | + | + | + | ||
“+" means that the cells have CPE changes, "-" means that the cells have no CPE changes or was in normal cell morphology, and "/" refers to cytotoxicity.
3.2. Network pharmacology and molecular docking
3.2.1. Compound-target network
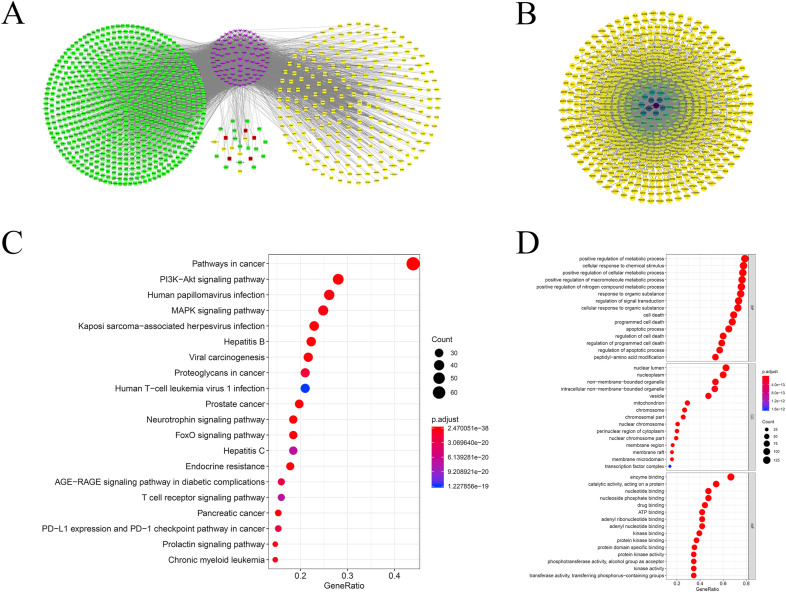
106 compounds that were mainly identified from two papers were adopted for analysis (Li et al., 2015; Yang et al., 2014). In a study from Li et al. (Li et al., 2015), compounds were obtained by ultra-performance liquid chromatography that were coupled to electrospray ionization tandem quadrupole time-of-flight mass spectrometry (UPLC-ESI-Q/TOF-MS) with the MSE (Edenotes collision energy) data acquisition mode. Yang et al. (Yang et al., 2014) identified compounds with good absorption, distribution, metabolism, and excretion (ADME). The structures of the representative components were shown in Fig. 1 . In the compound-target network (Fig. 2 A), 1069 compounds are connected by 4743 edges, including 93 compounds and 976 targets. 26 targets were related to the cytokine storm, while 5 of them were related to fever. The top three targets with the highest degrees were CA2, CA12 and CA1. In this network, 251 targets are co-expressed with ACE2. These results indicated that RDNI could potentially improve the symptoms caused by the disorder of ACE2 expression disorder that is associated with 2019-nCoV infection.
Fig. 1.
The representative components of Reduning injection.
Fig. 2.
Results of network pharmacology. (A) Compound-target gene interactions for RDNI. The purple nodes represent the compounds, the yellow nodes represent the targets co-expressed with ACE2, the red nodes represent the targets that are related to fever, and the green nodes represent the other targets. All the nodes were sorted by degree. The color round represents the targets of the cytokine storm. The radius of the point is proportional to the degree. More details of the network can be found in Supplementary file 2. (B) PPI network. All the nodes represent the targets of RDNI. The edges represent the interaction between targets. The higher the degree, the darker the color. More information on the network was list in Supplementary file 3. (C)Bubble charts of KEGG enrichment analysis. The dot size is proportional to the number of genes, and the color of the dot represents the p-value of each enrichment pathway. (D) Bubble charts of GO enrichment analysis. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2.2. PPI network of the targets
The PPI network consisted of a total of 617 nodes that were linked through 2480 edges (Fig. 2B). The average degree value of the nodes in the network was 8.03, and the target with the highest degree was HSP90AB1 (degree = 109). The degree values of a total of 167 targets were higher than the average degree value of these targets.
3.2.3. GO and KEGG analyze
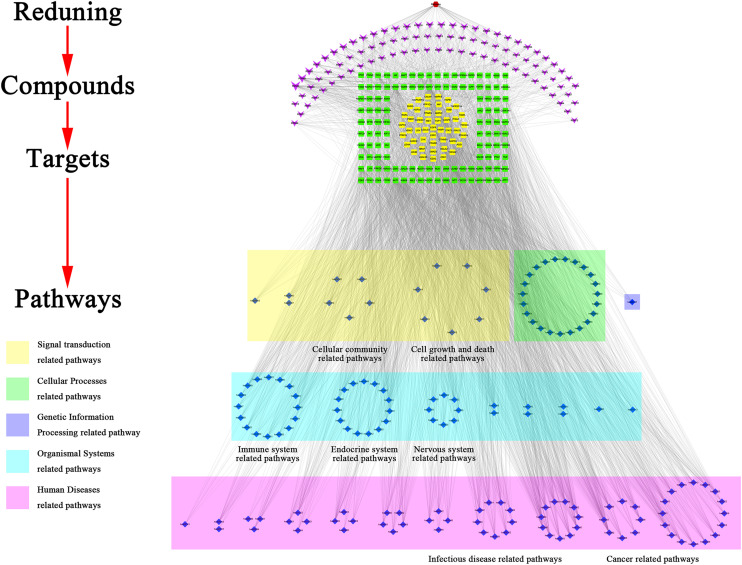
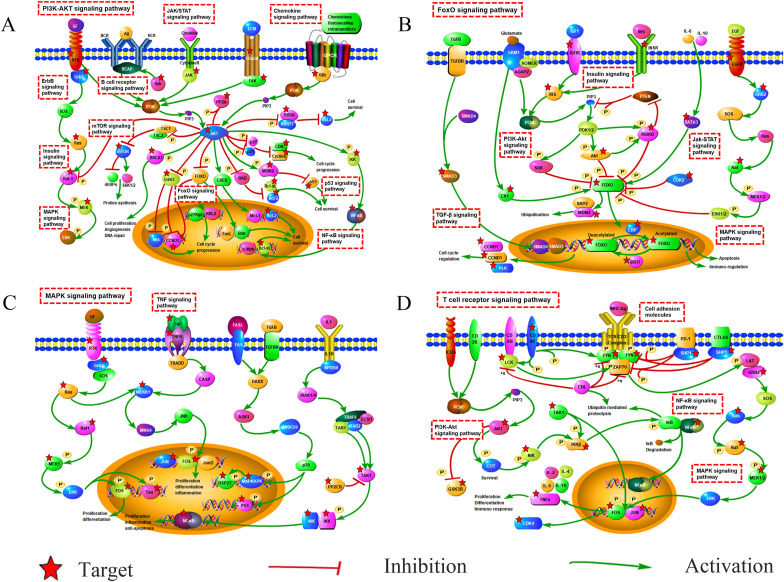
We conducted GO and KEGG enrichment analysis and disease functional clustering analysis on 167 targets was conducted with the degree value above average. A total of 1491 items with FDR<1*10−6 was obtained by enrichment of GO enrichment analysis. Including 1309 biological processes (BP) entries, 103 molecular functions (MF) entries and 79 cellular components (CC) entries. KEGG functional enrichment analysis contained 113 items with FDR<1*10−6, including 18 signal transduction pathways, 12 immune system pathways, 6 cell growth and death related pathways and 10 viral infectious disease pathways. The Enrichment results of KEGG and GO were shown in Fig. 2C and D. From the result of disease clustering analysis, the cluster with the highest enrichment levels (enrichment score: 21.30) contained 7 items, the first and third of them were lung cancer items and the fourth was related to a chronic obstructive pulmonary disease. The relationship between targets and pathways was shown in Fig. 3 . The pathway mechanism diagrams were shown in Fig. 4 . More details of the enrichment result can be found in supplementary file 5 and 6.
Fig. 3.
Drug-compound-target-pathway interactions for RDNI. Nodes in purple represent the compound; nodes in green and yellow represent the targets, nodes in yellow represent the targets that are co-expressed with ACE2, and the nodes in blue represent the potential pathways. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 4.
Schematic diagram of the critical path. (A) phosphatidylinositol-3-kinase (PI3K)-Akt signaling pathway; (B) FOXO signaling pathway; (C) mitogen-activated protein kinases (MAPK) signaling pathway; (D) T cell receptor signaling pathway. The dashed rectangle represent the pathways. The targets marked by a star represent the target of RDNI.
3.2.4. Molecular docking
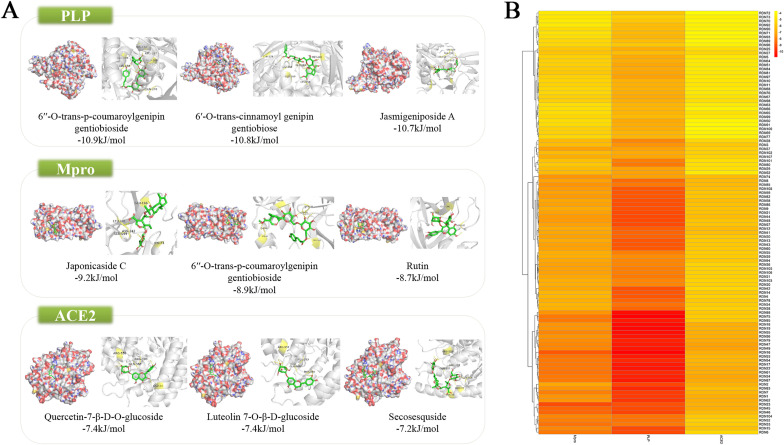
The mean binding energies of docking for the compounds with ACE2, PLP and Mpro were −5.32, −7.95, −6.4, respectively. The result of the molecular docking was shown in Fig. 5 . More detailed results of the docking simulation can be found in Supplementary file 7.
Fig. 5.
Result of molecular docking. (A) Compounds and targets with the lowest binding free energy. Green rectangles represent the targets. The diagrams on the left are the appearances of the surface, while the diagrams on the right are cartoon appearance. (B) Heat map of molecular docking. The deeper the color, the lower the binding energy. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Experimental verification
3.3.1. Western blot analysis
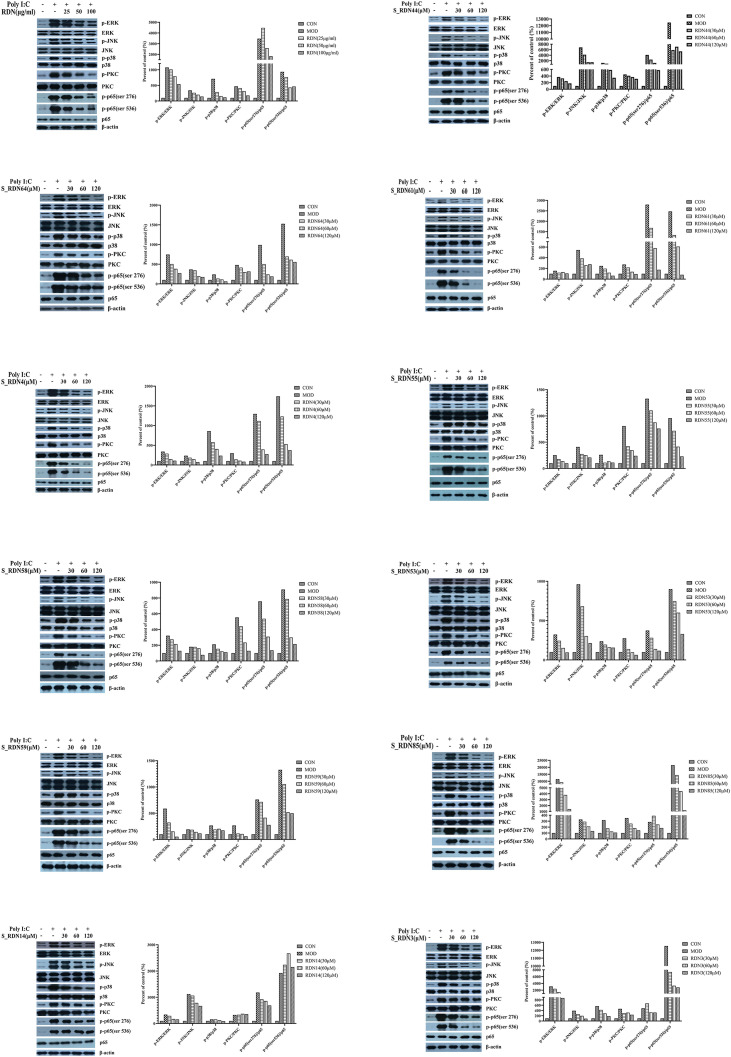
Western blotting was performed to determine the expressions of total and phosphorylated proteins of ERK, JNK, p38, PKC, and p65 NF-κB in BEAS-2B cells. The results showed that the expressions of phosphorylated proteins including ERK, JNK, p38, PKC, and p65 NF-κB, were significantly upregulated after being injected with PolyI:C (Fig. 6 ). There was no significant change in the phosphorylation of PKC and p65 (ser536) with geniposide acid-treated group, but RDNI and the 11 monomeric compoundscould effectively inhibit the expressions of these phosphorylated proteins, and the total proteins remained unchanged (Fig. 6).
Fig. 6.
Results of western blotting. Left shows representative Western blotting bands of p-ERK, ERK, p-JNK, JNK, p-p38, p38, p-PKC, PKC, p-p65(ser 276), p-p65(ser 536), p65 and β-actin. While right shows quantitative analysis of p-ERK/ERK, p-JNK/JNK, p-p38/p38, p-PKC/PKC, p-p65(ser 276)/p65 and p-p65(ser 536)/p65 by densitometry based on western blot images.
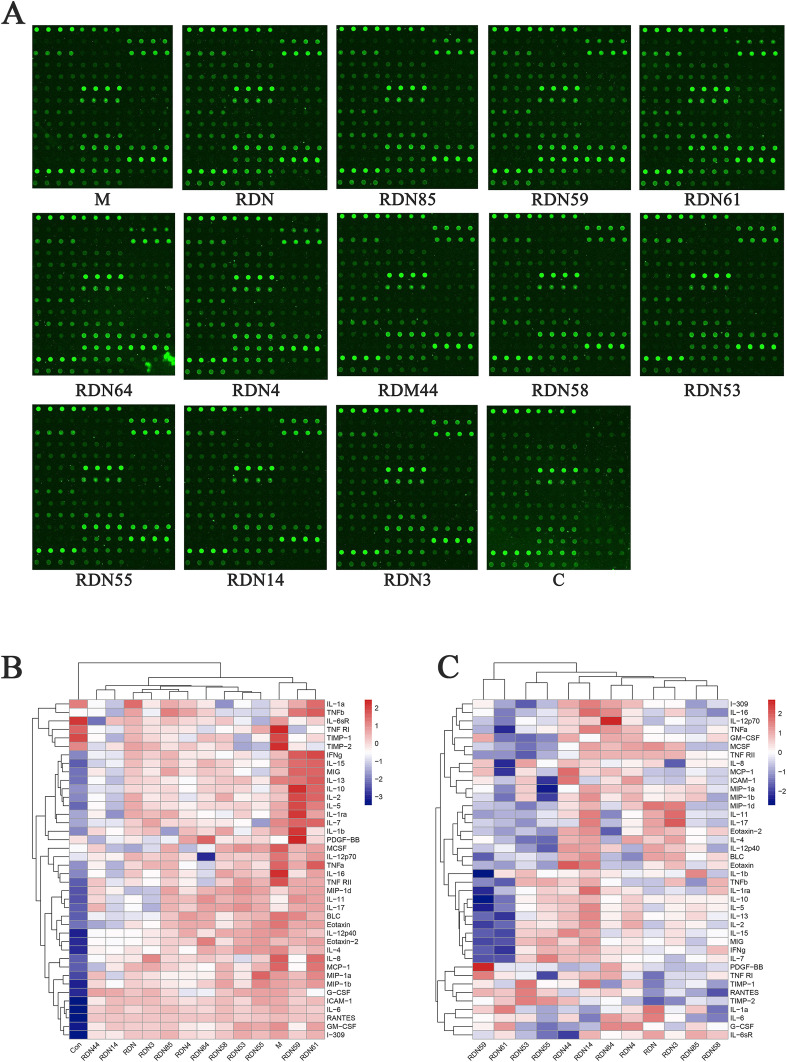
3.3.2. Cytokine microarray experiment
The results of the microarray analysis indicated that 35 out of 40 factors could be inhibited by RDNI treatment, including IL-1α, IL-1β, IL-4, IL-6, IL-8 and tumor necrosis factor (TNF-α). The inhibition of metallopeptidase inhibitor 1 (TIMP-1) was the most significant. After drug treatment, the expression of TIMP-1 was decreased by 36.53%. The results of the microarray analysis were listed in appendices at the end of this article and was shown in Fig. 7 .
Fig. 7.
The result from cytokine microarray. (A)Results of chip scan. (B) Heat map of chip signal value in each processing group. The colors are ranged from blue to red and indicate the expression are from low to high. (C) Heat map of the inhibition rate of Reduning injection and its compounds on cytokines levels. The colors are ranged from blue to red and indicate that the inhibition rate is from low to high. The red the color, the higher the inhibition rate, the lower the expression. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
COVID-19 brings a heavy economic burden to the society due to its properties of long cycle and highly infectious. Studies have found that compared with non-ICU patients, ICU patients who developed COVID-19 had higher plasma levels of IL2, IL7, IL10, colony stimulating factor 3 (CSF3, GCSF), C-X-C motif chemokine ligand 10 (CXCL10, IP10), C–C motif chemokine ligand 2 (CCL2, MCP1), C–C motif chemokine ligand 3 (CCL3, MIP1A), and TNFα (Huang et al., 2020). For intensive care survivors, these abnormal and excessive immune responses led to long-term lung damage and fibrosis, thereby resulting in dysfunction and reduced quality of life (Zumla et al., 2020). Chang et al. (Chang et al., 2015) reported that RDNI could markedly reduce the levels of IL-1β, TNF-α, IL-8, IL-10, and some other cytokines in a of lipopolysaccharide (LPS)-induced rat model of acute lung injury. However, many COVID-19 patients further developed ARDS, which caused pulmonary edema and lung failure, and liver, heart, and kidney damage. These symptoms are associated with cytokine storms (Mehta et al., 2020; Wu et al., 2020).
In the compound-target network, carbonic anhydrases (CAs) have the highest degree values, including CA2, CA12, CA1, and CA9. CAs are metalloenzymes that catalyze the hydration of carbon dioxide to bicarbonate and protons (Supuran, 2011). Studies have found that the inhibition of CA2 and CA4 in the proximal convoluted tubule could reduce bicarbonate reabsorption and causes metabolic acidosis, thereby would leading to the suppression of pro-inflammatory activation (Hudalla et al., 2019; Lan et al., 2017). CA inhibitors can be a base of anti-infectives (Supuran, 2010). This suggested that it can prevent from double infections. In addition, CA inhibitors are sometimes used as a respiratory stimulant for patients with a chronic obstructive pulmonary disease, itsgoal is to improve oxygenation (Adamson and Swenson (2017)). Besides, matrix metalloproteinase (MMP) family members, including MMP2, MMP9, MMP12, MMP13, MMP1 and MMP8, also have higher degree values. MMPs play prominent roles in pulmonary inflammation and remodeling and have recently emerged as promising novel therapeutic targets in lung disease (Summer et al., 2019; Golestani et al., 2017; Cui et al., 2017). Most of the MMPs promote (rather than inhibit) the development of pulmonary fibrosis, and for patients who developed COVID-19 the main symptoms of them were pneumonia and fibrosis in the later stage (Craig et al., 2015; Wu et al., 2020). This suggested that RDNI might influence pulmonary inflammation and remodeling and might reduce lung injury in critically ill patients. Furthermore, our results indicated that RDNI could potentially improve the symptoms caused by the disorder of ACE2 expression that is associated with COVID-19 and might influence the virus from entering into the body.
Among the targets pivotal in regulating COVID-19, we paid special attention to cytokine-related targets. Studies of SARS and MERS demonstrated that the levels of human coronaviruses could reach a high titer after infection and contain a variety of proteins that inhibit the interferon (IFN) response, suggesting that early resistance to the IFN response might delay or evade the innate immune response. Delayed IFN signal further coordinates with the inflammatory monocyte-macrophage (IMM) response and makes T cells to be more sensitive to apoptosis, thereby leading to the dysregulation of inflammatory response--cytokine storm. Rapid viral replication and a large number of pro-inflammatory cytokine/chemokine responses result in lung epithelial and endothelial cell apoptosis, which is acute lung injury (ALI) and ARDS (Channappanavar and Perlman (2017)). Therefore, the use of cytokine storm and immunoregulation therapy are necessary for the treatment of COVID-19. Several cytokines were involved in the target of RDNI treatment, which initially proved that RDNI had certain regulatory effects on cytokine storm (Tisoncik et al., 2012). In addition, cytokines also participate in several pathways that are related to fever induction, hence they are categorized as a group called pyrogenic cytokines. The major pyrogenic cytokines include interleukin-1 (IL-1α and IL-1β), TNF-α, IL-6, and interferon gamma, IL-8 and IFN-α, which possess pyrogenic activity all can be directly or indirectly and can be regulated by RDNI (Zampronio et al., 2015). This suggested that RDNI might be anti-pyretic.
From the analysis of PPI network, heat shock protein 90 alpha family class B member 1 (HSP90AB1) and heat shock protein 90 alpha family class A member 1 (HSP90AA1) have the highest degree values. TP53 and EGFR also showed high degree values. For heat shock protein 90 (HSP90), a 90-kDa and ATP-dependent molecular chaperone, can regulate inflammatory signaling networks. Its intracellular concentrations can be increased by stressors, e.g., increased temperature (fever), oxidative stress, ethanol, and infection that induce protein unfolding, misfolding, or aggregation (Tukaj and Węgrzyn (2016); Lilja et al., 2015). Importantly, an existing study proved that HSP90 inhibition could also prevent LPS-mediated inflammatory in human lung cells (Lilja et al., 2015). ALI and ARDS are clinical syndromes of noncardiogenic pulmonary edema that are caused primarily by increased permeability to proteins across the endothelial and epithelial barriers of the lung. Both are the clinical feature of COVID-19 (Huang et al., 2020). As for TP53, it is involved in the mediation of the barrier-protective effects of HSP90 inhibitors in human lung micro vascular endothelial cells and mouse lungs (Barabutis et al., 2016). HSP90 inhibition induces the upregulation of the wild-type p53, an effect that restores endothelial barrier function by disrupting LPS-induced ras homolog family member A (RHOA)/myosin light chain 2 (MLC2) pathway. p53 has significant anti-inflammatory effect on endothelial cells, and by carefully designing the response to repair the damaged endothelial monolayer. The first line of defense against inflammation is organized (Barabutis et al., 2019). With regard to EGFR, its inhibition can down-regulate the expression of p-p65 protein, block the activation of AKT and ERK1/2 signaling pathways and protect the lung from ALI (Tao et al., 2019). Taken together, these indicates that RDNI has regulatory effect on lung injury caused by COVID-19.
The disease clustering and functional enrichment analyses showed that RDNI is highly associated with pulmonary diseases. GO entries are closely related to the process of ALI and ARDS. It is mostly related to cell death and phosphorylation. Pathway enrichment analysis showed that 18 signal transduction pathways, 12 immune system pathways, 6 cell growth and death related pathways and 10 viral infectious disease pathways are related to RDNI treatment. MAPK/NF-κB signaling is associated with the pathogenesis and progression of ALI and inflammation, and is considered as major molecular pathways for ALI and ARDS (Wu et al., 2019; Liu et al., 2013; Cai et al., 2019; Tian et al., 2019). Forkhead box O (FOXOs) are negatively regulated by the canonical insulin signaling pathway through PI3K and AKT and activated in the presence of oxidative stress through JNK signaling (Eijkelenboom and Burgering, 2013). Studies have identified a correlation between increased FOXO expression and ARDS, ALI pathology (Sun et al., 2018; Artham et al., 2019). PI3K/AKT pathway is an important cellular signaling cascade in the cellular defense against inflammatory stimuli (Jiang et al., 2018). Regulation of apoptosis to protect against lung injury is mediated by activation of PI3K/AKT signaling (Shi et al., 2019; Wang et al., 2018; Luo et al., 2019). T cells also play an important role in the progress of lung damage and inflammation (Channappanavar, 2017; Wehrmann et al., 2016; DeBerge et al., 2013). By regulating the expression of IL-4, T cells can reduce the production of TNF-α from alveolar macrophages, thereby reducing the accumulation of M1 macrophages, inflammation and alveolar capillary leakage, and protecting the lung from LPS-induced lung injury (Wehrmann et al., 2016). In addition, JAK-STAT signaling pathway that plays a role in initiating the inflammatory response in the lung has also been enriched by the targets (Meletiadis et al., 2020; Song et al., 2015). The enriched pathways are closely associated with the pulmonary symptoms of COVID-19 and immune system, which are coincided with the efficacy of RDNI in clearing heat, detoxifying and regulating healthy qi. Another study showed that, immune-related genes are predominant in the Hot ZHENG network, which reflects the systemic and synergistic effect of Reduning for the treatment of diseases (Li et al., 2007).
From the results of molecular docking, the average binding energies between compounds and ACE2, PLP and Mpro were less than -5 kJ/mol. This indicated that the compounds have potential to inhibit the virus from entering into the human body and replicating. Some components with lower binding free energies are considered as potential antiviral active ingredients.
In experiment session, western blot experiment verified the regulation of key pathways by RDNI and its monomer compounds. ERK, p38 MAPK, and JNK are belonged to MAPK superfamily, and play an important role in the activation of signaling cascade of various inflammatory mediators including cytokines and chemical mediators (Kim and Choi (2010)). p38 inhibition had protective effect in a SARS-COV mouse model and p38 inhibitors are in the clinical stage for the treatment of COVID-19 (Grimes and Grimes (2020)). JNK plays an essential role in apoptosis regulation. It's inhibition significantly reduced inflammation in LPS-stimulated cells (Bin et al., 2019). Phosphorylated p65 (Ser536) migrates from the cytoplasm to the nucleus, activates multiple NF-kappa B target genes, and is one of the main mediators of ALI development. It is involved in the transcriptional regulation of proinflammatory cytokines, and the continuous elevation of proinflammatory cytokines in the lungs is associated with an increase in ALI mortality (Krupa et al., 2009). In addition, PKC plays an important role in lung permeability, contraction, migration, hypertrophy, proliferation, apoptosis and secretion (Dempsey et al., 2007). Our experimental results showed that RDNI could effectively inhibit the abnormal increase of these factors. This might be the mechanism by which RDNI protects the patients with COVID-19 from lung injury.
The results of protein microarray analysis preliminarily verified the regulation of cytokines by RDNI. It could effectively inhibit the expression of cytokines like IL-1α, IL-1β, IL-4, IL-6, IL-8 and TNF-α, thereby regulating cytokine storm. Therefore, RDNI could protect COVID-19 patients from lung injury and help to relieve fever and COVID-19 symptoms.
Anyway, limitations still exist in this article. The experiments we use to verify the efficacy and results are basic. More animal experiments and human experiments are needed to verify the results. Further experiments are needed to verify the docking between target and compound pairs with low binding energy. What's more, our molecular docking mainly focused on three targets, so other targets of COVID-19 were not well validated.
5. Conclusions
In summary, this study firstly performed anti- SARS-CoV-2 experiments in Vero E6 cells. Based on the results of cytotoxicity and antiviral tests of drugs, CC50 (dilution ratio) = 1:1280, CC50 = 2.031 mg crude drugs/mL (0.047 mg solid content/mL) and EC50 (diluted multiples) = 1:25140.3, EC50 = 103.420 μg crude drugs/mL (2.405 μg solid content/mL). Then, through constructing a network, this study found that RDNI could potentially improve the symptoms caused by the disorder of ACE2 expression that is associated with COVID-19 and might influence the virus from entering into the body. The level of 26 cytokines could be directly regulated by RDNI, including TNF-α, IL-8, IL-6, I1α, IL1β. This suggested that RDNI can regulate cytokine storm and reduce fever caused by COVID-19. MMPs, HSP90AA1, HSP90AB1, TP53 and EGFR have higher degree values from the compound-target network and PPI network, which indicated that RDNI might effectively reduce lung damage. Pathways like PI3K/AKT, FOXO, MAPK and T cell receptor signaling pathways enriched above 20 genes by RDNI treatment suggested that RDNI can regulate inflammation and protect COVID-19 patients from damage through these pathways. Molecular docking also indicated that the compounds of RDNI can spontaneously bind to PLP, Mpro and ACE2 inhibits SARS-CoV-2 from entering into the body and replicating. Finally, western blotting and a cytokine chip verified that RDNI can effectively inhibit the overexpression of MAPKs, PKC, p65 and cytokines. Although further experiments are needed to verify the underlying mechanisms of RDNI against COVID-19, this study provides a basis and guidance for further experiments.
Authors’ contributions
Conceptualization, Shanshan Jia. and Jiarui Wu.; Data curation, Xiaotian Fan, Yingying Liu and Shan Lu; Formal analysis, Xinzhuang Zhang, Shanshan Jia, Xinkui Liu and Xiaotian Fan; Funding acquisition, Jiarui Wu; Investigation, Shanshan Jia and Xinkui Liu; Methodology, Xinzhuang Zhang, Wei Zhou, and Xinkui Liu; Project administration, Jiarui Wu and Wei Xiao; Resources, Jiarui Wu, Wei Xiao and Liang Cao; Software, Jialin Li, Wei Zhou and Jingyuan Zhang; Supervision, Jingyuan Zhang and Siyu Guo; Validation, Siyu Guo, Zeyu Cao, Shan Lu and Xiaotian Fan; Visualization, Jialin Li, Yingying Liu; Writing - original draft, Shanshan Jia; Writing - review & editing, Hua Luo, Zeyu Cao, Liang Cao, Xinkui Liu. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Young Scientists Training Program of Beijing University of Chinese Medicine [grant number BUCM-QNLJ 2019001]; and the National Natural Science Foundation of China of China [grant number 81673829]. The funding body approved the design of the study, analysis, and interpretation of data, and publication of the manuscript.
Availability of data and materials
All data obtained or analyzed during this study are available from published articles and supplementary materials.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgements
We thank Wayen Biotechnologies (Shanghai), Inc. and Institute of Laboratory Animals Science for providing technical support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jep.2021.113871.
Abbreviation
- 3CLpro
3-chymotrypsin-like protease
- ACE2
angiotensin-converting enzyme-2
- AKT
serine/threonine kinase 1
- ALI
acute lung injury
- ARDS
acute respiratory distress syndrome
- BP
biological processe
- BSL-3
biosafety level-3 laboratory
- CA
carbonic anhydrases
- CC
cellular components
- CC50
half toxic concentration of drug
- CCL2
C–C motif chemokine ligand 2
- CCL3
C–C motif chemokine ligand 3
- COVID-19
Coronavirus Disease 2019
- CSF3
colony stimulating factor 3
- CT
computed tomography
- CXCL10
C-X-C motif chemokine ligand 10
- EC50
half effective concentration of drug
- EGFR
Epidermal growth factor receptor
- ERK
Extracellular signal-regulated kinase
- FOXO
Forkhead box O
- GO
gene ontology
- hCoVs
human coronaviruses
- HLMVECs
human lung microvascular endothelial cells
- HSP90
Heat shock protein 90
- HSP90AA1
heat shock protein 90 alpha family class A member 1
- HSP90AB1
heat shock protein 90 alpha family class B member 1
- HINT
High-qualityINTeractomes
- ICU
intensive care unit
- IFN
interferons
- IL1
interleukin-1 tumor necrosis factor α
- IL-6
interleukin-6
- IL-8
interleukin-8
- IMM
inflammatory monocyte-macrophage
- JNK
Jun N-terminal kinase
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LPS
lipopolysaccharide
- MAPK
mitogen-activated protein kinases
- MERS
Middle East respiratory syndrome
- MF
molecular functions
- MLC2
myosin light chain 2
- Mpro
aka main protease
- NF-κB
Nuclear factor-κB
- PF
pulmonary fibrosis
- PI3K
pathway through phosphatidylinositol-3-kinase
- PKB
protein kinase B
- PKC
protein kinase C
- PLP
papain-like protease
- PPI
protein-protein interaction
- RDNI
Reduning injection
- S
spike
- RHOA
ras homolog family member A
- SARS
severe acute respiratory syndrome
- SARS-CoV-2
SARS-coronavirus 2
- SI
selection index
- SMILES
simplified molecular input line entry specification
- TCID50/mL
50% tissue culture infective dose
- TCM
Traditional Chinese Medicine
- TNF-α
tumor necrosis factor
- TP53
Tumor protein P53
Appendices.
Appendice A.1. Concentrations of 40 factors in each treatment group (pg/mL)
| Names | M |
RDN |
RDN85 |
RDN59 |
RDN61 |
RDN64 |
RDN4 |
|---|---|---|---|---|---|---|---|
| Model set | Reduning injection | 3-O-caffeoylquinic acid | 4-O-caffeoylquinic acid | 3,4-di-O-caffeoylquinic acid | Trans-caffeic acid | Geniposide | |
| BLC | 1.63 | 0.51 | 0.95 | 1.51 | 1.09 | 1.19 | 1.36 |
| Eotaxin | 4.21 | 1.81 | 2.46 | 2.91 | 3.15 | 2.79 | 2.83 |
| Eotaxin-2 | 8.38 | 6.54 | 7.77 | 8.68 | 9.21 | 10.19 | 7.68 |
| G-CSF | 114.8 | 70.14 | 57.89 | 68.1 | 33.31 | 26.89 | 25.91 |
| GM-CSF | 37.06 | 22.73 | 24.8 | 17.04 | 32.21 | 17.11 | 18.28 |
| I-309 | 21.4 | 17.83 | 19.78 | 17.47 | 19.57 | 15.82 | 16.58 |
| ICAM-1 | 28379.92 | 24732.94 | 21601.71 | 21723.35 | 25236.22 | 21306.29 | 17881.68 |
| IFNg | 8.64 | 8.89 | 9.2 | 12.92 | 13.83 | 7.1 | 7.06 |
| IL-1a | 13.78 | 31.41 | 21.84 | 21.04 | 30.49 | 13.14 | 18.11 |
| IL-1b | 3.77 | 2.65 | 0.81 | 6.57 | 2.35 | 2.5 | 3.01 |
| IL-1ra | 110.73 | 113.25 | 112.62 | 165.68 | 118.16 | 77.38 | 86.14 |
| IL-2 | 88.71 | 96.55 | 82.77 | 123.8 | 114.84 | 81.12 | 66.82 |
| IL-4 | 9.8 | 4.98 | 5.87 | 5.98 | 7.14 | 7.37 | 4.09 |
| IL-5 | 28.55 | 28.61 | 22.82 | 41.57 | 34.55 | 24.25 | 21.04 |
| IL-6 | 2732.46 | 2183.47 | 2488.69 | 2354.09 | 2316.33 | 2226.42 | 2289.4 |
| IL-6sR | 26.79 | 21.91 | 21.01 | 14.45 | 18.26 | 16.32 | 17.13 |
| IL-7 | 28.6 | 22.1 | 21.59 | 31.82 | 31.77 | 20.09 | 21.47 |
| IL-8 | 607.35 | 270.41 | 296.32 | 252.86 | 536.43 | 338.74 | 303.05 |
| IL-10 | 1.43 | 1.17 | 1.2 | 3.15 | 2.38 | 1.27 | 0.89 |
| IL-11 | 6603.65 | 6146.92 | 9602.41 | 11530.19 | 10846.74 | 12087.45 | 8803.57 |
| IL-12p40 | 145.26 | 71.25 | 94.57 | 116.27 | 105.59 | 86.09 | 122.98 |
| IL-12p70 | 2.98 | 1.86 | 1.75 | 1.83 | 2.23 | 0 | 1.33 |
| IL-13 | 19.82 | 14.45 | 12.77 | 20.55 | 22.39 | 13.94 | 11.45 |
| IL-15 | 176.07 | 163.84 | 163.95 | 211.02 | 216.59 | 167.02 | 139.35 |
| IL-16 | 533.72 | 349.9 | 358.95 | 317.46 | 391.54 | 239.46 | 305.53 |
| IL-17 | 70.9 | 61.76 | 89.53 | 90.44 | 95.8 | 95 | 79.07 |
| MCP-1 | 174.89 | 123.82 | 112.83 | 91.47 | 162.43 | 90.25 | 99.34 |
| MCSF | 621.84 | 87.77 | 352.42 | 319.66 | 374.07 | 160.2 | 110.16 |
| MIG | 297.14 | 290.84 | 360.19 | 503.17 | 494.02 | 293.01 | 246.93 |
| MIP-1a | 123.1 | 69.21 | 41.4 | 78.1 | 145.19 | 38.49 | 39.4 |
| MIP-1b | 52.77 | 36.29 | 29.51 | 43.97 | 57.94 | 35.29 | 27.53 |
| MIP-1d | 46.76 | 15.16 | 78.72 | 73.81 | 78.75 | 67.33 | 40.87 |
| PDGF-BB | 44.33 | 51.8 | 39.73 | 274.81 | 61.27 | 162.98 | 99.35 |
| RANTES | 2258.4 | 2104 | 2078.93 | 1880.61 | 1906.76 | 1916.61 | 1974.55 |
| TIMP-1 | 19103.76 | 16252.61 | 14939.67 | 14517.99 | 15273.12 | 16054.85 | 13995.95 |
| TIMP-2 | 34985.02 | 24381.29 | 19675.03 | 18933.12 | 16431.14 | 18963.89 | 21244.41 |
| TNFa | 121.69 | 70.14 | 85.11 | 89.14 | 122.55 | 45.1 | 54.29 |
| TNFb | 66.75 | 135.64 | 181.31 | 189 | 228.49 | 87.71 | 135.55 |
| TNF RI | 69.58 | 42.12 | 36.73 | 27.53 | 33.71 | 30.3 | 36.13 |
| TNF RII | 14.66 | 0 | 2.39 | 5.9 | 5.66 | 0 | 0 |
Appendices A.2. Concentrations of 40 factors in each treatment group (pg/mL)
| Names | RDN44 |
RDN58 |
RDN53 |
RDN55 |
RDN14 |
RDN3 |
Con |
|---|---|---|---|---|---|---|---|
| Quercetin | 5-O-caffeoylquinic acid | 3,5-di-O-caffeoylquinic acid | 4,5-di-O-caffeoylquinic acid | Geniposidic acid | Secoxyloganin | Control | |
| BLC | 0.54 | 0.77 | 1.01 | 1.14 | 0.44 | 0.55 | 0.05 |
| Eotaxin | 0 | 1.68 | 2.51 | 3.18 | 0.46 | 1.28 | 0 |
| Eotaxin-2 | 6.56 | 7.14 | 8.73 | 9.47 | 6.19 | 6.86 | 1.93 |
| G-CSF | 105.64 | 73.74 | 100.76 | 116.95 | 96.23 | 91.2 | 3.01 |
| GM-CSF | 17.22 | 20.69 | 32.83 | 31 | 23.9 | 23.14 | 1.53 |
| I-309 | 16.02 | 18.5 | 20.77 | 19.09 | 14.95 | 17.75 | 0 |
| ICAM-1 | 15934.9 | 24655.82 | 19982.67 | 31195.54 | 24686.63 | 25198.62 | 1787.36 |
| IFNg | 5.9 | 5.95 | 7.41 | 5.59 | 5.36 | 7.86 | 3.1 |
| IL-1a | 9.76 | 0 | 10.06 | 5.13 | 4.42 | 13 | 27.79 |
| IL-1b | 2.52 | 3.63 | 1.71 | 2.85 | 2.02 | 2.98 | 0 |
| IL-1ra | 81 | 68.18 | 98.27 | 75.79 | 35.05 | 104.19 | 44.19 |
| IL-2 | 72.06 | 95.23 | 76.36 | 80.91 | 59.6 | 77.76 | 44.69 |
| IL-4 | 4.54 | 6.6 | 8.46 | 8.64 | 4.07 | 4.14 | 0.7 |
| IL-5 | 19.96 | 23.86 | 24.43 | 21.62 | 14.75 | 24.09 | 9.19 |
| IL-6 | 2303.77 | 2533.12 | 2324.76 | 2464.96 | 2519.36 | 2367.23 | 680.35 |
| IL-6sR | 5.81 | 19.67 | 12.63 | 8.62 | 20.72 | 17.06 | 50.64 |
| IL-7 | 16.82 | 19.88 | 18.49 | 12.54 | 16.84 | 25.65 | 13.17 |
| IL-8 | 233.86 | 298.96 | 200.27 | 319.26 | 270.91 | 480.76 | 107.35 |
| IL-10 | 0.72 | 1.22 | 1.18 | 0.77 | 0.64 | 1.11 | 0.33 |
| IL-11 | 7520.61 | 9793.19 | 10823.29 | 9520.3 | 4918.81 | 4339.98 | 2387.46 |
| IL-12p40 | 69.39 | 125.64 | 127.57 | 143.3 | 72.59 | 83.49 | 6.27 |
| IL-12p70 | 1.25 | 1.7 | 1.56 | 1.69 | 1.16 | 1.55 | 0.75 |
| IL-13 | 11.46 | 15.33 | 15.49 | 13.16 | 8.81 | 13.96 | 6.08 |
| IL-15 | 107.52 | 155.74 | 135.83 | 130.34 | 118.71 | 142.4 | 59.03 |
| IL-16 | 279.36 | 388.33 | 359.27 | 334.78 | 216.27 | 284.94 | 163.92 |
| IL-17 | 89.41 | 94.79 | 103.19 | 90.08 | 54.69 | 47.51 | 35.21 |
| MCP-1 | 64.67 | 123.45 | 101.02 | 123.77 | 97.88 | 128.79 | 34.71 |
| MCSF | 261.58 | 343.82 | 439.06 | 380.31 | 109.5 | 142.57 | 177.26 |
| MIG | 226.94 | 279.47 | 193.49 | 146.61 | 138.8 | 234.98 | 54.25 |
| MIP-1a | 79.73 | 92.37 | 43.36 | 209.56 | 15.73 | 53.52 | 0 |
| MIP-1b | 33.19 | 51.24 | 32.02 | 64.26 | 22.34 | 34.1 | 4.38 |
| MIP-1d | 49.36 | 77.79 | 93.43 | 76.14 | 34.64 | 16.66 | 0 |
| PDGF-BB | 24.18 | 51.78 | 59.69 | 62.23 | 38.25 | 32.06 | 56.64 |
| RANTES | 2027.35 | 2160.02 | 1812.26 | 1915.85 | 1945.14 | 2013.3 | 21.96 |
| TIMP-1 | 14593.53 | 15872.37 | 13000.38 | 14763.27 | 12951.41 | 15268.27 | 19025.71 |
| TIMP-2 | 16020.47 | 20550.94 | 13559.96 | 14337.84 | 19704.49 | 22892.11 | 26382.49 |
| TNFa | 60.38 | 69.55 | 79.82 | 68.74 | 36.41 | 83.48 | 29.98 |
| TNFb | 33.13 | 6.45 | 4.73 | 0 | 0 | 18.85 | 43.61 |
| TNF RI | 32.64 | 33.87 | 35.69 | 44.17 | 31.17 | 36.72 | 55.97 |
| TNF RII | 3.13 | 2.54 | 7.26 | 4.95 | 0 | 0 | 0 |
Note: The bold numbers in the table indicated that the values are below the minimum detection limit.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- Adamson R., Swenson E.R. Acetazolamide use in severe chronic obstructive pulmonary disease. Pros and cons. Annals of the American Thoracic Society. 2017;14(7):1086–1093. doi: 10.1513/AnnalsATS.201701-016FR. [DOI] [PubMed] [Google Scholar]
- Artham S., et al. Endothelial stromelysin1 regulation by the forkhead box-O transcription factors is crucial in the exudative phase of acute lung injury. Pharmacol. Res. 2019;141:249–263. doi: 10.1016/j.phrs.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabutis N., et al. Regulation of pulmonary endothelial barrier function by kinases. Am. J. Physiol. Lung Cell Mol. Physiol. 2016;311(5):L832–l845. doi: 10.1152/ajplung.00233.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabutis N., et al. Hsp90 inhibitors suppress P53 phosphorylation in LPS - induced endothelial inflammation. Cytokine. 2019;113:427–432. doi: 10.1016/j.cyto.2018.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bin W., et al. TRAF1 meditates lipopolysaccharide-induced acute lung injury by up regulating JNK activation. Biochem. Biophys. Res. Commun. 2019;511(1):49–56. doi: 10.1016/j.bbrc.2019.01.041. [DOI] [PubMed] [Google Scholar]
- Burley S.K., et al. Protein data bank (PDB): the single global macromolecular structure archive. Methods Mol. Biol. 2017;1607:627–641. doi: 10.1007/978-1-4939-7000-1_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., et al. Astaxanthin prevents against lipopolysaccharide-induced acute lung injury and sepsis via inhibiting activation of MAPK/NF-κB. Am. J. Tourism Res. 2019;11(3):1884–1894. [PMC free article] [PubMed] [Google Scholar]
- Cao B., et al. A trial of lopinavir-ritonavir in adults hospitalized with severe covid-19. N. Engl. J. Med. 2020;382(19):1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., et al. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell discovery. 2020;6:11. doi: 10.1038/s41421-020-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang X.J., et al. Mechanism of Reduning Injection on anti-acute lung injury in rats based on cytokine storm. Chin. Tradit. Herb. Drugs. 2015;46:236–239. 02. [Google Scholar]
- Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 2017;39(5):529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.W., et al. Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CL (pro)) structure: virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Research. 2020;9:129. doi: 10.12688/f1000research.22457.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coperchini F., et al. The cytokine storm in COVID-19: an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020;53:25–32. doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig V.J., et al. Matrix metalloproteinases as therapeutic targets for idiopathic pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 2015;53(5):585–600. doi: 10.1165/rcmb.2015-0020TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui N., et al. Biochemical and biological attributes of matrix metalloproteinases. Progress in Molecular Biology and Translational Science. 2017;147:1–73. doi: 10.1016/bs.pmbts.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W., et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science (New York, N.Y.). 2020;368(6497):1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das J., Yu H. HINT: high-quality protein interactomes and their applications in understanding human disease. BMC Syst. Biol. 2012;6:92. doi: 10.1186/1752-0509-6-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerge M.P., et al. ADAM17-mediated processing of TNF-α expressed by antiviral effector CD8+ T cells is required for severe T-cell-mediated lung injury. PloS One. 2013;8(11) doi: 10.1371/journal.pone.0079340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey E.C., et al. Lung disease and PKCs. Pharmacol. Res. 2007;55(6):545–559. doi: 10.1016/j.phrs.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Deng X., et al. Analysis of coronavirus temperature-sensitive mutants reveals an interplay between the macrodomain and papain-like protease impacting replication and pathogenesis. J. Virol. 2019;93(12) doi: 10.1128/jvi.02140-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S., et al. A guideline for homology modeling of the proteins from newly discovered betacoronavirus, 2019 novel coronavirus (2019-nCoV) J. Med. Virol. 2020;92:1542–1548. doi: 10.1002/jmv.25768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijkelenboom A., Burgering B.M. FOXOs: signalling integrators for homeostasis maintenance. Nat. Rev. Mol. Cell Biol. 2013;14(2):83–97. doi: 10.1038/nrm3507. [DOI] [PubMed] [Google Scholar]
- Ferner R.E., Aronson J.K. Remdesivir in COVID-19. BMJ (Clinical research ed.) 2020;369 doi: 10.1136/bmj.m1610. m1610. [DOI] [PubMed] [Google Scholar]
- Gao X., et al. Serum and urine metabolomics based on UPLC-Q-TOF/MS reveals the antipyretic mechanism of Reduning injection in a rat model. J. Ethnopharmacol. 2020;250:112429. doi: 10.1016/j.jep.2019.112429. [DOI] [PubMed] [Google Scholar]
- General office of the national health commission Diagnosis and treatment of novel coronavirus pneumonia (Trial Version Seventh) 2020. http://www.nhc.gov.cn/yzygj/s7653p/202003/46c9294a7dfe4cef80dc7f5912eb1989.shtml
- Geng T., et al. Influences of Re Du Ning Injection, a traditional Chinese medicine injection, on the CYP450 activities in rats using a cocktail method. J. Ethnopharmacol. 2015;174:426–436. doi: 10.1016/j.jep.2015.08.035. [DOI] [PubMed] [Google Scholar]
- Gfeller D., et al. SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014;42:W32–W38. doi: 10.1093/nar/gku293. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golestani R., et al. Matrix metalloproteinase-targeted imaging of lung inflammation and remodeling. J. Nucl. Med.: official publication, Society of Nuclear Medicine. 2017;58(1):138–143. doi: 10.2967/jnumed.116.176198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes J.M., Grimes K.V. p38 MAPK inhibition: a promising therapeutic approach for COVID-19. J. Mol. Cell. Cardiol. 2020;144:63–65. doi: 10.1016/j.yjmcc.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet (London, England) 2020;395(10223):497–506. doi: 10.1016/s0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., et al. Network pharmacology-based prediction and verification of the active ingredients and potential targets of zuojinwan for treating colorectal cancer. Drug Des. Dev. Ther. 2020;14:2725–2740. doi: 10.2147/dddt.S250991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudalla H., et al. Carbonic anhydrase inhibition ameliorates inflammation and experimental pulmonary hypertension. Am. J. Respir. Cell Mol. Biol. 2019;61(4):512–524. doi: 10.1165/rcmb.2018-0232OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K., et al. Barbaloin protects against lipopolysaccharide (LPS)-induced acute lung injury by inhibiting the ROS-mediated PI3K/AKT/NF-κB pathway. Int. Immunopharm. 2018;64:140–150. doi: 10.1016/j.intimp.2018.08.023. [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Sato Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020;29(1):28–35. doi: 10.1002/pro.3711. a publication of the Protein Society. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.K., Choi E.J. Pathological roles of MAPK signaling pathways in human diseases. Biochim. Biophys. Acta. 2010;1802(4):396–405. doi: 10.1016/j.bbadis.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Krupa A., et al. Anti-chemokine autoantibody: chemokine immune complexes activate endothelial cells via IgG receptors. Am. J. Respir. Cell Mol. Biol. 2009;41(2):155–169. doi: 10.1165/rcmb.2008-0183OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan C.C., et al. Carbonic anhydrase inhibitor attenuates ischemia-reperfusion induced acute lung injury. PloS One. 2017;12(6) doi: 10.1371/journal.pone.0179822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., et al. Understanding ZHENG in traditional Chinese medicine in the context of neuro-endocrine-immune network. IET Syst. Biol. 2007;1(1):51–60. doi: 10.1049/iet-syb:20060032. [DOI] [PubMed] [Google Scholar]
- Li H., et al. Chemical profiling of Re-Du-Ning injection by ultra-performance liquid chromatography coupled with electrospray ionization tandem quadrupole time-of-flight mass spectrometry through the screening of diagnostic Ions in MS(E) mode. PloS One. 2015;10(4) doi: 10.1371/journal.pone.0121031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilja A., et al. HSP90 inhibition suppresses lipopolysaccharide-induced lung inflammation in vivo. PloS One. 2015;10(1) doi: 10.1371/journal.pone.0114975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., et al. Angelicin regulates LPS-induced inflammation via inhibiting MAPK/NF-κB pathways. J. Surg. Res. 2013;185(1):300–309. doi: 10.1016/j.jss.2013.05.083. [DOI] [PubMed] [Google Scholar]
- Luo X., et al. Genipin attenuates mitochondrial-dependent apoptosis, endoplasmic reticulum stress, and inflammation via the PI3K/AKT pathway in acute lung injury. Int. Immunopharm. 2019;76:105842. doi: 10.1016/j.intimp.2019.105842. [DOI] [PubMed] [Google Scholar]
- Luo Y., et al. Application of Chinese medicine in acute and critical medical conditions. Am. J. Chin. Med. 2019;47(6):1223–1235. doi: 10.1142/s0192415x19500629. [DOI] [PubMed] [Google Scholar]
- Mehta P., et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet (London, England) 2020;395(10229):1033–1034. doi: 10.1016/s0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meletiadis J., et al. Interleukin-6 blocking vs. JAK-STAT inhibition for prevention of lung injury in patients with COVID-19. Infect. Dis. Ther. 2020;9(4):707–713. doi: 10.1007/s40121-020-00326-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickel J., et al. SuperPred: update on drug classification and target prediction. Nucleic Acids Res. 2014;42 doi: 10.1093/nar/gku477. (Web Server issue), W26-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H., et al. Repurposed antiviral drugs for COVID-19 –interim WHO SOLIDARITY trial results. MedRxiv. 2020 doi: 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders J.M., et al. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. Jama. 2020;323(18):1824–1836. doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- Shannon P., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., et al. PI3K/Akt pathway-mediated HO-1 induction regulates mitochondrial quality control and attenuates endotoxin-induced acute lung injury. Laboratory investigation; a journal of technical methods and pathology. 2019;99(12):1795–1809. doi: 10.1038/s41374-019-0286-x. [DOI] [PubMed] [Google Scholar]
- Sohrabi C., et al. World Health Organization declares global emergency: a review of the 2019 novel coronavirus (COVID-19) Int. J. Surg. 2020;76:71–76. doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., et al. Recombinant human brain natriuretic peptide ameliorates trauma-induced acute lung injury via inhibiting JAK/STAT signaling pathway in rats. The journal of trauma and acute care surgery. 2015;78(5):980–987. doi: 10.1097/ta.0000000000000602. [DOI] [PubMed] [Google Scholar]
- Summer R., et al. Matrix metalloproteinase activity in the lung is increased in Hermansky-Pudlak syndrome. Orphanet J. Rare Dis. 2019;14(1):162. doi: 10.1186/s13023-019-1143-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K., et al. Schisandrin attenuates lipopolysaccharide-induced lung injury by regulating TLR-4 and akt/FoxO1 signaling pathways. Front. Physiol. 2018;9:1104. doi: 10.3389/fphys.2018.01104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supuran C.T. Carbonic anhydrase inhibition/activation: trip of a scientist around the world in the search of novel chemotypes and drug targets. Curr. Pharmaceut. Des. 2010;16(29):3233–3245. doi: 10.2174/138161210793429797. [DOI] [PubMed] [Google Scholar]
- Supuran C.T. Bacterial carbonic anhydrases as drug targets: toward novel antibiotics? Front. Pharmacol. 2011;2:34. doi: 10.3389/fphar.2011.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., et al. Stitch 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016;44(D1):D380–D384. doi: 10.1093/nar/gkv1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao H., et al. Erlotinib protects LPS-induced acute lung injury in mice by inhibiting EGFR/TLR4 signaling pathway. Shock (Augusta, Ga.) 2019;51(1):131–138. doi: 10.1097/shk.0000000000001124. [DOI] [PubMed] [Google Scholar]
- Tian C., et al. The protective effect of the flavonoid fraction of Abutilon theophrasti Medic. leaves on LPS-induced acute lung injury in mice via the NF-κB and MAPK signalling pathways. Biomed. Pharmacother. 2019;109:1024–1031. doi: 10.1016/j.biopha.2018.10.197. [DOI] [PubMed] [Google Scholar]
- Tisoncik J.R., et al. Into the eye of the cytokine storm. Microbiol. Mol. Biol. Rev.: MMBR. 2012;76(1):16–32. doi: 10.1128/mmbr.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukaj S., Węgrzyn G. Anti-Hsp90 therapy in autoimmune and inflammatory diseases: a review of preclinical studies. Cell Stress Chaperones. 2016;21(2):213–218. doi: 10.1007/s12192-016-0670-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan S., et al. Clinical features and treatment of COVID-19 patients in northeast Chongqing. J. Med. Virol. 2020;92(7):797–806. doi: 10.1002/jmv.25783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., et al. Identification of the allergenic ingredients in reduning injection by ultrafiltration and high-performance liquid chromatography. Journal of immunology research. 2016:4895672. doi: 10.1155/2016/4895672. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., et al. Allicin attenuates lipopolysaccharide-induced acute lung injury in neonatal rats via the PI3K/Akt pathway. Mol. Med. Rep. 2018;17(5):6777–6783. doi: 10.3892/mmr.2018.8693. [DOI] [PubMed] [Google Scholar]
- Wang J., et al. ACE2 expression by colonic epithelial cells is associated with viral infection, immunity and energy metabolism. Med Rxiv. 2020 doi: 10.1101/2020.02.05.20020545. [DOI] [Google Scholar]
- Wehrmann F., et al. γδ T cells protect against LPS-induced lung injury. J. Leukoc. Biol. 2016;99(2):373–386. doi: 10.1189/jlb.4A0115-017RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Word Health Organization Coronavirus disease (COVID-19) outbreak situation. 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019
- Wrapp D., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (New York, N.Y.). 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., et al. Protostemonine alleviates heat-killed methicillin-resistant Staphylococcus aureus-induced acute lung injury through MAPK and NF-κB signaling pathways. Int. Immunopharm. 2019;77:105964. doi: 10.1016/j.intimp.2019.105964. [DOI] [PubMed] [Google Scholar]
- Wu J., et al. Novel coronavirus pneumonia (COVID-19) CT distribution and sign features. Chin. J. Tuberc. Respir. Dis. 2020;43(4):321–326. doi: 10.3760/cma.j.cn112147-20200217-00106. [DOI] [PubMed] [Google Scholar]
- Yang H., et al. A novel systems pharmacology model for herbal medicine injection: a case using Reduning injection. BMC Compl. Alternative Med. 2014;14:430. doi: 10.1186/1472-6882-14-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., et al. Traditional Chinese medicine in the treatment of patients infected with 2019-new coronavirus (SARS-CoV-2): a review and perspective. Int. J. Biol. Sci. 2020;16(10):1708–1717. doi: 10.7150/ijbs.45538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampronio A.R., et al. Central mediators involved in the febrile response: effects of antipyretic drugs. Temperature (Austin) 2015;2(4):506–521. doi: 10.1080/23328940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., et al. The clinical benefits of Chinese patent medicines against COVID-19 based on current evidence. Pharmacol. Res. 2020;157:104882. doi: 10.1016/j.phrs.2020.104882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A., et al. Reducing mortality from 2019-nCoV: host-directed therapies should be an option. Lancet (London, England) 2020;395(10224):e35–e36. doi: 10.1016/s0140-6736(20)30305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data obtained or analyzed during this study are available from published articles and supplementary materials.