Abstract
The year 2020 was a landmark year of a once-in-a-century pandemic of a novel coronavirus, SARS-CoV-2 virus, that led to a rapidly spreading coronavirus disease (COVID-19). The spectrum of disease with SARS-CoV-2 ranges from asymptomatic to mild upper respiratory illness, to moderate to severe disease with respiratory compromise to acute respiratory distress syndrome, multiorgan failure, and death. Early in the pandemic, risk factors were recognized that contributed to more severe disease, but it became evident that individuals and even young people could have severe COVID-19. As we started to understand the immunobiology of COVID-19, it became clearer that the immune responses to SARS-CoV-2 were variable, and in some cases, the excessive inflammatory response contributed to greater morbidity and mortality. In this review, we will explore some of the additional risk factors that appear to contribute to disease severity and enhance our understanding of why some individuals experience more severe COVID-19. Recent advances in genome-wide associations have identified potential candidate genes in certain populations that may modify the host immune responses leading to dysregulated host immunity. Genetic defects of the type I interferon pathway are also linked to a more clinically severe phenotype of COVID-19. Finally, dysregulation of the adaptive immune system may also play a role in the severity and complex clinical course of patients with COVID-19. A better understanding of the host immune responses to SARS-CoV-2 will hopefully lead to new treatment modalities to prevent the poor outcomes of COVID-19 in those individuals with pre-existing risk factors or genetic variants that contribute to the dysregulated host immune responses.
Abbreviations used: ACE2, Angiotensin-converting enzyme 2; ARDS, Acute respiratory distress syndrome; CI, Confidence interval; COVID-19, Coronavirus disease 2019; CVID, Common variable immune deficiency; GC, Germinal center; ICU, Intensive care unit; IFNAR1, IFN-α/β receptor 1; MIS-C, Multisystem inflammatory syndrome in children; N, Nucleocapsid protein; S, Spike protein; SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2; Th, T helper; TMPRSS2, Transmembrane serine protease 2
In the city of Wuhan in the Chinese province of Hubei at the end of 2019, clusters of patients with pneumonia of an unknown etiology were seen. In early January 2020, the cause of this unknown pneumonia was determined to be a novel coronavirus, later named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus responsible for causing coronavirus disease 2019 (COVID-19).1 Since early January and the identification of the first case of COVID-19 in the United States, the virus has rapidly spread throughout the world bringing about a global pandemic.2 There are more than 107 million confirmed cases in the world as of early February 2021 with more than 27 million confirmed cases in the United States and more than 472,000 deaths. The case fatality rate from this virus has been estimated to be approximately 2% to 3%, but is higher in certain ethnic groups, individuals older than 60, and in people with underlying risk factors.3 , 4 However, the transmissibility of this virus is high with an estimated basic reproduction number (R0) of 2.87 with increases up to 5 to 6 seen in densely populated areas, and the infectivity of asymptomatic individuals is thought to be responsible for the widespread COVID-19 disease.5 More complete details of the epidemiology and clinical aspects of COVID-19 disease can be found elsewhere.6 Although clinical disease is usually mild or even asymptomatic in children, a small percentage have serious inflammatory multiorgan disease process, multisystem inflammatory syndrome in children (MIS-C).7 , 8 Most adults also present with asymptomatic or mild-to-moderate symptoms (reviewed in the paper by Fang et al9). Recent data suggest that approximately 20% have a more progressive disease course with the development of pneumonia and respiratory failure progressing over days to weeks leading to intensive care admission. Approximately 10% of these individuals progress into a hyperinflammatory state, for example, acute respiratory distress syndrome (ARDS) with respiratory failure and death.10 In a CDC summary of the New York COVID-19 outbreak, the fatality rate among confirmed cases was 9.2% and 32.1% among hospitalized patients. Hospitalization and mortality were elevated among black and Hispanic persons and among residents of high-poverty neighborhoods.11 In this review, we will explore the possible explanations why some individuals have life-threatening COVID-19 disease whereas others have no or mild symptoms.
Risks Factors Predisposing to More Severe COVID-19 Infection
A number of sources have identified patient risk factors including race/ethnicity, cigarette smoking, age over 60, and the presence of other organ system diseases, for example, diabetes, obesity, hypertension, cardiovascular disease, chronic lung disease, and chronic renal disease.3 , 4 , 12 Surveillance studies also indicate that the black population and possibly the Hispanic population are disproportionally affected by COVID-19.12 Another factor that may play a role particularly in patients with darker skin is vitamin D. A number of studies have suggested that vitamin D has immune-modulating properties including viral replication.13 , 14 Vitamin D deficiency has been associated with an increased risk of respiratory infections such as respiratory syncytial virus infection and influenza.15 Szentpetery et al16 studied severe asthma exacerbations in 578 Puerto Rican children aged 6 to 14 years. Vitamin D insufficiency (plasma 25(OH)D <30 ng/mL) was consistently associated with a 5-fold increase in risk in those children without allergen sensitization for both severe exacerbations requiring systemic corticosteroids and 1 or more asthma hospitalizations. Meltzer et al17 reported that patients who were vitamin D deficient (<20 ng/mL) had an increased risk of COVID-19. COVID-19 rates in the deficient group were 21.6% (95% confidence interval [CI], 14.0%-29.2%) versus 12.2% (95% CI, 8.9%-15.4%) in the sufficient group. Similarly, Maghbooli et al18 reported that vitamin D levels >30 ng/mL reduced the risk of adverse clinical outcomes in patients with COVID-19 infection. Castillo et al19 evaluated the effects of calcifediol treatment on hospitalized Spanish patients on intensive care unit (ICU) admission and mortality from COVID-19. Of the 50 patients treated with calcifediol, 1 required admission to the ICU (2%), whereas 13 of the 26 untreated patients required admission to the ICU (50%; P < .001). African American and Hispanic populations that have high rates of vitamin D deficiency or insufficiency and bear a disproportionate burden of morbidity and mortality from SARS-CoV-2 are important populations to study to determine whether vitamin D supplementation can reduce the risk of severe COVID-19. Finally, in an interesting paper on the influence of temperature, humidity, and latitude on COVID-19 disease of 50 cities throughout the world, substantial community outbreaks of SARS-CoV-2 occurred along a narrow band between 30° N and 50° N, latitudes where vitamin D deficiency and insufficiency would be more common.20
Genetic Risk of Severe Life-Threatening COVID-19 Disease
The severe COVID-19 GWAS Group performed genotyping of patients with confirmed infection and receiving supplemental oxygen at Italian and Spanish epicenters.21 They identified a multigene locus at 3p21.31 and the ABO blood group locus at 9q34.2, which were associated with severe SARS-CoV-2 infection. Patients with blood type A had an increased risk for severe COVID-19, whereas those with blood group O had a decreased risk. Other groups have also implicated the ABO blood groups in the susceptibility to SARS-CoV-2.22 Six candidate genes in the 3p21.31 locus were associated with an increased risk of severe COVID-19. A highly significant variant (rs11385942) in the 3p21.31 locus was associated with respiratory failure and is associated with reduced expression of CXCR6 and increased expression of SLC6A20 and LZTFL1, both of which are strongly expressed in the lung. These gene loci may play an important role in host responses to the virus. SLC6A20 encodes a transporter protein that interacts with the angiotensin-converting enzyme 2 (ACE2) receptor, and the LZTFL1 gene is involved with T-cell activation. CXCR6 regulates lung resident memory CD8 T cells important in viral infections.23
Primary immune deficiencies have been an important model for characterizing the relationship between increased susceptibility to certain pathogens and monogenic inborn errors of immunity (reviewed in the paper by Notarangelo et al24). Perhaps the strongest data for the importance of the IFN pathway are the recent studies by the COVID Human Genetic Effort (www.covidhge.com) group. They sequenced the exome or genome of patients with life-threatening SARS-CoV-2 pneumonia and subjects with asymptomatic or mild infection, and identified an increase of loss-of-function variants in 13 candidate loci in individuals with severe COVID-19.25 Twenty-three patients (3.5%) carried 24 deleterious variants of 8 genes in the type I IFN pathway: autosomal-recessive deficiencies (IRF7 and IFNAR1) and autosomal dominant deficiencies (TLR3, UNC93B1, TICAM1, TBK1, IRF3, IRF7, IFNAR1, and IFNAR2). Liu et al26 applied a Mendelian randomization analysis and identified that the IFNAR2 gene (encoding the IFN-α/β receptor β chain) was associated with an increased risk and prognosis of COVID-19. The products of some of these genes are important as double-stranded RNA sensor products, whereas the other gene variants are important in the type I IFN pathway (Figure 1 ). In a related paper, Bastard et al27 identified neutralizing IgG autoantibodies against type I IFNs in 13.7% of patients with severe SARS-CoV-2 infection, whereas none of the individuals with asymptomatic or mild disease, and only 0.3% of healthy controls had these autoantibodies. Two of their patients with severe COVID-19 disease in this cohort had autoimmune polyendocrinopathy syndrome type I, a primary immune deficiency known to have autoantibodies to type I IFNs and other autoantibodies.28 Males accounted for 94% of the patients who had these autoantibodies and they tended to be older (49.5% were over age 65). IFN dysregulation represents a key part of SARS-CoV-2 susceptibility and may lead to new therapeutic modalities.29
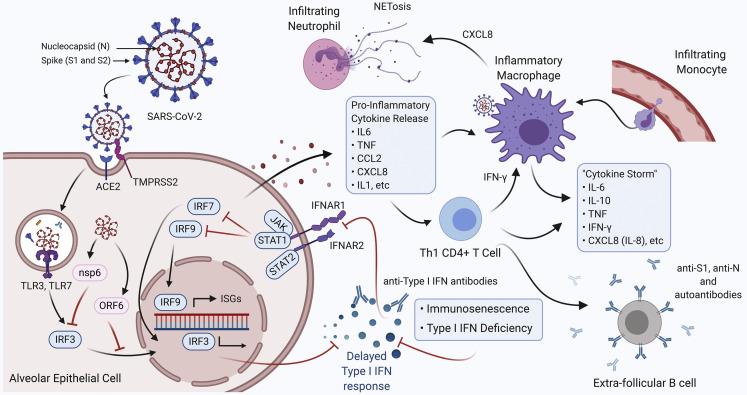
Figure 1.
Factors contributing to the hyperinflammatory immune response in severe SARS-CoV-2 infection. SARS-CoV-2 infects alveolar epithelial cells by binding to the angiotensin converting enzyme-2 (ACE2) receptor with the help of serine protease transmembrane serine protease 2 (TMPRSS2). The virus components (nsp6, ORF6) repress type I IFN responses by inhibiting the interferon regulatory factor 3 (IRF3) translocation to the nucleus. Repression of type I IFN responses may also be the result of pre-existing type I IFN monogenic variants, immunosenescence, or autoantibodies targeting type I IFN products (IFN α2, β, ω) preventing signaling through the IFNAR1/2 receptor complex. Delayed type I IFN responses result in the release of proinflammatory cytokines and chemokines, leading to the recruitment of monocyte-derived macrophages and T cells into the lungs. Proinflammatory signaling by activated, inflammatory macrophages leads to the recruitment of neutrophils into the lungs that undergo NETosis promoting further tissue damage. T cells recruited into the lungs appear to be of the CD4+ Th1 phenotype that promotes an inflammatory macrophage phenotype via IFN-γ signaling. In severe cases of COVID-19, humoral responses are derived from extrafollicular B cells that produce ineffective antibody responses coupled with the production of autoantibodies. This dysregulation and imbalance of the immune response leads to a hyperinflammatory state resulting in a “cytokine storm,” acute respiratory distress syndrome, and, in many cases, death.
(Figure created with Biorender.)
An international collaborative group of clinical immunologists reported on COVID-19 disease in 94 patients with inborn errors of immunity.30 Fifty-three patients (56%) had primary antibody deficiency, 9 (9.6%) had immune dysregulation syndrome, 6 (6.4%) a phagocyte defect, 7 (7.4%) an autoinflammatory disorder, 14 (15%) a combined immunodeficiency, 3 (3%) an innate immune defect, and 2 (2%) bone marrow failure. Ten had asymptomatic COVID-19 disease, 25 were treated as outpatients, 28 required admission, 13 required oxygen administration without invasive ventilation, whereas 23 patients required more intensive support in the ICU and 9 died. This latter group had predisposing risk factors similar to the general population for greater severity and mortality for COVID-19 disease. Patients with humoral immune deficiency, for example, x-linked agammaglobulinemia, hypogammaglobulinemia, and common variable immune deficiency (CVID), appeared to do better with asymptomatic or mild disease, perhaps by reducing the immune-mediated inflammatory response seen in severe COVID-19 disease.30 Cohen et al31 conducted a retrospective study of 135 patients with CVID, of whom 10 had COVID-19 infection. There was no mortality and low morbidity, reflecting a similar experience of Meyts et al.30 However, other groups have reported greater morbidity in patients with primary immune deficiency perhaps due to more severe risk factors and the types of immune deficiency in their patient population.32 , 33
Immune Responses Contributing to Severe COVID-19 Infection
Although the immune system response against SARS-CoV-2 for most patients is well regulated and functions similarly to infection with common seasonal coronaviruses, emerging evidence suggests that immune imbalance and dysregulation of innate and adaptive immune response results in severe COVID-19. As information on severe versus mild/asymptomatic patients is gathered, this imbalance becomes apparent at the earliest stages of infection leading to a signaling cascade triggering severe disease pathogenesis and ultimately resulting in immune failure. During initial stages of infection, SARS-CoV-2 Spike (S) protein binds to the ACE2 receptor expressed on human airway epithelial cells and, in concert with transmembrane serine protease 2 (TMPRSS2), enters these cells.34 , 35 Several studies have suggested the importance of the type I IFN-I pathway in mediating the innate immune responses in viral infections and influencing the development of adaptive immune responses.36 The IFN-I family consists of 13 IFN-α subtypes, IFN-β, IFN-ϖ, IFN-κ, and IFN-ε, that all signal through the heterodimeric IFN-I receptor, comprising IFN-α/β receptor 1 (IFNAR1) and IFNAR2. Type I IFNs are protective in acute viral infections but can have either protective or deleterious roles in bacterial infections and autoimmune diseases. Interestingly, Ziegler et al37 reported that the surface expression of ACE2 is regulated by IFN-I whereby an increase in IFN production induces surface expression of ACE2 that intuitively could facilitate cellular entry of SARS-CoV-2. However, this positive feedback loop contradicts evidence collected from plasma samples of patients with severe COVID-19 that are characterized by an initial, weak IFN-I response accompanied by an increase in proinflammatory cytokines such as tumor necrosis factor, IL-6, and CXCL10.38 , 39 Trouillet-Assant et al40 assessed the kinetics of plasma IFN-I production in patients with COVID-19 and found an impaired IFN-I response in approximately 1 in 5 COVID critically ill patients. Hadjadj et al41 studied 50 patients with COVID-19 with different disease severity and observed a distinct impaired IFN-I response phenotype in patients with severe COVID-19 characterized by no IFN-β and low IFN-α production. Furthermore, SARS-CoV-2 proteins nsp13, nsp6, and ORF6 have all been shown to contribute to decreased IFN-I production by antagonizing IFN regulatory factor 3 signaling and translocation to the nucleus in infected individuals (Figure 1).27
Severe COVID-19 disease is often found in the aging population, which indicates that immunosenescence, the generalized decline and remodeling of the immune system as a result of the aging process, may play a significant role in the early response to SARS-CoV-2 infection as seen in other upper respiratory infections.42 For instance, Kim et al43 showed that the aging population had attenuated IFN-I responses on influenza infection leading to an increased viral load. Similar IFN-I response impairment has been noted in obesity44 but not in diabetic individuals45 or recent kidney transplant patients on calcineurin inhibitors,46 both of which have increased levels of IFN-I, yet increased risk of severe COVID-19. However, there appears to be a temporal aspect to IFN-I responses in SARS-CoV-2 infection as Nienhold et al47 found increased IFN-I levels and enhanced IFN-stimulated gene products in postmortem analyses of lung tissue taken from COVID-19 victims and enhanced interferon-stimulated gene products. Thus, although the initial IFN-I response in those progressing to severe COVID-19 disease appears weak, this imbalance may only be temporary, possibly during rapid seeding of the viral reservoir in alveolar epithelial cells.48 The delayed or reduced production of type I IFNs may prolong the virus incubation time and persistence in the respiratory tract, and contribute to the increased production of proinflammatory cytokines resulting in a hyperinflammatory state. Because most of these studies have concentrated on hospitalized or convalescent patients, it is unclear if this impairment in the production of the type I IFNs is responsible for the susceptibility of individuals to severe disease.
The innate response to SARS-CoV-2 infection is largely regulated by macrophages and neutrophils. Similar to type 1 and 2 pneumocytes, alveolar tissue resident macrophages express ACE2 and can potentially be directly infected by SARS-CoV-2 but may also take up virus through phagocytic consumption of epithelial cell debris.49 In mild and asymptomatic COVID-19 cases, these alveolar macrophages predominate and act as effector cells to rapidly clear virally infected epithelial cells. However, age and accompanying immunosenescence shifts innate immune defenses toward a proinflammatory response driven by increased levels of inflammatory modulators such as IFN-γ as alveolar macrophages lose plasticity.50 It should be noted that these same shifts toward a more proinflammatory state of the innate immune response have not only been seen in older individuals, but also in individuals with hypertension, diabetes, and obesity, all of which are comorbidities in severe COVID-19 pathogenesis.51, 52, 53 Because of slowed immunoregulatory responses to control the inflammatory response in severe cases of COVID-19, infiltration of highly inflammatory, monocyte-derived macrophages inundates alveolar tissue as a consequence of an increase in the release of proinflammatory cytokines resulting in lung damage and subsequent ARDS.54 These inflammatory macrophages may polarize alveolar macrophages away from the M2 wound healing macrophage phenotype toward the highly proinflammatory M1 macrophage phenotype induced by the release of IFN-γ and IL-6.55 Infiltrating macrophages also release cytokines and chemoattractants, including IL-8, CXCL2, and CXCL8, that induce chemotaxis of neutrophils into the respiratory tract in severe cases of COVID-19. The resulting neutrophilia promotes the formation of neutrophil extracellular traps, exacerbating hyperinflammatory responses by facilitating further recruitment of neutrophils and other immune cells including T cells and monocytes to inflammatory sites.56 The influx of proinflammatory macrophages, neutrophils, and other immune cells into the respiratory tract invokes a powerful, uncontrollable proinflammatory response, driven by macrophage-activation syndrome presenting as a “cytokine storm” and culminating in often insurmountable lung injury and patient death (Figure 1).49 , 57, 58, 59, 60 The implications of early signaling events in SARS-CoV-2 infection and the uncoordinated innate immune response to these events paint a grim picture in severe COVID-19 pathogenesis wherein a patient's fate may be determined from the very outset of infection (Figure 1).
Whereas imbalance and dysfunction in the innate immune system are involved in inducing disease pathogenicity in patients with severe COVID-19, the adaptive immune system fairs no better in its response to SARS-CoV-2 infection. Although most patients with COVID-19 experiencing mild or asymptomatic disease generate what would be deemed a “normal” T-cell response, characterized by adequate CD4 and CD8 T-cell activation working in concert with the humoral immune system against a pathogen, patients with severe COVID-19 suffer from suboptimal CD8 T-cell responses coupled with pronounced, near universal lymphopenia.61 , 62 In older individuals, as well as individuals with obesity, lymphopenia may be a result of thymic involution, a process by which the thymic epithelial architecture is slowly replaced by adipose tissue deposits, leading to increased susceptibility to infection.63 , 64 Severe patients generally present with robust CD4 T-cell responses but suboptimal CD8 T-cell responses characterized by overactivation, impairment, or exhaustion. Studies by Habel et al65 indicate that the CD8 T-cell response may be limited to 2 HLA-A∗02:01-restricted SARS-CoV-2-specific CD8+ T-cell epitopes, A2/S269-277 and A2/Orf1ab3183-3191, which are biased toward naïve, stem cell memory rather than an effector phenotype and quickly undergo exhaustion. Schulien et al66 suggested that pre-existing CD8 T cells that cross-react with common cold coronaviruses may play a protective role in mild SARS-CoV-2 infection. Without such pre-existing cellular immunity, however, CD8 cytotoxic T-cell activity is impeded, greatly hindering SARS-CoV-2-infected cell clearance leading to sustained infection.
Whereas pre-existing CD8 T-cell immunity may lead to less severe disease, pre-existing CD4 memory T cells may play a detrimental role in severe SARS-CoV-2 infection.67 Although there are discrepancies as to the dynamics and imbalance of specific populations of CD4 T helper (Th) cells in severe versus mild/asymptomatic SARS-CoV-2 infection, the pragmatic view is that severe COVID-19 is driven by an increase in the Th1/Th2 ratio, which conforms to observed increases in levels of IFN-γ, contributing to macrophage M1 polarization and a further shift of naïve CD4 T cells toward the Th1 phenotype.68 These CD4 T-cell responses may derive from pre-existing memory T cells that are expanded in the elderly population, but bear low T-cell receptor avidity and impaired function.67 Contrary to older populations, observations of COVID-19 infection in children indicate a potentially protective effect of increased Th2 phenotype rather than a shift toward Th1 as seen in severe disease.69 Although counterintuitively associated with eosinophilic inflammation and increased TMPRSS2 expression, Th2 cytokine production downregulates ACE2 surface expression in epithelial cells of the respiratory tract. This downregulation of ACE2 before initial SARS-CoV-2 infection may play a role in greatly reducing symptoms in children.69 Lending credence to this theory of pre-emptive COVID-19 Th2-related protection is the observation that adolescents presenting with MIS-C express elevated levels of the Th1 cytokine IL-18 and Th1 chemokines CXCL10 and CXCL9.70 These results indicate that although patients with severe COVID-19 do mount a limited cellular-mediated immune response against SARS-CoV-2, dysfunction and imbalance in the helper CD4/CD8 T-cell response plays a significant role in determining severity and resolution of disease.
At first glance, the humoral immune response against SARS-CoV-2 appears to be strong in patients suffering from severe COVD-19. The antibody response to SAR-CoV-2 is directed against the viral envelope Spike (S) protein, comprising the S1 subunit that conceals the ACE2 receptor-binding domain and the S2 subunit, and the nucleocapsid protein (N). Serum IgG antibody titers directed against S and N correspond with disease severity with elevated titers seen in severe cases throughout disease pathogenesis in contrast to the lower levels found in mild and asymptomatic cases. Early IgM antibody responses against S1 and N appear to be short lived whether a patient experiences severe or mild disease progression.71 Counterintuitively, however, anti-S1 and anti-N IgG antibodies arise early in severe cases of COVID-19, and premature seroconversion may serve as a prognostic indicator of future disease severity.71 Although Imai et al72 suggest that these early IgG isotype antibodies may arise from immunological memory to cross-reactive antigens from previous exposure to other human coronaviruses, they may also result from the absence of germinal center (GC) formation in severe COVID-19 cases and reduction of Bcl-6-expressing GC B cells inducing extrafollicular B-cell activation.73 These extrafollicular B cells produce nonsomatically hypermutated IgG antibodies that may be of lower quality and of limited duration. They may also contribute to an autoantibody repertoire similar to that found in systemic lupus erythematous, acquiring prothrombotic antiphospholipid autoantibodies leading to the development of antiphospholipid syndrome with subsequent venous and arterial thrombosis.74 , 75 Furthermore, these extrafollicular B cells may themselves exhibit antibody-dependent and -independent pathogenesis through the secretion of inflammatory cytokines, further contributing to the hyperinflammatory state of severe COVID-19 (Figure 1).76 Although the mechanisms driving the loss of GCs is not yet fully understood, previous characterizations of extrafollicular responses have been associated with IL-6 and CXCL10, both of which are elevated in severe COVID-19.76 Chakraborty et al77 reported that patients with severe COVID-19 produced IgG1 receptor-binding domain antibodies with significantly reduced Fc fucosylation. These afucosylated Fc antibodies have enhanced interactions with the FcγRIIIa, leading to the increased production of inflammatory cytokines by monocytes. These observations in severe COVID-19 indicate that it may not simply be the quantity of the antibody response produced, but rather the quality of both the antibodies as well as the overall balance and health of the B-cell compartment.
The encompassing theme of the immune system in SARS-CoV-2 infection leading to severe COVID-19 is one of asymmetry and dysregulation eliciting a disproportionate and detrimental hyperinflammatory cascade. This immune disarray is particularly prominent in the elderly population, ascribable to initial failures in the immune defenses to viral infection brought about by immunosenescence. Although children and young/middle-aged adults tend to fair much better in their immune responses to SARS-CoV-2 infection, severe disease can still occur in these populations. Although specific mechanisms and/or genetic influences inducing severe COVID-19 in the younger demographic are currently unknown, it should be noted that hospitalized young adults often present with significant risk factors and comorbidities for severe COVID-19 such as obesity, diabetes, and hypertension.78 Although immunosenescence can explain some severe disease pathogenesis by restricting immune response and regulation of the hyperinflammatory state in older persons, it does not delineate why some older individuals may present as completely asymptomatic. Similarly, although diminished type I interferon responses are present in a large number of severe COVID-19 cases, these initial responses cannot fully explain all cases of severe COVID-19, and it is likely that a combination of multiple genetic and nongenetic factors contributes to an individual's unique immune response and susceptibility to SARS-CoV-2 infection.
Footnotes
No funding was received for this work.
Conflicts of interest: M. Ballow declares that he has no relevant conflicts of interest. C. L. Haga has received funding from the Schacknow Family Foundation, Inc.; and has previously served as a consultant for the purification of adenovirus-based therapeutics including vaccines for various companies.
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johns Hopkins University Coronavirus COVID-19 global cases by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU); 2020. https://coronavirus.jhu.edu/map.html Available from: Accessed January 11, 2021.
- 3.Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furukawa N.W., Brooks J.T., Sobel J. Evidence supporting transmission of severe acute respiratory syndrome coronavirus 2 while presymptomatic or asymptomatic. Emerg Infect Dis. 2020;26 doi: 10.3201/eid2607.201595. eid2607.201595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hosoki K., Chakraborty A., Sur S. Molecular mechanisms and epidemiology of COVID-19 from an allergist's perspective. J Allergy Clin Immunol. 2020;146:285–299. doi: 10.1016/j.jaci.2020.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.COVID-1RT C Coronavirus disease 2019 in children—United States, February 12-April 2, 2020. MMWR. 2020;69:422–426. doi: 10.15585/mmwr.mm6914e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felsenstein S., Hedrich C.M. SARS-CoV-2 infections in children and young people. Clin Immunol. 2020;220:108588. doi: 10.1016/j.clim.2020.108588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang F.C., Benson C.A., Del Rio C., Edwards K.M., Fowler V.G., Jr., Fredricks D.N. COVID-19—lessons learned and questions remaining. [published online ahead of print October 26, 2020]. Clin Infect Dis. [DOI] [PMC free article] [PubMed]
- 10.Sinha P., Matthay M.A., Calfee C.S. Is a “cytokine storm” relevant to COVID-19? JAMA Intern Med. 2020;180:1152–1154. doi: 10.1001/jamainternmed.2020.3313. [DOI] [PubMed] [Google Scholar]
- 11.Thompson C.N., Baumgartner J., Pichardo C., Toro B., Li L., Arciuolo R. COVID-19 outbreak—New York City, February 29-June 1, 2020. MMWR Morb Mortal Wkly Rep. 2020;69:1725–1729. doi: 10.15585/mmwr.mm6946a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garg S., Kim L., Whitaker M., O'Halloran A., Cummings C., Holstein R. Hospitalization rates and characteristics of patients hospitalized with laboratory-confirmed coronavirus disease 2019—COVID-NET, 14 states, March 1-30, 2020. MMWR Morb Mortal Wkly Rep. 2020;69:458–464. doi: 10.15585/mmwr.mm6915e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martineau A.R., Jolliffe D.A., Hooper R.L., Greenberg L., Aloia J.F., Bergman P. Vitamin D supplementation to prevent acute respiratory tract infections: systematic review and meta-analysis of individual participant data. BMJ. 2017;356:i6583. doi: 10.1136/bmj.i6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwalfenberg G.K. A review of the critical role of vitamin D in the functioning of the immune system and the clinical implications of vitamin D deficiency. Mol Nutr Food Res. 2011;55:96–108. doi: 10.1002/mnfr.201000174. [DOI] [PubMed] [Google Scholar]
- 15.Berry D.J., Hesketh K., Power C., Hyppönen E. Vitamin D status has a linear association with seasonal infections and lung function in British adults. Br J Nutr. 2011;106:1433–1440. doi: 10.1017/S0007114511001991. [DOI] [PubMed] [Google Scholar]
- 16.Szentpetery S.E., Han Y.Y., Brehm J.M., Acosta-Pérez E., Forno E., Boutaoui N. Vitamin D insufficiency, plasma cytokines, and severe asthma exacerbations in school-aged children. J Allergy Clin Immunol Pract. 2018;6:289–291.e2. doi: 10.1016/j.jaip.2017.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meltzer D.O., Best T.J., Zhang H., Vokes T., Arora V., Solway J. Association of vitamin D status and other clinical characteristics with COVID-19 test results. JAMA Netw Open. 2020;3:e2019722. doi: 10.1001/jamanetworkopen.2020.19722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maghbooli Z., Sahraian M.A., Ebrahimi M., Pazoki M., Kafan S., Tabriz H.M. Vitamin D sufficiency, a serum 25-hydroxyvitamin D at least 30 ng/mL reduced risk for adverse clinical outcomes in patients with COVID-19 infection. PLoS One. 2020;15:e0239799. doi: 10.1371/journal.pone.0239799. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Castillo M., Costa L., Barrios J., Diaz J., Miranda J., Bouillon R. Effect of calcifediol treatment and best available therapy versus best available therapy on intensive care unit admission and mortality among patients hospitalized for COVID-19: A pilot randomized clinical study. J Steroid Biochem Mol Biol. 2020;203:1–6. doi: 10.1016/j.jsbmb.2020.105751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sajadi M.M., Habibzadeh P., Vintzileos A., Shokouhi S., Miralles-Wilhelm F., Amoroso A. Temperature, humidity, and latitude analysis to estimate potential spread and seasonality of coronavirus disease 2019 (COVID-19) JAMA Network Open. 2020;3:e2011834. doi: 10.1001/jamanetworkopen.2020.11834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellinghaus D., Degenhardt F., Bujanda L., Buti M., Albillos A., Invernizzi P. Genomewide association study of severe Covid-19 with respiratory failure. N Engl J Med. 2020;383:1522–1534. doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao J., Yang Y., Huang H., Li D., Gu D., Lu X. Relationship between the ABO blood group and the COVID-19 susceptibility. [published online ahead of print August 4, 2020]. Clin Infect Dis. [DOI] [PMC free article] [PubMed]
- 23.Wein A.N., McMaster S.R., Takamura S., Dunbar P.R., Cartwright E.K., Hayward S.L. CXCR6 regulates localization of tissue-resident memory CD8 T cells to the airways. J Exp Med. 2019;216:2748–2762. doi: 10.1084/jem.20181308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Notarangelo L.D., Bacchetta R., Casanova J.L., Su H.C. Human inborn errors of immunity: an expanding universe. Sci Immunol. 2020;5:1–16. doi: 10.1126/sciimmunol.abb1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370:1–13. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu D., Yang J., Feng B., Lu W., Zhao C., Li L. Mendelian randomization analysis identified genes pleiotropically associated with the risk and prognosis of COVID-19. J Infect. 2021;82:126–132. doi: 10.1016/j.jinf.2020.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.H., Zhang Y. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:1–12. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo C.J., Leung P.S.C., Zhang W., Ma X., Gershwin M.E. The immunobiology and clinical features of type 1 autoimmune polyglandular syndrome (APS-1) Autoimmun Rev. 2018;17:78–85. doi: 10.1016/j.autrev.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 29.Monk P.D., Marsden R.J., Tear V.J., Brookes J., Batten T.N., Mankowski M. Safety and efficacy of inhaled nebulised interferon beta-1a (SNG001) for treatment of SARS-CoV-2 infection: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Respir Med. 2021;9:196–206. doi: 10.1016/S2213-2600(20)30511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyts I., Bucciol G., Quinti I., Neven B., Fischer A., Seoane E. Coronavirus disease 2019 in patients with inborn errors of immunity: an international study. J Allergy Clin Immunol. 2021;147:520–531. doi: 10.1016/j.jaci.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen B., Rubinstein R., Gans M.D., Deng L., Eisenberg R., Rubinstein A. COVID-19 infection in ten common variable immunodeficiency patients in New York City. J Allergy Clin Immunol Pract. 2020;9:504–507.e1. doi: 10.1016/j.jaip.2020.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho H.E., Mathew S., Peluso M.J., Cunningham-Rundles C. Clinical outcomes and features of COVID-19 in patients with primary immunodeficiencies in New York City. J Allergy Clin Immunol Pract. 2020;9:490–493.e2. doi: 10.1016/j.jaip.2020.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shields A.M., Burns S.O., Savic S., Richter A.G. COVID-19 in patients with primary and secondary immunodeficiency: the United Kingdom experience. J Allergy Clin Immunol. 2021;147 doi: 10.1016/j.jaci.2020.12.620. 870-5.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivashkiv L.B., Donlin L.T. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Acharya D., Liu G., Gack M.U. Dysregulation of type I interferon responses in COVID-19. Nat Rev Immunol. 2020;20:397–398. doi: 10.1038/s41577-020-0346-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Galani I.-E., Rovina N., Lampropoulou V., Triantafyllia V., Manioudaki M., Pavlos E. Untuned antiviral immunity in COVID-19 revealed by temporal type I/III interferon patterns and flu comparison. Nat Immunol. 2021;22:32–40. doi: 10.1038/s41590-020-00840-x. [DOI] [PubMed] [Google Scholar]
- 40.Trouillet-Assant S., Viel S., Gaymard A., Pons S., Richard J.C., Perret M. Type I IFN immunoprofiling in COVID-19 patients. J Allergy Clin Immunol. 2020;146:206–208.e2. doi: 10.1016/j.jaci.2020.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Domingues R., Lippi A., Setz C., Outeiro T.F., Krisko A. SARS-CoV-2, immunosenescence and inflammaging: partners in the COVID-19 crime. Aging (Albany NY) 2020;12:18778–18789. doi: 10.18632/aging.103989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim J.-A., Seong R.-K., Shin O.S. Enhanced viral replication by cellular replicative senescence. Immune Netw. 2016;16:286–295. doi: 10.4110/in.2016.16.5.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terán-Cabanillas E., Hernández J. Role of leptin and SOCS3 in inhibiting the type I interferon response during obesity. Inflammation. 2017;40:58–67. doi: 10.1007/s10753-016-0452-x. [DOI] [PubMed] [Google Scholar]
- 45.Newby B.N., Mathews C.E. Type I interferon is a catastrophic feature of the diabetic islet microenvironment. Front Endocrinol (Lausanne) 2017;8:1–15. doi: 10.3389/fendo.2017.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glowacka P., Rudnicka L., Warszawik-Hendzel O., Sikora M., Goldust M., Gajda P. The antiviral properties of cyclosporine. Focus on coronavirus, hepatitis C virus, influenza virus, and human immunodeficiency virus infections. Biology (Basel) 2020;9:192. doi: 10.3390/biology9080192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nienhold R., Ciani Y., Koelzer V.H., Tzankov A., Haslbauer J.D., Menter T. Two distinct immunopathological profiles in autopsy lungs of COVID-19. Nat Commun. 2020;11:5086. doi: 10.1038/s41467-020-18854-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xia H., Cao Z., Xie X., Zhang X., Chen J.Y.-C., Wang H. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 2020;33:1–12. doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang C., Xie J., Zhao L., Fei X., Zhang H., Tan Y. Alveolar macrophage dysfunction and cytokine storm in the pathogenesis of two severe COVID-19 patients. EBioMedicine. 2020;57:102833. doi: 10.1016/j.ebiom.2020.102833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stout R.D., Suttles J. Immunosenescence and macrophage functional plasticity: dysregulation of macrophage function by age-associated microenvironmental changes. Immunol Rev. 2005;205:60–71. doi: 10.1111/j.0105-2896.2005.00260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chiappetta S., Sharma A.M., Bottino V., Stier C. COVID-19 and the role of chronic inflammation in patients with obesity. Int J Obes (Lond) 2020;44:1790–1792. doi: 10.1038/s41366-020-0597-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang S., Wang J., Liu F., Liu J., Cao G., Yang C. COVID-19 patients with hypertension have more severe disease: a multicenter retrospective observational study. Hypertens Res. 2020;43:824–831. doi: 10.1038/s41440-020-0485-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lim S., Bae J.H., Kwon H.-S., Nauck M.A. COVID-19 and diabetes mellitus: from pathophysiology to clinical management. Nat Rev Endocrinol. 2021;17:11–30. doi: 10.1038/s41574-020-00435-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 55.Martinez F.O., Combes T.W., Orsenigo F., Gordon S. Monocyte activation in systemic Covid-19 infection: assay and rationale. EBioMedicine. 2020;59:102964. doi: 10.1016/j.ebiom.2020.102964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang J., Li Q., Yin Y., Zhang Y., Cao Y., Lin X. Excessive neutrophils and neutrophil extracellular traps in COVID-19. Front Immunol. 2020;11:2063. doi: 10.3389/fimmu.2020.02063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iwasaki M., Saito J., Zhao H., Sakamoto A., Hirota K., Ma D. Inflammation triggered by SARS-CoV-2 and ACE2 augment drives multiple organ failure of severe COVID-19: molecular mechanisms and implications. Inflammation. 2021;44:13–34. doi: 10.1007/s10753-020-01337-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jafarzadeh A., Chauhan P., Saha B., Jafarzadeh S., Nemati M. Contribution of monocytes and macrophages to the local tissue inflammation and cytokine storm in COVID-19: lessons from SARS and MERS, and potential therapeutic interventions. Life Sci. 2020;257:118102. doi: 10.1016/j.lfs.2020.118102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang Y., Liu J., Zhang D., Xu Z., Ji J., Wen C. Cytokine storm in COVID-19: the current evidence and treatment strategies. Front Immunol. 2020;11:1708. doi: 10.3389/fimmu.2020.01708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tomar B., Anders H.-J., Desai J., Mulay S.R. Neutrophils and neutrophil extracellular traps drive necroinflammation in COVID-19. Cells. 2020;9:1383. doi: 10.3390/cells9061383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Biasi S., Meschiari M., Gibellini L., Bellinazzi C., Borella R., Fidanza L. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat Commun. 2020;11:3434. doi: 10.1038/s41467-020-17292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Westmeier J., Paniskaki K., Karaköse Z., Werner T., Sutter K., Dolff S. Impaired cytotoxic CD8 T cell response in elderly COVID-19 patients. mBio. 2020;11:e02243–e02320. doi: 10.1128/mBio.02243-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aspinall R., Andrew D. Thymic involution in aging. J Clin Immunol. 2000;20:250–256. doi: 10.1023/a:1006611518223. [DOI] [PubMed] [Google Scholar]
- 64.Yang H., Youm Y.H., Vandanmagsar B., Rood J., Kumar K.G., Butler A.A. Obesity accelerates thymic aging. Blood. 2009;114:3803–3812. doi: 10.1182/blood-2009-03-213595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Habel J.R., Nguyen T.H.O., van de Sandt C.E., Juno J.A., Chaurasia P., Wragg K. Suboptimal SARS-CoV-2−specific CD8 T cell response associated with the prominent HLA-A∗02:01 phenotype. Proc Natl Acad Sci USA. 2020;117:24384–24391. doi: 10.1073/pnas.2015486117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schulien I., Kemming J., Oberhardt V., Wild K., Seidel L.M., Killmer S. Characterization of pre-existing and induced SARS-CoV-2-specific CD8+ T cells. Nat Med. 2021;27:78–85. doi: 10.1038/s41591-020-01143-2. [DOI] [PubMed] [Google Scholar]
- 67.Bacher P., Rosati E., Esser D., Rios Martini G., Saggau C., Schiminsky E. Pre-existing T cell memory as a risk factor for severe COVID-19 in the elderly. medRxiv 2020. Accessed February 2, 2021. [DOI]
- 68.Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369:eabc8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Steinman J.B., Lum F.M., Ho P.P.-K., Kaminski N., Steinman L. Reduced development of COVID-19 in children reveals molecular checkpoints gating pathogenesis illuminating potential therapeutics. Proc Natl Acad Sci USA. 2020;117:24620–24626. doi: 10.1073/pnas.2012358117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gruber C.N., Patel R.S., Trachtman R., Lepow L., Amanat F., Krammer F. Mapping systemic inflammation and antibody responses in multisystem inflammatory syndrome in children (MIS-C) Cell. 2020;183:982–995.e14. doi: 10.1016/j.cell.2020.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hu W., Howell J.C., Ozturk T., Benameur K., Bassit L., Ramonell R. Antibody profiles according to mild or severe SARS-CoV-2 infection, Atlanta, Georgia, USA, 2020. Emerg Infect Dis. 2020;26:2974. doi: 10.3201/eid2612.203334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Imai K., Kitagawa Y., Tabata S., Kubota K., Nagura-Ikeda M., Matsuoka M. Antibody response patterns in COVID-19 patients with different levels of disease severity—Japan. medRxiv 2020. Accessed February 2, 2021. [DOI] [PMC free article] [PubMed]
- 73.Kaneko N., Kuo H.-H., Boucau J., Farmer J.R., Allard-Chamard H., Mahajan V.S. Loss of Bcl-6-expressing T follicular helper cells and germinal centers in COVID-19. Cell. 2020;183:143–157.e13. doi: 10.1016/j.cell.2020.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zuniga M., Gomes C., Carsons S.E., Bender M.T., Cotzia P., Miao Q.R. Autoimmunity to the lung protective phospholipid-binding protein Annexin A2 predicts mortality among hospitalized COVID-19 patients. medRxiv 2021. Accessed February 2, 2021. [DOI] [PMC free article] [PubMed]
- 75.Zuo Y., Estes S.K., Ali R.A., Gandhi A.A., Yalavarthi S., Shi H. Prothrombotic autoantibodies in serum from patients hospitalized with COVID-19. Sci Transl Med. 2020;12:eabd3876. doi: 10.1126/scitranslmed.abd3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Woodruff M.C., Ramonell R.P., Nguyen D.C., Cashman K.S., Saini A.S., Haddad N.S. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat Immunol. 2020;21:1506–1516. doi: 10.1038/s41590-020-00814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chakraborty S., Gonzalez J., Edwards K., Mallajosyula V., Buzzanco A.S., Sherwood R. Proinflammatory IgG Fc structures in patients with severe COVID-19. Nat Immunol. 2021;22:67–73. doi: 10.1038/s41590-020-00828-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cunningham J.W., Vaduganathan M., Claggett B.L., Jering K.S., Bhatt A.S., Rosenthal N. Clinical outcomes in young US adults hospitalized with COVID-19. JAMA Intern Med. 2021;181:379–381. doi: 10.1001/jamainternmed.2020.5313. [DOI] [PMC free article] [PubMed] [Google Scholar]