Abstract
An effective early warning tool is of great administrative and social significance to the containment and control of an epidemic. Facing the unprecedented global public health crisis caused by COVID-19, wastewater-based epidemiology (WBE) has been given high expectations as a promising surveillance complement to clinical testing which had been plagued by limited capacity and turnaround time. In particular, recent studies have highlighted the role WBE may play in being a part of the early warning system. In this study, we briefly discussed the basics of the concept, the benefits and critical points of such an application, the challenges faced by the scientific community, the progress made so far, and what awaits to be addressed by future studies to make the concept work. We identified that the shedding dynamics of infected individuals, especially in the form of a mathematical shedding model, and the back-calculation of the number of active shedders from observed viral load are the major bottlenecks of WBE application in the COVID-19 pandemic that deserve more attention, and the sampling strategy (location, timing, and interval) needs to be optimized to fit the purpose and scope of the WBE project.
Keywords: COVID-19 surveillance, Epidemic early warning, Wastewater-based epidemiology, Fecal shedding, Virus genome recovery
Graphical abstract
1. Introduction
COVID-19 is an infectious respiratory disease caused by SARS-CoV-2 infection. Due to its highly contagious nature, following the initial cases reported in Wuhan, China at the end of 2019, COVID-19 has since swept the world, spreading to more than 200 countries and regions with a whopping worldwide case count of more than 61 million as of 30th November 2020 despite all the measures taken to control its transmission (WHO, 2020). The World Health Organization (WHO) had officially declared a global pandemic of COVID-19 on 11th March 2020.
Among the efforts to contain the COVID-19 pandemic and mitigate its adverse impact on the society in the absence of an effective vaccine, the ponderance of a reliable and timely epidemic surveillance system has been stressed. Conventionally, the epidemic surveillance relies heavily on clinical testing either conducted by existing healthcare facilities or temporarily established testing sites, and for COVID-19, using reverse transcription quantitative polymerase chain reaction (RT-qPCR) on nasopharyngeal swabs to detect RNA signal has been accepted as the standard testing procedure (D'Aoust et al., 2021). However, the high contagiousness and the presence of asymptomatic virus carriers have made the clinical testing capacity largely lag behind the demand, raising the concern about the grievous outcomes of underreporting, which has been suggested by both statistical analysis and seroprevalence surveys (Krantz and Rao, 2020; Medema et al., 2020a). Acknowledging the importance of filling the gap and lifting the pressure on testing facilities, recent studies have underlined the potential of wastewater-based epidemiology (WBE) as a solution complementary to clinical testing (Bivins et al., 2020b; Daughton, 2020; Thompson et al., 2020; Venugopal et al., 2020). Following its successful early applications of tracking illicit drug usage and lifestyle factors, WBE is gradually gaining popularity among researchers in the water-related field. Blessed with its community-wide coverage, ability to “see” the underreported and asymptomatic patients, and low-cost nature, WBE has been proposed to be a promising tool in infectious diseases surveillance, and unsurprisingly, high hopes are placed for its capability of helping combat COVID-19 as well (Daughton, 2020; Sims and Kasprzyk-Hordern, 2020; Thompson et al., 2020).
The basic concept of WBE centers around this principle: certain chemical or biological agents (also referred to as ‘biomarkers’) excreted by human bodies can be collected by the sewage network and end up entering the wastewater, making it a rich source of these substances. Via physicochemical methods, biomarkers can be recovered from wastewater and the measured concentration can then be used to infer the size of the shedding population and provide community-level health information (Xagoraraki and O'Brien, 2020). For SARS-CoV-2, although antigen testing is also emerging (Daughton, 2020), the viral genome has been widely accepted as the biomarker. To date, a handful of studies have reported the detection of the SARS-CoV-2 viral genome in sewage networks (Ahmed et al., 2020a; D'Aoust et al., 2021; Hata et al., 2020; La Rosa et al., 2020b; Medema et al., 2020b; Randazzo et al., 2020; Westhaus et al., 2021; Wurtzer et al., 2020).
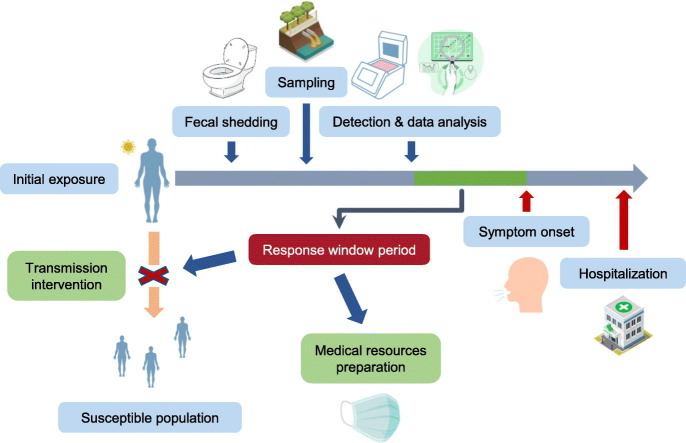
However, some believe that the true standout of WBE is the early warning capability. The term “early warning” can be interpreted in two ways in the context of COVID-19 surveillance: (1) signaling an early stage of an outbreak. Presymptomatic/asymptomatic transmission of COVID-19 is considered a key factor behind its rapid spread (Gandhi et al., 2020). Arons et al. (2020) recovered viable virus from 71% (17 in 24) of presymptomatic individuals 1 to 6 days prior to symptom onset, and He et al. (2020) estimated that 44% (95% CI 25–69%) of the secondary cases were infected when the index cases were in their presymptomatic stage. While asymptomatic and presymptomatic virus carriers can easily hide in the community due to the absence of appreciable symptoms such as fever and dry cough, by nature, WBE can indiscriminately detect their presence as long as they develop viral RNA shedding (Jones et al., 2020). Therefore, if a positive wastewater viral load is spotted in a region previously experiencing no or a low prevalence, it may indicate an unnoticed initial circulation of the virus in the community. This information can be made use by the local authority, who can take intervention by issuing warnings or administrative orders accordingly to inform the public of the potential threat and reduce the chance of invisible transmission. Also, as many countries and regions suffer from limited resources needed for a large-scale clinical testing program which greatly helps monitor the epidemic development and control the spread, getting a rough location of an initial circulation can help ease the burden and make the testing more efficient by guiding the valuable testing capacity to where it is most urgently needed; (2) foreshadowing an impending increase in infected individuals. The basic assumption behind this is: since infectiousness predates symptom onset, so can the viral shedding. Thus, if proper sampling tactics and quantification methods are adopted, an increased wastewater viral load may be observed and reported before the newly infected individuals develop symptoms and seek medical attention. Since there may be a correlation between wastewater viral load and the number of infected individuals, in addition to supporting the administrative and resource deployment measures previously described, from the perspective of disease treatment, the quantitative information also allows the healthcare facilities to take measures aimed at improving preparedness and coping with the anticipated new patients beforehand so that the facilities are less likely to be overwhelmed.
Both interpretations of early warning have been backed up by recent studies and events. Table 1 lists some selected recent wastewater surveillance studies that highlight the potential of WBE early warning of COVID-19. In terms of practical application, the University of Arizona made headlines in August 2020 when researchers there detected SARS-CoV-2 viral genome in the wastewater from a student dormitory, the university quickly took action and tested all 311 residents living and working in the said building and found two asymptomatic carriers among them, likely having prevented a potential outbreak and making it the first true application of WBE in COVID-19 early warning (Jaclyn Peiser, 2020).
Table 1.
Recent wastewater surveillance studies that indicate the potential of COVID-19 early warning via WBE.
| Region | Sample type | Primary concentration method | Sampling period | Population size in the WWTP catchment area | Major findings related to early warning potential | Reference |
|---|---|---|---|---|---|---|
| Murcia, Spain | Grab raw sewage | Al(OH)3 adsorption-precipitation | 2020/03/12–2020/04/14 | Multiple, from ~28,000 to ~530,000 | Wastewater samples from three WWTPs were tested positive 12–16 days before COVID-19 cases were reported in the respective catchment regions | (Randazzo et al., 2020) |
| Amersfoort, The Netherlands | Composite raw sewage | Ultrafiltration | 2020/02/05–2020/03/25 | ~234,000 | Sewage signaled virus circulation 6 days before the first cases were reported | (Medema et al., 2020b) |
| Milan, Italy | Composite raw sewage | PEG/dextran precipitation | 2020/02/03–2020/04/02 | ~1,050,000 | Samples were tested positive when the COVID-19 infections were very limited (29 in a larger area) | (La Rosa et al., 2020b) |
| Ishikawa, Japan | Grab raw sewage | PEG precipitation | 2020/03/05–2020/04/21 | Multiple, from ~31,000 to ~233,000 | Samples were tested positive when the prevalence was lower than one confirmed case per 100,000 people | (Hata et al., 2020) |
| Bozeman, MT, USA | Composite raw sewage | Ultrafiltration | 2020/03/30–2020/06/12 | ~50,000 | SARS-CoV-2 RNA levels in wastewater precede clinical PCR test results by 2–4 days | (Nemudryi et al., 2020) |
| New Haven, CT, USA | Primary sewage sludge | \ | 2020/03/19–2020/06/01 | ~200,000 | SARS-CoV-2 RNA concentrations in sludge predate hospital admissions by 1–4 days | (Peccia et al., 2020) |
However, despite the appealing side of WBE, a significant knowledge gap still exists regarding to what extent the promises regarding being an early warning system can be truly fulfilled, and potential bottlenecks are seldomly discussed in detail. To help readers fill the gap and draft a roadmap for further research, here in this study we would like to dive into the limiting factors in a real-world setting, discuss what has been done and made clear so far, what still awaits to be addressed, and where is the research frontier of these factors.
2. Limiting factors, current knowledge, and research needs
When used for detecting newly introduced virus carriers and initial virus circulation in a low-prevalence community, the viability of WBE and the confidence it offers largely lean on the lowest possible prevalence level that enables the detection of viral RNA in sewage. The practical value is governed by many factors including the sewage network layout and capacity, shedding profile of infected individuals, sewage characteristics, sampling strategy, the recovery efficiency of the concentration and quantification methods, and the detection limit of the instrument. A relatively reliable estimate requires the latest knowledge about the pathology of COVID-19, verified experimental method, as well as support from the local water agency. Some estimates have been given by previous studies, Hart and Halden (2020) performed a computational analysis with the City of Tempe, Arizona, USA being the studied region and estimated that a sensitivity of 1 in 144 to 2 million individuals can be achieved, depending on the assumptions used. Similarly, Ahmed et al. (2020a) reported an estimated prevalence level of 0.028% (95% CI 0.019–0.039%, 1 in 3571 individuals) based on viral RNA detection. However, these estimations may be too optimistic as some factors that can significantly affect the detection sensitivity are missing while others face significant uncertainty. For instance, neither of the two studies counted the recovery efficiency of the experimental method, the latter study also did not consider the natural degradation of the viral RNA. As for the example of the University of Arizona, despite a detection sensitivity of 0.64% (2 in 311) on paper, as further details (e.g., sampling strategy) remain undisclosed, it is unclear whether the same level of sensitivity can be expected under other conditions. Besides, in a quantitative sense, as an extension of calculating the lowest prevalence level that enables successful detection, a back-calculation model that projects the obtained wastewater viral load to the active shedding population is of foremost importance (Xagoraraki and O'Brien, 2020), yet so far, very few studies have challenged this issue.
Another measure of the viability of WBE in COVID-19 early warning is how responsive it can be. Even if the detection sensitivity is adequate for low prevalence detection, the value of detection can be seriously undermined, even nullified, if the result cannot reach the correct hands in time. Also, in regions where prevalence level is high enough to enable consistent viral RNA detection, WBE can still shine from its quantitative side; if the wastewater viral load is closely monitored and there is a surge in infections, as fecal shedding may predate symptom onset, an increase in viral load may appear before the newly infected individuals develop symptoms, seek medical attention, and be admitted to healthcare facilities after diagnosis, and the number of them may be inferred from the viral load. Different from the low prevalence detection which only gives a qualitative result, the quantitative outcome gives local healthcare facilities and their supervising agencies a response window period and an anticipated capacity demand. Just as in the case of testing capacity, in a time when many regions are having logistic difficulty handling the rapid increase in infections with limited resources (Kamerow, 2020), being able to forecast the demand may help get an upper hand and improve the preparedness as local healthcare facilities can make use of this time to (1) prepare necessary medical supplies and equipment including beds, ventilators, protective clothing, and masks; (2) arrange human resources to make sure there would be adequate health workers for the increased workload. In addition, from a higher angle, this community demand forecast may enable regional reallocation of available resources which can come in handy if there is an overall shortage.
However, both measures of the feasibility of WBE early warning face considerable uncertainty. In the previously mentioned studies regarding the lead of viral RNA in primary sewage sludge compared to local admissions, the analysis was performed in a retrospective way: the accumulated longitudinal SARS-CoV-2 quantification data were compared with the clinical reports during the same period. For WBE to be an active early warning tool, though, it needs to be performed in a timelier manner. One important index in the timeline of COVID-19 infection is the incubation period, which is the time gap between virus exposure to symptom onset. Some studies have reported very similar median or mean values of approximately 5 days (Lauer et al., 2020; Li et al., 2020; McAloon et al., 2020). After the symptom onset, there typically will be another period until the testing result comes out or the patient gets admitted to a hospital, Lauer et al. (2020) reported a mean value of 1.2 days, but it may vary greatly depending on the testing policy and the capacity of the hospital in question. Adding the two intervals up sets a reference for WBE; whether the workflow can be streamlined to beat this time largely affects its viability, although to what extent the outcome is useful also depends on how long is the response window period it leaves behind.
In the following sections, factors that may become bottlenecks, their significance, what previous and recent studies have revealed, and what awaits to be addressed and clarified by further studies are summarized and discussed to provide readers with a brief roadmap towards the final application of WBE as a part of the COVID-19 early warning system.
2.1. Shedding profile of infected individuals
The shedding profile of infected individuals directly determines the wastewater viral load and is hence regarded as one of the most critical factors in WBE. The shedding profile consists of three parts: the shedding rate, the beginning of shedding, and the shedding duration. When the shedding profile is relatively predictable and stable while showing finite between-person variation, it will greatly simplify the modeling process, but on the other hand, if the shedding profile bears significant stochastic fluctuations and between-person discrepancy, substantial extra efforts would be needed to handle the data noise and uncertainty.
So far, reports regarding shedding rate have mainly focused on hospitalized symptomatic patients due to the availability. Walsh et al. (2020) summarized in their review that while some studies reported little to no difference in the viral loads of symptomatic and asymptomatic patients, there are also studies that found the severity of symptoms can affect viral load, indicating substantial heterogeneity, and generally speaking, fecal viral shedding shows significant uncertainty and the overall pattern is more erratic than respiratory shedding. One of the earliest assessments of the rate and duration of fecal shedding was conducted by Wölfel et al. (2020), while the highest recorded viral load among 9 hospitalized patients reached 107 copies/g of stool sample, the results also show significant variation between cases; the viral load of one patient had stayed below 104 copies per gram of feces over the entire testing course. In a review by Parasa et al. (2020), the recorded viral load in stool samples also ranges from 550 to 1.21 × 105 copies per mL of feces. In addition to the variation in shedding rate, it has also been stated that not all infected individuals will develop fecal shedding. In the aforementioned study by Wölfel et al. (2020), the stool specimens of one patient were consistently negative. A meta-analysis by van Doorn et al. (2020) reported that 51.8% (95% CI 43.8–59.7%) of patients have their stool specimens tested positive while another systematic review by Gupta et al. (2020) reported a similar percentage (53.9%), but very limited information is available about the shedding ratio among asymptomatic virus carriers. Not only is this uncertain fecal shedding a hindrance to the estimation of achievable detection sensitivity, it also means that when the number of infected individuals is low, statistically, there is a chance that none of them sheds viral RNA into wastewater, making their presence undetectable by WBE no matter how sensitive the assays are.
Also, the timing of fecal shedding has a decisive role in determining how “early” the shedding can be detected. As the routine testing of fecal shedding typically only focuses on symptomatic patients after their hospitalization, solid evidence remains scarce as to the actual starting point of fecal shedding, especially among asymptomatic virus carriers. Alternatively, the timing of infectiousness development (respiratory shedding) may be used as a proxy. He et al. (2020) estimated that the infectious period begins at 2.3 days prior to symptom onset and peaks at 0.7 days before it. Nevertheless, it is important to keep in mind that respiratory shedding does not perfectly represent fecal shedding and may not exactly parallel it, related information hence must be interpreted and used prudently until more medical evidence becomes available.
As another critical aspect of the shedding profile, the persistency of shedding should also be given some consideration. It has been revealed that the shedding of SARS-CoV-2 in fecal specimens can outlast that in respiratory specimens (Jones et al., 2020; Wang et al., 2020; Wu et al., 2020; Xiao et al., 2020; Xu et al., 2020). The long-tailed fecal shedding may cause a masking effect on newly infected individuals, making their presence indistinguishable from the patients in their post-infection phase, especially when an infection peak has recently ended and the shedding population remains large. Although according to previous medical reports, the intensity of shedding steadily declines during the infection course, further clinical evidence is still needed to confirm whether the long-tailed shedding will become a concern for WBE application.
Because the shedding rate and duration determined from clinical case reports may be subjected to stochastic error and person-to-person variation, if possible, packing available data and biological explanation into a mathematical model for better generalization and easier extrapolation of the shedding dynamics is preferable. Currently, available information about this approach is very limited and further study is needed. Recently, Miura et al. (2020) fitted a shedding dynamics model (Eq. (1)) originally developed by Teunis et al. (2015) for norovirus fecal shedding.
| (1) |
This model assumes that virus particles first accumulate at an infection site and are then released from the intestinal tract into the environment. C(t|α, β) is the virus concentration in feces at time t, α and β are constants that are defined by the transport rate and effective volumes of the compartments within the intestinal tract, and C 0 is a constant controlling the height of peak virus concentration. The shedding curve features a rapid increase in the initial stage of infection, followed by a downward slide until the virus concentration falls below the detection limit. Other mathematical models have also been developed for virus shedding into saliva and blood based on the understanding of the infection process (Huynh and Rong, 2012; Osuna et al., 2016). However, although these previous models have provided some insights, as mentioned above, the fecal shedding of SARS-CoV-2 may have its own distinct characteristics and follow a different biological mechanism, it is unclear whether the same pathologic assumptions and consequently these mathematical models can be applied to SARS-CoV-2.
In conclusion, though much information has been made available, the current knowledge is still far from enough to support successful WBE application in absolute calculations. A heavy workload still lies ahead until the uncertainty in the fecal viral shedding can be properly addressed. However, it should be clearly stated that there is no guarantee that such a goal will finally be achieved therefore the worst scenario also needs to be considered: if the shedding profile is eventually found to be too erratic and unpredictable to be clearly described and properly modeled, as some existing literature suggests, the prospect of WBE will be critically impaired as it lacks the functionality to be a tool for absolute quantitative analysis. But to proceed from where we are now, a more comprehensive and holistic image of the shedding profile, including the rate, starting time, and duration is needed, which will benefit from further clinical evidence.
2.2. Recovery efficiency and instrument detection limit
Stable and efficient recovery and detection of the viral RNA is a decisive factor in wastewater surveillance. The recovery efficiency and instrument detection limit provide a critical reference when estimating the threshold prevalence level and back-calculating the shedding population from the viral load. For primary concentration and RNA extraction, several research articles and reviews have looked into this technical issue, focusing on either surrogates or other coronavirus strains (Ahmed et al., 2020d; La Rosa et al., 2020a; Rusiñol et al., 2020; Torii et al., 2020). Ahmed et al. (2020d) recently compared the recovery efficiency of some commonly used methods for wastewater virus concentration using murine hepatitis virus (MHV) as the surrogate for human coronavirus, the average recovery varied from 26.7 to 65.7%, with the method having the highest recovery efficiency being an adsorption-extraction method supplemented with MgCl2. Torii et al. (2020) conducted a similar study, in which Pseudomonas phage φ6 was used as the surrogate, and a method combining polyethylene glycol (PEG) precipitation and acid guanidinium thiocyanate-phenol-chloroform extraction achieved a mean recovery efficiency of 29.8 to 49.8%. From the standpoint of quantitative analysis, this means if the recovery efficiency is not considered, in other words assuming a 100% recovery, the estimated detection limit would be lower than the actual value, which may lead to a falsely high sense of security. However, even though both MHV and φ6 are enveloped viruses and may better resemble the behavior of SARS-CoV-2 than nonenveloped surrogates, discrepancy may still exist and the measured recovery efficiency should be used discreetly and only as a reference.
It is also worth mentioning that many established primary concentration methods were originally developed and validated for nonenveloped waterborne gastrointestinal viruses. Due to the distinct structure and surface property of enveloped viruses such as SARS-CoV-2, their behavior in the wastewater matrix may also be different, including the partitioning (Ye et al., 2016). This has been reflected by recent reports of the detection of SARS-CoV-2 in sewage sludge (D'Aoust et al., 2021; Kaplan et al., 2020; Peccia et al., 2020). However, it is important to point out that due to the sedimentation process, the viral load in primary sludge may be the result of an accumulation over several days and does not reflect the real-time change in the wastewater matrix. As for the wastewater solids, Kitamura et al. (2021) and Westhaus et al. (2021) recovered SARS-CoV-2 RNA from both the solid and liquid fractions of wastewater and the results suggest that wastewater solids may support more sensitive SARS-CoV-2 detection. Therefore, an extra step that helps release viral RNA from the solids (e.g., heat treatment and adsorption-elution) may improve recovery efficiency (Corpuz et al., 2020; Schwab et al., 1997), but additional research needs to be conducted to verify the efficacy for SARS-CoV-2.
The last barrier of the quantification assay is the detection and quantification limit. For RT-qPCR, a standard curve is necessary for converting the cycle threshold (Ct) value into virus titers, but if the signal intensity is below a certain Ct value, it would be indistinguishable from the potential noise. In practice, this Ct value limit is usually translated to gene copies per unit volume by referring to the standard curve. However, if the dilution series is not well configured, there could be a difference between the limit of detection (LoD) and the limit of quantification (LoQ). Attention should be paid to reduce or eliminate the gap between LoD and LoQ. PCR reaction inhibition is also a concern in wastewater surveillance, the introduction of process control, whether applied to the whole process, before RNA extraction and/or before RT-qPCR, has been proposed to help evaluate the extent of inhibition (Kitajima et al., 2020). In addition, the choice of RT-qPCR assay (Ahmed et al., 2020a; Hamouda et al., 2020; Hata et al., 2020; Kitamura et al., 2021) and nucleic acid extraction kit (Sidhu et al., 2013) can also affect the detection sensitivity. As in the case of the primary concentration method, at the current stage, a consensus of optimal recovery-detection assay has not been reached, researchers may need to conduct their experiments to determine the assay suitable for the lab condition and wastewater characteristics.
Some recent studies have employed droplet digital PCR (ddPCR) for the detection of SARS-CoV-2 RNA in clinical samples (Dang et al., 2020; Falzone et al., 2020; Suo et al., 2020; Yu et al., 2020) and suggested that ddPCR is a superior choice for clinical diagnosis for its higher sensitivity and other benefits such as not needing a standard curve for quantification. However, D'Aoust et al. (2021) compared RT-ddPCR and RT-qPCR using wastewater sludge samples and the results did not support the statement that RT-ddPCR performs better than RT-qPCR in the context of wastewater viral RNA detection. It is possible that the low detection limit offered by ddPCR can enhance the performance of the WBE approach, but related research needs to be further extended to investigate the effect of factors such as inhibition and optimize the assay.
2.3. Dilution factor and sampling strategy
Once the viral RNA is released from shedding individuals, it will enter the sewage network and get mixed with the rest of the wastewater. In its simplest form, the dilution factor can be determined by assuming a complete and homogeneous blend of the viral RNA shed by all shedding individuals in one day and the daily wastewater flowrate which is usually obtainable from the sewage network operator. But in practice, the mixing and dilution process is significantly influenced by the uneven diurnal wastewater flowrate and the timing of toilet flushing. There are two options of sampling: composite sample and grab sample. Composite samples are favored in recent detection reports and are generally considered more suitable for the task as multiple wastewater samples over a period of time are collected, this increases the success rate of detection given the high uncertainty of shedding, especially when the sampling period is set to 24 h and a flow-proportional sampler is used (Medema et al., 2020a; Michael-Kordatou et al., 2020; Thompson et al., 2020). However, considering the biological rhythm and living habits of human beings, the variance may, in turn, be beneficial and the grab sample may offer a higher chance of detection if the sampling time is optimized to capture the peak hours of toilet flushing. In the aforementioned study of Hata et al. (2020) in which a positive signal was detected when the catchment area had a low prevalence level (less than one confirmed case per 100,000 people), grab samples were used. However, due to the unevenly distributed in-sewer travel time in a large sewage network, this specific method may be more applicable to confined environments (e.g., dormitories and nursing homes). An early study on defecation concluded that defecations are more likely to occur in the early morning (Heaton et al., 1992), and Campisano and Modica (2015) reported that there are three toilet flushing peaks during a day, although it is necessary to point out that the result is merely based on a case study of a household and whether it also applies to a larger community needs further verification.
Previous studies have employed different sampling frequencies, from taking samples on discrete dates (Kumar et al., 2020; La Rosa et al., 2020b), to routine sampling with a relatively stable interval (D'Aoust et al., 2021; Hata et al., 2020; Randazzo et al., 2020) and daily sampling (Peccia et al., 2020). For retrospective analysis, frequent sampling over a long period (e.g., several months) may unnecessarily increase the total workload required for sample processing, therefore lower frequency is both acceptable and more realistic. But under the premise of using the measured viral load for early warning, especially considering the rapid progression of the COVID-19 epidemic and the potential social significance of the measures to be taken, daily or similarly frequent sampling is highly recommended as long as the laboratory capacity allows.
Apart from direct measuring, human fecal indicators may also help normalize the flowrate as well as help identify the peak flushing hours in a day if there are any. Some previous studies have opted for the use of pepper mild mottle virus (PMMoV) as an internal control because of its universal and stable presence in the wastewater matrix (Kitajima et al., 2014; Symonds et al., 2018), but more options including crAssphage, HAdV, JCPyV, human microbiome-specific HF183 Bacteroides 16S ribosomal rRNA, and eukaryotic 18S rRNA may also be used (Ballesté et al., 2019; Bofill-Mas et al., 2006; D'Aoust et al., 2021; Medema et al., 2020a), although it is worth mentioning that because their nature may be very distinct from that of the target biomarkers, these internal control targets cannot be used for signal normalization.
2.4. In-sewer travel time and degradation
Once the viral RNA is released from infected individuals and enters the wastewater, it will spend some time traveling in the sewer pipes until it reaches the designated sampling site. The in-sewer travel time is a function of many characteristics of the sewage system in question, such as the spatial configuration of the sewer network and wastewater flow rate in a given time, hence its value may vary greatly among sewage networks and is highly recommended to be determined for each WBE project individually if needed. Although its importance in WBE has been underlined (Thai et al., 2014), the in-sewer travel time of a given sewage network has rarely been reported, presumably due to the difficulty of performing an experiment or establishing a hydrological model. But according to limited existing estimates made for multiple purposes including WBE application, the mean value of in-sewer travel time, or wastewater residence time, typically falls within several hours. For instance, it has been estimated that the national median in-sewer travel time of the U.S. is 3.3 h (Kapo et al., 2017), and the approximate sewer transit times of the UK and Rome have also been estimated to be ~2 h and 3–5 h, respectively (D'Ascenzo et al., 2003; Holt et al., 1998). Also, in the case of grab sampling, the mean or median value does not consider the population heterogeneity, the wastewater produced by those living close to the sampling site will have significantly shorter in-sewer travel time compared to that from people living in the upstream area. To offset the potential impact, the demographic and geographic distributions of the population may also be considered a factor.
Wastewater is a complex matrix with a high concentration of microorganisms and substances that are either organic and inorganic, all of which may contribute to the natural degradation of viral RNA. Previous studies have investigated and reviewed the degradation of human coronaviruses including SARS-CoV and SARS-CoV-2 and their surrogates such as murine hepatitis virus (MHV) in the wastewater matrix (Ahmed et al., 2020c; Bivins et al., 2020a; Mandal et al., 2020). As the results suggest, the viral RNA of SARS-CoV-2 is more persistent than viable virus particles and can stay in wastewater for a relatively long period, and the decay rate increases as the wastewater temperature goes up. Bivins et al. (2020a) recorded a 26.2 days T 90 value in untreated wastewater at 20 °C, which is comparable to the study by Ahmed et al. (2020c) in which the T 90 values in untreated wastewater at 15 and 25 °C were 20.4 and 12.6 days, respectively. Because the typical residence time of wastewater is several hours, the effect of degradation may not be as pronounced as that of other factors, but it is still recommended to take the degradation kinetics into consideration for better accuracy. For instance, in a long-term monitoring project, the seasonal change in wastewater temperature may bring change to the prevalence estimation as the degradation during summer would be more significant and may reduce the measured viral load (Hart and Halden, 2020).
Although the majority of existing studies took samples from wastewater treatment plants (WWTPs) for better coverage and convenience, they are not the only option. If a better spatial resolution or a shorter response time is required, the strategy of ‘upstream sampling’ can also be employed, which means samples are taken from locations closer to the origins, such as sewer pumping stations and maintenance holes, to narrow down the coverage and shorten the in-sewer travel time (Medema et al., 2020a; Thompson et al., 2020).
2.5. Turnaround time for sample treatment and quantification
The turnaround time for sample treatment consists of sample transportation, virus concentration, quantification, and data analysis and organizing. Primary concentration methods can take anywhere from about an hour (ultrafiltration, electronegative membrane vortex) to overnight (PEG precipitation) (Ahmed et al., 2020d; Torii et al., 2020), the time required for subsequent steps varies depending on the reagent kits and instruments used but is also typically in the range of several hours if all steps are done consecutively. The time needed for sample transportation and data analysis depends heavily on real-life factors including the transportation method, the distance between the sampling site and laboratory, and the way of data processing, so far, very limited information about these two steps is available from existing literature. Due to the varied conditions, although it is possible for the WBE approach to predate symptom onset and hospital admission, the entire workflow is subject to significant uncertainty and different settings hence an estimated time cannot be given. We encourage future studies to include the information of the time required for each step to give a better image of the total turnaround time.
Worrying that the standard off-site RT-qPCR method may not fulfill the demand for rapid detection, recent studies have discussed alternative methods. Mao et al. (2020) and Bhalla et al. (2020) discussed the potential of paper-based analytical devices, which are easy-to-carry tools that can be deployed for rapid on-site nucleic acid testing. Yang et al. (2017) reported a paper-based “sample-to-answer” platform for the detection of human genomic DNA in untreated wastewater based on the loop-mediated isothermal amplification (LAMP), through which the result can be yielded within 45 min. Nguyen et al. (2020) shared similar optimism about LAMP, specifically stating that LAMP can be a potential candidate for COVID-19 early detection. While alluring on paper, compared to the conventional laboratory apparatus, the reliability of new methods and devices remains unverified, and very limited information is available regarding the practical application despite the strong interest. Although the development of novel devices and methods that can enable rapid and reliable detection of SARS-CoV-2 genome RNA is highly encouraged, considering that the derived information will be used to help make crucial decisions, the application must be proceeded with caution.
2.6. Data analysis
After obtaining the quantification result, there is a final step of data analysis. If the SARS-CoV-2 signal in a low prevalence region is positive for the first time, it indicates a high possibility that the predetermined threshold prevalence level has been exceeded, but for better dependability, especially considering the information will be used to support critical decisions that may leave a permanent impact on the society, an additional validation step may be employed at the cost of longer response time. As for long-term monitoring in a middle-to-high prevalence region where the viral load is consistently higher than LoQ, the concentration of viral RNA, or the active shedding population if a back-calculation model can be successfully established and validated, should be combined with previous results to assemble a longitudinal pattern.
Although theoretically, qualitative and semi-quantitative analysis can be performed without a back-calculation model that connects the viral load to the active shedding population (Daughton, 2020), the lack of quantitative projection would certainly impair the usefulness of the result. Although lagged correlation has been found in wastewater viral load and reported patient number (Nemudryi et al., 2020; Peccia et al., 2020), there is no model currently in existence that can infer the size of the total infected or shedding population, which could be much larger than reported cases due to undertesting and asymptomatic virus carriers. Previous WBE projects have used excretion-dilution-recovery mass balance models for the back-calculation of chemical biomarkers (Feng et al., 2018; Gracia-Lor et al., 2017; Lai et al., 2011), but in the case of COVID-19 surveillance, the same model may suffer greatly from the limited understanding and the variance of some parameters (mainly the shedding profile and dilution factor) as well as the potential data noise. One reason is that due to the persistent fecal shedding, the wastewater viral load is likely to be contributed by patients in different infection stages, this population may even include those who have the virus cleared in the respiratory tract. Thereby, not only modeling tools that help reduce the uncertainty (e.g., Bayesian inference and maximum likelihood estimation) are highly recommended, other mathematical models featuring different structures are also worth looking into as long as they offer a decent capability of capturing the correlation between viral load in wastewater and the shedding/infected population and dealing with the noisy data. Here, we not only encourage researchers who are already working on COVID-19 wastewater surveillance to reach out and look for potentially suitable modeling techniques but also call on experts from other disciplines, such as epidemiology and statistics, to join in and tackle the challenge together.
Knowing the de facto population size in the catchment area also helps reduce the uncertainty when doing quantitative analysis (Choi et al., 2018; Daughton, 2020; Medema et al., 2020a; O'Brien et al., 2014). Census data or estimation based on facility capacity can be used as an approximation, but they may deviate from reality. Another option is to use certain population biomarkers including exogenous markers such as nicotine and caffeine, and endogenous markers 5-hydroxyindoleacetic acid (5-HIAA) and ammonia (Choi et al., 2018), but it should be kept in mind that significance discrepancy may exist between regions and countries due to various lifestyles. Apart from the de facto population, in regions with high mobility (e.g., tourist attractions and transportation hubs), the frequent movements of people not only increase the risk of virus introduction but also introduce a new source of uncertainty for quantitative analysis (Lai et al., 2011). Further studies will need to employ appropriate methods to estimate and validate the population covered by the studied sewage network and consider how to incorporate the dynamics of the population into the result interpretation process.
As mentioned previously, WBE should not be considered as a standalone solution, but rather as a complementary data source in public health management (Ahmed et al., 2020b; D'Aoust et al., 2021; Thompson et al., 2020). Therefore, the result obtained from WBE should be viewed and assessed along with other supporting materials such as clinical reporting and estimation made with epidemic models before reaching any final decision.
3. Conclusions
While certainly holding a potential, the prospect of using wastewater-based epidemiology as an early warning system for COVID-19 surveillance still has many hurdles to overcome. As a result, we encourage experts from different disciplines to work together and share knowledge for the further refinement and validation of this novel approach as humanity continues to battle the ongoing COVID-19 pandemic. We summarized the main points that need to be addressed by further studies as follows:
-
•
The shedding profile of infected individuals, including the rate, ratio, timing, and consistency, is still subject to substantial uncertainty and needs to be better understood for WBE to be viable, more information needs to be extracted from existing and future clinical reports. Also, a shedding dynamics model needs to be established to help generalize the shedding profile.
-
•
The sampling strategy and experimental methods can be streamlined to shorten the total turnaround time and provide a longer response window period, future studies are encouraged to include related information for optimization. Also, the sampling strategy can significantly affect the representativeness of the sample and should be optimized according to the practical conditions and needs.
-
•
Back-calculation remains one of the biggest technical challenges, apart from continually updating the model with new information, auxiliary modeling tools and novel model structures are also worth looking into to improve its accuracy and robustness.
CRediT authorship contribution statement
Yifan Zhu: Investigation, Writing - original draft. Wakana Oishi: Validation, Writing - review & editing. Chikako Maruo: Validation. Mayuko Saito: Project administration. Rong Chen: Project administration. Masaaki Kitajima: Supervision, Writing - review & editing. Daisuke Sano: Conceptualization, Project administration, Funding acquisition, Supervision, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research was supported by the Japan Agency for Medical Research and Development (AMED) under Grant Number JPwm0125001. The graphical abstract was designed using Freepik.com resources.
Editor: Damia Barcelo
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728:138764. doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Angel N., Bibby K., Bivins A., Dierens L., Edson J., Ehret J., Gyawali P., Hamilton K.A., Hosegood I., Hugenholtz P., Jiang G., Kitajima M., Sichani H.T., Shi J., Shimko K.M., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Zaugg J., Mueller J.F. Detection of SARS-CoV-2 RNA in commercial passenger aircraft and cruise ship wastewater: a surveillance tool for assessing the presence of COVID-19 infected travellers. J. Travel Med. 2020;27:1–11. doi: 10.1093/jtm/taaa116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., Gyawali P., Korajkic A., Riddell S., Sherchan S.P., Simpson S.L., Sirikanchana K., Symonds E.M., Verhagen R., Vasan S.S., Kitajima M., Bivins A. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191:110092. doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., Korajkic A., McMinn B.R., Mueller J.F., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Kitajima M. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739:139960. doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arons M.M., Hatfield K.M., Reddy S.C., Kimball A., James A., Jacobs J.R., Taylor J., Spicer K., Bardossy A.C., Oakley L.P., Tanwar S., Dyal J.W., Harney J., Chisty Z., Bell M., Methner M., Paul P., Carlson C.M., McLaughlin H.P., Thornburg N., Tong S., Tamin A., Tao Y., Uehara A., Harcourt J., Clark S., Brostrom-Smith C., Page L.C., Kay M., Lewis J., Montgomery P., Stone N.D., Clark T.A., Honein M.A., Duchin J.S., Jernigan J.A. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N. Engl. J. Med. 2020;382:2081–2090. doi: 10.1056/NEJMoa2008457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesté E., Pascual-Benito M., Martín-Díaz J., Blanch A.R., Lucena F., Muniesa M., Jofre J., García-Aljaro C. Dynamics of crAssphage as a human source tracking marker in potentially faecally polluted environments. Water Res. 2019;155:233–244. doi: 10.1016/j.watres.2019.02.042. [DOI] [PubMed] [Google Scholar]
- Bhalla N., Pan Y., Yang Z., Payam A.F. Opportunities and challenges for biosensors and nanoscale analytical tools for pandemics: COVID-19. ACS Nano. 2020;14:7783–7807. doi: 10.1021/acsnano.0c04421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., Greaves J., Fischer R., Yinda K.C., Ahmed W., Kitajima M., Munster V.J., Bibby K. Persistence of SARS-CoV-2 in water and wastewater. Environ. Sci. Technol. Lett. 2020 doi: 10.1021/acs.estlett.0c00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., North D., Ahmad A., Ahmed W., Alm E., Been F., Bhattacharya P., Bijlsma L., Boehm A.B., Brown J., Buttiglieri G., Calabro V., Carducci A., Castiglioni S., Cetecioglu Gurol Z., Chakraborty S., Costa F., Curcio S., de los Reyes F.L., Delgado Vela J., Farkas K., Fernandez-Casi X., Gerba C., Gerrity D., Girones R., Gonzalez R., Haramoto E., Harris A., Holden P.A., Islam M.T., Jones D.L., Kasprzyk-Hordern B., Kitajima M., Kotlarz N., Kumar M., Kuroda K., La Rosa G., Malpei F., Mautus M., McLellan S.L., Medema G., Meschke J.S., Mueller J., Newton R.J., Nilsson D., Noble R.T., van Nuijs A., Peccia J., Perkins T.A., Pickering A.J., Rose J., Sanchez G., Smith A., Stadler L., Stauber C., Thomas K., van der Voorn T., Wigginton K., Zhu K., Bibby K. Wastewater-based epidemiology: global collaborative to maximize contributions in the fight against COVID-19. Environ. Sci. Technol. 2020 doi: 10.1021/acs.est.0c02388. [DOI] [PubMed] [Google Scholar]
- Bofill-Mas S., Albinana-Gimenez N., Clemente-Casares P., Hundesa A., Rodriguez-Manzano J., Allard A., Calvo M., Girones R. Quantification and stability of human adenoviruses and polyomavirus JCPyV in wastewater matrices. Appl. Environ. Microbiol. 2006;72:7894–7896. doi: 10.1128/AEM.00965-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisano A., Modica C. Appropriate resolution timescale to evaluate water saving and retention potential of rainwater harvesting for toilet flushing in single houses. J. Hydroinformatics. 2015;17:331–346. doi: 10.2166/hydro.2015.022. [DOI] [Google Scholar]
- Choi P.M., Tscharke B.J., Donner E., O’Brien J.W., Grant S.C., Kaserzon S.L., Mackie R., O’Malley E., Crosbie N.D., Thomas K.V., Mueller J.F. Wastewater-based epidemiology biomarkers: past, present and future. TrAC - Trends Anal. Chem. 2018;105:453–469. doi: 10.1016/j.trac.2018.06.004. [DOI] [Google Scholar]
- Corpuz M.V.A., Buonerba A., Vigliotta G., Zarra T., Ballesteros F., Campiglia P., Belgiorno V., Korshin G., Naddeo V. Viruses in wastewater: occurrence, abundance and detection methods. Sci. Total Environ. 2020;745:140910. doi: 10.1016/j.scitotenv.2020.140910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y., Liu N., Tan C., Feng Y., Yuan X., Fan D., Peng Y., Jin R., Guo Y., Lou J. Comparison of qualitative and quantitative analyses of COVID-19 clinical samples. Clin. Chim. Acta. 2020;510:613–616. doi: 10.1016/j.cca.2020.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aoust P.M., Mercier E., Montpetit D., Jia J.J., Alexandrov I., Neault N., Baig A.T., Mayne J., Zhang X., Alain T., Langlois M.A., Servos M.R., MacKenzie M., Figeys D., MacKenzie A.E., Graber T.E., Delatolla R. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Res. 2021;188:116560. doi: 10.1016/j.watres.2020.116560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ascenzo G., Di Corcia A., Gentili A., Mancini R., Mastropasqua R., Nazzari M., Samperi R. Fate of natural estrogen conjugates in municipal sewage transport and treatment facilities. Sci. Total Environ. 2003;302:199–209. doi: 10.1016/S0048-9697(02)00342-X. [DOI] [PubMed] [Google Scholar]
- Daughton C.G. Wastewater surveillance for population-wide Covid-19: the present and future. Sci. Total Environ. 2020;736:139631. doi: 10.1016/j.scitotenv.2020.139631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzone L., Musso N., Gattuso G., Bongiorno D., Palermo C.I., Scalia G., Libra M., Stefani S. Sensitivity assessment of droplet digital PCR for SARS-CoV-2 detection. Int. J. Mol. Med. 2020;46:957–964. doi: 10.3892/ijmm.2020.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L., Zhang W., Li X. Monitoring of regional drug abuse through wastewater-based epidemiology—a critical review. Sci. China Earth Sci. 2018;61:239–255. doi: 10.1007/s11430-017-9129-x. [DOI] [Google Scholar]
- Gandhi M., Yokoe D.S., Havlir D.V. Asymptomatic transmission, the achilles’ heel of current strategies to control Covid-19. N. Engl. J. Med. 2020;382:2158–2160. doi: 10.1056/NEJMe2009758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracia-Lor E., Rousis N.I., Zuccato E., Bade R., Baz-Lomba J.A., Castrignanò E., Causanilles A., Hernández F., Kasprzyk-Hordern B., Kinyua J., McCall A.K., van Nuijs A.L.N., Plósz B.G., Ramin P., Ryu Y., Santos M.M., Thomas K., de Voogt P., Yang Z., Castiglioni S. Estimation of caffeine intake from analysis of caffeine metabolites in wastewater. Sci. Total Environ. 2017;609:1582–1588. doi: 10.1016/j.scitotenv.2017.07.258. [DOI] [PubMed] [Google Scholar]
- Gupta S., Parker J., Smits S., Underwood J., Dolwani S. Persistent viral shedding of SARS-CoV-2 in faeces – a rapid review. Color. Dis. 2020;22:611–620. doi: 10.1111/codi.15138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamouda M., Mustafa F., Maraqa M., Rizvi T., Aly Hassan A. Wastewater surveillance for SARS-CoV-2: lessons learnt from recent studies to define future applications. Sci. Total Environ. 2020;759:143493. doi: 10.1016/j.scitotenv.2020.143493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart O.E., Halden R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020;730:138875. doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A., Hara-Yamamura H., Meuchi Y., Imai S., Honda R. Detection of SARS-CoV-2 in wastewater in Japan during a COVID-19 outbreak. Sci. Total Environ. 2020:143578. doi: 10.1016/j.scitotenv.2020.143578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., Lau Y.C., Wong J.Y., Guan Y., Tan X., Mo X., Chen Y., Liao B., Chen W., Hu F., Zhang Q., Zhong M., Wu Y., Zhao L., Zhang F., Cowling B.J., Li F., Leung G.M. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- Heaton K.W., Radvan J., Cripps H., Mountford R.A., Braddon F.E.M., Hughes A.O. Defecation frequency and timing, and stool form in the general population: a prospective study. Gut. 1992;33:818–824. doi: 10.1136/gut.33.6.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt M.S., Fox K.K., Burford M., Daniel M., Buckland H. UK monitoring study on the removal of linear alkylbenzene sulphonate in trickling filter type sewage treatment plants. Contribution to GREAT-ER project 2. Sci. Total Environ. 1998 doi: 10.1016/S0048-9697(98)00016-3. [DOI] [Google Scholar]
- Huynh G.T., Rong L. Modeling the dynamics of virus shedding into the saliva of Epstein-Barr virus positive individuals. J. Theor. Biol. 2012;310:105–114. doi: 10.1016/j.jtbi.2012.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.L., Baluja M.Q., Graham D.W., Corbishley A., McDonald J.E., Malham S.K., Hillary L.S., Connor T.R., Gaze W.H., Moura I.B., Wilcox M.H., Farkas K. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Sci. Total Environ. 2020;749:141364. doi: 10.1016/j.scitotenv.2020.141364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerow D. Covid-19: the crisis of personal protective equipment in the US. The BMJ. 2020;369:29–30. doi: 10.1136/bmj.m1367. [DOI] [PubMed] [Google Scholar]
- Kaplan E.H., Wang D., Wang M., Malik A.A., Zulli A., Peccia J. Aligning SARS-CoV-2 indicators via an epidemic model: application to hospital admissions and RNA detection in sewage sludge. Health Care Manag. Sci. 2020 doi: 10.1007/s10729-020-09525-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapo K.E., Paschka M., Vamshi R., Sebasky M., McDonough K. Estimation of U.S. sewer residence time distributions for national-scale risk assessment of down-the-drain chemicals. Sci. Total Environ. 2017;603–604:445–452. doi: 10.1016/j.scitotenv.2017.06.075. [DOI] [PubMed] [Google Scholar]
- Kitajima M., Iker B.C., Pepper I.L., Gerba C.P. Relative abundance and treatment reduction of viruses during wastewater treatment processes - identification of potential viral indicators. Sci. Total Environ. 2014;488–489:290–296. doi: 10.1016/j.scitotenv.2014.04.087. [DOI] [PubMed] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020;739:139076. doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura K., Sadamasu K., Muramatsu M., Yoshida H. Efficient detection of SARS-CoV-2 RNA in the solid fraction ofwastewater. Sci. Total Environ. 2021;763:144587. doi: 10.1016/j.scitotenv.2020.144587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantz S.G., Rao A.S.R.S. Level of underreporting including underdiagnosis before the first peak of COVID-19 in various countries: preliminary retrospective results based on wavelets and deterministic modeling. Infect. Control Hosp. Epidemiol. 2020;41:857–859. doi: 10.1017/ice.2020.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., Joshi C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746:141326. doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Bonadonna L., Lucentini L., Kenmoe S., Suffredini E. Coronavirus in water environments: occurrence, persistence and concentration methods - a scoping review. Water Res. 2020;179:115899. doi: 10.1016/j.watres.2020.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736:139652. doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F.Y., Ort C., Gartner C., Carter S., Prichard J., Kirkbride P., Bruno R., Hall W., Eaglesham G., Mueller J.F. Refining the estimation of illicit drug consumptions from wastewater analysis: co-analysis of prescription pharmaceuticals and uncertainty assessment. Water Res. 2011;45:4437–4448. doi: 10.1016/j.watres.2011.05.042. [DOI] [PubMed] [Google Scholar]
- Lauer S.A., Grantz K.H., Bi Q., Jones F.K., Zheng Q., Meredith H.R., Azman A.S., Reich N.G., Lessler J. The incubation period of coronavirus disease 2019 (CoVID-19) from publicly reported confirmed cases: Estimation and application. Ann. Intern. Med. 2020;172(9):577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J.…Liu M.Feng. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal P., Gupta A.K., Dubey B.K. A review on presence, survival, disinfection/removal methods of coronavirus in wastewater and progress of wastewater-based epidemiology. J. Environ. Chem. Eng. 2020;8:104317. doi: 10.1016/j.jece.2020.104317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao K., Zhang H., Yang Z. Can a paper-based device trace COVID-19 sources with wastewater-based epidemiology? Environ. Sci. Technol. 2020;54:3733–3735. doi: 10.1021/acs.est.0c01174. [DOI] [PubMed] [Google Scholar]
- McAloon C., Collins Á., Hunt K., Barber A., Byrne A.W., Butler F., Casey M., Griffin J., Lane E., McEvoy D., Wall P., Green M., O’Grady L., More S.J. Incubation period of COVID-19: A rapid systematic review and meta-analysis of observational research. BMJ Open. 2020;10(8):1–9. doi: 10.1136/bmjopen-2020-039652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Been F., Heijnen L., Petterson S. Implementation of environmental surveillance for SARS-CoV-2 virus to support public health decisions: opportunities and challenges. Curr. Opin. Environ. Sci. Heal. 2020;17:49–71. doi: 10.1016/j.coesh.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020 doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Michael-Kordatou I., Karaolia P., Fatta-Kassinos D. Sewage analysis as a tool for the COVID-19 pandemic response and management: the urgent need for optimised protocols for SARS-CoV-2 detection and quantification. J. Environ. Chem. Eng. 2020;8:104306. doi: 10.1016/j.jece.2020.104306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura F., Kitajima M., Omori R. Duration of SARS-CoV-2 viral shedding in faeces as a parameter for wastewater-based epidemiology: re-analysis of patient data using a shedding dynamics model. Sci. Total Environ. 2020:144549. doi: 10.1016/j.scitotenv.2020.144549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemudryi A., Nemudraia A., Wiegand T., Surya K., Buyukyoruk M., Cicha C., Vanderwood K.K., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep. Med. 2020;1:100098. doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T., Bang D.D., Wolff A. 2019 Novel coronavirus disease (COVID-19): paving the road for rapid detection and point-of-care diagnostics. Micromachines. 2020;11:1–7. doi: 10.3390/MI11030306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien J.W., Thai P.K., Eaglesham G., Ort C., Scheidegger A., Carter S., Lai F.Y., Mueller J.F. A model to estimate the population contributing to the wastewater using samples collected on census day. Environ. Sci. Technol. 2014;48:517–525. doi: 10.1021/es403251g. [DOI] [PubMed] [Google Scholar]
- Osuna C.E., Lim S.Y., Deleage C., Griffin B.D., Stein D., Schroeder L.T., Omange R., Best K., Luo M., Hraber P.T., Andersen-Elyard H., Ojeda E.F.C., Huang S., Vanlandingham D.L., Higgs S., Perelson A.S., Estes J.D., Safronetz D., Lewis M.G., Whitney J.B. Zika viral dynamics and shedding in rhesus and cynomolgus macaques. Nat. Med. 2016;22:1448–1455. doi: 10.1038/nm.4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parasa S., Desai M., Thoguluva Chandrasekar V., Patel H.K., Kennedy K.F., Roesch T., Spadaccini M., Colombo M., Gabbiadini R., Artifon E.L.A., Repici A., Sharma P. Prevalence of gastrointestinal symptoms and fecal viral shedding in patients with coronavirus disease 2019: a systematic review and meta-analysis. JAMA Netw. Open. 2020;3:e2011335. doi: 10.1001/jamanetworkopen.2020.11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L., Weinberger D.M., Arnold W., Omer S.B. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020 doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiser Jaclyn. 2020. University of Arizona Used Wastewater Testing to Detect Cases of Coronavirus in a Dorm [WWW Document] Washington Post. URL https://www.washingtonpost.com/nation/2020/08/28/arizona-coronavirus-wastewater-testing/ (accessed 9.7.20) [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 181. 2020 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusiñol M., Martínez-Puchol S., Forés E., Itarte M., Girones R., Bofill-Mas S. Concentration methods for the quantification of coronavirus and other potentially pandemic enveloped virus from wastewater. Curr. Opin. Environ. Sci. Heal. 2020;17:21–28. doi: 10.1016/j.coesh.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab K.J., Estes M.K., Neill F.H., Atmar R.L. Use of heat release and an internal RNA standard control in reverse transcription-PCR detection of Norwalk virus from stool samples. J. Clin. Microbiol. 1997;35:511–514. doi: 10.1128/jcm.35.2.511-514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu J.P.S., Ahmed W., Toze S. Sensitive detection of human adenovirus from small volume of primary wastewater samples by quantitative PCR. J. Virol. Methods. 2013;187:395–400. doi: 10.1016/j.jviromet.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Sims N., Kasprzyk-Hordern B. Future perspectives of wastewater-based epidemiology: monitoring infectious disease spread and resistance to the community level. Environ. Int. 2020;139:105689. doi: 10.1016/j.envint.2020.105689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo T., Liu X., Feng J., Guo M., Hu W., Guo D., Ullah H., Yang Y., Zhang Qiuhan, Wang X., Sajid M., Huang Z., Deng L., Chen T., Liu F., Xu K., Liu Yuan, Zhang Qi, Liu Yingle, Xiong Y., Chen G., Lan K., Chen Y. ddPCR: a more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg. Microbes Infect. 2020;9:1259–1268. doi: 10.1080/22221751.2020.1772678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds E.M., Nguyen K.H., Harwood V.J., Breitbart M. Pepper mild mottle virus: a plant pathogen with a greater purpose in (waste)water treatment development and public health management. Water Res. 2018;144:1–12. doi: 10.1016/j.watres.2018.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teunis P.F.M., Sukhrie F.H.A., Vennema H., Bogerman J., Beersma M.F.C., Koopmans M.P.G. Shedding of norovirus in symptomatic and asymptomatic infections. Epidemiol. Infect. 2015;143:1710–1717. doi: 10.1017/S095026881400274X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai P.K., Jiang G., Gernjak W., Yuan Z., Lai F.Y., Mueller J.F. Effects of sewer conditions on the degradation of selected illicit drug residues in wastewater. Water Res. 2014;48:538–547. doi: 10.1016/j.watres.2013.10.019. [DOI] [PubMed] [Google Scholar]
- Thompson J.R., Nancharaiah Y.V., Gu X., Lee W.L., Rajal V.B., Haines M.B., Girones R., Ng L.C., Alm E.J., Wuertz S. Making waves: Wastewater surveillance of SARS-CoV-2 for population-based health management. Water Res. 2020;184 doi: 10.1016/j.watres.2020.116181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii S., Furumai H., Katayama H. Applicability of polyethylene glycol precipitation followed by acid guanidinium thiocyanate-phenol-chloroform extraction for the detection of SARS-CoV-2 RNA from municipal wastewater. Sci. Total Environ. 2020:143067. doi: 10.1016/j.scitotenv.2020.143067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn A.S., Meijer B., Frampton C.M.A., Barclay M.L., de Boer N.K.H. Systematic review with meta-analysis: SARS-CoV-2 stool testing and the potential for faecal-oral transmission. Aliment. Pharmacol. Ther. 2020:1–13. doi: 10.1111/apt.16036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal A., Ganesan H., Sudalaimuthu Raja S.S., Govindasamy V., Arunachalam M., Narayanasamy A., Sivaprakash P., Rahman P.K.S.M., Gopalakrishnan A.V., Siama Z., Vellingiri B. Novel wastewater surveillance strategy for early detection of coronavirus disease 2019 hotspots. Curr. Opin. Environ. Sci. Heal. 2020;17:8–13. doi: 10.1016/j.coesh.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh K.A., Jordan K., Clyne B., Rohde D., Drummond L., Byrne P., Ahern S., Carty P.G., O’Brien K.K., O’Murchu E., O’Neill M., Smith S.M., Ryan M., Harrington P. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J. Infect. 2020;81:357–371. doi: 10.1016/j.jinf.2020.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zheng J., Guo L., Yao H., Wang L., Xia X.D., Zhang W. Fecal viral shedding in COVID-19 patients: clinical significance, viral load dynamics and survival analysis. Virus Res. 2020;289:198147. doi: 10.1016/j.virusres.2020.198147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhaus S., Weber F.A., Schiwy S., Linnemann V., Brinkmann M., Widera M., Greve C., Janke A., Hollert H., Wintgens T., Ciesek S. Detection of SARS-CoV-2 in raw and treated wastewater in Germany – suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021;751:141750. doi: 10.1016/j.scitotenv.2020.141750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2020. WHO Coronavirus Disease (COVID-19) Dashboard [WWW Document] URL https://covid19.who.int/ (accessed 11.30.20) [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., Yin H., Xiao Q., Tang Y., Qu X., Kuang L., Fang X., Mishra N., Lu J., Shan H., Jiang G., Huang X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020;5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.-M., Maday Y., Teyssou R., Richard E., Almayrac J.L., Moulin L. Evaluation of lockdown impact on SARS-CoV-2 dynamics through viral genome quantification in Paris wastewaters. Eurosurveillance. 2020;25 doi: 10.1101/2020.04.12.20062679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xagoraraki I., O’Brien E. Women in Water Quality. Springer; 2020. Wastewater-based epidemiology for early detection of viral outbreaks; pp. 75–97. [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833. doi: 10.1053/j.gastro.2020.02.055. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Li X., Zhu B., Liang H., Fang C., Gong Y., Guo Q., Sun X., Zhao D., Shen J., Zhang H., Liu H., Xia H., Tang J., Zhang K., Gong S. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020;26:502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Xu G., Reboud J., Kasprzyk-Hordern B., Cooper J.M. Monitoring genetic population biomarkers for wastewater-based epidemiology. Anal. Chem. 2017;89:9941–9945. doi: 10.1021/acs.analchem.7b02257. [DOI] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, Partitioning, and Recovery of Enveloped Viruses in Untreated Municipal Wastewater. Environ. Sci. Technol. 2016;50:5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Yu F., Yan L., Wang N., Yang S., Wang L., Tang Y., Gao G., Wang S., Ma C., Xie R., Wang F., Tan C., Zhu L., Guo Y., Zhang F. Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin. Infect. Dis. 2020;71:793–798. doi: 10.1093/cid/ciaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]