Abstract
The Coronavirus disease (Covid-19) has been declared a pandemic by World Health Organisation (WHO) and till date caused 585,727 numbers of deaths all over the world. The only way to minimize the number of death is to quarantine the patients tested Corona positive. The quick spread of this disease can be reduced by automatic screening to cover the lack of radiologists. Though the researchers already have done extremely well to design pioneering deep learning models for the screening of Covid-19, most of them results in low accuracy rate. In addition, over-fitting problem increases difficulties for those models to learn on existing Covid-19 datasets. In this paper, an automated Covid-19 screening model is designed to identify the patients suffering from this disease by using their chest X-ray images. The model classifies the images in three categories – Covid-19 positive, other pneumonia infection and no infection. Three learning schemes such as CNN, VGG-16 and ResNet-50 are separately used to learn the model. A standard Covid-19 radiography dataset from the repository of Kaggle is used to get the chest X-ray images. The performance of the model with all the three learning schemes has been evaluated and it shows VGG-16 performed better as compared to CNN and ResNet-50. The model with VGG-16 gives the accuracy of 97.67%, precision of 96.65%, recall of 96.54% and F1 score of 96.59%. The performance evaluation also shows that our model outperforms two existing models to screen the Covid-19.
Keywords: Covid-19, Transfer learning, VGG-16, CNN, ResNet-50, Pneumonia
1. Introduction
The Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV-2) is a novel virus causing respiratory illness known as Coronavirus disease (Covid-19). It has emerged in December 2019 in Wuhan, China [27] and spread rapidly by human-to-human transmission to other parts of the world [15], [21], [45]. Till date Covid-19 has affected 13,616,593 numbers of worldwide patients with 585,727 numbers of deaths across 216 countries according to the report of World Health Organization (WHO) [20]. On January 2020, WHO declared Covid-19 as international health concern [26]. The number of infected people is still increasing at a rapid rate on the day to day basis. To control the novel Coronavirus, quarantine of the patient seems to be the only way. Thus fast large scale screening for the virus is required.
The standard method used for the screening of Coronavirus is the detection of nucleic acid using reverse transcription polymerase chain reaction, in which the false negative rate is high and the screening needs to be repeated several times [46]. The clinical studies [7] show that chest X-ray is an effective screening technique as it can identify the similar features among the Corona cases and it outperformed lab testing using reverse transcription polymerase chain reaction. Therefore, chest X-ray is considered essential for the early diagnosis of Covid-19 positive patients. The number of Covid-19 positive cases is far greater than the number of radiologists. Thus the manual testing process should be replaced by automated screening of Covid-19. This will not only speed up the screening process, but also resolves the issues like cost of test, waiting time of result and unavailability of RT-PCR test kits.
The automatic diagnosis of diseases in medical field using machine learning have gained a lot of popularity by making the diagnosis faster using minimum man power. It enables the creation of the model that achieves promising results just by providing the input data, which do not need any manual feature extraction [18]. Machine learning techniques have been applied in various medical fields such as skin cancer classification [8], [11], detection of malaria [10,14,22], breast cancer detection [5,9], pneumonia detection using chest x-ray [31], brain disease classification [40], arrhythmia classification [1], [17], [47] lung segmentation [12,37] and fundus image segmentation [41]. Further Deep learning methods are involved in [4,7] to increase the accuracy of the test result. However the existing automated screening processes of Covid-19 have over-fitting problem due to smaller size of dataset and thus have lower accuracy rate. The Deep learning algorithms using chest CT images [16] have higher complexity.
The deficiencies of the existing works revealed from the state of the art study have motivated us to propose an automated Covid-19 screening model. In this paper, a model, called TLCoV, is proposed to screen the Coronavirus by using transfer learning algorithms on chest X-ray images. To learn the model we have used classical CNN and two transfer learning techniques VGG-16 and ResNet-50 individually. The chest X-ray images are collected from standard dataset of Kaggle. The TLCoV model is trained using 2905 chest x-ray images which are resized to 224 × 224 pixels. The horizontal offline augmentation technique is applied to the dataset as it enhances the accuracy to detect covid-19. Finally, the performance of TLCoV model is measured with all the three learning techniques separately. The experimental results show that TLCoV with VGG-16 outperforms other learning techniques in terms high accuracy, precision and recall value. The major contributions of the authors are listed as follows-
-
•
A novel automated Covid-19 screening model, called TLCoV, is proposed to fasten accurate screening of Coronavirus. The implementation of TLCoV model is available at https://github.com/chiranjeevbitm/Covid-19-detection-with-chest-scan.
-
•
The classical CNN and two transfer learning techniques VGG-16 and ResNet-50 are individually used with the proposed TLCoV model to find better combination in terms of higher accuracy.
-
•
Data augmentation is applied on the images so that it can effectively classify the covid-19 cases.
-
•
The chest X-ray images are collected from a standard dataset.
-
•
The performance of the proposed TLCoV model is compared with two existing automated screening model of Covid-19.
1.1. Roadmap
The remainder of the paper is organized as – Section 2 explains the state of the art study in related field, Section 3 describes the proposed work, Section 4 depicts the experimental results and analyses the performance, Section 5 describes the future research direction of our work and Section 6 concludes the paper followed by references.
2. Literature survey
The Coronavirus is originated in bats [36] and are transmitted to humans through various intermediary sources. It is transmitted with the close contact of infected person and its incubation period ranges between 2-14 days. Various computational intelligent techniques are used to detect Covid-19. Deep learning is introduced to detect the abnormal breathing pattern of the person [43]. It has proposed Respiratory Simulation Model to minimize the gap between the training data and the sparse real world data. The respiratory pattern is classified with accuracy of 94.5%, precision of 94.4%, recall of 95.1% and F1 score of 94.8%. A weakly supervised deep learning framework for classifying Covid-19 and localization of lesions by using 3D CT volumes was developed by [44]. They fed the segmented lung region to the 3D deep neural network for predicting the probability of Covid-19 infection which achieves 0.959 ROC AUC and 0.976 PR AUC.
An attention based deep 3D multiple instance learning (AD3D-MIL) scheme is proposed by [16] for the screening of covid-19, where the label named patient-level is marked in 3D chest CT that is considered as a bag of instances. AD3D-MIL generates deep 3D instances for applying an attention based pooling approach and further the bag of instance is converted into Bernoulli distribution to achieve more approachable learning. It yields the overall accuracy of 97.9%, AUC of 99% and Cohen kappa score of 95.7%. Another automated diagnosis of Covid-19 was developed by [25] using the features extracted from the CT images guarantee the completeness by using the backward neural network for each type of features. It achieves the accuracy of 95.5%, sensitivity of 96.6% and specificity of 93.2%.
A 3D deep learning method, named COVNet, is proposed for detection of Covid-19 cases with the accuracy of 96%. A combination of 3D CNN ResNet-18 and location-attention mechanism is designed by [4] to detect Covid-19 cases using pulmonary CT images. A drop weight-based Bayesian CNN is proposed by [13] for the detection of covid-19 using posterior–anterior chest radiography images, which achieves the accuracy of 89.92%. The modified inception transfer learning model by [44] detects Covid-19 using the CT images with the accuracy of 79.3% and sensitivity of 0.67.
A multilayer perceptron and LSTM model is applied by [2] on clinical data and achieves an AUC of 0.954. 2D CNN is used by [24] for the diagnosis of covid-19 using the Chest CT with an accuracy of 94.98% and AUC of 97.91%. Another method combines 3D UNet++ and ResNet-50 by [23] to identify Covid-19 using chest CT images and achieves the sensitivity of 0.974 and specificity of 0.922. CNN along with pre-trained transfer learning, called AlexNet, is applied on X-ray and chest CT images by [28] to achieve the accuracy of 98% and 94.1% respectively. The supervised machine learning is combined with digital signal processing (MLDSP) by [48] for the analysis of genomes and it achieves 100% accuracy for the classification of covid-19.
The authors of [42] have proposed an automated system for Covid-19 detection to reduce the load of radiologists. The system has used a Multi Scale Convolutional Neural Network (MSCNN) and evaluated on the dataset of Chest Tomography (CT) images. A Deep Convolutional Neural Network (CNN) model, viz. CVDNet [6], is proposed to classify Covid19, other pneumonia and normal patient by using their chest X-ray images. The residual neural network based architecture is proposed in this research. The global and local features of the input are captured by using two parallel levels that are constructed with different kernel sizes. The model is tested on a dataset that consists of 2905 number of chest x-ray images. The experimental results show that the model has achieved 96.69% of accuracy. In [34], the authors have proposed a deep feature plus support vector machine (SVM) based methodology to detect the Covid19 infected patient with the help of x-ray images. They have extracted the deep features from the fully connected layers and provided to SVM to classify the Covid19 infected patients from others. The deep features from 13 numbers of individual CNN models are fed to CNN. The performance analysis shows that ResNet-50 plus SVM achieves the highest accuracy of 98.66%.
Apart from the screening of Covid-19, machine learning algorithms have been used to make future prediction of the number of infections and deaths. Various supervised machine learning techniques namely SVM, LASSO, ES, and LR has been used by [33] to forecast the predictions such as number of new cases, number of deaths and number of recoveries for the upcoming 10 days. The implementation and evaluation shows that ES outperforms all the other three models. Another Machine Learning and Deep learning based model has proposed by [30] to forecast the transmission of Covid-19. It shows that polynomial regression yields minimum RMSE score as compared to other approaches.
In [29], support vector regression (SVR), ridge regression (RIDGE), autoregressive integrated moving average (ARIMA), random forest (RF) and cubist regression (CUBIST) are used to forecast for the Covid19 confirmed cases in Brazilian states in next one, three and six-days. The efficiencies of these models are evaluated based on the parameters like mean absolute error, improvement index and symmetric mean absolute percentage error. The performance analysis reveals that SVR achieves highest accuracy for the prediction of Covid19 among all the involved algorithms. Another model SEIR [39] is proposed to predict the future number of Covid19 in Indonesia. Isolation and vaccination parameters are also considered as model parameters. The simulation results show that isolation may take a big role to prevent from quick spread of this disease.
The authors of [32] have proposed a similar approach to forecast the probable Covid19 cases in next one, three and six days in five American and Brazilian states. The shallow machine learning techniques coupled with Variational Mode Decomposition (VMD) are used for forecasting and the performance evaluation shows that the model has the accuracy of 70%. In [3], the authors are interested in predict the number of possible Covid19 positive cases in the second wave of Iran. The proposed model considers most of the scenarios for the spread of Covid19 and their performance evaluation shows the higher accuracy.
The authors of [49] have proposed various regressor machine learning techniques to find the relationship between the spreading rate and the various factors of Covid19. The relationship between the weather variables and the number of confirmed cases are extracted to compute the impact of humidity and temperature on the transmission of Covid19. The experimental results show that the weather variables are more appropriate for predicting the mortality rate compared to other parameters such as age and population.
3. Proposed work
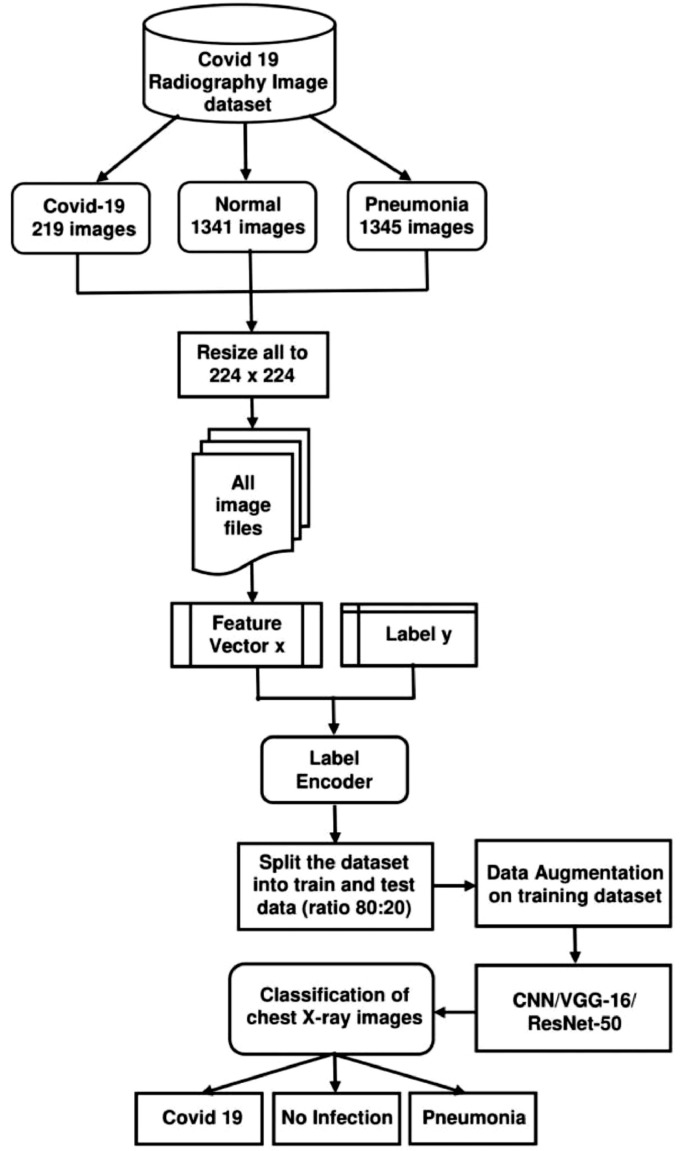
In this paper a deep learning model TLCoV is proposed for the automated screening of Covid-19. In TLCoV initially standard CNN is used, which works by extracting the relevant features from the convolution layers followed by passing from the pooling layer and then from a fully connected layer. Finally, two transfer learning algorithms VGG-16 and ResNet-50 is used on the same model to increase the accuracy. Transfer learning uses the pre trained model, i.e. the model is trained for one problem and is used in another relevant problem. The workflow diagram for the proposed TLCoV model is depicted in Fig. 1 .
Fig. 1.
Workflow diagram of TLCoV model.
3.1. Data preparation and preprocessing
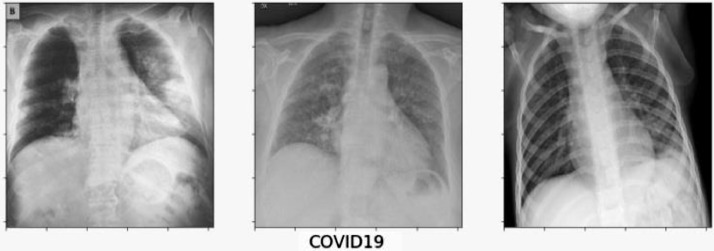
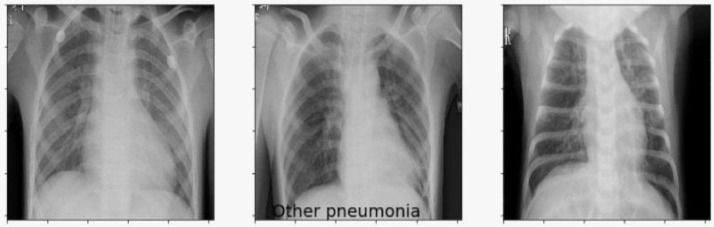
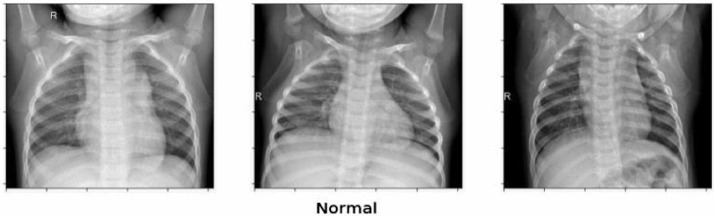
The dataset of Covid-19 radiography database is downloaded from the website of kaggle.com, which consists of three types of chest X-ray images - Covid-19 positive cases, viral pneumonia and no infection. There are 219 Covid-19 positive images, 1345 viral pneumonia images and 1341 no infection images that are depicted in Fig. 2, Fig. 3, Fig. 4 respectively. The behaviours of all the three types of chest x-ray images are analysed and all the three directories are joined together as a single directory. Each image is converted into equal size of pixel value 224 × 224 grids.
Fig. 2.
Corona positive chest X-ray images.
Fig. 3.
Viral pneumonia chest X-ray images.
Fig. 4.
Non Infected chest X-ray images.
Further the whole data is divided into training and test set in the ratio of 80:20. The offline augmentation technique is applied to the images that increase the size of the dataset by a factor equal to the number of the augmentation techniques. In TLCoV, horizontal flip is applied with rotation range 20 to increase the size of the dataset by the factor of 2.
3.2. Classification of Images
The images are classified as covid-19 positive cases, viral pneumonia and no infection using CNN, VGG-16 and ResNet-50 techniques. The following subsections describe all the techniques individually.
3.2.1. Classification using CNN
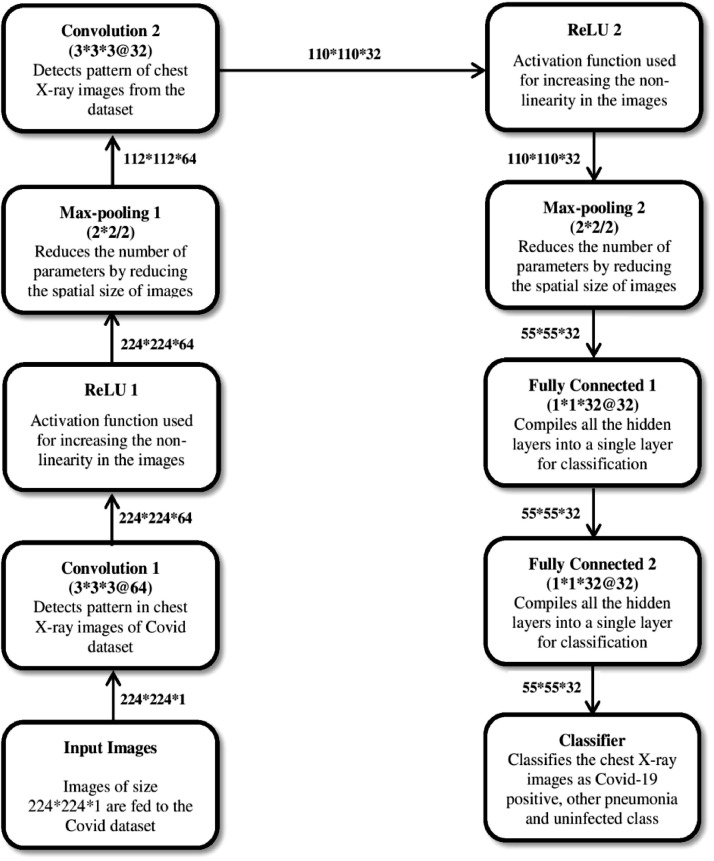
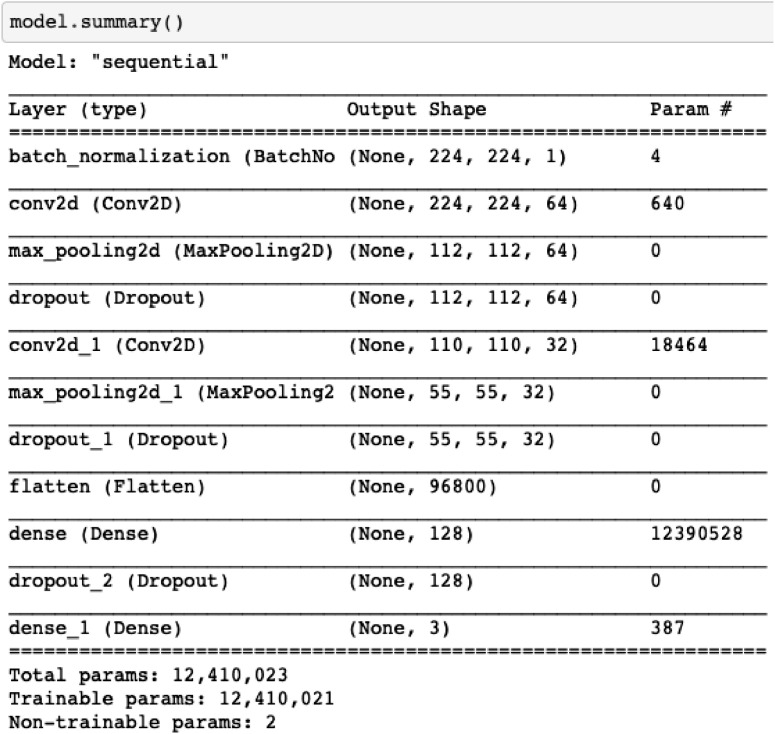
TLCoV-CNN model is used for the detection of covid-19 positive cases in which input layer reads the pre-processed images of chest x-ray from the dataset. The batch normalization layer is of shape 224 × 224 × 1 with 4 parameters, which is used for the standardization of inputs to the layers of each mini batch. It reduces the number of epochs required for the training of the model. The next layer, named convolution layer, has 640 parameters and output shape 224 × 224 × 64. It is used to detect the pattern in the chest X-ray images and retrieves the features from those images by using the set of filters with learning structures. The filters moved along with the images and the calculated activation feature map is provided as an output to the next layer of CNN.
Max pooling layer streamline the spatial size of network computation. It combines the parameter output of one layer into a single neuron and drops all the parameter in each stride having output shape of 112 × 112 × 64. The next layer dropout selects the value of outgoing edges of hidden unit randomly and set it as 0 to avoid the problem of overfitting. The value of rate parameter is taken as 0.2. It has output shape of 112 × 112 × 64 and does not have any parameters. These layers are followed by convolution of output shape 110 × 1110 × 32 with 18464 parameters which is followed by max pooling and dropout layer of output shape 55 × 55 × 32.
The next flatten layer with 96800 parameters is used to transform the n-dimension matrix of features into vector that is provided as an input to the dense layer. Dense layer has 128 filters with 12390528 activation and unit parameters. In this layer, the activation function ReLu is used, which replaces the negative valued pixels with zero in the computed convolved features for the generation of non-linearity map of features model. The dropout layer consists of 128 filters followed by dense layer having 3 filters to connect the neurons of this layer with the activation function of all the previous layers and classifies the convolved features of the chest X-ray images. The layers construction of TLCoV model using CNN is depicted in Fig. 5 .
Fig. 5.
Layers construction of TLCoV-CNN model.
It uses softmax activation function to interpret the probable values of the activation function results from the previous layer and calculates the probability distribution for 3 classes namely 0, 1 and 2. The output probability range is 0 to 1, while the sum of all the probabilities is equal to 1. The softmax function is defined in Eq. (1).
| (1) |
Where, is the normalized probability distribution, yj are the elements of input vector, m refers to the number of classes in multi-class classifier and is the normalization term that all the output values of the function will be equal to 1. Finally, the output layer labels the result as Covid-19 positive case, no infection and other viral pneumonia if the output value of the previous layer is 0, 1 and 2 respectively. The layer wise summary of TLCoV model is depicted in Fig. 6 .
Fig. 6.
Summary of TLCoV model using CNN.
3.2.2. Classification using transfer learning
Transfer learning leverage the knowledge gained from training of the previous model to train the new related model. Formally, a transfer learning includes – (i) a source domain ∂s and its corresponding source task ℑs and (ii) a target domain ∂t and its corresponding target task ℑt. A domain ∂ is defined by two components tuple that consists of feature space η and marginal probability P(Y), where Y = {y1, y2, y3,...., yn}. If two domains are different, then either of their feature space or marginal probability is different. A specific vector is represented by yi. A task ℑ is defined as two component tuple that consists of label space χ and an objective function γ which can be denoted as P(χ|Y) from the point of view of probability. The main motive of the transfer learning is to enable the model to learn the target conditional probability distribution P(χT|YT) in ∂t with the help of the information gained from the ∂s and ℑs, where ∂s ≠ ∂t and ℑs ≠ ℑt. Two transfer learning algorithms VGG-16 and ResNet-50 is used individually in the proposed TLCoV model.
3.2.2.1. VGG-16
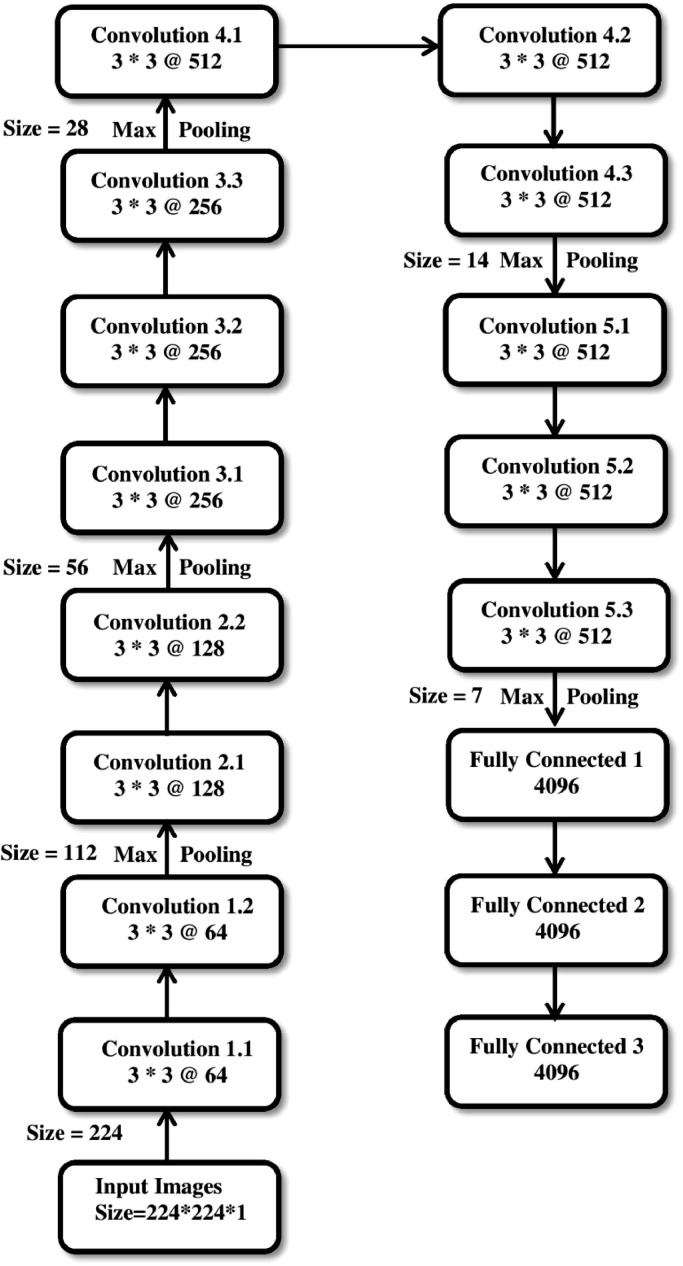
The pre trained transfer learning model VGG-16 is developed for computer vision benchmark dataset such as ImageNet image recognition task [35]. It is trained on millions of images using ImageNet in the range of many classes. The main motive of using transfer learning is weight, bias and features of the pre trained model can be transferred to our TLCoV scheme instead of starting from the scratch. This is achieved by applying these parameters while training on X-ray image dataset. Training a CNN model from scratch is a time consuming task as compared to training a pre trained model and it is computationally cheap if the dataset contains less number of images. VGG-16 model consists of 16 layers network that is built on the ImageNet database whose main aim is recognition and classification of the images. The chest X-ray images fed to the TLCoV model are unified and resized to 224 × 224. The set of parameters are configured for the model such as batch size is equal to 32, number of epoch is equal to 50 and learning rate is equal to 3e-4. The VGG-16 model consists of 13 convolution layer that uses 3 × 3 convolution filters, 5 max-pooling layers that is responsible for downsampling, 2 fully connected layers and 1 dense and flatten layer. The layers construction of TLCoV model using VGG-16 is depicted in Fig. 7 .
Fig. 7.
Layers construction of TLCoV-VGG-16 model.
If the input and output are represented as y and x and are in the form of usual maps, then they can be indexed as yk,l where k, l are the spatial coordinates. Let us consider a layer x = f(y). Sliding rectangular window field, also known as receptive field, is used to establish the relation that finds which component of y is influenced by which component of x. The output components x(k′,l′) depends only on the input component y(k,l), where (k,l) ∈ λ(k′,l′). The set λ(k′,l′) is a rectangular window field that can be defined using Eqs. (2) and (3).
| (2) |
| (3) |
Where, (αi,αj) is the stride, (βi, βj) is the offset and (δi, δj) is the receptive field size that measures the dependency of six network components. To calculate the receptive field filters size, it requires stride and padding for each layer, which is represented as i, s, p′ and sample point jo. The range of points that affect j0 in the input field can be represented as q, where q is defined as in Eq. (4), which is further simplified in Eqs. (5) and (6).
| (4) |
| (5) |
| (6) |
Where, p = and =.
The composing of receptive fields need to calculate the combination of two layers (i0, s0) and (i1, s1) as in Eqs. (7) and (8).
| (7) |
| (8) |
Eq. (9) is obtained by replacing value of j1 from Eq. (8) to Eq. (7).
| (9) |
Eq. (9) can further be simplified as in Eq. (10).
| (10) |
3.2.2.2. ResNet-50
ResNet-50 stands for Residual Network where 50 represent the number of layers. ResNet was introduced to solve exploding gradient and degradation problem that is faced while training a deep neural network model. It is pre trained on more than millions of images from ImageNet database [19]. This pre trained model is applied to train TLCoV on chest X-ray image dataset. ResNet 50 consists of 48 convolution layers, 1 average pool layer and 1 max pool layer and it consists of 3.8 × 109 floating point operations. The chest X-ray images are fed to the model and various parameters are configured like batch size is equal to 32, number of epoch is equal to 50 and learning rate is 3e-2. The work flow of TLCoV model using ResNet-50 is depicted in Fig. 8 .
Fig. 8.
Workflow of TLCoV-ResNet-50 model.
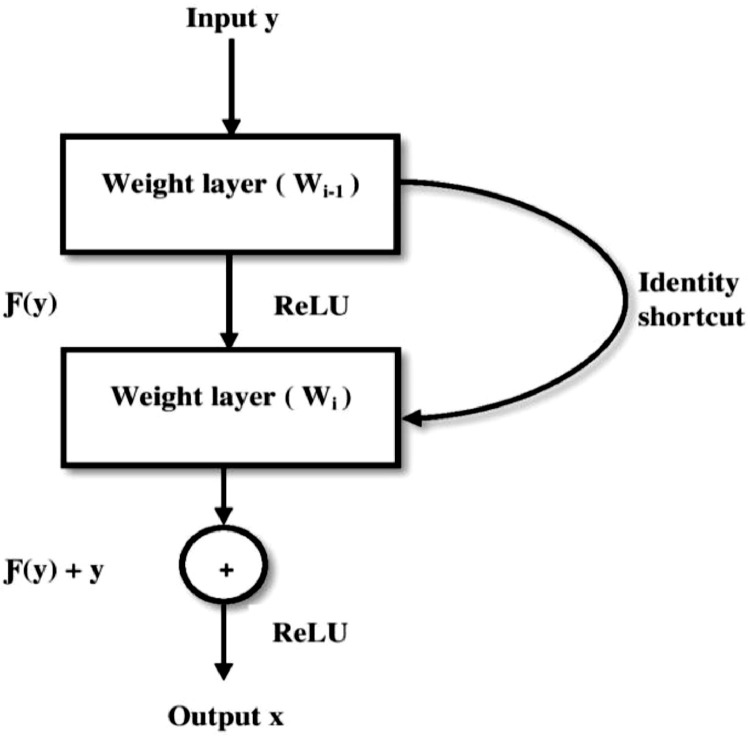
The identity shortcut can be used directly and the output function x is defined as in Eq. (11), when the input and output are of the same dimensions.
| (11) |
Where, y is the input to the residual block Ƒ(y, {Wj}) and Wj represents weight layers. In case of different dimensional input and output, the shortcut performs identity mapping by padding the extra zero entries with the dimension that is increased. The dimension is matched by using the projection shortcut as in Eq. (12).
| (12) |
Where, Ws is the extra parameter.
4. Experiments and results
This section describes the experimental setup, dataset, and performance metrics followed by analysing the performance of proposed TLCoV model by using CNN, VGG-16 and ResNet-50 separately and finds the best suite for our model. Finally, the performances of two existing schemes are also compared with our model TLCoV.
4.1. Experimental setup
The chest X-ray images are converted into 224 × 224 pixel value for achieving the requirement of the TLCoV model. It is trained and tested on Mac OS having Intel i5 1.6GHz dual core processor, 128GB PCI based SSD, 8GB of 2133MHz LPDDR3 on board RAM, boost upto 3.6 GHz and 4MB L3 cache. Online GPU from kaggle is used. Keras and tensorflow is used for the implementation and evaluation. The experiment is conducted to evaluate the performance of TLCoV model.
4.2. Dataset
The TLCoV scheme has used Covid-19 radiography dataset [19], which is downloaded from kaggle.com and consists of chest X-ray images of Covid-19 positive patients, normal patients and viral pneumonia patients. There are 219 corona positive images, 1345 viral pneumonia images and 1341 images for non-infected patients.
4.3. Performance metrics
The performance of the TLCoV model is evaluated based on various performance metrics such as accuracy, precision, recall and F-1 score which uses the following terminologies:
True Positive (TP): It signifies that a covid-19, normal and pneumonia case is correctly predicted as covid-19, normal and pneumonia respectively.
True Negative (TN): It signifies that a normal or pneumonia case is correctly predicted as normal or pneumonia.
False Positive (FP): It signifies that the case is normal or pneumonia infected case and is predicted as covid-19 case.
False Negative (FN): It signifies that the case is covid-19 and is predicted as normal or pneumonia case.
Accuracy is defined in Eq. (13) as the total number of records that is classified correctly to the total number of present records in the dataset.
| (13) |
Precision is defined in Eq. (14) as the ratio of positive records that is correctly classified to the total number predicted positive records in the dataset.
| (14) |
Recall is defined in Eq. (15) as the ratio of positive records that is correctly classified to the total number of positive records in the dataset. The higher rate of recall represents that the cases are correctly recognized.
| (15) |
High recall and low precision represents the positive records that are classified correctly though it contains large number of false positives, whereas low recall and high precision represents that the number of positive records may be missed, but either are predicted as positive or are truly positive.
F-1 score is the harmonic mean of precision and recall which is defined in Eq. (16). It reaches its best value at 1 and worst value at 0.
| (16) |
4.4. Performance evaluation of TLCoV using CNN, VGG-16 and ResNet-50
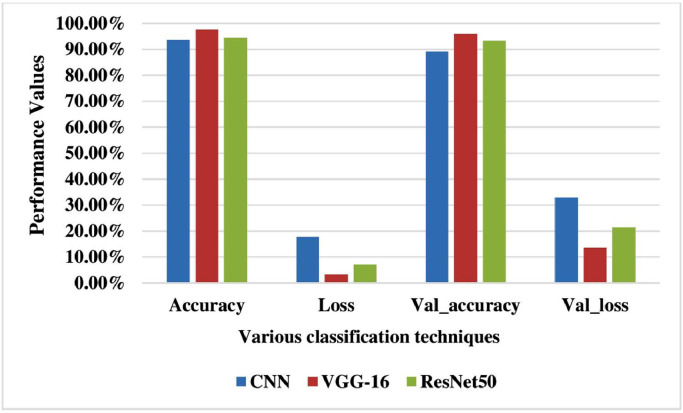
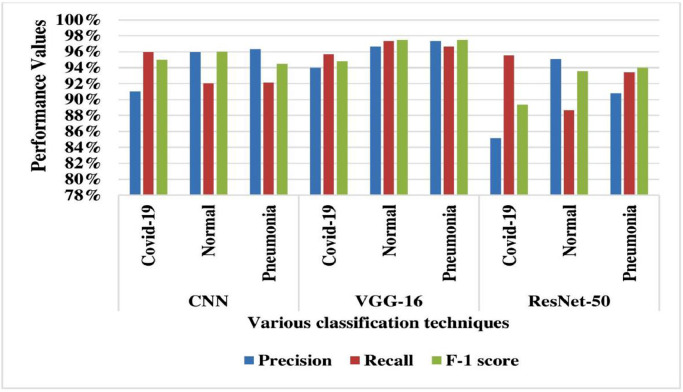
The performance of the TLCoV model is evaluated by performing the experiment using CNN, VGG-16 and ResNet-50. The performance metrics namely accuracy, loss, validation accuracy and validation loss are depicted in Table 1 and it -is represented graphically in Fig. 9 . The calculated value for precision, recall and F-1 score is presented in Table 2 and their graphical representation is depicted in Fig. 10 .
Table 1.
Performance comparison of different classifiers.
| Method | Accuracy | Loss | Val_accuracy | Val_loss |
|---|---|---|---|---|
| CNN | 93.67% | 17.62% | 89.16% | 32.86% |
| VGG-16 | 97.67% | 3.23% | 96.01% | 13.58% |
| ResNet-50 | 96.41% | 7.03% | 93.29% | 21.36% |
Fig. 9.
Performance comparison between different classifiers.
Table 2.
Multi class performance comparison of different classifiers.
| Classifier | Disease | Precision | Recall | F-1 score |
|---|---|---|---|---|
| CNN | Covid-19 | 91% | 95.97% | 95.27% |
| Normal | 95.95% | 92% | 95.81% | |
| Pneumonia | 96% | 92% | 94% | |
| VGG-16 | Covid-19 | 93.98% | 95.67% | 94.82% |
| Normal | 96.62% | 97.34% | 97.48% | |
| Pneumonia | 97.35% | 96.62% | 97.49% | |
| ResNet-50 | Covid-19 | 85.19% | 95.52% | 89.35% |
| Normal | 95.11% | 88.68% | 93.55% | |
| Pneumonia | 90.76% | 93.42% | 93.97% |
Fig. 10.
Multi class performance comparison of various classifiers.
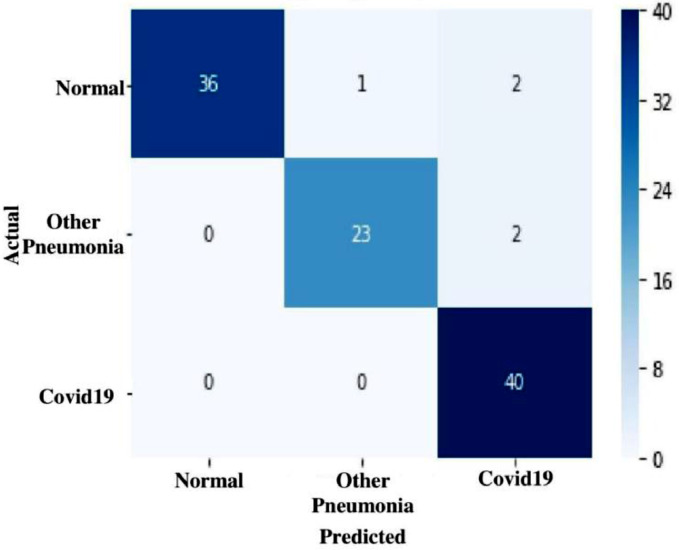
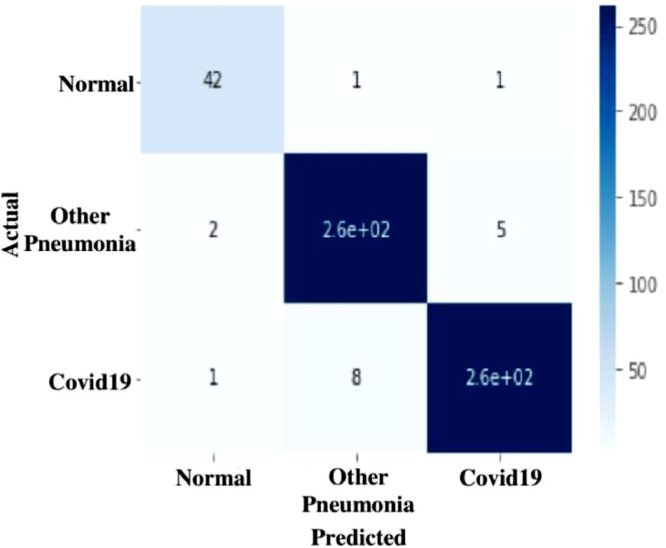
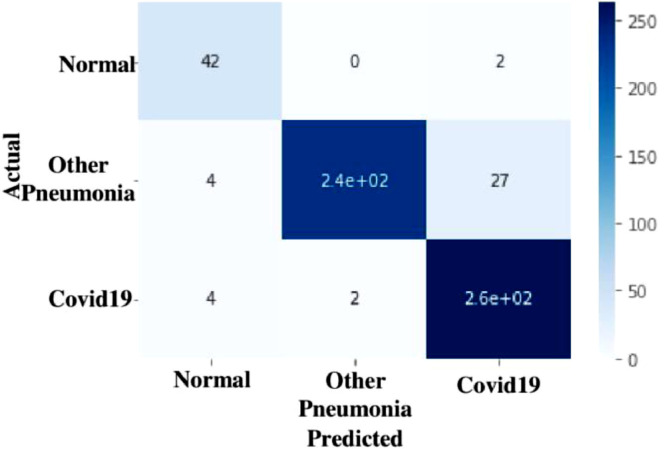
Confusion matrix for CNN, VGG-16 and ResNet-50 is depicted in Fig. 11, Fig. 12, Fig. 13 which gives evidence that the TLCoV model can screen coivid-19 cases correctly without missing any case.
Fig. 11.
Confusion matrix of TLCoV-CNN model.
Fig. 12.
Confusion matrix of TLCoV-VGG-16 model.
Fig. 13.
Confusion matrix of TLCoV-ResNet-50.
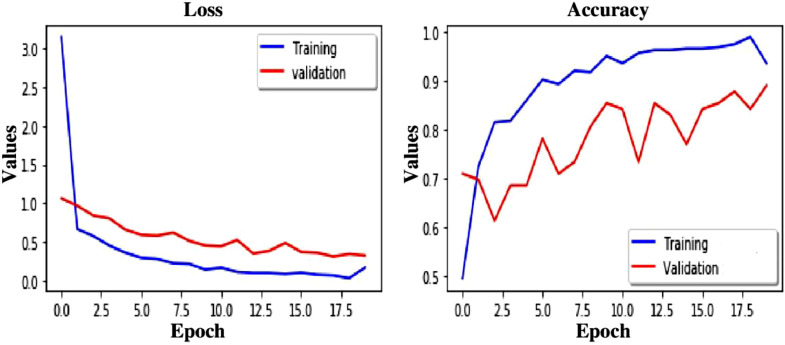
Accuracy and loss curve for TLCoV model using CNN, VGG-16 and ResNet-50 is depicted in Fig. 14, Fig. 15, Fig. 16 respectively.
Fig. 14.
Loss and Accuracy curve of TLCoV-CNN model.
Fig. 15.
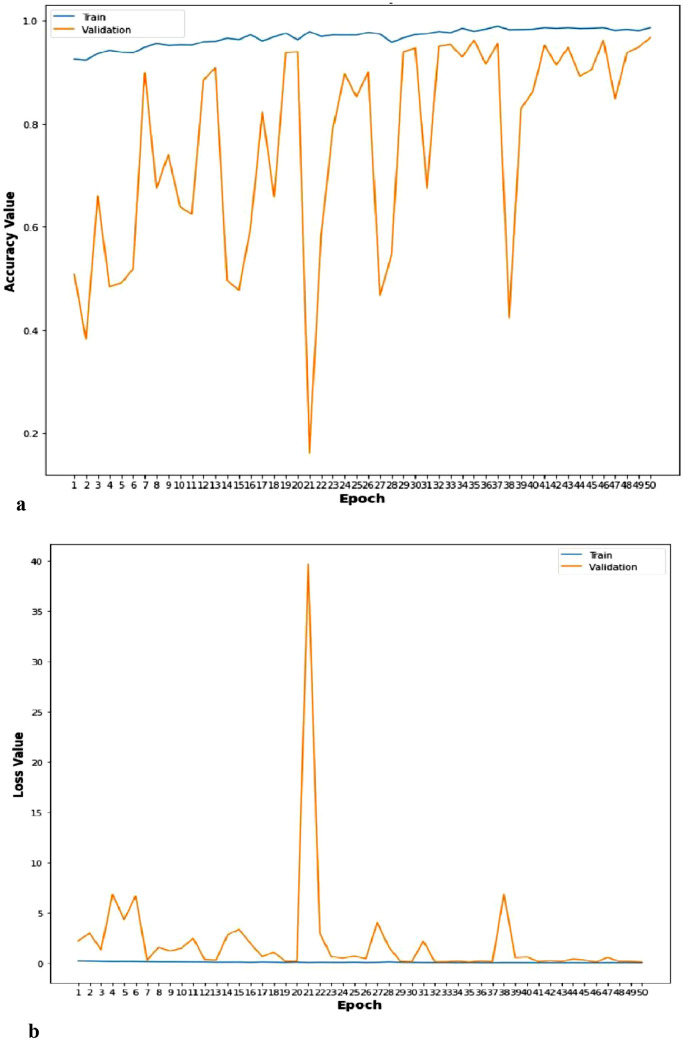
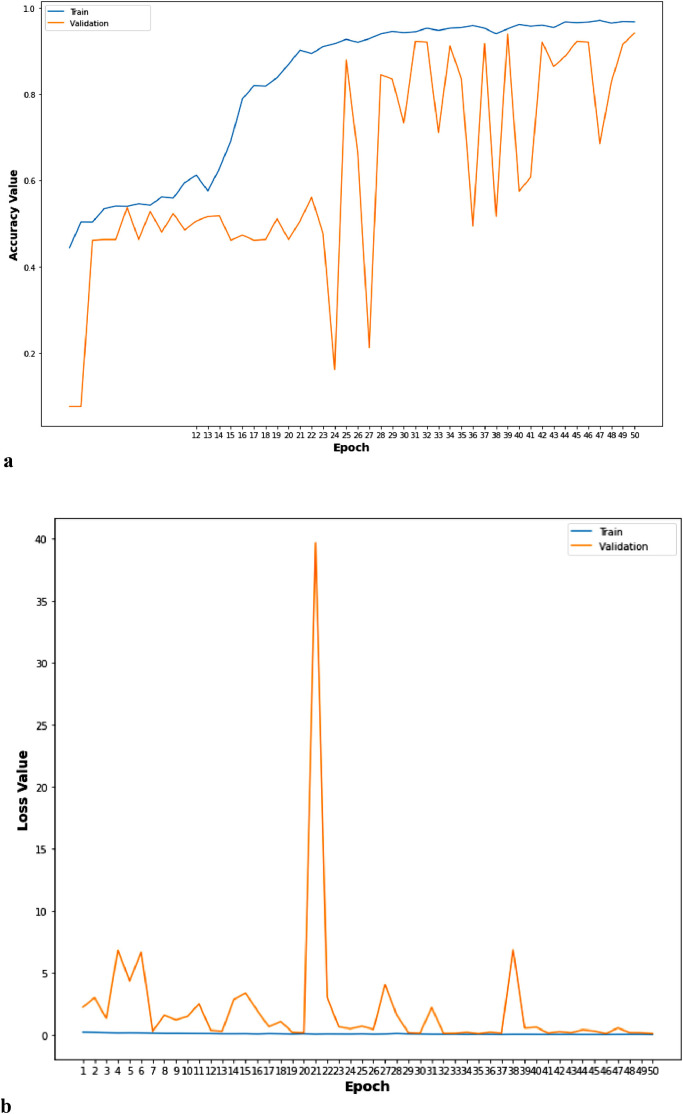
a. Accuracy curve of TLCoV-VGG-16 model. b. Loss curve of TLCoV-VGG-16 model.
Fig. 16.
a. Accuracy curve of TLCoV-ResNet-50 model. b. Loss curve of TLCoV-ResNet-50 model.
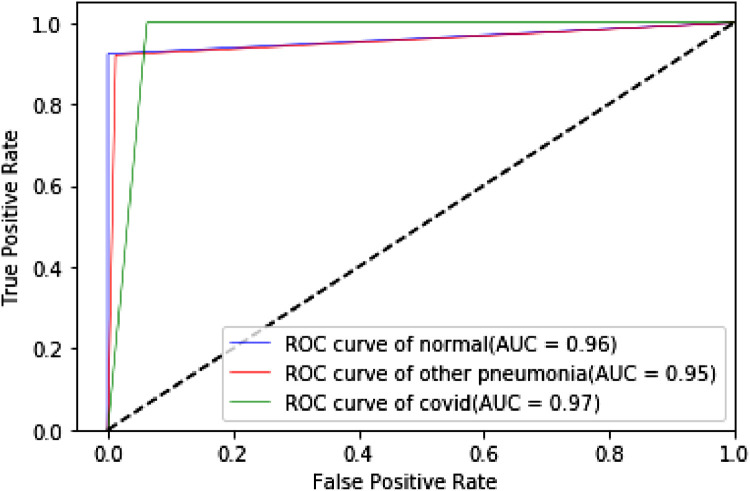
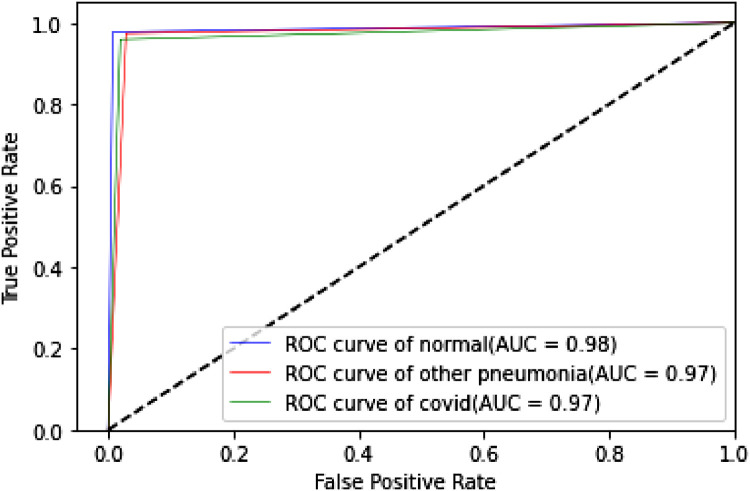
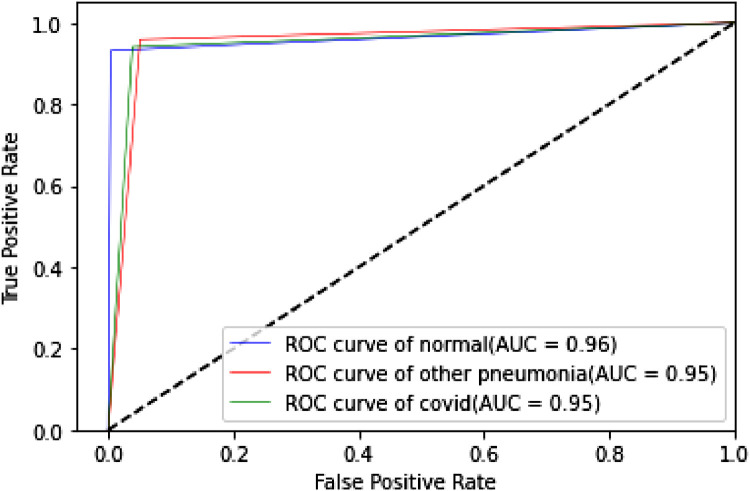
The proposed TLCoV model is solving a multiclass classification problem, in which the performance with various threshold settings can be measured by ROC (Receiver Operating Characteristics) curve and AUC (Area Under the Curve). The probability curve ROC depicts two parameters – True Positive Rate and false Positive Rate. AUC represents the entire two-dimensional area under the ROC curve from (0,0) to (1,1). Higher AUC means better model in classifying the patient as Covid-19 positive, viral pneumonia or with no infection. Roc curve of TLCoV model with CNN, VGG-16 and ResNet-50 is depicted in Fig. 17, Fig. 18, Fig. 19 respectively. The average ROC-AUC value for CNN is 0.96, for VGG-16 is 0.97 and for ResNet-50 is 0.95 indicates that VGG-16 is better suite with proposed TLCoV model.
Fig. 17.
ROC curve of TLCoV-CNN model.
Fig. 18.
ROC curve of TLCoV-VGG-16 model.
Fig. 19.
ROC curve of TLCoV-ResNet-50 model.
4.5. Performance comparison of TLCoV with the existing schemes
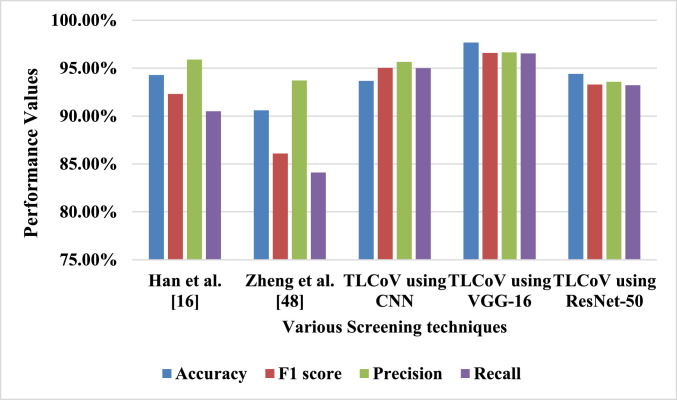
The previous subsection describes that TLCoV-VGG-16 is the best suite out of all the three combination. Thus the performance of TLCoV-VGG-16 is compared with two existing schemes as described in Table 3 and depicted in Fig. 20 , which show that the proposed model outperforms both the schemes.
Table 3.
Comparison between TLCoV-VGG-16 and existing schemes.
Fig. 20.
Comparision between TLCoV and existing schemes.
5. Future research
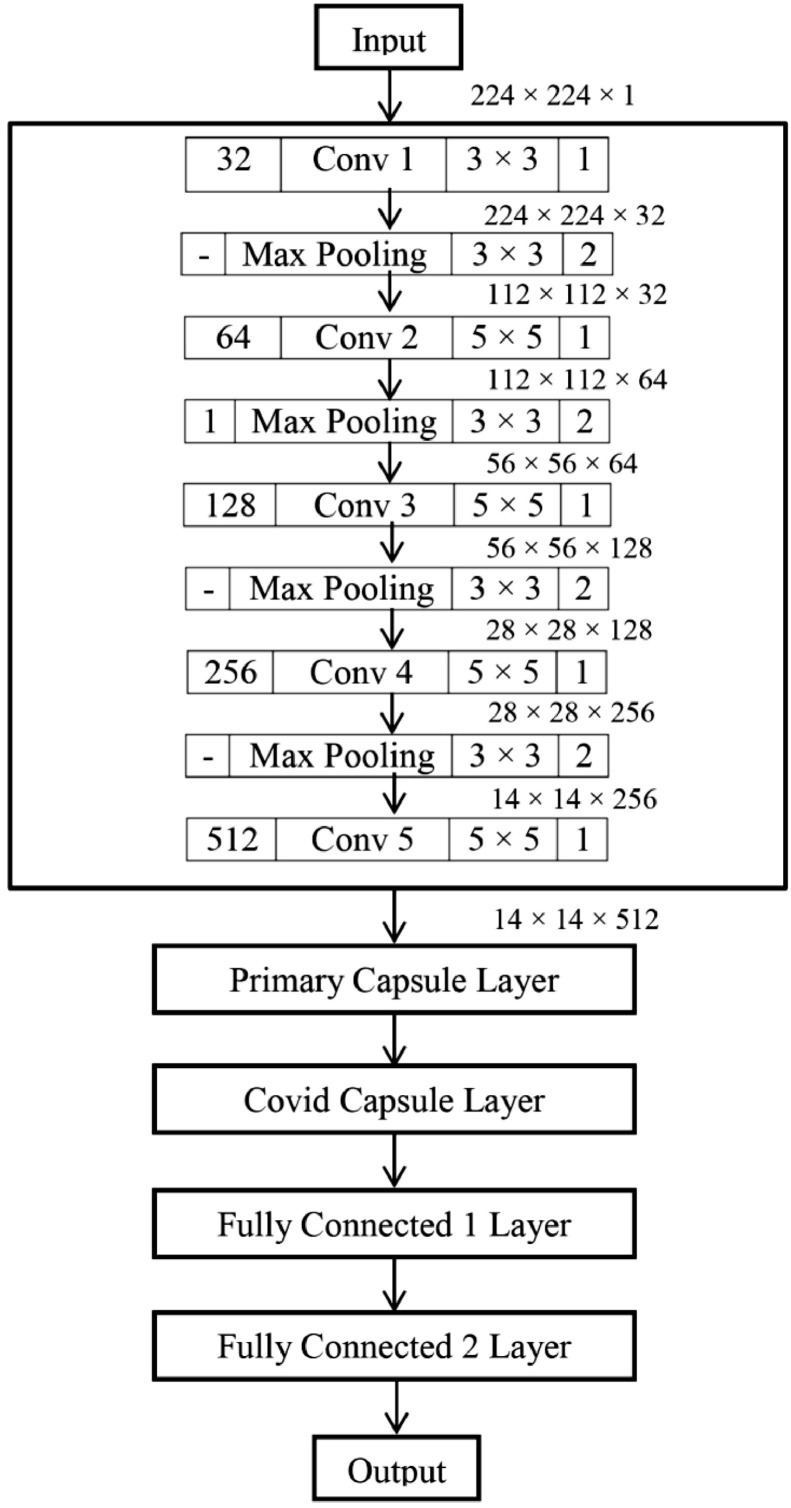
The main deficiency of standard convolutional neural network models is the use of successive pooling layers, which reduces the data dimension to achieve spatial invariance. Thus it is unable to recognize the object when direction is changed. Moreover, the pooling layer loses the required spatial information about the rotation, location, scale and different positional attributes of the object. This creates difficulties in object detection and segmentation. In CNN models, generally Max Pooling Layer is used as primitive type of routing mechanism. The most active feature in a local pool (say 4 × 4 grid) is routed to the higher layer and the higher-level detectors are not allowed to take the decision. This limitation of standard CNN models is motivated us to introduce Capsule Network (CapsNet) [38] for the screening of Covid19. In CapsNet, only those features that agree with high-level detectors will be allowed to be routed. We have designed a model for Covid19 screening as a future scope of this proposed work. The Capsule Network is used in our model for its superior dynamic routing mechanism.
Capsule Network (CapsNet) is a completely different approach than classical Neural Network. The CapsNet is more emphasized on creating a model of hierarchical relationships. A capsule is a group of neurons that stores different information about the identifiable object in a given image. The mostly stored information is about its position, rotation and scale in a high dimensional vector space. The dimensions are representing something special about the object than can be understood intuitively. The purpose of the capsule is to detect a feature and also to train the model to learn the variant such that the same capsule can detect the same object class with different orientations (for example, rotate clockwise). In order to screen Covid19, our model is using chest scan images as input images. Thus training on multiple convolutional layers will be beneficial. Hence, the novelty of our work is to use 5 convolutional layers in CapsNet to provide more deep analysis. Moreover, we have varied kernel size from 32 to 1024 to provide more accurate dot products for the pixels and lead to correct prediction. Kernel is a matrix that moves over the input data, performs the dot product with the sub-region of input data, and gets the output as the matrix of dot products. Kernel moves on the input data by the stride value. If the stride value is 2, then the kernel moves by 2 columns of pixels in the input matrix. Our proposed CapsNet architecture for the future implementation to detect the Covid19 disease from chest x-ray images is given in Fig. 21 .
Fig 21.
Proposed Capsule Network Architecture for Covid19 detection.
In the proposed architecture, two capsule layers is there, namely Primary Capsule Layer and Covid Capsule Layer. The output of the next layer j is predicted by each capsule i using the trainable weight matrix as in Eq. (17).
| (17) |
Where, is the instantiation parameter. The prediction is computed by Routing by Agreement process [38]. The agreement between the prediction and output is determined in Eq. (18).
| (18) |
The summation of agreements is calculated in Eq. (19).
| (19) |
The score that determines the contribution of prediction to the output is calculated in Eq. (20).
| (20) |
The actual output of Capsule j is determined by Eq. (21).
| (21) |
The CapsNet loss function associated with capsule n is computed in Eq. (22).
| (22) |
The value of Tn is 1 if the class n is present, otherwise it will be 0. , and represents the hyper parameter of the model.
6. Conclusion
In the proposed TLCoV model three learning techniques CNN, VGG-16 and ResNet-50 is used individually and the experimental results show that TLCoV-VGG-16 is the best suite for the detection and classification of Covid-19 positive cases from the chest X-ray images. The automated Covid-19 screening model TLCoV did multi class classification with accuracy of 97.67% and average ROC-AUC of 0.97. The performance evaluation of the proposed model proves that it outperforms two similar types of existing automated Covid-19 screening models. It helps in reducing the workload of radiologists and accelerates the screening process to identify Covid-19 positive patients.
CRediT authorship contribution statement
Ayan Kumar Das: Conceptualization, Data curation, Formal analysis, Methodology, Resources, Supervision, Validation, Writing - original draft, Writing - review & editing. Sidra Kalam: Conceptualization, Data curation, Formal analysis, Methodology, Resources, Validation, Visualization, Writing - original draft. Chiranjeev Kumar: Conceptualization, Data curation, Formal analysis, Methodology, Resources, Software, Validation, Visualization. Ditipriya Sinha: Conceptualization, Formal analysis, Methodology, Resources, Supervision, Validation, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no conflict of interest.
Acknowledgements
All persons who have made substantial contributions to the work reported in the manuscript (e.g., technical help, writing and editing assistance, general support), but who do not meet the criteria for authorship, are named in the Acknowledgements and have given us their written permission to be named. If we have not included an Acknowledgements, then that indicates that we have not received substantial contributions from non-authors.
References
- 1.Acharya U.R., Oh S.L., Hagiwara Y., Tan J.H., Adam M., Gertych A., San Tan R. A deep convolutional neural network model to classify heartbeats. Comput Biol Med. 2017;89:389–396. doi: 10.1016/j.compbiomed.2017.08.022. [DOI] [PubMed] [Google Scholar]
- 2.Bai, X., Fang, C., Zhou, Y., Bai, S., Liu, Z., Xia, L., & Xie, X. (2020). Predicting COVID-19 malignant progression with AI techniques. 10.1101/2020.03.20.20037325.
- 3.Ghanbari Behzad. On forecasting the spread of the COVID-19 in Iran: The second wave. (2020), On forecasting the spread of the COVID-19 in Iran: The second wave. Chaos Solitons Fract. 2020;140 doi: 10.1016/j.chaos.2020.110176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butt C., Gill J., Chun D., Babu B.A. Deep learning system to screen coronavirus disease 2019 pneumonia. Appl Intell. 2020;1 doi: 10.1007/s10489-020-01714-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Celik Y., Talo M., Yildirim O., Karabatak M., Acharya U.R. Automated invasive ductal carcinoma detection based using deep transfer learning with whole-slide images. Pattern Recognit Lett. 2020 doi: 10.1016/j.patrec.2020.03.011. [DOI] [Google Scholar]
- 6.Ouchicha C., Ammor O., Meknassi M. CVDNet: a novel deep learning architecture for detection of coronavirus (Covid-19) from chest x-ray images. Chaos Solitons Fract. 2020;140 doi: 10.1016/j.chaos.2020.110245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, J., Wu, L., Zhang, J., Zhang, L., Gong, D., Zhao, Y., Zhang, K. (2020). Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography: a prospective study. MedRxiv. 10.1101/2020.02.25.20021568. [DOI] [PMC free article] [PubMed]
- 8.Codella N.C., Nguyen Q.B., Pankanti S., Gutman D.A., Helba B., Halpern A.C., Smith J.R. Deep learning ensembles for melanoma recognition in dermoscopy images. IBM J Res Dev. 2017;61(4/5):5. doi: 10.1147/JRD.2017.2708299. -1. [DOI] [Google Scholar]
- 9.Cruz-Roa A., Basavanhally A., González F., Gilmore H., Feldman M., Ganesan S., Madabhushi A. Automatic detection of invasive ductal carcinoma in whole slide images with convolutional neural networks. Med Imag. 2014;9041 doi: 10.1117/12.2043872. [DOI] [Google Scholar]
- 10.Dong Y., Jiang Z., Shen H., Pan W.D., Williams L.A., Reddy V.V., Bryan A.W. Evaluations of deep convolutional neural networks for automatic identification of malaria infected cells. IEEE EMBS International Conference on Biomedical & Health Informatics (BHI); Orlando, FL, USA; 2017. [DOI] [Google Scholar]
- 11.Esteva A., Kuprel B., Novoa R.A., Ko J., Swetter S.M., Blau H.M., Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaál, G., Maga, B., & Lukács, A. (2020). Attention u-net based adversarial architectures for chest x-ray lung segmentation. arXiv preprint arXiv:2003.10304.
- 13.Ghoshal, B., & Tucker, A. (2020). Estimating uncertainty and interpretability in deep learning for coronavirus (COVID-19) detection. arXiv preprint arXiv:2003.10769.
- 14.Gopakumar G., Swetha M., Siva G.S., Subrahmanyam G.R.K. CNN based malaria diagnosis from focus-stack of blood smear images acquired using custom-built slide scanner. J Biophoton. 2018;11(3) doi: 10.1002/jbio.201700003. [DOI] [PubMed] [Google Scholar]
- 15.Gorbalenya, A. E., Baker, S. C., Baric, R., Groot, R. J. D., Drosten, C., Gulyaeva, A. A., Penzar, D. (2020). Severe acute respiratory syndrome-related coronavirus: The species and its viruses–a statement of the Coronavirus Study Group. 10.1101/2020.02.07.937862.
- 16.Han Z., Wei B., Hong Y., Li T., Cong J., Zhu X., Zhang W. Accurate Screening of COVID-19 using attention based deep 3D multiple instance learning. IEEE Trans Med Imaging. 2020 doi: 10.1109/TMI.2020.2996256. [DOI] [PubMed] [Google Scholar]
- 17.Hannun A.Y., Rajpurkar P., Haghpanahi M., Tison G.H., Bourn C., Turakhia M.P., Ng A.Y. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat Med. 2019;25(1):65. doi: 10.1038/s41591-018-0268-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE conference on computer vision and pattern recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [DOI] [Google Scholar]
- 19.https://www.kaggle.com/tawsifurrahman/covid19-radiography - database.
- 20.https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
- 21.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Cheng Z. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet North Am Ed. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hung J., Carpenter A. Proceedings of the IEEE conference on computer vision and pattern recognition workshops. 2017. Applying faster R-CNN for object detection on malaria images; pp. 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin C., Chen W., Cao Y., Xu Z., Zhang X., Deng L., Feng J. Development and evaluation of an AI system for COVID-19 diagnosis. medRxiv. 2020 doi: 10.1101/2020.03.20.20039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin S., Wang B., Xu H., Luo C., Wei L., Zhao W., Sun W. AI-assisted CT imaging analysis for COVID-19 screening: building and deploying a medical AI system in four weeks. medRxiv. 2020 doi: 10.1101/2020.03.19.20039354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang H., Xia L., Yan F., Wan Z., Shi F., Yuan H., Shen D. Diagnosis of coronavirus disease 2019 (covid-19) with structured latent multi-view representation learning. IEEE Trans Med Imaging. 2020 doi: 10.1109/TMI.2020.2992546. [DOI] [PubMed] [Google Scholar]
- 26.Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu H., Stratton C.W., Tang Y.W. Outbreak of pneumonia of unknown etiology in Wuhan China: the mystery and the miracle. J Med Virol. 2020;92(4):401–402. doi: 10.1002/jmv.25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maghdid H.S., Asaad A.T., Ghafoor K.Z., Sadiq A.S., Khan M.K. Diagnosing COVID-19 pneumonia from X-ray and CT images using deep learning and transfer learning algorithms. arXiv. 2020 preprint arXiv:2004.00038. [Google Scholar]
- 29.Ribeiro M.H.D.M., da Silva R.G., Mariani V.C., Coelho L.S. Short-term forecasting COVID-19 cumulative confirmed cases: perspectives for Brazil. Chaos Solitons Fract. 2020;135 doi: 10.1016/j.chaos.2020.109853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Punn N.S., Sonbhadra S.K., Agarwal S. COVID-19 epidemic analysis using machine learning and deep learning algorithms. medRxiv. 2020 doi: 10.1101/2020.04.08.20057679. [DOI] [Google Scholar]
- 31.Rajpurkar, P., Irvin, J., Zhu, K., Yang, B., Mehta, H., Duan, T., Ungren, M. P. (2017). Chexnet: radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv preprint arXiv:1711.05225.
- 32.da Silva R.G., Ribeiro M.H.D.M., Mariani V.C., Coelho Leandro dos Santos. Chaos Solitons Fract. 2020;139 doi: 10.1016/j.chaos.2020.110027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rustam F., Reshi A.A., Mehmood A., Ullah S., On B., Aslam W., Choi G.S. COVID-19 future forecasting using supervised machine learning models. IEEE Access. 2020 doi: 10.1109/ACCESS.2020.2997311. [DOI] [Google Scholar]
- 34.Sethy P.K., Behera S.K., Ratha P.K., Biswas P. Detection of coronavirus disease (COVID-19) based on deep features and support vector machine. Int. J. Math., Eng. Manage. Sci. 2020;5(4):643–651. [Google Scholar]
- 35.Simonyan, K., & Zisserman, A. (2014). Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556.
- 36.Singhal T. A review of coronavirus disease-2019 (COVID-19) Indian J. Pediatr. 2020:1–6. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Souza J.C., Diniz J.O.B., Ferreira J.L., da Silva G.L.F., Silva A.C., de Paiva A.C. An automatic method for lung segmentation and reconstruction in chest X-ray using deep neural networks. Comput Methods Programs Biomed. 2019;177:285–296. doi: 10.1016/j.cmpb.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 38.Toraman S., Alakus T.B., Turkoglu I. Convolutional capsnet: a novel artificial neural network approach to detect COVID-19 disease from X-ray images using capsule networks. Chaos Solitons Fract. 2020;140 doi: 10.1016/j.chaos.2020.110122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Annas S., Pratama M.I., Rifandi M, Sanusi W., Side S. Stability analysis and numerical simulation of SEIR model for pandemic COVID-19 spread in Indonesia. Chaos Solitons Fract. 2020;139 doi: 10.1016/j.chaos.2020.110072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Talo M., Yildirim O., Baloglu U.B., Aydin G., Acharya U.R. Convolutional neural networks for multi-class brain disease detection using MRI images. Comput Med Imaging Graph. 2019;78 doi: 10.1016/j.compmedimag.2019.101673. [DOI] [PubMed] [Google Scholar]
- 41.Tan J.H., Fujita H., Sivaprasad S., Bhandary S.V., Rao A.K., Chua K.C., Acharya U.R. Automated segmentation of exudates, haemorrhages, microaneurysms using single convolutional neural network. Inf Sci. 2017;420:66–76. doi: 10.1016/j.ins.2017.08.050. [DOI] [Google Scholar]
- 42.Yan T., Wong P.K., Ren H., Wang H., Li J.W.Y. Automatic distinction between COVID-19 and common pneumonia using multi-scale convolutional neural network on chest CT scans. Chaos Solitons Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Zhao Y. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, S., Kang, B., Ma, J., Zeng, X., Xiao, M., Guo, J., Xu, B. (2020). A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID-19). MedRxiv. 10.1101/2020.02.14.20023028. [DOI] [PMC free article] [PubMed]
- 45.Wang X., Deng X., Fu Q., Zhou Q., Feng J., Ma H., Zheng C. A Weakly-supervised framework for COVID-19 classification and lesion localization from chest CT. IEEE Trans Med Imaging. 2020 doi: 10.1109/TMI.2020.2995965. [DOI] [PubMed] [Google Scholar]
- 46.Wang, Y., Hu, M., Li, Q., Zhang, X. P., Zhai, G., & Yao, N. (2020). Abnormal respiratory patterns classifier may contribute to large-scale screening of people infected with COVID-19 in an accurate and unobtrusive manner. arXiv preprint arXiv:2002.05534.
- 47.Yıldırım Ö., Pławiak P., Tan R.S., Acharya U.R. Arrhythmia detection using deep convolutional neural network with long duration ECG signals. Comput Biol Med. 2018;102:411–420. doi: 10.1016/j.compbiomed.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 48.Zheng C., Deng X., Fu Q., Zhou Q., Feng J., Ma H., Wang X., Deep learning-based detection for COVID-19 from chest CT using weak label medRxiv. 2020 doi: 10.1101/2020.03.12.20027185. [DOI] [Google Scholar]
- 49.Malki Z., Atlam E.-S., Hassanien A.E., Dagnew G., Elhosseini M.A., Gad I. Association between weather data and COVID-19 pandemic predicting mortality rate: Machine learning approaches. Chaos Solitons Fract. 2020;138 doi: 10.1016/j.chaos.2020.110137. [DOI] [PMC free article] [PubMed] [Google Scholar]