Abstract
Mitigation of the ongoing coronavirus disease 2019 (COVID-19) pandemic requires reliable and accessible laboratory diagnostic services. In this study, the performance of one laboratory-developed test (LDT) and two commercial tests, cobas SARS-CoV-2 (Roche) and Amplidiag COVID-19 (Mobidiag), were evaluated for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA in respiratory specimens. A total of 183 specimens collected from suspected COVID-19 patients were studied with all three methods to compare their performance. In relation to the reference standard, which was established as the result obtained by two of the three studied methods, the positive percent agreement was highest for the cobas test (100%), followed by the Amplidiag test and the LDT (98.9%). The negative percent agreement was lowest for the cobas test (89.4%), followed by the Amplidiag test (98.8%), and the highest value was obtained for the LDT (100%). The dilution series of positive specimens, however, suggests significantly higher sensitivity for the cobas assay in comparison with the other two assays, and the low negative percent agreement value may be due to the same reason. In general, all tested assays performed adequately. Clinical laboratories need to be prepared for uninterrupted high-throughput testing during the coming months to mitigate the pandemic. To ensure no interruption, it is critical that clinical laboratories maintain several simultaneous platforms in their SARS-CoV-2 nucleic acid testing.
Mitigation of the ongoing coronavirus disease 2019 (COVID-19) pandemic requires reliable and accessible laboratory diagnostic services. The specific diagnosis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection relies on molecular methods, especially on reverse transcription PCR, although other technologies, including serologic immunoassays, are emerging.1 , 2 The first methods for SARS-CoV-2 detection were laboratory-developed reverse transcription PCR tests (LDTs), and one of the first methods published was described by Corman et al.3 This method was later endorsed by the World Health Organization and was widely implemented in clinical laboratories. Roche Molecular Systems (Branchburg, NJ) cobas SARS-CoV-2 test was the first commercial test to get emergency use authorization from the Food and Drug Administration on March 12, 2020. Since then (as of June 25, 2020), more than 100 commercial molecular in vitro diagnostic tests have been granted Food and Drug Administration or other national authorities' emergency use authorization and/or CE-mark (Kalorama Information, https://kaloramainformation.com/covid19diagnosticstracker, last accessed June 25, 2020). Both LDTs and commercial tests have been set up under high time pressure. Therefore, it is of great importance to evaluate the tests in the clinical laboratory settings. In this study, the performance of one LDT and two commercial tests, namely, cobas SARS-CoV-2 (Roche) and recently, CE/in vitro diagnostic–marked Amplidiag COVID-19 (Mobidiag, Espoo, Finland), were evaluated for the detection of SARS-CoV-2 RNA in respiratory specimens.
Materials and Methods
Specimens
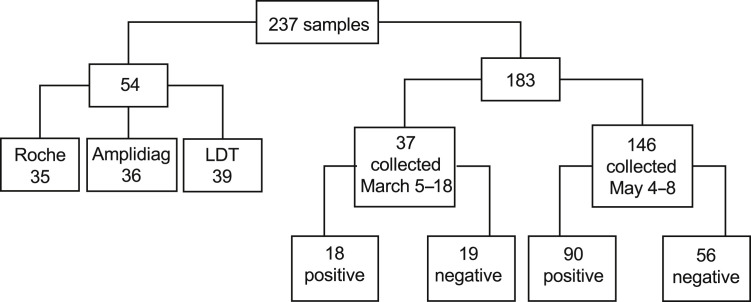
Altogether, 237 respiratory tract specimens referred to Helsinki University Hospital Laboratory (HUS Diagnostic Center, HUSLAB), Department of Virology and Immunology, Finland, were included in this study (Figure 1 ). The study was approved by the local review board (HUS/157/2020-29). Fifty-four specimens collected from patients with respiratory symptoms in 2018 to 2020 were used to verify the analytical specificity of the tests. The specimens from 2020 were negative for SARS-CoV-2 RNA by the LDT. A total of 183 specimens collected from suspected COVID-19 patients in 2020 were studied with all three methods to compare the performance of the tests in detecting SARS-CoV-2 RNA. Although most of the specimens tested were nasopharyngeal swabs (89%), oropharyngeal (9%) and nasal (2%) swabs also were included due to the global shortage of the swab sticks needed for nasopharyngeal sampling. The specimens were collected either in Copan UTM tubes (Copan, Brescia, Italy) or, due to the global shortage of Copan UTM tubes, in tubes containing 0.9% saline. The suitability of the 0.9% saline tubes as an alternative to the viral transport media in SARS-CoV-2 testing has been shown previously.4
Figure 1.
Flowchart of the samples analyzed in this study. Altogether, 237 samples were collected. Fifty-four samples that were positive for other respiratory viruses were used to evaluate the analytical specificity of the three methods. A total of 183 samples sent for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) analyses were collected, 37 in the early phase of the epidemic in Finland and a further 146 in the late phase of the epidemic. These 183 samples were tested with each of the three methods evaluated.
The specimens (n = 183) used to compare the performance of the three studied RT-PCR methods comprised two sets of specimens collected within two time frames. The first set of specimens (n = 37) was part of the material used for the initial verification of the cobas SARS-CoV-2 and Amplidiag COVID-19 tests. These specimens were collected between March 5 and March 18, 2020, and the SARS-CoV-2–positive specimens (n = 18) represent virus strains from the early epidemic. The second set of specimens (n = 146) were collected between May 4 and May 8, 2020, and the positive specimens (n = 90) thus represent strains from a declining phase of the epidemic in Finland. Specimens were stored at −20°C/−70°C after the initial analysis and were thawed upon analysis. Specimens were not thawed more than twice before RT-PCR.
In addition, QCMD 2020 Coronavirus Outbreak Preparedness EQA Pilot Study proficiency samples (Glasgow, Scotland, UK) were used to evaluate the performance of the methods.
Inactivation and Lysing of the Specimens
All specimens were inactivated in a biosafety cabinet in a biosafety level 2 laboratory that has negative pressure. At the beginning of the epidemic, an FFP3 mask, protective glasses, and protective clothing were worn when working in the laboratory. Later in the epidemic, a visor and a surgical mask replaced the FFP3 masks and protective glasses.
LDT, HUSLAB
The possible SARS-CoV-2 in the specimen was inactivated by adding 250 μL of MagNA Pure Lysis/Binding Buffer (Roche Diagnostics, Mannheim, Germany) to 250 μL of patient specimen. The lysates were incubated for a minimum of 10 minutes at room temperature before being processed further.
Cobas SARS-CoV-2
If needed, the specimens were first equilibrated to room temperature, after which the possible SARS-CoV-2 in the specimen was inactivated by adding 350 μL of MagNA Pure Lysis/Binding Buffer (Roche Diagnostics) to 350 μL of patient specimen. The lysates were incubated for a minimum of 10 minutes at room temperature before being processed further.
Amplidiag COVID-19
The possible SARS-CoV-2 in the specimen was inactivated either by adding 600 μL of the specimen to 1 mL of eNAT medium (Copan), or 360 μL to 1.2 mL of mNAT Medium (Mobidiag). The eNAT tubes were incubated for a minimum of 30 minutes, and the mNAT-tubes, for a minimum of 5 minutes at room temperature before being processed further.
Due to the different protocols, the inactivation step dilutes the specimens unevenly. The dilution factor for the LDT and cobas tests is 1:2, whereas it is 1:2.7 (eNAT tube) or 1:4.3 (mNAT tube) for the Amplidiag test.
Molecular Methods Evaluated in the Study
LDT, HUSLAB
The real-time LDT SARS-CoV-2 RT-PCR used in this study is a modification of the method published by Corman et al.3 The test is suitable for the detection of SARS-CoV-2 RNA from sputum; nasopharyngeal/tracheal aspirates; nasal, nasopharyngeal, and oropharyngeal swab specimens; and feces. Initially, all target genes (E, RdRP, and N) were included in the diagnostic assay. In addition, a PCR for the beta-globin gene5 was performed to verify successful sampling, extraction, and PCR. A full-length SARS-CoV in vitro transcript was used as a positive control. When the epidemic spread, and there was suddenly a high demand of testing in combination with the global shortage of supplies, the E gene PCR was first dropped out, because of the occasional unspecific positive signal obtained from negative controls/specimens.6 Later, RdRP also was excluded, and the diagnostics were continued with the N gene PCR only. The N gene PCR was chosen because a dilution series indicated better sensitivity for the N gene PCR over RdRP PCR, although earlier cycle threshold (CT) values were gained for RdRP PCR. Furthermore, findings from low-positive specimens suggested a better sensitivity for N gene RT-PCR (data not shown).
Nucleic acids were extracted from 450 μL of the respiratory specimen lysate using the MagNA Pure Viral NA Small Volume 2.0 Kit (Roche Diagnostics), with the MagNA Pure 96 instrument, and eluted in 50 μL of the elution buffer.
Real-time RT-PCR was performed using the SuperScript III Platinum One-Step qRT-PCR Kit (Invitrogen, Darmstadt, Germany) with 600 nmol/L of the forward primer 5′-CACATTGGCACCCGCAATC-3′, 800 nmol/L of the reverse primer 5′-GAGGAACGAGAAGAGGCTTG, and 200 nmol/L of the probe FAM-5′-ACTTCCTCAAGGAACAACATTGCCA-BBQ-3′.3 The RT-PCR reaction was performed on the Stratagene Mx3005P PCR instrument (Agilent Technologies, Santa Clara, CA). Positive and negative controls were included in each run. Five microliter of the extracted eluate was subjected to 25-μL PCR reaction with the following cycling conditions: 1 cycle of 55°C for 20 minutes, 1 cycle of 95°C for 15 minutes, followed by 45 cycles of 94°C for 15 seconds, and 58°C for 40 seconds.
Cobas SARS-CoV-2 Test
The cobas SARS-CoV-2 test (Roche Molecular Systems) is a qualitative test for fully automated cobas 6800/8800 platforms. The test is validated to detect SARS-CoV-2 RNA from nasal, nasopharyngeal, and oropharyngeal swab specimens. The test amplifies two SARS-CoV-2 targets: orf1ab (target 1), which is specific for SARS-CoV-2, and a conserved region of the E gene (target 2), which is pan-Sarbeco specific and detects also SARS-CoV and other Sarbecoviruses currently unknown to infect humans. In addition, the test includes an internal RNA control, which is added to the specimens before extraction. The test includes also positive and negative controls, which are processed the same way as the samples.
A total of 600 μL of the specimen lysate was subjected to the cobas 6800 system. The testing was performed according to the manufacturer's instructions, apart from the inactivation step.
Amplidiag COVID-19 Test
The Amplidiag COVID-19 test (Mobidiag) is a qualitative test for the detection of SARS-CoV-2 RNA in nasopharyngeal specimens. The test amplifies two SARS-CoV-2 targets: orf1ab and N gene, which are both specific for SARS-CoV-2. In addition, the test amplifies the human RNase P gene (RP), which serves as a sampling control. The test includes positive and negative controls.
The eNAT or the mNAT tubes were subjected to the Amplidiag Easy system, which extracts nucleic acids and does the PCR setup. The PCR plate was then transferred to a real-time PCR-machine (CFX96; Bio-Rad, Hercules, CA), which contains the Amplidiag Analyzer software version 1.8.0 (Mobidiag). The Amplidiag Analyzer software directly transfers data from the Amplidiag Easy system and automates the result interpretation. The testing was performed according to the manufacturer's instructions, apart from the specimen type (validated only for nasopharyngeal specimens) and the inactivation step with the eNAT-tubes.
A new version of the Amplidiag Easy Script Package software version 5.1.0 (Mobidiag) and Amplidiag COVID-19 Kit Configuration (v2-0-1) was introduced by the manufacturer during the evaluation. The new version does not collect data from the first 10 amplification cycles, and therefore, the obtained CT values are 10 cycles lower than with the old version. The new software version was introduced to get rid of the drift problems seen with the orf1ab target (false-positive interpretations of the old software). Because the new software did not allow analysis of the old data, 147 specimens from this evaluation were rerun from the same mNAT tubes within 5 days of the first run.
Workflow Evaluation and Relative Cost Efficiency of the Studied Tests
The turnaround time, hands-on time, and capacity of the tests were evaluated. An experienced lab technician performed all three tests and measured the time needed for each stage by a stopwatch. The specimen inactivation step was excluded from this analysis. The cost-efficiency ratio per an analyzed specimen was calculated using the test price and labor costs. Analytical instruments were not included in the calculation because some of the instruments are leased and others are owned by the laboratory.
Verification of the performance of the studied methods with specimens collected in Copan UTM versus 0.9% saline and using eNAT versus mNAT in the lysing/inactivation step in the Amplidiag COVID-19 test
When the epidemic started, the specimens were collected in Copan UTM tubes (3 mL), but later in tubes containing 1.5 mL of 0.9% saline, because of the global shortage in the supply of the Copan tubes. To verify that the used methods were performing at the same level, two independent dilution series were constructed: A patient specimen known to contain SARS-CoV-2 RNA originally collected in a Copan tube was diluted in pooled Copan-collected negative specimens. A patient specimen known to contain SARS-CoV-2 RNA originally collected in 0.9% saline was diluted in pooled 0.9% saline–collected negative specimens. The CT value for SARS-CoV-2 detection by RT-PCR was around 24 for both the Copan-collected and the saline-collected original positive specimens. A dilution series of 1:10 to 1:100,000 for both specimens (in Copan UTM or saline) in the respective specimen pools were constructed and analyzed with all three studied methods.
Due to the shortage of the eNAT tubes, a change to mNAT tubes was necessary. This change was evaluated with a small number of specimens. Eleven specimens defined positive by the cobas test were inactivated/lysed in parallel by pipetting the specimen into the mNAT and the eNAT tubes, and analyzed in the same PCR run.
Analytical Specificity
The analytical specificity of the tests was studied by analyzing patient specimens containing other respiratory viruses. Altogether, 54 specimens were included, but not all samples were run with all three methods. Some of the specimens contained several viruses (n = 11). In four specimens, the presence of respiratory virus nucleic acid was defined by xTAG RVP Fast (Luminex Diagnostics, Toronto, ON, Canada) assay, and in 50 specimens by the Allplex Respiratory Panel 1/2/3 (Seegene, Seoul, Republic of Korea). The specimens were chosen to contain moderate-to-high concentrations of viral nucleic acids (CT value <30 for specimens by the Seegene test or a high mean fluorescence intensity in the Luminex test). The specimens were collected during the 2018 to 2020 seasons, thus representing recent virus strains in Finland. The specimens are described in Supplemental Table S1.
Statistical Analysis
The positive percent agreement and negative percent agreement including two-sided 95% CIs were calculated with an online MEDCALC tool (MEDCALC, https://www.medcalc.org/calc/diagnostic_test.php, last accessed September 20, 2020). The overall agreement of the assays was evaluated by the kappa value, which was calculated (including a two-sided 95% CI) with an online QuickCalcs tool (GraphPad QuickCalcs, https://www.graphpad.com/quickcalcs/kappa1, last accessed September 20, 2020). Kappa values were interpreted as follows: no agreement (<0), slight agreement (0 to 0.20), fair agreement (0.21 to 0.40), moderate agreement (0.41 to 0.60), substantial agreement (0.61 to 0.80), and almost perfect agreement (0.81 to 1). All values were calculated relative to the reference standard, which was established as the result obtained by two of the three studied methods. The significance of the difference of the CT values obtained by Amplidiag test from the eNAT versus the mNAT tubes was estimated by two-tailed t-test. P < 0.05 was considered significant.
Results
Clinical Performance of the Evaluated Assays
Altogether, 183 specimens were analyzed by all three evaluated methods. One specimen gave an invalid result by the cobas SARS-CoV-2 test (0.5%), two specimens had an incorrect amount of specimen lysate in the LDT and were therefore considered invalid (1.1%), and seven specimens gave failed results by the Amplidiag COVID-19 test (3.8%). The failed/invalid specimens were excluded from the agreement analysis. In relation to the reference standard, the positive percent agreement was highest for the cobas test (100%), followed by the Amplidiag test (98.9%) and the LDT (98.9%). Negative percent agreement was lowest for the cobas test (89.4%), followed by the Amplidiag test (98.8%), and the best specificity value was obtained for the LDT (100%). The overall agreement as defined by kappa value was excellent for all studied tests (Table 1 ).
Table 1.
The PPA, NPA, and Concordance of the Evaluated Assays Relative to the Reference Values
| Test | Reference value∗ |
kappa | PPA (95% CI) | NPA (95% CI) | ||
|---|---|---|---|---|---|---|
| + | - | |||||
| LDT | + | 87 | 0 | 0.988 (0.966–1.0) | 98.9% (93.83–99.97) | 100% (95.75–100) |
| − | 1 | 85 | ||||
| Cobas | + | 88 | 9 | 0.896 (0.830–0.962) | 100% (95.89–100) | 89.4% (80.85–95.04) |
| − | 0 | 76 | ||||
| Amplidiag | + | 87 | 1 | 0.977 (0.945–1.0) | 98.9% (93.96–99.97) | 98.8% (93.62–99.97) |
| − | 1 | 84 | ||||
LDT, laboratory-developed test; NPA, negative percent agreement, PPA, positive percent agreement.
“Consensus result” was used as a reference value and was defined as the result obtained by at least two of the three studied methods.
Workflow Analysis and Relative Cost Efficiency of the Tests
The turnaround time for the cobas test was 3 hours 30 minutes for 80 samples; for the Amplidiag test, 3 hours 30 minutes for 48 samples; and for the LDT test, 4 hours 30 minutes for 93 samples. The hands-on time was shortest for the cobas test. To get a cost-efficiency ratio for the studied tests, the costs of the tests were divided by the cost of the cheapest test. The LDT was the most cost-efficient test for the laboratory with a cost-efficiency ratio of 1, followed by the cobas test with a cost-efficiency ratio of 1.25. The Amplidiag test was the least cost efficient with a cost-efficiency ratio of 1.38 (Table 2 ).
Table 2.
Workflow and Cost-Efficiency Evaluation
| Studied parameters | Cobas SARS-CoV-2 test | Amplidiag COVID-19 test | LDT |
|---|---|---|---|
| Throughput, one run | 93 (80∗) | 48 | 93 |
| Throughput in an 8-hour shift† | 372 | 96 | 186 |
| Hands-on time, one run | 50 min | 1 hour 25 min | 2 hours 30 min |
| Turnaround time | 3 hours 15 min | 3 hours 30 min | 4 hours 30 min |
| Cost-efficiency ratio‡ | 1.25 | 1.38 | 1.00 |
COVID-19, coronavirus disease 2019; LDT, laboratory-developed test; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Due to laboratory processing reasons, only 80 specimens were run during the time measurements, even though the maximum capacity is 93 specimens + 3 controls.
Throughput in an 8-hour shift was calculated with one operator and one set of instruments.
Cost-efficiency ratio includes test price and labor cost.
Performance Comparison of the Evaluated Methods for Specimens Collected in Copan UTM versus 0.9% Saline
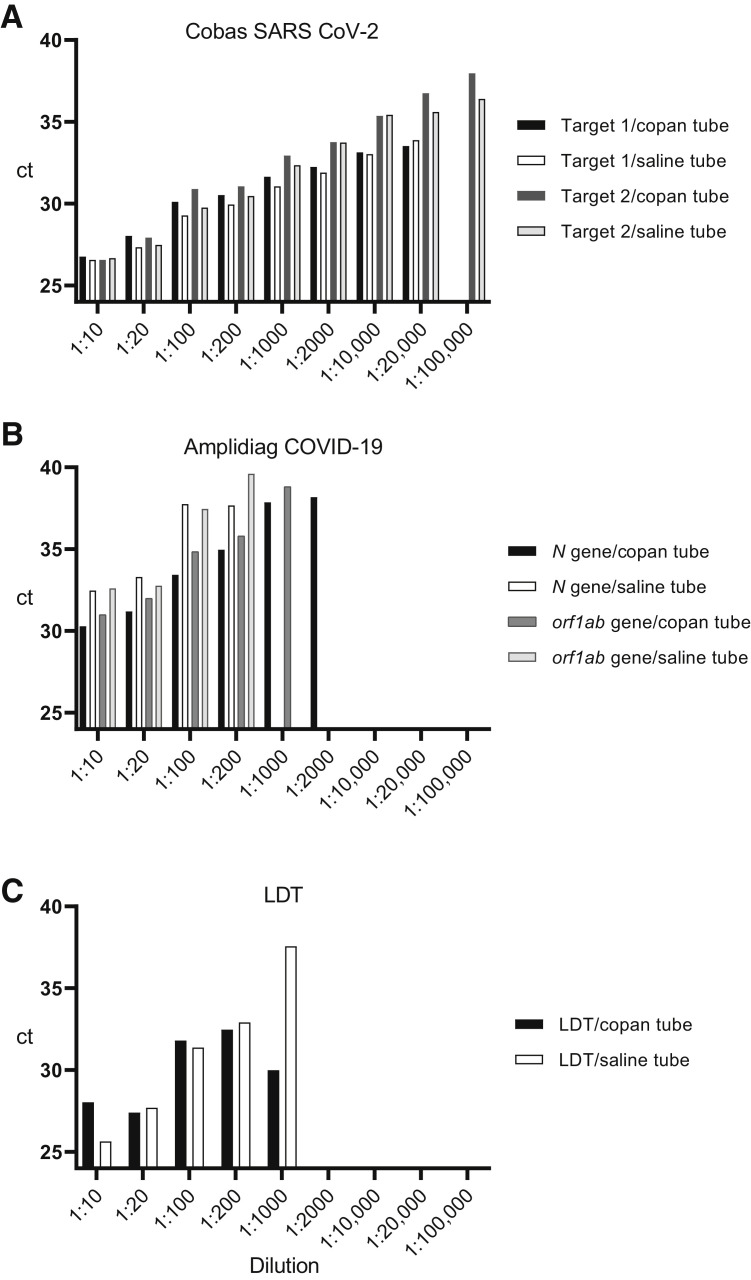
Two independent dilution series of two positive specimens were constructed in 0.9% saline and Copan UTM in a respiratory specimen matrix to verify that specimens collected in Copan and saline perform at the same level with the evaluated methods. The results indicate similar performance of all three methods independent of the collection medium used (Table 3 and Figure 2 ). However, the data suggest a higher overall sensitivity for the cobas test because the dilution 1:20,000 was still positive for both RT-PCR targets in the cobas test, whereas 1:200 to 1:1000 was the last dilution yielding positive test results in the LDT and in the Amplidiag test. The experiment was repeated with similar results. In the repeated experiment, the last dilution with a positive result with the LDT and the Amplidiag test was 1:100 (both for samples in saline and Copan), whereas a positive result was obtained for both targets in 1:10,000 dilution by the cobas test.
Table 3.
The Comparison of Results Obtained from Dilution Series of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in Copan UTM Tubes and Saline Tubes
| Dilution | Copan UTM |
Saline |
Copan UTM |
Saline |
Copan UTM |
Saline |
|---|---|---|---|---|---|---|
| Cobas result |
Amplidiag result |
LDT result |
||||
| (orf1ab CT/E gene CT) | (orf1ab CT/E gene CT) | (N gene CT/orf1ab CT) | (N gene CT/orf1ab CT) | (N gene CT) | (N gene CT) | |
| 1:10 | Pos (26.77/26.57) | Pos (26.57/26.67) | Pos (30.29/30.99) | Pos (32.47/32.6) | Pos (28.04) | Pos (25.65) |
| 1:20 | Pos (28.04/27.93) | Pos (27.34/27.48) | Pos (31.19/31.99) | Pos (33.3/32.77) | Pos (27.41) | Pos (27.70) |
| 1:100 | Pos (30.11/30.90) | Pos (29.28/29.77) | Pos (33.43/34.85) | Pos (37.76/37.46) | Pos (31.81) | Pos (31.38) |
| 1:200 | Pos (30.53/31.06) | Pos (29.95/30.47) | Pos (34.96/35.81) | Pos (37.67/39.61) | Pos (32.47) | Pos (32.91) |
| 1:1000 | Pos (31.64/32.94) | Pos (31.06/32.35) | Pos (37.86/38.82) | Neg (no CT/no CT) | Pos (30.00) | Pos (37.57) |
| 1:2000 | Pos (32.25/33.77) | Pos (31.90/33.74) | Pos (38.18/no CT) | Neg (no CT/no CT) | Neg (no CT) | Neg (no CT) |
| 1:10,000 | Pos (33.14/35.57) | Pos (33.04–35.43) | Neg (no CT/no CT) | Neg (no CT/no CT) | Neg (no CT) | Neg (no CT) |
| 1:20,000 | Pos (33.53/36.76) | Pos (33.88/35.61) | Neg (no CT/no CT) | Neg (no CT/no CT) | Neg (no CT) | Neg (no CT) |
| 1:100,000 | Pos (no CT/37.97) | Pos (no CT/36.41) | Neg (no CT/no CT) | Neg (no CT/no CT) | Neg (no CT) | Neg (no CT) |
LDT, laboratory-developed test.
Figure 2.
Comparison of the performance of the three studied methods using a dilution series in Copan UTM or 0.9% saline tubes. A known positive sample was diluted in either a pooled Copan-collected negative specimen or saline-collected negative specimen. A: Cobas SARS-CoV-2 test results. The test was positive for both targets at dilution 1:20,000 for both Copan and saline samples. Target 2 was detected with both tubes still in dilution 1:100,000. B: Amplidiag COVID-19 results. The Amplidiag test was positive for both targets in Copan and saline tubes at 1:200 dilutions, but at 1:1000 and 1:2000, only in Copan tubes (positive for both targets at 1:1000 and only N gene positive at 1:2000). No target was detected at dilutions 1:10,000 to 1:100,000. C: Laboratory-developed test (LDT) assay. No target was detected at dilutions 1:2000 to 1:100,000. The y axis is modified to show only CT values 24 to 40 to allow for a better visualization of the differences.
Verification of the eNAT versus the mNAT Tubes in the Lysing/Inactivation Step with the Amplidiag COVID-19 Test
Of the 11 specimens defined positive by the cobas test, eight were positive with the Amplidiag test independent of the tube used for the lysing/inactivation. Of the eight specimens positive by the Amplidiag test, the difference in N gene and orf1ab target CT values were calculated, and for the RNase P gene (sampling control), the difference was calculated from all 11 specimens. The mean difference in the obtained CT values was 0.77 for the N gene (P = 0.004), 0.87 (P = 0.189) for the orf1ab target, and 1.98 for the RNase P gene (P = 0.026).
Analysis of QCMD Proficiency Panel
QCMD 2020 Coronavirus Outbreak Preparedness EQA Pilot Study proficiency samples were used to assess the performance of the evaluated methods. The cobas test and the LDT were in 100% agreement with the QCMD expected results. The Amplidiag test failed to identify one SARS-CoV-2–positive result with a SARS-CoV-2 concentration of 3.3 log10 copies/mL (Table 4 ). However, the test identified correctly another sample containing SARS-CoV-2 RNA 2.3 log10 copies/mL. The Amplidiag test interpreted four samples as failed because the EQA samples lacked the human RNase P target needed for valid negative results with the test.
Table 4.
The Results of QCMD 2020 Coronavirus Outbreak Preparedness EQA Pilot Study Sample Panel with the Evaluated Methods
| Sample | Matrix | Sample content | Result with the cobas SARS-CoV-2 test | Result with the Amplidiag COVID-19 test | Result with the LDT | dPCR log10 copies/mL | Sample status |
|---|---|---|---|---|---|---|---|
| CVOP20S-01 | TM | SARS-CoV-2 | Positive | Positive | Positive | 4.30 | Positive |
| CVOP20S-02 | TM | Coronavirus-NL63 | Negative | Failed | Negative | 4.64 | Positive |
| CVOP20S-03 | TM | SARS-CoV-2 | Positive | Failed | Positive | 3.30 | Positive |
| CVOP20S-04 | TM | Coronavirus-OC43 | Negative | Failed | Negative | 4.03 | Positive |
| CVOP20S-05 | TM | Negative | Negative | Failed | Negative | Negative | |
| CVOP20S-06 | TM | SARS-CoV-2 | Positive | Positive | Positive | 4.30 | Positive |
| CVOP20S-07 | TM | SARS-CoV-2 | Positive | Positive | Positive | 5.30 | Positive |
| CVOP20S-08 | TM | SARS-CoV-2 | Positive | Positive | Positive | 2.30 | Borderline positive |
dPCR, digital PCR; TM, transport medium.
COVID-19, coronavirus disease 2019; LDT, laboratory-developed test; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Analytical Specificity
None of the studied SARS-CoV-2 tests gave positive results for the patient specimens containing other respiratory viruses (Supplemental Table S1).
Discrepant Analyses
Altogether, there were 12 discrepant results. In one specimen, which only gave a positive result with the Amplidiag test, the positive signal was obtained for the orf1ab target with a CT value of 40.8. However, in the amplification curve provided by the Amplidiag analyzer software no specific amplification could be seen. Instead of the expected logarithmic rise in the fluorescence signal, only a slow rise in the background signal was seen (data not shown). For one specimen, a positive result was obtained with the cobas and the Amplidiag tests, but a negative result with the LDT. In another specimen, positive results were obtained with the cobas test and the LDT, whereas the Amplidiag test gave a negative result. In that case, the specimen had gone through two freeze–thaw cycles before it was analyzed by the Amplidiag test, but only one or none when analyzed by the cobas or the LTD test, respectively. In nine specimens, a positive result was obtained only by the cobas test. However, a positive result was obtained for two of these specimens when rerun with the new Amplidiag COVID-19 software version. In all of the nine specimens, a positive signal was obtained for both amplification targets of the cobas test, but the CT values were high, >30 for eight of the nine specimens (Table 5 ).
Table 5.
Discrepant Analysis: Discrepancies in the Results between the Evaluated Methods and Discrepancies in the Results between Specimens Analyzed with the Old and the New Version of Amplidiag Software
| Number of specimens | Reference standard result | Cobas SARS-CoV-2 test result |
LDT result |
Amplidiag COVID-19 test result |
Comments |
|---|---|---|---|---|---|
| (orf1ab CT/E gene CT) | (N gene CT) | (N gene CT/orf1ab CT) | |||
| 1 | Neg | Neg (no CT) | Neg (no CT) | Pos (no CT/40.8) | Incorrect interpretation of the amplification curve by Amplidiag Analyzer |
| 1 | Pos | Pos (33.7/34.9) | Neg (no CT) | Pos (no CT/38.1) | |
| 1 | Pos | Pos (32.3/33.3) | Pos (30.46) | Neg (no CT) | The specimen had gone through two freezing–thawing cycles when analyzed by Amplidiag and only one with cobas test and none with LDT |
| 9 | Neg | Pos (28.6–34.4/29.1–37.9) | Neg (no CT) | Neg (no CT) | All specimens positive by cobas only had a late CT value. Apart from one specimen, all had a CT value > 30. Two of the specimens gave a positive result when specimens were rerun with the new Amplidiag software. |
| Number of specimens | Amplidiag result with old software |
Amplidiag result with new software |
Comments |
|---|---|---|---|
| (N gene CT/orf1ab CT) | (N gene CT/orf1ab CT) | ||
| 1 | Pos (no CT/40.8) | Neg (no CT/no CT) | Incorrect interpretation of the amplification curve by the old Amplidiag software version |
| 4 | Pos (no CT-38.1-no CT/38.5–40.8) | Neg (no CT/27.8–30.5) | New Amplidiag software version interprets the result as negative even though there is amplification from orf1ab gene |
| 2 | Neg (no CT/no CT) | Pos (27.5–28.0/no CT-27.5) | Specimens positive by cobas test |
| 2 | Pos (no CT/38.0–39.6) | Neg (no CT/no CT) |
Note that 10 CT cycles must be added to the CT value obtained with the new version of the software to compare the values to the CT values obtained by the old version.
LDT, laboratory-developed test.
Performance of the New Software Version of the Amplidiag COVID-19 Test
Of the 147 specimens run with both the new and the old software versions of the Amplidiag COVID-19 test, 11 specimens gave a failed result with version 1 or version 2 of the software or both (n = 2) and were excluded from the analysis. A total of 73 positive and 63 negative results were obtained by the old software, and 68 positive and 68 negative results by the new version. There were nine discrepant results. The specimen that gave a false-positive result by the earlier version of the Amplidiag software was negative in the new run with the new Amplidiag software. Four specimens that were positive with the old version gave negative results by the new version, even though there was some amplification of the orf1ab target. Additionally, two specimens were positive with the old version and negative with the new version without amplification of the orf1ab target. Two specimens that gave a negative result by the old software version gave a positive result when rerun with the new software version. In the comparison of the three methods, these two specimens were positive by the cobas assay only (Table 5).
Discussion
In general, all tested assays performed adequately. Both the time from the sample to the result and the hands-on time per sample were the shortest for the cobas test. Although the differences were not large, the LDT test was the most cost efficient for the laboratory, followed by the cobas test, whereas the Amplidiag test was the least cost efficient. The cobas and the Amplidiag assays allow for an automatic transfer of the results to the laboratory information system. On the other hand, both the LDT and the Amplidiag test allow for the evaluation of the actual amplification curves, which is very important for the quality assurance of the results, especially when the tests have been set up in a straining timetable.
Of the three RT-PCR methods evaluated in this study, the cobas SARS-CoV-2 test showed the best overall performance in the detection of SARS-CoV-2 RNA in the clinical respiratory specimens. In the absence of a gold standard, a reference value was created as a consensus of two methods. The agreement between the different methods tested was excellent. However, the positive percent agreement was highest for the cobas SARS-CoV-2 test (100%), whereas its negative percent agreement was the lowest (89.4%). All of the specimens positive only with the cobas SARS-CoV-2 test had high CT values (>30), with one exception, where CT values of 28.6 (orf1ab) and 29.1 (E gene) were obtained. The dilution series conducted for specimens collected in Copan UTM versus 0.9% saline, however, suggest a significantly higher sensitivity for the cobas assay in comparison with the other two assays, and the low negative percent agreement value may well be due to the same reason. Previous studies also suggest high sensitivity for the cobas SARS-CoV-2 test.7 , 8 The second commercial test evaluated, the Amplidiag COVID-19 test, obtained the same positive percent agreement (98.9%) as the LDT, but lower negative percent agreement (98.8% vs 100%, respectively). There was one specimen that was positive only by the Amplidiag COVID-19 test, but this seemed to be a false positive by the interpretation of the amplification curve of the positive target (orf1ab). This specimen was negative by the new Amplidiag software version, which did not give any false interpretations of negative results in this study. However, it seems that what is gained in specificity is lost in sensitivity: With the new software version, three specimens positive by the old version gave negative results, even though specific amplification from the orf1ab target could be seen. All things considered, the Amplidiag test seems to perform on the same level with the LDT, apart from the relatively high failure rate of the Amplidiag test (3.8%).
The comparison of the results from the eNAT tubes versus the mNAT tubes suggests that the Amplidiag test is for some reason less sensitive (later CT value) for the sampling control (RNase P gene) when using the mNAT tubes. The difference in the obtained CT values was greater for the RNase P gene (approximately 2 CT cycles) than for the specific virus targets (<1 CT cycle). This was also observed in the rising failure percent (from 3.8% to 8.4%, data not shown) when the change to the mNAT tubes was implemented. One limitation of this study is the inactivation step, which results in 0.46 of the specimen in the Amplidiag test in comparison to the cobas test and the LDT. This may affect the sensitivity of the Amplidiag test. However, the difference is not large, because this difference in the initial volume of the specimen results potentially in 1 CT cycle difference in the PCR result.
The QCMD 2020 Coronavirus Outbreak Preparedness EQA Pilot Study, samples were analyzed with all of the evaluated methods. The cobas test and the LDT identified all samples correctly. The Amplidiag test repeatedly missed one specimen with a digital PCR reference value of 3.3 log10 copies/mL, although it correctly identified another sample with a digital PCR reference value of 2.3 log10 copies/mL. In addition, the Amplidiag test yielded a failed test result for four samples because these samples lacked the human RNase P target needed for a valid negative result with the test. The lack of correct matrix in the EQA samples is a major drawback in general. The performance of the test identifying the target virus may be very different in the correct matrix in comparison to the virus alone. Human nucleic acids, if not yielding a false-positive result in a PCR, may weaken the amplification reaction (eg, if primers and/or the probe have binding sites in the human genome or transcriptome). For example, the LDT amplifies virus-positive specimens without human matrix more efficiently than in the specimens with human matrix included (data not shown).
It is common that RNA viruses accumulate mutations at high frequency because RNA polymerase lacks proofreading activity. Indeed, there is evidence that SARS-CoV-2 also is evolving over time, and mutations may occur in the target area of the molecular tests used. A previous report suggests that a mutation has already occurred in the target region of the cobas SARS-CoV-2 E gene test.9 In this study, all specimens gave a positive signal for both target genes by the cobas test. Fortunately, the test uses a dual-target approach, which means that the mutation proposed to be in the target area of the cobas E gene test does not compromise correct results. This highlights the importance of a dual-target approach.10, 11, 12
Many countries are now (in July 2020) moving from lockdown to test, trace, and isolate strategy in their COVID-19 mitigation, which requires a large capacity for sensitive and reliable SARS-CoV-2 laboratory testing. Clinical laboratories need to be prepared for uninterrupted high-throughput testing during the coming months. To achieve this, laboratories need to have optional testing methods in the event of instrumentation breakage, as well as prepare for the ongoing global shortage in testing supplies. Therefore, it is of critical importance for clinical laboratories to maintain several simultaneous platforms in their SARS-CoV-2 nucleic acid testing and continuously monitor the performance of the assays used.
Acknowledgment
We thank the laboratory personnel doing the SARS-CoV-2 diagnostics at HUSLAB for their input for this evaluation.
Footnotes
Supported by the Helsinki University Hospital, HUSLAB, Helsinki, Finland (M.L.).
H.J. and M.L. contributed equally to this work.
Disclosures: None declared.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2021.01.005.
Supplemental Data
References
- 1.Cheng M.P., Papenburg J., Desjardins M., Kanjilal S., Quach C., Libman M., Dittrich S., Yansouni C.P. Diagnostic testing for severe acute respiratory syndrome-related coronavirus-2: a narrative review. Ann Intern Med. 2020;172:726–734. doi: 10.7326/M20-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jääskeläinen A.J., Kekäläinen E., Kallio-Kokko H., Mannonen L., Kortela E., Vapalahti O., Kurkela S., Lappalainen M. Evaluation of commercial and automated SARS-CoV-2 IgG and IgA ELISAs using coronavirus disease (COVID-19) patient samples. Euro Surveill. 2020;25:2000603. doi: 10.2807/1560-7917.ES.2020.25.18.2000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodino K.G., Espy M.J., Buckwalter S.P., Walchak R.C., Germer J.J., Fernholz E., Boerger A., Schuetz A.N., Yao J.D., Binnicker M.J. Evaluation of saline, phosphate-buffered saline, and minimum essential medium as potential alternatives to viral transport media for SARS-CoV-2 testing. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00590-20. e00590-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nummi M., Mannonen L., Puolakkainen M. Development of a multiplex real-time PCR assay for detection of Mycoplasma pneumoniae, Chlamydia pneumoniae and mutations associated with macrolide resistance in Mycoplasma pneumoniae from respiratory clinical specimens. Springerplus. 2015;4:684. doi: 10.1186/s40064-015-1457-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Konrad R., Eberle U., Dangel A., Treis B., Berger A., Bengs K., Fingerle V., Liebl B., Ackermann N., Sing A. Rapid establishment of laboratory diagnostics for the novel coronavirus SARS-CoV-2 in Bavaria, Germany, February 2020. Euro Surveill. 2020;25:2000173. doi: 10.2807/1560-7917.ES.2020.25.9.2000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poljak M., Korva M., Knap Gašper N., Fujs Komloš K., Sagadin M., Uršič T., Avšič Županc T., Petrovec M. Clinical evaluation of the cobas SARS-CoV-2 test and a diagnostic platform switch during 48 hours in the midst of the COVID-19 pandemic. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00599-20. e00599-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pujadas E., Ibeh N., Hernandez M.M., Waluszko A., Sidorenko T., Flores V., Shiffrin B., Chiu N., Young-Francois A., Nowak M.D., Paniz-Mondolfi A.E., Sordillo E.M., Cordon-Cardo C., Houldsworth J., Gitman M.R. Comparison of SARS-CoV-2 detection from nasopharyngeal swab samples by the Roche cobas 6800 SARS-CoV-2 test and a laboratory-developed real-time RT-PCR test. J Med Virol. 2020;92:1695–1698. doi: 10.1002/jmv.25988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Artesi M., Bontems S., Göbbels P., Franckh M., Maes P., Boreux R., Meex C., Melin P., Hayette M.-P., Bours V., Durkin K. A recurrent mutation at position 26340 of SARS-CoV-2 is associated with failure of the E gene quantitative reverse transcription-PCR utilized in a commercial dual-target diagnostic assay. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.01598-20. e01598-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hokynar K., Rantakokko-Jalava K., Hakanen A., Havana M., Mannonen L., Jokela P., Kurkela S., Lappalainen M., Unemo M., Puolakkainen M. The Finnish new variant of Chlamydia trachomatis with a single nucleotide polymorphism in the 23S rRNA target escapes detection by the Aptima Combo 2 test. Microorganisms. 2019;7:227. doi: 10.3390/microorganisms7080227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mannonen L., Loginov R., Helanterä I., Dumoulin A., Vilchez R.A., Cobb B., Hirsch H.H., Lautenschlager I. Comparison of two quantitative real-time CMV-PCR tests calibrated against the 1st WHO international standard for viral load monitoring of renal transplant patients. J Med Virol. 2014;86:576–584. doi: 10.1002/jmv.23733. [DOI] [PubMed] [Google Scholar]
- 12.Beckmann C., Dumoilin A., Rinaldo C.H., Hirsch H.H. Comparison of a UL111a real-time PCR and pp65 antigenemia for the detection of cytomegalovirus. J Med Virol. 2011;83:2143–2150. doi: 10.1002/jmv.22232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.