Abstract
The COVID-19 pandemic has devastated the world, despite all efforts in infection control and treatment/vaccine development. Hospitals are currently overcrowded, with health statuses of patients often being hard to gauge. Therefore, methods for determining infection severity need to be developed so that high-risk patients can be prioritized, resources can be efficiently distributed, and fatalities can be prevented. Electrochemical prognostic biosensing of various biomarkers may hold promise in solving these problems as they are low-cost and provide timely results. Therefore, we have reviewed the literature and extracted the most promising biomarkers along with their most favourable electrochemical sensors. The biomarkers discussed in this paper are C-reactive protein (CRP), interleukins (ILs), tumour necrosis factor alpha (TNFα), interferons (IFNs), glutamate, breath pH, lymphocytes, platelets, neutrophils and D-dimer. Metabolic syndrome is also discussed as comorbidity for COVID-19 patients, as it increases infection severity and raises chances of becoming infected. Cannabinoids, especially cannabidiol (CBD), are discussed as a potential adjunct therapy for COVID-19 as their medicinal properties may be desirable in minimizing the neurodegenerative or severe inflammatory damage caused by severe COVID-19 infection. Currently, hospitals are struggling to provide adequate care; thus, point-of-care electrochemical sensor development needs to be prioritized to provide an approximate prognosis for hospital patients. During and following the immediate aftermath of the pandemic, electrochemical sensors can also be integrated into wearable and portable devices to help patients monitor recovery while returning to their daily lives. Beyond the COVID-19 pandemic, these sensors will also prove useful for monitoring inflammation-based diseases such as cancer and cardiovascular disease.
Keywords: COVID-19, Pathophysiology, Electrochemical sensors, Inflammatory biomarkers, Biofluids, Metabolic syndrome, Cannabinoids, Wearable and portable devices, Prognostic tools, Health monitoring
Graphical abstract
Abbreviations
- ACE2
Angiotensin-converting enzyme 2
- ALI
Acute lung injury
- ARDS
Acute respiratory distress syndrome
- CBD
Cannabidiol
- COPD
Chronic obstructive pulmonary disease
- CNS
Central nervous system
- CRP
C-reactive protein
- CRS
Cytokine release syndrome
- CV
Cyclic voltammetry
- CVD
Cardiovascular disease
- DPV
Differential pulse voltammetry
- EBC
Exhaled breath condensate
- ELISA
Enzyme-linked immunosorbent assay
- IFN
Interferon
- IL
Interleukin
- NLR
Neutrophil-lymphocyte ratio
- PAMP
Pathogen associated molecular pattern
- PLR
Platelet-lymphocyte ratio
- POC
Point of care
- PRR
Pattern Recognition Receptors
- S1
Spike protein
- subunit 1 S2
Spike protein, subunit 2
- THC
Δ9-tetrahydrocannabinol
- TNFa
Tumour necrosis factor alpha
- VOC
Volatile organic compounds
- WBC
White blood cells
- WHO
World Health Organization
1. Introduction
Viruses are nonliving entities that can replicate only in the presence of a host organism. This ability stems from their structure, which comprises a protein capsid containing DNA or RNA fragments. In December 2019, a contagious viral infection broke out in Wuhan, Hubei province, China [1]. The pathogen responsible has been identified as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is a part of the Coronaviridae family of viruses. Rapid global spread of the RNA-based SARS-CoV-2 has prompted extreme measures from the international community, including physical distancing, lockdowns, and travel restrictions causing severe social and economic disruptions around the globe. The World Health Organization (WHO) declared a global health pandemic on March 11th, 2020 [2]. The urgency of the COVID-19 pandemic necessitates the identification of informative biomarkers which can be used to forecast disease progression and alert clinicians to changes in patients’ health.
Mitigating the severe impact of COVID-19 on patients' health warrants an understanding of the disease's pathophysiology and clinical presentation. SARS-CoV-2 receptor binding domain (RBD) is well suited for binding angiotensin-converting enzyme 2 (ACE2) receptors which are highly expressed in lung tissues, nasal mucosa, kidneys, testes, the gastro-intestinal tract, oral, lung and intestinal tissues. The entry of SARS-CoV-2 into host cells increases inflammation through various mechanisms; serious cases are characterized by an uncontrollable release of proinflammatory cytokines. The “cytokine storm”, formally known as cytokine release syndrome (CRS), can cause serious damage to affected tissues, especially the lungs.
COVID-19 presents varying degrees of symptoms ranging from asymptomatic to fatal. A meta-analysis found 15% of COVID-19 infected individuals (n = 51,155) were asymptomatic [3]. The same meta-analysis found that half of asymptomatic individuals at the time of diagnosis developed symptoms at a later time [3]. Symptoms included fever, cough, myalgia, fatigue [4] and anosmia [5] while severe COVID-19 cases may progress to dyspnea (shortness of breath), bilateral interstitial pneumonia, acute respiratory distress syndrome (ARDS), sepsis and multiorgan dysfunction in addition to acute cardiac, kidney, and liver dysfunction [6,7]. Neurologically, COVID-19 impacts one's sense of smell by attacking olfactory cells [8,9]. Its ancestor, SARS-CoV has been known to attack the central nervous system (CNS), causing spine demyelinating and neurodegenerative diseases [10,11]. In addition, there are comorbidities that influence the onset and severity of these symptoms. For example, illnesses that are associated with metabolic syndrome (diabetes, obesity, etc.) have a 19.1% prevalence in Canada [12], and act as comorbidities by putting excess strain on the organ systems affected by COVID-19 [13]. It has been reported that COVID-19 patients with cardiovascular disease (CVD) and hypertension have a 6% and 10.5% mortality rate respectively, while patients without these comorbidities have a 2.3% mortality rate [13].
SARS-CoV-2 infection can be confirmed via reverse transcription-polymerase chain reaction (RT-PCR) [14]. There is an array of biomarkers related to COVID-19 which can be found in biofluids [15,16]. Therefore, there is an opportunity to pursue personalized diagnostic devices that can monitor patient health by rapidly and sensitively detecting biomarkers. This includes COVID-specific biomarkers (viral proteins and RNA) and non-specific biomarkers (CRP, ILs, D-dimer, TNFα, glutamate, pH, hematological biomarkers).
Although some biomarkers are not specific indicators of a COVID-19 diagnosis, they may be used along with RT-PCR [14] to indicate the severity of the infection. The current gold standard for protein biomarker measurement is the enzyme-linked immunosorbent assay (ELISA), which provides excellent sensitivity (<1 pg/mL) [17]. Western blot is also commonly used to provide great specificity and selectivity [17,18]. Alternatively, electrochemical sensing techniques involve detecting electroactive analytes in biofluids such as saliva, sweat, and blood [19]. They are easy to use and can reduce testing duration. Electrochemical sensors often involve the use of a simple technique called cyclic voltammetry (CV). The potential of the working electrode changes relative to a reference electrode until enough potential exists to drive a redox reaction of the analyte at the working electrode [20]. Faradaic current is generated and measured as electrons flow between the working and counter electrodes. A brief schematic illustrating this process is provided in Fig. 1 . Ideally, an electrochemical biosensor is a self-contained, integrated device which can analyze biofluids at high sensitivity and selectivity without the need for sample preparation. These attributes make electrochemical techniques ideal for continuous health monitoring in a pandemic where proper management of personnel and healthcare resources is critical [21]. There is an urgent need for a real-time, non-invasive sensor that can be used in point of care (POC) settings along with wearable devices that monitor biomarker levels outside of the hospital.
Fig. 1.
A) A schematic of a three-electrode system used in modern electrochemical potentiometry B) a plot of voltage at the working electrode (relative to the reference electrode) change over time during cyclic voltammetry C) A prototypical voltammogram, the current through the sample is measured at the voltage changes over time (as seen in B).
While biomarker sensing protocols are well established in lab-based techniques, miniaturized and portable sensors have been the beneficiaries of more recent exploration. This new attention is due to advancements in miniaturization, optimization and performance allowed by nanomaterials [22]. Decentralized testing methods using nanomaterials such as electrochemical sensors are cheaper, easier to mass produce and more accessible than centralized lab-based techniques (LBTs) like ELISA, Western blot and PCR. Centralized LBTs may be accessible to populations in big cities with well-established infrastructure. Populations in low to middle income countries may lack this infrastructure and any reliable alternatives — they would benefit from decentralized techniques like individualized electrochemical sensing [23]. Furthermore, electrochemical sensors allow for the creation of personalized multiplex or non-multiplex sensors that can be used by individuals to monitor their current biomarker levels at home. If abnormal levels (i.e. during a cytokine storm) are detected, individuals can be alerted to seek medical help. Considering the demand on hospitals during the COVID-19 pandemic, developing such sensors for personal use would help identify patients in need of further medical attention. In addition, electrochemical sensors are typically more portable, and faster than many LBTs (immunoassays, mass spectrometry) [24]. However, confounding variables when using electrochemical sensors, such as pH, can alter the performance of the sensor and must be considered when these sensors are designed [25]. Nevertheless, there are still further obstacles to face before these sensors can be used commercially.
The electrochemical sensors mentioned in this review have a wide variety of applications outside of COVID-19. Thus, the development of these sensors is an important endeavour to continue beyond the COVID-19 pandemic. This review pushes the field forward by highlighting current trends and promising sensors. Further applications include cancer prognosis and possibly even treatment [26]. For example, high IL-6 and IL-10 serum concentrations can prognose several cancers, independent of tumour heterogeneity [26]. Other conditions where similar sensors can be applied to prognosis are pancreatitis [27], malaria [28], acute ischemic stroke [29] and viral infections (such as respiratory syncytial virus [30] and HIV [31]). The use of multiplex sensors can differentiate between biomarker concentration profiles that are typical to specific diseases. Furthermore, multiplex sensors may be able to indicate what is physiologically occurring as a disease progresses [32]. However, one challenge to this may be the heterogeneity of baseline and diseased concentrations from patient to patient due to confounding variables [33,34]. This heterogeneity can be manifested in genetics, where mutations to various caspase proteases alter the cytokine response [34]. But this may be resolved by calibrating the reference values of a sensor with personalized normal values, detected prior to the incidence of a condition.
The purpose of this manuscript is to explore electrochemical detection of biomolecules affected by COVID-19 infection to monitor disease severity, especially when present with other diseases. While there are some excellent review papers reporting biomarkers’ value for COVID-19 prognosis [15,16], so far none of them have explored the advantages of electrochemical sensors for the detection of these biomolecules. Table 1 provides a summary of the biomarkers discussed in this paper. In this review paper, we discuss the progress, challenges and opportunities involved in electrochemical sensing of these molecules for researchers interested in developing prognostic tools to help address these urgent healthcare needs.
Table 1.
A summary of some biomarkers discussed in this paper. These biomarkers are prone to being influenced by a variety of variables. There is also a shortage of studies exploring Serum COVID-19 Concentrations.
| Name | Role | Approx. Healthy Concentration (μg/mL) | Serum Concentration in COVID-19 (μg/mL) |
|---|---|---|---|
| CRP | A protein which mediates inflammation in the body [35]. IL-6 is the main inducer for CRP production, with IL-1 enhancing it [36]. |
Serum median: | Severe: |
| 0.8 [35] | ≥41.8 [37] | ||
| IL-1β | A proinflammatory cytokine that helps with the production of synoviocytes [38] and proteases for inflammation [39]. | Serum: | Mild range: 1.729 × 10−5 – 3.422 × 10−5 Severe range: 2.589 × 10−5 – 4.718 × 10−5 [40] |
| 6.46 × 10−6 – 2.696 × 10−5 [40] | |||
| IL-4 | Immunoregulatory cytokines that help regulate antibody production, hematopoiesis, aids in the formation of the effector T-cell response [41]. | Serum: 3.486 × 10−5 ± 2.442 × 10−5 [40] |
Mild range: 3.251 × 10−5 – 5.550 × 10−5 Severe range: 3.646 × 10−5 – 1.0379 × 10−4 [40] |
| IL-6 | Most released cytokine by activated macrophages [42]. Induces an acute phase response | Serum: 9.0 × 10−7 – 1.0 × 10−5 [43,44] | Severe: ≥5.5 × 10−5 High likelihood of mortality: ≥8.0 × 10−5 [45] |
| lL-8 | A chemokine (CXL8) that is linked to the progression of ARDS [46]. Has a key role in neutrophil recruitment [47] |
Serum: 1.29 × 10−5 ± 1.393 × 10−5 [48] |
Severe: ≥5.0 × 10−5 [49] |
| IL-10 | Inhibits the activity of Th1, natural killer cells, and macrophages. All these are used for pathogen expulsion [50]. | Serum: 2.21 × 10−6 – 6.02 × 10−6 [40] |
Mild range: 3.32 × 10−6 – 4.16 × 10−6 Severe range: 6.30 × 10−6 – 8.05 × 10−6 [40] |
| D-dimer | A protein fragment produced by fibrinolysis (clot breakdown) [51]. | Serum: ≤0.5 [52] |
Survivors median: 1.02 Non-survivors median: 6.21 [52] |
| TNFα | A potent pleiotropic cytokine that participates in cell proliferation, differentiation and apoptosis [53,54]. | Serum: 1.12 × 10−5 ± 7.31 × 10−6 [48] | Not yet established |
| l-glutamate | An abundant excitatory neurotransmitter implicated in several neurodegenerative diseases [55]. | Plasma: 0.74–14.7 Saliva: 3.4 × 10-2 ± 2.6 × 10-2 Urine: 4.855 × 102–2.7072 × 103 creatinine [55] |
Not yet established |
2. Pathophysiology
In this section, we describe the physiological processes that lead to COVID-19 infection. One of the avenues SARS-CoV-2 uses to enter the body is ACE2, which can be found in the heart, lungs, kidneys, and endothelium [56]. ACE2 allows for the degradation of angiotensin II, affecting the constriction and dilation of vessels in the circulatory system in order to regulate blood pressure [57]. The virus' spike (S) protein, consisting of S1 and S2 subunits, facilitates viral entry [58]. S1 is responsible for viral attachment to the target cell's membrane via ACE2, while the S2 domain mediates the viral envelope's fusion with the host cell [58]. The viral genome then enters the host cell's cytosol, and the infection ultimately spreads to other cells.
SARS-CoV-2 infection first triggers the body's innate immunity, which serves to quickly and non-specifically prevent the spread of the virus within the body [59]. Innate immune cells (like macrophages, neutrophils, and mast cells) express Pattern Recognition Receptors (PRRs) that trigger host immune responses upon the binding of pathogen associated molecular patterns (PAMPs), for example surface glycoproteins or viral RNA [59,60]. These host responses include the release of pro-inflammatory cytokines such as IL-6 and TNF⍺ [61] which recruit leukocytes to help the body fight infection [62]. However, in severe cases, systemic inflammation can give rise to respiratory, cardiovascular, or neurological complications [63]. For instance, the IL-1β and IL-6 released during a COVID-19 infection can lead to lung inflammation, fever, and fibrosis [64].
The release of pro-inflammatory cytokines activates the adaptive immune system, which includes T- and B-lymphocytes. T-lymphocytes like Cluster of Differentiation 4+ and 8+ (CD4+ and CD8+) are memory cells that ‘remember’ antigens, allowing them to destroy infected host cells and subsequently recruit other immune cells to do the same [65]. B-lymphocytes facilitate the production of antibodies which attach to the antigens and mark them for destruction [65]. COVID-19 patients may present with lymphocytopenia (lymphocyte deficiency), which slows their immune response to the infection [66]. Lymphocytopenia has been reported in only 25% of mild COVID-19 cases [42] but in 80% of critical cases [57]. When T-cells are overstimulated with antigens they gradually enter a state of exhaustion in which they exhibit weaker effector function [66]. Since the pathogen is still present, pro-inflammatory cytokines are released in a positive-feedback loop. The resulting “cytokine storm” can lead to ARDS, sepsis, or multiple organ failure [63]. Since pro-inflammatory cytokines play a key role in the severity and progression of COVID-19, it is critical to investigate their validity as biomarkers of the disease.
3. Biological markers of inflammation
Inflammation is a complex biological process in which a symphony of markers and cells react to harmful stimuli. These markers include cytokines, immune cells, glutamate, pH and other proteins. Cytokines are signaling proteins that allow damaged tissues and immune cells to communicate [67]. Pro-inflammatory cytokines are a class of cytokines primarily released by macrophages and helper T-cells to upregulate inflammatory reactions [68]. IL-1, IL-6, TNFα, and IFN-γ belong to the pro-inflammatory class of cytokines [67,69]. Anti-inflammatory cytokines have the ability to inhibit the synthesis of pro-inflammatory cytokines. Two major cytokines in this class are IL-4 and IL-10 [68]. The aforementioned cytokines can be found in measurable quantities in various biofluids, indicating the health status of a patient. Such possible biofluids include whole blood, serum, urine, saliva and sweat. Non-cytokine protein biomarkers like CRP and D-dimer can also be found in some of these biofluids, and they also indicate infection in the context of COVID-19 [15,51]. Non-protein biomarkers like pH and glutamate have also been effective in assessing pulmonary inflammation (using breath) [[70], [71], [72]] and neurological status, respectively [11,73]. Hematological biomarkers, which can be cells or cell fragments, can indicate inflammation through the fluctuation of their concentrations in whole blood. COVID-19 specific biomarkers such as N-protein, S-protein, and viral RNA can specifically diagnose COVID-19 instead of simply indicating inflammation and these biomarkers can help indicate disease severity.
Out of the aforementioned biofluids, testing whole blood and its derivatives (serum, plasma, etc.) is considered invasive because it requires medical professionals to cut the skin or insert instruments into bodily openings. Whole blood is the gold standard of biofluids. It is a simple, well-defined broad appraisal of the human immune response. However, drawing blood would not provide a real-time assessment of health status and repeated blood harvesting leaves patients susceptible to infection by creating avenues for pathogen entry. This can be especially dangerous in an overcrowded hospital setting where bacteria and viruses tend to be strong and drug-resistant, as is the case during the COVID-19 pandemic. This could be overcome with implanted sensors, however, they are invasive and lack commercial viability [74]. Whole blood contains many potential interfering compounds, and tends to foul electrode surfaces, negatively impacting their long term performance [75]. This provides an opportunity for development and validation of anti-biofouling modifications [75]. Serum is derived from the centrifugation of invasively obtained whole blood with anticoagulants removed. Serum analysis does not suffer from the interference and fouling issues of whole blood analysis, but it is time-consuming to harvest and requires external lab equipment. Nevertheless, non-invasive biofluid alternatives like saliva, sweat, breath, and urine are also viable. In particular, saliva contains many of the constituents found in serum due to transcellular and paracellular routes between blood and saliva [76]. However, the concentration of various biomarkers is much lower in saliva compared to blood, providing a technological challenge for researchers [76]. Despite this, more sensitive materials are being used in biosensors, which has rendered the diagnostic/prognostic utility difference between saliva and blood less apparent [75,76]. Saliva can contaminate breath, which is another non-invasive biofluid that contains a variety of biomarkers that can be sensed. Breath provides insight into the health status of the lungs and is easy to harvest. Sweat is also a non-invasive matrix. Studies have found similar IL-1α, IL-1β, IL-6, IL-8, and TNFα concentrations between plasma and sweat [43]. One challenge with sweat analysis is the need for an adequate amount of fluid to be harvested; perspiration may vary among individuals and thus presents an obstacle to researchers [77]. Urine testing is a reliable and ubiquitous option, however its collection can also cause discomfort, risk disease transmission, and requires the handling of noxious fluids. Lab testing on urine is subject to time and temperature factors affecting particle stability [78]. Testing within 2 h of collection is considered optimal, however, POC testing is not subject to these limitations [79]. Despite the difficulties in collecting and measuring the aforementioned biofluids, they can quickly provide valuable information about patients’ health, especially when used in the context of electrochemical sensing. This section will describe some promising electrochemical sensors and the analytes they sense.
3.1. The biofouling challenge
Section Summary: Electrochemical sensors intended for repeated use need to be designed with biofouling taken into consideration because substances contacting the electrodes may diminish the sensor's performance over time. The mechanisms of biofouling and anti-biofouling methods are discussed below.
Biofouling has been identified as an important factor for prognostic sensors. Researchers designing sensors in the wearable or implanted sensor space will be prioritizing reusability in their design. The greatest obstacle these designers will face is biofouling at the surface of the electrode following repeated sensor use. This phenomenon blocks the transfer of electrons from the analyte to the electrode, diminishing the sensor's performance. Biofouling can occur by two mechanisms; it can affect the recognition components (including antibodies and aptamers), or the transducing components (including the electrode surface). The recognition element is affected by the non-specific adsorption of proteins present in the biofluid onto analyte receptors [80]. The transducing element is affected when an analyte in the sample participates in a redox reaction and forms electrochemically inactive products on the electrode surface [81].
Many anti-fouling strategies have been envisioned to treat the fouling of these components actively and passively. Passive anti-fouling takes advantage of material properties to promote the release of adsorbed proteins or render surfaces thermodynamically unfavourable for fouling material attachment [82]. Polyethylene glycol (PEG) and hydrogels form polymeric networks which retain water, thus forming a hydrophilic barrier on the electrode surface to which proteins have difficulty binding due to entropic unfavourability [82]. Active anti-fouling strategies utilize stimuli-responsive materials to reversibly change a material's properties. Mechanical stress, light, pH, temperature, acoustic waves or electrical/magnetic fields have been used in active anti-fouling sensors [83,84]. Temperature responsive polymers have a volume phase transition temperature (PVTT) property which marks the temperature at which the polymer drastically and discretely changes its physical arrangement. This property was exploited in the sensing of glucose with a nanocomposite hydrogel, the swelling of N-isopropylacrylamide (NIPAAm) and polysiloxane colloidal nanoparticles released adhered cells. The electrode was optimized for use in implanted glucose sensors; parameters such as temperature fluctuations, mechanical strength and swelling kinetics were all considered. This presented an active anti-fouling design that could take advantage of the body's own fluctuations in temperature to “clean” the electrode surface [84]. Ultimately, a similar demonstration of anti-fouling capabilities will be essential in real-sample analysis of complex matrices such as blood, urine, saliva and sweat.
3.2. C-reactive protein (CRP)
Section Summary:Electrochemical sensors can assess elevated CRP levels during COVID-19 infection, rising from healthy levels (around 0.8 μg/mL) to more than 41.8 μg/mL (severe cases), by using biofluids such as whole blood and serum. Many of the current viable label free electrochemical sensors lack the proper range of detection. However, two viable sensors were focused on: a EGOFET sensor with a linear range of 0.1 to 1 × 10 5 ng/mL and a wave-shaped microelectrode array with a range of detection of 0.01 to 10,000 ng/mL.
C-reactive protein (CRP) is a ring-shaped plasma protein produced in the liver [85]. Generally, CRP is a nonspecific indicator of tissue damage, infection, and other forms of acute inflammation [86]. Non-infected individuals have a median baseline concentration of 0.8 μg/L; up to 10 μg/L at the 99th percentile [35]. During inflammatory events, CRP can increase by more than 1000-fold, reaching between 100 μg/L and to 500 mg/L [86]. This large range makes changes in health status obvious as it provides an indication of inflammation along with an assessment of the intensity of the inflammation or infection [86]. During a COVID-19 infection, CRP levels can range between 1 μg/L (very mild cases) to more than 100 μg/L (severe cases) [15]. Future electrochemical sensors must operate in a wide linear range, ideally able to sense concentrations between 0.8 μg/L and 500 mg/L. There are a lack of studies indicating CRP presence in sweat, but there is evidence of CRP being present in blood [37]. Given the non-specific nature of CRP, biosensors targeting CRP will find many other applications such assessing autoimmune disease, the likelihood of developing CVD, amongst other conditions involving inflammation [86]. Evidently CRP is a viable biomarker for COVID-19 prognostics, meriting the exploration of various electrochemical sensors that measure this analyte.
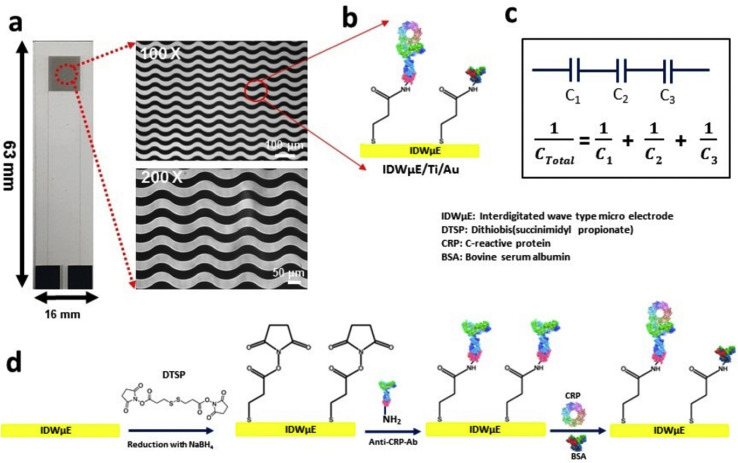
Some existing CRP electrochemical sensors are listed in Table 2 . Unfortunately, most approaches to sensing have failed to detect CRP outside of a narrow concentration range, while the clinical nature of the analyte necessitates a wide dynamic range (pM to μM) [87]. Additionally, most protocols are multistep and require extra processing, making them difficult to adapt to continuous or wearable devices [87]. One sensor that addressed both of these issues was based on an electrolyte-gated organic field-effect transistor (EGOFET) sensor that was fabricated through the absorption of anti-CRP monoclonal antibodies onto a poly-3-hexyl thiopene (P3HT) semi conductive organic surface [87]. Afterwards the modified semiconductor surface was treated with hydrophilic (N-[tris(hydroxy-methyl) methy] acrylamide-lipoic acid conjugate (pTHMMAA)) polymer to properly orient the anti-CRP antibodies and block non-specific binding. The linear range of this sensor was 0.1–1 × 105 ng/mL, and the limit of detection was 0.1 ng/mL, which is ideal for COVID-19. Beyond COVID-19, this range enables assessment of inflammation, cardiovascular health, and cancer [88]. This label-free sensor was compatible with disposable and flexible substrates while being adaptable to miniaturization and multianalyte arrays. An electrochemical capacitance immunosensor that used an interdigitated wave-shaped microelectrode array showed high performance (shown in Fig. 2 ) [89]. A self-assembled Dithiobis (succinimidyl propionate; DTSP) monolayer functionalized by NaBH4 was used to anchor anti-CRP antibodies onto the electrode surface. The sensor had a linear range of 0.01 ng/mL to 10,000 ng/mL (equivalent to 10 ng/L to 10 mg/L); this inadequately covered expected CRP concentrations in severe COVID-19 cases, but future developments may increase this range. The sensor was found to produce ideal electrochemical responses at a pH value of 7.4, a 50 μg/mL anti-CRP-antibody concentration and a 10-min incubation time. The sensor was able to detect CRP without significant effects from high concentrations of interfering agents. It had a miniaturized design, good storage stability (two weeks at 4°C) and was easily adaptable to a microcontroller-based potentiostat. Despite these advancements, there is a shortage of CRP electrochemical sensors with an optimal linear range for COVID-19 applications. Future research should focus on developing sensors with wide linear ranges and improved usability for POC applications.
Table 2.
A summary of electrochemical sensors targeting C-Reactive Protein analyte.
| Analyte | Electrode/Modification | Receptor | Matrix/Solvent | LOD (ng/mL) | Linear Range (ng/mL) | Ref. |
|---|---|---|---|---|---|---|
| CRP | Standard polycrystalline gold electrode modified with SAM immunosensor | Goat anti-human CRP polyclonal antibody | Pharmaceutical preparation, human serum | 0.135 | 0.384–38.4 | [90] |
| CRP | Wave shaped microelectrode array with a monolayer functionalized by NaBH4 to anchor antibodies | Human CRP antibody | PBS serum | 0.025–0.23 | 0.01 – 10 × 103 | [89] |
| CRP | Screen printed graphene electrode functionalized by l-cystine and gold to anchor Anti-1°Ab | Anti-1°Ab unlabeled capture antibody & Anthraquinone labelled anti-human CRP secondary antibody |
Human serum | 1.5 | 10 – 150 × 103 | [91] |
| CRP | Reduced graphene oxide (rGO)-modified screen-printed carbon electrode (SPCE) functionalized via 1-pyrenebutyric acid N hydroxy succinimide ester (PyNHS) | Anti-CRP antibody | Pharmaceutical preparation | – | 10 – 1 × 104 | [92] |
| CRP | Branched polyethylenimine functionalized with ferroced residues recognition layer anchored to electrode surface for the covalent anchoring of antibodies | Monoclonal IgG anti CRP antibodies | Rat blood | 0.5–2.5 | 1 – 5 × 104 | [93] |
| CRP | Gold printed screened electrode has affinity for sulfur and nitrogen, which allows for antibodies to covalently attach | Monoclonal anti-CRP antibody | Human serum | 780 | 6.25 × 103–5 × 104 | [94] |
| CRP | Disposable single walled carbon nanotubes field effect transistor immunosensor, able to absorb amino groups in anti-CRP antibodies | Goat anti-CRP | Pharmaceutical preparation | 0.1 | 0.1 – 1 × 105 | [95] |
Fig. 2.
A) Electrode array of the wave-like interdigitated electrode sensor, Low magnification at 100× and the high magnification shows 30 μm finger and spacing in detail B) DTSP is functionalized upon the e IDWμE modified wave shaped electrode, which allows for the immobilization of the anti-CRP antibody C) EGOFET sensor submerged in PBS where 1) anti CRP immobilized by P3HT surface 2) pTHMMAA absorbed by surface where anti-CRP is not present to prevent nonspecific binding 3) CRP binds to anti-CRP, producing an electrical current. D) The functionalization of DTSP, anti-CRP immobilization, and attachment between CRP and anti-CRP, respectively. A) B) C) and D) Reprinted and adapted with permission from Ref. [89]. Open Copyright 2019 Multidisciplinary Digital Publishing Institute (MDPI). C) Reprinted and adapted with permission from Ref. [70]. Copyright 2016 Springer.
3.3. Interleukins
Section Summary:The levels of many interleukins, such as IL-1, 4, 8, and 10, are affected by COVID19 inflammation. The most studied and promising interleukin is IL-6, whose levels in serum were found to be ≥ 5.5 × 10 −5 μg/mL in patients with risk of developing severe COVID-19 and ≥ 8.0 × 10 −5 μg/mL in patients with high likelihood of mortality. Detection of IL-6 within this range is achievable with existing electrochemical sensors, and detection in other biofluids is also possible. The sensor highlighted in this section utilizes rat breath to assess IL-6 levels through a silicon nanowire field effect transistor (SiNW-FET) and a Detection of Living Animal's Exhaled Breath Biomarker (dLABer) system.
Interleukins (IL) are a broad range of cytokines that have an essential role in intercellular communication, specifically among cells of the immune system and between immune cells and other tissue [96]. In inflammatory states, pro-inflammatory and anti-inflammatory interleukin levels fluctuate [96]. Pro-inflammatory interleukins such as IL-1, IL-6, and IL-8 were elevated in the sera of COVID-19 patients [[97], [98], [99], [100], [101], [102], [103], [104], [105]], while levels of anti-inflammatory interleukins like IL-10 and IL-4 were found to fluctuate [[103], [104], [105], [106]]. Out of these, IL-6 was most examined by the literature as a target for COVID-19 prognostics, and several IL-6 receptor inhibition therapies are currently being investigated [107,108]. In addition, interleukins can be detected in a variety of biofluids such as blood, saliva [109], sweat [110], and even breath [111]. IL-1α IL-1β, and IL-6 concentrations in sweat were not different from plasma, with healthy levels ranging between 1 and 10 pg/mL [43,44]. Meta-analysis of COVID-19 patients found IL-6 serum concentrations ≥5.5 × 10−5 μg/mL indicated a high risk for developing a severe COVID-19 infection, and concentrations ≥8.0 × 10−5 μg/mL indicated a high risk of mortality, although only one study examined this endpoint [45]. A meta-analysis found IL-6 serum concentrations were 2.9 times greater in severe cases compared to non-severe cases [112]. COVID-19 treatments like corticosteroids and drugs targeting pro-inflammatory signal pathways can be guided by interleukin biosensors to gauge efficacy and adjust treatment dosages [113]. Examples of these drugs include Tocilizumab (targeting IL-6) and Anakinra (targeting IL-1) [113]. Beyond their use in COVID-19 therapies and prognostics, interleukin biosensors will find applications in assessing conditions such as cancer [114], aneurysms [115], and chronic obstructive pulmonary disease (COPD) [116]. Electrochemical sensing developments for the measurement of interleukin levels are abundant due to their diverse applications, but performance differs between sensors.
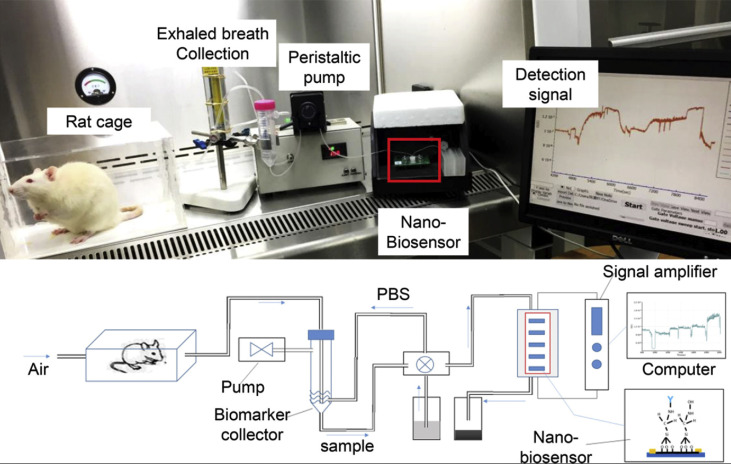
Table 3 presents some electrochemical interleukin biosensors. These sensors are highly specific due to their use of antibodies to bind to specific interleukins [17]. Sensors for IL-6 have achieved levels of detection which can distinguish between severe and non-severe cases of COVID-19. A fully integrated POC device, called the Detection of Living Animal's Exhaled Breath Biomarker (dLABer) is presented in Fig. 3 . It combined breath sampling, microfluidics, and a silicon nanowire field effect transistor (SiNW-FET) biosensor to measure the quantities of IL-6 in exhaled breath [117]. The breath exhaled by rats was absorbed with the collection liquid PBS via an airflow system, and the PBS was transported to the biosensing component for analysis. A commercial SiNW-FET (Vista Therapeutics, Inc., U.S.A.) was modified with rat-specific IL-6 antibody. IL-6 binding to the SiNW-FET antibody complex proportionally changed the conductance of the transistor, revealing IL-6 concentration. The dLABer had a signal-to-noise ratio 104 times higher than that of the ELISA when analyzing the same breath samples [117]. It was determined that the ELISA and sensor data were in agreement and the sensor demonstrated good repeatability and selectivity amongst ambient components. However, this method has only been tested on the exhaled breath of mice and further work is needed to adapt the breath collection component to human breath. Furthermore, the lower limit of the range of detection is 5 pg/mL which means it may be used in COVID-19 monitoring. However, this may restrict the applications of this sensor from conditions such as some forms of cancer [118]. This sensing technique can potentially be combined with ventilators to analyze patients' exhaled breath in intensive care settings to alert clinicians of alarming interleukin spikes. The use of breath to measure interleukin levels has proven to be possible and presents an opportunity for future research.
Table 3.
A summary of electrochemical sensors targeting Interleukins.
| Analyte | Electrode | Receptor | Matrix | LOD (pg/mL) | Linear Range (pg/mL) | Ref. |
|---|---|---|---|---|---|---|
| IL-1β, IL-10 |
Gold working microelectrode | Anti-human Il-6, IL-1, IL-10 antibody | Pharmaceutical preparation | 0.7, 0.3 |
1–15, 1–15 |
[119] |
| IL-6 | 8 × 50 μm gold needle electrodes | Anti-IL-6 antibody | 5% BSA solutions to simulate blood | – | – | [120] |
| IL-6 | Graphene based field effect transistor | IL-6 specific aptamers | Pharmaceutical preparation. Human saliva samples | 252 | – | [121] |
| IL-6 | Silicon nanowire field effect transistor | Rat IL-6 antibody | Rat exhale breath | 5 | 5 – 5 × 104 | [117] |
| IL-6 | Magnetic microparticles and planar graphite-screen printed electrodes | Anti-IL-6 antibody | Human serum | 0.3 | 1 – 1 × 106 | [122] |
| IL-6 | Carbon screen printed electrode modified with polypyrrole and gold nanoparticles. | IL-6 aptamer | Human serum | 0.33 | 1–15 | [123] |
| IL-6 | Functionalized single wall carbon nanotubes (SWCNT) microarrays | IL-6 aptamer | Spiked blood | 1.0 | 1 – 1 × 104 | [124] |
| IL-6 | Horizontally aligned single walled carbon nanotubes forms a liquid gate field effect transistor | Anti-IL-6 antibody | Rabbit serum | 1.37 | 1–100 | [17] |
Fig. 3.
A) Image of dLABer system include a live rat and the apparatus for detecting IL-6 in the exhaled breath of the rat, with future aspirations to be a device that measures human IL-6 levels B) Experimental schematic of the dLABer system setup in A. Describing breath sampling, microfluidics and the real-time sensing of IL-6 levels. Reprinted and adapted with permission from Ref. [117]. Copyright 2018 American Chemical Society.
3.4. Tumor necrosis factor alpha (TNFα) and interferon (IFN)
Section Summary: TNF and IFN levels are affected by COVID-19. The concentration in severe COVID-19 patients is not yet established but is likely to increase in the case of inflammation. Electrochemical sensors exist for TNF and IFN using both aptamers and antibodies. A multianalyte sensor using aptamers was able to simultaneously achieve a 47.6 and 48.8 pg/mL limit of detection for TNF and IFN respectively. Further development in multianalyte sensors is encouraged.
Tumor Necrosis Factor (TNF) is a potent pleiotropic cytokine playing important roles in cell proliferation, differentiation and apoptosis [53,54]. It is one of the most thoroughly studied cytokines and is considered the “master-regulator” of immune homeostasis by limiting the duration of the inflammatory process [125,126]. TNF-α is responsible for initiating strong inflammatory responses, inhibiting autoimmune diseases and impeding tumor growth [126]. Anti-TNF antibodies have been used to treat autoimmune diseases like rheumatoid arthritis (RA), inflammatory bowel disease (IBS), or ankylosing spondylitis [127]. TNF-α has also been found to increase during COVID-19 infection and the resulting cytokine storms, although exact concentrations have not yet been described [16,63]. These qualities make TNF a strong candidate for electrochemical sensing in COVID-19 applications.
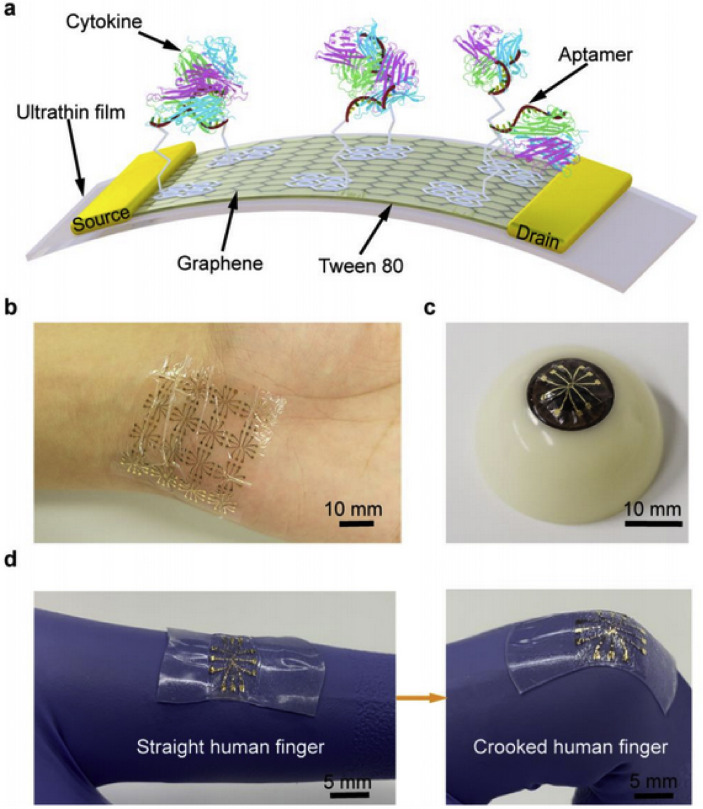
Many TNF-α electrochemical sensors have been described, using both aptamers [[128], [129], [130]] and anti-TNF antibodies [[131], [132], [133], [134]] as antigen receptors. A few are listed in Table 4 To address the challenges of the COVID-19 pandemic (for example, alerting clinicians to the onset of cytokine storms) a label-free, wearable sensor is desired. One study modified a glassy carbon electrode (GCE) with a [Fe(CN)6]4−-Glutaraldehyde-chitosan (KFe-GLA-CHIT) composite followed by Nafion and anti-TNF antibodies [133]. The KFe-GLA-CHIT was used as an electron transfer mediator. Nafion was used to protect the long-term stability of the sensor by blocking the interfering molecules from the sensor and to immobilize the anti-TNF antibodies. TNF present in the sample would bind to the capture antibodies and decrease the peak anodic current, this decrease has a linear relationship with concentration allowing the quantification of TNF in the sample. The sensor was highly sensitive, detecting 20–34,000 pg/mL of TNF with a limit of detection of 10 pg/mL while demonstrating excellent selectivity for TNF and long-term stability. Wearable TNF biosensors have been explored to yield an aptamer-based graphene field effect transistor built upon an ultrathin (2.5 μm) biocompatible mylar film shown in Fig. 4 . The substrate demonstrated good flexibility and durability over polymer-based substrates like polydimethylsiloxane (PDMS), polyester (PET) and polyethylene naphthalate (PEN) and was suitable for adhesion to non-planar, dynamic surfaces like the human wrist or eyeball. The sensor was a graphene monolayer modified with TNF-aptamer (the antigen receptor) and demonstrated sensitive, consistent, and timely detection of TNF in artificial tears [135]. Tween 80 was also used to protect the long-term stability of the sensor and block the potentially interfering molecules by passivating the unmodified graphene area. Although electrochemical TNF sensors can decrease assay time from several hours (ELISA) [117,136], future TNF sensors must emphasize timely/continuous detection.
Table 4.
A summary of electrochemical sensors targeting Tumor Necrosis Factor and Interferon.
| Analyte | Electrode/Modification | Receptor | Matrix/Solvent | LOD (pg/mL) | Linear Range (pg/mL) | Ref. |
|---|---|---|---|---|---|---|
| TNFα | Gold screen-printed electrode drop-casted with a modified Ag/Pt-Gr-aptamer nanocomposite | TNFα aptamer (RNA strand) | Human serum | 2.0 | 5.0–75 | [132] |
| TNFα | Carbon screen-printed electrode drop-casted with a modified Ag/Pt-Gr-aptamer nanocomposite | TNFα aptamer (RNA strand) | Human serum | 1.64 | 5–70 | [141] |
| TNFα IFN-γ |
Graphene field-effect transistor | TNFα and IFN-γ aptamers (RNaaA Strands) |
Artificial tears | 47.6, 48.8 |
– | [135] |
| TNFα | Screen printed electrode modified with Neutravidin-functionalized magnetic microbeads conjugated with capture antibodies | TNF Biotinylated capture antibodies | Human serum | 3 | 9.9 | [142] |
| TNFα | Glassy carbon electrode modified with anti-TNFα antibody | [Fe(CN)6]4−-Glutaraldehyde - chitosan probes | Pharmaceutical preparation | 10 | 20 – 3.4 × 104 | [133] |
| IFN-α2 | β-lactamase label-based pH sensitive field effect transistor | polyclonal anti-a-2 interferon antibodies | Pharmaceutical preparation | – | 1.0 × 107–1.0 × 108 | [143] |
Fig. 4.
A) A schematic of the TNF/IFN aptamer-based graphene FET built upon an ultrathin and flexible mylar substrate. A demonstration of sensor placement on the B) wrist and C) eyeball. D) A demonstration of the flexible nature of the constructed sensor. Reprinted and adapted with permission from Ref. [135]. Open Copyright 2020 Multidisciplinary Digital Publishing Institute (MDPI).
Interferons (IFNs) are a class of cytokines that interfere with viral and non-viral infections in the body by adapting cell functions [137]. IFN-α and IFN-β are typically produced by infected cells, while IFN-γ is produced by T-lymphocytes during cell division [138]. The administration of Type 1 interferons (IFN-α and -β) is being tested as a potential therapy for COVID-19. It can boost patients’ immune response and has been successful in treating other disorders such as multiple sclerosis (MS), MERS-CoV and SARS-CoV [139]. There is limited information about IFN concentrations in COVID-19 infected individuals. A small study examining the concentrations of various IFNs during COVID-19 progression found that patients who were IFN negative at diagnosis usually had higher viral load [140]. It was also found that IFN-α2 rose gradually after symptom onset and peaked approximately 8–10 days later. Patients without an increase of IFN-α all required ventilation and spent more time in the ICU. While IFN-α2 and IFN-α are relevant to COVID-19 prognosis, their relationship with regards to COVID-19 is not well documented and there is a shortage of electrochemical sensors targeting these two analytes. IFN-γ electrochemical sensors have been developed (listed in Table 4) but do not seem to be as relevant to COVID-19 prognosis. This presents an opportunity for future research and sensor development.
3.5. Glutamate
Section Summary: Glutamate has been linked to neurological diseases which have been observed in COVID-19 patients. However, the link between glutamate and COVID-19 is not well defined. Although electrochemical sensors can be used to detect glutamate current electrochemical sensors for glutamate do not adequately detect the low amount necessary for detection in biofluids. Functionalized nanomaterials can be used to increase sensitivity. Both enzymatic and non-enzymatic glutamate sensors are presented.
Glutamate is a nonessential amino acid acting as an excitatory neurotransmitter in the nervous system, making it responsible for neurocognitive functionality. Glutamate has been identified as a potential biomarker for disease conditions linked to neurological diseases such as stroke, epilepsy, Alzheimer's disease and amyotrophic lateral sclerosis (ALS) [11,73]. Relatives of SARS-CoV-2 have been known to impact the neurological system, so it is important to track glutamate concentrations in COVID-19 patients in case of complications [11,73].
The molecular virus-host interface is highly relevant to the neurological symptoms of COVID-19, which includes the loss of sense of smell [8,9]. Glutamate as the olfactory receptor cell neurotransmitter plays an important role in the virus-host interfaces [144]. Olfactory ionotropic glutamate receptors (iGluRs) are ligand gated ion channels regulated by G-protein-coupled receptors. iGluRs mediate chemical communication between neurons at synapses and can be modified to be used as genetically encoded chemical sensors [145]. Such chemical sensors could function as genetically encoded neuronal activators or inhibitors, as well as having broad practical applications, for example, in environmental pollutant detection or clinical diagnosis. The neurotransmitter glutamate plays the key role in neuronal communication and it is important to monitor glutamate level to understand iGluRs function and loss of sense of smell in SARS-CoV-2 infected people. Moreover, there is evidence of glutamate neurotoxicity involvement in coronavirus [146].
The presence of coronaviruses in the CNS may also result in long-term neurological diseases [9,147]. Coronaviruses can reach the smell center of the brain, primarily via vascular cells in the CNS [148]. Olfactory cells in the nasal cavity are most vulnerable to viral infection [148]. The first neurons infected are the neurons that detect odors in the nasal cavity; they can also signal when a virus is present to summon other brain and immune cells to help fight the infection [9,149]. The virus not only affects the respiratory system, but also has deleterious effects on the CNS [149]. During the SARS (severe acute respiratory syndrome) epidemic in 2003, the SARS coronavirus was also found to attack the CNS causing brain and spine demyelinating lesions and neurodegenerative diseases (e.g. multiple sclerosis, Alzheimer's disease, and Parkinson's disease) [10,11]. Brain stroke and seizures are already reported to be associated with coronavirus [73]. Neurological symptoms manifest in a notable proportion of patients with coronavirus disease [147]. As a possible explanation of neurotoxicity due to virus invasion, some coronaviruses have demonstrated the ability to spread via a synapse-connected route to the medullary cardiorespiratory center from the mechanoreceptors and chemoreceptors in respiratory system [149].
Since glutamate is the most abundant excitatory neurotransmitter in the CNS and neurotoxicity in COVID-19 evident, a glutamate-related biomarker can play an important role in detecting the severity of neurotoxicity. One major limitation, however, is that glutamate concentrations in some biofluids are lower than the detection limits of existing electrochemical sensors [55]. To address this, the functionalization of nanomaterials used for sensing (for example, NiO and Co3O4) can be improved with carbon nanotubes or β-cyclodextrin in order to improve sensing performance. Fig. 5 shows an example of one glutamate sensor made with Ni nanoparticle coated Pt electrode surfaces. A recent review identified the connection of glutamate to pathologies as a potential biomarker and emphasized development of electrochemical sensors to instantaneously detect glutamate in biofluids [55]. This review analyzed the performance of various materials for the working electrode of a non-enzymatic glutamate electrochemical sensor. Table 5 summarizes some important materials with modification approaches for electrochemical sensing of glutamate in different biofluids. Continuous and real-time monitoring of glutamate levels in biofluids can provide a means to identify the degree of neurological disorder or quantify the level of neurotoxicity due to COVID-19.
Fig. 5.
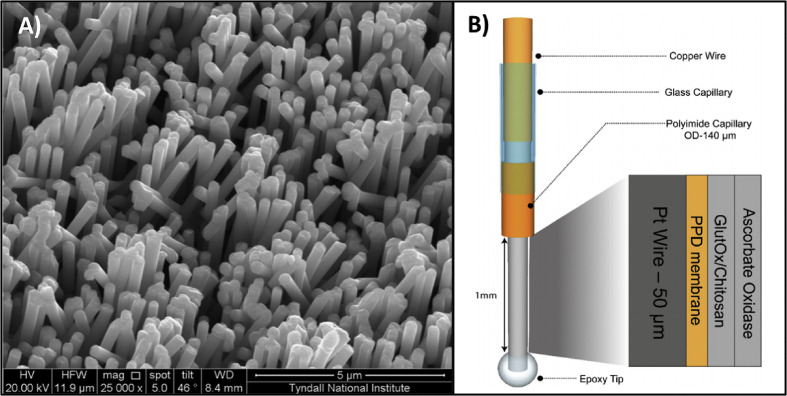
A) SEM images of a Pt coated Ni nanowire array electrode (non-enzymatic l-glutamate sensor). Reprinted and adapted with permission from Ref. [150]. Copyright 2013 Elsevier. B) Schematic of an enzymatic l-glutamate biosensor. The exposed copper wire makes contact with the potentiostat while the epoxy tip comes in contact with l-glutamate. Reprinted and adapted with permission from Ref. [151]. Copyright 2019 Elsevier.
Table 5.
A summary of electrochemical sensors targeting Glutamate.
| Analyte | Electrode/Modification | Receptor | Matrix/Solvent | LOD (ng/mL) | Linear Range (ng/mL) | Ref. |
|---|---|---|---|---|---|---|
| l-glutamic acid | Mesoporous Co3O4 nanosheets on glassy carbon electrodes | – | Serum and urine | 1.47 | 0.0147 – 1.47 × 107 | [152] |
| l-glutamate | Nickel-oxide nanoparticles on glassy carbon electrodes | – | Pharmaceutical preparation | 4.03 × 104 | 1.48 × 105–1.18 × 106 | [153] |
| l-glutamate | Ni-nanowire array electrode | – | Pharmaceutical preparation | 8.89 × 103 | 7.41 × 104–1.19 × 106 | [150] |
| l-glutamate | ZnO nanorods/polypyrrole modified pencil graphite electrode | l-glutamate oxidase | Chinese soup | 0.0267 | 2.96 – 7.41 × 104 | [154] |
| Glutamate | Ceramic-substrate enabled platinum microelectrode array | Glutamate oxidase | Pharmaceutical preparation | 23.7 | 1.48 × 103–8.44 × 103 | [155] |
| l-glutamate | Polyphenylenediamine, chitosan, ascorbate oxidase | Glutamate oxidase | In vivo and in vitro brain slices | 6.52 | 7.41 × 102–2.22 × 104 | [151] |
3.6. Exhaled breath pH
Section Summary: Currently no direct studies assessing breath pH levels during a COVID-19 infection have been conducted. However, breath pH levels increase with the incidence of lung injury, which is commonly caused by COVID-19. This section assesses two devices that collect and measure breath pH, one that attaches to a hospital ventilator and another that uses only a syringe, mouthpiece, cylinder, and ice as an exhaled breath condensate collection method.
It is difficult to assess pH as a biomarker at times due to the great number of variables that affect it in various biofluids [55]. Despite this difficulty, exhaled breath condensate (EBC), the condensed liquid form of exhaled breath, has been reliably observed to experience pH changes, resulting from inflammation caused by pulmonary diseases such as asthma, chronic obstructive pulmonary disease (COPD), and cystic fibrosis [70]. Such diseases cause oxygen limitations for lung cells, and this hypoxic state drives the cells into anaerobic respiration in which lactic acid is produced as a byproduct. In diseased states, EBC pH levels drop to a range of 4–6 while healthy patients' pH levels are around 7.4 or higher [156]. EBC pH can indicate inflammation even before clinical presentation of lung injury is observed [[157], [158], [159]]. A study examined patients requiring ventilation (with and without signs of acute lung injury (ALI) or ARDS and found that EBC pH in ventilated patients was lower and lactate levels were higher relative to a healthy group [157]. Furthermore, pro-inflammatory cytokine concentrations in breath correlated with both lower EBC pH and increased disease severity. To the best of our knowledge, no relationship between COVID-19 infection/severity and breath pH has been investigated. However, since viral infections commonly cause lung inflammation, breath pH may provide valuable insight into a patient's condition in the context of COVID-19. Thus, new research concerning this relationship is encouraged.
Unfortunately, collection of breath samples poses a risk for contamination. Most air contained in the respiratory tract does not contact the bloodstream at the alveoli; this is known as “dead-space air” [160]. During sample collection, dead-space air dilutes the sample containing clinically significant molecules. This issue can be overcome by exclusively sampling air exhaled at the end of breath [160]. Also, volatile organic compounds (VOCs) in atmospheric air may enter the respiratory tract during inhalation and contaminate the sample. In this case, correction for background concentrations of VOCs by parallel sampling of breath and external air is necessary [160]. Additionally, it is possible for breath samples to be contaminated by saliva, so collection techniques need to account for the separation of these matrices. They may also be harvested and analyzed in parallel to maximize the usability of the data collected. Various collection methods have been devised to avoid contaminants in EBC samples; a few of them are discussed below.
To capture exhaled breath for analysis, a simple condensation method is required [161]. In addition to a cooling system, gas standardization (de-aeration) may be used to address the issue of CO2 contamination, which can decrease the measured EBC pH values [162]. Two such collection devices are shown in Fig. 6 . A saliva filter is often used to ensure the breath condensate is not contaminated by saliva [161]. One proposed EBC collection mechanism was a system that attached to the exhaust port of a standard Servo-i hospital bedside ventilator [163]. This device included 2-chamber gas-standardization with a pH sensor that measured pH every 6 s. Extracting exhaled breath condensate during ventilation was also deemed to be non-detrimental to the patient and the apparatus [164]. This device would benefit from newer fabrication technologies, such as 3D printing, or new materials which could optimize cooling and hydrophobicity.
Fig. 6.
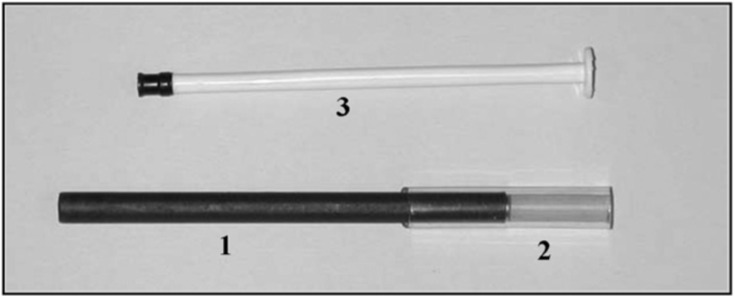
1) 88 mm long rubber covered stainless steel open cylinder, (2) 40 mm-long tube as a mouthpiece, (3) 1 mL syringe plunger. Reprinted and adapted with permission from Ref. [165]. Copyright 2006 American College of Allergy, Asthma & Immunology.
There are a variety of ventilator-independent commercial devices (Ecoscreen, RTube) used to collect EBC, however, their high costs restrict widespread use [165]. To address cost concerns, a device using only a stainless-steel open cylinder, a tube acting as a mouthpiece, and a syringe was developed [165]. The metal tube was placed in dry ice for 5 min and then patients exhaled into the tube. A sensor then measured pH levels despite not de-aerating the sample. CO2 present in exhaled breath lowers pH and necessitates the same collection-to-analysis time for consistent measurements [165]. This low-cost collection device is attractive to areas where healthcare may not be widely accessible due to costs. The two devices described here can be adapted for COVID-19 prognosis or assessing pulmonary injuries like ALI, COPD, and asthma.
3.7. Hematological biomarkers
Section Summary: Levels of hematological biomarkers such as lymphocytes, neutrophils, and thrombocytes represent the immune system's response to COVID-19 infection. T-lymphocytes are the most studied marker, and this section lists several electrochemical sensors that have already been developed to measure its levels in biofluids. A smartphone-compatible sensor is discussed in further detail.
Lymphocytopenia, neutrophilia and thrombocytopenia are present in severe and less pronounced in non-severe COVID-19 patients [166]. Therefore, lymphocyte, neutrophil, and platelet counts are all potential biomarkers that may aid in the prognosis of a COVID-19 infection. SARS-CoV-2 has an effect upon lymphocyte count by infecting T-cells directly, and the final result is a decrease in CD3+, CD4+, and CD8+ T lymphocytes accompanied by an increase in regulatory T-cells [167]. More lymphocyte apoptosis also results from the subsequent cytokine storm [167]. Lymphocyte count has already been combined with either platelet or neutrophil count, using ratios like neutrophil-lymphocyte ratio (NLR) and platelet-lymphocyte ratio (PLR); both of which are more predictive of disease state compared to sampling each biomarker alone [167]. Despite the potential of hematological biomarkers, white blood cell (WBC) counts are subject to change due to simultaneous viral or bacterial infections [168]. Steroids (like glucocorticoids) and intravenous immunoglobulin therapy can also alter WBC counts [168], both of which have been considered for COVID-19 treatment [169,170].
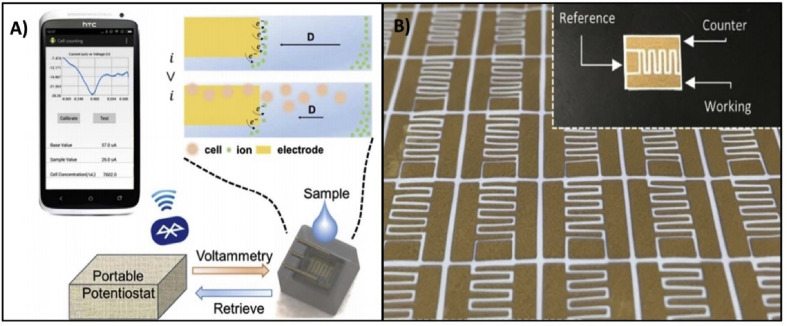
Several electrochemical sensors for immune cells have been listed in Table 6 . One publication documents a portable, label-free, paper-based smartphone compatible sensor with gold microelectrodes [171] as seen in Fig. 7 . A polyvinylidene fluoride (PVDF) membrane (paper) was used to physically trap WBCs, while interdigitated microelectrodes were used with ferrocyanide probes to quantify the trapped WBCs [171]. A portable potentiostat was also constructed with an Arduino Uno and two operational amplifiers; it was used to conduct differential pulse voltammetry (DPV) and send the collected data to a nearby smartphone via Bluetooth [171]. Furthermore, the dynamic range of this sensor was 150–15,000 cells/μL [171]. This covered most cases of leukocytopenia and leukocytosis, including severe leukocytopenia caused by COVID-19 (approx. 2000 cells/μL) [171,172]. This system's smartphone compatibility made data collection, storage, and analysis very quick, rendering it suitable for application in POC settings. The portable and cost-efficient nature of the Arduino potentiostat is also noteworthy; it paves the way for such electrochemical sensors to be used by patients at home in a similar fashion to blood-glucose testing in diabetics. Since the long-term effects of the COVID-19 infection are unknown, this feature could be especially useful for patients who have been discharged from the hospital and want to keep track of their recovery. This study takes steps to better sensor accessibility and ease-of-use while highlighting the opportunity to improve system design for sensors of other biomarkers mentioned in this review.
Table 6.
A summary of electrochemical sensors targeting Hematological Biomarkers.
| Analyte | Electrode/Modification | Receptor | Matrix/Solvent | LOD | Linear Range | Ref. |
|---|---|---|---|---|---|---|
| WBC | Microporous paper with pattered gold microelectrodes | Ferrocyanide electrochemical probes | Pharmaceutical preparation | 195 WBC/μL | 1.5 × 102–1.5 × 104 WBCs/μL WBC/μL |
[171] |
| CD4+ T lymphocyte | Magneto-actuated biosensor | AntiDC3 antibodies | Whole blood | 44 cells/μL | 89–912 cells/μL | [173] |
| CD4+ T lymphocyte | Densely packed working electrode pixels | Anti-human CD4+ antibody | Pharmaceutical preparation | – | – | [174] |
| CD4+ T lymphocyte | Single walled carbon nanotube electrodes | CD4 antibody | Pharmaceutical preparation | 100 cells/mL | 102–106 cells/mL | [175] |
| CD4+ lymphocyte | Gold-coated quartz crystal microbalance | Anti CD4+ antibody | Pharmaceutical preparation | – | – | [176] |
Fig. 7.
A) The system schematic for data collection from this paper-based WBC sensor B) The interdigitated design of the gold microelectrodes on the PVDF surface. Reprinted and adapted with permission from Ref. [171]. Copyright 2017 Elsevier.
3.8. D-dimer
Section Summary: D-dimer levels can be disrupted in the body as a result of clotting abnormalities that accompany a COVID-19 infection. 74.6% of patients have serum levels greater than 0.5 μg/mL, and high levels can be an indication of poor survival outcomes. Several existing D-dimer sensors are listed, and a low-cost, desk-top fabricated sensor is discussed in further detail.
D-dimer is a protein fragment produced as a by-product of fibrinolysis, the process of thrombus breakdown in the body [51]. It can be used to detect clotting abnormalities that arise when blood coagulation and fibrinolysis fall out of balance [51]. D-dimer is commonly measured in plasma, but techniques have been developed to allow measurement through saliva as well [177]. Although D-dimer can be elevated due to non-pathological factors such as race, age, or smoking, it is also elevated during pathologies such as stroke, atrial fibrillation, malignancy, or even COVID-19 [51]. One study (n = 248) found that 74.6% of COVID-19 patients had elevated D-dimer levels (≥0.5 μg/mL), and the median D-dimer levels were higher in non-survivors (6.21 μg/mL) than in survivors (1.02 μg/mL) [52].
D-dimer is a valuable biomarker to measure in COVID-19 patients as soon as the infection is diagnosed. One study has found that D-dimer levels greater than 2.0 μg/mL on hospital admission can predict mortality with a sensitivity of 92.3% and specificity of 83.3%, regardless of the patient's symptom severity [178]. Using electrochemical sensing techniques to monitor D-dimer early and frequently can allow health-care professionals to administer antithrombotic therapies if needed and prevent fatalities [179]. Furthermore, COVID-19 patients with metabolic syndrome are more likely to suffer cardiac events as the infection progresses [180], and D-dimer sensing could be valuable in predicting and intervening in those events. Monitoring D-dimer values after COVID-19 recovery may also be an important tool to preserve long term health for patients. It is possible that COVID-19, like its close relative SARS-CoV, leaves long lasting damage [181]. This can lead to cardiovascular events many years after recovery [182]. However, obtaining blood samples for D-dimer testing is an invasive procedure that may cause patients discomfort and increase chances of infection. Urine testing is reliable and ubiquitous but is subject to similar difficulties as blood, although it is less invasive. Despite these barriers, D-dimer's early elevation in response to infection, along with its strong predictive value make D-dimer electrochemical sensing an opportunity worth exploring.
Over the past decade, several electrochemical immunosensors have been developed to measure D-dimer levels in biofluids; some of these sensors are listed in Table 7 . One such sensor has an especially low-cost desktop fabrication method that could be valuable in areas where cleanrooms and expensive lab equipment are scarce [184]. The device was fabricated using a standard 4.7-inch polycarbonate compact disk (CD). After removing the protective layer from the CD with a knife, the disk was mounted onto an old CPU fan and spin-coated with photoresist (PR). The PR patterns were then exposed and developed, and electrodes were etched into the material with a gold etcher. The electrodes were functionalized by electrochemical deposition of polypyrrole (PPy) (see Fig. 8 ) and 12 h of incubation in a humidor with anti D-dimer antibody solution. This sensor measured D-dimer by treating its electrodes like an electric double-layer capacitor. When D-dimer in the test solution bound to the antibodies on the electrode surfaces, the properties of the capacitor's effective dielectric layer were altered, thus decreasing capacitance. This sensor's cost effectiveness and accessibility is an especially useful characteristic during today's COVID-19 climate, where medical equipment shortages occur frequently. Most of the equipment mentioned above can easily be acquired outside of a lab setting, and the cost of performing one test is approximately $1, which is much cheaper than the standard $495 ELISA kit (used for ninety-six tests). This sensor also saves time by providing results within minutes while an ELISA can take hours. Since this was just a preliminary study done with the intention of validating a low-cost fabrication method, the sensor's limit of detection and dynamic range are not optimized to COVID-19. Infected individuals typically have blood D-dimer levels greater than 0.5 μg/mL, but this sensor's dynamic range is 1 – 1 × 104 pg/mL. This sensor requires further testing and optimization before it can be applied to a COVID-19 patient. However, it provides a promising fabrication method that can make D-dimer prognosis technology available even in areas with funding limitations.
Table 7.
A summary of electrochemical sensors targeting D-dimer.
| Analyte | Electrode/Modification | Receptor | Matrix/Solvent | LOD (pg/mL) | Linear Range (pg/mL) | Reference |
|---|---|---|---|---|---|---|
| D-dimer | Silver nanoparticles decorated ZNO nanotubes | Mouse anti-human D-dimer antibody | Pharmaceutical preparation | 1 | 1 × 101–1 × 106 | [185] |
| D-dimer | Conducting polymer polypyrrole | D-dimer antibody 3B6 | Human plasma | 100 | 100 – 5 × 105 | [186] |
| D-dimer | Magnetic beads deposited on screen printed carbon electrodes (indirect competitive immunosensing) | Mouse monoclonal anti-D-dimer antibody | Human serum | 2.8 × 104 | 8.4 × 104–1.9 × 106 | [187] |
| D-dimer | Magnetic beads deposited on screen printed carbon electrodes (sandwich immunosensing) | Mouse monoclonal anti-D-dimer antibody | Human serum | 20 × 103 | 6 × 104–1 × 106 | [187] |
| D-dimer | Polycarbonate compact disk (CD) electrodes electropolymerized with polypyrrole | Anti D-dimer antibody | Pharmaceutical preparation | 1 | 1 – 1 × 104 | [184] |
| D-dimer | Functionalized carbon nanotubes (SWCNT-COOH) modified gold electrodes | Anti-D-dimer reduced antibody | Pharmaceutical preparation | 0.1 | 0.1 – 2 × 106 | [183] |
Fig. 8.
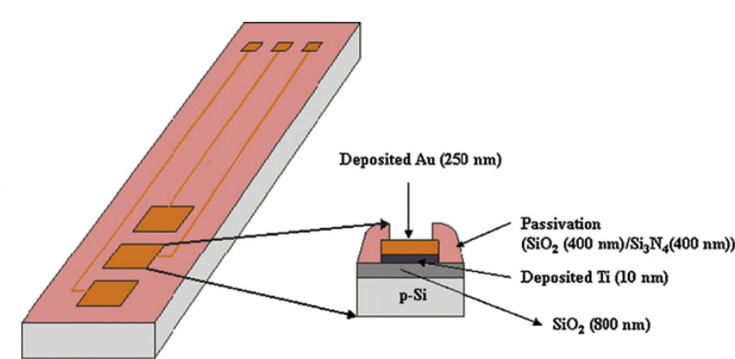
Schematic view of Impedimetric immunosensor with SWCNT-COOH modified gold microelectrodes. Sensor is used to detect D-dimer presence when deep venous thrombosis occurs. Reprinted and adapted with permission from Ref. [183]. Copyright 2017 Elsevier.
3.9. Existing electrochemical sensors related to COVID-19
Section Summary: In addition to the biomarkers mentioned previously, there have been advances in electrochemical sensing for biomarkers such as viral proteins and viral RNA specific to COVID-19. This section discusses these markers in detail, presents a few promising sensors, and identifies an opportunity for further investigation.
In addition to electrochemical sensors for the aforementioned biomarkers, there have been developments in sensing technology specific to SARS-CoV-2. These sensors involve the detection of molecules produced directly due to the presence of the virus; they would not exist in the body in large quantities unless an individual was infected. Some examples of these biomarkers are viral RNA and viral proteins such as spike protein and nucleocapsid phosphoprotein. As mentioned in the pathophysiology section (Section 2), the spike protein can be found on the surface of viral SAR-CoV-2 particles, allowing them to enter and infect host cells. The nucleocapsid protein, on the other hand, packages and assembles all the components of the virus within infected cells [188]. While other biomarkers like D-dimer, TNFα, and IL-6 indicate some form of inflammation/infection, viral proteins and RNA are COVID-19 specific, so they are great tools in the immediate diagnosis/prognosis of patients. Some sensors that have recently been developed for these purposes are listed in Table 8 (entries 1–4), and two such sensors are discussed in further detail below.
Table 8.
A summary of electrochemical sensors designed specifically for COVID-19.
| Analyte | Electrode/Modification | Receptor | Matrix/Solvent | LOD (copies/mL)a | Linear range (copies/mL)a | Ref. |
|---|---|---|---|---|---|---|
| Viral nucleocapsid phosphoprotein (N-gene) | Calixarene functionalized graphene oxide electrodes, super sandwich type sensor | Custom designed capture, signal, and auxiliary probes | Pharmaceutical preparation | 200 | – | [192] |
| Spike Receptor Binding Domain | Cobalt-functionalized TiO2 nanotubes (Co-TNTs) | Cobalt | SARS-CoV-2 Wuhan-Hu-1 sRBD with C-terminal hexa-histidine tag pharmaceutical preparation | 0.7 nM | 14 – 1.4 × 103 nM | [189] |
| COVID-specific viral RNA or the corresponding c-DNA | Electrode made of Gold nanoparticles on Ti surface | Complementary thiolated probes | – | – | – | [193] |
| Viral nucleocapsid phosphoprotein (N-gene) | Electrode made of AuNPs on graphene film | Highly specific antisense oligonucleotides (ssDNA) | Pharmaceutical Preparation | 6900 | 5.85 × 103 – 5.85 × 1010 | [194] |
| +Spike protein | Graphene FET-based sensor with Au/Cr electrodes | SARS-CoV-2 spike antibody | Nasopharyngeal secretions from COVID-19 infected patients | 242 | – |
[190] |
| Multiplex sensor for SARS-CoV-2 nucleocapsid protein (NP), S1-IgM, S1 IgG, and CRP | Laser-engraved graphene electrodes, sandwich/double-sandwich type assay (depending on the analyte) | Capture antigens and antibodies for NP, S1-IgM, S1-IgG and CRP immobilized onto the graphene electrode surface | Serum and saliva samples from RT PCR-confirmed COVID-19 positive patients and healthy patients | – | – | [191] |
The units used are copies/mL unless otherwise noted.
A non-enzymatic electrochemical sensor has been developed to rapidly detect the SARS-CoV-2-S-RBD (S Receptor Binding Domain, Spike protein) for diagnostic purposes [189]. This sensor is composed of cobalt functionalized TiO2 nanotubes which are relatively inexpensive to synthesize and have high sensitivity. A potentiostat was used to collect the data, and the limit of detection and sensitivity were found to be 0.7 nM and 14–1400 nM respectively. This test only took ∼30 s, making it a potential replacement for diagnostic tests that currently take several days. Furthermore, the metallic layer of this sensor can be redesigned for compatibility with other proteins, making it useful for diagnosing infections other than just COVID-19. Overall, this is a cost effective and sensitive sensor that can be adapted to other proteins, making it ideal for diagnostic applications during the current pandemic and potentially future ones as well. Another electrochemical sensor developed for sensing SARS-CoV-2 spike protein has even undergone trials in a clinical setting [190]. This graphene FET-based sensor is fabricated by first etching a graphene base, then forming an Au/Cr layer and immobilizing SARS-CoV-2 spike antibodies on top of it. The sensor's testing was done with secretions from nasopharyngeal swabs of both infected and non-infected individuals. In this medium, the limit of detection was discovered to be 242 copies/mL. Although this is greater than the limit of detection for current molecular diagnostic tests (∼50–100 copies), it is still suitable for practical use, and is one of the only current COVID-19-specific sensors that has undergone clinical trials. These two sensors are currently the most promising in the detection of viral proteins specific to COVID-19.
Despite the promising capabilities of the above-mentioned sensor, there remains a need for non-COVID-specific sensors that can be useful far beyond the end of the pandemic. At the present time, viral protein or RNA sensors will be a great help in diagnosing infected individuals. Post-infection, long term cardiological, neuropathological or inflammatory problems will be difficult to track using a COVID-specific sensor. Developing technologies for non-specific sensors will serve multiple purposes — inflammatory responses in patients can be monitored both throughout the infection and during recovery by continually tracking one or more inflammatory markers starting from the day of diagnosis. One such sensor is discussed in detail below.
A graphene-based multiplexed electrochemical sensor has been developed to diagnose COVID-19 and determine which stage of infection the patient is in Ref. [191]. It measures the concentration of four analytes: SARS-CoV-2 nucleocapsid protein (NP), CRP, and SARS-CoV-2 spike protein immunoglobulins S1-IgM and S1-IgG. The varying concentrations of these biomarkers in saliva or serum are used to distinguish between healthy, recovered, and infectious patients with varying degrees of symptom severity (presymptomatic, asymptomatic, symptomatic). Testing patient samples with this sensor can take as little as 1 min, and the materials used in its fabrication are also low cost. The design involves four laser-engraved electrodes functionalized with antibodies or coating proteins specific to each analyte. NP and CRP are detected via a sandwich-type assay, while S1-IgM and S1-IgG are detected via a double-sandwich type assay. Overall, this sensor utilizes inflammatory biomarkers and COVID-19 specific markers to diagnose the infection and determine its severity within minutes, making it an ideal device for use in POC settings. However, the use of this sensor requires the dilution of saliva and serum samples with PBS, which hinders the application of the system in at-home settings. This shortcoming is present in many of the other sensors discussed in this review as well. Moving forward, such multiplexed sensors can be designed to detect even more biomarkers such as ILs and IFNs, which may allow doctors to identify symptomatic patients in need of more direct attention or urgent care. Furthermore, the avenue of multiplexed sensors that measure COVID-19 specific and non-specific markers should be further explored to improve the simultaneous diagnosis and prognosis of COVID-19.
4. Metabolic syndrome
Section Summary:This section discusses metabolic syndrome as a comorbidity for COVID-19. In patients with metabolic syndrome, the markers mentioned in Section 3 can contribute to systemic inflammation; a COVID-19 infection can worsen this inflammation drastically, leading to a potentially fatal cytokine storm. It is possible that patients will face cardiovascular complications years after recovery from COVID-19. The electrochemical sensors mentioned in Section 3 can be useful for tracking this comorbidity as well.
Metabolic syndrome and its accompanying conditions are comorbidities that are often associated with COVID-19 [13]. Metabolic syndrome refers to a collection of risk factors such as obesity, insulin resistance/hyperglycemia, endothelial dysfunction, or hyperlipidemia that can increase a patient's chance of having type 2 diabetes or atherosclerotic cardiovascular disease (CVD), leading to a heart attack or stroke [195]. Since ACE2 is an enzyme involved in managing blood pressure, ACE2 expression is often elevated as a result of metabolic syndrome [196]. ACE2 also serves as SARS-CoV-2's primary target, so patients with metabolic syndrome are more frequently infected by COVID-19, and often suffer more severe infections than those without metabolic syndrome [13]. For instance, a study conducted in China showed that the case fatality rate for COVID-19 patients was 2.3%, but in patients with hypertension, diabetes, and CVD, it was 6%, 7.3%, and 10.5% respectively [197]. Since metabolic syndrome is a relatively well researched condition, knowledge about its symptoms, biomarkers, and management can be used to improve COVID-19 treatment outcomes for patients with metabolic syndrome.
Metabolic syndrome and its associated illnesses are commonly accompanied by chronic systemic inflammation, which overloads patients’ immune and cardiovascular systems [13,198]. In COVID-19 patients this is compounded by the inflammation from the infection, and the excess of inflammatory cytokines can lead to a fatal cytokine storm [199]. Several biomarkers can be used to monitor the progression of inflammation in metabolic syndrome, including pro-inflammatory cytokines such as IL-6, TNF⍺, and CRP [200]. IL-6 is a pro-inflammatory cytokine released upon any disruption of homeostasis in the body, but in the cardiovascular system, it is released primarily in response to vascular tissue remodeling. Initially, elevated levels of IL-6 have a protective effect by directing the immune system to injured tissue and facilitating healing [201]. However, chronically high levels of IL-6 induce systemic inflammation and are causally linked to CVD [201]. Similarly, TNF⍺ is a cytokine that contributes to heart contractility and peripheral resistance [202], but chronically elevated levels can promote atherosclerosis by inducing vascular dysfunction [203]. CRP is a biomarker that is very predictive of acute myocardial infarctions, and levels higher than 10 mg/L (in blood) give patients at least a 4% risk of experiencing a fatal cardiovascular event over the next ten years [204]. Recent investigations have proven that these biomarkers can be monitored in saliva as well [200,205]. During a COVID-19 infection, many of the same biomarkers are further elevated (CRP, IL-6, TNF⍺, etc.) [16], making them perfect targets for a biosensor that can be used to monitor concentrations of inflammatory cytokines as the infection progresses. Several examples of these sensors were discussed above in Section 3. This way, severe outcomes of the infection can be mitigated, especially in patients with metabolic syndrome which predisposes them to cardiac events and other fatalities.
Since the virus largely targets cells of the cardiorespiratory system it is possible that recovered patients are more likely to experience strokes or cardiac events long after they have recovered from COVID-19. This link has been found in a close relative of SARS-CoV-2, SARS-CoV [181]. In one study, patients were surveyed 12 years after recovery from a 2003 SARS-CoV outbreak [181]. It was determined that 68% had hyperlipidaemia, 60% had glucose metabolism disorders, and 44% had cardiovascular disorders. They were also more likely to contract lung infections and develop tumors. Although the mechanisms of these long-term health detriments are unknown, the correlations have been established. It is possible that this correlation applies to SARS-CoV-2 as well, and it can be especially damaging for patients with the metabolic syndrome comorbidity. Research is currently being conducted to develop therapies for COVID-19 in the short term, but as the pandemic fades, it will become important to monitor patients with comorbidities who can suffer fatal outcomes years after recovery. It is possible that tracking inflammatory biomarkers in COVID-19 patients now will give medical professionals insight into the long-term effects of the infection. Simultaneously, it will prepare them for outbreaks that may arise from similar viruses in the future.
5. Cannabinoids and the coronavirus
Section Summary: Cannabis-based medication (CBM) as a supplement to therapeutics has been discussed in the context of many complex diseases without conventional treatments (HIV, cancer, multiple sclerosis, chronic pain). Cannabinoids have also shown neuroprotective effects. These properties are being investigated to see if they could benefit COVID-19 patients. Cannabinoids sensors discussed here have high sensitivity on the ng/mL scale.
In this review we discuss unconventional approaches to deal with the COVID-19 pandemic, and cannabinoids are an unconventional therapeutic which can be monitored via electrochemical sensors simultaneously with inflammatory markers to determine efficacy. Cannabis (marijuana) is among the most consumed drugs worldwide [206]. Cannabinoids are a family of compounds derived from the Cannabis plant, the two most prominent of which are Δ9-tetrahydrocannabinol (THC) and cannabidiol (CBD) [207,208]. Δ9-THC is the psychoactive component responsible for somnolence and altered states of mind, while the non-psychoactive CBD is thought to be responsible for marijuana's sedating, analgesic and neuroprotective effects [[209], [210], [211], [212]].
Many of the effects of severe COVID-19 infection, such as cytokine storms [213], dyspnea (trouble-breathing) [214] and anosmia (loss of smell) [5] are related to inflammation, especially in the brain and lungs. There is extensive evidence in the scientific literature suggesting that an entourage of cannabinoids can decrease inflammation [[215], [216], [217], [218]]. These properties make cannabis well-suited for complex medical conditions which lack effective conventional treatments and their potential as an adjunct to conventional anti-viral treatments has been explored [219,220]. In addition, cannabis has been used in the management of chronic pain, neuropathy and loss of appetite associated with cancer [221,222] and HIV treatments [223,224], spasticity and muscle pain associated with multiple sclerosis [225] and mental illnesses like anxiety and depression [226]. They do this via four pathways involving the cannabinoid receptor 2 (CB2) which is highly expressed on the surface of immune cells [227]. The four pathways are apoptosis, inhibition of cell proliferation, cytokine/chemokine production inhibition and regulatory T cell induction [227]. In general, limited evidence exists concerning the influence of cannabinoids on patients with viral infections, and no investigations are available regarding SARS-CoV-2.
The novel interest in medicinal cannabis has led to a surge in evaluations and validations of therapeutic application for cannabis, including viral infections [228]. Although cannabis contains therapeutic potential for many diseases, it remains unclear whether THC and CBD would be advantageous or disadvantageous in the fight against COVID-19 [228]. Cannabinoids have been shown to inhibit immune cell recruitment and function in the lungs while having detrimental effects on lung function and decreasing respiratory pathogen clearance [229]. Understanding the role of cannabinoids in providing resistance against inflammatory damage may help clinicians with producing optimal health outcomes.
Deleterious neurological effects have been observed in a notable proportion of COVID-19 patients [147,149]. Anosmia (loss of smell) [230], necrotizing encephalopathy (brain damage) [231], stroke [73] and Guillain-Barré syndrome are neurological disorders associated with COVID-19 [232]. Although the exact mechanism is unknown, increasing evidence suggests some coronaviruses use synapse-connected routes to travel from the lungs and airways to the brainstem [149]. Despite a lack of information regarding the effects of CBD on COVID-19 recovery, cannabis has been used to guard against neurological developments caused by other viral infections. Studies have suggested cannabis exposure is associated with lower levels of neurocognitive impairment for individuals living with HIV, possibly due to its anti-inflammatory effects [233]. Other studies demonstrated CBD attenuated neuroinflammation induced by Theiler's Murine Encephalomyelitis Virus-Induced Demyelinating Disease (TMEV-IDD) in the brains of infected mice [234]. Treatment with CBD reduced the migration of activated immune cells through the blood-brain barrier (BBB) and decreased the induction of proinflammatory cytokines (CCL1, CCL2, IL-1β) and vascular cell adhesion molecule 1 (VCAM-1). Reducing leukocyte infiltration and microglia activation in the brain of TMEV-IDD mice improved their motor symptoms and neuroinflammation in the chronic phase of the infection [234]. Furthermore, in vitro studies have shown CBD to inhibit hepatitis C (HCV) replication by 86.4% at a 10 μM concentration with an EC50 (half maximal dose) of 3.163 μM, although no activity against hepatitis B (HBV) was exhibited [235]. Additionally, studies evaluating cannabis for the treatment of HBV and HCV found cannabis to be a useful adjunct to the standard interferon or ribavirin prescriptions by helping users adhere to challenging medication regimens [236]. These examples highlight the potential for cannabinoids, particularly CBD, as an adjunct to traditional antiviral therapies.
Electrochemical sensing of cannabinoids is well documented [81]. A summary of electrochemical sensors used to detect cannabinoids in aqueous solutions is presented in Table 9 . The biggest application of cannabinoid sensors is to determine impairment status of individuals. As a result, research has mostly targeted the psychoactive Δ9-THC and less commonly its metabolites 11-OH-THC and COOH-THC, while the development of electrochemical sensors for CBD has been completely neglected. Fig. 9 shows that Δ9-THC and CBD share a similar structure. The phenol which undergoes the redox reaction is common between the two [237]. This presents an opportunity to validate Δ9-THC biosensors for the analysis of CBD. The electrochemical behavior of CBD is largely unknown; a study of CBD's faradaic efficiency, redox site and transfer coefficient will be instrumental in developing CBD targeting sensors. Other opportunities are found in simultaneous sensing of multiple analytes. A multianalyte array simultaneously tracking cannabinoids and inflammatory markers may provide researchers new insights into medicinal cannabis and its effect on the immune system.
Table 9.
A summary of electrochemical sensors targeting Cannabinoids.
| Analyte | Electrode/Modification | Receptor/Mediator | Matrix/Solvent | LOD (ng/mL) | Linear Range (ng/mL) | Ref. |
|---|---|---|---|---|---|---|
| Δ9-THC | Screen-printed electrode | N-(4-amino-3-methoxyphenyl)-methanesulfonamide (OX0245) mediator | Real saliva | 25 | – | [238] |
| Δ9-THC 11-OH-THC COOH-THC |
Porous carbon paper | – | pH 10 buffer | 1.26 | – | [237] |
| 1.32 | ||||||
| 2.75 | ||||||
| Δ9-THC | Carbon screen printed electrode | – | Pharmaceutical preparation pH 7.4 | 314 | – | [239] |
| Δ9-THC | Glassy carbon immunosensor modified with gold nanoparticles | Anti-THC antibody | Rat serum pH 7.4 | 0.0033 | 0.01 – 1 × 103 | [240] |
| Δ9-THC | CNT molecularly imprinted polymer (MIP) | – | Pharmaceutical preparation | 0.18 | – | [241] |
| Δ9-THC | Carbon paste | Carbon paste; absorptive stripping voltammetry | Artificial saliva pH 10 | 157 | 157 – 2.2 × 103 | [242] |
Fig. 9.
The molecular structure of A) Δ9-Tetrahydrocannabinol (THC) and B) Cannabidiol (CBD).
When cannabinoids participate in redox reactions in aqueous solutions, radical phenols are formed. Phenol radicals readily participate in side-reactions which 1) result in surface fouling which quickly diminishes the reusability of these sensors and 2) decrease the faradaic efficiency of cannabinoids sensors thereby reducing their sensitivity. To address surface fouling issues, researchers use mediated (indirect) sensing arrays [81]. These sensors monitor a surrogate which changes concentration in the presence of a cannabinoid. For example, the mediator N-(4-amino-3-methoxyphenyl)-methanesulfonamide (OX0245) and supporting electrolytes were evaporated onto a porous electrode overlayer [238]. The overlayer's content dissolved into a sample when the two elements came into contact, allowing the sensor to be used without prior sample preparation. This sensor is presented in Fig. 10 b. This method achieved a limit of detection of 25 ng/mL and reported little surface fouling [238]. In this way the direct oxidation of cannabinoids is avoided, extending the lifespan of the sensor.
Fig. 10.
A) Molecularly imprinted polymer (MIP)-based electrochemical sensor Reprinted and adapted with permission from Ref. [241]. Copyright 2019 Elsevier. B) An electrochemical sensor in which an OX0245 mediator and supporting electrolytes are dissolved onto a porous overlayer, which eliminates sample preparation prior to electrochemical analysis. Reprinted and adapted with permission from Ref. [238]. Copyright 2016 Springer. C) A schematic fabrication a wearable sensor array, Ag/AgCl ink is used for contacts, carbon ink is used for working electrode which is then modified by MWCNT by drop-casting. D) The prototype for of the wearable ring sensor. C) and D) reprinted and adapted with permission from Ref. [248]. Copyright 2020 Elsevier.
Δ9-THC has very low solubility (1–2 μg/mL) in water [243]. In the past this has limited the use of electrochemical sensors in real bodily fluids because the sensitivity has not been adequate for detection [81]. Unmodified screen-printed carbon and glassy carbon electrodes have been documented with high sensitivity in non-aqueous solvents [[244], [245], [246]] or insufficient sensitivity in aqueous solutions [239]. Electrode surface modifications have been used to solve many problems in the electrochemical sensing space [247]. Multiwalled carbon nanotubes (MWCNT) drop-casted on a carbon ink electrode have been used in a wearable ring sensor to achieve a 157 ng/mL limit of detection [248]. This sensor can simultaneously sense THC and ethanol in the saliva of impaired drivers. The fabrication of this sensor is presented in Fig. 10c/d. Sensors based on absorptive stripping voltammetry [242], porous carbon paper [237], molecularly imprinted polymers (MIPs) (Fig. 10a) [241] have also produced highly sensitive electrodes capable of sensing concentrations of 31.5, 1.26 and 0.18 ng of THC/mL respectively.
6. Conclusion
In this review, the recent advances and current opportunities for electrochemical sensing of biomarkers related to COVID-19 and its comorbidities were discussed. We have emphasized advantages, disadvantages, and future opportunities in label-free and wearable technology to provide insight into disease progression for resource management and public health outcomes. Opportunities are present for targeting cytokines (ILs, TNFs, IFNs) and other biomarkers (CRP, NLR, PLR, D-dimer, pH and glutamate). Cannabinoids are also being explored as a possible supplement or adjunct to conventional antiviral therapies, as such continuous monitoring of cannabinoids in biofluids is desirable, particularly the non-psychoactive CBD. The development of sensors targeting these biomolecules is likely to progress rapidly as governments fund initiatives to minimize the impact of the COVID-19 pandemic. As mentioned in the respective sections, these biomolecules are involved in many diseases and continued work would improve health outcomes. Prior to publication, researchers should validate their sensors’ performance in biofluids (saliva, sweat, urine, tears, breath) to test performance in the presence of contaminants.
Multianalyte and simultaneous sensing is the newest frontier in electrochemical sensing. Multianalyte sensing could provide more rapid determination of disease progression and allow for an economical use of resources. We highlighted low-cost fabrication technologies throughout this paper and want to reiterate the importance of low-cost sensors. The WHO has developed the A.S.S.U.R.E.D criteria as a benchmark for identifying appropriate tools in areas where resources are limited. The A.S.S.U.R.E.D acronym stands for Affordable, Sensitive, Specific, User-friendly, Rapid and robust, Equipment-free and Deliverable to end-users. This is especially important during the COVID-19 pandemic where surges in cases may overwhelm developing or otherwise strained healthcare systems.
This COVID-19 pandemic has been the first-time humans have experienced the effects of the SARS-CoV-2 virus, so its long-term complications are largely unknown. Like its relative, SARS-CoV, it is likely that various cardiac and pulmonary complications of SARS-CoV-2 infection will become apparent over the next few years [181]. A study that assessed patients after 12 years of recovering from SARS-CoV infection suggested that 68%, 60%, and 40% had hyperlipidaemia, glucose metabolism disorders, and cardiovascular disorders, respectively; along with an increased risk of contracting lung infections and developing tumors [181]. Furthermore, virus-induced pneumonia survivors are at risk of having long term lung disability, with blood-gas diffusion abnormalities [249]. Lung fibrosis has also been suggested to be a long-term consequence of COVID-19 pneumonia [250]. Using portable, wearable, or POC electrochemical sensors to monitor inflammatory biomarkers after patients have recovered from COVID-19 can allow clinicians to observe residual injury and prevent any adverse effects from occurring in the future. Currently, POC testing can be made more efficient with the use of electrochemical sensors, providing real-time monitoring of various prognostic biomarkers. Wearable and portable sensors can be sent home with patients after discharge in order to monitor their recovery. In a post-COVID world, wearable and portable sensors will become very important as they will allow people returning to their daily lives to monitor any long-term complications of COVID-19. Apart from use during the COVID-19 pandemic, the sensors discussed in this paper have a myriad of use cases in other disorders involving inflammation, infection, and even cancer. Thus, these sensors should be developed further not only in the next few years with the purpose of post-COVID usage, but in the decades to come.
Credit author statement
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research is supported by Discovery Grant from the Natural Sciences and Engineering Research Council of Canada of Canada, an infrastructure grant from the Canada Foundation for Innovation, a FedDev of Southern Ontario grant, and McMaster Start-up Grant.
References
- 1.Lu H., Stratton C.W., Tang Y.W. Outbreak of pneumonia of unknown etiology in Wuhan, China: the mystery and the miracle. J. Med. Virol. 2020;92(4):401–402. doi: 10.1002/jmv.25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cucinotta D., Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020;91(1):157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He J., Guo Y., Mao R., Zhang J. Proportion of asymptomatic coronavirus disease 2019: a systematic review and meta-analysis. J. Med. Virol. 2021;93(2):820–830. doi: 10.1002/jmv.26326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singhal T. A review of coronavirus disease-2019 (COVID-19) Indian J. Pediatr. 2020:1–6. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han A.Y., Mukdad L., Long J.L., Lopez I.A. Anosmia in COVID-19: mechanisms and significance. Chem. Senses. 2020;45(6):423–428. doi: 10.1093/chemse/bjaa040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Z., Mcgoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China. Jama. 2020;323(13):1239. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 7.Cascella M., Rajnik M., Cuomo A., Dulebohn S.C., Di Napoli R. Statpearls [internet] StatPearls Publishing; 2020. Features, evaluation and treatment coronavirus (COVID-19) [PubMed] [Google Scholar]
- 8.Zubair A.S., Mcalpine L.S., Gardin T., Farhadian S., Kuruvilla D.E., Spudich S. Neuropathogenesis and neurologic manifestations of the coronaviruses in the age of coronavirus disease 2019. JAMA Neurol. 2020;77(8):1018. doi: 10.1001/jamaneurol.2020.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desforges M., Coupanec A.L., Dubeau P., Bourgouin A., Lajoie L., Dubé M., Talbot P.J. Human coronaviruses and other respiratory viruses: underestimated opportunistic pathogens of the central nervous system? Viruses. 2019;12(1):14. doi: 10.3390/v12010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zanin L., et al. SARS-CoV-2 can induce brain and spine demyelinating lesions. Acta Neurochir. (Wien). 2020;162:1491–1494. doi: 10.1007/s00701-020-04374-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cataldi M., Pignataro G., Taglialatela M. Neurobiology of coronaviruses: potential relevance for COVID-19. Neurobiol. Dis. 2020;143:105007. doi: 10.1016/j.nbd.2020.105007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prevalence of metabolic syndrome in the Canadian adult population. CMAJ (Can. Med. Assoc. J.) : Canad. Med. Asso. J. = J. de l'Asso. Med. Canad. 2019;191(5):E141. doi: 10.1503/cmaj.190063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costa F.F., Rosário W.R., Ribeiro Farias A.C., de Souza R.G., Duarte Gondim R.S., Barroso W.A. Metabolic syndrome and COVID-19: an update on the associated comorbidities and proposed therapies. Diab. Metabol. Syndr. 2020;14(5):809–814. doi: 10.1016/j.dsx.2020.06.016. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kubina R., Dziedzic A. Molecular and serological tests for COVID-19 a comparative review of SARS-CoV-2 coronavirus laboratory and point-of-care diagnostics. Diagnostics. 2020;10(6):434. doi: 10.3390/diagnostics10060434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kermali M., Khalsa R.K., Pillai K., Ismail Z., Harky A. The role of biomarkers in diagnosis of COVID-19 – a systematic review. Life Sci. 2020;254:117788. doi: 10.1016/j.lfs.2020.117788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ponti G., Maccaferri M., Ruini C., Tomasi A., Ozben T. Biomarkers associated with COVID-19 disease progression. Crit. Rev. Clin. Lab Sci. 2020;57(6):389–399. doi: 10.1080/10408363.2020.1770685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen H., Choo T.K., Huang J., Wang Y., Liu Y., Platt M., Tok A.I.Y. Label-free electronic detection of interleukin-6 using horizontally aligned carbon nanotubes. Mater. Des. 2016;90:852–857. [Google Scholar]
- 18.Chiswick E.L., Duffy E., Japp B., Remick D. Detection and quantification of cytokines and other biomarkers. Methods Mol. Biol. (Clifton, N.J.) 2012;844:15–30. doi: 10.1007/978-1-61779-527-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kissinger P.T., Heineman W.R. Cyclic voltammetry. J. Chem. Educ. 1983;60(9):702. doi: 10.1021/ed060p702. [DOI] [Google Scholar]
- 20.Abdulbari H.A., Basheer E.A. Electrochemical biosensors: electrode development, materials, design, and fabrication. ChemBioEng Rev. 2017;4(2):92–105. doi: 10.1002/cben.201600009. [DOI] [Google Scholar]
- 21.Jadon N., Jain R., Sharma S., Singh K. Recent trends in electrochemical sensors for multianalyte detection - a review. Talanta. 2016;161:894–916. doi: 10.1016/j.talanta.2016.08.084. [DOI] [PubMed] [Google Scholar]
- 22.Malik P., Katyal V., Malik V., Asatkar A., Inwati G., Mukherjee T.K. Nanobiosensors: concepts and variations. ISRN Nanomater. 2013;2013:1–9. doi: 10.1155/2013/327435. [DOI] [Google Scholar]
- 23.Peeling R.W., Wedderburn C.J., Garcia P.J., Boeras D., Fongwen N., Nkengasong J., Heymann D.L. Serology testing in the COVID-19 pandemic response. Lancet Infect. Dis. 2020;20(9) doi: 10.1016/s1473-3099(20)30517-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loo S.W., Pui T.S. Cytokine and cancer biomarkers detection: the dawn of electrochemical paper-based biosensor. Sensors. 2020;20(7):1854. doi: 10.3390/s20071854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alam A.U., Qin Y., Nambiar S., Yeow J.T., Howlader M.M., Hu N., Deen M.J. Polymers and organic materials-based pH sensors for healthcare applications. Prog. Mater. Sci. 2018;96:174–216. doi: 10.1016/j.pmatsci.2018.03.008. [DOI] [Google Scholar]
- 26.Lippitz B.E. Cytokine patterns in patients with cancer: a systematic review. Lancet Oncol. 2013;14(6):e218–e228. doi: 10.1016/S1470-2045(12)70582-X. [DOI] [PubMed] [Google Scholar]
- 27.Chen C.C., Wang S.S., Lee F.Y., Chang F.Y., Lee S.D. Proinflammatory cytokines in early assessment of the prognosis of acute pancreatitis. Am. J. Gastroenterol. 1999;94(1):213–218. doi: 10.1111/j.1572-0241.1999.00709.x. [DOI] [PubMed] [Google Scholar]
- 28.Day N.P., Hien T.T., Schollaardt T., Loc P.P., Chuong L.V., Hong Chau T.T., Ho M. The prognostic and pathophysiologic role of pro-and antiinflammatory cytokines in severe malaria. J. Infect. Dis. 1999;180(4):1288–1297. doi: 10.1086/315016. [DOI] [PubMed] [Google Scholar]
- 29.Li X., Lin S., Chen X., Huang W., Li Q., Zhang H., Shao B. The prognostic value of serum cytokines in patients with acute ischemic stroke. Aging Dise. 2019;10(3):544. doi: 10.14336/ad.2018.0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown P.M., Schneeberger D.L., Piedimonte G. Biomarkers of respiratory syncytial virus (RSV) infection: specific neutrophil and cytokine levels provide increased accuracy in predicting disease severity. Paediatr. Respir. Rev. 2015;16(4):232–240. doi: 10.1016/j.prrv.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keating S.M., Golub E.T., Nowicki M., Young M., Anastos K., Crystal H., Wu S. The effect of HIV infection and HAART on inflammatory biomarkers in a population-based cohort of US women. AIDS (London, England) 2011;25(15):1823. doi: 10.1097/QAD.0b013e3283489d1f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corwin E.J. Understanding cytokines part I: physiology and mechanism of action. Biol. Res. Nurs. 2000;2(1):30–40. doi: 10.1177/109980040000200104. [DOI] [PubMed] [Google Scholar]
- 33.Biancotto A., Wank A., Perl S., Cook W., Olnes M.J., Dagur P.K., McCoy J.P. Baseline levels and temporal stability of 27 multiplexed serum cytokine concentrations in healthy subjects. PloS One. 2013;8(12) doi: 10.1371/journal.pone.0076091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ulloa L., Tracey K.J. The ‘cytokine profile’: a code for sepsis. Trends Mol. Med. 2005;11(2):56–63. doi: 10.1016/j.molmed.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 35.Pepys M.B., Hirschfield G.M. C-reactive protein: a critical update. J. Clin. Invest. 2003;111(12):1805–1812. doi: 10.1172/JCI18921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szalai A.J., van Ginkel F.W., Dalrymple S.A., Murray R., McGhee J.R., Volanakis J.E. Testosterone and IL-6 requirements for human C-reactive protein gene expression in transgenic mice. J. Immunol. (Baltimore, Md: 1950) 1998;160(11):5294–5299. [PubMed] [Google Scholar]
- 37.Liu F., Li L., Xu M., Wu J., Luo D., Zhu Y., Li B., Song X., Zhou X. Prognostic value of interleukin-6, C-reactive protein, and procalcitonin in patients with COVID-19. J. Clin. Virol. : Off. Publ. Pan Am. Soc. Clin. Virol. 2020;127:104370. doi: 10.1016/j.jcv.2020.104370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brinckerhoff C.E., Guyre P.M. Increased proliferation of human synovial fibroblasts treated with recombinant interferon gamma. J. Immunol. 1985;134:3142–3146. [PubMed] [Google Scholar]
- 39.Dayer J.M., Beutler B., Cerami A. Cachectin/tumor necrosis factor stimulates collagenase and prostaglandin E2 production by human synovial cells and dermal fibroblasts. J. Exp. Med. 1985;162(6):2163–2168. doi: 10.1084/jem.162.6.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao Y., Qin L., Zhang P., Li K., Liang L., Sun J., Peng Y. Longitudinal COVID-19 profiling associates IL-1RA and IL-10 with disease severity and RANTES with mild disease. JCI Insight. 2020;5(13) doi: 10.1172/jci.insight.139834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown M.A., Hural J. Functions of IL-4 and control of its expression. Crit. Rev. Immunol. 1997;17(1):1–32. doi: 10.1615/critrevimmunol.v17.i1.10. [DOI] [PubMed] [Google Scholar]
- 42.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet (London, England) 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marques-Deak A., Cizza G., Eskandari F., Torvik S., Christie I.C., Sternberg E.M., Phillips T.M., Premenopausal, Osteoporosis Women, Alendronate, Depression Study Group Measurement of cytokines in sweat patches and plasma in healthy women: validation in a controlled study. J. Immunol. Methods. 2006;315(1–2):99–109. doi: 10.1016/j.jim.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 44.Thompson D.K., Huffman K.M., Kraus W.E., Kraus V.B. Critical appraisal of four IL-6 immunoassays. PloS One. 2012;7(2) doi: 10.1371/journal.pone.0030659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aziz M., Fatima R., Assaly R. Elevated interleukin-6 and severe COVID-19: a meta-analysis. J. Med. Virol. 2020 doi: 10.1002/jmv.25948. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Folkesson H.G., Matthay M.A., Hébert C.A., Broaddus V.C. Acid aspiration-induced lung injury in rabbits is mediated by interleukin-8-dependent mechanisms. J. Clin. Invest. 1995;96(1):107–116. doi: 10.1172/JCI118009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hammond M.E., Lapointe G.R., Feucht P.H., Hilt S., Gallegos C.A., Gordon C.A., Giedlin M.A., Mullenbach G., Tekamp-Olson P. IL-8 induces neutrophil chemotaxis predominantly via type I IL-8 receptors. J. Immunol. 1995;155(3):1428–1433. [PubMed] [Google Scholar]
- 48.Arican O., Aral M., Sasmaz S., Ciragil P. Mediators of inflammation; 2005. Serum Levels of TNF-α, IFN-γ, IL-6, IL-8, IL-12, IL-17, and IL-18 in Patients with Active Psoriasis and Correlation with Disease Severity. 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Del Valle D.M., Kim-Schulze S., Huang H.H., Beckmann N.D., Nirenberg S., Wang B., Marron T.U. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020:1–8. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Couper K.N., Blount D.G., Riley E.M. IL-10: the master regulator of immunity to infection. J. Immunol. 2008;180(9):5771–5777. doi: 10.4049/jimmunol.180.9.5771. [DOI] [PubMed] [Google Scholar]
- 51.Wakai A., Gleeson A., Winter D. Role of fibrin D-dimer testing in emergency medicine. Emerg. Med. J. : Eng. Manag. J. 2003;20(4):319–325. doi: 10.1136/emj.20.4.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yao Y., Cao J., Wang Q., Shi Q., Liu K., Luo Z., Hu B. D-dimer as a biomarker for disease severity and mortality in COVID-19 patients: a case control study. J. Inten. Care. 2020;8(1):1–11. doi: 10.1186/s40560-020-00466-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wajant H., Pfizenmaier K., Scheurich P. Tumor necrosis factor signaling. Cell Death Differ. 2003;10(1):45–65. doi: 10.1038/sj.cdd.4401189. [DOI] [PubMed] [Google Scholar]
- 54.Baud V., Karin M. Signal transduction by tumor necrosis factor and its relatives. Trends Cell Biol. 2001;11(9):372–377. doi: 10.1016/s0962-8924(01)02064-5. [DOI] [PubMed] [Google Scholar]
- 55.Schultz J., Uddin Z., Singh G., Howlader M.M. Glutamate sensing in biofluids: recent advances and research challenges of electrochemical sensors. The Analyst. 2020;145(2):321–347. doi: 10.1039/c9an01609k. [DOI] [PubMed] [Google Scholar]
- 56.Tikellis C., Thomas M.C. Angiotensin-converting enzyme 2 (ACE2) is a key modulator of the renin angiotensin system in health and disease. Inter. J. Peptide. 2012;2012:256294. doi: 10.1155/2012/256294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science (New York, N.Y.) 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muniyappa R., Gubbi S. COVID-19 pandemic, coronaviruses, and diabetes mellitus. Am. J. Physiol. Endocrinol. Metabol. 2020;318(5):E736–E741. doi: 10.1152/ajpendo.00124.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takeuchi O., Akira S. Innate immunity to virus infection. Immunol. Rev. 2009;227(1):75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saito T., Gale M. Principles of intracellular viral recognition. Curr. Opin. Immunol. 2007;19(1):17–23. doi: 10.1016/j.coi.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 61.Ragab D., Eldin H.S., Taeimah M., Khattab R., Salem R. The COVID-19 cytokine storm; what we know so far. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Turner M.D., Nedjai B., Hurst T., Pennington D.J. Cytokines and chemokines: at the crossroads of cell signalling and inflammatory disease. Biochim. Biophys. Acta. 2014;1843(11):2563–2582. doi: 10.1016/j.bbamcr.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 63.Tisoncik J.R., Korth M.J., Simmons C.P., Farrar J., Martin T.R., Katze M.G. Into the eye of the cytokine storm. Microbiol. Mol. Biol. Rev: MMBR (Microbiol. Mol. Biol. Rev.) 2012;76(1):16–32. doi: 10.1128/MMBR.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Conti P., Ronconi G., Caraffa A., Gallenga C.E., Ross R., Frydas I., Kritas S.K. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): anti-inflammatory strategies. J. Biol. Regul. Homeost. Agents. 2020;34(2):1. doi: 10.23812/CONTI-E. Advance online publication. [DOI] [PubMed] [Google Scholar]
- 65.Chaplin D.D. Overview of the immune response. J. Allergy Clin. Immunol. 2010;125(2 Suppl 2):S3–S23. doi: 10.1016/j.jaci.2009.12.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fathi N., Rezaei N. Lymphopenia in COVID-19: therapeutic opportunities. Cell Biol. Int. 2020 doi: 10.1002/cbin.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dmitrieva O.S., Shilovskiy I.P., Khaitov M.R., Grivennikov S.I. Interleukins 1 and 6 as main mediators of inflammation and cancer. Biochemistry (Mosc.) 2016;81(2):80–90. doi: 10.1134/S0006297916020024. [DOI] [PubMed] [Google Scholar]
- 68.Opal S.M., DePalo V.A. Anti-inflammatory cytokines. Chest. 2000;117(4):1162–1172. doi: 10.1378/chest.117.4.1162. [DOI] [PubMed] [Google Scholar]
- 69.Zhang C., Chen Z., Meng X., Li M., Zhang L., Huang A. The involvement and possible mechanism of pro-inflammatory tumor necrosis factor alpha (TNF-α) in thoracic ossification of the ligamentum flavum. PloS One. 2017;12(6) doi: 10.1371/journal.pone.0178986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carpagnano G.E., Barnes P.J., Francis J., Wilson N., Bush A., Kharitonov S.A. Breath condensate pH in children with cystic fibrosis and asthma: a new noninvasive marker of airway inflammation? Chest. 2004;125(6):2005–2010. doi: 10.1378/chest.125.6.2005. [DOI] [PubMed] [Google Scholar]
- 71.Kharitonov S.A., Barnes P.J. Exhaled biomarkers. Chest. 2006;130(5):1541–1546. doi: 10.1378/chest.130.5.1541. [DOI] [PubMed] [Google Scholar]
- 72.Aldakheel F.M., Thomas P.S., Bourke J.E., Matheson M.C., Dharmage S.C., Lowe A.J. Relationships between adult asthma and oxidative stress markers and pH in exhaled breath condensate: a systematic review. Allergy. 2016;71(6):741–757. doi: 10.1111/all.12865. [DOI] [PubMed] [Google Scholar]
- 73.Hess D.C., Eldahshan W., Rutkowski E. COVID-19-Related stroke. Transl. Stroke Res. 2020;11(3):322–325. doi: 10.1007/s12975-020-00818-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sokolov A., Hellerud B.C., Johannessen E.A., Mollnes T.E. Inflammatory response induced by candidate biomaterials of an implantable microfabricated sensor. J. Biomed. Mater. Res. 2012;100A(5):1142–1150. doi: 10.1002/jbm.a.34054. [DOI] [PubMed] [Google Scholar]
- 75.Jiang C., Alam M.T., Silva S.M., Taufik S., Fan S., Gooding J.J. Unique sensing interface that allows the development of an electrochemical immunosensor for the detection of tumor necrosis factor α in whole blood. ACS Sens. 2016;1(12):1432–1438. doi: 10.1021/acssensors.6b00532. [DOI] [Google Scholar]
- 76.Lee Y.H., Wong D.T. Saliva: an emerging biofluid for early detection of diseases. Am. J. Dent. 2009;22(4):241. [PMC free article] [PubMed] [Google Scholar]
- 77.Timmons B.W., Tarnopolsky M.A., Snider D.P., Bar-Or O. Immunological changes in response to exercise: influence of age, puberty, and gender. Med. Sci. Sports Exerc. 2006;38(2):293–304. doi: 10.1249/01.mss.0000183479.90501.a0. [DOI] [PubMed] [Google Scholar]
- 78.Dolan K., Rouen D., Kimber J. An overview of the use of urine, hair, sweat and saliva to detect drug use. Drug Alcohol Rev. 2004;23(2):213–217. doi: 10.1080/09595230410001704208. [DOI] [PubMed] [Google Scholar]
- 79.Delanghe J., Speeckaert M. Preanalytical requirements of urinalysis. Biochem. Med. 2014;24(1):89–104. doi: 10.11613/BM.2014.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gooding J.J. Finally, a simple solution to biofouling. Nat. Nanotechnol. 2019;14(12):1089–1090. doi: 10.1038/s41565-019-0573-0. [DOI] [PubMed] [Google Scholar]
- 81.Klimuntowski M., Alam M.M., Singh G., Howlader M.M. Electrochemical sensing of cannabinoids in biofluids: a noninvasive tool for drug detection. ACS Sens. 2020;5(3):620–636. doi: 10.1021/acssensors.9b02390. [DOI] [PubMed] [Google Scholar]
- 82.Xu J., Lee H. Anti-biofouling strategies for long-term continuous use of implantable biosensors. Chemosensors. 2020;8(3):66. [Google Scholar]
- 83.Sun C., Niu Y., Tong F., Mao C., Huang X., Zhao B., Shen J. Preparation of novel electrochemical glucose biosensors for whole blood based on antibiofouling polyurethane-heparin nanoparticles. Electrochim. Acta. 2013;97:349–356. [Google Scholar]
- 84.Gant R.M., Abraham A.A., Hou Y., Cummins B.M., Grunlan M.A., Coté G.L. Design of a self-cleaning thermoresponsive nanocomposite hydrogel membrane for implantable biosensors. Acta Biomater. 2010;6(8):2903–2910. doi: 10.1016/j.actbio.2010.01.039. [DOI] [PubMed] [Google Scholar]
- 85.Black S., Kushner I., Samols D. C-reactive protein. J. Biol. Chem. 2004;279(47):48487–48490. doi: 10.1074/jbc.R400025200. [DOI] [PubMed] [Google Scholar]
- 86.McBride J.D., Cooper M.A. A high sensitivity assay for the inflammatory marker C-Reactive protein employing acoustic biosensing. J. Nanobiotechnol. 2008;6:5. doi: 10.1186/1477-3155-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Magliulo M., De Tullio D., Vikholm-Lundin I., Albers W.M., Munter T., Manoli K., Palazzo G., Torsi L. Label-free C-reactive protein electronic detection with an electrolyte-gated organic field-effect transistor-based immunosensor. Anal. Bioanal. Chem. 2016;408(15):3943–3952. doi: 10.1007/s00216-016-9502-3. [DOI] [PubMed] [Google Scholar]
- 88.Palazzo G., De Tullio D., Magliulo M., Mallardi A., Intranuovo F., Mulla M.Y., Favia P., Vikholm-Lundin I., Torsi L. Detection beyond Debye's length with an electrolyte-gated organic field-effect transistor. Adv. Mater. (Deerfield Beach, Fla.) 2015;27(5):911–916. doi: 10.1002/adma.201403541. [DOI] [PubMed] [Google Scholar]
- 89.Chinnadayyala S.R., Park J., Kim Y.H., Choi S.H., Lee S.M., Cho W.W., Cho S. Electrochemical detection of C-reactive protein in human serum based on self-assembled monolayer-modified interdigitated wave-shaped electrode. Sensors. 2019;19(24):5560. doi: 10.3390/s19245560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bryan T., Luo X., Bueno P.R., Davis J.J. An optimised electrochemical biosensor for the label-free detection of C-reactive protein in blood. Biosens. Bioelectron. 2013;39(1):94–98. doi: 10.1016/j.bios.2012.06.051. [DOI] [PubMed] [Google Scholar]
- 91.Jampasa S., Siangproh W., Laocharoensuk R., Vilaivan T., Chailapakul O. Electrochemical detection of c-reactive protein based on anthraquinone-labeled antibody using a screen-printed graphene electrode. Talanta. 2018;183:311–319. doi: 10.1016/j.talanta.2018.02.075. [DOI] [PubMed] [Google Scholar]
- 92.Rajesh, Singal S., Kotnala R.K. Single frequency impedance analysis on reduced graphene oxide screen-printed electrode for biomolecular detection. Appl. Biochem. Biotechnol. 2017;183(2):672–683. doi: 10.1007/s12010-017-2510-8. [DOI] [PubMed] [Google Scholar]
- 93.Kowalczyk A., Sęk J.P., Kasprzak A., Poplawska M., Grudzinski I.P., Nowicka A.M. Occlusion phenomenon of redox probe by protein as a way of voltammetric detection of non-electroactive C-reactive protein. Biosens. Bioelectron. 2018;117:232–239. doi: 10.1016/j.bios.2018.06.019. [DOI] [PubMed] [Google Scholar]
- 94.Resende L.O., Castro A.C., Andrade A.O., Madurro J.M., Brito-Madurro A.G. Immunosensor for electrodetection of the C-reactive protein in serum. J. Solid State Electrochem. 2017;22(5):1365–1372. doi: 10.1007/s10008-017-3820-z. [DOI] [Google Scholar]
- 95.Justino C.I., Freitas A.C., Amaral J.P., Rocha-Santos T.A., Cardoso S., Duarte A.C. Disposable immunosensors for C-reactive protein based on carbon nanotubes field effect transistors. Talanta. 2013;108:165–170. doi: 10.1016/j.talanta.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 96.Akdis M., Burgler S., Crameri R., Eiwegger T., Fujita H., Gomez E., Klunker S., Meyer N., O'Mahony L., Palomares O., Rhyner C., Ouaked N., Schaffartzik A., Van De Veen W., Zeller S., Zimmermann M., Akdis C.A. Interleukins, from 1 to 37, and interferon-γ: receptors, functions, and roles in diseases. J. Allergy Clin. Immunol. 2011;127(3):701–721. doi: 10.1016/j.jaci.2010.11.050. e270. [DOI] [PubMed] [Google Scholar]
- 97.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., Tian D.S. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in wuhan, China. Clin. Infect. Dis: Off. Publ. Infect. Dise. Soc. Am. 2020;71(15):762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet (London, England) 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wong C.K., Lam C.W., Wu A.K., Ip W.K., Lee N.L., Chan I.H., Lit L.C., Hui D.S., Chan M.H., Chung S.S., Sung J.J. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin. Exp. Immunol. 2004;136(1):95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kim E.S., Choe P.G., Park W.B., Oh H.S., Kim E.J., Nam E.Y., Na S.H., Kim M., Song K.H., Bang J.H., Park S.W., Kim H.B., Kim N.J., Oh M.D. Clinical progression and cytokine profiles of Middle East respiratory syndrome coronavirus infection. J. Kor. Med. Sci. 2016;31(11):1717–1725. doi: 10.3346/jkms.2016.31.11.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. Jama. 2020;323(11):1061. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H., Zhang X., Zhang M., Wu S., Song J., Chen T., Han M., Li S., Luo X., Zhao J., Ning Q. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130(5):2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yang Y., Shen C., Li J., Yuan J., Yang M., Wang F., Liu Y. 2020. Exuberant Elevation of IP-10, MCP-3 and IL-1ra during SARS-CoV-2 Infection Is Associated with Disease Severity and Fatal Outcome. [DOI] [Google Scholar]
- 104.Chen X., Zhao B., Qu Y., Chen Y., Xiong J., Feng Y., Men D., Huang Q., Liu Y., Yang B., Ding J., Li F. Detectable serum SARS-CoV-2 viral load (RNAaemia) is closely correlated with drastically elevated interleukin 6 (IL-6) level in critically ill COVID-19 patients. Clin. Infect. Dise.: Off. Publ. Infect. Dise. Soc. Am. 2020:ciaa449. doi: 10.1093/cid/ciaa449. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dong T., Santos S., Yang Z., Yang S., Kirkhus N.E. Sputum and salivary protein biomarkers and point-of-care biosensors for the management of COPD. The Analyst. 2020;145(5):1583–1604. doi: 10.1039/c9an01704f. [DOI] [PubMed] [Google Scholar]
- 106.Wang W., He J., Lie P., Huang L., Wu S., Lin Y., Liu X. 2020. The definition and risks of cytokine release syndrome-like in 11 COVID-19-infected pneumonia critically ill patients: disease characteristics and retrospective analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Crisafulli S., Isgrò V., La Corte L., Atzeni F., Trifirò G. Potential role of anti-interleukin (IL)-6 drugs in the treatment of COVID-19: rationale, clinical evidence and risks. BioDrugs : Clin. Immunotherapeut. Biopharmaceut. Gene Ther. 2020;34(4):415–422. doi: 10.1007/s40259-020-00430-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Atal S., Fatima Z. IL-6 inhibitors in the treatment of serious COVID-19: a promising therapy? Pharmaceut. Med. 2020;34(4):223–231. doi: 10.1007/s40290-020-00342-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bagan L., Sáez G.T., Tormos M.C., Labaig-Rueda C., Murillo-Cortes J., Bagan J.V. Salivary and serum interleukin-6 levels in proliferative verrucous leukoplakia. Clin. Oral Invest. 2016;20(4):737–743. doi: 10.1007/s00784-015-1551-z. [DOI] [PubMed] [Google Scholar]
- 110.Hladek M.D., Szanton S.L., Cho Y.E., Lai C., Sacko C., Roberts L., Gill J. Using sweat to measure cytokines in older adults compared to younger adults: a pilot study. J. Immunol. Methods. 2018;454:1–5. doi: 10.1016/j.jim.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bucchioni E., Kharitonov S.A., Allegra L., Barnes P.J. High levels of interleukin-6 in the exhaled breath condensate of patients with COPD. Respir. Med. 2003;97(12):1299–1302. doi: 10.1016/j.rmed.2003.07.008. [DOI] [PubMed] [Google Scholar]
- 112.Coomes E.A., Haghbayan H. Interleukin-6 in Covid-19: a systematic review and meta-analysis. Rev. Med. Virol. 2020;30(6):1–9. doi: 10.1002/rmv.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Russell S.M., Alba-Patiño A., Barón E., Borges M., Gonzalez-Freire M., Rica R.D. Biosensors for managing the COVID-19 cytokine storm: challenges ahead. ACS Sens. 2020;5(6):1506–1513. doi: 10.1021/acssensors.0c00979. [DOI] [PubMed] [Google Scholar]
- 114.Smith P., Hobisch A., Lin D., Culig Z., Keller E. Interleukin-6 and prostate cancer progression. Cytokine Growth Factor Rev. 2001;12(1):33–40. doi: 10.1016/s1359-6101(00)00021-6. [DOI] [PubMed] [Google Scholar]
- 115.Harrison S.C., Smith A.J., Jones G.T., Swerdlow D.I., Rampuri R., Bown M.J., Humphries S.E. Interleukin-6 receptor pathways in abdominal aortic aneurysm. Eur. Heart J. 2012;34(48):3707–3716. doi: 10.1093/eurheartj/ehs354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chung K.F. Cytokines in chronic obstructive pulmonary disease. Eur. Respir. J. Suppl. 2001;34:50s–59s. [PubMed] [Google Scholar]
- 117.Chen H., Li J., Zhang X., Li X., Yao M., Zheng G. Automated in vivo nanosensing of breath-borne protein biomarkers. Nano Lett. 2018;18(8):4716–4726. doi: 10.1021/acs.nanolett.8b01070. [DOI] [PubMed] [Google Scholar]
- 118.Vainer N., Dehlendorff C., Johansen J.S. Systematic literature review of IL-6 as a biomarker or treatment target in patients with gastric, bile duct, pancreatic and colorectal cancer. Oncotarget. 2018;9(51):29820. doi: 10.18632/oncotarget.25661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Baraket A., Lee M., Zine N., Sigaud M., Bausells J., Errachid A. A fully integrated electrochemical biosensor platform fabrication process for cytokines detection. Biosens. Bioelectron. 2017;93:170–175. doi: 10.1016/j.bios.2016.09.023. [DOI] [PubMed] [Google Scholar]
- 120.Russell C., Ward A.C., Vezza V., Hoskisson P., Alcorn D., Steenson D.P., Corrigan D.K. Development of a needle shaped microelectrode for electrochemical detection of the sepsis biomarker interleukin-6 (IL-6) in real time. Biosens. Bioelectron. 2019;126:806–814. doi: 10.1016/j.bios.2018.11.053. [DOI] [PubMed] [Google Scholar]
- 121.Hao Z., Pan Y., Shao W., Lin Q., Zhao X. Graphene-based fully integrated portable nanosensing system for on-line detection of cytokine biomarkers in saliva. Biosens. Bioelectron. 2019;134:16–23. doi: 10.1016/j.bios.2019.03.053. [DOI] [PubMed] [Google Scholar]
- 122.Tertiş M., Melinte G., Ciui B., Şimon I., Ştiufiuc R., Săndulescu R., Cristea C. A novel label free electrochemical magnetoimmunosensor for human interleukin-6 quantification in serum. Electroanalysis. 2018;31(2):282–292. doi: 10.1002/elan.201800620. [DOI] [Google Scholar]
- 123.Tertiş M., Ciui B., Suciu M., Săndulescu R., Cristea C. Label-free electrochemical aptasensor based on gold and polypyrrole nanoparticles for interleukin 6 detection. Electrochim. Acta. 2017;258:1208–1218. doi: 10.1016/j.electacta.2017.11.176. [DOI] [Google Scholar]
- 124.Khosravi F., Loeian S.M., Panchapakesan B. Ultrasensitive label-free sensing of IL-6 based on PASE functionalized carbon nanotube micro-arrays with RNA-aptamers as molecular recognition elements. Biosensors. 2017;7(2):17. doi: 10.3390/bios7020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Parameswaran N., Patial S. Tumor necrosis factor-α signaling in macrophages. Crit. Rev. Eukaryot. Gene Expr. 2010;20(2):87–103. doi: 10.1615/critreveukargeneexpr.v20.i2.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Akdis M., Aab A., Altunbulakli C., Azkur K., Costa R.A., Crameri R., Duan S., Eiwegger T., Eljaszewicz A., Ferstl R., Frei R., Garbani M., Globinska A., Hess L., Huitema C., Kubo T., Komlosi Z., Konieczna P., Kovacs N., Kucuksezer U.C., Akdis C.A. Interleukins (from IL-1 to IL-38), interferons, transforming growth factor β, and TNF-α: receptors, functions, and roles in diseases. J. Allergy Clin. Immunol. 2016;138(4):984–1010. doi: 10.1016/j.jaci.2016.06.033. [DOI] [PubMed] [Google Scholar]
- 127.Gerriets V., Bansal P., Khaddour K. StatPearls [Internet] StatPearls Publishing; 2019. Tumor necrosis factor (TNF) inhibitors. [PubMed] [Google Scholar]
- 128.Liu Y., Zhou Q., Revzin A. An aptasensor for electrochemical detection of tumor necrosis factor in human blood. The Analyst. 2013;138(15):4321. doi: 10.1039/c3an00818e. [DOI] [PubMed] [Google Scholar]
- 129.Liu Y., Liu Y., Matharu Z., Rahimian A., Revzin A. Detecting multiple cell-secreted cytokines from the same aptamer-functionalized electrode. Biosens. Bioelectron. 2015;64:43–50. doi: 10.1016/j.bios.2014.08.034. [DOI] [PubMed] [Google Scholar]
- 130.Miao P., Yang D., Chen X., Guo Z., Tang Y. Voltammetric determination of tumor necrosis factor-α based on the use of an aptamer and magnetic nanoparticles loaded with gold nanoparticles. Microchimica Acta. 2017;184(10):3901–3907. [Google Scholar]
- 131.Filik H., Avan A.A. Electrochemical immunosensors for the detection of cytokine tumor necrosis factor alpha: a review. Talanta. 2020;211:120758. doi: 10.1016/j.talanta.2020.120758. [DOI] [PubMed] [Google Scholar]
- 132.Mazloum-Ardakani M., Hosseinzadeh L., Khoshroo A. Label-free electrochemical immunosensor for detection of tumor necrosis factor α based on fullerene-functionalized carbon nanotubes/ionic liquid. J. Electroanal. Chem. 2015;757:58–64. doi: 10.1016/j.jelechem.2015.09.006. [DOI] [Google Scholar]
- 133.Weng S., Chen M., Zhao C., Liu A., Lin L., Liu Q., Lin X. Label-free electrochemical immunosensor based on K3[Fe(CN)6] as signal for facile and sensitive determination of tumor necrosis factor-alpha. Sensor. Actuator. B Chem. 2013;184:1–7. doi: 10.1016/j.snb.2013.03.141. [DOI] [Google Scholar]
- 134.Belle J.T., Demirok U.K., Patel D.R., Cook C.B. Development of a novel single sensor multiplexed marker assay. The Analyst. 2011;136(7):1496. doi: 10.1039/c0an00923g. [DOI] [PubMed] [Google Scholar]
- 135.Wang Z., Hao Z., Yu S., Huang C., Pan Y., Zhao X. A wearable and deformable graphene-based affinity nanosensor for monitoring of cytokines in biofluids. Nanomaterials. 2020;10(8):1503. doi: 10.3390/nano10081503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pui T.S., Kongsuphol P., Arya S.K., Bansal T. Detection of tumor necrosis factor (TNF-α) in cell culture medium with label free electrochemical impedance spectroscopy. Sensor. Actuator. B Chem. 2013;181:494–500. doi: 10.1016/j.snb.2013.02.019. [DOI] [Google Scholar]
- 137.Sen G.C. Viruses and interferons. Annu. Rev. Microbiol. 2001;55(1):255–281. doi: 10.1146/annurev.micro.55.1.255. [DOI] [PubMed] [Google Scholar]
- 138.Morris A.G. Interferons. Immunology. 1988;64(Suppl 1):43. [PubMed] [Google Scholar]
- 139.Sallard E., Lescure F.X., Yazdanpanah Y., Mentre F., Peiffer-Smadja N., Florence A.D.E.R., Bouadma L. Type 1 Interferons as a Potential Treatment against COVID-19. Anti. Res. 2020:104791. doi: 10.1016/j.antiviral.2020.104791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Trouillet-Assant S., Viel S., Gaymard A., Pons S., Richard J.C., Perret M., Villard M., Brengel-Pesce K., Lina B., Mezidi M., Bitker L., Belot A., COVID HCL Study group Type I IFN immunoprofiling in COVID-19 patients. J. Allergy Clin. Immunol. 2020;146(1):206–208. doi: 10.1016/j.jaci.2020.04.029. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mazloum-Ardakani M., Hosseinzadeh L. A sensitive electrochemical aptasensor for TNF-α based on bimetallic Ag@ Pt core-shell nanoparticle functionalized graphene nanostructures as labels for signal amplification. J. Electrochem. Soc. 2016;163(5):B119. [Google Scholar]
- 142.Valverde A., Serafín V., Garoz J., Montero-Calle A., González-Cortés A., Arenas M., Pingarrón J.M. Electrochemical immunoplatform to improve the reliability of breast cancer diagnosis through the simultaneous determination of RANKL and TNF in serum. Sensor. Actuator. B Chem. 2020:128096. [Google Scholar]
- 143.Sergeyeva T., Soldatkin A., Rachkov A., Tereschenko M., Piletsky S., El`skaya A. β-Lactamase label-based potentiometric biosensor for α-2 interferon detection. Anal. Chim. Acta. 1999;390(1–3):73–81. doi: 10.1016/s0003-2670(99)00177-4. [DOI] [Google Scholar]
- 144.Berkowicz D.A., Trombley P.Q., Shepherd G.M. Evidence for glutamate as the olfactory receptor cell neurotransmitter. J. Neurophysiol. 1994;71(6):2557–2561. doi: 10.1152/jn.1994.71.6.2557. [DOI] [PubMed] [Google Scholar]
- 145.Abuin L., Bargeton B., Ulbrich M.H., Isacoff E.Y., Kellenberger S., Benton R. Functional architecture of olfactory ionotropic glutamate receptors. Neuron. 2011;69(1):44–60. doi: 10.1016/j.neuron.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Brison E., Jacomy H., Desforges M., Talbot P.J. Glutamate excitotoxicity is involved in the induction of paralysis in mice after infection by a human coronavirus with a single point mutation in its spike protein. J. Virol. 2011;85(23):12464–12473. doi: 10.1128/jvi.05576-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wu Y., Xu X., Chen Z., Duan J., Hashimoto K., Yang L., Yang C. Nervous system involvement after infection with COVID-19 and other coronaviruses. Brain Behav. Immun. 2020;87:18–22. doi: 10.1016/j.bbi.2020.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Brann D.H., Tsukahara T., Weinreb C., Lipovsek M., Van den Berge K., Gong B., Datta S.R. Non-neuronal expression of sars-cov-2 entry genes in the olfactory system suggests mechanisms underlying covid-19-associated anosmia. Sci. Adv. 2020;6(31) doi: 10.1126/sciadv.abc5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Li Y.C., Bai W.Z., Hashikawa T. The neuroinvasive potential of SARS-CoV2 may play a role in the respiratory failure of COVID-19 patients. J. Med. Virol. 2020;92(6):552–555. doi: 10.1002/jmv.25728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Jamal M., Hasan M., Mathewson A., Razeeb K.M. Disposable sensor based on enzyme-free Ni nanowire array electrode to detect glutamate. Biosens. Bioelectron. 2013;40(1):213–218. doi: 10.1016/j.bios.2012.07.024. [DOI] [PubMed] [Google Scholar]
- 151.Ganesana M., Trikantzopoulos E., Maniar Y., Lee S.T., Venton B.J. Development of a novel micro biosensor for in vivo monitoring of glutamate release in the brain. Biosens. Bioelectron. 2019;130:103–109. doi: 10.1016/j.bios.2019.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hussain M.M., Rahman M.M., Asiri A.M., Awual M.R. Non-enzymatic simultaneous detection of l-glutamic acid and uric acid using mesoporous Co3O4 nanosheets. RSC Adv. 2016;6(84):80511–80521. doi: 10.1039/c6ra12256f. [DOI] [Google Scholar]
- 153.Jamal M., Chakrabarty S., Shao H., Mcnulty D., Yousuf M.A., Furukawa H., Razeeb K.M. A non enzymatic glutamate sensor based on nickel oxide nanoparticle. Microsyst. Technol. 2018;24(10):4217–4223. doi: 10.1007/s00542-018-3724-6. [DOI] [Google Scholar]
- 154.Batra B., Yadav M., Pundir C.S. L-Glutamate biosensor based on l-glutamate oxidase immobilized onto ZnO nanorods/polypyrrole modified pencil graphite electrode. Biochem. Eng. J. 2016;105:428–436. doi: 10.1016/j.bej.2015.10.012. [DOI] [Google Scholar]
- 155.Scoggin J.L., Tan C., Nguyen N.H., Kansakar U., Madadi M., Siddiqui S., Arumugam P.U., DeCoster M.A., Murray T.A. An enzyme-based electrochemical biosensor probe with sensitivity to detect astrocytic versus glioma uptake of glutamate in real time in vitro. Biosens. Bioelectron. 2019;126:751–757. doi: 10.1016/j.bios.2018.11.023. [DOI] [PubMed] [Google Scholar]
- 156.Hunt J. Exhaled breath condensate pH assays. Immunol. Allergy Clin. 2007;27(4):597. doi: 10.1016/j.iac.2007.09.006. vi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gessner C., Hammerschmidt S., Kuhn H., Seyfarth H., Sack U., Engelmann L., Wirtz H. Exhaled breath condensate acidification in acute lung injury. Respir. Med. 2003;97(11):1188–1194. doi: 10.1016/s0954-6111(03)00225-7. [DOI] [PubMed] [Google Scholar]
- 158.Sack U., Scheibe R., Wötzel M., Hammerschmidt S., Kuhn H., Emmrich F., Hoheisel G., Wirtz H., Gessner C. Multiplex analysis of cytokines in exhaled breath condensate. Cytometr. Part A : J. Inter. Soc. Analyt. Cytol. 2006;69(3):169–172. doi: 10.1002/cyto.a.20231. [DOI] [PubMed] [Google Scholar]
- 159.Montuschi P. Analysis of exhaled breath condensate in respiratory medicine: methodological aspects and potential clinical applications. Ther. Adv. Respir. Dis. 2007;1(1):5–23. doi: 10.1177/1753465807082373. [DOI] [PubMed] [Google Scholar]
- 160.Buszewski B., Grzywinski D., Ligor T., Stacewicz T., Bielecki Z., Wojtas J. Detection of volatile organic compounds as biomarkers in breath analysis by different analytical techniques. Bioanalysis. 2013;5(18):2287–2306. doi: 10.4155/bio.13.183. [DOI] [PubMed] [Google Scholar]
- 161.Carter S.R., Davis C.S., Kovacs E.J. Exhaled breath condensate collection in the mechanically ventilated patient. Respir. Med. 2012;106(5):601–613. doi: 10.1016/j.rmed.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lin J.L., Bonnichsen M.H., Thomas P.S. Standardization of exhaled breath condensate: effects of different de-aeration protocols on pH and H₂O₂ concentrations. J. Breath Res. 2011;5(1) doi: 10.1088/1752-7155/5/1/011001. [DOI] [PubMed] [Google Scholar]
- 163.Walsh B.K., Mackey D.J., Pajewski T., Yu Y., Gaston B.M., Hunt J.F. Exhaled-breath condensate pH can be safely and continuously monitored in mechanically ventilated patients. Respir. Care. 2006;51(10):1125–1131. [PubMed] [Google Scholar]
- 164.Vaschetto R., Corradi M., Goldoni M., Cancelliere L., Pulvirenti S., Fazzini U., Navalesi P. Sampling and analyzing alveolar exhaled breath condensate in mechanically ventilated patients: a feasibility study. J. Breath Res. 2015;9(4) doi: 10.1088/1752-7155/9/4/047106. [DOI] [PubMed] [Google Scholar]
- 165.Prince P., Boulay M.E., Boulet L.P. A fast, simple, and inexpensive method to collect exhaled breath condensate for pH determination. Ann. Allergy Asthma Immunol. 2006;97(5):622–627. doi: 10.1016/S1081-1206(10)61091-5. [DOI] [PubMed] [Google Scholar]
- 166.Henry B.M., de Oliveira M., Benoit S., Plebani M., Lippi G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin. Chem. Lab. Med. 2020;58(7):1021–1028. doi: 10.1515/cclm-2020-0369. [DOI] [PubMed] [Google Scholar]
- 167.Chan A.S., Rout A. Use of neutrophil-to-lymphocyte and platelet-to-lymphocyte ratios in COVID-19. J. Clin. Med. Res. 2020;12(7):448–453. doi: 10.14740/jocmr4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yip T.T., Chan J.W., Cho W.C., Yip T.T., Wang Z., Kwan T.L., Law S.C., Tsang D.N., Chan J.K., Lee K.C., Cheng W.W., Ma V.W., Yip C., Lim C.K., Ngan R.K., Au J.S., Chan A., Lim W.W., Ciphergen SARS Proteomics Study Group Protein chip array profiling analysis in patients with severe acute respiratory syndrome identified serum amyloid a protein as a biomarker potentially useful in monitoring the extent of pneumonia. Clin. Chem. 2005;51(1):47–55. doi: 10.1373/clinchem.2004.031229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ye Z., Wang Y., Colunga-Lozano L.E., Prasad M., Tangamornsuksan W., Rochwerg B., Yao L., Motaghi S., Couban R.J., Ghadimi M., Bala M.M., Gomaa H., Fang F., Xiao Y., Guyatt G.H. Efficacy and safety of corticosteroids in COVID-19 based on evidence for COVID-19, other coronavirus infections, influenza, community-acquired pneumonia and acute respiratory distress syndrome: a systematic review and meta-analysis. CMAJ (Can. Med. Assoc. J.) : Canad. Med. Asso. J. = J. de l'Asso. Med. Canad. 2020;192(27):E756–E767. doi: 10.1503/cmaj.200645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sheianov M.V., Udalov Y.D., Ochkin S.S., Bashkov A.N., Samoilov A.S. Pulse therapy with corticosteroids and intravenous immunoglobulin in the management of severe tocilizumab-resistant COVID-19: a report of three clinical cases. Cureus. 2020;12(7) doi: 10.7759/cureus.9038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wang X., Lin G., Cui G., Zhou X., Liu G.L. White blood cell counting on smartphone paper electrochemical sensor. Biosens. Bioelectron. 2017;90:549–557. doi: 10.1016/j.bios.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 172.Fan B.E., Chong V., Chan S., Lim G.H., Lim K., Tan G.B., Mucheli S.S., Kuperan P., Ong K.H. Hematologic parameters in patients with COVID-19 infection. Am. J. Hematol. 2020;95(6):E131–E134. doi: 10.1002/ajh.25774. [DOI] [PubMed] [Google Scholar]
- 173.Carinelli S., Xufré Ballesteros C., Martí M., Alegret S., Pividori M.I. Electrochemical magneto-actuated biosensor for CD4 count in AIDS diagnosis and monitoring. Biosens. Bioelectron. 2015;74:974–980. doi: 10.1016/j.bios.2015.07.053. [DOI] [PubMed] [Google Scholar]
- 174.Jiang X., Spencer M.G. Electrochemical impedance biosensor with electrode pixels for precise counting of CD4 cells: a microchip for quantitative diagnosis of HIV infection status of AIDS patients. Biosens. Bioelectron. 2010;25(7):1622–1628. doi: 10.1016/j.bios.2009.11.024. [DOI] [PubMed] [Google Scholar]
- 175.Kim J., Park G., Lee S., Hwang S.W., Min N., Lee K.M. Single wall carbon nanotube electrode system capable of quantitative detection of CD4+ T cells. Biosens. Bioelectron. 2017;90:238–244. doi: 10.1016/j.bios.2016.11.055. [DOI] [PubMed] [Google Scholar]
- 176.Zhe J., Jagtiani A., Dutta P., Hu J., Carletta J. A micromachined high throughput Coulter counter for bioparticle detection and counting. J. Micromech. Microeng. 2007;17(2):304–313. doi: 10.1088/0960-1317/17/2/017. [DOI] [Google Scholar]
- 177.Wexels F., Seljeflot I., Pripp A.H., Dahl O.E. D-Dimer and prothrombin fragment 1+ 2 in urine and plasma in patients with clinically suspected venous thromboembolism. Blood Coagul. Fibrinolysis. 2016;27(4):396–400. doi: 10.1097/MBC.0000000000000461. [DOI] [PubMed] [Google Scholar]
- 178.Zhang L., Yan X., Fan Q., Liu H., Liu X., Liu Z., Zhang Z. D-dimer levels on admission to predict in-hospital mortality in patients with Covid-19. J. Thromb. Haemostasis : JTH. 2020;18(6):1324–1329. doi: 10.1111/jth.14859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lippi G., Favaloro E.J. D-Dimer is associated with severity of coronavirus disease 2019: a pooled analysis. Thromb. Haemostasis. 2020;120(5):876–878. doi: 10.1055/s-0040-1709650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Xie J., Zu Y., Alkhatib A., Pham T.T., Gill F., Jang A., Radosta S., Chaaya G., Myers L., Zifodya J.S., Bojanowski C.M., Marrouche N.F., Mauvais-Jarvis F., Denson J.L. Metabolic syndrome and COVID-19 mortality among adult black patients in new orleans. Diab. Care. 2020;44(1):188–193. doi: 10.2337/dc20-1714. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wu Q., Zhou L., Sun X., Yan Z., Hu C., Wu J., Xu L., Li X., Liu H., Yin P., Li K., Zhao J., Li Y., Wang X., Li Y., Zhang Q., Xu G., Chen H. Altered lipid metabolism in recovered SARS patients twelve years after infection. Sci. Rep. 2017;7(1):9110. doi: 10.1038/s41598-017-09536-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wu L., O'Kane A.M., Peng H., Bi Y., Motriuk-Smith D., Ren J. SARS-CoV-2 and cardiovascular complications: from molecular mechanisms to pharmaceutical management. Biochem. Pharmacol. 2020:114114. doi: 10.1016/j.bcp.2020.114114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Bourigua S., Hnaien M., Bessueille F., Lagarde F., Dzyadevych S., Maaref A., Bausells J., Errachid A., Jaffrezic Renault N. Impedimetric immunosensor based on SWCNT-COOH modified gold microelectrodes for label-free detection of deep venous thrombosis biomarker. Biosens. Bioelectron. 2010;26(4):1278–1282. doi: 10.1016/j.bios.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 184.Li S., Jiang Y., Eda S., Wu J.J. Low-cost and desktop-fabricated biosensor for rapid and sensitive detection of circulating D-dimer biomarker. IEEE Sensor. J. 2018;19(4):1245–1251. [Google Scholar]
- 185.Ibupoto Z., Jamal N., Khun K., Liu X., Willander M. A potentiometric immunosensor based on silver nanoparticles decorated ZnO nanotubes, for the selective detection of d-dimer. Sensor. Actuator. B Chem. 2013;182:104–111. doi: 10.1016/j.snb.2013.02.084. [DOI] [Google Scholar]
- 186.Chebil S., Hafaiedh I., Sauriat-Dorizon H., Jaffrezic-Renault N., Errachid A., Ali Z., Korri-Youssoufi H. Electrochemical detection of D-dimer as deep vein thrombosis marker using single-chain D-dimer antibody immobilized on functionalized polypyrrole. Biosens. Bioelectron. 2010;26(2):736–742. doi: 10.1016/j.bios.2010.06.048. [DOI] [PubMed] [Google Scholar]
- 187.Gamella M., Campuzano S., Conzuelo F., Reviejo A.J., Pingarrón J.M. Amperometric magnetoimmunosensors for direct determination of D-dimer in human serum. Electroanalysis. 2012;24(12):2235–2243. doi: 10.1002/elan.201200503. [DOI] [Google Scholar]
- 188.Chang C.K., Hou M.H., Chang C.F., Hsiao C.D., Huang T.H. The SARS coronavirus nucleocapsid protein-forms and functions. Antivir. Res. 2014;103:39–50. doi: 10.1016/j.antiviral.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Vadlamani B.S., Uppal T., Verma S.C., Misra M. Functionalized TiO2 nanotube-based electrochemical biosensor for rapid detection of SARS-CoV-2. Sensors. 2020;20(20):5871. doi: 10.3390/s20205871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Seo G., Lee G., Kim M.J., Baek S.H., Choi M., Ku K.B., Lee C.S., Jun S., Park D., Kim H.G., Kim S.J., Lee J.O., Kim B.T., Park E.C., Kim S.I. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 191.Torrente-Rodríguez R.M., Lukas H., Tu J., Min J., Yang Y., Xu C., Gao W. SARS-CoV-2 RapidPlex: a graphene-based multiplexed telemedicine platform for rapid and low-cost COVID-19 diagnosis and monitoring. Matter. 2020;3(6):1981–1998. doi: 10.1016/j.matt.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W., Zheng S. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020;58(6) doi: 10.1128/jcm.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Tripathy S., Singh S.G. Label-free electrochemical detection of DNA hybridization: a method for COVID-19 diagnosis. Transact. Indian Nat. Acad. Eng. 2020;5(2):205–209. doi: 10.1007/s41403-020-00103-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Alafeef M., Dighe K., Moitra P., Pan D. Rapid, ultrasensitive, and quantitative detection of SARS-CoV-2 using antisense oligonucleotides directed electrochemical biosensor chip. ACS Nano. 2020 doi: 10.1021/acsnano.0c06392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Huang P.L. A comprehensive definition for metabolic syndrome. Dise. Model Mech. 2009;2(5–6):231–237. doi: 10.1242/dmm.001180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.de Kloet A.D., Krause E.G., Woods S.C. The renin angiotensin system and the metabolic syndrome. Physiol. Behav. 2010;100(5):525–534. doi: 10.1016/j.physbeh.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Bansal M. Cardiovascular disease and COVID-19. Diab. Metabol. Syndr. 2020;14(3):247–250. doi: 10.1016/j.dsx.2020.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Marques P., Collado A., Martinez-Hervás S., Domingo E., Benito E., Piqueras L., Real J.T., Ascaso J.F., Sanz M.J. Systemic inflammation in metabolic syndrome: increased platelet and leukocyte activation, and key role of CX3CL1/CX3CR1 and CCL2/CCR2 axes in arterial platelet-proinflammatory monocyte adhesion. J. Clin. Med. 2019;8(5):708. doi: 10.3390/jcm8050708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.de Lucena T., da Silva Santos A.F., de Lima B.R., de Albuquerque Borborema M.E., de Azevêdo Silva J. Mechanism of inflammatory response in associated comorbidities in COVID-19. Diab. Metabol. Syndr. 2020;14(4):597–600. doi: 10.1016/j.dsx.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Abdul Rehman S., Khurshid Z., Hussain Niazi F., Naseem M., Al Waddani H., Sahibzada H.A., Sannam Khan R. Role of salivary biomarkers in detection of cardiovascular diseases (CVD) Proteomes. 2017;5(3):21. doi: 10.3390/proteomes5030021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Fontes J.A., Rose N.R., Čiháková D. The varying faces of IL-6: from cardiac protection to cardiac failure. Cytokine. 2015;74(1):62–68. doi: 10.1016/j.cyto.2014.12.024. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Meldrum D.R. Tumor necrosis factor in the heart. Am. J. Physiol. 1998;274(3):R577–R595. doi: 10.1152/ajpregu.1998.274.3.R577. [DOI] [PubMed] [Google Scholar]
- 203.Ferrari R. The role of TNF in cardiovascular disease. Pharmacol. Res. 1999;40(2):97–105. doi: 10.1006/phrs.1998.0463. [DOI] [PubMed] [Google Scholar]
- 204.Cozlea D.L., Farcas D.M., Nagy A., Keresztesi A.A., Tifrea R., Cozlea L., Carașca E. The impact of C reactive protein on global cardiovascular risk on patients with coronary artery disease. Curr. Health Sci. J. 2013;39(4):225–231. [PMC free article] [PubMed] [Google Scholar]
- 205.Khan R.S., Khurshid Z., Yahya Ibrahim Asiri F. Advancing point-of-care (PoC) testing using human saliva as liquid biopsy. Diagnostics. 2017;7(3):39. doi: 10.3390/diagnostics7030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Global overview of drug demand and supply . 2017. World Drug Report World Drug Report 2017. 1-66. [DOI] [Google Scholar]
- 207.Vemuri V.K., Makriyannis A. Medicinal chemistry of cannabinoids. Clin. Pharmacol. Therapeut. 2015;97(6):553–558. doi: 10.1002/cpt.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Grotenhermen F. Pharmacokinetics and pharmacodynamics of cannabinoids. Clin. Pharmacokinet. 2003;42(4):327–360. doi: 10.2165/00003088-200342040-00003. [DOI] [PubMed] [Google Scholar]
- 209.Mannucci C., Navarra M., Calapai F., Spagnolo E.V., Busardò F.P., Cas R.D., Calapai G. Neurological aspects of medical use of cannabidiol. CNS Neurol. Disord. - Drug Targets. 2017;16(5):541–553. doi: 10.2174/1871527316666170413114210. [DOI] [PubMed] [Google Scholar]
- 210.Mechoulam R., Peters M., Murillo-Rodriguez E., Hanuš L.O. Cannabidiol–recent advances. Chem. Biodivers. 2007;4(8):1678–1692. doi: 10.1002/cbdv.200790147. [DOI] [PubMed] [Google Scholar]
- 211.Russo E.B. Cannabidiol claims and misconceptions. Trends Pharmacol. Sci. 2017;38(3):198–201. doi: 10.1016/j.tips.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 212.Campos A.C., Fogaça M.V., Sonego A.B., Guimarães F.S. Cannabidiol, neuroprotection and neuropsychiatric disorders. Pharmacol. Res. 2016;112:119–127. doi: 10.1016/j.phrs.2016.01.033. [DOI] [PubMed] [Google Scholar]
- 213.Vaninov N. In the eye of the COVID-19 cytokine storm. Nat. Rev. Immunol. 2020;20(5) doi: 10.1038/s41577-020-0305-6. 277-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020:1–12. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Ryz N.R., Remillard D.J., Russo E.B. Cannabis roots: a traditional therapy with future potential for treating inflammation and pain. Cann. Cannabinoid Res. 2017;2(1):210–216. doi: 10.1089/can.2017.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Izzo A.A., Capasso R., Aviello G., Borrelli F., Romano B., Piscitelli F., Di Marzo V. Inhibitory effect of cannabichromene, a major non-psychotropic cannabinoid extracted from Cannabis sativa, on inflammation-induced hypermotility in mice. Br. J. Pharmacol. 2012;166(4):1444–1460. doi: 10.1111/j.1476-5381.2012.01879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Costa B., Colleoni M., Conti S., Parolaro D., Franke C., Trovato A.E., Giagnoni G. Oral anti-inflammatory activity of cannabidiol, a non-psychoactive constituent of cannabis, in acute carrageenan-induced inflammation in the rat paw. N. Schmied. Arch. Pharmacol. 2004;369(3):294–299. doi: 10.1007/s00210-004-0871-3. [DOI] [PubMed] [Google Scholar]
- 218.[a] Pellati F., Borgonetti V., Brighenti V., Biagi M., Benvenuti S., Corsi L. Cannabis sativa L. and nonpsychoactive cannabinoids: their chemistry and role against oxidative stress, inflammation, and cancer. BioMed Res. Int. 2018;2018 doi: 10.1155/2018/1691428. [DOI] [PMC free article] [PubMed] [Google Scholar]; [b] Abrams D.I., Guzman M. Cannabis in cancer care. Clin. Pharmacol. Therapeut. 2015;97(6):575–586. doi: 10.1002/cpt.108. [DOI] [PubMed] [Google Scholar]
- 219.Costiniuk C.T., Jenabian M.A. Acute inflammation and pathogenesis of SARS-CoV-2 infection: cannabidiol as a potential anti-inflammatory treatment? Cytokine Growth Factor Rev. 2020;53:63–65. doi: 10.1016/j.cytogfr.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Rossi F., Tortora C., Argenziano M., Di Paola A., Punzo F. Cannabinoid receptor type 2: a possible target in SARS-CoV-2 (CoV-19) infection? Int. J. Mol. Sci. 2020;21(11):3809. doi: 10.3390/ijms21113809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Abrams D.I., Guzman M. Cannabis in cancer care. Clin. Pharmacol. Ther. 2015;97(6):575–586. doi: 10.1002/cpt.108. [DOI] [PubMed] [Google Scholar]
- 222.Abrams D.I. Integrating cannabis into clinical cancer care. Curr. Oncol. 2016;23(Suppl 2):S8. doi: 10.3747/co.23.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Lutge E.E., Gray A., Siegfried N. The medical use of cannabis for reducing morbidity and mortality in patients with HIV/AIDS. Cochrane Database Syst. Rev. 2013;(4) doi: 10.1002/14651858.CD005175.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Woolridge E., Barton S., Samuel J., Osorio J., Dougherty A., Holdcroft A. Cannabis use in HIV for pain and other medical symptoms. J. Pain Symptom Manag. 2005;29(4):358–367. doi: 10.1016/j.jpainsymman.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 225.Corey-Bloom J., Wolfson T., Gamst A., Jin S., Marcotte T.D., Bentley H., Gouaux B. Smoked cannabis for spasticity in multiple sclerosis: a randomized, placebo-controlled trial. CMAJ (Can. Med. Assoc. J.) 2012;184(10):1143–1150. doi: 10.1503/cmaj.110837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Walsh Z., Gonzalez R., Crosby K., Thiessen M.S., Carroll C., Bonn-Miller M.O. Medical cannabis and mental health: a guided systematic review. Clin. Psychol. Rev. 2017;51:15–29. doi: 10.1016/j.cpr.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 227.Rieder S.A., Chauhan A., Singh U., Nagarkatti M., Nagarkatti P. Cannabinoid-induced apoptosis in immune cells as a pathway to immunosuppression. Immunobiology. 2010;215(8):598–605. doi: 10.1016/j.imbio.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Hill K.P. Cannabinoids and the coronavirus. Cannabis Cannabinoid Res. 2020;5(2):118–120. doi: 10.1089/can.2020.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Turcotte C., Blanchet M.R., Laviolette M., Flamand N. Impact of cannabis, cannabinoids, and endocannabinoids in the lungs. Front. Pharmacol. 2016;7:317. doi: 10.3389/fphar.2016.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Lechien J.R., Chiesa-Estomba C.M., De Siati D.R., Horoi M., Le Bon S.D., Rodriguez A., Dequanter D., Blecic S., El Afia F., Distinguin L., Chekkoury-Idrissi Y., Hans S., Delgado I.L., Calvo-Henriquez C., Lavigne P., Falanga C., Barillari M.R., Cammaroto G., Khalife M., Leich P., Saussez S. Olfactory and gustatory dysfunctions as a clinical presentation of mild-to-moderate forms of the coronavirus disease (COVID-19): a multicenter European study. Eur. Arch. Oto-Rhino-Laryngol. : Off. J. Europ. Feder. Oto-Rhino-Laryngol. Soc. (EUFOS) : affil. German Soc. Oto-Rhino-Laryngol. - Head Neck Surg. 2020;277(8):2251–2261. doi: 10.1007/s00405-020-05965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Poyiadji N., Shahin G., Noujaim D., Stone M., Patel S., Griffith B. COVID-19-associated acute hemorrhagic necrotizing encephalopathy: imaging features. Radiology. 2020;296(2):E119–E120. doi: 10.1148/radiol.2020201187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Toscano G., et al. Guillain–barré syndrome associated with SARS-CoV-2. N. Engl. J. Med. 2020 doi: 10.1056/nejmc2009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Watson C.W.M., Paolillo E.W., Morgan E.E., Umlauf A., Sundermann E.E., Ellis R.J., Grant I. Cannabis exposure is associated with a lower likelihood of neurocognitive impairment in people living with HIV. JAIDS J. Acqui. Immun. Def. Syndr. 2020;83(1):56–64. doi: 10.1097/QAI.0000000000002211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Mecha M., Feliú A., Iñigo P.M., Mestre L., Carrillo-Salinas F.J., Guaza C. Cannabidiol provides long-lasting protection against the deleterious effects of inflammation in a viral model of multiple sclerosis: a role for A2A receptors. Neurobiol. Dis. 2013;59:141–150. doi: 10.1016/j.nbd.2013.06.016. [DOI] [PubMed] [Google Scholar]
- 235.Lowe H.I., Toyang N.J., McLaughlin W. Potential of cannabidiol for the treatment of viral hepatitis. Pharmacogn. Res. 2017;9(1):116. doi: 10.4103/0974-8490.199780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Sylvestre D.L., Clements B.J., Malibu Y. Cannabis use improves retention and virological outcomes in patients treated for hepatitis C. Eur. J. Gastroenterol. Hepatol. 2006;18(10):1057–1063. doi: 10.1097/01.meg.0000216934.22114.51. [DOI] [PubMed] [Google Scholar]
- 237.Renaud-Young M., Mayall R.M., Salehi V., Goledzinowski M., Comeau F.J., MacCallum J.L., Birss V.I. Development of an ultra-sensitive electrochemical sensor for Δ9-tetrahydrocannabinol (THC) and its metabolites using carbon paper electrodes. Electrochim. Acta. 2019;307:351–359. [Google Scholar]
- 238.Wanklyn C., Burton D., Enston E., Bartlett C.A., Taylor S., Raniczkowska A., Murphy L. Disposable screen printed sensor for the electrochemical detection of delta-9-tetrahydrocannabinol in undiluted saliva. Chem. Cent. J. 2016;10(1):1. doi: 10.1186/s13065-016-0148-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Balbino M.A., Eleoterio I.C., de Oliveira M.F., McCord B.R. Electrochemical study of delta-9-tetrahydrocannabinol by cyclic voltammetry using screen printed electrode, improvements in forensic analysis. Sensors Transducers. 2016;207(12):73. [Google Scholar]
- 240.Lu D., Lu F., Pang G. A novel tetrahydrocannabinol electrochemical nano immunosensor based on horseradish peroxidase and double-layer gold nanoparticles. Molecules (Basel, Switzerland) 2016;21(10):1377. doi: 10.3390/molecules21101377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Zhang Q., Berg D., Mugo S.M. Molecularly imprinted carbon based electrodes for tetrahydrocannabinol sensing. Inorg. Chem. Commun. 2019;107:107459. doi: 10.1016/j.inoche.2019.107459. [DOI] [Google Scholar]
- 242.Nissim R., Compton R.G. Absorptive stripping voltammetry for cannabis detection. Chem. Cent. J. 2015;9:41. doi: 10.1186/s13065-015-0117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Jarho P., Pate D.W., Brenneisen R., Järvinen T. Hydroxypropyl-β-cyclodextrin and its combination with hydroxypropyl-methylcellulose increases aqueous solubility of Δ9-tetrahydrocannabinol. Life Sci. 1998;63(26):PL381–PL384. doi: 10.1016/s0024-3205(98)00528-1. [DOI] [PubMed] [Google Scholar]
- 244.Balbino M.A., Oiye É.N., Ribeiro M.F.M., Júnior J.W.C., Eleotério I.C., Ipólito A.J., de Oliveira M.F. Use of screen-printed electrodes for quantification of cocaine and Δ 9-THC: adaptions to portable systems for forensic purposes. J. Solid State Electrochem. 2016;20(9):2435–2443. [Google Scholar]
- 245.Balbino M.A., de Menezes M.M.T., Eleotério I.C., Saczk A.A., Okumura L.L., Tristão H.M., de Oliveira M.F. Voltammetric determination of Δ9-THC in glassy carbon electrode: an important contribution to forensic electroanalysis. Forensic Sci. Int. 2012;221(1–3):29–32. doi: 10.1016/j.forsciint.2012.03.020. [DOI] [PubMed] [Google Scholar]
- 246.Balbino M.A., Eleotério I.C., Oliveira L.S.D., de Menezes M.M., Andrade J.F.D., Ipólito A.J., Oliveira M.F.D. A comparative study between two different conventional working electrodes for detection of Δ9-tetrahydrocannabinol using square-wave voltammetry: a new sensitive method for forensic analysis. J. Braz. Chem. Soc. 2014;25(3):589–596. [Google Scholar]
- 247.Sandhyarani N. Electrochemical Biosensors. Elsevier; 2019. Surface modification methods for electrochemical biosensors; pp. 45–75. [Google Scholar]
- 248.Mishra R.K., Sempionatto J.R., Li Z., Brown C., Galdino N.M., Shah R., Wang J. Simultaneous detection of salivary Δ9-tetrahydrocannabinol and alcohol using a wearable electrochemical ring sensor. Talanta. 2020;211:120757. doi: 10.1016/j.talanta.2020.120757. [DOI] [PubMed] [Google Scholar]
- 249.Chen J., Wu J., Hao S., Yang M., Lu X., Chen X., Li L. Long term outcomes in survivors of epidemic Influenza A (H7N9) virus infection. Sci. Rep. 2017;7(1):17275. doi: 10.1038/s41598-017-17497-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Gentile F., Aimo A., Forfori F., Catapano G., Clemente A., Cademartiri F., Emdin M., Giannoni A. COVID-19 and risk of pulmonary fibrosis: the importance of planning ahead. Europ. J. Prevent. Cardiol. 2020;27(13):1442–1446. doi: 10.1177/2047487320932695. [DOI] [PMC free article] [PubMed] [Google Scholar]