Dear Editor,
In this Journal we previously reported the predicted SARS-CoV-2 spike-host cell receptor GRP78 binding site (1). New SARS-CoV-2 variant VUI 202,012/01 started in the UK and currently spreading in Europe and Australia during the last few days. The new variant bears about nine mutations in the spike protein (Δ69–70, Δ145, N501Y, A570D, D614G, P681H, T716I, S982A, and D1118H). The N501Y lies in the receptor-binding domain (RBD) of the spike and interacts with the host-cell receptor ACE2 responsible for viral recognition and entry. We tried to simulate the system of ACE2-SARS-CoV-2 spike RBD in the wildtype and mutated isoform of the RBD (N501Y). Additionally, the GRP78 association with the ACE2-SARS-CoV-2 spike RBD is modeled at the presence of this mutant variant of the viral spike.
Based on our previous study, the Heat Shock Protein A5 (HSPA5), also called, Glucose Regulated Protein 78 (GRP78) or Bip, is predicted to bind to the receptor-binding domain (RBD) of the SARS-CoV-2 Spike.1 The GRP78 is predicted to bind the Spike protein alongside the putative host-cell receptor, the Angiotensin-Converting Enzyme 2 (ACE2).2 , 3 The binding of GRP78 to the spike/ACE2 complex is predicted using HADDOCK 2.4 webserver4 (Fig. 1 A). PyMOL V2.2.2 was utilized to do a point mutation (N501Y) to resemble the RBD mutation found in the new variant of COVID-19.5 We docked GRP78 with both wild type SARS-CoV-2 Spike RBD-ACE2 complex (WT ACE2-RBD), and N501Y mutant SARS-CoV-2 Spike RBD-ACE2 complex (Mut ACE2-RBD). GRP78 and SARS-CoV-2 Spike RBD's active sites were T428, V429, V432, T434, F451, S452, V457 & I489 and C480-C488, respectively, and the rest of HADDOCK options were kept as default. The carbohydrate moieties (NAG) attached to the proteins were held in the structure.
Fig. 1.
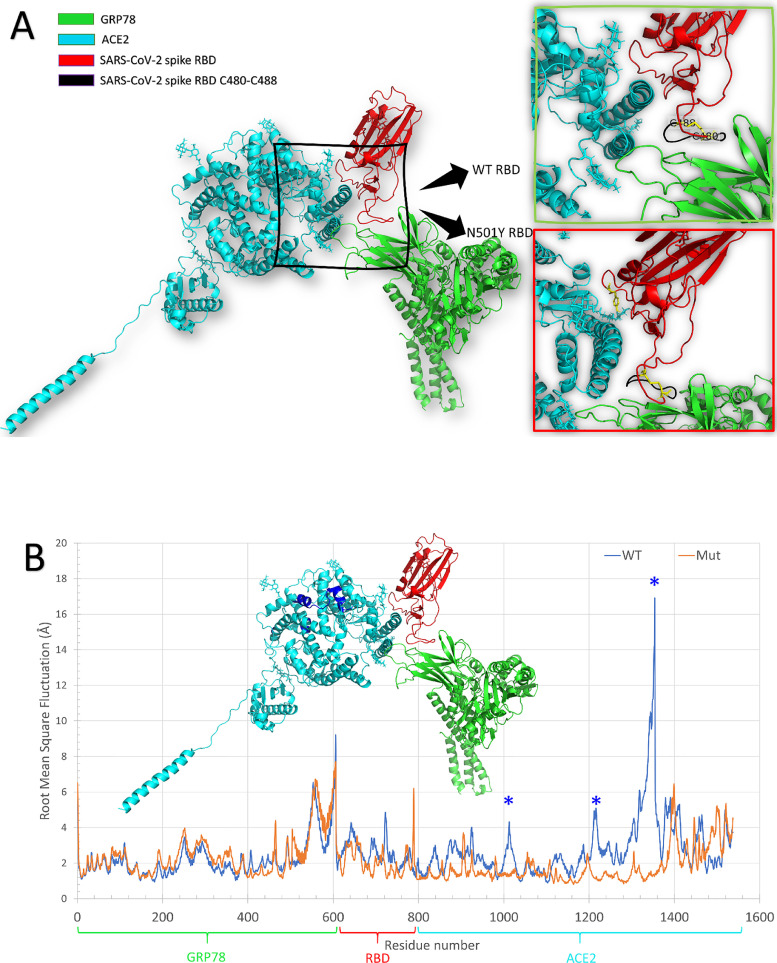
(A) The docking of GRP78 (5E84) to ACE2-SARS-CoV-2 Spike RBD complex (6M17). The enlarged panels on the right side show the docking pose in WT RBD (up) and the N501Y mutant variant (down). The C480-C488 of the RBD that was reported to bind GRP78 is shown in black cartoons. (B) The per -residue RMSF calculated for the 25 ns period MDS on the GRP78-RBD-ACE2 where both WT and N501Y mutant RBD is used. The proteins are represented with the same coloring scheme as Fig. 1. Blue asterisks denote the blue regions in the ACE2.
The HADDOCK score values for the GRP78 against WT ACE2-RBD and Mut ACE2-RBD are −74.3 ± 0.9 and −95.6 ± 1.0, respectively. This indicates better binding for the GRP78 against Mut ACE2-RBD than the WT ACE2-RBD complexes. There is a 28.7% increase in the HADDOCK score of GRP78 to the Mut ACE2-RBD form compared to the WT ACE2-RBD. The interactions between GRP78 and the two complexes are presented in Table 1 . GRP78 is tightly bound to the mutated complex with three H-bonds and five hydrophobic contacts instead of two H-bonds and three hydrophobic contacts in the case of WT ACE2-RBD, respectively. On the other hand, the docking scores and the interactions established upon docking of the ACE2 into SARS-CoV-2 spike RBD in wildtype and N501Y mutant isoforms are shown in Table 1.
Table 1.
The interaction patterns after docking GRP78 into the WT ACE2-RBD and Mut ACE2-RBD and ACE2 into the WT RBD and Mut RBD (N501Y). Bold residues are the interacting residues found in both WT and Mut ACE2-RBD complexes, while blue residues form Pi-stacking interactions.
| H-bonding | Hydrophobic interaction | Salt Bridge | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| complex | HADDOCK score | No. | Amino acids involved from RBD | Amino acids involved from GRP78 | No. | Amino acids involved from RBD | Amino acids involved from GRP78 | |||
| WT ACE2-RBD-GRP78 | −74.3 ± 0.9 | 2 | N481 and F486 | E427 and G454 | 3 | T478, P479, and V483 | V453 (2) and V457 | |||
| Mut ACE2-RBD-GRP78 | −95.6 ± 1.0 | 3 | E471, T478, and F486 | G430, S452, and G454 | 5 | P479, N481, V483 (2), and F486 | T428, V453 (2), V457, and V490 | |||
| complex | HADDOCK score | No. | Amino acids involved from RBD | Amino acids involved from ACE2 | No. | Amino acids involved from RBD | Amino acids involved from ACE2 | No. | Amino acids involved from RBD | Amino acids involved from ACE2 |
| WT ACE2-RBD | −126.1 ± 3.3 | 12 | K417, Y449, Y473, N487, Y489, Q493, S494, T500(3), G502, and V503 | E23, D30, H34(2), D38, Y41, Y83(2), T324, K353, D355, and R357 | 9 | F456(2), Y473, A475, F486(2), Y489, and T500(2) | Q24, T27(3), D30, Y41, M82, Y83, and D355 | 1 | E484 | K31 |
| Mut ACE2-RBD | −120.8 ± 1.7 | 13 | K417, G446, Y449, Y453, N487(3), Y489, F490, Q493, Q498(2), T500, and Y501 | Q24, T27, D30, K31(2), H34, Q42(4), Y83(2), K353, and R357 | 8 | F456(2), F486(3), F500, Y501, and Y505 | T27(2), K31, Y41, M82, Y83(2), and K353 | 1 | E484 | K31 |
As shown in the table, the HADDOCK score of ACE2 to both WT RBD and Mut RBD of SARS-CoV-2 spike protein is almost the same. The interactions are established through a dozen H-bonds, about eight hydrophobic contacts, and a salt bridge. The Y501 in the spike's mutant variant engaged in H-bond with K353 and formed π-stacking interaction with Y41 of the ACE2.
The best result from the two docking experiments (GRP78 against WT ACE2-RBD and Mut ACE2-RBD) was selected for Molecular Dynamic Simulation (MDS) using Nanoscale molecular dynamics software (NAMD) version 2.13.6 The necessary files for MDS were generated using the CHARMM-GUI webserver. The system's temperature and salt concentration were set to be 310 K and 0.154 M NaCl to resemble the physiological conditions. The system was minimized for 20,000 steps in a constant number of atoms, constant volume, constant temperature (NVT) ensemble using a conjugate gradient algorithm. The system was then equilibrated in a constant number of atoms, constant pressure, and constant temperature (NPT) ensemble for one nanosecond period. The pressure was controlled by the Nose-Hoover Langevin piston set to atmospheric pressure (1.01325 bar), while Langevin dynamics control the temperature. Finally, a production run of 25 ns was initialized in the NVT ensemble. The force field used was CHARMM36 force field parameters. TIP3P water model is used in the system simulation using NAMD 2.13 software.7 Different in-house scripts and the visualizing molecular dynamics (VMD) software tools are used to analyze data.8 , 9
Fig. 1B shows the superposition of the per-residue Root Mean Square Fluctuations in Å (calculated during 25 ns MDS) in the case of WT ACE2-RBD-GRP78 (blue line) and Mut ACE2-RBD-GRP78 (orange line). The WT complex show three highly flexible regions (blue asterisks) in the ACE2 (blue cartoon), while the other proteins show no significant differences. The systems need more in-depth analysis and calculations that require more time.
In this letter, we propose to shed light on the effect of the new variant mutation N501Y of the RBD on the viral recognition by the host cell-surface GRP78. This recognition can be targeted by peptides, antibodies, and phytochemicals (10).
Declaration of Competing Interest
All the authors declare no competing interest in this work.
Acknowledgments
Author contribution
A.E. own the research idea, wrote the manuscript, and draw figures, while I.I. performed the calculations and make the tables. All the authors approve the final version of the manuscript.
Acknowledgment
Cairo University supported this work through the COVID-19 grant.
References
- 1.Ibrahim I.M., Abdelmalek D.H., Elshahat M.E., Elfiky A.A. COVID-19 spike-host cell receptor GRP78 binding site prediction. J Infect. 2020;80(5):554–562. doi: 10.1016/j.jinf.2020.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elfiky A.A., Ibrahim I.M., Ismail A.M., Elshemey W.M. A possible role for GRP78 in cross vaccination against COVID-19. J Infect. 2020 doi: 10.1016/j.jinf.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elfiky A.A. SARS-CoV-2 spike-heat shock protein A5 (GRP78) recognition may be related to the immersed human coronaviruses. Front Pharmacol. 2020;11 doi: 10.3389/fphar.2020.577467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Dijk A.D., Bonvin A.M. Solvated docking: introducing water into the modelling of biomolecular complexes. Bioinformatics. 2006;22(19):2340–2347. doi: 10.1093/bioinformatics/btl395. [DOI] [PubMed] [Google Scholar]
- 5.2.4.1V . LLC; Schrödinger: 2020. The PyMOL Molecular Graphics System, Version 2.4.1. [Google Scholar]
- 6.Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mark P., Nilsson L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298K. J Phys Chem A. 2001;105(43):9954–9960. [Google Scholar]
- 8.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(1):27–28. doi: 10.1016/0263-7855(96)00018-5. 33-8. [DOI] [PubMed] [Google Scholar]
- 9.Elfiky A.A., Ismail A.M., Elshemey W.M. Recognition of gluconeogenic enzymes; Icl1, Fbp1, and Mdh2 by Gid4 ligase: a molecular docking study. J Mol Recognit. 2020;33(5):e2831. doi: 10.1002/jmr.2831. [DOI] [PubMed] [Google Scholar]
- 10.Elfiky A.A., Baghdady A.M., Ali S.A., Ahmed M.I. GRP78 targeting: hitting two birds with a stone. Life Sci. 2020;260 doi: 10.1016/j.lfs.2020.118317. [DOI] [PMC free article] [PubMed] [Google Scholar]