Abstract
Curtailing the Spring 2020 COVID-19 surge required sweeping and stringent interventions by governments across the world. Wastewater-based COVID-19 epidemiology programs have been initiated in many countries to provide public health agencies with a complementary disease tracking metric and non-discriminating surveillance tool. However, their efficacy in prospectively capturing resurgences following a period of low prevalence is unclear. In this study, the SARS-CoV-2 viral signal was measured in primary clarified sludge harvested every two days at the City of Ottawa's water resource recovery facility during the summer of 2020, when clinical testing recorded daily percent positivity below 1%. In late July, increases of >400% in normalized SARS-CoV-2 RNA signal in wastewater were identified 48 h prior to reported >300% increases in positive cases that were retrospectively attributed to community-acquired infections. During this resurgence period, SARS-CoV-2 RNA signal in wastewater preceded the reported >160% increase in community hospitalizations by approximately 96 h. This study supports wastewater-based COVID-19 surveillance of populations in augmenting the efficacy of diagnostic testing, which can suffer from sampling biases or timely reporting as in the case of hospitalization census.
Keywords: COVID-19, Wastewater-based epidemiology, SARS-CoV-2, Virus, Resurgence, Wave
Graphical abstract
1. Introduction
Wastewater has been used as a window in the habits and health of populations. A relatively novel epidemiological technique called wastewater-based epidemiology (WBE) has been successfully used by public health units and authorities for decades to track trends in the general population for numerous diverse substances: illicit and recreational drugs (Been et al., 2014; Feng et al., 2018; González-Mariño et al., 2016; Gushgari et al., 2018), toxic and harmful chemicals and pesticides (Du et al., 2018; Markosian and Mirzoyan, 2019; Rousis et al., 2017), as well as prevalence of disease and illness (Nordgren et al., 2009; Yang et al., 2017). An intense interest has developed in looking to sewage to seek out biomarkers, pathogens and viruses to further the understanding of the prevalence of diseases, diets and habits in the general population (Bowes and Halden, 2019; Lorenzo and Picó, 2019). The earliest occurrence of using sewage as a source of information was approximately 25 years ago, where scientists discovered that polio was in fact found and transmissible in sewage and human stool (Metcalf et al., 1995). One of the major advantages of WBE is that with well-designed sampling/testing regimes, it can be utilized to gain valuable, and sometimes otherwise unobtainable information for subsets of population, as small as a single building or neighborhood, or as large as a city or county (Choi et al., 2018; Tscharke et al., 2019).
Most recently, WBE has been applied to the quantification of COVID-19 disease signal. The COVID-19 global pandemic has had profound impacts on day-to-day life through sickness, death and the economic and social effects of lockdowns, closures and curfews put in place by local governments to control and limit the community spread of the disease. At the beginning of the pandemic, COVID-19 surveillance approaches invoked by nations around the world included case ascertainment of individual patients via nucleic acid-based tests, serological tests and contact tracing. As such, WBE has been investigated as a potential surveillance tool to obtain a rapid measure of the relative incidence rate of COVID-19 infections in the population at large, generating an entirely new epidemiological metric for public health units (PHU) to utilize. Fecal shedding of SARS-CoV-2 viral particles has been shown to occur before, during and after active COVID-19 infections, for periods ranging from a few days to several weeks (Chen et al., 2020; Cheung et al., 2020; Xing et al., 2020; Xu et al., 2020), with current literature suggesting that children may shed viral particles longer than adults (Li et al., 2020b; Ma et al., 2020). Additionally, fecal shedding of SARS-CoV-2 viral particles in stool occurs in patients infected with COVID-19 and appears to be independent of the presence of SARS-CoV-2 viral particles in the upper respiratory tract.
WBE has been applied at wastewater resource recovery facilities (WRRFs), with early studies demonstrating that viral signal is detected from both influent and primary clarified sludge, with recent studies identifying that at WRRFs, primary clarified sludge may provide more sensitive measurements than in influent (D'Aoust et al., 2021; Graham et al., 2020; Peccia et al., 2020). Additionally, recent studies outlined that viral particles of SARS-CoV-2 in wastewater could in fact be predominantly solid particle-bound (Canadian Water Network (CWN), 2020; Graham et al., 2020) due to the virus' lipid bilayer, which causes it to be lipophilic (Schoeman and Fielding, 2019). Furthermore, it was observed in several studies that SARS-CoV-2 viral signal in wastewater could be utilized as a tool to track epidemiological metric trends (Ahmed et al., 2020; Medema et al., 2020a; Nemudryi et al., 2020; Thompson et al., 2020). One of the recent developments to the field of WBE is the renewed interest in investigating biomarkers to normalize the SARS-CoV-2 viral signal. Previous WBE work identified several possible normalization biomarkers in wastewater such as 5-Hydroxyindoleacetic acid (Chen et al., 2014), caffeine and cotinine (Rico et al., 2017). However, due to the higher incidence of SARS-CoV-2 viral particles in feces as compared to urine (Collivignarelli et al., 2020; Jeong et al., 2020; Lodder and de Roda Husman, 2020; Zhurakivska et al., 2020), other biomarkers present in wastewater solids, such as pepper mild mottle virus (PMMoV) (Jafferali et al., 2020; Kitajima et al., 2018; Rosario et al., 2009; Symonds et al., 2014) and crAssphage (Crank et al., 2020; Polo et al., 2020) saw are being utilized for the normalization of SARS-CoV-2. Some recent studies have demonstrated that biomarker normalized SARS-CoV-2 viral signal may in fact track be capable of tracking the incidence of COVID-19 infections in the community (Gerrity et al., 2021; Kitamura et al., 2021) more effectively than non-biomarker-normalized viral signal (Jafferali et al., 2020; Wu et al., 2020), even if accounting for corrections due to flow or solids mass flux (D'Aoust et al., 2021).
To appropriately utilize epidemiological data obtained by measuring SARS-CoV-2 viral signal in wastewater, PHUs require an understanding of the predictive ability of this epidemiological metric to appropriately action obtained data (Hart and Halden, 2020; Hill et al., 2020; Thompson et al., 2020). Recent studies appear to demonstrate that WBE was able to identify the onset of community infection prior to clinical testing during the first wave, (D'Aoust et al., 2021; Kaplan et al., 2020; La Rosa et al., 2020; Medema et al., 2020b; Nemudryi et al., 2020; Peccia et al., 2020; Sherchan et al., 2020; Vallejo et al., 2020; Wu et al., 2020) however, there isn't a clear consensus on how early WBE can detect resurgences of the disease. Due to reporting limitations or absence of widespread clinical PCR or serological testing at the onset of the first wave of the COVID-19 pandemic, the predictive qualities of SARS-CoV-2 viral signal tracking in wastewater may not have been fully assessed, creating a critical and fundamental lack of understanding of intrinsic predictive qualities of wastewater-based SARS-CoV-2 surveillance, particularly for subsequent resurgences of COVID-19 infections in the community. As such, it remains unclear whether WBE can have a role in identifying COVID-19 resurgences before other, more traditional epidemiological metrics, such as clinical and serological testing results. As a result, the objective of this study is to establish a relationship between SARS-CoV-2 RNA signal in wastewater during a resurgence in a community with established clinical testing and (i) increases in SARS-CoV-2 RNA signal in wastewater, (ii) increases in the number of new COVID-19 positive patient cases in the community, and (iii) increases in the number of new hospitalizations of COVID-19 positive patients in the community.
2. Materials and methods
2.1. Characteristics of the City of Ottawa's water resource recovery facility
Primary clarified sludge samples were harvested from the City of Ottawa's Robert O. Pickard Environmental Centre, in Ottawa, ON, which services approximately 1 M residents of the national capital region. The hydraulic residence time of the Ottawa sewershed ranges from 2 h to 35 h, with an average residence time of approximately 12 h. The Ottawa water resource recovery facility (WRRF) has an average daily flow of 435,000 m3 per day. The facility is comprised of preliminary treatment consisting of coarse and fine screening and grit chambers for the removal of larger particles. The primary treatment units consist of rectangular primary clarifiers, where samples from this study are harvested (Fig. 1 ), and the secondary treatment is a conventional activated sludge system operating without nitrification. Finally, chlorination is used as a disinfection method followed by dechlorination prior to discharge to receiving waters.
Fig. 1.
Bird's-eye view of the Ottawa WRRF, with the primary clarifiers identified.
2.2. Sample collection, concentration, extraction and quantification
24-h composite samples of primary clarified sludge (PCS) were collected from the Ottawa WRRF at a frequency of every 2 days for a six-week period, from June 20th, 2020 until August 4th, 2020. Composite samples were collected at 6-h increments and stored at 4 °C with the four sample subsequently mixed and again stored at 4 °C for a maximum of 8 h until collected for processing. Processing of samples to concentrate, extract and quantify viral RNA was performed as per D'Aoust et al., (2021). To concentrate the samples, 32 mL of well homogenized PCS was first reacted with a sodium chloride (NaCl)/polyethylene glycol (PEG) solution with a final working concentration of 0.3 M NaCl and 80 mg/L PEG (Comelli et al., 2008; Hovi et al., 2001; Petterson et al., 2015). Samples were then well mixed on an orbital agitator set to a speed of 160 RPM for 12–17 h in a temperature-controlled chamber (4 °C). Following this, samples were centrifuged twice at 10,000 ×g in a refrigerated centrifuge set to 4 °C, for 45 min and 10 min, respectively. After each centrifugation cycle, the supernatant was decanted, preserving the pellet. The pellet was homogenized, and viral RNA was extracted using the RNeasy PowerMicrobiome Kit (Qiagen, Germantown, MD). The extraction was performed as per the manufacturer's recommendation, with the exception of the substitution of a chloroform-phenol solution with Trizol LS reagent (ThermoFisher, Ottawa, Canada). Quantification of the SARS-CoV-2 and PMMoV viral signals in wastewater was performed via singleplex one-step RT-qPCR with the Reliance One-Step Multiplex RT-qPCR Supermix (Bio-Rad, Hercules, CA) on a CFX Connect qPCR thermocycler (Bio-Rad, Hercules, CA). RT-qPCR reactions and the probe/primer sets utilized in this study are included in Supplemental Tables 1 and 2, respectively. It has been previously observed that normalization of SARS-CoV-2 viral copies with pepper mild mottle virus (copies SARS-CoV-2/copies PMMoV) is used to normalize viral SARS-CoV-2 signal for variations in wastewater physico-chemical characteristics, plant flows, wastewater solids organic/inorganic ratio and PCR amplification (D'Aoust et al., 2021). Evaluation of viral recovery efficiency was also performed as per D'Aoust et al. (2021); with the data in this study not being corrected for recovery efficiency. Dilution tests were periodically performed to determine if inhibition was present in the samples. The method's limit of detection for the N1 and N2 gene regions was accessed by determining the concentration at which a detection rate of ≥95% (<5% false negatives) was obtained, as per the MIQE recommendations (Bustin et al., 2009). Additionally, samples were discarded if they did not meet the following requirements: 1) standard curves are linear (R2 ≥ 0.95), 2) the copies/well are found in the linear range of the standard curve, and 3) the primer efficiency was between 90%–130%. Additionally, all samples were analyzed as technical triplicates and samples with values greater than 2 standard deviations away from the mean were discarded.
2.3. COVID-19 epidemiological data
During the study period, there were three main methods of COVID-19 surveillance used to compare to the wastewater RNA signal of SARS-CoV-2, with the three metrics being commonly communicated to the public. The first method was to observe the daily number of new confirmed COVID-19 infections, determined via laboratory-confirmed SARS-CoV-2 tests using reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR) that targeted the RNA-Dependent RNA polymerase for multiple genes. Tests were classified as positive for SARS-CoV-2 infection if at least one of any of the two gene “targets” were detected (Government of Ontario, 2020). There was a notable increase in COVID-19 testing in Ottawa during early June with an average daily testing increase from approximately 270 tests per day to approximately 800 tests per day. As a result of increased testing resources, additional testing was performed at dedicated community clinics, physician offices and in hospitals, retirement homes and long-term care facilities, ultimately improving the quality of clinical epidemiological data being generated daily.
A second method used for surveillance of COVID-19 infection is the test positivity rate of clinical testing within the health unit boundaries. COVID-19 test positivity is the proportion of positive COVID-19 cases of Ottawa residents reported as a percent of all individuals tested for COVID-19 on a specific date Test positivity is reported in this study on a daily basis and also as a seven-day mid-point average.
The third method used for COVID-19 surveillance was the daily hospital census counts, defined as the number of patients residing in Ottawa with confirmed COVID-19 infections as of midnight that day that were admitted to any of Ottawa's five acute care hospitals. The average length of stay during the study period was 19.5 days, interquartile range 22 days.
2.4. Statistical analysis
In order to test for significance and for strength of correlation between SARS-CoV-2 RNA signal and epidemiological metrics, a student's t-test and Pearson's correlation analyses were performed, with a p-value of 0.05 or lower signifying significance.
To evaluate if a lag existed between the appearance of increased SARS-CoV-2 RNA signal in wastewater and epidemiological metrics, time-step analyses were also performed where the correlations between viral RNA signal and epidemiological metrics were offset by a period of 1 to 7 days. The time-step analysis was performed across three discreet time-periods: i) pre-resurgence and resurgence, ending after the resurgence (June 21st to July 21st), ii) pre-resurgence and post-resurgence, extending to the end of the data set (June 21st to July 25th) and iii) full data period (June 21st to August 4th).
3. Results, discussion and implications
3.1. Trend and peak of wastewater SARS-CoV-2 RNA viral level in wastewater compared to other clinical COVID-19 surveillance metrics
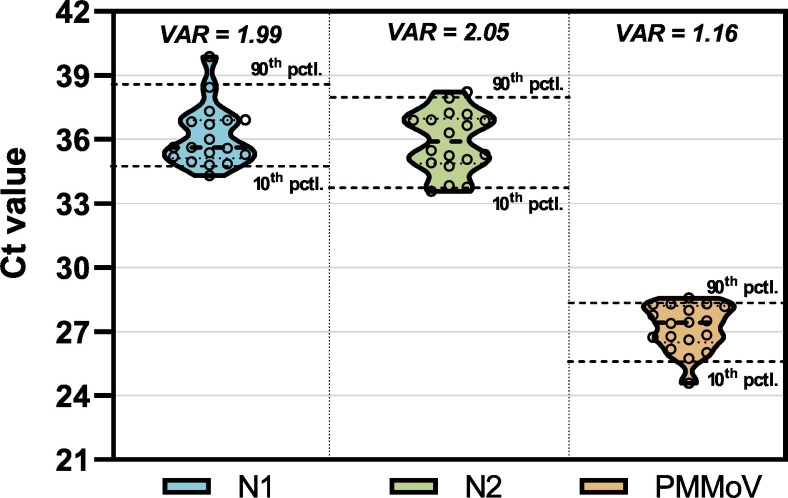
This study compares PMMoV normalized SARS-CoV-2 viral copies, a metric outlined in previous SARS-CoV-2 WBE work (D'Aoust et al., 2021; Kitamura et al., 2021; Wu et al., 2020) with clinical epidemiological metrics. This WBE-based metric normalizes SARS-CoV-2 viral copies to the number of copies of PMMoV in wastewater to account for variations inherent to wastewater samples; in particular variations related to the wastewaters water quality, solids and fecal matter concentrations. In addition, PMMoV normalizes for variations associated with sampling, storage, and testing. PMMoV is ubiquitous in both the liquid and solid fraction of wastewater and is effective due to its low temporal variability in wastewater (Fig. 2 ). The lower temporal variability of PMMoV is likely to be linked to the stability and hardiness of its virions (Sassi et al., 2018).
Fig. 2.
Ct values of the SARS-CoV-2 N1 and N2 gene regions, and the PMMoV normalization gene (1:10 dilution shown), outlining the low variability of PMMoV, justifying its use as a normalization gene. The 10th and 90th percentile are displayed, along with the median (dotted line inside shapes). The variance of the different targets is also shown at the top.
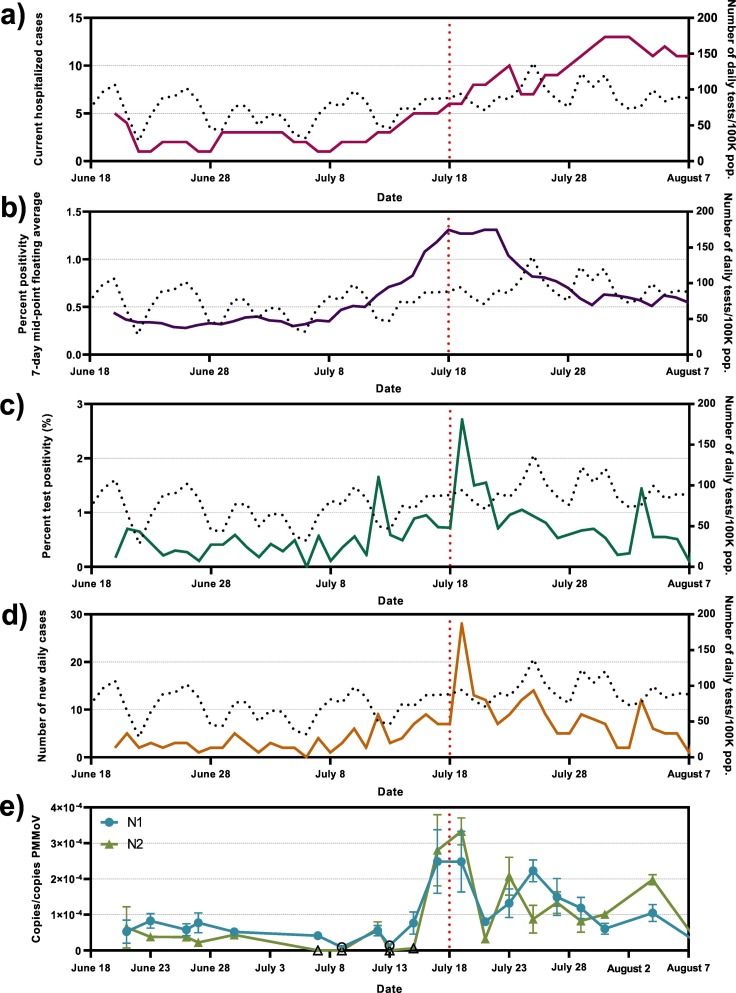
All conventional COVID-19 surveillance measures (# of hospitalized cases, 7-day mid-point floating average percent positivity, clinical test percent positivity and number of new daily cases; Fig. 3a–d) increase during the study period from a low level at the beginning of the study. The number of current hospitalized patient cases are at their highest on July 31-Aug 2 with a 168% increase compared to the week of July 13 (Fig. 3a–d). Meanwhile, percent positive cases and daily cases are at their highest on July 19, with increases of 215% and 326% respectively, relative to the preceding 7-day period (Fig. 3b–d).
Fig. 3.
Epidemiological metrics and SARS-CoV-2 viral concentrations over the study period; a) COVID-19-caused hospitalizations, b) percent test positivity (7-day mid-point floating average), c) percent test positivity and d) number of new daily cases, along with e) wastewater SARS-CoV-2 N1 and N2 gene copies/PMMoV gene copies. Open data points signify points which were below the limit of quantification. The dotted line in the background represents the number of daily clinical tests/100 K pop. Performed.
As observed with other epidemiological metrics, the PMMoV normalized viral signal in Ottawa's wastewater is also shown to be low at the beginning of the study period through to the week of July 13 with both N1 and N2 regions below 1.00 × 10−4 copies/copies PMMoV (Fig. 3e). There is a 450% relative increase (p < 0.05) in N1 and 440% increase (p < 0.05) in N2 viral signal on July 17 to 19 compared to July 13 to 15. An absolute increase of 2.03 × 10−4 and 3.01 × 10−4 copies/copies PMMoV is reported during this period of increase for N1 and N2 gene regions, respectively (Fig. 3e). The PMMoV normalized viral signal then decreases again following this peak, to levels higher than the previous baseline, but notably lower than the peak outlined between July 17 to 19.
Throughout the study, a good agreement is observed between N1 and N2 SARS-CoV-2 gene region targets (R = 0.852, r2 = 0.726), implying that some portions of the N1 and N2 gene regions may be preserved. This potentially indicates that RNA extracted from primary sludge and wastewater could be sequenced to track the transmission of new, and potentially more infectious mutations of the disease (Li et al., 2020a; Oude Munnink et al., 2020; Rockett et al., 2020; Volz et al., 2020)
This study also suggests that the PMMoV normalized viral signal peaks for a relatively short period of time before showing a subsequent rapid decrease in signal. This short peaking period may be an artifact of the inherent variation in wastewaters or associated with variations inherent to the assay. The short peak period may also be associated to fecal shedding patterns of the population contributing to the wastewater. It is noted that several current studies investigating fecal shedding timelines and intensity for SARS-CoV-2 may in fact be investigating the sickest of patients in a hospital setting (Chen et al., 2020; Fontana et al., 2020), and so patterns may not reflect occurrences in the general population, where a significant quantity of patients will likely suffer from milder symptoms, or no symptoms at all (Pan et al., 2020; Park et al., 2020). Furthermore, sicker or elderly patients may in fact “fall off” the wastewater monitoring efforts if bed pans or adult diapers are utilized as part of their treatment and convalescence in a hospital or retirement home (Coelho et al., 2015; Tsang et al., 2017).
Previous work on PMMoV identified that it is a useful environmental metric to track in surface waters and drinking waters as it correlated strongly with fecal contamination of said waters (Hamza et al., 2011; Haramoto et al., 2013; Rosiles-González et al., 2017). As a result, the use of PMMoV normalized data in SARS-CoV-2 is seeing increased utilization in several locales due to the metrics' ability to correct for the true quantity of fecal matter in wastewater (D'Aoust et al., 2021; Wu et al., 2020). It is hypothesized that due to the fact PMMoV signal in wastewater is likely diet based (due to consumption peppers, some tomatoes, or related foodstuff infected with PMMoV) (Jarret et al., 2008; Peng et al., 2015; Zhou et al., 2020), this approach will work best when performing a survey of a larger group of individuals or geographical area, where enough survey participants with varied enough diets will average out the signal.
3.2. Correlation between wastewater and clinical surveillance metrics
Positive correlations were observed (Fig. 3c, d and e) between the normalized viral RNA signal and both the number of new daily positive COVID-19 cases (N1: R = 0.673, p < 0.001; N2: R = 0.648, p < 0.001) and clinical testing percent positivity (N1: R = 0.468, p < 0.001; N2: R = 0.404, p < 0.001). These findings are in agreement with reported observations from previous studies (D'Aoust et al., 2021; Nemudryi et al., 2020; Peccia et al., 2020). This study, as opposed to an earlier study in the same region (D'Aoust et al., 2021), demonstrates that strong correlations do in fact exist between viral RNA signal and the epidemiological metric of clinical daily new COVID-19 cases, as supported by several other studies (Hart and Halden, 2020; Michael-Kordatou et al., 2020; Trottier et al., 2020; Wu et al., 2020). The moderate correlations between viral RNA signal and clinical daily new COVID-19 cases in previous work was largely attributed to inadequate resources to achieve a developed clinical testing framework early in the pandemic (lower daily # of tests). In this study, a strong testing regiment was deployed in Ottawa and hence produced a more reliable daily new COVID-19 cases metric across this study period. A weak positive correlation exists between N1 and N2 SARS-CoV-2 PMMoV-normalized RNA signal and COVID-19-caused hospitalizations (R = 0.347, p < 0.001 and R = 0.464, p < 0.001 for N1 and N2, respectively; Fig. 3a and e). This supports previous findings that WBE efforts to track the rate of incidence of COVID-19 infections in the community provides useful and confirmatory information.
3.3. Temporal association between surveillance metrics
A visual comparison of clinical testing percent positivity and the number of new daily positive COVID-19 cases compared to the PMMoV-normalized RNA signal shows that the viral RNA signal predates both the clinical testing percent positivity and new daily positive COVID-19 cases epidemiological data.
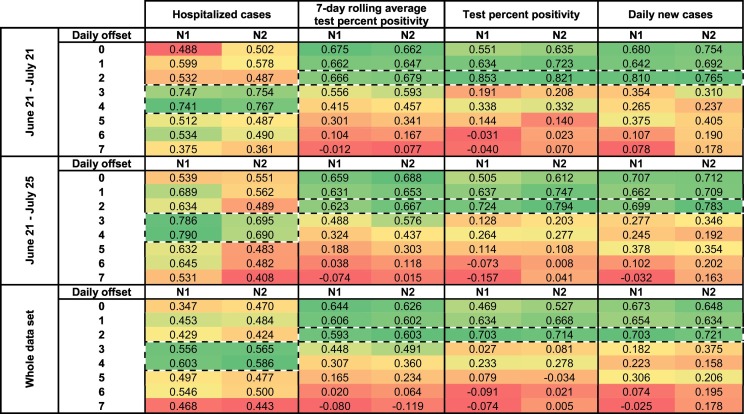
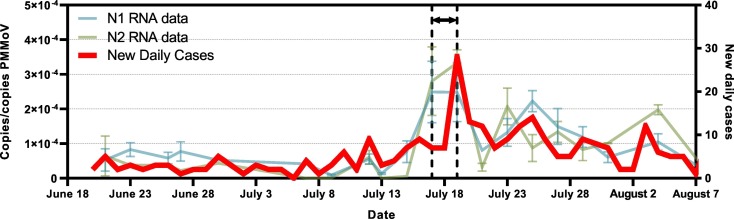
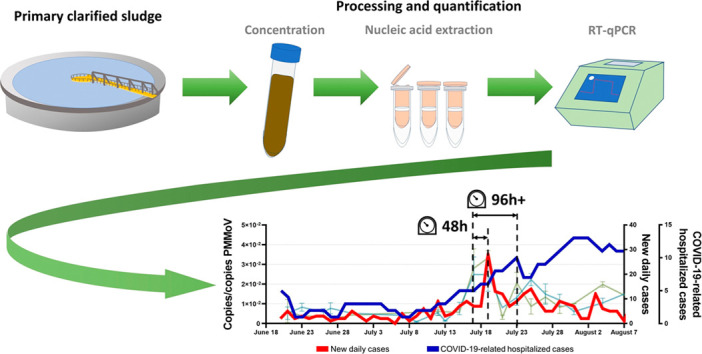
A times-step correlation analysis suggests that increases in SARS-CoV-2 signal in wastewater precedes increases in new daily positive COVID-19 cases and clinical testing percent positivity by two days (N1: R = 0.703, p < 0.001; N2: R = 0.721, p < 0.001; N1: R = 0.703, p < 0.001; N2: R = 0.714, p < 0.001) (Table 1 ). Furthermore, the same time-step correlation analysis also suggests that increases in SARS-CoV-2 signal in wastewater precedes increases in the number of hospitalized cases by four days (N1: R = 0.741, p < 0.001; N2: R = 0.767, p < 0.001). This is further confirmed visually (Fig. 4 ). The precedence of viral RNA signal vs. other epidemiological metrics has been reported elsewhere (Chavarria-Miró et al., 2020; Kumar et al., 2020; Peccia et al., 2020).
Table 1.
Time-step analyses of correlations (Pearson's R) between normalized SARS-CoV-2 viral RNA signal (copies/copies PMMoV) and 7-day rolling average percent positivity, test percent positivity and daily new cases epidemiological metrics.
Fig. 4.
Figure of the new daily cases epidemiological metric (in red) with SARS-CoV-2 viral RNA signal in Ottawa's primary clarified sludge samples, with the precedence of SARS-COV-2 viral signal in wastewater clearly outlined. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Recent studies proposed that wastewater viral SARS-CoV-2 RNA signal may in fact precede hospital admissions by 1 to 5 days in primary sludge (Fig. 4) (Kaplan et al., 2020; Peccia et al., 2020), and 6–14 days in raw wastewater (Kumar et al., 2020). This supports findings by Peccia et al. (2020), identifying that hospitalizations may begin peaking four days after an increased viral RNA signal, with the caveat that hospitalizations rates may be significantly higher when viral RNA signal is the result from shedding of an at-risk population group (young infants, elderly, immunocompromised, etc.). In addition, the RNA signal post-resurgence indicates that the resurgence events in viral RNA signal serves to weaken the correlation of subsequent peaks with clinical testing metrics. It is hypothesized that early and later infections and overlapping shedding events of duration and intensity convolute peaks within a period of time following the initial new peaks/surges in COVID-19 infections. Nonetheless, the RNA epidemiological metric of copies/copies PMMoV may become extremely valuable to public health units in their preparation for the next COVID-19 resurgences.
Earlier studies investigating the predictive capabilities of SARS-CoV-2 WBE (Kumar et al., 2020; Peccia et al., 2020) may have been performed at times where clinical metrics were dampened due to limited clinical testing resources, which may in turn have impacted the conclusions regarding predictive ability. This present study was performed during a period of relatively higher testing intensity in the community (avg. 80.9 tests per/100 K pop. performed) and confirms previous findings of early detection of SARS-CoV-2 WBE as compared to clinical metrics such as hospitalizations, new daily positives cases or percent positivity.
4. Conclusions
This study demonstrates that due to the general stability of SARS-CoV-2 RNA measurements in PCS, this fraction of wastewater can reliably be utilized to predict rapid increases or resurgences in COVID-19 cases in the community at large (Alpaslan-Kocamemi et al., 2020; D'Aoust et al., 2021; Peccia et al., 2020), and the results strongly suggest that PCS can be utilized as part of sentinel-type wastewater based epidemiology endeavors at WRRFs.
Advantages of this approach include: i) the analyses in water resource recovery facilities are high-enrollment surveys, which capture a majority of the urban population, regardless of individual's willingness to get tested clinically; ii) tests are anonymous and do not pose ethical challenges as opposed to mass clinical testing or targeted sewershed monitoring efforts; iii) data can be used as a confirmation to clinical tests, increases the effectiveness of local public health units and data remains independent of official testing strategies or media sentiment, and iv) provides an exit strategy/path forward for public health units as something to eventually transition to once immunization is available, to cut down on clinical testing costs, while maintaining broad surveillance capabilities. However, limitations to this WBE approach includes the following: i) data obtained only gives high-altitude view of the situation in the community, and it is currently not possible to correlate these data to an actual number of cases in the community; ii) testing protocols must be optimized and retain sensitivity at very-low levels of disease incidence to remain relevant; iii) widespread adoption of WBE requires scale-up of public health units' current capabilities and/or partnership with private laboratories or research institutions, and iv) increases in resolution (more localized, upstream of water resource recovery facilities, for example) of tests in watersheds may lead to ethical questions for public health unit if it wishes to act upon the data, due to the potential risk for identifying or singling out small subgroups of a population. It is noted however that these issues are not dissimilar to other ethical issues currently existing with other COVID-19 public health endeavors, such as COVID-19 potential exposure notification smartphone applications.
Funding
This research was supported by a CHEO (Children's Hospital of Eastern Ontario) CHAMO (Children's Hospital Academic Medical Organization) grant, awarded to Dr. Alex E. MacKenzie.
CRediT authorship contribution statement
Patrick M. D'Aoust: Investigation, Formal analysis, Writing – original draft. Tyson E. Graber: Validation, Supervision, Writing – review & editing. Elisabeth Mercier: Investigation, Formal analysis, Writing – original draft. Danika Montpetit: Investigation. Ilya Alexandrov: Validation, Writing – review & editing. Nafisa Neault: Validation, Writing – review & editing. Aiman Tariq Baig: Validation, Writing – review & editing. Janice Mayne: Validation, Writing – review & editing. Xu Zhang: Validation, Writing – review & editing. Tommy Alain: Validation, Writing – review & editing. Mark R. Servos: Methodology, Validation, Writing – review & editing. Nivetha Srikanthan: Methodology, Validation, Writing – review & editing. Malcolm MacKenzie: Validation, Writing – review & editing. Daniel Figeys: Validation, Writing – review & editing. Douglas Manuel: Validation, Writing – review & editing. Peter Jüni: Validation, Writing – review & editing. Alex E. MacKenzie: Methodology, Validation, Supervision, Writing – review & editing, Funding acquisition. Robert Delatolla: Methodology, Validation, Supervision, Writing – review & editing, Funding acquisition.
Declaration of competing interest
The authors declare that no known competing financial interests or personal relationships could appear to influence the work reported in this manuscript.
Acknowledgements
The authors wish to acknowledge the help and assistance of the University of Ottawa, the Ottawa Hospital, the Children's Hospital of Eastern Ontario, the Children's Hospital of Eastern Ontario's Research Institute, the City of Ottawa, Ottawa Public Health, Public Health Ontario and all their employees involved in the project during this study. Their time, facilities, resources and thoughts provided throughout the study helped the authors greatly. The authors also wish to specifically outline the assistance of Dr. Monir Taha at Ottawa Public Health.
Editor: Thomas Kevin V
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2021.145319.
Appendix A. Supplementary data
Supplementary tables
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728:138764. doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpaslan-Kocamemi B., Kurt H., Sait A., Sarac F., Saatci A.M., Pakdemirli B. SARS-CoV-2 detection in Istanbul wastewater treatment plant sludges. medRxiv. 2020 doi: 10.1101/2020.05.12.20099358. 2020.05.12.20099358. [DOI] [Google Scholar]
- Been F., Rossi L., Ort C., Rudaz S., Delémont O., Esseiva P. Population normalization with ammonium in wastewater-based epidemiology: application to illicit drug monitoring. Environ. Sci. Technol. 2014;48:8162–8169. doi: 10.1021/es5008388. [DOI] [PubMed] [Google Scholar]
- Bowes D.A., Halden R.U. Theoretical evaluation of using wastewater-based epidemiology to assess the nutritional status of human populations. Curr. Opin. Environ. Sci. Heal. 2019;9:58–63. doi: 10.1016/j.coesh.2019.05.001. [DOI] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., Wittwer C.T. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Canadian Water Network (CWN) Waterloo; Ontario, Canada: 2020. Phase I Inter-Laboratory Study: Comparison of Approaches to Quantify SARS-CoV-2 RNA in Wastewater. [Google Scholar]
- Chavarria-Miró G., Anfruns-Estrada E., Guix S., Paraira M., Galofré B., Sáanchez G., Pintó R., Bosch A. Sentinel surveillance of SARS-CoV-2 in wastewater anticipates the occurrence of COVID-19 cases. medRxiv. 2020 doi: 10.1101/2020.06.13.20129627. 2020.06.13.20129627. [DOI] [Google Scholar]
- Chen C., Kostakis C., Gerber J.P., Tscharke B.J., Irvine R.J., White J.M. Towards finding a population biomarker for wastewater epidemiology studies. Sci. Total Environ. 2014;487:621–628. doi: 10.1016/j.scitotenv.2013.11.075. [DOI] [PubMed] [Google Scholar]
- Chen Y., Chen L., Deng Q., Zhang G., Wu K., Ni L., Yang Y., Liu B., Wang W., Wei C., Yang J., Ye G., Cheng Z. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J. Med. Virol. 2020;92:833–840. doi: 10.1002/jmv.25825. [DOI] [PubMed] [Google Scholar]
- Cheung K.S., Hung I.F.N., Chan P.P.Y., Lung K.C., Tso E., Liu R., Ng Y.Y., Chu M.Y., Chung T.W.H., Tam A.R., Yip C.C.Y., Leung K.-H., Fung A.Y.-F., Zhang R.R., Lin Y., Cheng H.M., Zhang A.J.X., To, K.K.W, Chan K.-H., Yuen K.-Y., Leung W.K. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology. 2020:81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi P.M., Tscharke B.J., Donner E., O’Brien J.W., Grant S.C., Kaserzon S.L., Mackie R., O’Malley E., Crosbie N.D., Thomas K.V., Mueller J.F. Wastewater-based epidemiology biomarkers: past, present and future. TrAC - Trends Anal. Chem. 2018;105:453–469. doi: 10.1016/j.trac.2018.06.004. [DOI] [Google Scholar]
- Coelho M.F., de Castro D.F.A., Fumincelli L., Mazzo A. Characterization of adult patients who used disposable diaper during the hospital stay in a large teaching hospital. Int. J. Nurs. 2015;2:162–166. doi: 10.15640/ijn.v2n1a18. [DOI] [Google Scholar]
- Collivignarelli M.C., Collivignarelli C., Carnevale Miino M., Abbà A., Pedrazzani R., Bertanza G. SARS-CoV-2 in sewer systems and connected facilities. Process. Saf. Environ. Prot. 2020;143:196–203. doi: 10.1016/j.psep.2020.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comelli H.L., Rimstad E., Larsen S., Myrmel M. Detection of norovirus genotype I.3b and II.4 in bioaccumulated blue mussels using different virus recovery methods. Int. J. Food Microbiol. 2008;127:53–59. doi: 10.1016/j.ijfoodmicro.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Crank K., Li X., North D., Ferraro G.B., Iaconelli M., Mancini P., La Rosa G., Bibby K. CrAssphage abundance and correlation with molecular viral markers in Italian wastewater. Water Res. 2020;184:116161. doi: 10.1016/j.watres.2020.116161. [DOI] [PubMed] [Google Scholar]
- D’Aoust P.M., Mercier É., Montpetit D., Jia J., Alexandrov I., Neault N., Baig A.T., Mayne J., Zhang X., Alain T., Langlois M.-A., Servos M.R., MacKenzie M., Figeys D., MacKenzie A.E., Graber T.E., Delatolla R. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Res. 2021;188:116560. doi: 10.1016/j.watres.2020.116560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P., Zhou Z., Huang H., Han S., Xu Z., Bai Y., Li X. Estimating population exposure to phthalate esters in major Chinese cities through wastewater-based epidemiology. Sci. Total Environ. 2018;643:1602–1609. doi: 10.1016/j.scitotenv.2018.06.325. [DOI] [PubMed] [Google Scholar]
- Feng L., Zhang W., Li X. Monitoring of regional drug abuse through wastewater-based epidemiology—a critical review. Sci. China Earth Sci. 2018;61:239–255. doi: 10.1007/s11430-017-9129-x. [DOI] [Google Scholar]
- Fontana L., Villamagna A.H., Sikka M.K., McGregor J.C. Understanding viral shedding of SARS-CoV-2: review of current literature. Infect. Control Hosp. Epidemiol. 2020;2:1–10. doi: 10.1017/ice.2020.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrity D., Papp K., Stoker M., Sims A., Frehner W. Early-pandemic wastewater surveillance of SARS-CoV-2 in southern Nevada: methodology, occurrence, and incidence/prevalence considerations. Water Res. X. 2021;10:100086. doi: 10.1016/j.wroa.2020.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Mariño I., Gracia-Lor E., Rousis N.I., Castrignanò E., Thomas K.V., Quintana J.B., Kasprzyk-Hordern B., Zuccato E., Castiglioni S. Wastewater-based epidemiology to monitor synthetic Cathinones use in different European countries. Environ. Sci. Technol. 2016;50:10089–10096. doi: 10.1021/acs.est.6b02644. [DOI] [PubMed] [Google Scholar]
- Government of Ontario, 2020. Case definition for coronavirus disease 2019 (COVID-19).
- Graham K.E., Loeb S.K., Wolfe M.K., Catoe D., Sinnott-Armstrong N., Kim S., Yamahara K.M., Sassoubre L.M., Mendoza Grijalva L.M., Roldan-Hernandez L., Langenfeld K., Wigginton K.R., Boehm A.B. SARS-CoV-2 RNA in wastewater settled solids is associated with COVID-19 cases in a large urban sewershed. Environ. Sci. Technol. 2020 doi: 10.1021/acs.est.0c06191. acs.est.0c06191. [DOI] [PubMed] [Google Scholar]
- Gushgari A.J., Driver E.M., Steele J.C., Halden R.U. Tracking narcotics consumption at a southwestern U.S. university campus by wastewater-based epidemiology. J. Hazard. Mater. 2018;359:437–444. doi: 10.1016/j.jhazmat.2018.07.073. [DOI] [PubMed] [Google Scholar]
- Hamza I.A., Jurzik L., Überla K., Wilhelm M. Evaluation of pepper mild mottle virus, human picobirnavirus and torque Teno virus as indicators of fecal contamination in river water. Water Res. 2011;45:1358–1368. doi: 10.1016/j.watres.2010.10.021. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Kishida N., Konno Y., Katayama H., Asami M., Akiba M. Occurrence of pepper mild mottle virus in drinking water sources in Japan. Appl. Environ. Microbiol. 2013;79:7413–7418. doi: 10.1128/AEM.02354-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart O.E., Halden R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020;730:138875. doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K., Zamyadi A., Deere D., Vanrolleghem P.A., Crosbie N.D. SARS-CoV-2 known and unknowns, implications for the water sector and wastewater-based epidemiology to support national responses worldwide: early review of global experiences with the COVID-19 pandemic. Water Qual. Res. J. 2020:1–11. doi: 10.2166/wqrj.2020.100. [DOI] [Google Scholar]
- Hovi T., Stenvik M., Partanen H., Kangas A. Poliovirus surveillance by examining sewage specimens. Quantitative recovery of virus after introduction into sewerage at remote upstream location. Epidemiol. Infect. 2001;127:101–106. doi: 10.1017/S0950268801005787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafferali M.H., Khatami K., Atasoy M., Birgersson M., Williams C., Cetecioglu Z. Benchmarking virus concentration methods for quantification of SARS-CoV-2 in raw wastewater. Sci. Total Environ. 2020;755:142939. doi: 10.1016/j.scitotenv.2020.142939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarret R.L., Gillaspie A.G., Barkley N.A., Pinnow D.L. The occurrence and control of pepper mild mottle virus (PMMoV) in the USDA/ARS Capsicum germplasm collection. Seed Technol. 2008:26–36. [Google Scholar]
- Jeong H.W., Kim S.M., Kim H.S., Kim Y. Il, Kim J.H., Cho J.Y., Kim S. Hyung, Kang H., Kim S.G., Park S.J., Kim E.H., Choi Y.K. Viable SARS-CoV-2 in various specimens from COVID-19 patients. Clin. Microbiol. Infect. 2020;26:1520–1524. doi: 10.1016/j.cmi.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan E.H., Wang D., Wang M., Malik A.A., Zulli A., Peccia J. Aligning SARS-CoV-2 indicators via an epidemic model: application to hospital admissions and RNA detection in sewage sludge. Health Care Manag. Sci. 2020 doi: 10.1007/s10729-020-09525-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Sassi H.P., Torrey J.R. Pepper mild mottle virus as a water quality indicator. npj Clean Water. 2018:1. doi: 10.1038/s41545-018-0019-5. [DOI] [Google Scholar]
- Kitamura K., Sadamasu K., Muramatsu M., Yoshida H. Efficient detection of SARS-CoV-2 RNA in the solid fraction of wastewater. Sci. Total Environ. 2021;763:144587. doi: 10.1016/j.scitotenv.2020.144587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., Joshi C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;709:141326. doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736:139652. doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q., Wu, J., Nie, J., Zhang, Li, Hao, H., Liu, S., Zhao, C., Zhang, Q., Liu, H., Nie, L., Qin, H., Wang, M., Lu, Q., Li, Xiaoyu, Sun, Q., Liu, J., Zhang, Linqi, Li, Xuguang, Huang, W., Wang, Y., 2020a. The impact of mutations in SARS-CoV-2 Spike on viral infectivity and antigenicity. Cell 182, 1284–1294.e9. 10.1016/j.cell.2020.07.012 [DOI] [PMC free article] [PubMed]
- Li X., Xu W., Dozier M., He Y., Kirolos A., Theodoratou E. The role of children in transmission of SARS-CoV-2: a rapid review. J. Glob. Health. 2020;10:1–10. doi: 10.7189/JOGH.10.011101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W., de Roda Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020;5:533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo M., Picó Y. Wastewater-based epidemiology: current status and future prospects. Curr. Opin. Environ. Sci. Heal. 2019;9:77–84. doi: 10.1016/j.coesh.2019.05.007. [DOI] [Google Scholar]
- Ma X., Su L., Zhang Y., Zhang X., Gai Z., Zhang Z. Do children need a longer time to shed SARS-CoV-2 in stool than adults? J. Microbiol. Immunol. Infect. 2020;53:373–376. doi: 10.1016/j.jmii.2020.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markosian C., Mirzoyan N. Wastewater-based epidemiology as a novel assessment approach for population-level metal exposure. Sci. Total Environ. 2019;689:1125–1132. doi: 10.1016/j.scitotenv.2019.06.419. [DOI] [PubMed] [Google Scholar]
- Medema G., Been F., Heijnen L., Petterson S. Implementation of environmental surveillance for SARS-CoV-2 virus to support public health decisions: opportunities and challenges. Curr. Opin. Environ. Sci. Heal. 2020;17:49–71. doi: 10.1016/j.coesh.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Metcalf T.G., Melnick J.L., Estes M.K. Environmental virology: from detection of virus in sewage and water by isolation to identification by molecular biology - a trip of over 50 years. Annu. Rev. Microbiol. 1995;49:461–487. doi: 10.1146/annurev.mi.49.100195.002333. [DOI] [PubMed] [Google Scholar]
- Michael-Kordatou I., Karaolia P., Fatta-Kassinos D. Sewage analysis as a tool for the COVID-19 pandemic response and management: the urgent need for optimised protocols for SARS-CoV-2 detection and quantification. J. Environ. Chem. Eng. 2020;8:104306. doi: 10.1016/j.jece.2020.104306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemudryi A., Nemudraia A., Wiegand T., Surya K., Buyukyoruk M., Vanderwood K., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Reports Med. 2020;1:100098. doi: 10.2139/ssrn.3664367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordgren J., Matussek A., Mattsson A., Svensson L., Lindgren P.E. Prevalence of norovirus and factors influencing virus concentrations during one year in a full-scale wastewater treatment plant. Water Res. 2009;43:1117–1125. doi: 10.1016/j.watres.2008.11.053. [DOI] [PubMed] [Google Scholar]
- Oude Munnink B.B., Nieuwenhuijse D.F., Stein M., O’Toole Á., Haverkate M., Mollers M., Kamga S.K., Schapendonk C., Pronk M., Lexmond P., van der Linden A., Bestebroer T., Chestakova I., Overmars R.J., van Nieuwkoop S., Molenkamp R., van der Eijk A.A., GeurtsvanKessel C., Vennema H., Meijer A., Rambaut A., van Dissel J., Sikkema R.S., Timen A., Koopmans M., Oudehuis G.J.A.P.M., Schinkel J., Kluytmans J., Kluytmans-van den Bergh M., van den Bijllaardt W., Berntvelsen R.G., van Rijen M.M.L., Schneeberger P., Pas S., Diederen B.M., Bergmans A.M.C., van der Eijk P.A.V., Verweij J., Buiting A.G.N., Streefkerk R., Aldenkamp A.P., de Man P., Koelemal J.G.M., Ong D., Paltansing S., Veassen N., Sleven J., Bakker L., Brockhoff H., Rietveld A., Slijkerman Megelink F., Cohen Stuart J., de Vries A., van der Reijden W., Ros A., Lodder E., Verspui-van der Eijk E., Huijskens I., Kraan E.M., van der Linden M.P.M., Debast S.B., Al Naiemi N., Kroes A.C.M., Damen M., Dinant S., Lekkerkerk S., Pontesilli O., Smit P., van Tienen C., Godschalk P.C.R., van Pelt J., Ott A., van der Weijden C., Wertheim H., Rahamat-Langendoen J., Reimerink J., Bodewes R., Duizer E., van der Veer B., Reusken C., Lutgens S., Schneeberger P., Hermans M., Wever P., Leenders A., ter Waarbeek H., Hoebe C. Rapid SARS-CoV-2 whole-genome sequencing and analysis for informed public health decision-making in the Netherlands. Nat. Med. 2020;26:1405–1410. doi: 10.1038/s41591-020-0997-y. [DOI] [PubMed] [Google Scholar]
- Pan X., Chen D., Xia Y., Wu X., Li T., Ou X., Zhou L., Liu J. Asymptomatic cases in a family cluster with SARS-CoV-2 infection. Lancet Infect. Dis. 2020;20:410–411. doi: 10.1016/S1473-3099(20)30114-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Lee C.-W., Park D.-I., Woo H.-Y., Cheong H.S., Shin H.C., Ahn K., Kwon M.-J., Joo E.-J. Detection of SARS-CoV-2 in fecal samples from patients with asymptomatic and mild COVID-19 in Korea. Clin. Gastroenterol. Hepatol. 2020 doi: 10.1016/j.cgh.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L., Weinberger D.M., Arnold W., Omer S.B. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38:1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Shi B., Zheng H., Lu Y., Lin L., Jiang T., Chen J., Yan F. Detection of pepper mild mottle virus in pepper sauce in China. Arch. Virol. 2015;160:2079–2082. doi: 10.1007/s00705-015-2454-7. [DOI] [PubMed] [Google Scholar]
- Petterson S., Grøndahl-Rosado R., Nilsen V., Myrmel M., Robertson L.J. Variability in the recovery of a virus concentration procedure in water: implications for QMRA. Water Res. 2015;87:79–86. doi: 10.1016/j.watres.2015.09.006. [DOI] [PubMed] [Google Scholar]
- Polo D., Quintela-Baluja M., Corbishley A., Jones D.L., Singer A.C., Graham D.W., Romalde J.L. Making waves: wastewater-based epidemiology for COVID-19 – approaches and challenges for surveillance and prediction. Water Res. 2020;186:116404. doi: 10.1016/j.watres.2020.116404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico M., Andrés-Costa M.J., Picó Y. Estimating population size in wastewater-based epidemiology. Valencia metropolitan area as a case study. J. Hazard. Mater. 2017;323:156–165. doi: 10.1016/j.jhazmat.2016.05.079. [DOI] [PubMed] [Google Scholar]
- Rockett R.J., Arnott A., Lam C., Sadsad R., Timms V., Gray K.A., Eden J.S., Chang S., Gall M., Draper J., Sim E.M., Bachmann N.L., Carter I., Basile K., Byun R., O’Sullivan M.V., Chen S.C.A., Maddocks S., Sorrell T.C., Dwyer D.E., Holmes E.C., Kok J., Prokopenko M., Sintchenko V. Revealing COVID-19 transmission in Australia by SARS-CoV-2 genome sequencing and agent-based modeling. Nat. Med. 2020;26:1398–1404. doi: 10.1038/s41591-020-1000-7. [DOI] [PubMed] [Google Scholar]
- Rosario K., Symonds E.M., Sinigalliano C., Stewart J., Breitbart M. Pepper mild mottle virus as an indicator of fecal pollution. Appl. Environ. Microbiol. 2009;75:7261–7267. doi: 10.1128/AEM.00410-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosiles-González G., Ávila-Torres G., Moreno-Valenzuela O.A., Acosta-González G., Leal-Bautista R.M., Grimaldo-Hernández C.D., Brown J.K., Chaidez-Quiroz C., Betancourt W.Q., Gerba C.P., Hernández-Zepeda C. Occurrence of pepper mild mottle virus (PMMoV) in groundwater from a karst aquifer system in the Yucatan peninsula, Mexico. Food Environ. Virol. 2017;9:487–497. doi: 10.1007/s12560-017-9309-1. [DOI] [PubMed] [Google Scholar]
- Rousis N.I., Gracia-Lor E., Zuccato E., Bade R., Baz-Lomba J.A., Castrignanò E., Causanilles A., Covaci A., de Voogt P., Hernàndez F., Kasprzyk-Hordern B., Kinyua J., McCall A.K., Plósz B.G., Ramin P., Ryu Y., Thomas K.V., van Nuijs A., Yang Z., Castiglioni S. Wastewater-based epidemiology to assess pan-European pesticide exposure. Water Res. 2017;121:270–279. doi: 10.1016/j.watres.2017.05.044. [DOI] [PubMed] [Google Scholar]
- Sassi H.P., Tuttle K.D., Betancourt W.Q., Kitajima M., Gerba C.P. Persistence of viruses by qPCR downstream of three effluent-dominated Rivers in the Western United States. Food Environ. Virol. 2018;10:297–304. doi: 10.1007/s12560-018-9343-7. [DOI] [PubMed] [Google Scholar]
- Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., Ahmed W., Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 2020;743:140621. doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds E.M., Verbyla M.E., Lukasik J.O., Kafle R.C., Breitbart M., Mihelcic J.R. A case study of enteric virus removal and insights into the associated risk of water reuse for two wastewater treatment pond systems in Bolivia. Water Res. 2014;65:257–270. doi: 10.1016/j.watres.2014.07.032. [DOI] [PubMed] [Google Scholar]
- Thompson J.R., Nancharaiah Y.V., Gu X., Lee W.L., Rajal V.B., Haines M.B., Girones R., Ng L.C., Alm E.J., Wuertz S. Making waves: wastewater surveillance of SARS-CoV-2 for population-based health management. Water Res. 2020;184 doi: 10.1016/j.watres.2020.116181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trottier J., Darques R., Ait Mouheb N., Partiot E., Bakhache W., Deffieu M.S., Gaudin R. Post-lockdown detection of SARS-CoV-2 RNA in the wastewater of Montpellier, France. One Heal. 2020;10:0–3. doi: 10.1016/j.onehlt.2020.100157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang L.F., Sham S.Y.A., Chan S.K. Identifying reasons, gaps and prevalence of diaper usage in an acute hospital. Int. J. Urol. Nurs. 2017;11:151–158. doi: 10.1111/ijun.12143. [DOI] [Google Scholar]
- Tscharke B.J., O’Brien J.W., Ort C., Grant S., Gerber C., Bade R., Thai P.K., Thomas K.V., Mueller J.F. Harnessing the power of the census: characterizing wastewater treatment plant catchment populations for wastewater-based epidemiology. Environ. Sci. Technol. 2019;53:10303–10311. doi: 10.1021/acs.est.9b03447. [DOI] [PubMed] [Google Scholar]
- Vallejo, J., Rumbo-Feal, S., Conde, K., López-Oriona, Á., Tarrío, J., Reif, R., Ladra, S., Rodiño-Janeiro, B., Nasser, M., Cid, A., Veiga, M., Acevedo, A., Lamora, C., Bou, G., Cao, R., Poza, M., 2020. Highly predictive regression model of active cases of COVID-19 in a population by screening wastewater viral load 1–36. 10.1101/2020.07.02.20144865. [DOI]
- Volz E., Hill V., McCrone J.T., Price A., Jorgensen D., O’Toole Á., Southgate J., Johnson R., Jackson B., Nascimento F.F., Rey S.M., Nicholls S.M., Colquhoun R.M., da Silva Filipe A., Shepherd J., Pascall D.J., Shah R., Jesudason N., Li K., Jarrett R., Pacchiarini N., Bull M., Geidelberg L., Siveroni I., Goodfellow I., Loman N.J., Pybus O.G., Robertson D.L., Thomson E.C., Rambaut A., Connor T.R. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell. 2020:64–75. doi: 10.1016/j.cell.2020.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Poyet M., Moniz K., Washburne A.D., Erickson T.B., Chai P.R., Thompson J., Alm E.J. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5 doi: 10.1128/mSystems.00614-20. 2020.04.05.20051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y.H., Ni W., Wu Q., Li W.J., Li G.J., Di Wang W., Tong J.N., Song X.F., Wing-Kin Wong G., Xing Q.S. Prolonged viral shedding in feces of pediatric patients with coronavirus disease 2019. J. Microbiol. Immunol. Infect. 2020;53:473–480. doi: 10.1016/j.jmii.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Li X., Zhu B., Liang H., Fang C., Gong Y., Guo Q., Sun X., Zhao D., Shen J., Zhang H., Liu H., Xia H., Tang J., Zhang K., Gong S. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020;26:502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Xu G., Reboud J., Kasprzyk-Hordern B., Cooper J.M. Monitoring genetic population biomarkers for wastewater-based epidemiology. Anal. Chem. 2017;89:9941–9945. doi: 10.1021/acs.analchem.7b02257. [DOI] [PubMed] [Google Scholar]
- Zhou W.P., Li Y.Y., Li F., Tan G.L. First report of natural infection of tomato by pepper mild mottle virus in China. J. Plant Pathol. 2020;650201 doi: 10.1007/s42161-020-00688-y. [DOI] [Google Scholar]
- Zhurakivska K., Troiano G., Pannone G., Caponio V.C.A., Lo Muzio L. An overview of the temporal shedding of SARS-CoV-2 RNA in clinical specimens. Front. Public Health. 2020;8:1–9. doi: 10.3389/fpubh.2020.00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables