Abstract
Background and Objective
The new type of Coronavirus (2019-nCov) epidemic spread rapidly, causing more than 250 thousand deaths worldwide. The virus, which first appeared as a sign of pneumonia, was later called the SARS-COV-2 with Severe Acute Respiratory Syndrome by the World Health Organization. The SARS-COV-2 virus is triggered by binding to the Angiotensin-Converting Enzyme 2 (ACE 2) inhibitor, which is vital in cardiovascular diseases and the immune system, especially in conditions such as cerebrovascular, hypertension, and diabetes. This study aims to evaluate the prediction performance of death status based on the demographic/clinical factors (including COVID-19 severity) by data mining methods.
Methods
The dataset consists of 1603 SARS-COV-2 patients and 13 variables obtained from an open-source web address. The current dataset contains age, gender, chronic disease (hypertension, diabetes, renal, cardiovascular, etc.), some enzymes (ACE, angiotensin II receptor blockers), and COVID-19 severity, which are used to predict death status using deep learning and machine learning approaches (random forest, k-nearest neighbor, extreme gradient boosting [XGBoost]). A grid search algorithm tunes hyperparameters of the models, and predictions are assessed through performance metrics. Steps of knowledge discovery in databases are applied to obtain the relevant information.
Results
The accuracy rate of deep learning (97.15%) was more successful than the accuracy rate based on classical machine learning (92.15% for RF and 93.4% for k-NN), but the ensemble classifier XGBoost method gave the highest accuracy (99.7%). While COVID-19 severity and age calculated from XGBoost were the two most important factors associated with death status, the most determining variables for death status estimated from deep learning were COVID-19 severity and hypertension.
Conclusions
The proposed model (XGBoost) achieved the best prediction of death status based on the factors as compared to the other algorithms. The results of this study can guide patients with certain variables to take early measures and access preventive health care services before they become infected with the virus.
Keywords: SARS-COV-2, Data Mining, Deep Learning, Extreme Gradient Boosting, Machine Learning
1. Introduction
The new type of Coronavirus (2019-nCov) pandemic, which started in December 2019 in Wuhan, China, spread rapidly, causing more than 250 thousand deaths worldwide [1]. The virus, which first appeared as a sign of pneumonia, was later called the Coronavirus 2 (SARS-COV-2) with Severe Acute Respiratory Syndrome by the World Health Organization (WHO) [2]. Although pharmacological treatment has not been reported in the first studies on patients with severe symptoms; Some patients with Cardiovascular Disorders (CVD) have been shown to cause severe damage and an increased risk of death [3]. The SARS-COV-2 virus is triggered by binding to the Angiotensin-Converting Enzyme 2 (ACE 2) inhibitor, which is vital in CVD and the immune system, especially in conditions such as cerebrovascular, hypertension, and diabetes [4,5].
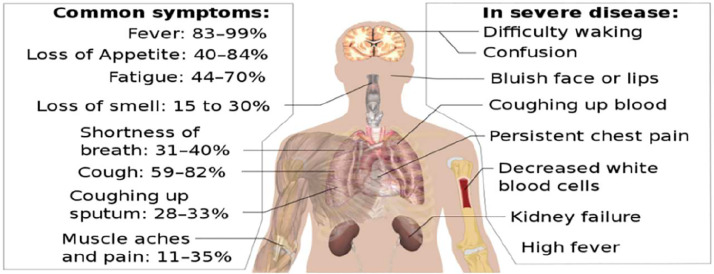
The most prevalent symptoms of the virus are high fever, dry cough, and weakness. Headache, pain in the muscles, sore throat and diarrhea, loss of taste and smell, and rash on the skin are less common. Dyspnea, shortness of breath, chest pain, loss of speech, and movement are severe symptoms [7,8].
According to the recommendation of the WHO, those who show severe symptoms should get medical help immediately and call by phone before visiting their doctor or health facility; People with mild symptoms and no other health problems should spend their treatment at home. People infected with the virus begin to show symptoms within an average of 5-6 days. However, this duration can take up to 14 days [6].
Data mining, which is also called the discovery of data in the systems, including databases, is a process of obtaining unearthed and potentially beneficial information from the data attained beforehand. The new-found data can be used in areas consisting of information management, inquiry work, and decision-making process control in this aspect. A great number of researchers working in the area of database systems, information base systems, artificial intelligence, machine learning, data-acquiring, statistic, spatial databases, and visualization of data show great interest in data mining [9,10]. Data mining has vital significance and potential, especially for the field of health [4,5].
The current study aims to evaluate the effects of age, gender, chronic disease (hypertension, diabetes, renal, cardiovascular, etc.), some enzymes (ACE, ARBs), and COVID-19 severity on the death status with deep learning and machine learning approaches.
2. Methods
2.1. Dataset
The current study included 1603 patients and 13 variables suffering from SARS-COV-2 disease and the public dataset was obtained from the related website (https://doi.org/10.1371/journal.pone.0235248.s001) on 16 July 2020 [11]. The related data on background pharmacological treatment up to the previous two years (January 1, 2018) were achieved from the National database of drug prescription and integrated with clinical chart information for hospitalized subjects. Data have been gathered on the subsequent drugs: angiotensin-converting enzyme (ACE) inhibitors (anatomical therapeutic chemical classes: C09A and C09B), angiotensin II receptor blockers (ARBs) (C09C and C09D), and other anti-diabetic or insulin medicines. Data on all subjects' age, gender, and pre-existing conditions was collected through data-linkage with hospital discharge abstracts (Italian SDO), which were queried from the day of diagnosis until January 1, 2015. Two physicians (LM and MEF) manually analyzed all admission data. They included the following conditions in the analysis: Malignant tumours, major cardiovascular disorders (heart failure, myocardial infarction, and stroke CVD), type II diabetes, renal disease, and chronic pulmonary obstructive disorders (COPD, bronchitis, pneumonia, asthma, and emphysema) [11]. Detailed explanations about the variables used in the current study are given in Table 1 .
Table 1.
The detailed explanation of the variables/attributes in the dataset.
| Abbreviation | Explanation | Role |
|---|---|---|
| Age | Birth year (year) | Input |
| Gender | Gender (0=female, 1=male) | Input |
| Diabetes | Diabetes (1= presence, 0= absence) | Input |
| Hypertension | Hypertension (1= presence, 0= absence) | Input |
| COPD | Chronic Obstructive Pulmonary Diseases (bronchitis, pneumonia, asthma, and emphysema) | Input |
| Cancer | Cancer Diseases (1= presence, 0= absence) | Input |
| Renal | Renal Diseases (1= presence, 0= absence) | Input |
| ACE | Angiotensin-Converting Enzyme (ATC classes: C09A and C09B) | Input |
| ARBs | Angiotensin II Receptor Blockers (C09C and C09D) | Input |
| CVD | Cardiovascular disorders (heart failure, myocardial infarction, and stroke-CVD) | Input |
| COVID Severity | SARS-COV-2 Severity (0=mild, 1= severe, 2= very severe) | Input |
| Death Status (DS) | (0=alive, 1=dead) | Output |
2.2. Knowledge discovery in databases (KDD)
In the process of KDD, data selection (output: DS; inputs: factors in Table 1), data preprocessing (outlier/extreme observation by local outlier factor (LOF) and missing value analyses by random forest), data transformation, statistical analyses, data mining (deep learning, random forest, k-nearest neighbor and extreme gradient boosting), evaluation (performance metrics), and interpretation of the results are performed throughout the study [12].
2.3. Data mining
2.3.1. Deep learning
Deep Learning can automatically extract feature representation from raw data, which is a new method of machine learning derived from artificial neural networks [13]. DL learns characteristic hierarchies with higher hierarchy features with a combination of low-level features. Thus, DL successfully solves complex and severe dimensional problems. It is used. Convolutional Neural Network (CNN) is one of the most successful deep learning models [14]. The value of the location of the layer l and its location in the k feature map (i, j), can be estimated as shown in Eq. (1).
| (1) |
Where = and are l th layers in the k property map is the weight vector and the bias. The activation value for the convolution feature can be expressed, as shown in Eq. (2).
| (2) |
Hyperparameters of the deep learning model are epsilon, rho, L1, L2, max w2, and dropout, which are tuned by a grid search optimization algorithm.
2.3.2. Machine learning methods
2.3.2.1. Random forest
Random Forest (RF) is a collection of tree-type classifiers. It can be considered an advanced type of method of bagging. The RF algorithm consists of the following steps. Each decision tree in RF, bootstrap re-sampling method technique (to create datasets of any size and quantity can be re-sampled by replacing observations from any size dataset. Thus, more information can be obtained from the dataset. The method described in this way is the “Bootstrap Re-sampling Method.”, and different samples are created by selection [15]. Forest brings together the class estimates made by the trees and reveals the best class estimation. The differentiating variable in RF is selected among m variables randomly determined from all variables. The number of m is constant for each tree, and it is generally predicted to be taken as (p: number of variables) [16]. Hyperparameters of the random forest model are minimal gain, minimal leaf size, minimal size for sp, and the number of preprun, which are calibrated by grid search optimization algorithm.
2.3.2.2. K-Nearest neighbor (K-NN)
K-Nearest Neighbor (K-NN) is a widely used machine learning method. Like the classifiers of the supervised learning paradigm, k-NN h (x) needs training data P⊆X containing data points x∈X whose values are known., The classifier is expected to estimate the class tag of the new sample using P information. K-NN is widely used due to its good performance as well as its simplicity. Also, k-NN, a nonparametric classifier, is not dependent on previously made assumptions about data distribution. However, for successful classification, the k-NN k value requires three essential factors: the distance metric used to determine neighbors and the sample size [17]. Hyperparameters of the k-NN model are k, measures type, mixed measures, which are optimized by a grid search algorithm.
2.3.2.3. Extreme Gradient Boosting (XGBoost)
The Extreme Gradient Boosting (XGBoost) by Chen and Guestrin [18] is a highly scalable end-to-end tree boosting system, a machine learning technique for classification and regression problems. XGBoost uses an ensemble of K classification and regression trees (CARTs), each of which has nodes [19]. The ultimate prediction is the sum of the prediction scores for each tree:
| (3) |
Where xi are members of the training set and yi are the corresponding class labels, fk is the leaf score for the k th tree, and F is the set of all K scores for all CARTs. Regularization is applied to improve the final result:
| (4) |
Hyperparameters of the XGBoost model are maximal depth, number of bins, and learning rate, which are tuned by grid search optimization algorithm.
2.4. Performance metrics
The 10-fold cross-validation method was used in the performance evaluation of all classifier methods to verify the quality of the models. Cross-validation is the re-sampling procedure used to evaluate machine learning models in a data sample. The procedure has a single parameter named k that expresses the number of groups to split a given data sample. In 10-fold cross-validation, the models are trained and tested ten different times, and then, mean performance metrics (i.e., accuracy, precision, and so on) are estimated at the end of the process [20]. All the model performances were calculated based on accuracy, precision, sensitivity, specificity, classification error, and kappa metrics [21].
2.5. Data analysis
Quantitative data were summarized as the arithmetic mean with standard deviation, median with min and max values, and qualitative data as the number by percentage. Differences among the groups with mild, severe, or very severe/fatal diseases were performed with Pearson chi-square test (cross-table) for categorical data and Kruskal Wallis H test for continuous data since the dataset did not show normal distribution given in Table 3. When significant differences in categorical data were determined among the groups (p<0.05), pairwise comparisons were performed by Bonferroni-adjusted Pearson chi-square test. Upon seeing significant differences (p<0.001) in the Kruskal Wallis H test, pairwise comparisons of the groups with significant differences were identified using the post-hoc Conover multiple comparison test. Data analyses were performed using “Statistical Analysis Software” [22], RStudio Version 1.1.463 [23], and RapidMiner Studio 8.1.001 [24] softwares. All p values < 0.05 were accept statistically significant.
Table 3.
The group comparisons of the variables.
| Variables | Mild | Severe | Very Severe |
Test Statistics |
|
|---|---|---|---|---|---|
| X2 | p-value | ||||
| Age (X̄±SD)/Med (Min-Max)) | 50.4 ± 20.2 / 51 (0-100)a |
66.4 ± 16.9 / 67.9 (19.1-100)b |
76.2 ± 12.9 / 78 (32.5-100)c |
355.1 | <0.001* |
| Diabetes (Count (%)) | 65a (34%) | 75b (38%) | 54c (28%) | 79.98 | <0.001** |
| Hypertension (Count (%)) | 207a (38%) | 207a (38%) | 129b (24%) | 186.9 | <0.001** |
| COPD (Count (%)) | 28a (29%) | 42b (43%) | 27a,c (28%) | 46.27 | <0.001** |
| Cancer (Count (%)) | 49a (40%) | 46b (38%) | 27b (22%) | 23.9 | <0.001** |
| CVD (Count (%)) | 66a (26%) | 122b (47%) | 70a,c (27%) | 157.9 | <0.001** |
| Renal (Count (%)) | 23a (27%) | 40b (46%) | 23a,c (27%) | 43.67 | <0.001** |
| ACE (Count (%)) | 107a (43%) | 88b (35%) | 56c (22%) | 45.79 | <0.001** |
| ARBs (Count (%)) | 86a (38%) | 90b (39%) | 52c (23%) | 59.18 | <0.001** |
| Gender (Count (%)) | Mild | Severe | Very Severe | X2 | p-value |
| Female | 550a (65%) | 213b (25%) | 82b (10%) | 22.523 | <0.001 |
| Male | 407a (54%) | 241b (32%) | 110b (14%) | ||
| Total | 957 (60%) | 454 (28%) | 192 (12%) | ||
The data are summarized as X̄±SD or median (min-max) and Count (Percent). Different superscripts in each row imply a significant difference between categories (Conover or Bonferroni-corrected Pearson chi-square tests for pairwise comparisons; p<0.05); *: Kruskal Wallis H test; **: Pearson chi-square test.
3. Results
Missing value analysis was applied to the variables in the data set first. Missing values were imputed with the random forest assignment method. Then, extreme and outlier values in the data set were examined. With the local outlier factor (LOF), object distances close to itself were calculated for each observation, and a total of 8 extreme or outlier values were deleted from the data set. Data transformation was applied to the quantitative variables in the data set. The transformation with the lowest test statistic was selected by calculating the Pearson P test statistic for each variable. Box-Cox transformation was applied for the age variable.
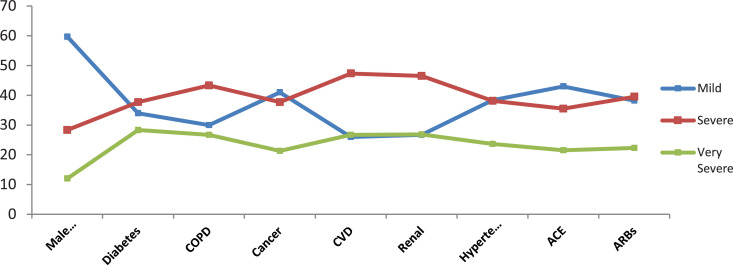
The average age of the sample consisting of 1603 people is 58.0, and 47.3% are males. In the overall sample, approximately 59.7% had mild symptoms, 28.3% severe symptoms, and 12% very severe or fatal illness. While the proportion of diabetic patients in the overall sample was 12.1%, approximately 34% of these patients had mild symptoms, 37.7% severe symptoms, and 28.3% very severe or fatal disease. Approximately 22.2% of diabetic patients died after failing to treatment. While the proportion of hypertensive patients in the overall sample was 33.9%, approximately 38.3% of these patients had mild symptoms, 38.1% severe symptoms, and 23.6% very severe or fatal disease. Approximately 20% of hypertensive patients died without treatment. While the proportion of cancer patients in the overall sample was 7.6%, 41% of these patients had mild symptoms, 37.7% severe symptoms, and 21.3% very severe or fatal disease. While the proportion of CVD patients in the overall sample was approximately 16.1%, 26% of these patients had mild symptoms, 47.3% severe symptoms, and 26.7% very severe or fatal disease. Approximately 25.2% of CVD patients died without treatment. While the proportion of COPD patients in the overall sample was 6%, 30% of these patients had mild symptoms, 43.3% severe symptoms, and 26.7% very severe or fatal disease. Approximately 26.8% of COPD patients died without treatment. While the proportion of renal patients in the overall sample was 5.4%, 26.7% of these patients had mild symptoms, 46.5% severe symptoms, and 26.8% very severe or fatal disease. Approximately 26.7% of renal patients died without treatment. The proportion of patients receiving antihypertensive treatment with ACE inhibitor enzyme was 15.7%, 43% of these patients had mild symptoms, 35.5% severe symptoms, and 21.5% very severe or fatal disease. Approximately 17.9% of the patients who received antihypertensive treatment with ACE inhibitor enzyme died after failing the treatment. The proportion of patients receiving antihypertensive treatment with ARBs inhibitor enzyme was 14.2%, 38.2% of these patients had mild symptoms, 39.5% severe symptoms, and 22.3% very severe or fatal disease. Approximately 20.1% of the patients who received antihypertensive treatment with the ARBs inhibitor enzyme died without a result of the treatment (Table 2 and Figure 3). The characteristics of the patients with respect to the variables are demonstrated in Figure 3 and Table 2.
Fig. 1.
SARS-COV-2 (yellow) from humans [6].
Fig. 2.
SARS-COV-2 symptoms [6].
Table 2.
The baseline characteristics of the sample.
| Variables | Overall Sample | Mild | Severe | Very Severe |
|---|---|---|---|---|
| n | 1603 | 957 | 454 | 192 |
| Mean age in years (X̄±SD) | 58.0±20.9 | 50.4± 20.2 | 66.4±16.9 | 76.2±12.9 |
| Male gender (Count (%)) | 758 (47.3) | 407 (53.7) | 241 (31.8) | 110 (14.5) |
| Diabetes (Count (%)) | 194 (12.1) | 65 (34) | 75 (37.7) | 54 (28.3) |
| COPD (Count (%)) | 97 (6.0) | 28 (28.9) | 42(43.3) | 27 (27.8) |
| Cancer (Count (%)) | 122 (7.6) | 49 (40.2) | 46 (37.7) | 27 (22.1) |
| CVD (Count (%)) | 258 (16.1) | 66 (25.6) | 122 (47.3) | 70 (27.1) |
| Renal Disease (Count (%)) | 86 (5.4) | 23 (26.7) | 40 (46.6) | 23 (26.7) |
| Hypertension (Count (%)) | 543 (33.9) | 207 (38.1) | 207 (38.1) | 129 (23.6) |
| ACE inhibitors (Count (%)) | 251 (15.7) | 107 (42.6) | 88 (35.1) | 56 (22.3) |
| ARBs (Count (%)) | 228 (14.2) | 86 (37.7) | 90 (39.5) | 52 (22.8) |
Fig. 3.
The characteristics of the sample.
According to Table 3 , a statistically significant difference was observed between the variables of age, diabetes, hypertension, COPD, CVD, cancer, renal, ACE, and ARBs and the severity of COVID (p<0.05). When significant differences were seen in the Kruskal Wallis H test for continuous variable (age), and cross tabulation chi-squared test for categorical ones. The groups with differences between them were determined with the Post Hoc Multiple comparison test. Significant differences were observed in all COVID-19 severity categories with age variable, diabetes, hypertensive disease, and antihypertensive treatment with ACE inhibitor enzyme. According to the ARBs inhibitor enzyme, renal, COPD, and, cancer disease, there was no difference in severe and very severe COVID-19 disease categories. In contrast, a significant difference was observed in mild and severe and mild and very severe COVID-19 disease categories. According to gender, there was no difference in severe and very severe COVID-19 disease categories in males and females. In contrast, a significant difference was observed in mild and severe, and mild and very severe COVID-19 disease categories.
Hyperparameters of the deep learning model were 1.0E-8 for epsilon, 0.99 for rho, 1.0E-5 for L1, 0.0 for L2, 10.0 for max w2, and 0.15 for dropout, respectively. Hyperparameter values related to the random forest model were 0.1 for minimal gain, 2 for minimal leaf size, 4 for minimal size for sp, and 3 for the number of preprun, fitted into the optimization algorithm for a grid search. The k-NN model’ hyperparameters were 1 for k, mixed measures for measures type, mixed Euclidean for composite measures, grid search optimization algorithm. In the same way, the XGBoost model had the hyperparameters, which were 5 for maximal depth, 20 for the number of bins, and 0.1 for learning rate hyperparameters, which are tuned by grid search optimization algorithm.
Figure 4 depicts the pseudo-codes of the XGBoost algorithm, which gives the best result of death status based on the demographic/clinical factors.
Fig. 4.
The pseudo-codes of the XGBoost algorithm.
Table 4 tabulates the importance levels of variables in SARS-COV-2 patients on the death status in the deep learning and XGBoost modeling. COVID-19 severity (1-10.3%), hypertension (0.98-10.1%), COPD (0.95-9.8%) and gender (0.92-9.5%) were calculated from deep learning. In comparison, the lowest relative significance was estimated for diabetes disease (0.77- 7.6%) from deep learning. COVID-19 severity (1-89.9%) and age (0.095-8.6%) provided the highest importance, while the lowest importance values were for cancer and COPD from the XGBoost technique.
Table 4.
Variable importance values for the deep learning and the XGBoost algorithms.
| Algorithms |
Deep learning |
XGBoost |
||
|---|---|---|---|---|
| Variable | Relative Importance | Percentage (%) | Relative Importance | Percentage (%) |
| Covid-19 severity | 1.00 | 10.3 | 1.00 | 89.9 |
| Hypertension | 0.98 | 10.1 | 0.004 | 0.35 |
| COPD | 0.95 | 9.8 | 0.000 | 0.00 |
| Gender | 0.92 | 9.5 | 0.002 | 0.15 |
| Renal | 0.89 | 9.2 | 0.001 | 0.11 |
| CVD | 0.89 | 9.1 | 0.004 | 0.4 |
| ARBs | 0.85 | 8.8 | 0.003 | 0.3 |
| Cancer | 0.84 | 8.7 | 0.000 | 0.00 |
| Age | 0.83 | 8.5 | 0.095 | 8.6 |
| ACE | 0.81 | 8.4 | 0.000 | 0.03 |
| Diabetes | 0.77 | 7.6 | 0.001 | 0.11 |
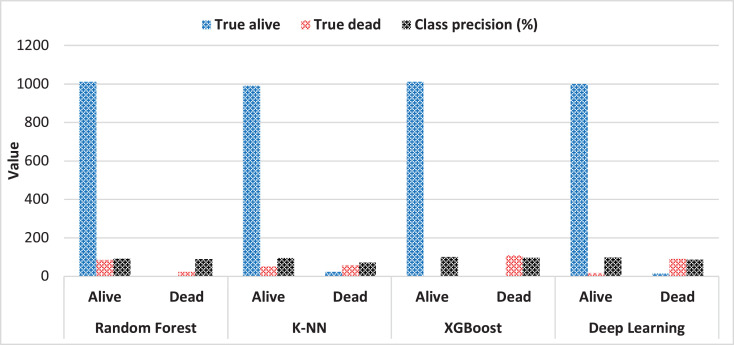
In the classification process performed with deep learning and machine learning approaches (random forest and k-NN), the correct positive rate in the deep learning algorithm, according to the confusion matrix indicated in Table 5 , is 92.2%. In comparison, the correct negative rate is 88.5%. According to the random forest algorithm, the correct positive rate is 98.3%, while the correct negative rate is 85.9%. In the k-NN algorithm, the true positive rate is 95.1%, while the rate of true negative is 71.2%. In the XGBoost algorithm, the true positive rate is 100%, while the rate of true negative is 97.3%.
Table 5.
Confusion matrix for the techniques.
| Random Forest | true alive | true dead | class precision |
| alive | 1011 | 85 | 92.24% |
| dead | 3 | 23 | 88.46% |
| class recall | 99.70% | 21.30% | |
| Deep Learning | |||
| alive | 999 | 17 | 98.33% |
| dead | 15 | 91 | 85.85% |
| class recall | 98.52% | 84.26% | |
| k-NN | |||
| alive | 991 | 51 | 95.11% |
| dead | 23 | 57 | 71.25% |
| class recall | 97.73% | 52.78% | |
| XGBoost | |||
| alive | 1011 | 0 | 100 |
| dead | 3 | 108 | 97.3 |
| class recall | 99.7 | 100 |
The graphical representation of the confusion matrix for the models is given in Figure 5 .
Fig. 5.
The graphical representation of the confusion matrix for the models.
According to the model performance metric results in Table 6 , the XGBoost classification algorithm gave the most successful result. The accuracy rate based on deep learning (97.15%) was more successful than the accuracy rate based on classic machine learning (RF 92.15% and k-NN 93.4%). Still, the ensemble classifier XGBoost method gave the highest accuracy. In kappa statistics, which measure the reliability of the statistical fit, the XGBoost and DL approaches represent a perfect fit with the values of 0.91 and 0.82. In the machine learning approach, the k-NN algorithm shows a good fit with a value of 0.58, while in the RF algorithm, it was observed that the fit with 0.19 value is insignificant.
Table 6.
Performance metrics of the models.
| Model | Accuracy | Precision | Sensitivity | Specificity | Class. Error | Kappa |
|---|---|---|---|---|---|---|
| DL | 97.15 | 98.5 | 92.2 | 88.5 | 2.85 | 0.82 |
| RF | 92.15 | 99.7 | 98.3 | 85.9 | 7.85 | 0.19 |
| k-NN | 93.4 | 97.7 | 95.1 | 71.2 | 6.6 | 0.58 |
| XGBoost | 99.7 | 99.7 | 99.7 | 1.00 | 0.03 | 0.91 |
4. Discussion
COVID-19 is an infectious disease caused in humans by a new virus never before described. With symptoms such as cough, fever, and, in extreme cases, pneumonia, this virus causes respiratory illness (for example, flu). The test is conducted on sputum or blood samples to detect the presence of this virus in humans, and the result is usually available within a few hours or, at most, days [25]. The current study intends to classify the effects of age, gender, chronic diseases (hypertension, diabetes, renal, cardiovascular, etc.), and some enzymes (ACE, ARBs) on the course of the COVID-19 pandemic in patients under treatment with deep learning and machine learning methods. Early diagnosis and prediction of COVID-19 are crucial in terms of saving people's lives and managing pandemic. If considered clinically, the COVID-19 causes illness in humans and creates severe damage in the lungs. COVID-19 has killed many people in the entire world, and chronic diseases, cancer, age, and gender are essential variables in the course of SARS-COV-2 disease. Cardiovascular disease, endocrine system disease, and respiratory system disease are the three most common chronic diseases coexisting. While imaging (CT) devices can follow the course of the disease, access to treatment is often difficult for COVID-19 patients. Applying to health centers in case of home quarantine and specific symptoms makes it difficult to monitor the course of the disease with imaging devices. Therefore, the steps to be followed in the early course of the disease are of vital importance. In the current study, we developed an early diagnosis and treatment method by classifying the effects of age, gender, chronic diseases, and some enzymes on the course of COVID-19 using artificial intelligence approaches.
The study conducted by Ahamad et al. (2020) [26] employed a machine learning model to identify early-stage symptoms of SARS-Cov-2 infected patients. They developed and tested a range of machine learning approaches and found the most significant clinical COVID-19 predictive features were (in descending order): lung infection, cough, pneumonia, runny nose, travel history, fever, isolation, age, muscle soreness, diarrhea, and gender. Their models predicted the stage of COVID-19 based on necessary patient information (age and gender), travel and isolation, and clinical symptoms (including fever, cough, and runny nose, and pneumonia). Similarly, the study of Banerjee et al. (2020) [27] used machine learning and artificial intelligence to forecast SARS-CoV-2 infection from full blood counts in a population. The authors mentioned the multiple independent models (statistical, random forest, and shallow learning) that can predict SARS-COV-2 with an AUC of up to 86% for community and 95% for regular ward patients, using only data collected from their normalized full blood counts. This situation provides an initial screen of SARS-CoV-2 positive from negative using biomarkers at an early stage in the disease presentation. This screen has been conducted on a set of data based on severity judged by the location of the patient in hospital (admitted to the regular ward compared to not admitted to hospital; ICU patients were excluded). Hence the models can distinguish from altered blood profiles in patients who were later diagnosed with other pathogens. In another paper, Brunese et al. (2020) [25] have suggested the adoption of deep learning for the detection of COVID-19 from X-rays to provide a fully automated and faster diagnosis. This experimental research study of 6,523 chest X-rays belonging to various institutions demonstrated the feasibility of the method proposed, with an average time of approximately 2.5 seconds for COVID-19 detection and an average accuracy of 0.97.
Based on biological criteria, Albari et al. (2020) [28] proposes a rescue framework for the transfusion of the best CP to the most critical patients with COVID-19 by using machine learning and novel multi-criteria decision-making approaches. They recommend that an intelligence-integrated concept is suggested to classify the most suitable convalescent plasma for corresponding COVID-19 priority patients to help doctors accelerate treatment. Considering another study of artificial intelligence, Pereira et al. (2020) [29] used only chest X-ray images to classify pneumonia caused by COVID-19 from other forms and even from healthy lungs. In the hierarchical classification scenario, the proposed solution tested in RYDLS-20 obtained a macro-average F1-Score of 0.65 using a multi-class solution and an F1-Score of 0.89 for the COVID-19 identification. In an unbalanced setting of more than three classes, the top identification rate obtained in this paper is the best nominal rate obtained for COVID-19 identification.
Several studies have been reported examining death outcomes from the COVID-19 pandemic in the past few months. In a study published in recent months, researchers aim to define baseline characteristics that predispose patients with COVID-19 to death in the hospital and evaluates retrospective analysis of 3,894 SARS-CoV-2 infected patients from 19 February to 23 May 2020 at 30 health centers across Italy. Random forest and Cox survival analysis are used for the specified prediction. After all, in a broad cohort of unselected patients with COVID-19, admitted to 30 separate clinical centers across Italy, impaired renal function, elevated C-reactive protein, and advanced age are significant predictors of in-hospital death [30]. Different regressor machine learning models are proposed in another work to extract the relationship between different factors and the COVID-19 spreading rate. By extracting the relationship between the number of reported cases and the weather variables in some regions, the machine learning algorithms used in the work estimate the effect of weather variables such as temperature and humidity on the transmission of COVID-19. From the experimental results, the researchers demonstrate that weather variables are more important in predicting the mortality rate, and thus, that temperature and humidity are essential characteristics for predicting the mortality rate of COVID-19 [31]. In order to find associations between these habits and the mortality rates caused by COVID-19 in another study, the eating habits of 170 countries are analyzed using machine learning techniques that group countries together according to the different distributions of fat, energy, and protein across 23 different types of food. The findings of the study show that obesity and high fat consumption occur in countries with the highest fat rate, while in countries with the lowest fat rate, higher cereal intake is correlated with a lower overall average intake [32].
Unlike the previously mentioned studies, our work presents a novel direction via the proposed model XGBoost, which attained the highest rate of predictive value, and the study outcomes can guide the related parties via the certain parameters so that the individuals can take the prompt measures and access to preventative health care service before getting infected by the COVID-19. Besides, our study successfully advocates the implementation of machine learning and deep learning in a thorough data mining context for the classification of survivability for people afflicted with COVID-19.
In brief, the SARS-COV-2 virus, which is effective worldwide, is very severe in people over 60 with chronic diseases. During the treatment process, the first amnesia is taken in patients with the disease, and the patient is examined. Vital signs are examined (heart rate, rhythm, respiratory rate, blood pressure, body temperature, and oxygen saturation are checked if the conditions are appropriate). The individual is admitted to the relevant service by providing respiratory support and circulatory support, and his examinations and chest radiography (CT) are taken. All of these processes are long, tiring, and risky processes. Due to the rapid spread of the virus in society and the rapid increase in the emergency density in the health center, it is necessary to take new and early measures in the fight against the virus. In our study, the proposed model (XGBoost) achieved the best prediction of death status based on the factors as compared to the other algorithms. The results of this study can guide patients with certain variables to take early measures and access preventive health care services before they become infected with the virus.
Acknowledgments
This study does not require ethical approval because the open-source data set is used. Also, there is no conflict of interest among the authors. Any institution or organization did not financially support this research.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.cmpb.2021.105951.
Appendix. Supplementary materials
References
- 1.W. Wang, J. Tang, F.J.J.o.m.v. Wei, Updated understanding of the outbreak of 2019 novel coronavirus (2019-nCoV) in Wuhan, China, 92 (2020) 441-447. [DOI] [PMC free article] [PubMed]
- 2.Bai Y., Yao L., Wei T., Tian F., Jin D.-Y., Chen L., Wang M.J.J. Presumed asymptomatic carrier transmission of COVID-19. 2020;323:1406–1407. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X.J.T.l. Clinical features of patients infected with 2019 novel coronavirus in. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.X. Yang, Y. Yu, J. Xu, H. Shu, H. Liu, Y. Wu, L. Zhang, Z. Yu, M. Fang, T.J.T.L.R.M. Yu, Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study, (2020). [DOI] [PMC free article] [PubMed]
- 5.J.-j. Zhang, X. Dong, Y.-y. Cao, Y.-d. Yuan, Y.-b. Yang, Y.-q. Yan, C.A. Akdis, Y.-d.J.A. Gao, Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China, (2020). [DOI] [PubMed]
- 6.Wikipedia, SARS-CoV-2, 2020.
- 7.Hui D.S., Azhar E.I., Madani T.A., Ntoumi F., Kock R., Dar O., Ippolito G., Mchugh T.D., Memish Z.A., Drosten C. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.W.M.H. Commission, Experts explain the latest bulletin of unknown cause of viral pneumonia.
- 9.Chen M.-S., Han J., Yu P.S., Engineering d. Data mining: an overview from a database perspective. 1996;8:866–883. [Google Scholar]
- 10.G. Piateski, W. Frawley, Knowledge discovery in databases, MIT press1991.
- 11.F. Bravi, M.E. Flacco, T. Carradori, C.A. Volta, G. Cosenza, A. De Togni, C.A. Martellucci, G. Parruti, L. Mantovani, L.J.m. Manzoli, Predictors of severe or lethal COVID-19, including Angiotensin Converting Enzyme Inhibitors and Angiotensin II Receptor Blockers, in a sample of infected Italian citizens, (2020). [DOI] [PMC free article] [PubMed]
- 12.U. Fayyad, Knowledge discovery in databases: An overview, Relational Data Mining, Springer2001, pp. 28-47.
- 13.Li Y., Nie X., Huang R. Web spam classification method based on deep belief networks. 2018;96:261–270. [Google Scholar]
- 14.Gu J., Wang Z., Kuen J., Ma L., Shahroudy A., Shuai B., Liu T., Wang X., Wang G., Cai J.J.P.R. Recent advances in convolutional neural networks. 2018;77:354–377. [Google Scholar]
- 15.Aktükün A. Asal bileşenler analizine bootstrap yaklaşımı. Ekonometri ve İstatistik e-Dergisi. 2005;1:1–11. [Google Scholar]
- 16.Breiman L. Random Forests, Machine Learning. 2001;45:5–32. [Google Scholar]
- 17.O.J.J.o.t.F.o.E. Yıldız, A.o.G. University, Melanoma detection from dermoscopy images with deep learning methods: Acomprehensive study, 34 (2019) 2241-2260.
- 18.Chen M.S., Han J., Yu P.S. Data mining: an overview from a database perspective. IEEE Transactions on Knowledge and data Engineering. 1996;8:866–883. [Google Scholar]
- 19.Chen T., Guestrin C. Xgboost: A scalable tree boosting system. Proceedings of the 22nd acm sigkdd international conference on knowledge discovery and data mining. 2016:785–794. [Google Scholar]
- 20.Yadav S., Shukla S. IEEE 6th International conference on advanced computing (IACC) IEEE; 2016. Analysis of k-fold cross-validation over hold-out validation on colossal datasets for quality classification; pp. 78–83. 2016. [Google Scholar]
- 21.Arlot S., Celisse A. A survey of cross-validation procedures for model selection. 2010;4:40–79. [Google Scholar]
- 22.Ş. YAŞAR, A. ARSLAN, C. Colak, S. Yoloğlu, A Developed Interactive Web Application for Statistical Analysis: Statistical Analysis Software, 6 227-239.
- 23.M. Campbell, RStudio Projects, Learn RStudio IDE, Springer2019, pp. 39-48.
- 24.M. Hofmann, R. Klinkenberg, RapidMiner: Data mining use cases and business analytics applications, CRC Press2016.
- 25.Brunese L., Mercaldo F., Reginelli A., Santone A. Explainable Deep Learning for Pulmonary Disease and Coronavirus COVID-19 Detection from X-rays. Computer Methods and Programs in Biomedicine. 2020;196 doi: 10.1016/j.cmpb.2020.105608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahamad M., Aktar S., Uddin S., Lió P., Xu H., Summers M.A., Quinn J.M., Moni M.A. A machine learning model to identify early stage symptoms of SARS-Cov-2 infected patients. 2020 doi: 10.1016/j.eswa.2020.113661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banerjee A., Ray S., Vorselaars B., Kitson J., Mamalakis M., Weeks S., Mackenzie L.S.J.I.i. Use of Machine Learning and Artificial Intelligence to predict SARS-CoV-2 infection from Full Blood Counts in a population. 2020 doi: 10.1016/j.intimp.2020.106705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O. Albahri, J.R. Al-Obaidi, A. Zaidan, A. Albahri, B. Zaidan, M.M. Salih, A. Qays, K. Dawood, R. Mohammed, K.H.J.C.m. Abdulkareem, p.i. biomedicine, Helping doctors hasten COVID-19 treatment: Towards a rescue framework for the transfusion of best convalescent plasma to the most critical patients based on biological requirements via ml and novel MCDM methods, 196 (2020) 105617. [DOI] [PMC free article] [PubMed]
- 29.Pereira R.M., Bertolini D., Teixeira L.O., Silla Jr C.N., Costa Y.M.J.C.M., Biomedicine P.i. COVID-19 identification in chest X-ray images on flat and hierarchical classification scenarios. 2020 doi: 10.1016/j.cmpb.2020.105532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Di Castelnuovo A., Bonaccio M., Costanzo S., Gialluisi A., Antinori A., Berselli N., Blandi L., Bruno R., Cauda R., Guaraldi G.J.N. Metabolism, C. Diseases, Common cardiovascular risk factors and in-hospital mortality in 3,894 patients with COVID-19: survival analysis and machine learning-based findings from the multicentre Italian CORIST Study. 2020;30:1899–1913. doi: 10.1016/j.numecd.2020.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malki Z., Atlam E.-S., Hassanien A.E., Dagnew G., Elhosseini M.A., Gad I.J.C. Solitons, Fractals, Association between weather data and COVID-19 pandemic predicting mortality rate: Machine learning approaches. 2020;138 doi: 10.1016/j.chaos.2020.110137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.García-Ordás M.T., Arias N., Benavides C., García-Olalla O., Benítez-Andrades J.A. Healthcare, Multidisciplinary Digital Publishing Institute; 2020. Evaluation of Country Dietary Habits Using Machine Learning Techniques in Relation to Deaths from COVID-19; p. 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.