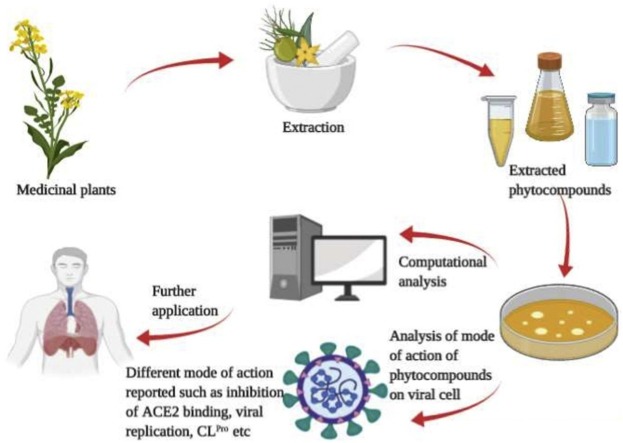
Graphical abstract
Keywords: Phytocompounds; SARS-CoV; MERS-CoV; Antiviral; SARS-CoV-2; COVID-19, mechanism
Abstract
Viral infections are one of the main cause of diseases worldwide due to the rising trends of migration, urbanization and global mobility of humans. The outbreak of corona virus diseases caused by SARS-CoV (year 2003), MERS-CoV (year 2012) and SARS-CoV-2 (year 2019) raised global health concerns. The side effects associated with the conventional drugs and increase in cases of anti-microbial resistance have led the researchers to switch to natural sources, especially plants, as they have immense potential to be used as antiviral agents. The aim of the article is to summarize the evidences of the bioactive phytocompounds from different plants as an effective alternative for the treatment of infections caused by coronaviruses. However, the use of most plant compounds succumbs to limitations due to lack of experimental evidences and safety studies. Therefore, further research and studies are required to validate their therapeutic uses for wide application of plant-based medicine, including anti-virals.
1. Introduction
The emerging trends in migration, urbanization and global travel have made viral outbreaks as an alarming threat for human health. Viral infections are of serious concern due to their complexity, diversity and limited availability of vaccines and antiviral therapies. As a result, they commonly lead to epidemic and pandemic events (Drexler, 2010; Neiderud, 2015). The treatment and control of viral infections largely depends on the availability of antiviral drugs, which are few in number and most of them do not act directly on virus, rather prevent their replication in the host. Viruses consist of a genome (either RNA or DNA) surrounded by a protein or lipid‐containing envelope, and the latter facilitates the viral entry into host cells resulting in several ailments like warts, colds, fever, or even death (Tapparel et al., 2013).
Viruses survive and replicate in the host by hijacking the metabolic pathways of the host cells. Therefore, it is difficult to design an appropriate drug to attack the virus without triggering any adverse effects on the host. Consequently, the unique features of viruses (specificity, affinity, and self‐defense mechanisms), and the limitations of antiviral chemotherapy necessitate the dire need to develop new antiviral agents, which possess efficient selectivity, strength, in vivo stability and safety profiles (Akram et al., 2018).
The Coronaviridae family comprises of enveloped single-stranded RNA viruses that are generally causative agents of common cold, upper respiratory disorders and lower respiratory infections, especially in elderly persons and children, who have weak immune system. Currently, there is an outbreak caused by a novel subtype of coronavirus named severe acute respiratory syndrome coronavirus 2 (SARS-Cov-2), which has resulted in a pandemic with millions of infections and deaths worldwide in humans. The novel coronavirus disease 2019 (COVID-19) caused by SARS-CoV-2 has become a global health concern since December 2019 till date. In this context, it is extremely important to understand the mechanism of the SARS-CoV-2 viral pathogenesis to develop specific and effective drugs. Currently, it has been reported that SARS-CoV-2 shows sequence homology with the SARS-CoV and a bat coronavirus (Gorbalenya et al., 2020). Despite its similarity to SARS-CoV, its transmission efficiency and diagnostic methods are very different, and SARS-CoV-2 is much more virulent compared to SARS-CoV-1. This could possibly be due to the nucleotide changes in the spike (S) protein and its receptor-binding domain (RBD) (Kannan et al., 2020; Coutard et al., 2020; Wan et al., 2020). Therefore, scientists worldwide are exploring the preventive methods and treatment for COVID-19, until a vaccine will be available (Balachandar et al., 2020).
As the world is awaiting curative remedies for COVID-19, there have been several attempts in the recent past towards repositioning of existing drugs to combat the spread of COVID-19. The World Health Organization (WHO) estimates that about 80 % of global population rely on traditional medicine to treat infectious diseases (Pan et al., 2013). Several in vitro and in vivo studies carried out on plants and their derived products have helped to develop effective antibiotic, antimitotic, and antiviral activities. In addition, the pharmaceutical companies began to develop new antimicrobial drugs from natural plant sources (Barreca et al., 2017). The evaluation of several medicinal plants revealed their potential to be used as therapeutic agents against different viruses (Akram et al., 2018). The available antiviral drugs act on specific enzymes involved in targeting the viral structure or in the replication cycle, making them effective targets. But the failure of several conventional drugs against viral infections and the rise in incidence of specific viral resistance has led to an interest in plants as an alternative source of effective antiviral agents (Irwin et al., 2016). Different plant components including essential oils and phytocompounds, such as phenolic acids, flavonoids, terpenes, lignans, coumarins, and alkaloids exhibit potential activity against viruses (Daglia, 2012). Thus, medicinal plants are a promising source for treatment viral diseases (Gomathi et al., 2020).
With the onset of COVID-19 pandemic, research has been initiated to screen the potential of several plant secondary metabolites in inhibiting the SARS-CoV-2 main protease (Mpro)/chymotrypsin-like protease (3CLpro) using molecular docking analysis to examine binding affinity. However, screening a large number of medicinal plants for phytocompounds with antiviral activity against SARS-CoV-2 will be a challenge in very short period of time. Drug discovery is a time consuming, slow and challenging process (Shaikh et al., 2013; Eweas et al., 2014). Thus, it is necessary to exploit computational tools for new drug development, which has made the process of drug discovery rapid and cost effective in the past (Eweas et al., 2014). For screening and searching phytocompounds, the ligand-based virtual screening tool/ molecular docking is very effective to identify most probable molecule with pharmacological activity (Guo et al., 2014; Banegas et al., 2018).
The aim of this review is to provide an update on the antiviral activity of different medicinal plants and their isolated bioactive phytocompounds, their mechanism of action and potential interactions with conventional drugs. The review focuses on the literature available on structure, immunological influence, mechanism of action of the phytocompounds, ongoing clinical trials, recent diagnostics and the potential use of certain medicinal herbs for the effective treatment of coronavirus. Based on the review of literature, we suggest that the traditional medicinal plants can be used as a beneficial and effective means to combat viruses like the SARS-CoV-1, MERS-CoV and SARS-CoV-2.
2. Overview of coronaviruses
There are total 39 species of coronaviruses under the realm of Riboviria, which belong to the family Coronaviridae, suborder Cornidovirineae and order Nidovirales (Gorbalenya et al., 2020). All the SARS-CoV viruses fall under the species severe acute respiratory syndrome-related coronavirus and genus Beta-coronavirus. Most of the species are enzootic and only few species infect humans (Schoeman and Fielding, 2019). So far, seven human CoVs (HCoVs) have been reported, which include, human coronavirus NL63 (HCoV-NL63) and human coronavirus 229E (HCoV-229E) of the alpha-coronavirus genus, and human coronavirus OC43 (HCoV−OC43), human coronavirus (HCoV-HKU1), SARS-CoV, SARS-CoV-2 and middle east respiratory syndrome coronavirus (MERS-CoV) of the beta-coronavirus genus. Human coronaviruses are mostly associated with upper respiratory tract infection, which ranges from mild to moderate, including common cold. It is believed that most of the people might have been infected with one of these viruses at some point in their lifetime (Killerby et al., 2018).
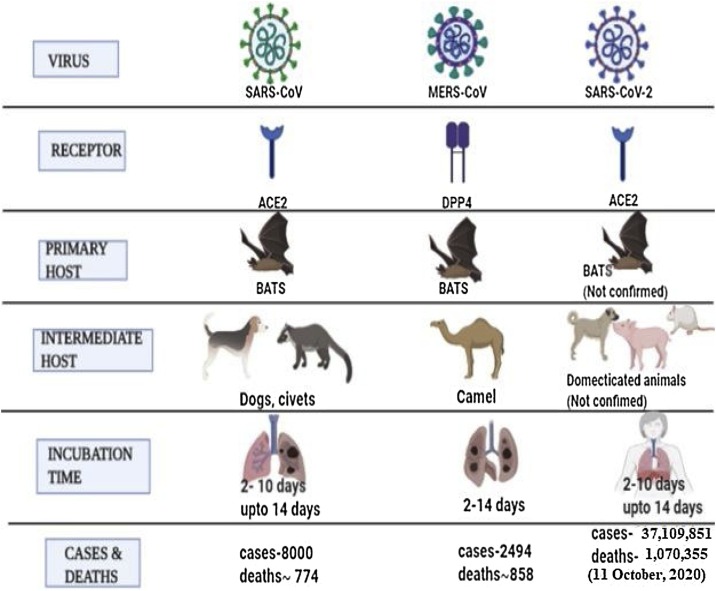
The SARS-CoV and MERS-CoV are the two major viruses that are responsible for severe pneumonia in humans (Song et al., 2019). The emergence of SARS-CoV was first reported in 2003, which led to more than 8000 infections and 774 deaths spread across 37 countries (Peiris et al., 2004). This was followed by the emergence of MERS-CoV in Saudi Arabia in 2012, which was responsible for 858 deaths out of 2494 infections (Zaki et al., 2012). Subsequently, an outbreak of SARS-CoV-2 emerged in December 2019 in the city of Wuhan, China from animal source and transmitted to human population, resulting in the COVID-19 pandemic with lakhs of deaths and millions of infected cases all over the world. As of October 11, 2020, the World Health Organization (WHO) has reported 37,109,851 infections in humans and 1,070,355 deaths worldwide due to COVID-19, and its spread to 214 countries and territories (World Health Organization (WHO, 2020a,b). To combat the battle against this dreadful virus, the WHO has strategized the following measures: to avoid human to human contact, maintaining social distancing and isolate patients at early stages, identify and stop its transmission from the animal source, finding out crucial aspects of the virus and speed up research activities for its prevention and cure, dissemination of appropriate directions to the public and minimize the social and economic impact.
The SARS-CoV-2 is a zoonotic virus that belongs to the coronaviridae family and can infect humans and several other animal species (Lu et al., 2020). The SARS-CoV-2 belongs to the subgenus sarbecovirus and mostly similar to bat coronavirus, with 96.2 % sequence homology (Chan et al., 2020a). The appearance of symptoms due to SARS-CoV-2 infection occurs slowly over an incubation period of around 2 weeks. During this incubation period, the virus replicates in the upper and lower respiratory tract, forming lesions (Chan et al., 2020b). The common symptoms reported in the infected individuals are loss of taste, fever, cough, dyspnea and lesion in the lungs (Huang et al., 2020). The key proteases of the SARS-CoV namely 3C- like Protease (3CLpro) and the Papain-like protease (PLpro) are potential targets for the development of inhibitors against SARS-CoV (Chou et al., 2003; Zhang and Yap, 2004; Anand et al., 2005). The 3CLpro and PLpro are the key enzymes involved in the processing of the non-structural proteins of the virus (nsps) required for the viral replication (Báez et al., 2015; Cascella et al., 2020). The surface of SARS-CoV is coated with the S proteins, which possess the potential to attach to human angiotensin converting enzyme 2 receptors (ACE2). ACE2 receptors are expressed in the lungs, kidney, endothelium, heart, and intestine (Zhang et al., 2020). An alternative way to block the activity of the SARS-CoV is to target its S protein or blocking the human ACE2 from binding to the S protein.
3. Structural and genomic organization of coronaviruses
The morphology of coronaviruses varies from spherical to pleomorphic shape, with a diameter of 80–120 nm (Masters, 2006). Coronaviruses possess the ability to form double membrane vesicles (DMVs) in the infected cells. They also exhibit recombination, high rates of mutation, and propensity to cross species. The coronavirus genome consists of 6–7 open reading frames (ORFs), as described below.
3.1. SARS-CoV
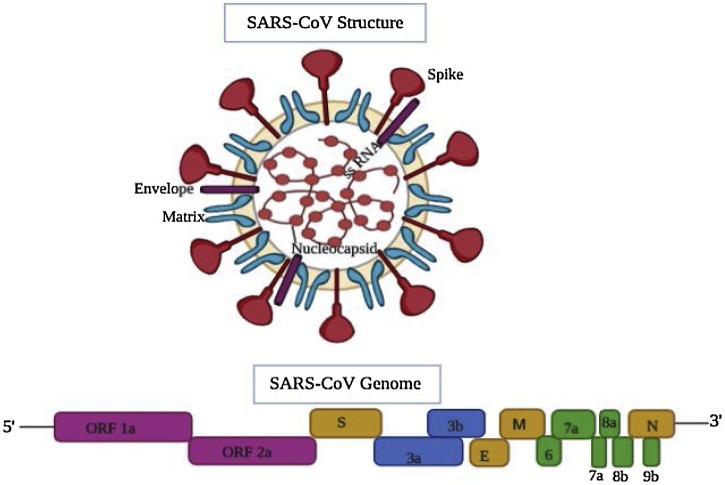
The genome of SARS-CoV encodes the structural spike (S) glycoprotein, membrane (M) glycoprotein, small envelope (E) protein and nucleocapsid (N) protein in 5ʹ- 3ʹ direction within the 3ʹ proximal 1/3rd region of the genome. All the SARS-CoV genomes harbor an extremely large gene 1 (separated into ORFs 1a and 1b and extending over two-thirds of the genome) encoding nonstructural proteins responsible for proteolytic processing of the gene 1 polyprotein products, virus genome replication, and sub-genomic (sg) mRNA synthesis (Fig. 1 ).
Fig. 1.
Schematic diagram of SARS-CoV structure and its genomic organization. The top panel depicts the structure of SARS- coronavirus, wherein the spike protein, envelope, and nucleocapsid are shown. Bottom panel: The genome organization map, which consists of genes encoding the structural spike (S) glycoprotein, membrane (M) glycoprotein, small envelope (E) protein and nucleocapsid (N) protein in 5ʹ-3ʹ direction within the 3ʹ proximal 1/3rd of the genome. A variable number of different ORFs (ORF 1a and 2a) appearing to be virus- or group-specific, encoding nonstructural proteins, are also present on the proximal end. The figure was made with the help of Biorender.com.
A variable number of ORFs appearing to be virus- or group-specific, encoding nonstructural proteins (nsps) are also present in the genome. These include ORF 3a (7.7 kDa protein), ORF 3b (27.7 kDa protein), and ORF 7 [0.7 kDa hydrophobic protein (HP)] in TGEV (Transmissible gastroenteritis coronavirus); ORF 3 (25.3 kDa protein) in PEDV(Porcine epidemic diarrhea coronavirus); ORF 4a (15.3 kDa protein) and ORF 4b (10.2 kDa protein) in HCoV-229E; ORF 2a (32 kDa protein), ORF 2b [65 kDa complete or 34.6 kDa truncated hemagglutinin esterase (HE) protein, depending on the strain], ORF 4 (17.8 kDa protein), ORF 5a (13.1 kDa protein), and an ORF internal to gene 7 [23 kDa internal (I) protein] in MHV (Mouse hepatitis coronavirus); ORF 2a (32 kDa protein), ORF 2b (65 kDa HE protein), ORF 4a (4.9 kDa protein), ORF 4b (4.8 kDa protein), ORF 5 (12.7 kDa protein), and an ORF internal to gene 7 (23 kDa I protein) in BCoV (Bovine coronavirus); and ORF 3a (6.7 kDa protein), ORF 3b (7.4 kDa protein), ORF 5a (7.5 kDa protein), and ORF 5b (9.5 kDa protein) in IBV (Avian coronavirus) (Fig. 1).
The 5’-untranslated region (UTRs) ranges in length from 209 to 528 nt and contains a similarly positioned short, AUG-initiated open reading frame (ORF) relative to the 5ʹ end (Morris and Geballe, 2000).
The 3ʹ UTRs vary in length from 288 to 506 nt, with some strains of IBV having 3’ UTRs of greater length because of internal sequence duplications (Williams et al., 1993. They possess an octameric sequence of GGAAGAGC upstream of the 3ʹ-terminal poly(A) tail.
3.2. MERS-CoV
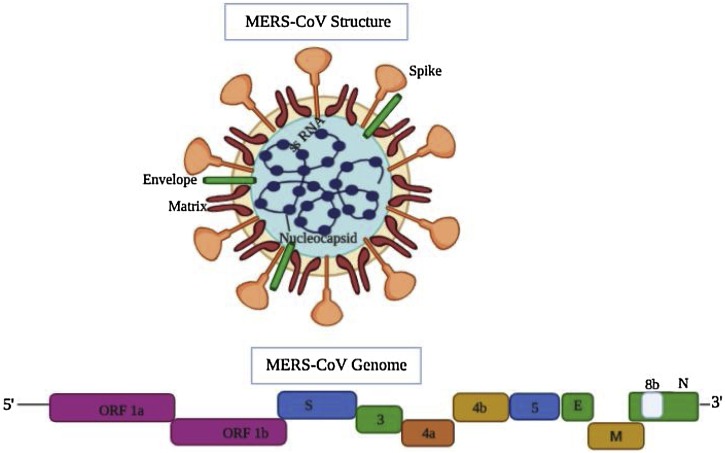
The genome of MERS-CoV strain was reported to be 30,114 nucleotides (nt), including the 3ʹ and 5ʹ UTRs. The MERS-CoV exhibits structural genomic organization of betacoronavirus with the following components: 5ʹ-untranslated region (UTR) (nt 1–272), replicase complex ORF1ab (nt 273–21508), S gene (nt 21450–25511), ORF3 (nt 25526–25837), ORF4a (nt 25846–26175), ORF4b (nt 26087–26827), ORF5 (nt26834 to 27,508), E gene (nt 27584–27832), M gene (nt 27847–28506), N gene (nt 28560–29801), ORF8b (nt 28756–29094), and 3ʹ UTR (nt 29094–30114) (Fig. 2 ). The two essential poly-proteins namely pp1ab and pp1a are cleaved into 15/16 non-structural proteins (nsp) by 3CLpro and PLpro. The nsps include nsp12, nsp13, nsp14, nsp15, and nsp16. The nsp14 protein functions as proofreading enzyme, thereby curtailing the mutation rates during replication of the genomic RNA of coronavirus (Ziebuhr et al., 2000; Snijder et al., 2003; Gorbalenya et al., 2006; Smith et al., 2013; Raj et al., 2014; Durai et al., 2015). Studies have revealed that the accessory ORF proteins play an important role in MERS-CoV infection and pathogenesis (Menachery et al., 2017).
Fig. 2.
MERS-CoV structure and genomic organization. The top panel shows the structure of MERS- coronavirus, wherein the spike protein, envelope, and nucleocapsid are shown. Bottom panel: The genome organization map of MERS-CoV, wherein the genes encoding the structural spike (S) glycoprotein, membrane (M) glycoprotein, small envelope (E) protein and nucleocapsid (N) protein in 5ʹ-3ʹ direction are shown within the 3ʹ proximal 1/3rd of the genome. The genome size of MERS-CoV strain was reported to be 30,114 nucleotide (nt) long, including the 3ʹ and 5ʹ UTRs. The MERS-CoV also contains 5ʹ-untranslated region (UTR), replicase complex ORF1ab, S gene, ORF3, ORF4a, ORF4b, ORF5, E gene, M gene, N gene, ORF8b gene and 3ʹ UTR. The figure was made with the help of Biorender.com.
The MERS-CoV has the potential to adjust and survive in new environments by acquiring various virulence factors and their transfer from one person to other during outbreak (Zumla et al., 2015). The phylogenetic analysis revealed that human and camel MERS-CoV were homologous to each other.
3.3. SARS-CoV-2
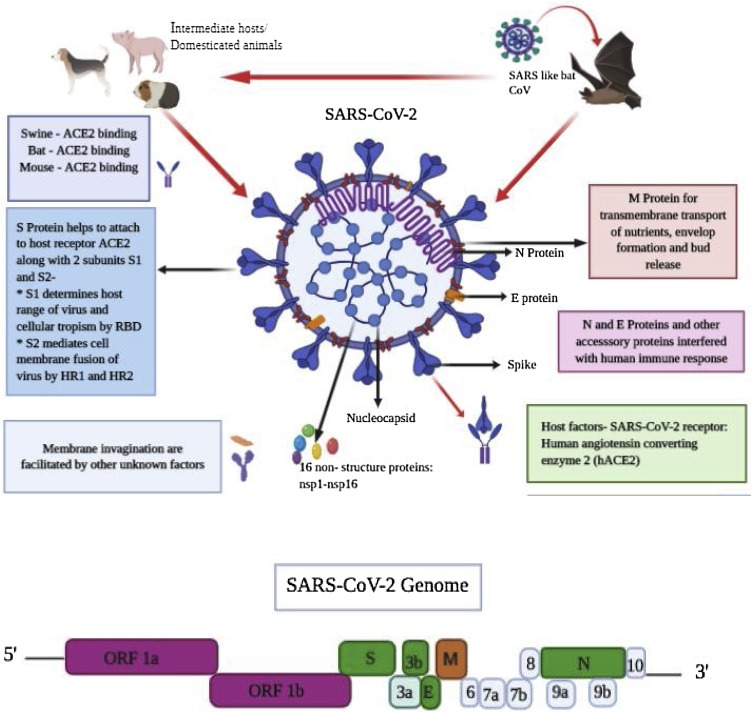
SARS‐CoV‐2 are spherical positive single‐stranded RNA viruses that are identified by the presence of S proteins projecting from the virion surface (Fig. 3 ) (Neuman et al., 2006; Bárcena et al., 2009). The spherical shape of SARS-CoV-2 along with the spike projections led to the name coronavirus from the Latin word “corona” which means “crown”, because of the appearance of the virus as a royal crown under the electron microscope (Neuman et al., 2006; Bárcena et al., 2009).
Fig. 3.
SARS-CoV-2 structural and genomic organization. S, M, and E proteins are located in the viral envelope but the N protein interacts with the viral RNA and is present in the core of the viral particle, forming the nucleocapsid. S protein exists as two subunits (S1 and S2) on the viral particle due to cleavage of S protein by host furin‐like proteases during viral replication. The SARS-CoV-2 genome has the following genes from 5’ to 3’: replicase open reading frame (ORF) 1ab; S; envelope (E); membrane (M); and N. The figure was made with the help of Biorender.com.
Recent studies have demonstrated that SARS-CoV-2 has a similar genomic organization to other beta-coronaviruses, containing 5′ UTR (265 nt), gene encoding a replicase complex (orf1ab), genes encoding non-structural proteins (nsps), S protein gene, E protein gene, M protein gene, N protein gene, 3′-UTR (229 nt), and other non-structural ORFs (Fig. 3) (Zhu et al., 2020). The S, ORF3a, E, M, and N genes of SARS-CoV-2 comprise 3822, 828, 228, 669, and 1260 nucleotides, respectively (Fig. 3). Similar to SARS-CoV, SARS-CoV-2 carries a predicted ORF8 gene (366 nt in length) found between the M and N ORF genes (Wu et al., 2020a,b). However, SARS-CoV-2 is more diverse from MERS-CoV and SARS-CoV. Recent studies highlighted that SARS-CoV-2 genes share < 80 % nucleotide identity and 89.10 % nucleotide homology with their corresponding genes of SARS-CoV (Zhou et al., 2020; Wu et al., 2020a,b).
The S, M, and E proteins are located in the viral envelope but the N protein interacts with the viral RNA and is present in the core of the viral particle, forming the nucleocapsid (Fehr and Perlman, 2015).
The S protein is a glycosylated protein that is responsible for homotrimeric spikes on the surface of the viral particle and mediates viral entry into host (Bosch et al., 2003). S protein exists as two subunits (S1 and S2) on the viral particle due to cleavage of S protein by host furin‐like proteases during viral replication (Bosch et al., 2003; Izaguirre, 2019). The N protein binds with the viral RNA and is involved in packaging of viral RNA into the viral particle during viral assembly (Chang et al., 2006; Hurst et al., 2009). The M protein is one of the most important proteins in the virion structure. It is present in higher quantities than any other protein in the viral particle, while E protein is found in small quantities within the virion (Nal et al., 2005).
Although the source of transmission of SARS-CoV-2 is as yet unclear, it is believed that humans were infected through an unidentified intermediary animal host (possibly bats) followed by intense and rapid spread from human-to humans (Fig. 3, Fig. 4 ). It is also believed that animals such as dogs, pigs might have served as intermediate host for SARS-CoV-2 (Fig. 4). In the advanced stages, the SARS-CoV-2 virus manifests the symptoms of pneumonia in the patients, which progresses to acute respiratory distress syndrome (ARDS), resulting in the need for life-support to sustain the patient's life (Gatera and Pavarini, 2020). The current pandemic caused by SARS-CoV-2 is maintaining a sustained progression throughout the world, thus calling for an emergency international alarm for finding an effective cure and vaccine for this infection.
Fig. 4.
Comparison of the receptor, hosts, number of cases and deaths (as per WHO data) in humans due to infection by SARS-CoV, MERS-CoV and SARS-CoV-2. The figure was made with the help of Biorender.com.
4. Current diagnostics and treatment for coronavirus infections
During SARS-CoV and MERS-CoV outbreaks, diagnostic tools were developed for their accurate detection but they are not effective for detection of SARS-CoV-2. The nucleic acid detection of the viral particle is primarily used in SARS-CoV-2 diagnosis (Wang et al., 2020a, 2020b, 2020c).
The collection of upper respiratory nasopharyngeal (NP) swabs for the diagnostic tests has been recommended (Centers for Disease Prevention and Control (CDC, 2020). The Charité algorithm has two steps: the first step involves two reverse transcriptase PCR (RT-PCR) assays for genes encoding E protein and RNA- dependent RNA polymerase (RdRp) of Sarbecovirus subfamily; if both the tests are positive, the sample proceeds to the second step, wherein it is tested for SARS-CoV-2 specific RdRp by RT-PCR (Loeffelholz and Tang, 2020). However, these methods are cost-intensive, and thus cheaper alternatives have been developed to track the symptoms of COVID-19 such as smart-phone surveillance (Dorigatti et al., 2020). Imaging techniques are also used as a diagnostic method in COVID-19, including chest CT scans, which have been commonly used to detect lung abnormalities caused by SARS-CoV-2 infection (Shi et al., 2020; Xu et al., 2020a, b).
The strategies used to halt the propagation of RNA viruses in host cells or tissues include inhibition of RNA transcription, RNA modification, virus packaging enzymes, and the capsid or surface proteins assisting the viral diffusion into the host cells (Dinesh et al., 2020). The current treatment regimen for COVID-19 includes drugs such as remdesivir, chloroquine, arbidol, favipiravir, anti-inflammatory drugs, anti-HIV drugs, and approaches such as interferon therapy, and monoclonal antibodies (Dinesh et al., 2020; Dong et al., 2020). However, there are no specific and completely effective medications against SARS-CoV-2. Although vaccine development against SARS-CoV-2 has been initiated, they are not available yet at the global level. Therefore, researchers are exploring the pool of natural and chemicals compounds that could inhibit the major proteases of the virus or downregulate the rate of propagation of the SARS-CoV-2 virus in the cells.
4.1. Current ongoing trials
Different research institutes and companies worldwide have been working on clinical trials to repurpose existing drugs as well as to develop vaccines and drugs to combat the fast spreading and fatal SARS-CoV-2 (Rudra et al., 2017). For repurposing the existing drugs, randomized controlled trials (RCTs) are being undertaken by various biotechnological companies and research organizations such as National Institutes of Health (NIH), USA to identify disease specific drugs. Clinical trials are underway for major drugs that have the potential to treat COVID-19. Research on traditional medicine is also being undertaken to utilize them in the treatment of COVID-19. Plants provide a natural source of anti-viral inhibitors that can be extended for treatment of diseases caused by SARS-CoV-2. Hence, by repurposing the compounds and extracts from medicinal plants, more innovative and effective drug options can be unveiled for eliminating this viral transmission.
5. Plants with antiviral properties for the treatment of SARS diseases
The Earth is a hub of more than 500,000 plant species, of which 10 % are utilized as food source and 10–15 % as source of drugs (Borris, 1996). Since ancient times, phytomedicine of ethnic communities used to be the basic method of treatment in China (Chang and But, 2014), India (Dev, 1999), Africa and many other countries (Schultes and Raffauf, 1990). Globally, a major proportion of the world’s populations rely on plant-based medications for primary health care and phytocompounds as ingredients of drugs (Farnsworth, 1990).
Plants can serve as a wide source of viral protein inhibitors for the treatment of SARS. Plants produce a wide array of secondary metabolites that can possess inhibitory effect on the enzymes, proteins and the propagation of viruses. Secondary metabolites are expressed in response to biotic and abiotic stresses. Some examples include plant active compounds such as flavonoids, carotenoids, and diarylheptanoids. Over centuries, medicinal herbs have been used as a treatment and preventive strategy for several diseases, including respiratory viral infections (Park et al., 2016; Kiran et al., 2020; ul Qamar et al., 2020,). The benefit of using these herbs in viral respiratory infections is due to their immune stimulating and inflammation modulating effects to manage the immune system. Although numerous studies have focused on the SARS-CoV and MERS-CoV, there are few studies on cure for COVID-19 disease, which are limited to in silico studies of the phytocompounds (Mohammadi and Shaghaghi, 2020; ul Qamar et al., 2020).
5.1. Antiviral activity of phytocompounds against SARS-CoV, MERS-CoV and SARS-CoV-2
Natural medicine is a valuable field of research to extract and establish curative properties. However, limited number of phytochemicals have been systematically reported for their therapeutic potential (De Clercq, 2005; Hostettmann et al., 2000). The antiviral properties of phytocompounds on SARS-CoV, MERS-CoV and SARS-CoV-2 are summarized below (Table 1 ):
Table 1.
List of medicinal plants, their habitat, target virus, the constituent antiviral phytocompounds, their classification, and mode of action (if reported) against coronaviruses.
| S No: | Plant | Habitat/ (wild/ cultivated) | Virus | Phyto-Compounds | Classification | Mode of action | References |
|---|---|---|---|---|---|---|---|
| 1. | Zingiber officinale | Asia, Hawaii, Europe (cultivated) | SARS- CoV-2 | 6-gingerol | Phenolics | In silico binding affinity to 5R7Y protease | Rathinavel et al. (2020) |
| 2. | Psorothamnus arborescens | Southwestern North America, desert and dry mountainous habitats (wild) | SARS- CoV-2 | 5,7,3′,4′-tetrahydroxy-2′-(3,3-dimethylallyl) isoflavone | Flavonoids | Inhibition of 3CLpro | ul Qamar et al. (2020) |
| 3. | Lycoris radiata (Stem cortex) | China, Korea, Nepal, Japan, United States (cultivated) | SARS- CoV | Lycorine | Alkaloids | Inhibition of virus replication and cellular entry | Li et al. (2005) |
| 4. | a) Gentiana scabra (dried rhizome) b) Dioscorea batatas (tuber) c) Cassia tora (dried seed) (d) Taxillus chinensis (leaf) |
United States and Japan (cultivated) Found in an apparently wild situation in valleys and on the slopes of hills in China, sunny slopes (wild) Central America (wild) Cambodia, Indonesia, Laos, Malaysia, Philippines, Thailand, Vietnam (wild+cultivated) |
SARS-CoV | ------- | ----------- | Inhibition of 3CLpro | Wen et al. (2011) |
| 5. | Glycyrrhiza glabra | Central and South-Western Asia, as well as to the Mediterranean region (wild) | SARS-CoV | Glycyrrhizin | Saponin | Inhibition of virus replication and cellular entry | Fiore et al. (2008) |
| 6. | a) Zingiber officinale b) Piper longum L. c) Syzygium aromaticum d) Tragia involucrata L e) Anacyclus pyrethrum f) Andrographis paniculata g) Hygrophila auriculata h) Terminalia chebula i) Justicia adhatoda L. j) Plectranthus amboinicus k) Saussurea lappa l) Tinospora cordifolia m) Clerodendrum serratum n) Sida acuta Burm. f. o) Cypreus rotundus L |
Asia, Europe, India (cultivated) India, Malaysia, Nepal, Sri Lanka and Vietnam (wild) Indonesia, India, Pakistan, Sri Lanka, Comoro Islands, Madagascar, Seychelles, and Tanzania (cultivated) Outer Himalayan ranges eastwards to Assam; southwards to Travancore, throughout warmer regions of India (wild) North Africa, Mediterranean region, Himalayas, North India, the Levant and in certain regions in the Arabian peninsula (wild+cultivated) India and Sri Lanka (wild+cultivated) Tropical Asia and Africa, India, Sri Lanka (cultivated) India and China (Yunnan), Sri Lanka, Malaysia, and Vietnam (wild) Indian subcontinent (Assam, Bangladesh, India, Nepal and Sri Lanka), Laos and Myanmar(wild) Southern and Eastern Africa, from South Africa (KwaZulu-Natal) and Swaziland to Angola and Mozambique and north to Kenya and Tanzania, India and Southeast Asia- (cultivated) North Asia- Kashmir and neighboring Himalaya region and also in Garhwal of Uttar Pradesh (wild) Indigenous to tropical regions of the Indian subcontinent (wild) Tropical and sub-tropical parts particularly in Bengal, Odisha and peninsular India (cultivated) Central America, northern Australia (wild) Africa, southern and central Europe to France and Austria), and southern Asia(wild) |
SARS-CoV-2 | β-sesquiphellandrene Piperine Eugenol; β-Caryophyllene Stigmosterol Squalene γ-Sitosterol Andrograpanin; 5-Hydroxy-7,8- dimethoxyflavanone Lupeol; Betulin Chebulagic acid Vasicinone Carvacrol Luteolin Tinosponone Bharangin Magnoflorine Cyperene |
Terpenoids Alkaloids Phenolics Terpenoids Lipids Terpenoids Lipids Terpenoids Flavonoids Terpenoids Terpenoids Phenolics Alkaloids Terpenoids Flavonoids Terpenoids Terpenoids Alkaloids Terpenoids |
In silico binding affinity to Spike (S) protein | Kiran et al. (2020) |
| 7. | a) Justicia adathoda L b) Carica Papaya c) Andrographis paniculata Burm d) Ocimum tenuiflorum e) Melia azedarach |
Asia, Indian subcontinent (Assam, Bangladesh, India, Nepal and Sri Lanka), Laos and Myanmar(wild+ cultivated) Mexico and northern South America, Caribbean Islands, Florida, Texas, California, Hawaii, and other tropical and subtropical regions of the world (cultivated) India and other Asian countries (cultivated) Indian subcontinent, Southeast Asian tropics (cultivated) Southeast Asia and northern Australia, USA (cultivated) |
SARS-CoV-2 | Vasicine Quercetin Andrographolide Ursolic acid Meliacine |
Alkaloids Flavonoids Terpenoids Terpenoids ------- |
In silico binding affinity to Spike (S) protein | Kiran et al. (2020) |
| 8. | a) Chamaecyparis obtuse b) Juniperus formosana c) Cryptomeria japonica |
Central Japan in East Asia, northern hemisphere (cultivated) China (from Tibet in the west to Zhejiang in the east) and in Taiwan (cultivated) Japan, China, India (cultivated) |
SARS-CoV | Ferruginol; Dehydroabieta-7-one; Sugiol; 8β-hydroxyabieta-9 (11); 6,7-dehydroroyleanone; Pinusolidic acid; α-cadinol; Hinokinin; Savinin 3β,12-diacetoxyabieta-681,113- Tetraene; Cedrane-312-diol; Betulonic acid; Cryptojaponol; 7β- Hydroxydeoxycryptojaponol; |
Terpenoids Terpenoids Terpenoids Terpenoids Terpenoids Terpenoids Phenolics Phenolics Terpenoids -------------- Terpenoids Terpenoids Terpenoids |
Inhibition of 3CLpro | Wen et al. (2007) |
| 9. | Utrica dioica | Europe, Asia and western North Africa, it is now found worldwide, including New Zealand and North America (wild+ cultivated) | SARS-CoV | Lectin | Inhibition of Spike (S) protein | Keyaerts et al. (2007); Van der Meer et al. (2007), Kumaki et al. (2011) | |
| 9. | Allium sativum | Central Asia and northeastern Iran (wild+cultivated) | SARS-CoV-2 | Allyl disulfide; Allyl trisulfide | Organosulfur | Inhibition of ACE2 receptor | Thuy et al. (2020) |
| 10. | Angelica keiskei | Japan, where it is found on the Pacific Coast (wild) | SARS-CoV | Chalcone | Flavonoids | Inhibition of 3CLpro amd PLpro | Park et al. (2016) |
| 11. | Betula pubescens | Northern Europe and northern Asia (wild) | SARS-CoV | Betulinic acid | Terpenoids | Inhibition of 3CLpro | Wen et al. (2007) |
| .12. | Galla chinensis | Asia (China) (wild) | SARS-CoV | Tetra-O-galloyl β-d-glucose | Phenolics | Inhibition of S protein | Yi et al. (2004) |
| 13. | Alnus japonica | Japan, Korea, Taiwan, eastern China, and Russia (cultivated) | SARS-CoV | Hirsutenone; Hirsutanonol; Oregonin; Rubranol; Rubranoside B; Rubranoside A |
Diarylheptanoids/ Phenolics | Inhibition of PLpro | Park et al. (2012) |
| 14. | Curcuma longa Isatis | Southern Asia especially India (cultivated) | SARS-CoV | Curcumin | Phenolics | Inhibition of 3CLpro | Wen et al. (2007) |
| 15. | Camellia sinensis (Black tea) | East Asia, and probably in the borderlands of north Burma and southwestern China (cultivated) | SARS-CoV | Tannic acid; Theaflavin-3,3ʹ-digallate |
Phenolics Phenolics |
Inhibition of 3CLpro | Chen et al. (2005) |
| 16. | Nicotiana benthamiana | Australia (cultivated) | SARS-CoV | NICTABA; Lectin | Viral growth inhibitor | Zheng et al. (2009); Demurtas et al. (2016) | |
| 17. | Houttuynia cordata | Southeast Asia (cultivated) | SARS-CoV | – | Inhibition of 3CLpro | Lau et al. (2008); Fung et al. (2011) | |
| 18. | Litchi chinensis | Native to the Guangdong and Fujian, India, other countries in Southeast Asia (cultivated) | MERS-CoV | Flavonoid | Flavonoids | Inhibition of 3CLpro | Kim et al., 2019 |
| 19. | Rhizoma Cibotii | China (wild) | SARS-CoV | – | Inhibition of 3CLpro | Wen et al. (2011) | |
| 20. | Polygonum multiflorum Thunb. | Central and southern China, Eastern Asia and the Russian Far East, Europe and North America (wild) | SARS-CoV | Emodin | Phenolics | Inhibition of ACE2 receptor, inhibition of S protein | Ho et al. (2007) |
| 21. | Rheum officinale | China (wild+cultivated) | SARS-CoV | Emodin | Phenolics | Inhibition of ACE2 receptor, inhibition of S protein | Ho et al. (2007) |
| 22. | Veronica linariifolia | Northern Hemisphere, though with some species from the Southern Hemisphere (cultivated) | SARS-CoV | Luteolin | Flavonoids | Inhibition of S protein | Yi et al. (2004) |
| 23. | Paulownia tomentosa | Central and western China, North America, Eastern USA (wild) | SARS-CoV | Tomentin A; Tomentin B; Tomentin C; Tomentin D; Tomentin E | Flavonoids | Inhibition of PLpro | Cho et al. (2013) |
| 24. | Stephania cepharantha Hayata | Eastern and southern Asia and Australia (wild) | SARS-CoV | Cepharanthine | Alkaloids | Viral Growth inhibitor | Chattopadhyay (2006) |
| 25. | Myrica faya | Macaronesia (the Azores, Madeira, and the Canary Islands), western coastal mainland Portugal (cultivated) | SARS-CoV | Myricetin | Flavonoids | Inhibition of Helicase, nsp13 | Yu et al. (2012) |
| 26. | Scutellaria lateriflora | North America (wild) | SARS-CoV | Scutellarein | Flavonoids | Inhibition of Helicase nsp13 | Yu et al. (2012) |
| 27. | Isatis indigotica | Europe, especially in Western and Southern Europe, China England, Germany and France (wild) |
SARS-CoV | Hesperetin; Sinigrin; Beta-sitosterol; Aloeemodin |
Flavonoids Glucosinolates Lipids Phenolics |
Inhibition of 3CLpro | Lin et al. (2005) |
| 28. | Torreya. nucifera | Southern Japan and to South Korea's Jeju Island (wild+cultivated) | SARS-CoV | Amentoflavone | Flavonoids | Inhibition of 3CLpro | Ryu et al. (2010) |
| 29. | Cinnamomum cassia | Southern China, South and Southeast Asia (India, Indonesia, Laos, Malaysia, Thailand, and Vietnam) (cultivated) | SARS-CoV | Procyanidin A2; Procyanidin B1; Cinnamtannin B1 |
Flavonoids | Viral growth inhibitor | Zhuang et al. (2009) |
| 30. | Linum usitatissimum | Syria, Switzerland and Germany, India, China (cultivated) | MERS‐CoV | Herbacetin | Flavonoids | Inhibition of 3CLpro | Kim et al., 2019 |
| 31. | Laurus nobilis | Mediterranean region (cultivated) | SARS-CoV | β-ocimene; 1,8-cineole; α-pinene; β-pinene |
Terpenoids | Viral Growth inhibitor | Loizzo et al. (2008) |
| 32. | Nicotiana tabacum | Caribbean, tropical and subtropical America(cultivated) | SARS-CoV | -------- | -------------- | Viral growth inhibitor | Zheng et al. (2009); Wang et al. (2005); Franconi et al. (2018) |
| 33. | Thymus vulgaris | Southern Europe from the western Mediterranean to southern Italy (cultivated) | SARS-CoV-2 | Ursolic acid | Terpenoids | In silico binding affinity to 6LU7 and 6Y2E proteases | Sampangi-Ramaiah et al. (2020) |
| 34. | Coriandrum sativum | Southern Europe and Northern Africa to Southwestern Asia(wild and cultivated) | SARS-CoV-2 | Coriandrin | Phenolics | In silico binding affinity to 6LU7 and 6Y2E proteases | Sampangi-Ramaiah et al. (2020) |
| 35. | Rosmarinus officinalis | Mediterranean region (wild and cultivated) | SARS-CoV-2 | Rosmarinic acid | Phenolics | In silico binding affinity to 6LU7 and 6Y2E proteases | Sampangi-Ramaiah et al. (2020) |
| 36. | Brassica juncea | Northwest Eastern Island, Midway Atoll, Hawaii, USA, India (wild and cultivated) | SARS-CoV-2 | Glucobrassicin | Glucosinolates | In silico binding affinity to 6LU7 and 6Y2E proteases | Sampangi-Ramaiah et al. (2020) |
Glycyrrhizin, a bioactive compound of Chinese liquorice (Glycyrrhiza uralensis Fisch), and lycorine isolated from Lycoris radiata L. showed strong anti-SARS-CoV activity (Li et al., 2005). Subsequently, Fiore et al. (2008) extracted glycyrrhizin from Glycyrrhiza glabra and reported that glycyrrhizin is a potent inhibitor of SARS-CoV virus replication, and adsorption and penetration of virus during the early steps of the replicative cycle.
The caffeine beverages like green and black tea from Camellia sinensis have bioflavonoids with several medicinal properties. A study reported that water soluble tannic acid and theaflavin-3, 3ʹ-digallate inhibit 3CLpro protease of SARS-CoV (Chen et al., 2005). Similarly, the phytocompounds namely hesperetin, sinigrin, beta-sitosterol, indigo and aloe emodin extracted from Isatis indigotica were found to have an inhibitory effect on the SARS-CoV 3CLpro (Lin et al., 2005).
Quercetin is a flavonoid that is abundant in several plants and food products with a multitude of medicinal and pharmacological properties (Massi et al., 2017). Both quercetin and quercetin 3-β- galactoside possess the ability to inhibit the activity of 3CLpro protease of SARS-CoV in vitro (Chen et al., 2006).
Wen et al. (2007) screened 221 phytocompounds isolated from Chamaecyparis obtuse, Juniperus formosana and Cryptomeria japonica against SARS-CoV and found that certain abietane type di-terpenoids and lignoids have best anti-viral effects. Similarly, bioactive compounds isolated from six herbal extracts namely, Gentiana scabra (dried rhizome), Dioscorea batatas (tuber), Cassia tora (dried seed), Taxillus chinensis (leaf), and Cibotium barometz (dried rhizome) exhibited anti-SARS-CoV activity in a cell-based assay on infected Vero E6 cells at concentrations between 25 and 200 μg/mL (Wen et al., 2011). Several independent studies have shown that the compounds such as curcumin, caffeic acid, chalcones, cinnamic acid and betulinic acid isolated from different medicinal plants are potent inhibitors of 3CLpro protease of SARS-CoV (Table 1). Similarly, Jo et al. (2019) showed that flavonoids of Litchi chinensis and herbacetin isolated and Linum usitatissimum showed inhibition of 3CLpro in MERS-CoV.
Urtica dioica (Stinging nettle) has been used in different countries as traditional medicine for years owing to its therapeutic effects on cardiovascular, immune, nervous and digestive systems (Dhouibi et al., 2020). Interestingly, lectins extracted from Nicotiana tabacum (tobacco agglutinin; NICTABA), Nicotiana benthamiana, and Urtica dioica exhibit strong inhibitory potential against proliferation of the SARS-CoV (Keyaerts et al., 2007; Zheng et al., 2009; Demurtas et al., 2016). Li et al. (2005) showed that extracts of Lycoris radiata possess anti-SARS-CoV activity with a significantly lower dose of effectiveness (about 2.1–2.4 u g/ml). The antiviral activity of the extracts was attributed to the presence of lycorine in L. radiata (Li et al., 2005).
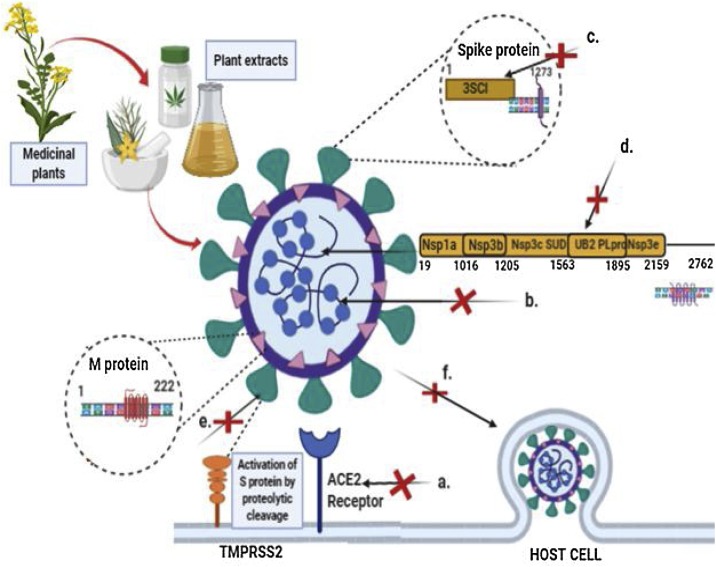
Phytocompounds targeting the second major protease PLpro include flavonoids (tomentins) from Paulownia tomentosa (Cho et al., 2013), chalcones from Angelica keiskei (Park et al., 2016), and diarylheptanoids extracted from Alnus japonica (Park et al., 2012), which were reported as potent inhibitors of PLpro of SARS-CoV (Fig. 5 ).
Fig. 5.
Summary of phytocompounds and their mode of action on coronaviruses. The targets of different phytocompounds is depicted as follows: a. Emodin, allyl disulfide, allyl trisulfide, and tetra-O-galloyl β-d-glucose inhibit ACE2 receptor binding to spike protein; b. Ferruginol, 3β,12-diacetoxyabieta-681,113- tetraene, and cryptojaponol inhibit virus replication; c. Flavonoids, caffeic acid, cinnamic acid, herbacetin, and hesperetin, and ursolic acid inhibit 3CLpro; d. Flavonoids, chalcones, and diarylheptanoids inhibit PLpro; e. Tetra-O-galloyl β-d-glucose, luteolin, terpenoids, carotenoids, ursolic acid and phytosterols target coronavirus spike protein; f. Quercetin and glycyrrhizin inhibit the cellular entry of SARS-CoV. The figure was made with the help of Biorender.com.
An alternative way to block the activity of the SARS-CoV is to target the spike protein (S) of SARS-CoV S proteins or blocking the human ACE2 receptor. Yi et al. (2004) reported that tetra-O-galloyl β-d-glucose from Galla chinensis and luteolin from Veronica linariifolia exhibit strong binding affinity to spike (S) protein of SARS-CoV. Emodin (1,3,8-trihydroxy-6-methylanthraquinone), rhein (1,8-dihydroxy-3-carboxyl-910-anthraquinone), and chrysin (5,7-dihydroxyflavone) are produced in high levels in plants of genus Rheum and Polygonum; these compounds were responsible for blocking the binding of S protein in SARS-CoV to ACE2 (Ho et al., 2007; Fig. 5). Emodin blocked the binding of S protein to ACE2 in a dose-dependent manner (Ho et al., 2007).
5.2. In silico studies on antiviral activity of phytocompounds against SARS-CoV-2
Owing to the recent occurrence of COVID-19 pandemic and advances in virtual tools and databases available for drug screening, several research groups worldwide have reported in silico screening of phytocompounds reported for activity against SARS-CoV and other viral diseases.
Zingiber officinale (ginger) is an herbaceous plant native to South Asia belonging to the Zingiberaceae family and has been used in various countries as traditional medicine for years. The phytocompounds of ginger have been screened for binding the proteins of SARS-CoV-2 (Rathinavel et al., 2020). The phytocompound 6-gingerol showed the highest binding affinity (-15.7591 kJ/mol) with 5R7Y SARS-CoV-2 main protease, which is essential for replication and propagation of SARS-CoV-2 (Rathinavel et al., 2020). Moreover, 6-gingerol possesses excellent drug likeliness with zero violations and very good pharmacokinetic properties, indicating its potential for treating COVID-19.
Organosulfur compounds are found in the plants of the Allium genus such as onions (e.g. Allium cepa), and garlic (Allium sativum). In a recent in silico study, the organosulfur materials such as allyl disulfide and allyl trisulfide from Allium sativum showed significant potential in binding to human ACE2, the target of SARS-CoV-2 (Thuy et al., 2020).
In another study ul Qamar et al. (2020) screened the Chinese medicinal plants library of 32,297 phytocompounds for potential binding with 3CLpro of SARS-CoV-2 generated by homology modeling. The isoflavone namely 5,7,3′,4′-tetrahydroxy-2′-(3,3-dimethylallyl) isoflavone extracted from Psorothamnus arborescens showed the highest binding affinity with 3CLpro of SARS-CoV-2 (Table 1; ul Qamar et al., 2020).
Sampangi-Ramaiah et al. (2020) explored the in-silico binding of 27 phytocompounds (present in spices and condiments used in Indian and other cuisines) to SARS-CoV-2 6LU7 protease (3CLpro) and 6Y2E protease, both of which are required for viral replication. Out of the 27 compounds, 15 showed binding affinity above threshold values for both the proteases. The key compounds with high binding affinity to 6LU7 and 6Y2E proteases include coriandrin (component of the essential oil of Coriandrum sativum widely in Indian cuisines across both north and south India), ursolic acid of Thymus vulgaris, rosmarinic acid of Rosmarinus officinalis and glucobrassicin of Brassica juncea (Sampangi-Ramaiah et al., 2020).
Siddha medicine is widely used in Southern India, which prescribes a concoction called Kabasura kudineer chooranam for cold, cough, and fever (Kiran et al., 2020). Molecular docking studies were performed for 32 phytochemical constituents of herbs present in Kabasura kudineer chooranam and 05 phytochemical constituents of herbal formulation named JACOM against spike protein (S) of SARS-CoV-2 (PDB ID: 6VSB) to observe the molecular interactions between target protein and ligands. All the phytochemical analogs were successfully docked with S protein of SARS-CoV-2 (Kiran et al., 2020; Table 1).
Recently, Rolta et al. (2020) reported molecular docking studies of 100 major phytocompounds of ten selected medicinal plants (Rheum emodi, Thymus serpyllum, Cymbopogon citratus, Moringa oleifera, Thalictrum foliolosum, Berberis aristata, Piper nigrum, Allium sativum, Myristica fragrans and Zanthoxylum armatum) for interaction with RNA binding domain (N-terminal Domain/ NTD; PDB 6VYO) of nucleocapsid phosphoprotein of SARS-CoV-2. The molecular docking study revealed that five phytocompounds namely emodin, anthrarufin, alizarine, aloe- emodin and dantron of Rheum emodi showed effective binding affinity and pharmacokinetic properties with NTD of RNA binding domain of nucleocapsid phosphoprotein of SARS- CoV-2 (Rolta et al., 2020).
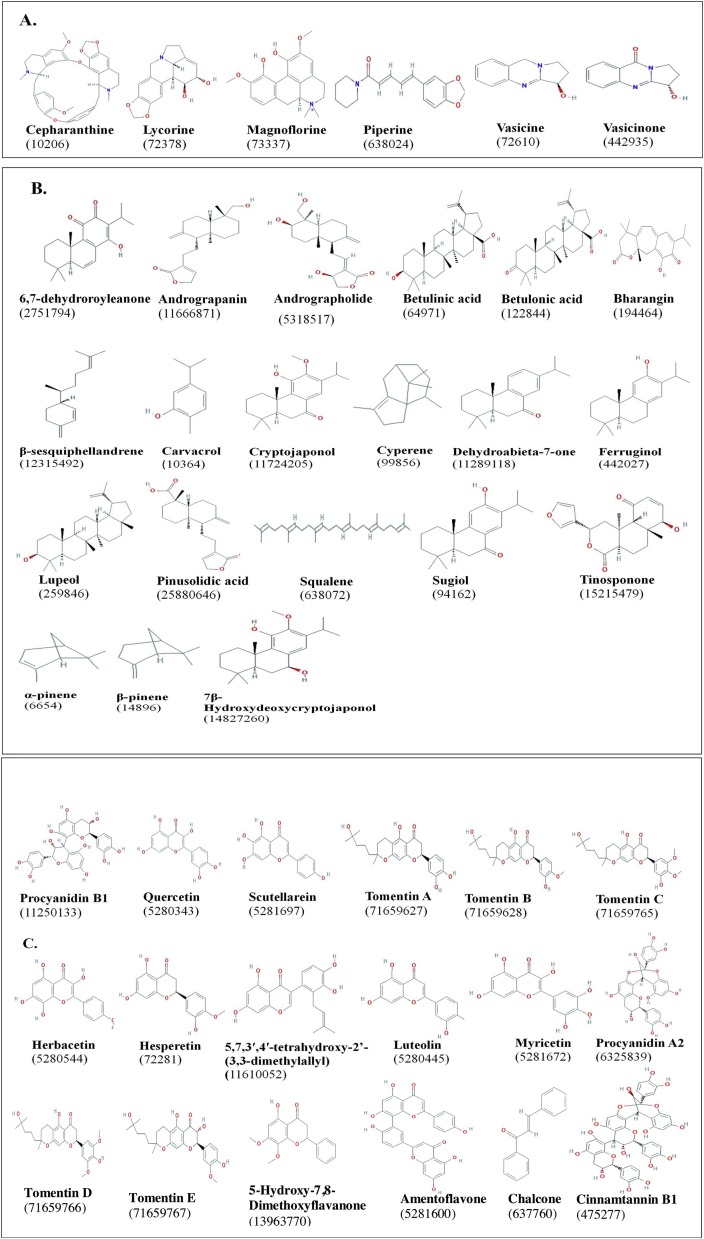
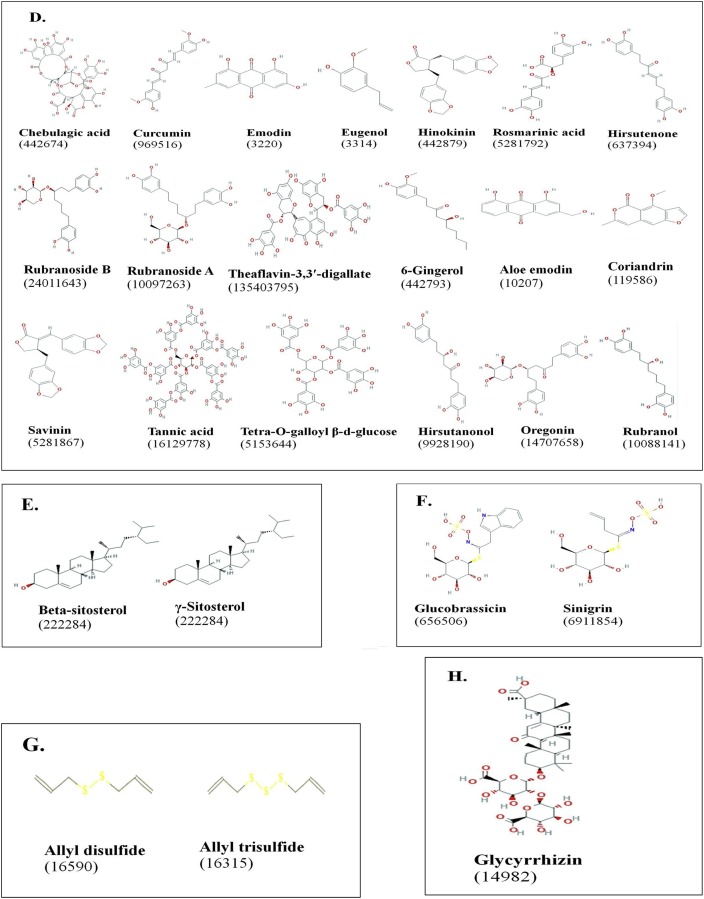
Based on the literature reviewed here, a total of 70 phytocompounds belonging to 8 classes from 62 different medicinal plants have been reported for anti-viral activities against coronaviruses, with focus on SARS-CoV (Fig. 6 ; Table 1). Most of these compounds have been used for treating other ailments in humans, revealing their multi-dimensional pharmacological properties. Analysis of the targets and mechanism of action of the phytocompounds indicates a broad spectrum of targets on both coronaviruses and the host receptors (Fig. 5). These include blocking of ACE2 receptor binding by spike protein, inhibition of viral proteases, inhibition of viral replication, targeting coronavirus spike protein, and blocking viral entry into host (Fig. 5; Table 1). With ongoing research for COVID-19 therapeutics, the list of anti-viral phytocompounds is certainly dynamic with more anticipated updates.
Fig. 6.
Structure of various classes of phytocompounds with anti-viral potential against coronaviruses studied by in vitro, in vivo, and in silico approaches. The PubChem CID of each compound is indicated in parenthesis. A: Alkaloids; B: Terpenoids; C: Flavonoids; D: Phenolics; E: phytosterols; F: Glucosinalates; G: Organosulfur compounds; H: Saponins (Source: Table 1). Structures were obtained from pubchem.ncbi.nlm.nih.gov.
6. Conclusions
Over the past few decades, there has been a huge demand to decipher the root of coronavirus infections not only in animals but also in humans. As a complementary approach, the search for new antiviral drugs of natural origin has gained momentum. Currently, COVID-19 has emerged as the most intense and petrifying viral infectious disease all over the world to be handled by the human race.
Based on the high degree of homology of the genome (> 96 %) and the receptors of SARS-CoV and SARS-CoV-2, this review was attempted to summarize the plant species and their bioactive compounds with anti-viral potential against coronaviruses, which can be used as anti-SARS-CoV-2 agents. The literature review suggested more than 70 compounds from 62 plant species with anti-viral activity against coronaviruses that might exhibit treatment potential for COVID-19 disease. Several of these anti-viral activities have been predicted from studies on molecular docking of bioactive compounds of different medicinal plants with proteases and spike protein of
SARS-CoV-2. Moreover, the anti-viral phytocompounds identified from in-silico studies also showed good drug-likeliness properties. Thus, the phytocompounds identified from in-silico studies can be validated by in vitro, in vivo and clinical studies against SARS-CoV-2. To assure public safety and control of infection in the event of a re-emergence of COVID-19, effective anti-SARS-CoV agents and treatments are highly desirable and imminent in the immediate and near future. The findings of this review will provide a reservoir of natural compounds for researchers world-wide focusing on drug discovery for COVID-19. The literature on anti-viral potential of phytocompounds will also pave way for experimental affirmation of several traditional and natural medicine in use for centuries in several sections of the world.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgement
We acknowledge Prem Kumar Khosla, Vice Chancellor, Shoolini University, and Yeast Biology lab for support and encouragement.
References
- Akram M., Tahir I.M., Shah S.M.A., Mahmood Z., Altaf A., Ahmad K., Mehboob H. Antiviral potential of medicinal plants against HIV, HSV, influenza, hepatitis, and coxsackievirus: a systematic review. Phytother. Res. 2018;32(5):811–822. doi: 10.1002/ptr.6024. [DOI] [PubMed] [Google Scholar]
- Anand K., Yang H., Bartlam M., Rao Z., Hilgenfeld R. Coronaviruses With Special Emphasis on First Insights Concerning SARS. Birkhäuser Basel; 2005. Coronavirus main proteinase: target for antiviral drug therapy; pp. 173–199. [Google Scholar]
- Báez-Santos Y.M., John S.E.S., Mesecar A.D. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandar V., Mahalaxmi I., Kaavya J., Vivekanandhan G., Ajithkumar S., Arul N., Devi S.M. COVID-19: emerging protective measures. Eur. Rev. Med. Pharmacol. Sci. 2020;24(6):3422–3425. doi: 10.26355/eurrev_202003_20713. [DOI] [PubMed] [Google Scholar]
- Banegas A.J., Cerón-Carrasco J.P., Pérez-Sánchez H. A review of ligand-based virtual screening web tools and screening algorithms in large molecular databases in the age of big data. Future Med. Chem. 2018;10(22):2641–2658. doi: 10.4155/fmc-2018-0076. [DOI] [PubMed] [Google Scholar]
- Bárcena M., Oostergetel G.T., Bartelink W., Faas F.G., Verkleij A., Rottier P.J., Bosch B.J. Cryo-electron tomography of mouse hepatitis virus: insights into the structure of the coronavirion. Proc. Natl. Acad. Sci. U.S.A. 2009;106(2):582–587. doi: 10.1073/pnas.0805270106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreca D., Gattuso G., Bellocco E., Calderaro A., Trombetta D., Smeriglio A., Nabavi S.M. Flavanones: Citrus phytochemical with health‐promoting properties. BioFactors. 2017;43(4):495–506. doi: 10.1002/biof.1363. [DOI] [PubMed] [Google Scholar]
- Borris R.P. Natural products research: perspectives from a major pharmaceutical company. J. Ethnopharmacol. 1996;51(1–3):29–38. doi: 10.1016/0378-8741(95)01347-4. [DOI] [PubMed] [Google Scholar]
- Bosch B.J., Van der Zee R., De Haan C.A., Rottier P.J. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77(16):8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascella M., Rajnik M., Cuomo A., Dulebohn S.C., Di Napoli R. Statpearls [internet] StatPearls Publishing; 2020. Features, evaluation and treatment coronavirus (COVID-19) [PubMed] [Google Scholar]
- Centers for Disease Prevention and Control (CDC) 2020. Centers for Disease Prevention and Control (CDC)https://www.cdc.gov/coronavirus/2019-ncov/index.html (Accessed on 25 March 2020) [Google Scholar]
- Chan J.F.W., Kok K.H., Zhu Z., Chu H., To K.K.W., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9(1):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.W., Yip C.C.Y., To K.K.W., Tang T.H.C., Wong S.C.Y., Leung K.H., et al. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J. Clin. Microbiol. 2020;58(5) doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.M., But P.P.H. Vol. I. 2014. (Pharmacology and Applications of Chinese Materia Medica). [Google Scholar]
- Chang C.K., Sue S.C., Yu T.H., Hsieh C.M., Tsai C.K., Chiang Y.C., Lin C.H. Modular organization of SARS coronavirus nucleocapsid protein. J. Biomed. Sci. 2006;13(1):59–72. doi: 10.1007/s11373-005-9035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay D. Role and scope of ethnomedical plants in the development of antivirals. Pharmacologyonline. 2006;3:64–72. [Google Scholar]
- Chen C.N., Lin C.P., Huang K.K., Chen W.C., Hsieh H.P., Liang P.H., Hsu J.T.A. Evidence-Based Complementary and Alternative Medicine. 2005. Inhibition of SARS-CoV 3C-like protease activity by theaflavin-3, 3’-digallate (TF3) p. 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Li J., Luo C., Liu H., Xu W., Chen G., Jiang H. Binding interaction of quercetin-3-β-galactoside and its synthetic derivatives with SARS-CoV 3CLpro: structure–activity relationship studies reveal salient pharmacophore features. Bioorg. Med. Chem. 2006;14(24):8295–8306. doi: 10.1016/j.bmc.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J.K., Curtis-Long M.J., Lee K.H., Kim D.W., Ryu H.W., Yuk H.J., Park K.H. Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia tomentosa. Bioorg. Med. Chem. 2013;21(11):3051–3057. doi: 10.1016/j.bmc.2013.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou K.C., Wei D.Q., Zhong W.Z. Binding mechanism of coronavirus main proteinase with ligands and its implication to drug design against SARS. Biochem. Biophys. Res. Commun. 2003;308(1):148–151. doi: 10.1016/S0006-291X(03)01342-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176 doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daglia M. Polyphenols as antimicrobial agents. Curr. Opin. Biotechnol. 2012;23(2):174–181. doi: 10.1016/j.copbio.2011.08.007. [DOI] [PubMed] [Google Scholar]
- De Clercq E. Recent highlights in the development of new antiviral drugs. Curr. Opin. Microbiol. 2005;8(5):552–560. doi: 10.1016/j.mib.2005.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demurtas O.C., Massa S., Illiano E., De Martinis D., Chan P.K., Di Bonito P., Franconi R. Antigen production in plant to tackle infectious diseases flare up: the case of SARS. Front. Plant Sci. 2016;7:54. doi: 10.3389/fpls.2016.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dev S. Ancient-modern concordance in Ayurvedic plants: some examples. Environ. Health Perspect. 1999;107(10):783–789. doi: 10.1289/ehp.99107783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhouibi R., Affes H., Salem M.B., Hammami S., Sahnoun Z., Zeghal K.M., Ksouda K. Screening of pharmacological uses of Urtica dioica and others benefits. Prog. Biophys. Mol. Biol. 2020;150:67–77. doi: 10.1016/j.pbiomolbio.2019.05.008. [DOI] [PubMed] [Google Scholar]
- Dinesh D.C., Tamilarasan S., Rajaram K., Bouřa E. Antiviral drug targets of single-stranded RNA viruses causing chronic human diseases. Curr. Drug Targets. 2020;21(2):105–124. doi: 10.2174/1389450119666190920153247. [DOI] [PubMed] [Google Scholar]
- Dong L., Hu S., Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19) Drug Discov. Ther. 2020;14(2020):58–60. doi: 10.5582/ddt.2020.01012. [DOI] [PubMed] [Google Scholar]
- Dorigatti I., Okell L., Cori A., Imai N., Baguelin M., Bhatia S., Fu H. Imperial College London; London: 2020. Report 4: Severity of 2019-novel Coronavirus (nCoV) [Google Scholar]
- Drexler M. 2010. What You Need to Know About Infectious Disease. [PubMed] [Google Scholar]
- Durai P., Batool M., Shah M., Choi S. Middle East respiratory syndrome coronavirus: transmission, virology and therapeutic targeting to aid in outbreak control. Exp. Mol. Med. 2015;47(8) doi: 10.1038/emm.2015.76. e181-e181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eweas A.F., Maghrabi I.A., Namarneh A.I. Advances in molecular modeling and docking as a tool for modern drug discovery. Der Pharma Chemica. 2014;6(6):211–228. [Google Scholar]
- Farnsworth N.R. The role of ethnopharmacology in drug development. Bioact. Compounds Plants. 1990;154:2–21. doi: 10.1002/9780470514009.ch2. [DOI] [PubMed] [Google Scholar]
- Fehr A.R., Perlman S. Coronaviruses. Humana Press; New York, NY: 2015. Coronaviruses: an overview of their replication and pathogenesis; pp. 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore C., Eisenhut M., Krausse R., Ragazzi E., Pellati D., Armanini D., Bielenberg J. Antiviral effects of Glycyrrhiza species. Phytotherapy Res. 2008;22(2):141–148. doi: 10.1002/ptr.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franconi R., Illiano E., Paolini F., Massa S., Venuti A., Demurtas O.C. Defence Against Bioterrorism. Springer; Dordrecht: 2018. Rapid and Low-cost tools derived from plants to face Emerging/Re-emerging infectious diseases and bioterrorism agents; pp. 123–139. [Google Scholar]
- Fung K.P., Leung P.C., Tsui K.W., Wan C.C., Wong K.B., Waye M.Y., et al. Immunomodulatory activities of the herbal formula Kwan Du Bu Fei Dang in healthy subjects: a randomised, double-blind, placebo-controlled study. Hong Kong Med. J.= Xianggang yi xue za zhi. 2011;17:41. [PubMed] [Google Scholar]
- Gatera G., Pavarini G. COVID-19: what is next for public health. Lancet. 2020;395:542–545. doi: 10.1016/S0140-6736(20)30374-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomathi M., Padmapriya S., Balachandar V. Drug studies on Rett syndrome: from bench to bedside. J. Autism Dev. Disord. 2020:1–25. doi: 10.1007/s10803-020-04381-y. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117(1):17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., et al. The species severe acute respiratory syndrome related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Yan Z., Zheng X., Hu L., Yang Y., Wang J. A comparison of various optimization algorithms of protein–ligand docking programs by fitness accuracy. J. Mol. Model. 2014;20(7):2251. doi: 10.1007/s00894-014-2251-3. [DOI] [PubMed] [Google Scholar]
- Ho T.Y., Wu S.L., Chen J.C., Li C.C., Hsiang C.Y. Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction. Antiviral Res. 2007;74(2):92–101. doi: 10.1016/j.antiviral.2006.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostettmann K., Marston A., Ndjoko K., Wolfender J.L. The potential of African plants as a source of drugs. Curr. Org. Chem. 2000;4(10):973–1010. [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Cheng Z. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst K.R., Koetzner C.A., Masters P.S. Identification of in vivo-interacting domains of the murine coronavirus nucleocapsid protein. J. Virol. 2009;83(14):7221–7234. doi: 10.1128/JVI.00440-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin K.K., Renzette N., Kowalik T.F., Jensen J.D. Antiviral drug resistance as an adaptive process. Virus Evol. 2016;2(1) doi: 10.1093/ve/vew014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaguirre G. The proteolytic regulation of virus cell entry by furin and other proprotein convertases. Viruses. 2019;11(9):837. doi: 10.3390/v11090837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S., Kim H., Kim S., Shin D.H., Kim M.S. Characteristics of flavonoids as potent MERS‐CoV 3C‐like protease inhibitors. Chem. Biol. Drug Des. 2019;94(6):2023–2030. doi: 10.1111/cbdd.13604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan S., Ali P.S.S., Sheeza A., Hemalatha K. COVID-19 (Novel Coronavirus 2019)-recent trends. Eur. Rev. Med. Pharmacol. Sci. 2020;24(4):2006–2011. doi: 10.26355/eurrev_202002_20378. [DOI] [PubMed] [Google Scholar]
- Keyaerts Els, Vijgen Leen, Pannecouque Christophe, Damme Els Van, Peumans Willy, Egberink Herman, Balzarini Jan, Ranst Marc Van. Plant lectins are potent inhibitors of coronaviruses by interfering with two targets in the viral replication cycle. Antiviral Res. 2007;75(3):179–187. doi: 10.1016/j.antiviral.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killerby M.E., Biggs H.M., Haynes A., Dahl R.M., Mustaquim D., Gerber S.I., Watson J.T. Human coronavirus circulation in the United States 2014–2017. J. Clin. Virol. 2018;101:52–56. doi: 10.1016/j.jcv.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiran G., Karthik L., Devi S., Sathiyarajeswaran P., Kanakavalli K., Kumar K.M., Kumar D.R. In silico computational screening of kabasura kudineer-official siddha formulation and JACOM against SARS-CoV-2 spike protein. J. Ayurveda Integr. Med. 2020;S0975–9476(20):30024–30033. doi: 10.1016/j.jaim.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau K.M., Lee K.M., Koon C.M., Cheung C.S.F., Lau C.P., Ho H.M., et al. Immunomodulatory and anti-SARS activities of Houttuynia cordata. J. Ethnopharmacol. 2008;118(1):79–85. doi: 10.1016/j.jep.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.Y., Chen C., Zhang H.Q., Guo H.Y., Wang H., et al. Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antiviral Res. 2005;67(1):18–23. doi: 10.1016/j.antiviral.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.W., Tsai F.J., Tsai C.H., Lai C.C., Wan L., Ho T.Y., Chao P.D.L. Anti-SARS coronavirus 3C-like protease effects of Isatis indigotica root and plant-derived phenolic compounds. Antiviral Res. 2005;68(1):36–42. doi: 10.1016/j.antiviral.2005.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffelholz M.J., Tang Y.W. Laboratory diagnosis of emerging human coronavirus infections–the state of the art. Emerg. Microbes Infect. 2020;9(1):747–756. doi: 10.1080/22221751.2020.1745095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loizzo M.R., Saab A.M., Tundis R., Statti G.A., Menichini F., Lampronti I., Doerr H.W. Phytochemical analysis and in vitro antiviral activities of the essential oils of seven Lebanon species. Chem. Biodivers. 2008;5(3):461–470. doi: 10.1002/cbdv.200890045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Bi Y. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massi A., Bortolini O., Ragno D., Bernardi T., Sacchetti G., Tacchini M., De Risi C. Research progress in the modification of quercetin leading to anticancer agents. Molecules. 2017;22(8):1270. doi: 10.3390/molecules22081270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Mitchell H.D., Cockrell A.S., Gralinski L.E., Yount B.L., Jr, Graham R.L., et al. MERS-CoV accessory ORFs play key role for infection and pathogenesis. mBio. 2017;8:e00665–17. doi: 10.1128/mBio.00665-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi N., Shaghaghi N. Inhibitory effect of eight secondary metabolites from conventional medicinal plants on COVID_19 virus protease by molecular docking analysis. Preprint. 2020 https://doi.org/10.26434/chemrxiv, 11987475, v1. [Google Scholar]
- Morris D.R., Geballe A.P. Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 2000;20(23):8635–8642. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nal B., Chan C., Kien F., Siu L., Tse J., Chu K., van der Werf S. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J. Gen. Virol. 2005;86(5):1423–1434. doi: 10.1099/vir.0.80671-0. [DOI] [PubMed] [Google Scholar]
- Neiderud C.J. How urbanization affects the epidemiology of emerging infectious diseases. Infect. Ecol. Epidemiol. 2015;5(1):27060. doi: 10.3402/iee.v5.27060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Adair B.D., Yoshioka C., Quispe J.D., Orca G., Kuhn P., Buchmeier M.J. Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. J. Virol. 2006;80(16):7918–7928. doi: 10.1128/JVI.00645-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan S.Y., Zhou S.F., Gao S.H., Yu Z.L., Zhang S.F., Tang M.K., et al. Evidence-Based Complementary and Alternative Medicine. 2013. New perspectives on how to discover drugs from herbal medicines: CAM’s outstanding contribution to modern therapeutics; p. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.Y., Jeong H.J., Kim J.H., Kim Y.M., Park S.J., Kim D., Ryu Y.B. Diarylheptanoids from Alnus japonica inhibit papain-like protease of severe acute respiratory syndrome coronavirus. Biol. Pharm. Bull. 2012:b12–00623. doi: 10.1248/bpb.b12-00623. [DOI] [PubMed] [Google Scholar]
- Park J.Y., Ko J.A., Kim D.W., Kim Y.M., Kwon H.J., Jeong H.J., et al. Chalcones isolated from Angelica keiskei inhibit cysteine proteases of SARS-CoV. J. Enzyme Inhib. Med. Chem. 2016;31(1):23–30. doi: 10.3109/14756366.2014.1003215. [DOI] [PubMed] [Google Scholar]
- Peiris J.S.M., Guan Y., Yuen K.Y. Severe acute respiratory syndrome. Nat. Med. 2004;10(12):S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Osterhaus A.D., Fouchier R.A., Haagmans B.L. MERS: emergence of a novel human coronavirus. Curr. Opin. Virol. 2014;5:58–62. doi: 10.1016/j.coviro.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinavel T., Palanisamy M., Palanisamy S., Subramanian A., Thangaswamy S. Phytochemical 6-Gingerol- A promising Drug of choice for COVID-19. Int. J. Adv. Sci. Eng. 2020;6(4):1482–1489. [Google Scholar]
- Rolta R., Yadav R., Salaria D., Trivedi S., Imran M., Sourirajan A., Baumler D.J., Dev K. In silicoscreening of hundred phytocompounds of ten medicinal plants as potential inhibitors of nucleocapsid phosphoprotein of COVID-19: an approach to prevent virus assembly. J. Biomol. Struct. Dyn. 2020:1–18. doi: 10.1080/07391102.2020.1804457. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra S., Kalra A., Kumar A., Joe W. Utilization of alternative systems of medicine as health care services in India: evidence on AYUSH care from NSS 2014. PLoS One. 2017;12(5) doi: 10.1371/journal.pone.0176916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu Y.B., Jeong H.J., Kim J.H., Kim Y.M., Park J.Y., Kim D., et al. Biflavonoids from Torreya nucifera displaying SARS-CoV 3CLpro inhibition. Bioorg. Med. Chem. 2010;18(22):7940–7947. doi: 10.1016/j.bmc.2010.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampangi-Ramaiah M.H., Vishwakarma R., Shaanker R.U. Molecular docking analysis of selected natural products from plants for inhibition of SARS-CoV-2 main protease. Curr. Sci. 2020;118(7):1087–1092. [Google Scholar]
- Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16(1):1–22. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultes R.E., Raffauf R.F. Dioscorides press; 1990. The Healing Forest: Medicinal and Toxic Plants of the Northwest Amazonia. [Google Scholar]
- Shaikh S.A., Jain T., Sandhu G., Soni A., Jayaram B. From drug target to leads-sketching a physico-chemical pathway for lead molecule design in silico. Front. Med. Chem. Online. 2013;6:324. [Google Scholar]
- Shi F., Yu Q., Huang W., Tan C. 2019 novel coronavirus (COVID-19) pneumonia with hemoptysis as the initial symptom: CT and clinical features. Korean J. Radiol. 2020;21(5):537. doi: 10.3348/kjr.2020.0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.C., Blanc H., Vignuzzi M., Denison M.R. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog. 2013;9(8) doi: 10.1371/journal.ppat.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., et al. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331(5):991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., Xu Y., Bao L., Zhang L., Yu P., Qu Y., Qin C. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11(1):59. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapparel C., Sobo K., Constant S., Huang S., Van Belle S., Kaiser L. Growth and characterization of different human rhinovirus C types in three-dimensional human airway epithelia reconstituted in vitro. Virology. 2013;446(1-2):1–8. doi: 10.1016/j.virol.2013.06.031. [DOI] [PubMed] [Google Scholar]
- ul Qamar M.T., Alqahtani S.M., Alamri M.A., Chen L.L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J. Pharm. Anal. 2020;10(4):313–319. doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Meer F.J.U.M., de Haan C.A.M., Schuurman N.M.P., Haijema B.J., Verheije M.H., Bosch B.J., Egberink H.F. The carbohydrate-binding plant lectins and the non-peptidic antibiotic pradimicin A target the glycans of the coronavirus envelope glycoproteins. J. Antimicrob. Chemother. 2007;60(4):741–749. doi: 10.1093/jac/dkm301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94(7) doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Chang Z., Ouyang J., Wei H., Yang R., Chao Y., Hung T. Profiles of IgG antibodies to nucleocapsid and spike proteins of the SARS-associated coronavirus in SARS patients. DNA Cell Biol. 2005;24(8):521–527. doi: 10.1089/dna.2005.24.521. [DOI] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Zhao Y. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. Jama. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.S., Wang Y.R., Ye D.W., Liu Q.Q. A review of the 2019 Novel Coronavirus (COVID-19) based on current evidence. Int. J. Antimicrob. Agents. 2020 doi: 10.1016/j.ijantimicag.2020.106137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen C.C., Kuo Y.H., Jan J.T., Liang P.H., Wang S.Y., Liu H.G., et al. Specific plant terpenoids and lignoids possess potent antiviral activities against severe acute respiratory syndrome coronavirus. J. Med. Chem. 2007;50(17):4087–4095. doi: 10.1021/jm070295s. [DOI] [PubMed] [Google Scholar]
- Wen C.C., Shyur L.F., Jan J.T., Liang P.H., Kuo C.J., Arulselvan P., et al. Traditional Chinese medicine herbal extracts of Cibotium barometz, Gentiana scabra, Dioscorea batatas, Cassia tora, and Taxillus chinensis inhibit SARS-CoV replication. J. Tradit. Complement. Med. 2011;1(1):41–50. doi: 10.1016/S2225-4110(16)30055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . World Health Organization; 2020. Middle East Respiratory Syndrome Coronavirus (MERS-CoV)https://www.who.int/emergencies/mers-cov/en/ (Accessed 1/4/2020) [Google Scholar]
- Williams A.K., Wang L., Sneed L.W., Collisson E.W. Analysis of a hypervariable region in the 3’non-coding end of the infectious bronchitis virus genome. Virus Res. 1993;28(1):19–27. doi: 10.1016/0168-1702(93)90086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO) 2020. Laboratory testing of 2019 Novel Coronavirus (2019-nCoV) in Suspected Human Cases.https://apps.who.int/iris/handle/10665/ [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Hu Y., Yuan M.L. 2020. Complete Genome Characterisation of a Novel Coronavirus Associated With Severe Human Respiratory Disease in Wuhan, China. bioRxiv. [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Yuan M.L. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Zhao S., Teng T., Abdalla A.E., Zhu W., Xie L., Guo X. Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses. 2020;12(2):244. doi: 10.3390/v12020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Yu C., Qu J., Zhang L., Jiang S., Huang D., Jiang R. Imaging and clinical features of patients with 2019 novel coronavirus SARS-CoV-2. Eur. J. Nucl. Med. Mol. Imaging. 2020:1–6. doi: 10.1007/s00259-020-04735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L., Li Z., Yuan K., Qu X., Chen J., Wang G., Chen L. Small molecules blocking the entry of severe acute respiratory syndrome coronavirus into host cells. J. Virol. 2004;78(20):11334–11339. doi: 10.1128/JVI.78.20.11334-11339.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M.S., Lee J., Lee J.M., Kim Y., Chin Y.W., Jee J.G., Jeong Y.J. Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13. Bioorg. Med. Chem. Lett. 2012;22(12):4049–4054. doi: 10.1016/j.bmcl.2012.04.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., Van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhang X.W., Yap Y.L. Old drugs as lead compounds for a new disease? Binding analysis of SARS coronavirus main proteinase with HIV, psychotic and parasite drugs. Bioorg. Med. Chem. 2004;12(10):2517–2521. doi: 10.1016/j.bmc.2004.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Penninger J.M., Li Y., Zhong N., Slutsky A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46(4):586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N., Xia R., Yang C., Yin B., Li Y., Duan C., Xie Q. Boosted expression of the SARS-CoV nucleocapsid protein in tobacco and its immunogenicity in mice. Vaccine. 2009;27(36):5001–5007. doi: 10.1016/j.vaccine.2009.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Chen H.D. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. BioRxiv. 2020 [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Niu P. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang M., Jiang H., Suzuki Y., Li X., Xiao P., Tanaka T., Qin C. Procyanidins and butanol extract of Cinnamomi Cortex inhibit SARS-CoV infection. Antiviral Res. 2009;82(1):73–81. doi: 10.1016/j.antiviral.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81(4):853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- Zumla A., Hui D.S., Perlman S. Middle east respiratory syndrome. Lancet. 2015;386(9997):995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]