Abstract
With surging global demand for SARS-CoV-2 testing capacity, laboratories seek automated, high-throughput molecular solutions, particularly for specimens not requiring specialized collection devices or viral transport media. Saliva specimens submitted from patients under investigation for COVID-19 from March to July 2020 were processed in the laboratory with sterile phosphate-buffered saline in a 1:2 dilution and tested using manual extraction and a commercial assay for detection of the SARS-CoV-2 E gene (LightMix®) in comparison to the Roche cobas® SARS-CoV-2 Test on the cobas® 6800 instrument. 34.4% (22/64) of saliva samples were positive for SARS-CoV-2. Positive and negative concordance between the LightMix® and cobas® assays were 100%. The overall invalid rate for saliva on the cobas® 6800 (1/128, 0.78%) was similar to the baseline invalid rate observed for nasopharyngeal swabs/viral transport media. Saliva is a feasible specimen type for SARS-CoV-2 testing on the cobas® 6800 platform, with potential to improve turnaround time and enhance testing capacity.
Keywords: SARS-Cov-2, COVID-19, saliva, cobas 6800, automated
1. Introduction
The global public health response to the COVID-19 pandemic highlighted the critical need for diagnostic testing which is sustainable, practical, and scalable (WHO 2020). With increasing worldwide demand for SARS-CoV-2 molecular testing, supply-chain issues for high-quality, flocked nasopharyngeal (NP) swabs have created significant challenges for testing capacity in clinical and public health laboratories. Alternate specimen types, such as saliva, have been reported in some studies to have nearly comparable sensitivity to nasopharyngeal swabs for the detection of SARS-CoV-2, and may be an appropriate supplemental or alternate diagnostic specimen (Jamal et al., 2020, Matic et al., 2020, To et al., 2020, Williams et al., 2020). Although a variety of methods for saliva collection have been described (Azzi et al., 2020, To et al., 2020, Wyllie et al., 2020), we have previously shown the utility of testing saliva in the absence of transport media (Matic et al., 2020), which enables a simple collection technique that avoids the introduction of potential inhibitors (Rodríguez and Vaneechoutte, 2019, Jiang et al., 2019) and dependence on supply of specialized saliva collection devices.
In addition to potentially obviating supply shortages, saliva has been increasingly described as a useful sample for the diagnosis of COVID-19 to overcome certain preanalytical collection challenges. Flocked NP swabs have been the preferred specimen type due to established sensitivity, but may occasionally result in false-negative test results due to poor specimen collection quality (Kinloch N et al., 2020) or timing of testing relative to symptom onset (He et al., 2020, Li et al., 2020, Wölfel et al., 2020). Lower respiratory tract specimens such as a bronchoalveolar lavage are often obtained from severely ill patients, but require aerosol-generating medical procedures and thus have the potential for aerosolization and transmission of SARS-CoV-2. Furthermore, only a minority of patients with COVID-19 are able to produce expectorated sputum (Huang C et al., 2020). Saliva is a convenient alternate sample to collect for SARS-CoV-2 detection, particularly for patients with high clinical suspicion for COVID-19 but repeatedly negative test results by NP swabs (Koven, 2020, Watson et al., 2020) or for individuals unwilling or unable to tolerate NP swab collection.
Within our clinical laboratory, processing and testing of saliva is currently a manual process requiring extraction (MagNA Pure Compact or MagNA Pure 96, Roche Molecular Diagnostics; Pleasanton, CA) followed by amplification (LightCycler® 480; Roche), which was previously validated in comparison to paired NP swabs (Matic et al., 2020). Postanalytical reporting into the electronic medical records system is also a manual process. As a result, capacity for saliva testing in our laboratory is limited, with delays in turnaround time compared to nasopharyngeal swabs which are processed entirely on the automated cobas® 6800 platform (Roche). We sought to evaluate the potential utilization of the cobas® 6800 platform for SARS-CoV-2 detection from saliva.
2. Materials and methods
From March to July 2020, saliva was ordered by clinicians from hospitalized patients, ambulatory patients, and long-term care residents for the diagnosis of COVID-19. Collection and processing of the saliva was previously described (Matic et al., 2020). Briefly, ≥1 mL of saliva was collected in a sterile screw-top container (Starplex Scientific Inc.; Etobicoke, Canada) without the addition of transport media, and then processed in the laboratory with phosphate-buffered saline (PBS) in a 1:2 dilution and vortexed with glass beads. Samples were extracted by the MagNA Pure 96 (extraction volume of 500 µL and elution volume of 50 µL) and tested using the LightMix® ModularDx SARS-CoV (COVID19) E-gene assay (TIB Molbiol; Berlin, Germany). The remaining volume of processed saliva samples was stored at –70°C, and subsequently tested with the cobas® SARS-CoV-2 Test (Roche Molecular Diagnostics, Laval, QC) on the cobas® 6800 platform (standard sample volume of 600 µL and processing volume of 400 µL). Prior to cobas® SARS-CoV-2 testing, a software upgrade (Assay Specific Analysis Package [ASAP]) was required on the cobas® 6800 to prevent viscous specimens from mistakenly being interpreted as clotted/invalid by the instrument. Samples known to be positive for SARS-CoV-2 based on results from the LightMix® assay were alternated with negative samples in a checkerboard pattern on the 96-well processing plate, and tested in duplicate. This study protocol was reviewed and approved by an institutional research ethics board, with waiver of consent obtained.
3. Results
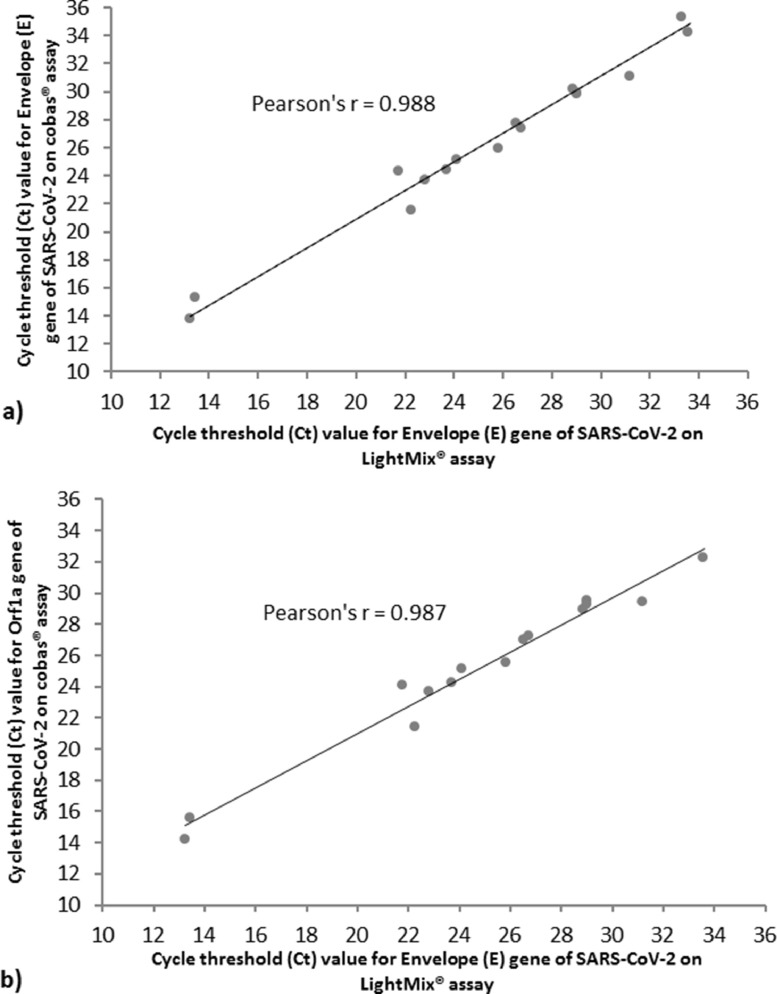
A total of 64 clinical saliva samples were included and tested in duplicate for SARS-CoV-2 on the cobas® 6800 platform. Twenty-2 (34.4%) of the samples were known to be positive for SARS-CoV-2 based on prior results from the LightMix® assay, and 42 (65.6%) had no detectable SARS-CoV-2. Compared to the LightMix® assay, positive percent agreement and negative percent agreement on the cobas® assay were 100%. Mean cycle threshold (Ct) values for the Envelope (E) gene showed close positive correlation between the LightMix® and cobas® assays with a Pearson's correlation coefficient of 0.988 (95% conflict of interest, 0.966 to 0.996), excluding 5 samples from the analysis which had required dilution prior to testing by the cobas® assay in order to maintain sufficient sample volume (Fig. 1a ). Correlation of the mean Ct values for the E gene on the LightMix® assay and Orf1a region on the cobas® assay was similar with a Pearson's correlation coefficient of 0.987 (95% conflict of interest, 0.960 to 0.996), although an additional 2 samples were excluded from the analysis where the Orf1a target was undetected by the cobas® assay (Fig. 1b). No carryover or cross-contamination of samples was observed, even with strongly positive samples (Ct values 13 to 24) directly adjacent to negative samples. One saliva sample produced an error which was reported by the cobas 6800® instrument as “Invalid”; this sample was successfully tested on the duplicate run. In total, the observed error rate for saliva samples tested on the cobas® 6800 was 0.78% (1/128).
Fig. 1.
(a) Mean cycle threshold (Ct) values for the Envelope (E) gene of SARS-CoV-2 in saliva samples showed close positive correlation between the LightMix® ModularDx SARS-CoV (COVID19) E-gene assay (TIB Molbiol; Berlin, Germany) and the cobas® SARS-CoV-2 Test (Roche Molecular Diagnostics; Laval, QC) on the cobas® 6800 platform. Five samples were excluded from analysis due to dilution that occurred prior to testing by the cobas® assay in order to increase sample volume. (b) Mean cycle threshold (Ct) values for the Envelope (E) gene of SARS-CoV-2 in saliva samples on the LightMix® assay and Orf1a gene on the cobas® assay showed close positive correlation. An additional 2 samples were excluded from analysis where the Orf1a target was undetected by the cobas® assay, but E gene was otherwise detected by both the LightMix® and cobas® assays.
4. Discussion
In our evaluation of the cobas® 6800 platform for saliva testing, we demonstrated complete concordance in comparison to the LightMix® assay, which is the current assay utilized in our laboratory for clinical testing of specimens other than NP swabs such as saliva.
Previous studies have evaluated the feasibility of saliva for SARS-CoV-2 detection on commercial instruments such as the Cepheid GeneXpert® System (McCormick-Baw et al., 2020) and cobas® 8800 (Nagura-Ikeda et al., 2020). However, there are technical challenges associated with processing saliva for automated, high-throughput testing. As a highly viscous sample compared to viral transport media, there is potential for pipetting errors or instrument contamination. In our laboratory, saliva is preprocessed with PBS and glass beads in order to decrease the viscosity of the sample, as previous attempts to perform nucleic acid extraction on neat saliva without these initial processing steps resulted in extraction failure on the MagNA Pure instruments or inhibition of internal controls in nearly all samples (Matic et al., 2020). Furthermore, we strived to minimize the potential for obstruction or contamination of the cobas® 6800 platform by highly viscous samples. Other studies have described the successful enzymatic or chemical treatment of sputum samples prior to nucleic acid extraction with use of agents such as proteinase K, dithiothreitol, or N-acetyl-L-cysteine (NaC), although NaC may not be optimal for targeting RNA by real-time RT-PCR (Centers for Disease Control and Prevention 2020, Sung et al., 2016, Yu et al., 2018). Use of PBS may ultimately be preferable in order for laboratories to maintain the ability to perform bacterial or viral culture of these specimens when needed. The samples utilized in the study were from a variety of clinical situations, including long-term care residents, inpatients at a tertiary care hospital, and outpatient contacts of known COVID-19 patients; these samples would be broadly representative of saliva ordered and collected for clinical testing in the future (Matic et al., 2020). Reassuringly, testing with this specimen type did not result in a significant number of invalid results (1/128, 0.78%), and was comparable to the rate of invalid results observed in our laboratory from nasopharyngeal swabs with the SARS-CoV-2 Test (0.20%) and plasma used with other cobas® 6800 assays (0.30%) (unpublished data). We also assessed the potential for carryover during pipetting on the cobas® and did not identify any cross-contamination of samples.
This study is limited by retrospective testing of a limited number of clinical saliva samples. Testing of retrospective samples was necessary to ensure a sufficient number of saliva samples with detectable SARS-CoV-2 RNA were included, and to allow time for cobas® software upgrades. Although testing was not performed in parallel, there was high concordance of testing despite sample storage. Due to global demand for cobas® SARS-CoV-2 Test reagents, it is not currently responsible to perform further research testing to attain a larger sample size in lieu of clinical testing. The use of clinical samples (rather than engineered or spiked) is considered a strength of this study, as the realistic performance of human saliva samples harboring live SARS-CoV-2 viral particles is reflected.
Due to limited remnant sample volume in some cases, 5 of the 25 saliva samples positive for SARS-CoV-2 (22.7%) underwent additional dilution in order to test on the cobas® 6800 platform. The Ct values in these cases cannot be directly compared to those from the LightMix® assay as a measure of assay performance; however, the dilution factor in these cases (1:3 to 1:5) would have had a negligible effect on the Ct values, and all 5 samples produced positive SARS-CoV-2 results on these qualitative assays.
Transitioning to an automated platform for saliva testing is critical for enhancing SARS-CoV-2 testing capacity, particularly in preparation for a potential resurgence of COVID-19 cases or mass testing of defined populations. Automated testing reduces errors in the preanalytical and postanalytical phases, and improves turnaround time by enabling saliva to be processed on multiple runs daily and overnight. Our evaluation confirmed the feasibility of saliva as a suitable specimen type for SARS-CoV-2 testing on the cobas® 6800 platform.
Author contributions statement
-
•
Nancy Matic: Conceptualization; Data curation; Formal analysis; Visualization; Writing - original draft.
-
•
Tanya Lawson: Data curation; Investigation.
-
•
Gordon Ritchie: Data curation; Investigation.
-
•
Aleksandra Stefanovic: Writing - review & editing.
-
•
Victor Leung: Writing - review & editing.
-
•
Sylvie Champagne: Writing - review & editing.
-
•
Marc G. Romney: Supervision; Writing - review & editing.
-
•
Christopher F. Lowe: Conceptualization; Supervision; Writing - original draft.
Funding
This study was supported by Roche Diagnostics for a software upgrade of the cobas® 6800, but no financial compensation was provided. Roche Diagnostics was not involved in the study design, implementation nor analysis of the data.
This study received approval from the Providence Health Care Research Ethics Board.
Potential competing interests
N.M. reports honoraria related to speaker engagement outside the submitted work for Roche Molecular Systems, Inc. All other authors report no relevant competing interests.
Acknowledgments
We are grateful to our medical laboratory technologists who are highly committed to patient care and laboratory quality improvement.
References
- Azzi L., Carcano G., Gianfagna F., Grossi P., Gasperina D.D., Genoni A., et al. saliva is a reliable tool to detect SARS-CoV-2. J Infect. 2020;81:E45–E50. doi: 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention, Processing of sputum specimens for nucleic acid extraction, (2020) 1. https://www.cdc.gov/coronavirus/2019-ncov/downloads/processing-sputum-specimens.pdf.
- He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- Huang C C.B., Wang Y, Li Z, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamal A.J., Mozafarihashjin M., Coomes E., Powis J., Li A.X., Paterson A., et al. Sensitivity of nasopharyngeal swabs and saliva for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin Infect Dis. 2020;27:9–11. doi: 10.1093/cid/ciaa848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Li L., Kang P., Yu H., Nie S.P., Xie M.Y., et al. Inappropriateness of RNA later to preserve Caenorhabditis elegans for RNA extraction. MethodsX. 2019;6:2460–2467. doi: 10.1016/j.mex.2019.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinloch N B.Z., Ritchie G, Brumme CJ, Dong W, Dong W, Lawson T, et al. Suboptimal biological sampling as a probable cause of false-negative COVID-19 diagnostic test results. J Infect Dis. 2020;222:899–902. doi: 10.1093/infdis/jiaa370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koven S. False negative tests for SARS-CoV-2 infection — challenges and implications. N Engl J Med. 2020;383(1-3):e38. doi: 10.1056/NEJMp2009027. [DOI] [PubMed] [Google Scholar]
- Li L., Lowe C.F., Ritchie G., Stefanovic A., Champagne S., Romney M.G., et al. SARS-CoV-2 molecular testing for the diagnosis of COVID-19: one test does not fit all. J Med Virol. 2020 doi: 10.1002/jmv.26532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matic N., Stefanovic A., Leung V., Lawson T., Ritchie G., Li L., et al. Practical challenges to the clinical implementation of saliva for SARS-CoV-2 detection. Eur J Clin Microbiol. 2020:1–4. doi: 10.1007/s10096-020-04090-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick-Baw C., Morgan K., Gaffney D., Cazares Y., Jaworski K., Byrd A., et al. Saliva as an alternate specimen source for detection of SARS-CoV-2 in symptomatic patients using cepheid Xpert Xpress SARS-CoV-2. J Clin Microbiol. 2020;58:2–3. doi: 10.1128/jcm.01109-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagura-Ikeda M., Imai K., Tabata S., Miyoshi K., Murahara N., Mizuno T., et al. Clinical evaluation of self-collected saliva by RT-qPCR, direct RT-qPCR, RT-LAMP, and a rapid antigen test to diagnose COVID-19. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.01438-20. e01438-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez A., Vaneechoutte M. Comparison of the efficiency of different cell lysis methods and different commercial methods for RNA extraction from Candida albicans stored in RNA later. BMC Microbiol. 2019;19:1–10. doi: 10.1186/s12866-019-1473-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H., Yong D., Ki C.S., Kim J.S., Seong M.W., Lee H., et al. Comparative evaluation of three homogenization methods for isolating middle east respiratory syndrome coronavirus nucleic acids from sputum samples for real-time reverse transcription PCR. Ann Lab Med. 2016;36:457–462. doi: 10.3343/alm.2016.36.5.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K.W., Tsang O.T.Y., Leung W.S., Tam A.R., Wu T.C., Lung D.C., et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;3099:1–10. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- W.H.O. (WHO) 2020. Laboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human cases. [Google Scholar]
- Watson J., Whiting P.F., Brush J.E. Interpreting a covid-19 test result. BMJ. 2020;369:m1808. doi: 10.1136/bmj.m1808. [DOI] [PubMed] [Google Scholar]
- Williams E., Bond K., Zhang B., Putland M., Williamson D.A. Saliva as a non-invasive specimen for detection of SARS-CoV-2. J Clin Microbiol. 2020;58 doi: 10.1128/jcm.00776-20. e00776-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P., et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N Engl J Med. 2020;383:13. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Qiu T., Zeng Y., Wang Y., Zheng S., Chen X., et al. Comparative evaluation of three preprocessing methods for extraction and detection of influenza A virus nucleic acids from sputum. Front. Med. 2018;5:7–10. doi: 10.3389/fmed.2018.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]