Abstract
Virus-specific humoral and cellular immunity act synergistically to protect the host from viral infection. We interrogate the dynamic changes of virological and immunological parameters in 12 patients with symptomatic acute SARS-CoV-2 infection from disease onset to convalescence or death. We quantify SARS-CoV-2 viral RNA in the respiratory tract in parallel with antibodies and circulating T cells specific for various structural (nucleoprotein [NP], membrane [M], ORF3a, and spike) and non-structural (ORF7/8, NSP7, and NSP13) proteins. Although rapid induction and quantity of humoral responses associate with an increase in disease severity, early induction of interferon (IFN)-γ-secreting SARS-CoV-2-specific T cells is present in patients with mild disease and accelerated viral clearance. These findings provide support for the prognostic value of early functional SARS-CoV-2-specific T cells with important implications in vaccine design and immune monitoring.
Keywords: T cell response, Longitudinal, Acute phase4, Convalescence, Humoral response, Antibodies
Graphical Abstract
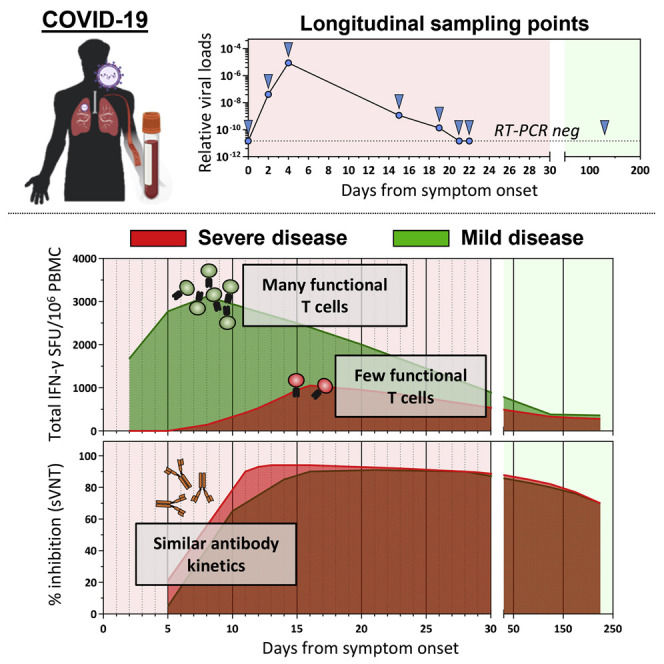
Tan et al. longitudinally analyzed the virological and immunological parameters in COVID-19 patients from disease onset until resolution or death. Early induction of functional SARS-CoV-2-specific T cells was observed in patients with mild disease and rapid viral clearance. This supports the prognostic value of detecting SARS-CoV-2-specific T cells.
Introduction
In December 2019, a new coronavirus was detected in Wuhan, China, in several patients with pneumonia and was later named SARS-CoV-2 (Zhou et al., 2020a). The illness (coronavirus disease 2019 [COVID-19]) resulting from SARS-CoV-2 infection is reported to be multifaceted with inflammation of the respiratory tract causing the leading symptoms of fever and dry cough (Chen et al., 2020). Both virus-specific humoral components (Long et al., 2020a; Gudbjartsson et al., 2020; Sun et al., 2020) and cellular components (Grifoni et al., 2020; Braun et al., 2020; Le Bert et al., 2020; Weiskopf et al., 2020) of adaptive immunity are induced in SARS-CoV-2-infected individuals, but their roles in viral control or disease pathogenesis need to be clarified. Viral clearance and reduced disease severity have been associated with coordinated activation of humoral and cellular anti-viral immunity (Rydyznski Moderbacher et al., 2020) and robust virus-specific T cell responses (Takahashi et al., 2020). A positive relationship between magnitude of SARS-CoV-2 antibodies (Hung et al., 2020; Long et al., 2020b; Wang et al., 2020) or T cells (Peng et al., 2020) and disease severity have also been reported. However, most of these studies have analyzed patients during the convalescent phase of infection, and only few studies have reported the dynamic changes of viral and virus-specific immunological parameters in severe COVID-19 patients during the initial phases of infection (Weiskopf et al., 2020; Rydyznski Moderbacher et al., 2020; Zhou et al., 2020b). To fill this gap, we longitudinally followed-up 12 patients (Table S1) with SARS-CoV-2 infection from symptom onset to convalescence or death. We quantified SARS-CoV-2 viral load in the upper respiratory tract and SARS-CoV-2-specific antibodies and T cells at multiple time points from acute disease until convalescence or death. Our data reveal a direct association between early induction of functional SARS-CoV-2-specific T cells and rapid control of viral infection.
Results
Dynamics of SARS-CoV-2 replication
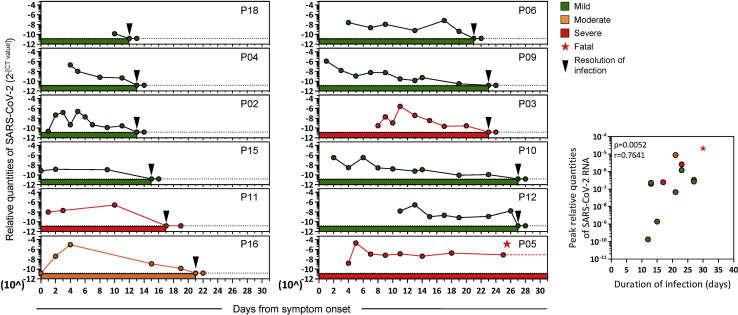
Relative quantities of SARS-CoV-2 in the respiratory tract and its persistence in each patient were calculated by using the number of RT-PCR cycles as a proxy of viral quantity (Figure 1 ). Duration of infection was defined from symptom onset until RT-PCR negativity (two negative SARS-CoV-2 RT-PCR tests 24 h apart). The patients displayed different profiles of virus quantity and persistence: short duration of infection was detected in four patients (P18, P04, P02, and P15) who became RT-PCR negative within 15 days of symptom onset. Five patients (P11, P16, P06, P09, and P03) were RT-PCR negative around day 17–24, whereas two patients (P10 and P12) became RT-PCR negative almost 1 month after onset of symptoms. One patient (P05) who succumbed to infection was persistently SARS-CoV-2 RT-PCR positive until day 31 after symptom onset, when he demised. Peak viral quantities was also positively correlated with duration of infection (Figure 1, insert). In addition, although patients who eliminated the virus within 15 days experienced mild respiratory symptoms (presence of fever or respiratory symptoms, but not requiring supplemental oxygen), patients with moderate symptoms (requiring oxygen supplementation of FiO2 < 0.5) and severe symptoms (requiring oxygen supplementation of FiO2 > 0.5, high-flow oxygen and/or mechanical ventilation) later eliminated the virus from the upper respiratory tract. Viral loads detected in patients with mild symptoms were also lower than those with moderate/severe symptoms (Figure S1A).
Figure 1.
Relative quantities of SARS-CoV-2 in the upper respiratory tract of symptomatic COVID-19 patients during acute infection
Longitudinal RT-PCR of SARS-CoV-2 RNA in the upper respiratory tract of COVID-19 patients (n = 12) with variable disease severity from symptom onset until RT-PCR negativity. Dotted lines denote positive cutoffs. Inserts show correlations between the peak relative quantities of SARS-CoV-2 and the duration of infection. The p value and the non-parametric Spearman correlation coefficient are indicated.
Dynamics of SARS-CoV-2-specific antibody response
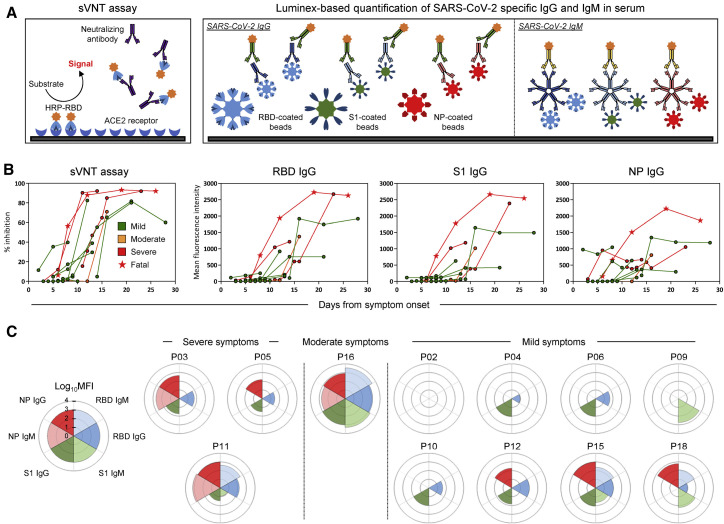
We then characterized the kinetics of anti-SARS-CoV-2 antibody appearance. Virus neutralization ability was tested longitudinally using the surrogate virus neutralization test (sVNT) that quantified the ability of serum antibodies to inhibit binding of Spike RBD (receptor binding domain) to the ACE2 receptor in vitro (Tan et al., 2020). We also quantified anti-RBD, anti-S1 (S1 domain of spike), and anti-NP (nucleoprotein) immunoglobulin G (IgG; Figure 2 ) and immunoglobulin M (IgM; Figure S2) antibodies with a Luminex-based quantification test that use beads coated with RBD, S1, and NP proteins, respectively (Figure 2A).
Figure 2.
Longitudinal analysis of SARS-CoV-2-specific antibody-related responses in acute COVID-19 patients
(A) Schematic representation of the surrogate virus neutralization assay and the Luminex-based assay to quantify SARS-CoV-2 RBD-, S1-, and NP-specific IgG and IgM antibodies. Cutoffs to define significant virus neutralization and antibody quantities were set at 20% inhibition for the sVNT assay (as defined in ref (Tan et al., 2020)) and MFI (mean fluorescence intensity) > 100 for the Luminex-based assay, respectively.
(B) SARS-CoV-2 neutralization and relative quantities of specific IgG and antibodies (n = 12).
(C) Rose plots represent the quantity of RBD-, S1-, and NP-specific IgG and IgM antibodies at first detectable antibody response (n = 12). Patient P02 had no detectable antibody response at 3, 4, 7, and 9 days after symptom onset for which samples were available.
Figure 2B shows that all COVID-19 patients developed neutralizing antibodies with the exception of patient P02, who nonetheless cleared SARS-CoV-2 at day 13 after symptom onset (Figure 1). The peak neutralizing activity was achieved within 9–15 days after symptom onset. Patients with moderate/severe symptoms exhibited stronger virus-specific antibody responses than those with mild disease (Figures S1B and S1C). The two patients who first reached 90% virus neutralization developed severe disease (patients P05, deceased, and P03, severe). The kinetics of anti-RBD, anti-S1, and anti-NP IgG also peaked around the 10- to 20-day period, similar to previous observations (Isho et al., 2020; Ripperger et al., 2020), with higher peak levels of antibodies against NP and Spike detected in patient P05, who succumbed to the infection. When we analyzed in parallel the kinetics of appearance of antibody responses against different proteins, we observed that patients with severe disease have an early NP-biased antibody response, whereas those with mild/moderate symptoms had either a spike-dominant or a balanced response (Figure 2C). These kinetics of appearance are consistent with recent data (Atyeo et al., 2020; Sun et al., 2020) showing that an anti-nucleocapsid humoral response is preferentially induced over a Spike IgG response in severe COVID-19 patients.
Dynamics of SARS-CoV-2-specific cellular responses
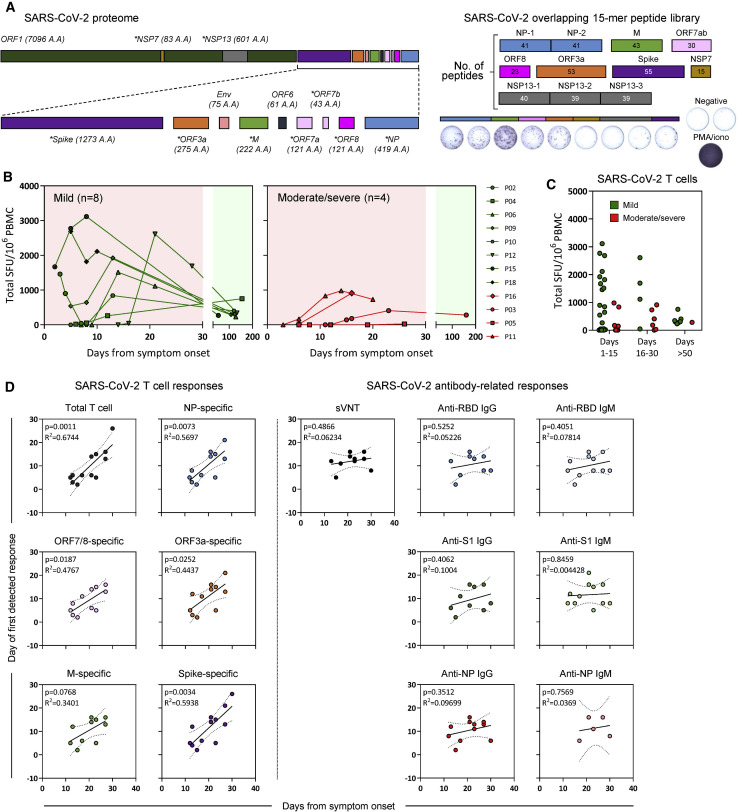
We next analyzed the kinetics of functional SARS-CoV-2-specific T cell appearance during the RT-PCR-positive phase of disease. Overlapping 15-mer peptide libraries (Table S2) covering the whole NP, membrane (M), ORF7ab, ORF8, ORF3a, the NSP7 and NSP13 of ORF1ab, and a pool of ∼40 peptides containing all confirmed T cell epitopes of Spike were used to stimulate PBMC (peripheral blood mononuclear cells) in an interferon (IFN)-γ ELISPOT (enzyme-linked immunospot) assay (Figure 3 A).
Figure 3.
Longitudinal analysis of SARS-CoV-2 T responses in COVID-19 patients during acute infection and at convalescence
(A) SARS-CoV-2 proteome organization. Analyzed proteins are marked by an asterisk. 15-mer peptides, which overlapped by 10 amino acids, comprising the NP, M, ORF7ab, ORF8, ORF3a, NSP7, and NSP13 were grouped into 10 pools with the indicated number of peptides in each pool. 15-mer predicted peptides previously shown to activate Spike-specific CD8 and CD4 T cells were grouped into a single pool. PMA+ionomycin was used as a positive control for all samples analyzed.
(B) Longitudinal analysis of the total SARS-CoV-2 T cell response in COVID-19 patients (n = 12) from onset of disease until convalescence. Individual lines represent single patients.
(C) Total SARS-CoV-2 T cell response detected in all COVID-19 patients (n = 12) during day 1–15, day 16–30, and >50 days after symptom onset. Patients with mild symptoms (n = 8) or moderate/severe symptoms (n = 4) are indicated.
(D) Linear regression analysis of the duration of infection and the number of days to the first detectable T cell response (total and NP-, ORF7/8-, ORF3a-, M-, or Spike-specific T cell response) or antibody-related response (sVNT and RBD-, S1-, or NP-specific IgG and IgM) are shown in the respective dotplots. The p values and the corresponding r2 values are shown.
During the initial phase of SARS-CoV-2 infection, the quantity of IFN-γ-secreting cells after stimulation by the different peptide pools increased progressively with the peak of frequency detected within ∼15 days after symptom onset in most tested patients (8 of 12, Figures 3B and 3C) in line with previous reports (Weiskopf et al., 2020; Rydyznski Moderbacher et al., 2020). In two patients, peak responses were detected beyond 20 days after symptom onset, whereas patient P05, who succumbed to the disease before viral clearance, had no detectable IFN-γ-secreting cells when stimulated with the different peptide pools until day 26, when stimulation with Spike peptides activated a weak response (Figure 3B). Importantly, production of IFN-γ was detected at all time points in all patients after stimulation with PMA+ionomycin (phorbol myristate acetate), showing that global cellular functionality was not fully compromised (Figure 4 A, insert). The frequency of IFN-γ-secreting cells reactive to all peptide pools was also quantified at least 1 month after resolution of infection. The magnitude of the IFN-γ response declined markedly in 7 of 8 patients tested (Figure 3B), consistent with waning of the cellular immune response that follows resolution of acute infection. The quantity and time of appearance of SARS-CoV-2 peptide-reactive cells were then analyzed in relation to the virological and clinical parameters.
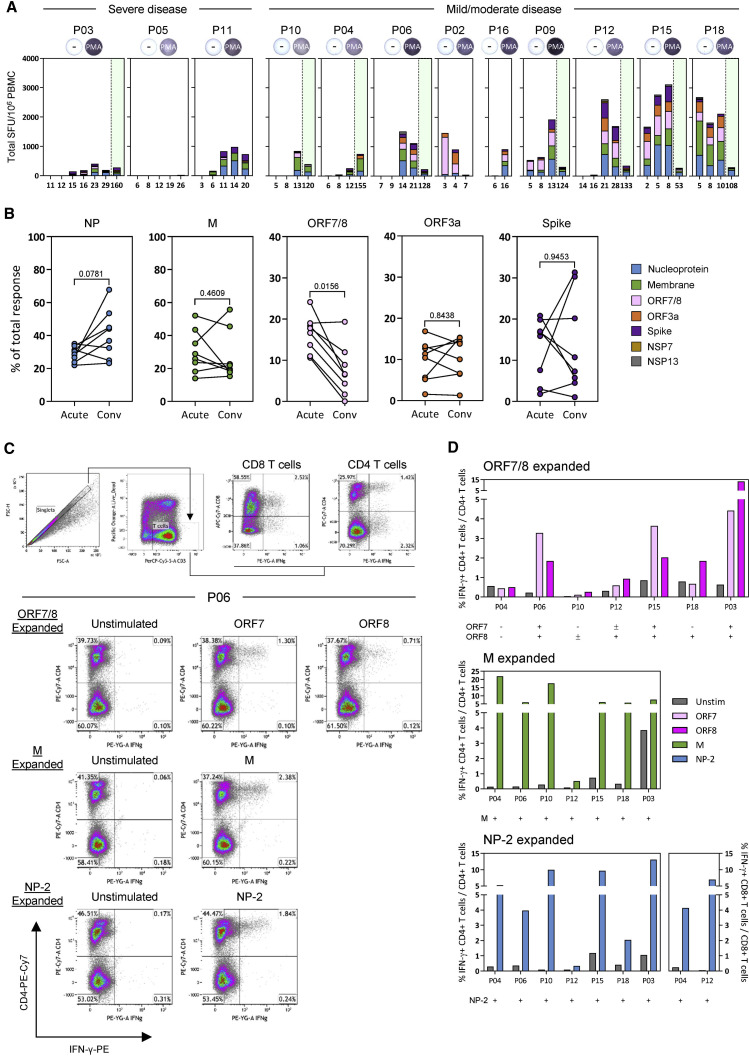
Figure 4.
Hierarchy of cellular responses toward different SARS-CoV-2 proteins
(A) Stacked bars denotes the frequency of peptide-reactive cells in all COVID-19 patients (n = 12) against the indicated SARS-CoV-2 protein at all time points tested. Green shaded areas denote the convalescence phase of the disease. Positive controls are inserted for each patient.
(B) Plots show the proportion of peptide-reactive cells attributed to the respective SARS-CoV-2 protein at the peak response during the acute phase and the convalescence phase of the disease (n = 8). A Wilcoxon matched-pairs test was used to evaluate the differences, and the p values are shown. Short-term T cell lines were also generated from the convalescent samples using the respective SARS-CoV-2 peptide pools. Each line was then stimulated with the corresponding peptide pool used for expansion, and the frequency of IFN-γ-producing T cells was quantified.
(C) Flow cytometry gating strategy is shown. Representative dotplots of ORF7/8-, M-, and NP-2-specific T cell lines generated from patient P06 are displayed.
(D) Frequencies of IFN-γ-producing CD4 or CD8 T cells of all in vitro-expanded T cell lines generated from the convalescent PBMCs of respective COVID-19 patients (n = 7).
First, we observed that in contrast to the antibody quantity, the overall magnitude of SARS-CoV-2 peptide-reactive cells was not proportional to the severity of disease. Figure 2B shows that although higher quantities of IgG were observed in patients with moderate/severe compared with mild COVID-19, an opposite pattern was detected when the total IFN-γ response detected after stimulation by all peptide pools was calculated. Higher frequencies of IFN-γ-secreting cells in both early stages (day 1–15) and late stages (day 15–30) were present in mild, but not in moderate/severe, COVID-19 patients (Figures 3B and 3C; Figure S1D). In addition, by analyzing the association between the time of SARS-CoV-2 T cell appearance and the length of infection, we observed a statistically significant direct correlation between early appearance of SARS-CoV-2 peptide-reactive cells (specific for NP, ORF7/8, ORF3a, M, and Spike) and shorter duration of infection (Figure 3D). In contrast, no correlation was observed when we analyzed the time of antibody and the length of infection. The temporal association of functional SARS-CoV-2-specific T cell appearance with reduced length of infection suggests that T cells play an essential role in the control of SARS-CoV-2 infection.
Hierarchy of T cell immunogenicity toward different SARS-CoV-2 proteins
Finally, we performed a granular analysis of the ability of different SARS-CoV-2 proteins to stimulate IFN-γ production in severe and mild cases of SARS-CoV-2 infection. Peptide pools covering all structural proteins (M, NP, ORF3a, and Spike) and the ORF7/8 accessory proteins stimulate IFN-γ production from the PBMCs of all the acute patients with the exception of patient P05 (Figure 4A). In line with our previous report, NSP7 and NSP13 pools were rarely able to trigger IFN-γ responses in COVID-19 patients (Le Bert et al., 2020). In most acute patients, we observed the simultaneous presence of IFN-γ-producing cells specific for all peptide pools both during the acute phase of infection and at convalescence (Figure 4A). We observed that the ORF7/8 peptide pool triggered a robust IFN-γ response preferentially in the early phases of infection but only in patients with mild disease (Figures 3B and 4A). When we calculated the proportion of IFN-γ-producing cells triggered by different SARS-CoV-2 proteins at the time of first detection and at convalescence (>30 days after viral clearance), we observed that ORF7/8 responses waned almost completely at convalescence. In contrast, the proportion of cells stimulated by the peptide pools covering other SARS-CoV-2 proteins remained unchanged (M, ORF3a, and Spike) or increased (NP) over time (Figure 4B). To demonstrate unequivocally that IFN-γ-secreting cells detected in our assays after peptide stimulation were indeed T cells, we performed a flow cytometry phenotypic analysis of IFN-γ-producing cells expanded after SARS-CoV-2 peptide stimulation of PBMCs. Unfortunately, such characterizations were performed only with PBMCs of SARS-CoV-2 patients at convalescence because of biosafety regulations that prevented us from analyzing PBMCs collected during active infection (RT-PCR positive) outside a biosafety level 3 (BSL3) laboratory. As expected and already demonstrated, CD3+ T cells produced IFN-γ after peptide pool stimulation (Figures 4C and 4D). Most peptide-responsive T cells were CD4 cells, but CD8 T cells specific to NP peptide pools were also detected (Figure 4D). We were also able to expand ORF7- or ORF8-specific T cells in 6 of 7 patients in whom we detected, in the early phases of infection, a robust population of IFN-γ-producing cells after activation with the combined ORF7/8 peptide pools. Phenotypic analysis showed that these cells were all CD4 T cells. Thus, even though we were unable to directly demonstrate that CD4 T cells were responsible for the IFN-γ response triggered by ORF7/8 pools during the early phases of infection, such interpretation was strongly supported by the exclusive expansion of ORF7- and ORF8-specific CD4 T cells in convalescent patients.
Discussion
Despite the small number of patients analyzed, our longitudinal analysis of the dynamics of virological and virus-specific immunological parameters during the acute phase of SARS-CoV-2 infection revealed a positive relation between early detection of IFN-γ-secreting SARS-CoV-2-specific T cells and early control of infection. In addition, the quantity of functional virus-specific T cells present during the acute phase of infection was not directly proportional to COVID-19 severity but was more robust in patients with mild disease. These data contrasted with the recent analysis of SARS-CoV-2-specific T cells in COVID-19 convalescent patients that reported a positive association of the frequency of SARS-CoV-2-specific T cells with disease severity (Peng et al., 2020). However, in this work, the T cell response was measured in patients who were already in the convalescent phase. Because our longitudinal analysis showed that the frequency of SARS-CoV-2-specific T cells rapidly waned after SARS-CoV-2 clearance, the time of T cell analysis can significantly influence the magnitude of T cell response detected. The most robust functional T cell response was detected in patients with mild symptoms who cleared the virus early, whereas for example, in the patient with severe disease who succumbed to infection, we could only detect a weak and monospecific IFN-γ-secreting cell response at 26 days after symptom onset. These observations are in line with other studies in which paucity of IFN-γ-producing SARS-CoV-2-specific T cells was observed in few patients with severe disease analyzed during the early phase of disease (Weiskopf et al., 2020; Rydyznski Moderbacher et al., 2020; Zhou et al., 2020b). This absence of IFN-γ-producing SARS-CoV-2-specific T cells in the peripheral blood of patients with severe disease could be caused by a defective induction of SARS-CoV-2 T cells. Alternatively, more robust sequestration of the cells in the highly inflamed lung environment (Zhao et al., 2016) or functional impairment of SARS-CoV-2 T cells could occur (Zhou et al., 2020b). The latter possibility is supported by data obtained with different methods (human leukocyte antigen [HLA] multimers or single-cell multiomic analysis) showing impairment of IFN-γ production by SARS-CoV-2-specific T cells in COVID-19 recovered patients (Rha et al., 2020) and the presence of a population of proliferating exhausted T cells during the early phases of severe COVID-19 (Su et al., 2020a).
Additional studies would have to be performed to confirm and/or differentiate these potential causes of the defect of SARS-CoV-2 T cells detected in more severe cases of COVID-19. Ultimately, combining these data with our observations stresses the importance of an early and functional SARS-CoV-2-specific T cell response in reducing infection and disease severity.
The opposite scenario was observed for antibodies. The two most severe COVID-19 patients studied here showed the most rapid and robust ability to achieve peak virus neutralization, and the overall quantities of SARS-CoV-2-specific antibodies were higher in severe than in milder COVID-19 cases. These data confirm the numerous observations that have linked virus-specific antibody production (Hung et al., 2020; Long et al., 2020b; Ripperger et al., 2020; Wang et al., 2020) and B cell hyperactivation (Woodruff et al., 2020) with increased disease severity. Because of the significant link between early induction of T cells and shorter duration of the infection, the demonstration of the early induction of ORF7/8-specific cellular immunity can be of particular significance in viral control. For example, a 382-nucleotide deletion that truncates ORF7b and ORF8, leading to the elimination of ORF8 transcription, had been reported at low frequencies in multiple countries (Su et al., 2020b). Although the mechanism leading to the acquisition of the genomic change is unresolved, the early induction of ORF7/8-specific T cells and a recent report of the robust early antibody response to ORF8 in SARS-CoV-2 infection would suggest that an immune-driven selection process could be involved (Hachim et al., 2020). Given that this viral variant was associated with a milder disease (Young et al., 2020), the early induction of ORF7/8 immunity is worthy of further investigation. It remains difficult to explain why ORF7/8-specific T cells were preferentially detected during the acute phase of infection. Findings show a corresponding increase in ORF8-specific antibodies during the early phases of SARS-CoV-2 infection (Hachim et al., 2020). However, there is no experimental evidence of preferential early expression of ORF7/8 proteins in SARS-CoV-2-infected cells that might contribute to increased immunogenicity of these accessory proteins in the early phases of SARS-CoV-2 infection. An alternative hypothesis is that pre-existing immunological memory to ORF7 or ORF8 might have caused selective accelerated expansion of ORF7/8 T cells, because ORF7/8-specific T cells can be detected occasionally in archived PBMC samples collected from healthy individuals before 2019 (Mateus et al., 2020). However, ORF7/8 is only expressed by SARS-CoV-1 and SARS-CoV-2 with little homology to other seasonal coronaviruses. Hence, the role of such peptide cross-reactive cells is puzzling and calls for a more detailed analysis of the effect of pre-existing immunity in the control or pathogenesis of SARS-CoV-2 infection and of the role of T cells specific for different antigens in SARS-CoV-2 protection.
Limitations of study
There are two main limitations of this study: sample size and use of a single functional assay (ELISPOT for IFN-γ production) for the detection of SARS-CoV-2 T cells during the PCR+ phase of infection.
The association of early SARS-CoV-2-specific T cell detection with accelerated SARS-CoV-2 clearance and milder symptoms is significant in our longitudinal analysis of 12 patients. Nevertheless, larger studies are needed to evaluate the potential influence of age, sex, and ethnicities in the obtained results and thus prove that measurement of IFN-γ-producing SARS-CoV-2-specific T cells in the heterogeneous populations of infected individuals might have prognostic value. Testing SARS-CoV-2 T cells with a method that only measures IFN-γ-producing cells prevents the possible detection of SARS-CoV-2 T cells producing different cytokines (i.e., interleukin [IL]-21, tumor necrosis factor alpha [TNF-α], and IL-17) or functionally exhausted T cells, information that would be important to better understand the mechanisms of COVID-19 severity and SARS-CoV-2 viral clearance. Similarly, the inability to perform a phenotypic analysis of IFN-γ-producing T cells does not clarify whether the preponderance of SARS-CoV-2 CD4 T cell response that we detected at the convalescent phase with flow cytometry is absolute or occurs only after the acute phase of disease. SARS-CoV-2 CD8 T cells are known to be induced in COVID-19 patients (Grifoni et al., 2020; Schulien et al., 2021). It will be important to understand whether the initial IFN-γ-producing cells detected in the early phase of infections are preferentially constituted by CD4 or CD8 T cells.
Despite these limitations, the direct association of early induction of IFN-γ-producing SARS-CoV-2-specific T cells with faster viral clearance and milder disease observed here supports the growing evidence of the protective role of T cells in SARS-CoV-2 infection and provides initial support for the prognostic value of SARS-CoV-2-specific T cell analysis in the management of COVID-19 patients.
STAR★methods
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-human IFN-γ coating antibody | Mabtech | Cat# 3420-3-1000; RRID:AB_907282 |
| anti-human IFN-γ biotin | Mabtech | Cat# 3420-6-1000; RRID:AB_907272 |
| anti-human CD3 PerCp Cy5.5 | BD Biosciences | Cat# 340949; RRID:AB_400190 |
| anti-human CD4 PE-Cy7 | BD Biosciences | Cat# 557852; RRID:AB_396897 |
| anti-human CD8 APC-Cy7 | BD Biosciences | Cat# 557834; RRID:AB_396892 |
| anti-human IFN-γ PE | R&D Systems | Cat# IC285P; RRID:AB_357309 |
| Goat anti-human IgG PE | eBioscience | Cat# 12-4998-82; RRID: AB_465926 |
| Goat anti-human IgM PE | SouthernBiotech | Cat# 2020-09; RRID; AB_2795606 |
| Biological samples | ||
| Peripheral blood mononuclear cells from COVID-19 patients | Recruited as part of the PROTECT and Novel Pathogens studies in Singapore | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Streptavidin-ALP | Mabtech | Cat# 3310-10-1000 |
| KPL BCIP/NBT phosphotase substrate | SeraCare | Cat# 5420-0038 |
| Recombinant human IL-2 | R&D Systems | Cat# 202-1L-050 |
| Yellow LIVE/DEAD fixable dead cell stain | Invitrogen | Cat# L34959 |
| 15-mer SARS-CoV-2 overlapping peptides | GL Biochem (Shanghai) | Amino acid sequences found in Table S2 |
| BD Cytofix/Cytoperm Fixation | BD Biosciences | Cat# 51-2090 KZ |
| HRP-conjugated SARS-CoV-2 RBD | Genscript | Custom synthesis |
| Human ACE2 Fc chimera | Genscript | Cat# Z03484 |
| SARS-CoV-2 RBD protein | Genscript | Cat# Z03479 |
| SARS-CoV-2 S1 protein | Genscript | Cat# Z03485 |
| SARS-CoV-2 N protein | Genscript | Cat# Z03480 |
| Critical commercial assays | ||
| xMap antibody coupling kit | Luminex | Cat# 40-50016 |
| MagPlex microspheres | Luminex | Cat# MC10072-01 |
| Software and algorithms | ||
| GraphPad Prism 7 | GraphPad | https://www.graphpad.com/scientific-software/prism |
| Kaluza software | Beckman Coulter | https://www.beckman.com/flow-cytometry/software/kaluza |
| Immunospot Software | Cellular Technology Limited | http://www.immunospot.com/ImmunoSpot-analyzers-software |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Antonio Bertoletti (antonio@duke-nus.edu.sg).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The published article includes all datasets generated or analyzed during this study. This study did not generate/analyze computer codes or algorithms.
Experimental model and subject details
Patients (n = 12) were enrolled in this study after being admitted to the hospital in Singapore and confirmed to be infected with SARS-CoV-2 based on a positive SARS-CoV-2 RT-PCR test as part of the PROTECT (National Healthcare Group Domain Specific Review Board reference number 2012/00917) and Novel Pathogens (CIRB ref. 2018/3045) studies. All participants provided written informed consent. None of the patients received immunomodulatory treatments during the study period. Six patients were male, six were female, their median age at time of admission was 52.5 years, ranging from 27 to 78 years. Additional patient information are found in Table S1.
Method details
Peripheral blood mononuclear cell isolation
Peripheral blood of acutely infected patients was collected in Mononuclear Cell Preparation tubes (CPT, BD Vacutainer) and transferred at 4°C to the biosafety level-3 (BSL3) facility for same-day processing. Blood from study participants at convalescent time points was obtained and processed in BSL2 laboratories. Peripheral blood mononuclear cells from all collected blood samples were isolated by Ficoll-Paque density gradient centrifugation.
Surrogate virus neutralization assay
sVNT assay to quantify the neutralizing antibody response were performed as previously described (Tan et al., 2020). Briefly, sera from acutely infected patients were prepared in BSL3 containment and heat-inactivated prior to sVNT assay. HRP-conjugated RBD (Genscript) were pre-incubated with 1:20 diluted serum at 37°C for 1h, followed by addition to the Fc-chimeric human ACE2-coated MaxiSORP ELISA plate (Nunc) for an hour at room temperature. Colorimetric signal was developed using TMB substrate (KPL) after extensive PBST washes and the reaction was stopped with 1M HCl. Absorbance reading at 450 nm and 570 nm were obtained using Hidex Sense microplate reader (Hidex).
Luminex analysis
SARS-CoV-2 RBD, S1 and N proteins (Genscript) were conjugated onto MagPlex microsphere (Luminex) using xMAP antibody coupling kit (Luminex). SARS-CoV-2 spike and N proteins specific antibodies were detected by pre-incubation of 100-fold diluted serum (in 1% BSA PBS) with conjugated microspheres (1250 beads/antigen) for 1h at room temperature, followed by 1:1000 diluted PE-conjugated anti-human IgG polyclonal antibody (eBioscience) or PE-conjugated anti-human IgM antibody (SouthernBiotech) for 1h at room temperature. The signal was detected using Luminex MAGPIX instrument.
IFN-γ ELISPOT assay
15-mer peptides that spanned the entire ORF of genes eight SARS-CoV-2 proteins (NP, M, ORF7, ORF8, ORF3, S, NSP7, NSP13) and antibody responses to two SARS-CoV-2 proteins (NP, S) were synthesized (GL Biochem, Shanghai, China) with 10 amino acids overlap and were grouped into pools of approximately 40 peptides (Table S2). CPT tubes were centrifuged at 1500 rcf. for 15 mins and approximately 2 mL of mononuclear cells located on top of the polyester gel were aliquoted and stored at −80°C. ELISPOT plates were prepared with anti-human IFN-γ coating antibody (MabTech) and peptides pools in 50 μL AIM-V medium (GIBCO; Thermo Fisher Scientific) supplemented with 2% AB human serum (GIBCO; Thermo Fisher Scientific) on the day of incubation with PBMCs. Vials containing PBMCs were thawed at RT, 2U Benzonase (SigmaAldrich) to remove remaining nucleic acids was added when fully thawed and incubated for 15 mins before centrifugation at 1000 rcf. for 10 mins. The cell pellet was dissolved in 1 mL of AIM-V medium (GIBCO; Thermo Fisher Scientific) supplemented with 2% AB human serum (GIBCO; Thermo Fisher Scientific) before quantification at undiluted and 1:10 dilution using a Scepter 2.0 cell counter (Millipore). Approximately 2 × 105 PBMCs in 100 μL per well were incubated in the presence of peptides overnight at 37°C and 5% CO2. After 24 hours, inoculum was removed and plates were washed six times with PBS. Biotinylated anti-human IFN-γ antibody (MabTech) at a 1:2000 dilution in PBS/0.5% FCS was incubated at RT for two hours, followed by six wash steps with PBS and incubation of Streptavidin-ALP (MabTech) at a 1:2000 dilution in PBS/0.5% FCS at RT for one hour. After another six PBS washes, 50 μL of KPL BCIP/NBT phosphatase substrate (SeraCare) was added and incubated at RT in the dark for 5-15 mins. The reaction was stopped by washing the plate with water extensively when the chromogenic reaction produced clearly visible spots. Subsequently, the plates were allowed to air-dry and spot forming units (SFU) were analyzed using an Immunospot reader and software (Cellular Technology Limited).
SARS-CoV-2 specific T cell lines
T cell lines were generated as follows: 20% of PBMC were pulsed with 10 μg/ml of the overlapping SARS-CoV-2 peptides (all pools combined) or single peptides for 1 hour at 37°C, subsequently washed, and co-cultured with the remaining cells in AIM-V medium (GIBCO; Thermo Fisher Scientific) supplemented with 2% AB human serum (GIBCO; Thermo Fisher Scientific). T cell lines were cultured for 10 days in the presence of 20 U/ml of recombinant IL-2 (R&D Systems).
Flow cytometry analysis
Expanded T cell lines were stimulated for 5h at 37°C with or without SARS-CoV-2 peptide pools (2 μg/ml) in the presence of 10 μg/ml brefeldin A (Sigma-Aldrich). Cells were stained with the yellow LIVE/DEAD fixable dead cell stain kit (Invitrogen) and anti-human CD3 PerCp Cy5.5 (clone SK7; 3:50), anti-human CD4 PE-Cy7 (clone SK3; 3:50), and anti-human CD8 APC-Cy7 (clone SK1; 3:50) antibodies. Cells were subsequently fixed and permeabilized using the Cytofix/Cytoperm kit (BD Biosciences-PharMingen) and stained with anti-human IFN-γ PE (clone 25723, R&D Systems; 1:25) and analyzed on a BD-LSR II FACS Scan. Data were analyzed by Kaluza (Beckman Coulter). Antibodies were purchased from BD Biosciences-PharMingen unless otherwise stated.
Quantification and statistical analysis
All statistical analysis were performed using GraphPad Prism v7. Where applicable, the statistical tests used and the definition of center were indicated in the figure legends. Statistical significance was defined as having a P value of less than 0.05. In all instances, “n” refers to the number of patients analyzed.
Acknowledgments
We express our gratitude to the study participants and personnel involved in ensuring the safety of the BSL3 laboratory operations. We also thank all clinical and nursing staff who provided care for the patients and staff in the Singapore Infectious Disease Clinical Research Network and Infectious Disease Research and Training Office, National Centre for Infectious Diseases, for coordinating patient recruitment. This study is supported by the Singapore Ministry of Health’s National Medical Research Council under its COVID-19 Research Fund (COVID19RF3-0060).
Author contributions
A.T.T., N.L.B., and A.B. designed the experiments. M.L., C.W.T., Y.Z., and G.J.D.S. performed the experiments in the BSL3 laboratory. W.N.C., C.W.T., and L.-F.W. performed the antibody analysis. K.K., C.Y.L.T., and A.C. performed all other experiments. A.T.T., M.L., N.L.B., and A.B. analyzed and interpreted the data. A.T.T. and A.B. prepared the figures and wrote the paper. B.Y., S.K., J.G.H.L., and D.L. recruited patients and provided all clinical data. A.B. designed and coordinated the study and provided funding.
Declaration of interests
A.B. is a cofounder of and A.T.T. consults for Lion TCR, a biotech company developing T cell receptors for treatment of virus-related diseases and cancers. None of the other authors has any competing interest related to the study.
Published: January 21, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.108728.
Supplemental information
References
- Atyeo C., Fischinger S., Zohar T., Slein M.D., Burke J., Loos C., McCulloch D.J., Newman K.L., Wolf C., Yu J. Distinct Early Serological Signatures Track with SARS-CoV-2 Survival. Immunity. 2020;53:524–532.e4. doi: 10.1016/j.immuni.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson D.F., Norddahl G.L., Melsted P., Gunnarsdottir K., Holm H., Eythorsson E., Arnthorsson A.O., Helgason D., Bjarnadottir K., Ingvarsson R.F. Humoral Immune Response to SARS-CoV-2 in Iceland. N. Engl. J. Med. 2020;383:1724–1734. doi: 10.1056/NEJMoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachim A., Kavian N., Cohen C.A., Chin A.W.H., Chu D.K.W., Mok C.K.P., Tsang O.T.Y., Yeung Y.C., Perera R.A.P.M., Poon L.L.M. ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection. Nat. Immunol. 2020;21:1293–1301. doi: 10.1038/s41590-020-0773-7. [DOI] [PubMed] [Google Scholar]
- Hung I.F.-N., Cheng V.C.-C., Li X., Tam A.R., Hung D.L.-L., Chiu K.H.-Y., Yip C.C.-Y., Cai J.-P., Ho D.T.-Y., Wong S.-C. SARS-CoV-2 shedding and seroconversion among passengers quarantined after disembarking a cruise ship: a case series. Lancet Infect. Dis. 2020;20:1051–1060. doi: 10.1016/S1473-3099(20)30364-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isho B., Abe K.T., Zuo M., Jamal A.J., Rathod B., Wang J.H., Li Z., Chao G., Rojas O.L., Bang Y.M. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci. Immunol. 2020;5:eabe5511. doi: 10.1126/sciimmunol.abe5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- Long Q.-X., Liu B.-Z., Deng H.-J., Wu G.-C., Deng K., Chen Y.-K., Liao P., Qiu J.-F., Lin Y., Cai X.-F. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Long Q.-X., Tang X.-J., Shi Q.-L., Li Q., Deng H.-J., Yuan J., Hu J.-L., Xu W., Zhang Y., Lv F.-J. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., Burger Z.C., Rawlings S.A., Smith D.M., Phillips E. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D., Dejnirattisai W., Rostron T., Supasa P., Liu C., Oxford Immunology Network Covid-19 Response T cell Consortium. ISARIC4C Investigators Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21:1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rha M.-S., Jeong H.W., Ko J.-H., Choi S.J., Seo I.-H., Lee J.S., Sa M., Kim A.R., Joo E.-J., Ahn J.Y. IFN-γ is Produced by Pd-1 + Cells Among SARS-CoV-2-Specific MHC-I Multimer+CD8 + T Cells in Acute and Convalescent COVID-19 Patients. Immunity. 2020 doi: 10.2139/ssrn.3684758. Published online September 15, 2020. [DOI] [Google Scholar]
- Ripperger T.J., Uhrlaub J.L., Watanabe M., Wong R., Castaneda Y., Pizzato H.A., Thompson M.R., Bradshaw C., Weinkauf C.C., Bime C. Orthogonal SARS-CoV-2 Serological Assays Enable Surveillance of Low-Prevalence Communities and Reveal Durable Humoral Immunity. Immunity. 2020;53:925–933.e4. doi: 10.1016/j.immuni.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., Belanger S., Abbott R.K., Kim C., Choi J. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell. 2020;183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulien I., Kemming J., Oberhardt V., Wild K., Seidel L.M., Killmer S., Sagar, Daul F., Salvat Lago M., Decker A. Characterization of pre-existing and induced SARS-CoV-2-specific CD8+ T cells. Nat. Med. 2021;27:78–85. doi: 10.1038/s41591-020-01143-2. [DOI] [PubMed] [Google Scholar]
- Su Y., Chen D., Lausted C., Yuan D., Choi J., Dai C., Voillet V., Scherler K., Troisch P., Duvvuri V.R. Multiomic Immunophenotyping of COVID-19 Patients Reveals Early Infection Trajectories. bioRxiv. 2020 doi: 10.1101/2020.07.27.224063. [DOI] [Google Scholar]
- Su Y.C.F., Anderson D.E., Young B.E., Linster M., Zhu F., Jayakumar J., Zhuang Y., Kalimuddin S., Low J.G.H., Tan C.W. Discovery and Genomic Characterization of a 382-Nucleotide Deletion in ORF7b and ORF8 during the Early Evolution of SARS-CoV-2. MBio. 2020;11:e01610–e01620. doi: 10.1128/mBio.01610-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B., Feng Y., Mo X., Zheng P., Wang Q., Li P., Peng P., Liu X., Chen Z., Huang H. Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg. Microbes Infect. 2020;9:940–948. doi: 10.1080/22221751.2020.1762515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., Ellingson M.K., Wong P., Israelow B., Lucas C., Klein J., Silva J., Mao T., Oh J.E., Tokuyama M., Yale IMPACT Research Team Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature. 2020;588:315–320. doi: 10.1038/s41586-020-2700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C.W., Chia W.N., Qin X., Liu P., Chen M.I.-C., Tiu C., Hu Z., Chen V.C.-W., Young B.E., Sia W.R. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat. Biotechnol. 2020;38:1073–1078. doi: 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang L., Sang L., Ye F., Ruan S., Zhong B., Song T., Alshukairi A.N., Chen R., Zhang Z. Kinetics of viral load and antibody response in relation to COVID-19 severity. J. Clin. Invest. 2020;130:5235–5244. doi: 10.1172/JCI138759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H., van den Akker J.P.C., Molenkamp R., Koopmans M.P.G., van Gorp E.C.M. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020;5:eabd2071. doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff M.C., Ramonell R.P., Nguyen D.C., Cashman K.S., Saini A.S., Haddad N.S., Ley A.M., Kyu S., Howell J.C., Ozturk T. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat. Immunol. 2020;21:1506–1516. doi: 10.1038/s41590-020-00814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young B.E., Fong S.-W., Chan Y.-H., Mak T.M., Ang L.W., Anderson D.E., Lee C.Y.-P., Amrun S.N., Lee B., Goh Y.S. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: an observational cohort study. Lancet. 2020;396:603–611. doi: 10.1016/S0140-6736(20)31757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhao J., Mangalam A.K., Channappanavar R., Fett C., Meyerholz D.K., Agnihothram S., Baric R.S., David C.S., Perlman S. Airway Memory CD4(+) T Cells Mediate Protective Immunity against Emerging Respiratory Coronaviruses. Immunity. 2016;44:1379–1391. doi: 10.1016/j.immuni.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R., To K.K.-W., Wong Y.C., Liu L., Zhou B., Li X., Huang H., Mo Y., Luk T.-Y., Lau T.T.-K. Acute SARS-CoV-2 Infection Impairs Dendritic Cell and T Cell Responses. Immunity. 2020;53:864–877.e5. doi: 10.1016/j.immuni.2020.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets generated or analyzed during this study. This study did not generate/analyze computer codes or algorithms.