Abstract
Spatiobehavioral characteristics are stable for, and hence predictive of, most cases of contagious diseases. They should be acknowledged as a formal way of defining the epidemiology of new contagious diseases at the early stage, enabling health authorities to implement precision control and prevention of the disease at the first moment possible.
Keywords: spatiobehavioral, spatial lifecourse epidemiology, COVID-19, contagious disease, infectious disease
The coronavirus disease 2019 (COVID-19) has visited over 200 countries and most, if not all, of them have not prepared well for it or realized the risk of COVID-19 until it landed on their territories [World Health Organization (2020) WHO Coronavirus Disease (COVID-19) Dashboard, https://covid19.who.int/]. It is essential to understand the epidemiology of an epidemic before we can model it in correct ways and curb it in its early stage. When COVID-19 was prevalent only in China, a call for studies to define its epidemiology and characterize its potential impact was launched before the number of detected cases outside China became unmanageable for public health authorities [1]. It was found vital for jurisdictions outside China to invest and prepare early to perform six types of studies (i.e., syndromic surveillance plus targeted viral testing, household studies, community studies, integration of multiple sources and data types, case–control studies, and viral shedding studies) as case numbers grew, in order to obtain the evidence needed for controlling the epidemic. However, it concluded that none of those studies could be done due to the small number of detected cases at that time [1]. Later, numbers went out of control rapidly; this was a shock, and, within a month, it burdened healthcare systems in over 200 countries. Therefore, those traditional ways of defining the epidemiology of such rapidly spreading contagious diseases (i.e., diseases of contact between bodies), in order to be effective and efficient, should have been conducted within a very short time window when the number of infected cases was transiently between being considerable and being manageable. Thus far, no country has succeeded within that time window in the panic of COVID-19.
Behind the slow or lack of response to the COVID-19 epidemic in most countries is the chicken-and-egg situation that, in traditional epidemiological study designs, the epidemiology of a new disease or epidemic cannot be well known until a large enough number of infected cases appear [1]. Also, traditional ways of defining the epidemiology of new diseases focus on the clinical and individual characteristics of infected cases, which are inherently retrospective and can only be summarized as afterthoughts. In addition, most, if not all, clinical and individual characteristics have varied among infected cases, which gives us uncertainties in prospectively identifying populations at (high) risk for early warning and timely control. If we could start over again, facing a forthcoming epidemic with high contagiousness and all these complexities, how could we be better prepared? Does defining its epidemiology have to occur prior to deciding to take statutory measures for preventing its spread? Can any aspect of the epidemiology of a new contagious disease be defined at its early stage and for all places once it has occurred in one place?
Defining one aspect of the epidemiology of COVID-19 – spatiobehavioral characteristics – has actually been practiced unconsciously or subconsciously during the COVID-19 epidemic. Facing new contagious diseases, spatiobehavioral characteristics may work better than other epidemiological characteristics in defining the epidemiology because they are stable for, and hence predictive of, most (if not all) infected cases; that is to say, most if not all infected cases have had close spatial contact with infectors for a certain amount of time. The spatial contact among human populations at a given moment is driven by human behaviors and can be monitored; hence, the contact at the next moment could be predicted. Thus, the spatiobehavioral characteristics of infected cases could be defined, or even precisely quantified, at the first moment possible on the basis of a small number of cases, and this would hold great potential for prospectively predicting populations at risk at a high spatial resolution [2]. Moreover, spatiobehavioral approaches – with the support of intelligent syndromic surveillance systems and smartphone-based alert systems that monitor the movement of anonymous infectors [3., 4., 5.] – could be used to identify potential asymptomatic infections on the basis of their movement and contact history, so that blanket testing can be better targeted to the populations and areas of greatest need. Spatiobehavioral approaches can also make blanket testing more cost-effective while maintaining the accuracy, for example, in a pooled screening strategy where how to pool samples could be determined based on their spatiobehavioral exposures [6].
Spatial lifecourse epidemiology has provided a family of advanced spatial and digital methods and tools for capturing one’s spatiobehavioral characteristics in high dimension, resolution, and frequency [7,8]. For example, long-distance (e.g., international, domestic) travel can be recorded by Geographic Information Systems; environmental conditions affecting human behaviors can be continually measured by remote sensing, also referred to as earth observation; daily human movements can be captured by Global Positioning Systems embedded in smartphone-based applications or other location-aware services (e.g., wearable devices); public understanding and responses to emergencies and interventions can be revealed by geo-tagged social media or other Internet-derived data, also called infodemiology; place-specific risk with exact date and time can be reported by citizens through mobile crowdsourcing apps, and those who have visited the same place at the same time could be alerted via Bluetooth; machine learning algorithms and spatiotemporally explicit agent-based simulation models could predict the progression of the place-specific risk over time, on the basis of real-time spatial big data and anonymous individual tracking data [9]. Therefore, a dose–response relationship could be established between the totality of spatiobehavioral exposures to infectors and infectious environments and the risk of infection, which could potentially be further developed into a future portable tool that can quantify one’s risk of infection on the basis of spatiobehavioral exposures. A smartphone-based app used at some places during the COVID-19 epidemic could be considered a crude prototype of such tools, which, for example, can show at the checkpoints whether one has traveled to high-risk places in the past 2 weeks in China. Furthermore, such precise quantification of spatiobehavioral exposures also enables speculation on the time of symptom onset, which is earlier than the currently recorded date of reporting (or diagnosis) and more important to predict for curbing the epidemic at the early stage.
Although spatiobehavioral characteristics have been to some extent involved in research on some contagious diseases (e.g., influenza, cholera), they have been used to understand retrospectively, rather than defining prospectively, the epidemiology of those diseases [10,11]. We are now in the digital era, where traditional epidemiological theories are insufficient to fully guide the definition of epidemiology of new diseases in a timely or cost-effective manner; instead, spatial technologies and big data, both historical and real-time, could be used to define, track, or even forecast the incidence, distribution, impact, and possible control strategies of many diseases at an early stage of the epidemic. However, the usefulness of the advanced technologies and big data for disease tracking would be compromised by privacy and other emerging concerns. Some frameworks and protocols with varying levels of privacy protection are also emerging in the context of the COVID-19 pandemic (e.g., Decentralized Privacy-Preserving Proximity Tracing, Pan-European Privacy-Preserving Proximity Tracing), aiming to minimize privacy risks for individuals and communities while notifying people of potential contact with infected patients.
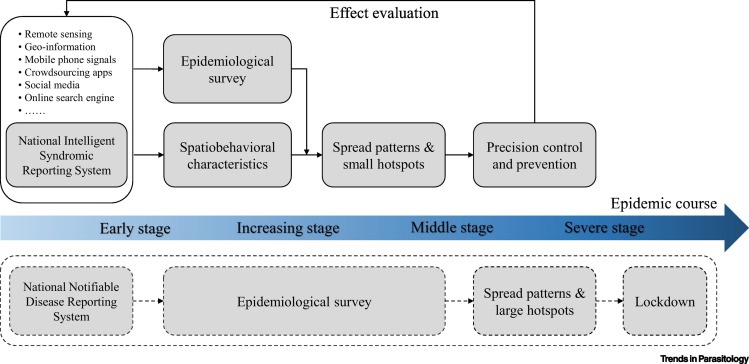
All in all, acknowledging spatiobehavioral characteristics as a formal way of defining the epidemiology of new contagious diseases in the digital era is vital and holds important public health implications. It could make privacy-ensured data-sharing mechanisms and infrastructures better prepared in normal times and ready to use at the early stage of new contagious diseases, which otherwise can work only during the important public health emergencies [12]. Moreover, it could provide valid early-stage evidence, perhaps statutory authority as well in the future, for implementing precise protection (e.g., wearing face masks in public) and prevention measures (e.g., lockdown) (Figure 1 ). Such measures have been demonstrated as the most effective medicine during a pandemic for equally protecting everyone, anytime, anywhere in the world. Therefore, using spatial lifecourse epidemiological theories, methods, and tools to better understand and characterize the spatiobehavioral characteristics of individual cases of new contagious diseases at the early stage would enable us to start prevention strategies compulsorily at the first moment possible for stopping disease transmission, and to outpace the epidemic eventually [2].
Figure 1.
Key Action Points for Implementing Precision Control and Prevention of the Disease over the Epidemic Course.
The traditional procedure is shown in the broken box.
Acknowledgments
Acknowledgements
We thank the International Institute of Spatial Lifecourse Epidemiology (ISLE) for research support.
References
- 1.Lipsitch M., et al. Defining the epidemiology of Covid-19 – studies needed. N. Engl. J. Med. 2020;382:1194–1196. doi: 10.1056/NEJMp2002125. [DOI] [PubMed] [Google Scholar]
- 2.Jia P., Yang S. Are we ready for a new era of high-impact and high-frequency epidemics? Nature. 2020;580:321. doi: 10.1038/d41586-020-01079-0. [DOI] [PubMed] [Google Scholar]
- 3.Budd J., et al. Digital technologies in the public-health response to COVID-19. Nat. Med. 2020;26:1183–1192. doi: 10.1038/s41591-020-1011-4. [DOI] [PubMed] [Google Scholar]
- 4.Jia P., Yang S. Early warning of epidemics: towards a national intelligent syndromic surveillance system (NISSS) in China. BMJ Glob. Health. 2020;5 doi: 10.1136/bmjgh-2020-002925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jia P., Yang S. China needs a national intelligent syndromic surveillance system. Nat. Med. 2020;26:990. doi: 10.1038/s41591-020-0921-5. [DOI] [PubMed] [Google Scholar]
- 6.Hogan C.A., et al. Sample pooling as a strategy to detect community transmission of SARS-CoV-2. JAMA. 2020;323:1967–1969. doi: 10.1001/jama.2020.5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jia P. Understanding the epidemic course in order to improve epidemic forecasting. Geohealth. 2020;4 doi: 10.1029/2020GH000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jia P. Spatial lifecourse epidemiology. Lancet Planet Health. 2019;3:e57–e59. doi: 10.1016/S2542-5196(18)30245-6. [DOI] [PubMed] [Google Scholar]
- 9.Jia P., et al. Spatial lifecourse epidemiology and infectious disease research. Trends Parasitol. 2020;36:235–238. doi: 10.1016/j.pt.2019.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Apolloni A., et al. Age-specific contacts and travel patterns in the spatial spread of 2009 H1N1 influenza pandemic. BMC Infect. Dis. 2013;13:176. doi: 10.1186/1471-2334-13-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bengtsson L., et al. Using mobile phone data to predict the spatial spread of cholera. Sci. Rep. 2015;5:8923. doi: 10.1038/srep08923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jia P., Yang S. Time to spatialise epidemiology in China. Lancet Glob. Health. 2020;8:e764–e765. doi: 10.1016/S2214-109X(20)30120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]