Abstract
Convalescent plasma (CP) has emerged as a treatment for COVID-19. However, the composition and mechanism of action are not fully known. Therefore, we undertook a two-phase controlled study in which, first the immunological and metabolomic status of recovered and severe patients were evaluated. Secondly, the 28-day effect of CP on the immune response in severe patients was assessed. Nineteen recovered COVID-19 patients, 18 hospitalized patients with severe disease, and 16 pre-pandemic controls were included. Patients with severe disease were treated with CP transfusion and standard therapy (i.e., plasma recipients, n = 9) or standard therapy alone (n = 9). Clinical and biological assessments were done on day 0 and during follow-up on days 4, 7, 14, and 28. Clinical parameters, viral load, total immunoglobulin (Ig) G and IgA anti-S1-SARS-CoV-2 antibodies, neutralizing antibodies (NAbs), autoantibodies, cytokines, T and B cells, and metabolomic and lipidomic profiles were examined. Total IgG and IgA anti-S1-SARS-CoV-2 antibodies were key factors for CP selection and correlated with NAbs. In severe COVID-19 patients, mostly interleukin (IL)-6 (P = <0.0001), IL-10 (P = <0.0001), IP-10 (P = <0.0001), fatty acyls and glycerophospholipids were higher than in recovered patients. Latent autoimmunity and anti–IFN–α antibodies were observed in both recovered and severe patients. COVID-19 CP induced an early but transient cytokine profile modification and increases IgG anti-S1-SARS-CoV-2 antibodies. At day 28 post-transfusion, a decrease in activated, effector and effector memory CD4+ (P < 0.05) and activated and effector CD8+ (P < 0.01) T cells and naïve B cells (P = 0.001), and an increase in non-classical memory B cells (P=<0.0001) and central memory CD4+ T cells (P = 0.0252) were observed. Moreover, IL-6/IFN-γ (P = 0.0089) and IL-6/IL-10 (P = 0.0180) ratios decreased in plasma recipients compared to those who received standard therapy alone. These results may have therapeutic implications and justify further post-COVID-19 studies.
Keywords: COVID-19, Convalescent plasma, Memory cells, Cytokines, Autoantibodies, Metabolomic profile
Abbreviations
- ACAs
Anti-cardiolipin antibodies
- β2GPI
Anti-β2glycoprotein antibodies
- BMI
Body mass index
- CCP3
Anti-cyclic citrullinated peptide third-generation antibodies
- CD
Cluster of differentiation
- CE
Cholesteryl
- COVID-19
Coronavirus disease 2019
- CP
Convalescent plasma
- CRP
C reactive protein
- DG
Diacylglycerols
- dsDNA
Anti-double-stranded DNA antibodies
- ED
Estimated difference
- ESI
Electrospray ionization
- ESR
Erythrocyte sedimentation rate
- FA
Fatty Acyls
- FiO2
Fraction of inspired oxygen
- FDA
Food and Drug Administration
- FDR
False discovery rate
- G-CSF
Granulocyte colony-stimulating factor
- GL
Global lipidomics
- GLMMs
Generalized linear mixed models
- GM
Global metabolomics
- GM-CSF
Granulocyte-macrophage colony-stimulating factor
- HILIC
Hydrophilic interaction liquid chromatography
- HODE
Hydroxyoctadecadienoic acid
- ICU
Intensive care unit
- IFN
Interferon
- Ig
Immunoglobulin
- IL
Interleukin
- IP-10
Interferon-γ induced protein 10
- IQR
Interquartile range
- JK
Jack-knife confident interval
- LDH
l-lactate dehydrogenase
- LPA
Lysophosphatidic acid
- LPC
Lysophosphatidylcholine
- LPE
Lysophosphatidylethanolamine
- MCP-1
Monocyte chemotactic protein-1
- NAbs
Neutralizing antibodies
- NSAIDS
Non-steroidal anti-inflammatory drugs
- OPLS-DA
Orthogonal partial least-squares discriminant analysis
- PaO2
Arterial partial pressure of oxygen
- PC
Phosphatidylcholine
- PCA
Principal-component analysis
- PE
Phosphatidylethanolamine
- PRNT
Plaque reduction neutralization test
- PS
Phosphoserine
- RANTES
Regulated on activation, normal T cell expressed and secreted
- RF
Rheumatoid factor
- RP-LC-QTOF-MS
Reverse phase-liquid chromatography-quadrupole time-of-flight mass spectrometry
- RT-PCR
Reverse transcriptase-polymerase chain reaction
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- SD
Standard deviation
- SE
Standard error
- SOFA
Sequential organ failure assessment score
- Tg
Anti-thyroglobulin antibodies
- Th
T helper
- TNF
Tumor necrosis factor
- TPO
Anti-thyroid peroxidase antibodies
- Tregs
Regulatory T cells
- TXB2
Thromboxane B2
- VIP
Variance important in projection
- WHO
World Health Organization
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causal agent of coronavirus disease 2019 (COVID-19), has infected over 98 million people around the world (https://coronavirus.jhu.edu/map.html) becoming a serious public health threat. Although most people with COVID-19 are asymptomatic or have mild disease, about 20% may present with severe illness [1,2].
Preventive and treatment options for COVID-19 remain under study. Therefore, traditional interventions have re-emerged as a new therapeutic alternative. CP has been used for decades for the prevention or treatment of infectious diseases when no specific treatment was available [3]. CP can provide passive immunity by transfusion of neutralizing antibodies (NAbs) and other immune components from patients who have recovered from an infection, to unexposed or infected individuals in order to reduce the risk of disease or to lessen its clinical impact [4]. This strategy has been used for the treatment of several viral infections, including Spanish influenza, parvovirus B19, H1N1, Ebola and other coronaviruses. However, conclusive data from randomized and controlled trials are lacking [[5], [6], [7], [8]].
Passive immunization with plasma from recovered COVID-19 patients to SARS-CoV-2 infected patients could help to control the infection before patients have generated a proper immune response [4]. The Food and Drug Administration (FDA) authorized the use of CP in COVID-19 patients as an investigational new drug for the treatment of this disease [9]. Up to date, several studies have published some promising benefits [[10], [11], [12], [13]]. The early administration of CP containing high IgG antibody titers against SARS-CoV-2 may not only decrease disease severity, but may also decrease mortality [14]. However, due to a high methodological variability in selection criteria for donors and recipients, dosage, NAbs concentration, disease severity, outcomes, among others, it is difficult to objectively assess the real therapeutic potential of CP [15].
COVID-19 is characterized by an overzealous activation of the innate and adaptive immune system leading to hyper-inflammation that are primarily a result of high levels of pro-inflammatory cytokines and cell dysregulation [16]. In this regard, transfusion of CP to infected patients may provide antibodies, cytokines, and other active factors that may modulate the immune response against SARS-CoV-2, thus attenuating the severe inflammatory response [3]. The main mechanism of action of CP involves, among others, the presence of NAbs against SARS-CoV-2 protein S, which are correlated with total IgG and IgA antibody levels [17]. Since its entire mechanisms of action are not fully understood, we aimed to evaluate the composition of the CP in terms of SARS-CoV-2 antibodies, autoantibodies, cytokines and metabolites, as well as its effect on the immune response in patients with severe COVID-19, up to 28 days after treatment.
2. Methods
This was a two-phase controlled study in which, first CP composition and the basal immunological and metabolomic status of recovered and severe patients were evaluated. Secondly, its longitudinal effect at 28-day follow-up period on the immune response in severe patients was determined (Fig. 1 A) (see section 2.7).
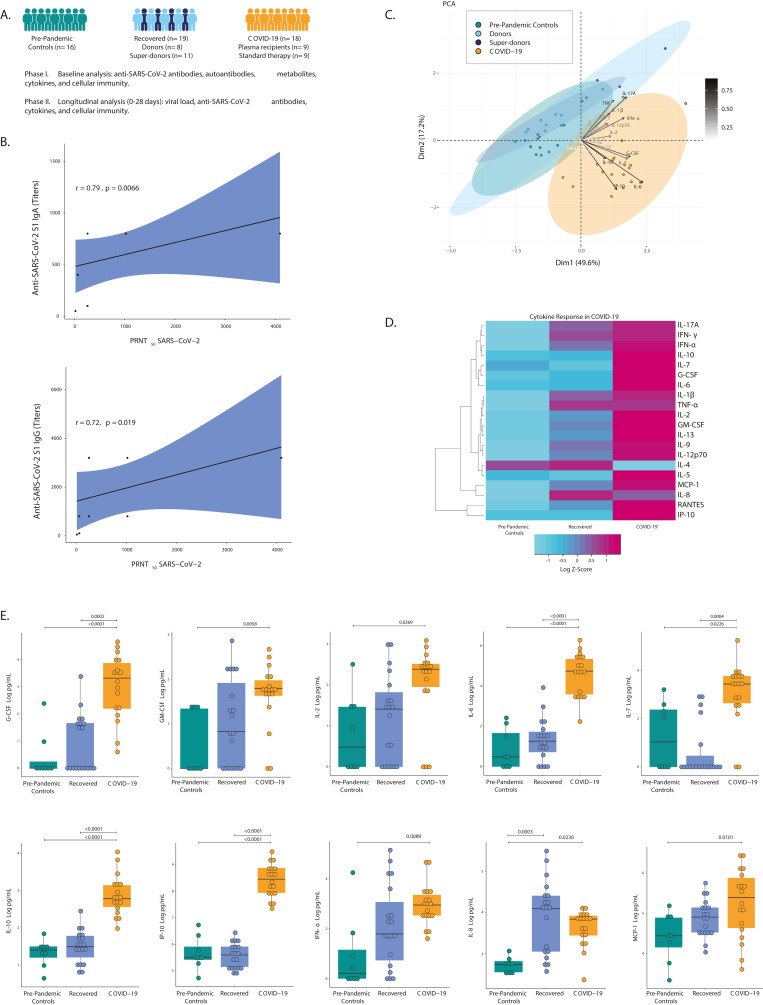
Fig. 1.
Immunological features of recovered and COVID-19 patients. A, Study summary flow diagram. Clinical analyses were done (see text for details, section 2.7). B, Strong Spearman correlation of IgA and IgG anti-SARS-CoV-2 titers by ELISA and neutralizing antibodies by PRNT50 in recovered patients. C, Analysis of cytokines by PCA shows that pre-pandemic controls, donors, and super-donors aggregate, whereas COVID-19 patient data segregates as an independent group. Color shows the data grouping. D, Heatmap of cytokine analysis among pre-pandemic controls, the recovered group, and COVID-19 patients. The color of the heatmap varies from blue, which indicates under-expression, to purple, which indicates over-expression. Row clustering was performed using the ward agglomeration method. The log-transformed cytokine concentration was used to construct the heatmap. The heatmap reflects the z scores of cytokines across groups, and it does not represent statistical significance. E, Representative box plots of cytokines that were plotted as log-transformed concentrations. Statistical analysis was performed using generalized linear mixed models that were adjusted for group, age, and sex. Abbreviations: G-CSF: Granulocyte colony-stimulating factor; GM -CSF: Granulocyte-macrophage colony stimulating factor; IFN: Interferon; Ig: Immunoglobulin; IL: Interleukin; IP-10: interferon-γ induced protein 10; MCP-1: Monocyte chemotactic protein-1; RANTES: Regulated on activation, normal T cell expressed and secreted; PCA, principal-component analysis; PRNT, plaque reduction neutralization test; TNF-α: Tumor necrosis factor-alpha.
2.1. Participants
The institutional review board at the Universidad del Rosario approved the study design. Written informed consent was obtained from all study participants. Participants were recruited from the following three medical centers: (1) Clínica del Occidente; (2) Clínica CES; and (3) Hospital Universitario Mayor Méderi in Colombia. Participants were recruited from July 1, 2020 to August 25, 2020. Follow-up was completed on September 22, 2020. Patients were selected using non-probabilistic sampling (i.e., convenience selection). Plasma recipients were selected from the CP pilot study (NCT04332380), whereas patients who received exclusively standard therapy were selected from a randomized controlled trial (NCT04332835). The latter group comprised the first nine consecutive controls who were included in that study. Consecutive inclusion of plasma donors was also performed.
Participants were divided into three groups, as follows: pre-pandemic control group (n = 16); patients who recovered from SARS-CoV-2 infection (n = 19), which included super-donors (n = 11) and donors (n = 8) on the basis of their antibody titers (see below); and patients who were hospitalized with severe COVID-19 (n = 18), whom after day 0 were split into two groups: plasma recipient (n = 9) and standard therapy (n = 9).
2.2. Procurement of convalescent plasma
Pre-donation process included the following steps/criteria: (1) signed informed consent; (2) aged between 18 and 65 years; (3) subjects with a laboratory-confirmed COVID-19 diagnosis by reverse transcriptase-polymerase chain reaction (RT-PCR) having been hospitalized but not at the intensive care unit (ICU), discharged and recovered between 14 and 30 days before the pre-donation assessment; (4) two consecutive negative RT-PCR results from nasopharyngeal swabs within 48 h before donation; (5) women were only accepted if they did not have pregnancy history or a current suspicion of pregnancy; (6) negativity for HIV, hepatitis B and C virus, HTLV 1 and 2, syphilis, and Trypanosoma cruzi infection. Potential donors were screened for IgG and IgA antibodies, and classified as super-donors and donors according to antibody levels. Subjects with IgG antibody titers ≥1:3200 and IgA antibody titers ≥1:800 to SARS-CoV-2 were considered super-donors and were chosen for plasmapheresis and further therapeutic transfusion. Subjects who did not reach those titers were considered as donors, and were discard for plasmapheresis, but its serum composition was analyzed in this study. Approximately, 800 mL of plasma were collected from super-donors. Prior freezing, pathogens inactivation with Riboflavin followed by UV light exposure was performed [18].
2.3. Inclusion criteria for COVID-19 patients
Inclusion criteria were the following: (1) signed informed consent; (2) aged at least 18 years; (3) COVID-19 diagnosis based on RT-PCR testing; (4) hospitalized patients; (5) Sequential Organ Failure Assessment score (SOFA) < 6; (6) severe cases according to Pneumonia Diagnosis and Treatment Scheme for Novel Coronavirus Infection (Trial Version 7). Severe COVID-19 was defined as respiratory distress (i.e., ≥30 breaths/min. in resting state, oxygen saturation of 90% or less on room air; or arterial partial pressure of oxygen (PaO2)/fraction of inspired oxygen (FiO2) of 300 or less). Subjects with life-threatening COVID-19 were not included.
2.4. Exclusion criteria for COVID-19 patients
Exclusion criteria included the following: (1) pregnancy or breast feeding; (2) patients with prior allergic reactions to transfusions; (3) critically ill patients in ICU; (4) patients with surgical procedures in the last 30 days; (5) subjects with active treatment for cancer (i.e., radiotherapy or chemotherapy); (6) diagnosis of HIV in subjects with viral failure (i.e., detectable viral load > 1000 copies/ml), two consecutive viral load measurements within a 3-month interval; (7) subjects with other confirmed infection that explains clinical manifestations; (8) end-stage kidney disease (i.e., glomerular filtration rate <15 ml/min/1.73 m2); (9) Child Pugh C stage liver cirrhosis; (10) high cardiac output diseases; (11) autoimmune diseases or immunoglobulin A nephropathy; (12) and subjects not willing to participate.
2.5. Convalescent plasma transfusion
Each transfusion dose of CP was 250 mL, patients received two doses for a total of 500 mL within 48 h after study inclusion. Each CP unit was kept separate from other super-donors’ units. The transfused CP ABO type was compatible with the recipient's ABO type in 8 out of 10 transfused patients. Each recipient received CP units from the same super-donor. CP transfusion was administered at 3 mL/min with close monitoring for the first 30 min, and regular monitoring over the following 6 h.
2.6. Standard therapy
Standard treatment consisted of symptomatic control and supportive care for COVID-19. This treatment was based upon recommendations from the Colombian Association of Infectology and institutional protocols, which included management with antibiotics, corticosteroids, oxygen, and anticoagulants [19]. Both plasma recipient and standard therapy groups received this treatment.
2.7. Patient monitoring and evaluation
Sociodemographic and pathological factors were evaluated on day 0. The biological baseline included cytokines, lymphocyte populations, IgA and IgG antibodies for SARS-CoV-2, viral load, blood gases, laboratory surrogate of possible thrombotic process (i.e., D-Dimer), hematological, inflammatory, hepatic and renal parameters. These measurements were repeated on days 4, 7, 14 and 28. In addition, the SOFA scale and the 4C mortality score (i.e., score for prediction of mortality 28 days after hospitalization) were evaluated on admission [20]. Clinical and paraclinical parameters were obtained using a standardized form. The former comprised all the variables that were included in the global COVID-19 clinical platform from the World Health Organization (WHO).
2.8. Biological parameters
2.8.1. Viral load
The viral load was measured using the Ampliphi ® RT-qPCR SARS-CoV-2 Viral Load Kit (www.ampliphi.co).
2.8.2. Antibody detection against SARS-CoV-2
The Euroimmun anti-SARS-CoV-2 ELISA (Euroimmun, Luebeck, Germany) was used for serological detection of human IgG and IgA antibodies against the SARS-CoV-2 S1 structural protein, in accordance with the manufacturer's instructions. The ratio interpretation was <0.8 = negative, ≥0.8 to <1.1 = borderline, ≥ 1.1 = positive [17,21,22]. Antibody titration was performed using serial dilutions of serum samples from 1:100 to 1:1,638,400.
2.8.3. Autoantibodies
Detection of IgM rheumatoid factor (RF), IgG anti-cyclic citrullinated peptide third-generation (CCP3), IgM and IgG anti-cardiolipin antibodies (ACAs), IgM and IgG anti-β2glycoprotein (β2GPI) antibodies, IgG anti-double-stranded DNA (dsDNA) antibodies, IgG anti-thyroglobulin (Tg) antibodies and anti-thyroid peroxidase (TPO) antibodies were quantified by ELISA as previously reported [23]. In addition, anti-nuclear antibodies were detected by IMTEC-ANA-LIA Maxx (Human Diagnostics Magdeburg, Germany) [24]. Assessment of anti–IFN–α antibodies was done by ELISA (Thermo Fisher Scientific, Waltham, MA) following manufacturer's specifications.
2.8.4. Plaque reduction neutralization test
A plaque reduction neutralization test (PRNT50) for SARS-CoV-2 was performed using Vero cells [17], with serum samples diluted from 1:16 to 1:4096.
2.8.5. Cytokine assay
Concentration of 20 human cytokines IL-2, IL-1β, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12p70, IL-13, IL-17A, granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), regulated on activation, normal T cell expressed and secreted (RANTES), monocyte chemotactic protein-1 (MCP-1), IFN-α, IFN-γ, tumor necrosis factor (TNF)-α, and interferon-γ induced protein 10 (IP-10) in serum samples from patients were assessed using the Cytometric Bead Array (BD Biosciences™, Franklin Lakes, New Jersey, U.S.) in a FACSCanto II™ flow cytometer (BD Biosciences™) [25].
2.8.6. Untargeted metabolomic and lipidomic analysis
To increase the metabolite coverage, serum samples were analyzed using an untargeted multiplatform metabolomic and lipidomic approach. The metabolomic analysis was performed using reverse phase-liquid chromatography-quadrupole time-of-flight mass spectrometry (RP-LC-QTOF-MS) with positive and negative modes electrospray ionization (ESI) and hydrophilic interaction liquid chromatography (HILIC)-LC-QTOF-MS with negative-ion mode ESI [[26], [27], [28]]. The significant features were selected by keeping only the variables that fulfilled: 1) UVA (p-value with a Benjamin-Hochberg False Discovery Rate (FDR) post hoc correction < 0.05); 2) multivariate analysis criteria (variance important in projection (VIP) > 2 with a Jack-knife confidence interval (JK) that did not include the zero value from orthogonal partial least-squares discriminant analysis (OPLS-DA) with CV-ANOVA < 0.05); and 3) percent change >50% (Supplementary methods).
2.8.7. Lymphocytes immunophenotype
For a detailed analysis of the cell phenotype, peripheral blood mononuclear cells were stained with fluorescent antibodies. A minimum of 100,000 lymphocytes per sample were acquired on a FACSCanto II™ flow cytometer (BD Biosciences™). Thirty cell subsets (Supplementary Table 1) were analyzed with FlowJo software version 9 (BD Biosciences™).
2.9. Statistical analyses
Briefly, in the univariate analysis, categorical variables were analyzed using frequencies, and quantitative continuous variables were expressed as the mean and standard deviation (SD) or the median and interquartile range (IQR). The Mann–Whitney U test, the chi-squared or the Fisher exact tests were used based on the results. Correlations among variables of interest were performed using the Spearman correlation test, and when necessary, corrected for multiple inference using Holm's method. Generalized linear mixed models (GLMMs) that were adjusted for age and sex were used to evaluate intergroup differences between pre-pandemic controls, donors, super-donors, and COVID-19 patients. Additionally, GLMMs were used to evaluate longitudinal changes in antibody ratios, cytokine delta, and lymphocyte population delta. A P value of <0.05 was set as significant. Analyses were performed using SAS Studio© (SAS® University Edition, Cary, North Carolina, U.S.) and R version 4.0.1.
3. Results
3.1. Composition of convalescent plasma
To establish the differential biological composition of CP from recovered COVID-19 patients, we compared the plasma of the following groups: samples collected prior to 2020 pandemic from healthy people (pre-pandemic controls n = 16); a second group consisted in hospitalized patients with severe COVID-19 who had confirmed SARS-CoV-2 infection (COVID-19 n = 18), and a third group consisted in 19 recovered patients of SARS-CoV-2 infection (super-donors n = 11 and donors n = 8) (Fig. 1A).
The mean age in the super-donor and donor groups was 42.18 years (SD, 6.71) and 39.63 years (SD, 12.08) respectively (Mann–Whitney U test, P = 0.3012). There were ten (90.9%) and seven (87.5%) men in the super-donor and donor groups, respectively (Chi-square, P = 0.8111). PRNT50 from both groups strongly correlated with total anti-SARS-CoV-2 S1 IgG (r = 0.72; P = 0.0185) and IgA titers (r = 0.79; P = 0.0066) (Fig. 1B). All super-donors had a PRNT50 ≥1:256. This guaranteed that super-donors exhibited higher neutralization capacity than donors which justified its therapeutic use.
Given the recent evidence of the autoimmune response in COVID-19, we evaluated whether recovered (i.e., donors and super-donors), hospitalized patients and pre-pandemic controls exhibited positivity for autoantibodies. This analysis revealed that recovered patients did not show an increased prevalence of autoimmunity as compared with pre-pandemic controls or hospitalized COVID-19 patients. However, some of them showed positivity for TPO antibodies, RF, anti–IFN–α, and ACAs. Antibodies against IFN-α were observed in 26.3% of recovered patients (Table 1 ). In patients transfused with CP containing autoantibodies including anti–IFN–α neither adverse events nor mortality was observed. On the other hand, two recipients positive for anti–IFN–α antibodies prior to transfusion deceased.
Table 1.
Autoantibodies in patients with COVID-19.
| Autoantibodies | Pre-Pandemic Controls (n: 8a) | Recovered (n: 19) | COVID-19 (n: 18) | P valueb |
|---|---|---|---|---|
| TPO | 1 (12.5%) | 9 (47.4%) | 6 (33.3%) | 0.2558 |
| RF | 2 (25.0%) | 7 (36.8%) | 4 (22.2%) | 0.6879 |
| IFN-α | 0 (0.0%) | 5 (26.3%) | 3 (16.7%) | 0.3494 |
| ACA IgM | 0 (0.0%) | 5 (26.3%) | 2 (11.1%) | 0.3082 |
| β2GPI IgM | 1 (12.5%) | 2 (10.5%) | 1 (5.6%) | 0.8244 |
| TG | 0 (0.0%) | 2 (10.5%) | 0 (0.0%) | 0.6545 |
| La | 0 (0.0%) | 2 (10.5%) | 0 (0.0%) | 0.6545 |
| dsDNA | 0 (0.0%) | 1 (5.3%) | 0 (0.0%) | 1.0000 |
| ACA IgG | 0 (0.0%) | 1 (5.3%) | 0 (0.0%) | 1.0000 |
| CENP-B | 0 (0.0%) | 0 (0.0%) | 1 (5.6%) | 0.5778 |
| β2GPI IgG | 1 (12.5%) | 0 (0.0%) | 0 (0.0%) | 0.1778 |
| CCP3 | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | – |
| Ro60 | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | – |
| Ro52 | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | – |
| Nucleosomes | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | – |
| Histones | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | – |
| SmD1 | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | – |
| PCNA | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | – |
| P0 | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | – |
| Scl70 | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | – |
| U1-snRNP | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | – |
| AMA-M2 | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | – |
| Jo1 | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | – |
| PM-Scl | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | – |
| Mi-2 | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | – |
| Ku | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | – |
When all the patients were taken together as a group (i.e., recovered and COVID-19) significant differences between this last group and pre-pandemic controls were not found.
Abbreviations: ACAs: anti-cardiolipin antibodies; β2GPI: anti-β2glycoprotein antibodies; CCP3: anti-cyclic citrullinated peptide third-generation; dsDNA: anti-double-stranded DNA antibodies; IFN: Interferon; Ig: Immunoglobulin; RF: Rheumatoid factor; RNP: ribonucleoprotein; Sm: Smith; Tg: anti-thyroglobulin.
Only 8 samples were available for autoantibody evaluation.
P value by Fisher exact test.
Next, a panel of 20 cytokines was evaluated. This is of interest since transfusion of pro-inflammatory or anti-inflammatory mediators could influence the recipient response. Cytokines were gathered using principal-component analysis (PCA) (Fig. 1C). This analysis revealed, that the only difference between donors and super-donors relied on anti-SARS-CoV-2 S1 levels. Thus, since both donors and super-donors disclosed a similar cytokine profile, it was appropriate to combine the data of these two groups as a unique recovered group for subsequent analyses.
The heatmap analysis of cytokines suggest a different profile among groups (Fig. 1D). G-CSF (ED = 1.7871; P = 0.0002), IL-6 (ED = 3.2240; P = <0.0001), IL-7 (ED = 1.9736; P = 0.0004), IL-10 (ED = 1.3119; P=<0.0001), and IP-10 (ED = 2.9264; P=<0.0001) levels were higher in COVID-19 than in recovered patients (Fig. 1E). Although IL-4, IL-8, TNF-α and IFN-γ z-score was slightly increased in recovered patients (Fig. 1D), no differences were found at the statistical level.
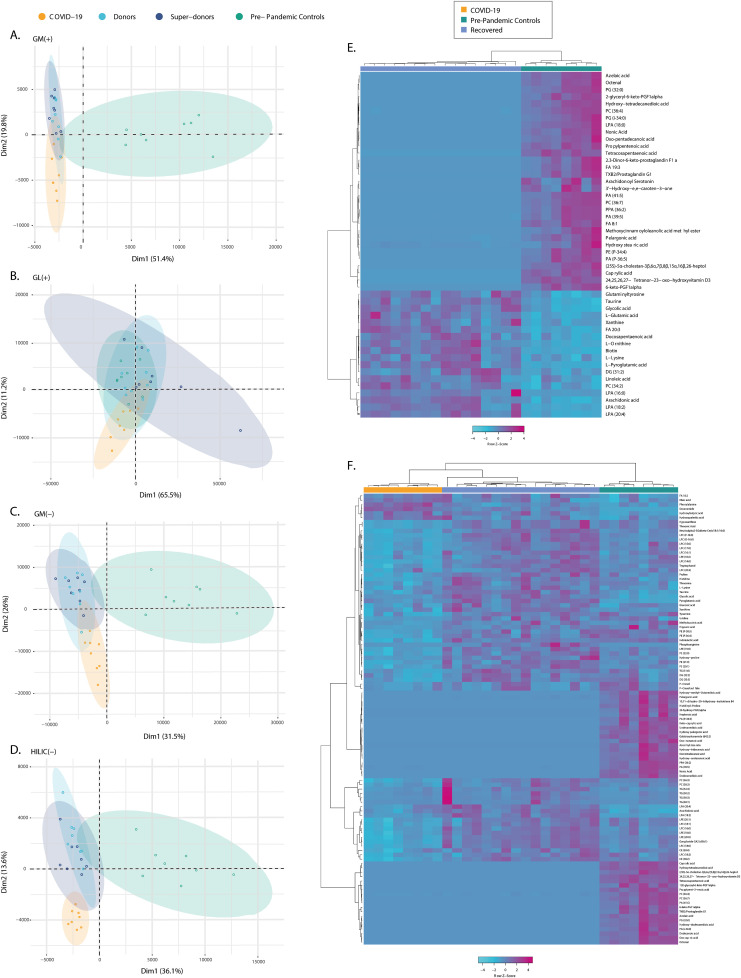
Non-targeted metabolomic and lipidomic profile was evaluated to assess additional components of CP. The total coverage of molecular features from the global metabolomics (GM) and lipidomics (GL) after data processing and filtering comprised 845, 651, 452, and 542 features by GM-RP-LC-QTOF-MS(+), GM-RP-LC-QTOF-MS(−), GM-HILIC-LC-QTOF-MS(−), and GL-LC-QTOF-MS(+), respectively. PCA and OPLS-DA models showed that there was no separation between donors and super-donors on all platforms (Fig. 2 A–D). This confirmed that donors and super-donors were only distinguished based on anti-SARS-CoV-2 S1 levels and therefore were gathered again as a single recovered group for all subsequent OPLS-DA models.
Fig. 2.
PCA score plots for global lipidomics (GL) and metabolomics (GM) and heatmaps of significant metabolites. A, GL by RP-LC-QTOF-MS (+): R2 (cum): 0.92, Q2 (cum): 0.798. B, GM by LC-QTOF-MS (+): R2 (cum): 0.832, Q2 (cum): 0.72. C, GM by LC-QTOF-MS (−): R2 (cum): 0.697, Q2 (cum): 0.475. D, GM by HILIC-LC-QTOF-MS (−): R2 (cum): 0.704, Q2 (cum): 0.444. Color shows the data grouping. COVID-19 (n = 8); donors (n = 8); super-donors (n = 8); and pre-pandemic controls (n = 8). E, Heatmap of recovered vs. pre-pandemic controls comparison. F, Heatmap of COVID-19 vs. recovered and pre-pandemic controls comparison. The significant features were selected by: 1) UVA (p-value with a Benjamin–Hochberg false discovery rate (FDR) post hoc correction < 0.05); and 2) multivariate analysis (MVA) criteria VIP>2 with JK not including the zero value from orthogonal partial least-squares discriminant analysis (OPLS-DA) with CV-ANOVA < 0.05); and 3) Percent change >50%. Abbreviations: CE: Cholesteryl; DG: Diacylglycerols; FA: Fatty acyls; GM: Global metabolomics; GL: Global lipidomics; HILIC: Hydrophilic interaction liquid chromatography; LPA: Lysophosphatidic acid; LPC: Lysophosphatidylcholine; LPE: Lysophosphatidylethanolamine; PC: Phosphatidylcholine; PE: Phosphatidylethanolamine; PS: Phosphoserine; TXB2: Thromboxane B2. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
For recovered and pre-pandemic controls, 50 metabolites were found to be statistically significant (Supplementary Table 2). The heatmaps showed that the proportion of most statistically significant metabolite levels were lower in recovered patients, which corresponds to fatty acyls and glycerophospholipids (Fig. 2E). When the COVID-19 group was compared with the pre-pandemic control and recovered groups (Supplementary Table 3), the largest class of differential metabolites corresponded to a dysregulation in fatty acids, glycerophospholipids, and glycerolipids, and in a lower proportion of sterol lipids and sphingolipids (Fig. 2F). However, the phenotype of recovered patients did not return to a similar phenotype of pre-pandemic controls, and it was characterized by continued altered levels of unsaturated fatty acids, such as arachidonic and linoleic acid (Fig. 2).
3.2. Clinical and immunological features of COVID-19 patients
3.2.1. Clinical characteristics
After the evaluation of CP composition, we aimed to evaluate the immune response in hospitalized patients with COVID-19. Inter-group analysis revealed that plasma recipients and standard therapy patients were similar in age, sex, WHO scale, SOFA score, 4C score, PaO2-FiO2, and viral load on inclusion. In addition, comorbidities and body mass index were similar between groups. Patients who underwent plasma transfusion received CP within the first 72 h after hospitalization. None of the patients presented adverse reactions nor acute exacerbation of the disease after CP transfusion (Table 2 ).
Table 2.
General characteristics of patients with COVID-19.
| Variable | Standard therapy (n = 9) | Plasma recipients (n = 9) | P value |
|---|---|---|---|
| Sex (%) | 0.3428 | ||
| Female | 5 (55.6%) | 3 (33.3%) | |
| Male | 4 (44.4%) | 6 (66.7%) | |
| Age (Mean - SD) | 53.667 (6.708) | 47.889 (9.688) | 0.0768 |
| BMI (Mean - SD) | 29.284 (3.926) | 31.351 (4.841) | 0.4015 |
| WHO scale (%) | 1.0000 | ||
| 4 points | 7 (77.8%) | 7 (77.8%) | |
| 5 points | 2 (22.2%) | 2 (22.2%) | |
| Therapy (%) | |||
| Antimalarials | 0 (0%) | 0 (0%) | 1.0000 |
| Corticosteroids | 8 (88.9%) | 4 (44.4%) | 0.0455 |
| Antibiotics | 7 (77.8%) | 9 (100.0%) | 0.1336 |
| NSAIDs | 0 (0%) | 0 (0%) | 1.0000 |
| Heparin | 6 (66.7%) | 9 (100.0%) | 0.0578 |
| Log viral load (Mean - SE) | 11.70 (2.05) | 13.75 (1.94) | 0.9993 |
| 4C Mortality score (Mean - SD) | 6.889 (2.028) | 5.667 (2.500) | 0.2647 |
| SOFA on inclusion (Median - IQR) | 2 (2–2) | 2 (2–2) | 0.6535 |
| PaO2-FiO2 on inclusion (Median - IQR) | 199.55 (128.375–258.35) | 289.6 (197.9–293.3) | 0.3862 |
| Time from symptoms onset to inclusion (Days - Mean - SD) | 7.000 (2.398) | 7.667 (2.693) | 0.4939 |
| Time from symptoms onset to plasma transfusion (Days - Mean - SD) | – | 8.667 (2.693) | – |
| Time from hospital admission to plasma transfusion (Days - Mean - SD) | – | 2.333 (0.707) | – |
| Total days of hospitalization (Days - Mean - SD) | 17.222 (10.244) | 9.333 (3.937) | 0.1953 |
| ICU admission (%) | 4 (44.4%) | 2 (22.2%) | 0.3173 |
| ICU total days of management (Mean - SD) | 12.250 (3.594) | 6.500 (7.778) | 0.3545 |
| Requirement of mechanical ventilation (%) | 3 (33.3%) | 2 (22.2%) | 0.5987 |
| Days on mechanical ventilation (Mean - SD) | 11.667 (6.110) | 6.500 (7.778) | 0.2482 |
| Mortality (%) | 1 (11.1%) | 2 (22.2%) | 0.5271 |
| Comorbidities (%) | |||
| Hypertension | 1 (11.1%) | 1 (11.1%) | 1.0000 |
| Dyslipidemia | 5 (55.6%) | 1 (11.1%) | 0.0455 |
| Asthma | 0 (0.0%) | 1 (11.1%) | 0.3035 |
| CKD | 1 (11.1%) | 0 (0.0%) | 0.3035 |
| Acid peptic disease | 5 (55.6%) | 1 (11.1%) | 0.0455 |
| Diabetes | 3 (33.3%) | 0 (0.0%) | 0.0578 |
| Current smoker | 0 (0.0%) | 1 (11.1%) | 0.3035 |
| Former smoker | 4 (44.4%) | 2 (22.2%) | 0.3173 |
| Admission laboratories (Median - IQR) | |||
| Platelets | 225,000 (180,000–266,000) | 207,000 (186,000–228,000) | 0.6911 |
| Leucocytes | 9480 (8110–13,950) | 8830 (5990–9110) | 0.2004 |
| Lymphocytes | 790 (580–1100) | 800 (680 -1370) | 0.5078 |
| Neutrophils | 7670 (7230–12,830) | 7110 (4640–7820) | 0.1451 |
| C reactive protein (mg/L) | 123.100 (78.21–166.69) | 229.150 (83.54–255.04) | 0.7573 |
| Erythrocyte sedimentation rate (mm/hr) | 25.000 (8–39) | 28.500 (20–38.25) | 0.5309 |
| Albumin (g/dL) | 3.500 (3.32–3.9) | 3.505 (3.482–3.598) | 1.0000 |
| Total bilirubin (μmol/L) | 0.470 (0.36–0.62) | 0.550 (0.43–0.73) | 0.5074 |
| Urea (mg/dL) | 14.2 (11–17.8) | 13.7 (11.7–14.4) | 0.5363 |
| Creatinine (mg/dL) | 0.7 (0.63–0.89) | 0.86 (0.75–0.98) | 0.3538 |
| Creatine kinase (U/L) | 133 (80–656) | 78 (64–105) | 0.1331 |
| D dimer (mg/L) | 1.490 (1.13–1.94) | 0.420 (0.33–0.68) | 0.0170 |
| Ferritin (ng/mL) | 1199 (634.8–1921) | 1193 (798.6–1548) | 0.9233 |
| Lactate dehydrogenase (U/L) | 360 (308–490) | 337 (305–367) | 0.5660 |
| Procalcitonin (ng/mL) | 0.219 (0.086–0.33) | 0.112 (0.073–0.226) | 0.4414 |
| Troponin T (ng/mL) | 0.006 (0.006–0.008) | 0.004 (0.003–0.005) | 0.0308 |
Abbreviations: BMI: Body mass index; ICU: Intensive care unit; NSAIDs: Non-steroidal anti-inflammatory drugs; SD: Standard deviation; SE: Standard error; SOFA: Sequential organ failure assessment; WHO: World health organization.
3.2.2. Immune response in COVID-19
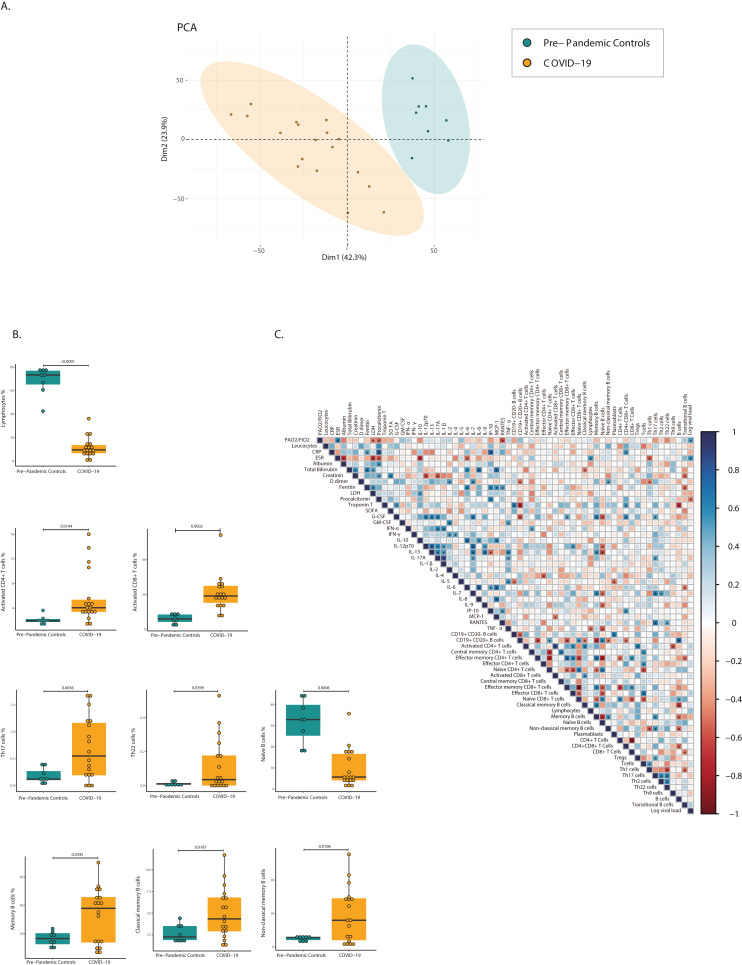
To characterize the T- and B-cell signature in COVID-19 patients’ prior transfusion, a cellular analysis was performed before plasma or standard therapy treatment. PCA of 30 cell subsets clearly distinguished between COVID-19 patients and pre-pandemic controls (Fig. 3 A). Marked lymphopenia in COVID-19 patients was accompanied by higher activated CD4+ (ED = 1.1861; P = 0.0144) and CD8+ (ED = 1.6467; P = 0.0002) T cell levels. Within CD4+ Th cells, Th17 (ED = 1.5658; P = 0.0056) and Th22 (ED = 2.6618; P = 0.0399) subsets were higher in COVID-19 patients. For B cells, memory (ED = 1.0325; P = 0.0345), classical memory (ED = 0.8466; P = 0.0187), and non-classical memory B (ED = 1.6689; P = 0.0108) cell levels were higher in COVID-19 patients. Additionally, naïve B cell levels were lower in COVID-19 compared to pre-pandemic controls (ED = −1.7522; P = 0.0006) (Fig. 3B).
Fig. 3.
Cellular immune profile and correlation between biological and clinical parameters in COVID-19. A, Principal component analysis of 30 cell subsets that were analyzed in pre-pandemic controls and COVID-19 patients. Color shows the data grouping for pre-pandemic controls (n = 8) and COVID-19 (n = 18). B, Significant differences in immune cell subsets between pre-pandemic controls and COVID-19 patients are plotted as percentages. Each dot represents a subject. For all box plots, the center is the median of the measurement, and the lower and upper lines of the box correspond to the first and third percentiles. Statistical analysis was performed using generalized linear mixed models adjusted for group, age, and sex. C, Correlation matrix of cytokines, cell subsets, and clinical features of COVID-19 patients at day 0. Spearman correlation coefficient is visualized by color intensity. Only significant correlations (P < 0.05) and those adjusted by Holm's correction are presented with an asterisk. Abbreviations: CD: Cluster of differentiation; CRP: C reactive protein; ESR: Erythrocyte sedimentation rate; G-CSF: Granulocyte colony-stimulating factor; GM-CSF: Granulocyte-macrophage colony stimulating factor; IFN: Interferon; IL: Interleukin; IP-10: interferon-γ induced protein 10; LDH: Lactate dehydrogenase; MCP-1: Monocyte chemotactic protein-1; RANTES: Regulated on activation, normal T cell expressed and secreted; SOFA: Sequential organ failure assessment; Th: T helper; TNF- α: Tumor necrosis factor-alpha; Tregs: Regulatory T cells. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2.3. Immunological and clinical correlates in COVID-19
A correlation matrix among clinical and biological parameters was constructed (Fig. 3C). Negative correlations between erythrocyte sedimentation rate (ESR) with Th1 cells (r = −0.56; P = 0.0201); ferritin and creatinine with transitional B cells (r = −0.52; P = 0.0316; r = −0.52; P = 0.0273, respectively) and bilirubin with classical memory B cells (r = −0.49; P = 0.0377) were observed. Moreover, PaO2/FiO2 and ESR were positively correlated with viral load (r = 0.51; P = 0.0322; r = 0.56; P = 0.0163, respectively). Furthermore, the following positive correlations were observed: between ferritin and IL-6 (r = 0.61; P = 0.0093) and IP-10 (r = 0.58; P = 0.0140); between bilirubin and IL-6 (r = 0.58; P = 0.0121); between C reactive protein and IP-10 (r = 0.78; P = 0.0001), between D-dimer and IL-7 (r = 0.55; P = 0.0182); between lactate dehydrogenase and IL-10 (r = 0.51; P = 0.0311), and between procalcitonin and IP-10 (r = 0.61; P = 0.0093). ESR was negatively correlated with IL-6 (r = −0.35; P = 0.0289) and IL-10 (r = −0.7; P = 0.0019) (Supplementary Table 4).
Prior longitudinal analysis, a baseline analysis (i.e., Day 0) between the plasma receptor group (i.e., recipients) and the standard therapy group looking for cytokines and immune cell signature was performed. Since this analysis revealed differences for some cell populations (Supplementary Table 5) and cytokines (Supplementary Table 6), we conducted a longitudinal analysis [delta (Δ) changes] to evaluate the effect of CP.
3.3. Longitudinal changes on immune response after CP transfusion
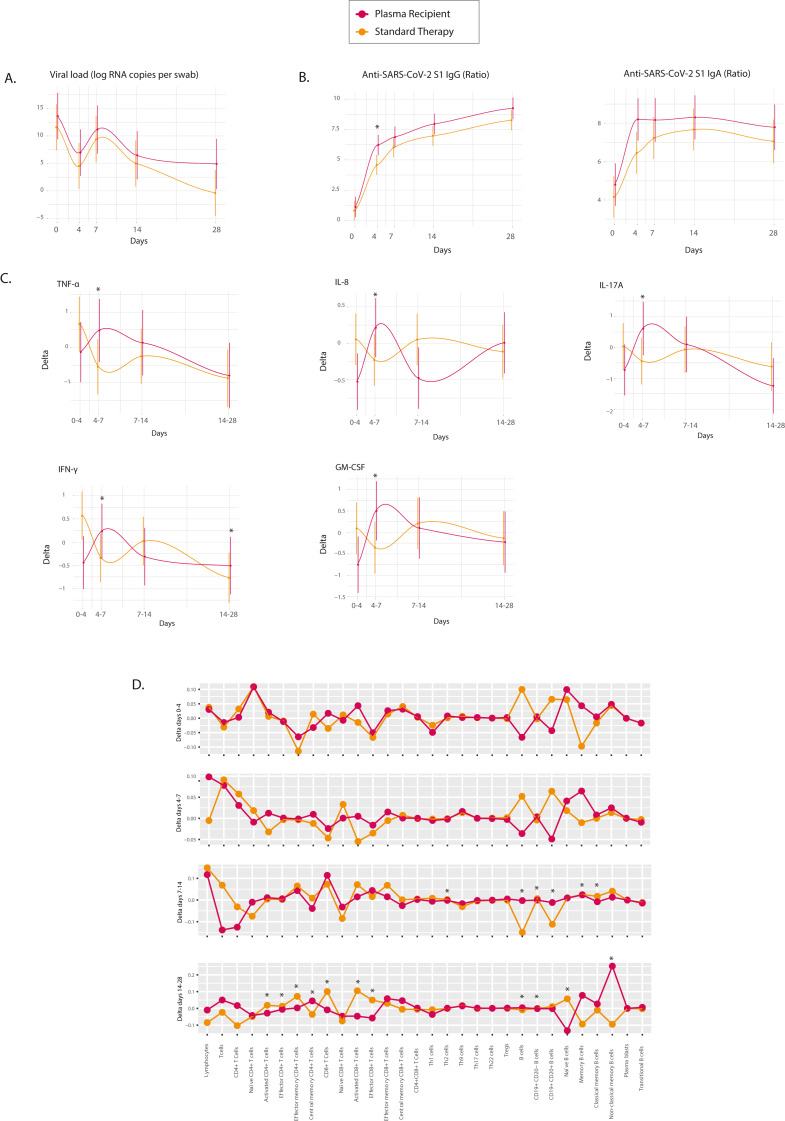
Based on the results of the previous section, we evaluated the immune response induced by CP through longitudinal follow-up in patients with COVID-19 after treatment with CP or standard therapy. No significant differences in clinical outcomes or viral load were found between groups during the follow-up (Fig. 4 A, Table 2). However, the IgG anti-S1-SARS-CoV-2 ratio value was higher on day 4 in the plasma recipient group (dilution 1:200 βinter = 1.3301; P = 0.0371) (Fig. 4B, Supplementary Table 7). This difference was observed from a titer of 1:200 to 1:800. No significant differences were observed in IgA antibodies between groups.
Fig. 4.
Longitudinal immune profile of standard therapy vs. convalescent plasma recipients. A, Temporal delta changes in the viral load were plotted as log RNA copies per swab against days of follow-up for standard therapy (n = 9) and plasma recipient (n = 9) groups. B, Dynamics of the SARS-CoV-2 antibody response and the change in anti-SARS-CoV-2 IgA and IgG ratios (OD sample/OD calibrator) were plotted against days of follow-up. Estimated ratios from generalized linear mixed models are presented. C, Estimated delta of significant cytokine concentrations in standard therapy vs. the plasma recipient group. D, Estimated delta of all the lymphocyte subsets in standard therapy vs. the plasma recipient group. All significant β interactions for delta change in day × therapy are depicted with an asterisk. Abbreviations: CD: Cluster of differentiation; GM-CSF: Granulocyte-macrophage colony stimulating factor; IFN: Interferon; Ig: Immunoglobulin; IL: Interleukin; Th: T helper; TNF- α: Tumor necrosis factor-alpha; Tregs: Regulatory T cells.
The plasma recipient group had a different cytokine profile over time than the standard therapy group. CP transfusion induced an early but transient increase in ΔD4 to D7 for TNF-α (βinter = 1.8174; P = 0.0176), IL-8 (βinter = 1.0210; P = 0.0032), IL-17A (βinter = 1.8041; P = 0.0148), GM-CSF (βinter = 1.7074; P = 0.0039), and IFN-γ (βinter = 1.5836; P = 0.0020). For IFN-γ the change was persistent, since was also higher in ΔD14 to D28 (βinter = 1.2757; P = 0.0159) (Fig. 4C). On day 28, the ratios IL-6/IFN-γ (βinter = −0.8536; P = 0.0089) and IL-6/IL-10 (βinter = −0.5105; P = 0.0180) decreased in the plasma recipient group compared with the standard therapy group (Supplementary Table 7).
For cellular sub-phenotypes, most of the between-group differences were observed at ΔD14 to D28. Plasma recipients showed a positive Δchange in central memory CD4+ T cells (βinter = 0.1270; P = 0.0252), B cells (βinter = 0.1790; P = 0.0026), and non-classical memory B cells (βinter = 0.3425; P=<0.0001), whereas negative Δchange were observed for activated, effector and effector memory CD4+ (βinter = −0.0614; P = 0.0444; βinter = −0.0156; P = 0.0239; βinter = −0.1189; P = 0.0441, respectively), and activated and effector CD8+ (βinter = −0.2099; P = 0.0063; βinter = −0.1238; P = 0.0004, respectively) T cells, as well as naïve B cells (βinter = −0.2249; P = 0.0010) (Fig. 4D).
4. Discussion
Our study showed an ongoing and dynamic immune response differential between treatments. Although antibody-independent mechanisms of action have been proposed to explain the effect of CP [3], no difference between donors and super-donors in terms of cytokines and metabolomic profile was observed. Super-donors had NAbs titers ≥1:256, which exceeded the FDA recommendation of ≥1:160 [9,29]. Thus, the main difference between super-donors and donors was in their antibody levels, which highly correlated with NAbs [17].
We found a differential metabolomic profile of recovered COVID-19 patients compared with pre-pandemic controls and severe COVID-19 patients. Consistent with previous reports, ours confirmed that COVID-19 patients are characterized by lipid dysregulation in fatty acids, glycerophospholipids, glycerolipids, and sphingolipids, which has been associated with dyslipidemia and oxidative stress [30,31]. Recovered patients regulated the most prominent metabolic perturbations that were associated with SARS-CoV-2 infection on metabolic function, such as a decrease in lipid levels, along with an increase in ketone bodies. However, the phenotype of recovered patients did not return to a similar phenotype of pre-pandemic controls, and it was characterized by continued altered levels of unsaturated fatty acids, such as arachidonic and linoleic acid.
COVID-19 patients disclose latent rheumatologic, thyroid and antiphospholipid autoimmunity (Table 1) [32]. Several emerging reports have shown that SARS-CoV-2 can lead to autoimmune and auto-inflammatory diseases [[33], [34], [35]]. The pathogenic mechanisms shared between these diseases suggest that SARS-CoV-2 could act as a triggering factor for the development of autoimmune phenomena in susceptible individuals [16,36]. Post-COVID-19 long-term studies are warranted to determine whether this latent autoimmunity persists and may evolve towards overt autoimmunity (i.e., clinical disease) [37]. Furthermore, anti–IFN–α antibodies were confirmed in COVID-19 patients. A recent study, showed that some infected individuals with SARS-CoV-2, mainly men produce autoantibodies against IFN-α due to inborn errors of type I IFN immunity [38]. These autoantibodies are associated with high risk of developing life-threatening pneumonia [39]. However, in this small study, clinical worsening following CP transfusion containing such antibodies was not observed, suggesting that allogenic antibodies anti–IFN–α do not influence clinical outcomes.
We observed that transfusion of CP with high NAbs titers did not significantly modify viral load kinetics. Although early administration of CP to hospitalized COVID-19 patients could reduce severity and mortality [14], this phenomenon is not exclusively attributable to viral load reduction. Severe COVID-19 patients exhibit a higher viral shedding in a variety of tissues for 20–40 days after onset of disease than mildly ill patients, indicating that severity of illness influences viral load kinetics [40]. All together, data indicate that the level of antibodies does not guarantee viral clearance.
Patients with COVID-19 showed a correlation between hematological and clinical parameters and pro-inflammatory cytokines, as previously reported for SARS-CoV-2 infection [[41], [42], [43], [44]]. COVID-19 patients showed a hyper-inflammatory response that was characterized by high levels of IP-10, GM-CSF, G-CSF, MCP-1, IFN-α, IL-6, IL-7, IL-8, and IL-10 (Fig. 5 ). Accumulating evidence indicate that the severity of COVID-19 is associated with an increased level of inflammatory mediators including the cytokines and chemokines mentioned above [[45], [46], [47]]. Among these cytokines, IL-6 is one of the key factors associated with lethal complications of COVID-19 patients [48]. IL-6 amplifier is activated by coactivation of nuclear factor (NF)-κB and STAT3, which are potential regulators of the COVID-19-mediated cytokine storm [47]. High IFN-α levels were observed in COVID-19 patients, as previously shown [46]. However, opposite results for IFN-α have been published [38,39,49]. Cytokines release in COVID-19 is influenced by disease duration and severity, sex, age, genetic background including ancestry, and autoimmune phenomena [38,39,50].
Fig. 5.
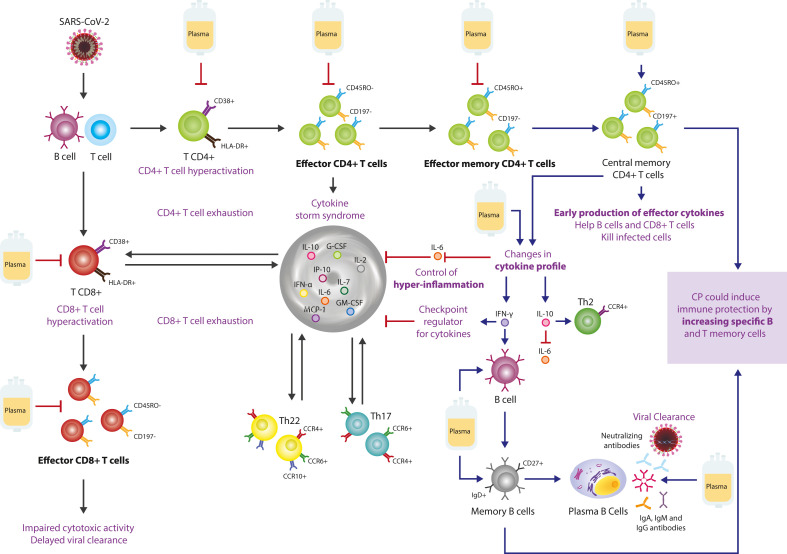
Possible mechanisms of action of convalescent plasma in severe COVID-19 patients. Upon infection with SARS-CoV-2, T cells are activated by viral proteins presented by B cells, then they undergo clonal expansion. Enrichment of activated CD38+ HLA-DR + CD4+ and CD8+ T cells leads to dysfunctional effector cells with an ineffective state of differentiation, preceding T cell exhaustion. Th17 and Th22 perpetuates inflammation. CP blocks the hyperactivation of CD4+ and CD8+ T cells, and decreases effector T cells. Most effector T cells die, but a subset persists, transitioning to memory T cells. CP decreases IL-6 and increases IFN-γ and IL-10, and increases central memory CD4+ T cells, which can be located in secondary lymphoid tissues, to be reactivated by exposure to antigen. CP also increases B and memory B cells, which can differentiate into long-lived plasma cells to maintain long-term antibody production. Black lines indicate the effect induced by SARS-CoV-2 infection. Blue lines indicate the stimulation effect generated by the CP. Red lines indicate the inhibitory effect generated by CP. Abbreviations: CCR: C–C chemokine receptor; CD: Cluster of differentiation; CP: Convalescent plasma; HLA: human leukocyte antigen, Ig: Immunoglobulin; IL: Interleukin; IP-10: interferon-γ induced protein 10; IFN: Interferon; G-CSF: Granulocyte colony-stimulating factor; GM-CSF: Granulocyte-macrophage colony stimulating factor; MCP-1: Monocyte Chemotactic Protein-1; Th: T helper. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
High levels of IL-6 and IL-10 can directly block the expansion of lymphoid progenitors [48,51], causing lymphopenia. The hyper-inflammatory state (i.e., cytokine storm syndrome) drives the exhaustion of T cell subsets, characterized by perpetuated activation of CD4+ and CD8+ T cells [[52], [53], [54], [55]]. IL-6 also induces Th17 and Th22 differentiation and favors inflammation [56].
The inflammatory profile of cytokines in COVID-19 differed from that of recovered patients. CP transfusion early regulates the inflammatory balance among the cytokines evaluated in this study, which have been associated with COVID-19 severity [57]. An increase in IL-8 was observed in recovered patients compared to pre-pandemic controls. Similar results were reported by Hähnel et al. [58], who observed a slight increase in IL-8 in plasma donors, indicating that some pro-inflammatory cytokines may still be present in the plasma of recovered COVID-19 patients.
CP decreased the IL-6/IL-10 and IL-6/IFN-γ ratios on day 28 post-transfusion. Recent reports have shown that an increase in the IL-6/IL-10 ratio is related to a poor prognosis in COVID-19 [59]. IL-10 is an immunoregulatory cytokine that can attenuate inflammatory responses through its Th2 activity and by blocking NF-κB, which negatively regulates IL-6 production [60,61]. However, a large amount of IL-10 inhibits immune cell function and proliferation, which delays viral clearance and can consequently cause lung damage [62]. The decrease in the IL-6/IFN-γ ratio in the plasma recipient group indicates another mechanism of action of CP. Lagunas-Rangel et al. [63], showed that an increase in this ratio could be associated with COVID-19 severity. Interaction between these two cytokines contributes to the recruitment and adequate elimination of neutrophils which favors the timely resolution of the infection and prompt transition between innate and adaptive immunity [64,65]. Moreover, CP induced an early but transient increase of TNF-α, IL-8, IL-17A, GM-CSF, and IFN-γ, although the latter was persistently increased at day 28. The understanding of the inflammatory profiles in COVID-19 have demonstrated a dual role of IFN-γ. Some have shown increased IFN-γ in severe COVID-19 patients [66], while others have shown opposite results [67]. Our results indicated that CP induces a temporary increase in inflammatory cytokines (e.g., IFN-γ and TNF-α) that counteracts the infection by controlling the cytokine storm syndrome, as observed in the decrease in the IL-6/IL-10 and IL-6/IFN-γ ratios.
Our findings also demonstrate that the use of CP attenuates the exhausted phenotype observed in patients before transfusion, since CP decreases activated and effector CD4+ and CD8+ T cells (Fig. 5). Kalfaoglu et al. [68], demonstrated in patients with severe disease that hyperactivated T cells differentiate into multiple effector T cell lineages. These hyperactivated T cells secrete proteins (e.g., furin) that facilitate viral entry and damage to the lung. In addition, CP induces immune memory through an increase in central memory CD4+ T cells and memory B cells, specifically non-classical memory B cells. Rodda et al. [69], found that SARS-CoV-2-specific memory lymphocytes have antiviral function: memory T cells produce cytokines and expand upon antigen re-encounter, while memory B cells express receptors capable of neutralizing the virus. Previous reports have shown that recovery is associated with the increase of T cell memory as suggested by the lack of central memory in severe COVID-19 patients [70]. Likewise, the frequency of memory B cells is a critical indicator of the resolution of the disease in SARS-CoV-2 [71]. CP induces a robust CD4+ and CD8+ T cell response, along with a memory T and B phenotype that could provide long-term immunity.
5. Strengths and limitations
Our study has several strengths. We included patients in the first 72 h of admission, and in the first 14 days after symptoms onset, which allowed to evaluate the effects of CP in the early stages of disease. The timing of inclusion guaranteed the comparability in clinical progression and staging of disease. All included patients fulfilled the definition for severe COVID-19. In addition, transfused patients received high quality CP that surpassed the FDA NAbs recommendation. None of the included patients received experimental treatments that may had influenced the immune response.
Limitations must be acknowledged. A longitudinal analysis of the effect of CP on metabolic profiles and NAbs was not done, which would have been informative and complementary to the immunological assessment. A broader panel of B and T cells, as well as their functional analyses was not possible owing to the small number of cells. Genetic evaluation was not investigated. Another potential limitation of the present study is that the observed results might be from chance alone or the moderate sample size. However, this is unlikely because of the highly significant results that were seen after adjustments and corrections as well as their consistent direction and magnitude within the different analyses.
6. Conclusions
Total IgG and IgA anti-S1-SARS-CoV-2 antibodies are key factors for CP selection and correlate with NAbs. Serum from recovered patients discloses a different cytokine and metabolite composition than that from patients with severe disease. Latent autoimmunity and anti–IFN–α antibodies are observed in COVID-19. COVID-19 CP induces an early but transient effect on the antibody and cytokine profile of patients with severe disease, attenuates the exhausted phenotype and increases memory T and B-lymphocytes at day 28 post-transfusion together with a reduction of IL-6/IFN-γ and IL-6/IL-10 ratios. Our results may have therapeutic implications and justify further post-COVID-19 studies.
7. CP-COVID-19 group
Paula Andrea Gaviria García (Instituto Distrital de Ciencia Biotecnología e Investigación en Salud, IDCBIS, Bogota, Colombia), Adriana Rojas-Villarraga (Fundación Universitaria de Ciencias de la Salud (FUCS), Bogota, Colombia), Juan Camilo Díaz (Internal Medicine Department, Universidad CES, Medellin, Colombia), Verónica Posada Vélez (Internal Medicine Department, Universidad CES, Medellin, Colombia), Lina Marcela Acevedo Landinez (Instituto Distrital de Ciencia Biotecnología e Investigación en Salud, IDCBIS, Bogota, Colombia), Luisa Paola Duarte Correales (Instituto Distrital de Ciencia Biotecnología e Investigación en Salud, IDCBIS, Bogota, Colombia), Oscar Gómez (GenomaCES, Universidad CES, Medellin, Colombia), Gustavo Andrés Salguero López (Instituto Distrital de Ciencia Biotecnología e Investigación en Salud, IDCBIS, Bogota, Colombia), Jeser Santiago Grass Guaqueta (Instituto Distrital de Ciencia Biotecnología e Investigación en Salud, IDCBIS, Bogota, Colombia), Cristian Alejandro Ricaurte Pérez (Instituto Distrital de Ciencia Biotecnología e Investigación en Salud, IDCBIS, Bogota, Colombia), Jorge Carrillo (Hospital Universitario Mayor Méderi, Universidad del Rosario, Bogota, Colombia), José Alejandro Daza Vergara (Hospital Universitario Mayor Méderi, Universidad del Rosario, Bogota, Colombia), Sandra Landinez (Laboratorio Clínico Compensar, Hospital Universitario Mayor Méderi, Universidad del Rosario, Bogota, Colombia), Rubén D. Mantilla (Center for Autoimmune Diseases Research (CREA), School of Medicine and Health Sciences, Universidad del Rosario, Bogota, Colombia), Jairo David Torres Yepes (Internal Medicine Department, Hospital Universitario Mayor Méderi, Bogotá, Colombia), Oscar Andrés Briceño Ricaurte (Internal Medicine Department, Universidad CES, Medellin, Colombia), Carlos E. Pérez-Díaz (Infectious diseases, Clínica de Marly, Bogota, Colombia), Norma Montoya García (Clínica del Occidente, Bogota, Colombia), Yady Nataly Mateus (Clínica del Occidente, Bogota, Colombia), and Laura Mancera Navarro (Clínica del Occidente, Bogota, Colombia).
Ethical statement
The authors declare that they have no competing interests.
Funding
The CP-COVID-19 group was supported by grants from Universidad del Rosario (ABN011) and IDCBIS, Bogota, Colombia, and ISA group and Suramericana, Medellin, Colombia.
Role of the funder/sponsor
The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Author statement
This work has not been submitted elsewhere for consideration. All figures are original. No permissions are required.
Author contribution
Yeny Acosta-Ampudia: Acquisition, analysis, or interpretation of data, Statistical analysis, Writing and Editing, Revision of the manuscript and final approval. Diana M. Monsalve: Statistical analysis, Writing and Editing, Revision of the manuscript and final approval. Manuel Rojas: Acquisition, analysis, or interpretation of data, Statistical analysis, Writing and Editing, Revision of the manuscript and final approval. Yhojan Rodríguez: Revision of the manuscript and final approval. Juan Esteban Gallo: Acquisition, analysis, or interpretation of data, Revision of the manuscript and final approval. Juan Carlos Salazar-Uribe: Acquisition, analysis, or interpretation of data, Statistical analysis, Revision of the manuscript and final approval. María José Santander: Acquisition, analysis, or interpretation of data, Revision of the manuscript and final approval. Mónica P. Cala: Revision of the manuscript and final approval. Wildeman Zapata: Acquisition, analysis, or interpretation of data, Revision of the manuscript and final approval. María Isabel Zapata: Acquisition, analysis, or interpretation of data, Revision of the manuscript and final approval. Rubén Manrique: Acquisition, analysis, or interpretation of data, Obtained funding, Revision of the manuscript and final approval. Juan Mauricio Pardo-Oviedo: Acquisition, analysis, or interpretation of data, Revision of the manuscript and final approval. Bernardo Camacho: Acquisition, analysis, or interpretation of data, Obtained funding, Revision of the manuscript and final approval. Carolina Ramírez-Santana: had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis, Conceptualization, Acquisition, analysis, or interpretation of data, Statistical analysis, Writing and Editing, Revision of the manuscript and final approval. Juan-Manuel Anaya: had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis, Conceptualization, Acquisition, analysis, or interpretation of data, Statistical analysis, Writing and Editing, Obtained funding, Revision of the manuscript and final approval
Declaration of competing interest
None.
Acknowledgments
We thank Jodi Smith, PhD, from Edanz Group (https://en-author-services.edanzgroup.com/ac) for editing and reviewing this manuscript for English language.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jaut.2021.102598.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Guan W.-J., Ni Z.-Y., Hu Y., Liang W.-H., Ou C.-Q., He J.-X., Liu L., Shan H., Lei C.-L., Hui D.S.C., Du B., Li L.-J., Zeng G., Yuen K.-Y., Chen R.-C., Tang C.-L., Wang T., Chen P.-Y., Xiang J., Li S.-Y., Wang J.-L., Liang Z.-J., Peng Y.-X., Wei L., Liu Y., Hu Y.-H., Peng P., Wang J.-M., Liu J.-Y., Chen Z., Li G., Zheng Z.-J., Qiu S.-Q., Luo J., Ye C.-J., Zhu S.-Y., Zhong N.-S. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berenguer J., Ryan P., Rodríguez-Baño J., Jarrín I., Carratalà J., Pachón J., Yllescas M., Arriba J.R. vol. 26. The Official Publication of the European Society of Clinical Microbiology and Infectious Diseases; 2020. pp. 1525–1536. (Characteristics and Predictors of Death Among 4035 Consecutively Hospitalized Patients with COVID-19 in Spain., Clinical Microbiology and Infection). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rojas M., Rodríguez Y., Monsalve D.M., Acosta-Ampudia Y., Camacho B., Gallo J.E., Rojas-Villarraga A., Ramírez-Santana C., Díaz-Coronado J.C., Manrique R., Mantilla R.D., Shoenfeld Y., Anaya J.-M. Convalescent plasma in Covid-19: possible mechanisms of action. Autoimmun. Rev. 2020;19:102554. doi: 10.1016/j.autrev.2020.102554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wood E.M., Estcourt L.J., McQuilten Z. How should we use convalescent plasma therapies for COVID-19? Blood. 2020 doi: 10.1182/blood.2020008903. blood.2020008903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Griensven J., Edwards T., de Lamballerie X., Semple M.G., Gallian P., Baize S., Horby P.W., Raoul H., Magassouba N., Antierens A., Lomas C., Faye O., Sall A.A., Fransen K., Buyze J., Ravinetto R., Tiberghien P., Claeys Y., De Crop M., Lynen L., Bah E.I., Smith P.G., Delamou A., De Weggheleire A., Haba N. Evaluation of convalescent plasma for Ebola virus disease in Guinea. N. Engl. J. Med. 2016;374:33–42. doi: 10.1056/NEJMoa1511812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marano G., Vaglio S., Pupella S., Facco G., Catalano L., Liumbruno G.M., Grazzini G. Convalescent plasma: new evidence for an old therapeutic tool? Blood Transf.Trasf. Del Sangue. 2016;14:152–157. doi: 10.2450/2015.0131-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hung I.F., To K.K., Lee C.-K., Lee K.-L., Chan K., Yan W.-W., Liu R., Watt C.-L., Chan W.-M., Lai K.-Y., Koo C.-K., Buckley T., Chow F.-L., Wong K.-K., Chan H.-S., Ching C.-K., Tang B.S., Lau C.C., Li I.W., Liu S.-H., Chan K.-H., Lin C.-K., Yuen K.-Y. vol. 52. Clinical Infectious Diseases : An Official Publication of the Infectious Diseases Society of America; 2011. pp. 447–456. (Convalescent Plasma Treatment Reduced Mortality in Patients with Severe Pandemic Influenza A (H1N1) 2009 Virus Infection). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng Y., Wong R., Soo Y.O.Y., Wong W.S., Lee C.K., Ng M.H.L., Chan P., Wong K.C., Leung C.B., Cheng G. Use of convalescent plasma therapy in SARS patients in Hong Kong. Eur. J. Clin. Microbiol. Infect. Dis. 2005;24:44–46. doi: 10.1007/s10096-004-1271-9. Official Publication of the European Society of Clinical Microbiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Recommendations for investigational COVID-19 convalescent plasma, food and drug administration. 2020. https://www.fda.gov/vaccines-blood-biologics/investigational-new-drug-ind-or-device-exemption-ide-process-cber/recommendations-investigational-covid-19-convalescent-plasma
- 10.Hegerova L., Gooley T.A., Sweerus K.A., Maree C., Bailey N., Bailey M., Dunleavy V., Patel K., Alcorn K., Haley R., Johnsen J.M., Konkle B.A., Lahti A.C., Alexander M.L., Goldman J.D., Lipke A., Lim S.-J., Sullivan M.D., Pauk J.S., Pagel J.M. Use of convalescent plasma in hospitalized patients with COVID-19: case series. Blood. 2020;136:759–762. doi: 10.1182/blood.2020006964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joyner M., Wright R.S., Fairweather D., Senefeld J., Bruno K., Klassen S., Carter R., Klompas A., Wiggins C., Shepherd J.R., Rea R., Whelan E., Clayburn A., Spiegel M., Johnson P., Lesser E., Baker S., Larson K., Ripoll Sanz J., Andersen K., Hodge D., Kunze K., Buras M., Vogt M., Herasevich V., Dennis J., Regimbal R., Bauer P., Blair J., van Buskirk C., Winters J., Stubbs J., Paneth N., Casadevall A. The Preprint Server for Health Sciences; 2020. Early Safety Indicators of COVID-19 Convalescent Plasma in 5,000 Patients. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duan K., Liu B., Li C., Zhang H., Yu T., Qu J., Zhou M., Chen L., Meng S., Hu Y., Peng C., Yuan M., Huang J., Wang Z., Yu J., Gao X., Wang D., Yu X., Li L., Zhang J., Wu X., Li B., Xu Y., Chen W., Peng Y., Hu Y., Lin L., Liu X., Huang S., Zhou Z., Zhang L., Wang Y., Zhang Z., Deng K., Xia Z., Gong Q., Zhang W., Zheng X., Liu Y., Yang H., Zhou D., Yu D., Hou J., Shi Z., Chen S., Chen Z., Zhang X., Yang X. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. U.S.A. 2020;117:9490–9496. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J., Wang F., Li D., Yang M., Xing L., Wei J., Xiao H., Yang Y., Qu J., Qing L., Chen L., Xu Z., Peng L., Li Y., Zheng H., Chen F., Huang K., Jiang Y., Liu D., Zhang Z., Liu Y., Liu L. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. J. Am. Med. Assoc. 2020;323:1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salazar E., Christensen P.A., Graviss E.A., Nguyen D.T., Castillo B., Chen J., V Lopez B., Eagar T.N., Yi X., Zhao P., Rogers J., Shehabeldin A., Joseph D., Leveque C., Olsen R.J., Bernard D.W., Gollihar J., Musser J.M. Treatment of coronavirus disease 2019 patients with convalescent plasma reveals a signal of significantly decreased mortality. Am. J. Pathol. 2020;190:2290–2303. doi: 10.1016/j.ajpath.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rojas M., Anaya J.-M. Why will it never be known if convalescent plasma is effective for COVID-19. J. Trans. Autoimmun. 2020;3:100069. doi: 10.1016/j.jtauto.2020.100069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodríguez Y., Novelli L., Rojas M., De Santis M., Acosta-Ampudia Y., Monsalve D.M., Ramírez-Santana C., Costanzo A., Ridgway W.M., Ansari A.A., Gershwin M.E., Selmi C., Anaya J.-M. Autoinflammatory and autoimmune conditions at the crossroad of COVID-19. J. Autoimmun. 2020;114:102506. doi: 10.1016/j.jaut.2020.102506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okba N.M.A., Müller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., Lamers M.M., Sikkema R.S., de Bruin E., Chandler F.D., Yazdanpanah Y., Le Hingrat Q., Descamps D., Houhou-Fidouh N., Reusken C.B.E.M., Bosch B.-J., Drosten C., Koopmans M.P.G., Haagmans B.L. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg. Infect. Dis. 2020;26:1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tonn T., Corman V.M., Johnsen M., Richter A., Rodionov R.N., Drosten C., Bornstein S.R. Stability and neutralising capacity of SARS-CoV-2-specific antibodies in convalescent plasma. Lancet Microbe. 2020;1:e63. doi: 10.1016/S2666-5247(20)30037-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saavedra Trujillo C.H. Consenso colombiano de atención, diagnóstico y manejo de la infección por SARS-COV-2/COVID 19 en establecimientos de atención de la salud. Recomendaciones basadas en consenso de expertos e informadas en la evidencia, Infection. 2020;24:1. doi: 10.22354/in.v24i3.851. [DOI] [Google Scholar]
- 20.Knight S.R., Ho A., Pius R., Buchan I., Carson G., Drake T.M., Dunning J., Fairfield C.J., Gamble C., Green C.A., Gupta R., Halpin S., Hardwick H.E., Holden K.A., Horby P.W., Jackson C., Mclean K.A., Merson L., Nguyen-Van-Tam J.S., Norman L., Noursadeghi M., Olliaro P.L., Pritchard M.G., Russell C.D., Shaw C.A., Sheikh A., Solomon T., Sudlow C., V Swann O., Turtle L.C., Openshaw P.J., Baillie J.K., Semple M.G., Docherty A.B., Harrison E.M. Risk stratification of patients admitted to hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: development and validation of the 4C Mortality Score. BMJ (Clinic., Res. Ed). 2020;370 doi: 10.1136/bmj.m3339. m3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Theel E.S., Harring J., Hilgart H., Granger D. Performance characteristics of four high-throughput immunoassays for detection of IgG antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.01243-20. e01243-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weidner L., Gänsdorfer S., Unterweger S., Weseslindtner L., Drexler C., Farcet M., Witt V., Schistal E., Schlenke P., Kreil T.R., Jungbauer C. Quantification of SARS-CoV-2 antibodies with eight commercially available immunoassays. J. Clin. Virol. 2020;129:104540. doi: 10.1016/j.jcv.2020.104540. The Official Publication of the Pan American Society for Clinical Virology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Franco J.-S., Amaya-Amaya J., Molano-González N., Caro-Moreno J., Rodríguez-Jiménez M., Acosta-Ampudia Y., Mantilla R.D., Rojas-Villarraga A., Anaya J.-M. Autoimmune thyroid disease in Colombian patients with systemic lupus erythematosus. Clin. Endocrinol. 2015;83:943–950. doi: 10.1111/cen.12662. [DOI] [PubMed] [Google Scholar]
- 24.Pacheco Y., Monsalve D.M., Acosta-Ampudia Y., Rojas C., Anaya J.-M., Ramírez-Santana C. Antinuclear autoantibodies: discordance among four different assays. Ann. Rheum. Dis. 2020;79:e6. doi: 10.1136/annrheumdis-2018-214693. [DOI] [PubMed] [Google Scholar]
- 25.Pacheco Y., Barahona-Correa J., Monsalve D.M., Acosta-Ampudia Y., Rojas M., Rodriguez Y., Saavedra J., Rodriguez-Jimenez M., Mantilla R.D., Ramirez-Santana C., Molano-Gonzalez N., Anaya J.-M. Cytokine and autoantibody clusters interaction in systemic lupus erythematosus. J. Transl. Med. 2017;15:239. doi: 10.1186/s12967-017-1345-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ciborowski M., Javier Rupérez F., Martínez-Alcázar M.P., Angulo S., Radziwon P., Olszanski R., Kloczko J., Barbas C. Metabolomic approach with LC-MS reveals significant effect of pressure on diver's plasma. J. Proteome Res. 2010;9:4131–4137. doi: 10.1021/pr100331j. [DOI] [PubMed] [Google Scholar]
- 27.Whiley L., Godzien J., Ruperez F.J., Legido-Quigley C., Barbas C. In-vial dual extraction for direct LC-MS analysis of plasma for comprehensive and highly reproducible metabolic fingerprinting. Anal. Chem. 2012;84:5992–5999. doi: 10.1021/ac300716u. [DOI] [PubMed] [Google Scholar]
- 28.Contrepois K., Jiang L., Snyder M. Optimized analytical procedures for the untargeted metabolomic profiling of human urine and plasma by combining hydrophilic interaction (HILIC) and reverse-phase liquid chromatography (RPLC)-Mass spectrometry. Mol. Cell. Proteomics: MCP. 2015;14:1684–1695. doi: 10.1074/mcp.M114.046508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salazar E., V Kuchipudi S., Christensen P.A., Eagar T.N., Yi X., Zhao P., Jin Z., Long S.W., Olsen R.J., Chen J., Castillo B., Leveque C., Towers D.M., Lavinder J., Gollihar J.D., Cardona J., Ippolito G.C., Nissly R.H., Bird I.M., Greenawalt D., Rossi R.M., Gontu A., Srinivasan S., Poojary I.B., Cattadori I.M., Hudson P.J., Joselyn N., Prugar L., Huie K., Herbert A., Bernard D.W., Dye J., Kapur V., Musser J.M. bioRxiv [Preprint]; 2020. Relationship between Anti-spike Protein Antibody Titers and SARS-CoV-2 in Vitro Virus Neutralization in Convalescent Plasma. 2020.06.08.138990. [DOI] [Google Scholar]
- 30.Shen B., Yi X., Sun Y., Bi X., Du J., Zhang C., Quan S., Zhang F., Sun R., Qian L., Ge W., Liu W., Liang S., Chen H., Zhang Y., Li J., Xu J., He Z., Chen B., Wang J., Yan H., Zheng Y., Wang D., Zhu J., Kong Z., Kang Z., Liang X., Ding X., Ruan G., Xiang N., Cai X., Gao H., Li L., Li S., Xiao Q., Lu T., Zhu Y., Liu H., Chen H., Guo T. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182 doi: 10.1016/j.cell.2020.05.032. 59–72.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bruzzone C., Bizkarguenaga M., Gil-Redondo R., Diercks T., Arana E., García de Vicuña A., Seco M., Bosch A., Palazón A., San Juan I., Laín A., Gil-Martínez J., Bernardo-Seisdedos G., Fernández-Ramos D., Lopitz-Otsoa F., Embade N., Lu S., Mato J.M., Millet O. SARS-CoV-2 infection dysregulates the metabolomic and lipidomic profiles of serum. iScience. 2020;23:101645. doi: 10.1016/j.isci.2020.101645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Botello A., Herrán M., Salcedo V., Rodríguez Y., Anaya J.-M., Rojas M. Prevalence of latent and overt polyautoimmunity in autoimmune thyroid disease: a systematic review and meta-analysis. Clin. Endocrinol. 2020;93:375–389. doi: 10.1111/cen.14304. [DOI] [PubMed] [Google Scholar]
- 33.Mateu-Salat M., Urgell E., Chico A. SARS-COV-2 as a trigger for autoimmune disease: report of two cases of Graves' disease after COVID-19. J. Endocrinol. Invest. 2020;43:1527–1528. doi: 10.1007/s40618-020-01366-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y., Xiao M., Zhang S., Xia P., Cao W., Jiang W., Chen H., Ding X., Zhao H., Zhang H., Wang C., Zhao J., Sun X., Tian R., Wu W., Wu D., Ma J., Chen Y., Zhang D., Xie J., Yan X., Zhou X., Liu Z., Wang J., Du B., Qin Y., Gao P., Qin X., Xu Y., Zhang W., Li T., Zhang F., Zhao Y., Li Y., Zhang S. Coagulopathy and antiphospholipid antibodies in patients with covid-19. N. Engl. J. Med. 2020;382:e38. doi: 10.1056/NEJMc2007575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mantovani Cardoso E., Hundal J., Feterman D., Magaldi J. Concomitant new diagnosis of systemic lupus erythematosus and COVID-19 with possible antiphospholipid syndrome. Just a coincidence? A case report and review of intertwining pathophysiology. Clin. Rheumatol. 2020;39:2811–2815. doi: 10.1007/s10067-020-05310-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ehrenfeld M., Tincani A., Andreoli L., Cattalini M., Greenbaum A., Kanduc D., Alijotas-Reig J., Zinserling V., Semenova N., Amital H., Shoenfeld Y. Covid-19 and autoimmunity. Autoimmun. Rev. 2020;19:102597. doi: 10.1016/j.autrev.2020.102597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lerma L.A., Chaudhary A., Bryan A., Morishima C., Wener M.H., Fink S.L. Prevalence of autoantibody responses in acute coronavirus disease 2019 (COVID-19) J. Trans. Autoimmun. 2020;3:100073. doi: 10.1016/j.jtauto.2020.100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., Ogishi M., Sabli I.K.D., Hodeib S., Korol C., Rosain J., Bilguvar K., Ye J., Bolze A., Bigio B., Yang R., Arias A.A., Zhou Q., Zhang Y., Onodi F., Korniotis S., Karpf L., Philippot Q., Chbihi M., Bonnet-Madin L., Dorgham K., Smith N., Schneider W.M., Razooky B.S., Hoffmann H.-H., Michailidis E., Moens L., Han J.E., Lorenzo L., Bizien L., Meade P., Neehus A.-L., Ugurbil A.C., Corneau A., Kerner G., Zhang P., Rapaport F., Seeleuthner Y., Manry J., Masson C., Schmitt Y., Schlüter A., Le Voyer T., Khan T., Li J., Fellay J., Roussel L., Shahrooei M., Alosaimi M.F., Mansouri D., Al-Saud H., Al-Mulla F., Almourfi F., Al-Muhsen S.Z., Alsohime F., Al Turki S., Hasanato R., van de Beek D., Biondi A., Bettini L.R., D'Angio’ M., Bonfanti P., Imberti L., Sottini A., Paghera S., Quiros-Roldan E., Rossi C., Oler A.J., Tompkins M.F., Alba C., Vandernoot I., Goffard J.-C., Smits G., Migeotte I., Haerynck F., Soler-Palacin P., Martin-Nalda A., Colobran R., Morange P.-E., Keles S., Çölkesen F., Ozcelik T., Yasar K.K., Senoglu S., Karabela Ş.N., Rodríguez-Gallego C., Novelli G., Hraiech S., Tandjaoui-Lambiotte Y., Duval X., Laouénan C., Snow A.L., Dalgard C.L., Milner J.D., Vinh D.C., Mogensen T.H., Marr N., Spaan A.N., Boisson B., Boisson-Dupuis S., Bustamante J., Puel A., Ciancanelli M.J., Meyts I., Maniatis T., Soumelis V., Amara A., Nussenzweig M., García-Sastre A., Krammer F., Pujol A., Duffy D., Lifton R.P., Zhang S.-Y., Gorochov G., Béziat V., Jouanguy E., Sancho-Shimizu V., Rice C.M., Abel L., Notarangelo L.D., Cobat A., Su H.C., Casanova J.-L. Science; New York, NY: 2020. Inborn Errors of Type I IFN Immunity in Patients with Life-Threatening COVID-19; p. 370. eabd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.-H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Béziat V., Manry J., Shaw E., Haljasmägi L., Peterson P., Lorenzo L., Bizien L., Trouillet-Assant S., Dobbs K., de Jesus A.A., Belot A., Kallaste A., Catherinot E., Tandjaoui-Lambiotte Y., Le Pen J., Kerner G., Bigio B., Seeleuthner Y., Yang R., Bolze A., Spaan A.N., Delmonte O.M., Abers M.S., Aiuti A., Casari G., Lampasona V., Piemonti L., Ciceri F., Bilguvar K., Lifton R.P., Vasse M., Smadja D.M., Migaud M., Hadjadj J., Terrier B., Duffy D., Quintana-Murci L., van de Beek D., Roussel L., Vinh D.C., Tangye S.G., Haerynck F., Dalmau D., Martinez-Picado J., Brodin P., Nussenzweig M.C., Boisson-Dupuis S., Rodríguez-Gallego C., Vogt G., Mogensen T.H., Oler A.J., Gu J., Burbelo P.D., Cohen J.I., Biondi A., Bettini L.R., D'Angio M., Bonfanti P., Rossignol P., Mayaux J., Rieux-Laucat F., Husebye E.S., Fusco F., Ursini M.V., Imberti L., Sottini A., Paghera S., Quiros-Roldan E., Rossi C., Castagnoli R., Montagna D., Licari A., Marseglia G.L., Duval X., Ghosn J., Tsang J.S., Goldbach-Mansky R., Kisand K., Lionakis M.S., Puel A., Zhang S.-Y., Holland S.M., Gorochov G., Jouanguy E., Rice C.M., Cobat A., Notarangelo L.D., Abel L., Su H.C., Casanova J.-L. vol. 370. Science; New York, NY: 2020. (Autoantibodies against Type I IFNs in Patients with Life-Threatening COVID-19). abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y., Zhang L., Sang L., Ye F., Ruan S., Zhong B., Song T., Alshukairi A.N., Chen R., Zhang Z., Gan M., Zhu A., Huang Y., Luo L., Mok C.K.P., Al Gethamy M.M., Tan H., Li Z., Huang X., Li F., Sun J., Zhang Y., Wen L., Li Y., Chen Z., Zhuang Z., Zhuo J., Chen C., Kuang L., Wang J., Lv H., Jiang Y., Li M., Lin Y., Deng Y., Tang L., Liang J., Huang J., Perlman S., Zhong N., Zhao J., Malik Peiris J.S., Li Y., Zhao J. Kinetics of viral load and antibody response in relation to COVID-19 severity. J. Clinic. Investig. 2020;130:5235–5244. doi: 10.1172/JCI138759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Velavan T.P., Meyer C.G. Mild versus severe COVID-19: laboratory markers. Int. J. Infect. Dis. 2020;95:304–307. doi: 10.1016/j.ijid.2020.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leisman D.E., Ronner L., Pinotti R., Taylor M.D., Sinha P., Calfee C.S., V Hirayama A., Mastroiani F., Turtle C.J., Harhay M.O., Legrand M., Deutschman C.S. Cytokine elevation in severe and critical COVID-19: a rapid systematic review, meta-analysis, and comparison with other inflammatory syndromes. Lancet Respir. Med. 2020 doi: 10.1016/S2213-2600(20)30404-5. S2213-2600–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ponti G., Maccaferri M., Ruini C., Tomasi A., Ozben T. Biomarkers associated with COVID-19 disease progression. Crit. Rev. Clin. Lab Sci. 2020;57:389–399. doi: 10.1080/10408363.2020.1770685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goel H., Harmouch F., Garg K., Saraiya P., Daly T., Kumar A., Hippen J.T. Publish Ah; 2020. The Liver in COVID-19: Prevalence, Patterns, Predictors, and Impact on Outcomes of Liver Test abnormalities., European Journal of Gastroenterology & Hepatology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Costela-Ruiz V.J., Illescas-Montes R., Puerta-Puerta J.M., Ruiz C., Melguizo-Rodríguez L. SARS-CoV-2 infection: the role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020;54:62–75. doi: 10.1016/j.cytogfr.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M., Ellingson M.K., Mao T., Oh J.E., Israelow B., Takahashi T., Tokuyama M., Lu P., Venkataraman A., Park A., Mohanty S., Wang H., Wyllie A.L., Vogels C.B.F., Earnest R., Lapidus S., Ott I.M., Moore A.J., Muenker M.C., Fournier J.B., Campbell M., Odio C.D., Casanovas-Massana A., Herbst R., Shaw A.C., Medzhitov R., Schulz W.L., Grubaugh N.D., Dela Cruz C., Farhadian S., Ko A.I., Omer S.B., Iwasaki A. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hojyo S., Uchida M., Tanaka K., Hasebe R., Tanaka Y., Murakami M., Hirano T. How COVID-19 induces cytokine storm with high mortality. Inflamm. Regen. 2020;40:37. doi: 10.1186/s41232-020-00146-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., Damoraki G., Gkavogianni T., Adami M.-E., Katsaounou P., Ntaganou M., Kyriakopoulou M., Dimopoulos G., Koutsodimitropoulos I., Velissaris D., Koufargyris P., Karageorgos A., Katrini K., Lekakis V., Lupse M., Kotsaki A., Renieris G., Theodoulou D., Panou V., Koukaki E., Koulouris N., Gogos C., Koutsoukou A. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27 doi: 10.1016/j.chom.2020.04.009. 992–1000.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C., Breillat P., Carlier N., Gauzit R., Morbieu C., Pène F., Marin N., Roche N., Szwebel T.-A., Merkling S.H., Treluyer J.-M., Veyer D., Mouthon L., Blanc C., Tharaux P.-L., Rozenberg F., Fischer A., Duffy D., Rieux-Laucat F., Kernéis S., Terrier B. vol. 369. Science; New York, NY: 2020. Impaired Type I Interferon Activity and Inflammatory Responses in Severe COVID-19 Patients; pp. 718–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takahashi T., Ellingson M.K., Wong P., Israelow B., Lucas C., Klein J., Silva J., Mao T., Oh J.E., Tokuyama M., Lu P., Venkataraman A., Park A., Liu F., Meir A., Sun J., Wang E.Y., Casanovas-Massana A., Wyllie A.L., Vogels C.B.F., Earnest R., Lapidus S., Ott I.M., Moore A.J., Shaw A., Fournier J.B., Odio C.D., Farhadian S., Dela Cruz C., Grubaugh N.D., Schulz W.L., Ring A.M., Ko A.I., Omer S.B., Iwasaki A. Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature. 2020 doi: 10.1038/s41586-020-2700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taga K., Tosato G. IL-10 inhibits human T cell proliferation and IL-2 production. J. Immunol. (Baltimore, Md? 1950;148:1143–1148. 1992. [PubMed] [Google Scholar]
- 52.Chen Z., John Wherry E. T cell responses in patients with COVID-19. Nat. Rev. Immunol. 2020;20:529–536. doi: 10.1038/s41577-020-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moon C. Fighting COVID-19 exhausts T cells. Nat. Rev. Immunol. 2020;20:277. doi: 10.1038/s41577-020-0304-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., Chen L., Li M., Liu Y., Wang G., Yuan Z., Feng Z., Zhang Y., Wu Y., Chen Y. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., Kuri-Cervantes L., Pampena M.B., D'Andrea K., Manne S., Chen Z., Huang Y.J., Reilly J.P., Weisman A.R., Ittner C.A.G., Kuthuru O., Dougherty J., Nzingha K., Han N., Kim J., Pattekar A., Goodwin E.C., Anderson E.M., Weirick M.E., Gouma S., Arevalo C.P., Bolton M.J., Chen F., Lacey S.F., Ramage H., Cherry S., Hensley S.E., Apostolidis S.A., Huang A.C., Vella L.A., Betts M.R., Meyer N.J., Wherry E.J. vol. 369. Science; New York, NY: 2020. Deep Immune Profiling of COVID-19 Patients Reveals Distinct Immunotypes with Therapeutic Implications. eabc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu D., Yang X.O. TH17 responses in cytokine storm of COVID-19: an emerging target of JAK2 inhibitor Fedratinib. J. Microbiol. Immunol. Infect. 2020;53:368–370. doi: 10.1016/j.jmii.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Del Valle D.M., Kim-Schulze S., Huang H.-H., Beckmann N.D., Nirenberg S., Wang B., Lavin Y., Swartz T.H., Madduri D., Stock A., Marron T.U., Xie H., Patel M., Tuballes K., Van Oekelen O., Rahman A., Kovatch P., Aberg J.A., Schadt E., Jagannath S., Mazumdar M., Charney A.W., Firpo-Betancourt A., Mendu D.R., Jhang J., Reich D., Sigel K., Cordon-Cardo C., Feldmann M., Parekh S., Merad M., Gnjatic S. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hähnel V., Peterhoff D., Bäuerlein V., Brosig A.-M., Pamler I., Johnson C., Bica A., Totir M., Ossner T., Stemmer B., Toelge M., Schütz A., Niller H.-H., Schmidt B., Wagner R., Gessner A., Burkhard R., Offner R. Manufacturing of convalescent plasma of COVID-19 patients: aspects of quality. PloS One. 2020;15 doi: 10.1371/journal.pone.0243967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McElvaney O.J., Hobbs B.D., Qiao D., McElvaney O.F., Moll M., McEvoy N.L., Clarke J., O'Connor E., Walsh S., Cho M.H., Curley G.F., McElvaney N.G. A linear prognostic score based on the ratio of interleukin-6 to interleukin-10 predicts outcomes in COVID-19. EBioMedicine. 2020;61:103026. doi: 10.1016/j.ebiom.2020.103026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tang Y., Liu J., Zhang D., Xu Z., Ji J., Wen C. Cytokine storm in COVID-19: the current evidence and treatment strategies. Front. Immunol. 2020;11:1708. doi: 10.3389/fimmu.2020.01708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rojas J.M., Avia M., Martín V., Sevilla N. IL-10: a multifunctional cytokine in viral infections. J. Immunol. Res. 2017;(2017):6104054. doi: 10.1155/2017/6104054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.He S., Zhou C., Lu D., Yang H., Xu H., Wu G., Pan W., Zhu R., Jia H., Tang X., Chen X., Wu X. Relationship between chest CT manifestations and immune response in COVID-19 patients. Int. J. Infect. Dis. 2020;98:125–129. doi: 10.1016/j.ijid.2020.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lagunas-Rangel F.A., Chávez-Valencia V. High IL-6/IFN-γ ratio could be associated with severe disease in COVID-19 patients. J. Med. Virol. 2020 doi: 10.1002/jmv.25900. 10.1002/jmv.25900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McLoughlin R.M., Witowski J., Robson R.L., Wilkinson T.S., Hurst S.M., Williams A.S., Williams J.D., Rose-John S., Jones S.A., Topley N. Interplay between IFN-gamma and IL-6 signaling governs neutrophil trafficking and apoptosis during acute inflammation. J. Clinic. Investig. 2003;112:598–607. doi: 10.1172/JCI17129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Acharya D., Liu G., Gack M.U. Dysregulation of type I interferon responses in COVID-19. Nat. Rev. Immunol. 2020;20:397–398. doi: 10.1038/s41577-020-0346-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu J., Li S., Liu J., Liang B., Wang X., Wang H., Li W., Tong Q., Yi J., Zhao L., Xiong L., Guo C., Tian J., Luo J., Yao J., Pang R., Shen H., Peng C., Liu T., Zhang Q., Wu J., Xu L., Lu S., Wang B., Weng Z., Han C., Zhu H., Zhou R., Zhou H., Chen X., Ye P., Zhu B., Wang L., Zhou W., He S., He Y., Jie S., Wei P., Zhang J., Lu Y., Wang W., Zhang L., Li L., Zhou F., Wang J., Dittmer U., Lu M., Hu Y., Yang D., Zheng X. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020;55:102763. doi: 10.1016/j.ebiom.2020.102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H., Zhang X., Zhang M., Wu S., Song J., Chen T., Han M., Li S., Luo X., Zhao J., Ning Q. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clinic. Investig. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kalfaoglu B., Almeida-Santos J., Tye C.A., Satou Y., Ono M. T-cell hyperactivation and paralysis in severe COVID-19 infection revealed by single-cell analysis. Front. Immunol. 2020;11:589380. doi: 10.3389/fimmu.2020.589380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rodda L.B., Netland J., Shehata L., Pruner K.B., Morawski P.A., Thouvenel C.D., Takehara K.K., Eggenberger J., Hemann E.A., Waterman H.R., Fahning M.L., Chen Y., Hale M., Rathe J., Stokes C., Wrenn S., Fiala B., Carter L., Hamerman J.A., King N.P., Gale M., Campbell D.J., Rawlings D.J., Pepper M. 2020. Functional SARS-CoV-2-specific Immune Memory Persists after Mild COVID-19. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Odak I., Barros-Martins J., Bošnjak B., Stahl K., David S., Wiesner O., Busch M., Hoeper M.M., Pink I., Welte T., Cornberg M., Stoll M., Goudeva L., Blasczyk R., Ganser A., Prinz I., Förster R., Koenecke C., Schultze-Florey C.R. Reappearance of effector T cells is associated with recovery from COVID-19. EBioMedicine. 2020;57:102885. doi: 10.1016/j.ebiom.2020.102885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Newell K.L., Clemmer D.C., Cox J.B., Kayode Y.I., Zoccoli-Rodriguez V., Taylor H.E., Endy T.P., Wilmore J.R., Winslow G. The Preprint Server for Health Sciences; 2020. Switched and Unswitched Memory B Cells Detected during SARS-CoV-2 Convalescence Correlate with Limited Symptom Duration. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.