Abstract
Coronavirus Disease 2019 (COVID-19) is an acute respiratory syndrome, reported at the end of 2019 in China originally and immediately spread affecting over ten million world population to date. This pandemic is more lethal for the older population and those who previously suffered from other ailments such as cardiovascular diseases, respiratory disorders, and other immune system affecting abnormalities including cancers. Lung cancer is an important comorbidity of COVID-19. In this review, we emphasized the impact of lung tumor microenvironment (TME) on the possibility of enhanced severity of infection caused by the SARS-Co-V2. The compromised lung TME is further susceptible to the attack of viruses. The lung cells are also abundant in the virus entry receptors. Several SARS-Co-V2 proteins can modulate the lung TME by disrupting the fragile immune mechanisms contributing to cytokine storming and cellular metabolic variations. We also discussed the impact of medication used for lung cancer in the scenario of this infection. Since other respiratory infections can be a risk factor for lung cancer, COVID-19 recovered patients should be monitored for tumor development, especially if there is genetic susceptibility or it involves exposure to other risk factors.
Keywords: Covid-19, Lung cancer, Tumor microenvironment, Cytokines, Risk factors
Lung cancer
Lung cancer is the most frequent malignancy worldwide, afflicting 2.09 million new cancer cases, and 1.76 million deaths annually, thereby the leading cause of cancer-related deaths [1]. Predisposing factors for the lung cancer development include tobacco smoking, genetic polymorphisms, family history, alcohol, dietary preferences, carcinogen exposure (asbestos, silica, and polycyclic aromatic hydrocarbons), air pollution, radiations, chronic inflammation because of infections like tuberculosis and pneumonia [2].
Lung cancer is of two major types; small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). SCLC accounts for 15% of all lung cancer cases and more common in men. It is an aggressive form which originates in the bronchi and metastasizes to other body organs. NSCLC is the most common lung cancer and accounts for 80–85% cases of lung cancer cases. It originates in the mucus, squamous, or any other cell in the lungs [3]. The dilemma with lung cancer is that the symptoms rarely appear until the situation gets complicated and the cancer is already at a progressive stage. The symptoms like persistent coughing, chest pain and fatigue are mistaken for other causes, such as smoking effects or any other infection. This may delay the diagnosis, which should be within 2–4 weeks after the presentation of symptoms, hence leading to metastasis [4].
Metastasis of lung cancer occurs to the contralateral lung, adjacent lymph nodes, adrenal glands, bones, brain, and liver [5]. The process of metastasis starts with the migration of tumor cells through blood or lymphatic system to a distinct site. Upon extravasation, the tumor cell ends up in any adjacent or distant organ and starts growing there upon finding a suitable environment [6]. Different signaling pathways and epigenetic factors regulate tumor metastasis [7]. A successful metastasis depends on the imbalance between cell survival and apoptotic signals.
Tumor microenvironment in lung cancer
The effective development of tumors at primary and metastatic sites depends on the surrounding environment called tumor microenvironment (TME). A variety of stromal cells including T-cells, B-cells, Natural Killer (NK) cells, fibroblasts, adipocytes, vascular endothelial cells, and pericyte surround the growing tumor. These cells secrete signals involved in tumor survival, growth, invasion, migration, and change the behavior of cancer cells, also called oncomodulation [8]. Oncomodulation is induced by the effect of various factors that change the components of TME and alter the mechanisms acting on cancer cells and associated stromal cells such as suppressing immunity, halting normal apoptosis mechanisms, altering metabolic pathways, triggering inflammation, inducing angiogenesis, proliferation, invasion, and migration signals in cancer cells [9].
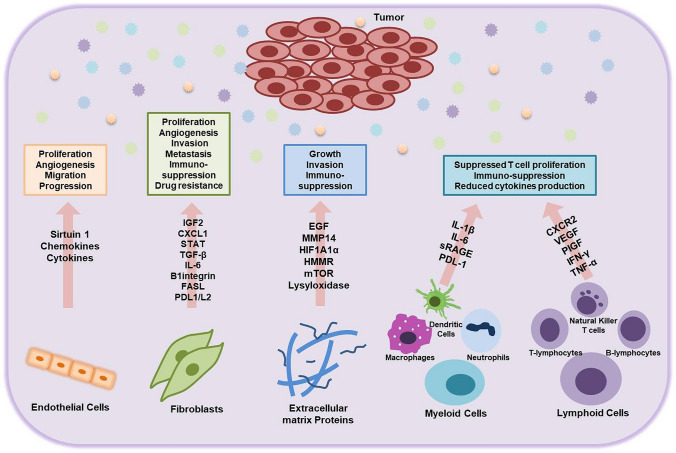
In lung cancer, the TME is significantly active. Invading cancer cells can reprogram the components of lung TME promoting carcinogenesis. Different TME components like cancer activated fibroblast, extracellular matrix, endothelial cells, immune cells that include myeloid cells (macrophages, neutrophils, dendritic cells, natural killer cells) and lymphoid cells (T and B lymphocytes) get modulated under the effect of signals from infiltrating tumor cells and further activate the cascades leading to survival and growth of tumor cell [10]. Figure 1 depicts the remodulation of the surrounding environment in lung cancer.
Fig. 1.
Tumor microenvironment (TME) in lung cancer. The TME components support the tumor growth and secrete signals and initiate cascades that facilitate the proliferation, growth, migration, invasion, and stabilization of tumor cells. They also suppress immune responses and confer drug resistance. IGF2 (insulin-like growth factor-2), CXCL1 (C-X-C Motif Chemokine Ligand 1), STAT (Signal Transducer And Activator Of Transcription), TGF-β (Transforming Growth Factor Beta), IL-6 (Interleukin-6), FASL (Fas Ligand), PDL1/L2 (Programmed cell death protein ligand 1 and 2), EGF (Epidermal growth factor), MMP14 (Matrix metalloproteinase 14), HIF1A1α (Hypoxia-inducible factor 1A1alpha), HMMR (Hyaluronan mediated motility receptor), mTOR (mammalian target of rapamycin), IL-1β (Interleukin-1beta), sRAGE (soluble Advanced Glycosylation End-Product Specific Receptor), CXCR2 (C-X-C Motif Chemokine Receptor 2), VEGF (Vascular endothelial growth factor), PIGF (Placental growth factor), IFN-γ (Interferon-gamma), TNF-α (Tumor necrosis factor-alpha)
Upon prolonged exposure to risk factors, the redesigned lung TME becomes more supportive of tumor growth. Persistent exposure to smoking leads to hypoxic conditions in the blood and other organs, including lungs leading to impaired gaseous exchange and increased inflammation [11]. This hypoxic environment induces Hypoxia-inducible factor-1 (HIF-1) expression and aggravates the malignant potential of the tumor. Increased HIF-1 expression leads to the overexpression of the genes associated with angiogenesis, cell survival, and migration in cancer cells [12, 13]. Hypoxia also causes resistance in the lung cancer cells of NSCLC against chemotherapeutic drugs such as gefitinib [14].
Lung infections cause chronic inflammation and further lead to accumulating pro-inflammatory cytokines, changing TME, and resulting in metastasis. The macrophages secrete pro-inflammatory cytokines (such as TGF-β, IL-6, IL-10, and TNF-α) and further induce stem cell like characteristics in the tumor cells, which enable them to initiate growth and sustainability [15]. The chronic exposure of these cytokines further activate several inflammatory pathways in lung malignancies such as NF-κB pathway activation by TNF-α [16], STAT3 pathway by over-expressed IL-6 [17] and enhanced TGF-β signaling pathway [18]. Prolonged inflammation also increases the E-selectin expression, which promotes hyper-permeability supporting extravasation and facilitates the homing of lung tumor cells in an unfamiliar environment [6]. Another important candidate in the lung TME cross talk is the chemokine receptor CXCR4, which creates a “pro TME” and promotes distant metastasis [19]. CXCR4 antagonists are being considered as an important therapeutic target in NSLC [20].
Taken together, this information suggests that TME plays an important factor in tumor initiation and progression. Several factors such as smoking, inflammation mediators, and infections can modulate the TME and make it more tumor friendly. Here we discuss different scenarios that COVID-19 infection can influence lung cancer patients because of TME modification.
Impact of Covid-19 on lung cancer: how TME support this infection
What is Covid-19 infection
COVID-19 is an outbreak reported in China at the end of 2019. World Health Organization (WHO) on 11 February 2020 named the novel coronavirus-induced pneumonia as Coronavirus Disease 2019 (COVID-19). The disease causative agent was named as Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by the International Virus Classification Commission [21].
The COVID-19 transmission is reported by both symptomatic and asymptomatic individuals, according to WHO. The origin of this virus is zoonotic as bats are reported as the reservoir hosts of this virus [22]. The primary mode of transmission is by close contact with the infected individual and contaminated areas [23]. The infection primarily affects the respiratory system of the human host. The incubation period of this virus is approximately five days. At this time the viral load in the upper respiratory tract is very high. The symptoms may appear after 14 days of infection [24]. Because of which SARS-CoV-2 has the potential to spread massively before being detected symptomatically.
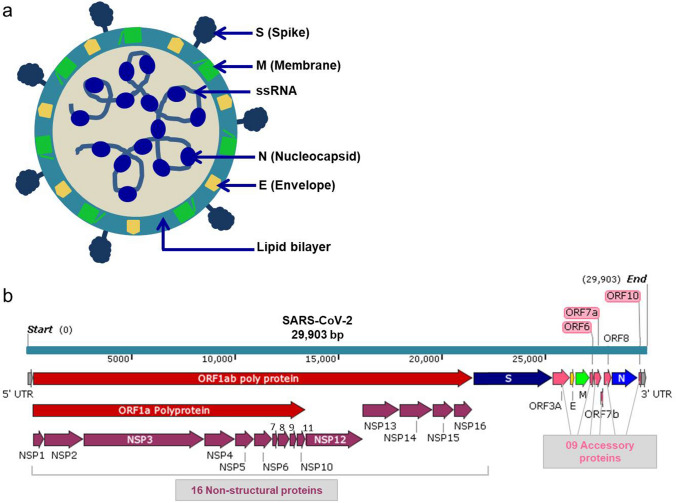
SARS-CoV-2 belongs to the genus Betacronavirus and structurally comprises the RNA genome of 29.8 kb which encodes 29 proteins including structural (n = 4), non-structural (n = 16) and accessory (n = 9) proteins. Structural proteins maintain the virus physical characters and include spike (S) protein, envelop (E) protein, membrane (M) and nucleocapsid (N) protein (Fig. 2a). S protein (YP 009724390.1) is a 141 kilo Dalton (kDa) protein and binds to the surface receptors of the host cell, mediates the entry of virus inside the host cell and starts its life-cycle by taking control of the host system. E protein (YP 009724392.1) is an 8.4 kDa hydrophobic protein and forms the outer covering of virus and essential for virus assembly or release. M protein (YP 009724393.1) is a 25 kDa highly expressed protein of SARS-CoV-2 and associated with the production of viral particles and mediate inflammatory pathways. N protein (YP 009724397.2) is a 45.6 kDa highly expressed protein in the host at the early stages of infection [25]. It binds to RNA of the virus and forms ribonucleoprotein, which facilitates in interaction with cellular processes after the entry of virus into the host. Figure 2b illustrates various regions in the genome of SARS-CoV-2 encoding 29 important proteins.
Fig. 2.
Structure of SARS-CoV-2. a A virus particle showing structural proteins and single-stranded RNA genome; b A schematic figure of viral 29.9 kb genome showing 5′UTR, location of the polyprotein, 04 structural proteins, 16 non-structural proteins, 09 accessory proteins, and 3′UTR
Along with the four structural proteins, 16 non-structural proteins (NSP) degrade the host RNA and take hold of its transcription machinery. These proteins are synthesized as two long polypeptides, one of which is auto-proteolytically processed into 16 proteins (NSP1-NSP16; with a molecular weight ranging from 1.3 to 217 kDa). Of these proteins, RNA polymerase (NSP12; YP 009725307.1) and the main protease (NSP3; YP 009725299.1) are the significant proteins which are involved in virus replication processes, promote cytokine expression, and block innate immune responses [26].
The remaining nine accessory proteins aid in the inflammation, induce apoptosis and halt antiviral responses of the host. These accessory proteins include ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF8, ORF9b, ORF9c and ORF10; with a molecular weight ranging from 4.4 to 31 kDa.
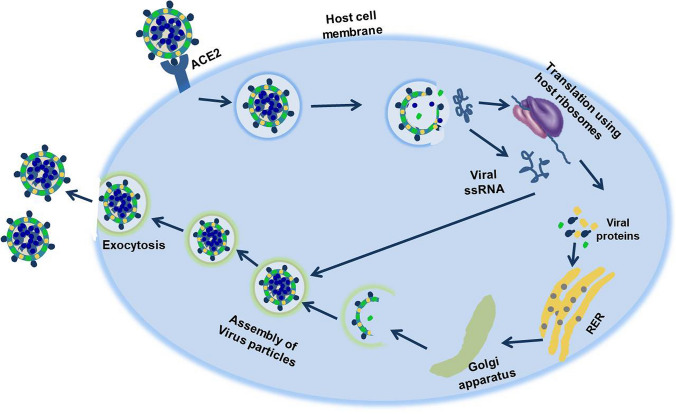
SARS-CoV-2 gets entry into the host cell by binding to the angiotensin-converting enzyme 2 (ACE2) receptors through its S protein [27, 28]. These receptors are abundantly present on the alveolar type II (AT2) epithelial cells of the respiratory tract [29]. The AT2 cells abundance is also the hallmark of lung tumor. SARS-CoV-2 infects the lower respiratory tract and can cause severe respiratory failure that may lead to a high death rate among lung cancer patients [30]. ACE2 knockout mouse model has strongly supported this idea for SARS-CoV-2 entry in pulmonary cells [31]. Another protein, TMPRSS2, involved in propagation of several viruses, including influenza virus, as previously reported, and can also mediate the SARS-CoV-2 entry [32]. The respiratory tract except the alveolar epithelium significantly expresses TMPRSS2 [33] and Matsuyama et al., [32] have shown its vulnerability towards SARS-CoV-2 entry. Recently, Meng et al. [34] have showed that the co-expression of ACE2 and TMPRSS2 and has a propensity for development of SARS-CoV-2 infection. Another preliminary study reported that the elevated expression of CD147 in tumor cells facilitates virus entry by binding to S protein [35]. Moreover, integrins are being investigated for its role in SARS-CoV-2 entry, but a solid confirmation is still lacking [36]. Following the binding of host receptor to virus S protein, the proteolytic activity in S-protein by host proteases fuses the virus envelope to the host membrane. Once inside, the virus gets hold of the cell translation machinery and starts translating its replicase gene and other non-structural components. This translation is followed by the synthesis of genomic and sub-genomic virus RNA. Sub-genomic RNA translates into the structural proteins and going through host cytoplasmic compartments leads to the assembly of the mature virus, which are then transported to host cell surface and are exocytosed (Fig. 3) [37, 38]. SARS-CoV-2 genome like other coronaviruses has a tendency for recombination and this makes it excessively adaptable to different conditions [39]. In two independent studies, SARS-CoV-2 is shown to use neuropilin-1 (NRP1) for getting entry into the host cell by binding through spike protein.
Fig. 3.
SARS-CoV-2 binds to ACE2 receptors on the host cell surface and gains entry. Once inside it can start its life cycle by using components of the host cell. The virus makes multiple copies that leave by exocytosis to infect other cells
Possible effects of Covid-19 infection on lung cancer patients
Cancer patients are more susceptible to COVID-19 infection than individuals without cancer. This could be because of systemic reduced immunity or anticancer therapy being offered to these cancer patients. The situation could be worse in lung cancer patients as they already have chronic pulmonary inflammation because of the underlying TME and lung pathology [40]. The already hostile lung environment may offer suitability for the virus entry, growth, and infection. So, lung cancer can have significant comorbidity for the SARS-CoV-2 infection. The similarity between the symptoms of lung cancer and SARS-CoV-2 infection can also delay the diagnosis of infection in these individuals which can lead to further deterioration of lungs [41]. Overall, cancer patients are more prone to acquire this infection and have poor survival. In a Chinese study, lung cancer was the most reported type of cancer patients carrying COVID-19 infection [42]. A multicenter retrospective study was performed on 205 cancer patients with SARS-CoV-2 infection from China. Lung cancer was one of the most common solid tumors and reported in 12% of all cases [43]. Another multicenter study during the COVID-19 pandemic was performed on 105 cancer patients and 536 age-matched non-cancer patients confirmed with COVID-19 from China [44]. Lung cancer was the most frequent cancer and reported in 21% of all cancer cases. Lung cancer patients reported the highest frequency of severe events including death rate (18.18%), ICU admission rate (27.27%), risks of critical symptoms (50.00%), and the chance of use of invasive mechanical ventilation (18.18%) of reported lung cancer patients. The increased risk of infection in lung cancer can also be because of the abundance of viral spike protein binding receptors to the host cells in the lungs. The ACE2 receptors are expressed on the lung capillaries [45] and can make the situation worse by facilitating infection in such patients. Recently two lung cancer patients with COVID-19 infection underwent lung lobectomies. Pathological examination of lung tissues showed edema, proteinaceous exudate, patchy inflammatory infiltrates, and multinucleated giant cells [46].
The TME in the lung could also be very supportive of the viral proteins, which can act as the signals for activation of several inflammatory pathways. These possibilities are discussed further in this review.
Oncomodulation due to Covid-19 infection
Several studies have shown how a virus can alter the processes in the tumor and its surrounding stromal cells. The signaling cascades will be affected and lead to altered metabolism, immunosuppression, decreased apoptosis, transformed cellular communications, and increased angiogenesis. The crosstalk between the viral proteins and inflammatory mediators oncomodulates the TME resulting in increased tumor growth and survival chances [47, 48].
SARS-CoV-2 proteins might interact with the components of TME and may support tumor growth or cause resistance to the ongoing therapy. A detailed proteomic analysis of host cell (SARS-CoV-2 infected colon carcinoma cell line) after the infection was performed [49]. The SARS-CoV-2 infection affected several pathways. On closer observation of the proteins whose expression is upregulated or downregulated in response to infection give clues about the effects on the TME. Tumor cell metabolism utilizes glucose and glutamine more efficiently than normal cells which help tumor growth. Excessive utilization of glucose by growing tumor cells deprive TME of glucose and competition between tumor and T cells for glucose starts [50] resulting in decreased IFN-γ hindering T cell antitumor activity. Increased glycolysis in tumor cells results in lactate accumulation in TME making it acidic and decrease T cell proliferation and INF-gamma levels. In lung cancer, it is also demonstrated that lactate upregulates the expression of “Programmed cell death ligand-1 (PDL-1)” starting PD-1 signaling in T-cells and suppressing the immune system [51].
The increased extracellular glutamine levels also make tumor cells more aggressive, and with lesser survival period for patients. This increased utilization of glutamine is supported by viral infections [52]. SARS-CoV-2 infected colon cancer cells have shown that a set of genes involved in the metabolic pathways get altered by this infection [49]. Also, the TME is usually hypoxic and nutrient deprived which supports suppressed immune response providing a favorable environment for viral growth.
Cytokines rush is the phenomenon characterized by increased production of cytokines leading to an elevated inflammatory response. It can occur in tumor cells because of several underlying factors and with immune based therapeutics [53]. SARS-CoV-2 infection can fuel up this process, causing the top mortality rates in cancer patients. When the viruses attack the respiratory tract cells, the cytokine response is triggered in a variety of ways. Pro-inflammatory chemokines are released in response to infection and further recruit immune system cells including lymphocytes, macrophages, and neutrophils to the site and increase inflammation and immune suppression. Along with chemokines, interleukins, tumor necrosis factors, and interferon-gamma are rushed to the site to release cytokines. These excessive cytokines which initially affect the TME, get leaked from the site of infection in the respiratory tract and end up in other organs where they produce tumor promoting effects [54]. However, host susceptibility to this storm depends on the underlying genetics of the host innate immune system [55].
SARS-CoV-2, NSP3 affects the host innate immune mechanisms and promotes cytokine production which may lead to the storming of cells with pro-inflammatory cytokines. ORF6 and ORF9b, the accessory peptides in the SARS-CoV-2 genome, act as type I IFNs antagonists. Type I interferon has a protective function for tumor cells as they activate the JAK-STAT pathway leading to the expression of transcription factors and genes with antiviral, antiproliferative, and immunomodulatory functions. These viral peptides (ORF6 and ORF9b) can block IFN and may lead to increased proliferation and tumor survival. SARS-CoV-2 can also modify the host immune system through another accessory peptide, ORF3a which can activate the NLRP3 (nucleotide-binding domain (NOD)-like receptor protein 3) inflammasome. This activation produces excessive IL-1β and IL-18 in response to infection, but increased pro-inflammatory signals can make the situation worse. It is noteworthy that the activity of this inflammasome is regulated by type I IFN which is also affected by other viral proteins discussed before so a shift in type I IFN level has the potential to change the scenario in TME of the lung.
TNF-α mediated NF-κB pathway activation and TGF- β signaling in TME are crucial as these cytokines regulate growth, proliferation and differentiation, and apoptosis [56]. However, the destruction of cellular mRNA by viral proteins (such as NSP1) can lead to the destruction of the inhibitory molecules for these pathways triggering their activation, thus promoting inflammation of lung tissues.
Similarly, ORF7 peptide can induce apoptosis in host cells and NSP2 can bind to a host protein “Prohibitin” which is a regulator of cell proliferation, several transcription factors, and mitochondrial processes. The functions of several other SARS-CoV-2 proteins in the host cells still need to be determined.
Ongoing therapy and fighting COVID-19 infection
As the SARS-CoV-2 dampens down the immune system of the host, a question arises if the individuals are on immune supportive therapy can better fight with this infection than those without such therapy. Because of the symptoms resemblance between lung cancer and COVID-19, it may affect similar mechanisms and cascades. Based on this, we have assessed different chemotherapeutic and immunotherapeutic drugs being administered to lung cancer patients.
The immunotherapy in lung cancer patients targets either the Programmed cell Death-1 receptor and its ligand (PD-1/PDL-1) [57] such as pembrolizumab, nivolumab, atezolizumab, and durvalumab or cytotoxic T-lymphocyte associated protein 4 (CTLA-4) [58] such as Ipilimumab. Conflicting data reported effective, adverse, or with no effect of anti-PD-1/PDL-1 or anti-CTLA-4 therapies to lung cancer patients. This variation can be attributed to the diverse innate immune system of the studied participants. However, a conclusion cannot be drawn from these studies because of the small sample size of available data. Also, the hyperactivation of the immune response in lung cancer patients or the cytokine storm because of immunotherapy may lead to injury in other body organs and increase the severity of damage leading to death.
The chemotherapeutic drugs given for cancer treatment may also provide additional benefits against SARS-CoV-2 peptides. Gordon et al., [26] developed an interactome of human proteins and SARS-CoV-2 peptides. Further, they sought the ligands interacting with those human proteins and then used viral peptides as bait to determine their interaction with the ligands. From the data, several chemotherapeutic compounds can be spotted, which might be the candidates against SARS-CoV-2 peptides. The Casein Kinase 2 inhibitor (Silmitasertib), HDAC2 inhibitor (Valproic Acid), RIPK1 inhibitor (Ponatinib), Protein kinase inhibitor MARK1/3 (Midostaurin), Topoisomerase Inhibitor (Daunorubicin) and NEK9 inhibitor (Dabrafenib) interacted with N protein, NSP5, NSP12, ORF9b, ORF9c, and NSP9 when used as baits. However, verifiable data from patients is still missing and the clinical manifestation for these medications against COVID-19 infection is also required.
Prospects of COVID-19 leading to lung cancer
The chances of pulmonary infections to develop into lung cancer are very high. Pneumonia, tuberculosis, and influenza infections are potential risk factors for developing lung cancer [2]. As SARS-CoV-2 infects the respiratory tract and leads to similar symptoms, it might trigger some cascades which facilitate the modification of the lung environment for tumor appearance and growth. The situation could be more alarming for the SARS-CoV-2 infected cases where genetic predisposition or other risk factors like smoking are also involved.
At a later stage in COVID-19 with respiratory distress, a hyper-activated immune system is observed. This can cause cytokine rush leading to multiple organ damage. It is previously shown that acute lung injury (ALI) or acute respiratory distress syndrome (ARDS) caused by infections can lead to inflammatory response and can also predispose for lung cancer development [59]. ALI represents the most severe outcome of SARS-CoV-2 infection [60]. The precise mechanism of ARDS/ALI in COVID-19 is still unclear. The predominant factor noted in several studies is age of the patient [61]. Two hospital-based studies from China and Singapore investigated the clinical features of typical ARDS and COVID-19 related ARDS cases. The lung pathology was similar in both groups, but thrombosis and mortality were more common in cases of COVID-19 ARDS [62]. A similar study from Italy reported the same findings among COVID-19 related and unrelated ARDS cases [63]. Being heterogeneous, the management of ARDS is being tailored in context to the over whelming pressure on health care systems because of COVID.
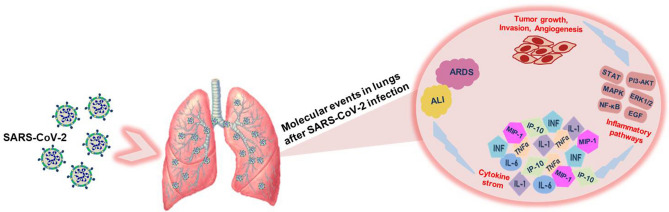
The symptoms of ARDS and ALI are overlapping with the lung cancer and it can be predicted that SARS-CoV-2 infection can lead to cancer. The early players of the inflammatory pathway after acute lung damage are TNF-α, IL-1β, IL-8, and IL-6 which are very active mediators of inflammatory pathways, whereas the anti-inflammatory cytokines appear late [64]. Figure 4 explains the connection between the lung infections and cancer. The monoclonal anti-IL-6 receptor antibody, tocilizumab emerges to be an effective treatment choice in COVID-19 patients who are at risk of a cytokine storm. Significantly improved clinical, biological, and radiological outcomes are noted within 5–7 days after tocilizumab treatment [65].
Fig. 4.
Molecular events predicting the development of lung tumor in response to SARS-CoV-2 infection. ALI (acute lung injury), ARDS (acute respiratory distress syndrome), IFN (Interferons), TNF-α (tumor necrosis factor-alpha), IL-6 (interleukin-6), IL-1 (interleukin-1), MIP-1 (macrophage inflammatory protein-1), IP-10 (interferon gamma-induced protein 10), STAT (signal transducers and activators of transcription), MAPK (mitogen-activated protein kinase), NF-κB (nuclear factor kappa B), ERK1/2 (extracellular receptor kinase), EGF (epidermal growth factor), PI3-AKT (phosphatidylinositol 3 kinase- protein kinase B)
Conclusion
This review summarizes that TME has an important role in determining the route of tumor development. The lung tumor environment supports for infectious agents like SARS-CoV-2 and can expedite infection. We conclude from the evidence to date, the immune and chemotherapeutic drugs administered during lung cancer have no convincing evidence of being supportive in case of accompanying SARS-CoV-2 infection. We propose that the COVID-19 recovered patients should pay unusual attention to the symptoms of lung cancer, as the viral infection may have elicited any tumor initiation mechanisms.
Acknowledgements
None.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Research involving human and/or animals participants
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Malhotra J, Malvezzi M, Negri E, La Vecchia C, Boffetta P. Risk factors for lung cancer worldwide. Eur Respir J. 2016;48:889–902. doi: 10.1183/13993003.00359-2016. [DOI] [PubMed] [Google Scholar]
- 3.Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, Chirieac LR, Dacic S, Duhig E, Flieder DB, et al. The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10:1243–1260. doi: 10.1097/JTO.0000000000000630. [DOI] [PubMed] [Google Scholar]
- 4.Ellis PM, Vandermeer R. Delays in the diagnosis of lung cancer. J Thorac Dis. 2011;3:183–188. doi: 10.3978/j.issn.2072-1439.2011.01.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rami-Porta R, Call S, Dooms C, Obiols C, Sánchez M, Travis WD, Vollmer I. Lung cancer staging: a concise update. Eur Respir J. 2018;51:1800190. doi: 10.1183/13993003.00190-2018. [DOI] [PubMed] [Google Scholar]
- 6.Popper HH. Progression and metastasis of lung cancer. Cancer Metastasis Rev. 2016;35:75–91. doi: 10.1007/s10555-016-9618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fares J, Fares MY, Khachfe HH, Salhab HA, Fares Y. Molecular principles of metastasis: a hallmark of cancer revisited. Signal Transduct Target Ther. 2020;5:28. doi: 10.1038/s41392-020-0134-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baghban R, Roshangar L, Jahanban-Esfahlan R, Seidi K, Ebrahimi-Kalan A, Jaymand M, Kolahian S, Javaheri T, Zare P. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun Signal. 2020;18:59. doi: 10.1186/s12964-020-0530-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whiteside TL. The tumor microenvironment and its role in promoting tumor growth. Oncogene. 2008;27:5904–5912. doi: 10.1038/onc.2008.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altorki NK, Markowitz GJ, Gao D, Port JL, Saxena A, Stiles B, McGraw T, Mittal V. The lung microenvironment: an important regulator of tumor growth and metastasis. Nat Rev Cancer. 2019;19:9–31. doi: 10.1038/s41568-018-0081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fricker M, Goggins BJ, Mateer S, Jones B, Kim RY, Gellatly SL, Jarnicki AG, Powell N, Oliver BG, Radford-Smith G, Talley NJ, Walker MM, Keely S, Hansbro PM. Chronic cigarette smoke exposure induces systemic hypoxia that drives intestinal dysfunction. JCI Insight. 2018;3:e94040. doi: 10.1172/jci.insight.94040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mittal V, El Rayes T, Narula N, McGraw TE, Altorki NK, Barcellos-Hoff MH. The microenvironment of lung cancer and therapeutic implications. Adv Exp Med Biol. 2016;890:75–110. doi: 10.1007/978-3-319-24932-2_5. [DOI] [PubMed] [Google Scholar]
- 13.Wood SL, Pernemalm M, Crosbie PA, Whetton AD. The role of the tumor-microenvironment in lung cancer-metastasis and its relationship to potential therapeutic targets. Cancer Treat Rev. 2014;40:558–566. doi: 10.1016/j.ctrv.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 14.Murakami A, Takahashi F, Nurwidya F, Kobayashi I, Minakata K, Hashimoto M, Nara T, Kato M, Tajima K, Shimada N, Iwakami S, Moriyama M, Moriyama H, Koizumi F, Takahashi K. Hypoxia increases gefitinib-resistant lung cancer stem cells through the activation of insulin-like growth factor 1 receptor. PLoS ONE. 2014;9:e86459. doi: 10.1371/journal.pone.0086459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y, Tan W, Wang C. Tumor-associated macrophage-derived cytokines enhance cancer stem-like characteristics through epithelial-mesenchymal transition. Onco Targets Ther. 2018;11:3817–3826. doi: 10.2147/OTT.S168317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen W, Li Z, Bai L, Lin Y. NF-kappaB in lung cancer, a carcinogenesis mediator and a prevention and therapy target. Front Biosci. 2011;16:1172–1185. doi: 10.2741/3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, Luo F, Xu Y, Wang B, Zhao Y, Xu W, Shi L, Lu X, Liu Q. Epithelial-mesenchymal transition and cancer stem cells, mediated by a long non-coding RNA, HOTAIR, are involved in cell malignant transformation induced by cigarette smoke extract. Toxicol Appl Pharmacol. 2015;282:9–19. doi: 10.1016/j.taap.2014.10.022. [DOI] [PubMed] [Google Scholar]
- 18.Saito A, Horie M, Nagase T. TGF-β signaling in lung health and disease. Int J Mol Sci. 2018;19:2460. doi: 10.3390/ijms19082460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang JX, Gao W, Liang Y, Zhou XM. Chemokine receptor CXCR4 expression and lung cancer prognosis: a meta-analysis. Int J Clin Exp Med. 2015;8:5163–5174. [PMC free article] [PubMed] [Google Scholar]
- 20.Wald O. CXCR4 based therapeutics for non-small cell lung cancer (NSCLC) J Clin Med. 2018;7:303. doi: 10.3390/jcm7100303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA, Haagmans BL, Lauber C, Leontovich AM, Neuman BW, Penzar D. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mackenzie JS, Smith DW. COVID-19: a novel zoonotic disease caused by a coronavirus from China: what we know and what we don't. Microbiol Aust. 2020 doi: 10.1071/MA20013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malik YA. Properties of Coronavirus and SARS-CoV-2. Malays J Pathol. 2010;42:3–11. [PubMed] [Google Scholar]
- 24.Lauer SA, Grantz KH, Bi Q, Jones FK, Zheng Q, Meredith HR, Azman AS, Reich NG, Lessler J. The Incubation Period of Coronavirus Disease 2019 (COVID-19) From publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020;172:577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dutta NK, Mazumdar K, Gordy JT. The nucleocapsid protein of SARS–CoV-2: a target for vaccine development. J Virol. 2020 doi: 10.1128/JVI.00647-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, O'Meara MJ, Rezelj VV, Guo JZ, Swaney DL, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020 doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santos RAS. Angiotensin-(1–7): a comprehensive review. Cham: Springer Publications; 2019. p. 132. [Google Scholar]
- 30.Ferone G, Lee MC, Sage J, Berns A. Cells of origin of lung cancers: lessons from mouse studies. Genes Dev. 2020;34:1017–1032. doi: 10.1101/gad.338228.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuba K, Imai Y, Rao S, Gao H, Guo F, Guan B, Huan Y, Yang P, Zhang Y, Deng W, Bao L. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus–induced lung injury. Nat Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuyama S, Nao N, Shirato K, Kawase M, Saito S, Takayama I, Nagata N, Sekizuka T, Katoh H, Kato F, Sakata M. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. PNAS. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matusiak M, Schürch CM. Expression of SARS-CoV-2 entry receptors in the respiratory tract of healthy individuals, smokers and asthmatics. Respir Res. 2020;21:1–6. doi: 10.1186/s12931-020-01521-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meng T, Cao H, Zhang H, Kang Z, Xu D, Gong H, Wang J, Li Z, Cui X, Xu H, Wei H. The insert sequence in SARS-CoV-2 enhances spike protein cleavage by TMPRSS. BioRxiv. 2020 doi: 10.1101/2020.02.08.926006. [DOI] [Google Scholar]
- 35.Xia P, Dubrovska A. Tumor markers as an entry for SARS-CoV-2 infection? FEBS J. 2020;287:3677–3680. doi: 10.1111/febs.15499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sigrist CJ, Bridge A, Le Mercier P. A potential role for integrins in host cell entry by SARS-CoV-2. Antiviral Res. 2020;177:104759. doi: 10.1016/j.antiviral.2020.104759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schoeman D, Fielding BC. Coronavirus envelope protein: current knowledge. Virol J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang YZ, Holmes EC. A genomic perspective on the origin and emergence of SARS-CoV-2. Cell. 2020;181:223–227. doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ballaz S, Mulshine JL. The potential contributions of chronic inflammation to lung carcinogenesis. Clin Lung Cancer. 2003;5:46–62. doi: 10.3816/CLC.2003.n.021. [DOI] [PubMed] [Google Scholar]
- 40.Moujaess E, Kourie HR, Ghosn M. Cancer patients and research during COVID-19 pandemic: a systematic review of current evidence. Crit Rev Oncol Hematol. 2020;150:102972. doi: 10.1016/j.critrevonc.2020.102972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang W, Guan W, Chen R, Wang W, Li J, Xu K, Li C, Ai Q, Lu W, Liang H, Li S, He J. Cancer patients in SARS-CoV-2 infection: a nationwide analysis in China. Lancet Oncol. 2020;21:335–337. doi: 10.1016/S1470-2045(20)30096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang K, Sheng Y, Huang C, Jin Y, Xiong N, Jiang K, Lu H, Liu J, Yang J, Dong Y, et al. Clinical characteristics, outcomes, and risk factors for mortality in patients with cancer and COVID-19 in Hubei, China: a multicentre, retrospective, cohort study. Lancet Oncol. 2020;21:904–913. doi: 10.1016/S1470-2045(20)30310-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dai M, Liu D, Liu M, Zhou F, Li G, Chen Z, Zhang Z, You H, Wu M, Zheng Q, et al. Patients with cancer appear more vulnerable to SARS-CoV-2: a multicenter study during the COVID-19 outbreak. Cancer Discov. 2020;10:783–791. doi: 10.1158/2159-8290.CD-20-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riordan JF. Angiotensin-I-converting enzyme and its relatives. Genome Biol. 2003;4:225. doi: 10.1186/gb-2003-4-8-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tian S, Hu W, Niu L, Liu H, Xu H, Xiao SY. Pulmonary pathology of early-phase 2019 novel coronavirus (COVID-19) pneumonia in two patients with lung cancer. J Thorac Oncol. 2020;15:700–704. doi: 10.1016/j.jtho.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blaylock RL. Viruses and tumor cell microenvironment: a brief summary. Surg Neurol Int. 2019;10:160. doi: 10.25259/SNI_351_2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Vries CR, Kaufman HL, Lattime EC. Oncolytic viruses: focusing on the tumor microenvironment. Cancer Gene Ther. 2015;22:169–171. doi: 10.1038/cgt.2015.11. [DOI] [PubMed] [Google Scholar]
- 48.Bojkova D, Klann K, Koch B, Widera M, Krause D, Ciesek S, Cinatl J, Münch C. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020 doi: 10.1038/s41586-020-2332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roy M, Finley SD. Metabolic reprogramming dynamics in tumor spheroids: Insights from a multicellular, multiscale model. PLoS Comput Biol. 2019;15:e1007053. doi: 10.1371/journal.pcbi.1007053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de la Cruz-López KG, Castro-Muñoz LJ, Reyes-Hernández DO, García-Carrancá A, Manzo-Merino J. Lactate in the regulation of tumor microenvironment and therapeutic approaches. Front Oncol. 2019;9:1143. doi: 10.3389/fonc.2019.01143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shimabukuro-Vornhagen A, Gödel P, Subklewe M, Stemmler MJ, Schlößer HA, Schlaak M, Kochanek M, Böll B, von Bergwelt-Baildon MS. Cytokine release syndrome. J Immunother Cancer. 2018;6:56. doi: 10.1186/s40425-018-0343-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woo YL, Kamarulzaman A, Augustin Y, Staines H, Altice F, Krishna S. A genetic predisposition for cytokine storm in life-threatening COVID-19 infection. OSF Preprints. 2020 doi: 10.31219/osf.io/mxsvw. [DOI] [Google Scholar]
- 54.Tisoncik JR, Korth MJ, Simmons CP, Farrar J, Martin TR, Katze MG. Into the eye of the cytokine storm. Microbiol Mol Biol Rev. 2012;76:16–32. doi: 10.1128/MMBR.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.D'Elia RV, Harrison K, Oyston PC, Lukaszewski RA, Clark GC. Targeting the "cytokine storm" for therapeutic benefit. Clin Vaccine Immunol. 2013;20:319–327. doi: 10.1128/CVI.00636-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schulze AB, Schmidt LH. PD-1 targeted Immunotherapy as first-line therapy for advanced non-small-cell lung cancer patients. J Thorac Dis. 2017;9:E384–E386. doi: 10.21037/jtd.2017.03.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mooradian MJ, Gainor JF. Putting the brakes on CTLA-4 inhibition in lung cancer? Transl Lung Cancer Res. 2018;7:S35–S38. doi: 10.21037/tlcr.2018.01.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Young AY, Shannon VR. Acute respiratory distress syndrome in cancer patients. Oncol Crit Care. 2019 doi: 10.1007/978-3-319-74588-6_48. [DOI] [Google Scholar]
- 59.Luke JJ, Ascierto PA. Biology confirmed but biomarkers elusive in melanoma immunotherapy. Nat Rev Clin Oncol. 2020;17:198–199. doi: 10.1038/s41571-020-0328-8. [DOI] [PubMed] [Google Scholar]
- 60.Cai A, McClafferty B, Benson J, Ramgobin D, Kalayanamitra R, Shahid Z, Groff A, Aggarwal CS, Patel R, Polimera H, Vunnam R. COVID-19: catastrophic cause of acute lung injury. S D Med. 2020;73:252–260. [PubMed] [Google Scholar]
- 61.Acosta MA, Singer BD. Pathogenesis of COVID-19-induced ARDS: implications for an ageing population. Eur Respir J. 2020 doi: 10.1183/13993003.02049-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gibson PG, Qin L, Puah SH. COVID-19 acute respiratory distress syndrome (ARDS): clinical features and differences from typical pre-COVID-19 ARDS. Med J Aust. 2020;213:54–56. doi: 10.5694/mja2.50674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grasselli G, Tonetti T, Protti A, Langer T, Girardis M, Bellani G, Laffey J, Carrafiello G, Carsana L, Rizzuto C, Zanella A. Pathophysiology of COVID-19-associated acute respiratory distress syndrome: a multicentre prospective observational study. Lancet Respir Med. 2020;8:1201–1208. doi: 10.1016/S2213-2600(20)30370-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stroud CR, Hegde A, Cherry C, Naqash AR, Sharma N, Addepalli S, Cherukuri S, Parent T, Hardin J, Walker P. Tocilizumab for the management of immune mediated adverse events secondary to PD-1 blockade. J Oncol Pharm Pract. 2019;25:551–557. doi: 10.1177/1078155217745144. [DOI] [PubMed] [Google Scholar]