Abstract
In recent years, a total of seven human pathogenic coronaviruses (HCoVs) strains were identified, i.e., SARS-CoV, SARS-CoV-2, MERS-CoV, HCoV-OC43, HCoV-229E, HCoV-NL63, and HCoV-HKU1. Here, we performed an analysis of the protease recognition sites and antigenic variation of the S-protein of these HCoVs. We showed tissue-specific expression pattern, functions, and a number of recognition sites of proteases in S-proteins from seven strains of HCoVs. In the case of SARS-CoV-2, we found two new protease recognition sites, each of calpain-2, pepsin-A, and caspase-8, and one new protease recognition site each of caspase-6, caspase-3, and furin. Our antigenic mapping study of the S-protein of these HCoVs showed that the SARS-CoV-2 virus strain has the most potent antigenic epitopes (highest antigenicity score with maximum numbers of epitope regions). Additionally, the other six strains of HCoVs show common antigenic epitopes (both B-cell and T-cell), with low antigenicity scores compared to SARS-CoV-2. We suggest that the molecular evolution of structural proteins of human CoV can be classified, such as (i) HCoV-NL63 and HCoV-229E, (ii) SARS-CoV-2, and SARS-CoV and (iii) HCoV-OC43 and HCoV-HKU1. In conclusion, we can presume that our study might help to prepare the interventions for the possible HCoVs outbreaks in the future.
Keywords: Human coronavirus, Protease recognition sites, Antigenic variation, S-protein, Molecular phylogenetics
1. Introduction
Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) gained universal attention in December 2019 after the onset of a respiratory outbreak in China. It now poses a global threat as confirmed cases spread over 191 countries (Chakraborty et al., 2020a; Chakraborty et al., 2020b; Wang et al., 2020a). Being a member of β-coronavirus (CoV) group, SARS-CoV-2 is presumed to be linked with the previous two outbreaks of SARS-CoV in China (in the year 2002 and 2003), and Middle East respiratory syndrome coronavirus (MERS-CoV) in the Middle East (in the year 2012) (Chakraborty et al., 2020a). In addition to SARS-CoV-2, SARS-CoV, and MERS-CoV, other CoVs known to cause infection in humans are HCoV-OC43, HCoV-229E, HCoV-NL63, and HCoV-HKU1. It has been established that these four CoVs strains do not impose major health threats; however, they might cause respiratory illnesses, including pneumonia (Zeng et al., 2018). Therefore, there are a total of seven CoVs strains that can infect the human species. Understanding the molecular similarities and variations among SARS-CoV-2 with different strains of CoVs can provide insights into their pathogenesis that might further help in the identification of appropriate targets for intervention (Pal et al., 2020).
Members of Coronaviridae show the remarkable ability for interspecies transmission and zoonotic outbreaks (Bosch and Rottier, 2007; Cutler et al., 2010; Woo et al., 2009). It is possible that their zoonotic potential is due to the viral entry process, which might help to infect the target cell (Belouzard et al., 2012). It is well observed that enveloped viruses can show infectivity only when they can invade the target host cells and incorporate their genome in the host cell. Viral entry is mediated by one or more surface proteins displayed on the viral envelope that induce specific attachment to one or more host cell receptors, followed by fusion of the viral envelope with the cell membrane. Following receptor adherence, a dedicated viral fusion protein induces fusion of membrane and allows internalization of the virus via endocytosis (Simons et al., 1982).
It is known that an envelope-anchored spike (S) protein with a dual function of mediating receptor binding and fusion directs CoV entry into the host cells (Li, 2016; Mocarski et al., 2013). An excess signal, such as proteolytic activation, receptor binding, and low pH, is employed by CoV, which allows the liberation of the fusion peptide into target membranes at a particular time (Bosch and Rottier, 2007). S-protein is a class I fusion viral protein that is stimulated for fusion by a variety of proteases of the host cell (Belouzard et al., 2012; White et al., 2008). The S-protein has two types of ectodomains (Mocarski et al., 2013) that are further separated into two functional domains, i.e., S1 subunit for ACE2 receptor-binding and S2 subunit for fusion mechanism. Host cellular proteases are responsible for the cleavage and activation of the S-protein that enables the S2 unit for the fusion. As a result, SARS-CoV-2 takes entrance into the susceptible cells by the combined action of proteolytic processing as well as receptor-binding of the S-protein to support host cell-virus fusion (Walls et al., 2020). The membrane fusion of the CoV S-protein needs two initial cleavages by proteases of the host at the portion of the S1-S2 boundary. The fusion peptide, located adjacent to the S2′ site, comprises a highly conserved motif called I-E-D-L-L-F in all the seven strains under study (Burkard et al., 2014; Millet and Whittaker, 2015; Mocarski et al., 2013). Apart from these well-characterized cleavage sites, the S-protein can also be cleaved at several other less-characterized locations by a multitude of proteases. Based on CoV's strain and the type of infected host cell, the cleavage phenomena occur at several sites and different stages of the CoV infection cycle. It may be significant in cell/tissue tropism, modulating pathogenicity, and host range of the virus (Li, 2016; Mocarski et al., 2013). A large number of cellular proteases, namely, proprotein convertase (e.g., furin and furin-like proteases), trypsin, cathepsins, elastase, plasmin, TMPRSS2 (transmembrane protease, serine 2), and several others have recognition sites in variable number and at various locations in different CoV strains (Belouzard et al., 2010; Gierer et al., 2013; Kam et al., 2009; Matsuyama et al., 2005; Shirato et al., 2013; Simmons et al., 2013; Williamson et al., 2013). However, all of them may not be functionally relevant as their efficiency in cleavage scores is low. Therefore, there is an urgent need to identify the putative protease recognition sites of seven CoVs along with this pandemic virus. Additionally, presumed protease recognition sites of SARS-CoV-2 can help us to understand its pathomechanism.
It has been noted that S-protein has several antigenic sites, which are the most important target for vaccine development (Bhattacharya et al., 2020b; Giurgea et al., 2020). The epitopes in S-protein in both SARS-CoV-2 and SARS-CoV can help develop antibodies (Tian et al., 2020; Yuan et al., 2020). Therefore, it is necessary to identify the variations among the antigenic sites in the S-protein of seven CoVs pathogenic to humans. Moreover, data on the evolutionary relationships of the human pathogenic strains of CoVs is limited, and further research work is required in this direction.
Therefore, we first identified the protease recognition sites of seven CoVs, including SARS-CoV-2. We also predicted S-protein's antigenicity and identified the antigenic sites in S-protein of seven human coronaviruses (HCoVs) strains via identifying the multi-epitopic regions of B-cells and T-cells using immunoinformatics tools. Further, we compared the antigenicity of SARS-CoV-2 with other HCoVs. Finally, we mapped the evolutionary relationships, sequence similarity, align positions (align blocks) analysis of spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins among human pathogenic CoVs.
2. Materials and methods
2.1. Retrieval of proteomic data
The S-protein sequences of the seven different strains of HCoVs were collected from the NCBI (Coordinators, 2017). For protease recognition sites, the S-protein of the seven different strains of HCoVs information (numbers of amino acids, accession ID, and version) were retrieved, which are shown in supplementary table S1. For the prediction of antigenicity and identification of antigenic sites of the S-protein, the respective retrieved protein information (numbers of amino acids, accession ID, and version) are shown in supplementary table S2. Furthermore, we have collected amino acid sequences for all structural proteins (S-, M-, E-, N-proteins) of seven HCoVs pathogens from NCBI to analyze the evolutionary relationships, sequence similarity, and align positions (Supplementary table S3 to S9).
2.2. Mapping of the protease recognition sites and tissue-specific expression pattern analysis of the proteases
A segment of S-protein was selected (varying in length from 66 to 210 amino acids) encompassing the S1/S2 and S2′ junctions, along with a portion of subunit 2 in between for comparative assessment of protease recognition sites in the seven strains of CoVs.
The target amino acid sequences from each of the seven strains were then scanned using Procleave and PROSPER: Protease substrate specificity web server (Li et al., 2020; Song et al., 2012). In Procleave, all possible protease recognition sites were identified with cleavage scores ranging from 0.02 to 0.9. In contrast, in PROSPER, there was a rigidity in the number of proteases identified, and their putative recognition sites showed a cleavage score from 0.6 to 1.3. Since a higher score indicates a strong prediction of cleavage, proteases that were expressed in human epithelium with cleavage score > 0.7 were considered for further analysis from both the web servers.
For tissue-specific expression pattern analysis, an extensive literature survey complemented by data from GeneCards ((Safran et al., 2010; Stelzer et al., 2011) and The Human Protein Atlas ((Thul and Lindskog, 2018; Uhlen et al., 2010) was used to validate the tissue-specific expression pattern and function of the proteases. A putative map of the protease recognition site in the target amino acid sequence of SARS-CoV-2 was generated to understand the virus's high infectivity and disease severity compared to the other six related strains.
2.3. Estimation of location and solvent accessibility of protease recognition sites in S glycoprotein
With reference to the atomic model of closed SARS-CoV-2 S glycoprotein ectodomain trimer (PDB 6VXX) (Walls et al., 2020) the solvent accessibility of the candidate protease recognition sites were cross-examined in one of the three monomers (monomer A) constituting the protein structure using PDBePISA server (Krissinel and Henrick, 2007). It is a bioinformatics tool that explores the interface of biological macromolecules. A similar assessment was done for the open S-protein trimer (PDB 6VYY). The accessibility was further validated together with allocating the cleavage sites in the protein structure using PyMOL software (DeLano, 2002).
2.4. Identification and selection of B-cell epitopes
The amino acid sequences of selected seven different strains of HCoVs were explored for accurate identification and selection of B-cell epitopes using the Immune Epitope Database (IEDB) prediction server. The IEDB cooperated BepiPred Linear Epitope Prediction algorithm was used for the B-cell epitope identification from the targeted S-proteins sequences of CoV strains (Jespersen et al., 2017; Vita et al., 2015). This method requires a single FASTA file containing amino acid sequences to access the B-cell epitopes.
2.5. Identification and selection of T-cell epitopes within selected B-cell epitopes and their antigenicity prediction
T-cell epitopes are required to trigger the cell-mediated immunity in the host cell. This analysis used ProPred and Propred-I servers to predict the MHC-II and MHC-I binding epitopes, respectively (Singh and Raghava, 2003). These servers predict epitopes with high efficiency and might propose the interaction with the allelic forms (a probability with any of 47 MHC-I and 51 MHC-II alleles). The present study has considered those T-cell epitopes that have a potent interacting affinity towards the B-cells. The 9mer B-cell epitopes were further processed in the Proped and Proped-I server to identify T-cell and select known common epitopes. Hence we predicted the antigenic score to recognize the antigenicity of the common epitopes (both B-cells and T-cells) within the S-protein of seven CoVs strains. Additionally, the VaxiJen 2.0- Drug Design server was used for the calculation of antigenicity of these common epitopes within the HCoVs S-protein (Doytchinova and Flower, 2007).
2.6. Immunogenicity and conservancy analysis of common B-cell and T-cell epitopes
The IEDB class I immunogenicity tool is used to predict the immunogenicity of a peptide MHC (pMHC) complex. This method was validated only on 9mers presented on the HLA class I molecules (Calis et al., 2013). Subsequently, the IEDB conservancy analysis tool was employed to analyze the degree of conservation of B-cell and T-cell epitopes of seven HCoV strains. This tool calculates the degree of the conservancy of an epitope within a given protein sequence set at different degrees of sequence identity (Bui et al., 2007).
2.7. Mapping of molecular phylogenetics
Bioinformatics tool was used to construct the phylogenetic tree of seven HCoVs strains using different structural proteins. We constructed the phylogenetic tree with bootstrap values for bootstrap analysis using software that employs different signature algorithms, such as PhyML, TreeDyn MUSCLE, and Gblocks (Dereeper et al., 2008). PhyML (for tree building), TreeDyn (for tree depiction), MUSCLE (for multiple alignments), and Gblocks are the highly used algorithms. This tool utilizes the Maximum-likelihood (ML) algorithm for tree building along with the Bayesian algorithm for the analysis of molecular phylogenetics.
2.8. Analysis of sequence similarity
ClustalW calculates the best match for sequences, their likeness, and differences, and it can be detected and evaluated. ClustalW is a well-accepted algorithm for Multiple Sequence Alignment (MSA). This program applies a progressive MSA method, which consists of an algorithm for 3 phases: distance matrix, neighbor-joining, and progressive alignment. The basic algorithm to describe the particular pairwise alignment scores on the ground of its significant algorithm is Needleman and Wunsch algorithm (Needleman and Wunsch, 1970), where gaps have no penalty (d = 0). We utilized Clustal Omega to understand the similarity and elucidate respective pairwise alignment scores for sequence similarity analysis. The Clustal omega tool has a graphical and user-friendly interface. This server was developed based on a ‘progressive algorithm’ (Hogeweg and Hesper, 1984). Clustal Omega is designed on the platform of ClustalW. We used Clustal Omega to understand the similarity and explain the specific pairwise alignment of all structural proteins (S-, M-, E-, N-proteins) of seven human CoVs pathogens.
2.9. Creation of align positions
Previously, we used several sequences of insulin (n = 60) to find the aligned blocks using a modern method (Gblocks server), which is a significant server for removing divergent and obscurely aligned blocks (Chakraborty et al., 2012). In this study, we also utilized the Gblocks server to understand the aligned blocks between all structural proteins (S-, M-, E-, N-proteins) of seven human CoVs pathogens. The Gblocks server explains a set of conserved parts from MSA (Talavera and Castresana, 2007).
3. Result
3.1. Mapping of the protease recognition sites and tissue-specific expression pattern analysis of the proteases
In the search for protease candidates for this study, the proteases' tissue-specific expression pattern was created from an extensive literature survey. This was then validated with data available from GeneCards and The Human Protein Atlas. For quantification of gene expression of proteases, we referred to the tissue-specific confidence scores provided by TISSUES 2.0 in GeneCards. The proteases with significantly higher expression in human epithelial tissues and cleavage scores >0.7 were selected for further analysis. Details of the candidate proteases with potential recognition sites in all of the seven strains of CoVs have been summarized in table 1 . SARS-CoV-2 evades the human immune surveillance more effectively than SARS-CoV and other CoVs strains for which their pathogenicity is much higher. So we elaborated our study by mapping the putative protease recognition sites of SARS-CoV-2 spike glycoprotein in an attempt to address the pathogenicity of the virus (Fig. 1 ).
Table 1.
Summary of tissue-specific expression pattern, functions and number of recognition sites of proteases under study in S-proteins from seven strains of HCoVs.
| Name of proteases | Tissue-specific expression | Number of recognition site(s) of proteases in Coronavirus |
Functions | ||||||
|---|---|---|---|---|---|---|---|---|---|
| SARS-CoV-2 | SARS- COV-1 | MERS-COV | HCOV-OC43 | HCOV-229E | HCOV-NL63 | HCO- HKU1 | |||
| Elastase-2 (ELANE) | Blood, Lung, Liver, Bone marrow, Eye, Heart, Nervous system, Skin, Intestine, Kidney | 7 | 10 | 7 | 6 | 8 | 11 | 11 | Collagen degradation, neutrophil degranulation, ECM organization and remodelling, MMP activation, perpetuate airway inflammatory immune response, disrupt innate immunity, incite inflammatory response |
| MMP-2 | Nervous system, Lung, Bone, Muscle, Skin, Heart, Intestine, Kidney, Blood, Gall bladder, Eye, Lymph node, Liver, Pancreas, Stomach, Spleen, Thyroid gland, Bone marrow | 0 | 1 | 2 | 2 | 1 | 3 | 2 | Regulator in matrix remodelling and molecules involved in signal transduction, particularly expressed in lung, placenta, ovary, pancreas, spleen and intestine |
| MMP-3 | Lung, Blood, Skin, Intestine, Bone, Heart, Muscle, Nervous system | 0 | 0 | 2 | 2 | 0 | 2 | 1 | Breakdown of ECM in normal physiological processes as well as in disease processes. |
| MMP-9 | Blood, Lung, Bone marrow, Intestine, Lymph node, Heart, Nervous system, Liver, Muscle, Spleen, Skin, Bone, Kidney, Eye, Stomach, Pancreas | 7 | 10 | 9 | 1 | 8 | 13 | 7 | Overexpression in human respiratory epithelial healing, ECM organization and remodelling |
| Cathepsin- G | Blood, Skin, Bone marrow, Spleen, Lung, Lymph node, Heart | 5 | 6 | 8 | 4 | 4 | 5 | 7 | Activation of secretion from epithelia of airway submucosal glands, Regulation of innate immunity and inflammation, stimulates neutrophils to respond to chemotactic signals, increase pulmonary immune response by PLTP degradation. |
| Cathepsin- K | Bone, Skin, Blood, Spleen, Kidney, Lung, Nervous system, Pancreas, Bone marrow, Gall bladder, Muscle, Eye, Lymph node, Heart | 1 | 2 | 5 | 2 | 3 | 3 | 7 | Collagen degradation, ECM organization and remodelling, bone remodelling and resorption, affects innate immunity, apoptosis, TLR cascade. |
| Calpain-1 | Pancreas, Nervous system, Kidney, Liver, Intestine, Lung, Muscle, Blood, Eye, Heart, Stomach, Skin | 2 | 1 | 1 | 0 | 0 | 0 | 2 | Stomach and muscle specific cysteine protease |
| Calpain-2 | Pancreas, Skin, Nervous system, Liver, Lung, Lymph node, Blood, Kidney, Intestine, Muscle, Gall bladder | 2 | 0 | 0 | 0 | 0 | 0 | 0 | Stomach and muscle specific cystine protease |
| Pepsin-A | Stomach, Nervous system | 2 | 0 | 0 | 0 | 0 | 0 | 0 | Stomach specific protease |
| Caspase-6 | Blood, Lung, Intestine, Nervous system | 1 | 0 | 0 | 0 | 0 | 0 | 0 | Apoptosis |
| Caspase-8 | Blood, Intestine, Lung, Bone marrow, Liver, Lymph node, Nervous system, Heart, Kidney, Skin, Muscle, Stomach, Pancreas | 2 | 0 | 0 | 0 | 0 | 0 | 0 | Apoptosis. |
| Caspas-3 | Nasal epithelium, Peripheral blood mononuclear cells, Lungs, Heart, Blood | 1 | 0 | 0 | 0 | 0 | 0 | 0 | Apoptosis. |
| Furin | Liver, Blood, Lung, Nervous system, Kidney, Pancreas, Intestine, Muscle, Heart, Skin, Spleen | 1 | 0 | 0 | 0 | 0 | 0 | 0 | Secretory protease acting as proproteinconvertase, Activation of HIV envelope glycoproteins gp160 and gp140, probable role in tumor progression |
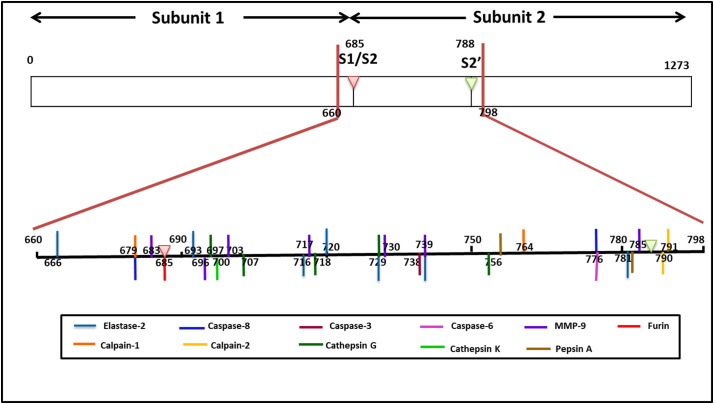
Fig. 1.
A representative map of putative protease recognition sites in target sequence in S-protein encompassing the S1/S2 site and S2’ site. The coloured bars corresponds to cleavage site of individual proteases.
The variation in host proteases responsible for S-protein activation determines its host range, tissue tropism, and viral transmissibility. S-protein of SARS-CoV-2 possesses one furin cleavage site in between the amino acid residues 682 and 685 (RRAR/SVAS), within the S1/S2 domain (Fig. 1), which is distinct from all other CoVs. A similar furin cleavage site was also found in MERS-CoV, but its cleavage score (0.1) was found much lesser than that of SARS-CoV-2 (0.92). A number of cleavage sites for elastase-2, cathepsin-K, and cathepsin-G in the S-protein of all the seven viral strains were observed at different locations. One out of ten recognition sites for elastase-2 in SARS-CoV1(DIPI/GAGI) and one out of eleven in HCOV-HKU1 (AIPT/NFTI) are identical with one from seven sites in SARS-CoV-2 each. Out of the six cathepsin G cleavage sites in SARS-CoV-1, two sites (AYTM/SLGA and LLQY/GSFC) are indistinguishable from SARS-CoV-2. In contrast, another site exhibited a slight deviation in the amino acid sequence (SVAY/SNNS). MMP9 also has multiple recognition sites in the S-protein of all the strains of Coronavirideae under study. Two sites from SARS-CoV-1 (VDCT/MYIC and (FAQV/KQIY) and another from HCOV-229E (IPTN/FTIS) showed similarity with recognition sequence in SARS-CoV-2. The recognition site for calpain-1 was noted in SARS-CoV-1, SARS-CoV-2, and MERS-CoV-2, whereas only SARS-CoV-2 possessed a cleavage site in calpain-2. Two cleavage sites for pepsin-A were exclusive only for SARS-CoV-2. Cleavage sites for caspase-3, caspase-6 and caspase-8 were identified in the S2 subunit of SARS-CoV-2. The position and recognition sequence of cleavage sites for all the candidate proteases in the S-protein of SARS-CoV-2 have been described in supplementary table S10.
3.2. Allocation of protease cleavage sites and assessment of their solvent accessibility
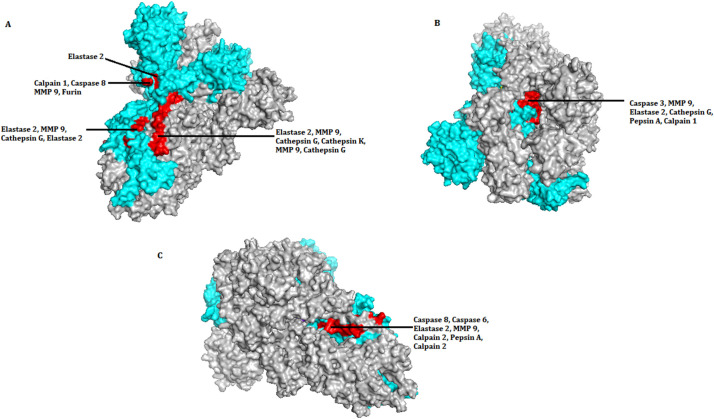
Among the multiple conformational states available for SARS-CoV-2 S glycoprotein, one with a prefusion ectodomain trimer in both close and open state was used to determine the position of the cleavage sites in the protein structure. For that, corresponding structure data represented by PDB formats 6VXX (closed state) and 6VYY (open state) were considered for analysis using PDBePISA software (Krissinel and Henrick, 2007). Solvent accessibility of individual amino acid residue located in the three chains of the S-protein was cross-examined. The target sites for 31 proteolytic cleavages were classified as either accessible or inaccessible, as shown in table 2 . Out of 31 sites, twenty-eight were accessible while three were inaccessible. The same observation was recorded for the open conformation also. The structure information provided in the respective PDB files were used to obtain the protein structure with three chains using PyMOL (Krissinel and Henrick, 2007). From there, the location of recognition sites for each of the proteases under study was determined, and the accessibility was validated. The analysis was done for all the three S- protein chains in both closed and open states, and the same observation was recorded in both of the two states. In Fig. 2 , a representative model has been generated to show the location and the accessibility of the cleavage sites in one chain only but from different positions. The representation of every site is mutually exclusive without any repetition. The model obtained corroborated with the observation from the PDBePISA platform (Wu et al., 2020).
Table 2.
Summary of accessibility of protease cleavage sites on SARS-CoV-2 spike protein (600–798 amino acid residues).
| Serial Number | Position of cleavage site | Name of protease | Cleavage site | Solvent accessibility |
|---|---|---|---|---|
| 1 | 666 | Elastase 2 | DIPI/GAGI | accessible |
| 2 | 679 | Calpain 1 | TQTN/SPRR | accessible⁎ |
| 3 | 679 | Caspase 8 | TQTN/SPRR | accessible⁎ |
| 4 | 683 | MMP9 | SPRR/ARSV | accessible⁎ |
| 5 | 685 | Furin | RRAR/SVAS | accessible⁎ |
| 6 | 693 | Elastase 2 | QSII/AYTM | accessible |
| 7 | 696 | MMP9 | IAYT/MS | accessible |
| 8 | 697 | Cathepsin G | AYTM/SLGA | accessible |
| 9 | 700 | Cathepsin K | MSLG/AENS | inaccessible |
| 10 | 703 | MMP9 | GAEN/SVAY | inaccessible |
| 11 | 707 | Cathepsin G | SVAY/SNNS | inaccessible |
| 12 | 716 | Elastase 2 | AIPT/NFTI | accessible |
| 13 | 717 | MMP9 | IPTN/FTIS | accessible |
| 14 | 718 | Cathepsin G | PTNF/TISV | accessible |
| 15 | 720 | Elastase 2 | NFTI/SVTT | accessible |
| 16 | 729 | Elastase 2 | ILPV/SMTK | accessible |
| 17 | 729 | Cathepsin G | ILPV/SMTK | accessible |
| 18 | 730 | MMP9 | LPVS/MTKT | accessible |
| 19 | 738 | Caspase 3 | SVDC/TMYI | accessible |
| 20 | 739 | MMP9 | VDCT/MYIC | accessible |
| 21 | 739 | Elastase 2 | VDCT/MYIC | accessible |
| 22 | 756 | Cathepsin G | LLQY/GSFC | accessible |
| 23 | 759 | Pepsin A | SFCT/QLN | accessible |
| 24 | 764 | Calpain 1 | TQLN/RALT | accessible |
| 25 | 776 | Caspase 6 | EQDK/NTQE | accessible |
| 26 | 776 | Caspase 8 | EQDK/NTQE | accessible |
| 27 | 778 | Pepsin A | QEVF/AQVK | accessible |
| 28 | 781 | Elastase 2 | TQEV/FAQV | accessible |
| 29 | 785 | MMP9 | FAQV/KQIY | accessible |
| 30 | 790 | Calpain 2 | QIYK/TPPI | accessible |
| 31 | 791 | Calpain 2 | KQIY/ KTTP | accessible |
‘/’ indicates cleavage site.
Accessibility predicted based on their positions as the residues are not present in crystal structure.
Fig. 2.
SARS-CoV-2 spike glycoprotein showing location of recognition sites for proteases from three orientations, A, B and C in both closed (6VXX) and open (6VYY) states. The positions are mutually exclusive. Out of the three chains in Spike protein structure, one chain was selected (marked in cyan) to mark the location of protease cleavage site. In the chain, red coloured patches indicate the presence of individual and overlapping positions of recognition sites. In orientation (A) proteolytic cleavage sites are shown in between amino acid sequence from 600 to 720. In orientations (B) and (C) cleavage sites are shown in between amino acid sequences 738–761 and 776 to 798 respectively. Only the sites accessible to solvent have been shown. Three sites that were inaccessible have not been shown (see Table 2).
3.3. Identification and selection of B-cell epitopes
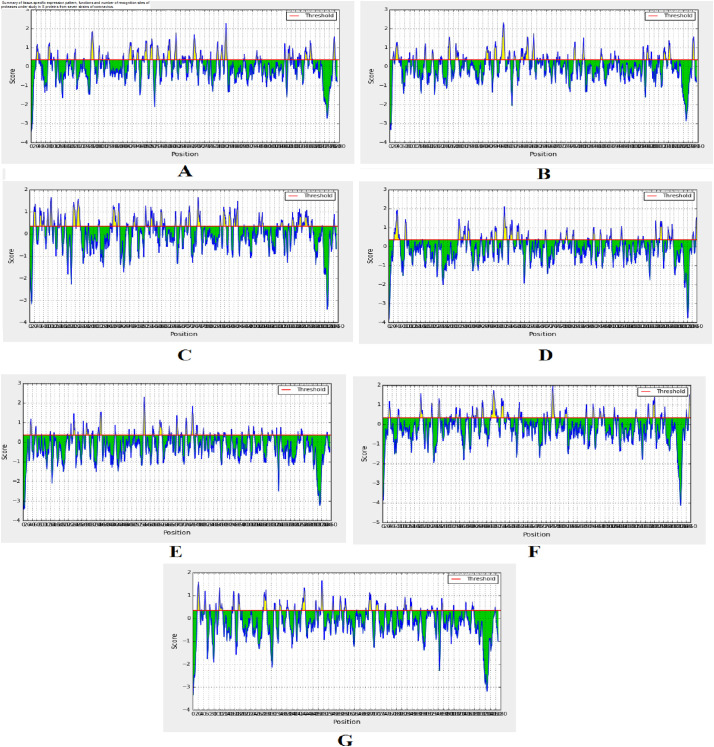
The linear B-cell epitopes were identified within various strains of CoV. The epitopic prediction of IEDB applying the BepiPred prediction method is shown in the graphical presentation (Fig. 3 ). The yellow portions represent the B-cell epitopes, and the green colors represent the non-epitopic area.
Fig. 3.
Graphical representations of B-cell epitopic region from seven strains of HCoVs S-protein using Bepipred prediction portal of IEDB server. (A: SARS-CoV-2, B: SARS-CoV, C: MERS-CoV, D: HCoV-229E, E: HCoV-NL63, F: HCoV-OC43 G: HCoV-HKU1). A red line is drawn in the score versus residue position plot at the chosen score threshold value to predict epitopes. The yellow peaks show the peptide sequences that are potential epitopes whereas the green peaks show the peptides that are not epitopic in nature.
3.4. Identification and selection of T-cell epitopes within selected B-cell epitopes and their antigenicity prediction
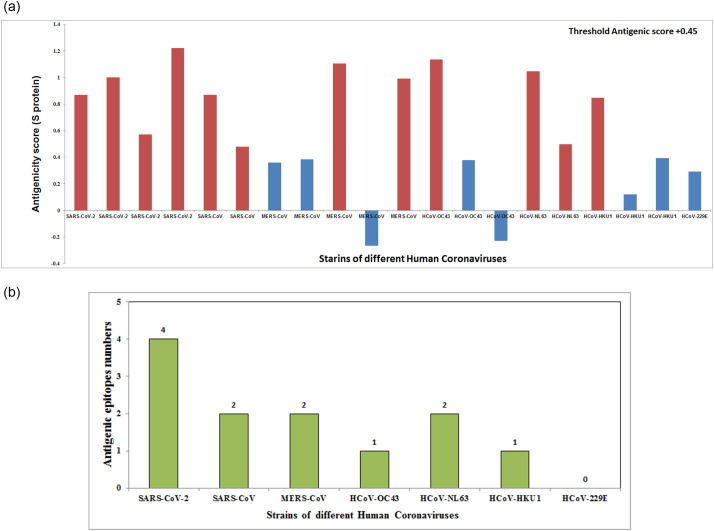
T-cell mediated immune response and its cascade relay after binding to peptide-MHC complexes are pivotal for specific cellular immunogenicity. As these epitopes are selected from the B-cell epitopic part of the S-proteins, these can serve as both B-cell and T-cell epitopes that interact with the human target receptor. Here, seven CoV strains were characterized according to their antigenicity and their interacting affinity towards MHC-I and MHC-II alleles (supplementary table S11). Among them, only the SARS-CoV-2 strain showed the most potent antigenic epitopes (highest antigenicity score with maximum numbers of epitope regions) (Fig. 4A). Our analysis also demonstrates that the S-protein of SARS-CoV-2 contains four epitopic regions (VRQIAPGQT: 0.8675, YQAGSTPCN: 0.4992, FQPTNGVGF: 0.5711, ILPDPSKPS: 1.2217) that bear the maximum antigenicity score (>threshold score 0.45) and are considered as the probable potent antigen for both B-cell and T-cell (supplementary table S11). Other six strains of HCoVs showed less numbers of antigenic epitopes (common B-cell and T-cell) and low antigenicity scores compared to SARS-CoV-2 (Fig. 4B).
Fig. 4.
(A) Graphical representations shows the antigenicity score of common B-cell and T -cell epitopes from S-protein of seven strain HCoVs. (B) Nos of epitopes (common for B-cell and T-cell) present in seven HCoVs strains.
3.5. Immunogenicity and conservancy analysis of common B-cell and T-cell epitopes
Immunogenicity prediction of peptide epitope is typically based on amino acid sequence configuration. The enrichment of an amino acid in an immunogenic peptide is determined by the importance of the position at which it was found to calculate the score of HLA class I within the presented peptide. In prediction results, the higher score indicates a greater probability of eliciting an immune response. Out of the total 20 epitope sequences (common for B-cell and T-cell) from HCoVs analyzed, the SARS-CoV-2 strain epitopes bear the highest immunogenicity (table 3 ) compared to all other CoV strains epitopes. Whereas the epitope conservancy analysis is applied to analyze the variability or conservation of studied epitopes. The calculated of conservancy degree and the matching between minimum identity andmaximum identity levels within the protein sequence set of each HCoVs S-protein is presented in table 4 . Different epitopes were identified from the protein data set, showing the positions and the matching protein sub-sequences. The corresponding matching identity of the epitope in every protein sequence is also depicted. In our current study, 16 identified epitopes from the S-protein have more than 5% of protein sequence matches. Likewise, the minimum identity was determined from the given seven pathogenic HCoVs S-protein sequences.
Table 3.
Immunogenicity (HLA class I) analysis of S-protein epitopes.of seven HCoVs.
| Sl. no | Human CoVs strain name | Common B and T-cell epitopes sequence | Epitope (peptide) length | Immunogenicity (HLA class I) score |
|---|---|---|---|---|
| 1 | SARS-CoV-2 | VRQIAPGQT | 9 | 0.0849 |
| 2 | YQAGSTPCN | 9 | −0.11862 | |
| 3 | FQPTNGVGF | 9 | 0.1157 | |
| 4 | ILPDPSKPS | 9 | −0.33629 | |
| 5 | SARS-CoV | VRQIAPGQT | 9 | 0.0849 |
| 6 | FSPDGKPCT | 9 | −0.19214 | |
| 7 | MERS-CoV | LLSGTPPQV | 9 | −0.06928 |
| 8 | YGTDTNSVC | 9 | −0.04887 | |
| 9 | LTPRSVRSV | 9 | −0.12674 | |
| 10 | YQNISTNLP | 9 | −0.00468 | |
| 11 | LGNSTGIDF | 9 | 0.02641 | |
| 12 | HCoV-OC43 | YRRKPDLPN | 9 | −0.20596 |
| 13 | FKPQPAGVF | 9 | −0.04141 | |
| 14 | FTGPYKCPQ | 9 | −0.25874 | |
| 15 | HCoV-NL63 | VKSGSPGDS | 9 | −0.14958 |
| 16 | VRPRNSSDN | 9 | −0.24021 | |
| 17 | HCoV-HKU1 | ITAYDPRSC | 9 | −0.03284 |
| 18 | YNSPSSSSS | 9 | −0.61797 | |
| 19 | YYYPEPISD | 9 | 0.09036 | |
| 20 | HCoV-229E | LPRSGSRVA | 9 | −0.2046 |
Table 4.
Epitope conservancy analysis of S-protein epitopes of seven HCoVs.
| Sl. no | Human CoVs strain name | Common B and T-cell epitopes sequence | Epitope (peptide) length | Percent of protein sequence matches at identity ≤ 100% | Minimum identity | Maximum identity |
|---|---|---|---|---|---|---|
| 1 | SARS-CoV-2 | VRQIAPGQT | 9 | 5.26% (1/19) | 11.11% | 100.00% |
| 2 | YQAGSTPCN | 9 | 5.26% (1/19) | 11.11% | 100.00% | |
| 3 | FQPTNGVGF | 9 | 0.00% (0/19) | 22.22% | 88.89% | |
| 4 | ILPDPSKPS | 9 | 5.26% (1/19) | 11.11% | 100.00% | |
| 5 | SARS-CoV | VRQIAPGQT | 9 | 5.56% (1/18) | 22.22% | 100.00% |
| 6 | FSPDGKPCT | 9 | 5.56% (1/18) | 22.22% | 100.00% | |
| 7 | MERS-CoV | LLSGTPPQV | 9 | 5.00% (1/20) | 22.22% | 100.00% |
| 8 | YGTDTNSVC | 9 | 5.00% (1/20) | 22.22% | 100.00% | |
| 9 | LTPRSVRSV | 9 | 5.00% (1/20) | 22.22% | 100.00% | |
| 10 | YQNISTNLP | 9 | 5.00% (1/20) | 11.11% | 100.00% | |
| 11 | LGNSTGIDF | 9 | 5.00% (1/20) | 22.22% | 100.00% | |
| 12 | HCoV-OC43 | YRRKPDLPN | 9 | 0.00% (0/20) | 22.22% | 88.89% |
| 13 | FKPQPAGVF | 9 | 0.00% (0/20) | 11.11% | 77.78% | |
| 14 | FTGPYKCPQ | 9 | 0.00% (0/20) | 22.22% | 88.89% | |
| 15 | HCoV-NL63 | VKSGSPGDS | 9 | 5.00% (1/20) | 22.22% | 100.00% |
| 16 | VRPRNSSDN | 9 | 5.00% (1/20) | 22.22% | 100.00% | |
| 17 | HCoV-HKU1 | ITAYDPRSC | 9 | 5.00% (1/20) | 11.11% | 100.00% |
| 18 | YNSPSSSSS | 9 | 5.00% (1/20) | 11.11% | 100.00% | |
| 19 | YYYPEPISD | 9 | 5.00% (1/20) | 11.11% | 100.00% | |
| 20 | HCoV-229E | LPRSGSRVA | 9 | 0.00% (0/17) | 22.22% | 88.89% |
3.6. Mapping of molecular phylogenetics
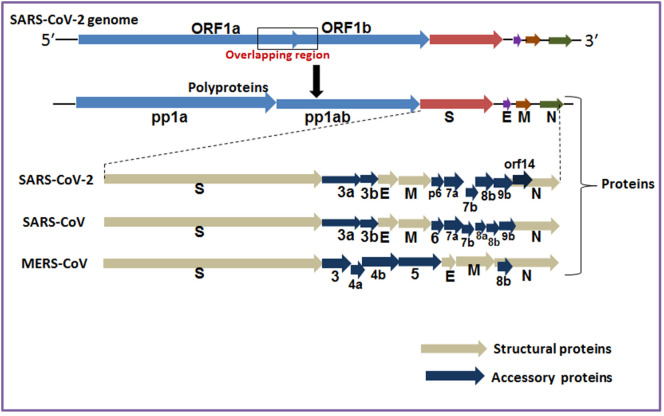
The genome of SARS-CoV-2 encodes polyproteins (pp1a and pp1b) and structural proteins (Fig. 5 ). The structural proteins of CoVs are the building blocks of their capsid that encase different proteins, i.e., E-, M-, N- and S-proteins (Supplementary table S12). Our molecular evolution study involves different SPs of the seven HCoVs pathogens (Supplementary table S12). We have collected amino acid sequences for all structural proteins for seven HCoVs pathogens from NCBI. We used the latest computational biology tools to generate data on the structural proteins of seven recognized human pathogenic CoVs. We specifically analyzed four critical points in the phylogenetic tree.
Fig. 5.
The genomes and structural proteins of different human pathogenic CoVs. The polyprotein includes pp1a and pp1b. Along with the structural proteins 3′ terminus SARS-CoV- 2 genome contain accessory protein (3a, 3b,p6, 7a, 7b, 8b,9b, orf14). Similarly 3′ terminus SARS-CoV genome contain accessory protein (3a, 3b, p6, 7a, 7b, 8a,8b, 9b). Likewise 3′ terminus MERS-CoV genome contain accessory protein (3, 4a, 4b, 5, 8b) include spike (S), membrane (M), envelope (E), and nucleocapsid (N) (Walls et al., 2019; Walls et al., 2020).
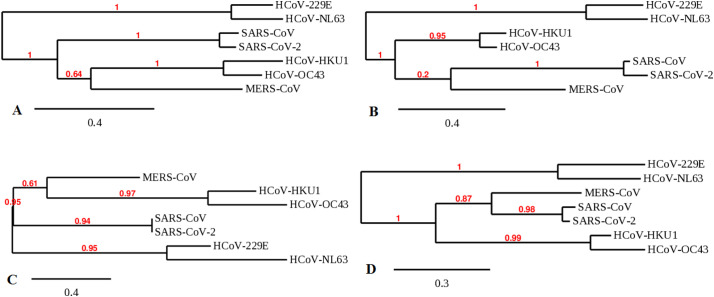
First, the S-protein of CoVs showed origin from an ancestral node for HCoV-NL63 and HCoV-229E. Similarly, SARS-CoV, as well as SARS-CoV-2, expresses the same line and point of origin. Besides, HCoV-OC43 and HCoV-HKU1 are associated with MERS-CoV as they showed the origin point from the ancestral node (Fig. 6A). Secondly, the M-protein of different viral strains showed HCoV-229E and HCoV-NL63 having the same point of origin from the ancestral node. Likewise, HCoV-HKU1 and HCoV-OC43 are also expressed at the same origin point. It was also observed that the SARS-CoV-2 and SARS-CoV originated from the ancestral node that links MERS-CoV (Fig. 6B). Thirdly, the E-protein of different strains has illustrated that HCoV-HKU1, HCoV-OC43 originated from the ancestral node associated with MERS-CoV. Also, SARS-CoV-2 and SARS-CoV originated from ancestral nodes similar to HCoV-NL63 and HCoV-229E (Fig. 6C). Fourth, the N-protein showed HCoV-NL63 and HCoV-229E with a similar point of origin. Therefore, it can be suggested that SARS-CoV-2 and SARS-CoV are linked and share the same point and line of origin as MERS-CoV. HCoV-HKU1 and HCoV-OC43 also showed the same origin point (Fig. 6D).
Fig. 6.
Molecular evolution of structural proteins of seven human pathogenic CoVs ((A) evolution of spike protein in human pathogenic CoV, (B) evolution of membrane protein in human pathogenic CoV, (C) evolution of envelope protein in human pathogenic CoV, and (D) evolution of nucleocapsid protein of human pathogenic CoV.
3.7. Analysis of sequence similarity
To understand the sequence similarity of structural proteins of seven HCoVs, we performed multiple sequence alignment (MSA). We found a high level of sequence similarity between different human pathogenic CoVs strains in S-, M-, E- and N-proteins (Supplementary Fig. S1-S4).
3.8. Creation of HCoVs align positions
To understand the align positions of structural proteins of HCoVs, we performed a G-Block analysis. Several align positions of different pathogenic HCoVs have been noted in S-proteins (Supplementary Fig. S5), M- protein (Supplementary Fig. S6), E-protein (Supplementary Fig. S7), and N-protein (Supplementary Fig. S8). More number of align positions are found in S-proteins because it may be a long protein compare to other proteins.
4. Discussion
The SARS-CoV infection in China occurred in 2002–03, and the MERS-CoV outbreak followed in the Middle East a decade later (Chakraborty et al., 2020a; Zaki et al., 2012). Although the human pathogenic CoVs, namely HCoV-HKU1, HCoV-NL63, HCoV-229E, and HCoV-OC43 have been well-established, they didn't create significant health threats. But, they were associated with respiratory illnesses, including pneumonia (Zeng et al., 2018). Therefore, we focused on the seven human pathogenic CoVs.
In CoVs, S-proteins' tethering to the viral envelope is essential for receptor binding, followed by membrane fusion and viral entry in host cells. Therefore, it is a significant determinant of its infectivity (Ou et al., 2016; Xia et al., 2020). The two subunits, i.e., S1 and S2, of the trimeric S-protein, assist binding and membrane fusion, respectively (Colman and Lawrence, 2003; Lamb and Jardetzky, 2007). S-glycoprotein with a metastable perfusion conformation following translation is a typical representative of class I fusion protein. It needs to be activated by proteolytic cleavage at S1/S2 and/or S2’ sites by proteases of the host cell followed by a configuration change in the S2 subunit that facilitates the release of spike fusion peptide to promote the fusion reaction and viral entry (Hulswit et al., 2016; Wang et al., 2020b). Therefore, we restricted our search for candidate protease recognition site to a segment of the S-protein starting from 25 amino acid upstream of S1/S2 junction, encompassing the S2 subunit up to 10 amino acid downstream of S2’ site in each of the seven strains of CoVs. The S-protein size is highly variable among different CoVs species ranging from approximately 1100 to 1600 residues in length (Hoffmann et al., 2020). An in silico study was performed using the target sequences of SARS-CoV-1 (159 aa), SARS-CoV-2 (137 aa), MERS-CoV (167 aa), HCoV-HKU1 (134 aa), HCoV-OC43 (66 aa), HCoV-229E (147 aa), and HCoV-NL63 (210 aa). In the present study, an in silico analysis was performed using the target sequences of these seven HCoVs pathogens, wherefrom a number of proteases with a significant level of expression in human epithelium and high level of cleavage efficiency were selected for our analysis (Table 1). SARS-CoV-2 evades the human immune surveillance more effectively than SARS-CoV and other CoV strains for which their pathogenicity is much higher. Therefore, our study concentrated on mapping the putative protease recognition sites of SARS-CoV-2 S-glycoprotein (Fig. 1).
S-protein activation is a complex process that involves multiple cleavage events at discrete locations by a multitude of host proteases, as reported in SARS-CoV, MERS-CoV, and other CoVs (Hoffmann et al., 2020). Moreover, the host proteases that can prime S-protein activation vary among different CoVs. This diversity is a deciding factor for the virus's epidemiological and pathological properties, like host range, tissue tropism, transmissibility, and mortality (Wang et al., 2020b). S-protein of SARS-CoV-2 possesses a unique multi basic furin cleavage site (−RRAR–) in between the amino acid residues 682 and 685, within the S1/S2 domain (Fig. 1), discrete from SARS-CoV and other related CoVs which contain only trypsin or TMPRSS2 cleavage site at R667 (Wang et al., 2020b). The presence of a similar furin cleavage site has also been reported in MERS-CoV (Hoffmann et al., 2020; Millet and Whittaker, 2014). However, our analyses revealed that the efficiency of cleavage for MERS-CoV is much lesser than that of SARS-CoV-2, indicating a higher basic reproductive rate. Hence, the greater infectivity transmissibility of the later (Wang et al., 2020b). The overexpression of furin in saliva and the salivary gland may expedite the process of mucous membrane invasion by SARS-CoV-2 in the upper respiratory tract (Zupin et al., 2020). The ubiquitous expression of furin in multiple organs and tissues like the lung, gastrointestinal tract, brain, pancreas, liver, and reproductive tissues allows SARS-CoV-2 to access and infect these organs or tissues that are impervious to other CoVs, causing systematic infection humans (Hoffmann et al., 2020).
There are multiple recognition sites for cysteine proteases elastase-2, cathepsin-K, and serine protease cathepsin-G in the S-protein of all the seven viral strains. However, the number and positions are variable. Following receptor binding, lysosomal cathepsins, elastase, or TMPRSS-2 are reported to activate S-protein for viral entry either alone or through cumulative effect with furin. The efficiency and magnitude of this protease-mediated activation step of the target cells may regulate cellular tropism and viral pathogenesis (Coutard et al., 2020). One of the ten recognition sites for elastase in SARS-CoV-1 is identical with one from seven sites in SARS-CoV-2. Out of the six cathepsin G cleavage sites in SARS-CoV-1, two sites are indistinguishable from SARS-CoV-2, while two other sites are similar with minor alteration in amino acid sequences. Considering elastase-2, only HCOV-HKU1 has a recognition site similar to SARS-CoV-2.
The majority of these protease cleavage sites (28 out of 31) were found to be accessible in protein structure analysis. Our observation corroborated with findings in several reports that advocated that coronaviruses use conformational masking and glycan shielding to limit recognition by the immune system of affected individuals (Walls et al., 2019). As most of the cleavage sites are accessible, most of the host proteases available can cleave the S-protein to facilitate the fusion of viral membrane with the host cell membrane in a tissue-specific manner (Walls et al., 2020).
Pathogenesis of acute lung injury is caused by inflammatory damage in the epithelium of alveoli and endothelial capillary membrane and subsequent basement membrane degradation. Matrix metallopeptidases (MMPs) have a significant role in a wide range of such pulmonary pathologies by degrading extracellular matrix parts, which are non-matrix mediators of lung injury (cell surface receptors and chemokines) and the basement membrane (Davey et al., 2011). All of the seven strains of CoV were found to have several recognition sites for MMP-9, while the recognition sites for MMP-2 and MMP-3 were found in all strains except for SARS-CoV-2.
Calpains are calcium-dependent cysteine proteases that cleave cytoskeletal and cytoplasmic proteins leading to apoptosis and necrosis. Calpain-1 and calpain-2 are stomach and muscle-specific enzymes. In this study, we identified calpain-1 cleavage site in all of the CoV strains that cause a severe form of respiratory ailments, whereas only SARS-CoV-2 S- protein could be cleaved by calpain-2. It has been previously reported that conventional calpains' elevated activity may aggravate cardiovascular diseases, such as cardiac hypertrophy, cardiac infarction, diabetes-related myocardial hypertrophy, and hyperglycemia (Sorimachi and Ono, 2012). A severe form of COVID-19 can be associated with similar symptoms in patients with cardiovascular disorders, which may be attributed to calpain activity in S-protein priming.
Two cleavage sites were identified for pepsin-A in SARS-CoV-2 only out of all the strains studied. Aspiration of gastric fluids damages airway epithelial cells leading to chronic lung diseases. Apart from low pH, the presence of pepsin A induces the release of inflammatory mediators, like IL-6 and IL-8 (Bathoorn et al., 2011). These mediators are an essential component of the cytokine storm that complements the severe form of COVID-19. So in patients with gastroesophageal reflux disease, Pepsin-A activity may trigger an elevated cytokine storm in the case of COVID-19 co-infection.
In the process of apoptosis, caspase-8 acts as initiator while caspase-3 and 6 act as executioner molecules. In most viral infections characterized by host cell death, the viruses can utilize apoptotic caspase activity to facilitate their proliferation through the formation of cleavage products with novel activities. While caspase-3 mediated, cleavage of Amdovirus protein NS1 and Hepatitis C virus protein NS5A promotes nuclear entry, nucleoprotein of Influenza A activation by caspase-3 facilitates the transport of ribonucleoprotein complexes into the cytoplasm. All these mechanisms enhance the replicative proficiency of the viruses (Connolly and Fearnhead, 2017). So the S2 subunit cleavage by caspases-3, caspases-6, caspases-8 upstream of the S2’ site may play a significant role in the release of fusion peptide and hence in viral replication and proliferation.
In the second part of our analysis, advanced immunoinformatic approaches were applied to understand the viral antigens of seven HCoVs strains. It was required to predict the multiepitopes and to assess their antigenicity. In this work, common B-cell and T-cell epitopes were identified from the S-protein of seven HCoVs strains for characterizing the antigenic infections on the host body. The S-protein of SARS-CoV-2 consisting of four epitopic regions (VRQIAPGQT: 0.8675, YQAGSTPCN: 0.4992, FQPTNGVGF: 0.5711, ILPDPSKPS: 1.2217) bearing the maximum antigenicity score (>threshold score 0.45) were considered as the potential potent antigens for both B-cell and T-cell (Fig. 3 and Fig. 4)(Table 3). Our previous work identified different epitopic regions of S-protein (SARS-CoV-2) that demonstrated a potential of activating the immune system (Bhattacharya et al., 2020a; Bhattacharya et al., 2020b; Bhattacharya et al., 2020c). The six strains of HCoVs show antigenic epitopes (common B-cell and T-cell) fewer in numbers than SARS-CoV-2. Our analysis also demonstrates that the SARS-CoV-2 is more infectious and can inflict excessive respiratory infection on the human respiratory tract. Our study may help towards the vaccine development of these seven human pathogenic CoVs.
The immunogenicity analysis reveals that higher immunogenicity is present among the common B-cell and T-cell epitopes of SARS-CoV-2 S-protein compare to all other HCoV strains. From conservancy analysis of S-protein, we found that the MERS-CoV has maximum conserved epitopes, whereas HCoV-OC43 epitopes have a minimum level of conservancy pattern. Therefore, the maximum variations (multivalent) and high immunogenic epitopes might be suitable for the peptide-based vaccine development.
The third part of our study shows that the molecular evolution of structural proteins of HCoVs can be classified into some groups: (i) HCoV-NL63 and HCoV-229E, (ii) SARS-CoV-2 and SARS-CoV, and (iii) HCoV-OC43 and HCoV-HKU1 (Fig. 6). The HCoV-229E and HCoV-NL63 group appear to possess the same pathogenic profile reported for seroconversion in children (Dijkman et al., 2008). As shown in our analysis, SARS-CoV-2 and SARS-CoV share the same evolutionary path and point of origin for all four structural proteins as predicted in the recent report that named SARS-CoV-2 (C.S.G of the International, 2020). A previous study has already pointed out that HCoV-HKU1 and HCoV-OC43 have common ancestors (Al-Khannaq et al., 2016).
5. Conclusion
This current study is highly significant because it will increase our understanding level in several novel directions. First, the putative protease recognition sites of seven CoVs can help to understand their pathomechanism. Second, the expression of different proteases in different organs and tissues like the brain, liver, lung, pancreas, gastrointestinal tract, and reproductive tissues allows us to comprehend the infectivity of seven human pathogenic CoVs with the SARS-CoV-2 to access and infect those organs or tissues. Third, epitopic variations of the S-protein of seven human pathogenic CoVs and the SARS-CoV-2 will help us assist in peptide-based vaccine development. Fourth, our analysis reaffirms that the three groups of CoVs came into existence due to cross-species transmission. The co-evolution of human pathogenic viral strains occurs in host animals in a million years to create distinct patterns and different structural proteins evolutionarily. This study provides scientific evidence for the evolutionary grouping of human pathogenic CoVs using computational biology. Finally, we can conclude that our research will contribute to future preparedness to encounter possible outbreaks of infectious diseases.
Author contributions
All authors contributed equally and agreed to the published version of the manuscript.
Ethics approval
Not required.
Declaration of Competing Interest
The authors have no conflicts of interest.
Acknowledgments
This study was supported by Hallym University Research Fund and by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF -2020R1C1C1008694 & NRF-2020R1I1A3074575 ).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2021.104729.
Appendix A. Supplementary data
Supplementary Figures (S1-S8)
Supplementary Tables (S1-S12)
References
- Al-Khannaq M.N., Ng K.T., Oong X.Y., Pang Y.K., Takebe Y., Chook J.B., Hanafi N.S., Kamarulzaman A., Tee K.K. Molecular epidemiology and evolutionary histories of human coronavirus OC43 and HKU1 among patients with upper respiratory tract infections in Kuala Lumpur, Malaysia. Virol. J. 2016;13:33. doi: 10.1186/s12985-016-0488-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bathoorn E., Daly P., Gaiser B., Sternad K., Poland C., Macnee W., Drost E.M. Cytotoxicity and induction of inflammation by pepsin in acid in bronchial epithelial cells. Int. J. Inflamm. 2011;2011:569416. doi: 10.4061/2011/569416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Madu I., Whittaker G.R. Elastase-mediated activation of the severe acute respiratory syndrome coronavirus spike protein at discrete sites within the S2 domain. J. Biol. Chem. 2010;285:22758–22763. doi: 10.1074/jbc.M110.103275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya M., Sharma A.R., Mallick B., Sharma G., Lee S.-S., Chakraborty C. Immunoinformatics approach to understand molecular interaction between multi-epitopic regions of SARS-CoV-2 spike-protein with TLR4/MD-2 complex. Infect. Genet. Evol. 2020;85:104587. doi: 10.1016/j.meegid.2020.104587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya M., Sharma A.R., Patra P., Ghosh P., Sharma G., Patra B.C., Lee S.S., Chakraborty C. Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): Immunoinformatics approach. J. Med. Virol. 2020;92:618–631. doi: 10.1002/jmv.25736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya M., Sharma A.R., Patra P., Ghosh P., Sharma G., Patra B.C., Saha R.P., Lee S.-S., Chakraborty C. A SARS-CoV-2 vaccine candidate: in silico cloning and validation. Inform. Med. Unlocked. 2020;100394 doi: 10.1016/j.imu.2020.100394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., Rottier P.J. Nidovirus entry into cells. Nidoviruses. 2007:157–178. [Google Scholar]
- Bui H.-H., Sidney J., Li W., Fusseder N., Sette A. Development of an epitope conservancy analysis tool to facilitate the design of epitope-based diagnostics and vaccines. BMC Bioinform. 2007;8:361. doi: 10.1186/1471-2105-8-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard C., Verheije M.H., Wicht O., van Kasteren S.I., van Kuppeveld F.J., Haagmans B.L., Pelkmans L., Rottier P.J., Bosch B.J., de Haan C.A. Coronavirus cell entry occurs through the endo−/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C.S.G of the International The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calis J.J., Maybeno M., Greenbaum J.A., Weiskopf D., De Silva A.D., Sette A., Keşmir C., Peters B. Properties of MHC class I presented peptides that enhance immunogenicity. PLoS Comput. Biol. 2013;9 doi: 10.1371/journal.pcbi.1003266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty C., Roy S.S., Hsu M.J., Agoramoorthy G. Can computational biology improve the phylogenetic analysis of insulin? Comput. Methods Prog. Biomed. 2012;108:860–872. doi: 10.1016/j.cmpb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Chakraborty C., Sharma A., Sharma G., Bhattacharya M., Lee S. SARS-CoV-2 causing pneumonia-associated respiratory disorder (COVID-19): diagnostic and proposed therapeutic options. Eur. Rev. Med. Pharmacol. Sci. 2020;24:4016–4026. doi: 10.26355/eurrev_202004_20871. [DOI] [PubMed] [Google Scholar]
- Chakraborty C., Sharma A.R., Bhattacharya M., Sharma G., Lee S.-S. The 2019 novel coronavirus disease (COVID-19) pandemic: a zoonotic prospective. Asian Pac J Trop Med. 2020;13:242. [Google Scholar]
- Colman P.M., Lawrence M.C. The structural biology of type I viral membrane fusion. Nat. Rev. Mol. Cell Biol. 2003;4:309–319. doi: 10.1038/nrm1076. [DOI] [PubMed] [Google Scholar]
- Connolly P.F., Fearnhead H.O. Viral hijacking of host caspases: an emerging category of pathogen-host interactions. Cell Death Differ. 2017;24:1401–1410. doi: 10.1038/cdd.2017.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coordinators N.R. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2017;45:D12–D17. doi: 10.1093/nar/gkw1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S.J., Fooks A.R., van der Poel W.H. Public health threat of new, reemerging, and neglected zoonoses in the industrialized world. Emerg. Infect. Dis. 2010;16:1–7. doi: 10.3201/eid1601.081467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey A., McAuley D.F., O’Kane C.M. Matrix metalloproteinases in acute lung injury: mediators of injury and drivers of repair. Eur. Respir. J. 2011;38:959–970. doi: 10.1183/09031936.00032111. [DOI] [PubMed] [Google Scholar]
- DeLano W.L. Pymol: an open-source molecular graphics tool. CCP4 newsletter on protein crystallography. 2002;40:82–92. [Google Scholar]
- Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.F., Guindon S., Lefort V., Lescot M., Claverie J.M., Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkman R., Jebbink M.F., El Idrissi N.B., Pyrc K., Müller M.A., Kuijpers T.W., Zaaijer H.L., Van Der Hoek L. Human coronavirus NL63 and 229E seroconversion in children. J. Clin. Microbiol. 2008;46:2368–2373. doi: 10.1128/JCM.00533-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doytchinova I.A., Flower D.R. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007;8:4. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierer S., Bertram S., Kaup F., Wrensch F., Heurich A., Kramer-Kuhl A., Welsch K., Winkler M., Meyer B., Drosten C., Dittmer U., von Hahn T., Simmons G., Hofmann H., Pohlmann S. The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J. Virol. 2013;87:5502–5511. doi: 10.1128/JVI.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giurgea L.T., Han A., Memoli M.J. Universal coronavirus vaccines: the time to start is now. NPJ Vaccin. 2020;5:43. doi: 10.1038/s41541-020-0198-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Pohlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78(779–784) doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogeweg P., Hesper B. The alignment of sets of sequences and the construction of phyletic trees: an integrated method. J. Mol. Evol. 1984;20:175–186. doi: 10.1007/BF02257378. [DOI] [PubMed] [Google Scholar]
- Hulswit R.J., de Haan C.A., Bosch B.J. Coronavirus spike protein and tropism changes. Adv. Virus Res. 2016;96:29–57. doi: 10.1016/bs.aivir.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespersen M.C., Peters B., Nielsen M., Marcatili P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017;45:W24–W29. doi: 10.1093/nar/gkx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam Y.W., Okumura Y., Kido H., Ng L.F., Bruzzone R., Altmeyer R. Cleavage of the SARS coronavirus spike glycoprotein by airway proteases enhances virus entry into human bronchial epithelial cells in vitro. PLoS One. 2009;4 doi: 10.1371/journal.pone.0007870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Lamb R.A., Jardetzky T.S. Structural basis of viral invasion: lessons from paramyxovirus F. Curr. Opin. Struct. Biol. 2007;17:427–436. doi: 10.1016/j.sbi.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, function, and evolution of coronavirus spike proteins. Ann. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Leier A., Liu Q., Wang Y., Xiang D., Akutsu T., Webb G.I., Smith A.I., Marquez-Lago T., Li J., Song J. Procleave: predicting protease-specific substrate cleavage sites by combining sequence and structural information. Genomics Proteomics Bioinform. 2020;18:52–64. doi: 10.1016/j.gpb.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Ujike M., Morikawa S., Tashiro M., Taguchi F. Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proc. Natl. Acad. Sci. U. S. A. 2005;102:12543–12547. doi: 10.1073/pnas.0503203102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. U. S. A. 2014;111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski, E., Shenk, T., Griffiths, P., Pass, R., 2013. Cytomegaloviruses in: Knipe DM, Howley PM, ed. Fields Virology. Philadelphia, PA, USA: Wolters Kluwer Lippincott Williams & Wilkins. 1960-2014.
- Needleman S.B., Wunsch C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Ou X., Zheng W., Shan Y., Mu Z., Dominguez S.R., Holmes K.V., Qian Z. Identification of the fusion peptide-containing region in betacoronavirus spike glycoproteins. J. Virol. 2016;90:5586–5600. doi: 10.1128/JVI.00015-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal M., Berhanu G., Desalegn C., Kandi V. Severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2): an update. Cureus. 2020;12 doi: 10.7759/cureus.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safran M., Dalah I., Alexander J., Rosen N., Iny Stein T., Shmoish M., Nativ N., Bahir I., Doniger T., Krug H., Sirota-Madi A., Olender T., Golan Y., Stelzer G., Harel A., Lancet D. GeneCards Version 3: the human gene integrator. Database: J. Biol. Databases Curation 2010. 2010:1–16. doi: 10.1093/database/baq020. baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Kawase M., Matsuyama S. Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J. Virol. 2013;87:12552–12561. doi: 10.1128/JVI.01890-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Zmora P., Gierer S., Heurich A., Pohlmann S. Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antivir. Res. 2013;100:605–614. doi: 10.1016/j.antiviral.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K., Garoff H., Helenius A. How an animal virus gets into and out of its host cell. Sci. Am. 1982;246:58–66. doi: 10.1038/scientificamerican0282-58. [DOI] [PubMed] [Google Scholar]
- Singh H., Raghava G.P. ProPred1: prediction of promiscuous MHC class-I binding sites. Bioinformatics. 2003;19:1009–1014. doi: 10.1093/bioinformatics/btg108. [DOI] [PubMed] [Google Scholar]
- Song J., Tan H., Perry A.J., Akutsu T., Webb G.I., Whisstock J.C., Pike R.N. PROSPER: an integrated feature-based tool for predicting protease substrate cleavage sites. PLoS One. 2012;7 doi: 10.1371/journal.pone.0050300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorimachi H., Ono Y. Regulation and physiological roles of the calpain system in muscular disorders. Cardiovasc. Res. 2012;96:11–22. doi: 10.1093/cvr/cvs157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer G., Dalah I., Stein T.I., Satanower Y., Rosen N., Nativ N., Oz-Levi D., Olender T., Belinky F., Bahir I., Krug H., Perco P., Mayer B., Kolker E., Safran M., Lancet D. In-silico human genomics with GeneCards. Human Genom. 2011;5:709–717. doi: 10.1186/1479-7364-5-6-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G., Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Thul P.J., Lindskog C. The human protein atlas: a spatial map of the human proteome. Protein Sci. 2018;27:233–244. doi: 10.1002/pro.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y., Ying T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M., Oksvold P., Fagerberg L., Lundberg E., Jonasson K., Forsberg M., Zwahlen M., Kampf C., Wester K., Hober S., Wernerus H., Bjorling L., Ponten F. Towards a knowledge-based human protein atlas. Nat. Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- Vita R., Overton J.A., Greenbaum J.A., Ponomarenko J., Clark J.D., Cantrell J.R., Wheeler D.K., Gabbard J.L., Hix D., Sette A., Peters B. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015;43:D405–D412. doi: 10.1093/nar/gku938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Xiong X., Park Y.-J., Tortorici M.A., Snijder J., Quispe J., Cameroni E., Gopal R., Dai M., Lanzavecchia A. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell. 2019;176:1026–1039. doi: 10.1016/j.cell.2018.12.028. e1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(281–292) doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Qiu Y., Li J.-Y., Zhou Z.-J., Liao C.-H., Ge X.-Y. A unique protease cleavage site predicted in the spike protein of the novel pneumonia coronavirus (2019-nCoV) potentially related to viral transmissibility. Virol. Sin. 2020:1–3. doi: 10.1007/s12250-020-00212-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.M., Delos S.E., Brecher M., Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson D.M., Elferich J., Ramakrishnan P., Thomas G., Shinde U. The mechanism by which a propeptide-encoded pH sensor regulates spatiotemporal activation of furin. J. Biol. Chem. 2013;288:19154–19165. doi: 10.1074/jbc.M112.442681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Huang Y., Yuen K.Y. Coronavirus diversity, phylogeny and interspecies jumping. Exp. Biol. Med. (Maywood). 2009;234:1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Lan Q., Su S., Wang X., Xu W., Liu Z., Zhu Y., Wang Q., Lu L., Jiang S. The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Signal Trans. Target. Therapy. 2020;5:1–3. doi: 10.1038/s41392-020-0184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zeng Z.-Q., Chen D.-H., Tan W.-P., Qiu S.-Y., Xu D., Liang H.-X., Chen M.-X., Li X., Lin Z.-S., Liu W.-K. Epidemiology and clinical characteristics of human coronaviruses OC43, 229E, NL63, and HKU1: a study of hospitalized children with acute respiratory tract infection in Guangzhou, China. Eur. J. Clin. Microbiol. Infect. Dis. 2018;37:363–369. doi: 10.1007/s10096-017-3144-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupin L., Pascolo L., Crovella S. Is FURIN gene expression in salivary glands related to SARS-CoV-2 infectivity through saliva? J. Clin. Pathol. 2020:1–3. doi: 10.1136/jclinpath-2020-206788. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures (S1-S8)
Supplementary Tables (S1-S12)