Abstract
Antagonistic coevolution between host and parasite drives species evolution. However, most of the studies only focus on parasitism adaptation and do not explore the coevolution mechanisms from the perspective of both host and parasite. Here, through the de novo sequencing and assembly of the genomes of giant panda roundworm, red panda roundworm, and lion roundworm parasitic on tiger, we investigated the genomic mechanisms of coevolution between nonmodel mammals and their parasitic roundworms and those of roundworm parasitism in general. The genome-wide phylogeny revealed that these parasitic roundworms have not phylogenetically coevolved with their hosts. The CTSZ and prolyl 4-hydroxylase subunit beta (P4HB) immunoregulatory proteins played a central role in protein interaction between mammals and parasitic roundworms. The gene tree comparison identified that seven pairs of interactive proteins had consistent phylogenetic topology, suggesting their coevolution during host–parasite interaction. These coevolutionary proteins were particularly relevant to immune response. In addition, we found that the roundworms of both pandas exhibited higher proportions of metallopeptidase genes, and some positively selected genes were highly related to their larvae’s fast development. Our findings provide novel insights into the genetic mechanisms of coevolution between nonmodel mammals and parasites and offer the valuable genomic resources for scientific ascariasis prevention in both pandas.
Keywords: coevolution, comparative genomics, parasitism, pandas
Introduction
Coevolution is an important biological driver of species evolution that affects speciation, population differentiation, phenotype evolution, and biodiversity maintenance (Woolhouse et al. 2002). Coevolution includes mutualistic and antagonistic coevolution. Coevolution between plants and pollinating insects and that between insects and symbiotic fungi are classic examples of mutualistic coevolution, whereas cases of coevolution between bacteria and bacteriophages, plants and pathogenic fungi, and humans and parasites are well-known examples of antagonistic coevolution. Because the above coevolution systems are experimentally manipulatable, the genetic mechanisms of their coevolution have been extensively studied, including gene-for-gene interactions, genotype-by-genotype interactions, and multilocus gene-for-gene interactions (Shan et al. 2007; Brockhurst and Koskella 2013; Cogni et al. 2016). In contrast, studies on the genetic mechanisms of coevolution between nonmodel animals and parasites are rare (Kirkness et al. 2010), because nonmodel coevolution systems are difficult to be studied under controlled and temporal dynamic conditions. With the rapid development of next-generation sequencing technology, de novo whole-genome assemblies and innovative bioinformatics methods are enabling genome-wide studies for detecting the molecular mechanisms of coevolution. However, although the genomes of an increasing number of parasites are being sequenced (Jex et al. 2011; Tsai et al. 2013; Tang et al. 2014; Schwarz et al. 2015; Zhu et al. 2015; Wang et al. 2016; Zheng et al. 2019), these genome-level studies have mainly focused on the genetic bases of parasitism but have not explored coevolution between parasites and their hosts from the perspective of both sides.
The giant panda roundworm (Baylisascaris schroederi) is an obligate parasite of giant pandas (Sprent 1968). Wild giant pandas are infected by B. schroederi at an infection rate of >50% (Zhang et al. 2011). Research showed that the deaths of 50% of the examined dead wild giant pandas were due to roundworm infection, suggesting that roundworm infection is a major cause of wild giant panda deaths (Zhang et al. 2008). The red panda roundworm (Baylisascaris ailuri) is an obligate parasite of red pandas and is also an important cause of wild red panda deaths (Yang and Wang 2000). Although the phylogeny and genetic diversity of the giant panda roundworm and red panda roundworm have been studied based on mitochondrial DNA markers (Zhou et al. 2013; Liu et al. 2014; Xie et al. 2014), the genetic bases of the adaptation of the roundworms from both pandas to parasitism remain unclear. More importantly, the coevolution mechanisms between pandas and their obligate roundworms remain to be investigated.
Here, we investigated the genome-level mechanisms of coevolution between nonmodel mammals and their parasitic roundworms. We performed the de novo whole-genome sequencing and assembly of the giant panda roundworm, the red panda roundworm, and the lion roundworm Toxascaris leonina parasitic on tiger and identified the genomic signatures of roundworm parasitism using a comparative genomics method. Then, combined with the host gene expression profiles through simulating the parasite’s life cycle within the host, we investigated the protein interaction networks between host and roundworm. Finally, on the basis of gene tree topology between host and roundworm, we identified the genetic signatures of coevolution between nonmodel mammals and parasitic roundworms. Our findings provide novel insights into the genomic mechanisms of coevolution and contribute to understanding the effects of parasitism on host genome evolution. In addition, this study provides important genomic resources for scientific ascariasis prevention in both pandas.
Results
Genome Assembly and Annotation of Three Roundworm Species
We sequenced the genomes of three roundworm species (i.e., the giant panda roundworm B. schroederi, the red panda roundworm B. ailuri, and the lion roundworm T. leonina parasitic on tiger) using the Illumina HiSeq 4000 platform with a whole-genome shotgun sequencing strategy. In total, we sequenced the genomes of B. schroederi, B. ailuri, and T. leonina at average depths of 449.3×, 256.3×, and 229.8×, respectively (supplementary tables S1–S3, Supplementary Material online). In particular, to obtain a better genome assembly, we performed PacBio RS II sequencing of the B. schroederi genome at an average depth of 13.5× (supplementary table S4, Supplementary Material online). For the B. schroederi genome assembly, we applied a combined assembly strategy for second-generation and third-generation sequencing reads and obtained a de novo genome assembly with a size of 281.6 Mb and a scaffold N50 of 888.9 kb (supplementary table S5, Supplementary Material online). For B. ailuri, we performed the de novo assembly of a genome with a size of 266.9 Mb and a scaffold N50 of 50.6 kb, whereas for T. leonina, we produced a de novo-assembled genome with a size of 284.8 Mb and a scaffold N50 of 35.5 kb (supplementary tables S6 and S7, Supplementary Material online). Benchmarking Universal Sing-Copy Orthologs (BUSCO) assessment (Waterhouse et al. 2018) of the genome assemblies showed that 87.9%, 80.9%, and 60.4% of the 982 conserved Nematoda genes were assembled in complete form for B. schroederi, B. ailuri, and T. leonina, respectively (supplementary table S8, Supplementary Material online). Compared with previously published genomes of nematode species (https://wormbase.org/), the genome assemblies of B. schroederi and B. ailuri were high quality, whereas the assembly quality of T. leonina was middle level (supplementary table S9, Supplementary Material online).
The MAKER2 gene annotation pipeline (Holt and Yandell 2011) predicted 13,284, 12,252, and 16,087 protein-coding genes in the B. schroederi, B. ailuri, and T. leonina genomes, respectively (supplementary table S10, Supplementary Material online). The BUSCO assessment of the gene annotations showed that 87.0%, 81.1%, and 62.0% of the 982 conserved Nematoda genes were annotated as complete for B. schroederi, B. ailuri, and T. leonina, respectively (supplementary table S11, Supplementary Material online). Gene functional annotation demonstrated that 93.64%, 93.76%, and 72.00% of the respective gene set could be functionally annotated (supplementary table S12, Supplementary Material online).
Genome Functions Related to Parasitism
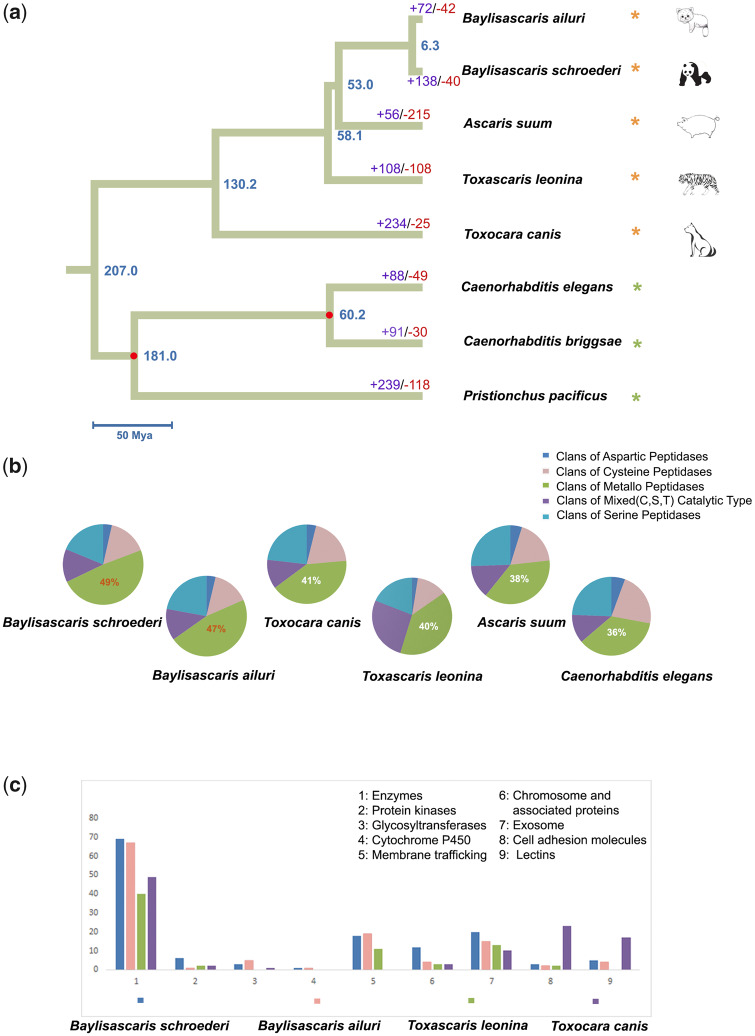
Proteases play a critical role in parasitism including the involvement in invading the host by parasite migration through tissue barriers, degradation of hemoglobin and other blood proteins, immune evasion, and activation of inflammation (McKerrow et al. 2006). Protease inhibitors create a safer environment in the host by inhibiting and regulating protease activity and immune regulation (Ranasinghe and McManus 2017). Thus, in order to explore the adaptation of roundworms to parasitism, we identified the protease and protease inhibitor gene families in the B. schroederi, B. ailuri, and T. leonina genomes (supplementary tables S13 and S14, Supplementary Material online). To achieve a better comparison, we also identified the protease and protease inhibitor gene families in the dog roundworm (Toxocara canis), pig roundworm (Ascaris suum), and Caenorhabditis elegans genomes. The results showed that these six nematode species exhibited similar numbers of protease genes and gene families (supplementary table S13, Supplementary Material online). However, in terms of the relative proportions of protease gene families, the B. schroederi and B. ailuri genomes presented higher proportions of metallopeptidases (49% and 47%, respectively) (fig. 1b). Metallopeptidases are considered to be involved in host tissue invasion (Haffner et al. 1998), ecdysis (Rhoads et al. 1997), and nutrient digestion (Rhoads and Fetterer 1998). For instance, in Strongyloides stercoralis, it has been shown that a zinc-endometallopeptidase secreted by infective larva is used to penetrate the skin of mammalian hosts (McKerrow et al. 1990). In addition, in A. suum female adults, the intestine exhibits 3- to 8-fold higher metallopeptidase activity than other tissues and fluids, suggesting that this enzyme shows digestive activity (Rhoads and Fetterer 1998). Therefore, the higher levels of metallopeptidase found in B. schroederi and B. ailuri may be utilized to degrade host tissues and digest special bamboo metabolites. In contrast, the T. leonina genome presented the highest proportion of the T03 peptidase gene family (i.e., gamma-glutamyltransferase 1) (supplementary table S13, Supplementary Material online), which is closely related to glutathione metabolism and amino acid absorption (Lieberman et al. 1996; Zhang and Forman 2009). For the protease inhibitor gene families, the B. schroederi, B. ailuri, and T. leonina genomes exhibited fewer protease inhibitor genes and gene families than other nematodes (supplementary table S14, Supplementary Material online).
Fig. 1.
Genome-wide phylogenetic tree, protease gene family composition, and annotated KEGG pathways for the secretomes of parasitic roundworms. (a) Phylogenomic tree, divergence times, and gene family expansion and contraction for five parasitic roundworms and three free-living nematodes. A yellow asterisk indicates a parasitic roundworm and its corresponding host, and a green asterisk indicates a free-living nematode. Two divergence times (red node) were used as the calibration points for estimating divergence times. (b) The proportions of different clans of proteases. The mixed type indicates the mixture of the cysteine, serine, and threonine catalytic types. (c) Annotated KEGG pathways for secretomes. The x-coordinate shows the types of annotated KEGG pathways, and the y-coordinate shows the number of genes for the corresponding KEGG pathways.
Excretory/secretory peptides are central to understanding parasite–host interactions, because the release of them is associated with contacting host cytoplasm and interference with the host immune system (Gahoi et al. 2019). We predicted the secretomes of three roundworm species sequenced in this study and then compared their annotated KEGG pathways together with that of Toxo. canis (Zhu et al. 2015). The results showed that more genes related to the glycosyltransferase and cytochrome P450 pathways presented in B. schroederi and B. ailuri than in other roundworms (fig. 1c and supplementary table S15, Supplementary Material online). These two pathways play a key role in neutralizing the toxic effects of xenobiotics, especially in the detoxification mechanism for plant secondary metabolites (PSMs; Tiwari et al. 2016). It has been shown that an ethanolic extract from leaves of Phyllostachys edulis upregulates the activities of the cytochrome P450 enzymes CYP1A2 and CYP3A11 and uridine diphosphate glucuronosyltransferase in the mouse liver (Koide et al. 2011). Accordingly, the glycosyltransferase and cytochrome P450 pathways in the secretomes of B. schroederi and B. ailuri may represent a coping strategy to detoxify secondary metabolites of the host’s bamboo diet.
Secreted proteases play important roles in host tissue degradation, feeding, and larval migration in a range of helminths (McKerrow et al. 2006). Forty-nine, 34, and 37 secreted proteases were found among the secretomes of B. schroederi, B. ailuri, and T. leonina, respectively (supplementary table S16, Supplementary Material online, see http://merops.sanger.ac.uk/ for clan definitions), including relatively high numbers of metalloprotease clans and serine protease clans.
Phylogenomics and Divergence Time
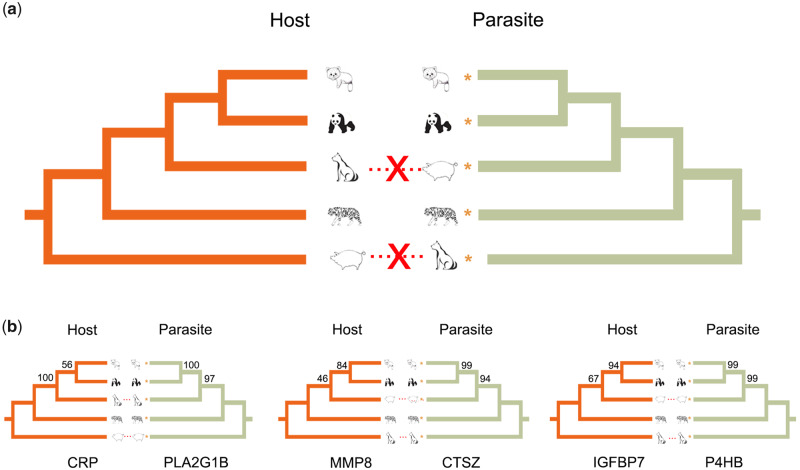
To understand the evolutionary relationships of these nematode species and their coevolution with host phylogeny, we constructed a genome-wide phylogenetic tree for eight nematode species: B. schroederi, B. ailuri, and T. leonina sequenced in this study, and A. suum, Toxo. canis, C. elegans, C. briggsae, and Pristionchus pacificus whose genomes have been published (supplementary table S17, Supplementary Material online). A total of 129,940 protein-coding genes from these eight species were subjected to gene family analysis, and 24,903 gene families were identified, including 1,444 single-copy orthologous genes across all eight species. Based on these single-copy protein-coding genes, the phylogenomic tree (fig. 1a) showed that B. schroederi and B. ailuri clustered together, whereas T. leonina and Toxo. canis were phylogenetically adjacent, and A. suum was located in the middle of the tree. This phylogenetic tree was basically consistent with that based on mitochondrial genomes (Liu et al. 2014). However, the phylogenetic topology of these parasitic roundworms was not consistent with their host phylogeny (fig. 4a), suggesting that these species have not phylogenetically coevolved with their hosts.
Fig. 4.
Species-level and gene-level phylogenetic relationships between hosts and roundworms, reflecting gene-level coevolution. (a) The species-level phylogenetic tree for hosts and roundworms. The yellow asterisk indicates the corresponding parasite. Symbol X shows inconsistency between two phylogenetic topologies. (b) Consistent gene trees for three pairs of interactive proteins, suggesting their molecular coevolution. The yellow asterisk indicates the corresponding parasite. The number on the branches represents the bootstrap value using Neighbor-Joining method.
A divergence time of 6.3 Ma between B. schroederi and B. ailuri was estimated using two divergence calibration points (fig. 1a). This divergence time was much later than that between the giant panda and red panda (47.5 Ma; Hu et al. 2017), suggesting that the speciation of the roundworms of both pandas occurred after their hosts’ species divergence. This result may indicate that the parasitic roundworms have evolved slowly because of the relatively stable internal intestinal environment.
Selection Signatures in Obligate Panda Roundworms
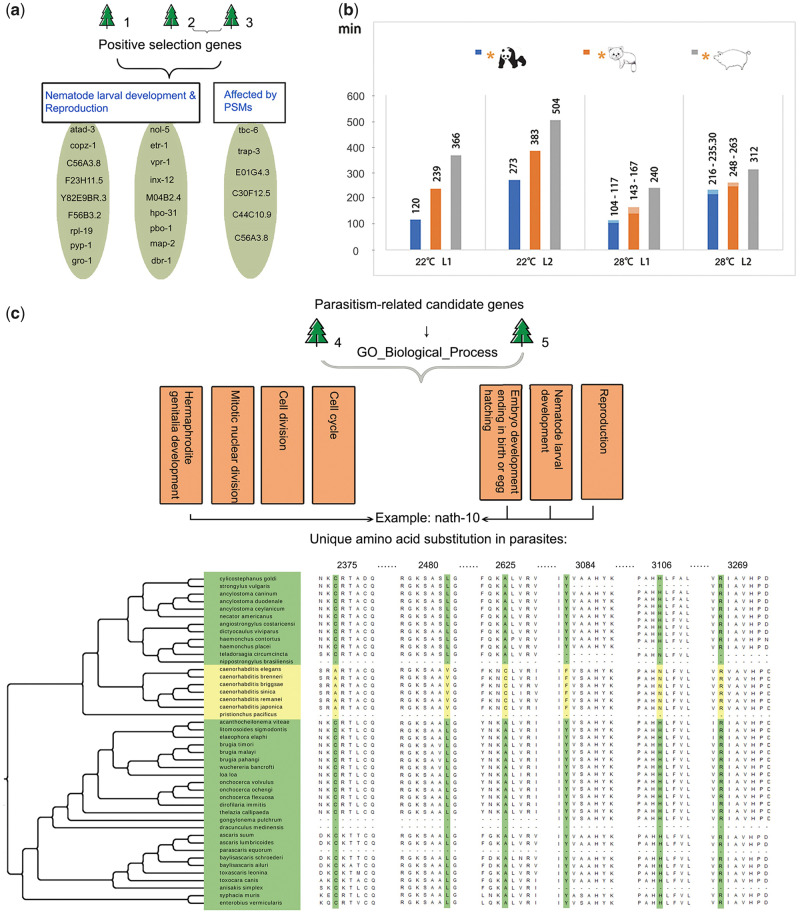
The giant panda and red panda are almost exclusive bamboo feeders, so their obligate parasitic roundworms may have evolved adaptability to cope with the hosts’ specialized diet. Based on the previous experimental observations of panda roundworms under in vitro culture (Wu et al. 1985; Wu 1988), we compared the development time of different roundworms (fig. 2b). Baylisascaris schroederi exhibited particularly short times of development into L1 and L2 larvae in vitro, of only 120 and 273 h at 22°, respectively, whereas B. ailuri took 239 and 383 h to develop into L1 and L2 larvae in vitro. Under the same conditions, A. suum exhibited relatively longer times of 366 and 504 h (Wu et al. 1985; Wu 1988). Therefore, signatures of common mechanisms related to faster development rates might be observed.
Fig. 2.
Positive selection related to the nutrient utilization and development of the roundworms from giant and red pandas, and positive selection and unique amino acid substitutions related to parasitism. (a) Two functional categories of positively selected genes in the roundworms from giant and red pandas. Trees 1, 2, and 3 indicate the first three positive selection analysis strategies, including both panda roundworms, only the giant panda roundworm, and only the red panda roundworm as the foreground branches, respectively. PSM, plant secondary metabolites. (b) In vitro development time of L1- and L2-stage larvae of three parasitic roundworms. The x-coordinate shows different temperature conditions and larval development stages, and the y-coordinate represents the development time (measured in minutes). A yellow asterisk indicates the host of the corresponding parasite. A lighter color in the column shows the fluctuation range of the development time. (c) GO biological process term enrichment of positively selected genes and unique amino acid substitutions related to parasitism. Trees 4 and 5 indicate the fourth and fifth positive selection analysis strategies, which both set all the parasitic roundworms as the foreground branches. Tree 4 only considered the positive selection of the most recent common ancestor lineage of five roundworms, and Tree 5 considered the positive selection of five roundworm lineages and their common ancestor lineages. Taking the nath-10 gene as an example, the arrow shows the enriched GO terms related to nath-10, and the multiple sequence alignments for 44 nematodes were ordered based on their phylogenetic relationships. Yellow represents free-living nematodes, and green represents parasites. Six amino acid substitutions were unique to all the parasites, with the number in the alignment being the location of unique amino acid substitutions.
Based on the phylogenomic tree that we constructed, three positive selection analyses were performed, which included both panda roundworms, only the giant panda roundworm, or only the red panda roundworm as the foreground branches (supplementary table S18, Supplementary Material online). A total of 5,587, 5,963, and 5,948 orthologous genes were included in these analyses; consequently, 69, 62, and 97 positively selected genes were identified, respectively. Among these genes, 42, 36, and 55 exhibited homologous gene annotations to C. elegans (supplementary tables S19–S21, Supplementary Material online). Only the annotated genes were included in further analysis. Moreover, since we focused on the common selection signatures of both panda roundworms, the positively selected genes obtained from the second and third analysis strategies were intersected and then combined with those from the first analysis strategy. As a result, a total of 49 positively selected genes were obtained (supplementary table S22, Supplementary Material online). The functional enrichment results showed that some genes were significantly enriched in gene ontology (GO) terms related to nematode larval development (GO:0002119) and reproduction (GO:0000003) (fig. 2a and supplementary table S23, Supplementary Material online).
Specifically, the atad-3 protein plays an important role in upregulating mitochondrial activity during the transition to higher larval stages. Its null mutant in C. elegans was found to arrest at developmental stages with low mitochondrial activity (Hoffmann et al. 2009). Additionally, the null mutant of pyp-1 exhibits developmental arrest at an early larval stage in C. elegans, and the larval arrest phenotype is successfully rescued by reintroduction (Ko et al. 2007). Thus, the positive selection of atad-3 and pyp-1 in panda roundworms might facilitate larval development. In addition, the daf-7 protein is a regulatory growth factor for dauer larva development and functions as a gauge of environmental conditions to regulate energy balance through a neural circuit. In daf-7 mutants of C. elegans, the perception of depleted food resources leads to fat accumulation without increasing the feeding rate (Greer et al. 2008). The positive selection of daf-7 in panda roundworms might be beneficial for sensing food conditions and modulating feeding behavior. In conclusion, these positively selected genes that are critical for larval development and reproduction may be involved in the faster larval development of B. schroederi and B. ailuri. Ascaris worms have access to partially digested nutrients in their hosts since they live in their upper small intestine (Beames and King 1972). Both giant and red pandas feed on high-fiber bamboo but have retained a short carnivoran alimentary tract, resulting in a very low digestive efficiency (Dierenfeld et al. 1982; Wei et al. 1999, 2015). Selective pressure on genes that increase larval development speed and nutrient utilization may come from their hosts’ adverse food sources. Only larvae that present stronger viability are able to survive under food shortage conditions. Accordingly, we inferred that the rapid development of panda roundworms may be an adaptation to survive in a nutritionally deficient host environment. Additionally, studies have indicated that six positively selected genes in our data set (fig. 2a) are affected by PSMs such as rotenone, quercetin, and hydrolyzable tannins, based on RNA-seq and microarray analyses (from https://wormbase.org/), which may be related to their hosts’ specialized bamboo diet.
Evolution of Roundworm Parasitism
To identify common selection signatures related to parasitism adaptation among roundworm genomes, we conducted positive selection analyses using the fourth and fifth strategies (supplementary table S18, Supplementary Material online), in which the parasitic roundworms were set the foreground branches and free-living nematodes as the background branches. As a result, we identified 186 and 12 positively selected genes, respectively. Furthermore, we detected the specific amino acid substitutions of positively selected genes that are probably related to parasitism. We used the reciprocal best-hit method to expand the orthologs of the above 196 positively selected genes (supplementary table S24, Supplementary Material online) from eight species to 87 nematode species (75 parasitic and 12 free-living species; supplementary table S9, Supplementary Material online). Then, considering possible sequencing errors in these genomic variations, we identified significantly different amino acid substitutions by calculating the frequency of each amino acid between the parasitic and free-living nematodes based on the multiple sequence alignment of each gene and requiring that a candidate amino acid substitution is shared by at least ten free-living species and at least 20 parasitic species. Consequently, we detected 57 genes with 114 such amino acid substitutions. To identify amino acid substitutions that stringently discriminate parasitic from free-living nematodes, we focused on 44 closely related nematodes (fig. 2c) on the basis of the above analysis of 87 nematode species, and the multiple sequence alignment of the above candidate amino acid substitutions was manually checked. Finally, a total of 34 positively selected genes with 60 unique amino acid substitutions were identified in the parasitic nematodes (supplementary table S25, Supplementary Material online). Five of these unique substitutions were predicted to be possibly damaging and one of them was probably damaging to the protein structure (supplementary table S25, Supplementary Material online). GO analyses revealed significant terms involved in embryo development ending in birth or egg hatching (GO:0009792), reproduction (GO:0000003), nematode larval development (GO:0002119), hermaphrodite genitalia development (GO:0040035), and mitotic nuclear division (GO:0007067) (fig. 2c and supplementary table S26, Supplementary Material online).
Some of these positively selected genes were related to parasitism adaptation. Parasites often increase their reproductive potential through the production of great numbers of eggs. For example, a single Ascaris lumbricoides individual can produce more than 200,000 eggs per day for several months (Schmidt et al. 1977). Two genes, smc4 and nath-10, are involved in increasing the offspring number of parasites. The smc4 protein is involved in mitotic sister chromatid segregation and localizes to the centromeric region and condensed chromosomes. It has been suggested that smc4 plays essential roles in parasite proliferation and transmission (Pandey et al. 2020). The nath-10 protein was also shown to be responsible for slight increases in the egg-laying rate and the total number of sperm, affecting the tradeoff between fertility and minimal generation time (Duveau and Félix 2012). In addition, the ran-3 protein is required for infection virulence and efficient nuclear trafficking (Frankel et al. 2007). A unique amino acid substitution in this gene in parasites may improve the efficiency of infection, which is the key step for parasite invasion. On the whole, these positively selected genes may play a vital role during parasitism adaptation.
Protein Interaction between Nonmodel Mammals and Their Parasitic Roundworms
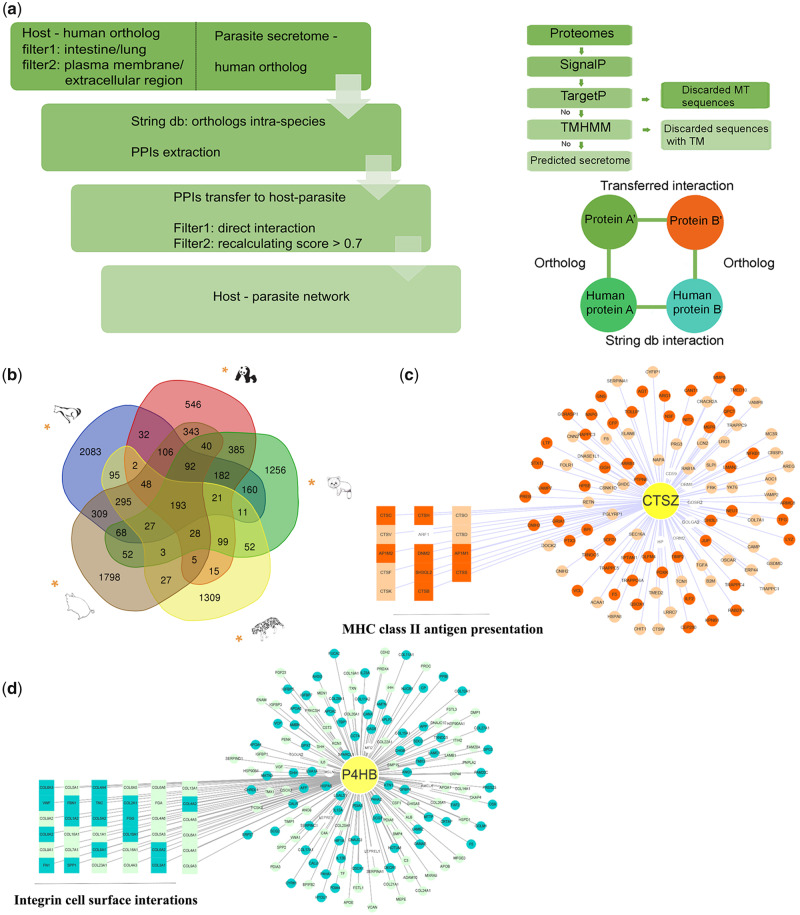
It is generally believed that host–parasite coevolution is based on antagonism. The host reacts to the presence of symbionts, mounting a defense against foreign invaders, and successful symbionts must evolve coping strategies to evade host defense (Schmidt et al. 1977). Host–parasite protein–protein interactions (PPIs) can facilitate the understanding of molecular-level coevolution between hosts and parasites. In our study, we constructed PPI networks of five host–parasite coevolution systems using genome-wide filtered proteomes, including the giant panda–B. schroederi (GB), red panda–B. ailuri (RB), tiger–T. leonina (TT), dog–Toxo. canis (DT), and pig–A. suum (PA) systems. In total, we identified 2,137, 2,669, 3,724, 2,230, and 3,434 PPIs in the GB, RB, TT, DT, and PA systems, respectively. Among these interactions, 193 PPIs were identified in all five systems, whereas GB exhibited 546 unique PPIs, RB exhibited 1,256 unique PPIs, TT exhibited 1,309 unique PPIs, DT exhibited 2,083 unique PPIs, and PA exhibited 1,798 unique PPIs (fig. 3b). According to the network topology results, the degree was used as a measure to rank the importance of proteins in the networks of the five host–parasite systems (supplementary tables S27–S31, Supplementary Material online). For example, in the GB system (supplementary table S27, Supplementary Material online), the first-ranking protein was ADAM10, which contains a metalloprotease domain and is highly involved in immune system (Gibb et al. 2011). The 11th-ranking protein, ERP44, is related to parasite-induced endoplasmic reticulum stress and contributes to the success of infection (Inácio et al. 2015). Additionally, we found that the ninth-ranking protein COL4A6, an important extracellular matrix structural constituent, from the host in the GB system was a positively selected gene from Hu et al. (2017).
Fig. 3.
PPI network between hosts and parasites. (a) The pipeline for building the host–parasite PPI network, the steps for predicting the secretome, and the homology-based PPI identification method. (b) Venn diagram for the number of PPIs in five host–parasite systems. The yellow asterisk indicates the host of the corresponding parasite. (c) The PPI network of the parasite protein CTSZ with host proteins. Darker orange indicates that this protein appears in all five systems, lighter orange indicates that this protein appears in more than one system, and white indicates that this protein appears in only one system. Fourteen host proteins displayed in squares were enriched in the MHC-II antigen presentation pathway. (d) The PPI network of parasite protein P4HB with host proteins. Darker blue indicates that the protein appears in all four systems, lighter blue indicates that the protein appears in more than one system, and white indicates that the protein appears in only one system. Thirty-six host proteins shown in squares were enriched in the integrin cell surface interaction pathway.
We also identified the specific interaction networks within the GB and RB interaction systems and the TT and DT interaction systems. The GB- and RB-specific networks presented 425 interactions, whereas the TT- and DT-specific networks exhibited 402 interactions. There were five parasite proteins (degree > 1) specifically involved in the GB and RB networks and not in TT and DT networks (supplementary table S32, Supplementary Material online). Only two parasite proteins (degree > 1) were specifically involved in TT and DT networks and not in the GB and RB networks (supplementary table S33, Supplementary Material online).
Moreover, we found that all five host–parasite interaction systems included the CTSZ and prolyl 4-hydroxylase subunit beta (P4HB) proteins among the top ten proteins ranked by the degree (table 1). The CTSZ protein (cathepsin Z) is a lysosomal cysteine protease and a member of the peptidase C1 family. The corresponding gene was also under positive selection (supplementary table S24, Supplementary Material online). We performed functional enrichment analysis for all the proteins from the host proteomes that interacted with parasite CTSZ and discovered significant reactome pathways, including MHC class II antigen presentation pathway (fig. 3c and supplementary table S34, Supplementary Material online). The long-term survival of parasites in the host is related to modifying and downregulating augmented host immune responses (Zandman-Goddard and Shoenfeld 2009). CTSZ presents a strong interaction with IL-1β and is involved in B/T-cell proliferation and adhesion of macrophages (Bernhardt et al. 2010). CTSZ-deficient mice show a reduced efficiency of IL-1β secretion by antigen-presenting cells, leading to a reduction in Th17 responses (Allan et al. 2017). So, roundworms may utilize the CTSZ protein to regulate MHC class II antigen presentation and immune response of the host.
Table 1.
The List of Top Ten Proteins Based on the Degree Ranking among All Five Host–Parasite Interaction Systems.
| Ranking | Degree | Gene Name | Degree | Gene Name | Degree | Gene Name | Degree | Gene Name | Degree | Gene Name |
|---|---|---|---|---|---|---|---|---|---|---|
| GB | RB | DT | TT | PA | ||||||
| 1 | 203 | ADAM10 | 217 | ITGB1 | 169 | P4HB | 339 | PIK3CA | 226 | ADAM10 |
| 2 | 150 | P4HB | 165 | ACTB | 153 | LAMB1 | 186 | PSAP | 159 | P4HB |
| 3 | 137 | ACTB | 165 | P4HB | 125 | HSP90B1 | 144 | CSNK1D | 141 | LAMC1 |
| 4 | 116 | LAMC1 | 122 | SNW1 | 107 | FBXO11 | 133 | P4HB | 124 | NOTCH1 |
| 5 | 110 | HSP90B1 | 120 | HSP90B1 | 103 | RET | 98 | UBE2A | 121 | UBE2V2 |
| 6 | 92 | CTSZ * | 98 | CTSZ * | 102 | CTSZ * | 94 | CTSZ * | 114 | STUB1 |
| 7 | 92 | RALA | 98 | EIF2S2 | 98 | EIF2S3 | 88 | CTSD | 99 | CTSZ * |
| 8 | 74 | RAB11B | 93 | EFNB1 | 96 | CTSD | 83 | HSPG2 | 98 | MBTPS1 |
| 9 | 63 | COL4A6* | 86 | RBM5 | 96 | CALU | 76 | TTR | 97 | PCF11 |
| 10 | 59 | C6orf120 | 82 | GUSB | 96 | RALA | 75 | TMED7 | 96 | RPL37 |
Note.—GB, giant panda–Baylisascaris schroederi system; RB, red panda–Baylisascaris ailuri system; DT, dog–Toxocara canis system; TT, tiger–Toxascaris leonina system; PA, pig–Ascaris suum system; P4HB and CTSZ (bold) occurred in all five interaction systems.
indicates positively selected gene.
P4HB has also been indicated as a key protein in other host–parasite PPI studies (Cuesta-Astroz et al. 2019), consistent with our results. Enrichment analysis identified significant reactome pathways such as integrin cell surface interaction, immunoregulatory interactions between a lymphoid and nonlymphoid cells, and antigen presentation, including the folding, assembly, and peptide loading of class I MHC (fig. 3d and supplementary table S35, Supplementary Material online). Previous studies have shown that P4HB plays a key role in internalization of certain pathogens (Ali Khan and Mutus 2014) and induction of phagocytosis (Santos et al. 2009). P4HB is also associated with other pathogens by inhibiting entry of viruses such as HIV, dengue virus, or rotavirus (Calderon et al. 2012; Wan et al. 2012). Therefore, in host–parasite interactions, P4HB may be utilized by roundworms to interfere with the host phase of internalizing the exogenous parasites.
Coevolution between Nonmodel Mammals and Their Parasitic Roundworms
In host and parasite systems, any similarity of phylogenetic trees must be due to some type of evolutionary interaction (Lovell and Robertson 2010). For the 193 common PPIs within the above five interaction systems, only 186 PPIs showed clear sources of hosts and roundworms for each protein (supplementary table S36, Supplementary Material online). We constructed gene trees of 186 pairs of interactive proteins between hosts and roundworms separately. The results showed that only seven pairs of gene trees for PPIs had identical tree topology, that is, C-reactive protein (CRP)–PLA2G1B, PLA2G7–PLA2G1B, PLA2R1–PLA2G1B, PLB1–PLA2G1B, IGFBP7–P4HB, MMP8–CTSZ, and QPCT–CTSZ (fig. 4b and supplementary fig. S1, Supplementary Material online), among which four pairs had gene tree topology identical to that of the hosts and three pairs had topology identical to that of the roundworms. Specifically, for the CRP–PLA2G1B interaction, CRP from the host is an acute-phase protein that serves as an early marker of inflammation or infection (Paul et al. 2012). It can bind to the component of surface of damaged cells such as phosphocholine and activate the immune system and phagocytosis (World Health Organization 2014). PLA2G1B protein from the roundworm encodes a secreted member of the phospholipase A2 (PLA2) class of enzymes. Phospholipases are a group of enzymes that hydrolyze phospholipids into fatty acids and other lipophilic molecules. Thus, coevolution may occur because CRP binds PLA2G1B in a way and it induces the host resistance to the roundworm during the long-term infection. For the IGFBP7–P4HB interaction, IGFBP7 is a protein secreted by monocytes in response to parasite stimulation. In malaria, it can trigger infected erythrocytes to form the type II rosettes and escape phagocytosis. The plasmodium can also sense the protein and escape phagocytosis through attaching to the infected erythrocytes (Lee et al. 2020). MMP8 in the MMP8–CTSZ interaction is proved to play a role in the trafficking of lymphocytes into central nervous system during chronic infection with the parasite Toxoplasma gondii (Clark et al. 2011). Taken together, these coevolutionary PPIs may be particularly relevant to the immune response during the antagonistic coevolution of hosts and roundworms.
Discussion
Species interactions between hosts and parasites are typical examples of antagonistic coevolution. In this study, we performed the de novo sequencing of the genomes of three parasitic roundworms from wild animals. Comparative genomics analysis showed that some genes related to larval development and detoxification were under positive selection in both the giant panda roundworm and red panda roundworm and that some genes were involved in parasitism evolution. In particular, we constructed protein interaction networks between hosts and roundworms through simulating the parasite’s life cycle within the host in five interaction systems and found that seven pairs of gene trees for PPIs had the same topologies for hosts and roundworms. These candidate key coevolutionary proteins were involved in immune regulation, providing novel insights into the molecular mechanisms of antagonistic host–parasite coevolution. Finally, these genome resources, secretomes, and candidate genes related to parasitism and coevolution will be useful for the effective development of specific antiparasitic drugs and vaccines, which will aid in the conservation of these high-profile threatened mammals.
Materials and Methods
Sample Collection and Species Identification
We collected parasitic roundworm individuals from the fresh feces of captive giant panda, Chinese red panda, and Amur tiger individuals. After sample collection, we quickly stored the roundworms in a refrigerator. To ensure the validity of the roundworm species used, we performed polymerase chain reaction amplification of the mitochondrial 12S rRNA gene for these samples (Niu et al. 2012) and then conducted BLAST alignment against the NCBI database for species identification. The BLAST results showed that the roundworms collected from the fresh feces of captive giant panda, Chinese red panda, and Amur tiger were B. schroederi, B. ailuri, and T. leonina, respectively. We subsequently used these roundworm individuals for de novo genome sequencing.
De Novo Whole-Genome Sequencing
The genomes of three roundworm species were sequenced using a whole-genome shotgun strategy. Genomic DNA was extracted from the whole body tissue of a single roundworm individual after eliminating the outer cuticle. For the genome sequencing of the giant panda roundworm, B. schroederi, we used a combined strategy of second-generation and third-generation high-throughput sequencing. First, paired-end and mate-pair libraries with insert sizes of 250–10 kb were constructed and then sequenced on the Illumina HiSeq 4000 platform (supplementary table S1, Supplementary Material online). Second, a 10-kb genome library was constructed and then sequenced on the PacBio RS II platform (supplementary table S4, Supplementary Material online). Because of the low quantity of the genomic DNA derived from a single roundworm individual, we used genomic DNA that was directly extracted to construct the paired-end libraries, but we used genomic DNA obtained from whole-genome amplification to construct the mate-pair libraries and PacBio libraries. For the genome sequencing of the red panda roundworm B. ailuri and the lion roundworm T. leonina from the tiger, we only used a second-generation sequencing strategy. Paired-end and mate-pair libraries with insert sizes of 235–5 kb were constructed and then sequenced on the Illumina HiSeq 4000 platform (supplementary tables S2 and S3, Supplementary Material online).
Genome Assembly and Annotation
For the genome assemblies of B. schroederi, B. ailuri, and T. leonina, we used SOAPdenovo v2.04 (Luo et al. 2012) to assemble the genome with Illumina sequencing reads. Then, we used the GapCloser module within SOAPdenovo2 to fill gaps in the scaffolded assembly. Next, we used BLAST to align the assembled scaffolds against the NR database to filter the DNA sequences from mitochondria, indigenous microbes, and hosts. In particular, for the genome assembly of B. schroederi, we combined the PacBio long sequencing reads. The PacBio long reads were error corrected by the LSC method (Au et al. 2012) using the Illumina short reads from the same individual. Then, we used the PBJelly (English et al. 2012) and SSPACE-LongRead (Boetzer and Pirovano 2014) methods to obtain the final genome based on the PacBio long reads and the above Illumina read-based scaffolds. We used the BUSCO method to assess the completeness of these genome assemblies.
We used the MAKER2 annotation pipeline (Holt and Yandell 2011) to perform protein-coding gene annotation according to two strategies: protein homology and ab initio gene prediction. The protein homology input consisted of all proteins of the pig roundworm A. suum, the dog roundworm Toxo. canis, and C. elegans from the NCBI database. Ab initio gene predictions were produced by three programs (SNAP, Augustus, and GeneMark) within the MAKER2 pipeline. Then, we assessed the completeness of the gene annotations using the BUSCO method. Finally, we used the BlastP method to perform functional annotations of predicted protein-coding genes against the NR, GO, KEGG, COG-KOG, and SwissProt databases.
Prediction of the Protease and Protease Inhibitor Gene Families and Secretome
Proteases (also called peptidases or proteinases) are enzymes that catalyze proteolysis and breakdown proteins into smaller polypeptides or single amino acids. They are involved in a large class of biological functions and are important in host–parasite interactions (Malagón et al. 2013). These proteases are divided into five major classes (aspartic, cysteine, metallo, serine, and threonine proteases). In our study, proteases and protease inhibitors were identified and classified into families by using BlastP against the MEROPS peptidase database (Rawlings et al. 2016, http://www.ebi.ac.uk/merops), with at least 60 amino acids matched for each protein. Excretory/secretory proteins (i.e., secretome) were predicted by the programs SignalP (Petersen et al. 2011), TargetP (Emanuelsson et al. 2000), and TMHMM (Krogh et al. 2001). Proteins with a signal peptide sequence but without a transmembrane region were identified as secretome proteins, excluding the mitochondrial sequences.
Gene Family Expansion and Contraction, Phylogenomic Tree, and Divergence Time Estimation
We used the OrthoMCL v2.0.9 pipeline (Li et al. 2003) to build gene families for eight nematode species. In addition to the gene sets of the three roundworm species sequenced in this study, protein-coding genes from five other nematode species, including A. suum, Toxo. canis, C. elegans, C. briggsae, and P. pacificus, were downloaded from the NCBI database. First, BlastP was used for the pairwise alignment of protein sequences between species. Then, OrthoMCL was used to identify gene families including single-copy orthologous protein-coding genes. Gene family data from OrthoMCL were introduced into CAFÉ v3.0 (De Bie et al. 2006) to identify the expansion and contraction of gene families.
Each single-copy orthologous gene sequence was aligned and concatenated by the MUSCLE method (Edgar 2004). ModelTest (Posada and Crandall 1998) was used to estimate the best-fit substitution model (the GTRGAMMA model). Then, a maximum-likelihood phylogenomic tree was constructed by using RAxML software (Stamatakis 2014) with 1,000 bootstrap replicates. Free-living nematode species were used as the outgroup. Furthermore, we used the r8s program (Sanderson 2003) to estimate divergence times between roundworm species with two calibration points applied from the TimeTree database (www.timetree.org): the divergence time of C. elegans and C. briggsae (60.2 Ma) and the divergence time of C. elegans and P. pacificus (181.0 Ma).
Positive Selection Analysis
Based on the reconstructed phylogenomic tree (fig. 1a), we performed a positive selection analysis using the branch-site model of codon evolution with model = 2 and NSsites = 2 in PAML v4.8 (Yang 2007). We aligned 1:1:1 orthologous genes using Prank (Löytynoja 2014) and obtained conservative information sites with Gblocks (Castresana 2000). We performed five positive selection analyses (supplementary table S18, Supplementary Material online). The first three strategies were used to identify the adaptation signatures of panda roundworms to their almost exclusively bamboo-eating hosts: 1) the giant and red panda roundworms were set as the foreground branches, and the pig roundworm, dog roundworm, and tiger roundworm were set as the background branches; 2) only the giant panda roundworm was set as the foreground branch, and the other three species (pig/dog/tiger roundworms) were set as the background branches; or 3) only the red panda roundworm was set as the foreground branch, and the other three species (pig/dog/tiger roundworms) were set as the background branches. The fourth and fifth strategies were used to analyze the selection signals related to the parasitic lifestyle: 4) “#” was used to label the common ancestor of five parasites (giant_panda/red_panda/pig/dog/tiger roundworms) in the phylogenetic tree file used by PAML, which means that only the selection of the common ancestor lineage was considered, and C. elegans, C. briggsae, and P. pacificus were used as the background branches; 5) “$” was used to label the common ancestor of the five parasites, and the other settings were identical to (4), which means that the selection of the common ancestor and the five parasite branches was considered. Finally, the functional enrichment of these positively selected genes according to GO terms was performed by the DAVID method (Huang et al. 2009), with the 1,444 orthologous genes of eight species as the reference gene set. The significantly enriched category included at least two genes, and the hypergeometric test was used to estimate significance (P < 0.05). The FDR method was further used to correct for multiple testing and estimate q values, with q < 0.1 being considered significant.
Identification of Unique Amino Acid Substitutions Related to Parasitism
We focused on the positively selected gene sets obtained from the fourth and fifth strategies in the positive selection analysis and applied two steps to screen for unique amino acid substitutions shared by the parasitic nematodes. First, 87 currently released nematode proteomes (including those of 75 parasitic nematodes and 12 free-living nematodes) were obtained (supplementary table S9, Supplementary Material online). Then, the orthologs of positively selected genes in these nematodes were obtained by the reciprocal best-hit BLAST approach. Briefly, with these orthologous genes of C. elegans as a reference, the protein set of each collected species was BLAST searched against the reference, and reciprocal best-hit pairs were considered as orthologs. Next, multiple sequence alignment was performed for each orthologous gene using Prank (Löytynoja 2014). To avoid the impact of high-throughput sequencing errors on the identification of unique amino acid substitutions, we calculated the frequency of an amino acid substitution that is dominant in parasitic roundworm species (i.e., the dominant amino acid substitution was shared by at least 20 species) and its corresponding frequency in free-living nematode species (i.e., the dominant amino acid was shared by at least ten species). The frequency difference was examined by Fisher’s exact test. An amino acid substitution with a significant frequency difference (P < 0.001) was considered a candidate unique amino acid substitution.
Second, to find the most stringent amino acid substitutions that discriminate parasitic from free-living nematodes, the candidate unique amino acid substitutions identified in the first step were further manually checked in 44 nematode species that are phylogenetically closely related, including 37 parasitic and seven free-living nematodes, and the phylogenetic tree of these species was derived from International Helminth Genomes Consortium (2019). The multiple sequence alignment results were displayed in accordance with the order of their evolutionary relationships using MEGA (Kumar et al. 2016), and the accuracy of the BLAST results was manually checked for the sequences near the candidate amino acid substitutions. Therefore, the positively selected genes related to parasitism were selected only if they contained amino acid substitutions that met the following conditions: 1) the amino acid substitutions at the site were identical among all parasitic species and 2) among all free-living species, the amino acid substitutions at the site were different from those among all parasitic species. We used PolyPhen-2 to predict the functional significance of the amino acid substitution for each homologous gene to that of C. elegans (Adzhubei et al. 2010). The functional enrichment of the filtered gene set for GO terms was tested by the DAVID method, with the 1,444 orthologous genes of eight species as the reference gene set. The significantly enriched category included at least two genes, and the hypergeometric test was used to estimate significance (P < 0.05). The FDR method was further used to correct for multiple testing and estimate q value, with q < 0.1 being considered significant.
Genome-Wide Host–Parasite Protein Interaction Analysis
Coevolution allows proteins to change with a similar magnitude while maintaining their interactions (Bitbol et al. 2016; Cong et al. 2019). Thus, the functional interactions between proteins can help to understand the coevolution between host and parasite. The homology-based method (Soyemi et al. 2018) (fig. 3a) has been applied to build host–parasite interactions. For the hosts, we implemented the reciprocal best-hit BLAST method to find the orthologs of five hosts (giant panda, red panda, dog, pig, and tiger) with the proteome of human as the reference. Moreover, proteins were filtered to build host–parasite interaction network since not all proteins are likely to interact with the parasite’s proteins. According to the life cycle information for Ascaris presented at https://www.cdc.gov/parasites/, only proteins expressed in the small intestine and lungs of the host and those located in the plasma membrane and extracellular region were considered. The gene expression and amino acid site information were obtained from the TISSUES and COMPARTMENTS databases, which provide high-confidence information on tissue expression and cellular localization, respectively (Binder et al. 2014; Santos et al. 2015).
For the parasites, we predicted the secretomes of five parasitic roundworms (the giant panda roundworm B. schroederi, the red panda roundworm B. ailuri, the dog roundworm Toxo. canis, the pig roundworm A. suum, and the lion roundworm T. leonina parasitic on tiger) (fig. 3a). The human orthologous genes corresponding to the secretomes of the parasites were obtained using the reciprocal BLAST best-hit method. These genes were used for the construction of host–parasite PPI networks.
The interaction files in the STRING database (Jensen et al. 2009) allow the filtering of PPIs by confidence scores; these files include several individual evidence channels (neighborhood, gene fusion, cooccurrence, coexpression, experiments, databases, and text mining) and are further subdivided into direct and transferred evidence. The transferred evidence is derived from ortholog transfer performed by the STRING database itself. To obtain high-credibility PPIs, we excluded those that only exhibited the transferred scores; moreover, we retained the high-confidence PPIs (combined score > 0.7) using the recalculated method (Cuesta-Astroz et al. 2019).
Once we obtained the predicted host–parasite PPIs, we used the degree index (number of connections) to reveal relevant proteins that may play a key role in host–parasite coevolution. Functional enrichment analysis of Reactome pathways was performed using the GeneTrail2 method (Stöckel et al. 2016) and took the combined filtered proteomes of the hosts and parasites as background. The significantly enriched category included at least two genes, and the hypergeometric test was used to estimate significance (P < 0.05).
Host–Parasite Coevolution Analysis
Congruence of gene-level phylogenetic trees was used as an indicator of PPI in two different biological systems such as hosts and parasites (Lovell and Robertson 2010), thus phylogenetic tree test was performed for the common protein interactions within the five interactive systems. First, we identified the source of each protein following two rules. For A–B interaction, one is that if A comes from the filtered proteome of host, then B must come from the filtered proteome of roundworm. The other is that A must be present in the filtered proteomes of the hosts in all five systems, and B must be present in the filtered proteomes of the roundworms in all five systems. After identifying the respective sources of the two proteins in the interactions, we extracted amino acid sequences and used the Neighbor-Joining method (bootstrap value of 500) and maximum-likelihood method in MEGA to construct the phylogenetic tree of the targeted interactive proteins from the hosts and roundworms separately.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This study is supported by the Strategic Priority Research Program of Chinese Academy of Sciences (XDB31000000), the National Natural Science Foundation of China (31822050 and 31821001), the Frontier Key Project of Chinese Academy of Sciences (QYZDY-SSW-SMC019), and the Youth Innovation Promotion Association, CAS (2016082). We thank Yixiang Shi and Long Huang for help with genomic data analysis and thank Chenglin Zhang, Hongshuai Shang, Yanqiang Yin, and Yunfang Xiu for help with sample collection.
Author Contributions
F.W. conceived and supervised this study. Y.H. performed the sample collection. Y.H., Lij.Y., and H.F. conducted the genomic data analyses. F.W., G.H., Q.W., Y.N., S.L., and Li Y. were involved in the project implementation and data interpretation. Y.H., Lij.Y., and F.W. wrote the manuscript with input from other authors.
Data Availability
The sequence data and the genome assembly of Baylisascaris schroederi have been deposited at NCBI under BioProject ID PRJNA612649. The sequence data and the genome assembly of Baylisascaris ailuri have been deposited at NCBI under BioProject ID PRJNA612650. The sequence data and the genome assembly of Toxascaris leonina have been deposited at NCBI under BioProject ID PRJNA612651.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR.. 2010. A method and server for predicting damaging missense mutations. Nat Methods. 7(4):248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali Khan H, Mutus B.. 2014. Protein disulfide isomerase a multifunctional protein with multiple physiological roles. Front Chem. 2:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan ERO, Campden RI, Ewanchuk BW, Tailor P, Balce DR, McKenna NT, Greene CJ, Warren AL, Reinheckel T, Yates RM.. 2017. A role for cathepsin Z in neuroinflammation provides mechanistic support for an epigenetic risk factor in multiple sclerosis. J Neuroinflammation 14(1):103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au KF, Underwood JG, Lee L, Wong WH.. 2012. Improving PacBio long read accuracy by short read alignment. PLoS One 7(10):e46679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beames CG Jr, King GA.. 1972. Factors influencing the movement of materials across the intestine of Ascaris. In: Comparative biochemistry of parasites. New York: Academic Press. p. 275–282. [Google Scholar]
- Bernhardt A, Kuester D, Roessner A, Reinheckel T, Krueger S.. 2010. Cathepsin X-deficient gastric epithelial cells in co-culture with macrophages. J Biol Chem. 285(44):33691–33700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder JX, Pletscher-Frankild S, Tsafou K, Stolte C, O’Donoghue SI, Schneider R, Jensen LJ.. 2014. COMPARTMENTS: unification and visualization of protein subcellular localization evidence. Database 2014:bau012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitbol AF, Dwyer RS, Colwell LJ, Wingreen NS.. 2016. Inferring interaction partners from protein sequences. Proc Natl Acad Sci U S A. 113(43):12180–12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetzer M, Pirovano W.. 2014. SSPACE-LongRead: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinf. 15(1):211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockhurst MA, Koskella B.. 2013. Experimental coevolution of species interactions. Trends Ecol Evol (Amst). 28(6):367–375. [DOI] [PubMed] [Google Scholar]
- Calderon MN, Guerrero CA, Acosta O, Lopez S, Arias CF.. 2012. Inhibiting rotavirus infection by membrane-impermeant thiol/disulfide exchange blockers and antibodies against protein disulfide isomerase. Intervirology 55(6):451–464. [DOI] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. J Mol Evol. 17(4):540–552. [DOI] [PubMed] [Google Scholar]
- Clark RT, Nance JP, Noor S, Wilson EH.. 2011. T-cell production of matrix metalloproteinases and inhibition of parasite clearance by TIMP-1 during chronic Toxoplasma infection in the brain. ASN Neuro 3(1):e00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogni R, Cao C, Day JP, Bridson C, Jiggins FM.. 2016. The genetic architecture of resistance to virus infection in Drosophila. Mol Ecol. 25(20):5228–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Q, Anishchenko I, Ovchinnikov S, Baker D.. 2019. Protein interaction networks revealed by proteome coevolution. Science 365(6449):185–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta-Astroz Y, Santos A, Oliveira G, Jensen LJ.. 2019. Analysis of predicted host–parasite interactomes reveals commonalities and specificities related to parasitic lifestyle and tissues tropism. Front Immunol. 10:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bie T, Cristianini N, Demuth JP, Hahn MW.. 2006. CAFE: a computational tool for the study of gene family evolution. Bioinformatics 22(10):1269–1271. [DOI] [PubMed] [Google Scholar]
- Dierenfeld ES, Hintz HF, Robertson JB, Van Soest PJ, Oftedal OT.. 1982. Utilization of bamboo by the giant panda. J Nutr. 112(4):636–641. [DOI] [PubMed] [Google Scholar]
- Duveau F, Félix MA.. 2012. Role of pleiotropy in the evolution of a cryptic developmental variation in Caenorhabditis elegans. PLoS Biol. 10(1):e1001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G.. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 300(4):1005–1016. [DOI] [PubMed] [Google Scholar]
- English AC, Richards S, Han Y, Wang M, Vee V, Qu J, Qin X, Muzny DM, Reid JG, Worley KC, et al. 2012. Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS One 7(11):e47768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel MB, Mordue DG, Knoll LJ.. 2007. Discovery of parasite virulence genes reveals a unique regulator of chromosome condensation 1 ortholog critical for efficient nuclear trafficking. Proc Natl Acad Sci U S A. 104(24):10181–10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahoi S, Singh S, Gautam B.. 2019. Genome-wide identification and comprehensive analysis of Excretory/Secretory proteins in nematodes provide potential drug targets for parasite control. Genomics. 111(3):297–309. [DOI] [PubMed] [Google Scholar]
- Gibb DR, Saleem SJ, Chaimowitz NS, Mathews J, Conrad DH.. 2011. The emergence of ADAM10 as a regulator of lymphocyte development and autoimmunity. Mol Immunol. 48(11):1319–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer ER, Pérez CL, Van Gilst MR, Lee BH, Ashrafi K.. 2008. Neural and molecular dissection of a C. elegans sensory circuit that regulates fat and feeding. Cell Metab. 8(2):118–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffner A, Guilavogui AZ, Tischendorf FW, Brattig NW.. 1998. Onchocerca volvulus: microfilariae secrete elastinolytic and males nonelastinolytic matrix-degrading serine and metalloproteases. Exp Parasitol. 90(1):26–33. [DOI] [PubMed] [Google Scholar]
- Hoffmann M, Bellance N, Rossignol R, Koopman WJ, Willems PH, Mayatepek E, Bossinger O, Distelmaier F.. 2009. C. elegans ATAD-3 is essential for mitochondrial activity and development. PLoS One 4(10):e7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C, Yandell M.. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinf. 12(1):491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Wu Q, Ma S, Ma T, Shan L, Wang X, Nie Y, Ning Z, Yan L, Xiu Y, et al. 2017. Comparative genomics reveals convergent evolution between the bamboo-eating giant and red pandas. Proc Natl Acad Sci U S A. 114(5):1081–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA.. 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inácio P, Zuzarte-Luís V, Ruivo MT, Falkard B, Nagaraj N, Rooijers K, Mann M, Mair G, Fidock DA, Mota MM.. 2015. Parasite‐induced ER stress response in hepatocytes facilitates Plasmodium liver stage infection. EMBO Rep. 16(8):955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Helminth Genomes Consortium. 2019. Comparative genomics of the major parasitic worms Nat. Genet. 51(1):163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. 2009. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 37(Database issue):D412–D416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jex AR, Liu S, Li B, Young ND, Hall RS, Li Y, Yang L, Zeng N, Xu X, Xiong Z, et al. 2011. Ascaris suum draft genome. Nature 479(7374):529–533. [DOI] [PubMed] [Google Scholar]
- Kirkness EF, Haas BJ, Sun W, Braig HR, Perotti MA, Clark JM, Lee SH, Robertson HM, Kennedy RC, Elhaik E, et al. 2010. Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proc Natl Acad Sci U S A. 107(27):12168–12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko KM, Lee W, Yu JR, Ahnn J.. 2007. PYP-1, inorganic pyrophosphatase, is required for larval development and intestinal function in C. elegans. FEBS Lett. 581(28):5445–5453. [DOI] [PubMed] [Google Scholar]
- Koide CL, Collier AC, Berry MJ, Panee J.. 2011. The effect of bamboo extract on hepatic biotransforming enzymes—findings from an obese–diabetic mouse model. J Ethnopharmacol. 133(1):37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL.. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305(3):567–580. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WC, Russell B, Sobota RM, Ghaffar K, Howland SW, Wong ZX, Maier AG, Dorin-Semblat D, Biswas S, Gamain B, et al. 2020. Plasmodium-infected erythrocytes induce secretion of IGFBP7 to form type II rosettes and escape phagocytosis. Elife 9:e51546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ Jr, Roos DS.. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13(9):2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman MW, Wiseman AL, Shi ZZ, Carter BZ, Barrios R, Ou CN, Chévez-Barrios P, Wang Y, Habib GM, Goodman JC, et al. 1996. Growth retardation and cysteine deficiency in gamma-glutamyl transpeptidase-deficient mice. Proc Natl Acad Sci U S A. 93(15):7923–7926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GH, Zhou DH, Zhao L, Xiong RC, Liang JY, Zhu XQ.. 2014. The complete mitochondrial genome of Toxascaris leonina: comparison with other closely related species and phylogenetic implications. Infect Genet Evol. 21:329–333. [DOI] [PubMed] [Google Scholar]
- Lovell SC, Robertson DL.. 2010. An integrated view of molecular coevolution in protein–protein interactions. Mol Biol Evol. 27(11):2567–2575. [DOI] [PubMed] [Google Scholar]
- Löytynoja A. 2014. Phylogeny-aware alignment with PRANK. Methods Mol Biol. 1079:155 –170. [DOI] [PubMed] [Google Scholar]
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience 1(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malagón D, Benítez R, Kašný M, Adroher FJ.. 2013. Peptidases in parasitic nematodes: a review. In: Parasites: ecology, diseases and management. Hauppauge (NY: ): Nova Science Publishers. p. 61–102. [Google Scholar]
- McKerrow JH, Brindley P, Brown M, Gam AA, Staunton C, Neva FA.. 1990. Strongyloides stercoralis: identification of a protease that facilitates penetration of skin by the infective larvae. Exp Parasitol. 70(2):134–143. [DOI] [PubMed] [Google Scholar]
- McKerrow JH, Caffrey C, Kelly B, Loke P, Sajid M.. 2006. Proteases in parasitic diseases. Annu Rev Pathol Mech Dis. 1(1):497–536. [DOI] [PubMed] [Google Scholar]
- Niu L, Li Y, Gu X, Yu H, Deng J, Yan H, Yu X, Chen W, Wang S, Yang G.. 2012. Phylogenetic analysis of ascarids from Ailuropoda melanoleuca and five other species of wild mammals based on the cytochrome b gene. Chin Vet Sci. 42:895–899. [Google Scholar]
- Pandey R, Abel S, Boucher M, Wall RJ, Zeeshan M, Rea E, Freville A, Lu XM, Brady D, Daniel E, et al. 2020. Plasmodium condensin core subunits (SMC2/SMC4) mediate atypical mitosis and are essential for parasite proliferation and transmission. Cell Rep. 30(6):1883–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul R, Sinha PK, Bhattacharya R, Banerjee AK, Raychaudhuri P, Mondal J.. 2012. Study of C reactive protein as a prognostic marker in malaria from Eastern India. Adv Biomed Res. 1(1):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H.. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 8(10):785. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA.. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14(9):817–818. [DOI] [PubMed] [Google Scholar]
- Ranasinghe SL, McManus DP.. 2017. Protease inhibitors of parasitic flukes: emerging roles in parasite survival and immune defence. Trends Parasitol. 33(5):400–413. [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Finn R.. 2016. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 44(D1):D343–D350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads ML, Fetterer RH.. 1998. Purification and characterisation of a secreted aminopeptidase from adult Ascaris suum. Int J Parasitol. 28(11):1681–1690. [DOI] [PubMed] [Google Scholar]
- Rhoads ML, Fetterer RH, Urban JF Jr.. 1997. Secretion of an aminopeptidase during transition of third- to fourth-stage larvae of Ascaris suum. J Parasitol. 83(5):780–784. [PubMed] [Google Scholar]
- Sanderson MJ. 2003. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics 19(2):301–302. [DOI] [PubMed] [Google Scholar]
- Santos A, Tsafou K, Stolte C, Pletscher-Frankild S, O’Donoghue SI, Jensen LJ.. 2015. Comprehensive comparison of large-scale tissue expression datasets. PeerJ. 3:e1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos CX, Stolf BS, Takemoto PV, Amanso AM, Lopes LR, Souza EB, Goto H, Laurindo FR.. 2009. Protein disulfide isomerase (PDI) associates with NADPH oxidase and is required for phagocytosis of Leishmania chagasi promastigotes by macrophages. J Leukoc Biol. 86(4):989–998. [DOI] [PubMed] [Google Scholar]
- Schmidt GD, Roberts LS, Janovy J.. 1977. Foundations of parasitology. St Louis (MO: ): Mosby. [Google Scholar]
- Schwarz EM, Hu Y, Antoshechkin I, Miller MM, Sternberg PW, Aroian RV.. 2015. The genome and transcriptome of the zoonotic hookworm Ancylostoma ceylanicum identify infection-specific gene families. Nat Genet. 47(4):416–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan LB, He P, Sheen J.. 2007. Endless hide-and-seek: dynamic co-evolution in plant–bacterium warfare. J Integr Plant Biol. 49(1):105–111. [Google Scholar]
- Soyemi J, Isewon I, Oyelade J, Adebiyi E.. 2018. Inter-species/host–parasite protein interaction predictions reviewed. Curr Bioinf. 13(4):396–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprent JFA. 1968. Notes on Ascaris and Toxascaris, with a definition of Baylisascaris gen. nov. Parasitology 58(1):185–198. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöckel D, Kehl T, Trampert P, Schneider L, Backes C, Ludwig N, Gerasch A, Kaufmann M, Gessler M, Graf N, et al. 2016. Multi-omics enrichment analysis using the GeneTrail2 web service. Bioinformatics 32(10):1502–1508. [DOI] [PubMed] [Google Scholar]
- Tang YT, Gao X, Rosa BA, Abubucker S, Hallsworth-Pepin K, Martin J, Tyagi R, Heizer E, Zhang X, Bhonagiri-Palsikar V, et al. 2014. Genome of the human hookworm Necator americanus. Nat Genet. 46(3):261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari P, Sangwan RS, Sangwan NS.. 2016. Plant secondary metabolism linked glycosyltransferases: an update on expanding knowledge and scopes. Biotechnol Adv. 34(5):714–739. [DOI] [PubMed] [Google Scholar]
- Tsai IJ, Zarowiecki M, Holroyd N, Garciarrubio A, Sánchez-Flores A, Brooks KL, Tracey A, Bobes RJ, Fragoso G, Sciutto E, et al. 2013. The genomes of four tapeworm species reveal adaptations to parasitism. Nature 496(7443):57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan SW, Lin CF, Lu YT, Lei HY, Anderson R, Lin YS.. 2012. Endothelial cell surface expression of protein disulfide isomerase activates β1 and β3 integrins and facilitates dengue virus infection. J Cell Biochem. 113(5):1681–1691. [DOI] [PubMed] [Google Scholar]
- Wang S, Wang S, Luo Y, Xiao L, Luo X, Gao S, Dou Y, Zhang H, Guo A, Meng Q, et al. 2016. Comparative genomics reveals adaptive evolution of Asian tapeworm in switching to a new intermediate host. Nat Commun. 7:12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, Seppey M, Simão FA, Manni M, Ioannidis P, Klioutchnikov G, Kriventseva EV, Zdobnov EM.. 2018. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol. 35(3):543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F, Feng Z, Wang Z, Zhou A, Hu J.. 1999. Use of the nutrients in bamboo by the red panda (Ailurus fulgens). J Zool. 248(4):535–541. [Google Scholar]
- Wei F, Hu Y, Yan L, Nie Y, Wu Q, Zhang Z.. 2015. Giant pandas are not an evolutionary cul-de-sac: evidence from multidisciplinary research. Mol Biol Evol. 32(1):4–12. [DOI] [PubMed] [Google Scholar]
- Woolhouse ME, Webster JP, Domingo E, Charlesworth B, Levin BR.. 2002. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat Genet. 32(4):569–577. [DOI] [PubMed] [Google Scholar]
- World Health Organization. 2014. C-reactive protein concentrations as a marker of inflammation or infection for interpreting biomarkers of micronutrient status. No. WHO/NMH/NHD/EPG/14.7.
- Wu J. 1988. Red panda ascariasis. Chin J Wildl. 2:24. [Google Scholar]
- Wu J, Jiang Y, Wu G, He G, Zhang D, Yang W, Hu H.. 1985. Observation on the egg development period of Baylisascaris schroederi. Chin Vet Sci. 10:9–12. [Google Scholar]
- Xie Y, Zhou X, Zhang Z, Wang C, Sun Y, Liu T, Gu X, Wang T, Peng X, Yang G.. 2014. Absence of genetic structure in Baylisascaris schroederi populations, a giant panda parasite, determined by mitochondrial sequencing. Parasit Vectors 7:606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang GY, Wang CD.. 2000. Research advances of parasite and parasitosis in red pandas. Chin Vet J. 26:36–38. [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Zandman-Goddard G, Shoenfeld Y.. 2009. Parasitic infection and autoimmunity. Lupus 18(13):1144–1148. [DOI] [PubMed] [Google Scholar]
- Zhang H, Forman HJ.. 2009. Redox regulation of gamma-glutamyl transpeptidase. Am J Respir Cell Mol Biol. 41(5):509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JS, Daszak P, Huang HL, Yang GY, Kilpatrick AM, Zhang S.. 2008. Parasite threat to panda conservation. EcoHealth 5(1):6–9. [DOI] [PubMed] [Google Scholar]
- Zhang L, Yang X, Wu H, Gu X, Hu Y, Wei F.. 2011. The parasites of giant pandas: individual-based measurement in wild animals. J Wildl Dis. 47(1):164–171. [DOI] [PubMed] [Google Scholar]
- Zheng L, Jiang N, Sang X, Zhang N, Zhang K, Chen H, Yang N, Feng Y, Chen R, Suo X, et al. 2019. In-depth analysis of the genome of Trypanosoma evansi, an etiologic agent of surra. Sci China Life Sci. 62(3):406–419. [DOI] [PubMed] [Google Scholar]
- Zhou X, Xie Y, Zhang ZH, Wang CD, Sun Y, Gu XB, Wang SX, Peng XR, Yang GY.. 2013. Analysis of the genetic diversity of the nematode parasite Baylisascaris schroederi from wild giant pandas in different mountain ranges in China. Parasit Vectors 6(1):233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XQ, Korhonen PK, Cai H, Young ND, Nejsum P, von Samson-Himmelstjerna G, Boag PR, Tan P, Li Q, Min J, et al. 2015. Genetic blueprint of the zoonotic pathogen Toxocara canis. Nat Commun. 6:6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequence data and the genome assembly of Baylisascaris schroederi have been deposited at NCBI under BioProject ID PRJNA612649. The sequence data and the genome assembly of Baylisascaris ailuri have been deposited at NCBI under BioProject ID PRJNA612650. The sequence data and the genome assembly of Toxascaris leonina have been deposited at NCBI under BioProject ID PRJNA612651.