Abstract
DNA cytosine methylation is central to many biological processes, including regulation of gene expression, cellular differentiation, and development. This DNA modification is conserved across animals, having been found in representatives of sponges, ctenophores, cnidarians, and bilaterians, and with very few known instances of secondary loss in animals. Myxozoans are a group of microscopic, obligate endoparasitic cnidarians that have lost many genes over the course of their evolution from free-living ancestors. Here, we investigated the evolution of the key enzymes involved in DNA cytosine methylation in 29 cnidarians and found that these enzymes were lost in an ancestor of Myxosporea (the most speciose class of Myxozoa). Additionally, using whole-genome bisulfite sequencing, we confirmed that the genomes of two distant species of myxosporeans, Ceratonova shasta and Henneguya salminicola, completely lack DNA cytosine methylation. Our results add a notable and novel taxonomic group, the Myxosporea, to the very short list of animal taxa lacking DNA cytosine methylation, further illuminating the complex evolutionary history of this epigenetic regulatory mechanism.
Keywords: methylome evolution, whole-genome bisulfite sequencing (WGBS), Cnidaria, parasite, cytosine methylation
Introduction
DNA methylation is a chemical modification of genomic DNA present in both prokaryotes and eukaryotes that affects gene regulation. DNA methylation of cytosines (cytosine methylation) has been extensively studied. An important functional consequence of DNA cytosine methylation is the suppression of gene expression when methylation is present in promoter regions (Schübeler 2015). This regulation affects a wide variety of key processes in animals including gametogenesis, embryonic development, cellular differentiation, X-chromosome inactivation, and transposon repression (Richa and Sinha 2014). Cytosine methylation also plays an important role in genome evolution. Cytosine methylation is mutagenic, as methylated cytosines can spontaneously deaminate to thymines (Bird 1980; Mendizabal et al. 2014). As a result, cytosine–guanine dinucleotides (CpGs) tend to change to thymine–guanine dinucleotides in genomic DNA over the course of evolution (Bird 1980; Mendizabal et al. 2014). Recent methods have allowed researchers to use CpG to thymine–guanine conversion rates to reconstruct past DNA methylation patterns (Mendizabal et al. 2014; Pedersen et al. 2014). Methylated genes tend to be more conserved than nonmethylated genes, even between distantly related species (Sarda et al. 2012). Such conservation attests to the importance of methylation at the genome level.
Similarly, the fact that cytosine methylation has been described in a wide variety of organisms, including viruses, prokaryotes, and eukaryotes, suggests a key biological importance (e.g., Zemach et al. 2010). Most of what we know about DNA cytosine methylation in animals comes from studies in bilaterians. Nevertheless, it has also been described in a number of early branching animal lineages, including sponges, ctenophores, and cnidarians (e.g., Hassel et al. 2010; Zemach et al. 2010; Dabe et al. 2015; Li et al. 2018; de Mendoza et al. 2019; Liew et al. 2020), and it is widely assumed that DNA methylation likely existed in the common ancestor of animals (Zemach and Zilberman 2010; Yi 2012). Strikingly, despite the biological importance of cytosine methylation and its high degree of conservation, a handful of animals are known to have secondarily lost this epigenomic modification. For example, cytosine methylation has been completely lost in the free-living nematode Caenorhabditis elegans, indicating that it is not crucial for gene regulation and development in this organism (Greer et al. 2015). It may have been also lost in the helminth parasite Schistosoma mansoni, although data are presently contradictory (Geyer et al. 2013; Raddatz et al. 2013). There is evidence that cytosine methylation has been completely lost in Diptera (flies), and in some hymenopterans (Bewick et al. 2017). However, the loss of cytosine methylation in the model dipteran Drosophila melanogaster is disputed (Raddatz et al. 2013; Capuano et al. 2014; Wang et al. 2018). Some species of lepidopterans, coleopterans, hemipterans, and blattodeans also show extremely low levels of methylation, although due to the lack of genome assemblies, whether DNA methyltransferases (DNMTs) exist in these species is currently unresolved (Bewick et al. 2017). Adenine methylation is also known to occur in diverse species, including D. melanogaster and Ca. elegans, although the function of this type of DNA methylation in eukaryotes remains poorly understood (Low et al. 2001; Ratel et al. 2006; Greer et al. 2015; Zhang et al. 2015; Wu et al. 2016; Liang et al. 2018).
Cytosine-methylation-mediated gene regulation involves a network of proteins working to methylate nucleotides, bind to methylated regions, and subsequently alter gene expression. DNMTs are highly conserved proteins responsible for DNA methylation (Fuks et al. 2000; Jin and Robertson 2013; Edwards et al. 2017). Three DNMTs have been inferred to be present in the common ancestor of animals (DNMT1, DNMT2/TRDMT1, and DNMT3) (Albalat et al. 2012). Studies in mammalian model systems demonstrated that patterns of cytosine methylation are first established by DNMT3, and then maintained by DNMT1 (Goll and Bestor 2005). The third DNMT, DNMT2/TRDMT1, has been implicated in the methylation of tRNA rather than DNA. Despite a very high degree of evolutionary conservation of DNMTs (e.g., Ponger and Li 2005), some DNMTs have been duplicated, or lost, in specific animal lineages (e.g., Yokomine et al. 2006; Barau et al. 2016; Bewick et al. 2017; Alvarez-Ponce et al. 2018). Cytosine methylation can trigger the binding of methyl-CpG-binding domain (MBD) proteins to methylated DNA, resulting in the recruitment of histone methyltransferases and histone deacetylases. These enzymes, in turn, modify the histone tails and affect gene expression (Fuks et al. 2000; Du et al. 2015). Two MBD proteins binding to methylated DNA have been inferred to be present in the common ancestor of animals and in cnidarians: MBD1/2/3 and MBD4/MeCP2 (Albalat et al. 2012). Two additional proteins with an MBD domain, MBD5 and MBD6, exist in animals but their function is unclear since, in mammals, they do not bind to methylated DNA (Laget et al. 2010); for this reason, they were not considered in our analyses. Methylated cytosines can be converted into hydroxymethylated cytosines (5hmC), and then to c5-formylcytosine (5fC) and 5-carbozylcytosine (5caC) by proteins known as Ten-eleven translocation methylcytosine dioxygenases (TETs) (Kriaucionis and Heintz 2009; Tahiliani et al. 2009; Guo et al. 2011). The resulting 5hmC, 5fC, and 5caC can be subsequently converted to unmethylated cytosine via base excision repair or replication-dependent dilution (reviewed by Wu and Zhang [2017]). Consequently, TETs can actively demethylate methylated cytosines. Since their first discovery, the role of TETs in regulating methylation and demethylation has been shown to be critical in development and gene regulation (Lu et al. 2015). Although three copies of TET are present in vertebrates, a single copy of the gene is assumed to have been present in the ancestor of animals (Liu et al. 2020). Similar proteins are involved in adenine methylation, with methyltransferase like 4 (METTL4) having an analogous function for adenine as DNMTs do for cytosine (Iyer et al. 2016). At present, DNA N6-methyl methyltransferase (DAMT-1), the Ca. elegans ortholog of METTL4, is the only animal protein that has been confirmed to act as an adenine DNMT (Greer et al. 2015; Luo and He 2017). Collectively, these DNA methylation-related proteins lay the foundation for DNA methylation driven regulation.
DNA methylation enzymes, and methylomes, are relatively understudied in nonmodel organisms, including cnidarians (see fig. 1) (Fuchs et al. 2014; Putnam et al. 2016; Li et al. 2018; Eirin-Lopez and Putnam 2019). Here, we present the first investigation of DNA methylation in a wide variety of cnidarians, including both free-living and parasitic cnidarians. Myxozoans are microscopic and parasitic cnidarians that typically infect fish and annelids (Kent et al. 2001; Chang et al. 2015; Atkinson et al. 2018). They are divided into two classes: Myxosporea, which encompass the vast majority of myxozoans; and the less-studied Malacosporea, with fewer than 10 species (Fiala et al. 2015). Myxosporeans are further divided into a “marine/polychaete–host” and “freshwater/oligochaete–host” clades (Fiala et al. 2015). They exhibit highly reduced genomes (22–175 Mb) compared with other cnidarians (256–1,260 Mb), consistent with the pattern of parasitic organisms having smaller genomes than their closest free-living relatives (Dieterich and Sommer 2009; Chang et al. 2015).
Fig. 1.
Presence/absence of methylation-related genes across cnidarians. The tree was assembled from published trees (Kayal et al. 2018; Yahalomi et al. 2020). Black and white squares represent the presence and absence of each gene, respectively. Predicted MBD proteins were characterized as part of our phylogenetic analyses. According to this tree 1) DNMT1, DNMT2/TRDMT1, and MBD4/MeCP2 would have been lost in an ancestor of Myxozoa (red circle); 2) DNMT3 and TET proteins would have been lost in a common ancestor of Myxozoa and Polypodium hydriforme (orange circle); 3) MBD1/2/3 would have been lost in a common ancestor of Myxosporea (black circle); and 4) METLL4 would have been lost in an ancestor of P. hydriforme and in an ancestor of Buddenbrockia plumatellae. It should be noted, however, that only transcriptomic data are available for P. hydriforme, and that only sparse EST data are available for Malacosporea (asterisks represent species with only transcriptomic or EST data), and thus it is possible that all relevant genes may be present in these species and that DNMTs, MBDs, and TETs would have been lost in the branch preceding the diversification of Myxosporea. Question marks indicate uncertainties regarding the absence of genes in that species due to incomplete genome data. The “2” indicates that the gene encoding MBD1/2/3 is duplicated in Dendronephthya gigantea. The last column corresponds to the presence and absence of DNA cytosine methylation in each species (or in another species of the same genus); the absence of a square indicates that neither a methylome nor the absence thereof has been reported. Cytosine methylation data were obtained from Zemach et al. (2010) (Nematostella vectensis), Hassel et al. (2010) (Hydra), Putnam et al. (2016) (Pocillopora damicornis), Dixon et al. (2016) (Acropora millepora), Liew et al. (2018) (Stylophora pistillata), Li et al. (2018) (Exaiptasia pallida), and the current study (Ceratonova shasta and Henneguya salminicola).
We searched for the presence of known methylation-associated proteins (DNMTs, MBDs, and TETs) in available genomic and transcriptomic data from 29 cnidarians, including eight myxozoans representative of the myxosporean diversity (representing the “marine/polychaete–host” lineage: Ceratonova shasta, Kudoa iwatai, and Enteromyxum leei; and the “freshwater/oligochaete–host” lineage: Sphaeromyxa zaharoni, Henneguya salminicola, Thelohanellus kitauei, Myxobolus cerebralis, and Myxobolus squamalis; Atkinson et al. 2018; Holzer et al. 2018; Yahalomi et al. 2020). Although homologs of genes encoding methylation-associated proteins are present in all nonmyxozoan cnidarians with available complete genomes, we did not find any of these genes in any of the studied myxosporean genomes. We thus hypothesized that myxosporeans had lost DNA cytosine methylation. To test this hypothesis, we then performed whole-genome bisulfite sequencing (WGBS, a technique that allows determining which DNA cytosines are methylated) on two highly divergent myxosporeans (C. shasta and H. salminicola) to determine the level of DNA cytosine methylation in these species. Our sequencing results confirmed that these myxosporeans completely lack cytosine methylation. We thus add a new animal group, the Myxosporea, to the very short list of species known to lack DNA cytosine methylation.
Results
DNA Cytosine Methylation Proteins Were Lost in an Ancestor of Myxosporea
We obtained the sequences of the DNMT, MBD, TET, and METTL4 proteins of the cnidarians Orbicella faveolata (Anthozoa) and Hydra vulgaris (Hydrozoa). Using these sequences, we performed BLAST searches against the genome assemblies, annotated protein sequences, and/or transcriptomes, of 29 cnidarian species. These included eight myxosporeans, Polypodium hydriforme (the most closely related cnidarian species to myxozoans; Chang et al. 2015), and 20 other nonmyxozoan cnidarians including both anthozoans (corals and sea anemone) and medusozoans (jellies and hydras). We detected homologs of all these proteins in all of the nonmyxozoan cnidarians, except for P. hydriforme, which lacked transcripts encoding TET, DNMT3, and METTL4 (fig. 1). We detected METTL4-predicted proteins in all eight myxosporeans included in our study but did not detect any DNMT, MBD, or TET protein (fig. 1).
The fast rate of evolution of myxozoans might have hindered the detection of the proteins of interest using BLAST searches. To address this possibility, we designed seven Hidden Markov Model (HMM) profiles (see Materials and Methods) for the DNMT1, DNMT2/TRDMT1, DNMT3, MBD1/2/3, MBD4/MeCP2, TET, and METTL4 proteins to search against predicted protein databases deposited in the NCBI or generated from published transcriptomes of six myxosporeans (C. shasta, H. salminicola, K. iwatai, T. kitauei, M. cerebralis, and M. squamalis). We manually evaluated the homology of each HMM hit with reciprocal BlastP searches against the nr database (NCBI Resource Coordinators 2016). Phylogenetic trees were constructed for each protein data set including the database of proteins representative of the animal and cnidarian diversity selected for the HMM construction and the hit found in Myxosporea and P. hydriforme, with the sponge Amphimedon queenslandica as outgroup (supplementary figs. 1–7, Supplementary Material online). Once again, we detected METTL4-predicted proteins for the myxosporeans included in our study but did not detect any DNMT, MBD, or TET protein in myxosporeans. Thus, our HMM searches corroborate our BLAST searches.
Mapping the presence or absence of these genes onto a cnidarian phylogenetic tree (fig. 1) allowed us to infer that 1) the most recent common ancestor of cnidarians harbored the same number of methylation genes as the ancestor of animals; 2) DNMT1, DNMT2/TRDMT1, and MBD proteins were lost in a common ancestor of Myxosporea; 3) DNMT3 and TET were lost in a common ancestor of P. hydriforme and Myxozoa; and 4) METTL4 were lost in an ancestor of P. hydriforme. It should be noted, however, that no complete genome is available for P. hydriforme, and because transcriptomic data sets do not represent all genes, it is possible that DNMT, MBD, and TET proteins might have been lost in an ancestor of Myxosporea (fig. 1).
Ceratonova shasta and H. salminicola Lack DNA Cytosine Methylation
The absence of genes related to cytosine methylation in myxosporeans led us to hypothesize that they lacked DNA cytosine methylation. To test this hypothesis, we sequenced the methylomes of two distantly related myxosporeans, C. shasta (representative of the “marine/polychaete” lineage) and H. salminicola (representative of the “freshwater/oligochaete” lineage). To directly measure cytosine methylation in these myxosporeans, we obtained total genomic DNA from the intestine of a rainbow trout (Oncorhynchus mykiss) infected with C. shasta and from a muscle cyst of a Chinook salmon (Oncorhynchus tshawytscha) infected with H. salminicola. We added lambda phage DNA to both samples (as a negative control for bisulfite conversion rate, since it is unmethylated), and then we performed WGBS. Therefore, our raw WGBS data for C. shasta contained sequences from C. shasta, rainbow trout and lambda phage genomic DNA (supplementary table 1, Supplementary Material online). Similarly, our raw WGBS data for H. salminicola contained H. salminicola genomic DNA sequences, as well as trace amounts of Chinook salmon and lambda phage DNA sequences (supplementary table 2, Supplementary Material online).
We first aligned both WGBS data sets to the lambda phage genome using Bismark to calculate the bisulfite nonconversion rates (Krueger and Andrews 2011). In addition to the CpG context, we also examined CHG and CHH cytosine methylation (where H represents A, T, or C nucleotides), which occurs in eukaryotic genomes (Zemach et al. 2010). For the C. shasta WGBS data set, we determined the bisulfite nonconversion rates to be 0.1%, 0.2%, and 0.1% for the CpG, CHG, and CHH methylation contexts, respectively (table 1). For the H. salminicola WGBS data set, we determined the bisulfite nonconversion rates to be 0.1% for each of the CpG, CHG, and CHH methylation contexts, respectively (table 1).
Table 1.
Methylation Analysis with Bismark Showed That Ceratonova shasta and Henneguya salminicola Have No DNA Cytosine Methylation.
| Bisulfite Sequence Data | Reference Genome Assembly | C Methylated in CpG Context (%) | C Methylated in CHG Context (%) | C Methylated in CHH Context (%) |
|---|---|---|---|---|
| Ceratonova shasta + rainbow trout + lambda phage | Lambda phage | 0.1 | 0.2 | 0.1 |
| Ceratonova shasta + rainbow trout | Rainbow trout | 73.8 | 0.2 | 0.2 |
| Ceratonova shasta | Ceratonova shasta | 0.1 | 0.1 | 0.1 |
| Henneguya salminicola + Chinook salmon + lambda phage | Lambda phage | 0.1 | 0.1 | 0.1 |
| Henneguya salminicola + Chinook salmon | Chinook salmon | 49.0 | 11.9 | 16.2 |
| Henneguya salminicola | H. salminicola | 0.1 | 0.1 | 0.1 |
Note.—The table lists the percent cytosine methylation for the CpG, CHG, and CHH methylation contexts for each of the six bisulfite sequence alignments performed using Bismark. The first row summarizes the methylation results for the alignment of the raw WGBS sequence reads for C. shasta to the lambda phage genome. The second row summarizes the results for the alignment of the C. shasta filtered reads from the previous alignment to the rainbow trout genome. The third row summarizes the results for the alignment of the C. shasta final filtered reads from the previous alignment to the C. shasta genome excluding cytosine sites overlapping SNPs from the reference genome. The fourth row summarizes the methylation results for the alignment of the raw WGBS sequence reads for H. salminicola to the lambda phage genome. The fifth row summarizes the results for the alignment of the H. salminicola filtered reads from the previous alignment to the Chinook salmon genome. The sixth row summarizes the results for the alignment of the H. salminicola final filtered reads from the previous alignment to the H. salminicola genome excluding cytosine sites overlapping SNPs from the reference genome. The percent methylation data shown here are from after deduplication and extraction using deduplicate_bismark and bismark_methylation_extractor.
We then aligned the remaining WGBS reads (those that did not align to the lambda phage genome) in the C. shasta and H. salminicola data sets to the rainbow trout and Chinook salmon genomes (Berthelot et al. 2014; Christensen et al. 2018), respectively. These alignments served as positive control, as fish genomes are known to be highly methylated (Jabbari et al. 1997). We detected 73.8% CpG methylation for the rainbow trout in the C. shasta WGBS data set (table 1) and 49.0% CpG methylation for the Chinook salmon in the H. salminicola WGBS data set (table 1). The lower methylation levels measured for the Chinook salmon were most likely due to the low number of sequence reads for this species contained in the H. salminicola data set (supplementary table 2, Supplementary Material online).
We also performed deep whole-genome sequencing (WGS) to identify polymorphic sites that occur at cytosines between the reference genome and the sequenced samples. We observed a 2.8% single-nucleotide polymorphism (SNP) difference between the C. shasta genome assembly and our C. shasta WGS data set, and a 0.5% SNP difference between the H. salminicola genome assembly and our H. salminicola WGS data set (supplementary table 3, Supplementary Material online). These sites only impacted between 3.5–4.8% and 0.58–0.72% of CpG, CHG, and CHH sites for C. shasta and H. salminicola respectively and we removed them from downstream analyses to avoid errors due to SNPs.
We aligned the remaining WGBS reads (those that did not align to either the lambda phage genome or the fish host genomes) to the genome assemblies of C. shasta and H. salminicola (see Materials and Methods). By virtue of extremely high sequencing depth, the average coverages of the mapped cytosines were very high: ∼400× for C. shasta and ∼1,600× for H. salminicola (figs. 2 and 3; supplementary tables 4 and 5, Supplementary Material online). For both species, we determined the CpG, CHG, and CHH methylation rates to be 0.1% for each of these cytosine contexts (table 1). These methylation rates were nearly equal to the bisulfite nonconversion rates, indicating that C. shasta and H. salminicola lack DNA cytosine methylation.
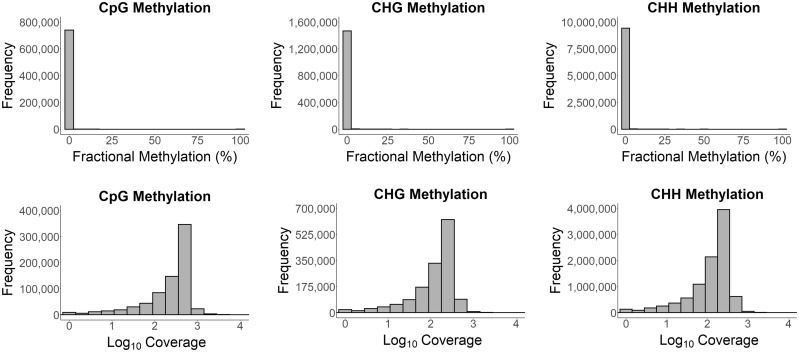
Fig. 2.
Cytosine methylation in Ceratonova shasta. Histograms of fractional methylation (ratio of the number of methylated reads to the total number of methylated and unmethylated reads) for the CpG, CHG, and CHH methylation contexts for the alignment of the deduplicated final filtered C. shasta reads to the C. shasta assembly (top row). Histograms of read coverage per base for three different methylation contexts for the alignment of the final filtered C. shasta reads to the C. shasta assembly (bottom row).
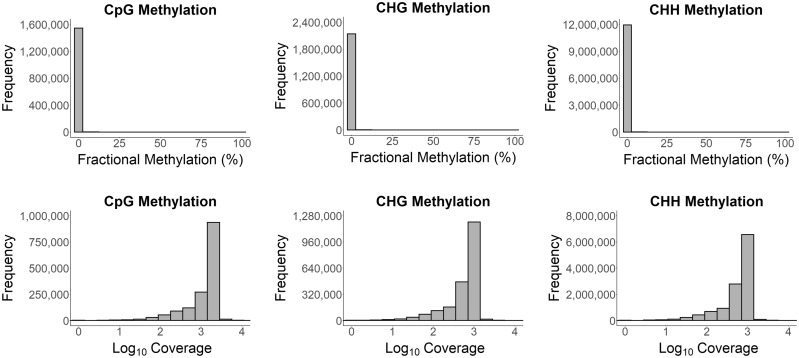
Fig. 3.
Cytosine methylation in Henneguya salminicola. Histograms of fractional methylation (ratio of the number of methylated reads to the total number of methylated and unmethylated reads) for the CpG, CHG, and CHH methylation contexts for the alignment of the deduplicated final filtered H. salminicola reads to the H. salminicola assembly (top row). Histograms of read coverage per base for three different methylation contexts for the alignment of the final filtered H. salminicola reads to the H. salminicola assembly (bottom row).
We calculated the fractional methylation (number of methylated reads/total reads per cytosine site) for all three cytosine contexts (CpG, CHG, and CHH) for the final sequence alignments of our WGBS data sets. The histograms of fractional methylation frequency showed only one peak at the zero end of the x-axis for each of the three methylation contexts, thus indicating no cytosine methylation for either C. shasta or H. salminicola (figs. 2 and 3).
Discussion
We searched the genomes, proteomes, and/or transcriptomes of 29 cnidarians, including eight myxosporeans, for the presence of genes encoding core proteins involved in DNA methylation. Our BLAST and HMMER searches revealed that DNMTs, MBDs, and TET are present in most of the studied nonmyxozoan cnidarians, in agreement with previous reports of DNA cytosine methylation in a wide range of cnidarians (Hassel et al. 2010; Zemach et al. 2010; Dixon et al. 2016; Putnam et al. 2016; Li et al. 2018; Liew et al. 2018) (fig. 1). However, we could not find these genes in any of the eight myxosporeans with available genomes or transcriptomes (fig. 1). Placing this information into a phylogenetic context suggests that proteins encoding these genes were lost in a common ancestor of Myxosporea (fig. 1).
The only nonmyxozoan cnidarian that appears to lack some of these proteins is P. hydriforme, an enigmatic parasite of paddlefish and sturgeon oocytes which is the sister taxon to Myxozoa (Chang et al. 2015). This species appears to lack DNMT3 and TET proteins, which would indicate that these proteins would have been lost in a common ancestor of Myxozoa and P. hydriforme. It should be noted, however, that our searches against P. hydriforme relied on transcriptome data, as a complete genome assembly is currently unavailable. As transcriptomes do not capture the whole set of genes of an organism, it is possible that these genes are present in P. hydriforme, in which case they would have been most likely lost in an ancestor of Myxosporea, subsequent to divergence from the common ancestor with P. hydriforme. Future analyses using a whole-genome assembly of this species will allow to draw a definitive conclusion.
Our analyses are based on eight myxozoan species that belong to the subclass Myxosporea (a clade that contains the vast majority of myxozoan species). Myxozoa, however, encompass a second distant subclass, the Malacosporea, which include only two genera (Buddenbrockia and Tetracapsuloides) and less than ten species (Fiala et al. 2015). Unfortunately, in public databases, genomic data are limited to <1,500 EST for Malacosporea, which precluded the inclusion of this subclass in our analyses. Preliminary BLAST searches and phylogenetic analyses, however, revealed the presence of MBD1/2/3 in Buddenbrockia plumatellae (sequence accession: ES599526). This suggests that the loss of cytosine methylation is limited to Myxosporea and not a characteristic shared by all Myxozoa. Future studies should determine if Malacosporea possess a complete set of methylation enzymes or represent an intermediate stage in the loss of cytosine methylation.
In agreement with our observation that myxosporeans lack DNMTs, MBDs, and TET proteins, our newly generated WGBS data show that two highly divergent myxosporeans, C. shasta and H. salminicola (representatives from the freshwater and the marine lineages, respectively; Fiala et al. 2015; Holzer et al. 2018), completely lack DNA cytosine methylation (table 1 and figs. 2 and 3). Thus, we have identified a new taxonomic group with no cytosine methylation, placing Myxosporea among the very few animal clades known to lack this form of epigenetic modification.
Given that DNMT1, DNMT2/TRDMT1, and MBDs (and potentially DNMT3 and TET proteins too, as discussed above) were lost in the same branch of the cnidarian tree (the branch predating the diversification of Myxosporea), it is possible that the loss of DNMTs in an ancestor of Myxosporea may have led to a subsequent loss of MBD and TET proteins, as without DNA cytosine methylation, these proteins would have been rendered nonfunctional. Alternatively, MBDs could have been lost first, which would have rendered cytosine methylation (and thus DNMTs) nonfunctional. The sequencing of Malacosporea may help to answer this question.
Given the importance of cytosine methylation in regulation of gene expression, development, and genome defense, it is a pressing question to understand how the loss of cytosine methylation impacts these processes. In terms of gene expression, other epigenetic mechanisms, notably histone modification, may play a major role of regulation, as in Ca. elegans and D. melanogaster, which also lack cytosine modification (Chang and Liao 2017). Similar molecular mechanisms may exist in myxosporeans. Interestingly, we found METLL4 orthologs in all the studied cnidarians (including all myxosporeans), except P. hydriforme (for which, as discussed above, we lack a complete genome). Thus, cnidarians, including myxozoans, might harbor adenine DNA methylation (even though the functional relevance of this modification needs to be resolved; Flusberg et al. 2010).
Our study adds to a growing list of species where DNA methylation has been lost. Evolutionary forces that facilitate loss of DNA methylation remain unclear (Takuno et al. 2016; Yi 2017; Bewick et al. 2019; Schmitz et al. 2019). An interesting observation is that myxozoans generally have very small genomes, and thus are potentially relatively free from the burden of active genome defense. In plant genomes, there is a strong negative correlation between genome size and the degree of genomic CHG DNA methylation (Niederhuth et al. 2016; Vidalis et al. 2016), which is the primary DNA methylation of transposable elements in plants. In comparison, factors affecting genomic DNA methylation in animal lineages are still relatively little understood. Future studies should determine the significance of genome defense and genomic DNA methylation in cnidarians.
The current understanding of the evolutionary loss of DNA methylation in animals comes primarily from studies using model species and human parasites. Our work offers an example of a DNA methylation survey of a group of nonmodel organisms using straightforward and highly replicable molecular and computational techniques. Given that it is becoming less prohibitive to generate WGS data and WGBS data of nonmodel organisms, an investigative framework similar to the method used here could be applied to a variety of eukaryotic species in a near future. Such data will allow evolutionary biologists to generate a more complete understanding of the intricate evolutionary dynamics of DNA methylation. On one hand, the pervasiveness of DNA methylation across a broad variety of taxa, as well as the high level of conservation of the methylation machinery, is a testament to the biological importance of this epigenetic modification in genomic regulation. On the other hand, despite this paradigm of regulation, our findings suggest that a speciose group of parasitic cnidarians—the Myxosporea—are among a short but notable list of animal taxa that have lost DNA cytosine methylation and the enzymatic machinery responsible for it. With future data on genomic DNA methylation surveys from diverse species, we will soon be able to illuminate the factors that determine the tradeoffs between conservation and loss of DNA methylation across the tree of life.
Materials and Methods
BLAST Searches
We performed online BlastP searches (Altschul et al. 1997) using as query the Orbicella faveolata and Hy. vulgaris DNMT1, DNMT2/TRDMT1, DNMT3, MBD1/2/3, MBD4/MeCP2, TET, and METTL4 protein sequences against all cnidarians with protein sequences in the NCBI nr database and with a scaffold N50 >90,000 on the NCBI Genome database (NCBI Resource Coordinators 2016). The latter criterion was chosen to ensure that the absence of a protein could not be explained by a low assembly quality (low N50 values result in many genes being spread across multiple scaffolds, which may lead to incomplete and/or erroneous protein sequence predictions). The list of cnidarian species meeting these criteria is provided in bold in supplementary table 6, Supplementary Material online. In addition, we performed TBlastN searches using the same query sequences against the unannotated genomes and transcriptomes of additional nonmyxozoans representative of the cnidarian diversity; all cnidarians with an available genome assembly in the NCBI Genome database (as per June 2019) were included in our analyses (fig. 1 and supplementary table 6, Supplementary Material online). For each cnidarian species considered, the orthology of the first five hits was assessed by performing reciprocal blast searches against the Hy. vulgaris proteome and reconstructing phylogenetic trees (see below).
We next performed local TBlastN searches (BLAST+ v2.9.0) (Camacho et al. 2009), using all the nonmyxozoan cnidarian protein sequences identified in supplementary tables 7–13, Supplementary Material online, against the genome assemblies or transcriptomes of eight myxosporeans (C. shasta, H. salminicola, K. iwatai, E. leei, S. zaharoni, T. kitauei, M. cerebralis, and M. squamalis), and the transcriptome of P. hydriforme. Accession numbers of the assemblies used are provided in supplementary table 6, Supplementary Material online. For each myxosporean hit obtained, a reciprocal BlastX search was performed against the NCBI’s nr database (last accessed in February 2020) to verify the identity of the protein sequences identified by our local TBlastN. These BLAST searches ensured that the myxosporean hits were indeed cnidarian sequences rather than host contamination or erroneous BLAST hits.
All BLAST searches were conducted using default parameters. The accession numbers for all protein sequences used for and found during our BLAST searches can be found in supplementary tables 7–13, Supplementary Material online.
HMM Profile Construction and Searches
For HMM profile construction, all cnidarian sequences identified in our previous BLAST searches were included together with representatives of the main animal lineages (see supplementary tables 7–13, Supplementary Material online). Multiple sequence alignments of each group of proteins were created using MAFFT v7.450 with default parameters (“auto”) (Katoh and Standley 2013), and the alignments were manually inspected in Geneious Prime software version 2019.2.3 (Kearse et al. 2012). The accession numbers of all proteins used in HMM profile construction can be found in supplementary tables 7–13, Supplementary Material online. HMM profiles were built based on the MAFFT alignments using hmmbuild from the HMMER package v3.1b2 (Eddy 2011). Alignments and HMM profiles are provided in supplementary files 1–15, Supplementary Material online.
The T. kitauei, M. squamalis, and H. salminicola proteomes were retrieved from the NCBI Database (supplementary table 6, Supplementary Material online). Proteins sequences of the other three myxosporean species (C. shasta, K. iwatai, and M. cerebralis) and P. hydriforme were predicted from transcriptome sequences using Transdecoder version v5.5.0 (https://github.com/TransDecoder; last accessed: April 30, 2020) with default parameters. For each species, the protein database generated or downloaded from NCBI was searched using hmmsearch from the HMMER package v3.1b2 under the HMM built as described above (Eddy 2011). Because there is no transcriptome data available for E. leei or S. zaharoni, these two species were excluded from our HMM analyses.
Finally, to confirm the orthology of the proteins identified using BLAST and HMM searches, phylogenetic trees were reconstructed for each gene considered in this study (DNMT1, DNMT2/TRDMT1, DNMT3, MBD2, MBD4, TET, and METTL4) using the amino acid alignments used previously for HMM build. Specifically, the GUIDANCE2 server version 2.02 (Sela et al. 2015) was used to remove ambiguously aligned positions, with the following parameters: MAFFT program, 100 bootstrap, “–maxiterate 1000,” and “–localpair.” Unreliable column positions with a reliability score below 0.5 were excluded, as well as columns which included more than 50% of gaps. The different alignments are provided in supplementary file 1, Supplementary Material online. Maximum-likelihood phylogenies were inferred using IQ-TREE version 1.6.12 (Nguyen et al. 2015) with automatic model detection using ModelFinder Plus and 1,000 bootstrap replicates (supplementary figs. 1–7, Supplementary Material online).
Genome Assemblies
The H. salminicola genome assembly was obtained from Yahalomi et al. (2020). We used our draft C. shasta genome assembly (Version Velvet2015-93; S.D. Atkinson, unpublished data; 15,423 sequences, average contig length: 6,823, N50: 47,028, assembled size: 105.2 Mb, average coverage: ∼250×, filtered of rainbow trout and bacterial contamination). The genome assembly IDs for all genomes used in our study are listed in supplementary table 6, Supplementary Material online.
Biological Material
Fresh H. salminicola was obtained in September 2016 from the skeletal muscle of an adult Chinook salmon (Oncorhynchus tshawytscha) from the Willamette River, Oregon, USA. Parasite material, predominantly mature myxospores, was aspirated from a single pseudocyst, and then dried in a SpeedVac (Savant), before being stored at −20 °C until processing for sequencing. Fresh C. shasta was obtained in February 2014 from the intestine of a juvenile rainbow trout (Oncorhynchus mykiss) that had been infected during a cage exposure in the Willamette River, Oregon. Fresh intestine was dissected from an infected fish, macerated with scissors, and suspended parasite spores washed through a 70-µm cell sieve using phosphate-buffered saline (PBS). The filtered material was washed twice, by pelleting by centrifugation (∼2,000 × g, 10 min), with the supernatant exchanged with fresh PBS after each centrifugation. The semipure myxospores were then layered on top of a Percoll (Sigma-Aldrich) gradient (layers with concentrations of 25%, 50%, and 75%) and centrifuged for ∼2,500 × g for 15 min. The almost pure myxospores formed a band on top of the 75% layer. The spores were aspirated from the gradient, then again washed by centrifugation in PBS to remove Percoll. The spores were then dried in a SpeedVac (Savant), before being stored at −20 °C until processing for sequencing.
Whole-Genome Sequencing
WGS libraries were generated from DNA extracted using the DNeasy Blood and Tissue DNA kit (Qiagen). From each sample, 500 ng to 1 µg of DNA was sheared on a Covaris ultrasonicator (Covaris, Woburn, MA) to 200–600 bp at the Emory Integrated Genomics Core. The DNA fragment ends were repaired with the End-It DNA End-Repair Kit (#ER81050, Epicentre, Madison, WI) and A-overhangs were added (#M0202, New England Biolabs, Ipswich, MA) before Nextera barcode adaptors were ligated to the DNA fragments overnight. Finally, the libraries were polymerase chain reaction (PCR) amplified to increase concentration and enrich for adaptor-ligated DNA fragments. WGS libraries were sequenced using Illumina HiSeq X 150 with paired-end reads at the Macrogen Clinical Laboratory.
Variant Calling for Whole-Genome Sequences
We used FastQC (v0.11.7) and Trim Galore (v0.5.0) to examine the quality and remove the adaptor sequences in our raw, paired-end WGS data for C. shasta and H. salminicola. The raw WGS data for C. shasta contained C. shasta and rainbow trout genomic DNA sequences. The raw WGS data for H. salminicola contained H. salminicola and Chinook salmon genomic DNA sequences. We used BBSplit, part of the BBMap package (v38.62) (Bushnell 2014), to remove all fish contamination from both sets of WGS data. Next, we aligned the WGS reads to the respective C. shasta and H. salminicola reference genome assemblies using BWA (v0.7.17) (Li and Durbin 2009) under default parameters. We then used samtools (v1.9) to convert the SAM files produced by the alignments to BAM files. Then, we used samtools (v1.9) again to extract correctly paired reads, remove duplicate reads, and extract all reads with a map quality of 30 or higher, and acquire statistics to calculate the average genome coverage for the C. shasta and H. salminicola WGS data.
To perform variant calling, we used bcftools (v1.9) (Li et al. 2009). We then used SnpSift (v4.0) (Cingolani et al. 2012) to extract all SNPs where the sequence depth was ≥20. These high-quality SNPs were used for a comprehensive variant calling analysis performed with bcftools for the C. shasta and H. salminicola WGS data. The SNP density was computed with vcftools (v1.9) (Danecek et al. 2011), which allowed to calculate average numbers of SNPs per kb. Afterward, we used ANGSD (v0.928) (Korneliussen et al. 2014) to convert the BAM files to consensus FASTA files. Finally, we used Mauve (February 13, 2015 build) (Darling et al. 2004) to compare the reference genome assembly FASTA files and consensus FASTA files for C. shasta and H. salminicola and the outputted statistics to calculate percent differences between the genome assembly and consensus FASTA files. All the relevant statistical information obtained from our analysis are presented in supplementary table 3, Supplementary Material online.
Whole-Genome Bisulfite Sequencing
To generate WGBS libraries, genomic DNA was pooled with 1–5% lambda phage DNA as a test to control for bisulfite reaction efficiency. The DNA samples were then sheared on a Covaris ultrasonicator to 200–600 bp. The DNA fragment ends were repaired and A-overhangs were added before bisulfite compatible adaptors were ligated to the DNA fragments overnight. Next, the DNA fragments were bisulfite converted using the MethylCode Bisulfite Conversion Kit (#MECOV50, ThermoFisher). Purified gDNA was treated with CT conversion reagent in a thermocycler for 10 min at 98 °C, followed by 2.5 h at 640 °C. Bisulfite-treated DNA fragments remain single stranded as they are no longer complementary. Low-cycle (4–8) PCR amplification was performed with Kapa HiFi Uracil Hotstart polymerase enzyme (#KK2801, KAPA Biosystems, Wilmington, MA), which can tolerate uracil residues. The final library fragments contain thymines and cytosines in place of the original unmethylated cytosine and methylated cytosines, respectively. WGBS libraries were sequenced using Illumina HiSeq X 150 with paired-end reads at the Macrogen Clinical Laboratory.
Whole-Genome Bisulfite Sequence Quality Control
The raw WGBS data for C. shasta contained C. shasta, rainbow trout, and lambda phage genomic DNA sequences. The raw WGBS data for H. salminicola contained H. salminicola, Chinook salmon, and lambda phage genomic DNA sequences. We first used FastQC (v0.11.7) (Andrews 2010) to analyze the quality of the reads. Next, we used Trim Galore (v0.5.0) (Martin 2011; Krueger and Andrews 2012; Krueger 2015) to remove the Illumina adaptor sequences and very low quality reads. Finally, we used FastQC again to analyze the quality of the trimmed reads.
Whole-Genome Bisulfite Sequence Alignment with Bismark
We aligned and analyzed the trimmed reads for the C. shasta and H. salminicola data sets using Bismark (v0.20.0) (Krueger and Andrews 2011, 2012) with Bowtie 1 (v1.0.0) (Langmead et al. 2009). First, we aligned the reads to the lambda phage genome to calculate the bisulfite conversion rate (Wreczycka et al. 2017). As part of this step, we used Bismark’s “–un” flag to retain the myxosporean and host fish reads, as these reads do not align to the lambda phage genome. Note that the term “host fish” refers to the vertebrate hosts of C. shasta and H. salminicola, which are the rainbow trout and the Chinook salmon, respectively. Next, we aligned the unmapped reads (myxosporean and host fish sequences) from the previous step to the host fish genomes, while using the “–un” flag to retain all myxosporean reads. Finally, we aligned the unmapped reads (myxozoan sequences) from the previous step to the respective C. shasta and H. salminicola reference genome assemblies. Afterward, we used deduplicate_bismark on the BAM output files produced in the previous steps to remove PCR duplication bias. Then, we used bismark_methylation_extractor on these deduplicated BAM files to acquire the final methylation data (Krueger and Andrews 2012; Wreczycka et al. 2017). Additionally, we used bam2nuc on the deduplicated BAM output files produced from the final C. shasta and H. salminicola alignments to assess the nucleotide coverage of the alignments (supplementary tables 4 and 5, Supplementary Material online). Finally, we used bismark2report to create six HTML data summary report files from the six sets of files created from the previous steps. All codes used for the quality control and sequence analysis are available upon request. The methylation data we obtained from each of the six alignments were used to construct table 1.
DNA Methylation Analysis
We utilized Bismark’s bismark_methylation_extractor, bismark2bedGraph, and coverage2cytosine in series (v0.20.0) (Krueger and Andrews 2011, 2012) to generate reports for cytosine in three nucleotide contexts, CpG, CHG, and CHH. To counteract inaccurate methylation calls that can arise due to polymorphisms between the reference genomes and WGBS samples (i.e., C-to-T polymorphisms would always be designated unmethylated irrespective of reference methylation status), all cytosine sites overlapping identified variants were removed. We calculated the fractional methylation (ratio of the number of methylated cytosine reads to the total number of methylated and unmethylated reads) for each analyzed site. Finally, we plotted the frequencies of fractional methylation in all three contexts for both C. shasta and H. salminicola.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors are grateful to Tom Parchman for providing access to his server and Richard Tillett for helpful advice on data analysis. R.K., A.L.-N., S.P., and D.A.-P. were supported by a grant from the National Science Foundation (MCB 1818288) and a Pilot Grant from the Smooth Muscle Plasticity COBRE of the University of Nevada, Reno, funded by the National Institutes of Health (Grant No. 5P30GM110767-04). T.L., D.S., and S.V.Y. were supported by a grant from the National Science Foundation (MCB 1615664). D.H. was supported by a grant from the Tel-Aviv University Vice President’s fund for research support (Grant No. 30003072000) and by a grant from the Israel Science Foundation (Grant No. 652/20). S.D.A. and J.L.B. were supported in part by the Bureau of Reclamation, US Department of Interior through Inter-agency Agreement #R15PG00065, as part of its mission to manage, develop, and protect water and related resources in an environmentally and economically sound manner in the interest of the American public. The views in this report are the authors’ and do not necessarily represent the views of Bureau of Reclamation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Mention of trade names does not imply U.S. Government endorsement.
Data Availability
Sequencing data have been deposited to the BioProject database (accession numbers PRJNA623035 and PRJNA623156).
References
- Albalat R, Martí-Solans J, Cañestro C.. 2012. DNA methylation in amphioxus: from ancestral functions to new roles in vertebrates. Brief Funct Genomics. 11(2):142–155. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ.. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25(17):3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Ponce D, Torres-Sánchez M, Feyertag F, Kulkarni A, Nappi T.. 2018. Molecular evolution of DNMT1 in vertebrates: duplications in marsupials followed by positive selection. PLoS One 13(4):e0195162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed April 30, 2020.
- Atkinson SD, Bartholomew JL, Lotan T.. 2018. Myxozoans: ancient metazoan parasites find a home in phylum Cnidaria. Zoology 129:66–68. [DOI] [PubMed] [Google Scholar]
- Barau J, Teissandier A, Zamudio N, Roy S, Nalesso V, Hérault Y, Guillou F, Bourc’his D.. 2016. The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Science 354(6314):909–912. [DOI] [PubMed] [Google Scholar]
- Berthelot C, Brunet F, Chalopin D, Juanchich A, Bernard M, Noël B, Bento P, Da Silva C, Labadie K, Alberti A, et al. 2014. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat Commun. 5(1):36–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewick AJ, Vogel KJ, Moore AJ, Schmitz RJ.. 2017. Evolution of DNA methylation across insects. Mol Biol Evol. 34(3):654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewick AJ, Zhang Y, Wendte JM, Zhang X, Schmitz RJ.. 2019. Evolutionary and experimental loss of gene body methylation and its consequence to gene expression. G3 (Bethesda) 9(8):2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AP. 1980. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 8(7):1499–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell B. 2014. BBMap: a fast, accurate, splice-aware aligner. Berkeley (CA: ): Lawrence Berkeley National Lab. (LBNL; ). [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL.. 2009. BLAST+: architecture and applications. BMC Bioinf. 10(1):421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capuano F, Mülleder M, Kok R, Blom HJ, Ralser M.. 2014. Cytosine DNA methylation is found in Drosophila melanogaster but absent in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and other yeast species. Anal Chem. 86(8):3697–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AY, Liao BY.. 2017. Recruitment of histone modifications to assist mRNA dosage maintenance after degeneration of cytosine DNA methylation during animal evolution. Genome Res. 27(9):1513–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang ES, Neuhof M, Rubinstein ND, Diamant A, Philippe H, Huchon D, Cartwright P.. 2015. Genomic insights into the evolutionary origin of Myxozoa within Cnidaria. Proc Natl Acad Sci U S A. 112(48):14912–14917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen KA, Leong JS, Sakhrani D, Biagi CA, Minkley DR, Withler RE, Rondeau EB, Koop BF, Devlin RH.. 2018. Chinook salmon (Oncorhynchus tshawytscha) genome and transcriptome. PLoS One 13(4):e0195461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Patel VM, Coon M, Nguyen T, Land SJ, Ruden DM, Lu X.. 2012. Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Front Genet. 3:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabe EC, Sanford RS, Kohn AB, Bobkova Y, Moroz LL.. 2015. DNA methylation in basal metazoans: insights from ctenophores. Integr Comp Biol. 55(6):1096–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 27(15):2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AC, Mau B, Blattner FR, Perna NT.. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14(7):1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mendoza A, Hatleberg WL, Pang K, Leininger S, Bogdanovic O, Pflueger J, Buckberry S, Technau U, Hejnol A, Adamska M, et al. 2019. Convergent evolution of a vertebrate-like methylome in a marine sponge. Nat Ecol Evol. 3(10):1464–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich C, Sommer RJ.. 2009. How to become a parasite—lessons from the genomes of nematodes. Trends Genet. 25(5):203–209. [DOI] [PubMed] [Google Scholar]
- Dixon GB, Bay LK, Matz MV.. 2016. Evolutionary consequences of DNA methylation in a basal metazoan. Mol Biol Evol. 33(9):2285–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q, Luu PL, Stirzaker C, Clark SJ.. 2015. Methyl-CpG-binding domain proteins: readers of the epigenome. Epigenomics 7(6):1051–1073. [DOI] [PubMed] [Google Scholar]
- Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol. 7(10):e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JR, Yarychkivska O, Boulard M, Bestor TH.. 2017. DNA methylation and DNA methyltransferases. Epigenet Chromatin 10:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirin-Lopez JM, Putnam HM.. 2019. Marine environmental epigenetics. Annu Rev Mar Sci. 11(1):335–368. [DOI] [PubMed] [Google Scholar]
- Fiala I, Bartošová-Sojková P, Whipps CM.. 2015. Classification and phylogenetics of Myxozoa. In: Okamura B, Gruhl A, Bartholomew JL, editors. Myxozoan evolution, ecology and development. New York: Springer International Publishing. p. 85–110. [Google Scholar]
- Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, Korlach J, Turner SW.. 2010. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 7(6):461–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs B, Wang W, Graspeuntner S, Li Y, Insua S, Herbst EM, Dirksen P, Böhm AM, Hemmrich G, Sommer F, et al. 2014. Regulation of polyp-to-jellyfish transition in Aurelia aurita. Curr Biol. 24(3):263–273. [DOI] [PubMed] [Google Scholar]
- Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T.. 2000. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 24(1):88–91. [DOI] [PubMed] [Google Scholar]
- Geyer KK, Chalmers IW, Mackintosh N, Hirst JE, Geoghegan R, Badets M, Brophy PM, Brehm K, Hoffmann KF.. 2013. Cytosine methylation is a conserved epigenetic feature found throughout the phylum Platyhelminthes. BMC Genomics. 14(1):462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Bestor TH.. 2005. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 74(1):481–514. [DOI] [PubMed] [Google Scholar]
- Greer EL, Blanco MA, Gu L, Sendinc E, Liu J, Aristizábal-Corrales D, Hsu CH, Aravind L, He C, Shi Y.. 2015. DNA methylation on N6-adenine in C. elegans. Cell 161(4):868–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H.. 2011. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 145(3):423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassel M, Cornelius MG, Vom Brocke J, Schmeiser HH.. 2010. Total nucleotide analysis of Hydra DNA and RNA by MEKC with LIF detection and 32P-postlabeling. Electrophoresis 31(2):299–302. [DOI] [PubMed] [Google Scholar]
- Holzer AS, Bartošová‐Sojková P, Born‐Torrijos A, Lövy A, Hartigan A, Fiala I.. 2018. The joint evolution of the Myxozoa and their alternate hosts: a cnidarian recipe for success and vast biodiversity. Mol Ecol. 27(7):1651–1666. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Zhang D, Aravind L.. 2016. Adenine methylation in eukaryotes: apprehending the complex evolutionary history and functional potential of an epigenetic modification. BioEssays 38(1):27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbari K, Cacciò S, Païs de Barros JP, Desgrès J, Bernardi G.. 1997. Evolutionary changes in CpG and methylation levels in the genome of vertebrates. Gene 205(1–2):109–118. [DOI] [PubMed] [Google Scholar]
- Jin B, Robertson KD.. 2013. DNA methyltransferases, DNA damage repair, and cancer. Adv Exp Med Biol. 754:3–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT—multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:72–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayal E, Bentlage B, Sabrina Pankey M, Ohdera AH, Medina M, Plachetzki DC, Collins AG, Ryan JF.. 2018. Phylogenomics provides a robust topology of the major cnidarian lineages and insights on the origins of key organismal traits. BMC Evol Biol. 18(1):68. [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent ML, Andree KB, Bartholomew JL, El-Matbouli M, Desser SS, Devlin RH, Feist SW, Hedrick RP, Hoffmann RW, Khattra J, et al. 2001. Recent advances in our knowledge of the Myxozoa. J Eukaryotic Microbiol. 48(4):395–413. [DOI] [PubMed] [Google Scholar]
- Korneliussen TS, Albrechtsen A, Nielsen R.. 2014. ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinf. 15(1):356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N.. 2009. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324(5929):929–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F. 2015. A wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files. Cambridge, UK: Babraham Institute.
- Krueger F, Andrews SR.. 2011. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27(11):1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR.. 2012. Quality control, trimming and alignment of Bisulfite-Seq data (Prot 57). In: Department of Medicine, Hematology and Oncology, Domagkstr. 3, 48149. Münster: Epigenesys; p. 1–13. Available from: https://www.epigenesys.eu/images/stories/protocols/pdf/20120720103700_p57.pdf.
- Laget S, Joulie M, Le Masson F, Sasai N, Christians E, Pradhan S, Roberts RJ, Defossez PA.. 2010. The human proteins MBD5 and MBD6 associate with heterochromatin but they do not bind methylated DNA. PLoS One 5(8):e11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL.. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10(3):R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liew YJ, Cui G, Cziesielski MJ, Zahran N, Michell CT, Voolstra CR, Aranda M.. 2018. DNA methylation regulates transcriptional homeostasis of algal endosymbiosis in the coral model. Sci Adv. 4(8):eaat2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Shen L, Cui X, Bao S, Geng Y, Yu G, Liang F, Xie S, Lu T, Gu X, et al. 2018. DNA N6-adenine methylation in Arabidopsis thaliana. Dev Cell 45(3):406–416. [DOI] [PubMed] [Google Scholar]
- Liew YJ, Howells EJ, Wang X, Michell CT, Burt JA, Idaghdour Y, Aranda M.. 2020. Intergenerational epigenetic inheritance in reef-building corals. Nat Climate Change 10:254–259. [Google Scholar]
- Liew YJ, Zoccola D, Li Y, Tambutté E, Venn AA, Michell CT, Cui G, Deutekom ES, Kaandorp JA, Voolstra CR, et al. 2018. Epigenome-associated phenotypic acclimatization to ocean acidification in a reef-building coral. Sci Adv. 4(6):eaar8028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Hu H, Panserat S, Marandel L.. 2020. Evolutionary history of DNA methylation related genes in chordates: new insights from multiple whole genome duplications. Sci Rep. 10(1):970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low DA, Weyand NJ, Mahan MJ.. 2001. Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect Immun. 69(12):7197–7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Zhao BS, He C.. 2015. TET family proteins: oxidation activity, interacting molecules, and functions in diseases. Chem Rev. 115(6):2225–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo GZ, He C.. 2017. DNA N6-methyladenine in metazoans: functional epigenetic mark or bystander? Nat Struct Mol Biol. 24(6):503–506. [DOI] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17 (1):10–12. [Google Scholar]
- Mendizabal I, Keller TE, Zeng J, Yi SV.. 2014. Epigenetics and evolution. Integr Comp Biol. 54(1):31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI Resource Coordinators. 2016. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 44:D7–D19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, Haeseler V, Minh BQ A. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederhuth CE, Bewick AJ, Ji L, Alabady MS, Kim KD, Li Q, Rohr NA, Rambani A, Burke JM, Udall JA, et al. 2016. Widespread natural variation of DNA methylation within angiosperms. Genome Biol. 17(1):194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen JS, Valen E, Velazquez AMV, Parker BJ, Rasmussen M, Lindgreen S, Lilje B, Tobin DJ, Kelly TK, Vang S, et al. 2014. Genome-wide nucleosome map and cytosine methylation levels of an ancient human genome. Genome Res. 24(3):454–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponger L, Li WH.. 2005. Evolutionary diversification of DNA methyltransferases in eukaryotic genomes. Mol Biol Evol. 22(4):1119–1128. [DOI] [PubMed] [Google Scholar]
- Putnam HM, Davidson JM, Gates RD.. 2016. Ocean acidification influences host DNA methylation and phenotypic plasticity in environmentally susceptible corals. Evol Appl. 9(9):1165–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raddatz G, Guzzardo PM, Olova N, Fantappié MR, Rampp M, Schaefer M, Reik W, Hannon GJ, Lyko F.. 2013. Dnmt2-dependent methylomes lack defined DNA methylation patterns. Proc Natl Acad Sci U S A. 110(21):8627–8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratel D, Ravanat JL, Berger F, Wion D.. 2006. N6‐methyladenine: the other methylated base of DNA. BioEssays 28(3):309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richa R, Sinha RP.. 2014. Hydroxymethylation of DNA: an epigenetic marker. EXCLI J. 13:592–610. [PMC free article] [PubMed] [Google Scholar]
- Sarda S, Zeng J, Hunt BG, Yi SV.. 2012. The evolution of invertebrate gene body methylation. Mol Biol Evol. 29(8):1907–1916. [DOI] [PubMed] [Google Scholar]
- Schmitz RJ, Lewis ZA, Goll MG.. 2019. DNA methylation: shared and divergent features across eukaryotes. Trends Genet. 35(11):818–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schübeler D. 2015. Function and information content of DNA methylation. Nature 517(7534):321–326. [DOI] [PubMed] [Google Scholar]
- Sela I, Ashkenazy H, Katoh K, Pupko T.. 2015. GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 43(W1):W7–W14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324(5929):930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takuno S, Ran J-H, Gaut BS.. 2016. Evolutionary patterns of genic DNA methylation vary across land plants. Nat Plants 2(2):15222. [DOI] [PubMed] [Google Scholar]
- Vidalis A, Živković D, Wardenaar R, Roquis D, Tellier A, Johannes F.. 2016. Methylome evolution in plants. Genome Biol. 17(1):264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Minakhina S, Tran H, Changela N, Kramer J, Steward R.. 2018. Tet protein function during Drosophila development. PLoS One 13(1):e0190367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wreczycka K, Gosdschan A, Yusuf D, Grüning B, Assenov Y, Akalin A.. 2017. Strategies for analyzing bisulfite sequencing data. J Biotechnol. 261:105–115. [DOI] [PubMed] [Google Scholar]
- Wu TP, Wang T, Seetin MG, Lai Y, Zhu S, Lin K, Liu Y, Byrum SD, Mackintosh SG, Zhong M, et al. 2016. DNA methylation on N6-adenine in mammalian embryonic stem cells. Nature 532(7599):329–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhang Y.. 2017. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat Rev Genet. 18(9):517–534. [DOI] [PubMed] [Google Scholar]
- Yahalomi D, Atkinson SD, Neuhof M, Chang ES, Philippe H, Cartwright P, Bartholomew JL, Huchon D.. 2020. A cnidarian parasite of salmon (Myxozoa: Henneguya) lacks a mitochondrial genome. Proc Natl Acad Sci U S A. 117(10):5358–5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi SV. 2012. Birds do it, bees do it, worms and ciliates do it too: DNA methylation from unexpected corners of the tree of life. Genome Biol. 13(10):174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi SV. 2017. Insights into epigenome evolution from animal and plant methylomes. Genome Biol Evol. 9(11):3189–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomine T, Hata K, Tsudzuki M, Sasaki H.. 2006. Evolution of the vertebrate DNMT3 gene family: a possible link between existence of DNMT3L and genomic imprinting. Cytogenet Genome Res. 113(1–4):75–80. [DOI] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D.. 2010. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328(5980):916–919. [DOI] [PubMed] [Google Scholar]
- Zemach A, Zilberman D.. 2010. Evolution of eukaryotic DNA methylation and the pursuit of safer sex. Curr Biol. 20(17):R780–785. [DOI] [PubMed] [Google Scholar]
- Zhang G, Huang H, Liu D, Cheng Y, Liu X, Zhang W, Yin R, Zhang D, Zhang P, Liu J, et al. 2015. N6-methyladenine DNA modification in Drosophila. Cell 161(4):893–906. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been deposited to the BioProject database (accession numbers PRJNA623035 and PRJNA623156).