Abstract
Metallothioneins (MTs) are proteins devoted to the control of metal homeostasis and detoxification, and therefore, MTs have been crucial for the adaptation of the living beings to variable situations of metal bioavailability. The evolution of MTs is, however, not yet fully understood, and to provide new insights into it, we have investigated the MTs in the diverse classes of Mollusks. We have shown that most molluskan MTs are bimodular proteins that combine six domains—α, β1, β2, β3, γ, and δ—in a lineage-specific manner. We have functionally characterized the Neritimorpha β3β1 and the Patellogastropoda γβ1 MTs, demonstrating the metal-binding capacity of the new γ domain. Our results have revealed a modular organization of mollusk MT, whose evolution has been impacted by duplication, loss, and de novo emergence of domains. MTs represent a paradigmatic example of modular evolution probably driven by the structural and functional requirements of metal binding.
Keywords: α, β1, β2, β3, γ, δ, domains, bi- and multimodular metallothioneins, cysteine motifs, de novo evolution, metal-binding capacity and preference
Introduction
Metallothioneins (MTs) are a superfamily of intracellular, cysteine-rich (≈15–30%), and mostly low-molecular-weight (<100 amino acids) proteins present across eukaryotes, from various protists to plants, fungi, and animals (Capdevila and Atrian 2011; Blindauer 2014). Their cysteine (Cys, C) residues are arranged in distinctive motifs (i.e., CxC, CC, and CCC), whose number and distribution led to the original definition of the two functional domains in vertebrate MTs (Braun et al. 1986) designated as α domain (with 11–12 cysteines at the C-terminal region) and β domain (with nine cysteines at the N-terminal region), joined by a linker sequence. In this bimodular structure made of two domains, the cysteine motifs of each domain are able to form metallic clusters, thus conferring the capacity of binding both essential and nonessential metals (Nielson and Winge 1985). Notice that the α and β nomenclature has been also used for referring to N-terminal (α) or C-terminal (β) domains in some gastropod MTs (Baumann et al. 2017; Niederwanger, Calatayud, et al. 2017; Palacios et al. 2017; Schmielau et al. 2019). In order to avoid confusion, however, we will use here the α/β nomenclature in its primary meaning for classifying domains based on the number and distribution of cysteine motifs (Jenny et al. 2016; Nam and Kim 2017), and not on their N- or C-terminal position.
Thanks to their metal-binding capacity, MTs are directly involved in metal homeostasis and detoxification, but also in radical scavenging, oxidative stress protection, and antiapoptotic defense (Capdevila et al. 2012), leading to their role as a model system for the investigation of the genetic mechanisms by which organisms adapt to diverse metal bioavailabilities or stress. For instance, duplications of MT genes (Maroni et al. 1987; Adamo et al. 2012) or expansions of MT domains (Tanguy and Moraga 2001; Jenny et al. 2016; Pedrini-Martha et al. 2020), along with elevated levels of MT expression (Timmermans et al. 2005; Janssens et al. 2008, 2009; Costa et al. 2012; Catalan et al. 2016; de Francisco et al. 2018) or changed metal specificity (Tio et al. 2004; Palacios et al. 2011; de Francisco et al. 2017) have been considered adaptive events contributing to increase the metal and stress tolerance of the organisms in different environments. In particular, a previous thorough analysis investigating the role of cadmium (Cd) on the evolution of gastropod MTs has led us to suggest that lineage-specific changes of metal-selectivity features might have been important during the recurrent colonization of marine gastropods to terrestrial and freshwater habitats, where they had to face the challenge of adapting to ecosystems with different levels of metal bioavailability (Dallinger et al. 2020). In this context, studies on MT evolution have been of interest to evolutionary ecologists (Janssens et al. 2009; Faddeeva-Vakhrusheva et al. 2016; Zhang et al. 2018, 2019; Purać et al. 2019) who have associated environmental factors—that is, concentrations of heavy metals—with the evolution of diverse MTs.
Gastropoda is composed of five distinctive lineages ranked as subclasses, whose interrelationships are still controversial: Patellogastropoda, Vetigastropoda, Neritimorpha, Caenogastropoda, and Heterobranchia (Zapata et al. 2014; Cunha and Giribet 2019). Although several gastropod MTs have been previously investigated, these studies have an uneven phylogenetic distribution. MT identification and metal-binding selectivity have been determined for 15 MTs from seven heterobranch species (with the most extensive studies), for four MTs from three caenogastropod species, and for a single vetigastropod MT. In order to fully understand the evolution of gastropod MTs and their metal-binding specificities, it was therefore necessary to extend studies to include particular species of Patellogastropoda (true limpets) and Neritimorpha (snails known as nerites and their allies). To add both patellogastropods and neritimorphs here, we have conducted an exhaustive survey of gastropod MTs in public databases, including raw sequencing data from transcriptomic and genomic high-throughput sequencing projects. We have been able to reconstruct and identify new MTs from limpet and neritid species. We have characterized the metal-binding properties of new MTs for each, selecting Lottia gigantea and Nerita peloronta MTs as representatives of each taxon. Our data set extends the knowledge of metal-binding preferences to MTs of previously unstudied gastropod taxa, broadening the scope of known diversification of metal-binding selectivity in MTs for gastropods, the most species-rich, and arguably the most morphologically and ecologically diverse, class of Mollusca.
By extending what we have learned with new genomic searches, we have added novel MTs from the other three gastropod clades (Vetigastropoda, Caenogastropoda, and Heterobranchia) and from all mollusk classes, identifying and classifying new MTs from other conchiferan (i.e., Gastropoda, Bivalvia, Cephalopoda, Monoplacophora, and Scaphopoda) and aculiferan (i.e., Polyplacophora, Solenogastres, and Caudofoveata classes) mollusks. In summary, by collecting 272 MTs from 189 different species, we have created the most comprehensive catalog of mollusk MTs compiled so far, and we have exposed patterns of modular organization across molluskan MTs.
Results
Identification of Patellogastropoda MTs
We surveyed the genome project of the L. gigantea (Patellogastropoda: Lottiidae). This search yielded an automatically predicted MT (Gene ID: 20249168; hypothetical protein: XP_009056965.1), whose sequence, structure, and size suspiciously differed from other Gastropoda MT genes. The low-quality sequence of the genomic region containing the putative MT gene (NW_008709190.1) prompted us to reanalyze this region by polymerase chain reaction (PCR) amplifying, cloning, and resequencing >12 kb of L. gigantea genome (supplementary fig. S1A, Supplementary Material online). Sequence analysis identified two MT genes tandem repeated in this region (accession number MK795721), which we named LgiMT1 and LgiMT2. Our predictions were further supported by PCR amplification of the corresponding cDNAs (MK770430 and MK770431) and by nine L. gigantea expressed sequence tags (supplementary table S1, Supplementary Material online), corroborating the misassembly of the MT genomic region in the genome project, and revealing some degree of polymorphism at the amino acid level (S/T and S/A at amino acid positions 3 and 68, respectively, supplementary fig. S1B, Supplementary Material online). Comparison of the coding region (CDS) of LgiMT1 and LgiMT2 genes revealed that they were 97.78% identical at the nucleotide level (220 out of 225 nt) and both encoded for a 75 amino acid protein with 19 cysteines (25.7%). LgiMT1 and LgiMT2 proteins only differed at positions 6, 20, and 49: P, L, and P residues in LgiMT1, and A, S, and S in LgiMT2 (supplementary fig. S1B, Supplementary Material online).
We extended the limpet MT listing to three families and eight additional species of Patellogastropoda (supplementary fig. S2A and table S2, Supplementary Material online, and Dallinger et al. 2020). These include five species of Lottiidae: Lottia digitalis (LdiMT1 partial), Lottia kogamogai (LkoMT1), Lottia scutum (LscMT1 partial), Nipponacmea fuscoviridis (NfuMT1), and Patelloida pygmaea (PpyMT1); two species of Nacellidae: Cellana rota (CroMT1) and Nacella concinna (NcoMT1); and one species of Patellidae: Patella vulgata (PvuMT1 and PvuMT2). Amino acid comparison of the 11 Patellogastropoda MTs revealed high sequence similarity (from 69% to 97% identity) with the distinctive 19 cysteines fully conserved. Interestingly, the 19 cysteines were organized into two domains: a novel MT domain at the N-terminal region with ten cysteines arranged in five CC pairs (CCx5CCx4CCx6CCx7CC) that we named γ domain (notice that this γ domain is not related with the N-terminal, six Cys domain of plant Ec-1 MTs, also named γ domain [Loebus et al. 2011]), which was connected by a linker of four to six amino acids to an archetypal 9-Cys β1 domain ([CxC]x3[CxC]x3Cx5[CxC]x3[CxC]) (Jenny et al. 2016; Nam and Kim 2017) at the C-terminal region (table 1 and supplementary fig. S2A, Supplementary Material online). We concluded therefore that the structure of Patellogastropoda MTs was of γ/β1 domains (fig. 1).
Table 1.
Cysteine Motifs of Mollusca Bimodular MTs.
| Class | Clade | Domain | Cysteine Motifs |
||||
|---|---|---|---|---|---|---|---|
| N-Terminal | Linker | C-Terminal | |||||
| Conchifera | Gastropoda | Patellogastropoda | γ | β1 | CCx5CCx4CCx6CCx7CC | X5–6 | [CxC]x3[CxC]x3Cx5[CxC]x3[CxC] |
| Vetigastropoda | β3 | β1 | Cx3Cx4[CxC]x3[CxC]x4[CxC]x2C | X3–6 | [CxC]x3[CxC]x3Cx5[CxC]x3[CxC] | ||
| Neritimorpha | β3 | β1 | Cx3Cx5[CxC]x3[CxC]x3[CxC]x2C | X3–4 | [CxC]x4[CxC]x3Cx5[CxC]x3[CxC] | ||
| Caenogastropoda | β3 | β1 | Cx3Cx4[CxC]x5[CxC]x3[CxC]x2C | X2–3 | [CxC]x4[CxC]x3Cx6[CxC]x3[CxC] | ||
| Heterobranchia | β3 | β1 | Cx3Cx4[CxC]x3[CxC]x3[CxC]x2C | X2 | [CxC]x4[CxC]x3Cx5[CxC]x3[CxC] | ||
| Scaphododa | β2 | β1 | [CxC]x5[CxC]x3[CxC]x4[CxC]x4C | X8–10 | [CxC]x4[CxC]x3Cx5[CxC]x3[CxC] | ||
| Bivalvia | α | β1 | [CxC]x5[CxC]x3Cx5[CxC]x3[CxC]x3[CxC]x2C | X3–4 | [CxC]x6[CxC]x3Cx5[CxC]x3[CxC] | ||
| β2 | β2 | [CxC]x5[CxC]x3[CxC]x4[CxC]x2C | X2–3 | Cx2Cx4[CxC]x3[CxC]x3[CxC]x2C | |||
| β2 like | β1 | Cx4Cx4[Cx0–1C]x4[Cx0–1C]x3[CxC]x2C | X1–3 | [CxC]x4[CxC]x3Cx5[CxC]x3[CxC] | |||
| Cephalopoda | α | β1 | [CxC]x6[CxC]x3Cx4[CxC]x3[CxC]x3[CxC]x2C | X3–4 | [CxC]x4[CxC]x3Cx5[CxC]x3[CxC] | ||
| Monoplacophora | β2 | β1 | [CxC]x6[CxC]x3[CxC]x2Cx[CxC]x2C | X3 | [CxC]x3[CxC]x3Cx5[CxC]x3[CxC] | ||
| Aculifera | Polyplacophora | α | β1 | [CxC]x4[CxC]x3Cx4[CxC]x3[CxC]x3[CxC]x2C | X3 | [CxC]x4[CxC]x3Cx5[CxC]x3[CxC] | |
| Solenogastres | β2 | β1 | [CxC]x5[CxC]x3[CxC]x4[CxC]x4C | X6 | [CxC]x4[CxC]x3Cx5[CxC]x3[CxC] | ||
| α | β1 | [CxC]x5[CxC]x3Cx5[CxC]x3[CxC]x3[CxC]x2C | X7 | [CxC]x3[CxC]x3Cx5[CxC]x3[CxC] | |||
| Caudofoveata | δ | β1 | [CxC]x3[CxC]CCx4Cx3[CxC]x3CCx4[CxC]C | X3 | [CxC]x4[CxC]x3Cx5[CxC]x3[CxC] | ||
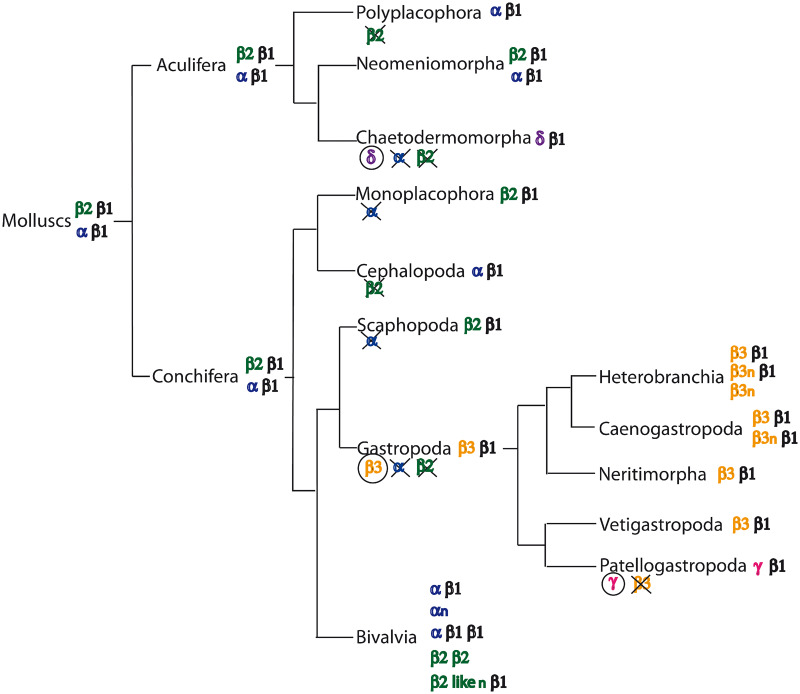
Fig. 1.
Structural evolution of MTs along the Mollusca phylum. Most mollusk MTs have a bimodular structure made of a variable taxon-specific N-terminal domain—α, β2, β3, γ or δ—, and a conserved C-terminal β1 domain. Domains are classified based on the number and configuration of the cysteine motifs. Based on the distribution of domains among the different clades, the most parsimonious evolutionary scenario would be that α, β1, and β2 domains are ancient, already present in the MTs of mollusk ancestor. In contrast, β3, γ, and δ domains would be de novo domains arose in Gastropoda, Patellogastropoda, and Caudofoveata, respectively. Domain gains (circled) and losses (crossed) are indicated under each clade. Deviations of the standard bimodular structure are found in MTs of Bivalvia and Gastropoda, including multimodular MTs with more than two domains (αn [Jenny et al. 2004, 2016]; β1n and β2-liken [Nam and Kim 2017] in Bivalvia; and β3n in Heterobranchia and Caenogastropoda [Niederwanger, Calatayud, et al. 2017; Palacios et al. 2017]), and MTs lacking the conserved β1 domain (αn and β2β2 MTs in Bivalvia; β3n MTs in Heterobranchia). Phylogenetic relationships within Mollusca are based on Zapata et al. (2014), Cunha and Giribet (2019), and Kocot et al. (2020)
Metal-Binging Capacity of Patellogastropoda MTs
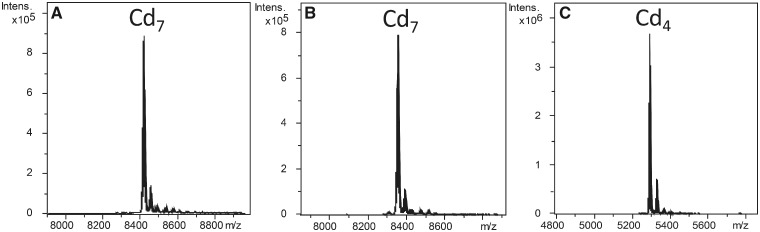
In order to demonstrate the MT nature and to explore the metal-selectivity features of the two L. gigantea MTs, we studied the formation of metal-LgiMT1 and metal-LgiMT2 complexes by the proteins heterologously expressed in Escherichia coli and grown in media supplemented with copper (Cu), cadmium (Cd), or zinc (Zn) salts by inductively coupled plasma atomic emission spectrometer (ICP-AES) and electrospray ionization mass spectrometry (ESI-MS) analyses. ICP-AES is an analytical technique that allows for protein quantification and metal-to-protein stoichiometry determination through the measurement of element composition of the samples (S, Zn, Cd, and Cu) (Bongers et al. 1988), and ESI-MS is used to determine the molecular mass of the species formed, that is, the speciation of the samples (Capdevila et al. 2012). The ICP-AES and native ESI-MS (recorded at neutral pH in order to allow the observation of unaltered species) analyses (data not shown) of the recovered samples showed that LgiMT1 and LgiMT2 rendered a mixture of metallated species both in Zn(II)- as well as in Cu(II)-supplemented cultures, thus indicating the absence of preference for either of those metal ions (Zn(II) and Cu(II) stand for divalent Zn2+ and Cu2+ ions). On the contrary, unique Cd7-LgiMT1 and Cd7-LgiMT2 species, that is, MTs loaded with seven Cd(II) ions, were recovered from Cd-enriched culture media (fig. 2A and B). The identification of single species after recombinant metal-supplemented productions normally reveals the existence of a preferred thermodynamically favored metal cluster, whereas in the absence of any specific species, the formation of a variety of species with similar but not identical metal content is observed (Palacios et al. 2014). The observation of single Cd7-LgiMT species, together with the observation by acid ESI-MS (recorded at pH 2.4 that lead to partial protonation of thiol Cys groups, which normally results on the release of Zn and Cd while bound Cu is maintained) that both proteins are reluctant to release Cd(II) by acidification at pH 2.4 giving rise to Cd4-LgiMT species, demonstrated the Cd-thionein character of both Patellogastropoda MTs.
Fig. 2.
Deconvoluted ESI-MS spectra LgiMT1 (A), LgiMT2 (B), and γLgiMT2 (C) recombinantly produced by Escherichia coli in Cd-enriched media.
Metal-Binding Functionality of the New γ Domain
In order to determine the independent metal-binding capacity of the new γ domain, we analyzed its ability to form metal complexes when expressed alone. Thus, we heterologously expressed the 10-Cys γ domain of LgiMT2 (from Met1 to Gln45), hereafter as γLgiMT2, in E. coli grown in medium supplemented with Cu, Cd, or Zn salts. The election of the γ domain of LgiMT2 relied on the fact that it was more conserved than that of LgiMT1 when compared across the γ domains of other Patellogastropoda MTs (supplementary fig. S2A, Supplementary Material online). The distinct metal-γLgiMT2 preparations obtained after purification were characterized by ICP-AES and ESI-MS. The results showed a clear preference of γLgiMT2 for divalent Zn(II) and Cd(II) ions, as major M4-γLgiMT2 species were identified for both metal ions by ESI-MS at pH 7. Interestingly, formation of Cd4-γLgiMT2 complexes was more favored than Zn4-γLgiMT2 complexes, as suggested by the fact that the former one was obtained as a single species (fig. 2C), whereas the latter one coexisted with other minor species (data not shown). This Cd4-γLgiMT2 cluster also exhibited a high resistance against demetallation at acidic pH levels, as this required an acidification down to pH 1 to obtain the apo-protein. Conversely to the Zn- and Cd-cultures, the preparations obtained from Cu(II)-enriched media yielded mixtures of multiple Cux-γLgiMT2 complexes (x ranging from 5 to 10), confirming the poor preference of this domain for Cu(I).
Identification of Neritimorpha MTs
We collected 12 MTs from two families and six species of Neritimorpha, including five species of Neritidae: Clithon retropictum (CretrMT1 and CretrMT2), Nerita albicilla (NalMT1 and NalMT2), Nerita melanotragus (NmeMT1 and NmeMT2), N. peloronta (NpeMT1 and NpeMT2), and Neritina pulligera (NpuMT1 and NpuMT2); one species of Neritopsidae: Titiscania limacina (TliMT1 and TliMT2 partial) (supplementary fig. S2B and table S2, Supplementary Material online, and Dallinger et al. 2020). All Neritimorpha species possessed two MTs, and this appears to be a lineage-specific gene duplication shared by at least two of the four extant superfamilies within Neritimorpha. As for patellogastropods, amino acid comparison of the 12 Neritimorpha MTs showed high sequence conservation (from 65% to 96% identity) with 19–20 cysteines organized in two MT domains: at the N-terminal region, a 9-Cys β3 domain (Cx3Cx4–5[CxC]x3[CxC]x3[CxC]x2C, formerly known as α1/2 domain in non-Neritimorpha species [Baumann et al. 2017; Niederwanger, Calatayud et al. 2017; Palacios et al. 2017; Schmielau et al. 2019] with an extra cysteine in Neritimorpha MT1s), connected by a linker of three to four amino acids to a 9-Cys β1 domain at the C-terminal region ([CxC]x3–4[CxC]x3Cx5[CxC]x3[CxC]) (table 1 and supplementary fig. S2B, Supplementary Material online). The structure of Neritimorpha MTs was therefore of β3/β1 domains (fig. 1).
Metal-Binding Capacity of Neritimorpha MTs
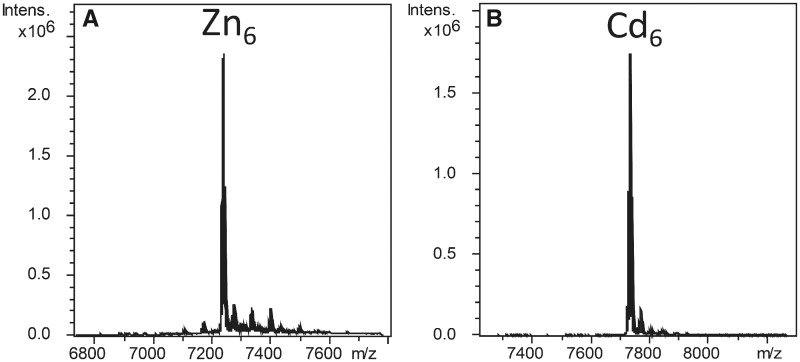
In order to analyze the metal-binding abilities of Neritimorpha MTs, we studied the formation of metal-MT complexes of NpeMT1 and NpeMT2 heterologously expressed in E. coli grown in medium supplemented with Cu, Cd, or Zn salts. Metal-NpeMT complexes were purified and analyzed by ICP-AES and ESI-MS. Our data showed that both Neritimorpha MTs presented similar specificities for divalent metal ions, Zn(II) and Cd(II). However, their binding preference was not exactly the same, since NpeMT1 was more specific for Zn(II) than NpeMT2, whereas NpeMT2 was more specific for Cd(II) than NpeMT1. Consequently, Zn6-NpeMT1 and Cd6-NpeMT2 complexes could be recovered as single species (fig. 3). The Zn-thionein character of NpeMT1 and the Cd-thionein nature of NepMT2 were also supported by the recombinant productions of this protein in Cu-enriched media, which rendered mixtures of heterometallic Zn, Cu-MT complexes for NpeMT1, but mixtures of homometallic Cu-MT species for NpeMT2 (data not shown).
Fig. 3.
Deconvoluted ESI-MS spectra of the in vivo preparations of Escherichia coli recombinant NpeMT1 obtained from a Zn-enriched medium (A) and NpeMT2 synthesized in Cd-enriched cultures (B).
MTs from Other Gastropoda Clades
We extended the surveys of MTs to gastropod species belonging to Caenogastropoda, Heterobranchia, and Vetigastropoda, considered separately below. We collected a total of 163 MT sequences, including newly identified MTs and previously reported ones (supplementary table S2, Supplementary Material online).
Caenogastropoda MTs
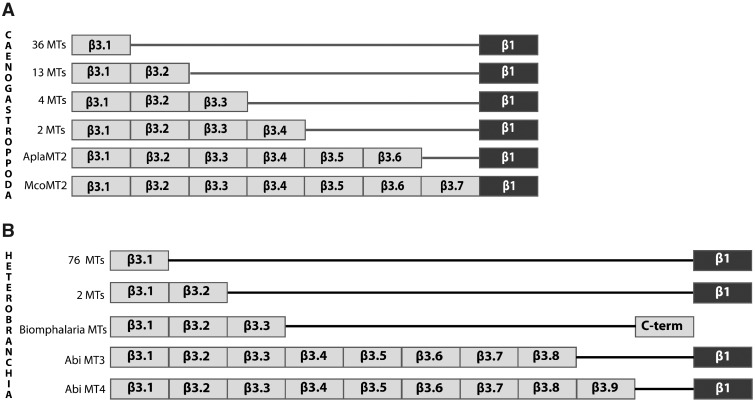
We collected 55 MT sequences (43 new) of 43 Caenogastropoda species (supplementary fig. S2C and table S2, Supplementary Material online). MT multiplicity was observed in 11 species (ten species with two, and Pomacea canaliculata with three MTs). Size diversity within Caenogastropoda was high, with MTs ranging from 66 amino acids (BacMT1) to 251 amino acids (McornMT2), with a cysteine content of 17 (25.8%) and 72 cysteines (28.7%), respectively. Overall, Caenogastropoda MTs were organized in an N-terminal 9-Cys β3 domain (Cx3Cx4[CxC]x3–5[CxC]x3[CxC]x2C) linked by two to three residues to a C-terminal 9-Cys β1 domain ([CxC]x3–4[CxC]x3Cx5–6[CxC]x3[CxC]) (table 1 and supplementary fig. S2C, Supplementary Material online). The archetypal structure of Caenogastropoda MTs was therefore of β3/β1 domains (fig. 1). Remarkably, β3 domain duplications were observed in a number of Caenogastropoda MTs: one single duplication (i.e., β3.1/β3.2) in 13 MTs; two duplications (i.e., β3.1/β3.2/β3.3) in JjaMT1, PbrMT2, and PcanMT2 sequences; three duplications (from β3.1 to β3.4) in partial EheMT2 and PcanMT1; five duplications (from β3.1 to β3.6) in AplaMT2; and six duplications (from β3.1 to β3.7) in McornMT2 (fig. 4A and supplementary fig. S3A, Supplementary Material online). Since these multi-β3 MTs were unevenly distributed among the Littorinimorpha, Neogastropoda, and “architaenioglossan” representatives, independent events of internal domain duplications were the most plausible origin of such MTs (Schmielau et al. 2019). Also noteworthy, ten MTs (most of them duplicated copies) of the Architaenioglossa order showed an additional H3–4C4 motif (HxHHHx2Cx3Cx6–9Cx0–1C) at the N-terminal region (supplementary fig. S2C, Supplementary Material online). Since these MTs belonged to the same taxonomic group, a lineage-specific event that added the H3–4C4 motif to the N-terminus of an ancestral Architaenioglossa MT duplicate would be the most parsimonious explanation.
Fig. 4.
Schematic representation of multimodular Gastropoda MTs. Internal duplications of the N-terminal β3 domain (gray box) generated multimodular MTs in Caenogastropoda (A) and Heterobranchia (B) (see supplementary fig. S2 and table S2, Supplementary Material online, for further details of species). A variable number of β3 domains, ranging from 1 (top) to 9 (bottom), is followed the by a conserved β1 domain (black box). In Heterobranchia, Biomphalaria MTs have three β3 domains followed by a C-terminal tail of five cysteines (C-term) but lack the conserved β1 domain.
Heterobranchia
From an MT perspective, this was the most studied Gastropoda clade. We collected 86 MT sequences (61 new) of 55 Heterobranchia species. MT multiplicity was observed in 22 species with two (14 species), three (seven species), or four (one species) MTs (supplementary fig. S2D and table S2, Supplementary Material online). Size diversity of Heterobranchia MTs was the highest within the Gastropoda class, with MTs ranging from 58 (AbaMT1, GscMT1, GcuMT1, GtrMT1, and LstMT1) to 319 (AbiMT4) amino acids, with a cysteine content of 15 (25%) and 85 cysteines (26.2%), respectively. Overall, Heterobranchia MTs were organized in a N-terminal 9-Cys β3 domain (Cx3Cx4[CxC]x3[CxC]x3[CxC]x2C) linked by two residues to a C-terminal 9-Cys β1 domain ([CxC]x4[CxC]x3Cx5[CxC]x3[CxC]) (table 1 and supplementary fig. S2D, Supplementary Material online). The structure of Heterobranchia MTs was therefore of β3/β1 domains (fig. 1). There were, however, several exceptions to this structure. Species of Lymnaeidae, for instance, lacked the last [CxC] motif of the β1 domain, MTs of Bradybaena similaris and Fiona pinnata had an additional β3 domain (i.e., β3.1/β3.2) (supplementary fig. S2D, Supplementary Material online), and Alinda biplicata AbiMT3 and AbiMT4 had eight and nine β3 domains, respectively (Pedrini-Martha et al. 2020). But the most deviant Heterobranchia MTs were those of the Biomphalaria species, B. glabrata and B. pfeifferi. These MTs lacked the β1 domain and had three β3 domains (β3.1/β3.2/β3.3) followed by a C-terminal tail with five cysteines (Cx6Cx5Cx4CC) (fig. 4B and supplementary fig. S3B, Supplementary Material online) (Niederwanger, Calatayud, et al. 2017). The fact that both species shared the same structure suggested that it could be a general MT feature for Biomphalaria.
Vetigastropoda
We collected 22 MT sequences (18 new) of 18 Vetigastropoda species (supplementary fig. S2E and table S2, Supplementary Material online). MT multiplicity was found in four species: Clanculus pharaonius (CphMT1 and CphMT2 partial), Perotrochus lucaya (PluMT1 and PluMT2), Phasianella ventricosa (PveMT1 and PveMT2), and Prothalotia lehmanni (PleMT1 and PleMT2). Overall, Vetigastropoda MTs were 65–71 amino acid long with 18 cysteines (25.4–27.7%) organized in a N-terminal 9-Cys β3 domain (Cx3Cx4[CxC]x3[CxC]x3–4[CxC]x2C) linked by 3–6 residues to a C-terminal 9-Cys β1 domain ([CxC]x3[CxC]x3Cx5[CxC]x3[CxC]) (table 1 and supplementary fig. S2E, Supplementary Material online). The last common ancestor for Vetigastropoda thus likely had MTs with β3/β1 domains (fig. 1).
Comparisons across all of the phylogenetically diverse Gastropoda have suggested that the ancestral Gastropoda already had a β3/β1 MT (fig. 1), which underwent diverse lineage-specific modifications during evolution: 1) gene duplications leading to parallel MT multiplicity in divergent gastropod lineages, 2) internal domain duplications and losses in the Caenogastropoda and Heterobranchia taxa, and 3) acquisitions of novel modules such the H3–4-C4 motif in certain Caenogastropoda, and the new γ domain in Patellogastropoda.
New MTs in Other Mollusk Classes
Recent phylogenomic analyses have split Mollusks into two major clades (see fig. 1) (Kocot et al. 2011, 2020; Smith et al. 2011): Conchifera (including Gastropoda, Bivalvia, Cephalopoda, Monoplacophora, and Scaphopoda) and Aculifera (including Polyplacophora, Solenogastres, and Caudofoveata). We used publicly available molluskan Sequence Read Archives (SRA) projects to assemble new MTs that, together with previously reported ones, represented an extensive evolutionary list of mollusk MTs. Thus, our analysis has led us to identify for the first time MTs in Scaphopoda, Cephalopoda, and Monoplacophora classes, and the first MTs for chiton, solenogastres and caudofoveatan representatives of Aculifera.
Conchifera Clade
Scaphopoda
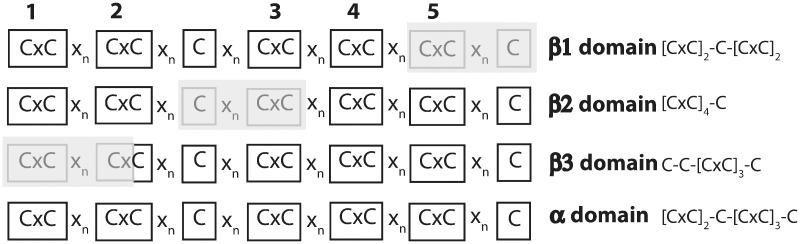
This molluskan class has been considered the sister group of Gastropods (Smith et al. 2011), though this phylogenetic relationship is still under debate. We identified two Scaphopoda MTs from Antalis entalis (AenMT1) and Graptacme eborea (GebMT1) species (supplementary fig. S2F and table S2, Supplementary Material online). Scaphopoda MTs were 68–72 amino acid long with 18–19 cysteines (26.4%) organized in a N-terminal 9-Cys β domain ([CxC]x5[CxC]x3[CxC]x2–4[CxC]x4C) linked by 8–10 residues to a C-terminal 9-Cys β1 domain ([CxC]x4[CxC]x3Cx5[CxC]x3[CxC]) (table 1 and supplementary fig. S2F, Supplementary Material online). Interestingly, the organization of the [CxC] motifs in the Scaphopoda N-terminal β domain, [CxC]4-C, resembled that of β2 domain defined in the unconventional bivalve MTIIIs (Jenny et al. 2016), whereas it differed from that of Gastropoda β1 and β3 domains—[CxC]2-C-[CxC]2 and C-C-[CxC]3-C, respectively—(table 1 and fig. 5). The structure of Scaphopoda MTs was therefore of β2/β1 domains (fig. 1).
Fig. 5.
Hypothetical relationship between α and β domains. Based on the configuration of the C and [CxC] motifs, a putative connection between α and β domains is hypothesized. The five [CxC] motifs and the two additional Cs distributed as [CxC]2-C-[CxC]3-C in the α domain appear to have been trimmed (shadowed) at the C-terminal end in β1 domain: [CxC]2-C-[CxC]2; at the middle in β2: [CxC]4-C; or at the N-terminal end in β3: C-C-[CxC]3-C.
Bivalvia
In order to have a broad perspective of the MTs within Bivalvia, we analyzed 62 MTs from 36 species (supplementary fig. S2G and table S2, Supplementary Material online). For clarity, and since the structural diversity of MTs in oysters and mussels has been extensively described elsewhere (Mackay et al. 1993; Jenny et al. 2004, 2016; Aceto et al. 2011), we focused our analysis on the MTI and MTIV within available species of Ostreidae, and on MT10, MT10B, and MT20 within Mytilidae, which have been considered to have the plesiomorphic condition for bivalve MTs (Nam and Kim 2017). Most Bivalvia MTs were 66–78 amino acid long with 21–25 cysteines (26.9–32%) organized in an N-terminal 12-Cys α domain ([CxC]x5[CxC]x3Cx4–5[CxC]x3[CxC]x3–4[CxC]x2C) linked by three residues to a C-terminal 9-Cys β1 domain ([CxC]x3–6[CxC]x3Cx5[CxC]x3–5[CxC]) (table 1 and supplementary fig. S2G, Supplementary Material online). The likely ancestral structure of these Bivalvia MTs was therefore of α/β1 domains (fig. 1). Other Bivalvia MTs (e.g., Crassostrea MTIIIs in the order Ostreoidea and PmarMT2 of Pinctada martensii in the order Pterioida) were organized in two β2 domains. These MTs probably derived from an ancestral β2/β1 form (fig. 1). Deviations from these structures were found, ranging from loss or gain of some cysteines or small protein fragments (e.g., MquMT1 and CgiMTIV), to significant structural modifications, such as those previously described in a sphaeriid clam (Pisidium coreanum) PcorMT1 and the oyster (Alectryonella plicatula) ApliMT1: α/β1/β1; in Crassostrea MTIIs: αn or α/β1/β1; or in the scallop (Argopecten irradians) AirMT1 and AirMT2: β2-like/β2-like/β2-like/β1 and β2-like/β2-like/β1, respectively (notice that organization of the [CxC] motifs in the β2-like domain was more similar to β3 than to β2 domain) (Tanguy and Moraga 2001; Baek et al. 2009; Jenny et al. 2016; Nam and Kim 2017).
Cephalopoda
We identified the MTs of four Cephalopoda species: Nautilus pompilius (NpoMT1), Octopus vulgaris (OvuMT1), Octopus bimaculoides (ObiMT1), and Sepia esculenta (SesMT1) (supplementary fig. S2H and table S2, Supplementary Material online). Cephalopoda MTs were 69–73 amino acid long with 19–21 cysteines (27.5–28.7%) organized in α/β1 domains more similar to those of other molluskan classes in the Nautilus MT than in the other species. The N-terminal 12-Cys α domain ([CxC]x5–6[CxC]x3Cx4[CxC]x3[CxC]x3[CxC]x2C) in the Nautilus MT, but slightly divergent in the coleoid cephalopods ([CxC]x5–6[CxC]x3Cx4Cx3[CxC]x3[CxC]x2C in Sepia and Octopus MTs), was linked by three or four residues to a C-terminal 9-Cys β1 domain ([CxC]x4[CxC]x3Cx5[CxC]x3[CxC] in Nautilus and Sepia MTs, but somewhat modified in Octopus MTs; table 1 and supplementary fig. S2H, Supplementary Material online). Based on Nautilus and Sepia MTs, we concluded that the prototypical structure of Cephalopoda MTs was of α/β1 domains (fig. 1).
Monoplacophora
We identified the first MT in a Monoplacophora species, Laevipilina hyalina (LhyMT1) (supplementary fig. S2I and table S2, Supplementary Material online). LhyMT1 was 63 amino acid long with 19 cysteines (30.2%) organized in an N-terminal β2 domain ([CxC]x6[CxC]x3[CxC]x2[CxC]xCx2C) linked by three residues to a C-terminal 9-Cys β1 domain ([CxC]x3[CxC]x3Cx5[CxC]x3[CxC]) (table 1 and supplementary fig. S2I, Supplementary Material online). The structure of the Monoplacophora MT was therefore of β2/β1 domains (fig. 1).
Aculifera Clade
Polyplacophora
We identified five MTs in Polyplacophora species: Acanthochitona crinita (AcriMT1), Chaetopleura apiculata (CapMT1), Chiton olivaceus (ColMT1), Tonicella lineata (TliMT1), and Leptochiton rugatus (LruMT1) (supplementary fig. S2J and table S2, Supplementary Material online). Polyplacophora MTs were 70–73 amino acid long with 21 cysteines (28.8–30.0%) organized in a N-terminal 12-Cys α domain ([CxC]x5[CxC]x3Cx4[CxC]x3[CxC]x3[CxC]x2C) linked by three residues to a C-terminal 9-Cys β1 domain ([CxC]x3–4[CxC]x3Cx5[CxC]x3[CxC]) (table 1 and supplementary fig. S2J, Supplementary Material online). The structure of Polyplacophora MTs was therefore of α/β1 domains (fig. 1).
Solenogastres
We identified eight MTs of Solenogastres species: Alexandromenia crassa (AcraMT1 partial), Amphimeniidae sp. (AspMT1 partial), Micromenia fodiens (MfoMT1 partial), Neomenia carinata (NcaMT1), Neomenia megatrapezata (NmegMT1 and NmegMT2), and Neomeniomorpha sp. (NspMT1 and NspMT2 partial) (supplementary fig. S2K and table S2, Supplementary Material online). Full-length Solenogastres MTs (i.e., NcaMT1, NmegMT1, and NspMT1) were 65–66 amino acid long with 18–19 cysteines (27.3–29.3%) organized in an N-terminal 9-Cys β2 domain ([CxC]x5[CxC]x3[CxC]x2–4[CxC]x3–4C) linked by six residues to a C-terminal 9-Cys β1 domain ([CxC]x3–4[CxC]x3Cx5[CxC]x3[CxC]) with an additional cysteine in NmegMT1 and NspMT1 sequences (table 1 and supplementary fig. S2K, Supplementary Material online). The structure of these MTs was therefore of β2/β1 domains (fig. 1). Interestingly, Neomenia megatrapezata species had a second MT, NmegMT2, whose N-terminal domain was a 12-Cys α domain ([CxC]x5[CxC]x3Cx5[CxC]x3[CxC]x3[CxC]x2C) linked by seven residues to a C-terminal 9-Cys β1 domain ([CxC]x3[CxC]x3Cx5[CxC]x3[CxC]) (table 1 and supplementary fig. S2L, Supplementary Material online). The NmegMT2 structure was, therefore, of the α/β1 type, meaning that two structurally different MTs coexisted in the same Solenogastres species (fig. 1).
Caudofoveata
We identified four MTs in Caudofoveata species: Chaetoderma nitidulum (CniMT1), Falcidens caudatus (FcaMT1), Falcidens sagittiferus (FsaMT1), and Scutopus ventrolineatus (SveMT1) (supplementary fig. S2M and table S2, Supplementary Material online). The Caudofoveata MTs were 73–78 amino acid long with 23 cysteines (29.5–31.5%) organized in a novel N-terminal 14-Cys domain ([CxC]x3[CxC]CCx4Cx3–7[CxC]x3CCx4[CxC]C) that we named δ domain, linked by three residues to a C-terminal 9-Cys β1 domain ([CxC]x3–4[CxC]x3Cx5[CxC]x3[CxC]) (table 1 and supplementary fig. S2M, Supplementary Material online). The structure of Caudofoveata MTs was therefore of δ/β1 domains (fig. 1).
In summary, we have identified more than 270 MTs in 189 different species distributed across the eight molluskan classes, ≈64% as single copy sequences (supplementary table S2, Supplementary Material online; the existence of additional MTs in some species cannot be excluded because some sequence databases are still in progress). Our data showed that a single MT with a two-domain (i.e., bimodular) structure was the predominant type of MTs in most Mollusca species (fig. 1 and table 1). Our results revealed, however, many exceptions to this situation, with MT multiplicity and/or multimodular MTs in many Mollusca species, which denoted an intricate and dynamic evolutionary history of mollusk MTs.
Discussion
Functional Evolution of Mollusk MTs
Evolution of the Metal-Binding Capacity
Most mollusk MTs are bimodular proteins with 18 (in β2–3/β1 MTs), 19 (in γ/β1), 21 (in α/β1), or 23 (in δ/β1) cysteines. Neritimorpha β3/β1 MTs render homometallic complexes with six divalent ions, either Zn(II) or Cd(II) (fig. 3), meaning that each 9-Cys domain is designed to allocate three divalent metal ions. This capacity is similar to those reported for other bimodular gastropod MTs (Perez-Rafael et al. 2012, 2014; Palacios et al. 2014; Dvorak et al. 2018) and agrees with 3D structural analysis of Littorina littorea (Baumann et al. 2017) and Helix pomatia MTs (Beil et al. 2019). The metal-binding capacity of Patellogastropoda γ/β1 LgiMTs, both with 19 cysteines, is slightly higher since they bind seven divalent metal ions (fig. 2A and B), similar to the 21-Cys α/β1 MTs of Bivalvia (Munoz et al. 2002; Orihuela et al. 2008). The successful synthesis of the γ domain of LgiMT2, with ten Cys residues, and the characterization of the species produced in Cd(II)-enriched media indicate that the LgiMTs render metal-aggregates containing seven Cd(II) ions because these MTs are capable of binding three Cd(II) ions in their β1 domain, whereas their γ domain allocates four Cd(II) ions (fig. 2C). The metal-binding capacity of MTs appears, therefore, to rely on the number and position of the cysteines in the different domains and, as expected, the higher the cysteine content, the higher the metal-binding capacity.
In that sense, an effective evolutionary strategy for increasing the metal-binding capacity of MTs has been the design of multimodular forms with high cysteine content and a high capacity of metal binding (Niederwanger, Dvorak et al. 2017; Palacios et al. 2017; Calatayud et al. 2018). Multimodular MTs had been identified in a few Bivalve and Gastropoda species (Tanguy and Moraga 2001; Jenny et al. 2004, 2016; Baumann et al. 2017; Nam and Kim 2017; Niederwanger, Calatayud et al. 2017; Palacios et al. 2017; Schmielau et al. 2019; Pedrini-Martha et al. 2020), and our results have increased this list with 16 new proteins containing a variable number of repeated domains. Sequence comparisons (fig. 4 and supplementary fig. S3, Supplementary Material online) and structural analysis (Baumann et al. 2017) indicate that these multimodular MTs originated by N-terminal duplications of the β3 domain, in agreement with the idea that proteins tend to increase in length mainly by the gain of sequences at the 5′-end of their genes (Toll-Riera and Alba 2013). The evolution of such multimodular MTs in some mollusk species suggests that diverse lineages have had to adapt to different conditions of metal bioavailability and stress, despite the ecophysiological determinants that have favored them in only some species remain, however, unknown.
Evolution of the Metal-Binding Preference
The evolution of MTs with different metal preferences has usually been associated with scenarios of MT multiplicity, in which neofunctionalization processes yielded MT duplicates with new metal-binding selectivities. Sixty-seven species patchily distributed across the mollusk phylogeny possess at least two MTs (supplementary table S2, Supplementary Material online), and their sequence (supplementary fig. S2, Supplementary Material online) and domain conservation (fig. 1 and table 1) suggest that most of them originated after the splitting of the main Mollusk groups by lineage-specific duplications. For instance, a gene duplication in the ancestor of the Neritimorpha class resulted in two MTs that according to the analysis of N. peloronta NpeMT1 and NpeMT2 diverged in their metal preferences (i.e., a Zn-thionein character for NpeMT1 and a Cd-thionein nature for NpeMT2; fig. 3). In contrast, the duplication found in L. gigantea appears more recent since both duplicates are 96% identical (supplementary fig. S1, Supplementary Material online) and still share a Cd binding preference (fig. 2). Our results support the idea that metal-binding preference and hence, functional specificity, does not mainly rely on the number and position of the cysteines (>94% identical between NpeMT1 and NpeMT2) but on the nature of the noncoordinating amino acids (≈70% different between NpeMT1 and MpeMT2) distributed along the protein sequence (Palacios et al. 2011; Perez-Rafael et al. 2014; Dallinger et al. 2020), although we are still far from being able to predict metal preference based on the analysis of the noncoordinating amino acids.
We do not know the biological determinants that favored the evolution of MTs with different metal preferences in certain mollusk lineages, but the colonization of new habitats (Dallinger et al. 2020) and the emergence of physiological novelties (Dallinger et al. 2005; Höckner et al. 2011) have been proposed as significant evolutionary factors. We do not know either the binding selectivity of the ancestral mollusk MT, but the widespread cadmium-binding capacity of many MTs—not only in mollusk but in diverse marine animals (Narula et al. 1995; Riek et al. 1999; Valls et al. 2001; Guirola et al. 2012; Calatayud et al. 2018)—along with the ancient origin α and β domains (see below), lead us to speculate that ancestral MTs might have been a detoxification system that was later co-opted for homeostatic functions for essential metals. Other scenarios are possible but since Cd is a highly toxic metal because it competes for Zn-dependent cellular processes, and Cd is frequently found with Zn in ore deposits of the earth crust, an early evolution of Cd-detoxifying MTs could have conferred a significant physiological advantage to marine organisms. This advantage would be especially important after increased Cd levels during Paleozoic era (Dallinger et al. 2020), concomitantly with the Cambrian explosion and the emergence of most animal phyla. From the ancestral MT, different metal-selective MTs would have independently evolved in different molluskan lineages: Cu-selective MTs in Heterobranchia species (Höckner et al. 2011; Perez-Rafael et al. 2011; Palacios et al. 2014), Zn-selective forms in Neritimorpha gastropods (this work) and bivalves (Orihuela et al. 2008), and metal-unselective MTs in Vetigastropoda and Heterobranchia lineages (Höckner et al. 2011; Perez-Rafael et al. 2011, 2012, 2014; Niederwanger, Calatayud, et al. 2017). This functional diversification might be related to the extraordinary evolutionary success of the phylum, with species that have colonized and adapted to very diverse habitats around the world.
Structural Evolution of Mollusca MTs
Bimodular Structure of Mollusca MTs
Vertebrate MTs have a bimodular structure made of two independent functional domains—an 11–12-Cys α domain and a 9-Cys β domain—each one capable to bind metal ions (Braun et al. 1986; Capdevila et al. 1997; Cols et al. 1999). By comparison with vertebrates, the bimodular structure has been extended to gastropod (Palacios et al. 2011; Perez-Rafael et al. 2012, 2014; Dvorak et al. 2018; Beil et al. 2019) and bivalve MTs (Jenny et al. 2004, 2016; Nam and Kim 2017; Yingprasertchai et al. 2019). Our results spread the bimodular structure to the MTs of all molluskan classes since 90% of their MTs are two-domain proteins. In these bimodular MTs, an N-terminal domain that it is variable depending on the taxon (an α domain in Bivalvia, Cephalopoda, and Polyplacophora MTs; a β2 domain in Scaphopoda, Monoplacophora, and Solenogastres MTs; a δ domain in Caudofoveata MTs; and a β3 domain in Gastropoda MTs, with the exception of the γ domain in Patellogastropoda MTs) is linked to a conserved β1 domain at the C-terminal region (fig. 1 and table 1). The pervasiveness of the C-terminal β1 domain suggests a conserved role for this domain, probably related to the stabilization of the 3D cluster structure of the entire protein (Dallinger et al. 2020).
Modular Evolution of MTs: Ancient and Recent Domains
The origin and the evolutionary relationships of the distinct α, β1, β2, β3, γ, and δ domains were intriguing. The finding of α, β1, and β2 domains in diverse molluskan classes together with the presence of α/β1 and β2/β1 MTs in species of both Conchifera and Aculifera (fig. 1) supports that the origin of the domains predated the diversification of the phylum, which has estimated at more than 545 Ma (Kocot et al. 2020), and suggests that an ancient MT multiplicity was subsequently maintained or lost in a lineage-specific manner. In addition, a possible connection between α and β domains might be envisaged based on the number and configuration of the cysteine motifs (fig. 5). Thus, the three β domains might have derived from an ancestral α domain trimmed at the C-terminal end (β1), at the middle (β2), and at the N-terminal end (β3), respectively. This possibility challenges the classical view that β domains represent ancestral forms, and that α domains evolved later to provide detoxification capacity in front of toxic metals such as cadmium (Cols et al. 1999).
In contrast to the ancient origin of α, β1, and β2 domains, the restricted distribution of β3, γ, and δ domains in some mollusk lineages suggests a more recent origin, probably concomitant with the appearance of these taxonomic groups (fig. 1). Novel N-terminal domains would have replaced former and older ones specifically in some mollusk lineages: the β3 domain in Gastropoda, the γ domain in Patellogastropoda, and the δ domain in Caudofoveata. Other scenarios of domain evolution cannot be ruled out, but they would require assuming complex processes of parallel or convergent evolution to justify the current distribution of the different domains throughout Mollusca.
De Novo Domain Evolution
Whereas there might be an evolutionary relationship between α and β domains, any link among these domains and the novel γ and δ domains is obscured by their differences. Although it cannot be ruled out that γ and δ domains derived from α or β domains that have diverged too much for homology to be recognized, the sequence and cysteine motifs in the γ and δ domains are so divergent from those in α and β domains that they seem to have evolved de novo. Recent analyses have shown that, indeed, de novo evolution is more frequent than previously thought (Neme and Tautz 2013; Toll-Riera and Alba 2013; Weisman and Eddy 2017; Levy 2019). Studies about the emergence of novel domains in human proteins, for instance, have revealed more than 400 “young” domains, 164 of which are found combined with older ones and preferentially located at the N-terminus of the proteins (Toll-Riera and Alba 2013). These new domains are rich in low-complexity sequences (Toll-Riera et al. 2012) and tend to be structurally disordered (Moore and Bornberg-Bauer 2012). Such structural features match well those of MTs, which are considered as low complexity and intrinsically disordered proteins. Thus, de novo emergence of MT domains might be relatively easy under an evolutionary perspective because the only requirement for a peptide to function as a metal ion chelator would be a high content of coordinating residues (e.g., cysteines) and a relative small length that favored the polypeptide folding (Capdevila and Atrian 2011). De novo evolution of MTs has been, indeed, implicitly stated from diverse evolutionary studies concluding that MTs likely evolved more than once in different animal phyla (Capdevila and Atrian 2011; Blindauer 2014; Isani and Carpene 2014; Ziller and Fraissinet-Tachet 2018).
In summary, the evolution of the mollusk MTs is intriguing. At the short term (at low taxonomic ranks), it appears to have followed the habitual evolutionary patterns based on progressive accumulation of changes in the sequence, and on duplications or losses of internal domains or genes. At the long term (at high taxonomic ranks), in contrast, MT evolution seems to have been mainly impacted by emergence of new structural domains. The modular structure of mollusk MTs makes the analyses of their domain organization and cysteine motifs more informative for inferring their evolution during the diversification of the phylum than the classic comparisons of sequences, which may be biased toward a general cysteine-richness due to the structural and functional requirements of metal binding.
Materials and Methods
Database Searches and MT Identification
Molluskan MT sequences were identified from public databases by Entrez searches using “metallothionein” and “Mollusca” as queries. Retrieved MT sequences were then used as queries in TBlastN searches in expressed sequence tags and genomic NCBI and eSnail (http://soft.bioinfo-minzhao.org/esnail/index.html) databases. In addition, RNA-SRA for each mollusk species deposited in NCBI were Blast searched using as queries MT sequences from the nearest phylogenetically species as well as from different mollusk species covering all the major clades. Raw sequence data were retrieved from the SRA and assembled using SeqMan 8.0.2 (Pro Assembler) software from the DNASTAR Lasergene package, and manually inspected in order to reconstruct new MT sequences. The MT nature of each new identified sequence was evaluated by BlastX searches against metazoan NCBI nonredundant protein sequence database. The amino acid sequences and the accession numbers of the retrieved MTs are provided in supplementary table S2, Supplementary Material online.
Characterization of L. gigantea MT Genes
Collection of three L. gigantea specimens, dissection of their hepatopancreas, RNA extraction, and storage in RNA later (Thermo Fisher Scientific, Waltham, CA) were performed by one of us (D.J.E.). Genomic DNA was obtained from hepatopancreatic tissue disrupted with the TissueLyser II (Qiagen, Hilden, Germany) procedure, and following the manufacturer’s instructions of DNeasy Plant Mini Kit (Qiagen) for DNA extraction. The DNA concentration, purity, and integrity were checked using Tecan Infinite M200 (Tecan Group Ltd, Switzerland) measuring the absorbance at 260 and 280 nm. For the RNA extraction, tissue of the midgut gland was homogenized with glass beads using the Precellys homogenizer (Bertin Instruments, Montigny-le-Bretonneux, France). Total RNA was isolated applying the RNeasy Plant Mini Kit (Qiagen) including on-column DNase I digestion (Qiagen) according to manufacturer’s constructions. RNA integrity was checked by visualization on a 1.5% agarose gel (Biozym, Hessisch Oldendorf, Germany). For cDNA synthesis, 450 ng total RNA was used applying the RevertAid Reverse Transcriptase (Fermentas by Thermo Fisher Scientific).
The genomic region containing the putative MT genes of L. gigantea was PCR amplified, cloned PCR primers (supplementary table S3, Supplementary Material online) were designed based on the L. gigantea genome project (Scaffold and resequenced. 35, NW_008709190.1) in order to amplify overlapping fragments of different sizes (from 500 bp to 4 kb), covering the entire putative MT-containing genomic region. For each PCR reaction, 1 ng of genomic DNA was amplified using selected pairs of primers and the Phusion High-Fidelity DNA Polymerase (Invitrogen, Thermo Fisher Scientific) in a final 25 μl reaction. PCR conditions were 98 °C 30 s (s); 35 cycles of 98 °C 10 s, 58 °C 30 s, and 72 °C 3 min; and 72 °C 10 min. PCR products were visualized in 0.7% agarose gels, isolated with the GelElute Plasmid Miniprep Kit (Sigma-Aldrich, Merck KGaA, Darmstadt, Germany), and cloned with TOPO TA Cloning Kit (Invitrogen, Thermo Fisher Scientific). Plasmid DNA was purified from bacteria using the GeneElutet Plasmid Miniprep Kit (Sigma-Aldrich), screened for insert presence by digestion with EcoRI (EcoRI Fast Digest Restriction Enzyme, Invitrogen, Thermo Fisher Scientific), and sequenced at the Scientific and Technological Centers of the University of Barcelona using the Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) in an automatic sequencer (ABIPRISM 310, Applied Biosystems). Several internal primers (supplementary table S3, Supplementary Material online) were used to fully sequence the overlapping PCR fragments, which were manually assembled in order to reconstruct the entire genomic region.
The prediction of the L. gigantea MT genes was corroborated by the PCR amplification of the corresponding cDNAs using gene specific primers (supplementary table S3, Supplementary Material online) designed with CLC main workbench (Version 6.9), and Advantage 2 polymerase (Clontech, Takara Bio Europe, Saint-Germain-en-Laye, France) in a final 50 μl reaction. Cycling conditions were 95 °C 1 min; 30 cycles 95 °C 30 s, 53 °C (for LgiMT2)/55.5 °C (for LgiMT1) 30 s, and 68 °C 40 s; and 68 °C 5 min. PCR products were visualized on a 1.5% agarose gels, purified using the QIAquick gel extraction kit (Qiagen) and cloned with the TOPO TA Cloning Kit (Invitrogen, Thermo Fisher Scientific). Insert containing plasmids were isolated using the QIAprep Spin Miniprep Kit (Qiagen) and sent for sequencing to Microsynth (Balgach, Switzerland). Sequences were analyzed via CLC main workbench (Version 6.9).
Production and Purification of Recombinant of Metal-MT Complexes
Synthetic cDNAs codifying the selected MTs (i.e., LgiMT1, LgiMT2, LgiMT2-γ domain, NpeMT1, and NpeMT2) were provided by Synbiotech (Monmouth Junction, NJ, USA) cloned in the pGEX-4T-1 expression vector (GE Healthcare). Recombinant plasmids were transformed in E. coli BL21 strain, a protease-deficient strain used for heterologous protein expression. For heterologous protein production, 500 ml of Luria-Bertani (LB) medium with 100 μg/ml ampicillin was inoculated with E. coli BL21 cells transformed with the corresponding recombinant plasmid. After overnight growth at 37 °C/250 rpm, the cultures were used to inoculate 5 l of fresh LB-100 μg/ml ampicillin medium. Gene expression was induced with 100 μM isopropyl-β-d-thiogalactopyranoside for 3 h. After the first 30 min of induction, cultures were supplemented with ZnCl2 (300 μM), CdCl2 (300 μM), or CuSO4 (500 μM) in order to generate metal-MT complexes. Cells were harvested by centrifugation for 5 min at 9,100 × g (7,700 rpm), and bacterial pellets were suspended in 125 ml of ice-cold phosphate-buffered saline (PBS) (1.4 M NaCl, 27 mM KCl, 101 mM Na2HPO4, 18 mM KH2PO4, and 0.5% v/v β-mercaptoethanol). Resuspended cells were sonicated (Sonifier Ultrasonic Cell Disruptor) 8 min at voltage 6 with pulses of 0.6 s, and then centrifuged for 40 min at 17,200 × g (12,000 rpm) and 4 °C. Soluble protein extracts containing GST-MT fusion proteins were incubated with glutathione sepharose beads (GE Healthcare) for 1 h at room temperature with gentle rotation. GST-MT fusion proteins bound to the sepharose beads were washed with 30 ml of cold 1× PBS bubbled with argon to prevent oxidation. After three washes, GST-MT fusion proteins were digested with thrombin (GE Healthcare, 25 U/l of culture or SERVA, 25 U/l of culture) overnight at 17 °C, thus enabling separation of the metal-MT complexes from the GST that remained bound to the sepharose matrix. The eluted metal-MT complexes were concentrated with a 3-kDa Centripep Low Concentrator (Amicon, Merck) and fractionated on a Superdex-75 FPLC column (GE Healthcare) equilibrated with 20 mM Tris–HCl, pH 7.0. The protein-containing fractions, identified by their absorbance at 254 nm, were pooled and stored at −80 °C until use.
Analysis of Metal-MT Complexes
Protein quantification and element composition of all the samples were achieved by ICP-AES measurements performed in a Optima 4300DV (Perkin-Elmer, MA, USA) apparatus (S, 182.040 nm; Zn, 213.856 nm; Cd, 228.802 nm; and Cu, 324.803 nm) under conventional conditions following an already established method (Bongers et al. 1988).
Molecular weights were determined by ESI-MS, in a MicroTof-Q instrument (Bruker Daltonics Gmbh, Bremen, Germany) connected to a Series 1100 HPLC pump (Agilent Technologies) controlled by the Compass Software. The instrument was calibrated with ESI-L Low Concentration Turning Mix (Agilent Technologies, Santa Clara, CA). Metallated forms were detected under native conditions: 20 µl of sample injected through a PEEK tube at 30–50 µl min−1 in a 3.5–5.0-kV capillary-counter voltage, at 90–110 °C of desolvation temperature, and with dry gas at 6 l min−1. Spectra were recorded between a m/z range from 800 to 3,000. The liquid carrier was a 90:10 mixture of 15 mM ammonium acetate and acetonitrile at pH 7.0. All molecular masses were calculated according to the bibliography (Fabris et al. 1996).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
Parts of this study were financed by a cooperation grant to R.D. from the Austrian Science Fund (DACH grant No I 1482-N28). Researchers from Barcelona want to acknowledge the Spanish Ministerio de Ciencia e Innovación and FEDER for the projects BIO2015-67358-C2-2-P (M.C. and O.P.) and BIO2015-67358-C2-1-P (R.A.). M.C. and O.P. are members of the “Grup de Recerca de la Generalitat de Catalunya,” ref. 2017SGR-864, and R.A. of ref. 2017SGR-1665. M.G.-R. acknowledges to the UAB the PIF grant. D.J.E. acknowledges support from the United States National Science Foundation (Grant No. DEB-1355230). We also thank the Servei d’Anàlisi Química (SAQ) at the Universitat Autònoma de Barcelona (ICP-AES, ESI-MS) for allocating instrument time, the Centres Científics i Tecnològics (CCiT) at the Universitat de Barcelona for DNA sequencing, and to Sebastian Artime for the experimental support.
Data Availability
The data underlying this article are available in NCBI (https://www.ncbi.nlm.nih.gov/) and eSnail (http://soft.bioinfo-minzhao.org/esnail/index.html) databases. The amino acid sequences and the accession numbers of the mollusk MTs are available in supplementary table S2, Supplementary Material online.
References
- Aceto S, Formisano G, Carella F, De Vico G, Gaudio L.. 2011. The metallothionein genes of Mytilus galloprovincialis: genomic organization, tissue expression and evolution. Mar Genomics. 4(1):61–68. [DOI] [PubMed] [Google Scholar]
- Adamo GM, Lotti M, Tamas MJ, Brocca S.. 2012. Amplification of the CUP1 gene is associated with evolution of copper tolerance in Saccharomyces cerevisiae. Microbiology 158(9):2325–2335. [DOI] [PubMed] [Google Scholar]
- Baek M-K, Lee J-S, Kang S-W, Lee J-B, Kang H-J, Jo YH, Noh M-Y, Han Y, Choi S-H, Chae S-H, et al. 2009. Phylogenetic analysis based on metallothionein gene sequence of an indigenous species Pisidium (Neopisidium) coreanum in Korea. Korean J Malacol. 25(2):153–160. [Google Scholar]
- Baumann C, Beil A, Jurt S, Niederwanger M, Palacios O, Capdevila M, Atrian S, Dallinger R, Zerbe O.. 2017. Structural adaptation of a protein to increased metal stress: NMR structure of a marine snail metallothionein with an additional domain. Angew Chem Int Ed Engl. 56(16):4617–4622. [DOI] [PubMed] [Google Scholar]
- Beil A, Jurt S, Walser R, Schonhut T, Guntert P, Palacios O, Atrian S, Capdevila M, Dallinger R, Zerbe O.. 2019. The solution structure and dynamics of Cd-metallothionein from Helix pomatia reveal optimization for binding Cd over Zn. Biochemistry 58(45):4570–4581. [DOI] [PubMed] [Google Scholar]
- Blindauer CA. 2014. Metallothioneins. In: Binding, transport and storage of metal ions in biological cells. Cambridge, United Kingdom: The Royal Society of Chemistry. p. 594–653. [Google Scholar]
- Bongers J, Walton CD, Richardson DE, Bell JU.. 1988. Micromolar protein concentrations and metalloprotein stoichiometries obtained by inductively coupled plasma atomic emission spectrometric determination of sulfur. Anal Chem. 60(24):2683–2686. [DOI] [PubMed] [Google Scholar]
- Braun W, Wagner G, Worgotter E, Vasak M, Kagi JH, Wuthrich K.. 1986. Polypeptide fold in the two metal clusters of metallothionein-2 by nuclear magnetic resonance in solution. J Mol Biol. 187(1):125–129. [DOI] [PubMed] [Google Scholar]
- Calatayud S, Garcia-Risco M, Rojas NS, Espinosa-Sanchez L, Artime S, Palacios O, Cañestro C, Albalat R.. 2018. Metallothioneins of the urochordate Oikopleura dioica have Cys-rich tandem repeats, large size and cadmium-binding preference. Metallomics 10(11):1585–1594. [DOI] [PubMed] [Google Scholar]
- Capdevila M, Atrian S.. 2011. Metallothionein protein evolution: a miniassay. J Biol Inorg Chem. 16(7):977–989. [DOI] [PubMed] [Google Scholar]
- Capdevila M, Bofill R, Palacios Ò, Atrian S.. 2012. State-of-the-art of metallothioneins at the beginning of the 21st century. Coord Chem Rev. 256(1–2):46–62. [Google Scholar]
- Capdevila M, Cols N, Romero-Isart N, Gonzalez-Duarte R, Atrian S, Gonzalez-Duarte P.. 1997. Recombinant synthesis of mouse Zn3-beta and Zn4-alpha metallothionein 1 domains and characterization of their cadmium(II) binding capacity. Cell Mol Life Sci. 53(8):681–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalan A, Glaser-Schmitt A, Argyridou E, Duchen P, Parsch J.. 2016. An indel polymorphism in the MtnA 3′ untranslated region is associated with gene expression variation and local adaptation in Drosophila melanogaster. PLoS Genet. 12(4):e1005987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cols N, Romero-Isart N, Bofill R, Capdevila M, Gonzalez-Duarte P, Gonzalez-Duarte R, Atrian S.. 1999. In vivo copper- and cadmium-binding ability of mammalian metallothionein beta domain. Protein Eng. 12(3):265–269. [DOI] [PubMed] [Google Scholar]
- Costa D, Marien J, Janssens TK, van Gestel CA, Driessen G, Sousa JP, van Straalen NM, Roelofs D.. 2012. Influence of adaptive evolution of cadmium tolerance on neutral and functional genetic variation in Orchesella cincta. Ecotoxicology 21(7):2078–2087. [DOI] [PubMed] [Google Scholar]
- Cunha TJ, Giribet G.. 2019. A congruent topology for deep gastropod relationships. Proc Biol Sci. 286(1898):20182776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallinger R, Chabicovsky M, Hödl E, Prem C, Hunziker P, Manzl C.. 2005. Copper in Helix pomatia (Gastropoda) is regulated by one single cell type: differently responsive metal pools in rhogocytes. Am J Physiol Regul Integr Comp Physiol. 289(4):R1185–R1195. [DOI] [PubMed] [Google Scholar]
- Dallinger R, Zerbe O, Baumann C, Egger B, Capdevila M, Palacios Ò, Albalat R, Calatayud S, Ladurner P, Schlick-Steiner BC, et al. 2020. Metallomics reveal a persisting impact of cadmium on the evolution of metal-selective snail metallothioneins. Metallomics 12(5):702–720. [DOI] [PubMed] [Google Scholar]
- de Francisco P, Martin-Gonzalez A, Turkewitz AP, Gutierrez JC.. 2017. Extreme metal adapted, knockout and knockdown strains reveal a coordinated gene expression among different Tetrahymena thermophila metallothionein isoforms. PLoS One 12(12):e0189076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Francisco P, Martin-Gonzalez A, Turkewitz AP, Gutierrez JC.. 2018. Genome plasticity in response to stress in Tetrahymena thermophila: selective and reversible chromosome amplification and paralogous expansion of metallothionein genes. Environ Microbiol. 20(7):2410–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak M, Lackner R, Niederwanger M, Rotondo C, Schnegg R, Ladurner P, Pedrini-Martha V, Salvenmoser W, Kremser L, Lindner H, et al. 2018. Metal binding functions of metallothioneins in the slug Arion vulgaris differ from metal-specific isoforms of terrestrial snails. Metallomics 10(11):1638–1654. [DOI] [PubMed] [Google Scholar]
- Fabris D, Zaia J, Hathout Y, Fenselau C.. 1996. Retention of thiol protons in two classes of protein zinc ion coordination centers. J Am Chem Soc. 118(48):12242–12243. [Google Scholar]
- Faddeeva-Vakhrusheva A, Derks MF, Anvar SY, Agamennone V, Suring W, Smit S, van Straalen NM, Roelofs D.. 2016. Gene family evolution reflects adaptation to soil environmental stressors in the genome of the collembolan Orchesella cincta. Genome Biol Evol. 8(7):2106–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirola M, Perez-Rafael S, Capdevila M, Palacios O, Atrian S.. 2012. Metal dealing at the origin of the Chordata phylum: the metallothionein system and metal overload response in amphioxus. PLoS One 7(8):e43299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höckner M, Stefanon K, de Vaufleury A, Monteiro F, Pérez-Rafael S, Palacios O, Capdevila M, Atrian S, Dallinger R.. 2011. Physiological relevance and contribution to metal balance of specific and non-specific metallothionein isoforms in the garden snail. Biometals 24(6):1079–1092. [DOI] [PubMed] [Google Scholar]
- Isani G, Carpene E.. 2014. Metallothioneins, unconventional proteins from unconventional animals: a long journey from nematodes to mammals. Biomolecules 4(2):435–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens TKS, Lopéz RDR, Mariën J, Timmermans MJTN, Montagne-Wajer K, van Straalen NM, Roelofs D.. 2008. Comparative population analysis of metallothionein promoter alleles suggests stress-induced microevolution in the field. Environ Sci Technol. 42(10):3873–3878. [DOI] [PubMed] [Google Scholar]
- Janssens TKS, Roelofs D, Van Straalen NM.. 2009. Molecular mechanisms of heavy metal tolerance and evolution in invertebrates. Insect Sci. 16(1):3–18. [Google Scholar]
- Jenny MJ, Payton SL, Baltzegar DA, Lozier JD.. 2016. Phylogenetic analysis of molluscan metallothioneins: evolutionary insight from Crassostrea virginica. J Mol Evol. 83(3–4):110–125. [DOI] [PubMed] [Google Scholar]
- Jenny MJ, Ringwood AH, Schey K, Warr GW, Chapman RW.. 2004. Diversity of metallothioneins in the American oyster, Crassostrea virginica, revealed by transcriptomic and proteomic approaches. Eur J Biochem. 271(9):1702–1712. [DOI] [PubMed] [Google Scholar]
- Kocot KM, Cannon JT, Todt C, Citarella MR, Kohn AB, Meyer A, Santos SR, Schander C, Moroz LL, Lieb B, et al. 2011. Phylogenomics reveals deep molluscan relationships. Nature 477(7365):452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocot KM, Poustka AJ, Stoger I, Halanych KM, Schrodl M.. 2020. New data from Monoplacophora and a carefully-curated dataset resolve molluscan relationships. Sci Rep. 10(1):101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A. 2019. How evolution builds genes from scratch. Nature 574(7778):314–316. [DOI] [PubMed] [Google Scholar]
- Loebus J, Peroza EA, Bluthgen N, Fox T, Meyer-Klaucke W, Zerbe O, Freisinger E.. 2011. Protein and metal cluster structure of the wheat metallothionein domain gamma-E(c)-1: the second part of the puzzle. J Biol Inorg Chem. 16(5):683–694. [DOI] [PubMed] [Google Scholar]
- Mackay EA, Overnell J, Dunbar B, Davidson I, Hunziker PE, Kagi JH, Fothergill JE.. 1993. Complete amino acid sequences of five dimeric and four monomeric forms of metallothionein from the edible mussel Mytilus edulis. Eur J Biochem. 218(1):183–194. [DOI] [PubMed] [Google Scholar]
- Maroni G, Wise J, Young JE, Otto E.. 1987. Metallothionein gene duplications and metal tolerance in natural populations of Drosophila melanogaster. Genetics 117(4):739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AD, Bornberg-Bauer E.. 2012. The dynamics and evolutionary potential of domain loss and emergence. Mol Biol Evol. 29(2):787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz A, Forsterling FH, Shaw CF 3rd, Petering DH.. 2002. Structure of the (113)Cd(3)beta domains from Homarus americanus metallothionein-1: hydrogen bonding and solvent accessibility of sulfur atoms. J Biol Inorg Chem. 7(7–8):713–724. [DOI] [PubMed] [Google Scholar]
- Nam YK, Kim EJ.. 2017. Diversification and domain evolution of molluskan metallothioneins: a mini review. Fish Aquatic Sci. 20(1):8. [Google Scholar]
- Narula SS, Brouwer M, Hua Y, Armitage IM.. 1995. Three-dimensional solution structure of Callinectes sapidus metallothionein-1 determined by homonuclear and heteronuclear magnetic resonance spectroscopy. Biochemistry 34(2):620–631. [DOI] [PubMed] [Google Scholar]
- Neme R, Tautz D.. 2013. Phylogenetic patterns of emergence of new genes support a model of frequent de novo evolution. BMC Genomics. 14(1):117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederwanger M, Calatayud S, Zerbe O, Atrian S, Albalat R, Capdevila M, Palacios O, Dallinger R.. 2017. Biomphalaria glabrata metallothionein: lacking metal specificity of the protein and missing gene upregulation suggest metal sequestration by exchange instead of through selective binding. Int J Mol Sci. 18(7):1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederwanger M, Dvorak M, Schnegg R, Pedrini-Martha V, Bacher K, Bidoli M, Dallinger R.. 2017. Challenging the metallothionein (MT) gene of Biomphalaria glabrata: unexpected response patterns due to cadmium exposure and temperature stress. Int J Mol Sci. 18(8):1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielson KB, Winge DR.. 1985. Independence of the domains of metallothionein in metal binding. J Biol Chem. 260(15):8698–8701. [PubMed] [Google Scholar]
- Orihuela R, Domenech J, Bofill R, You C, Mackay EA, Kagi JH, Capdevila M, Atrian S.. 2008. The metal-binding features of the recombinant mussel Mytilus edulis MT-10-IV metallothionein. J Biol Inorg Chem. 13(5):801–812. [DOI] [PubMed] [Google Scholar]
- Palacios O, Jimenez-Marti E, Niederwanger M, Gil-Moreno S, Zerbe O, Atrian S, Dallinger R, Capdevila M.. 2017. Analysis of metal-binding features of the wild type and two domain-truncated mutant variants of Littorina littorea metallothionein reveals its Cd-specific character. Int J Mol Sci. 18(7):1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios O, Pagani A, Pérez-Rafael S, Egg M, Höckner M, Brandstätter A, Capdevila M, Atrian S, Dallinger R.. 2011. Shaping mechanisms of metal specificity in a family of metazoan metallothioneins: evolutionary differentiation of mollusc metallothioneins. BMC Biol. 9(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios O, Perez-Rafael S, Pagani A, Dallinger R, Atrian S, Capdevila M.. 2014. Cognate and noncognate metal ion coordination in metal-specific metallothioneins: the Helix pomatia system as a model. J Biol Inorg Chem. 19(6):923–935. [DOI] [PubMed] [Google Scholar]
- Pedrini-Martha V, Koll S, Dvorak M, Dallinger R.. 2020. Cadmium uptake, MT gene activation and structure of large-sized multi-domain metallothioneins in the terrestrial door snail Alinda biplicata (Gastropoda, Clausiliidae). Int J Mol Sci. 21(5):1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Rafael S, Atrian S, Capdevila M, Palacios O.. 2011. Differential ESI-MS behaviour of highly similar metallothioneins. Talanta 83(3):1057–1061. [DOI] [PubMed] [Google Scholar]
- Perez-Rafael S, Mezger A, Lieb B, Dallinger R, Capdevila M, Palacios O, Atrian S.. 2012. The metal binding abilities of Megathura crenulata metallothionein (McMT) in the frame of gastropoda MTs. J Inorg Biochem. 108:84–90. [DOI] [PubMed] [Google Scholar]
- Perez-Rafael S, Monteiro F, Dallinger R, Atrian S, Palacios O, Capdevila M.. 2014. Cantareus aspersus metallothionein metal binding abilities: the unspecific CaCd/CuMT isoform provides hints about the metal preference determinants in metallothioneins. Biochim Biophys Acta 1844(9):1694–1707. [DOI] [PubMed] [Google Scholar]
- Purać J, Nikolić TV, Kojić D, Ćelić AS, Plavša JJ, Blagojević DP, Petri ET.. 2019. Identification of a metallothionein gene in honey bee Apis mellifera and its expression profile in response to Cd, Cu and Pb exposure. Mol Ecol. 28(4):731–745. [DOI] [PubMed] [Google Scholar]
- Riek R, Precheur B, Wang Y, Mackay EA, Wider G, Guntert P, Liu A, Kagi JH, Wuthrich K.. 1999. NMR structure of the sea urchin (Strongylocentrotus purpuratus) metallothionein MTA. J Mol Biol. 291(2):417–428. [DOI] [PubMed] [Google Scholar]
- Schmielau L, Dvorak M, Niederwanger M, Dobieszewski N, Pedrini-Martha V, Ladurner P, Pedregal JR, Marechal JD, Dallinger R.. 2019. Differential response to cadmium exposure by expression of a two and a three-domain metallothionein isoform in the land winkle Pomatias elegans: valuating the marine heritage of a land snail. Sci Total Environ. 648:561–571. [DOI] [PubMed] [Google Scholar]
- Smith SA, Wilson NG, Goetz FE, Feehery C, Andrade SC, Rouse GW, Giribet G, Dunn CW.. 2011. Resolving the evolutionary relationships of molluscs with phylogenomic tools. Nature 480(7377):364–367. [DOI] [PubMed] [Google Scholar]
- Tanguy A, Moraga D.. 2001. Cloning and characterization of a gene coding for a novel metallothionein in the Pacific oyster Crassostrea gigas (CgMT2): a case of adaptive response to metal-induced stress? Gene 273(1):123–130. [DOI] [PubMed] [Google Scholar]
- Timmermans MJ, Ellers J, Roelofs D, van Straalen NM.. 2005. Metallothionein mRNA expression and cadmium tolerance in metal-stressed and reference populations of the springtail Orchesella cincta. Ecotoxicology 14(7):727–739. [DOI] [PubMed] [Google Scholar]
- Tio L, Villarreal L, Atrian S, Capdevila M.. 2004. Functional differentiation in the mammalian metallothionein gene family: metal binding features of mouse MT4 and comparison with its paralog MT1. J Biol Chem. 279(23):24403–24413. [DOI] [PubMed] [Google Scholar]
- Toll-Riera M, Alba MM.. 2013. Emergence of novel domains in proteins. BMC Evol Biol. 13(1):47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toll-Riera M, Rado-Trilla N, Martys F, Alba MM.. 2012. Role of low-complexity sequences in the formation of novel protein coding sequences. Mol Biol Evol. 29(3):883–886. [DOI] [PubMed] [Google Scholar]
- Valls M, Bofill R, Gonzalez-Duarte R, Gonzalez-Duarte P, Capdevila M, Atrian S.. 2001. A new insight into metallothionein (MT) classification and evolution. The in vivo and in vitro metal binding features of Homarus americanus recombinant MT. J Biol Chem. 276(35):32835–32843. [DOI] [PubMed] [Google Scholar]
- Weisman CM, Eddy SR.. 2017. Gene evolution: getting something from nothing. Curr Biol. 27(13):R661–R663. [DOI] [PubMed] [Google Scholar]
- Yingprasertchai T, Yu RMK, Tran TKA, Chong Kong RY, O’Connor WA, MacFarlane GR.. 2019. Characterisation of the metallothionein gene in the Sydney rock oyster and its expression upon metal exposure in oysters with different prior metal exposure histories. Mar Environ Res. 151:104775. [DOI] [PubMed] [Google Scholar]
- Zapata F, Wilson NG, Howison M, Andrade SC, Jorger KM, Schrodl M, Goetz FE, Giribet G, Dunn CW.. 2014. Phylogenomic analyses of deep gastropod relationships reject Orthogastropoda. Proc Biol Sci. 281(1794):20141739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Jansen M, De Meester L, Stoks R.. 2018. Thermal evolution offsets the elevated toxicity of a contaminant under warming: a resurrection study in Daphnia magna. Evol Appl. 11(8):1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Jansen M, De Meester L, Stoks R.. 2019. Rapid evolution in response to warming does not affect the toxicity of a pollutant: insights from experimental evolution in heated mesocosms. Evol Appl. 12(5):977–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller A, Fraissinet-Tachet L.. 2018. Metallothionein diversity and distribution in the tree of life: a multifunctional protein. Metallomics 10(11):1549–1559. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in NCBI (https://www.ncbi.nlm.nih.gov/) and eSnail (http://soft.bioinfo-minzhao.org/esnail/index.html) databases. The amino acid sequences and the accession numbers of the mollusk MTs are available in supplementary table S2, Supplementary Material online.