Abstract
Despite significant advances in invertebrate phylogenomics over the past decade, the higher-level phylogeny of Pycnogonida (sea spiders) remains elusive. Due to the inaccessibility of some small-bodied lineages, few phylogenetic studies have sampled all sea spider families. Previous efforts based on a handful of genes have yielded unstable tree topologies. Here, we inferred the relationships of 89 sea spider species using targeted capture of the mitochondrial genome, 56 conserved exons, 101 ultraconserved elements, and 3 nuclear ribosomal genes. We inferred molecular divergence times by integrating morphological data for fossil species to calibrate 15 nodes in the arthropod tree of life. This integration of data classes resolved the basal topology of sea spiders with high support. The enigmatic family Austrodecidae was resolved as the sister group to the remaining Pycnogonida and the small-bodied family Rhynchothoracidae as the sister group of the robust-bodied family Pycnogonidae. Molecular divergence time estimation recovered a basal divergence of crown group sea spiders in the Ordovician. Comparison of diversification dynamics with other marine invertebrate taxa that originated in the Paleozoic suggests that sea spiders and some crustacean groups exhibit resilience to mass extinction episodes, relative to mollusk and echinoderm lineages.
Keywords: arthropods, Pycnogonida, mitogenome, ultraconserved, diversification
Introduction
Pycnogonida (sea spiders), the sister group to the remaining Chelicerata, are exclusively marine arthropods ranging from 1 to 750 mm in leg span (fig. 1). The morphological architecture of sea spiders is unusual, with a typically small body that is dwarfed by the much longer legs (hence, the alternate name “Pantopoda,” or “all legs”), into which diverticula of major organ systems emanate. Sea spiders are found throughout the world’s oceans from the intertidal zone to abyssal depths but are especially abundant and diverse in polar benthic communities. In contrast to many invertebrate groups that flourish in the tropics, the peak of sea spider diversity is concentrated in the Southern Ocean, which also harbors multiple cases of gigantism in distantly related species (Arnaud and Bamber 1987; Griffiths et al. 2011).
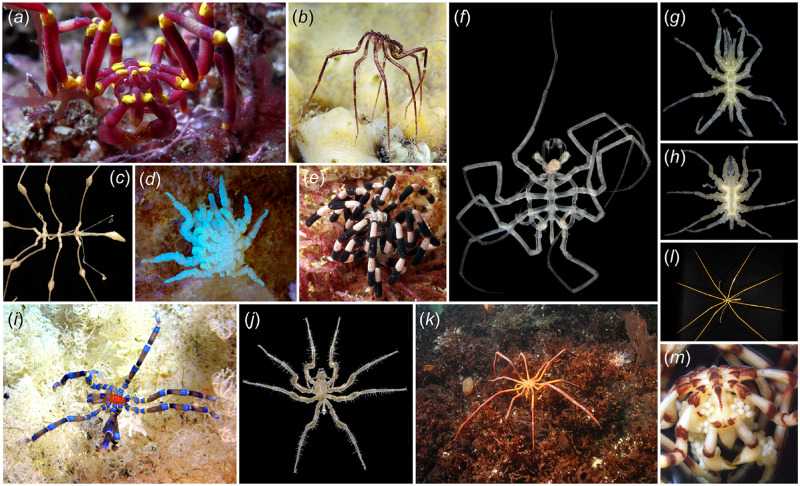
Fig. 1.
Exemplars of sea spider diversity. (a) Pallenella harrisi (Callipallenidae). (b) Nymphon grossipes (Nymphonidae). (c) Rhopalorhynchus magdalenae (Colossendeidae). (d) Copulating pair of Pycnogonum litorale (Pycnogonidae) with UV illumination. (e) Stylopallene sp. (Callipallenidae), photograph by Iain Gray. (f) Nymphonella tapetis (Ascorhynchidae sensu lato). (g) Austrodecus glaciale (Austrodecidae). (h) Rhynchothorax australis (Rhynchothoracidae). (i) Anoplodactylus evansi (Phoxichilidiidae). (j) Cilunculus armatus (Ammotheidae), (k) Decolopoda australis (Colossendeidae), photograph by Andrei Utevsky. (l) Colossendeis megalonyx (Colossendeidae). (m) Male of Pallenella sp. (Callipallenidae) with egg clutch.
Pycnogonids typically have four pairs of walking legs attached to the small body; the cephalon bears an anterior proboscis, composed of three longitudinal antimeres with the triradiate pharynx lumen in their center; and three pairs of cephalic appendages: the chelifores, palps, and ovigers. Extant Pycnogonida lack a segmented opisthosoma (abdomen or posterior tagma), and thus also lack segmentally iterated opisthosomal respiratory organs that are found in other chelicerates. Sea spiders respire instead via cuticular gas exchange, with peristaltic contractions of the gut facilitating oxygen transport through the body (Woods et al. 2017; Lane et al. 2018). Ovigers, a type of modified leg unique to Pycnogonida, are used for grooming and by the males to carry egg masses (fig. 1M) (Arnaud and Bamber 1987). A remarkable exception from the conserved body architecture are genera with supernumerary body segments (resulting in ten-legged species), which occur in three families (Colossendeidae, Pycnogonidae, and Nymphonidae), and two genera (Dodecolopoda and Sexanymphon) are even characterized by 12 legs (Arango and Wheeler 2007). Beyond this, the cephalic appendages show generally a high degree of variation. Families are often distinguishable by the number of articles in the palps and ovigers (Arango 2002); and in some lineages, adults may lack one or more cephalic appendage types altogether.
Early conceptions of sea spider phylogeny envisioned a gradual reductive trend characterized by unidirectional, stepwise losses of appendage types (Hedgpeth 1947; Fry 1978; Munilla León 1999). Phylogenetic investigations of sea spider relationships based on anatomical data (Arango 2002) or combined analyses of morphology and molecular sequence data (Arango and Wheeler 2007) suggested instead that reduction of appendages occurred independently across the phylogeny, but tree topologies were highly discordant between data partitions. Subsequent approaches to infer sea spider relationships under model-based approaches (Nakamura et al. 2007; Arabi et al. 2010; Sabroux et al. 2018) were repeatedly frustrated by the instability of basal relationships, which are attributable to two possible causes.
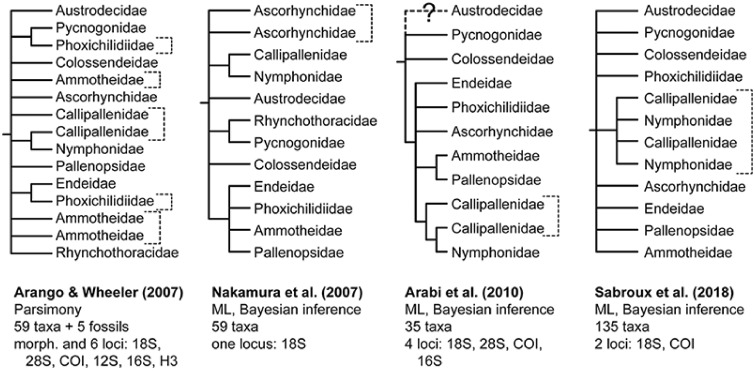
First, efforts to infer the phylogeny of Pycnogonida have been based on a small number of loci (one to six genes) that evolve at high rates (e.g., mitochondrial genes cytochrome c oxidase subunit I and 16S rRNA) or those that evolve at uninformatively low rates (e.g., nuclear ribosomal genes) (fig. 2) (Arango and Wheeler 2007; Nakamura et al. 2007; Arabi et al. 2010; Sabroux et al. 2018). Separately, mitochondrial genes of sea spiders exhibit well-known lineage-specific compositional biases (Arabi et al. 2010). Data sets based on fast-evolving mitochondrial genes have exhibited limited utility in resolving Paleozoic relationships of various invertebrate groups, and the placement of sea spiders within Chelicerata specifically (Masta et al. 2010; Rota-Stabelli et al. 2010).
Fig. 2.
Historical hypotheses of higher-level sea spider relationships based on molecular sequence data. Nodes lacking nodal support (<70% bootstrap support; <0.90 posterior probability) or conflicting across analyses have been collapsed. Brackets correspond to paraphyletic lineages. In Sabroux et al. (2018), both Nymphonidae and Callipallenidae were recovered as polyphyletic. 12S, 12S rRNA; 16S, 16S rRNA; 18S, 18S rRNA; 28S, 28S rRNA; COI, cytochrome c oxidase subunit I; H3, histone H3.
Second, previous phylogenetic studies have poorly sampled (with respect to number of loci), or omitted entirely, two small-bodied families of sea spiders, Austrodecidae and Rhynchothoracidae (Arabi et al. 2010; Sabroux et al. 2018). Austrodecidae (fig. 1G) are distinguished from other sea spiders by the annulation of the proboscis. Little is known about their biology, as most austrodecids are small-bodied species (<5 mm) that are infrequently encountered (Arnaud and Bamber 1987; Griffiths et al. 2011). Even less understood are species of Rhynchothoracidae (fig. 1H), which typically do not exceed 1 mm in length (Arnaud and Bamber 1987; Arango 2002).
The lack of a robust sea spider phylogeny has hindered inferences of major macroevolutionary trends in the group (Brenneis and Scholtz 2015; Brenneis et al. 2017, 2018). Although phylotranscriptomic approaches have proved effective for resolving relationships of various chelicerate groups (Sharma, Kaluziak, et al. 2014; Santibáñez-López et al. 2019), the inaccessibility of rare sea spider lineages has obviated RNA-Seq-based approaches, as cryptic and small-bodied species are often not identified in ethanol-preserved samples until weeks to years after their initial collection. Moreover, sea spider specimens are often covered with epibionts and the sea spider digestive system extends into all but the two most distal podomeres of the legs. Thus, RNA-Seq-based approaches carry high risks of contaminations from epibionts and gut contents, especially for small-bodied species.
Resolving sea spider relationships thus requires an approach that can overcome several challenges to on-target data collection. To surmount these challenges, we undertook a target capture sequencing approach toward generating a robustly resolved phylogenetic backbone for Pycnogonida. We present here the first phylogenomic tree of sea spiders sampling all extant families. To assess whether whole-genome duplications (WGDs) discovered in some chelicerate orders have affected orthology inference in sea spiders, we separately generated the first developmental transcriptomes of this group and investigated the incidence of Hox duplications across Pycnogonida. To place this branch of the tree of life in a temporal context, we inferred for the first time the age of the crown group Pycnogonida using a node-dating approach under a Bayesian inference framework.
Results
Phylogenomic Relationships of Pycnogonida
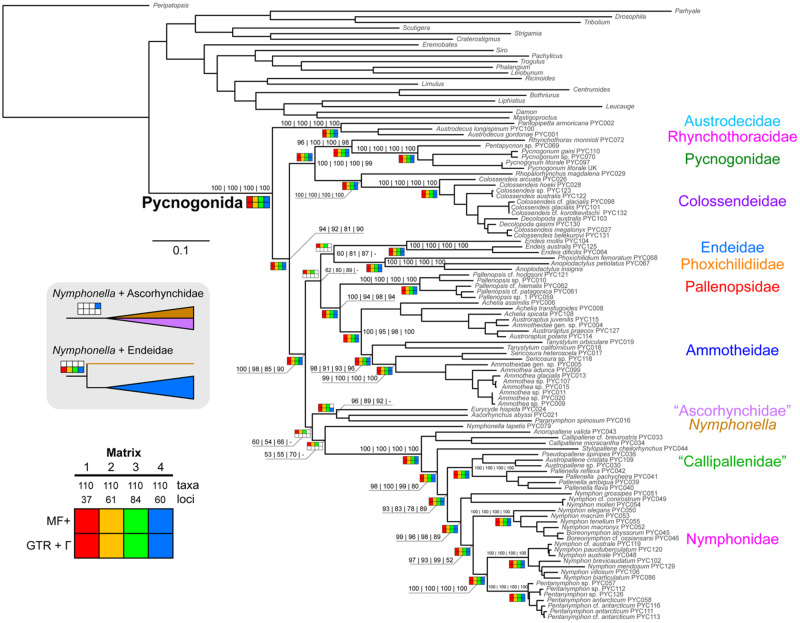
Maximum likelihood analyses of Matrices 1–4 consistently recovered the monophyly of Pycnogonida with maximal nodal support (bootstrap resampling frequency [BS]) (fig. 3). Austrodecidae was recovered in all analyses as the sister group to the remaining sea spiders with support (BS = 81–94%). At the next internal node, we recovered a clade comprised of Colossendeidae, Pycnogonidae, and Rhynchothoracidae (BS = 100%), with unambiguous support for the sister group relationship of the latter two families (BS = 96–100%). All other families were consistently recovered as a clade (BS = 85–100%).
Fig. 3.
Phylogenomic relationships of Pycnogonida based on maximum likelihood analysis of Matrix 3 (ln L = −472,525.59). Colors of branches correspond to families (right). Numbers on nodes indicate bootstrap resampling frequencies for Matrices 1–4 with model fitting using ModelFinder. Quotation marks on Ascorhynchidae reflect the inclusion of the putative ammotheid genus Paranymphon. Bottom left: Sensitivity plot indicating design of matrices and phylogenomic analyses. Inset (gray background): Alternative placements of Nymphonella tapetis.
As shown in figure 3, maximum likelihood analyses of Matrices 1–4 with model fitting using ModelFinder recovered Endeidae and Phoxichilidiidae as sister taxa (BS = 60–87%). Endeidae + Phoxichilidiidae was in turn sister group (BS = 62–89%) to a clade comprised of the reciprocally monophyletic Pallenopsidae and Ammotheidae (BS = 94–100%). The remaining sea spider lineages consisted of a monophyletic Nymphonidae nested within Callipallenidae (BS = 80–100%), and this lineage in turn subtended with weak support by a grade comprised of Nymphonella tapetis (BS = 53–70%) and a paraphyletic Ascorhynchidae sensu stricto (BS = 54–66%). Ascorhynchidae consistently included the putative ammotheid genus Paranymphon (BS = 89–96%).
Model-fitting strategy did not affect the backbone tree topology, save for the placement of Nymphonella. Assigning a unique GTR (general time reversible) + Γ model to each locus for Matrices 1–4 consistently recovered Nymphonella as the sister group of Endeidae. Matrix 4 alternatively recovered Nymphonella as nested within Ascorhynchidae when the partition models were developed using ModelFinder (fig. 3). Barring the placement of Nymphonella and its attendant topological instability, all analyses congruently resolved the monophyly of all families, excepting Rhynchothoracidae (one terminal only), Ammotheidae (due to the placement of Paranymphon), and Callipallenidae (consistently recovered as paraphyletic with respect to Nymphonidae). To assess support for nonmonophyly of Callipallenidae, we performed approximately unbiased tests, using a constraint tree that enforced the mutual monophyly of Callipallenidae and Nymphonidae. The monophyly of Callipallenidae was rejected under both Matrix 1 (Δln L = 101.92, P = 1.67 × 10−3) and Matrix 4 (Δln L = 132.34, P = 3.27 × 10−4).
Most genera sampled with two or more taxa were also recovered as monophyletic. Exceptions included Decolopoda (a result attributable to the lack of any loci that include both Decolopoda species) and Achelia (sampled with three terminals). Our results corroborated the nonmonophyly of Colossendeis due to the nested placement of species with supernumerary segments (e.g., the genera Decolopoda and Dodecolopoda; Dietz et al. 2019) and additionally revealed here the nonmonophyly of Nymphon due to the nested placement of Boreonymphon and the ten-legged genus Pentanymphon.
Comparative Assessment of Phylogenetic Data Classes
Performance measures for phylogenetic data classes consisted of number of taxa sampled, alignment length, GC content (see Arabi et al. 2010), Robinson–Foulds (RF) distance, weighted Robinson–Foulds (wRF) distance, and evolutionary rate (mean percent pairwise sequence identity; MPSI). Properties of individual loci and matrices are separately provided in supplementary tables S4 and S5, Supplementary Material online.
The mean number of taxa per locus was highest for the mitochondrial and nuclear ribosomal genes (68.2 and 85.7, respectively) and comparable for targeted exons, albeit with high variance (mean = 50.9, σ2 = 20.9). Ultraconserved element (UCE) loci bore the most missing data, with an average of 22.2 terminals per locus. In addition, after trimming, the targeted exons and UCEs had comparable alignment lengths and were shorter on average than mitochondrial and nuclear ribosomal genes. GC content of nuclear exons and UCEs was also comparable, with the average values intermediate between mitochondrial genes and nuclear ribosomal genes (supplementary fig. S1, Supplementary Material online).
Assessment of congruence between gene trees and the pruned species trees showed that targeted exons and UCEs had comparable distributions of RF and wRF distances (supplementary fig. S2, Supplementary Material online). Average values of both metrics were intermediate for targeted exons and UCEs, with respect to the lower values of mitochondrial genes and the higher values of nuclear ribosomal genes. Paralleling these distributions, MPSI values of targeted exons and UCEs were intermediate between mitochondrial genes and nuclear ribosomal genes (supplementary fig. S2, Supplementary Material online).
Inference of tree topologies based on each of the four data partitions was performed in IQ-TREE with identical heuristics as for the species tree analyses. Comparison of tree topologies reflected the performance metrics measured above; mitochondrial genes and targeted exons retained many of the higher-level relationships recovered by the species tree, such as the monophyly of various families, and the sister group relationships of Callipallenidae + Nymphonidae and Pallenopsidae + Ammotheidae (supplementary fig. S3, Supplementary Material online). However, by themselves, none of these partitions could resolve basal relationships with support. Reflecting the high proportion of missing data, the UCE tree topology was largely discordant with the species tree and other partitions, although some higher-level relationships were consistently recovered (e.g., the monophyly of some families; Callipallenidae + Nymphonidae; Pallenopsidae + Ammotheidae). The nuclear ribosomal partition retained some higher-level relationships, but branch lengths subtending interfamilial relationships were close to zero and were not supported (supplementary fig. S3, Supplementary Material online).
Assessment of WGD and Paralogy in Pycnogonida
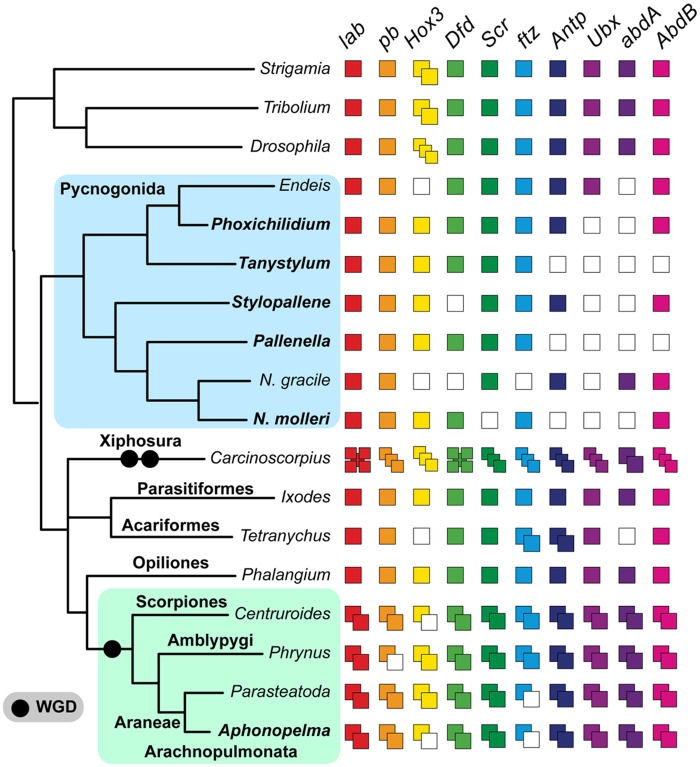
It has recently been shown that WGDs have independently occurred in at least two different groups of Chelicerata, the horseshoe crabs (Kenny et al. 2016; Shingate et al. 2020) and the arachnopulmonates (e.g., spiders and scorpions; Sharma, Schwager, et al. 2014; Schwager et al. 2017; Leite et al. 2018). Due to the unavailability of genomes sampling all chelicerate orders, it is presently unknown whether WGD has also affected Pycnogonida and whether putative orthologs of sea spiders may in fact be paralogous copies. To assess this possibility, we generated the first developmental transcriptomes of five sea spider species, sampling four families. We surveyed these transcriptomes for the presence of Hox genes, which reliably exhibit duplicates in embryonic transcriptomes of WGD taxa (Sharma, Schwager, et al. 2014; Gainett and Sharma 2020) and a subset of which has been shown to be expressed in canonical Hox-like domains in the protonymphon stage of sea spiders (Jager et al. 2006). Between six and eight Hox genes were recovered per species from the five sea spider transcriptomes, with gene tree analysis inferring all these to be single-copy (fig. 4). By comparison, an embryonic transcriptome of the mygalomorph spider Aphonopelma hentzi (an exemplar of Arachnopulmonata) that was sequenced in parallel with the embryonic sea spiders, revealed a complement of 18 Hox genes (two copies of all ten Hox genes except for Hox3 and fushi tarazu), consistent with the pattern across arachnopulmonates. Other genes surveyed for potential shared duplication included appendage patterning genes (e.g., dachshund, homothorax; Nolan et al. 2020) and members of the retinal determination gene network (e.g., sine oculis; Optix); these were also found to be single copy across all sea spiders. Broadly, these patterns suggest that Pycnogonida is not part of the WGD events exhibited by Xiphosura and Arachnopulmonata.
Fig. 4.
Summary of Hox gene complements of surveyed taxa. Boldface text indicates new embryonic transcriptomes generated for this study. Hox genes for Carcinoscorpius rotundicauda are drawn from Shingate et al. (2020). Hox genes for Endeis spinosa and Nymphon gracile were reported by Manuel et al. (2006) using RACE-PCR. Phylogenetic relationships within Chelicerata are based on Ballesteros and Sharma (2019) and this study. Multiple sequence alignment, gene tree topology, and transcriptomic assemblies are provided in the Dryad Digital Repository.
As a separate approach to validating orthology, we annotated our multiple sequence alignments of targeted exons de novo by identifying the best BLAST hit to the proteome of the common fruit fly (Drosophila melanogaster). All recovered annotations were consistent with the gene IDs of the single-copy orthologs initially selected for probe design (from Sharma, Kaluziak, et al. 2014; supplementary table S6, Supplementary Material online). Taken together with the contents of sea spider embryonic transcriptomes, these results suggest that sea spiders are not included among the chelicerate orders that exhibit WGD and thus do not exhibit the same patterns of paralogy.
Age and Tempo of Sea Spider Diversification
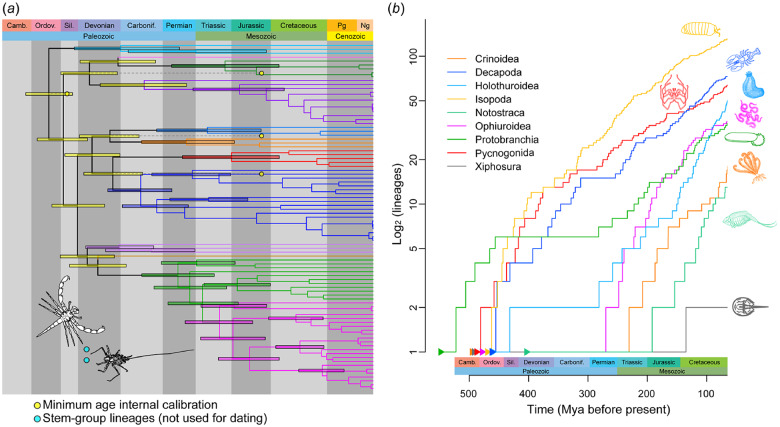
Phylogenomic estimation of divergence times was performed using 11 outgroup and four ingroup fossil calibrations; fossil taxa and justification are provided in supplementary text S2, Supplementary Material online. Dating was performed under a correlated rates clock model and the most complete matrix (Matrix 1) recovered a Late Cambrian to Early Silurian age for the crown group of Pycnogonida (median: 457 Ma [Ordovician]; 95% highest posterior density [HPD] interval: 423–489 Ma) (fig. 5A). Comparable ages were recovered for the same data set under an uncorrelated rates clock model (supplementary fig. S4, Supplementary Material online). Diversification of sea spider families (with Nymphonidae considered as a derived lineage of Callipallenidae) was estimated to have occurred during the Paleozoic under both clock models. Separately, we reran molecular dating analyses under both clock models with two alternative calibration schemes: 1) all calibrations except for the fossil Haliestes dasos and 2) outgroup calibrations only. Results varied little between analyses (supplementary fig. S4, Supplementary Material online). Bayesian analysis of macroevolutionary mixtures (BAMM) of sea spider diversification revealed no evidence of rate shifts within Pycnogonida, recovering instead a monotonic and near-constant process of diversification (supplementary fig. S5, Supplementary Material online).
Fig. 5.
(a) Phylogenomic dating of sea spiders based on the most complete data matrix and a correlated rates molecular clock model. Colors of branches and 95% HPD intervals correspond to families, as in figure 2. Line drawings (inset) correspond to stem-group fossils Palaeoisopus problematicus (top) and Flagellopantopus blocki (bottom). (b) Log lineage through time trajectories for selected Paleozoic aquatic taxa (sources in text). Branching times are truncated at the Cenozoic to mitigate undersampling of recent diversity and/or oversampling of intraspecific terminals.
Discussion
A Phylogenomic View of Higher-Level Sea Spider Relationships
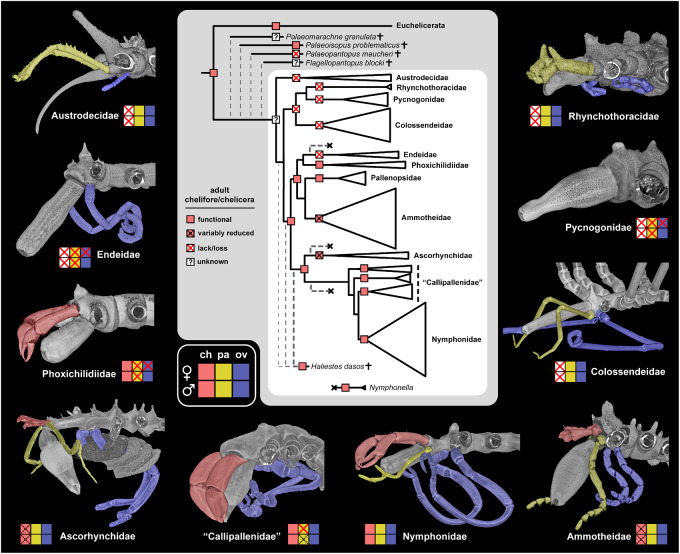
Traditional views of sea spider evolution suggested that the sister group of the remaining sea spiders was Nymphonidae (Munilla and de Haro 1981), based on its generalized appendage morphology (Hedgpeth 1947), or possibly Ammotheidae (previously thought to include Eurycyde; Stock 1994), based on anatomical similarities between ammotheids and fossils like Palaeoisopus problematicus. The species tree that we produced decisively recovered the enigmatic family Austrodecidae as the sister group to the remaining sea spiders. Earlier classifications had already pointed to the distinctness of Austrodecidae, segregating the family as sole member of the suborder Stiripasterida (Fry 1978; Bamber 2007). The recovery of a sister group relationship of Pycnogonidae and Rhynchothoracidae has been previously proposed based on morphology (Munilla León 1999) and an analysis of 18S rRNA (Nakamura et al. 2007). Potential morphological apomorphies of Pycnogonidae and Rhynchothoracidae include the general habitus (compact body, relatively short legs), shape and configuration of the proboscis, presence of female gonopores on the last leg pair only, and the egg-carrying behavior of males (single pancake-shaped mass into which both ovigers insert). Overall, our data set established a stable backbone topology for the remaining families (fig. 3). Only the position of N. tapetis engendered topological discordance across the data sets.
Establishing the sequence of basal branching families facilitates reconstruction of evolutionary transformation series for major character systems. Upon reconstructing adult appendage characteristics on our tree topology, we found that adult chelifores are absent in a grade of families at the base of extant Pycnogonida (fig. 6). Given some ambiguity in the state of the chelifore in some fossil taxa, as well as uncertainty about their phylogenetic placement, our results suggest that the presence of an adult chelifore may not be the unambiguous ancestral condition for crown-group sea spiders. In any case, a strictly gradual reduction of cephalic appendages (sensu Munilla León [1999]) has to be refuted for the crown group, as the traditionally advocated early branching families Nymphonidae and Ammotheidae with full sets of adult appendages are resolved in derived positions (a result consistent with previous studies; Arango 2002; Arango and Wheeler 2007; Arabi et al. 2010). The sister group relationship of Endeidae and Phoxichilidiidae accords with the shared loss of palps and female ovigers in these families. We also consistently obtained the nonmonophyly of Callipallenidae, recapitulating a result that has been suggested, albeit with weak support, in previous analyses of one to six loci (Arango and Wheeler 2007; Nakamura et al. 2007; Arabi et al. 2010). In contrast to previous topologies, we recovered the monophyly of Nymphonidae as a derived lineage within the callipallenids across all data sets and analyses. This result calls for systematic revision of these two families and illustrates another instance of cephalic appendage regain instead of reduction during crown-group evolution. Although functional chelifores are shared by both families, adult palps are lacking in the majority of callipallenid taxa (fig. 6), with the exception of few genera where they are present but largely reduced in males only (e.g., Anoropallene). By contrast, all nymphonids possess well-differentiated five-articled adult palps in both sexes (fig. 6; Brenneis et al. 2011). Moreover, this callipallenid–nymphonid relationship also suggests the reemergence of the pycnogonid-typical protonymphon larva in the nymphonid lineage, having been lost in the more direct development of callipallenids (Brenneis et al. 2011).
Fig. 6.
Cephalic appendage evolution in sea spiders, with emphasis on the chelifore. Reconstruction of adult chelifores is mapped on the topology obtained herein, with addition of fossil stem group and crown group representatives. Ancestral state reconstruction is based on equally weighted parsimony. Note the omission of functional adult chelifores in colossendeids with supernumerary segments (a derived state within the genus Colossendeis). Specimens in counterclockwise order from top left: male Austrodecus glaciale, male Endeis spinosa, female Phoxichilidium femoratum, egg-bearing male Ascorhynchus ramipes, male Stylopallene cheilorhynchus, male Nymphon gracile, female Ammothella longipes, female Colossendeis angusta, female Pycnogonum litorale, female Rhynchothorax australis.
Although genera were largely recovered as monophyletic, those defined to accommodate the condition of supernumerary segments (e.g., Decolopoda, Pentanymphon) were nested within larger genera of eight-legged species (e.g., Colossendeis, Nymphon). The exception was Pentapycnon, although our sampling of the family Pycnogonidae remains too sparse to test the monophyly of Pycnogonum (∼100 described species). Lineages with supernumerary segments are remarkable in an evolutionary context; in other Chelicerata, segment number tends to be fixed within a given extant order, as exemplified by such groups as scorpions and harvestmen. Changes in the segmentation of the prosoma (the appendage-bearing tagma of chelicerates) are especially rare (e.g., the synziphosurines Offacolus kingi and Weinbergina opitzi; Dunlop and Lamsdell 2017), yet all sea spiders are distinguished from the remaining Chelicerata by the presence of an additional oviger-bearing segment (the post-tritocerebral segment, which bears a walking leg in all other chelicerates). The mechanistic basis of supernumerary segment addition is currently an enigma, due to limited developmental genetic resources for sea spiders (Jager et al. 2006; Brenneis et al. 2008). Future work should prioritize the gap segmentation genes of sea spiders, especially those functionally linked to gap segmentation phenotypes in chelicerate model species (Schwager et al. 2009; Pechmann et al. 2011; Setton and Sharma 2018).
Nymphonella and the Composition of Ascorhynchidae
Surprisingly, the position of the putative ascorhynchid N. tapetis was unstable to analytical treatment of the data set, a result that cannot be attributed to missing data alone (N. tapetis was represented in 47–59% of loci across Matrices 1–4). Nymphonella is clearly distinguished from other sea spiders by anterolaterally projecting chelifores, annulated distal podomeres of the first walking leg, and a bulbous proboscis (fig. 1F). Previous efforts to infer the placement of Nymphonella have recovered limited support for its placement or for the monophyly of Ascorhynchidae, though Nymphonella has only been represented by 18S rRNA in such studies (Miyazaki et al. 2015; Sabroux et al. 2018). Nymphonella may therefore constitute a rogue taxon, an inference corroborated by its variable placement across data partitions generated in this study (supplementary fig. S3, Supplementary Material online).
By contrast, we recovered support for a nested placement of the putative ammotheid genus Paranymphon within Ascorhynchidae sensu stricto (represented by the genera Eurycyde and Ascorhynchus). Paranymphon has never been previously sequenced, but like Nymphonella, this terminal exhibited topological incongruence across data classes, being recovered within ammotheids by the mitochondrial data partition (albeit being represented therein by only three mitochondrial genes), and with ascorhynchids by UCEs and exons (13 and 17 loci, respectively).
Given the uncertainty surrounding the composition of Ascorhynchidae, future systematic efforts should target deeper sequencing of the genera Nymphonella (three described species) and Paranymphon (six described species).
Combining Data Classes Overcomes Limitations of Individual Partitions
Historical efforts to infer higher-level relationships of sea spiders have leaned heavily on the Sanger-sequenced nuclear ribosomal and mitochondrial markers, but these have not yielded a stable sea spider phylogeny to date (Arango and Wheeler 2007; Arabi et al. 2010; Sabroux et al. 2018). Ideally, new markers should exhibit evolutionary rates intermediate between slow-evolving nuclear ribosomal genes and fast-evolving mitochondrial markers. Targeted exons and UCEs sequenced in this study exhibit rates that propitiously fall exactly within this desired range (supplementary fig. S2, Supplementary Material online).
Problematically for the UCE data set, capture efficiency of UCE probes was much lower than other data partitions (supplementary fig. S1, Supplementary Material online). This phenomenon appears to be partly linked amount and quality of genomic DNA in a given extraction, as reported in a previous study on spiders with the same UCE probe set, resulting in low recovery rates for extractions with small quantities of DNA (Wood et al. 2018). Low quantity of DNA is unavoidable for such minute and rare lineages as Rhynchothorax and austrodecids. However, coverage inefficiency of the arachnid UCE probe set was systemic for sea spiders, even when ample DNA was available for large-bodied species, suggesting that the Arachnida 1.1Kv1 probe set is not universally effective across chelicerate orders (e.g., a single mite exemplar in Starrett et al. [2017] similarly exhibited low yields of UCE loci, by comparison to other arachnid taxa). As a result, of an initial 230 UCEs amplified, we discarded 56% of alignments where fewer than six sea spider terminals were obtained. The remaining 101 UCE loci bore high proportions of missing data, in contrast to the targeted exons. Similar outcomes have been reported for palpimanoid spiders, a group that had the advantage of a well-annotated theridiid spider genome for validation of probe design (Wood et al. 2018).
Nevertheless, we observed high informativeness of the UCE loci despite missing data, as inferred from distributions of RF and weighted RF distances. The concatenated UCE tree, while incongruent with the species tree, recovered such higher-level groupings as Nymphonidae nested within Callipallenidae, and Colossendeidae sister group to Rhynchothoracidae + Pycnogonidae. Given the promise of this phylogenetic data class, efforts to improve the recovery of UCE data sets in sea spiders should target the generation of high-quality sea spider genomes, with downstream improvements in the design of sea spider-specific UCE probes. Such strategies have been shown to overcome limitations inherent to the arachnid UCE bait set for spiders (Kulkarni et al. 2020).
The First Molecular Dating of Pycnogonida Reveals Ancient Diversification and Monotonic Evolutionary Rates
Generally, the fossil record for many invertebrate groups (especially marine arthropods) is scarce. The appearance of a Cambrian fossil resembling a sea spider early developmental instar, together with the exquisite preservation of the fossil Haliestes dasos, clearly point to an ancient origin of sea spiders before the Silurian (supplementary text S1, Supplementary Material online). However, a fossil record dating to the Paleozoic is not dispositive of ancient diversification of the crown group. Evolutionary relicts like Xiphosura (horseshoe crabs) and Nautiloidea appeared early in the fossil record (oldest fossils belonging to the Ordovician for Xiphosura; Late Cambrian for Nautiloidea) but survived to the present as a small number of species that diverged relatively recently (Obst et al. 2012; Combosch et al. 2017). Molecular divergence time estimation for such groups as Crinoidea (feather stars) and Ophiuroidea (brittle stars) has shown that both these echinoderm groups diversified in the wake of the end-Permian mass extinction (Rouse et al. 2013; O’Hara et al. 2014), having survived this extinction episode as a single lineage that subsequently recovered some fraction of its diversity (a “revenant” taxon sensu Sharma and Wheeler [2013]). By contrast, marine invertebrate groups like Holothuroidea (sea cucumbers) (Miller et al. 2017) and Protobranchia (protobranch bivalves) (Sharma et al. 2013) diversified in the Paleozoic but retain the signature of the end-Permian mass extinction in their dated phylogenies, which manifests as a low diversification rate (the plateau of an antisigmoidal curve in a log-lineage through time plot) until the beginning of the Mesozoic (fig. 5B) (Crisp and Cook 2009; Sharma et al. 2013).
To date, molecular divergence time estimation has never been performed for sea spiders. It was previously postulated that sea spiders diversified relatively recently (in the Mesozoic), but this speculation was based solely on the branch length (i.e., substitutions per site) subtending Pycnogonida in a molecular phylogeny, rather than a parametric molecular dating approach or the use of fossil calibrations (Arabi et al. 2010). Our divergence time estimation unambiguously recovered Cambrian to Ordovician ages of sea spider diversification, a result that is independent of the choice of clock model or use of ingroup fossil calibrations. Ancient diversification of Pycnogonida during the Ordovician is consistent with their fossil record (e.g., Jurassic sea spiders that are assignable to families; supplementary text S1, Supplementary Material online), and further suggests that Devonian sea spiders with opisthosomal segments (e.g., Flagellopantopus, Palaeoisopus) constitute stem lineages that diverged from extant Pycnogonida before the Ordovician and thereafter went extinct. A parallel evolutionary history has been reconstructed for spiders and their extinct sister group Uraraneida, which was recently shown to have survived at least until the Cretaceous (Huang et al. 2018; Wang et al. 2018).
Comparison of lineage accumulation through time for selected marine invertebrate groups reveals the marked difference between the evolutionary history of sea spiders and other Paleozoic fauna (fig. 5B) (Lins et al. 2012; Obst et al. 2012; Korn et al. 2013; Rouse et al. 2013; Sharma et al. 2013; O’Hara et al. 2014; Miller et al. 2017; Wolfe et al. 2019). In contrast to groups like Protobranchia and Crinoidea, Pycnogonida exhibited a static diversification regime, with a monotonic process of slowing diversification rate since initial divergence, and no evidence of rate shifts (fig. 5 and supplementary fig. S5, Supplementary Material online), by comparison to other Paleozoic taxa. Notably, the use of log-lineage through time plots as the sole window for examining evolutionary history has been recently criticized for nonidentifiability; it has been shown mathematically that myriad combinations of cladogenetic and extinction rate regimes can engender the same net diversification rate curves (Louca and Pennell 2020). Fossil data sets, when available and densely sampled, were proposed as more reliable arbiters of the accuracy of log-lineage through time trajectories, and thus potential solutions to this impasse (Louca and Pennell 2020). Fortuitously, such direct comparisons between net diversification curves and the fossil record are available for a subset of taxa that exhibit a detailed fossil record (fig. 5B). Specifically, dynamics of generic diversity of fossil echinoderms substantiate the signature of susceptibility to the end-Permian extinction event in dated phylogenies, with coincident accumulation of lineages in extant phylogenies and the fossil record in the wake of the mass extinction episodes (figure 3 of Rouse et al. 2013; figure 5 of Miller et al. 2017). Taken together with the absence of inferred rate shifts for sea spider in BAMMs (supplementary fig. S6, Supplementary Material online), our results indicate the absence of evidence for radical changes in diversification dynamics of Pycnogonida during most of the Phanerozoic.
The lack of any signature of major mass extinction events in the early (pre-Mesozoic) evolutionary history of extant sea spider and crustacean lineages is unexpected. In contrast to groups like bivalves and echinoderms, sea spiders do not form large calcified hard parts (valves or tests) whose deposition was severely affected by cataclysmic historical environmental changes (e.g., during the end-Permian event) (Krishnan 1955). However, this differing composition of hard parts does not account for the Paleozoic origin and post-Permian diversification of aquatic arthropod groups like Xiphosura and Notostraca (Obst et al. 2012; Korn et al. 2013). Moreover, the cuticle of decapods is indeed biomineralized, with denser cuticle associated with higher calcification in benthic Decapoda (Amato et al. 2008). Apropos, the log-lineage through time trajectory for Decapoda does indeed exhibit a small decline in lineage accumulation rate immediately preceding the Triassic (fig. 5B), but this result was only observed under divergence time estimation under one of two clock models (Wolfe et al. 2019).
We therefore postulate that the inferred resilience of sea spiders is not exclusively due to their lack of a calcified exoskeleton. Patterns of sea spider species richness observed today could be attributable to higher diversification rates in cooler regions, as has previously been shown in other marine arthropod groups like Anomura (Bracken-Grissom et al. 2013; Davis et al. 2016). In deep-sea isopods, diversification in the deep sea occurred in parallel with anoxic events, with the earliest radiation dated to the early Permian and subsequent episodes of rapid colonization and radiation (Lins et al. 2012). Such processes could partly explain the oddities of sea spider biogeography (such as the concentration of their diversity in Antarctica), but the role that the Southern Ocean has played as a potential center of endemism is not clear based on our results.
In the present study, which is focused on higher-level phylogenetic relationships, we lack sufficiently broad geographic sampling to assess the biogeography of sea spiders and a putative role for the polar regions as the driver of sea spider diversity in lower latitudes. Population genomic works have begun tackling such questions at shallower taxonomic scales (within species or species groups), with high levels of precision (Dietz et al. 2015, 2019; Dömel et al. 2015, 2019; Harder et al. 2016; Soler-Membrives et al. 2017; Collins et al. 2018), in tandem with challenging comparative physiological experiments in Southern Ocean species that address the dynamic evolution of body size in Pycnogonida (Woods et al. 2009; Shishido et al. 2019). Denser sampling of extant sea spider diversity in future studies will augment the predictive power of such approaches in deciphering the evolutionary history of this ancient group.
Materials and Methods
Species Sampling and Locus Selection
Specimens of all sea spider families were obtained from field expeditions and museum collections, as well as material collected during multiple deep sea cruises. A list of taxa sampled is provided in supplementary table S1, Supplementary Material online. DNA was extracted using the Qiagen DNeasy Blood and Tissue Kit (Valencia, CA) from one to five specimens, prioritizing tissue from the propodus and main claw (gut-free leg podomeres) where possible. Taxonomic sampling consisted of 89 sea spiders; outgroups consisted of 14 Arachnida (including one Xiphosura, which has recently been shown to be possibly nested within the arachnids; Ballesteros and Sharma 2019; Ballesteros et al. 2019), 3 Myriapoda, 3 Pancrustacea, and 1 Onychophora.
Probes were designed using available references and databases, given the absence of a published sea spider genome, following the pipeline established by Breinholt et al. (2018). Probes for mitochondrial genomes were designed using a newly sequenced mitogenome of Pycnogonum litorale and five published sea spider mitogenomes: Tanystylum orbiculare (GU370074.1), Ammothea hilgendorfi (GU370075.1), Achelia bituberculata (NC_009724.1), Nymphon unguiculatum-charcoti complex sp. (GU370076.1), and Nymphon gracile (NC_008572.1). From previous work on the phylogeny of Chelicerata (Sharma, Kaluziak, et al. 2014), we identified 56 nuclear exons that 1) occurred in single copy across Panarthropoda, 2) were present for at least one of two sea spider Illumina transcriptomes used as templates for probe design (Anoplodactylus insignis and Pycnogonum litorale; Ballesteros and Sharma [2019]); and 3) exhibited sufficient conservation of intron–exon boundaries in the genomes of arthropods for 125-bp probe design with 3× tiling. Probes design was guided by conservation of intron–exon boundaries in the genomes of D. melanogaster, Strigamia maritima, and Parasteatoda tepidariorum. For UCE sequencing, we deployed probes from the UCE Arachnida 1.1Kv1 bait set (Starrett et al. 2017). Probe design, synthesis, automated library preparation, and paired-end sequencing (2 × 150 bp) on the Illumina Hi-Seq 2500 platform were performed through RAPiD Genomics (Gainesville, FL). Sequencing efficiency is provided in supplementary table S2, Supplementary Material online; details of locus selection and orthology validation are provided in supplementary text S1, Supplementary Material online. All probe sequences are made available in the Dryad Digital Repository.
Bioinformatic and Phylogenomic Operations
Raw reads were assembled using Trinity v.2.7 (Grabherr et al. 2011; Haas et al. 2013) using a path reinforcement distance of 75. Reference sequences of sea spiders (mitogenomes and nuclear exons) and/or euchelicerates (UCEs) were used to separate loci into alignments. We included the nuclear ribosomal genes 5.8S rRNA, 18S rRNA, and 28S rRNA, which were obtained as bycatch; on-target amplification of these bycatch loci was verified using BlastN against available sea spider ribosomal sequences in GenBank. Outgroup taxa were subsequently added into the alignments using available transcriptomes from our previous works (for nuclear exons; Sharma, Kaluziak, et al. 2014), from GenBank-accessioned data (mitochondrial genes/genomes and nuclear ribosomal genes), and from a subset of the taxa in chelicerate UCE alignments (Starrett et al. 2017). Due to the unavailability of the same phylogenetic data classes among the genomic resources of some data-poor outgroups, we created chimeric terminals for such terminals as Eremobates and Ricinoides. The nature of these congeneric chimeras is that sequence data for nuclear exons and ribosomal genes are drawn from the transcriptomes of one species (e.g., Eremobates cf. transfugoides; Ricinoides atewa), but the mitochondrial genome is drawn from a congener (e.g., Eremobates cf. palpisetulosus; Ricinoides karschii). The list of chimeric outgroup terminals is detailed in supplementary table S3, Supplementary Material online.
Multiple sequence alignment was performed using MUSCLE v.3.8.31 (Edgar 2004). For ribosomal genes, alignments were performed on nucleotide sequences. For nuclear and mitochondrial exons, alignments were performed using peptide translations and subsequently reverted to nucleotides. We discarded the mitochondrial genes 12S rRNA, 16S rRNA, and tRNAs, due to their high rate of evolution, as well as off-target capture of non-sea spider sequence for these regions, which we presumed to have resulted from sequencing of gut content and/or epibionts.
Alignments were trimmed using trimal v.1.2 (Capella-Gutiérrez et al. 2009) to cull overhanging ends. Each alignment was filtered to ensure the inclusion of a minimum of six sea spider terminals and four outgroup terminals (resulting in the exclusion of the mitochondrial protein ATP8 and 129 out of 230 UCE alignments). Gene trees were inferred using IQ-TREE v.1.6 (Nguyen et al. 2015) with the best-fitting model selected by ModelFinder (-m MFP) (Kalyaanamoorthy et al. 2017). Gene tree topologies were visually inspected for potential paralogs, which were summarily discarded from alignments (principally found in the UCE alignments previously generated for arachnids). Orthology was validated using BLAST-based annotation; the annotations for all sequenced loci are provided in supplementary table S6, Supplementary Material online.
Four matrices were constructed for our main analyses and all data sets were treated as nucleotide alignments. Matrices 1–3 were constructed using taxon occupancy thresholds of 50%, 33%, and 25%, respectively. Matrix 4 consisted of loci stipulated to sample at least one Austrodecidae (the putative sister group to the remaining sea spiders). The final alignments were analyzed as concatenated supermatrices using IQ-TREE v.1.6 with best-fitting models per locus. Due to ongoing debate over the benefits of model fitting, we additionally analyzed our data sets using a unique GTR + Γ4 model for each locus as well. Nodal support was estimated using bootstrap resampling frequency with 1,000 ultrafast bootstrap replicates in IQ-TREE. In addition, approximately unbiased tests of monophyly were performed using in-built tools in IQ-TREE for selected phylogenetic hypotheses using Matrices 1 and 4.
For assessment of data set performance, we inferred the normalized RF and wRF distances between the species tree inferred using the most complete matrix (Matrix 1) and each gene tree. To overcome missing data, the species tree was recursively pruned of tips to match the terminals of each gene tree prior to calculation of RF and wRF distances, and the distances were normalized using the maximal possible distance between each gene tree and the pruned species tree. Other metrics of performance consisted of locus length, taxon sampling, GC content, and MPSI (a proxy for evolutionary rate). Tree manipulations and calculations of performance metrics were performed using R packages ape v.5.3 (Paradis and Schliep 2019) and phytools v.0.6-99 (Revell 2012).
Bayesian analyses were trialed using PhyloBayes-mpi v.1.7 (Lartillot and Philippe 2004), but these analyses failed to reach convergence after several months of computation and were ultimately discarded.
Embryonic Transcriptomes
Ovigerous sea spider males with prominent egg masses were hand collected from field sites between 2015 and 2017 for five sea spider species: Phoxichilidium cf. femoratum (Phoxichilidiidae), Tanystylum orbiculare (Ammotheidae), Pallenella flava (Callipallenidae), Stylopallene cheilorhynchus (Callipallenidae), and Nymphon molleri (Nymphonidae). Egg masses were separated from the males with fine forceps and immediately transferred to RNAlater (ThermoFisher). Due to asynchrony of development within egg masses, these samples represented a range of embryonic stages, from early embryogenesis through protonymphon larvae. As a control sample, we included a range of embryonic stages from the tarantula A. hentzi; field collection and processing of embryos of this spider are described in Setton et al. (2019). All samples were removed from RNAlater and transferred to TRIZOL TriReagent for RNA extraction, following our previous procedures (Sharma, Kaluziak, et al. 2014). Library preparation, paired-end Illumina sequencing, and transcriptome assembly with Trinity also followed our previous approach (Sharma, Kaluziak, et al. 2014).
We used the conserved homeodomain sequences of all Hox genes of the spider Parasteatoda tepidariorum to retrieve Hox sequences from new libraries, using tBLASTn searches. Putative A. hentzi and sea spider Hox homologs were added to multiple sequence alignments recently generated by us (Gainett and Sharma 2020), in addition to all available sea spider Hox sequences previously generated using RACE PCR (Manuel et al. 2006). Gene tree inference was performed using IQ-TREE, treating sequence data as peptides and model fitting using ModelFinder. The Hox gene tree and the multiple sequence alignment used for Hox orthology inference are provided in the Dryad Digital Repository.
Phylogenomic Dating
Phylogenomic estimation of divergence times was estimated using a node-dating approach with MCMCTree (Yang 2007) on two data sets (Matrices 1 and 3), implementing a likelihood approximation of branch lengths using a multivariate normal distribution (dos Reis and Yang 2019). Fossils used to inform the dating consisted of 11 outgroup and 4 ingroup node calibrations. All calibrations were implemented as soft minimum and soft maximum ages.
The oldest unequivocal fossil of crown group Pycnogonida, Haliestes dasos, dates to the Silurian (424 Ma) (Siveter et al. 2004), whereas the oldest fossil assigned to the pycnogonid (stem) lineage is an early developmental instar from the Upper Cambrian (Cambropycnogon klausmuelleri) (Waloszek and Dunlop 2002). Haliestes dasos superficially resembles Nymphonidae, clearly lacks an annulated or segmented proboscis, and has been recovered at the base of Pycnogonida in some morphological cladistic analyses (Siveter et al. 2004). As a conservative measure, we calibrated the age of crown group Pycnogonida using a soft minimum age of 424 Ma and a soft maximum age of 501 Ma. Various other fossils (Flagellopantopus blocki, Palaeothea devonica, Palaeoisopus problematicus, Palaeopantopus maucheri, Palaeomarachne granulata, Pentapantopus vogteli), some of which exhibit morphologies attributable to stem-group lineages (e.g., a flagellum; opisthosomal segments), were not considered usable for node dating, due to poor preservation and/or an age younger than Haliestes dasos.
Sea spider crown group fossils from the Late Jurassic and La-Voulte-sur-Rhône were used to calibrate ingroup nodes: Palaeopycnogonides gracilis (Ammotheidae), Colossopantopodus boissinensis and Colossopantopodus nanus (Colossendeidae), and Palaeoendeis elmii (Endeidae) (Charbonnier et al. 2007; Sabroux et al. 2019). Each of these fossils was treated as a minimum age calibration for the corresponding family, with a maximum soft bound age of 501 Ma. We abstained from using Palaeothea devonica, Pentapantopus vogteli, and Eurycyde golem in this study, as the poor preservation and lack of characteristic details of these likely crown group fossils preclude reliable placement in any extant genus or family.
For Arachnida, we constrained the crown group of Opiliones using the soft minimum age of 411 Ma, based on the harvestman fossil Eophalangium sheari, and the divergence of Eupnoi harvestmen with a soft minimum age of 305 Ma, based on the age of Macrogyion cronus (Garwood et al. 2011). The divergence of spiders from the remaining Tetrapulmonata was constrained to at least 386 Ma (age of Attercopus fimbriunguis) and the divergence of Liphistius at 305 Ma (age of Palaeothele montceauensis) (Selden et al. 2008; Wang et al. 2018). Pedipalpi (Amblypygi + Uropygi) was set to a minimum age of 319 Ma, based on the age of the fossil Parageralinura naufraga (Tetlie and Dunlop 2008). The divergence of Xiphosura from Riniculei was constrained to a minimum of 445 Ma (age of Lunataspis aurora) (Rudkin et al. 2008). The stem-group age of scorpions was constrained to a soft minimum age of 435 Ma and a soft maximum of 514 Ma (based on the ages of Parioscorpio venator and Eramoscorpius brucensis) (Waddington et al. 2015; Wendruff et al. 2020). The crown group age of scorpions was constrained between 112.6 and 313.7 Ma (based on the ages of Protoischnurus axelrodurum and Compsoscorpius buthiformis) (Dunlop et al. 2016).
For Mandibulata, we constrained the basal diversification of Chilopoda between 416 and 521 Ma, and of Pleurostigmophora between 382.7 and 521 Ma. Altocrustacea (the node comprising the most recent common ancestor of the malacostracan exemplar Parhyale hawaiensis and the hexapods in this analysis) was constrained between 429.8 and 521 Ma. The root of the tree (split of Onychophora and Arthropoda) was constrained between 550 and 636 Ma. Mandibulate and root calibrations are based on various fossils reviewed by Wolfe et al. (2016).
Both the independent rates and correlated rates clock models were used to infer node ages with Matrix 1 (the most complete matrix), under the maximum likelihood tree topology inferred for this matrix with model selection using ModelFinder. We constrained the monophyly of Mandibulata and Phalangida (the non-Cyphophthalmi Opiliones) for node calibration. Four independent chains were run for 106 generations for each analysis, sampling every 100th generation. Convergence diagnostics were assessed using Tracer v.1.7 (Rambaut et al. 2018) and inbuilt tools in MCMCTree, ensuring minimum effective sample sizes >200 for summary statistics. As a conservative measure, 1,000 trees (10%) were discarded as burn-in. Separately, we ran two other dating analyses to assess the impact of sea spider fossils as calibration points: 1) using all fossil calibration points except for Halieses dasos and 2) using only outgroup calibration points.
Analyses of diversification rates through time were performed using BAMM v.2.5.0 (Rabosky 2014). The chronogram inferred using all calibration points and the correlated rates clock model was used as the input for BAMM. Outgroups were removed prior to analysis using phytools. Priors were iteratively tested for the expected number of rate shifts (one to five). Rate shifts were permitted on all branches. Sampling frequency was set to 6.54%, using the number of described species as a proxy for extant diversity. As an exploration of the effect of this parameter, we additionally reran the analysis with sampling frequency set to 1.308% and 0.654% (i.e., inferring total extant diversity to equal five times or ten times the current number of described species). MCMC (Markov chain Monte Carlo) chains were run for 500,000 generations, sampling event data every 10,000 generations, and discarding 25% of the run as burn-in.
Microcomputed Tomography
Fixed specimens were dehydrated via an ascending ethanol series, incubated in solution of 2% iodine (resublimated; Carl RothGmbH&Co. KG, Karlsruhe, Germany; cat. #X864.1) in 99.5% ethanol for ∼48 h at ambient temperature, briefly rinsed in 99.5% ethanol (3–4× 10 min) and either transferred into a vial in ethanol (wet scan) or critical point-dried with a Leica EM CPD300 and glued with the posterior body pole on plastic welding rods (dry scan). Scans were performed with an Xradia MicroXCT-200 (Carl Zeiss Microscopy GmbH). Scan settings were individually optimized for each specimen, including objective choice (0.39×, 4×, 10×) according to size. Scans were performed under 40 kV/200 µA/8W, exposure times ranged from 0.4 to 1.5 s. Tomography projections were reconstructed using the XMReconstructor software (Carl Zeiss Microscopy GmbH) with TIFF format image stacks as output. All scans were performed with Binning 2 to reduce noise and subsequently reconstructed with Binning 1 (=full resolution) to avoid information loss. Processing and 3D visualization of the image stacks (including highlighting of cephalic appendages and removal of nontarget structures) were performed with Imaris (version 7.0.0., Bitplane AG, Switzerland) as described previously (Scholtz and Brenneis 2016).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work is dedicated to the memory of Roger Norman Bamber. We are indebted to Katsumi Miyazaki, Anna Soler-Membrives, and Julia D. Sigwart for providing specimens for study. We thank the crews of many research vessels for assistance with fieldwork, and the Australian and Paris Museums for loans of materials for study. Sequencing and assembly of the Pycnogonum litorale mitogenome were performed by Anke Braband and Jakob Machner. Chronograms of selected marine invertebrate taxa were kindly provided by Heather D. Bracken-Grissom, Simon Y.W. Ho, Tim D. O’Hara, Greg W. Rouse, and Jo M. Wolfe. Access to computing nodes for intensive tasks was facilitated by the Bioinformatics Resource Center (BRC) and Christina Koch at the Center for High Throughput Computing (CHTC) of the University of Wisconsin-Madison. Comments from three anonymous reviewers and the Associate Editor improved a previous draft of this work. J.A.B. was supported by the M. Guyer postdoctoral fellowship. E.V.W.S. was supported by a National Science Foundation Graduate Research Fellowship. C.E.S.L. was supported by postdoctoral CONACYT (Grant Reg. 207146/454834). Morphological data collection was supported by Deutsche Forschungsgemeinschaft (Grant No. BR5039/3-1 to G.B.). Fieldwork in Antarctica was supported by National Science Foundation (Grant Nos. ANT-0551969 to A.L.M. and ANT-0440577 to H.A.W.). Collection under projects POL2006-06399/CGL (CLIMANT) and CTM2012-39350-C02-01 (ECOWED) were funded by the Spanish Comisión Interministerial de Ciencia y Tecnología and the Ministerio de Economía y Competitividad. Revolta Program 1124, led by Guillaume Lecointre, was supported by Institut Polaire Français Paul Émile Victor (IPEV). C.P.A was supported by ABRS grant 204-61 and Antarctic Science Grant AA3010. This work was supported by intramural funds of the American Museum of Natural History (W.C.W.) and intramural funds of the University of Wisconsin-Madison and National Science Foundation Grant No. IOS-1552610 (P.P.S).
Permitting
Specimens from multiple deep sea cruises were collected under the auspices of the United States Antarctic Program, in compliance with the US Antarctic Conservation Act. All diplomatic formulas for sampling the Icelandic economic zone were given for cruises M85/3 (IceAGE1) and POS456 (IceAGE2). Specimens from Australia were collected under permit no. 15111 of the Tasmanian Department of Primary Industries, Parks, Water and Environment and Marine Parks permit G08/27858.1.
Author Contributions
C.P.A., W.C.W., and P.P.S. conceived the study. Specimens were collected and/or contributed by C.P.A., G.B., S.B., E.C.-S., G.F.D., M.P.E., C.G., L.M., A.L.M., R.M., G.G., H.A.W., P.J.L.G., and P.P.S. Tissue for developmental transcriptomes was collected by G.B. and P.P.S. A new mitogenome of Pycnogonum litorale was contributed by G.S. for bait design. Coordination of sample sorting from the DIVA3 and IceAGE cruises was performed at the DZMB in Hamburg by S.B. Taxonomic identification from IceAGE and DIVA3 collections was performed by C.P.A., G.B., E.V.W.S., and P.P.S. E.V.W.S. and P.P.S. performed molecular work. E.V.W.S. performed imaging of specimens used for extraction. G.B. performed µCT scans and their subsequent processing and visualization. J.A.B., C.E.S.L., M.D., S.M., C.W., and P.P.S. performed bioinformatic and phylogenetic analyses. K.F.C., J.T.Z., E.V.W.S., and P.P.S. analyzed embryonic transcriptome data. W.C.W. and P.P.S. funded the work. P.P.S. drafted the manuscript, and all authors edited and approved the final content.
Data Availability
The complete data set, including sequence alignments, annotated tree files, chronograms, and embryonic transcriptomic assemblies, have been deposited at Dryad Digital Repository under URL https://datadryad.org/stash/share/HsFKzSdD00rwa5BQSEf9km6dXuURuiAtOwv0w0QVDUg. Raw read data were deposited at NCBI Sequence Read Archive. Loci from targeted sequencing were deposited in NCBI GenBank.
References
- Amato CG, Waugh DA, Feldmann RM, Schweitzer CE.. 2008. Effect of calcification on cuticle density in decapods: a key to lifestyle. J Crustac Biol. 28(4):587–595. [Google Scholar]
- Arabi J, Cruaud C, Couloux A, Hassanin A.. 2010. Studying sources of incongruence in arthropod molecular phylogenies: sea spiders (Pycnogonida) as a case study. C R Biol. 333(5):438–453. [DOI] [PubMed] [Google Scholar]
- Arango CP. 2002. Morphological phylogenetics of the sea spiders (Arthropoda: Pycnogonida). Org Divers Evol. 2(2):107–125. [Google Scholar]
- Arango CP, Wheeler WC.. 2007. Phylogeny of the sea spiders (Arthropoda, Pycnogonida) based on direct optimization of six loci and morphology. Cladistics 23(3):255–293. [DOI] [PubMed] [Google Scholar]
- Arnaud F, Bamber RN.. 1987. The biology of Pycnogonida. Adv Mar Biol. 24:1–95. [Google Scholar]
- Ballesteros JA, Santibáñez López CE, Kováč Ľ, Gavish-Regev E, Sharma PP.. 2019. Ordered phylogenomic subsampling enables diagnosis of systematic errors in the placement of the enigmatic arachnid order Palpigradi. Proc R Soc B. 286(1917):20192426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros JA, Sharma PP.. 2019. A critical appraisal of the placement of Xiphosura (Chelicerata) with account of known sources of phylogenetic error. Syst Biol. 68(6):896–917. [DOI] [PubMed] [Google Scholar]
- Bamber RN. 2007. A holistic re-interpretation of the phylogeny of the Pycnogonida Latreille, 1810 (Arthropoda). Zootaxa 1668(1):295–312. [Google Scholar]
- Bracken-Grissom HD, Cannon ME, Cabezas P, Feldmann RM, Schweitzer CE, Ahyong ST, Felder DL, Lemaitre R, Crandall KA.. 2013. A comprehensive and integrative reconstruction of evolutionary history for Anomura (Crustacea: Decapoda). BMC Evol Biol. 13(1):128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breinholt JW, Earl C, Lemmon AR, Lemmon EM, Xiao L, Kawahara AY.. 2018. Resolving relationships among the megadiverse butterflies and moths with a novel pipeline for anchored phylogenomics. Syst Biol. 67(1):78–93. [DOI] [PubMed] [Google Scholar]
- Brenneis G, Arango CP, Scholtz G.. 2011. Morphogenesis of Pseudopallene sp. (Pycnogonida, Callipallenidae) I: embryonic development. Dev Genes Evol. 221(5–6):309–328. [DOI] [PubMed] [Google Scholar]
- Brenneis G, Bogolomova EV, Arango CP, Krapp F.. 2017. From egg to “no-body”: an overview and revision of developmental pathways in the ancient arthropod lineage Pycnogonida. Front Zool. 14:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenneis G, Scholtz G.. 2015. Serotonin-immunoreactivity in the ventral nerve cord of Pycnogonida – support for individually identifiable neurons as ancestral feature of the arthropod nervous system. BMC Evol Biol. 15:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenneis G, Scholtz G, Beltz B.. 2018. Comparison of ventral organ development across Pycnogonida (Arthropoda, Chelicerata) provides evidence for a plesiomorphic mode of late neurogenesis in sea spiders and myriapods. BMC Evol Biol. 18(1):47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenneis G, Ungerer P, Scholtz G.. 2008. The chelifores of sea spiders (Arthropoda, Pycnogonida) are the appendages of the deutocerebral segment. Evol Dev. 10(6):717–724. [DOI] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T.. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25(15):1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonnier S, Vannier J, Riou B.. 2007. New sea spiders from the Jurassic La Voulte-sur-Rhône Lagerstätte. Proc R Soc B. 274(1625):2555–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins EE, Galaska MP, Halanych KM, Mahon AR.. 2018. Population genomics of Nymphon australe Hodgson, 1902 (Pycnogonida, Nymphonidae) in the Western Antarctic. Biol Bull. 234(3):180–191. [DOI] [PubMed] [Google Scholar]
- Combosch DJ, Lemer S, Ward PD, Landman NH, Giribet G.. 2017. Genomic signatures of evolution in Nautilus—an endangered living fossil. Mol Ecol. 26(21):5923–5938. [DOI] [PubMed] [Google Scholar]
- Crisp MD, Cook LG.. 2009. Explosive radiation or cryptic mass extinction? Interpreting signatures in molecular phylogenies. Evolution 63(9):2257–2265. [DOI] [PubMed] [Google Scholar]
- Davis KE, Hill J, Astrop TI, Wills MA.. 2016. Global cooling as a driver of diversification in a major marine clade. Nat Commun. 7:13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz L, Arango CP, Dömel JS, Halanych KM, Harder AM, Held C, Mahon AR, Mayer C, Melzer RR, Rouse GW, et al. 2015. Regional differentiation and extensive hybridization between mitochondrial clades of the Southern Ocean giant sea spider Colossendeis megalonyx. R Soc Open Sci. 2(7):140424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz L, Dömel JS, Leese L, Mahon AR, Mayer C.. 2019. Phylogenomics of the longitarsal Colossendeidae: the evolutionary history of an Antarctic sea spider radiation. Mol Phylogenet Evol. 136:206–214. [DOI] [PubMed] [Google Scholar]
- Dömel JS, Convey P, Leese F.. 2015. Genetic data support independent glacial refugia and open ocean barriers to dispersal for the Southern Ocean sea spider Austropallene cornigera (Möbius, 1902). J Crustac Biol. 35(4):480–490. [Google Scholar]
- Dömel JS, Macher T-H, Dietz L, Duncan S, Mayer C, Rozenberg A, Wolcott K, Leese F, Melzer RR.. 2019. Combining morphological and genomic evidence to resolve species diversity and study speciation processes of the Pallenopsis patagonica (Pycnogonida) species complex. Front Zool. 16:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis M, Yang Z.. 2019. Bayesian molecular clock dating using genome-scale datasets. In: Anisimova M, editor. Evolutionary genomics. Methods in Molecular Biology. Vol. 1910. New York: Humana. p. 309–330. [DOI] [PubMed] [Google Scholar]
- Dunlop JA, Lamsdell JC.. 2017. Segmentation and tagmosis in Chelicerata. Arthropod Struct Dev. 46(3):395–418. [DOI] [PubMed] [Google Scholar]
- Dunlop JA, Legg DA, Selden PA, Fet V, Schneider JW, Rößler R.. 2016. Permian scorpions from the Petrified Forest of Chemnitz, Germany. BMC Evol Biol. 16:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry WG. 1978. A classification within the pycnogonids. Zool J Linn Soc. 63(1–2):35–38. [Google Scholar]
- Gainett G, Sharma PP.. 2020. Genomic resources and toolkits for developmental study of whip spiders (Amblypygi) provide insights into arachnid genome evolution and antenniform leg patterning. EvoDevo 11(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garwood RJ, Dunlop JA, Giribet G, Sutton MD.. 2011. Anatomically modern Carboniferous harvestmen demonstrate early cladogenesis and stasis in Opiliones. Nat Commun. 2:444. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29(7):644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths HJ, Arango CP, Munilla T, McInnes SJ.. 2011. Biodiversity and biogeography of Southern Ocean pycnogonids. Ecography 34(4):616–627. [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 8(8):1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder AM, Halanych KM, Mahon AR.. 2016. Diversity within the sea spider genus Pallenopsis (Pycnogonida: Chelicerata) in the Western Antarctic. Polar Biol. 39(4):677–688. [Google Scholar]
- Hedgpeth JW. 1947. On the evolutionary significance of the Pycnogonida. Smiths Misc Coll. 106:1–53. [Google Scholar]
- Huang D, Hormiga G, Cai C, Su Y, Yin Z, Xia F, Giribet G.. 2018. Origin of spiders and their spinning organs illuminated by mid-Cretaceous amber fossils. Nat Ecol Evol. 2(4):623–627. [DOI] [PubMed] [Google Scholar]
- Jager M, Murienne J, Clabaut C, Deutsch J, Guyader HL, Manuel M.. 2006. Homology of arthropod anterior appendages revealed by Hox gene expression in a sea spider. Nature 441(7092):506–508. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny NJ, Chan KW, Nong W, Qu Z, Maeso I, Yip HY, Chan TF, Kwan HS, Holland PWH, Chu KH, et al. 2016. Ancestral whole-genome duplication in the marine chelicerate horseshoe crabs. Heredity 116(2):190–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn M, Rabet N, Ghate HV, Marrone F, Hundsdoerfer AK.. 2013. Molecular phylogeny of the Notostraca. Mol Phylogenet Evol. 69(3):1159–1171. [DOI] [PubMed] [Google Scholar]
- Krishnan G. 1955. Nature of the cuticle of Pycnogonida. Nature 175(4464):904. [Google Scholar]
- Kulkarni S, Wood H, Lloyd M, Hormiga G.. 2020. Spider-specific probe set for ultraconserved elements offers new perspectives on the evolutionary history of spiders (Arachnida, Araneae). Mol Ecol Resour. 20(1):185–203. [DOI] [PubMed] [Google Scholar]
- Lane SJ, Moran AL, Shishido CM, Tobalske BW, Woods HA.. 2018. Cuticular gas exchange by Antarctic sea spiders. J Exp Biol. 221(8):jeb177568. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Philippe H.. 2004. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 21(6):1095–1109. [DOI] [PubMed] [Google Scholar]
- Leite DJ, Baudouin-Gonzalez L, Iwasaki-Yokozawa S, Lozano-Fernandez J, Turetzek N, Akiyama-Oda Y, Prpic N-M, Pisani D, Oda H, Sharma PP, et al. 2018. Homeobox gene duplication and divergence in arachnids. Mol Biol Evol. 35(9):2240–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lins LSF, Ho SYW, Wilson GDF, Lo N.. 2012. Evidence for Permo-Triassic colonization of the deep sea by isopods. Biol Lett. 8(6):979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louca S, Pennell MW.. 2020. Extant timetrees are consistent with a myriad of diversification histories. Nature 580(7804):502–505. [DOI] [PubMed] [Google Scholar]
- Manuel M, Jager M, Murienne J, Clabaut C, Le Guyader H.. 2006. Hox genes in sea spiders (Pycnogonida) and the homology of arthropod head segments. Dev Genes Evol. 216(7–8):481–491. [DOI] [PubMed] [Google Scholar]
- Masta SE, McCall A, Longhorn SJ.. 2010. Rare genomic changes and mitochondrial sequences provide independent support for congruent relationships among the sea spiders (Arthropoda, Pycnogonida). Mol Phylogenet Evol. 57(1):59–70. [DOI] [PubMed] [Google Scholar]
- Miller AK, Kerr AM, Paulay G, Reich M, Wilson NG, Carvajal JI, Rouse GW.. 2017. Molecular phylogeny of extant Holothuroidea (Echinodermata). Mol Phylogenet Evol. 111:110–131. [DOI] [PubMed] [Google Scholar]
- Miyazaki K, Tomiyama T, Yamada K, Tamaoki M.. 2015. 18S analysis of the taxonomic position of an endoparasitic pycnogonid, Nymphonella tapetis (Arthropoda: Pycnogonida: Ascorhynchidae). J Crustac Biol. 35(4):491–494. [Google Scholar]
- Munilla León T. 1999. Evolución y filogenia de los picnogónidos. In: Melic A, de Haro JJ, Mendez M, Ribera I, editors. Evolución y filogenia de Arthropoda. Sociedad Entomológica Aragonesa (SEA), Zaragoza. p. 273–279. [Google Scholar]
- Munilla T, de Haro A.. 1981. An electrophoretical and immunological study of Pycnogonida, with phylogenetic considerations. Bijdr Dierkd. 51:191–198. [Google Scholar]
- Nakamura K, Kano Y, Suzuki N, Namatame T, Kosaku A.. 2007. 18S rRNA phylogeny of sea spiders with emphasis on the position of Rhynchothoracidae. Mar Biol. 153(2):213–223. [Google Scholar]
- Nguyen L-T, Schmidt HA, Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan ED, Santibáñez-López CE, Sharma PP.. 2020. Developmental gene expression as a phylogenetic data class: support for the monophyly of Arachnopulmonata. Dev Genes Evol. 230(2):137–153. [DOI] [PubMed] [Google Scholar]
- O’Hara TD, Hugall AF, Thuy B, Moussalli A.. 2014. Phylogenomic resolution of the class Ophiuroidea unlocks a global microfossil record. Curr Biol. 24(16):1874–1879. [DOI] [PubMed] [Google Scholar]
- Obst M, Faurby S, Bussarawit S, Funch P.. 2012. Molecular phylogeny of extant horseshoe crabs (Xiphosura, Limulidae) indicates Paleogene diversification of Asian species. Mol Phylogenet Evol. 62(1):21–26. [DOI] [PubMed] [Google Scholar]
- Paradis E, Schliep K.. 2019. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35(3):526–528. [DOI] [PubMed] [Google Scholar]
- Pechmann M, Khadjeh S, Turetzek N, McGregor AP, Damen WGM, Prpic N-M.. 2011. Novel function of Distal-less as a gap gene during spider segmentation. PLoS Genet. 7(10):e1002342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabosky DL. 2014. Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS One 9(2):e89543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA.. 2018. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst Biol. 67(5):901–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol. 3(2):217–233. [Google Scholar]
- Rota-Stabelli O, Kayal E, Gleeson D, Daub J, Boore JL, Telford MJ, Pisani D, Blaxter M, Lavrov DV.. 2010. Ecdysozoan mitogenomics: evidence for a common origin of the legged invertebrates, the Panarthropoda. Genome Biol Evol. 2:425–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse GW, Jermiin LS, Wilson NG, Eeckhaut I, Lanterbecq D, Oji T, Young CM, Browning T, Cisternas P, Helgen LE, et al. 2013. Fixed, free, and fixed: the fickle phylogeny of extant Crinoidea (Echinodermata) and their Permian-Triassic origin. Mol Phylogenet Evol. 66(1):161–181. [DOI] [PubMed] [Google Scholar]
- Rudkin DM, Young GA, Nowlan GS.. 2008. The oldest horseshoe crab: a new xiphosurid from Late Ordovician Konservat-Lagerstätten deposits, Manitoba, Canada . Palaeontology 51(1):1–9. [Google Scholar]
- Sabroux R, Audo D, Charbonnier S, Corbari L, Hassanin A.. 2019. 150-million-year-old sea spiders (Pycnogonida: Pantopoda) of Solnhofen. J Syst Palaeontol. 17(22):1927–1938. [Google Scholar]
- Sabroux R, Corbari L, Krapp F, Bonillo C, Le Prieur S, Hassanin A.. 2018. Biodiversity and phylogeny of Ammotheidae (Arthropoda: Pycnogonida). Eur J Taxon. 286:1–33. [Google Scholar]
- Santibáñez-López CE, González E, Monod L, Sharma PP.. 2019. Phylogenomics facilitates stable scorpion systematics: reassessing the relationships of Vaejovidae and a new higher-level classification of Scorpiones (Arachnida). Mol Phylogenet Evol. 135:22–30. [DOI] [PubMed] [Google Scholar]
- Scholtz G, Brenneis G.. 2016. The pattern of a specimen of Pycnogonum litorale (Arthropoda, Pycnogonida) with a supernumerary leg can be explained with the “boundary model” of appendage formation. Sci Nat. 103:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwager EE, Pechmann P, Feitosa NM, McGregor AP, Damen WGM.. 2009. hunchback functions as a segmentation gene in the spider Achaearanea tepidariorum. Curr Biol. 19(16):1333–1340. [DOI] [PubMed] [Google Scholar]
- Schwager EE, Sharma PP, Clarke T, Leite DJ, Wierschin T, Pechmann M, Akiyama-Oda Y, Esposito L, Bechsgaard J, Bilde T, et al. 2017. The house spider genome reveals an ancient whole-genome duplication during arachnid evolution. BMC Biol. 15(1):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selden PA, Shear WA, Sutton MD.. 2008. Fossil evidence for the origin of spider spinnerets, and a proposed arachnid order. Proc Natl Acad Sci U S A. 105(52):20781–20785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setton EVW, Hendrixson BE, Sharma PP.. 2019. Embryogenesis in a Colorado population of Aphonopelma hentzi (Girard, 1852) (Araneae: Mygalomorphae: Theraphosidae): establishing a promising system for the study of mygalomorph development. J Arachnol. 47(2):209–216. [Google Scholar]
- Setton EVW, Sharma PP.. 2018. Cooption of an appendage-patterning gene cassette in the head segmentation of arachnids. Proc Natl Acad Sci U S A. 115(15):E3491–E3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma PP, Kaluziak ST, Pérez-Porro AR, González VL, Hormiga G, Wheeler WC, Giribet G.. 2014. Phylogenomic interrogation of Arachnida reveals systemic conflicts in phylogenetic signal. Mol Biol Evol. 31(11):2963–2984. [DOI] [PubMed] [Google Scholar]
- Sharma PP, Schwager EE, Extavour CG, Wheeler WC.. 2014. Hox gene duplications correlate with posterior heteronomy in scorpions. Proc Biol Sci. 281(1792):20140661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma PP, Wheeler WC.. 2013. Revenant clades in historical biogeography: the geology of New Zealand predisposes endemic clades to root age shifts. J Biogeogr. 40(8):1609–1618. [Google Scholar]
- Sharma PP, Zardus JD, Boyle EE, González VL, Jennings RM, McIntyre E, Wheeler WC, Etter RJ, Giribet G.. 2013. Into the deep: a phylogenetic approach to the bivalve subclass Protobranchia. Mol Phylogenet Evol. 69(1):188–204. [DOI] [PubMed] [Google Scholar]
- Shingate P, Ravi V, Prasad A, Tay B-H, Garg KM, Chattopadhyay B, Yap L-M, Rheindt FE, Venkatesh B.. 2020. Chromosome-level assembly of the horseshoe crab genome provides insights into its genome evolution. Nat Commun. 11(1):2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shishido CM, Woods HA, Lane SJ, Toh MWA, Tobalske BW, Moran AL.. 2019. Polar gigantism and the oxygen–temperature hypothesis: a test of upper thermal limits to body size in Antarctic pycnogonids. Proc R Soc B. 286(1900):20190124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siveter DJ, Sutton MD, Briggs DEG, Siveter DJ.. 2004. A Silurian sea spider. Nature 431(7011):978–980. [DOI] [PubMed] [Google Scholar]
- Soler-Membrives A, Linse K, Miller KJ, Arango CP.. 2017. Genetic signature of Last Glacial Maximum regional refugia in a circum-Antarctic sea spider. R Soc Open Sci. 4(10):170615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starrett J, Derkarabetian S, Hedin M, Bryson RW Jr, McCormack JE, Faircloth BC.. 2017. High phylogenetic utility of an ultraconserved element probe set designed for Arachnida. Mol Ecol Resour. 17(4):812–823. [DOI] [PubMed] [Google Scholar]
- Stock JH. 1994. Indo-West Pacific Pycnogonida collected by some major oceanographic expeditions. Beaufortia 1:17–77. [Google Scholar]
- Tetlie OE, Dunlop JA.. 2008. Geralinura carbonaria (Arachnida: Uropygi) from Mazon Creek, Illinois, USA, and the origin of subchelate pedipalps in whip scorpions. J Paleontol. 82(2):299–312. [Google Scholar]
- Waddington J, Rudkin DM, Dunlop JA.. 2015. A new mid-Silurian aquatic scorpion - one step closer to land? Biol Lett. 11(1):20140815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waloszek D, Dunlop JA.. 2002. A larval sea spider (Arthropoda: Pycnogonida) from the Upper Cambrian ‘Orsten’ of Sweden, and the phylogenetic position of pycnogonids. Palaeontology 45(3):421–446. [Google Scholar]
- Wang B, Dunlop JA, Selden PA, Garwood RJ, Shear WA, Müller P, Lei X.. 2018. Cretaceous arachnid Chimerarachne yingi gen. et sp. nov. illuminates spider origins. Nat Ecol Evol. 2(4):614–622. [DOI] [PubMed] [Google Scholar]
- Wendruff AJ, Babcock LE, Wirkner CS, Mukulic DG.. 2020. A Silurian ancestral scorpion with fossilised internal anatomy illustrating a pathway to arachnid terrestrialisation. Sci Rep. 10:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe JM, Breinholt JW, Crandall KA, Lemmon AR, Lemmon EM, Timm LE, Siddall ME, Bracken-Grissom HD.. 2019. A phylogenomic framework, evolutionary timeline and genomic resources for comparative studies of decapod crustaceans. Proc R Soc B. 286(1901):20190079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe JM, Daley AC, Legg DA, Edgecombe GD.. 2016. Fossil calibrations for the arthropod tree of life. Earth Sci Rev. 160:43–110. [Google Scholar]
- Wood HM, González VL, Lloyd M, Coddington J, Scharff N.. 2018. Next-generation museum genomics: phylogenetic relationships among palpimanoid spiders using sequence capture techniques (Araneae: Palpimanoidea). Mol Phylogenet Evol. 127:907–918. [DOI] [PubMed] [Google Scholar]
- Woods HA, Lane SJ, Shishido C, Tobalske BW, Arango CP, Moran AL.. 2017. Respiratory gut peristalsis by sea spiders. Curr Biol. 27:R623–R641. [DOI] [PubMed] [Google Scholar]
- Woods HA, Moran AL, Arango CP, Mullen L, Shields C.. 2009. Oxygen hypothesis of polar gigantism not supported by performance of Antarctic pycnogonids in hypoxia. Proc R Soc B. 276(1659):1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete data set, including sequence alignments, annotated tree files, chronograms, and embryonic transcriptomic assemblies, have been deposited at Dryad Digital Repository under URL https://datadryad.org/stash/share/HsFKzSdD00rwa5BQSEf9km6dXuURuiAtOwv0w0QVDUg. Raw read data were deposited at NCBI Sequence Read Archive. Loci from targeted sequencing were deposited in NCBI GenBank.