Abstract
The exocrine-gland secreting peptide (ESP)gene family encodes proteinaceous pheromones that are recognized by the vomeronasal organ in mice. For example, ESP1 is a male pheromone secreted in tear fluid that regulates socio-sexual behavior, and ESP22 is a juvenile pheromone that suppresses adult sexual behavior. The family consists of multiple genes and has been identified only in mouse and rat genomes. The coding region of a mouse ESP gene is separated into two exons, each encoding signal and mature sequences. Here, we report the origin and evolution of the ESP gene family. ESP genes were found only in the Muridea and Cricetidae families of rodents, suggesting a recent origin of ESP genes in the common ancestor of murids and cricetids. ESP genes show a great diversity in number, length, and sequence among different species as well as mouse strains. Some ESPs in rats and golden hamsters are expressed in the lacrimal gland and the salivary gland. We also found that a mature sequence of an ESP gene showed overall sequence similarity to the α-globin gene. The ancestral ESP gene seems to be generated by recombination of a retrotransposed α-globin gene with the signal-encoding exon of the CRISP2 gene located adjacent to the ESP gene cluster. This study provides an intriguing example of molecular tinkering in rapidly evolving species-specific proteinaceous pheromone genes.
Keywords: pheromone, gene family evolution, globin, rodents, molecular tinkering
Introduction
Pheromones are chemical signals that have evolved for the communication between individuals of the same species and to trigger a specific reaction, such as an innate behavior or an endocrine or emotional change (Liberles 2014; Wyatt 2014a). Not only small volatile molecules but also nonvolatile peptides and proteins are used as pheromones. Many invertebrates and vertebrates, on land as well as in water, utilize peptides and proteins as pheromones (Wyatt 2014b).
The exocrine-gland secreting peptide 1 (ESP1) is the first proteinaceous pheromone identified in mammals (Kimoto et al. 2005). The ESP1 is a 7 kDa protein secreted from the extraorbital lacrimal gland (ELG) into tear fluid of male mice. It stimulates the vomeronasal organ (VNO) in female mice via a specific receptor Vmn2r116 (also named V2Rp5) and enhances lordosis, a female sexual receptive behavior upon male mounting (Haga et al. 2010). Moreover, ESP1 enhances male aggressiveness in conjunction with unfamiliar male urine, and further acts as an autostimulatory factor that enhances male aggressiveness by self-exposure (Hattori et al. 2016). ESP1 is also a key factor involved in the Bruce effect, in which pregnancy of a recently pregnant female mouse is blocked upon exposure to unfamiliar males (Hattori et al. 2017). Among laboratory mouse strains, the pregnancy block is observed in between a particular combination of different strains but is not observed within the same strain. Some strains secrete ESP1 but others do not (Kimoto et al. 2007; Haga et al. 2010); hence, it was proposed that ESP1 not only functions as a pheromone but also may serve as a signature molecule that conveys the strain information (Hattori et al. 2017).
ESP22 is another proteinaceous pheromone in mice, the gene of which has a sequence similarity to ESP1. ESP22 is released into tear fluid of 2- to 3-week-old mice of both sexes and inhibits adult sexual behavior via the specific receptor Vmn2r115 (also named V2Rp4), which is closely related (87% identical) to the ESP1 receptor, Vmn2r116 (Ferrero et al. 2013; Osakada et al. 2018).
In 2007, by using the genome sequence available at that time, we reported that the mouse ESP multigene family consists of 38 genes including 24 putatively functional genes and 14 pseudogenes (Kimoto et al. 2007). Moreover, 10 ESP genes were found in rats but no ESP gene was found in the human genome. ESP genes consist of 3–5 exons and the coding sequence is present within the last two exons; the initiation codon and a coding region of a putative signal peptide are located in the second from the last exon, and the mature peptide is encoded in the last exon. Out of the 24 intact ESP genes, 15 were expressed in the ELG, Harderian gland (HG), and/or submaxillary gland (SMG) of sexually mature BALB/c mice. Moreover, recombinant proteins for all the 15 expressed ESPs evoked an electrical response in the VNO. Therefore, although detailed functions of ESPs other than ESP1 and ESP22 are unknown, it can be said that the members of the ESP multigene family in mice may possess pheromonal functions.
It has neither been known which other species have ESP genes in their genomes, nor has it yet been clear whether ESPs have a pheromonal function in species other than mice. In this study, we performed extensive homology searches of ESP genes against over 100 available mammalian genomes. We found that the origin of ESP genes could be traced back to the common ancestor of the family Muridae (including mice and rats) and the family Cricetidae (including hamsters). We also examined the expression of these nonmouse ESPs in the lacrimal and salivary glands. Moreover, we propose a possible evolutionary scenario for the origin of the ESP gene family.
Results
Identification of ESP Genes in Rodents
In mice, the coding sequence of an ESP gene is separated into two exons by a phase 0 intron. The boundary between the two exons nearly corresponds to the boundary between signal and mature peptides of an ESP, though the signal peptide is one amino acid shorter than the sequence encoded by the first of the two exons (Kimoto et al. 2007). In this paper, we call the coding sequence of each of the two exons as “signal sequence” and “mature sequence,” respectively. We identified a signal sequence and a mature sequence separately, because the combination of signal and mature sequences was uncertain (see below).
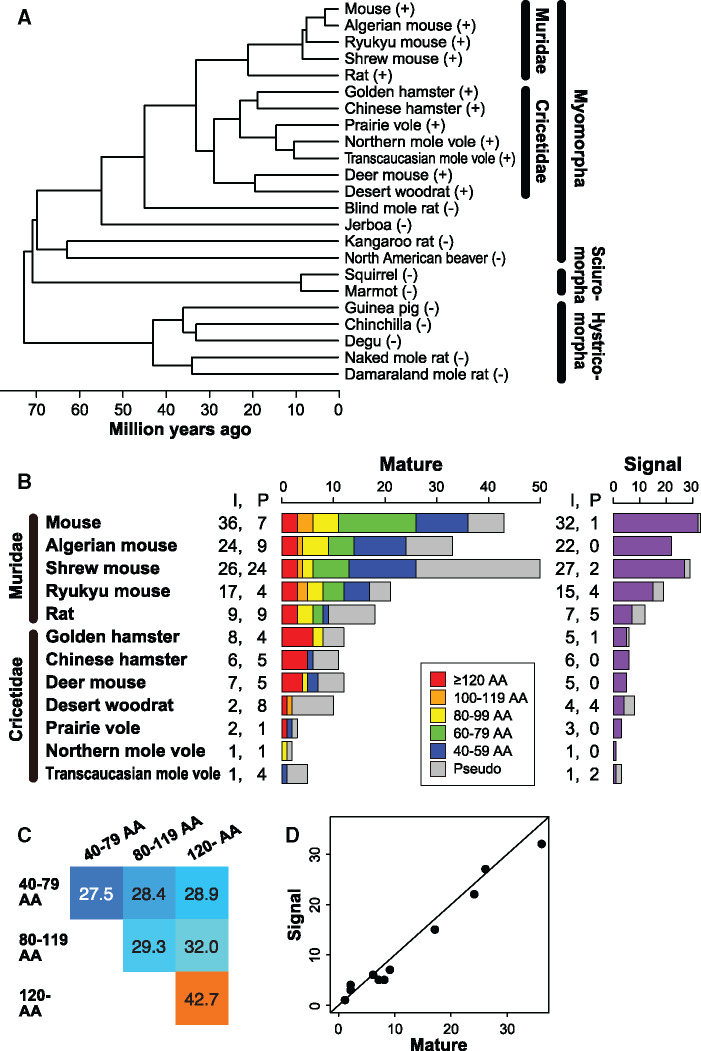
We first examined the presence or absence of mature sequences in the whole genome sequences of 23 rodents (supplementary table S1, Supplementary Material online) and 87 nonrodent mammals (supplementary table S2, Supplementary Material online). The phylogeny of the 23 rodent species examined is shown in figure 1A. As a result, we found mature sequences in all species of the families Muridae and Cricetidae, both of which belong to the suborder Myomorpha in the order Rodentia. However, no mature sequence was identified in the genomes of any other rodent species or nonrodent species. It is therefore suggested that the origin of ESP genes can be traced back to the common ancestor of murids and cricetids that was present ∼33 Ma (fig. 1A).
Fig. 1.
ESP genes in the Muridae and Cricetidae genomes. (A) Presence and absence of ESP genes in 23 rodent species. The species with and without ESP genes are shown with a plus and a minus sign, respectively. Divergence times were obtained from TimeTree (Kumar et al. 2017). (B) Number of mature and signal sequences identified from five Muridae and seven Cricetidae species. “I” and “P” indicate intact sequences and pseudogenes, respectively. The left bar graph is colored according to the length of intact mature sequences. AA, amino acids. (C) Mean amino acid sequence identities (%) among mature sequences according to their lengths. For example, there are 32 mature sequences containing 120 or more amino acids, and the mean amino acid sequence identity based on all possible pairwise comparisons is 42.7%. Each pairwise alignment was constructed by using ClustalW2 (Larkin et al. 2007), and the amino acid sequence identity was calculated after excluding all alignment gaps to exclude the effect of the difference in length between the two compared sequences. (D) Strong correlation between the number of intact mature sequences and that of intact signal sequences in each species (r = 0.989, P < 10−8).
The number of mature sequences identified from 12 murid and cricetid species is shown in figure 1B. In total, we identified 139 intact mature sequences from the 12 rodent species (supplementary fig. S1A, Supplementary Material online). Each sequence was named by using an abbreviation of the species name and the gene number (supplementary table S3, Supplementary Material online). Among the 12 species, the mouse Mus musculus has the largest number of intact mature sequences (36). This number is considerably larger than the one in the previous report (24) (Kimoto et al. 2007). Each of the 43 mature sequences including pseudogenes was named MmESP1–37 according to a previous study (Kimoto et al. 2007). Some mature sequences (e.g., MmESP13a–13d) were newly identified in this study from the latest versions of the mouse and rat genomes and were depicted by a suffix such as a, b, etc.
The lengths of mature sequences were highly variable, ranging from 42 to 131 amino acids. Murids tend to have a larger number of mature sequences than cricetids. However, the lengths of mature sequences are significantly longer in cricetids (mean ± SD, 104.3 ± 31.2 amino acids) than in murids (76.9 ± 26.0 amino acids; P < 0.001 by the Wilcoxon rank-sum test; supplementary fig. S2A, Supplementary Material online). We also found that longer mature sequences tend to be more conserved in amino acid sequence than shorter mature sequences (fig. 1C). In fact, although the overall amino acid sequences are not well conserved among all the mature sequences identified, the multiple alignment of mature sequences that were 120 amino acids or longer in length showed relatively high conservation in amino acid sequence except for MmESP12 (supplementary fig. S1B, Supplementary Material online).
We also identified signal sequences in 12 murid and cricetid species (fig. 1B and supplementary fig. S3, Supplementary Material online). The lengths of signal sequences were 16–23 amino acids. Although the signal sequences were identified separately from the mature sequences, the number of signal sequences found in each species is nearly the same as that of the mature sequences (fig. 1D), and most of the signal sequences are located near to the mature sequences.
Cluster Organization
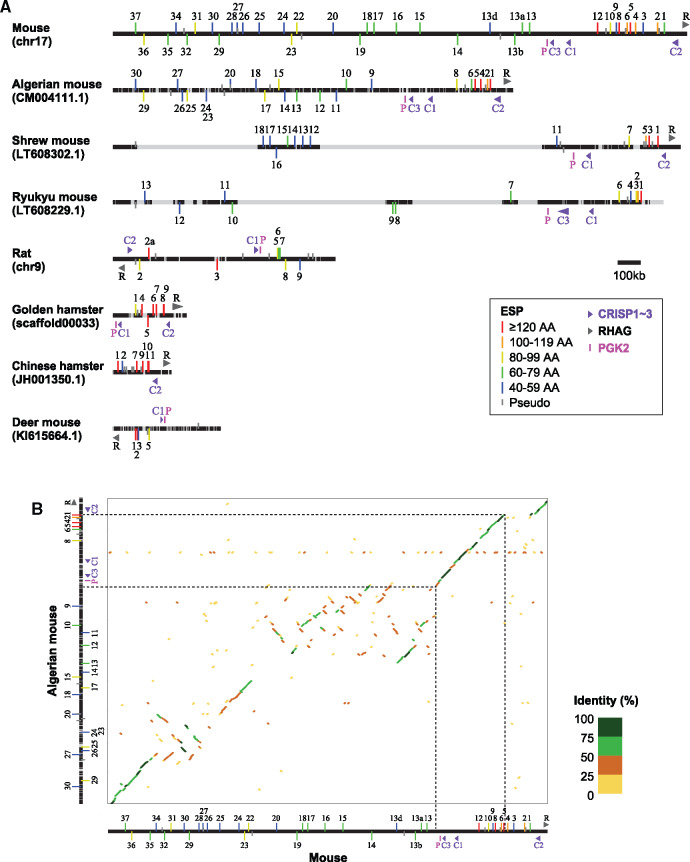
We compared the organization of ESP gene clusters among eight species having three or more intact mature sequences (fig. 2A). In mice, all of the 43 mature sequences including pseudogenes are located in one cluster spanning a ∼2.4 Mb region on chromosome 17. In shrew mice and Ryukyu mice, there are large sequencing gaps within the ESP gene clusters, suggesting that these species may have more ESP genes than those shown in figure 1B. In rats, eight out of nine intact mature sequences are located in one genomic cluster on chromosome 9. One intact mature sequence (RnESP15) is on chromosome 14, but its nucleotide sequence is 100% identical to that of RnESP3 on chromosome 9, suggesting a possibility of genome misassembly. In Chinese hamsters, all ESP genes are located on one scaffold named JH001350.1, whereas in golden hamsters, all intact mature sequences but one are located on one scaffold (scaffold00033). In deer mice, ESP genes reside in several scaffolds; the scaffold KI615664.1 contains four out of seven intact mature sequences.
Fig. 2.
Organization of ESP gene cluster in eight rodent species. (A) For each species, a chromosome or a scaffold name containing the ESP gene cluster is shown in parentheses. The black horizontal line represents the DNA sequence including the entire ESP gene cluster with 100-kb sequences in both directions. The horizontal line in gray indicates a region in which the nucleotide sequences are undetermined. The vertical line attached to the black horizontal line represents a mature sequence of an ESP gene, and the vertical lines above and below the black horizontal line indicate a sequence encoded in plus and minus strands, respectively. The gray vertical line represents a pseudogene, whereas the vertical line in red, orange, yellow, green, and blue represents an intact sequence (a color code indicates the length of a sequence). Each intact mature sequence is shown with a gene number (e.g., MmESP1 is depicted as “1”). The locations of CRISP1–3, RHAG, and PGK2 genes are also shown. (B) Dot plot for the comparison of ESP gene clusters between mice and Algerian mice constructed by D-Genies (Cabanettes and Klopp 2018).
Within the mouse ESP gene cluster, there is a large interval (∼300 kb) between MmESP12 and MmESP13. The mouse ESP gene cluster can be separated into two parts at the interval, and we named them as upstream part (MmESP13–37) and downstream part (MmESP1–12). The intact mature sequences in the downstream part (mean ± SD, 95.2 ± 29.3 amino acids) are significantly longer than those in the upstream part (65.5 ± 13.5 amino acids; P < 0.01 by the Wilcoxon rank-sum test; supplementary fig. S2B left, Supplementary Material online), whereas the intervals between intact mature sequences are significantly shorter in the downstream part (mean ± SD, 32.7 ± 17.2 kb) than in the upstream part (69.9 ± 46.3 kb; P < 0.02 by the Wilcoxon rank-sum test; supplementary fig. S2B right, Supplementary Material online). However, the lengths of intact mature sequences and the intervals of intact mature sequences in the downstream part are not significantly different from those in the clusters of three cricetid species as shown in figure 2A (P > 0.05 by the Wilcoxon rank-sum test; supplementary fig. S2B, Supplementary Material online): The lengths of intact mature sequences in the three cricetids are 111.8 ± 26.7 amino acids, and the intervals of intact mature sequences are 22.8 ± 17.2 kb (note that this value is inaccurate because of the presence of undetermined nucleotides in the genome sequences of these species).
There are three non-ESP genes within the interval between the upstream part and the downstream part in the mouse ESP gene cluster: phosphoglycerate kinase 2 (PGK2), cysteine-rich secretory proteins 3 (CRISP3), and CRISP1. CRISP genes form a multigene family, and there are four CRISP genes in the mouse genome, CRISP1–4. CRISP2 is located to the immediate downstream of MmESP1. Therefore, the downstream part (MmESP1–12) is surrounded by two CRISP genes, CRISP1 and CRISP2. Rh-associated glycoprotein (RHAG) gene is located next to the CRISP2 gene.
We identified CRISP, PGK2, and RHAG genes in eight rodent species as shown in figure 2A. We found that these genes are located close to the ESP gene cluster in all the species examined. A phylogenetic analysis showed that the gene duplication between CRISP1 and CRISP3 occurred before the divergence between mice and Ryukyu mice, and probably after the divergence between mice and shrew mice (supplementary fig. S4, Supplementary Material online). Therefore, both CRISP1 and CRISP3 genes in mice are orthologous to a CRISP1 gene in rats and cricetids. In cricetid species, the entire ESP gene cluster is surrounded by PGK2/CRISP1 and CRISP2/RHAG (fig. 2A). Therefore, the downstream part in mice (MmESP1–12) appears to correspond to the entire ESP genes clusters in cricetids. From these observations, it is suggested that the downstream part in the mouse ESP gene cluster is the “prototype” of an ESP gene cluster, whereas the upstream part is murid-specific.
Consistent with the above notion, the downstream part is more conserved than the upstream part. The dot plot for the comparison of ESP gene clusters between mice and Algerian mice demonstrates that the downstream part (including PGK2, CRISP1–3, and RHAG genes) is well conserved between the two species, though the genomic region of MmESP1–4 is absent from the Algerian mouse genome (fig. 2B). However, the upstream part, especially the central region including MmESP13–20, is poorly conserved, and it is likely that many genomic rearrangements have occurred in this region. We also found that there were two mouse-specific segmental duplications in the central region: One is in the genomic region containing MmESP13–13b and that containing MmESP17–19, and the other is in the genomic region containing MmESP15 and that containing MmESP16 (supplementary fig. S5, Supplementary Material online). The amino acid sequences for the corresponding mature sequences are highly similar (supplementary fig. S6, Supplementary Material online). This observation suggests that the upstream part was recently generated in a mouse-specific manner.
ESP Genes in Various Mouse Strains
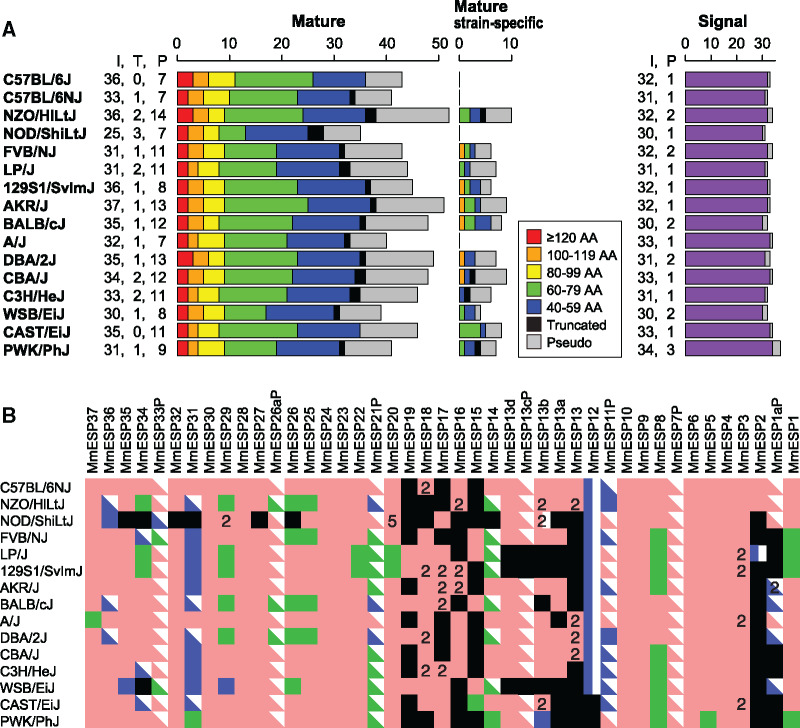
To see the diversity of ESP gene repertoires among different mouse strains, we identified ESP genes from de novo assemblies for 15 inbred mouse strains in addition to the reference genome for C57BL/6J (Lilue et al. 2018). The results showed that the number of intact mature sequences varied among different strains (fig. 3A). Because the quality of genome assemblies for these 15 strains is much lower (N50 < 25,000) as compared with that of the reference genome (supplementary table S4, Supplementary Material online), it is possible that some ESP genes are missing due to incompleteness of genome assembly. In fact, some mature sequences are truncated at the coding sequence.
Fig. 3.
Comparison of ESP gene repertoires among 16 mouse inbred strains. (A) Number of mature and signal sequences identified in the reference mouse genome (C57BL/6J) and other 15 inbred strains. “I,” “T,” and “P” indicate intact sequences, truncated sequences, and pseudogenes, respectively. The bar graphs on the left and in the middle are colored according to the length of intact mature sequences. AA, amino acids. (B) ESP mature sequence repertoires in 15 inbred mouse strains were compared with that of the reference genome (C57BL/6J). The square, rectangle, and right triangle represent an intact sequence, truncated sequence, and pseudogene, respectively. Pink, green, and blue colors indicate sequences that are identical to those in the C57BL/6 genome, sequences containing nonsynonymous SNPs, and sequences containing indels, respectively. The black box indicates a sequence that is missing in a given strain. The numbers on the pink squares show that the sequences are encoded in multiple locations in the genome.
We found that mature sequences of ESP genes are diverse in amino acid sequence among different strains (fig. 3B and supplementary table S5, Supplementary Material online). Of the 36 intact mature sequences in the reference mouse genome, only nine sequences (MmESP3, 4, 6, 9, 10, 23, 24, 28, and 30) are identical in amino acid sequence among all the strains examined. Thirteen sequences (MmESP1, 5, 8, 14, 15, 20, 22, 25, 26, 29, 31, 34, and 37) including ESP1 and ESP22 contain nonsynonymous single nucleotide polymorphisms (SNPs) (supplementary fig. S7, Supplementary Material online). Four sequences (MmESP14, 31, 34, and 36) are segregating pseudogenes; they are intact in the reference genome but are pseudogenes in some other strains. In addition, MmESP11P is another segregating pseudogene: it is a pseudogene in the reference genome but is intact in some strains.
Several mature sequences are missing in the central region in some strains (fig. 3B), which is consistent with the observation that the extent of overall conservation is relatively low in the central region of the mouse ESP gene cluster. However, it should be noted that ESP genes may be missing in the central region due to relatively low quality of genome assembly in this region.
Some mature sequences are strain-specific (fig. 3A). In all, we identified additional 10 mature sequences that are present in nonreference strains but are absent from the reference genome. These sequences were named MmESP39–48P. Six out of the 10 sequences are intact in at least one strain (supplementary fig. S8, Supplementary Material online).
Origin of ESP Genes
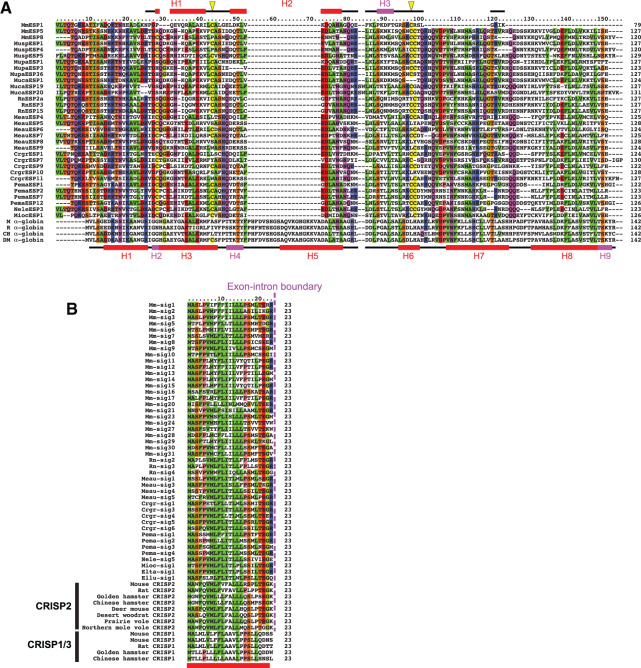
During the process of homology searches, we found that a mature ESP sequence named CrgrESP7 that was newly identified in this study from the Chinese hamster genome showed a weak (Blast E value is 2e−04) similarity to the α-globin gene encoded in Chinese hamster scaffold JH017478.1. In fact, a BlastP search (Altschul et al. 1997) using CrgrESP7 as a query against the nonredundant protein sequences in GenBank database suggested that CrgrESP7 was similar to both known ESP genes and α-globin genes: for example, the E values to RnESP3 in rats, MmESP5 in mice, marmot α-globin, and mouse α-globin are 2e−23, 1e−16, 1e−08, and 2e−06, respectively. Note that mouse and rat ESP genes that have been reported so far did not show any significant similarity to α-globin genes (supplementary fig. S9, Supplementary Material online). However, when we used only mature sequences that are 120 amino acids or longer in length, an overall similarity to α-globin genes became evident in a multiple alignment (fig. 4A).
Fig. 4.
Similarity of ESP genes to α-globin and CRISP genes. (A) Multiple alignment of MmESP1, 31 long (≥120 amino acids) mature sequences of ESP genes, and four α-globin genes from the mouse (M; GenBank accession number, NP_001077424.1), rat (R; NP_001013875.1), Chinese hamster (CH; ERE82205.1), and deer mouse (DM; XP_006978909.1). All long (≥120 amino acids) mature sequences were used except MmESP12 which shows an exceptionally low sequence conservation as compared with the other long mature sequences (supplementary fig. S1B, Supplementary Material online). The diagrams above and below the multiple alignment represent secondary structures of MmESP1 (Yoshinaga et al. 2013) and mouse α-globin (PDB code, 3HRW), respectively. Red and magenta rectangles indicate α-helix and 310-helix, respectively. The horizontal line shows the portion for which the 3D structures were determined; for MmESP1, the NMR structure of 55-amino-acid-long C-terminal portion was previously reported (Yoshinaga et al. 2013). Yellow triangles in the upper diagram indicate two cysteine residues involved in a disulfide bond in MmESP1, which are well conserved across ESP genes. Multiple alignment was constructed by MAFFT (Katoh et al. 2005). The amino acids that are conserved among half or more of all sequences, at a given site, have been shown in color according to their chemical properties. (B) Multiple alignment of 45 signal sequences consisting of 23 amino acids in nine rodent species along with the first 23 amino acid sequences of CRISP1–3 in several rodent species. A magenta dotted line represents the exon–intron boundary, which is common for ESP and CRISP2 genes. A red bar below the alignment indicates a signal sequence.
The 3D structure of the C-terminal portion of an MmESP1 protein has been determined by NMR (Yoshinaga et al. 2013). The structure contains three helices: two α-helices (H1 and H2) and one 310-helix (H3; fig. 4A). An α-globin protein has also a helix-rich structure with six α-helices and three 310-helices. The locations of the helices in MmESP1 are generally in good agreement with those in α-globin (fig. 4A). Altogether, these observations suggest that mature sequences of ESP genes may have originated from α-globin gene.
We also found that the signal sequence of ESP genes was similar to the signal of a CRISP2 gene (fig. 4B) which is located adjacent to the ESP gene cluster (fig. 2A). In the process of identification of ESP signal sequences, homology searches detected CRISP2 signal sequences as well as genuine ESP signal sequences, and thus the CRISP2 signal sequences were eliminated from our data set of the signal sequences of ESP genes in the final step of our process (see Materials and Methods section). In fact, the CRISP2 signal sequences are indistinguishably similar to the ESP signal sequences, whereas CRISP1/3 signals are less similar to ESP signals (fig. 4B). In addition, the exon–intron boundary is located at exactly the same position for ESP and CRISP2 genes, whereas it is different for CRISP1/3 genes (fig. 4B). These observations strongly suggest that the signal sequences of ESP genes were originated from the signal-encoding exon of a CRISP2 gene.
Molecular Evolution of the ESP Gene Family
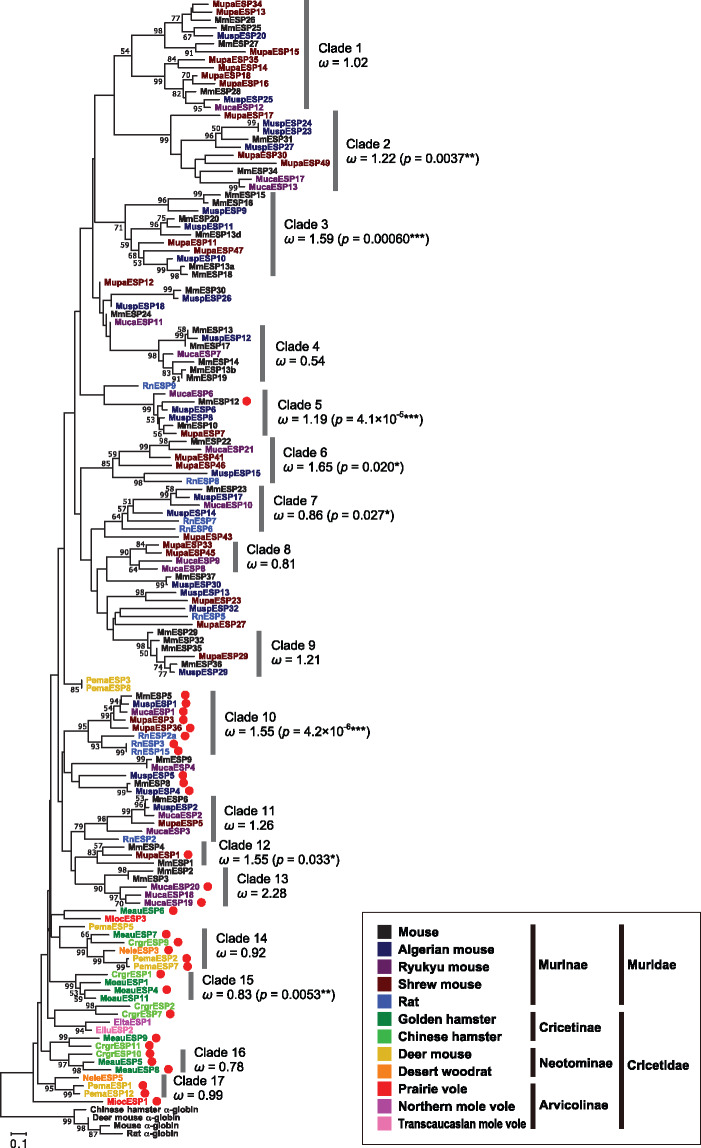
Under the assumption that the mature sequence has originated from α-globin gene, we can root the phylogenetic tree of mature sequences by using α-globin genes as the outgroup. Although the bootstrap values are generally low because mature sequences are short, the phylogenetic tree showed the following clear tendency (fig. 5): Cricetid mature sequences are located the closest to the root, followed to long (≥120 amino acids) mature sequences in murids, and short mature sequences in murids are located at the most outside of the tree. Among long mature sequences, MmESP12 is exceptionally located to the outside of the tree, but it has relatively low amino acid sequence conservation as compared with the other long mature sequences (supplementary fig. S1B, Supplementary Material online). Overall, the results suggest that mature sequences of ESP genes in cricetids tend to retain the ancestral sequence, whereas short mature sequences in murids appear to have rapidly diverged by recent gene duplications.
Fig. 5.
Neighbor-joining phylogenetic tree (Saitou and Nei 1987) constructed by MEGA7 (Kumar et al. 2016) for the amino acid sequences of 139 ESP mature sequences in 12 rodent species together with four α-globin genes used to determine the root. The evolutionary distances were computed using the Poisson correction method after all of the alignment gaps were eliminated for each sequence pair. Each sequence name was colored according to the color code for the species. The red filled circle represents a long (≥120 amino acids) mature sequence. Bootstrap values obtained from 500 replicates are shown for nodes with >50% bootstrap supports. Phylogenetic clades each of which is supported with a > 50% bootstrap value and contains three or more genes is numbered 1–17. The overall ω value calculated by PAML (Yang 2007) under model M0 is presented for each clade. The P value of the χ2 test for rejecting the null model of no positive selection (M7) over the alternative model of positive selection (M8) is also shown in parentheses for P < 0.05.
To evaluate selective constraints working on ESP genes, we calculated the ratio of the rate of nonsynonymous substitutions (dN) to that of synonymous substitutions (dS), ω (=dN/dS). Because ESP genes are highly divergent in sequence, we identified phylogenetic clades each of which is supported with a > 50% bootstrap value and contains three or more sequences and analyzed the ESP mature sequences in each clade separately. There are 17 such clades (fig. 5). The overall ω value in each clade is generally high, suggesting relaxation of selective constraints or diversifying selection, or both in the evolution of ESP genes. We then performed likelihood tests implemented in the PAML package (Yang 2007) to see the possibility of the occurrence of positive selection. Signals of positive selection were detected for eight of the 17 clades examined with a statistical significance (P < 0.05, fig. 5 and supplementary table S6, Supplementary Material online). Amino acid sites that were suggested to be under positive selection are distributed among the entire mature sequence (supplementary fig. S10 and table S6, Supplementary Material online).
Expression of Nonmouse ESPs
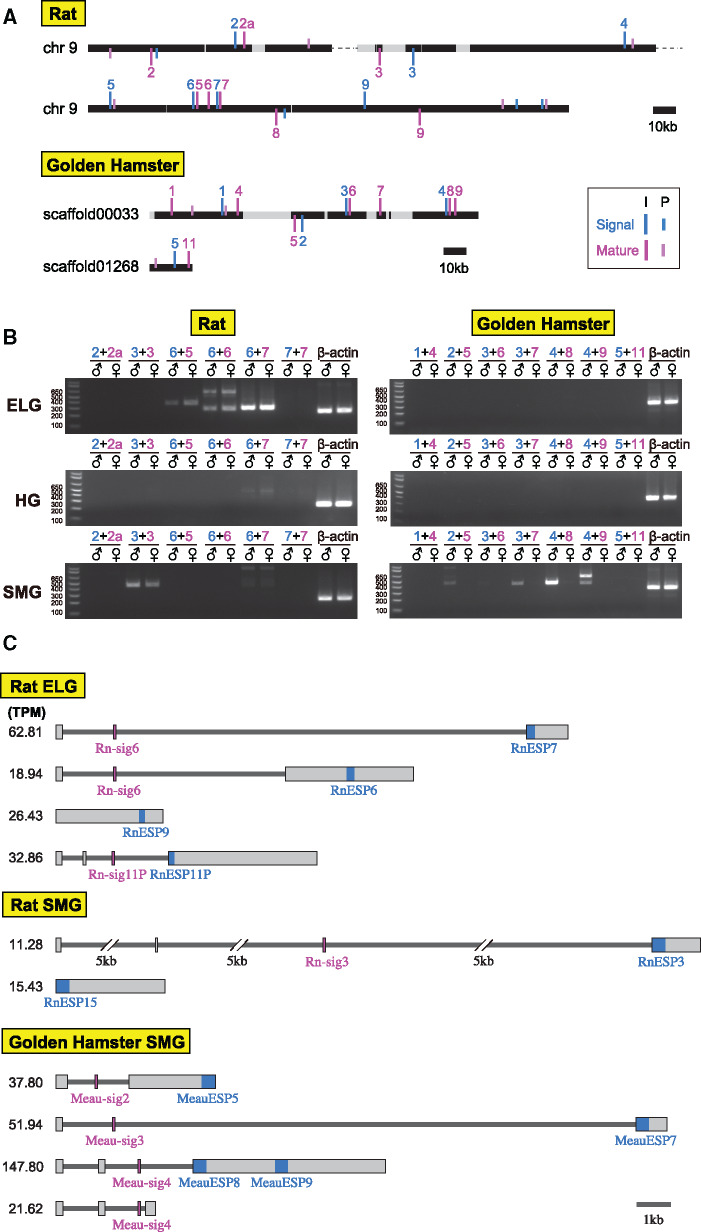
We performed expression analyses of ESP genes in nonmouse rodents. We first conducted reverse transcriptase-polymerase chain reaction (RT-PCR) for the ELG and HG that secrete tear fluid and the SMG that secretes saliva in rats and golden hamsters. Forward and reverse primers were designed on the signal and mature sequences, respectively (supplementary table S7, Supplementary Material online). Because a combination of signal and mature sequences in an ESP gene is unclear, we considered all possible combinations of signal and mature sequences that are encoded in the same strand and are located within a short distance (<20 kb; fig. 6A).
Fig. 6.
Expression profiles of ESP genes in rats and golden hamsters. (A) Genomic locations of ESP single and mature sequences on chromosome 9 in rats (upper) and on scaffold00033 and scaffold01268 in golden hamsters (lower). Signal and mature sequences are shown in sky blue and magenta vertical lines, respectively, along with the names (e.g., “3” indicates RnESP3 and Rn-sig3; see supplementary table S3, Supplementary Material online). I and P indicate intact sequences and pseudogenes, respectively. (B) Expression profile of ESP genes in the ELG, HG, and SMG of male and female rats (left) and golden hamsters (right) determined by RT-PCR. (C) Results of RNA-seq analyses for the ELG and SMG of the male rat and the male golden hamster. Reconstructed ESP transcript models with the TPM values >10 are shown. All transcript models mapped to genomic regions encoding ESP genes are summarized in supplementary table S8, Supplementary Material online. No ESP transcripts were obtained for the golden hamster ELG. Mature and signal sequences are depicted in sky blue and magenta, respectively.
The results demonstrate that RnESP5–7 were expressed in the ELG of both male and female rats (fig. 6B). Interestingly, we found that one signal sequence, Rn-sig6, is combined with three different mature sequences (RnESP5–7), whereas the expression for the combination of Rn-sig7 with RnESP7 was not detected. RnESP3 was expressed in the SMG of both male and female rats. RnESP9 gene was amplified by RT-PCR using both of the primers designed within a mature sequence (supplementary fig. S11, Supplementary Material online), but it was not amplified using primers on a signal sequence and on a mature sequence. In golden hamsters, MeauESP5, 7–9 were specifically expressed in the male SMG. The signal sequence Meau-sig4 is commonly used for both MeauESP8 and 9 (fig. 6B). No ESP gene expression was found in the ELG and HG of both male and female hamsters. We also examined the expression of ESP genes in various tissues (eye, brain, lung, heart, liver, kidney, and testis) of rats and golden hamsters, but we did not detect expression from any tissues (supplementary fig. S12, Supplementary Material online).
Interestingly, we found a clear relationship between the expression of ESP genes and their lengths. All ESP genes with a mature sequence of 100 amino acids or less are exclusively expressed in the ELG, whereas those with a mature sequence of 120 amino acids or more are exclusively expressed in the SMG with the exception of MmESP5. ESP genes with a mature sequence of 100–120 amino acids are expressed in both the ELG and SMG (supplementary table S9, Supplementary Material online). The lengths of mature sequences exclusively expressed in SMG (mean ± SD, 125.3 ± 3.1 amino acids) are significantly longer than those in ELG (71.3 ± 17.8 amino acids; P < 0.001 by the Wilcoxon rank-sum test; supplementary fig. S13, Supplementary Material online).
To further investigate the expression of ESP genes in nonmouse species, we conducted RNA-seq analyses for the ELG and SMG of the male rat and the male golden hamster. Transcript models mapped to the genomic regions encoding EPS genes are shown in figure 6C and supplementary table S8, Supplementary Material online. The results are generally in good agreement with those obtained by the RT-PCR. For example, Rn-sig6 is expressed together with RnESP6 and RnESP7 in the rat ELG; Meau-sig3 is expressed together with MeauESP7, and Meau-sig4 is expressed with MeauESP8 and MeauESP9 in the golden hamster SMG. No ESP transcripts were detected in the golden hamster ELG, which is consistent with the results of RT-PCR. Interestingly, some mature sequences (e.g., RnESP6 and MeanESP9) are encoded at the middle of an exon, suggesting that they are transcribed, but not translated. RnESP9 and RnESP15 are transcribed without signal sequences. RnESP11P is also expressed, though it contains an interrupting stop codon. In summary, at least some ESP mature sequences are expressed together with a signal sequence in the ELG and SMG of rats and golden hamsters, suggesting that these ESPs are secreted into tear and saliva.
We next assessed whether rat and golden hamster ESP peptides could stimulate the vomeronasal sensory neurons in each species. We produced a recombinant protein for each ESP in Escherichia coli (supplementary fig. S14, Supplementary Material online) and tested their activities by c-Fos in situ hybridization. However, the number of c-Fos positive cells in the AOB mitral/tufted cells of both male and female rats stimulated with recombinant rat ESPs and those of both male and female golden hamsters stimulated with recombinant golden hamster ESPs were not significantly different from the control (supplementary fig. S15, Supplementary Material online). Although we observed a small increase in the number of c-Fos positive cells when male rats were stimulated with recombinant RnESP3 and RnESP5 proteins, the activities, if any, were much weaker than those for mice ESPs (Haga et al. 2010). We also examined the possibility of interspecific activity of the ESPs, because some proteins secreted by rats, such as major urinary protein 13 (MUP13) and cystatin-related protein 1, activate the mouse VNO (Papes et al. 2010; Tsunoda et al. 2018). We exposed rat ESPs and golden hamster ESPs to mice and investigated their vomeronasal-stimulating activity. There was no responsive cell observed in the VNO of mice upon their stimulation with either the mixture of rat ESPs or the mixture of golden hamster ESPs (supplementary fig. S16, Supplementary Material online).
Discussion
In this study, we report the following novel findings: 1) ESP genes were identified only in the genomes of the families Muridae and Cricetidae, suggesting that the ESP gene family had originated in the common ancestor of murids and cricetids. 2) Murids tend to have a larger number of ESP genes than cricetids, whereas the length of a mature sequence is significantly shorter in murids than in cricetids. 3) Longer mature sequences tend to be more conserved in amino acid sequence than shorter mature sequences. 4) The mature sequence of ESP genes shows an overall weak similarity in amino acid sequence to the α-globin gene. 5) The signal sequence of ESP genes is indistinguishably similar to the signal-encoding exon of the CRISP2 gene. 6) Some ESPs in rats and golden hamsters are expressed in the lacrimal gland and the salivary gland; however, they did not induce an obvious vomeronasal-stimulating activity.
α-Globin as a Possible Origin of the ESP Mature Sequence
We found that the mature sequences of ESP genes show a weak amino acid sequence similarity to the α-globin gene. Moreover, both ESP and α-globin proteins are helix-rich, and the secondary structures of the two proteins roughly correspond to each other. Here, we argue a possibility that the mature sequence of ESP genes might have originated from the α-globin gene.
The mouse α-globin gene cluster is located on chromosome 11, which is different from the location of the ESP gene cluster (chromosome 17). Moreover, the rodent α-globin gene is interrupted by two introns, whereas an ESP gene is intronless within a mature sequence. These features suggest that an α-globin-like gene was integrated into the genomic region close to the ESP gene cluster via retrotransposition of a processed mRNA. Intronless retrotransposed gene copies usually become pseudogenes due to the lack of regulatory elements (called processed pseudogenes); however, a substantial number of them can be transcribed and translated into functional proteins by “hitch-hiking” on the preexisting regulatory machinery of other genes (called retrogenes) (Ding et al. 2006; Kaessmann et al. 2009). In fact, it has been known for more than 35 years that one processed pseudogene (ψα3) in mice arose from the α-globin gene (Vanin 1984). It is therefore plausible that an α-globin gene created a retrogene.
Retrotransposition needs to occur in the germ line cells. In this regard, it has been reported that both α- and β-globin mRNAs and proteins are found at high levels in mouse periovulatory cumulus cells surrounding the egg and they function as gas transporter molecules (Brown et al. 2015). Moreover, interestingly, the PGK2 gene located within the ESP gene cluster (fig. 2A) is one of the most well-known examples of a retrogene (Boer et al. 1987; McCarrey and Thomas 1987). There are two PGK genes found in most mammals. The X-linked PGK1 gene is ubiquitously expressed in all somatic cells and contains ten introns. However, the PGK2 gene is intronless; it is expressed in a tissue-, cell type-, and developmental stage-specific manner during spermatogenesis and is required for normal sperm motility and fertility (Danshina et al. 2010). Retrotransposed gene copies would preferentially be inserted into open and actively transcribing chromatin (Kaessmann et al. 2009). The ESP gene cluster is located close to the germ line-expressed PGK2 gene. Therefore, the genomic region containing the ESP gene cluster might be a “hot spot” of retrotransposition.
Chimeric Origin of the Ancestral ESP Gene
We found that 1) the signal sequence of ESP genes is indistinguishably similar to the signal sequence of the CRISP2 gene, 2) the exon–intron boundaries of a signal-encoding exon of ESP genes and those of CRISP2 genes are exactly the same, and 3) the CRISP2 gene is located adjacent to the ESP gene cluster in almost all the species examined. These observations strongly suggest that the signal sequence of ESP genes has originated from the CRISP2 gene.
In mammals, CRISP genes are primarily expressed in the reproductive tract (Roberts et al. 2007; Gibbs et al. 2008), and some CRISP proteins are involved in fertilization (Ernesto et al. 2015; Carvajal et al. 2018). One study showed that mouse CRISPs are expressed in various tissues; for example, CRISP1 and CRISP3 are expressed in the salivary gland, and CRISP1–3 are expressed in the lacrimal gland (Reddy et al. 2008). Interestingly, in various snakes and lizards, CRISP proteins are secreted in the venom gland and function as a venom. They block the cyclic nucleotide-gated ion channels and inhibit smooth muscle contraction (Yamazaki and Morita 2004). It is hypothesized that the CRISPs were present in the ancestral salivary tissue and venom-secreting CRISPs are derivations of the previously existing salivary CRISPs (Fry 2005). Therefore, it is likely that the CRISP2 signal sequence has an ability of secreting a protein into the salivary and/or lacrimal glands.
The first coding exon of the CRISP2 gene nearly perfectly corresponds to the signal peptide. Owing to this feature, if an α-globin-like sequence was by chance inserted immediately downstream of the CRISP2 first coding exon, it is likely that the α-globin-like protein would be secreted in saliva or other secretions of exocrine glands with the aid of the signal peptide.
On the basis of the above argument, we propose that ESP genes have originated from a retrotransposed α-globin-like sequence combined with a signal-encoding exon of the CRISP2 gene. This study provides an intriguing example of “molecular tinkering” (Jacob 1977) in rapidly evolving species-specific proteinaceous pheromone genes, the ESP multigene family.
Coevolution of ESP and V2R Genes
As aforementioned, ESP1 and ESP22 specifically bind to Vmn2r116 and Vmn2r115, respectively. Both Vmn2r116 and Vmn2r115 belong to the subfamily A3 in the phylogenetic tree of V2R genes (supplementary fig. S17, Supplementary Material online) (Francia et al. 2015). According to Francia et al. (2015), genes of subfamily A3 were identified only in murids and cricetids. Therefore, the subfamily A3 and the ESP gene family are probably evolutionarily correlated with each other.
We recently showed that β-globin (but not α-globin) acted as a chemosignal to elicit the digging behavior in lactating female mice via a specific vomeronasal receptor, Vmn2r88, which belongs to the subfamily A1 (Osakada T, et al., unpublished data). Isogai et al. (2018) also reported that β-globin is a ligand of Vmn2r88. Francia et al. (2015) showed that genes in subfamilies A1/A2 are widely present in the infraorder Myodonta, whereas the subfamily A3 is specific to murids/cricetids and the subfamily A4 is specific to only murids. It is therefore likely that the subfamilies A3/A4 have originated from the ancestral gene of the subfamilies A1/A2 in the common ancestor of murid and cricetids. This evolutionary relationship of V2R subfamilies is in parallel to our hypothesis that ESPs (the ligands of V2Rs in the subfamily A3) have been originated from globins (the ligands of V2Rs in the subfamily A1) in the murids/cricetids ancestor.
To further investigate coevolution of ESP and V2R genes, we identified subfamilies A3/A4 V2R genes from 23 rodent genomes examined in this study (supplementary table S1, Supplementary Material online). However, because a V2R gene is encoded in multiple exons, and the quality of the genome assembly is not very high except for mice and rats, it is difficult to reconstruct full-length sequences of V2R genes. For this reason, we focused on a single exon, exon3, of a V2R gene, which is ∼270 amino acids long. The rationale is that 1) exon3 encompasses most of the ligand-binding domain of an V2R, and 2) the phylogeny of exon3 sequences shows a very similar topology with that of full-length V2R genes, as suggested in Francia et al. (2015). As a result, we identified exon3 of subfamily A3 V2R genes from all murid and cricetid species examined except for the Northern mole vole, whereas no such sequences were detected from nonmurid/cricetid rodents (supplementary figs. S18 and S19A, Supplementary Material online). Exon3 of subfamily A4 V2R genes were found only from murid species. These observations are consistent with Francia et al. (2015). We also found that the numbers of exon3 of the subfamily A3/A4 V2R genes are positively correlated with those of ESP mature sequences in 12 murid/cricetid species (Spearman’s correlation coefficient rS = 0.783; supplementary fig. S19B, Supplementary Material online).
Evolutionary Scenario of the ESP Gene Family
In this study, we proposed an evolutionary scenario for the origin of ESP genes, in which the ancestral ESP gene was originated from the α-globin gene combined with the signal-encoding exon of the CRISP gene. As aforementioned, the signal peptide of the ancestral CRISP protein may have had an ability to secret proteins into saliva. Our findings suggested that long (≥120 amino acids) mature sequences appear to be the “prototype” of ESP genes. We also found an intriguing clear tendency that SMG-expressing ESP genes are longer than ELG-expressing ESP genes (supplementary fig. S13 and table S9, Supplementary Material online). It is therefore reasonable to speculate that the ancestral ESP was secreted into saliva.
Although we confirmed that rat and golden hamster ESPs were expressed in the SMG and ELG, we did not detect an obvious vomeronasal-stimulating activity of rat or golden hamster ESPs. It is therefore possible that the pheromone activity of ESPs was acquired in the Mus lineage after the divergence from rats. It is, however, also possible that c-Fos signals might be more difficult to observe for rat/hamster ESPs than for mouse ESPs due to poorer folding of recombinants and/or rarity of some cognate proteins. To conclude the presence/absence of vomeronasal-stimulating activity of ESPs in nonmouse rodents, further analyses should be necessary.
In the Mus lineage, ESP genes became shorter and diversified rapidly. At the same time, these ESPs would have begun to be secreted into tear fluid to gain the function of communicating with other individuals. We also found that ESP genes are highly variable even among different mouse strains. In fact, the ESP gene loci are one of the most highly diversified genomic regions among different mouse strains along with the major histocompatibility complex and the olfactory receptor gene loci (Lilue et al. 2018). It was proposed that the expression levels of ESPs might convey some information of individuals (Hattori et al. 2017). Our study suggests that the sequences of ESPs might also be used as signals for recognizing individuals. In this regard, it would be interesting to note that MUP proteins, which are secreted in mouse urine and were often proposed to be used as chemical signatures for individual recognition, show an unexpectedly low inter-individual variation (Thoß et al. 2016).
Several questions remain elusive. It is essential to investigate the function of a long-type ESP, which appears to be the prototype of ESPs, for the further understanding of the evolution of ESP gene family. The presence/absence of the pheromone activity of ESPs in nonmouse rodents should be examined more thoroughly. The functional difference among different ESP alleles in various mouse strains is also an open question. Finally, deorphanization of V2Rs is critical to elucidate the coevolution of ESP and V2R gene families in more detail, because most V2Rs remain orphaned yet.
Materials and Methods
Genome Data
The latest genome assemblies for 23 rodent species and 16 inbred mouse strains (Lilue et al. 2018) were obtained from GenBank (https://www.ncbi.nlm.nih.gov/genbank/) or UCSC Genome Browser (http://genome.ucsc.edu/) (see supplementary tables S1 and S4, Supplementary Material online). We also examined the whole genome sequences of 87 nonrodent mammals (supplementary table S2, Supplementary Material online), though we did not find any significant hits.
Identification of Mature Sequences
To identify ESP genes in the whole genome sequences, we first performed TBlastN searches (Altschul et al. 1997) against each of the genome sequences using known ESP gene sequences as queries with the cutoff E value of 0.001 (see supplementary fig. S20, Supplementary Material online). We used the mature sequences of 37 mouse ESP genes and eight rat ESP genes as queries (Kimoto et al. 2007) (GenBank Accession numbers: NP_001033589.1 and AB306980–AB307028). Because all of the query sequences were similar to one another, multiple query sequences may hit the same genomic region. We therefore extracted the “best-hit,” showing the lowest E value among all Blast hits corresponding to a given genomic region. MmESP12, MmESP06, and MmESP02 matched numerous genomic regions of the Muridae genomes; probably because these mature sequences have a weak similarity to some Muridae-specific repetitive sequences. We, therefore, eliminated the best-hits that matched to only either of MmESP12, MmESP06, and MmESP02 (criterion 1 in supplementary fig. S20, Supplementary Material online). We also eliminated the best-hits that were less than 40 amino acids long, because the shortest mature sequence (MmESP30) is 42 amino acids long (criterion 2). All of the remaining best-hit sequences were regarded as mature sequences. At this stage, we did not achieve any hits for nonmurid/cricetid rodent genomes and nonrodent genomes.
Among the remaining best-hit sequences of murid/cricetid genomes, intact mature sequences were identified by the following filtering processes. The sequences other than intact mature sequences were regarded as pseudogenes. For each of the best-hit sequences, if the codon at the end was not a stop codon, they were extended in the 3′ direction along the genomic DNA sequence up to a stop codon (criterion 3). Some sequences contained interrupting stop codons or frameshifts. We therefore extracted the longest coding sequence that contained no interrupting stop codons or frameshifts and ended at a stop codon. If the extracted sequence was shorter than 40 amino acids in length, then it was discarded (criterion 4). We then constructed a multiple alignment for all of the remaining sequences by MAFFT (Katoh et al. 2005). If a sequence had gaps at five or more amino acid sites in the N-terminal conserved region of a mature sequence, it was excluded (criterion 5). To determine the exon–intron boundary for each of the remaining sequences, the codon that was present immediately after an AG dinucleotide and was located to the most upstream within the sequence was identified, and the sequence starting from that codon was extracted. We again constructed a multiple alignment for all of the sequences obtained above using MAFFT, and any sequence having gaps at five or more amino acid sites in the N-terminal region was discarded (criterion 6). The remaining sequences were considered as intact mature sequences. We then performed a second-round TBlastN search against genome sequences using the intact mature sequences identified above as queries with the E value of 0.001, and we conducted the same filtering processes described above to identify additional mature sequences. These processes were iteratively performed until no new mature sequences were identified. The coordinates of mature sequences identified in this study are provided in supplementary tables S3 and S5, Supplementary Material online.
Identification of Signal Sequences
A signal sequence of an ESP gene is short in length (16–23 amino acids long). Therefore, to avoid missing any sequences with a weak similarity to queries, we performed both TBlastN and BlastN searches with the E value of 1 against the whole genome sequence using known signal sequences as queries (see supplementary fig. S21, Supplementary Material online). We used signal sequences of 24 mouse ESP genes with an intact coding sequence as queries (Kimoto et al. 2007). From the Blast hits obtained by TBlastN and BlastN searches, we extracted a best-hit sequence for a given genomic region in a similar manner as explained in the section “Identification of mature sequences.” From these best-hit sequences, we extracted intact signal sequences by the following filtering processes. The sequences other than intact signal sequences were regarded as pseudogenes. First, a best-hit including a frameshift (detected by a BlastN search) was regarded as a pseudogene (criterion 1). We then constructed a multiple alignment for all the remaining sequences together with the queries. Many of the query sequences (known signal sequences of mouse ESP genes) are 23 amino acids long. Let us denote the amino acid positions in the multiple alignment as position 1 to position 23. Unless a given best-hit sequence was aligned to the region from position 1 to position 23 without any gaps, the sequence was extended in both directions, the 3′ direction and the 5′ direction, along the genomic DNA sequence up to position 1 and position 23 (criterion 2). After extension, we examined the position of first methionine and the dinucleotide sequence immediately after the position 23 for each best-hit sequence. If the sequence did not meet to either of the following two conditions, it was regarded as a signal pseudogene (criterion 3): 1) the first methionine is located between position 1 and position 8, and 2) the dinucleotide after position 23 is GT (an exon–intron boundary). Among the remaining sequences, the sequence containing an interrupting stop codon was regarded as a pseudogene (criterion 4). The remaining sequences were considered as intact signal sequences. These processes were repeated until no new signal sequences were identified by using identified signal sequences as queries (criterion 5). Finally, to exclude CRISP2 signal sequences, we performed a BLASTP search using 32 intact signal sequences in mice identified after performing the process described above as well as CRISP2 signal sequences in six species (mice, Algerian mice, shrew mice, rats, Chinese hamsters, and golden hamsters) as queries against all of the identified signal sequences. If a sequence showed a lower E value for a CRISP2 signal sequence as compared with an ESP signal sequence, it was eliminated (criterion 6). The coordinates of signal sequences identified in this study are provided in supplementary tables S3 and S5, Supplementary Material online.
Identification of CRISP, PGK2, and RHAG Genes
CRISP gene sequences of mice, rats, golden hamsters, and Chinese hamsters were downloaded from GenBank (see supplementary fig. S4 for their accession numbers, Supplementary Material online). The exon–intron structures of CRISP genes of eight rodent species (fig. 2A) were predicted by using the GENEWISEDB program of the WISE2 package (Birney et al. 2004). We first constructed multiple alignments for CRISP1/3 and CRISP2 separately by using MAFFT (Katoh et al. 2005). The profile Hidden Markov Model (HMM) was constructed from each alignment by using the HMMBUILD program of the HMMER package version 2.3.2 (Eddy 2011). For each species, we extracted a genomic DNA sequence containing the entire ESP gene cluster with 100 kb elongated sequences from the mature ESP sequence at the end of the cluster in the both directions. The profile HMM, created as described above, was aligned to the extracted genomic DNA sequence for each species by using GENEWISEDB. GENEWISEDB analyses were also performed by using each query sequence in place of a profile HMM. We then compiled the results and extracted the exon–intron structure with the largest bits value for a given genomic region. The genomic locations of PGK2 and RHAG genes were identified in the same way.
Comparison among 16 Mouse Strains
Mature and signal sequences of ESP genes were identified in the de novo genome assemblies of 15 nonreference mouse strains (Lilue et al. 2018) in the same way as described in the sections “Identification of mature sequences” and “Identification of signal sequences.” The quality of genome assemblies for nonreference mouse strains is much lower than that of the reference genome, and the assembly of the genomic region of the ESP gene cluster in the nonreference strains is incomplete. Therefore, we assigned each of the mature sequences identified in the 15 strains to MmESP1–37 in the reference genome in the following way. First, pairwise amino acid sequence alignments were constructed for all combinations between the mature sequences identified in the nonreference strain and MmESP1–37 by using ClustalW (Larkin et al. 2007), and the amino acid identities were calculated. We then identified the reciprocal best hits of mature sequences between the reference genome and each of the nonreference strains. In all, 97 mature sequences of the nonreference genomes were not assigned to any of MmESP1–37 in the reference genome. We also identified the reciprocal best hits of mature sequences between two nonreference genomes. The remaining 97 mature sequences were classified into 10 groups on the basis of reciprocal best hits (two sequences that were a reciprocal best hit to each other were assigned into the same group), and the sequences assigned to the same group were considered to be at the same allele in the mouse genome. These 10 groups were named as MmESP39–48P. We did not use the name “ESP38” in Kimoto et al. (2007), because its sequence is missing in the latest version of the reference genome. The sequences that did not show reciprocal best hits to any sequence in other strains, that is, the sequences that were found in only one strain, were discarded. There were 10 such sequences.
Selection Tests
The overall ω (= dN/dS) value was calculated by codeml implemented in the PAML 4.8a package (Yang 2007) under model M0 for each clade separately. A codon alignment of ESP mature sequences was generated by MAFFT (Katoh et al. 2005). We compared two models, the null model of no positive selection (M1a and M7) which assumes no sites with ω > 1 and the alternative model of positive selection (M2a and M8, respectively) which assumes the presence of sites with ω > 1, and calculated P values by the χ2 tests with the degree of freedom of two for the twice difference in log-likelihood values between the two models (Nielsen and Yang 1998; Yang et al. 2000). For the clades in which the presence of positively selected sites was suggested with P < 0.05, the Bayes Empirical Bayes approach was used to calculate the posterior probability distribution of ω for each site. We ran codeml program with “CodonFreq = 2” (F3 × 4 codon frequency model) and “cleandata = 1” (complete deletion).
Animals
Brown Norway rats (Rattus norvegicus), golden hamsters (Mesocricetus auratus), and BALB/c mice were purchased from SLC (Shizuoka, Japan). They were maintained under a 12-h dark/light cycle. Food and water were provided ad libitum. Experiments were carried out in accordance with animal experimentation protocols approved by the animal care and use committees at the University of Tokyo.
Cloning of the Rat and Golden Hamster ESP Genes
Ten-week-old Brown Norway rats and golden hamsters were sacrificed and their ELG, HG, SMG, and some tissues (eye, brain, lung, heart, liver, kidney, and testis) were collected. Total RNA samples were prepared by using TRIzol reagent (Invitrogen, USA). One microgram of DNaseI-treated RNA isolated from each tissue was used to prepare oligo (dT) adaptor-primed cDNAs by SuperScript III (Invitrogen, USA), according to the manufacturer’s protocol. Amplification was carried out by using gene-specific primers for 35 cycles at 94 °C for 15 s, 55 °C for 30 s, 68 °C for 30 s. See supplementary table S7, Supplementary Material online for the sequences of primers utilized in this study.
RNA-seq Analyses
Total RNA was extracted from the ELG and the SMG of a male rat and a male golden hamster treated with RNAlater (Ambion) using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. RNA quantity and integrity were checked using agarose gel electrophoresis and NanoDrop (Thermo Fisher Scientific). The RNA samples were used to construct RNA-seq libraries using TruSeq Stranded mRNA Library Kit (Illumina) according to manufacturer’s instruction. Subsequently, paired-end high-throughput sequencing was performed using Illumina NovaSeq6000 platform (151 bp × 2). Library construction and RNA sequencing were done by Macrogen Japan (Kyoto). As a result, the following sizes of RNA-seq reads were obtained: 12.2 Gb for the rat ELG, 14.9 Gb for the rat SMG, 12.2 Gb for the golden hamster ELG, and 14.2 Gb for the golden hamster SMG. Low-quality sequences and adapters were removed using the Trimmomatic version 0.39 (Bolger et al. 2014) with the following parameters: ILLUMINACLIP:TruSeq3-PE.fa:2:30:10, LEADING:20, TRAILING:20, SLIDINGWINDOW:5:25, and MINLEN:36. Trimmed RNA-seq reads were mapped to the rat and golden hamster genome assemblies using HISAT2 version 2.2.0 (Kim et al. 2019) with –dta option. Reconstruction of transcript models and estimate of gene expression levels in fragments per kilobase of exon per million reads mapped and in transcripts per million (TPM) were performed using StringTie version 2.1.3 (Pertea et al. 2015, 2016).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the Exploratory Research for Advanced Technology (ERATO) Touhara Chemosensory Signal Project from Japan Science and Technology Agency (JPMJER1202 to K.T.) and by a Grant-in-Aid for Scientific Research (C) from Japan Society for the Promotion of Science (JSPS KAKENHI Grant Number JP18K06359 to Y.N.).
Author Contributions
Y.N. and K.T. conceived the project and designed the research. Y.N, S.K., and S.S. performed the bioinformatic analyses. M.T., S.K., K.M., and T.Y. performed experiments. Y.N., M.T., and K.T. wrote the manuscript.
Data Availability
Nucleotide sequence data for 139 mature sequences of ESP genes from 12 rodent species and those for the CRISP genes newly identified in this study (for Algerian mice, Shrew mice, Ryukyu mice, and deer mice) are provided in supplementary data S1 and S2, Supplementary Material online, respectively. Amino acid sequences for 122 exon3 sequences of subfamilies A3/A4 V2R genes are provided in supplementary data S3, Supplementary Material online. RNA-seq data for the ELG and the SMG of a male rat and a male golden hamster have been deposited to GenBank (BioSample accession numbers: SAMD00238582, golden hamster ELG; SAMD00238583, golden hamster SMG; SAMD00238584, rat ELG; SAMD00238585, rat SMG).
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ.. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25(17):3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R.. 2004. GeneWise and Genomewise. Genome Res. 14(5):988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer PH, Adra CN, Lau YF, McBurney MW.. 1987. The testis-specific phosphoglycerate kinase gene pgk-2 is a recruited retroposon. Mol Cell Biol. 7(9):3107–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown HM, Anastasi MR, Frank LA, Kind KL, Richani D, Robker RL, Russell DL, Gilchrist RB, Thompson JG.. 2015. Hemoglobin: a gas transport molecule that is hormonally regulated in the ovarian follicle in mice and humans. Biol Reprod. 92(1):26. [DOI] [PubMed] [Google Scholar]
- Cabanettes F, Klopp C.. 2018. D-GENIES: dot plot large genomes in an interactive, efficient and simple way. PeerJ 6:e4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal G, Brukman NG, Weigel Munoz M, Battistone MA, Guazzone VA, Ikawa M, Haruhiko M, Lustig L, Breton S, Cuasnicu PS.. 2018. Impaired male fertility and abnormal epididymal epithelium differentiation in mice lacking CRISP1 and CRISP4. Sci Rep. 8(1):17531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danshina PV, Geyer CB, Dai Q, Goulding EH, Willis WD, Kitto GB, McCarrey JR, Eddy EM, O'Brien DA.. 2010. Phosphoglycerate kinase 2 (PGK2) is essential for sperm function and male fertility in mice. Biol Reprod. 82(1):136–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding W, Lin L, Chen B, Dai J.. 2006. L1 elements, processed pseudogenes and retrogenes in mammalian genomes. IUBMB Life 58(12):677–685. [DOI] [PubMed] [Google Scholar]
- Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol. 7(10):e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernesto JI, Weigel Munoz M, Battistone MA, Vasen G, Martinez-Lopez P, Orta G, Figueiras-Fierro D, De la Vega-Beltran JL, Moreno IA, Guidobaldi HA, et al. 2015. CRISP1 as a novel CatSper regulator that modulates sperm motility and orientation during fertilization. J Cell Biol. 210(7):1213–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrero DM, Moeller LM, Osakada T, Horio N, Li Q, Roy DS, Cichy A, Spehr M, Touhara K, Liberles SD.. 2013. A juvenile mouse pheromone inhibits sexual behaviour through the vomeronasal system. Nature 502(7471):368–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francia S, Silvotti L, Ghirardi F, Catzeflis F, Percudani R, Tirindelli R.. 2015. Evolution of spatially coexpressed families of type-2 vomeronasal receptors in rodents. Genome Biol Evol. 7(1):272–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry BG. 2005. From genome to “venome”: molecular origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences and related body proteins. Genome Res. 15(3):403–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs GM, Roelants K, O'Bryan MK.. 2008. The CAP superfamily: cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins—roles in reproduction, cancer, and immune defense. Endocr Rev. 29(7):865–897. [DOI] [PubMed] [Google Scholar]
- Haga S, Hattori T, Sato T, Sato K, Matsuda S, Kobayakawa R, Sakano H, Yoshihara Y, Kikusui T, Touhara K.. 2010. The male mouse pheromone ESP1 enhances female sexual receptive behaviour through a specific vomeronasal receptor. Nature 466(7302):118–122. [DOI] [PubMed] [Google Scholar]
- Hattori T, Osakada T, Masaoka T, Ooyama R, Horio N, Mogi K, Nagasawa M, Haga-Yamanaka S, Touhara K, Kikusui T.. 2017. Exocrine gland-secreting peptide 1 is a key chemosensory signal responsible for the Bruce effect in mice. Curr Biol. 27(20):3197–3201.e3. 3197-3201.e3193. [DOI] [PubMed] [Google Scholar]
- Hattori T, Osakada T, Matsumoto A, Matsuo N, Haga-Yamanaka S, Nishida T, Mori Y, Mogi K, Touhara K, Kikusui T.. 2016. Self-exposure to the male pheromone esp1 enhances male aggressiveness in mice. Curr Biol. 26(9):1229–1234. [DOI] [PubMed] [Google Scholar]
- Isogai Y, Wu Z, Love MI, Ahn MH, Bambah-Mukku D, Hua V, Farrell K, Dulac C.. 2018. Multisensory logic of infant-directed aggression by males. Cell 175(7):1827–1841.e17. 1827-1841.e1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F. 1977. Evolution and tinkering. Science 196(4295):1161–1166. [DOI] [PubMed] [Google Scholar]
- Kaessmann H, Vinckenbosch N, Long M.. 2009. RNA-based gene duplication: mechanistic and evolutionary insights. Nat Rev Genet. 10(1):19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T.. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33(2):511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL.. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 37(8):907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimoto H, Haga S, Sato K, Touhara K.. 2005. Sex-specific peptides from exocrine glands stimulate mouse vomeronasal sensory neurons. Nature 437(7060):898–901. [DOI] [PubMed] [Google Scholar]
- Kimoto H, Sato K, Nodari F, Haga S, Holy TE, Touhara K.. 2007. Sex- and strain-specific expression and vomeronasal activity of mouse ESP family peptides. Curr Biol. 17(21):1879–1884. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB.. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 34(7):1812–1819. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. ClustalW and ClustalX version 2.0. Bioinformatics 23(21):2947–2948. [DOI] [PubMed] [Google Scholar]
- Liberles SD. 2014. Mammalian pheromones. Annu Rev Physiol. 76(1):151–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilue J, Doran AG, Fiddes IT, Abrudan M, Armstrong J, Bennett R, Chow W, Collins J, Collins S, Czechanski A, et al. 2018. Sixteen diverse laboratory mouse reference genomes define strain-specific haplotypes and novel functional loci. Nat Genet. 50(11):1574–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarrey JR, Thomas K.. 1987. Human testis-specific PGK gene lacks introns and possesses characteristics of a processed gene. Nature 326(6112):501–505. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Yang Z.. 1998. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148(3):929–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakada T, Ishii KK, Mori H, Eguchi R, Ferrero DM, Yoshihara Y, Liberles SD, Miyamichi K, Touhara K.. 2018. Sexual rejection via a vomeronasal receptor-triggered limbic circuit. Nat Commun. 9(1):4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papes F, Logan DW, Stowers L.. 2010. The vomeronasal organ mediates interspecies defensive behaviors through detection of protein pheromone homologs. Cell 141(4):692–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL.. 2016. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 11(9):1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL.. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 33(3):290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy T, Gibbs GM, Merriner DJ, Kerr JB, O'Bryan MK.. 2008. Cysteine-rich secretory proteins are not exclusively expressed in the male reproductive tract. Dev Dyn. 237(11):3313–3323. [DOI] [PubMed] [Google Scholar]
- Roberts KP, Johnston DS, Nolan MA, Wooters JL, Waxmonsky NC, Piehl LB, Ensrud-Bowlin KM, Hamilton DW.. 2007. Structure and function of epididymal protein cysteine-rich secretory protein-1. Asian J Androl. 9(4):508–514. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M.. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4(4):406–425. [DOI] [PubMed] [Google Scholar]
- Thoß M, Enk V, Yu H, Miller I, Luzynski KC, Balint B, Smith S, Razzazi-Fazeli E, Penn DJ.. 2016. Diversity of major urinary proteins (MUPs) in wild house mice. Sci Rep. 6(1):38378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoda M, Miyamichi K, Eguchi R, Sakuma Y, Yoshihara Y, Kikusui T, Kuwahara M, Touhara K.. 2018. Identification of an intra- and inter-specific tear protein signal in rodents. Curr Biol. 28(8):1213–1223.e6. [DOI] [PubMed] [Google Scholar]
- Vanin EF. 1984. Processed pseudogenes. Characteristics and evolution. Biochim Biophys Acta 782(3):231–241. [DOI] [PubMed] [Google Scholar]
- Wyatt TD. 2014. a. Pheromones and animal behavior: chemical signals and signatures. Cambridge: Cambridge University Press. [Google Scholar]
- Wyatt TD. 2014. b. Proteins and peptides as pheromone signals and chemical signatures. Anim Behav. 97:273–280. [Google Scholar]
- Yamazaki Y, Morita T.. 2004. Structure and function of snake venom cysteine-rich secretory proteins. Toxicon 44(3):227–231. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Goldman N, Pedersen AM.. 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga S, Sato T, Hirakane M, Esaki K, Hamaguchi T, Haga-Yamanaka S, Tsunoda M, Kimoto H, Shimada I, Touhara K, et al. 2013. Structure of the mouse sex peptide pheromone ESP1 reveals a molecular basis for specific binding to the class C G-protein-coupled vomeronasal receptor. J Biol Chem. 288(22):16064–16072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Nucleotide sequence data for 139 mature sequences of ESP genes from 12 rodent species and those for the CRISP genes newly identified in this study (for Algerian mice, Shrew mice, Ryukyu mice, and deer mice) are provided in supplementary data S1 and S2, Supplementary Material online, respectively. Amino acid sequences for 122 exon3 sequences of subfamilies A3/A4 V2R genes are provided in supplementary data S3, Supplementary Material online. RNA-seq data for the ELG and the SMG of a male rat and a male golden hamster have been deposited to GenBank (BioSample accession numbers: SAMD00238582, golden hamster ELG; SAMD00238583, golden hamster SMG; SAMD00238584, rat ELG; SAMD00238585, rat SMG).