Abstract
In the last 240,000 years, males of the Drosophila simulans species clade have evolved striking differences in the morphology of their epandrial posterior lobes and claspers (surstyli). These appendages are used for grasping the female during mating and so their divergence is most likely driven by sexual selection. Mapping studies indicate a highly polygenic and generally additive genetic basis for these morphological differences. However, we have limited understanding of the gene regulatory networks that control the development of genital structures and how they evolved to result in this rapid phenotypic diversification. Here, we used new D. simulans/D. mauritiana introgression lines on chromosome arm 3L to generate higher resolution maps of posterior lobe and clasper differences between these species. We then carried out RNA-seq on the developing genitalia of both species to identify the expressed genes and those that are differentially expressed between the two species. This allowed us to test the function of expressed positional candidates during genital development in D. melanogaster. We identified several new genes involved in the development and possibly the evolution of these genital structures, including the transcription factors Hairy and Grunge. Furthermore, we discovered that during clasper development Hairy negatively regulates tartan (trn), a gene known to contribute to divergence in clasper morphology. Taken together, our results provide new insights into the regulation of genital development and how this has evolved between species.
Keywords: evolution, development, Drosophila, sexual selection, morphology, gene regulatory networks
Introduction
To understand the evolution of animal morphology, we need to better link genotypic and phenotypic changes. This requires identifying the causative genes, how they are integrated into gene regulatory networks, and how changes in these interactions alter developmental processes and consequently the phenotype (Stern 2011; Nunes et al. 2013; Kittelmann et al. 2018). There has been great progress in identifying genes that cause changes in animal morphology (reviewed in Martin and Orgogozo [2013]). However, we still lack information on the genes that contribute to changes in quantitative traits, such as organ size, and how they combine to achieve this.
The size and shape of male genital organs evolve rapidly among species, driven by sexual selection (Eberhard 1985, 2010; Hosken and Stockley 2004; House et al. 2013; Simmons 2014). For example, the epandrial posterior lobes and claspers (surstyli) have changed dramatically in size in the Drosophila simulans species clade in the last 240,000 years (Garrigan et al. 2012) (fig. 1A). Both the claspers and posterior lobes play important roles during copulation. The claspers open the female oviscapt through interdigitization of bristles, and help achieve correct copulatory positioning (Robertson 1988; Acebes et al. 2003; Jagadeeshan and Singh 2006; Kamimura and Mitsumoto 2011; Yassin and Orgogozo 2013; Masly and Kamimura 2014; Mattei et al. 2015), whereas the posterior lobes also contribute to stability during mating by inserting into grooves on the female tergites (Robertson 1988; Kamimura and Mitsumoto 2011; Yassin and Orgogozo 2013).
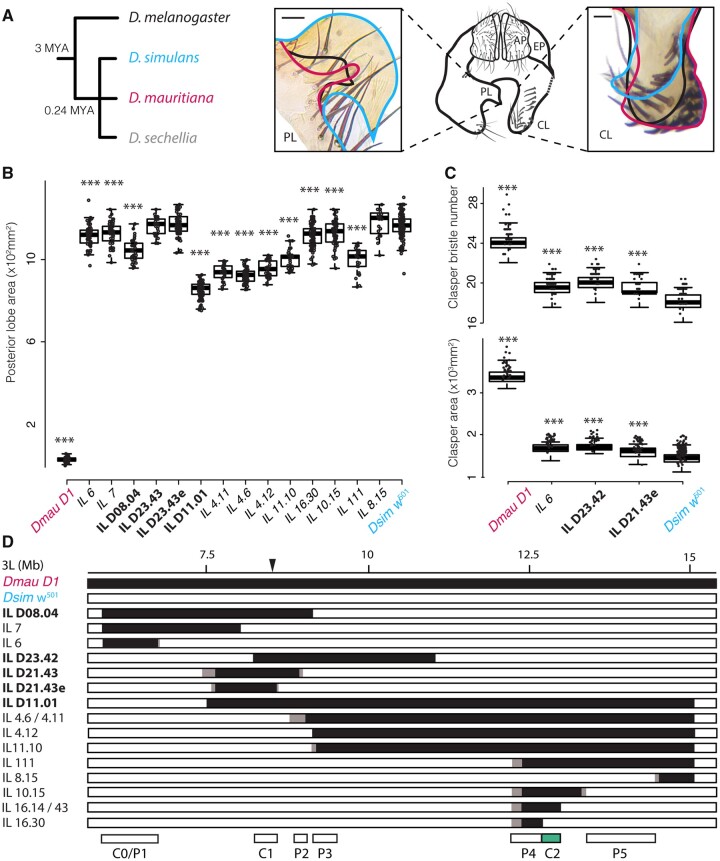
Fig. 1.
Differences in external male-terminal structures among Drosophila melanogaster subgroup species and introgression mapping between D. simulans and D. mauritiana. (A) Relationships of D. melanogaster and species of the D. simulans clade (left). The central diagram depicts a schematic of male genital arch morphology of D. melanogaster (posterior view). The posterior lobes (left-hand box) typically obscure visualization of the claspers (right-hand box), and therefore they are shown here dissected away on the right-hand side of the central schematic. The relative size and shape of the lobes and the claspers of D. melanogaster (black), D. simulans (blue), and D. mauritiana (red) are illustrated in the left and right schematics and images, respectively. D. simulans has much larger posterior lobes and smaller claspers, with fewer, thinner, and shorter bristles than D. mauritiana and D. melanogaster. Posterior lobes (PL), claspers (CL), anal plates (AP), and epandrium (EP). Scale bars = 20 µm. (B–D) Mapping and phenotypic effect of candidate regions on posterior lobe size (B), clasper bristle number (C, upper plot), and clasper area (C, lower plot). Boxes indicate the range, upper and lower quartiles, and median for each sample. Asterisks indicate significant comparisons with Dsim w501 where P < 0.001*** (Dunnett’s test for posterior lobe and clasper size, Dunn’s test for clasper bristle number, supplementary file 2D–F, Supplementary Material online). Differences in the effect of the introgressed regions (supplementary file 3 and supportive text, Supplementary Material online) on posterior lobe size (B) and clasper size/bristle number (C), allowed refinement of candidate regions P1–P5, C0, and C1 (D). The previously identified C2 region is shown in green (Hagen et al. 2019) (D). Black bars indicate Dmau D1 DNA, white bars indicate Dsim w501 DNA, and gray boxes regions containing break points that have not been precisely determined. The black triangle indicates the position of P-element insertion originally used for generating the introgressions. New introgression lines are shown in nonbold font.
The posterior lobes are a novelty of the D. melanogaster species subgroup (Kopp and True 2002; Jagadeeshan and Singh 2006; Glassford et al. 2015). In D. mauritiana, they are small, thin, finger-like projections in comparison to the much larger, helmet-shaped lobes of D. simulans (fig. 1A). D. melanogaster has intermediate-sized lobes, which are trapezoid shaped (fig. 1A), whereas the D. sechellia lobes are also intermediate in size and resemble “boots.” It is important to note that there is some variation within species but the extremes of intraspecific variation do not overlap with the differences observed between species (McNeil et al. 2011; Hackett et al. 2016).
The claspers lie beneath the posterior lobes, and about twice as large in D. mauritiana compared with D. simulans, with a third more bristles (True et al. 1997; Tanaka et al. 2015) (fig. 1A). The morphology of these bristles also differs between the species, with the D. mauritiana bristles being generally shorter and thicker than those of D. simulans (True et al. 1997; Tanaka et al. 2015). D. sechellia male claspers have very similar morphology to those of D. simulans, whereas the claspers of D. melanogaster appear to be intermediate between D. mauritiana, and D. simulans/D. sechellia (fig. 1A).
Genetic mapping of changes to posterior lobe and clasper morphology among D. melanogaster subgroup species have shown that these differences are polygenic and generally additive (Coyne et al. 1991; Liu et al. 1996; Laurie et al. 1997; True et al. 1997; Macdonald and Goldstein 1999; Zeng et al. 2000; Tanaka et al. 2015, 2018). For example, up to 19 QTL have been identified for the difference in posterior lobe size between D. mauritiana and D. simulans, and QTL have been mapped to all major autosomal arms for the differences in clasper size between these species (Laurie et al. 1997; True et al. 1997; Zeng et al. 2000; Tanaka et al. 2015, 2018). Therefore, it appears that many loci contribute to these differences in genital organ size.
We previously used an introgression-based approach to fine-scale map QTL on chromosome arm 3L underlying posterior lobe and clasper size differences between D. mauritiana and D. simulans (True et al. 1997; Zeng et al. 2000; Tanaka et al. 2015; Hagen et al. 2019). The genomes of these lines were D. simulans, apart from introgressed regions of D. mauritiana DNA on 3L (Tanaka et al. 2015; Hagen et al. 2019). The regions that we found to contribute to posterior lobe and clasper size differences were mutually exclusive; suggesting that different genes underlie divergence in these two structures (Tanaka et al. 2015). Furthermore, this approach revealed that sequence divergence in tartan (trn), which encodes a leucine-rich repeat transmembrane protein, contributes to the larger claspers of D. mauritiana compared with D. simulans (Hagen et al. 2019). This is likely due to more extensive and persistent expression of trn in the developing claspers in D. mauritiana (Hagen et al. 2019). However, since trn does not appear to contribute to posterior lobe size differences and explains only 16% of the clasper size difference between the species (Hagen et al. 2019), there must be additional loci involved in posterior lobe and clasper size differences on chromosome arm 3L.
To try to identify other causative genes on 3L, we generated new introgression lines to further refine existing candidate regions (Tanaka et al. 2015). We complemented this approach with RNA-seq on the developing genitalia of both species to identify genes expressed and differentially expressed both genome-wide and in the mapped regions. Subsequent functional testing of positional and expression candidate genes in D. melanogaster identified novel players involved in genital development, including the transcription factors (TFs) Grunge (Gug) and Hairy (H), which appear to positively and negatively regulate clasper size, respectively. Furthermore, we found that H represses trn expression in the developing claspers suggesting that changes in this regulatory interaction may contribute to interspecific differences in this structure. Taken together our findings provide new insights into the genetic interactions that underlie genital development, as well as the divergence of genital morphology between Drosophila species.
Results
Mapping Genomic Regions Underlying Male Genital Divergence between D. simulans and D. mauritiana
Previously, we resolved the C2 candidate region for clasper size divergence between D. simulans and D. mauritiana by successfully identifying trn as the causative gene in this region (Hagen et al. 2019). In order to increase the resolution of other candidate regions contributing to male genitalia divergence (Tanaka et al. 2015), we generated 23 new introgression lines with smaller introgressed D. mauritiana regions in a D. simulans background (fig. 1B–D and supplementary file 1, Supplementary Material online). We mapped clasper size and clasper bristle number to two regions that collectively explain 16.8% of clasper size differences between these species (table 1 and supplementary supportive text, Supplementary Material online). We confirmed the location and effect size of the previously identified C1 region (Tanaka et al. 2015) and identified a new region, C0, which explains 11% of the divergence in clasper morphology (fig. 1D, table 1, and supplementary supportive text, Supplementary Material online). We mapped posterior lobe size to five regions that collectively explain 29.3% of posterior lobe size differences between D. mauritiana and D. simulans, two of which, P4 and P5, are new (fig. 1D, table 1, and supplementary supportive text, Supplementary Material online). In total, these regions contain 380 protein-coding genes (as annotated in D. melanogaster, table 1).
Table 1.
Summary of Candidate Regions Underlying Clasper and Posterior Lobe Divergence.
| Candidate Region | Phenotypic Effect Sizea (%) | Number of Expressed Genes |
||||||
|---|---|---|---|---|---|---|---|---|
| Total Number of Genesb | Total | Diffc | Upregulatedc |
Genes Tested by RNAi in D. mel |
||||
| Dsim w 501 | Dmau D1 | Total | Developmental Candidatesd | |||||
| C0 | 11 (20) | 99 | 69 | 14 | 5 | 9 | 7 | sgl |
| C1 | 6 (21) | 58 | 35 | 6 | 4 | 2 | 32 | hairy, Cpr66D, Gug, Mcm7, foi |
| P1 | 4 | 99 | 69 | 14 | 5 | 9 | 7 | — |
| P2 | 6 | 7 | 2 | 1 | 1 | 0 | 7 | — |
| P3 | 6 | 71 | 49 | 10 | 5 | 5 | 5 | — |
| P4 | 5 | 52 | 38 | 5 | 3 | 2 | 2 | — |
| P5 | 9 | 93 | 67 | 13 | 7 | 6 | 0 | — |
The phenotypic effect size is calculated as a percentage of the difference in phenotype between the parental strains. Brackets in C regions indicate effect size for clasper bristle number.
Protein-coding orthologs in Drosophila melanogaster R6.24.
Differentially expressed between Dmau D1 and Dsim w501, Padj (FDR) < 0.05.
Genes that significantly affect either clasper size, bristle number, or posterior lobe size compared with both UAS and driver controls (P < 0.05) after RNAi knock-down are considered developmental candidates.
Analysis of Genes Expressed in Developing Male Genitalia
We next carried out RNA-seq on stages 2 and 4.5 of male genital development in D. mauritiana strain D1 (Dmau D1) and D. simulans strain w501 (Dsim w501) (Hagen et al. 2019). This allowed us to assay the genes expressed in the developing genitalia and those differentially expressed between these two species genome-wide and in our mapped regions.
We detected expression of 8,984 and 8,458 genes above the threshold value of 1 transcripts per million (TPM) in all biological replicates in the developing genital arches of Dsim w501 and Dmau D1, respectively (supplementary file 4A, Supplementary Material online). A total of 760 genes are only expressed in Dsim w501, whereas 264 genes are only expressed in Dmau D1. However, many of these genes (114 and 121 genes, respectively) have low expression in the species where they are detected (<2 TPM on an average between replicates) and therefore are less likely to underlie functional expression differences between species. Gene ontology (GO) analysis of the remaining 676 detected genes in Dsim w501 indicated the most significant enrichment is in genes involved in heme binding (supplementary file 4B, Supplementary Material online) such as Cyp4d14, Cyp9b2, Cyp6d5, Cyp6t1, Cyp4g1, Cyp12a5, Cyt-c-d, Cyp6d2, glob2, Cyp6a20, and Cyp4aa1. The remaining 143 detected genes exclusive to Dmau D1 were enriched for ion transmembrane transporters (supplementary file 4B, Supplementary Material online), the majority of which were ionotropic receptors (IRs) (IR76b, IR7g, IR60b, IR7f, IR25a) as well as the ionotropic glutamate receptor eye-enriched kainate receptor (Ekar).
Of the 8,194 genes detected in both species, 1,169 were significantly differentially expressed between Dsim w501 and Dmau D1, with 547 upregulated in the former and 622 in the latter, respectively (Padj < 0.05, supplementary file 4C, Supplementary Material online). Using the Kyoto Encyclopedia of Genes and Genomes (KEGG), we determined that 14/547 and 16/622 of these differentially expressed genes encode proteins in signaling pathways (supplementary file 4D, Supplementary Material online). This includes components of the mTOR, Notch, Hippo, Toll, and Imd pathways that are upregulated in Dsim w501, and members of the mTOR, MAPK, Wnt, FOX0, Toll, and Imd pathways that are upregulated in Dmau D1 (supplementary file 4D, Supplementary Material online). However, note that none of these genes is located in the introgressed regions we have analyzed.
To further explore divergence in gene regulation in the developing male genitalia of these species, we next assessed the expression of TF-encoding genes. We found 802 out of 994 genes-encoding TFs and cofactors are expressed in the developing genitalia of Dmau D1 and Dsim w501 according to our RNA-seq data set (supplementary file 4E, Supplementary Material online). We identified eight TF genes that appear to be exclusively expressed in the developing male genitalia of Dmau D1, whereas 16 appear to be exclusive to Dsim w501. However, three and ten of these TFs, respectively, were detected at relatively low levels (TPM < 2) and are therefore not likely to contribute to functional regulatory differences in genital development between species. Of the 778 TF genes expressed in both species (supplementary file 4E, Supplementary Material online), 49 are differentially expressed with 33 upregulated in Dmau D1, and 16 upregulated in Dsim w501 (supplementary fig. 1, Supplementary Material online). This includes five of the TF genes whose spatial expression in the developing genitalia of D. melanogaster was recently characterized (hinge3, Myb oncogene-like, single stranded-binding protein c31A, Sox21b and enhancer of split m3, helix-loop-helix) (Vincent et al. 2019).
We then focused on which of the genes in our mapped introgression regions are expressed in the developing genitalia. We found that 260 of the 380 protein-coding genes in the introgression-mapped regions could be detected in our RNA-seq data, including 31 TFs (supplementary file 5, Supplementary Material online). 49 of the expressed candidate genes are differentially expressed between Dsim w501 and Dmau D1, with about half the genes being upregulated in each (table 1). This includes one TF that is upregulated in Dsim w501 (mirror [P4]) and four in Dmau D1 (meiotic central spindle, Sox21b, CG17359 [all P5], and CG10147 [C0/P1]).
Identifying Developmental Candidate Genes
We next sought to test if the positional candidate genes that are expressed in the genitalia according to our RNA-seq data have a role in the development of either the posterior lobes or the claspers. To do this, we performed RNAi in D. melanogaster to knockdown candidate genes in the smallest posterior lobe (P2) and clasper (C1) candidate regions, as well as a selection of promising genes from the other regions based on their expression profiles (table 1 and supplementary files 5 and 6, Supplementary Material online). RNAi knockdown of the two expressed genes within P2 had no significant effect on posterior lobe size (nor on clasper size) (supplementary file 6, Supplementary Material online).
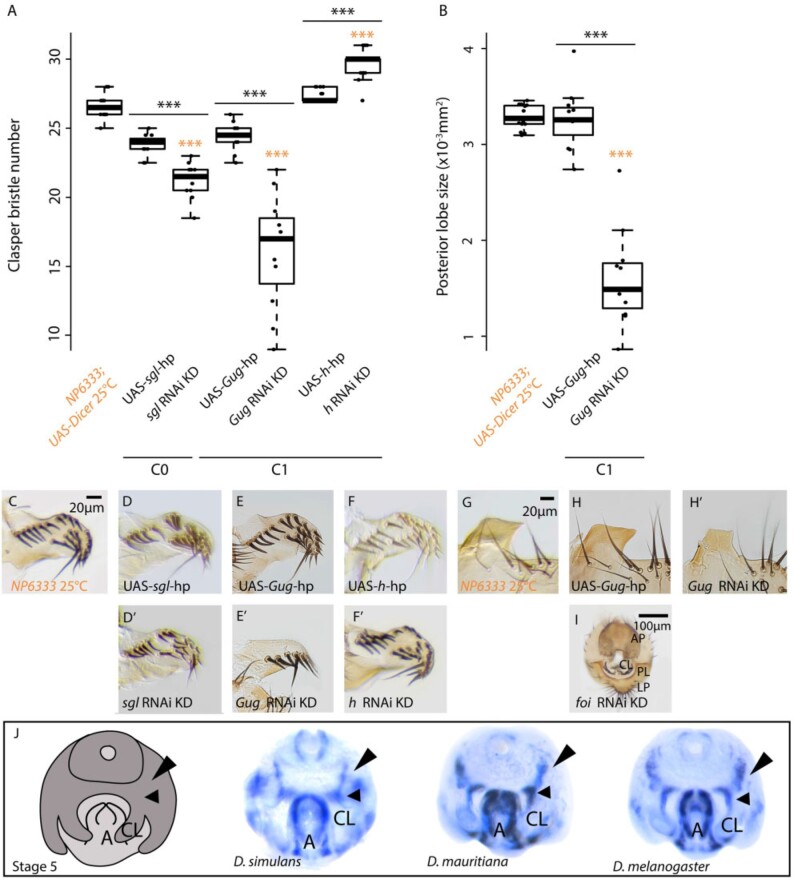
In combination with our previous study (Tanaka et al. 2015), we have now carried out RNAi for all expressed C1 candidate genes with available UAS lines (32 out of 35, supplementary file 6, Supplementary Material online). We previously observed that RNAi knockdown of cuticular protein 66D (Cpr66D) and minichromosome maintenance 7 (Mcm7) results in larger and smaller claspers, respectively (Tanaka et al. 2015). In addition to these two genes, we have now found that knocking down hairy (h), Grunge (Gug), and fear of intimacy (foi) significantly affects clasper bristle number and clasper morphology (fig. 2A, E′, F′, and I; supplementary file 6, Supplementary Material online). Knockdown of h results in larger claspers with more bristles (fig. 2A, F and F′), whereas reducing Gug expression gives rise to smaller claspers with fewer bristles (fig. 2A, E and E′). This implies that the H and Gug TFs play opposite roles in the regulation of clasper size. Interestingly, Gug also appears to positively regulate posterior lobe size; since knocking down this gene significantly reduces the size of these structures (fig. 2B, H and H′; supplementary file 6, Supplementary Material online). foi knockdown results in severe developmental defects, with fusion of the appendages of the male external genitalia including the claspers (fig. 2I).
Fig. 2.
Functional analysis of positional candidate genes in D. melanogaster male genitalia. (A) Knocking down C0 candidate gene sgl (Tanaka et al. 2015) and C1 candidate gene Gug resulted in significantly fewer clasper bristles compared with both the UAS-Gug-hp (black asterisks, P < 0.001) and NP6333 driver controls (orange asterisks, P < 0.001). In contrast, knocking down C1 candidate gene h resulted in significantly more clasper bristles compared with the NP6333 driver (P < 0.001, orange asterisks) and UAS-h-hp controls (P < 0.001, black lines and black asterisks). (B) In addition, knocking down C1 candidate gene Gug resulted in the development of significantly smaller posterior lobes compared with the UAS-Gug-hp (orange asterisks) and NP6333 driver controls (P < 0.001, black asterisks). Asterisks indicate significant differences detected with Tukey’s pairwise comparisons, where P < 0.001*** and P > 0.05 = “ns” (supplementary file 6, Supplementary Material online). Orange indicates comparisons between the NP6333 driver control and UAS-gene-hp controls/gene knockdowns, whereas comparisons between UAS controls and knockdowns are indicated by black lines and black asterisks. Boxes indicate the range, upper and lower quartiles, and median for each sample. hp, hairpin; KD, knockdown. (C–I) Morphology of claspers and posterior lobes in NP6333 driver controls, UAS controls, and gene knockdowns (D–F′ and H, H′). (J) An illustration of stage 5 male genitalia (excluding the posterior lobes) and in situ hybridizations of Cpr66D in Dsim w501, Dmau D1, and Dmel w1118. Cpr66D transcripts were detected in a wider domain along the clasper inner edge (small arrowheads) and in bands extending toward the anal plates (large arrowheads) in the two species with larger clasper. Crpr66D is also expressed in the aedeagus of all three species. CL, clasper primordia; A, aedeagus (internal genitalia). Note that sgl RNAi knockdown data were generated in Tanaka et al. (2015) and reanalysed here.
Therefore, the C1 region contains five promising clasper developmental candidate genes. However, none of these genes is differentially expressed between Dsim w501 and Dmau D1 (upregulated in the latter, supplementary file 5, Supplementary Material online). Analysis of the spatial expression of Cpr66D during genital development revealed that this gene is expressed in a wider domain along the inner clasper edge in D. melanogaster and D. mauritiana compared with D. simulans, and in bands extending from this region toward the lateral edge of the anal plates (fig. 2J).
Region C0/P1 encompasses 99 genes (table 1). About 69 of these genes are expressed in the developing genitalia according to our RNA-seq data, with 14 exhibiting significantly differential expression between Dmau D1 and Dsim w501 (table 1 and supplementary file 5, Supplementary Material online). Together with our previous study (Tanaka et al. 2015), we have now carried out RNAi against two of these differentially expressed genes (SP1173 and CG9953), and five other nondifferentially expressed genes (sugarless [sgl], CG32388, ventral veins lacking [vvl], CG10064, and lactate dehydrogenase [ImpL3]) in addition to sugarless [sgl] which we had previously knocked down (Tanaka et al. 2015). Only RNAi against sgl produced a phenotype (Tanaka et al. 2015). sgl appears to have a role in clasper development because RNAi knockdown of this gene led to significantly smaller claspers, but had no effect on the posterior lobes (fig. 2A, D, and D′; supplementary file 6, Supplementary Material online) (as shown previously in Tanaka et al. 2015).
Interactions between Genes Underlying Clasper Divergence
trn is the only gene identified so far that has been shown to contribute to clasper differences between D. simulans and D. mauritiana. The D. mauritiana allele of trn generates larger claspers with more bristles than the D. simulans allele (Hagen et al. 2019). This is likely achieved through the expanded and/or more enduring expression of trn in the developing claspers in D. mauritiana compared with D. simulans (Hagen et al. 2019). It was previously shown that the transcriptional corepressor h, represses trn expression during embryogenesis in D. melanogaster (Chang et al. 1993; Kok et al. 2015) and in Drosophila Kc cells (Bianchi-Frias et al. 2004). Since we found that h RNAi in D. melanogaster results in significantly larger claspers with more bristles (fig. 2), we hypothesized that this gene might negatively regulate clasper size through repression of trn.
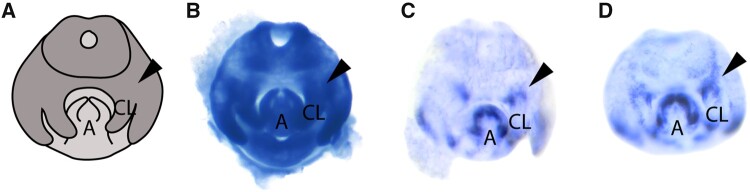
Consistent with a previous study (Vincent et al. 2019), we found that h is ubiquitously expressed throughout the male genitalia of D. melanogaster, including the clasper primordia (fig. 3B). We then analyzed the expression of trn in the developing genitalia of h RNAi knockdowns (fig. 3C and D; supplementary fig. 2, Supplementary Material online). Upon h RNAi knockdown trn expression at the base of the developing claspers at 46 hAPF/stage 5 appears to be expanded and the edges of the domain are less well defined compared with controls, with the bands of expression extending in wisps dorsally (fig. 3C and D; supplementary fig. 2, Supplementary Material online). The extreme differences in trn expression upon h RNAi are shown in supplementary figure 2, Supplementary Material online, and may explain the range of clasper bristle count data generated by this treatment (fig. 2A and supplementary file 6, Supplementary Material online). This ectopic expression indicates that the larger claspers produced upon h RNAi knockdown in D. melanogaster are likely due to increased trn expression at the base of the claspers, and that h acts upstream of trn in the clasper GRN. Despite being ubiquitously expressed throughout the male genitalia (fig. 3B), the selective targeting of trn by H may explain the role of this TF in regulating clasper but not posterior lobe development; since trn is not expressed in the developing posterior lobes (fig. 3C). However, h is expressed throughout the genital arch, and so its role in the development of other genital structures is still unclear.
Fig. 3.
trn expression expands in the clasper primordia upon h RNAi knockdown in D. melanogaster. (A) A schematic of genital development based on DAPI at stage 5 (Hagen et al. 2019). (B) h mRNA in situ hybridization in D. melanogaster w1118. (C) trn mRNA in situ hybridization on UAS parental control and (D) trn mRNA in situ hybridization upon h RNAi knockdown at stage 5. Both trn in situ hybridizations were carried out in parallel and allowed to develop for the same length of time. h RNAi results in a distortion of trn expression at the base of the claspers compared with the UAS control (black arrows). CL, claspers; A, aedeagus. N = 5 for each experiment.
Discussion
Regions on Chromosome Arm 3L Contributing to Inter- and Intraspecific Variation in Posterior Lobe and Clasper Size
As found previously, all regions identified through our introgression approach affect the claspers and/or posterior lobes consistently in the direction of their differences between the two species: Dmau D1 DNA resulted in larger claspers and smaller posterior lobes than Dsim w501 and vice versa (Zeng et al. 2000; Tanaka et al. 2015; Hagen et al. 2019).
Also consistent with previous studies, we have shown clasper area and clasper bristle number map to the same genomic locations, which suggests that the same genes may influence both traits (Tanaka et al. 2015; Hagen et al. 2019). This could at least in part be explained by the process of bristle formation through lateral inhibition (Heitzler and Simpson 1991) and consequently large claspers developing more bristles than small claspers. It is not clear, therefore, whether selection drove changes in clasper bristle number, and clasper size changed as a by-product, or vice versa. However, the interdigitization of clasper bristles with those of the female oviscapt would perhaps argue for the former scenario (Mattei et al. 2015).
Apart from C0/P1, all regions identified only affected either the claspers or the posterior lobes, which suggests different genes underlie the diversification in size of these two structures between D. simulans and D. mauritiana (fig. 1). The effects observed for C0/P1 could be explained by a single evolved locus that is able to affect growth of the claspers and posterior lobes in opposite directions with D. mauritiana C0/P1 alleles generating smaller posterior lobes and larger claspers (fig. 1B and C). Alternatively, since C0/P1 is still a relatively large region, it is possible that further mapping would resolve this region into distinct clasper and posterior lobe loci.
Interestingly, genes within region C0/P1 may underlie intraspecific variation as well as interspecific differences in posterior lobe size. This region overlaps with the 3L QTL peak observed in other interspecific mapping studies of differences in posterior lobe size between D. simulans and D. mauritiana or D. sechellia (Liu et al. 1996; Macdonald and Goldstein 1999; Zeng et al. 2000; Masly et al. 2011), as well as QTL peaks found in studies that mapped genetic variation underlying differences in posterior lobe size between D. melanogaster strains (McNeil et al. 2011; Takahara and Takahashi 2015; Hackett et al. 2016). Several other studies have also found cases where intraspecific variation maps to the same genomic region as interspecific variation (Nuzhdin and Reiwitch 2000; Gleason et al. 2002; Tatsuta and Takano-Shimizu 2006). Therefore, P1 genes represent excellent candidates for contributing to variation in the size of this structure both within and between species.
Genome-Wide Gene Expression During Genital Development in D. mauritiana and D. simulans
We carried out RNA-seq to identify and compare genes expressed in the developing genitalia between D. mauritiana and D. simulans. We were able to filter out positional candidates and also obtain a genome-wide perspective of gene activity during genital development as well as inferring differential expression between species.
In both species, all the key genes known to pattern the genital disc are expressed, such as homeotic genes and sex-determination genes (Casares et al. 1997; Chen and Baker 1997; Estrada and Sanchez-Herrero 2001; Keisman and Baker 2001; Sanchez and Guerrero 2001; Estrada et al. 2003) and signaling genes, like wingless, decapentaplegic, and hedgehog (Casares et al. 1997; Chen and Baker 1997; Abdelilah-Seyfried et al. 2000; Keisman and Baker 2001; Sanchez and Guerrero 2001). We also detected expression of 80% TF-encoding genes, including those previously shown to pattern the male genital disc, such as cubitus interruptus, engrailed (Kornberg et al. 1985; Eaton and Kornberg 1990; Simmons and Garcia-Gonzalez 2011), dachshund (Keisman and Baker 2001), distal-less (Estrada and Sanchez-Herrero 2001), and Drop (Chatterjee et al. 2011).
We found that a small proportion of genes (<10%) are exclusively expressed in the developing genitalia of either Dsim w501 or Dmau D1. The Dsim w501 male genital-specific genes are enriched for iron ion binding proteins, whereas the Dmau D1 genes are enriched for multiple IRs. IRs are a conserved family of chemosensory receptors best known for their role in olfaction (Benton et al. 2009; Grosjean et al. 2011; Silbering et al. 2011; Min et al. 2013; Ziegler et al. 2013). Interestingly, some IRs, for example, IR52c and IR52d, are candidate taste and pheromone receptors (Koh et al. 2014) expressed in a sexually dimorphic manner on the sensilla of the D. melanogaster male foreleg, which makes contact with the female during courtship (Koh et al. 2014). The neurons in which these IRs are expressed in D. melanogaster males are only activated upon contact with females of the same species (Koh et al. 2014). Therefore, the striking differences in IR expression between male Drosophila species’ genitalia may be an evolved mechanism to prevent conspecific mating.
Of the genes that are expressed in both Dsim w501 and Dmau D1, we found that 1,169 were differentially expressed. This includes 30 signaling pathway components and 49 TF-encoding genes. This suggests that the regulatory landscape of developing genitalia is generally conserved between D. mauritiana and D. simulans. However, the differentially expressed TFs will help to better understand the gene regulatory networks involved in genital development and evolution, and represent excellent candidate genes for further investigation.
Functional Analysis of Expressed Positional Candidates on Chromosome 3L During Genital Development
We have now analyzed the function of 57 of the expressed genes by RNAi knockdown in D. melanogaster, including 32 out of the 35 genes expressed in C1 (including those we studied previously in Tanaka et al. [2015]), as well as all expressed genes in P2 (table 1). Note that we did not just focus on differentially expressed genes because genes can exhibit localized differences in expression during genital development that may contribute to morphological differences (Hagen et al. 2019).
RNAi against the expressed P2 genes did not have any significant effect on the posterior lobes (supplementary file 6, Supplementary Material online). RNAi against some of these genes simply may not have worked for various reasons, including when partial knock-down of the gene may not be sufficient to result in a phenotype (although numerous P2 genes were tested with multiple RNAi constructs, supplementary file 6, Supplementary Material online). Given this caveat of potential false negatives from RNAi, this approach allows us to prioritize genes for downstream analysis rather than completely exclude them as candidates. It remains possible that a nonprotein coding element in region P2 may explain the phenotypic effect of this region on posterior lobe size. Indeed, P2 encompasses a microRNA, mir-4940, as well as a long noncoding RNA CR45408 (Thurmond et al. 2019). Therefore, the causative element in P2 could be either of these factors, or a long-range enhancer responsible for the differential regulation of a gene outside P2 between these two species.
Our functional analysis of region C0/P1 identified one excellent candidate gene, sgl. sgl has been implicated in boundary formation and may interact with Wnt signaling (Hacker et al. 1997). RNAi against sgl resulted in smaller claspers (fig. 2A and D′; Tanaka et al. 2015), but this gene is not differentially expressed between D. mauritiana and D. simulans. However, since C0/P1 is a large region that is likely to contain many other developmental candidates, higher resolution mapping, and functional analysis of genes in C0/P1 is needed.
RNAi against C1 genes revealed five interesting genes for clasper development and evolution: Gug, foi, Mcm7, Cpr66D, and h (this study; Tanaka et al. 2015). Cpr66D expression is more extensive along the inner edge of the claspers and in bands extending toward the anal plates in Dmau D1 compared with Dsim w501 (fig. 2K). Cpr66D encodes a structural protein that forms chitin-based cuticle (Ren et al. 2005; Chandran et al. 2014; Stahl et al. 2017) and its role in genital development merits further study.
We also found evidence for potential interactions between other genes in mapped regions during genital development. Repression of trn by H has been predicted (Bianchi-Frias et al. 2004; Kok et al. 2015) or shown (Chang et al. 1993) in different developmental contexts. We found that H also negatively regulates trn expression in the developing claspers of D. melanogaster; with larger claspers generated by h RNAi likely being caused by expansion of the trn expression domain (figs. 2A and F′ and 3D; supplementary fig. 2, Supplementary Material online). H also negatively regulates trn expression during embryogenesis to help define compartmental boundaries (Chang et al. 1993; Pare et al. 2019). Therefore, this regulatory interaction could represent a more general mechanism for coordinating the correct positioning of cells during development. However, h is not differentially expressed between Dsim w501 and Dmau D1 and appears to be ubiquitously expressed in the developing genitalia of D. melanogaster (fig. 3B) (Vincent et al. 2019). Although it is possible that there could also be localized differences in h expression in the developing genitalia, these observations suggest that the differences in trn expression between Dsim w501 and Dmau D1 could be the result of protein-coding changes that affect the DNA-binding efficiency of H, or variation in the number and/or sensitivity of H binding sites in trn regulatory elements. Indeed, there are several predicted H binding sites across the trn locus, but identification of trn genital enhancers and further analyses of H binding sites between D. mauritiana and D. simulans is needed to test this further.
In addition to trn, H may regulate multiple genes during clasper development including candidates revealed by our mapping and functional analyses. For example, H is also predicted to negatively regulate the C1 candidate gene, Gug (Yeung et al. 2017). Indeed, Gug itself is predicted to regulate the C0 candidate gene sgl, as this gene contains a Gug binding site in its intron (Yeung et al. 2017). However, since Gug acts as a transcriptional corepressor, and RNAi against both Gug and sgl reduces clasper size, it is unclear at this stage if there is a regulatory interaction between these genes in the developing claspers. It will be interesting to test these predictions in the future to learn more about the architecture of the gene regulatory network for clasper development and how this evolved during the rapid diversification of these structures.
Materials and Methods
Introgression Line Generation and Phenotyping
We generated new recombinants in our candidate regions by backcrossing virgin IL D11.01/Dsim w501 heterozygous females, and virgin IL D08.04/Dsim w501 heterozygous females to Dsim w501 males. IL D1101 is an introgression line with D. mauritiana D1 DNA in the genomic location 3L: 7527144.15084689 Mb and encompasses the candidate regions C1, P2, and P3 (Tanaka et al. 2015). IL D08.04 is an introgression line with D. mauritiana w− DNA on 3L: 5911371.9167745 Mb (R2.02 D. simulans) and includes candidate regions P1 and C1 (Tanaka et al. 2015). New recombinants were detected by selecting for the loss of the visible marker D1 (True et al. 1997; Tanaka et al. 2015) (fig. 1D), restriction fragment length polymorphisms, and sequencing markers (see supplementary file 7, Supplementary Material online for primer list). New introgression lines (supplementary file 1, Supplementary Material online) were all maintained as homozygous stocks.
Male genitalia were phenotyped from flies cultured under controlled growth conditions. All males used were progeny of ten females and five males that were transferred every 2 days, and allowed to develop at 25 °C in a 12-h light–12-h dark cycle incubator unless otherwise stated. All adult males were maintained on a standard cornmeal diet at 25 °C for at least 3 days before collection and storage in 70% EtOH.
Where possible, two or three replicates of ILs were phenotyped. Replicates are defined as introgression lines derived from the same recombination event and therefore containing the same introgressed region of D. mauritiana DNA. The abdominal tip and T1 leg were dissected for each fly in 70% EtOH, and transferred to Hoyer’s medium. Using entomological pins, the posterior lobes were then dissected away from the claspers and anal plates. The claspers, posterior lobes, and T1 tibia were mounted in Hoyer’s medium for imaging.
Images were taken using a Zeiss Axioplan light microscope at 250× magnification for the claspers and lobes and 160× for the T1 tibia, using a DFC300 camera. Clasper area, posterior lobe size, and tibia length were measured manually using ImageJ (Schneider et al. 2012), and bristle number was counted for each clasper (supplementary file 2A, Supplementary Material online). T1 tibia length was used as a proxy for body size, in order to assess consistency in rearing conditions and to ensure genital differences were not a result of general differences in size. Most introgression lines showed no significant difference in T1 tibia length compared with Dsim w501 (supplementary file 2G, Supplementary Material online), and since genitalia are hypoallometric (Coyne et al. 1991; Liu et al. 1996; Macdonald and Goldstein 1999; Eberhard 2009; Shingleton et al. 2009; Masly et al. 2011), the phenotypic data were not corrected for body size. A detailed description of statistical methods and the comparisons used to map candidate regions based on these data can be found in the supplementary supportive text, Supplementary Material online.
RNA Sequencing and Differential Expression Analysis
Three independent RNA-seq library replicates were generated for Dsim w501 and Dmau D1 developing male genitalia. Flies were reared under the above conditions, and white prepupae collected. Males were selected using gonad size and allowed to develop in a humid container at 25 °C until either stage 2 or stage 4.5 (Hagen et al. 2019). Between these stages, the claspers develop from a ridge structure to a distinct appendage separate from the surrounding tissue, and the posterior lobe has begun to extend outward from the lateral plate primordia (Hagen et al. 2019). The anterior of pupae were impaled with a needle onto a charcoal agar plate and submerged in 1×PBS. Dissection scissors were used to remove the distal tip of the pupal case and the outer membrane, and pressure applied to the abdomen to allow the developing genitalia to be quickly expelled from the pupal case and dissected away from the abdomen. Note that the entire genital arch, including internal genital organs (but not including abdominal tissue), was isolated for RNA extraction. The genitalia from 15 males from each stage were collected and then combined in TRIzol (ThermoScientific). RNA was then extracted using standard procedures. Quality and quantity of RNA were verified using a Qubit fluorometer. Samples were sequenced by the NERC Biomolecular Analysis Facility (NBAF) at the Centre for Genomic Research, University of Liverpool, where dual-indexed, strand-specific RNA-seq libraries were prepared using NEBNext polyA selection and Ultra Directional RNA preparation kits. Samples were then sequenced using Illumina HiSeq 4000 (paired-end, 2×150-bp sequencing). These RNA-seq data have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-9465 (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-9465). Ribosomal reads were filtered out using default settings in SortMeRNA version 4.2.0 (Kopylova et al. 2012), and Trimmomatic version 0.38 (Bolger et al. 2014) was used to trim low-quality reads using default parameters. The remaining D. simulans reads were mapped against the reannotated transcriptomes of D. simulans, “GSE76252_ReanDsim_with_ ReanDmau_GeneSet_1to1orth,” and the remaining D. mauritiana reads were mapped against the reannotated transcriptome of D. mauritiana, “GSE76252_PubDmau_ with_ReanDsim_exoutput_1to1orth” (Torres-Oliva et al. 2016), using Bowtie2 version 2.3.5 with the –very-sensitive-local option (Langmead and Salzberg 2012). Reads mapped to reannotated genomes with an overall alignment rate of 68–69%. The SAM files were then converted to BAM files, sorted by coordinate, and index files created using Samtools version 1.10 (Li et al. 2009). Duplicate reads were marked but left in the data set. These data were then used with HTSeq-count version 0.11.1 in order to generate raw read counts for each gene (Anders et al. 2015). TPM was calculated using these counts in order to quantify gene expression, and the DEseq2 R package version 1.28.1 was used to determine differential expression between species using the default parameters (Love et al. 2014). Genes were considered to be expressed if TPM > 1 in all three biological replicates. Genes were only considered differentially expressed in comparisons where Padj (FDR) < 0.05.
Gene Ontology Analysis
In order to investigate the nature of the expressed, not expressed, and differentially expressed genes in our RNA-seq data set, we determined their ontology using PANTHER version 15.0 (Thomas et al. 2003). We conducted overrepresentation tests (released 09/11/2019) of GO (released December 9, 2019) for the positional genes against the D. melanogaster reference list using the Fisher test (Thomas et al. 2006). Genes were considered significantly overrepresented when Padj (FDR) < 0.05.
Pathway Database Analysis
To identify potential differences in signaling pathway gene expression between Dsim w501 and Dmau D1 developing male genitalia, we searched for differentially expressed genes in the KEGG pathway database (Kanehisa and Goto 2000). Those annotated as signaling pathway components are reported in supplementary file 4D, Supplementary Material online.
Annotation of TFs Present in RNA-Seq Data
In order to extract the genes encoding TFs from the RNA-seq data set, we used the databases of genes from Flymine (https://www.flymine.org/flymine/begin.do;Lyne et al. 2007), amiGO (http://amigo.geneontology.org/;Carbon et al. 2009), and Flybase (Thurmond et al. 2019), and bioinformatic analysis and manual curation from Hens et al. (2011). We filtered the genes in our data set corresponding to TFs by their GO terms and gene groups in molecular function using the previously mentioned sources. The GO terms used were the following: “FlyTF_putativeTFs” from Flymine (Lyne et al. 2007), “transcription factor regulator activity” and “transcription factor coregulator activity” from amiGO (Carbon et al. 2009), “transcription factor gene group” and “transcription coregulator activity” from Flybase (Thurmond et al. 2019) and the data set of TFs from Hens et al. (2011). Genes that were annotated with these terms in any of the four resources were considered TF genes and used for downstream analysis.
RNAi Knockdown of Candidate Genes
The developmental role of genes was tested using RNAi in D. melanogaster. UAS-RNAi lines for these genes were provided by the Vienna Drosophila RNAi Center and the Bloomington Drosophila Stock Center (see supplementary file 6, Supplementary Material online for stock numbers). UAS males of candidate genes were crossed to NP6333–Gal4 (“NP6333”) driver virgins (P[GawB]PenNP6333) (Chatterjee et al. 2011) carrying UAS-Dicer-2 P[UAS-Dcr-2.D] (Stieper et al. 2008). RNAi knockdown was conducted at either 25 or 28 °C (supplementary file 6, Supplementary Material online) (Tanaka et al. 2015), under identical rearing conditions, and dissection, imaging, and analysis were carried out as described above (supplementary file 6, Supplementary Material online). To assess the role of a gene during genitalia development, we compared the phenotype of genital structures of gene knockdowns against the respective NP6333 driver controls using a Dunnett’s test (supplementary file 6, Supplementary Material online). If the gene knockdown phenotype differed significantly from the NP6333 driver control, we then assessed whether or not this significant effect is a result of genetic background (e.g., an effect of the UAS-parental phenotype), or reflects a role of the gene in genital development. To do this, we compared all three experimental groups of males using an ANOVA (supplementary file 6, Supplementary Material online). If this was significant, we then analyzed where these differences arise from using a Tukey’s test, and only concluded genes have a developmental role in the genitalia if the RNAi knockdown males were significantly different in phenotype compared with both parental controls.
In Situ Hybridization
Sample collection, RNA extraction, cDNA synthesis, and probe synthesis were conducted as described in Hagen et al. (2019). We performed in situ hybridization to detect expression of Cpr66D in D. mauritiana, D. simulans, and D. melanogaster, h in Dmel w1118 and trn in UAS-h Bloomington TRiP 27738, NP6333-Gal4; UAS-Dicer x UAS-h Bloomington TRiP 27738 using species-specific probes. Probes were generated using the following oligos (forward followed by reverse) with the addition of T7 linker sequences added to the 5′ end of each primer; trn (514 bp) ATCGAGGAGCTGAATCTGGG and TCCAGGTTACCATTGTCGCT (Hagen et al. 2019), Cpr66D (314 bp) CTCCTCGTATCAGTTTGGCTTC and CTGGTGGTACT GTGGCTGCT. Antisense h probes were generated by amplification using T7 primers from a BLUESCRIBE plasmid that contained sequences for all three h coding exons (a gift from B. Jennings, Oxford Brookes University). In situ hybridizations were based on the Carroll lab “Drosophila abdominal in situ” protocol (http://carroll.molbio.wisc.edu/methods.html) with minor modifications. All in situ hybridizations were conducted at least twice, with n = 5–10 in each experiment.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was funded by a Natural Environment Research Council grant (NE/M001040/1) to A.P.M., a Japanese Society for the Promotion of Science KAKENHI grant (15J05233) to K.M.T., and a Deutsche Forschungsgemeinschaft Fellowship (FR 3929/1-1) to F.A.F. J.F.D.H. and J.F.J. were funded by Nigel Groome studentships from Oxford Brookes University, and S.R.B. and A.M.R. by Biotechnology and Biological Sciences Research Council DTP studentships. We thank the NERC Biomolecular Analysis Facility (NBAF) at the Centre for Genomic Research, University of Liverpool for sequencing.
Author Contributions
J.F.D.H., C.C.M., A.P.M., and M.D.S.N. designed this project. J.F.D.H., C.C.M., J.F.J., F.A.F., L.B.G., S.R.B., K.M.T., A.M.R., and S.A. performed the experimental work. J.F.D.H., C.C.M., A.P.M., K.M.T., and M.D.S.N. analyzed data. J.F.D.H., A.P.M., and M.D.S.N. wrote the article.
Contributor Information
Joanna F D Hagen, Department of Biological and Medical Sciences, Oxford Brookes University, Oxford, United Kingdom.
Cláudia C Mendes, Department of Biological and Medical Sciences, Oxford Brookes University, Oxford, United Kingdom.
Shamma R Booth, Department of Biological and Medical Sciences, Oxford Brookes University, Oxford, United Kingdom.
Javier Figueras Jimenez, Department of Biological and Medical Sciences, Oxford Brookes University, Oxford, United Kingdom.
Kentaro M Tanaka, Department of Biological and Medical Sciences, Oxford Brookes University, Oxford, United Kingdom.
Franziska A Franke, Department of Biological and Medical Sciences, Oxford Brookes University, Oxford, United Kingdom.
Luis Baudouin-Gonzalez, Department of Biological and Medical Sciences, Oxford Brookes University, Oxford, United Kingdom.
Amber M Ridgway, Department of Biological and Medical Sciences, Oxford Brookes University, Oxford, United Kingdom.
Saad Arif, Department of Biological and Medical Sciences, Oxford Brookes University, Oxford, United Kingdom; Centre for Functional Genomics, Oxford Brookes University, Oxford, United Kingdom.
Maria D S Nunes, Department of Biological and Medical Sciences, Oxford Brookes University, Oxford, United Kingdom; Centre for Functional Genomics, Oxford Brookes University, Oxford, United Kingdom.
Alistair P McGregor, Department of Biological and Medical Sciences, Oxford Brookes University, Oxford, United Kingdom; Centre for Functional Genomics, Oxford Brookes University, Oxford, United Kingdom.
Data Availability
The data underlying this article have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-9465 (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-9465) or are available in its Supplementary Material online.
References
- Abdelilah-Seyfried S, Chan YM, Zeng C, Justice NJ, Younger-Shepherd S, Sharp LE, Barbel S, Meadows SA, Jan LY, Jan YN.. 2000. A gain-of-function screen for genes that affect the development of the Drosophila adult external sensory organ. Genetics 2: 733–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acebes A, Cobb M, Ferveur JF.. 2003. Species-specific effects of single sensillum ablation on mating position in Drosophila. J Exp Biol. 206(17):3095–3100. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W.. 2015. HTSeq – a Python framework to work with high-throughput sequencing data. Bioinformatics 31(2):166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB.. 2009. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136(1):149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi-Frias D, Orian A, Delrow JJ, Vazquez J, Rosales-Nieves AE, Parkhurst SM.. 2004. Hairy transcriptional repression targets and cofactor recruitment in Drosophila. PLoS Biol. 2(7):E178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S, AmiGO Hub, Web Presence Working Group. 2009. AmiGO: online access to ontology and annotation data. Bioinformatics 25(2):288–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casares F, , Sánchez L, , Guerrero I, , Sánchez-Herrero E. 1997. The genital disc of Drosophila melanogaster. I. Segmental and compartmental organization. Dev Genes Evol. 207(4):216–228. [DOI] [PubMed] [Google Scholar]
- Chandran RR, Iordanou E, Ajja C, Wille M, Jiang L.. 2014. Gene expression profiling of Drosophila tracheal fusion cells. Gene Expr Patterns. 15(2):112–123. [DOI] [PubMed] [Google Scholar]
- Chang Z, Price BD, Bockheim S, Boedigheimer MJ, Smith R, Laughon A.. 1993. Molecular and genetic characterization of the Drosophila tartan gene. Dev Biol. 160(2):315–332. [DOI] [PubMed] [Google Scholar]
- Chatterjee SS, Uppendahl LD, Chowdhury MA, Ip PL, Siegal ML.. 2011. The female-specific doublesex isoform regulates pleiotropic transcription factors to pattern genital development in Drosophila. Development 138(6):1099–1109. [DOI] [PubMed] [Google Scholar]
- Chen EH, Baker BS.. 1997. Compartmental organization of the Drosophila genital imaginal discs. Development 1: 205–218. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Rux J, David JR.. 1991. Genetics of morphological differences and hybrid sterility between Drosophila–sechellia and its relatives. Genet Res. 57(2):113–122. [DOI] [PubMed] [Google Scholar]
- Eaton S, Kornberg TB.. 1990. Repression of ci-D in posterior compartments of Drosophila by engrailed. Genes Dev. 6: 1068–1077. [DOI] [PubMed] [Google Scholar]
- Eberhard WG. 1985. Sexual selection and animal genitalia. Cambridge: Harvard University Press. [Google Scholar]
- Eberhard WG. 2009. Static allometry and animal genitalia. Evolution 63(1):48–66. [DOI] [PubMed] [Google Scholar]
- Eberhard WG. 2010. Evolution of genitalia: theories, evidence, and new directions. Genetica 1:5–18. [DOI] [PubMed] [Google Scholar]
- Estrada B, Casares F, Sánchez-Herrero E.. 2003. Development of the genitalia in Drosophila melanogaster. Differentiation 6: 299–310. [DOI] [PubMed] [Google Scholar]
- Estrada B, Sanchez-Herrero E.. 2001. The Hox gene Abdominal-B antagonizes appendage development in the genital disc of Drosophila. Development 3: 331–339. [DOI] [PubMed] [Google Scholar]
- Garrigan D, Kingan SB, Geneva AJ, Andolfatto P, Clark AG, Thornton KR, Presgraves DC.. 2012. Genome sequencing reveals complex speciation in the Drosophila simulans clade. Genome Res. 22(8):1499–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glassford WJ, Johnson WC, Dall NR, Smith SJ, Liu Y, Boll W, Noll M, Rebeiz M.. 2015. Co-option of an ancestral Hox-regulated network underlies a recently evolved morphological novelty. Dev Cell. 34(5):520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason JM, Nuzhdin SV, Ritchie MG.. 2002. Quantitative trait loci affecting a courtship signal in Drosophila melanogaster. Heredity 89(1):1–6. [DOI] [PubMed] [Google Scholar]
- Grosjean Y, Rytz R, Farine JP, Abuin L, Cortot J, Jefferis GS, Benton R.. 2011. An olfactory receptor for food-derived odours promotes male courtship in Drosophila. Nature 478(7368):236–240. [DOI] [PubMed] [Google Scholar]
- Hacker U, Lin X, Perrimon N.. 1997. The Drosophila sugarless gene modulates Wingless signaling and encodes an enzyme involved in polysaccharide biosynthesis. Development 18:3565–3573. [DOI] [PubMed] [Google Scholar]
- Hackett JL, Wang X, Smith BR, Macdonald SJ.. 2016. Mapping QTL contributing to variation in posterior lobe morphology between strains of Drosophila melanogaster. PLoS One 11(9):e0162573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen JFD, Mendes CC, Blogg A, Payne A, Tanaka KM, Gaspar P, Figueras Jimenez J, Kittelmann M, McGregor AP, Nunes MDS.. 2019. tartan underlies the evolution of Drosophila male genital morphology. Proc Natl Acad Sci U S A. 116(38):19025–19030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzler P, Simpson P.. 1991. The choice of cell fate in the epidermis of Drosophila. Cell 64(6):1083–1092. [DOI] [PubMed] [Google Scholar]
- Hens K, Feuz JD, Isakova A, Iagovitina A, Massouras A, Bryois J, Callaerts P, Celniker SE, Deplancke B.. 2011. Automated protein-DNA interaction screening of Drosophila regulatory elements. Nat Methods. 8(12):1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosken DJ, Stockley P.. 2004. Sexual selection and genital evolution. Trends Ecol Evol. 19(2):87–93. [DOI] [PubMed] [Google Scholar]
- House CM, Lewis Z, Hodgson DJ, Wedell N, Sharma MD, Hunt J, Hosken DJ.. 2013. Sexual and natural selection both influence male genital evolution. PLoS One 8(5):e63807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadeeshan S, Singh RS.. 2006. A time-sequence functional analysis of mating behaviour and genital coupling in Drosophila: role of cryptic female choice and male sex-drive in the evolution of male genitalia. J Evol Biol. 19(4):1058–1070. [DOI] [PubMed] [Google Scholar]
- Kamimura Y, Mitsumoto H.. 2011. Comparative copulation anatomy of the Drosophila melanogaster species complex (Diptera: Drosophilidae). Entomol Sci. 14(4):399–410. [Google Scholar]
- Kanehisa M, Goto S.. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28(1):27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keisman EL, Baker BS.. 2001. The Drosophila sex determination hierarchy modulates wingless and decapentaplegic signaling to deploy dachshund sex-specifically in the genital imaginal disc. Development 9: 1643–1656. [DOI] [PubMed] [Google Scholar]
- Kittelmann S, Buffry AD, Franke FA, Almudi I, Yoth M, Sabaris G, Couso JP, Nunes MDS, Frankel N, Gomez-Skarmeta JL, et al. 2018. Gene regulatory network architecture in different developmental contexts influences the genetic basis of morphological evolution. PLoS Genet. 14(5):e1007375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh TW, He Z, Gorur-Shandilya S, Menuz K, Larter NK, Stewart S, Carlson JR.. 2014. The Drosophila IR20a clade of ionotropic receptors are candidate taste and pheromone receptors. Neuron 83(4):850–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok K, Ay A, Li LM, Arnosti DN.. 2015. Genome-wide errant targeting by Hairy. Elife 4: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp A, True JR.. 2002. Evolution of male sexual characters in the oriental Drosophila melanogaster species group. Evol Dev. 4(4):278–291. [DOI] [PubMed] [Google Scholar]
- Kopylova E, Noé L, Touzet H.. 2012. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28(24):3211–3217. [DOI] [PubMed] [Google Scholar]
- Kornberg T, Siden I, O’Farrell P, Simon M.. 1985. The engrailed locus of Drosophila: in situ localization of transcripts reveals compartment-specific expression. Cell 1: 45–53. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie CC, True JR, Liu J, Mercer JM.. 1997. An introgression analysis of quantitative trait loci that contribute to a morphological difference between Drosophila simulans and D. mauritiana. Genetics 2:339–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Mercer JM, Stam LF, Gibson GC, Zeng ZB, Laurie CC.. 1996. Genetic analysis of a morphological shape difference in the male genitalia of Drosophila simulans and D. mauritiana. Genetics 4:1129–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyne R, Smith R, Rutherford K, Wakeling M, Varley A, Guillier F, Janssens H, Ji W, McLaren P, North P, et al. 2007. FlyMine: an integrated database for Drosophila and Anopheles genomics. Genome Biol. 8(7):R129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald SJ, Goldstein DB.. 1999. A quantitative genetic analysis of male sexual traits distinguishing the sibling species Drosophila simulans and D. sechellia. Genetics 4:1683–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Orgogozo V.. 2013. The loci of repeated evolution: a catalog of genetic hotspots of phenotypic variation. Evolution 5:1235–1250. [DOI] [PubMed] [Google Scholar]
- Masly JP, Dalton JE, Srivastava S, Chen L, Arbeitman MN.. 2011. The genetic basis of rapidly evolving male genital morphology in Drosophila. Genetics 189(1):357–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masly JP, Kamimura Y.. 2014. Asymmetric mismatch in strain-specific genital morphology causes increased harm to Drosophila females. Evolution 8:2401–2411. [DOI] [PubMed] [Google Scholar]
- Mattei AL, Riccio ML, Avila FW, Wolfner MF.. 2015. Integrated 3D view of postmating responses by the Drosophila melanogaster female reproductive tract, obtained by micro-computed tomography scanning. Proc Natl Acad Sci U S A. 27:8475–8480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil CL, Bain CL, Macdonald SJ.. 2011. Multiple quantitative trait loci influence the shape of a male-specific genital structure in Drosophila melanogaster. G3 (Bethesda) 5:343–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min S, Ai M, Shin SA, Suh GS.. 2013. Dedicated olfactory neurons mediating attraction behavior to ammonia and amines in Drosophila. Proc Natl Acad Sci U S A. 14:1321–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes MD, Arif S, Schlotterer C, McGregor AP.. 2013. A perspective on micro-evo-devo: progress and potential. Genetics 195(3):625–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuzhdin SV, Reiwitch SG.. 2000. Are the same genes responsible for intra- and interspecific variability for sex comb tooth number in Drosophila? Heredity 84(1):97–102. [DOI] [PubMed] [Google Scholar]
- Pare AP, Naik P, Shi J, Mirman Z, Palmquist KH, Zallen JA.. 2019. An LRR receptor-teneurin system directs planar polarity at compartment boundaries. Dev Cell. 51(2):208–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren N, Zhu C, Lee H, Adler PN.. 2005. Gene expression during Drosophila wing morphogenesis and differentiation. Genetics 171(2):625–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM. 1988. Mating asymmetries and phylogeny in the Drosophila melanogaster species complex. Pac Sci. 42(1–2):72–80. [Google Scholar]
- Sanchez L, Guerrero I.. 2001. The development of the Drosophila genital disc. Bioessays 8: 698–707. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW.. 2012. NIH image to ImageJ: 25 years of image analysis. Nat Methods. 9(7):671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingleton AW, Estep CM, Driscoll MV, Dworkin I.. 2009. Many ways to be small: different environmental regulators of size generate distinct scaling relationships in Drosophila melanogaster. Proc R Soc B. 276(1667):2625–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silbering AF, Rytz R, Grosjean Y, Abuin L, Ramdya P, Jefferis GS, Benton R.. 2011. Complementary function and integrated wiring of the evolutionarily distinct Drosophila olfactory subsystems. J Neurosci. 31(38):13357–13375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons LW. 2014. Sexual selection and genital evolution. Aust Entomol. 53(1):1–17. [Google Scholar]
- Simmons LW, Garcia-Gonzalez F.. 2011. Experimental coevolution of male and female genital moprhology. Nat Commun. 374: 1–6. [DOI] [PubMed] [Google Scholar]
- Stahl AL, Charlton-Perkins M, Buschbeck EK, Cook TA.. 2017. The cuticular nature of corneal lenses in Drosophila melanogaster. Dev Genes Evol. 227(4):271–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL. 2011. Evolution, development, & the predictable genome. Colorado: Roberts and Company Publishers. [Google Scholar]
- Stieper BC, Kupershtok M, Driscoll MV, Shingleton AW.. 2008. Imaginal discs regulate developmental timing in Drosophila melanogaster. Dev Biol. 321(1):18–26. [DOI] [PubMed] [Google Scholar]
- Takahara B, Takahashi KH.. 2015. Genome-wide association study on male genital shape and size in Drosophila melanogaster. PLoS One 10(7):e0132846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka KM, Hopfen C, Herbert MR, Schlotterer C, Stern DL, Masly JP, McGregor AP, Nunes MD.. 2015. Genetic architecture and functional characterization of genes underlying the rapid diversification of male external genitalia between Drosophila simulans and Drosophila mauritiana. Genetics 200(1):357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka KM, Kamimura Y, Takahashi A.. 2018. Mechanical incompatibility caused by modifications of multiple male genital structures using genomic introgression in Drosophila. Evolution 72(11):2406–2418. [DOI] [PubMed] [Google Scholar]
- Tatsuta H, Takano-Shimizu T.. 2006. Genetic architecture of variation in sex-comb tooth number in Drosophila simulans. Genet Res. 87(2):93–107. [DOI] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A.. 2003. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13(9):2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Kejariwal A, Guo N, Mi H, Campbell MJ, Muruganujan A, Lazareva-Ulitsky B.. 2006. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 1:645–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurmond J, Goodman JL, Strelets VB, Attrill H, Gramates LS, Marygold SJ, Matthews BB, Millburn G, Antonazzo G, Trovisco V, et al. 2019. FlyBase 2.0: the next generation. Nucleic Acids Res. 47(D1):D759–D765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Oliva M, Almudi I, McGregor AP, Posnien N.. 2016. A robust (re-)annotation approach to generate unbiased mapping references for RNA-seq-based analyses of differential expression across closely related species. BMC Genomics 17(1):1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True JR, Liu J, Stam LF, Zeng Z-B, Laurie CC.. 1997. Quantitative genetic analysis of divergence in male secondary sexual traits between Drosophila simulans and Drosophila mauritiana. Evolution 51(3):816–832. [DOI] [PubMed] [Google Scholar]
- Vincent BJ, Rice GR, Wong GM, Glassford WJ, Downs KI, Shastay JL, Charles-Obi K, Natarajan M, Gogol M, Zeitlinger J, et al. 2019. An atlas of transcription factors expressed in male pupal terminalia of Drosophila melanogaster. G3 (Bethesda) 12:3961–3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassin A, Orgogozo V.. 2013. Coevolution between male and female genitalia in the Drosophila melanogaster species subgroup. PLoS One 8(2):e57158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung K, Boija A, Karlsson E, Holmqvist P, Tsatskis Y, Nisoli I, Yap D, Lorzadeh A, Moksa M, Hirst M, et al. 2017. Atrophin controls developmental signaling pathways via interactions with Trithorax-like. eLife 6:e23084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng ZB, Liu JJ, Stam LF, Kao CH, Mercer JM, Laurie CC.. 2000. Genetic architecture of a morphological shape difference between two drosophila species. Genetics 1:299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler AB, Berthelot-Grosjean M, Grosjean Y.. 2013. The smell of love in Drosophila. Front Physiol. 72:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-9465 (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-9465) or are available in its Supplementary Material online.