Abstract
Embryonic stem cells (ESCs) can self-renew indefinitely and maintain their pluripotency status. The pluripotency gene regulatory network is critical in controlling these properties and particularly chromatin remodeling complexes. In this review, we summarize the research progresses of the functional and mechanistic studies of BAF complex in mouse ESCs and early embryonic development. A discussion of the mechanistic bases underlying the distinct phenotypes upon the deletion of different BAF subunits in ESCs and embryos will be highlighted.
1. Introduction
Embryonic stem cells (ESCs) are derived from the inner cell mass of blastocysts in early embryos [1–3]. With the remarkable abilities to indefinitely self-renew and differentiate to all types of cells in the body, ESCs become an ideal model to study cell fate determination and lineage differentiation, therefore having broad applications in the fields of regenerative medicine and translational medicine.
Since their isolation, the mechanism underlying the self-renewal and pluripotency of ESCs has been the focus of intensive research in the field of stem cell biology [4]. Numerous studies demonstrate that the identity of ESCs is controlled by a core transcriptional regulatory network composed of signaling pathways such as the LIF/STAT3 pathway [4–6], pluripotent transcription factors such as OCT4, SOX2, NANOG, and KLF4 [7–9], protein complexes [10–12], microRNAs [13], and chromatin remodeling complexes [12].
2. Chromatin Remodeling Complexes
Specific transcriptomes expressed in different types of mammalian cells are controlled partly by their unique chromatin states. The regulation of chromatin states selectively causes gene expression or silencing via controlling the access of transcriptional factors to gene regulatory elements. This variation of transcriptional activity according to the chromatin structural changes is called chromatin remodeling [14]. There are two main types of chromatin remodeling: one is covalent histone modification, which includes acetylation, methylation, phosphorylation, and ubiquitination; the other is ATP-dependent physical modification, which is achieved mainly through ATP-dependent protein complexes [14].
The ATP-dependent protein complexes with ATPase activity, termed chromatin remodeling complex, use the energy generated by hydrolysis of ATP to make the four changes in the nucleosomes structure and thereby regulate gene expression (Figure 1) [15].
Figure 1.
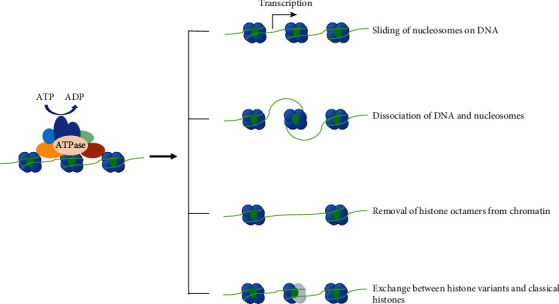
Schematic of the chromatin remodeling complex functional mode. Chromatin remodeling complex, which has ATPase activity, could change the structure of nucleosomes with the energy generated by hydrolyzing ATP, to regulate the accessibility of chromatin and further affect gene expression.
According to the difference in structure and composition of ATPase, chromatin remodeling complexes are divided into four categories: switching (SWI)/sucrose nonfermenting (SNF) [16, 17], INO80 [18], ISWI (imitation SWI) [19], and CHD (chromodomain helicase DNA binding) [20].
3. Structure and Function of SWI/SNF
The SWI/SNF complex is first discovered in yeast [16] and later in Drosophila [21] and mammals [22, 23]. The mammalian SWI/SNF complex, also named BAF (BRG1/BRM-associated factor) complex, is a multi-subunit protein complex of about 2 MDa, which is composed of 12-15 subunits encoded by 29 genes [24]. According to the different composition of subunits, BAF complexes are divided into canonical BAF (cBAF), PBAF, and noncanonical (ncBAF) [25]. The structural characteristics of the three types of SWI/SNF complexes are shown in Figure 2. Recent studies reveal the assembly process of these three types of BAF complexes (Figure 2) [25, 26] In different developmental stages and different tissues, the composition of the BAF complex also changes to regulate distinct gene expression, thereby performing different functions [27].
Figure 2.

Three types of SWI/SNF assembly processes. (a) BAF: ARID1 and BAF core modules form a subcomplex (ARID1/BAF core), then combine with DPF2, and recruit ATPase modules (including SS18) to complete BAF assembly; (b) PBAF: ARID2 first combines with BAF core to form a subcomplex (ARID2/PBAF core), then combines BRD7 and PHF10, followed by the recruitment of ATPase module (excluding SS18), and finally combines with PBRM1 to complete PBAF assembly; (c) ncBAF: GLTSCAR1/1L BRD9 combines with BAF core module to form the core module of ncBAF and combines BRD9 with ATPase module (containing SS18) to form the ncBAF complex [25, 26].
4. The Role of BAF Complex in mESCs
esBAF, a specific BAF complex in ESCs, consists of 9-11 subunits, which includes the ATPase subunit BRG1 not BRM, BAF250a instead of BAF200, BAF60a/b instead of BAF60c, and BAF155 dimer instead of BAF155 and BAF170 (Figure 3) [28]. Numerous studies reveal the functional importance of the BAF complex in ESCs and embryonic development [28–30]. Here, we summarize the roles of various subunits of the esBAF complex in ESCs (Table 1) and embryonic development (Table 2).
Figure 3.
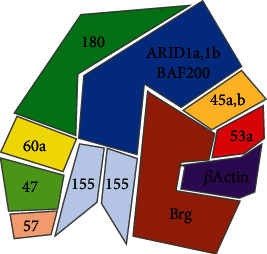
The subunits constituent of the esBAF complex.
Table 1.
The role of BAF subunits in mESCs.
| Subunit | Phenotypes | References |
|---|---|---|
| BRG1 | Knockdown or knockout of Brg1 resulted in ESC differentiation and downregulation of self-renewal and pluripotency genes such as Oct4 and Sox2. | [28, 31, 32] |
| BAF47 | Knockdown of Baf47 blocks differentiation; overexpression of Baf47 enhances differentiation; knockdown of Baf57 upregulates Cdx2 expression. | [33, 34] |
| BAF155 | Depletion of BAF155 resulted in decreased proliferation, decreased Oct4 expression, and increased apoptosis of ESCs. | [28] |
| BAF250a | The self-renewal ability of mESCs decreases after knocking out BAF250a, and the differentiation of ES cells into the mesoderm and endoderm is inhibited. | [35, 36] |
| BAF45d | Knockout of BAF45d perturbs ESC self-renewal and impairs its differentiation to three lineages. | [30] |
| BAF53a | Knockdown of Baf53a reduces the expression of pluripotent genes in ESCs. Baf53a protects mESCs from differentiating into primitive endoderm; knockout of Baf53a represses cell proliferation and induces cell apoptosis. | [37, 38] |
| BRD9 | Preserving the naive pluripotency of ESCs | [39] |
Table 2.
The role of BAF subunits in early mouse embryonic development.
| Subunit | Phenotypes | References |
|---|---|---|
| BRG1 | Brg1 null embryos die during implantation, and mice heterozygous for Brg1 are prone to cause tumor formation and anencephaly. | [32, 40] |
| BAF47 | Baf47 null mice die during embryo implantation. Baf47 heterozygous mice are prone to cause anencephaly. | [41, 42] |
| BAF155 | Baf155 knockout embryos are lethal during implantation. | [29, 34] |
| BAF250a | Baf250a knockout embryos die on E6.5. Baf250a regulates heart development. | [35, 43] |
4.1. BRG1
As the core catalytic subunit of the esBAF complex, BRG1 alone can reshape nucleosomes in vitro, but the efficiency is very low. The smallest complex of four subunits, BAF155, BAF170, Baf47, and BRG1, can exert catalytic activity efficiently [44].
BRG1 participates in chromatin remodeling to maintain ESC self-renewal and pluripotency [28, 31]. The absence of Brg1 results in the impairment of ESC self-renewal and pluripotency [28, 31, 32]. Deletion of Brg1 leads to the decreased expression of Oct4 and Sox2 and increased expression of lineage-specific genes, indicating its function in ESC differentiation [28, 31]. The BRG1 null embryos die at the blastocyst stage. ES cells cannot be isolated from Brg1-deficient embryos [40, 45].
BRG1 directly binds the promoter regions of Oct4, Sox2, and Nanog genes, indicating its regulatory roles on the expression of core pluripotency genes. Consistently, BRG1 interacts directly with NANOG, OCT4, and SOX2 and binds with many of their common target genes [30, 46].
In addition, BRG1 also regulates the expression of ESC-related genes by participating in LIF/STAT3 signaling pathways [47]. Leukemia Inhibitory Factor (LIF) is required to maintain the pluripotency of mESCs and naïve human ESCs [5, 6, 48]. In mESCs, the binding of BRG1 and STAT3 colocalize extensively on the genome [30, 47]. The binding of STAT3 to genes associated with pluripotency depends on the presence of the catalytic subunit BRG1 in the esBAF complex, which loosens the chromatin structure at the target gene of STAT3 and thus responds to the LIF signal [47]. BRG1 can enhance the LIF-STAT3 signaling pathway by antagonizing the PcG complex [47]. On the other hand, BRG1 and the PRC complex bind together to four Hox loci, thereby inhibiting the differentiation of ESCs [47].
Recently, YY1 was reported to interact with BRG1 to promote proliferation and pluripotency of mouse ESCs. The knockdown of Yy1 gene downregulates Nanog and upregulates differentiation marker genes Pax3 and Cdx2 [49].
The interaction between BRG1 and TOP2 is required for the initial stage of accessibility induction. Top2 can make the chromatin more accessible for chromatin remodelers as well as transcription factors [50], suggesting that TOP2 may work together with the BAF complex to remodel chromatin and optimize BAF-mediated recruitment of transcriptional factors.
4.2. BAF47
BAF47 (also known as SMARCB1/SNF5/INI1) is involved in the differentiation of stem cells. The knockdown of BAF47 enhances cell pluripotency and prevents differentiation [33]. Overexpressing BAF47 promotes ESC differentiation. BAF47 can fine-tune the level of OCT4 and affect the nucleosome occupation at the regulatory region of OCT4 target genes, thus breaking the balance between pluripotency and differentiation and determining the fate of cells [33]. In contrast, a recent report indicates the upregulated Cdx2 expression in Baf47 KO ESCs [34]. Therefore, further study to clarify the function of Baf47 in ESCs is needed.
The BAF47 null blastocysts do not hatch and cannot implant into the uterus for further development [41, 42], which may cause death of Baf47 null embryos during implantation [41, 42].
4.3. BAF155 and BAF170
BAF155 (also known as SRG3) shares 61.7% amino acid homology with BAF170, but they have different functions [28]. The esBAF complex contains a homodimer of two BAF155 without BAF170 [28]. The deletion of BAF155 resulted in the defects of ESC self-renewal and pluripotency [28]. As expected, overexpression of BAF170 cannot restore the defects of Brg155 KO ESCs [28]. Similarly, knockdown of Baf155 expression also resulted in inhibited ESC proliferation, decreased expression of the pluripotent gene Oct4, and increased apoptosis [28]. Consistently, deletion of BAF155 fails to form inner cell mass [51].
In contrast to mESCs, esBAF in hESCs contains heterodimers composed of BAF155 and BAF170. The contents of BAF155 and BAF170 in the BAF complex seem to determine the fate of the cell [52].
The deletion of BAF155 prevented mouse embryos from developing properly and died during implantation [29]. Depletion of BAF155 leads to increased expression of Nanog in the ICM and its ectopic expression in TE. However, the overexpression of BAF155 leads to the development arrested at the E3.5 to E4.5 transition and upregulation of Cdx2 and Sox17 at E4.5 embryos [29].
4.4. BAF53a
BAF53a (also known as ACTL6a or ARP4) is expressed in a variety of stem/progenitor cells, including neural progenitor cells, hematopoietic stem cells, epidermal progenitor cells, and ES cells [37, 38, 53]. The knockdown of BAF53a in ESCs reduces the expression of pluripotent genes such as Oct4 and Nanog and induces ESC differentiation towards the original endoderm [37]. It is interesting that another report indicates that knockout of Baf53a increases the expression of Oct4 and Nanog. Deletion of Baf53a repressed cell proliferation and induced apoptosis [54].
4.5. BAF45
BAF45 has two PHD domains, which can recruit the BAF complex to specific histone modification sites [55]. BAF45 includes four subunits: BAF45a, BAF45b, BAF45c, and BAF45d [30]. Only BAF45a and BAF45d are contained in esBAF [30]. BAF45a plays an important role in the maintenance of hematopoietic stem cells [56], but its role in mESCs is not clear. BAF45d, also known as Dpf2, is widely expressed in a variety of cells [30]. Deletion of Dpf2 in mESCs leads to the differentiation defects, which cannot be restored by BAF45a and BAF45c [30]. Further study demonstrates that Dpf2 regulates ESC differentiation by regulating Tbx3 expression [30].
4.6. BAF250a
BAF250a (ARID1A) is a unique subunit of esBAF, which belongs to the trithorax group (TrxG) family [57, 58]. BAF250a is abundantly expressed in early mouse embryos and ESCs [35, 36]. Deletion of BAF250a inhibits ESC self-renewal and upregulates the expression of the primitive endoderm marker genes in ESCs [35, 36]. The lack of BAF250a prevents ESCs from developing into mesoderm-derived cardiomyocytes, adipocytes, and skeletal muscle cells, but can differentiate into ectoderm-derived nerve cells [35, 43].
BAF250a is necessary for the development of early embryos. The loss of BAF250a caused the development of early embryos (E6.5) of mice to stagnate, and the lack of mesoderm prevented further development of gastrulation embryos [35].
4.7. ncBAF in ESCs
Gatchalian and colleagues found the existence of ncBAF in mESCs that puts BRD9 as the core [39]. Compared with esBAF, ncBAF lacks BAF47, BAF57, and ARID1A subunits. The knockdown of BRD9, the core subunit of the ncBAF complex, inhibited the proliferation of ESCs [39]. Although both esBAF and ncBAF are involved in ESC self-renewal and pluripotency maintenance, ChIP-seq analysis showed that esBAF and ncBAF complexes target distinct sites in the genome and cobound with different pluripotent transcription factors (TFs) [30, 39, 45]. esBAF tends to bind to active enhancers rich in h3k4me1 modification [30], while ncBAF is more likely to bind to promoter regions rich in h3k4me3 [39]. Different from esBAF, ncBAF tends to cobind with KLF4 and CTCF, indicating its distinct mechanisms from cBAF in ESCs [39].
In summary, different components of the BAF complex function differentially in ESC maintenance and differentiation. Deletion of core subunits such as Brg1, Baf155, or Baf250a reduced the expression of Oct4, Sox2, and Nanog, the key pluripotency genes of ES cells [28, 31]. On the contrary, Baf47 negatively regulated Oct4 expression in ESCs [33]. Deletion of Baf250a promoted the expression of endoderm marker genes Gata4 and Gata6 [35], while deletion of Baf45d decreased Tbx3 expression and impaired mesoendoderm differentiation [30]. During embryonic development, knockout of Brg1, Baf155, or Baf250a led to embryo death during peri-implantation [40–42, 51]. Deletion of Baf250a resulted in embryo death in later embryonic development stage [35].
Consistently, BAF complexes also play important roles in the reprogramming of somatic cells to induced pluripotent stem cells (iPSCs). Depletion of Brg1 leads to the failures in reprogramming [59, 60]. Overexpression of Brg1 and Baf155 increases the reprogramming efficiency of MEFs to iPSCs [61], whereas downregulation of Brm and Baf170 improves reprogramming efficiency [62]. Therefore, similar to the distinct roles of different BAF subunits for the maintenance and differentiation of ES cells, different BAF components also play different roles in the reprogramming.
5. Conclusion
BAF complexes are functionally important for the self-renewal and development of ESCs and mouse embryonic development. Deletion of different subunits in ESCs and embryos results in distinct phenotypes in ESC maintenance and differentiation and embryonic development, while the underlying mechanisms are far from clear. Schick et al.'s work reveals that the loss of a single subunit of the BAF complex did not destroy the entire complex, but will change the composition of the BAF complex [24]. Consistently, a recent study shows that deletion of Dpf2 only affects about 8% of BRG1 binding sites on the genome [30]. Therefore, it is attractive to propose that distinct BAF subunit controls the integrity of a part of the BAF complex on the genome, and therefore, its deletion only affects the binding of a part of the BAF complex, which directly changes the expression of distinct pluripotency TFs in both ESCs and differentiating cells with other TFs and chromatin modifiers. It is intriguing to extend the proposed mechanism further to other chromatin remodeling complexes. To confirm the proposal, future works are required to study the deletion of specific subunits on the binding of BRG1 and some other core factors of BAF and other chromatin remodeling complexes.
Contributor Information
Ronghua Shi, Email: rhsh@ustc.edu.cn.
Ying Ye, Email: yye@suda.edu.cn.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
Heyao Zhang, Xuepeng Wang, and Jinsheng Li are co-first authors.
References
- 1.Evans M. J., Kaufman M. H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292(5819):154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 2.Martin G. R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proceedings of the National Academy of Sciences of the United States of America. 1981;78(12):7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomson J. A., Itskovitz-Eldor J., Shapiro S. S., et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 4.Surani M. A., Hayashi K., Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128(4):747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 5.Matsuda T., Nakamura T., Nakao K., et al. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. The EMBO Journal. 1999;18(15):4261–4269. doi: 10.1093/emboj/18.15.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niwa H., Burdon T., Chambers I., Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes & Development. 1998;12(13):2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loh K. M., Lim B. A precarious balance: pluripotency factors as lineage specifiers. Cell Stem Cell. 2011;8(4):363–369. doi: 10.1016/j.stem.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 8.Thomson M., Liu S. J., Zou L. N., Smith Z., Meissner A., Ramanathan S. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell. 2011;145(6):875–889. doi: 10.1016/j.cell.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim J., Chu J., Shen X., Wang J., Orkin S. H. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132(6):1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyer L. A., Plath K., Zeitlinger J., et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441(7091):349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 11.Lee T. I., Jenner R. G., Boyer L. A., et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125(2):301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tee W. W., Reinberg D. Chromatin features and the epigenetic regulation of pluripotency states in ESCs. Development. 2014;141(12):2376–2390. doi: 10.1242/dev.096982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marson A., Levine S. S., Cole M. F., et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134(3):521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harikumar A., Meshorer E. Chromatin remodeling and bivalent histone modifications in embryonic stem cells. EMBO Reports. 2015;16(12):1609–1619. doi: 10.15252/embr.201541011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hasan N., Ahuja N. The emerging roles of ATP-dependent chromatin remodeling complexes in pancreatic cancer. Cancers. 2019;11(12) doi: 10.3390/cancers11121859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neigeborn L., Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics. 1984;108(4):845–858. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stern M., Jensen R., Herskowitz I. Five _SWI_ genes are required for expression of the _HO_ gene in yeast. Journal of Molecular Biology. 1984;178(4):853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- 18.Conaway R. C., Conaway J. W. The INO80 chromatin remodeling complex in transcription, replication and repair. Trends in Biochemical Sciences. 2009;34(2):71–77. doi: 10.1016/j.tibs.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 19.Klement K., Luijsterburg M. S., Pinder J. B., et al. Opposing ISWI- and CHD-class chromatin remodeling activities orchestrate heterochromatic DNA repair. The Journal of Cell Biology. 2014;207(6):717–733. doi: 10.1083/jcb.201405077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murawska M., Brehm A. CHD chromatin remodelers and the transcription cycle. Transcription. 2014;2(6):244–253. doi: 10.4161/trns.2.6.17840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kennison J. A., Tamkun J. W. Trans-regulation of homeotic genes in Drosophila. The New Biologist. 1992;4(2):91–96. [PubMed] [Google Scholar]
- 22.Wolffe A. P. Transcriptional activation: switched-on chromatin. Current Biology. 1994;4(6):525–528. doi: 10.1016/S0960-9822(00)00114-7. [DOI] [PubMed] [Google Scholar]
- 23.Owen-Hughesand T., Workman J. L. Experimental analysis of chromatin function in transcription control. Critical Reviews in Eukaryotic Gene Expression. 1994;4(4):403–441. [PubMed] [Google Scholar]
- 24.Schick S., Rendeiro A. F., Runggatscher K., et al. Systematic characterization of BAF mutations provides insights into intracomplex synthetic lethalities in human cancers. Nature Genetics. 2019;51(9):1399–1410. doi: 10.1038/s41588-019-0477-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mashtalir N., D'Avino A. R., Michel B. C., et al. Modular organization and assembly of SWI/SNF family chromatin remodeling complexes. Cell. 2018;175(5):1272–1288.e20. doi: 10.1016/j.cell.2018.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He S., Wu Z., Tian Y., et al. Structure of nucleosome-bound human BAF complex. Science. 2020;367(6480):875–881. doi: 10.1126/science.aaz9761. [DOI] [PubMed] [Google Scholar]
- 27.Sonand E. Y., Crabtree G. R. The role of BAF (mSWI/SNF) complexes in mammalian neural development. American Journal of Medical Genetics. Part C, Seminars in Medical Genetics. 2014;166C(3):333–349. doi: 10.1002/ajmg.c.31416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho L., Ronan J. L., Wu J., et al. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(13):5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panamarova M., Cox A., Wicher K. B., et al. The BAF chromatin remodelling complex is an epigenetic regulator of lineage specification in the early mouse embryo. Development. 2016;143(8):1271–1283. doi: 10.1242/dev.131961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang W., Chronis C., Chen X., et al. The BAF and PRC2 complex subunits Dpf2 and Eed antagonistically converge on Tbx3 to control ESC differentiation. Cell Stem Cell. 2019;24(1):138–152.e8. doi: 10.1016/j.stem.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kidder B. L., Palmer S., Knott J. G. SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells. 2009;27(2):317–328. doi: 10.1634/stemcells.2008-0710. [DOI] [PubMed] [Google Scholar]
- 32.Singhal N., Esch D., Stehling M., Schöler H. R. BRG1 is required to maintain pluripotency of murine embryonic stem cells. Biores Open Access. 2014;3(1):1–8. doi: 10.1089/biores.2013.0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.You J. S., De Carvalho D. D., Dai C., et al. SNF5 is an essential executor of epigenetic regulation during differentiation. PLoS Genetics. 2013;9(4, article e1003459) doi: 10.1371/journal.pgen.1003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakakura M., Ohta S., Yagi M., et al. Smarcb1 maintains the cellular identity and the chromatin landscapes of mouse embryonic stem cells. Biochemical and Biophysical Research Communications. 2019;519(4):705–713. doi: 10.1016/j.bbrc.2019.09.054. [DOI] [PubMed] [Google Scholar]
- 35.Gao X., Tate P., Hu P., Tjian R., Skarnes W. C., Wang Z. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(18):6656–6661. doi: 10.1073/pnas.0801802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lei I., Tian S., Chen V., Zhao Y., Wang Z. SWI/SNF component BAF250a coordinates OCT4 and WNT signaling pathway to control cardiac lineage differentiation. Frontiers in Cell and Developmental Biology. 2020;7:p. 358. doi: 10.3389/fcell.2019.00358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu W., Fang L., Ouyang B., et al. Actl6a protects embryonic stem cells from differentiating into primitive endoderm. Stem Cells. 2015;33(6):1782–1793. doi: 10.1002/stem.2000. [DOI] [PubMed] [Google Scholar]
- 38.Krasteva V., Buscarlet M., Diaz-Tellez A., Bernard M. A., Crabtree G. R., Lessard J. A. The BAF53a subunit of SWI/SNF-like BAF complexes is essential for hemopoietic stem cell function. Blood. 2012;120(24):4720–4732. doi: 10.1182/blood-2012-04-427047. [DOI] [PubMed] [Google Scholar]
- 39.Gatchalian J., Malik S., Ho J., et al. A non-canonical BRD9-containing BAF chromatin remodeling complex regulates naive pluripotency in mouse embryonic stem cells. Nature Communications. 2018;9(1):p. 5139. doi: 10.1038/s41467-018-07528-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bultman S., Gebuhr T., Yee D., et al. A _Brg1_ null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Molecular Cell. 2000;6(6):1287–1295. doi: 10.1016/S1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 41.Klochendler-Yeivin A., Fiette L., Barra J., Muchardt C., Babinet C., Yaniv M. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Reports. 2000;1(6):500–506. doi: 10.1093/embo-reports/kvd129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guidi C. J., Sands A. T., Zambrowicz B. P., et al. Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Molecular and Cellular Biology. 2001;21(10):3598–3603. doi: 10.1128/MCB.21.10.3598-3603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lei I., West J., Yan Z., et al. BAF250a protein regulates nucleosome occupancy and histone modifications in priming embryonic stem cell differentiation. Journal of Biological Chemistry. 2015;290(31):19343–19352. doi: 10.1074/jbc.M115.637389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phelan M. L., Sif S., Narlikar G. J., Kingston R. E. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Molecular Cell. 1999;3(2):247–253. doi: 10.1016/S1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 45.Bultman S. J., Herschkowitz J. I., Godfrey V., et al. Characterization of mammary tumors from _Brg1_ heterozygous mice. Oncogene. 2008;27(4):460–468. doi: 10.1038/sj.onc.1210664. [DOI] [PubMed] [Google Scholar]
- 46.Ho L., Jothi R., Ronan J. L., Cui K., Zhao K., Crabtree G. R. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(13):5187–5191. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ho L., Miller E. L., Ronan J. L., Ho W. Q., Jothi R., Crabtree G. R. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nature Cell Biology. 2011;13(8):903–913. doi: 10.1038/ncb2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buecker C., Srinivasan R., Wu Z., et al. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell. 2014;14(6):838–853. doi: 10.1016/j.stem.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang J., Wu X., Wei C., et al. YY1 positively regulates transcription by targeting promoters and super- enhancers through the BAF complex in embryonic stem cells. Stem Cell Reports. 2018;10(4):1324–1339. doi: 10.1016/j.stemcr.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller E. L., Hargreaves D. C., Kadoch C., et al. TOP2 synergizes with BAF chromatin remodeling for both resolution and formation of facultative heterochromatin. Nature Structural & Molecular Biology. 2017;24(4):344–352. doi: 10.1038/nsmb.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim J. K., Huh S. O., Choi H., et al. Srg3, a mouse homolog of yeast SWI3, is essential for early embryogenesis and involved in brain development. Molecular and Cellular Biology. 2001;21(22):7787–7795. doi: 10.1128/MCB.21.22.7787-7795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang X., Li B., Li W., et al. Transcriptional repression by the BRG1-SWI/SNF complex affects the pluripotency of human embryonic stem cells. Stem Cell Reports. 2014;3(3):460–474. doi: 10.1016/j.stemcr.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bao X., Tang J., Lopez-Pajares V., et al. ACTL6a enforces the epidermal progenitor state by suppressing SWI/SNF- dependent induction of KLF4. Cell Stem Cell. 2013;12(2):193–203. doi: 10.1016/j.stem.2012.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu B., Ueda A., Song X., Horike S. I., Yokota T., Akagi T. Baf53a is involved in survival of mouse ES cells, which can be compensated by Baf53b. Scientific Reports. 2017;7(1):p. 14059. doi: 10.1038/s41598-017-14362-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kadoch C., Crabtree G. R. Mammalian SWI/SNF chromatin remodeling complexes and cancer: mechanistic insights gained from human genomics. Science Advances. 2015;1(5, article e1500447) doi: 10.1126/sciadv.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krasteva V., Crabtree G. R., Lessard J. A. The BAF45a/PHF10 subunit of SWI/SNF-like chromatin remodeling complexes is essential for hematopoietic stem cell maintenance. Experimental Hematology. 2017;48:58–71.e15. doi: 10.1016/j.exphem.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 57.Nie Z., Xue Y., Yang D., et al. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Molecular and Cellular Biology. 2000;20(23):8879–8888. doi: 10.1128/MCB.20.23.8879-8888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vazquez M., Moore L., Kennison J. A. The trithorax group gene Osa encodes an ARID-domain protein that genetically interacts with the brahma chromatin-remodeling factor to regulate transcription. Development. 1999;126(4):733–742. doi: 10.1242/dev.126.4.733. [DOI] [PubMed] [Google Scholar]
- 59.Hansis C., Barreto G., Maltry N., Niehrs C. Nuclear reprogramming of human somatic cells by Xenopus egg extract requires BRG1. Current Biology. 2004;14(16):1475–1480. doi: 10.1016/j.cub.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 60.Egli D., Eggan K. Recipient cell nuclear factors are required for reprogramming by nuclear transfer. Development. 2010;137(12):1953–1963. doi: 10.1242/dev.046151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singhal N., Graumann J., Wu G., et al. Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell. 2010;141(6):943–955. doi: 10.1016/j.cell.2010.04.037. [DOI] [PubMed] [Google Scholar]
- 62.Jiang Z., Tang Y., Zhao X., Zhang M., Donovan D. M., Tian X. C. Knockdown of Brm and Baf170, components of chromatin remodeling complex, facilitates reprogramming of somatic cells. Stem Cells and Development. 2015;24(19):2328–2336. doi: 10.1089/scd.2015.0069. [DOI] [PMC free article] [PubMed] [Google Scholar]


