Abstract
Diversity-generating retroelements (DGRs) vary protein sequences to the greatest extent known in the natural world. These elements are encoded by constituents of the human microbiome and the microbial ‘dark matter’. Variation occurs through adenine-mutagenesis, in which genetic information in RNA is reverse transcribed faithfully to cDNA for all template bases but adenine. We investigated the determinants of adenine-mutagenesis in the prototypical Bordetella bacteriophage DGR through an in vitro system composed of the reverse transcriptase bRT, Avd protein, and a specific RNA. We found that the catalytic efficiency for correct incorporation during reverse transcription by the bRT-Avd complex was strikingly low for all template bases, with the lowest occurring for adenine. Misincorporation across a template adenine was only somewhat lower in efficiency than correct incorporation. We found that the C6, but not the N1 or C2, purine substituent was a key determinant of adenine-mutagenesis. bRT-Avd was insensitive to the C6 amine of adenine but recognized the C6 carbonyl of guanine. We also identified two bRT amino acids predicted to nonspecifically contact incoming dNTPs, R74 and I181, as promoters of adenine-mutagenesis. Our results suggest that the overall low catalytic efficiency of bRT-Avd is intimately tied to its ability to carry out adenine-mutagenesis.
INTRODUCTION
Adaptation by organisms to novel selective pressures requires variation. While this usually occurs over multiple generations and lengthy time scales, there are two examples of instantaneous adaptation that take place within a single generation. These are the variation of antigen receptors by the vertebrate adaptive immune system and of select proteins belonging to diversity-generating retroelements (DGRs) (1). DGRs are prevalent in the human virome and microbiome, and the microbial ‘dark matter’, which appears to constitute a major fraction of microbial life (2–7). The level of DGR variation greatly exceeds the 1014–16 variation of the vertebrate immune system (8). A DGR variable protein with 1020 possible sequences has been structurally characterized (9), and one with 1030 possible sequences has been identified (10). In the adaptive immune system, variation enables the recognition of novel targets and consequent adaptation to dynamic environments. A similar benefit appears to be provided by DGRs, as documented for the prototypical DGR of Bordetella bacteriophage (1). This DGR encodes the bacteriophage's receptor-binding protein Mtd. Variation in Mtd enables the bacteriophage to adapt to the loss of potential surface receptors by its host Bordetella, which happens because of environmental changes or immune pressure (11,12).
DGRs vary protein sequences through a mechanism that is fundamentally different from that of the adaptive immune system and indeed any biological system. Sequence variation by DGRs arises from adenine-mutagenesis, in which genetic information is transmitted faithfully for all bases but adenine (Figure 1). This occurs during reverse transcription of an RNA transcript that contains the intergenic template region (TR), which is nearly identical in sequence to a variable region (VR) within a DGR variable protein gene. Misincorporation at adenines during reverse transcription of TR-RNA results in adenine-mutagenized TR-cDNA, which homes to and replaces VR, giving rise to a new protein variant. As shown for the Bordetella bacteriophage DGR, misincorporation at TR-RNA adenines occurs at an astonishingly high frequency of 50% (1,13,14).
Figure 1.
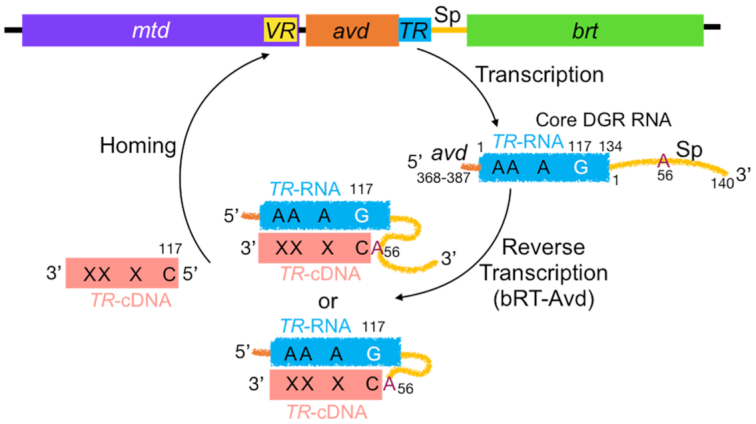
Diversity-generating retroelement. The Bordetella bacteriophage DGR consists of the variable protein gene mtd, which contains a variable region (VR); avd (Accessory Variability Determinant); an intergenic template region (TR); an intergenic spacer (Sp) region; and a reverse transcriptase (brt). An RNA transcript that contains TR and 5′ and 3′ sequences from avd and Sp, respectively, is reverse transcribed and adenine-mutagenized by the bRT-Avd complex. Synthesis of TR-cDNA is primed by the RNA from Sp A56, either through an internal 2′-OH or a terminal 3′-OH following cleavage of the RNA. TR G117 is the first position reverse transcribed, and cDNAs extend to TR 22–24 or just into avd. TR-cDNA homes to and replaces VR, involving either the covalently linked RNA-cDNA molecule or the cDNA alone.
We recently reconstituted adenine-mutagenesis in vitro (14). This reconstitution showed that a complex formed by the Bordetella bacteriophage DGR reverse transcriptase bRT and the associated pentameric Accessory Variability Determinant (Avd) protein (15), along with a specific DGR RNA, are necessary and sufficient for adenine-mutagenesis (Figure 1). The DGR RNA contains the 134-nucleotide (nt) TR flanked by two functional elements, a short 20-nt segment from avd at the 5′ end and a longer 140-nt spacer (Sp) region separating TR and brt at the 3′ end (14). This RNA is identical to the ‘core’ DGR RNA described previously (14). The mechanistic role of the 5′ avd sequence is unknown, but the Sp region has been shown to provide an essential binding site for Avd and to supply the site from which reverse transcription is primed, Sp A56 (13,14). A number of pieces of evidence indicate that the 2′-OH of Sp 56 provides the priming nucleophile, resulting in a branched, covalently-linked RNA-cDNA molecule (14) (Figure 1). An alternative model involving RNA cleavage at Sp 56 to expose its 3′-OH for priming has been proposed as well (13). The first nucleotide reverse transcribed is TR G117, and cDNAs typically extend from there to TR 22–24 (∼90 nt) or just further into avd (∼120 nt); the shorter cDNA includes all of the adenines in TR whose substitution leads to a coding change.
The bRT-Avd complex is also capable of synthesizing cDNAs from non-DGR RNA templates (14). For this, an exogenous primer is required and only short cDNAs (∼5–35 nt) are synthesized. These results indicate that template-priming and processive polymerization are both specific properties of the DGR RNA. Evidence suggests that this is because bRT-Avd and the DGR RNA combine to form a structured ribonucleoprotein (RNP) particle that aligns the priming site at Sp 56 with the reverse transcription start site at TR 117, and also maintains interaction between bRT-Avd and the RNA that is conducive to processive polymerization (14). Avd is still required for cDNA synthesis from non-DGR RNA templates, indicating that Avd has a role in catalysis that is independent of its role in binding Sp in the DGR RNA. Notably, cDNAs produced from non-DGR RNA templates are adenine-mutagenized, indicating that adenine-mutagenesis is an intrinsic property of the bRT-Avd complex and independent of the RNA template and the mechanism of priming.
To understand the determinants of adenine-mutagenesis, we characterized reverse transcription of the DGR RNA by bRT-Avd in vitro. We found that the catalytic efficiency (kcat/Km) of bRT-Avd for correct incorporation was strikingly low across all template bases, as generally observed for low fidelity polymerases (16), with the lowest occurring for adenine. The catalytic efficiency of misincorporation across a template adenine was only somewhat lower than correct incorporation. Using nucleobase analogs, we identified the C6 position of the purine ring as a key determinant of adenine-mutagenesis. An amine at C6, as in adenine, had no effect on misincorporation, neither increasing nor decreasing it. In contrast, a carbonyl at C6, as in guanine, decreased misincorporation. bRT-Avd was able to incorporate dNTPs across an abasic template site, albeit with a significant incidence of deletions and with only a partial preference for adenine as compared to A-rule polymerases (16). We also found that two bRT amino acids, Arg 74 and Ile 181, promoted adenine-mutagenesis. These amino acids are predicted by an in silico model to have counterparts in HIV reverse transcriptase (RT) that nonspecifically stabilize incoming dNTPs. These results provide the first detailed characterization of the nucleobase and protein determinants of adenine-mutagenesis in DGRs.
MATERIALS AND METHODS
Protein and RNA
The bRT-Avd complex was expressed in Escherichia coli and purified, and the core DGR RNA (avd 368 – TR 140) was produced through in vitro transcription with T7 polymerase and gel purified, both as previously described (14). Mutants of bRT were generated using QuikChange mutagenesis (Agilent), and expressed and purified as bRT-Avd complexes. Mutants of the DGR RNA were also generated through QuikChange mutagenesis.
RNA with nucleobase analogs
RNA oligonucleotides spanning avd 368 to TR 26 were chemically synthesized (Dharmacon), with adenine, guanine, or base analogs at TR 23 and 24 (or just 24), and gel purified. RNA spanning TR 27 to Sp 140 was in vitro transcribed, gel purified, and treated with alkaline phosphatase (NEB) according to the manufacturer's directions at 37°C for 2 h to remove the triphosphate from the 5′ end. The dephosphorylation reaction was quenched at 80°C for 5 min to inactivate the phosphatase. A phosphate group was added to the 5′ end of the RNA using T4 polynucleotide kinase and ATP, according to the manufacturer's directions (NEB). The RNA was then purified by phenol:chloroform extraction followed by a G-25 desalting column. The chemically synthesized RNA oligonucleotide (1.1 μM) and the in vitro transcribed RNA (2.2 μM) were annealed in the presence of 1.7 μM splint oligodeoxynucleotide P1 (Supplementary Table S1), 8% DMSO, and 0.2× T4 RNA Ligase 1 (T4Rnl1) buffer in 250 μl. The splint was annealed to the RNA by heating at 95°C for 3 min and cooling at 0.2°C/min to 20°C. A 250 μl solution consisting of 0.8× T4Rnl1 buffer, 2 mM ATP, 4% DMSO and 540 units T4Rnl1 (NEB) were mixed with the annealing reaction. The resulting mixture was incubated for 8 h at 37°C. The sample was then extracted with phenol:chloroform and ethanol–glycogen precipitated. The pellet was resuspended in water and gel purified.
Reverse transcription reactions and next-generation sequencing
Reverse transcription reactions were carried out with wild-type or mutant bRT-Avd at 37°C for 12 h, and resulting cDNA was purified, both as previously described (14). The region from TR 21 to TR 98 was PCR amplified using Pfu polymerase from purified cDNA using primers P2 and P3, which have partial Illumina adapter sequences at their 5′ ends (Supplementary Table S1). For amplification of the region from TR 114 to TR 117, reverse transcription of RNA–cDNA molecules with primer P2 was first carried out, as previously described (14), and the resulting cDNA was PCR amplified with Pfu polymerase and primers P2 and P4, which likewise had partial Illumina adapter sequences at their 5′ ends (Supplementary Table S1). The reverse transcription step was necessary as TR 117 is at the 5′ end of the cDNA and attached covalently to the core DGR RNA at Sp 56. The amplified PCR product was subjected to short-read next-generation sequencing (Amplicon EZ, Genewiz). The quality scores for sequencing reactions were Q30, which is equivalent to an error probability of 1 in 1000. Fastq files generated from next-generation sequencing containing paired-end reads were aligned with the TR reference sequence using bowtie2, and output files were sorted and indexed using samtools (parameters in Supplementary Data) (17–19). (Mis)incorporation frequencies in the template region were calculated using IGV (integrative genomics viewer) (20).
Reverse transcription reactions with Moloney Murine Leukemia Virus (MMLV) RT (BioBharati Life Sciences Pvt. Ltd) and HIV RT (Worthington Biochemical) were carried out according to the manufacturer's directions using primer P5 (Supplementary Table S1). PCR amplification of cDNAs was carried out with Pfu polymerase and primers P2 and P3 (Supplementary Table S1).
Quantitative PCR
Quantitative PCR (qPCR) was carried out on cDNA reverse transcribed by bRT-Avd from the core DGR RNA (25 μL reaction). The cDNA was purified as previously described (14) and amplified in the presence of 1x SYBR Green 1 dye (Thermo Fisher) using primers P6 and P7 (Supplementary Table S1). The reaction was performed on a Bio-Rad CFX Connect Real Time System apparatus. A standard curve was generated using chemically synthesized single-stranded DNA (TR 1 to TR 117, HPLC purity of 99.9%), with template concentrations in the range of 100 fg–1 ng. qPCR reactions were performed in triplicate, and the cycle threshold (Ct, the cycle number at which the fluorescence due to the reaction crosses the fluorescence background threshold) for each reaction was determined using CFX Maestro Software. The average of Ct was plotted against the log of the DNA mass, and a linear fit from the plot was used to calculate the quantity and concentration of cDNA generated in the bRT-Avd reverse transcription reaction.
Single deoxynucleotide primer extension assay
Oligodeoxynucleotide P117 (Supplementary Table S1) was 5′-[32P]-labeled as previously described (14). Wild-type or mutant core DGR RNA (0.5 μM) was mixed with primer P117 (0.5 μM), 5′-[32P]-labeled P117 (0.05 μM), varying concentration of dNTPs, and 20 units RNase inhibitor (NEB) in 75 mM KCl, 3 mM MgCl2, 10 mM DTT, 50 mM HEPES, pH 7.5 and 10% glycerol in a 20 μl final volume. The mixture was incubated at 37°C, and then wild-type or mutant 1 μM bRT-Avd was added. A 2.5 μl aliquot of the reaction was removed at various time points, and quenched by addition to 7.5 μl of an ice-cold solution of proteinase K (1.3 mg/ml) followed by incubation at 50°C for 20 min. The quenched reactions were incubated with 0.5 μl RNase A/T1 mix (Ambion) at room temperature for 20 min in a final volume of 20 μl. The samples were then ethanol-glycogen precipitated overnight at −20°C. Samples were centrifuged the next day, and pellets were air dried and resuspended in 20 μl of RNA loading dye. Five μL of the reaction sample was loaded on an 8% denaturing sequencing gel to resolve unreacted and extended primers. The radiolabeled products were visualized by autoradiography using a Typhoon Trio (GE Healthcare Life Sciences), and band densities were quantified using ImageQuant TL 8.1 (GE Healthcare Life Sciences). Background values determined from band densities prior to any reaction were subtracted. The steady-state initial velocity with respect to substrate concentration was fit to the Michaelis-Menten equation using nonlinear regression analysis in GraphPad Prism.
RESULTS
Adenine-mutagenesis of TR
To characterize adenine-mutagenesis of TR by bRT-Avd at fine-scale, we pursued next-generation sequencing (NGS) of cDNAs. The NGS read count of ∼100 000 enabled conclusions to be drawn about the distribution of adenine-mutagenesis that were not possible due to the small number of sequences previously available from single clones (∼30) (1). The template was the ∼300-nt core DGR RNA (14), consisting of the TR (134 nt) flanked by upstream avd and downstream Sp regions that are functionally essential (Figure 1) (14). As described above, reverse transcription was template-primed by the core DGR RNA from Sp A56 and initiated from TR G117 (14).
To determine the baseline detection level of our methodology, we carried out NGS on cDNA that had not been adenine-mutagenized. For this, we used chemically synthesized, purified single-stranded (ss) DNA that corresponded to TR 1–117. During NGS analysis we set the concentration of this ssDNA to be equivalent to that of the cDNA produced by bRT-Avd. The latter was determined through quantitative PCR (Supplementary Figure S1a), which showed that 1 × 1011 cDNA molecules were synthesized from 6 × 1012 DGR RNA molecules. NGS of the non-mutagenized ssDNA yielded an average 0.13% misincorporation frequency, with a range of 0.1–0.7% (Supplementary Figure S1b). As a further control, we synthesized TR-cDNA from the DGR RNA template using an exogenous primer with the high fidelity reverse transcriptase MMLV RT, which has a reported error rate of ∼10−5 (21). NGS analysis revealed an average 0.36% misincorporation frequency, with a range of 0.1–2.6% (Supplementary Figure S1c). Interestingly, TR adenines had the highest average misincorporation frequency (0.8%).
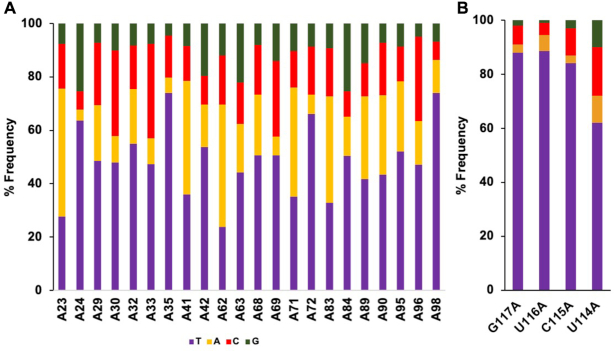
We then carried out NGS analysis on cDNAs synthesized by bRT-Avd. We found through three independent experiments that the average misincorporation frequency across the 22 adenines in TR that lead to coding changes was 51.6 ± 2.3% (Figure 2A and Supplementary Figure S2a), similar to the level previously observed in vitro and in vivo (1,13,14). The most frequently misincorporated base across a template adenine was adenine itself (22.2 ± 0.7%), followed by cytosine (17.8 ± 2.8%) and guanine (11.6 ± 0.2%) (Supplementary Figure S2a). By comparison, the misincorporation frequency across template uracils, cytosines, and guanines in TR was at or near baseline detection levels (0.5 ± 0.2%, 0.3 ± 0.2% and 1.6 ± 1.2%, respectively) (Supplementary Figure S2b).
Figure 2.
Adenine-mutagenesis. (A) Frequency of deoxynucleotides incorporated (thymine, purple) or misincorporated (adenine, orange; cytosine, red; guanine, green) by bRT-Avd across template adenines in TR. Results from one of three independent experiments is shown. This color coding is used throughout. (B) Frequency of deoxynucleotides (mis)incorporated across template adenines individually substituted at TR 114–117.
The misincorporation frequency of bRT-Avd varied widely across individual template adenines (Figure 2A and Supplementary Figure S3). TR A23 and A62 were especially prone to misincorporation, with frequencies of 72.3 ± 1.9% and 76.3 ± 0.9%, respectively. Notably these adenines are the first bases in AAC codons (for Mtd 344 and 357, respectively), indicating an enhanced potential for amino acid substitution at these positions (22). In contrast, TR A35 and A98 were especially resistant to misincorporation, with frequencies of only 28.0 ± 2.8% and 29.9 ± 5.2%, respectively. Equally notably, these adenines are the only ones in their codons (ACG for Mtd 348 and ATC for Mtd 369, respectively), indicating a curtailed potential for amino acid substitution at these codons (22). The differences in misincorporation frequencies were not attributable to any obvious RNA primary sequence patterns.
To determine whether misincorporation also occurred at artificially introduced adenines, we substituted adenines at the initial four positions that are reverse transcribed (TR 117–114). These positions are normally occupied by G, C or U. Misincorporation was evident at all four positions when substituted by adenine. Positions 117, 116 and 115 were somewhat more resistant to misincorporation than the most resistant adenines positions naturally occurring in TR, while 114 was within the range observed for naturally occurring adenine positions (Figure 2B). Thus adenine-mutagenesis can occur outside naturally occurring positions, and similar to naturally occurring positions, the misincorporation frequency is variable.
Enzymatic parameters of bRT-Avd
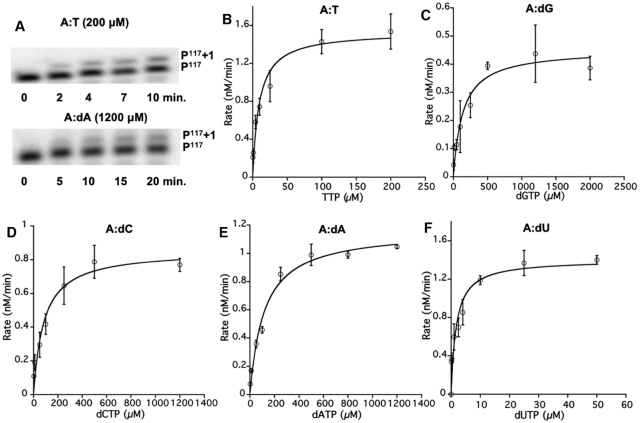
We next examined the rate of single deoxynucleotide addition by bRT-Avd. As misincorporation occurred at a detectable level across TR G117A, we used oligodeoxynucleotide P117 (Supplementary Table S1) to prime synthesis from TR G117A of the core DGR RNA. We have previously shown that P117 primes cDNA synthesis from the natural start site of reverse transcription, TR G117, and concurrently inhibits template-primed cDNA synthesis (14). The addition of a single deoxynucleotide to the radiolabeled P117 primer was unambiguously detectable (Figure 3A). In the case of extension with dATP, the template contained both TR U116G and G117A substitutions, so as to avoid incorporation of a second dATP across TR U116. Using this single deoxynucleotide primer extension assay, we determined the steady-state enzymatic parameters of bRT-Avd for correct incorporation and misincorporation across a template adenine.
Figure 3.
Kinetics of single deoxynucleotide (mis)incorporation. (A) Single deoxynucleotide primer extension by bRT-Avd of [32P]-labeled P117 by TTP (top) or dATP (bottom), as templated by the core DGR RNA containing TR G117A. The extended product (P117 +1) was resolved from the reactant (P117) with an 8% sequencing gel. (B–F) Steady-state kinetic characterization of single deoxynucleotide (mis)incorporation of TTP, dGTP, dCTP, dATP and dUTP, across the core DGR RNA containing TR G117A (and TR U116G, in the case of dATP) by bRT-Avd. The Michaelis-Menten fit is shown, and error bars represent standard deviations from three independent measurements.
This analysis showed that the kcat varied little (only up to three-fold) between correct incorporation of TTP and misincorporation of the other dNTPs across the adenine at TR 117 (Figure 3B–F and Table 1). Similarly, the Km for the incoming dNTP varied over a very small range, with the greatest difference being ∼17-fold between dGTP and TTP (Table 1). The most favorable Km was for dUTP, reflecting the previously noted preference in bRT-Avd for dUTP over TTP (14). The catalytic efficiencies (kcat/Km) for the misincorporation of dGTP, dCTP, and dATP were on average 23-fold lower than for the correct incorporation of TTP. The misincorporation ratios (catalytic efficiency for misincorporation to correct incorporation) for dCTP and dATP were 0.05, and 0.02 for dGTP (Table 1). These values corresponded closely to the misincorporation frequencies at TR G117A determined by NGS from template-primed reverse transcription (Figure 2B, 7%, 3% and 2%, respectively). Thus, template- and P117-primed synthesis resulted in similar misincorporation frequencies, which indicated that the enzymatic parameters determined here for oligodeoxynucleotide-primed synthesis were likely to be applicable to template-primed synthesis.
Table 1.
Steady-state enzymatic parameters for (mis)incorporation by bRT-Avd at TR G117A
| WT | K m (μM) | k cat (min−1) x 10−3 | kcat/Km (μM−1 min−1) x 10−3 | Misincorporation ratiob |
|---|---|---|---|---|
| A:Ta | 7.97 ± 2.34 | 11.64 ± 0.71 | 1.46 | - |
| A:dG | 138.03 ± 48.33 | 3.62 ± 0.32 | 0.03 | 0.02 |
| A:dC | 84.35 ± 26.10 | 6.85 ± 0.56 | 0.08 | 0.05 |
| A:dA | 115.25 ± 22.31 | 9.33 ± 0.46 | 0.08 | 0.05 |
| A:dU | 1.73 ± 0.64 | 11.20 ± 0.96 | 6.47 | 4.43 |
aTemplate base:(mis)incorporated base.
bMisincorporation ratio = [kcat (incorrect)/Km (incorrect)]/[kcat (correct)/Km (correct)].
We then examined the enzymatic parameters for correct dNTP incorporation across the other template bases at TR 117. For template guanine, the wild-type DGR RNA template was used, and for template cytosine and uracil, mutated DGR RNA templates were used (TR G117C for the former, and TR U116C/G117U for the latter to avoid incorporation of a second dATP). The experiment showed that the kcat for correct incorporation varied little among template guanine, cytosine, uracil, and adenine (Tables 1 and 2, Supplementary Figure S4). However, there was a major difference in Km. While the Km was similar for template guanine, cytosine, and uracil, this value was on average ∼36-fold lower than for a template adenine. Similarly, the catalytic efficiencies were nearly identical for these first three template bases but 28-fold greater than for a template adenine.
Table 2.
Steady-state enzymatic parameters for incorporation by bRT-Avd at TR G117, G117C and G117U
| WT | K m (μM) | k cat (min−1) x 10−3 | k cat/Km (μM−1 min−1) x 10−3 | Efficiencya |
|---|---|---|---|---|
| G:dC | 0.26 ± 0.09 | 10.41 ± 0.62 | 40.04 | 27.42 |
| C:dG | 0.17 ± 0.06 | 6.92 ± 0.41 | 40.71 | 27.88 |
| U:dA | 0.24 ± 0.09 | 9.95 ± 0.62 | 41.46 | 28.40 |
aEfficiency = [kcat (correct)/Km (correct)]/[kcat (A:T)/Km (A:T)].
Nucleobase determinants of adenine-mutagenesis
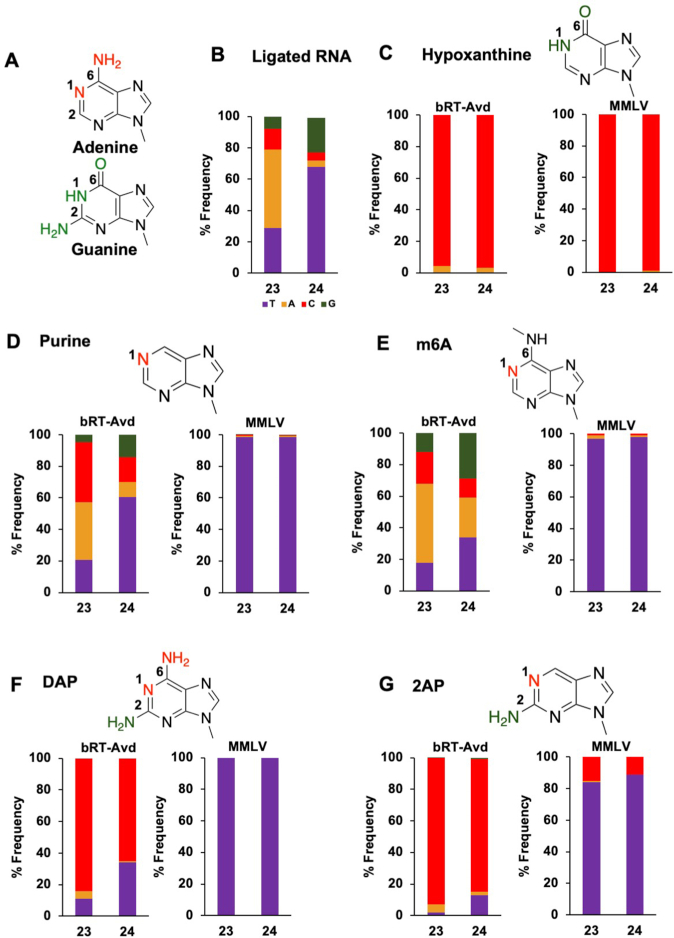
We next asked which features of adenine promoted misincorporation. To address this, we compared the nearly isosteric features of infidelity-promoting adenine and fidelity-promoting guanine. The differences between A and G primarily occur at the N1, C2 and C6 positions (Figure 4A). We sought to probe these ring positions using base analogs. For this, short RNA oligonucleotides corresponding to the 5′ portion of the core DGR RNA were chemically synthesized with nucleobase analogs at TR 23 and 24, which have misincorporation frequencies of 72.3 ± 1.9% and 39.7 ± 3.3%, respectively. The oligonucleotide was ligated to a longer in vitro transcribed RNA corresponding to the rest of the core DGR RNA. To validate this method, we first constructed the ligated core DGR RNA using an oligonucleotide that had adenines rather than analogs at TR 23 and 24. The cDNAs produced by bRT-Avd from the ligated core DGR RNA template had a near identical misincorporation frequency as the fully in vitro transcribed core DGR RNA template (Figures 2A and 4B). As further validation, we used the same ligation method to introduce guanines at TR 23 and 24, and found almost exclusive incorporation of cytosine by bRT-Avd (Supplementary Figure S5), indicating that there is no barrier to correct incorporation at these positions.
Figure 4.
Nucleobase determinants of adenine-mutagenesis. (A) Adenine and guanine differ at N1, C2 and C6 positions (red for A, green for G). (B) Frequency of deoxynucleotides (mis)incorporated by bRT-Avd at TR 23 and TR 24 using the core DGR RNA template that had been ligated from chemically synthesized and in vitro transcribed sections. The chemically synthesized section contained adenosines at TR 23 and 24. (C) Top, structure of hypoxanthine. Bottom, (mis)incorporation frequencies of bRT-Avd (left) and MMLV RT (right) with hypoxanthine at TR 23 and 24. (D) Top, structure of purine. Bottom, (mis)incorporation frequencies of bRT-Avd (left) and MMLV RT (right) with purine at TR 23 and 24. (E) Top, structure of N6-methyladenine (m6A). Bottom, (mis)incorporation frequencies of bRT-Avd (left) and MMLV RT (right) with m6A at TR 23 and 24. (F) Top, structure of DAP. Bottom, (mis)incorporation frequencies of bRT-Avd (left) and MMLV RT (right) with DAP at TR 23 and 24. (G) Top, structure of 2,6-diaminopurine (2AP). Bottom, (mis)incorporation frequencies of bRT-Avd (left) and MMLV RT (right) with 2AP at TR 23 and 24.
N1 and C6
The first nucleobase analog we examined was hypoxanthine, in which the N1 and C6 groups of adenine (N with a lone electron pair and amine, respectively) are substituted with those of guanine (NH and carbonyl, respectively). Hypoxanthine preferentially forms a Watson-Crick base pair with cytosine, and a less stable wobble base pair with adenine (23,24). This preference was verified through reverse transcription with MMLV RT (Figure 4C). Cytosine was almost exclusively incorporated across template hypoxanthines by MMLV RT. Reverse transcription was then carried out with bRT-Avd. Significantly, bRT-Avd correctly incorporated cytosine with nearly the same frequency, 96–97% (Figure 4C), as MMLV RT. Adenines constituted the rest. This result showed that an NH at N1 or a carbonyl at C6, or both, decrease misincorporation relative to adenine.
We next asked if the amine at C6 in adenine has an impact on misincorporation. To do so, we used purine (i.e. nebularine), which lacks a substituent at C6, as the base analog. Purine preferentially base pairs with thymine (25), which we verified with MMLV RT (Figure 4D). With bRT-Avd, template purines led to misincorporation with very similar frequencies as those observed for template adenines (Figure 4D). This result indicated that the C6 amine of adenine was functionally equivalent to no substituent at this position. Thus, we concluded that an amine at C6 had no impact on misincorporation, neither decreasing nor increasing it.
To probe the C6 position further, we used N6-methyladenine (m6A). As previously reported, reverse transcription of m6A by MMLV results in the incorporation of thymine (Figure 4E) (26). However, for bRT-Avd, m6A resulted in greater misincorporation, most notably at TR 24, where misincorporation increased from 40% to 66% (Figure 4E). Misincorporation also increased at TR 23, although less strikingly, from 72% to 82%. Thus, while an amino group at the C6 position did not impact misincorporation, the bulkier methylamino group at the same position increased misincorporation. Importantly, these results also suggested that the N1 position had little if any role in modulating misincorporation. This is because while adenine and m6A are identical at the N1 position, they differed in misincorporation frequency.
Taken together, these results indicated that the C6 but not the N1 purine position was a major determinant of adenine-mutagenesis, and that the C6 carbonyl of guanine decreased misincorporation while the C6 amine was equivalent to having no substituent at this position.
C2
The C2 position was probed using 2,6-diaminopurine (DAP), which is identical to adenine except containing an amine at C2 as well (as does guanine). DAP preferentially base pairs with thymine but can also form a wobble base pair with cytosine (27,28). In the case of MMLV RT, exclusive incorporation of thymine was observed (Figure 4F). However for bRT-Avd, the major species incorporated was cytosine, followed to a lesser extent by thymine and adenine (Figure 4F). This was the case even though the Watson-Crick base pair between DAP and thymine involves three hydrogen bonds, while the wobble base pair between DAP and cytosine involves only one.
A similar trend was seen for 2-amino purine (2AP), which is identical to DAP but lacks an amine at C6. 2AP forms a base pair with thymine but can also form a wobble or protonated base pair with cytosine (28–30). Both forms involve two hydrogen bonds. MMLV RT preferentially incorporated thymine but also a substantial level of cytosine. In the case of bRT-Avd, incorporation of cytosine was greatly preferred, followed by thymine and adenine (Figure 4G), similar to the results with DAP (Figure 4F). The similarity of misincorporation frequencies between DAP and 2-AP (which differ only in an amine at C6) confirmed that an amine at C6 was equivalent to having no substituent at this position.
These results showed that an amine at C2 led to a more homogeneous distribution of incorporated deoxynucleotide, but with those forming a wobble base pair being favored over those forming a Watson–Crick base pair. The exception to this was when the C6 position was occupied by a carbonyl, that is when the template base was a guanine, in which case a Watson–Crick base pair was favored. Thus, these results further emphasized the importance of the C6 position to adenine-mutagenesis.
Abasic site and the A-rule
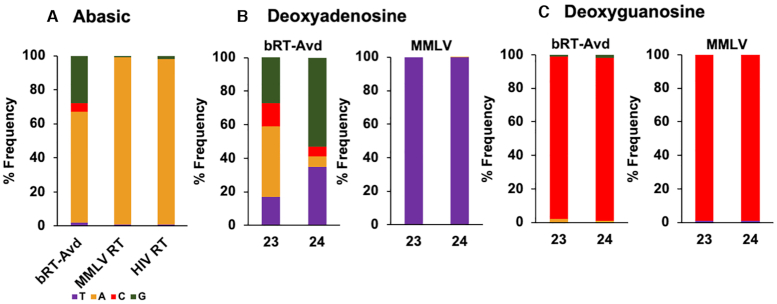
We noticed that adenine was the most frequently misincorporated base across the 22 sites in TR (Figure 2A and Supplementary Figure S2a). A number of nucleotide polymerases have the tendency to insert an adenine across an abasic template site, which is called the A-rule (31). We investigated whether bRT-Avd follows the A-rule by constructing a core DGR RNA template with abasic sites at TR 23 and 24. HIV RT has been documented to tolerate abasic sites (32), and thus we used HIV RT as a positive control. However, tandem abasic sites led to a high incidence of deletions at these and surrounding sites with HIV RT, as well as with MMLV RT and bRT-Avd (Supplementary Figure S6a). Thus, we limited the abasic site to TR 23. HIV RT incorporated adenine almost exclusively (98%) across the abasic site (Figure 5a), and while deletions still occurred, they occurred much less frequently than with tandem abasic sites (Supplementary Figure S6b). MMLV RT has been reported to be intolerant to abasic sites (32), and indeed the incidence of deletion was ∼3-fold higher at abasic TR 23 for MMLV RT than for HIV RT (Supplementary Figure S6b). Nevertheless, for those cDNAs lacking deletions, MMLV RT incorporated adenine almost exclusively (99%) across the abasic site (Figure 5a). A significant level of deletion at and around the abasic site was also seen with bRT-Avd, with an incidence that resembled that of MMLV RT (Supplementary Figure S6b). However, the near exclusive preference for incorporating adenine seen for HIV and MMLV RTs was not observed for bRT-Avd, and instead at best a strong preference for adenine was evident (65%) (Figure 5A). Guanine was also incorporated at an appreciable frequency by bRT-Avd across the abasic site (Figure 5A). These results indicated that bRT-Avd did not follow the A-rule as strictly as HIV RT.
Figure 5.
Abasic and deoxy template sites. (A) Frequency of deoxynucleotides (mis)incorporated across an abasic site at TR 23 for bRT-Avd, MMLV RT, and HIV RT. (B) Frequency of deoxynucleotides (mis)incorporated by bRT-Avd (left) and MMLV RT (right) with deoxyadenosines at TR 23 and 24. (C) Frequency of deoxynucleotides (mis)incorporated by bRT-Avd (left) and MMLV RT (right) with deoxyguanosines at TR 23 and 24.
2′-OH
We also asked whether adenine-mutagenesis occurred when a deoxynucleotide was reverse transcribed. We therefore instituted deoxyadenosine at TR 23 and 24 in the core DGR RNA template. As expected, thymine was incorporated exclusively by MMLV RT across TR dA23 and dA24 (Figure 5B). In contrast, misincorporation increased for bRT-Avd. It went from 40% to 65% at TR 24 and from 72% to 83% at TR 23 (Figure 5B). To ascertain whether selectivity to adenine was maintained, TR 23 and 24 were instituted with deoxyguanosine. MMLV RT incorporated cytosine (99%) almost exclusively across these sites. Similarly, bRT-Avd incorporated cytosine across deoxyguanosine with nearly the same frequency as across guanosine (97% versus 98%) (Figure 5C). These results indicated that the 2′-OH of a template adenosine promoted correct incorporation and therefore countered adenine-mutagenesis.
bRT amino acids that modulate adenine-mutagenesis
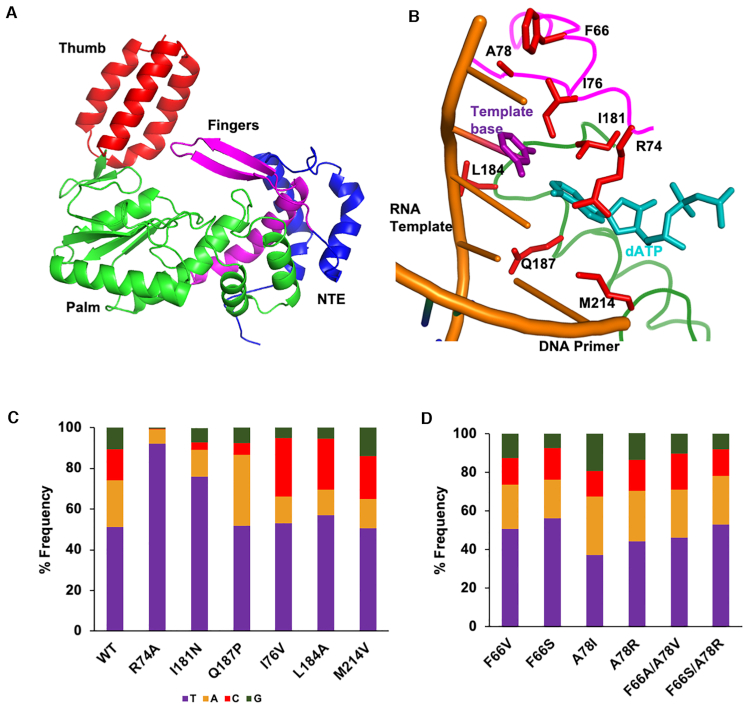
We next sought to identify bRT amino acids that modulated adenine-mutagenesis. We pursued this through structure-guided mutagenesis using a high-confidence model of bRT (100% confidence level for 95% of the amino acid sequence) that was generated using Phyre2 (33) (Supplementary Data). This bRT model was based in part on the structures of group II intron maturases (34,35), including the high fidelity GsI-IIc RT (36). This latter structure also contains a bound RNA template-DNA primer heteroduplex and an incoming dATP. The in silico bRT model consisted of all the important functional elements of RTs—the canonical fingers, palm, and thumb domain (Figure 6A). In addition to this, we relied on the extensive literature on the fidelity of HIV RT, and superposed the structure of HIV RT (37) with the in silico model of bRT to guide the choice of substitution sites.
Figure 6.
bRT amino acids that modulate selective fidelity. (A) In silico model of bRT in cartoon representation, showing the N-terminal extension (NTE, blue), Fingers (magenta), Palm (green), and Thumb (red) subdomains. (B) In silico model of bRT with amino acids subjected to mutagenesis shown as red bonds. The main chain is shown as a coil, colored by subdomains as in panel a. For reference, the RNA template-DNA primer heteroduplex (gold) and incoming dATP (cyan) from the structure of the GsI-IIc group II intron maturase is shown. The template base (thymine) is purple. (C) Average frequency of deoxynucleotides (mis)incorporated across the 22 TR adenines for wild-type bRT and bRT containing substitutions at amino acids implicated in modulating selective fidelity. (D) Average frequency of deoxynucleotides (mis)incorporated across the 22 TR adenines for bRT containing substitutions at amino acids predicted to be proximal to the templating base.
Based on this superposition, bRT Arg 74 was predicted to form a part of the binding pocket for the incoming dNTP (Figure 6b). Its putative homolog in HIV RT is Arg 72, which when substituted by Ala decreases misincorporation (38,39). We found a similar effect for bRT(R74A)-Avd. Substitution of bRT Arg 74 with Ala led to a marked decrease in misincorporation frequency across the 22 template adenines in TR from 52% to 8%, with some sites reaching as low as 1% misincorporation (Figure 6C and Supplementary Figure S7a). Misincorporation at other template bases (0.4%, 0.4% and 1.7% for uracil, cytosine and guanine, respectively) were at the levels within error seen for wild-type bRT (Supplementary Figure S7b). Thus, bRT Arg74 is a promoter of adenine-mutagenesis. Single deoxynucleotide primer extension analysis was carried out with bRT R74A to determine the basis for the decreased misincorporation. While there was almost no change in the Km for the correct incorporation of TTP across a template adenine at TR 117, a 40% decrease in kcat was observed (Table 3). The catalytic efficiency of bRT(R74A)-Avd for proper incorporation was 68% of that of wild-type bRT. Efforts to quantify misincorporation of dATP, the most frequently misincorporated dNTP, by primer extension analysis were unsuccessful, with no misincorporation evident for bRT(R74A)-Avd.
Table 3.
Steady-state enzymatic parameters for incorporation by mutant bRT-Avd
| bRT mutant | K m (μM) | k cat (min−1) x 10−3 | k cat/Km (μM−1 min−1) x 10−3 | Efficiencya |
|---|---|---|---|---|
| R74A, A:T | 7.09 ± 4.68 | 7.03 ± 0.78 | 0.99 | 0.68 |
| I181N, A:T | 218.85 ± 55.94 | 11.13 ± 1.04 | 0.05 | 0.03 |
| I181N, A:dA | 259.10 ± 144.40 | 1.37 ± 0.17 | 0.01 | 0.01 |
aEfficiency = [kcat (mutant)/Km (mutant)]/[kcat (wt)/Km (wt)].
Ile 181 of bRT was also predicted to be proximal to the incoming dNTP, and corresponds to HIV RT Gln 151, whose substitution by Asn decreases misincorporation (40–42). Ile 181 is the first amino acid of the signature DGR RT motif [I/V/L]GxxxSQ (1). In most retroviral RTs, non-LTR retrotransposon RTs, and group II intron maturases, this motif is instead QGxxxSP. We found that bRT I181N decreased the average misincorporation frequency across template adenines from 52% to 24% (Figure 6C and Supplementary Figure S7c). The misincorporation frequencies at other template bases were similar within error to those of wild-type bRT-Avd (Supplementary Figure S7d, 1.2%, 0.5% and 3% for uracil, cytosine, and guanine respectively). These results indicated that, like Arg 74, Ile 181 promoted misincorporation across template adenines. Single deoxynucleotide primer extension analysis of bRT(I181N)-Avd for correct TTP incorporation across a template adenine at TR 117 showed a large increase in Km as compared to wild-type bRT-Avd (Table 3, 27-fold). Notably, the Km for misincorporation of dATP across a template adenine by bRT(I181N)-Avd was similar to that of correct incorporation (Table 3). However, there was a large difference in kcat. While the kcat’s for correct incorporation by bRT(I181N)-Avd and wild-type bRT-Avd were similar, the kcat for misincorporation of dATP by bRT(I181N)-Avd was ∼8-fold slower (Table 3). The catalytic efficiency for correct incorporation of TTP by bRT(I181N)-Avd was 3% of wild-type bRT-Avd, and for misincorporation of dATP 13% of wild-type.
We also substituted the last amino acid in the signature DGR RT motif [I/V/L]GxxxSQ, bRT Gln 187, with the proline from the QGxxxSP motif. Proline at this position in HIV RT, P157, contacts a base in the template strand (37). bRT(Q187P)-Avd was unaltered in misincorporation frequency across template adenines compared to wild-type bRT, although this complex showed an increased bias towards misincorporating adenines over other bases (Figure 6C).
Substitutions were made in three bRT amino acids with putative equivalents in HIV RT that interact with the template or primer strand proximal to the catalytic site (37). These were bRT I176V, L184A and M214V (Figure 6B, C). The predicted structural equivalents in HIV RT and the misincorporation-decreasing substitutions are, respectively, Leu 74 substituted with Val (43); Lys 154 substituted with Ala (40); and Met 184 substituted with Val (44,45). While none of these substitutions in bRT changed the misincorporation frequency, they all curiously changed the misincorporation bias towards cytosine and away from adenine (Figure 6C).
Lastly, we probed two bRT amino acids predicted to be proximal to the template base: Phe 66 and Ala 78 (Figure 6B). The size and hydrophobicity of these positions were altered through F66V, F66S, A78V and A78R substitutions, as well as F66A/A78V and F66S/A78R double substitutions. None of these resulted in a significant decrease in misincorporation frequency, and indeed, the misincorporation frequency increased substantially for bRT(A78I)-Avd and somewhat for bRT(A78R)-Avd (Figure 6D).
DISCUSSION
DGRs bring about massive protein sequence variation through the unique mechanism of adenine-mutagenesis. This results in variability being restricted to adenine-encoded amino acids, with non-adenine-encoded amino acids remaining conserved. As seen in a number of DGR variable proteins (9,22,46–48), adenine-encoded amino acids are organized by the C-type lectin-fold of the variable protein into a solvent-exposed binding site. Non-adenine-encoded amino acids form the invariable structural scaffolding for the variable binding site. AAY (Y = pyrimidine) codons are especially prevalent in DGR variable proteins, and as previously noted, adenine-mutagenesis of AAY codons captures the gamut of amino acid chemistry but precludes a stop codon (22). Adding a layer of complexity to adenine-mutagenesis is the distribution of misincorporation frequencies documented here. For example, this makes some amino acid positions more variable (e.g. Mtd 357 with A62 in its codon, 76% misincorporation frequency) and some less (e.g. Mtd 369 with A98 in its codon, 30% misincorporation frequency), and thereby shapes the repertoire of ligands functionally bound by DGR variable proteins. These positional effects on misincorporation frequencies are also seen in cDNAs synthesized in vivo (13) and mtd sequences that have undergone variation in the absence of selection (49). This is most noticeable for TR A62, which consistently has a high misincorporation frequency (∼70%). No obvious relationship was evident between positional variation in misincorporation frequency and primary sequence. It is possible that the secondary or even tertiary structure of the template plays a role in this.
Extensive work has shown that fidelity in nucleotide polymerases depends both on Watson–Crick hydrogen bonding and shape complementarity between base pairs (50–56). Indeed, shape complementarity appears to be the dominant discriminator (57), as supported by several lines of evidence, including the observation that hydrogen bonding is dispensable for fidelity in DNA polymerase I (58). The general consensus is that pairing between the template and incoming base is sterically evaluated by polymerases (31,52). If the pairing is correct, polymerases undergo an open to closed transition, which places catalytic groups in the right positions for chemistry to proceed. If the pairing is incorrect, the open to closed transition fails to occur, providing time enough for the incorrect dNTP to dissociate before chemistry can occur.
High and low fidelity nucleotide polymerases are alike when it comes to misincorporation but differ crucially with respect to correct incorporation (16). Both types of polymerases display similarly low catalytic efficiencies (kcat/Km) for misincorporation, but high fidelity polymerases have high catalytic efficiencies for correct incorporation while low fidelity polymerases remain at low catalytic efficiencies. High fidelity polymerases have a ∼105-fold difference between catalytic efficiencies of correct incorporation versus misincorporation, but low fidelity polymerases have only a ∼102-fold difference. While low fidelity polymerases are inefficient enzymes, they have evolved to be inefficient for specific purposes. For example, members of the Y family of DNA polymerases are low fidelity enzymes that are responsible for replicating through DNA lesions (51). Synthesis through a lesion, effectively a non-standard template site, requires low fidelity. Reminiscent of bRT-Avd, the Y family DNA polymerase iota (pol ι) misincorporates dGTP at a frequency of 0.72 across template thymines and has a low catalytic efficiency for incorporating correct base pairs, ranging between 10−1 to 10−4 μM−1min−1 (59–61). Notably, the 1–40 × 10−4 μM−1min−1 catalytic efficiency of bRT-Avd for correct incorporation falls within this range (Table 1), and is ∼104 lower than the efficiency of high fidelity polymerases (16). DNA pol ι and other Y family DNA polymerases synthesize only short stretches of DNA to repair lesions (62), and indeed DNA pol ι tends to terminate synthesis after incorporating a dGTP across a template thymine (60). This is also reminiscent of bRT-Avd, which synthesizes only short cDNAs (5–35 nt) with non-DGR RNA templates. However, bRT-Avd becomes processive with the DGR RNA as its template and synthesizes extended cDNAs (90- and 120-nt), likely due to the formation of a structured RNP. These observations suggest that the low catalytic efficiency of bRT-Avd is intimately tied to its ability to carry out adenine-mutagenesis, and while the inherent tendency of bRT-Avd is to synthesize short cDNAs, the DGR RNA provides a means to synthesize longer cDNAs. It is possible that a low catalytic efficiency is tolerated by DGRs because the target for variation (e.g. the gene encoding the variable protein) usually exists in single copy number and thus requires only a single cDNA molecule to effect sequence variation. While the efficiency of cDNA synthesis by the Bordetella bacteriophage DGR in vivo requires further study, it is worth noting that the overall efficiency of sequence variation in vivo by this DGR is quite low (10−6) (15).
To understand the nucleobase determinants that modulate adenine-mutagenesis, we took advantage of the near isosteric features of infidelity-promoting adenine and fidelity-promoting guanine. Using nucleobase analogs that have adenine- or guanine-like groups, we found that the substituent at the C6 position, but not the N1 or C2 position, had a major effect on misincorporation frequency. An amine at the C6 position, which in adenine acts as a hydrogen bond donor in a Watson-Crick base pair, was functionally equivalent to having no substituent at this position. In contrast, a carbonyl at C6, which in guanine acts as a hydrogen bond acceptor in a Watson-Crick base pair, greatly lowered the misincorporation frequency. Thus, it appears that bRT-Avd is sensitive to the mispositioning of a C6 carbonyl but not a C6 amine.
We also explored the possibility that adenine flips out of the catalytic site, leaving it empty. This has been suggested for DNA pol ι, in which incorporation is more efficient across an abasic rather than a pyrimidine template site (60,61). However, in the case of bRT-Avd an abasic site resulted in a substantial level of deletions, and the misincorporation pattern at the abasic site was not the same as with adenine. While both abasic and adenine sites led predominantly to adenine misincorporation, an abasic site led to preferential misincorporation of guanine over cytosine. In contrast for a template adenine the preference was cytosine over guanine. Thus, the adenine-mutagenesis pattern is not explained by the flipping out of adenine from the catalytic site. In addition, these results indicated that the catalytic site of bRT-Avd is not as predisposed towards an incoming adenine as has been suggested for A-rule polymerases (31).
We sought to identify amino acids in bRT that have a role in modulating adenine-mutagenesis, and relied on a three-dimensional in silico model based primarily on group II intron maturases (34–36). We probed eight amino acids predicted to be located at or near the catalytic site, and identified two that modulated adenine-mutagenesis: R74 and I181. Based on an in silico model of bRT, Arg 74 is predicted to correspond to Arg 72 of HIV RT. This HIV RT amino acid contacts the base and phosphate of the incoming dNTP (37). Substitution of HIV RT Arg 72 with Ala leads to a significant decrease in misincorporation (about three-fold on average but up to 25-fold at specific sites) with a significant decrease in kcat (30- to 100-fold) (38,39). Similarly, we observed that substitution of bRT R74 with Ala led to a decrease in misincorporation accompanied by a decrease in kcat. However, the effects for bRT-Avd were much more modest (1.8-fold decrease in misincorporation and 1.7-fold decrease in kcat). The second amino acid, Ile 181 of bRT, is predicted to correspond to HIV RT Q151. This amino acid in HIV RT contacts the ribose of the incoming dNTP (37). Substitution of HIV RT Gln 151 with Asn decreases misincorporation by 8- to 27-fold (41), and decreases the affinity for the correct incoming dNTP by 120-fold and for incorrect ones to levels that are not measurable (42). A similar but smaller 1.5-fold decrease in misincorporation across a template was seen for bRT I181N, as well as a 27-fold increase in Km for correct incorporation of TTP. However, the decrease in misincorporation in bRT(I181N)-Avd was not due to a further increase in Km for the incorrect deoxynucleotide but instead a marked decrease in kcat. In HIV RT, Arg 74 and Gln 151 provide contacts that stabilize both correct and incorrect incoming dNTPs. In the absence of the nonspecific contacts provided by Arg 74 and Gln 151, the contributions of correct hydrogen bonding and shape complementarity become more consequential and thereby increase discrimination between pairings, leading to a decrease in misincorporation. Similarly, bRT Arg 74 and Ile 181 are likely to provide nonspecific stabilization of base pairs and thereby promote adenine-mutagenesis.
In summary, our results provide evidence that bRT-Avd is a catalytically inefficient enzyme, a property that is likely intimately tied to its ability to carry out adenine-mutagenesis. We found that the C6, but not the N1 or C2, purine substituent was a key determinant of adenine-mutagenesis, and that Arg 74 and Ile 181 have significant roles in promoting adenine-mutagenesis. Our results provide the first detailed characterization of the nucleobase and protein determinants of adenine-mutagenesis in DGRs.
DATA AVAILABILITY
The alignment files for NGS data are available at the NIH Sequence Read Archive with accession number PRJNA676165.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Simpson Joseph and Yitzhak Tor for valuable conversations about aspects of this work.
Notes
Present address: Andres Reyna, Department of Chemistry, University of Washington, Seattle, WA 98195, USA.
Contributor Information
Sumit Handa, Department of Chemistry & Biochemistry, 9500 Gilman Drive, La Jolla, CA, 92093-0375, USA.
Andres Reyna, Department of Chemistry & Biochemistry, 9500 Gilman Drive, La Jolla, CA, 92093-0375, USA.
Timothy Wiryaman, Department of Chemistry & Biochemistry, 9500 Gilman Drive, La Jolla, CA, 92093-0375, USA.
Partho Ghosh, Department of Chemistry & Biochemistry, 9500 Gilman Drive, La Jolla, CA, 92093-0375, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01 GM132720 to P.G.]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Liu M., Deora R., Doulatov S.R., Gingery M., Eiserling F.A., Preston A., Maskell D.J., Simons R.W., Cotter P.A., Parkhill J. et al.. Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science. 2002; 295:2091–2094. [DOI] [PubMed] [Google Scholar]
- 2. Minot S., Bryson A., Chehoud C., Wu G.D., Lewis J.D., Bushman F.D.. Rapid evolution of the human gut virome. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:12450–12455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ye Y. Identification of diversity-generating retroelements in human microbiomes. Int. J. Mol. Sci. 2014; 15:14234–14246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Paul B.G., Bagby S.C., Czornyj E., Arambula D., Handa S., Sczyrba A., Ghosh P., Miller J.F., Valentine D.L.. Targeted diversity generation by intraterrestrial archaea and archaeal viruses. Nat. Commun. 2015; 6:6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Paul B.G., Burstein D., Castelle C.J., Handa S., Arambula D., Czornyj E., Thomas B.C., Ghosh P., Miller J.F., Banfield J.F. et al.. Retroelement-guided protein diversification abounds in vast lineages of Bacteria and Archaea. Nat. Microbiol. 2017; 2:17045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Benler S., Cobian-Guemes A.G., McNair K., Hung S.H., Levi K., Edwards R., Rohwer F.. A diversity-generating retroelement encoded by a globally ubiquitous Bacteroides phage. Microbiome. 2018; 6:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yan F., Yu X., Duan Z., Lu J., Jia B., Qiao Y., Sun C., Wei C.. Discovery and characterization of the evolution, variation and functions of diversity-generating retroelements using thousands of genomes and metagenomes. BMC Genomics. 2019; 20:595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boehm T., McCurley N., Sutoh Y., Schorpp M., Kasahara M., Cooper M.D.. VLR-based adaptive immunity. Annu. Rev. Immunol. 2012; 30:203–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Le Coq J., Ghosh P. Conservation of the C-type lectin fold for massive sequence variation in a Treponema diversity-generating retroelement. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:14649–14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu L., Gingery M., Abebe M., Arambula D., Czornyj E., Handa S., Khan H., Liu M., Pohlschroder M., Shaw K.L. et al.. Diversity-generating retroelements: natural variation, classification and evolution inferred from a large-scale genomic survey. Nucleic Acids Res. 2018; 46:11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Melvin J.A., Scheller E.V., Miller J.F., Cotter P.A.. Bordetella pertussis pathogenesis: current and future challenges. Nat. Rev. Microbiol. 2014; 12:274–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guo H., Arambula D., Ghosh P., Miller J.F.. Diversity-generating retroelements in phage and bacterial genomes. Microbiol. Spectr. 2014; 2:doi:10.1128/microbiolspec.MDNA3-0029-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Naorem S.S., Han J., Wang S., Lee W.R., Heng X., Miller J.F., Guo H.. DGR mutagenic transposition occurs via hypermutagenic reverse transcription primed by nicked template RNA. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E10187–E10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Handa S., Jiang Y., Tao S., Foreman R., Schinazi R.F., Miller J.F., Ghosh P.. Template-assisted synthesis of adenine-mutagenized cDNA by a retroelement protein complex. Nucleic Acids Res. 2018; 46:9711–9725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Alayyoubi M., Guo H., Dey S., Golnazarian T., Brooks G.A., Rong A., Miller J.F., Ghosh P.. Structure of the essential diversity-generating retroelement protein bAvd and its functionally important interaction with reverse transcriptase. Structure. 2013; 21:266–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Beard W.A., Shock D.D., Vande Berg B.J., Wilson S.H.. Efficiency of correct nucleotide insertion governs DNA polymerase fidelity. J. Biol. Chem. 2002; 277:47393–47398. [DOI] [PubMed] [Google Scholar]
- 17. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Proc G.P.D.. The sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Miller J.R., Koren S., Sutton G.. Assembly algorithms for next-generation sequencing data. Genomics. 2010; 95:315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P.. Integrative genomics viewer. Nat. Biotechnol. 2011; 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sebastian-Martin A., Barrioluengo V., Menendez-Arias L.. Transcriptional inaccuracy threshold attenuates differences in RNA-dependent DNA synthesis fidelity between retroviral reverse transcriptases. Sci. Rep. 2018; 8:627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McMahon S.A., Miller J.L., Lawton J.A., Kerkow D.E., Hodes A., Marti-Renom M.A., Doulatov S., Narayanan E., Sali A., Miller J.F. et al.. The C-type lectin fold as an evolutionary solution for massive sequence variation. Nat. Struct. Mol. Biol. 2005; 12:886–892. [DOI] [PubMed] [Google Scholar]
- 23. Case-Green S.C., Southern E.M.. Studies on the base pairing properties of deoxyinosine by solid phase hybridisation to oligonucleotides. Nucleic Acids Res. 1994; 22:131–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martin F.H., Castro M.M., Aboulela F., Tinoco I.. Base-Pairing involving Deoxyinosine - Implications for probe design. Nucleic Acids Res. 1985; 13:8927–8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rahman M.S., Humayun M.Z.. Nebularine (9-2′-deoxy-beta-D-ribofuranosylpurine) has the template characteristics of adenine in vivo and in vitro. Mutat. Res. 1997; 377:263–268. [DOI] [PubMed] [Google Scholar]
- 26. Harcourt E.M., Ehrenschwender T., Batista P.J., Chang H.Y., Kool E.T.. Identification of a selective polymerase enables detection of N-6-Methyladenosine in RNA. J. Am. Chem. Soc. 2013; 135:19079–19082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cheong C., Tinoco I., Chollet A.. Thermodynamic studies of base-pairing involving 2,6-diaminopurine. Nucleic Acids Res. 1988; 16:5115–5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Patro J.N., Urban M., Kuchta R.D.. Role of the 2-Amino group of purines during dNTP polymerization by human DNA polymerase α. Biochemistry. 2009; 48:180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Watanabe S.M., Goodman M.F.. Kinetic measurement of 2-aminopurine.cytosine and 2-aminopurine.thymine base-pairs as a test of DNA-polymerase fidelity mechanisms. Proc. Natl. Acad. Sci. 1982; 79:6429–6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reha-Krantz L.J., Hariharan C., Subuddhi U., Xia S.L., Zhao C., Beckman J., Christian T., Konigsberg W.. Structure of the 2-aminopurine-cytosine base pair formed in the polymerase active site of the RB69 Y567A-DNA polymerase. Biochemistry. 2011; 50:10136–10149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Strauss B.S. The “A” rule revisited: polymerases as determinants of mutational specificity. DNA Repair (Amst.). 2002; 1:125–135. [DOI] [PubMed] [Google Scholar]
- 32. Kupfer P.A., Crey-Desbiolles C., Leumann C.J.. Trans-lesion synthesis and RNaseH activity by reverse transcriptases on a true abasic RNA template. Nucleic Acids Res. 2007; 35:6846–6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015; 10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhao C., Pyle A.M.. Crystal structures of a group II intron maturase reveal a missing link in spliceosome evolution. Nat. Struct. Mol. Biol. 2016; 23:558–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Qu G., Kaushal P.S., Wang J., Shigematsu H., Piazza C.L., Agrawal R.K., Belfort M., Wang H.W.. Structure of a group II intron in complex with its reverse transcriptase. Nat. Struct. Mol. Biol. 2016; 23:549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stamos J.L., Lentzsch A.M., Lambowitz A.M.. Structure of a thermostable group II intron reverse transcriptase with Template-Primer and its functional and evolutionary implications. Mol. Cell. 2017; 68:926–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huang H., Chopra R., Verdine G.L., Harrison S.C.. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998; 282:1669–1675. [DOI] [PubMed] [Google Scholar]
- 38. Sarafianos S.G., Pandey V.N., Kaushik N., Modak M.J.. Site-directed mutagenesis of arginine 72 of HIV-1 reverse transcriptase. Catalytic role and inhibitor sensitivity. J. Biol. Chem. 1995; 270:19729–19735. [DOI] [PubMed] [Google Scholar]
- 39. Lewis D.A., Bebenek K., Beard W.A., Wilson S.H., Kunkel T.A.. Uniquely altered DNA replication fidelity conferred by an amino acid change in the nucleotide binding pocket of human immunodeficiency virus type 1 reverse transcriptase. J. Biol. Chem. 1999; 274:32924–32930. [DOI] [PubMed] [Google Scholar]
- 40. Weiss K.K., Isaacs S.J., Tran N.H., Adman E.T., Kim B.. Molecular architecture of the mutagenic active site of human immunodeficiency virus type 1 reverse transcriptase: roles of the beta 8-alpha E loop in fidelity, processivity, and substrate interactions. Biochemistry. 2000; 39:10684–10694. [DOI] [PubMed] [Google Scholar]
- 41. Kaushik N., Talele T.T., Pandey P.K., Harris D., Yadav P.N., Pandey V.N.. Role of glutamine 151 of human immunodeficiency virus type-1 reverse transcriptase in substrate selection as assessed by site-directed mutagenesis. Biochemistry. 2000; 39:2912–2920. [DOI] [PubMed] [Google Scholar]
- 42. Weiss K.K., Bambara R.A., Kim B.. Mechanistic role of residue Gln151 in error prone DNA synthesis by human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT). J. Biol. Chem. 2002; 277:22662–22669.Pre-steady state kinetic study of the Q151N HIV-1 RT mutant with increased fidelity. [DOI] [PubMed] [Google Scholar]
- 43. Jonckheere H., De Clercq E., Anne J.. Fidelity analysis of HIV-1 reverse transcriptase mutants with an altered amino-acid sequence at residues Leu74, Glu89, Tyr115, Tyr183 and Met184. Eur. J. Biochem. 2000; 267:2658–2665. [DOI] [PubMed] [Google Scholar]
- 44. Pandey V.N., Kaushik N., Rege N., Sarafianos S.G., Yadav P.N.S., Modak M.J. Role of methionine 184 of human immunodeficiency virus type-1 reverse transcriptase in the polymerase function and fidelity of DNA synthesis. Biochemistry. 1996; 35:2168–2179. [DOI] [PubMed] [Google Scholar]
- 45. Wainberg M.A., Drosopoulos W.C., Salomon H., Hsu M., Borkow G., Parniak M.A., Gu Z.X., Song Q.B., Manne J., Islam S. et al.. Enhanced fidelity of 3TC-selected mutant HIV-1 reverse transcriptase. Science. 1996; 271:1282–1285. [DOI] [PubMed] [Google Scholar]
- 46. Miller J.L., Coq J.L., Hodes A., Barbalat R., Miller J.F., Ghosh P.. Selective ligand recognition by a Diversity-Generating retroelement variable protein. PLoS Biol. 2008; 6:e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Handa S., Paul B.G., Miller J.F., Valentine D.L., Ghosh P.. Conservation of the C-type lectin fold for accommodating massive sequence variation in archaeal diversity-generating retroelements. BMC Struct. Biol. 2016; 16:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Handa S., Shaw K.L., Ghosh P.. Crystal structure of a Thermus aquaticus diversity-generating retroelement variable protein. PLoS One. 2019; 14:e0205618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Doulatov S., Hodes A., Dai L., Mandhana N., Liu M., Deora R., Simons R.W., Zimmerly S., Miller J.F.. Tropism switching in Bordetella bacteriophage defines a family of diversity-generating retroelements. Nature. 2004; 431:476–481. [DOI] [PubMed] [Google Scholar]
- 50. Beard W.A., Wilson S.H.. Structural insights into the origins of DNA polymerase fidelity. Structure. 2003; 11:489–496. [DOI] [PubMed] [Google Scholar]
- 51. Kunkel T.A. Evolving views of DNA replication (in)fidelity. Cold Spring Harb. Symp. Quant. Biol. 2009; 74:91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Freudenthal B.D., Beard W.A., Wilson S.H.. New structural snapshots provide molecular insights into the mechanism of high fidelity DNA synthesis. DNA Repair (Amst.). 2015; 32:3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Echols H., Goodman M.F.. Fidelity mechanisms in DNA replication. Annu. Rev. Biochem. 1991; 60:477–511. [DOI] [PubMed] [Google Scholar]
- 54. Kim T.W., Delaney J.C., Essigmann J.M., Kool E.T.. Probing the active site tightness of DNA polymerase in subangstrom increments. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15803–15808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sydow J.F., Cramer P.. RNA polymerase fidelity and transcriptional proofreading. Curr. Opin. Struct. Biol. 2009; 19:732–739. [DOI] [PubMed] [Google Scholar]
- 56. Menendez-Arias L. Molecular basis of fidelity of DNA synthesis and nucleotide specificity of retroviral reverse transcriptases. Prog. Nucleic Acid Res. Mol. Biol. 2002; 71:91–147. [DOI] [PubMed] [Google Scholar]
- 57. Kool E.T. Hydrogen bonding, base stacking, and steric effects in dna replication. Annu. Rev. Biophys. Biomol. Struct. 2001; 30:1–22. [DOI] [PubMed] [Google Scholar]
- 58. Moran S., Ren R.X.F., Kool E.T. A thymidine triphosphate shape analog lacking Watson–Crick pairing ability is replicated with high sequence selectivity. Proc. Natl. Acad. Sci. 1997; 94:10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bebenek K., Tissier A., Frank E.G., McDonald J.P., Prasad R., Wilson S.H., Woodgate R., Kunkel T.A.. 5′-Deoxyribose phosphate lyase activity of human DNA polymerase iota in vitro. Science. 2001; 291:2156–2159. [DOI] [PubMed] [Google Scholar]
- 60. Zhang Y., Yuan F., Wu X., Wang Z.. Preferential incorporation of G opposite template T by the low-fidelity human DNA polymerase iota. Mol. Cell. Biol. 2000; 20:7099–7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Johnson R.E., Washington M.T., Haracska L., Prakash S., Prakash L.. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature. 2000; 406:1015–1019. [DOI] [PubMed] [Google Scholar]
- 62. McCulloch S.D., Kunkel T.A.. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008; 18:148–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The alignment files for NGS data are available at the NIH Sequence Read Archive with accession number PRJNA676165.