Abstract
The PAQosome is a large complex composed of the HSP90/R2TP chaperone and a prefoldin-like module. It promotes the biogenesis of cellular machineries but it is unclear how it discriminates closely related client proteins. Among the main PAQosome clients are C/D snoRNPs and in particular their core protein NOP58. Using NOP58 mutants and proteomic experiments, we identify different assembly intermediates and show that C12ORF45, which we rename NOPCHAP1, acts as a bridge between NOP58 and PAQosome. NOPCHAP1 makes direct physical interactions with the CC-NOP domain of NOP58 and domain II of RUVBL1/2 AAA+ ATPases. Interestingly, NOPCHAP1 interaction with RUVBL1/2 is disrupted upon ATP binding. Moreover, while it robustly binds both yeast and human NOP58, it makes little interactions with NOP56 and PRPF31, two other closely related CC-NOP proteins. Expression of NOP58, but not NOP56 or PRPF31, is decreased in NOPCHAP1 KO cells. We propose that NOPCHAP1 is a client-loading PAQosome cofactor that selects NOP58 to promote box C/D snoRNP assembly.
INTRODUCTION
Non-coding RNPs play essential functions in cellular processes such as transcription, splicing and translation, and their assembly often involves complex mechanisms requiring chaperones and cofactors (1–4). Ribosomes are among the best studied molecular machines built from non-coding RNAs and proteins. Their biogenesis is an ordered process that assembles numerous proteins on ribosomal RNAs (rRNAs) in a highly regulated manner, and it involves nearly two hundred assembly factors that are not maintained in the mature functional particle (5). These factors play many roles during the assembly process, including a quality-control of the particles produced.
Ribosomal RNAs also contain many nucleotide modifications that are important for ribosome biogenesis and for accurate translation of proteins (6,7). Most of them are catalyzed by snoRNPs, which are another important class of non-coding RNPs involved in both nucleotide modification and rRNA processing (3,8). SnoRNPs are divided in two groups, C/D and H/ACA, which guide 2′-O-methylation of the ribose or isomerization of uridine into pseudouridine, respectively. The position of the modified nucleotide is determined by base pairing between the snoRNA and its RNA target, and nucleotide modification is carried out by the methyltransferase Fibrillarin (FBL) for C/D snoRNPs and the pseudouridine synthase Dyskerin (DKC1) in H/ACA snoRNPs. These enzymes are stable components of the snoRNPs, which also contain several others core proteins. These play important role in the maintenance and function of the RNP particles: SNU13, NOP56 and NOP58 for C/D snoRNPs; NHP2, NOP10 and GAR1 for H/ACA snoRNPs (9,10).
Each core protein is positioned in a similar manner in the various snoRNP particles, irrespective of the snoRNA it contains (11–16). In the C/D snoRNPs, SNU13 directly recognizes a K-turn in the C/D and C′/D′ motifs, which are located at the basal and apical part of the snoRNAs, respectively (17). NOP56 and NOP58 are two paralog proteins containing NOP and coiled-coil (CC) domains (18). In these proteins, the NOP domain is an RNP binding module specific for SNU13:RNA complexes (19), while their CC domain allows their hetero-dimerization across the C/D and C′/D′ motifs, thereby creating a pseudo dimeric structure (11,15,20). NOP56 and NOP58 also bind each a copy of Fibrillarin through their N-terminal domains (11). Interestingly, the U4 snRNA forms an RNP that displays similarities with C/D snoRNPs. U4 RNA contains a K-turn that binds SNU13, and it also associates with PRPF31, which is another paralog of NOP58 (17,21). As in C/D snoRNPs, the PRPF31 NOP domain binds SNU13:RNA complexes (19). Its CC domain however hetero-dimerizes with PRPF6, which belongs to U5 snRNP and helps to form the U4/U6.U5 tri-snRNP particle (19).
Studies in the last ten years have shown that the HSP90/R2TP chaperone is a key player in C/D snoRNP biogenesis (22–24). The R2TP is composed of RPAP3, PIH1D1 and the AAA+ ATPases RUVBL1/RUVBL2. These highly related ATPases form hetero-hexamers and hetero-dodecamers, and they are proposed to have chaperone activity on their own (25). RPAP3 and PIH1D1 form a stable heterodimer and are believed to function as specific adaptors and regulators for HSP90 and RUVBL1/2 (26–28). RPAP3 binds HSP90 and HSP70 through its TPR domains (29–31), and it binds RUVBL1/2 through its C-terminal domain (26,32). PIH1D1 recruits R2TP clients through an N-terminal phospho-binding pocket (31,33). The R2TP chaperone is conserved from yeast to human but in metazoans, it further associates with a prefoldin-like module to form the PAQosome (34). A fascinating property of the PAQosome is that it seems specialized in the assembly of multi-subunit complexes. Several clients of this chaperone have been identified besides the C/D snoRNPs. They are essential macro-molecular complexes such as the nuclear RNA polymerases, the PIKK containing complexes like mTORC1, and other RNP complexes including U4 and U5 snRNPs, or H/ACA snoRNPs (34).
The precise mechanism of action of R2TP remains incompletely characterized. With regards to C/D snoRNP biogenesis, RUVBL1/2 were proposed to promote an important remodeling event on immature C/D snoRNPs (22,35). It is also known that NOP58 and SNU13 are clients of HSP90 and make ATP-dependent interaction with RUVBL1/2 (36). SNU13 also forms a stable complex with the NUFIP1/ZNHIT3 heterodimer, which is another assembly factor that interacts with R2TP via PIH1D1 (23,37). Bcd1 is also involved in C/D snoRNP assembly. It was discovered in yeast and found to play an essential role during C/D snoRNA biogenesis (38). Its human homolog, called ZNHIT6, was found to be required for maintenance of C/D snoRNA level and to make ATP-dependent interaction with RUVBL1/2 (36,39). Interestingly, ZNHIT6 and ZNHIT3 have a very similar zf–HIT domain, which is common to other ZNHIT family members and may be a RUVBL1/2 interaction domain (40,41). By proteomic experiments, a protein-only complex containing SNU13, NOP58, RUVBL/2, ZNHIT6 and the NUFIP1/ZNHIT3 heterodimer was identified (22,39). This complex is believed to form by the independent recruitment of several factors to R2TP (i.e. NOP58, ZNHIT6 and a ZNHIT3/NUFIP1/SNU13 module), followed by a rearrangement of these proteins, possibly in an ATP-dependent manner (22,39).
Here, we characterized C12ORF45, which we rename NOPCHAP1, as a new assembly factor for C/D snoRNPs. We show that it binds directly NOP58 and RUVBL1/2, and that it can bridge them in absence of snoRNAs. NOP58 is destabilized in NOPCHAP1 knock-out cells, suggesting that it chaperones NOP58 during early step of C/D snoRNP biogenesis.
MATERIALS AND METHODS
Cell culture and cell line construction
HeLa Flp-In cells were a gift of S. Emiliani (Institut Cochin, Paris and 42). HEK 293T cells were from the ATCC collection. All cells were grown at 37°C, 5% CO2 and plasmid transfections were done with JetPrime (Ozyme). HeLa Flp-In and HEK 293T cells were grown in Dulbecco's modified eagle medium (DMEM) containing 10% fetal bovine serum (FBS), glutamine (2.9 mg/ml), and penicillin/streptomycin (10 U/ml). For SILAC, after plasmid recombination in FRT site of HeLa Flp-In by co-transfection of FlpO expression plasmid, clones were selected in hygromycin B (150 μg/ml), picked individually and characterized by Western Blot and fluorescence microscopy. NOPCHAP1 CRISPR/Cas9 knock-out cells (NOPCHAP1 KO cells) were generated by transfection of HEK 293T cells expressing Flag-Cas9 with Sanger Lentiviral CRISPR vector plasmid U6-gRNA/PGK-puro-2A-BFP and guide RNA to the NOPCHAP1 or control locus according to the manufacturer's protocol (Sigma-Aldrich). Sequences of RNA guides are as following: NOPCHAP1-CGAACTGTTTGAAGAGTGGAGG and Ct-AGCACGTAATGTCCGTGGAT. After selection with puromycin and blasticidin, one single-cell per well was sorted into 96-well plates by FACS, and clones KO for NOPCHAP1 were screened by Western Blot and PCR and sequencing on genomic DNA.
Plasmids and cloning
DNA cloning was performed using standard techniques or with the Gateway™ system (InVitrogen). For mutation in NOP58, PRPF31, RUVBL1 and RUVBL2 ORFs, mutagenesis was performed on the pDon vectors with the QuickStrand mutagenesis kit (Agilent) following the manufacturer's protocol. NOP58 fragments were generated by PCR and cloned in pDon vector by BP reaction (Gateway). To generate GFP fusions for SILAC-IPs, pcDNA5-FRT-GFP-3xFLAG-Rf was recombined with pDon vectors by LR reaction (Gateway). For the LUMIER-IP assays, recombination was done with pcDNA5-FRT-3xFLAG-FL-Rf for the bait, and L30-HA-RL-Rf for the prey. For the bridged LUMIER IP assay, the third plasmid is a pcDNA5-myc-Rf recombined with pDon-NOPCHAP1 or pDon-PHAX. Most of cDNAs were from human origin except for RUVBL1, RUVBL2 and PHAX which were from mouse. Plasmids for in vitro expression in Escherichia coli are as follow. RUVBL1 and RUVBL2 were cloned in the pETDuet vector (Novagen) by manufacturer (GenScript) between NcoI and HindIII, and NdeI and XhoI, respectively, then the RUVBL1-RUVBL2 fragment of this construct was subcloned using standard technics in pnCS vector between NdeI and BamHI. The sequences of NOP58-CC-NOP, yNOP58(1–447), yNOP58-CC-NOP and NOPCHAP1 are codon optimized sequences for E. coli expression. They were cloned between NdeI and BamHI in expression vectors (pnEA-3CH for N-His6 tagged protein, pnCS and pnYK for native proteins) compatible for co-transformation and co-expression in E. coli. Detailed maps and sequences are available upon request.
Antibodies
Antibodies and dilutions for Western Blots were the following: rabbit polyclonal anti-NOP58 (Bethyl laboratories, A302-719A) at 1:1000, mouse monoclonal anti-GAPDH (Abcam, ab8245) at 1:20 000, rabbit polyclonal anti-RUVBL1 (Proteintech Group, 10210-2-AP) at 1:1000, rabbit polyclonal anti-NOPCHAP1 (Bethyl laboratories, A304-707A) at 1:1000. Secondary antibodies were goat anti-rabbit and anti-mouse conjugated to horseradish peroxidase (HRP) (Sigma-Aldrich) at 1:20000 and 1:10000 respectively. Antibodies and dilutions for immunolabelling were the following: rabbit polyclonal anti-NOP58 (Bethyl laboratories, A302-719A) at 1:200, mouse monoclonal antibody anti-Fibrillarin (72B9, personal gift of J. Cavaillé) at 1:20, rabbit polyclonal anti-Coilin (personal gift of A. Lamond) at 1:400, rabbit polyclonal anti-dyskerin (Santacruz, sc48794) at 1:100 and rabbit polyclonal anti-B23 (Santacruz, sc5564) at 1:200. Secondary antibodies were goat anti-rabbit and goat anti-mouse coupled to FITC (Jackson laboratories) used at 1:200.
Sequence analyses
Sequences were retrieved from the NCBI annotated databases (nr and EST, http://www.ncbi.nlm.nih.gov), using Annotation search tools for specific domains (pfam 15370/CDD:292014 for NOPCHAP1, pfam 1043/CDD:313644 for NUFIP, pfam 13877/CDD:258144 for RPAP3-Cter, pfam 08156/CDD:311881 for NOP56-58), and when necessary NCBI PHI-BLAST as well as BLAST, available in the Geneious 11.1.5 software package (Biomatters, http://www.geneious.com/). Orthology was determined by reciprocal BLAST analysis and domain architecture. The 2301 accession numbers used for the Figure 3F are listed in Supplementary Table S4. The phylogenetic tree was drawn according to (43).
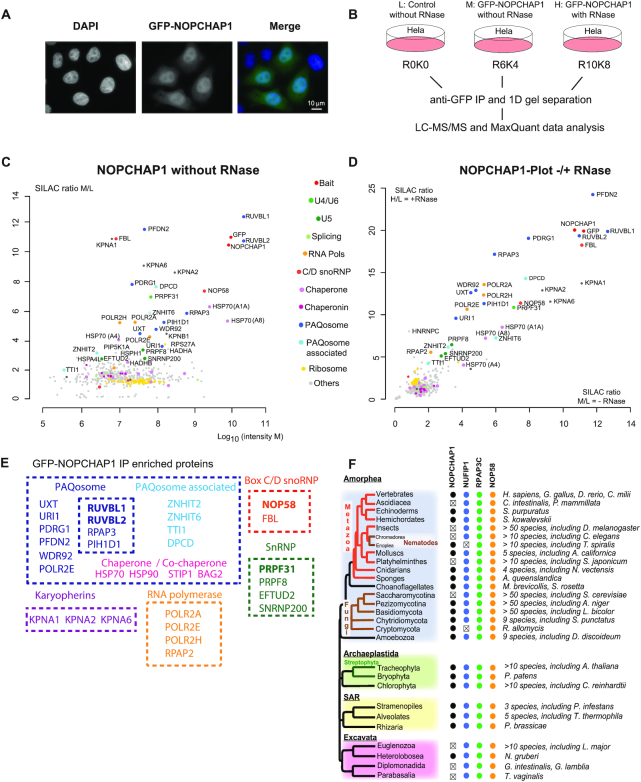
Figure 3.
Characterization of NOPCHAP1. (A) Epifluorescence microscopy images of HeLa cells expressing GFP-NOPCHAP1. Blue/left panel: DAPI staining; Green/middle panel: GFP. Scale bar is 10 μm. (B) Schematic representation of SILAC IP experiments shown in C and D. (C) Proteomic analyses of the partners of GFP-NOPCHAP1. This IP is done without RNase treatment. Full hit list with Significance B values are given in Supplementary Table S3. Legend as in Figure 1 for graphic representation. (D) Comparison of GFP-NOPCHAP1 interactome in presence or in absence of RNase. SILAC ratios from condition with RNase (y axis, shown in Figure S2A) or without RNase (x axis, shown in C) were plotted against each other. (E) Scheme showing groups of NOPCHAP1 interactants found in our SILAC IPs and grouped according to the known complexes in which they belong. (F) Conservation of NOPCHAP1, NUFIP1, RPAP3-Cter (Cterminal domain) and NOP58 across Eukaryotes. Members that were not found are indicated by ‘x’. In Alveolates, NOPCHAP1 and NUFIP1 were not found in Perkinsea.
Immunofluorescence staining (IF), RNA fluorescence in situ hybridization (FISH) and image acquisition
IF experiments were carried out on cells grown on coverslips. Cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100 in PBS. Coverslips were incubated with primary antibodies in 3% BSA for 1 h at room temperature. Coverslips were washed three times with PBS and incubated with secondary conjugated antibodies for 45 min. Coverslips were wash 3 times in PBS and fixed again before doing RNA FISH experiments. SmiFISH method was done as previously described (44) using Cy3-labeled DNA fragments specific to U3 and U85 snoRNAs. After incubation with the hybridization mix for one night, coverslips were all washed twice in 10% formamide in 2XSSC, once in PBS and mounted in Vectashield solution containing DAPI (Vector Laboratories). Samples were observed at RT using an upright epifluorescence microscope (LEICA DM6000) with a ×63 oil objective (NA 1.3). Images were captured with a CCD camera (Coolsnap HQ2 from Photometrics) using MetaMorph (Molecular Devices) and processed with Photoshop (Adobe). Deconvolution was proceeded with Huygens Professional (Scientific Volume Imaging).
Co-immunoprecipitation and Western Blot
Cells transfected with pcDNA5-GFP-3xFLAG-NOPCHAP1 were lyzed in HNTG buffer (20 mM HEPES, pH 7.9, 150 mM NaCl, 1% Triton X-100, 10% glycerol, 1 mM MgCl2, 1 mM EGTA, and protease inhibitors) and incubated for 20 min at 4°C. Cellular debris were removed by centrifugation (10 min at 9000 ×g). For control IP, untransfected cells were used. Extracts were put on GFP-TRAP beads for 1.5 h at 4°C (ChromoTek) and then were washed 4 times with HNTG before resuspending in 2× Laemmli buffer. Inputs and pellets were loaded on 12% SDS-PAGE and transferred to a nitrocellulose membrane (Protean, Amersham). Membranes were blocked with 5% nonfat milk (weight/volume) in PBST (0.05% Tween-20 in PBS) and incubated with appropriate primary antibody diluted in 1% nonfat milk followed by incubation with secondary antibody conjugated to HRP. Enzymatic activity was detected using the ECL kit (Roche) or SuperSignal West Pico Chemiluminescence Substrate (Thermo Fischer Scientific).
Luciferase assays
HEK 293T cells were grown on 24-well plates and co-transfected with 50 ng of plasmid expressing a 3xFLAG-tagged Firefly luciferase (3xFLAG-FL) in fusion with the protein of interest, and 450 ng of plasmid coding Renilla luciferase alone (RL) with 1 μl of JetPrime (Ozyme). After 48 h, cells were extracted in 100 μl of PLB buffer (Promega) and incubated at 4°C for 15 min. RL and FL activities were measured on 96-well plates using 4 μl of cell extract and the dual-luciferase assay kit (Promega). Values obtained for FL were normalized to RL values. Experiments were done at least in triplicate. When treated with Geldanamycin (GA), drug was added 24 h before extraction to 2 μM final.
LUMIER IPs
HEK 293T cells were grown on 24-well plates and co-transfected with 450 ng of the RL fusion and 50 ng of the 3xFLAG-FL fusion. Each IP was performed in duplicate by transfecting twice the same plasmids. After 48 h, cells were lyzed in 450 μl of HNTG containing protease inhibitor cocktail (Roche), incubated for 15 min at 4°C and spun down at 4°C and at 20 000 × g for 15 min. 100 μl of the extract were dispatched in two wells of a 96-well plate, with one well being coated with anti-FLAG antibody (10 μg/ml in PBS; F1804 Sigma-Aldrich), and one control well without antibodies. Plates were incubated for 3 h at 4°C, and then washed 5 times with 300 μl of ice-cold HNTG, for 10 min at 4°C for each wash. After the last wash, 10 μl of PLB buffer was added in each well. To measure the signal in the input, 2 μl of extract and 8 μl of PLB buffer was put in empty remaining wells. Plates were then incubated 5 min at RT, and FL and RL luciferase activities were measured in IP and input wells, using the dual luciferase kit (Promega). Every transfection was performed at least twice as independent replicates. Co-IP efficiency was defined as the RL/FL ratio in the pellet, divided by the RL/FL ratio in the input. Unless otherwise stated, statistical significance was evaluated using Z-test assaying whether the co-IP efficiency in the anti-FLAG IP was more than 6 times higher than the mean values obtained in the control IP, done without antibodies (32).
Production and purification of recombinant proteins
N-His6 tagged protein and its putative protein partners were co-expressed in E. coli BL21(DE3) strain (NEB) supplemented with pRARE2 plasmids (Novagen) and co-transformed with the corresponding plasmids, in 100 ml Luria-Bertani medium, O/N at 20°C after induction with 0.2 mM IPTG when OD600 reaches 0.7. Then the cells were harvested by centrifugation, 15 min at 4200 ×g at 4°C. The cell pellet was resuspended in 4 ml lysis buffer (25 mM HEPES pH 7.5; 300 mM NaCl; 10 mM Imidazole; 5% glycerol) and sonicated (corresponds to So in Supplementary Figure S3). Then the lysate was centrifuged 30 min at 16 100 ×g at 4°C and the supernatant (corresponds to SN in Figure 7 and Supplementary Figure S3) was incubated with 200 μl of 50% slurry talon beads for 30 min at 4°C for binding step. The pellet is resuspended in 4 ml of lysis buffer. Then, after 5 min centrifugation at 700 ×g at 4°C, the supernatant was discarded (corresponds to the FT in Figure 7 and Supplementary Figure S3) and the resin was washed 3 times with 500 μl of lysis buffer (corresponds to B in Figure 7 and Supplementary Figure S3). The complexes were eluted from the resin using 500 μl elution buffer (25 mM HEPES pH 7.5; 300 mM NaCl; 300 mM Imidazole; 5% Glycerol) (corresponds to E in Figure 7 and Supplementary Figure S3). An additional step was to incubate the elution with in house 3C protease, O/N at 4°C, to remove the N-His6 tag (corresponds to Figure 7H–I). A size exclusion chromatography carried out in gel filtration buffer (10 mM HEPES pH 7.5; 150 mM NaCl; 2% Glycerol; 1 mM DTT) on a Superose® 6 10/300 analytical column permitted to isolate the protein complex of interest.
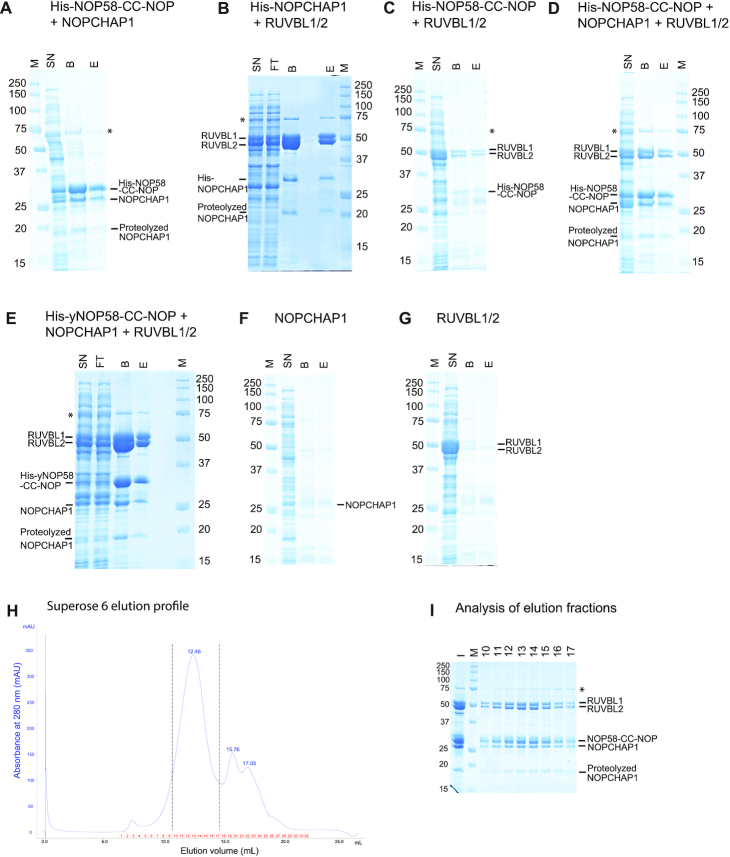
Figure 7.
In vitro characterization of a NOP58:NOPCHAP1:RUVBL1/2 complex. (A) Coomassie-stained SDS-PAGE 10% of His-NOP58-CC-NOP co-expressed with NOPCHAP1 in E. coli and purified on TALON resin. From left to right: M: size of the markers in kDa; SN: soluble fraction; B: bound on resin; E: imidazole eluate. *: non-specific protein (Chaperone protein DnaK) from E. coli extract. (B) Coomassie-stained SDS-PAGE 10% of His-NOPCHAP1 co-expressed with RUVBL1/2 in E. coli and purified on TALON resin. Legend as in A. FT: flow through. All labeled bands are identified by LC-MALDI. (C) Coomassie-stained SDS-PAGE 10% of His-NOP58-CC-NOP co-expressed with RUVBL1/2 in E. coli and purified on TALON resin. Legend as in A. (D) Coomassie-stained SDS-PAGE 10% of His-NOP58-CC-NOP co-expressed with NOPCHAP1 and RUVBL1/2 in E. coli and purified on TALON resin. Legend as in A. All labeled bands are identified by LC-MALDI. (E) Coomassie-stained SDS-PAGE 10% of His-yNOP58-CC-NOP (amino acids 159 to 412 from S. cerevisiae NOP58) co-expressed with NOPCHAP1 and RUVBL1/2 in E. coli and purified on TALON resin. Legend as in A. (F) Coomassie-stained SDS-PAGE 10% of NOPCHAP1 expressed in E. coli and purified on TALON resin. Legend as in A. (G) Coomassie-stained SDS-PAGE 10% of RUVBL1/2 expressed in E. coli and purified on TALON resin. Legend as in A. (H) Gel filtration profile of His-NOP58-CC-NOP+NOPCHAP1+RUVBL1/2 eluted and cleaved by 3C protease. The chromatogram shows the absorbance (y axis) as a function of the elution volume (x axis) of the purified NOP58-CC-NOP:NOPCHAP1:RUVBL1/2 complex on an Superose® 6 10/300 analytical column. The fractions of the majority peak between the two dashed lines were analyzed by SDS-PAGE. (I) Coomassie-stained SDS-PAGE 10% of the various gel filtration fractions from H. The gel shows the major complex eluted from the column. From left to right: I: injection; M: size of the markers in kDa; Fraction numbers from 10 to 17. *: non-specific protein (Chaperone protein DnaK) from E. coli extract.
After purification, the different fractions were loaded on a 10% SDS-polyacrylamide gel for PAGE run (45 min run at 200V in 1× Laemmli buffer), and the complexes were revealed by Coomassie blue staining.
Production of RUVBL1/2 and RUVBL1-ΔDII/RUVBL2-ΔDII
RUVBL1 carries an N-terminal 6 × His followed by thrombin cleavage site, while RUVBL2 has a C-terminal FLAG-FH8 preceded by a Human Rhino 3C cleavage site (HRV-3C). The RUVBL1/RUVBL2 complex was expressed in Escherichia coli (DE3) (Novagen, 71400), with 100 μM IPTG overnight at 18°C in a New Brunswick™ (Innova®) 44R Shaker at 225 rpm. The complex was immobilized in a 5 ml HisTrap™ HP (GE Healthcare), previously equilibrated in Buffer A (20 mM HEPES pH 8.0, 250 mM NaCl, 2 mM MgCl2, 0.5 mM TCEP), and eluted with 300 mM imidazole. Peak fractions collected from the HisTrap were incubated with 5 mM CaCl2 during 1 hr and loaded onto an HiPrep™ Octyl FF 16/10 column (GE Healthcare) equilibrated in Buffer B (20 mM HEPES pH 8.0, 200 mM NaCl, 5 mM CaCl2, 0.5 mM TCEP). Bound proteins were eluted using Buffer C (Buffer B without NaCl and CaCl2 and supplemented with 5 mM EDTA). FLAG-FH8 removal of collected samples was performed by incubating 18 h at 4°C with 1% (w/w) HRV-3C protease (Thermo Fisher Scientific). To separate oligomeric species, a Superose® 6 column was used, equilibrated with Buffer D (20 mM HEPES pH 8.0, 150 mM NaCl, 0.5 mM TCEP), allowing the isolation of the major dodecameric specie. The peak fractions were pooled and concentrated to 17.3 mg/ml using a 30 kDa Cut-off Amicon Ultra centrifugal filter (Millipore). RUVBL1-ΔDII/RUVBL2-ΔDII expression and purification is performed as described above. All purification steps were carried out at RT and were monitored by NuPage Bis–Tris gels (Invitrogen, NP0302).
Cloning, expression and biotinylation of NOPCHAP1 for surface plasmon resonance (SPR) experiments
The coding sequences for BirA-FLAG and Avi-NOPCHAP1-6xHis were synthetized and cloned into pRSF-Duet™-1 by GenScript. BirA protein was cloned on Multiple Cloning Site (MCS) 1 while the NOPCHAP1 protein on MCS 2. The two proteins were co-expressed in E. coli Tuner™ (DE3) (Novagen, 70623) in LB medium, with 500 μM IPTG overnight at 18°C in a New Brunswick™ (Innova®) 44R Shaker at 160 rpm. To promote cellular biotinylation of BirA on NOPCHAP1, 5 μg/ml of D-Biotin (Merck, B4501) was supplied to the culture upon IPTG addition. The cells were resuspended in BugBuster™ Protein Extraction Reagent (Novagen, 70584) supplemented with 0.1 mg/ml Lysozyme, 5 U/ml Benzonase and 1 mM PMSF and placed on ice for 15 min. The clarified extract was injected onto a PD-10 Desalting column equilibrated in Buffer D supplemented with 1 mM PMSF. Protein expression and biotinylation was assessed by Western blotting using Streptavidin, Alkaline Phosphate Conjugate (Invitrogen, S921).
SPR experiments
The interaction between the three versions of RUVBLs and the cellular biotinylated NOPCHAP1 was assessed by SPR at 25°C using a Biacore T200 instrument (Cytiva). The surface of a CM5 sensor chip (Series S) was activated with 400 mM EDC and 100 mM NHS for 10 min and coated with NeutrAvidin™ Biotin-Binding Protein (Thermo Scientific, 31000) at 50 μg/ml in 10 mM sodium citrate pH 4.5. The cellular biotinylated NOPCHAP1 at 15 μg/ml in HBS-P+ (10 mM HEPES pH 7.4, 150 mM NaCl and 0.05% Tween® 20) was coupled to the NeutrAvidin™ Biotin-Binding Protein coated surface for 100 s at a flow rate of 10 μl/min to reach 90 RU. The background buffer used during immobilization was HBS-P+.
RUVBL1/2 and RUVBL1-ΔDII/RUVBL2-ΔDII (analytes) were directly dissolved in running buffer (20 mM NaKPi pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT and 0.05% Tween® 20) and injected at 10 different concentrations using a 2-fold dilution series, with the highest concentrations tested being 1 and 10 nM, respectively. Interaction analysis cycles consisted of a 220 s sample injection (association phase) followed by 600 s of buffer flow (dissociation phase) at a flow rate of 30 μl/min. To assess the impact of ATP-γ-S (Jena Bioscience, NU-406) in the interaction between the RUVBL1/2 and NOPCHAP1, RUVBL1/2 was directly dissolved in running buffer supplemented with saturating concentrations of ATP-γ-S (100 μM). Following the ATP-γ-S assay, NOPCHAP1 surface activity was validated in the absence of nucleotide (data not shown).
All sensorgrams were processed by first subtracting the binding response recorded from the nearest buffer blank injection and all datasets were fit with a heterogeneous ligand model to determine the interaction affinity and kinetic rate constants, using the provided Biacore T200 evaluation software.
Nuclear magnetic resonance
For NMR analysis, we produced a 15N-labeled sample of NOPCHAP1 by growing E. coli BL21(DE3) cells transformed with a pnEA plasmid encoding the full-length protein fused to a N-terminal 6xHIS tag. Bacteria were grown in a minimal M9 media containing 15NH4Cl and d6-Glucose as sole source of nutriments. Overexpression of the protein was induced by adding 0.2 mM IPTG to a culture with an OD600 value of 0.6. After one night at 20°C under agitation, bacteria were harvested by centrifugation, then sonicated in lysis buffer (25 mM HEPES, pH7.5, 300 mM NaCl, 0.5 mM TCEP, 10 mM imidazole). The recombinant NOPCHAP1 was isolated from the supernatant after binding to TALON beads, elution (by lysis buffer with imidazole adjusted to 300 mM), and 3C-cleavage step. A final gel filtration performed in 10 mM NaPi, pH 6.4, 150 mM NaCl, 0.5 mM TCEP provided a NMR sample concentrated at 55 μM. A 1H-15N HSQC spectrum was recorded at 300 K on a 600 MHz spectrometer equipped with a TXI probe. Data were processed with TOPSPIN-3 (Bruker).
Mass spectrometry
Gel bands were processed by successive washes at room temperature under agitation in a 50 μl volume at all steps: for cysteine reduction/alkylation, bands were incubated once in AB (ammonium bicarbonate, 100 mM), once in AB containing 50 mM DTT for 45 min, once in AB, once in AB containing 50 mM IAA (IodoAcetAmide) for 45 min. then, they were washed through two cycles as follows: 5 min in AB/ACN (Acetonitrile), 1:1, 15 min in AB. Finally, they were dehydrated twice in ACN and dried in a speed-vac for one hour. Bands were digested with 50 ng trypsin (sequencing grade, Promega) overnight in 10 μl Tris, pH 8.0, 50 mM, 1 mM CaCl2. The next day, peptides were extracted twice in 10 μl ACN, 80%, TFA (triFluoroacetic acid) 1% for 7 min under sonication. Extracts were pooled and dried in a speed vac, resuspended in 10 μl 2% ACN, 0.1% TFA and processed for fractionation by nano-HPLC on an Ultimate3000 system equipped with a 20 μl sample loop, a pepMap 100 C18 desalting precolumn and a 15 cm pepMap RSLC C18 fractionation column (all from Dionex). Samples (5 μl) were injected using the μLPickUp mode and eluted by a 2–45% ACN gradient over 30 min at 300 nl/min. Fractions (170, 9 s each) were collected on a ProteineerFcII (Bruker) over 25.5 min and elutions were directly mixed on MTP-1536 TF target (Bruker) spots to α-cyano-4-hydroxycinnamic acid (Bruker). LC-MALDI runs were processed using dedicated automatic methods piloted by WARP-LC software on an Autoflex speed MALDI-TOF/TOF mass spectrometer (Bruker) in the 700–4500 mass range, using next-neighbour external calibration for all MALDI spots, using 2000 random laser shots per spot at a 2000 Hz frequency. Masses detected with S/N above 50 were selected for TOF/TOF fragmentation in LIFT mode. Peptide assignments were performed from TOF/TOF spectra by Mascot interrogation (Matrix Science) of the full Swissprot database piloted and compiled by Proteinscape with a mass tolerance of 50 ppm in TOF mode and 0.8 Da in TOF/TOF mode, with optional cysteine carbamidomethylation, methionine oxidation and trypsin cut with one optional miss-cleavage. Only peptides with Mascot scores above 20 were taken into account and proteins were considered as identified with certainty if they obtained a score >80.
SILAC labeling and proteomic analysis
For SILAC experiments, HeLa Flp-In cells were grown for 15 days in each isotopically labeled media (CIL/Eurisotop), to ensure complete incorporation of isotopically labeled arginine and lysine (light label (R0K0, L) or semi-heavy label l-lysine-2HCl (2H4, 96–98%)/l-arginine–HCl (13C6, 99%) (R6K4, M) or l-lysine–2HCl (13C6, 99%; 15N2, 99%)/l-arginine–HCl (13C6, 99%; 15N4, 99%) heavy label (R10K8, H) (percentages represent the isotopic purity of the labeled amino acids). Eight 15-cm diameter plates were used per SILAC condition. Cells were rinsed with PBS, trypsinized and cryogrinded (45) and powder was resuspended in HNT lysis buffer (20 mM HEPES, pH 7.4, 150 mM NaCl, 0.5% triton X-100, protease inhibitor cocktail (cOmplete, Roche)). Extracts were incubated 20 min at 4°C and clarified by centrifugation for 10 min at 20 000 ×g. For RNase treated extract, RNase A was added in the lysis buffer at 10 μg/ml final concentration before incubation. For all IP experiments, extracts were pre-cleared by incubation with Protein G Sepharose beads (GE healthcare) for 1 h at 4°C. The control was extracted from the SILAC light condition prepared from parental HeLa cells that did not express the GFP fusion. Each extract was then incubated with 50 μl of GFP-Trap beads (Chromotek) for 1.5 h at 4°C, washed 5 times with HNT buffer, and beads from the different isotopic conditions were finally pooled. Bound proteins were eluted by adding 1% SDS to the beads and boiling for 10 min. Proteomic analysis was performed as previously described (32).
RNA-Seq analysis
Triplicates of RNA samples were prepared from 3 plates of HEK control cells and three plates of HEK KO NOPCHAP1 cells by adding 1 ml of Trizol reagent (Thermo Fisher) and 200 μl of Chloroform. Aqueous phase was precipitated with Isopropanol in presence of glycogen. RNAs were treated with RQ1 DNAse (Promega) and depleted for ribosomal RNA (Thermofisher). Libraries were prepared with TruSeq Stranded total RNA Sample Preparation kit (Illumina).
Image analyses and base calling were performed using the Illumina NovaSeq Control Software and Real-Time Analysis component. Demultiplexing was performed using Illumina's conversion software (bcl2fastq 2.20). The quality of the raw data was assessed using FastQC (v0.11.8) from the Babraham Institute and the Illumina software SAV (Sequencing Analysis Viewer). Potential contaminants were investigated with the FastQ Screen software from the Babraham Institute (0.14.0). All samples were aligned to human genome (UCSC hg38) using Bowtie2. Final read alignments having more than six mismatches were discarded. Samtools (1.9) was used to sort the alignment files. Then, the counting was performed with Featurecounts. The data is from a strand-specific assay, the read has to be mapped to the opposite strand of the gene. Before statistical analysis, genes with <15 reads (cumulating all the analysed samples) were filtered and thus removed. Differentially expressed genes were identified using the R (v3.6.2) Bioconductor package DESeq2 1.26.0. Data were normalized using the DESeq2 normalization method. Genes with adjusted P-value <5% (according to the FDR method from Benjamini-Hochberg) were declared differentially expressed.
RESULTS
Identification of early C/D snoRNP assembly intermediates using NOP58 mutants
To characterize early steps in C/D snoRNP biogenesis, we decided to generate mutant forms of NOP58 that could not be incorporated in mature snoRNPs. PRPF31 is a NOP58 paralog that belongs to the U4 snRNP, and we previously used mutant versions of PRPF31 to characterize U4 assembly pathway (46). We had used two mutants of PRPF31, a double mutant in the NOP domain that prevents association to SNU13:RNA complexes, and a single one at the basis of the CC domain that prevents association with other U4 components. As NOP and CC domains are well conserved between PRPF31 and NOP58 (30% identity and 50% similarity over these domains, Supplementary Figure S1C), we generated similar mutations in NOP58 (K310A/A313R, thereafter referred to as NOP58-KA/AR; and NOP58-A283P, Supplementary Figure S1B). GFP fusions of the mutants and wild-type NOP58 were transiently expressed in HeLa cells. While GFP-NOP58-WT localized to the nucleolus with a minor part present in the nucleoplasm, GFP-NOP58-A283P and GFP-NOP58-KA/AR were restricted to nucleoplasm and did not accumulate in nucleolus (Figure 1A). This confirmed that these NOP58 mutants are not properly assembled into mature C/D snoRNPs.
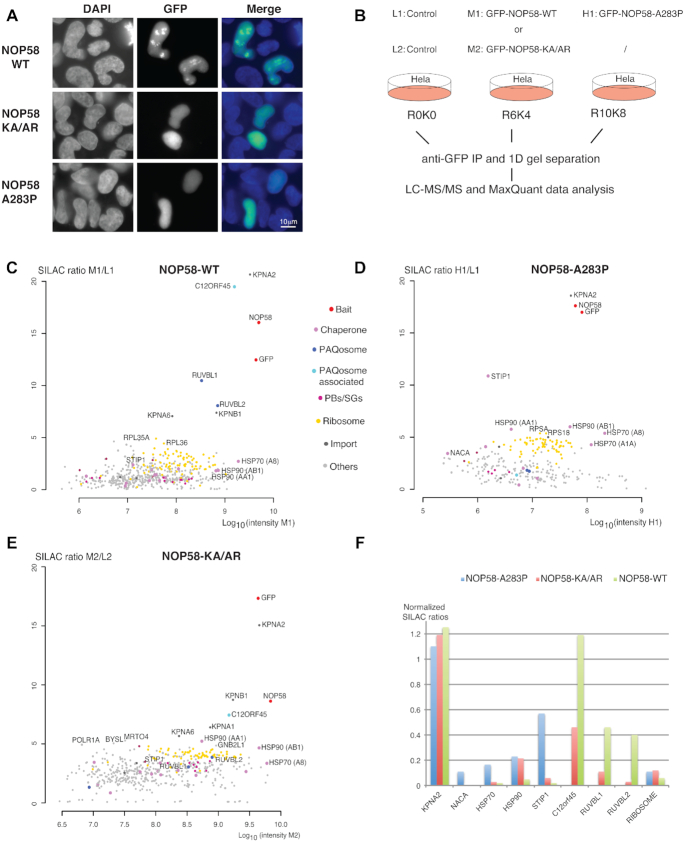
Figure 1.
Identification of early assembly intermediates with NOP58 mutant proteins. (A) Epifluorescence microscopy images of HeLa cells expressing wild type (WT) or mutant GFP fusions of NOP58 (NOP58-KA/AR or NOP58-A283P). Blue/left panels: DAPI staining; Green/middle panels: GFP. Scale bar is 10 μm. (B) Schematic representation of SILAC IP experiments shown in C, D and E. First experiment was done with three conditions (L1: light label for control done with parental HeLa cells; M1: medium label for GFP-NOP58-WT; H1: heavy label for GFP-NOP58-A283P) and second with two conditions (L2: light label for control; M2: medium label for GFP-NOP58-KA/AR). (C–E) Proteomic analyses of the partners of GFP-NOP58-WT (C), GFP-NOP58- A283P (D) and GFP-NOP58-KA/AR (E). Graphs display SILAC ratios (y axis, specific versus control IP) as a function of signal abundance (x axis, log10(intensity)). Each dot represents a protein and is color encoded according to the classification shown between panels C and D. The labeled dots highlight proteins relevant to this study. Full hit list with Significance B values are given in Supplementary Table S1. (F) Bar plot comparing SILAC ratios shown in C, D and E for a few selected partners. In each experiment, the normalization is done by dividing the SILAC ratio of the protein of interest with that of NOP58. Blue bars are for NOP58-A283P, red bars for NOP58-KA/AR and green bars for NOP58-WT.
To characterize early assembly intermediates in C/D snoRNP biogenesis, we performed proteomic experiments using stable isotope labeling by amino acids in cell culture (SILAC) followed by immunoprecipitation (SILAC-IP), using mutant NOP58 proteins as baits. We generated stable HeLa clones expressing GFP-NOP58-WT, GFP-NOP58-A283P and GFP-NOP58-KA/AR. The two mutant fusion proteins were expressed at a level similar to that of wild-type GFP-NOP58 and also endogenous NOP58 (Supplementary Figure S1A). Our SILAC-IPs were performed on total extract prepared after cryogrinding of pelleted cells (45). This method depletes nucleoli and thus enriches for snoRNP assembly intermediates. Each extract was incubated with anti-GFP antibodies and immunoprecipitated proteins were analyzed by quantitative mass spectrometry (MS) and compared with a control purification performed simultaneously with parental HeLa cells not expressing the GFP fusion (Figure 1B). Wild-type and mutants NOP58 fusions were found with high SILAC ratio (Figure 1C–E), and their partners were considered according to their B Significance (47 and Supplementary Table S1). After normalizing of the SILAC ratio to the NOP58 value found in each IP, we directly compared several of the proteins found in the three IPs to know to which proportion they associate to NOP58 mutants as compared to the wild type (Figure 1F). We found a high SILAC ratio for KPNA2 in all the three IPs (normalized value of ≈1.2). KPNA2 is an importin and it could be important for nuclear import of newly synthesized NOP58, and/or to chaperone its long positively charged C-terminal domains that was previously shown to function as a nuclear/nucleolar localization signal (48,49). This suggested that our procedure isolated newly synthesized NOP58. C12ORF45, thereafter referred to as NOPCHAP1 (NOP protein chaperone 1), was the second most enriched factor found with GFP-NOP58-WT (with a normalized SILAC ratio of ≈1.2). It was also associated with GFP-NOP58-KA/AR, however to a lesser extent, and it was not found with GFP-NOP58-A283P. The R2TP factors RUVBL1 and RUVBL2 associated to GFP-NOP58-WT, but showed only insignificant enrichment levels with GFP-NOP58-KA/AR (Figure 1F), and were not detected with GFP-NOP58-A283P. In contrast, the two NOP58 mutants showed stronger interaction with HSP90 than wild-type NOP58, and GFP-NOP58-A283P additionally associated with the HSP90 co-chaperone STIP1 and the ribosome associated chaperone NACA. This indicated that GFP-NOP58-A283P is blocked at an early assembly stage just after its translation. GFP-NOP58-KA/AR appears to progress further in the assembly process because it associates with NOPCHAP1, but it is still blocked early since it does not bind RUVBL1/2 as does wild-type GFP-NOP58.
Identification of early U4 assembly intermediates using PRPF31 mutants
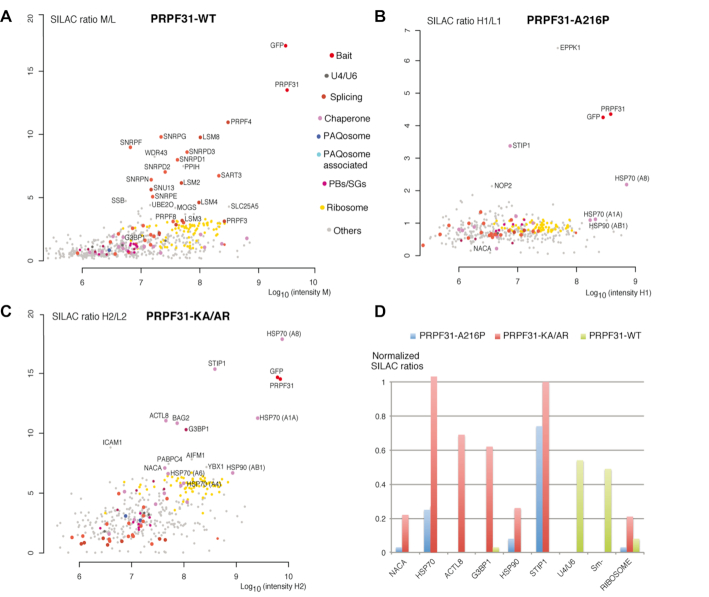
PRPF31 is structurally similar to NOP58 and we previously showed that its assembly in U4 snRNP also involves C/D snoRNP assembly factors such as NUFIP1 and ZNHIT3 (46). We thus set out to compare the proteins associated with NOP58 and PRPF31 mutant proteins. GFP-PRPF31-KA/AR (K243A and A246R) and GFP-PRPF31-A216P were detected in the cytoplasm while GFP-PRPF31-WT was nucleoplasmic as previously reported (46). HeLa cells stably expressing the three PRPF31 fusion proteins were prepared and used for SILAC-IP on total extract from each clone (Figure 2A–C). After normalization to PRPF31 level, we directly compared the enrichment levels of the various proteins (Figure 2D and Supplementary Table S2). Proteins of the U4/U5/U6 tri-snRNP particle associated with the wild-type PRPF31 protein but not with the mutants, confirming that they are deficient for assembly. In contrast, GFP-PRP31-A216P and GFP-PRPF31-KA/AR were found enriched with HSP70 and STIP1, while these proteins were not detected with wild-type GFP-PRPF31, most likely because most of the protein is assembled with the U4 snRNP in the nucleus. GFP-PRPF31-KA/AR also associated with ACTL8, G3BP1 and BAG2, which function as HSP70 co-chaperones (50), and also at a low level with HSP90. As in the case of NOP58, the two PRPF31 mutants were thus blocked in assembly shortly after PRPF31 translation and before their association with other U4 proteins. Interestingly, GFP-PRPF31WT or mutants did not associate with NOPCHAP1, suggesting that either the association was too weak to be detected, or that PRPF31 does not associate with this factor during assembly.
Figure 2.
Identification of early assembly intermediates with PRPF31 mutant proteins. (A–C) Proteomic analyses of the partners of PRPF31-WT (A), GFP-PRPF31-A216P (B) and PRPF31-KA/AR (C). First and second experiments were done with three conditions (L1/L2: light label for control done with parental HeLa cells; M1/M2: medium label for GFP-PRPF31-WT; H1: heavy label for GFP-PRPF31-A216P in the first experiment; H2: heavy label for GFP-PRPF31-KA/AR in the second experiment). Full hit list with Significance B values are given in Supplementary Table S2. Legends as in Figure 1 for the graphic representation. SILAC ratio shown in A is mean of the two experiments. (D) Bar plot comparing SILAC ratios shown in A, B and C for a few selected partners. In each experiment, the normalization is done by dividing the SILAC ratio of the protein of interest with that of PRPF31. Blue bars are for PRPF31-A216P, red bars for PRPF31-KA/AR and green bars for PRPF31-WT.
NOPCHAP1 associates with PAQosome and PAQosome clients
Identification of NOPCHAP1 with RUVBL1/2 in the NOP58 IP raised the possibility of a new intermediate in C/D snoRNP biogenesis, involving other factors than those already known such as NUFIP1/ZNHIT3 and ZNHIT6. The NOPCHAP1 protein has been described as a small protein (188 amino acids) conserved in S. pombe and X. laevis but lacking in the yeast S. cerevisiae. Interestingly, exploration of the phylogenic conservation of NOPCHAP1 indicates that it is a highly conserved protein, present in most phyla including primitive eukaryotes such as N. gruberi (Figure 3F). We first characterized the localization of a GFP-NOPCHAP1 fusion protein stably expressed in HeLa cells (Figure 3A). The fusion protein was diffusely localized in the nucleoplasm with a faint signal in the cytoplasm. To better characterize its function, we first performed a proteomic identification of its partners. In this experiment, we used a triple SILAC encoding scheme to compare RNase-treated and untreated extracts and to determine the contribution of RNA in the observed complexes (Figure 3B). Plotting the two conditions showed that most partners of GFP-NOPCHAP1 were equally enriched with or without RNase supporting the idea that the complexes observed are not RNA dependent (Figure 3D and Supplementary Table S3). The most enriched partners of GFP-NOPCHAP1 were RUVBL1 and RUVBL2 (Figure 3C and Supplementary Figure S2A), and this interaction was confirmed by immuno-precipitating GFP-NOPCHAP1 and performing western blot against endogenous RUVBL1 (Supplementary Figure S2B). RPAP3 and PIH1D1 were also found together with prefoldins and prefoldin-like proteins, confirming the association of the entire PAQosome with GFP-NOPCHAP1. Interestingly, a number of well-known R2TP clients were also found in the IPs. The core C/D snoRNP proteins NOP58 and Fibrillarin were identified with high SILAC ratios, as well as PRPF31. Of these proteins, NOP58 seemed by far the most abundant one. Moreover, other R2TP clients were detected, such as the U5 snRNP proteins PRPF8, EFTUD2, SNRNP200 and the RNA polymerase subunits POLR2A, POLR2E, POLR2H. Finally, some known PAQosome co-factors such as ZNHIT2, ZNHIT6 and TTI1 also bound NOPCHAP1 (Figure 3E). Thus, NOPCHAP1 associates with PAQosome, PAQosome-related factors and a series of clients, raising the question of which interaction is direct and which is indirect.
NOPCHAP1 binds to RUVBL1/2 domain II in an ATP-dependent manner
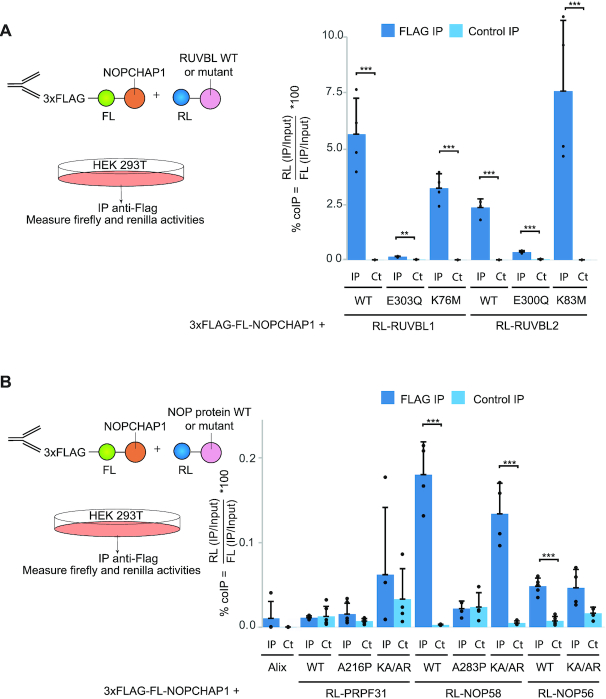
To further analyze protein-protein interactions, we used LUMIER IP, which is a quantitative immunoprecipitation assay using pairs of over-expressed proteins (Figure 4A and 51). RUVBL1/2 and NOPCHAP1 were fused to Renilla luciferase (RL) and FLAG-tagged Firefly luciferase (3xFLAG-FL), respectively. We then measured RL and FL activities in the input and pellet of an anti-FLAG IP, or an IP without antibody as control. Co-IP efficiency was defined as the IP/input ratio of RL relative to that of FL. WT forms of RL-RUVBL1 and RL-RUVBL2 were efficiently co-precipitated with 3xFLAG-FL-NOPCHAP1 (6.9% and 2.5% of co-IP efficiency respectively). RUVBL1 and RUVBL2 are known to form hetero-hexamer or dodecamer and they occur in different forms that can be loaded or not with ATP. To test whether the interaction of RUVBL1/2 with NOPCHAP1 could change depending on their ATP association state, we measured the binding of NOPCHAP1 to RUVBL1/2 mutants known to preferentially associate with distinct nucleotide forms (Figure 4A). We first tested E to Q substitutions in their Walker B domain, known to prevent nucleotide hydrolysis (52). For these mutants (RL-RUVBL1-E303Q and RL-RUVBL2-E300Q), we found a strongly decreased co-IP efficiency (0.16% and 0.35% respectively). Second, we tested K to M mutation in their Walker A domain (RL-RUVBL1-K76M and RL-RUVBL2-K83M), known to prevent nucleotide binding (53). These mutants showed an interaction with NOPCHAP1 similar to wild type RUVBL1/2 (3.7% and 7.5% respectively). Thus, NOPCHAP1 interacts with RUVBL1 and RUVBL2 that are empty or that can bind ADP, and much less with mutants that cannot hydrolyze ATP.
Figure 4.
NOPCHAP1 interacts with WT and mutant forms of RUVBL1/2 and NOP58. (A) Left panel: schematic representation of the LUMIER-IP assay between NOPCHAP1 and RUVBL proteins. Right panel: graph plotting the % of co-IP efficiency for the interaction between co-expressed 3xFLAG-FL-NOPCHAP1 and RL-RUVBL1 or RL-RUVBL2, either WT or mutants as indicated below the graph. % of co-IP was calculated from the co-precipitated RL fusion protein (IP/Input ratios of RL activity), normalized with IP/Input ratio of FL activity obtained in the anti-FLAG IP. The values are means of four experiments, each represented by a dot. Error bars: standard deviation. Stars: values significantly greater than six-times the mean value obtained in the control IPs without anti-FLAG antibody (Control IP: light blue bars). *** P-value ≤ 0.001, ** P-value ≤ 0.01 and * P-value ≤ 0.05. (B) Left panel: schematic representation of the LUMIER-IP assay between NOPCHAP1 and NOP proteins. Right panel: graph plotting the % of co-IP efficiency for the interaction between 3xFLAG-FL-NOPCHAP1 and RL-PRPF31, RL-NOP58 or RL-NOP56 either WT or mutants as indicated below the graph. Alix is an alias name for human protein PDCD6IP and is used as a negative control. Legend as in (A). *** P-value ≤ 0.01.
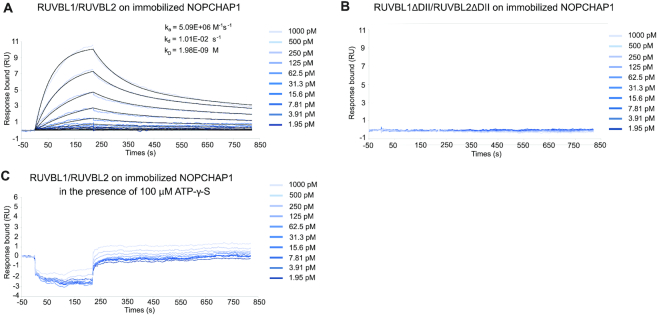
To confirm and expand these results, we turned to in vitro approaches and used SPR to characterize the interaction between NOPCHAP1 and RUVBL1/2. We observed a very strong interaction between NOPCHAP1 and RUVBL1/2 WT with a calculated KD of 1.98 nM (Figure 5A), confirming the results previously obtained by LUMIER IP. RUVBL domain II is a unique regulatory domain within the AAA+ ATPases family. In order to map the NOPCHAP1-RUVBL1/2 interaction interface, we used a domain II-truncated form of RUVBL1/2 and no interaction was observed in SPR (Figure 5B; compare RUVBL1/2 WT and RUVBL1-ΔDII/RUVBL2-ΔDII), suggesting domain II to be pivotal for complex formation. To evaluate the nucleotide binding effect on NOPCHAP1-RUVBL1/2 interaction, the same experiment was performed in presence of 100 μM ATP-γ-S, a non-hydrolysable ATP-form. Interestingly, NOPCHAP1 and RUVBL interaction was completely abolished in the presence of this nucleotide (Figure 5C), suggesting that NOPCHAP1 binds RUVBL domain II only in an ATP-depleted state.
Figure 5.
SPR binding assays between NOPCHAP1 and RUVBL1/2. The graphs depict the immobilized NOPCHAP1 response upon injection of RUVBL1/2 in the presence and absence of 100 μM ATP-γ-S (A and C, respectively) and RUVBL1-ΔDII/RUVBL2-ΔDII (B) (t = 0 s) at different concentrations or upon washing (t = 220 s). X-axis: time (s); Y-axis: response (arbitrary units). All assays were performed in triplicates.
NOPCHAP1 binds to NOP58
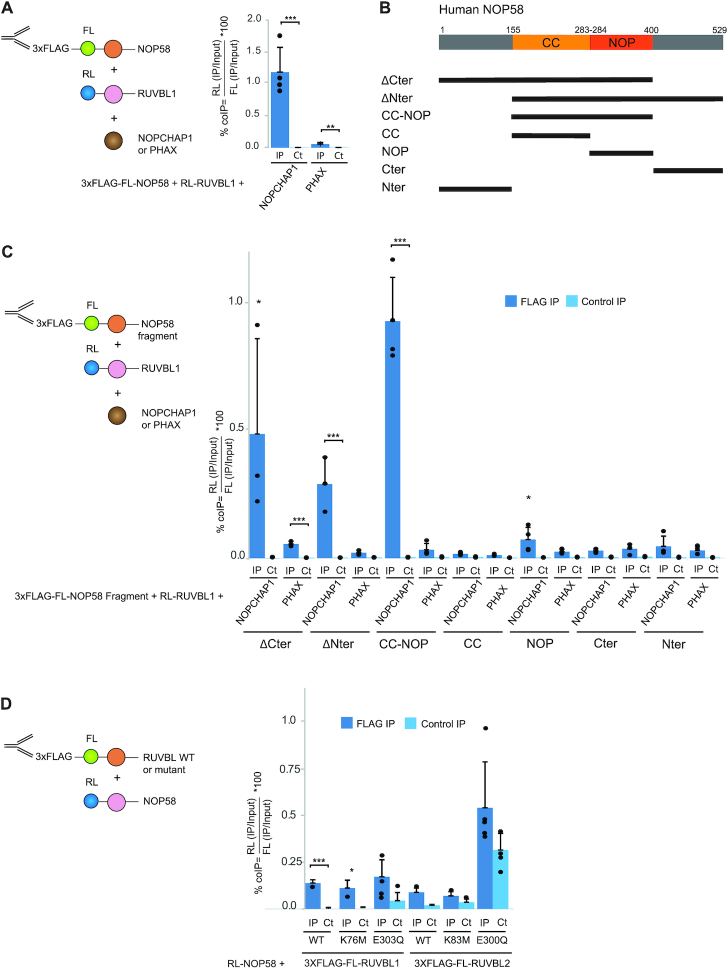
Next, we used LUMIER-IPs to test the ability of NOPCHAP1 to bind the different NOP proteins and their mutants (Figure 4B). Binding of 3xFLAG-FL-NOPCHAP1 with WT and mutants RL-PRPF31 was tested but no interaction was detected. RL-NOP58 showed an interaction 64-fold higher than in the control IP and demonstrating a specific interaction. This interaction was however much weaker than the binding of RUVBL1/2 with NOPCHAP1 (0.18% versus 3–8% co-IP efficiency). Interestingly, NOP58-KA/AR interacted to a similar level than WT NOP58, but NOP58-A283P did not bind NOPCHAP1, in agreement with the results obtained in the proteomic experiments. Thus, in contrast to NOP58-A283P, NOP58-KA/AR keeps the ability to associate with NOPCHAP1. Due to the high similarity of NOP56 with NOP58, we also wondered whether NOP56 could bind NOPCHAP1. We tested both RL-NOP56-WT and RL-NOP56-KA/AR (K326A and A329R; similar to the NOP58 KA/AR mutant). NOP56-KA/AR did not interact with NOPCHAP1, while NOP56-WT displayed a significant but very weak binding (<0.05%; Figure 4B). We concluded that NOPCHAP1 specifically binds NOP58 as compared to the structurally related proteins PRPF31 and NOP56. The association of NOPCHAP1 with PRPF31 seen in the NOPCHAP1 proteomic experiments (Figure 3) is thus likely indirect and mediated by other proteins, for instance the RUVBLs, as seen for the other PAQosome clients detected in this IP.
NOPCHAP1 bridges NOP58 to RUVBL1/2 in vivo
NOPCHAP1 interacts with both NOP58 and RUVBL1/2, we thus hypothesized that it could make a bridge between these proteins. To test this hypothesis, we performed LUMIER IPs to measure the interaction between 3xFLAG-FL-NOP58 and RL-RUVBL1, while simultaneously overexpressing either NOPCHAP1 or the unrelated protein PHAX as control. Co-expression of NOPCHAP1 increased the NOP58-RUVBL1 interaction by 20-fold as compared to PHAX (Figure 6A), and moreover NOP58 bound only very weakly to wild-type and mutant forms of RUVBL1 and RUVBL2 (<0.1% co-IP efficiency; Figure 6D). These data suggest that NOPCHAP1 may act as a tether to bridge NOP58 to the RUVBL1/2 complex.
Figure 6.
NOPCHAP1 bridges NOP58 CC-NOP domain to RUVBL1. (A) Left panel: schematic representation of the LUMIER-IP assay. Right panel: graph plotting the % of co-IP efficiency for the interaction between 3xFLAG-FL-NOP58 and RL-RUVBL1 using NOPCHAP1 as a third co-expressed fusion or PHAX (unrelated protein for a negative control). The values are means of four experiments that are all represented by a dot. Error bars: standard deviation. Stars: values significantly greater than six-times the mean value obtained in the control IPs without anti-FLAG antibody (Control IP: light blue bars). *** P-value ≤ 0.001, ** P-value ≤ 0.01 and * P-value ≤ 0.05. (B) Schematic representation of human NOP58 protein sequence. CC and NOP domains are represented. Below are the truncation mutants used for the LUMIER IPs shown in C. (C) Left panel: schematic representation of the LUMIER-IP assay. Right panel: graph plotting the % of co-IP efficiency for the interaction between 3xFLAG-FL-NOP58 fragments and RL-RUVBL1 using NOPCHAP1 as a third co-expressed fusion or PHAX as a negative control. The 3xFLAG-FL-NOP58 fragments used in the assay are indicated below the graph. Legend as in A. (D) Left panel: schematic representation of the LUMIER-IP assay. Right panel: graph plotting the % of co-IP efficiency for the interaction between RL-NOP58 and 3xFLAG-FL-RUVBL1 or RUVBL2 (WT or mutants, as indicated below the graph). Legend as in A.
In order to determine the minimal domain of NOP58 that is involved in the formation of this complex, we generated various truncation mutants of 3xFLAG-FL-NOP58 (Figure 6B). These mutants were then used for LUMIER IPs with RL-RUVBL1, in presence of NOPCHAP1 or PHAX as control (Figure 6C). The NOP58-CC-NOP fragment, which contains the conserved coiled-coil and NOP domains (amino acids 154–400 in NOP58), gave similar co-IP efficiency as full-length NOP58, and binding was also enhanced by NOPCHAP1. We also observed a NOPCHAP1-dependent interaction for NOP58-ΔCter (amino acids 1–400) and NOP58 ΔNter (amino acids 154–529), but not for the others NOP58 fragments. These data show that together, the CC and NOP domains of NOP58 are necessary and sufficient to form a complex with NOPCHAP1 and RUVBL1/2.
In vitro characterization of NOPCHAP1
To explore the structural properties of NOPCHAP1, we employed solution-state NMR spectroscopy recorded on a 15N-labeled sample of the protein. Remarkably, the 1H–15N HSQC spectrum of NOPCHAP1 was characteristic of an intrinsically disordered protein (IDP; Supplementary Figure S3A). Indeed, 1H resonances of backbone amide groups are tightly centered around 8.1 ppm, whereas 15N resonances spread between 107 and 131 ppm, and resonances in the region of side-chain -NH2 groups are strongly overlapped. More importantly, we could unambiguously count 150 peaks in the backbone amide region over the 173 expected peaks for this protein, these peaks being rather thin for a 20 kDa entity. These features differentiate the 1H–15N HSQC spectrum of NOPCHAP1 from a spectrum recorded on an aggregated protein. Considering that resonance overlap very frequently occurs in IDPs (as no stable 3D structure in the protein can promote variations of the mean chemical shift values), we could assume that almost all residues of NOPCHAP1 give a signal on the spectrum. As example, we can focus on the Glycine region in the spectrum (Supplementary Figure S3A). We could count there 10 narrow peaks for 11 glycines in the sequence, with one of those peaks being more intense than the others (and thus probably gathering two residues). From our NMR analysis, we concluded that NOPCHAP1 is an IDP, almost fully disordered in its free state.
NOP58, NOPCHAP1 and RUVBL1/2 make a quaternary complex in vitro
In order to confirm that NOPCHAP1 is sufficient to make a bridge between NOP58 and RUVBL1/2, we reconstituted the complex in vitro (Figure 7). We generated a His-tagged NOP58-CC-NOP soluble fragment (amino acids 156–400) and co-expressed it with untagged NOPCHAP1 in E. coli. NOPCHAP1 was retained on Cobalt beads and co-eluted with His-NOP58-CC-NOP, but it did not bind the resin when expressed alone, therefore showing a direct interaction between these proteins (Figure 7A). Next, we co-expressed His-tagged NOPCHAP1 with untagged RUVBL1/RUVBL2. We found that these proteins also co-purified on Cobalt beads and co-eluted with imidazole treatment (Figure 7B), while RUVBL1/2 did not exhibit non-specific binding to the resin (Figure 7F and G respectively). The co-expression of His-NOP58-CC-NOP with RUVBL1/2 showed a weak binding of RUVBL1/2 (Figure 7C). However, when His-NOP58-CC-NOP was expressed with both NOPCHAP1 and RUVBL1/RUVBL2, we found that all these proteins co-purified and co-eluted together, suggesting that they formed a quaternary complex (His-NOP58-CC-NOP/NOPCHAP1/RUVBL1/2; Figure 7D). To validate this hypothesis, we performed a gel filtration on analytical Superose 6 in buffer without nucleotide (Figure 7H and I). The chromatogram showed a major peak whose analysis on SDS-PAGE and mass spectrometry demonstrated the formation of the RUVBL1/2:NOPCHAP1:NOP58-CC-NOP complex.
NOPCHAP1 is not present in budding yeast. However, given the high degree of conservation of NOP58 between human and yeast (46.8% of identity and 68% of similarity), we repeated the same co-expression experiments using S. cerevisiae NOP58 instead of the human protein. Remarkably, we found very similar results as with the human protein: human NOPCHAP1 co-purified with yeast His-yNOP581–447 (amino acids 1–447) and His-yNop58-CC-NOP (amino acids 159–412; Supplementary Figure S3B and C), and a quaternary complex with human RUVBL1/2 could also form (Figure 7E). Again, when yeast His-yNop58-CC-NOP was co-expressed with RUVBL1/2 in absence of NOPCHAP1, only limited binding of RUVBL1/2 could be observed (Supplementary Figure S3D), indicating that NOPCHAP1 also promotes the interaction of yeast NOP58-CC-NOP with RUVBL1/2. Taken together, these data demonstrate that a quaternary complex can be formed between NOP58, NOPCHAP1 and RUVBL1/2, and that NOPCHAP1 functions as a bridge between NOP58 and the RUVBLs.
NOPCHAP1 promotes NOP58 expression
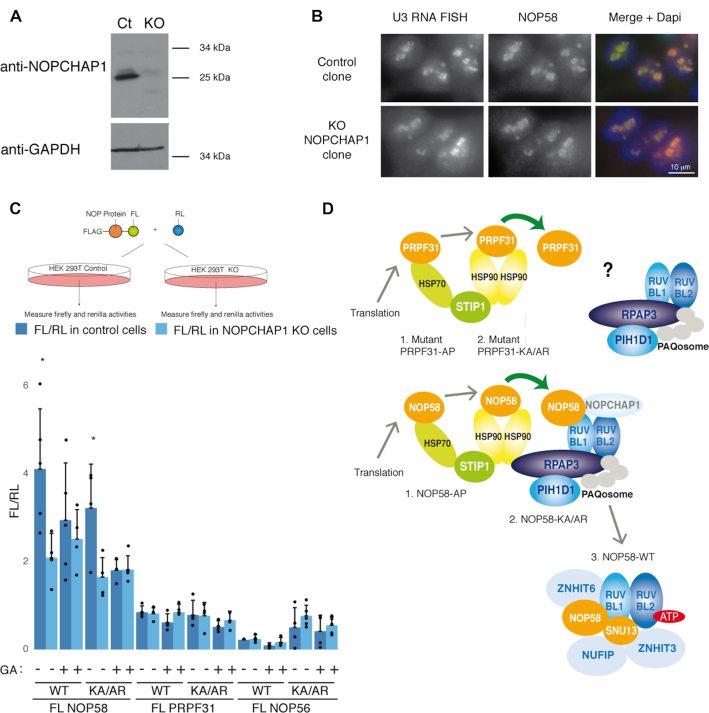
To further study the function of NOPCHAP1, we knocked out the gene in HEK 293T human cells using the CRISPR/Cas9 gene editing method. Individual clones were genotyped and the absence of NOPCHAP1 was confirmed by immunoblotting (Figure 8A). In control cells, we observed a predicted 25 kDa protein that was absent in KO cells. Cell cycle analysis with propidium iodide labeling and cytometry measurement showed no significant difference in the G1, S and G2 phase distribution between KO and control cells (Supplementary Figure S4A). In absence of cell growth change, we wondered if the absence of NOPCHAP1 changed some morphological aspects of the cells and especially nucleoli as they would be impacted by changes in C/D snoRNP biogenesis. We labeled U3 C/D snoRNA by RNA FISH and immunolabelled NOP58 and Fibrillarin. Nucleoli had similar aspects in control and NOPCHAP1 KO clones with a punctuated distribution of U3, NOP58 and Fibrillarin as expected for constituents of the dense fibrillar component of nucleoli (Figure 8B and Supplementary Figure S4B). We also looked at Cajal body (CB), because snoRNAs also localize to CB during their biogenesis and some scaRNAs, which localize in CB, contain a C/D fold and assemble with C/D core proteins. We tested the localization of scaRNA U85 that possess both box C/D and H/ACA motifs, and FISH combined with Coilin immuno-labelling confirmed that U85 scaRNA was still localized in CBs of KO cells. CBs were also similar in size and number in presence or absence of NOPCHAP1 (Supplementary Figure S4C). We also labeled two other nucleolar proteins, dyskerin, which is a constituent of H/ACA snoRNAs and scaRNAs, and B23 that is involved in late steps of rRNA processing. Both were localized in the nucleolus with similar distribution in control and NOPCHAP1 KO clones (Supplementary Figure S5A and B). Finally, RNAseq analysis in NOPCHAP1 KO cells did not show any difference for C/D and H/ACA snoRNA levels as compared to WT cells (Supplementary Figure S5C). Messenger mRNA coding for C/D and H/ACA core proteins were also not affected, indicating no gross alteration of snoRNP and nucleolar metabolism.
Figure 8.
NOPCHAP1 enhances expression of NOP58. (A) Characterization of a NOPCHAP1 KO clone generated in human HEK 293T cells. Western Blot with anti-NOPCHAP1 antibody using Laemmli extracts from NOPCHAP1 KO clone (KO) or control clone (Ct). Anti-GAPDH antibody was used as loading control. (B) The nucleolar structure is not affected in the NOPCHAP1 KO clone. Epifluorescence microscopy images of HEK 293T control and NOPCHAP1 KO clones. Red/left panels: RNA FISH with U3 specific probes. Green/ middle panels: IF with an anti-NOP58 antibody. Right panels: merge with DAPI staining. Scale bar is 10 μm. (C) Top panel: schematic representation of the assay. Bottom panel: graph showing the ratio FL/RL representing the expression of indicated FL-fusion protein in control cells (dark blue) and NOPCHAP1 KO cells (light blue), in normal conditions or after Geldanamycin treatment (GA) for 12 hr. The WT or mutant forms of protein used in the assay is indicated below the graph. A co-transfected plasmid expressing RL alone was used to normalize the value obtained for FL-fusion proteins. The values are means of four experiments that are each represented by a dot. Error bars: standard deviation. * P-value ≤ 0.05 according to a Student test. (D) Model for the assembly of PRPF31 and NOP58 and specific role of NOPCHAP1 in box C/D snoRNP biogenesis. Newly-synthesized NOP58 associates with HSP70 and HSP90, and is transferred to R2TP by NOPCHAP1. A SNU13/ZNHIT3/NUFIP1 module binds R2TP via PIH1D1, and this leads to the assembly of NOP58 with SNU13, maintained by ATP-loaded RUVBL1/2, ZNHIT6 and NUFIP1/ZNHIT3. In the case of PRPF31, it is unclear how this protein associates with R2TP. The steps where NOP58 and PRPF31 mutants accumulate are indicated.
Next, we tested whether NOP58 biogenesis could be affected. As steady state NOP58 did not seem impacted in NOPCHAP1 KO cells based on its subcellular localization and signal intensity (Figure 8B), we analyzed newly synthesized NOP58. To this end, we transiently transfected a fusion of NOP58 with Firefly luciferase (FL-NOP58) and measured luciferase activity in KO and control cells (Figure 8C). Both WT and NOP58-KA/AR mutants were tested as they both associated with NOPCHAP1. Following normalization with a RL expression vector to normalize transfection efficiencies, we found that FL-NOP58 levels were two-fold decreased in NOPCHAP1 KO cells, for both the WT and KA/AR mutant. We next tested the effect of Geldanamycin (GA), a competitive inhibitor of HSP90 ATPase activity that often triggers destabilization of HSP90 clients. Using the same assay, we found that GA treatment decreased the level of FL-NOP58 as previously described (23). Interestingly, GA had no effect on the levels of FL-NOP58 in KO cells, and the levels of FL-NOP58 in KO cells were similar to that of WT cells treated with GA (Figure 8C). NOPCHAP1 and HSP90 are thus both required for NOP58 expression, suggesting that they act on the same pathway. Finally, we did the same experiments using PRPF31 and NOP56 in both their WT and mutant KA/AR forms (Figure 8C). Knocking out NOPCHAP1 and/or treating cells with GA had no significant effect on their expression levels, suggesting that NOPCHAP1 acted only on NOP58 in agreement with our interaction data, and that NOP56 and PRPF31 were not HSP90 clients, in agreement with previous results (23). Taken together, these data suggest that the role of NOPCHAP1 is restricted towards NOP58 and could be to chaperone the newly synthesized protein.
DISCUSSION
NOPCHAP1 binds and stabilizes newly synthesized NOP58
The PAQosome is composed of the HSP90/R2TP chaperone and a prefoldin-like module. The role of HSP90/R2TP in the assembly of C/D snoRNPs has been extensively demonstrated (22–24,34). While the role of prefoldins in C/D snoRNP assembly is currently unknown, the fact they co-purify with C/D assembly factors and core C/D proteins suggests that the entire PAQosome is mobilized in this process. HSP90 stabilizes newly-synthesized NOP58 and SNU13, and these two proteins are assembled into a protein-only complex containing the RUVBL1/2 AAA+ ATPases, the assembly factor ZNHIT6 and the NUFIP1/ZNHIT3 heterodimer (22). Here, we described the assembly factor NOPCHAP1, for NOP-protein chaperone 1, formerly known as C12ORF45. This factor directly interacts with NOP58 and RUVBL1/2 hetero-multimers. Remarkably, in vitro experiments demonstrate that a stable complex containing NOP58-CC-NOP, NOPCHAP1 and RUVBL1/2 can be formed, while little interactions are seen between NOP58-CC-NOP and RUVBL1/2 without NOPCHAP1. Thus, NOPCHAP1 can bridge NOP58 to RUVBL1/2. In addition, transiently expressed NOP58 is produced at lower levels in NOPCHAP1 KO than in wild-type cells. NOPCHAP1 thus stabilizes newly synthesized NOP58 and channels it to RUVBL1/2. Because NOPCHAP1 is not essential in HeLa cells, its function can probably be bypassed, at least in normal culture conditions. Its high degree of conservation nevertheless suggests that it as a selective advantage.
Several partners of RUVBL1 or RUVBL2 have been suggested to favor interaction with specific clients. This is notably the case for proteins of the ZNHIT family, which interact with RUVBL1 and RUVBL2 via their zf-HIT domain (40,41). ZNHIT2 is specific for the U5 snRNP particle and was shown to bridge EFTUD2 with RUVBL1 (40,54). ZNHIT1 and ZNHIT4 belong to the SRCAP and INO80 remodeling complex, respectively, and they mediate contacts of SWR1 and INO80 with RUVBL1/2 (55–59). ZNHIT3 and ZNHIT6 are, like NOPCHAP1, involved in C/D snoRNP biogenesis. Interestingly, ZNHIT6 preferentially binds RUVBL1/2 when they are ATP-loaded (36), in contrast to NOPCHAP1 that only interacts with RUVBL1/2 in an ATP-free state, as demonstrated by our in vitro SPR results in presence of ATP-γ-S. Given that ZNHIT6 is also involved in the assembly of NOP58 (22,36,38), this suggests a switch whereby NOP58 interacts with RUVBL1/2 via NOPCHAP1 in their empty or ADP state, and via ZNHIT6 in their ATP state. In agreement with this possibility, two recent studies reported the discovery of RUVBL1/2 ATPase inhibitors, and one showed that in presence of one such inhibitor, RUVBLs associate less with NOPCHAP1 and HSPs, but more with ZNHIT6 (52,60; see model in Figure 8D). Because NOPCHAP1 associates with the entire PAQosome, we propose that NOPCHAP1 is a PAQosome cofactor involved in client recognition with a specific role for NOP58 and C/D snoRNP biogenesis. In the future, it will be interesting to test whether NOPCHAP1 regulates the conformation and ATPase activity of the RUVBLs, akin the client-loading co-chaperones of HSP90 (61).
NOPCHAP1 discriminates NOP58 from related proteins via its CC-NOP domain
PRPF31, NOP56 and NOP58 are paralog proteins that share a very similar structure, and we previously showed that the HSP90/R2TP chaperones are involved not only in C/D snoRNP biogenesis but also in the assembly of PRPF31 on U4 snRNP (46). PRPF31, NOP56 and NOP58 share a common CC-NOP fold where the NOP domain is involved in binding SNU13:RNA complexes while the CC domain binds additional partners: NOP56 for C/D snoRNPs (11) and PRPF6 for U4 snRNP (19). Here, we show that NOPCHAP1 binds tightly the NOP58-CC-NOP domain but not the CC or NOP domains separately. Interestingly, the NOP58-A283P mutant does not bind NOPCHAP1, suggesting that the relative orientation of the NOP and CC domains might be important for binding. In the future, it will be interesting to determine precisely where NOPCHAP1 binds and whether it prevents NOP58 from associating with NOP56 and/or SNU13:RNA complexes, thereby eliminating non-functional/non-specific premature assembly as was observed for a number of other RNP chaperones (22,62,63). Most interestingly, NOPCHAP1 appears to be specific for NOP58 as it binds weakly or not at all to NOP56 and PRPF31 in LUMIER-IPs. NOPCHAP1 could thus enhance the specificity of RNP assembly by directing NOP58 to a dedicated pathway. It is interesting to note that NOPCHAP1 is an ancient protein well conserved across evolution, as it is present even in primitive eukaryotes. A comparative analysis with other easily traceable C/D assembly factors such as NUFIP1 and the RPAP3-Cter domain (32) shows that these have similar conservation and therefore form an ancient module likely devoted to assembly of C/D snoRNPs. Since Archaea have C/D sRNPs but appears to lack these factors, they may have evolved in primitive eukaryotes. NOPCHAP1 has been lost in S. cerevisiae. This echoes the lack of SMN complex in S. cerevisiae and suggests that biogenesis of sn/snoRNPs may follow simplified pathways in budding yeast. In this organism, Nop58p recruitment on R2TP has been proposed to be mediated by Pih1p, which may have evolved to compensate the absence of NOPCHAP1 (28,64).
HSP70/90 chaperones function early during NOP58 biogenesis and hand over the protein to NOPCHAP1 and the PAQosome
HSP70 and HSP90 chaperones work sequentially to fold newly synthesized proteins (61). HSP70 provides a platform for folding nascent polypeptides and misfolded proteins (65), while the co-chaperone STIP1 promotes the transfer of some clients from HSP70 to HSP90, which works at final steps in folding pathways (66,67). These three proteins are major partners of the NOP58 and PRPF31 mutants that we studied here. These mutants were designed to prevent assembly into their respective RNP complexes and depending on the mutation, the associated factors are not the same or vary quantitatively. NOP58-KA/AR is a double mutant that was designed to prevent association with SNU13 without affecting the overall protein structure, while NOP58-A283P mimics a PRPF31 mutation occurring in retinitis pigmentosa patients and likely affects the relative orientation of the NOP and CC domain (19,68). In this pathology, PRPF31 mutants cannot integrate in their mature particle and are unstable (69). In agreement, GFP-PRPF31-A216P and GFP-NOP58-A283P are found mostly associated with HSP70. In contrast, GFP-NOP58-KA/AR mutant associates more with HSP90 than HSP70, and also binds NOPCHAP1. We thus propose a model in which, after translation, NOP58 is sequentially taken in charge by the chaperones HSP70 and HSP90 and then associates with NOPCHAP1 and PAQosome (Figure 8D). Moreover, wild-type GFP-NOP58 associates more strongly with the RUVBLs than the KA/AR mutant. Since this mutant does not bind SNU13 and since we previously identified a complex containing NOP58, SNU13, RUVBL1/2, ZNHIT6 and ZNHIT3/NUFIP1, we can speculate that NOP58 may be transferred from NOPCHAP1/PAQosome to SNU13/ZNHIT3/NUFIP1/ZNHIT6/RUVBL1/2. As described above, this could occur concomitantly with ATP loading of the RUVBLs. Thus, NOP58 would be chaperoned all the way from its synthesis to its productive assembly on C/D snoRNPs (Figure 8D).
We demonstrated that NOPCHAP1 is an intrinsically disordered protein in its free state. One remarkable feature of IDPs is their surprising stability in vivo despite their lack of 3D fold. IDPs play crucial roles in many cellular functions such as transcription, translation and signaling. IDPs have also been shown to be involved in the assembly of large macromolecular protein complexes (70), and to interact with chaperones to promote their assembly with their biological partners (71). Thus, the IDP status of NOCHAP1 is in good agreement with its addressing and bridging function in a pathway involving HSP and RUVBL1/2 chaperones.
DATA AVAILABILITY
For RNAseq data, see GEO Submission (GSE161155) [NCBI tracking system #21422403]. For proteomic data, see Pride Submission (2020-05-26| PXD019390; 2020-05-27| PXD019414; 2020-05-27| PXD019416; 2020-05-27| PXD019415; 2020-05-26| PXD019389).
Supplementary Material
ACKNOWLEDGEMENTS
We thank P.P.F., M.R.I., M.G.X. and M.G.C. facilities of UMS Montpellier BioCampus. M.R.I. is a member of the national France-BioImaging infrastructure supported by the French National Research Agency (ANR-10-INBS-04, «Investments for the future»). We thank Remy Bordonné, S. Boulon, B. Pradet-Balade, J. Henry for critical reading of the manuscript and Ali Akl for its help in characterization of NOPCHAP1 KO cells. We thank J. Cavaillé and A. Lamond for the gift of antibodies, and C. Goujon for the gift of HEK 293T cells expressing Cas9. We acknowledge IBSLor (UMS 2008/US40 Université de Lorraine, CNRS, INSERM, F-54000 Nancy France) for NMR and Proteomics core facilities.
Contributor Information
Yoann Abel, IGMM, CNRS, Univ Montpellier, Montpellier, France; Equipe labellisée Ligue Nationale Contre le Cancer, Montpellier, France; IGH, CNRS, Univ Montpellier, Montpellier, France.
Ana C F Paiva, iBET, Instituto de Biologia Experimental e Tecnológica, Apartado 12, Oeiras, 2781-901, Portugal; Instituto de Tecnologia Química e Biológica António Xavier, Universidade Nova de Lisboa, Av. da República, Oeiras, 2780-157, Portugal.
Jonathan Bizarro, IGMM, CNRS, Univ Montpellier, Montpellier, France; Equipe labellisée Ligue Nationale Contre le Cancer, Montpellier, France.
Marie-Eve Chagot, Université de Lorraine, CNRS, IMoPA, F-54000 Nancy, France.
Paulo E Santo, iBET, Instituto de Biologia Experimental e Tecnológica, Apartado 12, Oeiras, 2781-901, Portugal; Instituto de Tecnologia Química e Biológica António Xavier, Universidade Nova de Lisboa, Av. da República, Oeiras, 2780-157, Portugal.
Marie-Cécile Robert, IGMM, CNRS, Univ Montpellier, Montpellier, France; Equipe labellisée Ligue Nationale Contre le Cancer, Montpellier, France; IGH, CNRS, Univ Montpellier, Montpellier, France.
Marc Quinternet, Université de Lorraine, CNRS, INSERM, IBSLor, Biophysics and Structural Biology Core Facility, F-54000, Nancy, France.
Franck Vandermoere, IGF, CNRS, INSERM, Univ Montpellier, Montpellier, France.
Pedro M F Sousa, iBET, Instituto de Biologia Experimental e Tecnológica, Apartado 12, Oeiras, 2781-901, Portugal; Instituto de Tecnologia Química e Biológica António Xavier, Universidade Nova de Lisboa, Av. da República, Oeiras, 2780-157, Portugal.
Philippe Fort, CRBM, CNRS, Univ Montpellier, Montpellier, France.
Bruno Charpentier, Université de Lorraine, CNRS, IMoPA, F-54000 Nancy, France.
Xavier Manival, Université de Lorraine, CNRS, IMoPA, F-54000 Nancy, France.
Tiago M Bandeiras, iBET, Instituto de Biologia Experimental e Tecnológica, Apartado 12, Oeiras, 2781-901, Portugal; Instituto de Tecnologia Química e Biológica António Xavier, Universidade Nova de Lisboa, Av. da República, Oeiras, 2780-157, Portugal.
Edouard Bertrand, IGMM, CNRS, Univ Montpellier, Montpellier, France; Equipe labellisée Ligue Nationale Contre le Cancer, Montpellier, France; IGH, CNRS, Univ Montpellier, Montpellier, France.
Céline Verheggen, IGMM, CNRS, Univ Montpellier, Montpellier, France; Equipe labellisée Ligue Nationale Contre le Cancer, Montpellier, France; IGH, CNRS, Univ Montpellier, Montpellier, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centre National de la Recherche Scientifique (CNRS); Université de Montpellier (UM); Université de Lorraine (UL); Agence Nationale de la Recherche [ANR-16-CE11-0032-04]; Ligue Nationale Contre le Cancer (‘équipe labellisée’); Institut National du Cancer [INCa PLBio 2016-161]; Portuguese national funds through FCT – Fundação para a Ciência e a Tecnologia, I.P. [PTDC/BBB-BEP/1463/2014]; iNOVA4Health – UIDB/04462/2020; Fundação para a Ciência e Tecnologia/Ministério da Educação e Ciência, through national funds; Y.A. was supported by INCa and ANR; J.B. had a fellowship from Ministère de l’Enseignement supérieur et de la Recherche and Fondation ARC. Funding for open access charge: Ligue contre le cancer.
Conflict of interest statement. None declared.
REFERENCES
- 1. Battle D.J., Kasim M., Yong J., Lotti F., Lau C.-K., Mouaikel J., Zhang Z., Han K., Wan L., Dreyfuss G.. The SMN complex: an assembly machine for RNPs. Cold Spring Harb. Symp. Quant. Biol. 2006; 71:313–320. [DOI] [PubMed] [Google Scholar]
- 2. Lafontaine D.L.J. Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat. Struct. Mol. Biol. 2015; 22:11–19. [DOI] [PubMed] [Google Scholar]
- 3. Massenet S., Bertrand E., Verheggen C.. Assembly and trafficking of box C/D and H/ACA snoRNPs. RNA Biol. 2017; 14:680–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Matera A.G., Wang Z.. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014; 15:108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Peña C., Hurt E., Panse V.G.. Eukaryotic ribosome assembly, transport and quality control. Nat. Struct. Mol. Biol. 2017; 24:689–699. [DOI] [PubMed] [Google Scholar]
- 6. Bohnsack M.T., Sloan K.E.. Modifications in small nuclear RNAs and their roles in spliceosome assembly and function. Biol. Chem. 2018; 399:1265–1276. [DOI] [PubMed] [Google Scholar]
- 7. Sloan K.E., Warda A.S., Sharma S., Entian K.-D., Lafontaine D.L.J., Bohnsack M.T.. Tuning the ribosome: the influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2017; 14:1138–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Watkins N.J., Bohnsack M.T.. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip. Rev. RNA. 2012; 3:397–414. [DOI] [PubMed] [Google Scholar]
- 9. Reichow S.L., Hamma T., Ferré-D’Amaré A.R., Varani G.. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007; 35:1452–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tran E., Brown J., Maxwell E.S.. Evolutionary origins of the RNA-guided nucleotide-modification complexes: from the primitive translation apparatus. Trends Biochem. Sci. 2004; 29:343–350. [DOI] [PubMed] [Google Scholar]
- 11. Aittaleb M., Rashid R., Chen Q., Palmer J.R., Daniels C.J., Li H.. Structure and function of archaeal box C/D sRNP core proteins. Nat. Struct. Mol. Biol. 2003; 10:256–263. [DOI] [PubMed] [Google Scholar]
- 12. Cahill N.M., Friend K., Speckmann W., Li Z.-H., Terns R.M., Terns M.P., Steitz J.A.. Site-specific cross-linking analyses reveal an asymmetric protein distribution for a box C/D snoRNP. EMBO J. 2002; 21:3816–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chaker-Margot M., Barandun J., Hunziker M., Klinge S.. Architecture of the yeast small subunit processome. Science. 2017; 355:eaal1880. [DOI] [PubMed] [Google Scholar]
- 14. Lin J., Lai S., Jia R., Xu A., Zhang L., Lu J., Ye K.. Structural basis for site-specific ribose methylation by box C/D RNA protein complexes. Nature. 2011; 469:559–563. [DOI] [PubMed] [Google Scholar]
- 15. Qu G., van Nues R.W., Watkins N.J., Maxwell E.S.. The Spatial-Functional coupling of box C/D and C′/D′ RNPs is an evolutionarily conserved feature of the eukaryotic box C/D snoRNP nucleotide modification complex. Mol. Cell. Biol. 2011; 31:365–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rashid R., Liang B., Baker D.L., Youssef O.A., He Y., Phipps K., Terns R.M., Terns M.P., Li H.. Crystal structure of a Cbf5-Nop10-Gar1 complex and implications in RNA-Guided pseudouridylation and dyskeratosis congenita. Mol. Cell. 2006; 21:249–260. [DOI] [PubMed] [Google Scholar]
- 17. Watkins N.J., Ségault V., Charpentier B., Nottrott S., Fabrizio P., Bachi A., Wilm M., Rosbash M., Branlant C., Lührmann R.. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000; 103:457–466. [DOI] [PubMed] [Google Scholar]
- 18. Gautier T., Bergès T., Tollervey D., Hurt E.. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell. Biol. 1997; 17:7088–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu S., Li P., Dybkov O., Nottrott S., Hartmuth K., Lührmann R., Carlomagno T., Wahl M.C.. Binding of the human Prp31 nop domain to a composite RNA-Protein platform in U4 snRNP. Science. 2007; 316:115–120. [DOI] [PubMed] [Google Scholar]
- 20. Watkins N.J., Dickmanns A., Lührmann R.. Conserved Stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol. Cell. Biol. 2002; 22:8342–8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schultz A., Nottrott S., Watkins N.J., Lührmann R.. Protein-protein and protein-RNA contacts both contribute to the 15.5K-mediated assembly of the U4/U6 snRNP and the box C/D snoRNPs. Mol. Cell. Biol. 2006; 26:5146–5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bizarro J., Charron C., Boulon S., Westman B., Pradet-Balade B., Vandermoere F., Chagot M.-E., Hallais M., Ahmad Y., Leonhardt H. et al.. Proteomic and 3D structure analyses highlight the C/D box snoRNP assembly mechanism and its control. J. Cell Biol. 2014; 207:463–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Boulon S., Marmier-Gourrier N., Pradet-Balade B., Wurth L., Verheggen C., Jády B.E., Rothé B., Pescia C., Robert M.-C., Kiss T. et al.. The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. J. Cell Biol. 2008; 180:579–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhao R., Kakihara Y., Gribun A., Huen J., Yang G., Khanna M., Costanzo M., Brost R.L., Boone C., Hughes T.R. et al.. Molecular chaperone Hsp90 stabilizes Pih1/Nop17 to maintain R2TP complex activity that regulates snoRNA accumulation. J. Cell Biol. 2008; 180:563–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jeganathan A., Leong V., Zhao L., Huen J., Nano N., Houry W.A., Ortega J.. Yeast Rvb1 and Rvb2 proteins oligomerize as a conformationally variable dodecamer with low frequency. J. Mol. Biol. 2015; 427:1875–1886. [DOI] [PubMed] [Google Scholar]
- 26. Martino F., Pal M., Muñoz-Hernández H., Rodríguez C.F., Núñez-Ramírez R., Gil-Carton D., Degliesposti G., Skehel J.M., Roe S.M., Prodromou C. et al.. RPAP3 provides a flexible scaffold for coupling HSP90 to the human R2TP co-chaperone complex. Nat. Commun. 2018; 9:3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Muñoz-Hernández H., Pal M., Rodríguez C.F., Fernandez-Leiro R., Prodromou C., Pearl L.H., Llorca O.. Structural mechanism for regulation of the AAA-ATPases RUVBL1-RUVBL2 in the R2TP co-chaperone revealed by cryo-EM. Sci. Adv. 2019; 5:eaaw1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tian S., Yu G., He H., Zhao Y., Liu P., Marshall A.G., Demeler B., Stagg S.M., Li H.. Pih1p-Tah1p puts a lid on hexameric AAA+ ATPases Rvb1/2p. Structure. 2017; 25:1519–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Back R., Dominguez C., Rothé B., Bobo C., Beaufils C., Moréra S., Meyer P., Charpentier B., Branlant C., Allain F.H.-T. et al.. High-Resolution structural analysis shows How Tah1 tethers Hsp90 to the R2TP complex. Structure. 2013; 21:1834–1847. [DOI] [PubMed] [Google Scholar]
- 30. Henri J., Chagot M.-E., Bourguet M., Abel Y., Terral G., Maurizy C., Aigueperse C., Georgescauld F., Vandermoere F., Saint-Fort R. et al.. Deep structural analysis of RPAP3 and PIH1D1, two components of the HSP90 co-chaperone R2TP complex. Structure. 2018; 26:1196–1209. [DOI] [PubMed] [Google Scholar]
- 31. Pal M., Morgan M., Phelps S.E.L., Roe S.M., Parry-Morris S., Downs J.A., Polier S., Pearl L.H., Prodromou C.. Structural basis for Phosphorylation-Dependent recruitment of Tel2 to Hsp90 by Pih1. Structure. 2014; 22:805–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maurizy C., Quinternet M., Abel Y., Verheggen C., Santo P.E., Bourguet M., Paiva A.C.F., Bragantini B., Chagot M.-E., Robert M.-C. et al.. The RPAP3-Cterminal domain identifies R2TP-like quaternary chaperones. Nat. Commun. 2018; 9:2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hořejší Z., Stach L., Flower T.G., Joshi D., Flynn H., Skehel J.M., O’Reilly N.J., Ogrodowicz R.W., Smerdon S.J., Boulton S.J.. Phosphorylation-dependent PIH1D1 interactions define substrate specificity of the R2TP cochaperone complex. Cell Rep. 2014; 7:19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Houry W.A., Bertrand E., Coulombe B.. The PAQosome, an R2TP-based chaperone for quaternary structure formation. Trends Biochem. Sci. 2018; 43:4–9. [DOI] [PubMed] [Google Scholar]
- 35. Watkins N.J., Lemm I., Ingelfinger D., Schneider C., Hoßbach M., Urlaub H., Lührmann R.. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol. Cell. 2004; 16:789–798. [DOI] [PubMed] [Google Scholar]
- 36. McKeegan K.S., Debieux C.M., Watkins N.J.. Evidence that the AAA+ proteins TIP48 and TIP49 bridge interactions between 15.5K and the related NOP56 and NOP58 proteins during box C/D snoRNP biogenesis. Mol. Cell. Biol. 2009; 29:4971–4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rothé B., Saliou J.-M., Quinternet M., Back R., Tiotiu D., Jacquemin C., Loegler C., Schlotter F., Peña V., Eckert K. et al.. Protein Hit1, a novel box C/D snoRNP assembly factor, controls cellular concentration of the scaffolding protein Rsa1 by direct interaction. Nucleic Acids Res. 2014; 42:10731–10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Paul A., Tiotiu D., Bragantini B., Marty H., Charpentier B., Massenet S., Labialle S.. Bcd1p controls RNA loading of the core protein Nop58 during C/D box snoRNP biogenesis. RNA. 2019; 25:496–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McKeegan K.S., Debieux C.M., Boulon S., Bertrand E., Watkins N.J.. A dynamic scaffold of Pre-snoRNP factors facilitates human box C/D snoRNP assembly. Mol. Cell. Biol. 2007; 27:6782–6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cloutier P., Poitras C., Durand M., Hekmat O., Fiola-Masson É., Bouchard A., Faubert D., Chabot B., Coulombe B.. R2TP/prefoldin-like component RUVBL1/RUVBL2 directly interacts with ZNHIT2 to regulate assembly of U5 small nuclear ribonucleoprotein. Nat. Commun. 2017; 8:15615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Verheggen C., Pradet-Balade B., Bertrand E.. SnoRNPs, ZNHIT proteins and the R2TP pathway. Oncotarget. 2015; 6:41399–41400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tantale K., Mueller F., Kozulic-Pirher A., Lesne A., Victor J.-M., Robert M.-C., Capozi S., Chouaib R., Bäcker V., Mateos-Langerak J. et al.. A single-molecule view of transcription reveals convoys of RNA polymerases and multi-scale bursting. Nat. Commun. 2016; 7:12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Adl S.M., Bass D., Lane C.E., Lukeš J., Schoch C.L., Smirnov A., Agatha S., Berney C., Brown M.W., Burki F. et al.. Revisions to the classification, nomenclature, and diversity of eukaryotes. J. Eukaryot. Microbiol. 2019; 66:4–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tsanov N., Samacoits A., Chouaib R., Traboulsi A.-M., Gostan T., Weber C., Zimmer C., Zibara K., Walter T., Peter M. et al.. smiFISH and FISH-quant – a flexible single RNA detection approach with super-resolution capability. Nucleic Acids Res. 2016; 44:e165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Domanski M., Molloy K., Jiang H., Chait B.T., Rout M.P., Jensen T.H., LaCava J.. Improved methodology for the affinity isolation of human protein complexes expressed at near endogenous levels. BioTechniques. 2012; 0:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bizarro J., Dodré M., Huttin A., Charpentier B., Schlotter F., Branlant C., Verheggen C., Massenet S., Bertrand E.. NUFIP and the HSP90/R2TP chaperone bind the SMN complex and facilitate assembly of U4-specific proteins. Nucleic Acids Res. 2015; 43:8973–8989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cox J., Mann M.. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008; 26:1367–1372. [DOI] [PubMed] [Google Scholar]
- 48. Jäkel S., Mingot J.-M., Schwarzmaier P., Hartmann E., Görlich D.. Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J. 2002; 21:377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pradet-Balade B., Girard C., Boulon S., Paul C., Azzag K., Bordonné R., Bertrand E., Verheggen C.. CRM1 controls the composition of nucleoplasmic pre-snoRNA complexes to licence them for nucleolar transport. EMBO J. 2011; 30:2205–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Taipale M., Tucker G., Peng J., Krykbaeva I., Lin Z.-Y., Larsen B., Choi H., Berger B., Gingras A.-C., Lindquist S.. A quantitative chaperone interaction network reveals the architecture of cellular protein homeostasis pathways. Cell. 2014; 158:434–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Barrios-Rodiles M., Brown K.R., Ozdamar B., Bose R., Liu Z., Donovan R.S., Shinjo F., Liu Y., Dembowy J., Taylor I.W. et al.. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005; 307:1621–1625. [DOI] [PubMed] [Google Scholar]
- 52. Assimon V.A., Tang Y., Vargas J.D., Lee G.J., Wu Z.Y., Lou K., Yao B., Menon M.-K., Pios A., Perez K.C. et al.. CB-6644 is a selective inhibitor of the RUVBL1/2 complex with anticancer activity. ACS Chem. Biol. 2019; 14:236–244. [DOI] [PubMed] [Google Scholar]
- 53. Afanasyeva A., Hirtreiter A., Schreiber A., Grohmann D., Pobegalov G., McKay A.R., Tsaneva I., Petukhov M., Käs E., Grigoriev M. et al.. Lytic water dynamics reveal evolutionarily conserved mechanisms of ATP hydrolysis by TIP49 AAA+ ATPases. Structure. 2014; 22:549–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Malinová A et al.. Assembly of the U5 snRNP component PRPF8 is controlled by the HSP90/R2TP chaperones. J. Cell Biol. 2017; 216:1579–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ayala R., Willhoft O., Aramayo R.J., Wilkinson M., McCormack E.A., Ocloo L., Wigley D.B., Zhang X.. Structure and regulation of the human INO80–nucleosome complex. Nature. 2018; 556:391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chen L., Cai Y., Jin J., Florens L., Swanson S.K., Washburn M.P., Conaway J.W., Conaway R.C.. Subunit organization of the human INO80 chromatin remodeling complex: an evolutionarily conserved core complex catalyzes ATP-dependent nucleosome remodeling. J. Biol. Chem. 2011; 286:11283–11289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Eustermann S., Schall K., Kostrewa D., Lakomek K., Strauss M., Moldt M., Hopfner K.-P.. Structural basis for ATP-dependent chromatin remodelling by the INO80 complex. Nature. 2018; 556:386–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sardiu M.E., Cai Y., Jin J., Swanson S.K., Conaway R.C., Conaway J.W., Florens L., Washburn M.P.. Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc. Natl. Acad. Sci. 2008; 105:1454–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tosi A., Haas C., Herzog F., Gilmozzi A., Berninghausen O., Ungewickell C., Gerhold C.B., Lakomek K., Aebersold R., Beckmann R. et al.. Structure and subunit topology of the INO80 chromatin remodeler and its nucleosome complex. Cell. 2013; 154:1207–1219. [DOI] [PubMed] [Google Scholar]
- 60. Yenerall P., Das A.K., Wang S., Kollipara R.K., Li L.S., Villalobos P., Flaming J., Lin Y.-F., Huffman K., Timmons B.C. et al.. RUVBL1/RUVBL2 ATPase activity drives PAQosome maturation, DNA replication and radioresistance in lung cancer. Cell Chem. Biol. 2020; 27:105–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kirschke E., Goswami D., Southworth D., Griffin P.R., Agard D.A.. Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles. Cell. 2014; 157:1685–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Neuenkirchen N., Englbrecht C., Ohmer J., Ziegenhals T., Chari A., Fischer U.. Reconstitution of the human U snRNP assembly machinery reveals stepwise Sm protein organization. EMBO J. 2015; 34:1925–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Walbott H., Machado-Pinilla R., Liger D., Blaud M., Réty S., Grozdanov P.N., Godin K., van Tilbeurgh H., Varani G., Meier U.T. et al.. The H/ACA RNP assembly factor SHQ1 functions as an RNA mimic. Genes Dev. 2011; 25:2398–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yu G., Zhao Y., Tian S., Rai J., He H., Spear J., Sousa D., Fan J., Yu H.-G., Stagg S.M. et al.. Yeast R2TP interacts with extended termini of client protein Nop58p. Sci. Rep. 2019; 9:20228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rosenzweig R., Nillegoda N.B., Mayer M.P., Bukau B.. The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 2019; 20:665–680. [DOI] [PubMed] [Google Scholar]
- 66. Luengo T.M., Mayer M.P., Rüdiger S.G.D.. The Hsp70–Hsp90 chaperone cascade in protein folding. Trends Cell Biol. 2019; 29:164–177. [DOI] [PubMed] [Google Scholar]
- 67. Mayer M.P., Le Breton L.. Hsp90: Breaking the symmetry. Mol. Cell. 2015; 58:8–20. [DOI] [PubMed] [Google Scholar]
- 68. Vithana E.N., Abu-Safieh L., Allen M.J., Carey A., Papaioannou M., Chakarova C., Al-Maghtheh M., Ebenezer N.D., Willis C., Moore A.T. et al.. A human homolog of yeast Pre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11). Mol. Cell. 2001; 8:375–381. [DOI] [PubMed] [Google Scholar]
- 69. Deery E.C., Vithana E.N., Newbold R.J., Gallon V.A., Bhattacharya S.S., Warren M.J., Hunt D.M., Wilkie S.E.. Disease mechanism for retinitis pigmentosa (RP11) caused by mutations in the splicing factor gene PRPF31. Hum. Mol. Genet. 2002; 11:3209–3219. [DOI] [PubMed] [Google Scholar]
- 70. Wright P. E., Dyson H. J.. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015; 16:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hegyi H., Tompa P.. Intrinsically disordered proteins display no preference for chaperone binding in vivo. PLoS Comput. Biol. 2008; 4:e1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
For RNAseq data, see GEO Submission (GSE161155) [NCBI tracking system #21422403]. For proteomic data, see Pride Submission (2020-05-26| PXD019390; 2020-05-27| PXD019414; 2020-05-27| PXD019416; 2020-05-27| PXD019415; 2020-05-26| PXD019389).