Significance
While human genes underlying monogenic disorders are expected to undergo negative selection, the factors that impact the intensity of negative selection remain unknown. We find pervasive negative selection at genes underlying both autosomal dominant and the most severe autosomal recessive inborn errors. Among genes underlying dominant disorders, we show that loss of function entails stronger selection than gain of function, and haploinsufficiency than negative dominance. We develop a method that predicts whether an autosomal gene is more likely to underlie a recessive or a dominant disease. These results have evolutionary implications for studies of the drivers of negative selection, and practical implications in the search for genes underlying life-threatening, heritable conditions.
Keywords: immunodeficiency, selection, evolution, genetics, method
Abstract
Genetic variants underlying life-threatening diseases, being unlikely to be transmitted to the next generation, are gradually and selectively eliminated from the population through negative selection. We study the determinants of this evolutionary process in human genes underlying monogenic diseases by comparing various negative selection scores and an integrative approach, CoNeS, at 366 loci underlying inborn errors of immunity (IEI). We find that genes underlying autosomal dominant (AD) or X-linked IEI have stronger negative selection scores than those underlying autosomal recessive (AR) IEI, whose scores are not different from those of genes not known to be disease causing. Nevertheless, genes underlying AR IEI that are lethal before reproductive maturity with complete penetrance have stronger negative selection scores than other genes underlying AR IEI. We also show that genes underlying AD IEI by loss of function have stronger negative selection scores than genes underlying AD IEI by gain of function, while genes underlying AD IEI by haploinsufficiency are under stronger negative selection than other genes underlying AD IEI. These results are replicated in 1,140 genes underlying inborn errors of neurodevelopment. Finally, we propose a supervised classifier, SCoNeS, which predicts better than state-of-the-art approaches whether a gene is more likely to underlie an AD or AR disease. The clinical outcomes of monogenic inborn errors, together with their mode and mechanisms of inheritance, determine the levels of negative selection at their corresponding loci. Integrating scores of negative selection may facilitate the prioritization of candidate genes and variants in patients suspected to carry an inborn error.
Negative (or purifying) selection is the natural process by which deleterious alleles are selectively purged from the population (1). In diploid species, the strength of negative selection at a given locus is predicted to increase with decreasing fitness and increasing dominance of the genetic variants controlling traits: Variation causing early death in the heterozygous state are the least likely to be transmitted to the next generation, as their carriers have fewer offspring than noncarriers (2). Human genetic variants that cause severe diseases are, thus, expected to be the primary targets of negative selection, particularly for diseases affecting heterozygous individuals. In humans, several studies have ranked protein-coding genes according to their levels of negative selection (3–5). Nevertheless, the extent to which negative selection affects human disease-causing genes, and the factors determining its strength, remain largely unknown, particularly because our knowledge of the severity, mode, and mechanism of inheritance of the corresponding human diseases remains incomplete (3, 6–8).
The strength of negative selection at a given gene has been traditionally approximated by comparing the coding sequence of the gene in a given species with that of one or several closely related species; it depends on the proportion of amino acid changes that have accumulated during evolution (9–11). With the advent of high-throughput sequencing, intraspecies metrics have been developed, based on the comparison of the probability of predicted loss-of-function (pLOF) mutations for a gene under a random model with the frequency of pLOF mutations observed in population databases (5, 12, 13), which capture the species-specific evolution of genes. Using an interspecies-based method and a hand-curated version of the Online Mendelian Inheritance in Man (hOMIM) database, a previous study elegantly showed that most human genes for which mutations cause highly penetrant diseases, including autosomal dominant (AD) diseases in particular, evolve under stronger negative selection than genes associated with complex disorders (6). However, other studies based on OMIM genes have reported conflicting results (3, 14–17), probably due to the incompleteness and heterogeneity of the datasets used. Moreover, no study has yet addressed this problem with intraspecies metrics, even though it has been suggested that the choice of the reference species for interspecies metrics contributes to discrepancies across studies (6).
We aimed to improve the identification of the drivers of negative selection acting on human disease-causing genes, by developing a negative selection score combining several informative intraspecies and interspecies statistics, focusing on inborn errors of immunity (IEI). IEI, previously known as primary immunodeficiencies (18), are genetic diseases that disrupt the development or function of human immunity. They form a large and expanding group of genetic diseases that has been widely studied, and they are well characterized physiologically (immunologically) and phenotypically (clinically) (19–21). IEI are often symptomatic in early childhood, and at least until the turn of the 20th century and the introduction of antibiotics, most individuals with IEI probably died before reaching reproductive maturity. Accordingly, IEI genes have probably been under strong negative selection from the dawn of humankind until very recently. In this study, we investigated whether the severity of IEI and their mode and mechanism of inheritance have left signatures of negative selection of various intensities in the corresponding human genes. Furthermore, we validated our model on genes underlying inborn errors of neurodevelopment (IEND), another group of well-characterized severe genetic diseases.
Results
CoNeS Is a Consensus-Based Measure of Negative Selection.
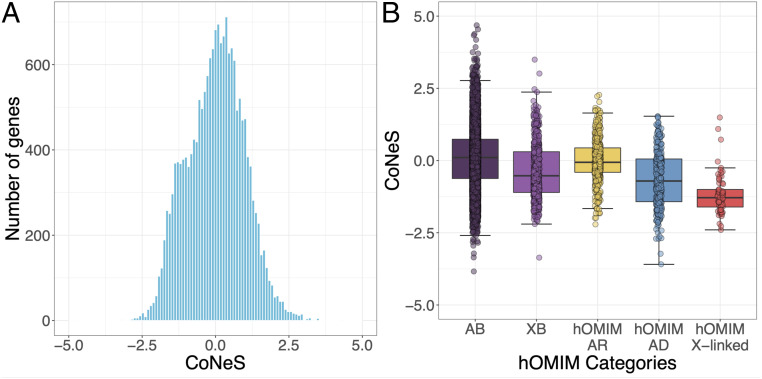
We developed a score, consensus negative selection (CoNeS), to take into account information from both interspecies [the f parameter from SnIPRE (11), lofTool (13), and evoTol (22)] and intraspecies [RVIS (5), LOEUF (23), pLI (12), and SIS (24)] statistics to approximate the strength of negative selection. The correlation between these different statistics is shown in SI Appendix, Fig. S1. Although these scores are different, they can be grouped into three correlated categories: scores that integrate structural protein conservation measures (lofTool and evoTol), those that treat heterozygotes and homozygotes differently (pLI and LOEUF), and the others (SIS, f parameter, and RVIS). We did not include in the computation gene-level metrics that are not aimed at measuring the strength of negative selection [such as the GDI (25) or pRecessive (12)] or that are unavailable for more than 25% of the genes [such as Sel (3)]. CoNeS was obtained through a standardized (i.e., mean of 0 and SD of 1) projection of these seven methods on the first principal component, which captures 81.8% of the total variance. The CoNeS distribution for 18,026 autosomal protein-coding genes is shown in Fig. 1A. The distribution is slightly bimodal due to the inclusion of bimodal metrics (pLI and LOEUF) in the calculation. Low CoNeS values are associated with stronger selection constraints (i.e., low f, lofTool, evoTol, LOEUF, and RVIS; high SIS and pLI). As expected, CoNeS values were significantly lower for X-linked genes than for autosomal genes (median on X, −0.718; Wilcoxon one-tailed test between CoNeS values for the genes on X and CoNeS values for autosomal genes, P = 1.89 × 10−51; SI Appendix, Fig. S2), as negative selection acts on deleterious X-linked variants underlying recessive disease in both homozygous females and hemizygous males. We therefore considered X chromosome genes separately from autosomal genes in all subsequent analyses. We assessed the dependence to each of the individual scores by calculating CoNeS after the removal of each of the seven metrics contributing to the combined score. The resulting scores were strongly correlated with CoNeS (0.955 < Spearman’s R2 < 0.993; SI Appendix, Fig. S3), indicating that CoNeS is not particularly affected by a single statistic.
Fig. 1.
The distribution of CoNeS for human genes. (A) The distribution of CoNeS for 18,037 autosomal human genes. (B) The distribution of CoNeS for genes causing Mendelian diseases with complete penetrance (hOMIM), according to their dominant (AD), recessive (AR), or X-linked mode of inheritance, relative to autosomal (AB) and X chromosome (XB) background genes.
Mendelian Disease-Causing Genes Show Stronger Negative Selection Scores than Background Genes.
We sought to replicate previous observations based on the hOMIM database (6). We compared the CoNeS of 833 autosomal and 66 X-linked genes annotated by hOMIM as Mendelian disease causing, to that of 15,219 “autosomal background” (AB) and 650 “X background” (XB) genes, respectively, these background genes not being known to be essential [i.e., indispensable for the survival (26)] or to underlie any severe genetic disorders (see Methods for details). The CoNeS was significantly lower for hOMIM autosomal genes than for the AB group (Wilcoxon one-tailed test, P = 5.34 × 10−16; resampling test accounting for the size of coding genes, P < 10−5; Fig. 1B), indicating that the hOMIM genes were subject to stronger selection constraints. However, this result was dependent of the mode of disease inheritance: The difference in CoNeS between hOMIM genes causing AD diseases and the AB group was highly significant (Wilcoxon one-tailed test, P = 2.05 × 10−31; resampling test, P < 10−5; SI Appendix, Table S1), whereas this difference was not significant for hOMIM genes causing autosomal recessive (AR) diseases (SI Appendix, Table S2) (Fig. 1B). Furthermore, X-linked hOMIM genes showed significantly stronger levels of negative selection (median, −1.28) than XB genes (Wilcoxon one-tailed test, P = 1.08 × 10−12; resampling test, P < 10−5; Fig. 1B and SI Appendix, Table S3). CoNeS, together with lofTool and evoTol, were the metrics that were the most significantly different between hOMIM genes and background genes. Overall, these results confirm that genes underlying known monogenic disorders, especially for AD and X-linked disorders, are under stronger negative selection constraints than the rest of the coding genome. This result is probably conservative, as the rest of the coding genome probably comprises hitherto-unknown disease-causing loci.
Levels of Negative Selection at Autosomal IEI Genes Depend on the Mode of Inheritance.
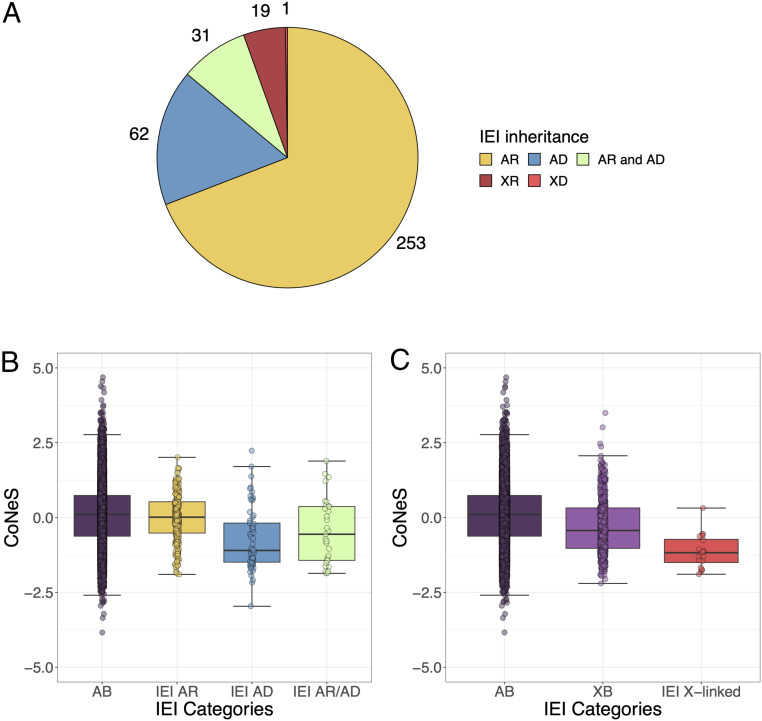
We leveraged the unique information collected for genes underlying autosomal IEI to determine how the mode and mechanism of disease inheritance, together with disease severity, impact levels of negative selection on human disease-causing genes. There are 416 known IEI, caused by defects of 370 genes, in the latest International Union of Immunological Societies Committee classification (18). Historically, IEI were considered to be Mendelian disorders, with both complete clinical penetrance and detectable immunological abnormalities. More recently, IEI with incomplete penetrance and/or without detectable immunological phenotypes have been described (19, 21). We obtained negative selection metrics for 366 out of 370 IEI genes (Methods). Out of the remaining 366 IEI genes, more than two-thirds (253/366) underlie IEI that are AR; a smaller number of genes (62/366) underlie IEI that are AD; an even smaller number underlie IEI that are X-recessive (XR) (19/366); and only one gene (WAS) underlies an IEI that is X-dominant (XD). A small number of loci underlie IEI diseases with both AR and AD inheritance patterns (31/366) (Fig. 2A). Consistent with the results obtained for hOMIM genes, CoNeS was significantly lower for IEI AD and IEI AR/AD genes, relative to AB genes (medians, −1.08 and −0.55; Wilcoxon test, P = 1.71 × 10−9 and 3.02 × 10−3; resampling test, P < 10−5 and P = 8.19 × 10−3, respectively), whereas that for IEI AR genes was not (median, 2.09 × 10−2; Wilcoxon test, P = 0.101; resampling test, P = 0.180) (Fig. 2B, Table 1, and SI Appendix, Tables S4 and S5). Most individual statistics (with the exception of evoTol) supported IEI AD genes being under significantly stronger negative selection than AB genes, but the difference was the most significant for CoNeS (Table 1). These results suggest that genes underlying AD IEI are under stronger negative selection than genes underlying AR IEI or both AR and AD IEI, which is expected because deleterious mutations causing dominant disease decrease the fitness of both heterozygous and homozygous carriers. This is consistent with the notion that disease dominance has a strong impact on the levels of negative selection on human disease-causing genes.
Fig. 2.
The distribution of CoNeS for genes underlying inborn errors of immunity (IEI). (A) The number of genes underlying IEI, according to the mode of disease inheritance. (B) The distribution of CoNeS for autosomal genes underlying IEI, according to their dominant (AD), recessive (AR), or both dominant and recessive (AR/AD) mode of inheritance, relative to autosomal background (AB) genes. (C) The distribution of CoNeS for X-linked genes underlying IEI, relative to autosomal (AB) and X chromosome (XB) background genes.
Table 1.
Statistical significance of differences in negative selection scores between genes underlying AD IEI and AB genes
| Score | Wilcoxon-based test | Resampling-based test |
| evoTol | 0.209 | 0.767 |
| f parameter | 5.16 × 10−7 | 5 × 10−4 |
| LOEUF | 1.29 × 10−7 | <10−5 |
| lofTool | 3.25 × 10−5 | 2.70 × 10−3 |
| pLI | 1.59 × 10−5 | <10−5 |
| RVIS | 3.29 × 10−6 | 2.42 × 10−2 |
| SIS | 4.48 × 10−5 | 7.60 × 10−3 |
| CoNeS | 1.71 × 10−9 | <10−5 |
The P values for a one-tailed Wilcoxon test and a resampling-based test (Methods) assessing the difference between IEI AD genes and AB genes are shown.
Negative Selection Scores Are Stronger at X-Linked IEI Genes than at Other X-Linked Genes.
Men carry only one copy of the X chromosome. Variants underlying XR diseases are therefore expected to decrease fitness in both homozygous women and hemizygous men, and thus to be more rapidly purged from the population than variants underlying AR diseases. Consistent with this hypothesis, we found that the CoNeS for XB genes is significantly lower than that for AB genes (Fig. 2C). Furthermore, IEI XR genes showed stronger evidence for negative selection than XB genes, as indicated by their lower CoNeS (median, −1.17; Wilcoxon test, P = 1.19 × 10−5; resampling-based test, P = 2.21 × 10−2; SI Appendix, Table S6). CoNeS, pLI, and evoTol were the statistics yielding the most significant difference between IEI XR and XB (SI Appendix, Table S6). At the individual gene level, only one IEI XR gene has a positive CoNeS value: CSF2RA (0.324). Human CSF2RA deficiency causes juvenile pulmonary alveolar proteinosis, a disease that was lethal until very recently (27). However, CSF2RA lies in the pseudoautosomal region of the X and Y chromosomes, accounting for pseudoheterozygous boys not developing disease. Serving as a natural control, this gene was, therefore, unsurprisingly under weaker selection than the genes underlying truly XR IEI. Collectively, these results suggest that X chromosome genes underlying IEI are, like autosomal IEI genes, under stronger selective constraints than the rest of the coding genome, a trend that may become stronger as new IEI are being discovered.
Negative Selection Scores Are Stronger in Genes Underlying Early-Onset, Highly Penetrant Recessive IEI.
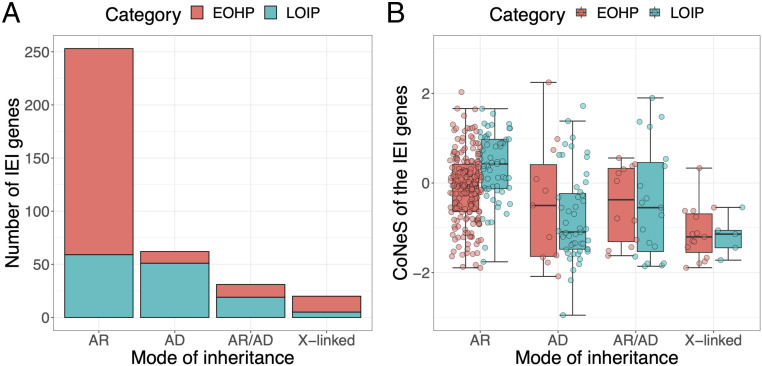
We investigated whether the genes underlying diseases that decrease fitness the most were under the strongest selective constraints, by classifying genes underlying IEI into two categories: 231 genes that, when mutated, underlie severe disease and prevent patients from reaching reproductive age (i.e., early-onset, highly penetrant diseases [EOHP]) in the absence of modern treatment, and 134 genes, comprising all the other genes causing diseases with incomplete penetrance and/or a more moderate impact (i.e., later-onset, incompletely penetrant diseases [LOIP]), as demonstrated by the findings for at least one reported multigenerational multiplex family (whether dominant or recessive). Genes underlying AR and XR IEI are enriched in EOHP genes (76.5% and 75.0%, respectively), whereas AD diseases are typically associated with LOIP genes (80.9%) (χ2 test, P = 4.56 × 10−17; Fig. 3A). This observation suggests that variants underlying AR IEI decrease fitness more than variants underlying AD IEI, consistent with the negative relationship observed between fitness and the dominance coefficient in Drosophila, yeast, and thale cress (28–30). However, we caution this result, because the observed enrichment may also be due to a bias in the IEI database (e.g., severe dominant diseases are more difficult to discover and study). Interestingly, we observed that the IEI AR genes of the EOHP group show significantly lower negative selection scores than those of the LOIP group (medians, −0.0742 and 0.413, respectively; Wilcoxon one-tailed test, P = 1.72 × 10−5) or AB genes (Wilcoxon one-tailed test, P = 2.13 × 10−3; SI Appendix, Table S7). The observation was not replicated for IEI AD, IEI AR/AD, and IEI XR genes (Wilcoxon one-tailed test, P = 0.771, 0.602, and 0.466, respectively; Fig. 3B), possibly because of a lack of power. Together, our findings suggest that genes underlying severe AR IEI are under stronger selective constraints than genes causing milder AR IEI. They also provide evidence that age of disease onset and clinical penetrance affect the levels of negative selection at human disease-causing genes.
Fig. 3.
The distribution of CoNeS for genes underlying inborn errors of immunity (IEI), according to disease, age of onset, and penetrance. (A) The number of IEI genes underlying early-onset, highly penetrant (EOHP) and later-onset, incompletely penetrant (LOIP) IEI, according to the mode of disease inheritance. (B) The distribution of CoNeS for the IEI genes, as a function of age of onset, penetrance, and mode of inheritance.
The Mechanism of Dominance Affects Negative Selection Scores.
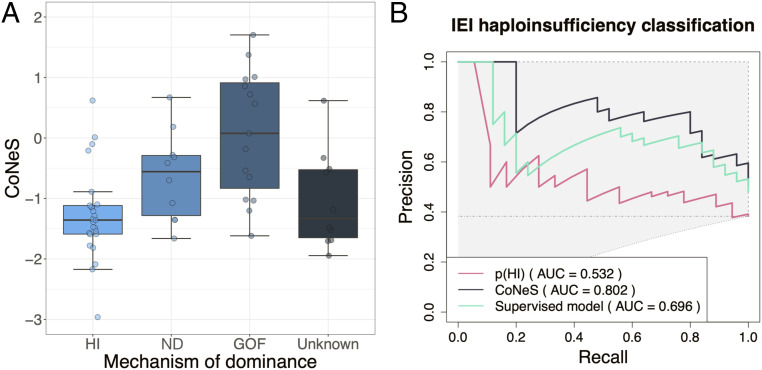
Dominance can operate by negative dominance (ND), haploinsufficiency (HI), or gain of function (GOF) (31). In AD disorders due to ND, the AD cellular and clinical deficiencies are caused by the interference of the mutant gene product with the activity of the wild-type (WT) product, whatever the molecular mechanism. In AD disorders due to HI, the mutant copy is not functional and does not interfere with the WT product, and the single functional WT copy produces too little protein to fulfill the function required by the whole organism. HI is more commonly associated with loss-of-expression alleles and ND with normally or highly expressed alleles, but rare examples have been reported of HI with normal levels of the mutant protein (32), and of ND with a lack of detectable mutant protein (33). Autosomal dominance by GOF defines a third category, in which the mutant protein is produced at various levels yet results in abnormally enhanced biological activity (34). We hypothesized that genes causing dominant disease through HI mechanisms are under stronger negative selection than those in which the underlying mechanism is ND or GOF, because any loss-of-expression mutation at HI loci is likely to be LOF and potentially disease-causing (35). Dominant forms of IEI have been reported to be caused by variants acting by HI (20 genes), GOF (17 genes), and ND (10 genes) (SI Appendix, supplementary files). RAC2 is the only AD gene to have been shown to be associated with two different mechanisms (ND and GOF), while TLR3 and IKZF1 are associated with two similar mechanisms (HI and ND) (36); we classified these three genes as having “unknown” modes of dominance. As predicted, the CoNeS of genes operating by HI was lower than that of ND and GOF genes (medians of −1.36, −0.554, and 0.323, respectively) (Fig. 4A). Despite the small number of genes in each group, the difference between AD by LOF and AD by GOF, as well as between HI and ND, are significant (P = 1.06 × 10−4 and 3.45 × 10−2; SI Appendix, Table S8). The difference between LOF and GOF IEI AD genes was the most significant for CoNeS, pLI, and LOEUF, the latter two metrics reflecting the strength of selection acting on heterozygotes (23, 37) (SI Appendix, Table S8). Furthermore, CoNeS, LOEUF, and RVIS are the scores that provide the most significant separation between HI and ND IEI AD genes. Accordingly, differences in negative selection scores between genes underlying AR and AD IEI were stronger when focusing on genes underlying IEI AD by LOF (SI Appendix, Table S9). Collectively, these findings indicate that the mechanism of dominance (HI, ND, or GOF) affects the levels of negative selection on human genes underlying AD IEI.
Fig. 4.
Negative selection scores of genes with different mechanisms of disease dominance. (A) The distribution of CoNeS for genes causing autosomal dominant IEI, according to mechanism of disease dominance. (B) Precision–recall curves for p(HI), CoNeS, and the best supervised model when trying to distinguish between haploinsufficient and haplosufficient (gain-of-function or dominant-negative) IEI genes. The dashed line is the maximum precision–recall curve, the dotted line the minimum precision–recall curve, and the mixed line a random classifier precision–recall curve.
CoNeS and Most Individual Scores Predict Haploinsufficiency of IEI Genes.
We attempted to predict whether a given AD IEI gene can be HI or not by using negative selection scores as predictors. All individual scores performed better at this task than P(HI), a supervised model that combines genomic, evolutionary, and functional features (38), but includes genes underlying recessive disorders as negative controls (rather than genes underlying dominant disorders by means other than HI), with CoNeS displaying the best results. We then trained supervised models by using the IEI AD genes with a unique and known mode of dominance as training set and the individual scores as features. We used support vector machines (SVMs) with Euclidian, polynomial, or radial kernels as well as random forests and k-nearest neighbors and adjusted their parameters with nested cross-validation (Fig. 4B, SI Appendix, Table S10, and Methods for details). An SVM with a polynomial kernel was the best supervised model, but was outperformed by pLI, SIS, LOEUF, and CoNeS. We tested versions of CoNeS with each of the individual selection scores removed one by one, but none of these versions improved the performance of the original CoNeS (SI Appendix, Table S11). Out of all the tested models, CoNeS is the most appropriate score to predict whether a gene that causes an AD IEI does so by HI or by another mechanism.
Disease Severity, Mode of Inheritance, and Mechanism of Dominance Independently Affect the Strength of Negative Selection on IEI Genes.
Recessive IEI tend to be more severe than dominant IEI (Fig. 3A). We therefore investigated whether dominance and disease severity affect the measured levels of negative selection on IEI genes in an independent manner. We fitted a linear regression model to all IEI autosomal genes, predicting CoNeS and using as covariates the mode of inheritance (coded as AR, AD, or both), a measure of the severity of the associated disease (coded as EOHP or LOIP), coding sequence length, and gene GC content (coded as a percentage) (SI Appendix, Tables S12 and S13). This multiple linear regression model predicted CoNeS with significant performance (P = 8.3 × 10−7). The mode of inheritance and severity predicted CoNeS better than they would predict any of the individual scores, with the exception of pLI, whereas coding sequence length and GC content did not improve predictive performance (P = 0.63 and 0.45, respectively). GC content significantly improved the prediction of evoTol, lofTool, and RVIS (P = 3.5 × 10−2, 3.0 × 10−3, and 4.5 × 10−4, respectively), these relationships probably being due to computational artifacts in these methods (see SI Appendix, Table S12 for a full comparison). Coding sequence length did not improve the prediction score for any method. When IEI AD genes were considered separately, the mechanism of dominance improved prediction even further for most methods (ANOVA, P = 8.12 × 10−5 for CoNeS; see SI Appendix, Tables S14 and S15 for complete results). These results demonstrate that mode of inheritance, mechanism of dominance, and clinical severity of IEI are three independent determinants of the strength of negative selection, as measured by CoNeS, on disease-causing genes.
The Mode and Mechanism of Inheritance Affect the Levels of Negative Selection on Genes Underlying IEND.
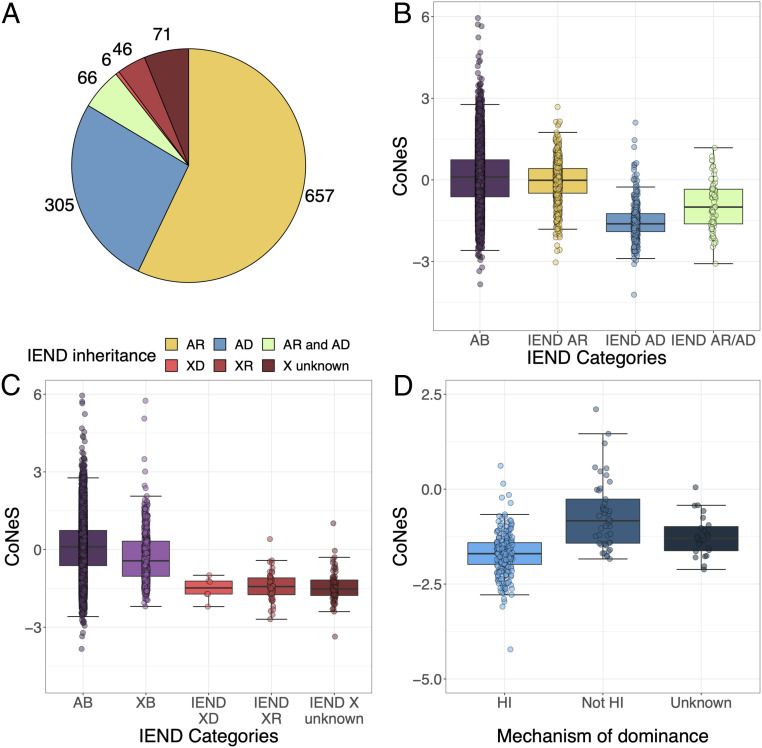
To validate the results obtained for IEI genes, we compared the CoNeS values of genes underlying IEND, a group of severe, early-onset diseases with well-characterized genetic etiologies (39). We classified the 1,140 genes underlying IEND according to their mode of inheritance: 650 genes underlie AR IEND, 303 underlie AD IEND, and 65 underlie both AR and AD IEND, whereas 46 and 6 X-linked genes underlie XR and XD IEND, respectively, and 70 X-linked IEND genes have an unknown mode of inheritance (Fig. 5A). Consistent with the results obtained for IEI, negative selection scores were significantly lower for AD IEND genes (median, −1.61) than AB genes (Wilcoxon one-tailed test, P = 4.54 × 10−133, and resampling test, P < 10−5) (Fig. 5B and SI Appendix, Table S16), whereas the scores for AR IEND genes were not different (median, −1.44 × 10−2; resampling-based P = 0.192; SI Appendix, Table S17). In contrast to the results obtained for IEI, AR/AD IEND genes showed stronger evidence for negative selection than AB genes (median, −1.01; Wilcoxon test, P = 4.55 × 10−14; Fig. 5B and SI Appendix, Table S18), although to a lesser degree than IEND AD genes. Stronger evidence for negative selection was also found for X-linked IEND genes, relative to XB genes (Wilcoxon one-tailed test, P = 3.39 × 10−37; Fig. 5C and SI Appendix, Table S19). We found no significant difference between X-linked genes underlying diseases with unknown modes of inheritance, recessive or dominant diseases (medians, −1.54, −1.48, and −1.43, respectively). Finally, CoNeS was lower for the 237 IEND AD acting by HI (median, −1.69), than for the 44 IEND AD genes not acting by HI (median, −0.857) (P = 1.12 × 10−13) (Fig. 5D and SI Appendix, Table S20). These results confirm that the levels of negative selection on disease-causing genes depend on the mode of inheritance and mechanism of dominance of disease.
Fig. 5.
The distribution of CoNeS for genes underlying inborn errors of neurodevelopment (IEND), according to disease mode of inheritance and mechanism of dominance. (A) The number of genes underlying IEND, according to the mode of disease inheritance. (B) The distribution of CoNeS for autosomal genes underlying IEND, according to their dominant (AD), recessive (AR), or both recessive and dominant (AR/AD) mode of inheritance, relative to autosomal background (AB) genes. (C) The distribution of CoNeS for genes on the X chromosome, relative to AB genes. (D) The distribution of CoNeS for IEND AD genes, according to the mechanism of dominance.
Negative Selection Scores Predict Autosomal Dominant and Recessive Inheritance.
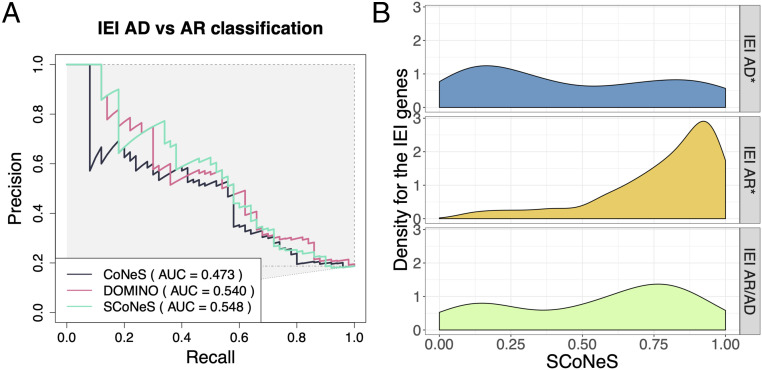
We attempted to predict whether an autosomal candidate gene is likely to underlie a dominant or recessive disease. We compared our performance with that of DOMINO, a supervised method that integrates numerous gene-level metrics that capture conservation and protein structure features in order to predict whether a gene is likely to underlie a dominant or recessive disease (40). pLI and DOMINO are the methods that perform the best on hOMIM, while CoNeS and DOMINO are those that perform the best on IEI genes (SI Appendix, Fig. S5), and DOMINO and LOEUF on IEND genes (SI Appendix, Table S21). We improved the performance of CoNeS by removing some individual scores (SI Appendix, Table S22), but its performance did not reach that of DOMINO on either hOMIM or IEND genes. We then trained a supervised model to distinguish between AR and AD genes on a dataset combining hOMIM, IEI, and IEND that included as features both DOMINO and the seven individual selection scores previously described. We evaluated its performance through leave-one-out cross-validation. Our supervised metric, which we denote as SCoNeS (supervised CoNeS), is the predicted probability of a given gene being AR. We chose a random forest classifier but also tested SVM (with Euclidian, polynomial, and radial kernels) and k-nearest neighbors (Methods and SI Appendix, Table S23). SCoNeS outperformed DOMINO as well as every other metric on IEI (Fig. 6A), IEND (SI Appendix, Fig. S6), and hOMIM genes (SI Appendix, Fig. S7) that are not part of the DOMINO training set. After DOMINO, LOEUF and pLI were the components that contributed the most to SCoNeS (SI Appendix, Fig. S8). The distribution of SCoNeS for genes underlying AR IEI and AD IEI have largely different modes (Fig. 6B, in leave-one-out for the IEI AR and IEI AD genes; see SI Appendix, Fig. S9 for the IEND genes and SI Appendix, Fig. S10 for the hOMIM genes). Only 7 out of 245 IEI AR genes are misclassified as AD (SCoNeS < 0.25), when removed from the training set (SI Appendix, supplementary file). All these genes underlie early-onset diseases with complete penetrance. By contrast, 20 out of 59 IEI AD genes are misclassified as IEI AR (SCoNeS > 0.75) when removed from the training set (SI Appendix, supplementary file). More than two-thirds of these genes (14/20) are either dominant through GOF or with an unknown mode of dominance, a quarter (5/20) are ND with incomplete penetrance, while another, CD46, is classified as HI, but has a complex mode of inheritance, with some AR cases reported in the literature (41). The false-negative rate of SCoNeS is therefore lower for genes underlying AR than AD diseases, which may be due to the heterogeneous levels of negative selection observed for genes underlying AD diseases. These results demonstrate that SCoNeS provides an improvement over state-of-the-art methods when predicting the mode of transmission of genes that cause IEI.
Fig. 6.
The performance of SCoNeS to predict disease mode of inheritance on genes underlying inborn errors of immunity (IEI). (A) The precision–recall curves for CoNeS, DOMINO, and SCoNeS when trying to distinguish between genes underlying autosomal dominant (AD) IEI and autosomal recessive (AR) IEI. SCoNeS values are calculated through leave-one-out. (B) The distribution of SCoNeS for genes underlying AD IEI, AR IEI, and AR/AD IEI. SCoNeS for genes underlying IEI AD and for genes underlying IEI AR was calculated through leave-one-out.
Discussion
With recent advances in high-throughput DNA sequencing, genes underlying inborn errors are increasingly discovered and characterized. Detailed studies of disease transmission and functional impacts of disease-causing variants provide the unique opportunity to evaluate how disease inheritance, severity, and mechanism of dominance translate into various levels of negative selection on the corresponding human protein-coding genes. We quantified the levels of selective constraints on known disease-causing genes by developing and using the CoNeS score, which combines interspecies and intraspecies measurements of negative selection. We first focused on genes responsible for IEI and then replicated our approach on genes underlying IEND. We demonstrated that CoNeS approximates the strength of negative selection acting on human genes, as it is lower for genes on the X chromosome than for those on autosomes, for loci underlying Mendelian diseases than for other genes, and at loci underlying dominant diseases than those underlying recessive diseases. Importantly, we showed that the CoNeS score was significantly lower at loci underlying recessive IEI with a high degree of clinical severity than for other genes underlying recessive IEI. This result contrasts with the nonsignificant difference observed in a previous study based on highly penetrant OMIM genes (6) but confirms a more recent study based on the Bayesian estimation of the selection coefficient of heterozygotes for HI genes (8). This discrepancy probably reflects differences in power and in the annotations of disease databases. Together, these results indicate that the effects of negative selection on genetic variation depend on both the mode of inheritance and the clinical outcome of human diseases.
We found stronger evidence for negative selection at genes underlying inborn errors that are dominant by HI, relative to genes that have other mechanisms of dominance. The significantly lower CoNeS score for HI genes than for other AD genes is not confounded by other factors such as gene size or GC content and is to be accounted for principally by the inclusion of the LOEUF and pLI statistics (SI Appendix, Table S14). pLI was originally described as “the probability of being loss-of-function intolerant” and has been used for the classification of HI genes (12). However, it was recently argued that pLI cannot be used to infer the HI status of genes directly, because it reflects only the strength of selection acting on heterozygotes (37). Here, we showed that negative selection scores are significantly different between HI and other genes, for both pLI and CoNeS. Three nonmutually exclusive mechanisms can explain the increased negative selection on HI genes: 1) diseases caused by mutations in HI genes are more severe than dominant diseases caused by mutations in other genes (i.e., their selection coefficient s is more negative); 2) heterozygotes for mutations in HI genes may have a clinical presentation that is as severe as that of homozygotes (i.e., their dominance coefficient h is higher); 3) and/or the probability of a LOF variant to affect the molecular function of the protein is higher for a HI gene, whereas most variants of GOF or ND genes are not disease causing, whether isomorphic, hypomorphic, or even LOF, because they do not create GOF or ND (35). We hypothesize that this translates quantitatively into stronger evolutionary constraints on HI genes than on other autosomal genes underlying dominant conditions.
The search for candidate genes for a specific Mendelian or non-Mendelian monogenic disease requires to study disease transmission and determine its mode of inheritance. This information is usually leveraged to increase the power of linkage analyses. We suggest that candidate genes can be further prioritized based on their mode of inheritance predicted from negative selection scores. Our study shows that, although all scores are informative for this, some metrics do perform differently. For example, lofTool and evoTol are the only scores able to distinguish hOMIM AR disease-causing genes from AB genes. One of the reasons could be their internal use of FATHMM (42), which includes known disease mutations as a training set (43) and may overfit well-known disease genes included in hOMIM. An overfitted gene score would increase error to predict the mode of inheritance of novel disease-causing genes, whose mutation patterns do not fit existing databases. CoNeS and SCoNeS show two paths to avoid overfitting: unsupervised classification for CoNeS and supervised classification for SCoNeS by leave-one-out cross-validation. Both scores perform similarly to, or better than, individual scores on the independent datasets of genes underlying IEI and IEND, indicating good performance.
Two related limitations of our study are the assumption that all mutations at a given locus cause diseases with the same severity and mode of inheritance, and that the strength of negative selection is equal all along a given gene. Several genes, such as C3 (44) and STAT1 (45), were found to be under strong negative selection (CoNeS of −1.64 and −1.84, respectively), but to be associated with several diseases of different severities, modes of inheritance, and/or incomplete penetrance. The additive effects of multiple small constraints on most of the sequence result in strong overall constraints on these genes. Conversely, a small number of genes are under weak negative selection, whereas their variants underlie severe diseases. For instance, some heterozygous variants of TCF3 (CoNeS of 0.420) underlie an AD deficiency of the E47 transcription factor (46), a very severe disease, as well as to a severe AR hypogammaglobulinemia (47). However, all reported heterozygous patients share an identical mutation in the small bHLH domain of the TCF3 gene, suggesting that there may be heterogeneity in the selective constraints on the gene. We tested this hypothesis with subRVIS (48), a domain-level version of RVIS (5) (SI Appendix, Fig. S11). Our findings confirmed that most domains of the gene were not particularly constrained (subRVIS = 83.8; i.e., 83.8% of the domains of all human proteins are under stronger constraints), while the bHLH domain was under relatively strong negative selection (subRVIS = 18.4). These examples suggest that future studies should take such heterogeneity into account and integrate local measurements of selective constraints (48, 49).
In summary, our results indicate that genes underlying inborn errors show various levels of negative selection according to disease mode of inheritance, disease severity, and mechanism of dominance, in good agreement with expectations from population genetics models. The observation of stronger negative selection acting on HI genes, relative to other genes underlying dominant diseases, calls for further theoretical and experimental testing. Our study shows that negative selection scores, including the consensus CoNeS metric, can provide valuable information to predict whether a gene is likely to underlie a recessive or dominant disease. By integrating negative selection scores with other gene-specific metrics, such as pathway centrality (50) and epigenetic marks (51), future studies based on supervised machine learning (52, 53) may help predicting the dominance and pathogenicity of candidate variants for severe disorders, ultimately facilitating the dissection of genetic etiologies of human diseases.
Methods
Gene and Disease Annotations.
The lists of hOMIM and IEI genes and their modes of inheritance were obtained from previous publications (6, 54). Each IEI gene was manually annotated for severity and (when AD) for mode of dominance (SI Appendix, supplementary file 1). The IEND gene list was assembled from the SysID reference database (39) (SI Appendix, supplementary file 1). The AB and XB gene groups included all human genes not listed in the OMIM, IEI, or IEND lists or in the list of essential mouse and human genes defined in a previous study (26).
Computation of the Scores.
EvoTol (22), lofTool (13), and SIS (24) statistics were downloaded from the corresponding publications. Values for pLI (12) and LOEUF (23) were obtained from gnomAD, version 2.1. For RVIS (5), we downloaded the values calculated with ExAC v2 from the RVIS website. For the f parameter from SNIPRE (11), we used the values calculated in a previous study (55). We unified the gene names through the checkGeneSymbols function of the HGNChelper package, version 0.8.1 (56). For each of these scores, we computed the missing values with the imputePCA function of the missMDA package, version 1.14 (57). We then used the PCA function from FactoMineR, version 1.41 (58), and the first component, which we standardized through the scale function, as the CoNeS score. In total, we computed the individual statistics and CoNeS for 18,801 genes (SI Appendix, supplementary file 1). For the calculation of subRVIS (48) for TCF3, we used the subRVIS website with the options domain-level and quantile values. We used R, version 3.5.2.
Comparison with Random Groups of Genes.
For comparisons of negative selection statistics for a test group of autosomal (or X chromosome) genes with the AB background (or XB) group, we created 100,000 groups of randomly sampled genes with a coding sequence length in the same decile of the genome-wide distribution as those of the test group. P values were estimated as the proportion of random groups with a median for negative selection statistics below that of the test group. Based on the number of random samples, the lowest nonzero P value possible is 1/100,000 = 10−5. When the proportion equaled 0, we therefore noted P < 10−5 .
Supervised Classification.
In order to build supervised classification models, we trained SVM with linear, radial, and polynomial kernels from the e1071 R package. We trained random forest with the randomForest function from the RandomForest R package. We used the knn function from the class package for k-nearest neighbors. When a gene was present in more than one dataset among IEI, IEND, and hOMIM, we annotated the gene as AD (respectively AR) if the gene was considered AD (respectively AR) in all the datasets in which it was present. Otherwise, we annotated the gene as AR/AD. We adjusted the parameters of the SVMs using an internal cross-validation loop (function tune.svm) and chose the parameters of the best model to test the performance. The performance is displayed with a precision–recall curve (function pr.curve from PRROC package).
Supplementary Material
Acknowledgments
This work was supported in part by the Rockefeller University, the St. Giles Foundation, Institut National de la Santé et de la Recherche Médicale, Paris Descartes University, the National Center for Research Resources and the National Center for Advancing Sciences, NIH Clinical and Translational Science Award program (Grant UL1TR001866), the Integrative Biology of Emerging Infectious Diseases Laboratory of Excellence (Grant ANR-10-LABX-62-IBEID), and the French National Research Agency (ANR) (Grants ANR-10-IAHU-01, ANR-15-CE17-0014). The laboratory of L.Q.-M. and E.P. is supported by the Institut Pasteur, the Collège de France, the Investissement d’Avenir program, Laboratoires d’Excellence “Milieu Intérieur” (Grant ANR-10-LABX-69-01), Fondation pour la Recherche Médicale (Grant FRM DEQ20180339214), Fondation Allianz–Institut de France, and Fondation de France. We thank members of the Laboratory of Human Genetics of Infectious Diseases for helpful discussions and critical reading.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001248118/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Loewe L., Negative selection. Nat. Educ. 1, 59 (2008). [Google Scholar]
- 2.Kacser H., Burns J. A., The molecular basis of dominance. Genetics 97, 639–666 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bustamante C. D., et al. , Natural selection on protein-coding genes in the human genome. Nature 437, 1153–1157 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Wang E. T., Kodama G., Baldi P., Moyzis R. K., Global landscape of recent inferred Darwinian selection for Homo sapiens. Proc. Natl. Acad. Sci. U.S.A. 103, 135–140 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petrovski S., Wang Q., Heinzen E. L., Allen A. S., Goldstein D. B., Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 9, e1003709 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blekhman R., et al. , Natural selection on genes that underlie human disease susceptibility. Curr. Biol. 18, 883–889 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nielsen R., et al. , Darwinian and demographic forces affecting human protein coding genes. Genome Res. 19, 838–849 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cassa C. A., et al. , Estimating the selective effects of heterozygous protein-truncating variants from human exome data. Nat. Genet. 49, 806–810 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kimura M., Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature 267, 275–276 (1977). [DOI] [PubMed] [Google Scholar]
- 10.Li W. H., Wu C. I., Luo C. C., A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol. Biol. Evol. 2, 150–174 (1985). [DOI] [PubMed] [Google Scholar]
- 11.Eilertson K. E., Booth J. G., Bustamante C. D., SnIPRE: Selection inference using a Poisson random effects model. PLoS Comput. Biol. 8, e1002806 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lek M. et al.; Exome Aggregation Consortium , Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fadista J., Oskolkov N., Hansson O., Groop L., LoFtool: A gene intolerance score based on loss-of-function variants in 60 706 individuals. Bioinformatics 33, 471–474 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Smith N. G., Eyre-Walker A., Human disease genes: Patterns and predictions. Gene 318, 169–175 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Huang H., et al. , Evolutionary conservation and selection of human disease gene orthologs in the rat and mouse genomes. Genome Biol. 5, R47 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondrashov F. A., Ogurtsov A. Y., Kondrashov A. S., Bioinformatical assay of human gene morbidity. Nucleic Acids Res. 32, 1731–1737 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas P. D., Kejariwal A., Coding single-nucleotide polymorphisms associated with complex vs. Mendelian disease: Evolutionary evidence for differences in molecular effects. Proc. Natl. Acad. Sci. U.S.A. 101, 15398–15403 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tangye S. G., et al. , Human inborn errors of immunity: 2019 update on the classification from the International Union of Immunological Societies Expert Committee. J. Clin. Immunol. 40, 24–64 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Casanova J. L., Abel L., Primary immunodeficiencies: A field in its infancy. Science 317, 617–619 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Casanova J. L., Abel L., Human genetics of infectious diseases: Unique insights into immunological redundancy. Semin. Immunol. 36, 1–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Notarangelo L. D., Bacchetta R., Casanova J. L., Su H. C., Human inborn errors of immunity: An expanding universe. Sci. Immunol. 5, eabb1662 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rackham O. J., Shihab H. A., Johnson M. R., Petretto E., EvoTol: A protein-sequence based evolutionary intolerance framework for disease-gene prioritization. Nucleic Acids Res. 43, e33 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karczewski K. J. et al., Genome Aggregation Database Consortium , The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443, 10.1038/s41586-020-2308-7, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aggarwala V., Voight B. F., An expanded sequence context model broadly explains variability in polymorphism levels across the human genome. Nat. Genet. 48, 349–355 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Itan Y., et al. , The human gene damage index as a gene-level approach to prioritizing exome variants. Proc. Natl. Acad. Sci. U.S.A. 112, 13615–13620 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartha I., di Iulio J., Venter J. C., Telenti A., Human gene essentiality. Nat. Rev. Genet. 19, 51–62 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Hildebrandt J., et al. , Characterization of CSF2RA mutation related juvenile pulmonary alveolar proteinosis. Orphanet J. Rare Dis. 9, 171 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phadnis N., Fry J. D., Widespread correlations between dominance and homozygous effects of mutations: Implications for theories of dominance. Genetics 171, 385–392 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agrawal A. F., Whitlock M. C., Inferences about the distribution of dominance drawn from yeast gene knockout data. Genetics 187, 553–566 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huber C. D., Durvasula A., Hancock A. M., Lohmueller K. E., Gene expression drives the evolution of dominance. Nat. Commun. 9, 2750 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rieux-Laucat F., Casanova J. L., Immunology. Autoimmunity by haploinsufficiency. Science 345, 1560–1561 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Lim H. K., et al. , Severe influenza pneumonitis in children with inherited TLR3 deficiency. J. Exp. Med. 216, 2038–2056 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pérez de Diego R., et al. , Human TRAF3 adaptor molecule deficiency leads to impaired Toll-like receptor 3 response and susceptibility to herpes simplex encephalitis. Immunity 33, 400–411 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boisson B., Quartier P., Casanova J. L., Immunological loss-of-function due to genetic gain-of-function in humans: Autosomal dominance of the third kind. Curr. Opin. Immunol. 32, 90–105 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson A. F., Nguyen H. T., Veitia R. A., Causes and effects of haploinsufficiency. Biol. Rev. Camb. Philos. Soc. 94, 1774–1785 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Zhang S. Y., et al. , TLR3 deficiency in patients with herpes simplex encephalitis. Science 317, 1522–1527 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Fuller Z. L., Berg J. J., Mostafavi H., Sella G., Przeworski M., Measuring intolerance to mutation in human genetics. Nat. Genet. 51, 772–776 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang N., Lee I., Marcotte E. M., Hurles M. E., Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 6, e1001154 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kochinke K., et al. , Systematic phenomics analysis deconvolutes genes mutated in intellectual disability into biologically coherent modules. Am. J. Hum. Genet. 98, 149–164 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quinodoz M., et al. , DOMINO: Using machine learning to predict genes associated with dominant disorders. Am. J. Hum. Genet. 101, 623–629 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richards A., et al. , Mutations in human complement regulator, membrane cofactor protein (CD46), predispose to development of familial hemolytic uremic syndrome. Proc. Natl. Acad. Sci. U.S.A. 100, 12966–12971 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greif P. A., et al. , Evolution of cytogenetically normal acute myeloid leukemia during therapy and relapse: An exome sequencing study of 50 patients. Clin. Cancer Res. 24, 1716–1726 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Stenson P. D., et al. , The human gene mutation database: Towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum. Genet. 136, 665–677 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sfyroera G., et al. , Rare loss-of-function mutation in complement component C3 provides insight into molecular and pathophysiological determinants of complement activity. J. Immunol. 194, 3305–3316 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chapgier A., et al. , A partial form of recessive STAT1 deficiency in humans. J. Clin. Invest. 119, 1502–1514 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boisson B., et al. , A recurrent dominant negative E47 mutation causes agammaglobulinemia and BCR(−) B cells. J. Clin. Invest. 123, 4781–4785 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ben-Ali M., et al. , Homozygous transcription factor 3 gene (TCF3) mutation is associated with severe hypogammaglobulinemia and B-cell acute lymphoblastic leukemia. J. Allergy Clin. Immunol. 140, 1191–1194.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Gussow A. B., Petrovski S., Wang Q., Allen A. S., Goldstein D. B., The intolerance to functional genetic variation of protein domains predicts the localization of pathogenic mutations within genes. Genome Biol. 17, 9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davydov E. V., et al. , Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLOS Comput. Biol. 6, e1001025 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Asif M., Martiniano H. F. M. C. M., Vicente A. M., Couto F. M., Identifying disease genes using machine learning and gene functional similarities, assessed through Gene Ontology. PLoS One 13, e0208626 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kalayci S., et al. , ImmuneRegulation: A web-based tool for identifying human immune regulatory elements. Nucleic Acids Res. 47, W142–W150 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boudellioua I., Kulmanov M., Schofield P. N., Gkoutos G. V., Hoehndorf R., DeepPVP: Phenotype-based prioritization of causative variants using deep learning. BMC Bioinformatics 20, 65 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rentzsch P., Witten D., Cooper G. M., Shendure J., Kircher M., CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 47, D886–D894 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Picard C., et al. , International Union of Immunological Societies: 2017 primary immunodeficiency diseases committee report on inborn errors of immunity. J. Clin. Immunol. 38, 96–128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deschamps M., et al. , Genomic signatures of selective pressures and introgression from archaic hominins at human innate immunity genes. Am. J. Hum. Genet. 98, 5–21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oh S., Abdelnabi J., Al-Dulaimi R., Davis S., Riester M., Waldron L., HGNChelper: Identification and correction of invalid gene symbols for human and mouse. 10.1101/2020.09.16.300632 (18 September 2020). [DOI] [PMC free article] [PubMed]
- 57.Josse J., Husson F., missMDA: A package for handling missing values in multivariate data analysis. J. Stat. Softw. 70, (2016). [Google Scholar]
- 58.Le S., Josse J., Husson F., FactoMineR: An R package for multivariate analysis. J. Stat. Softw. 25, 1–18 (2008). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.