Significance
Eczema, hives, and allergic contact dermatitis affect millions of people every year, particularly children. Many skin allergies are caused by exaggerated production of type-2 cytokines, such as IL-4 and IL-13, by T helper (Th) cells in response to various substances when they contact the skin. Here, we illustrate that maladaptive Th2 responses to a cutaneous allergen can be recapitulated in a transgenic mouse model in which a subset of CD326+ dendritic cells (DCs) that migrate from the skin to the draining lymph nodes are artificially depleted. This depletion perturbs the balance of DC subsets in lymph nodes, which biases T cell responses toward type-2 inflammation. Thus, preservation of lymph node CD326+ DCs is essential to prevent or attenuate skin allergies.
Keywords: plasmacytoid DC, allergy, skin, contact hypersensitivity, Th2
Abstract
Plasmacytoid dendritic cells (pDCs) specialize in the production of type I IFN (IFN-I). pDCs can be depleted in vivo by injecting diphtheria toxin (DT) in a mouse in which pDCs express a diphtheria toxin receptor (DTR) transgene driven by the human CLEC4C promoter. This promoter is enriched for binding sites for TCF4, a transcription factor that promotes pDC differentiation and expression of pDC markers, including CLEC4C. Here, we found that injection of DT in CLEC4C-DTR+ mice markedly augmented Th2-dependent skin inflammation in a model of contact hypersensitivity (CHS) induced by the hapten fluorescein isothiocyanate. Unexpectedly, this biased Th2 response was independent of reduced IFN-I accompanying pDC depletion. In fact, DT treatment altered the representation of conventional dendritic cells (cDCs) in the skin-draining lymph nodes during the sensitization phase of CHS; there were fewer Th1-priming CD326+ CD103+ cDC1 and more Th2-priming CD11b+ cDC2. Single-cell RNA-sequencing of CLEC4C-DTR+ cDCs revealed that CD326+ DCs, like pDCs, expressed DTR and were depleted together with pDCs by DT treatment. Since CD326+ DCs did not express Tcf4, DTR expression might be driven by yet-undefined transcription factors activating the CLEC4C promoter. These results demonstrate that altered DC representation in the skin-draining lymph nodes during sensitization to allergens can cause Th2-driven CHS.
Plasmacytoid dendritic cells (pDCs) are a subset of bone-marrow–derived DCs that produce substantial amounts of type I interferon (IFN-I; i.e., IFN-α, IFN-β) upon recognition of viral and microbial nucleic acids through toll-like receptor (TLR) 7 and 9 (1–4). Secreted IFN-I binds to the IFN-I receptor (IFNAR) on pDCs, completing an autocrine loop that sustains IFN-I production (5). pDC secretion of IFN-I peaks at early time points during viral infections and mediates immediate containment of viral replication, but wanes and becomes less important later during infection as pDCs undergo cell-intrinsic IFNAR-mediated apoptosis (6) and other host cells become more dominant producers of IFN-I. In addition to a transient IFN-I response against viral infections, pDCs have been proposed to contribute to naive CD4+T cell differentiation. Since pDCs express major histocompatibility complex (MHC) class II molecules as well as the costimulatory molecules CD40, CD80, and CD86, they can present antigens to CD4+ T cells, although not as efficiently as conventional DCs (cDCs) (7). pDCs have been reported to prime Th1 (8), Th2 (9), or tolerogenic T cells (10–14), depending on the cell surface and soluble signals that they provide to CD4+ T cells together with the MHC-peptide complex.
To further address the impact of pDCs in CD4+ T cell responses in vivo, we used a transgenic mouse that expresses the diphtheria toxin receptor (DTR) under the control of the promoter of the human CLEC4C gene, which encodes the pDC-specific C-type lectin CLEC4C (also known as BDCA2). The DNA sequence of this promoter contains four binding sites for E2-2, the transcription factor encoded by the TCF4 gene that drives pDC development (15). Thus, DTR is preferentially expressed in pDCs, and injection of DT depletes pDCs (16). In this study, we found that injection of DT in CLEC4C-DTR+ mice markedly augmented Th2-dependent skin inflammation in a model of contact hypersensitivity (CHS) induced by the hapten fluorescein isothiocyanate (FITC). However, the exaggerated Th2 response was not due to a diminished IFN-I or IFN-λ accompanying pDC depletion. In fact, DT treatment impacted the priming phase of FITC CHS by reducing not only pDCs, but also a subset of cDCs expressing CD103 and CD326, which migrate from the inflamed skin into the draining lymph nodes (DLN). Single-cell RNA-sequencing (scRNA-seq) of DCs isolated from the skin DLN of Zbtb46GFP cDC reporter mice crossed with CLEC4C-DTR+ mice revealed that DTR was expressed not only on pDCs but also on migrating CD326+ cDCs, which were acutely depleted together with pDCs after DT injection, creating a DC environment prone to prime Th2. These results suggest that a defective representation of CD103+ CD326+ migrating DCs during CD4+ T cell priming facilitates the development of skin allergic pathologies.
Results
DT Treatment of CLEC4C-DTR+ Mice Exacerbates FITC-Induced CHS.
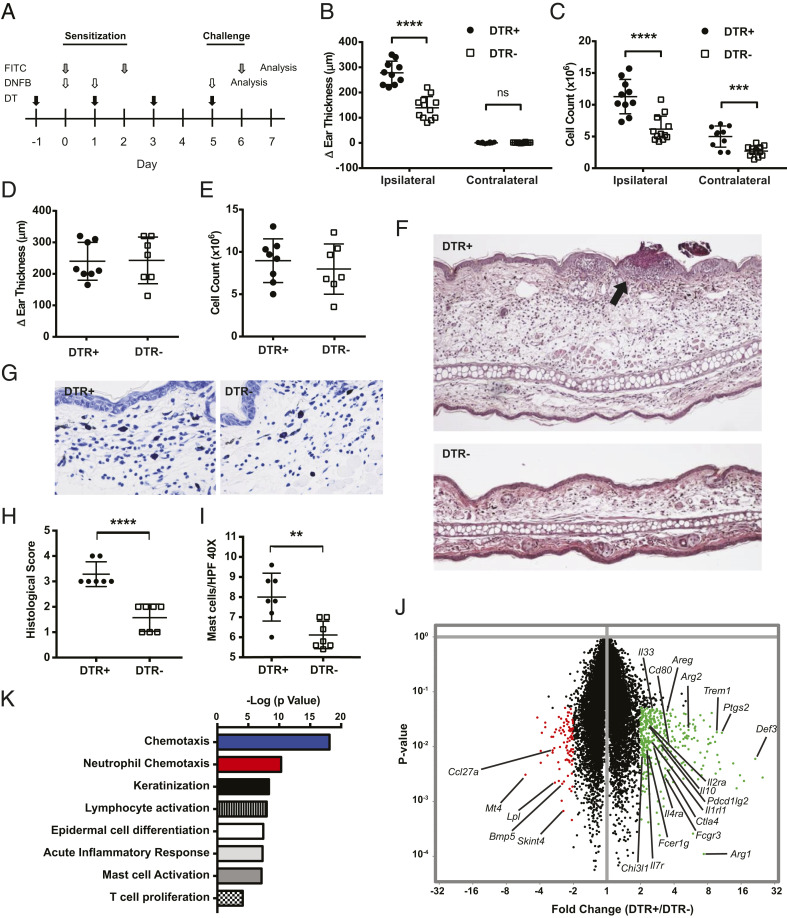
This study was initially prompted by the observation that CLEC4C-DTR+ mice treated with DT developed noticeable skin inflammation when felt-tip markers were repeatedly used to mark their tails (SI Appendix, Fig. S1A), suggesting that pDCs might control skin CHS. To directly test the impact of pDCs on CHS, we performed CHS assays in DT-treated CLEC4C-DTR+ mice and transgene-negative littermate controls (DTR–) using FITC and 2,4-dinitrofluorobenzene (DNFB) as haptens. These CHS models trigger Th2 and Th1 CD4+ T cell responses, respectively (17, 18), allowing us to determine whether pDCs uniquely control Th1- or Th2-mediated skin pathology. Mice were primed by applying haptens on the abdomen and then challenged on the ears 5 to 6 d later to induce skin inflammation (Fig. 1A). Mice were administered DT every other day throughout the experiment, resulting in >90% pDC depletion in the spleen and skin DLN in CLEC4C-DTR+ mice, while no depletion was observed in DTR– littermates (SI Appendix, Fig. S1B). In the FITC model, CLEC4C-DTR+ mice exhibited increased ear swelling and DLN cellularity 24 h post challenge compared to DTR– mice (Fig. 1 B and C). However, in the DNFB model, we found no difference between CLEC4C-DTR+ and DTR– mice in either ear swelling or DLN cellularity post challenge (Fig. 1 D and E). Histologic examination of FITC challenged ears from CLEC4C-DTR+ mice revealed markedly enhanced dermal edema, granulocytic and lymphocytic infiltration, and epidermal crusting (Fig. 1 F and H). Toluidine blue staining showed mast cell hyperplasia in CLEC4C-DTR+ mice (Fig. 1 G and I).
Fig. 1.
DT treatment of CLEC4C-DTR+ mice enhances FITC-induced CHS. (A) CLEC4C-DTR+ mice and DTR– littermate controls were subjected to two different CHS models. For FITC CHS, FITC was applied to the shaved abdomen on day 0 and day 2 (sensitization), and FITC was applied to the ear on day 6 (challenge). For DNFB CHS, 20 μL of 0.5% DNFB was applied to the shaved abdomen on day 0 and 1 (sensitization), and 20 μL of 0.2% DNFB was applied to the ear on day 5 (challenge). DT was administered on day –1, 1, 3, and 5 in each model. (B) Ear swelling measured 24 h post challenge in the FITC-treated ear (ipsilateral) and solvent-treated ear (contralateral). Data represent three pooled experiments (n = 10 CLEC4C-DTR+, n = 12 DTR– littermates). (C) Cellularity of the ipsilateral ear DLN and contralateral DLN 24 h post challenge with FITC. Numbers reflect cell yield from three pooled superior cervical nodes digested with collagenase D. Data represent three pooled experiments (n = 10 CLEC4C-DTR+, n = 12 DTR– littermates). (D) Ear swelling measured 24 h post challenge in the DNFB model. Data represent three pooled experiments (n = 8 CLEC4C-DTR+, n = 7 DTR– littermates). (E) Ipsilateral DLN cellularity 24 h post challenge in the DNFB model. Numbers reflect cell yield from three pooled superior cervical nodes digested with collagenase D. Data represent three pooled experiments (n = 8 CLEC4C-DTR+, n = 7 DTR– littermates). (F) Hematoxylin and eosin histology of the ear pinna 24 h post challenge with FITC at 10× magnification. Epidermal crusting is indicated by the black arrow. Representative of two experiments (n = 7 CLEC4C-DTR+, n = 7 DTR– littermates). (G) Toluidine blue staining 24 h post challenge with FITC at 40× magnification. Data are representative of two experiments (n = 7 CLEC4C-DTR+, n = 7 DTR– littermates). (H) Histological scoring of ear pinna 24 h post challenge based on dermal edema, leukocyte infiltration, and epidermal crusting. Data representative of two experiments (n = 7 CLEC4C-DTR+, n = 7 DTR– littermates). (I) Quantification of mast cells per high-power field (HPF) at 40× via toluidine blue staining of the FITC-treated ear 24 h post challenge. Data represent two experiments, and each data point represents an average of 15 high-power-fields per sample (n = 7 CLEC4C-DTR+, n = 7 DTR– littermates). (J) Volcano plot comparing gene expression by microarray in ear skin 1 d post FITC challenge. Data represent fold change and P value calculated from three RNA samples per group; each sample is RNA derived from skin of one mouse (n = 3 CLEC4C-DTR+, n = 3 DTR– littermates). Transcripts in green or red indicate P < 0.05 and fold change ≥ 2. (K) Most highly significant pathways derived from Gene Set Enrichment Analysis (Broad) after input of transcripts significantly increased in DT-treated mice (FC > 2; P < 0.05). **P < 0.01, ***P < 0.001, ****P < 0.0001; statistics by unpaired two-tailed Student’s t test. Data in B–E, H, and I are mean ± SD, and each symbol represents an individual mouse.
To further characterize the enhanced pathology in CLEC4C-DTR+ mice, we analyzed the transcriptome of a bulk preparation of the ear skin after FITC challenge. Microarray analysis of the FITC-challenged skin showed evidence of an enhanced type 2 immune response in CLEC4C-DTR+ mice, with elevation of Il33, IL4ra, Fcer1g, Arg1, Arg2, and Ptgs2 messenger RNAs (mRNAs) (Fig. 1J). Pathway analysis showed evidence of enhanced lymphocyte and granulocyte infiltration and keratinocyte and mast cell activation (Fig. 1K). Concordantly, flow cytometric analysis of the skin confirmed an increased amount of infiltrating CD45+ cells, dominated by granulocytes in CLEC4C-DTR+ mice (SI Appendix, Fig. S1 C and D). The percentages of neutrophils among CD45+ cells were significantly elevated in CLEC4C-DTR+ mice with an additional trend toward an increased percentage of eosinophils (SI Appendix, Fig. S1D). CD11c+ FITC+ cells were increased in the DLN post challenge, reflecting increased inflammation in the skin (SI Appendix, Fig. S1E). Together, these data demonstrated that DT treatment of CLEC4C-DTR+ mice exacerbated CHS reaction to the FITC hapten and Th2-driven skin pathology, but did not affect the DNFB-triggered Th1-mediated disease.
Since FITC-induced contact hypersensitivity depends on keratinocyte release of thymic stromal lymphopoietin (TSLP) (19), we tested whether CLEC4C-DTR+ mice treated with DT were susceptible to another model of TSLP-dependent atopic dermatitis, which is driven by the vitamin D analog calcipotriol (MC903) (SI Appendix, Fig. S1F). Upon application to the skin for 21 d, calcipotriol acts to relieve tonic transcriptional repression on the TSLP locus, resulting in robust epidermal TSLP production that drives type 2 immunopathology (20). In this model, DT-treated CLEC4C-DTR+ mice had exacerbated gross pathology, increased ear swelling, and robust pathological changes to the skin including epidermal thickening and architectural distortion compared to DTR– mice (SI Appendix, Fig. S1 G–I). These data corroborated that DT treatment in CLEC4C-DTR+ mice exacerbates type 2 allergic pathologies of the skin induced by allergens and/or adjuvants that elicit TSLP.
DT Treatment of CLEC4C-DTR+ Mice Biases CD4+ T Cell Response to FITC toward Th2.
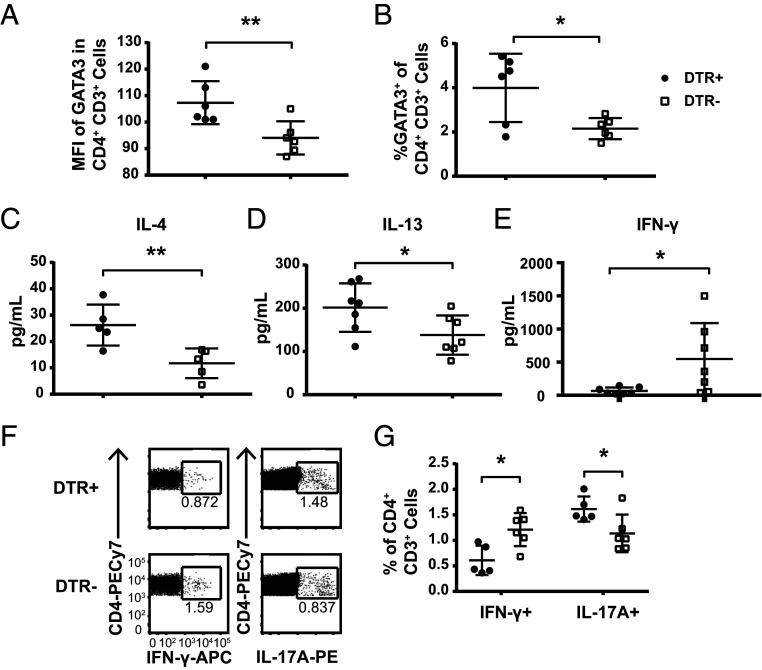
We sought to directly evaluate the effector T cell response underlying the exacerbated FITC-driven CHS. There was a significantly higher level of GATA3 expression within the entire CD4+ T cell compartment in the DLN of CLEC4C-DTR+ mice compared to DTR– controls on day 7 (Fig. 2A), as well as a higher percentage of GATA3+ cells within CD4+ T cells (Fig. 2B), suggesting a bias toward Th2 responses. Assessment of cytokine production ex vivo in the DLN revealed that whole lymph node cultures from CLEC4C-DTR+ mice produced greater amounts of IL-4 and IL-13, while those from DTR– controls produced more IFN-γ (Fig. 2 C–E). We further evaluated cytokine production on a per-cell basis by intracellular cytokine staining, which revealed that a higher percentage of CD4+ T cells from DTR– mice made IFN-γ (Fig. 2 F and G), while the IFN-γ production from the CD8+ T cells was similar in both groups (SI Appendix, Fig. S2 A and B). We also noted that a slightly higher percentage of CD4+ and CD8+ T cells from CLEC4C-DTR+ mice produced IL-17A (Fig. 2 F and G and SI Appendix, Fig. S2 A and B). Type 2 cytokines were difficult to detect by intracellular staining, most likely due to limitations in the reagents currently available.
Fig. 2.
DT treatment of CLEC4C-DTR+ mice during FITC CHS increases Th2 polarization and ILC2 in DLN. Cells from the ipsilateral superior cervical lymph nodes were isolated from CLEC4C-DTR+ and littermate control mice 24 h post FITC challenge. (A) Median fluorescence intensity of GATA3 in CD4+CD3+ cells and (B) percentage of GATA3+ of CD4+CD3+ cells. Data show one experiment, representative of two individual experiments (depicted: n = 6 CLEC4C-DTR+, n = 6 DTR– littermates; total: n = 12 CLEC4C-DTR+, n = 12 DTR– littermates). (C–E) Cells from DLN were stimulated with Phorbol 12-myristate 13-acetate (PMA)/ionomycin for 12 h at a concentration of 1 × 106 cells/mL, and supernatants were analyzed for cytokine concentrations by CBA (IFN-γ and IL-4) or ELISA (IL-13). Data in C were pooled from two experiments (n = 5 CLEC4C-DTR+, n = 5 DTR– littermates). Data in D were pooled from two experiments (n = 7 CLEC4C-DTR+, n = 7 DTR– littermates). Data in E were pooled from two experiments (n = 7 CLEC4C-DTR+, n = 7 DTR– littermates). Each data point reflects the mean of two technical replicates from the same sample. Each sample is derived from one mouse. (F) Representative staining of intracellular cytokine staining of CD4+ cells after stimulation with PMA/ionomycin for 6 h with Brefeldin A and Monensin in culture for the final 4 h. (G) Quantification of the percentage of IFN-γ+ or IL-17A+ cells within the CD4+CD3+ population. Data are representative of one of two experiments (depicted: n = 5 CLEC4C-DTR+, n = 6 DTR– littermates; total: n = 10 CLEC4C-DTR+, n = 10 DTR– littermates). *P < 0.05, **P < 0.01; statistics by unpaired two-tailed Student’s t test. Data in A–E and G are mean SD, and each symbol represents an individual mouse.
Since many studies have indicated that pDCs induce tolerance to allergens and other antigens by inducing Tregs through a variety of mechanisms (14, 21–24), we assessed the frequency and number of Tregs in the DLN at day 7 after ear challenge. Unexpectedly, CLEC4C-DTR+ mice had enhanced rather than reduced frequencies and numbers of Tregs compared to DTR– controls (SI Appendix, Fig. S2 C–F). Moreover, a higher percentage of Tregs expressed the ST2 subunit of the IL-33R (SI Appendix, Fig. S2F), which triggers an increased expression of Foxp3 (25, 26). Thus, enhanced CHS in CLEC4C-DTR+ mice was not due to reduced immune regulation. Overall, these data indicate that DT treatment of CLEC4C-DTR+ mice biases T cell responses toward Th2 while reducing Th1 immunity.
Exacerbated FITC CHS Occurs Independently of IFN-I and IFN-λ.
It has been shown that IFN-I and pDC-produced IFN-I promote Th1 responses (8, 27). Recent studies have further demonstrated that IFN-I inhibits proliferation and survival of type 2 innate lymphoid cells (ILC2s), thereby tempering skin allergy (28, 29). Moreover, pDC-produced IFN-I was shown to limit ILC2s and airway hyperreactivity in lung (30). Accordingly, we found that DT-treated CLEC4C-DTR+ mice had more ILC2s (numbers and percentages) in the DLN after FITC challenge than DTR– controls (SI Appendix, Fig. S3 A–C). Thus, we hypothesized that enhanced CHS in CLEC4C-DTR+ mice was due to a deficit of IFN-I resulting in Th1 reduction and/or ILC2 increase. To test this hypothesis, we assessed FITC CHS in Ifnar−/− and wild-type (WT) mice. Like CLEC4C-DTR+ mice, Ifnar−/− mice had increased numbers and percentages of ILC2s in the DLN after FITC challenge (SI Appendix, Fig. S3 A–C), but ear swelling was similar in Ifnar−/− and WT mice (SI Appendix, Fig. S3D). Thus, we concluded that reduction of pDC-derived IFN-I and consequent increase of ILC2s is not sufficient to explain exacerbated FITC CHS in CLEC4C-DTR+ mice. Because pDCs also produce IFN-λ (31), we evaluated the impact of IFN-λ on FITC CHS by examining mice that lack the receptor for IFN-λ, IL-28R. Il28ra−/− and WT mice were equally susceptible to FITC CHS (SI Appendix, Fig. S3D), excluding the possibility that pDCs limit type 2 responses through IFN-λ. Altogether, these results indicate that exacerbated Th2 response to FITC is most likely independent of the defect of interferons that accompanies pDC depletion.
DT Treatment of CLEC4C-DTR+ Mice Alters Lymph Node cDC Subsets in Steady State and during CHS.
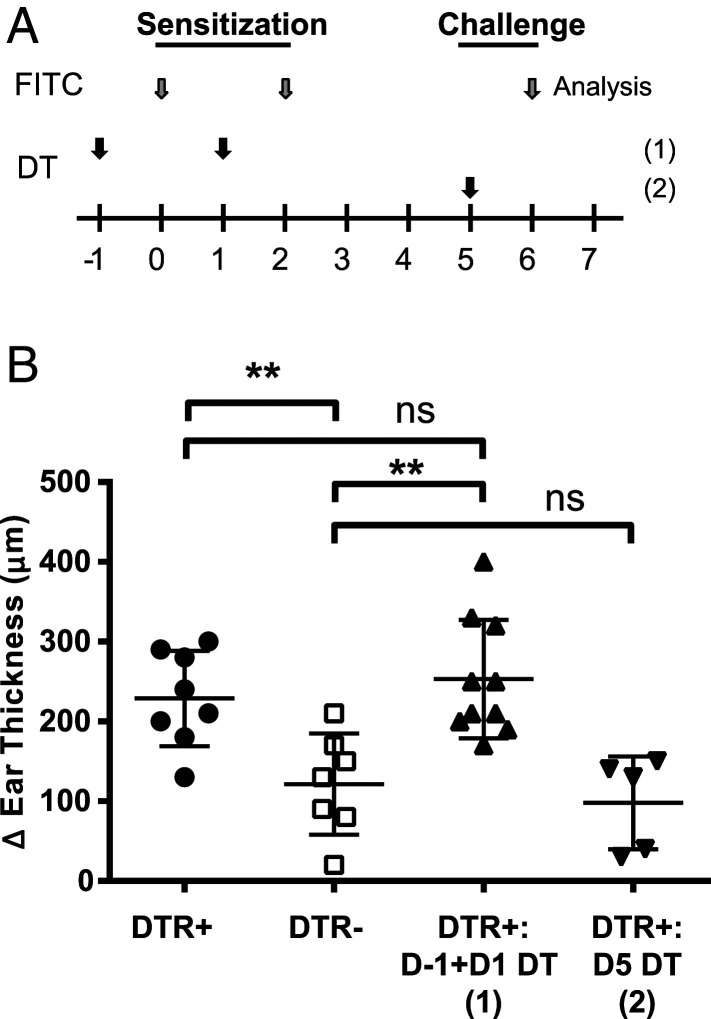
The FITC-induced model of CHS encompasses priming and challenge phases. To evaluate the impact of DT treatment on each phase, we treated mice with either two doses of DT on days –1 and 1 prior to FITC sensitization on the abdomen or with a single dose of DT the day prior to FITC challenge on the ear (Fig. 3A). DT treatment during the sensitization phase was sufficient to exacerbate ear swelling at challenge (Fig. 3B). We concluded that DT treatment in CLEC4C-DTR+ mice promotes the priming of the Th2 response.
Fig. 3.
DT administration during the FITC CHS sensitization phase results in exacerbated pathology. (A) Mice were administered a modified FITC CHS protocol with DT administered on day –1 and 1 [D–1 + D1 depletion (1)] or administered on day 5 [D5 depletion (2)]. (B) Ear swelling was measured 24 h post challenge in the FITC-treated ear. Data are pooled from three experiments (n = 8 CLEC4C-DTR+, n = 7 DTR– littermates, n = 10 D–1 and D1 depleted CLEC4C-DTR+, n = 5 D5-depleted CLEC4C-DTR+). **P < 0.01; statistics by Student’s t test. Data in B are mean ± SD, and each symbol represents an individual mouse; ns, nonsignificant.
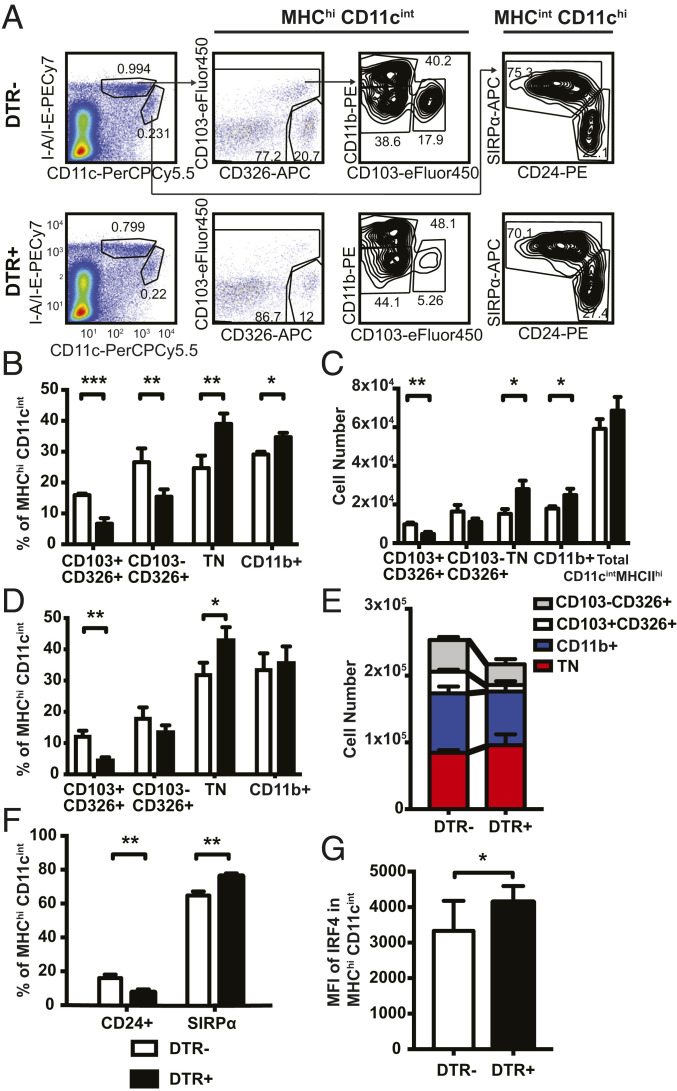
Given the established role of cDCs in priming T helper responses (32), we next evaluated whether DT treatment affected cDCs in addition to pDCs in skin DLN. We characterized the CD11chiMHCIIint resident and CD11cintMHCIIhi migratory populations in the skin DLN in DT-treated CLEC4C-DTR+ and DTR– mice at steady state and 48 h after FITC priming, when cDC migration from skin peaks (SI Appendix, Fig. S4). Within the resident cDCs, two subsets could be identified using CD24 and Sirpα, corresponding to resident cDC1 and cDC2, respectively. Within the migratory cDCs we could define four separate populations: CD103–CD326+ Langerhans cells (LC), CD103+CD326+ migrating cDC1, CD11b+ cDC2, and triple negative (TN) cDC2, as previously described (33, 34) (Fig. 4A). In the steady state, DT treatment caused a reduction in the frequency and number of CD103+CD326+ cDC1 and CD103–CD326+ LCs, as well as an increase in CD11b+ and TN cDC2 within migratory cDCs of skin DLN (Fig. 4 B and C); no effect was noted on the resident cDC populations. After FITC sensitization, DT caused a similar subset skewing in CLEC4C-DTR+ mice with significantly more migratory TN cDC2 and fewer CD326+CD103+ cDC1 in the inguinal and axillary lymph nodes (Fig. 4 D and E).
Fig. 4.
DT-treated mice have more migratory cDC2 and fewer migratory cDC1 in skin DLN. (A) Representative flow plot for migratory and resident DC populations in the axillary and inguinal lymph nodes 48 h post FITC sensitization. Cells pregated on live FSChi SSChi. (B) Frequency and (C) number of migratory cDC subsets in DT-treated mice at steady state (no FITC). Data are representative of one of two separate experiments (depicted: n = 3 CLEC4C-DTR+, n = 4 DTR– littermates, total: n = 6 CLEC4C-DTR+, n = 7 DTR– littermates). (D and F) Percentages and (E) numbers of indicated migratory DC populations. Data represent one of three experiments (depicted: n = 3 CLEC4C-DTR+, n = 3 DTR– littermates; total: n = 12 CLEC4C-DTR+, n = 12 DTR– littermates). (G) Mean fluorescence intensity (MFI) of IRF4 in migratory cDCs (MHChiCD11cint) in inguinal and axillary lymph nodes. Data are pooled from two separate experiments (n = 9 CLEC4C-DTR+, n = 9 DTR– littermates). *P < 0.05, **P < 0.01, ***P < 0.001; statistics by unpaired two-tailed Student’s t test. Data in B–G are mean ± SD.
This result was confirmed with an additional gating strategy using Sirpα and CD24 within CD11cintMHCIIhi cDCs to distinguish migratory cDC2 and cDC1, respectively. Here, CLEC4C-DTR+ mice had a significantly greater percentage of migratory Sirpα+ cDC2 and a corresponding decrease in the percentage of CD24+ cDC1 (Fig. 4F). Furthermore, we observed an increase in the IRF4 expression within the entire migratory cDC population in CLEC4C-DTR+ mice, supporting the skewing in the cDC1:cDC2 ratio (Fig. 4G). We conclude that DT treatment of CLEC4C-DTR+ mice not only depletes pDCs but also reduces the ratio between migratory cDC1 and cDC2. This altered cDC1:cDC2 ratio may have a major impact on susceptibility to FITC CHS, as suggested by a previous report showing that lack of cDC1 in Batf3−/− mice is associated with increased Th2 responses and IL-4 production during FITC CHS (33). In contrast, Batf3−/− mice do not exhibit exacerbated skin pathology in the Th1-driven DNFB model of CHS (35), similar to DT-treated CLEC4C-DTR+ mice.
scRNA-seq Reveals DTR Expression on pDCs and cDC Subsets in Skin DLN of CLEC4C-DTR+ Mice.
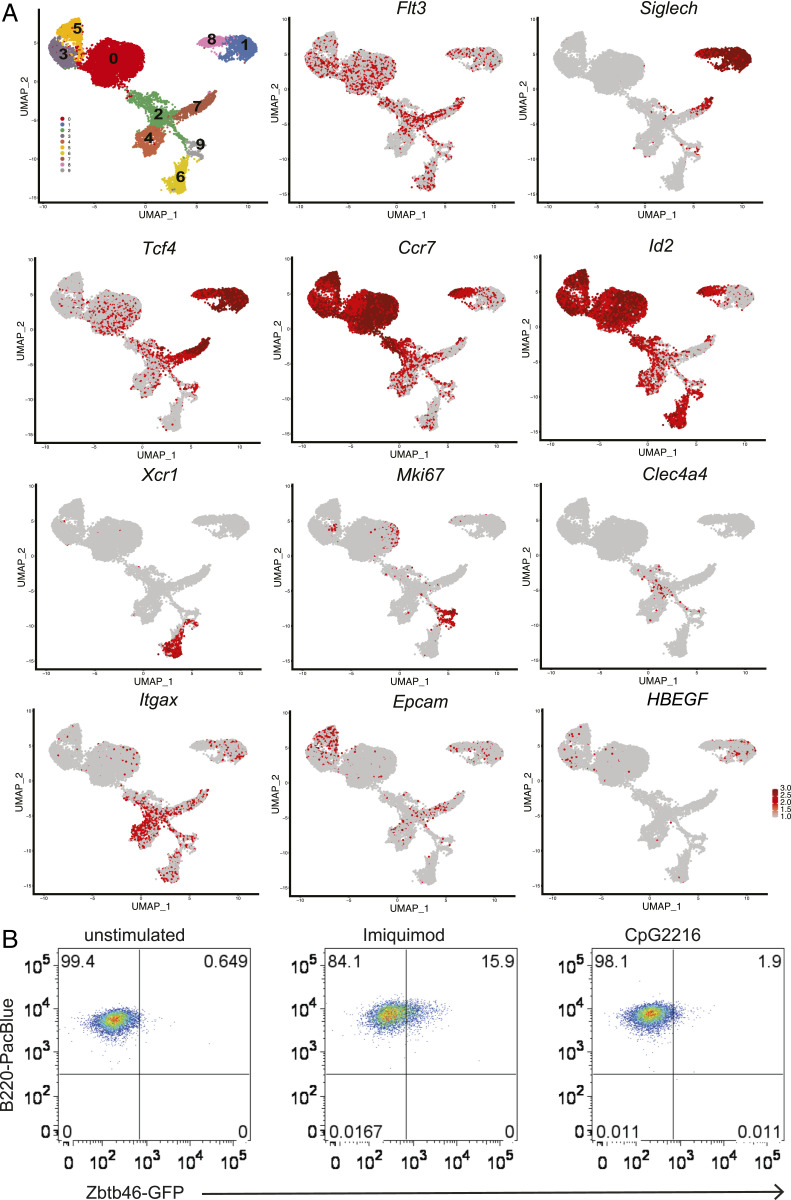
To identify a mechanism that could explain why DT depletion in CLEC4C-DTR+ mice led to an altered cDC1:cDC2 ratio in skin DLN at steady state and during CHS, we resorted to an unbiased approach and performed scRNA-seq. To facilitate the isolation of cDCs from skin DLN, CLEC4C-DTR+ transgenic mice were crossed with the Zbtb46GFP reporter mice, in which all cDCs express GFP (36, 37). We sorted SIGLECH+GFP– cells, which encompass pDCs, as well as SIGLECH+GFPlow/int and SIGLECH–GFPhigh cells, which capture cDCs. Unsupervised clustering by Uniform Manifold Approximation and Projection (UMAP) identified 10 distinct populations at steady state (Fig. 5A). All populations expressed high levels of Flt3 mRNA, with the exception of cluster 4, which corresponded to macrophages expressing high levels of Csf1r, Adgre1 (encoding the scavenger receptor F4/80), and the macrophage-associated transcription factor Mafb (Fig. 5A and SI Appendix, Fig. S5). Three distinct clusters expressed the pDC markers Siglech and Tcf4. The most abundant cluster 1 had low levels of costimulatory and adhesion molecules and lacked Zbtb46. The smaller cluster 8 expressed high levels of Cd40, Cd86, Icam1, and Ccr7, indicative of activation, and, unexpectedly, the cDC markers Zbtb46 and Id2. To validate that pDC activation also induces some cDC markers, we sorted SIGLECH+GFP– pDCs from spleens of Zbtb46GFP mice and activated them in vitro with different stimuli. Approximately 10 to 15% of pDCs cultured with the TLR7 agonist imiquimod acquired intermediate GFP expression (Fig. 5B), whereas CpG-A ODN2216 had no effect (Fig. 5B). Thus, pDCs acquire de novo expression of Zbtb46 in response to some activation stimuli. The third Siglech+Tcf4+ cluster 7, which segregated away from the pDC clusters 1 and 8, expressed pDC markers such as Cd209a, Lifr, and Lag3, as well as unique markers such as Cd200r1, Cd209e, Ccr1, and Ngfr, and some macrophage/DC2-related markers such as Cx3cr1, Mgl2, CD300a, Ms4a6c, and Csf1r. This cluster also expressed higher levels of Zbtb46 than cluster 8. Based on this expression profile, we determined that cluster 7 corresponded to the previously described CX3CR1+ noncanonical DCs (38, 39) and the recently appreciated pDC-like cells (40, 41). Clusters 6 and 9 represented resident cDC1, as shown by expression of Xcr1, Clec9a, Itgae, and Wdyf4. Cluster 9 further expressed several cyclins and Mki67, indicating a proliferating fraction within resident cDC1. Cluster 2 represented resident cDC2 marked by expression of Clec4a4. Clusters 0, 3, and 5 represented migrating cDCs, as indicated by the low expression of Itgax (encoding CD11c). Cluster 0 corresponded to migrating cDC2, as suggested by expression of Ccl22, Ccl17, and Nrp2, although neither of the two transcription factors required for cDC2 commitment, Irf4 and Klf4, was more expressed in this subset compared to the remaining clusters (SI Appendix, Fig. S5). Cells in the right portion of cluster 0 expressed Mki67, suggesting active proliferation. Clusters 3 and 5 expressed markers of dermal cDC1 and LCs, such as Epcam (encoding CD326), Cd207, and Itgb8 (SI Appendix, Fig. S5), which encodes a subunit of the avb8 integrin that facilitates activation of latent TGFβ, a cytokine known to drive LC development.
Fig. 5.
scRNA-seq analysis of pDCs and cDCs isolated from skin DLN reveals diverse lineages of resident and migrating DCs. (A) SIGLECH+GFP–, SIGLECH+GFPlow/int and SIGLECH–GFPhigh cells from inguinal and axillary skin DLN of Zbtb46GFP x CLEC4C-DTR+ mice were submitted for scRNA-seq via the 10× Genomics platform. UMAP plots of scRNA-seq data show DC subsets and distinguishing markers. Pooled cells from two individual mice are shown. (B) Induction of GFP expression in SIGLECH+GFP– pDCs sorted from Zbtb46GFP spleens after stimulation with imiquimod; CpG-A ODN2216 had no effect. One experiment representative of two is depicted.
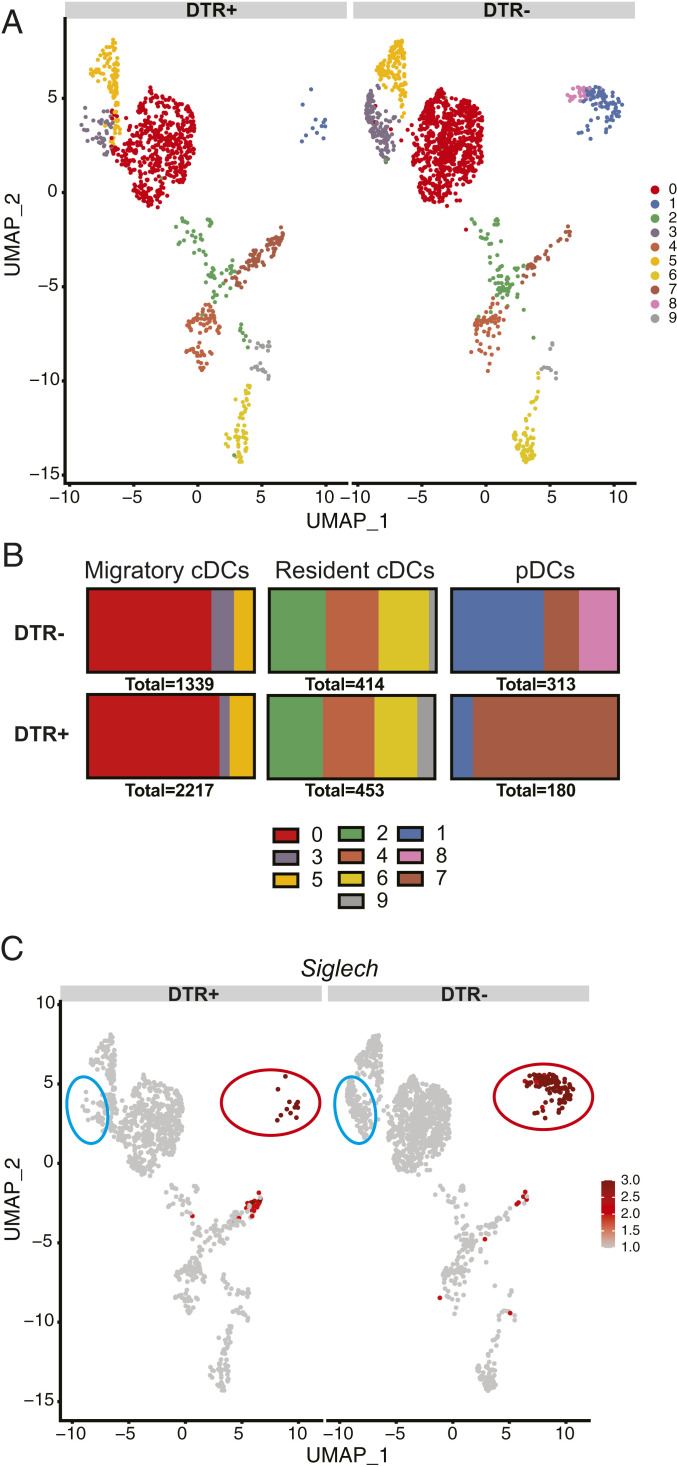
By including the nucleotide sequence of simian HBEGF encoding DTR in the mouse sequence database used for aligning mRNA sequences, we were able to determine the expression of DTR in each cluster (Fig. 5A). As expected, we detected HBEGF mRNA in pDC clusters 1 and 8. However, we did not detect HBEGF in cluster 7 encompassing CX3CR1+ noncanonical DCs and pDC-like cells, despite high expression of Tcf4 in this cluster. Importantly, we detected unexpected HBEGF expression in clusters 3 and 5 of Epcam+Cd207+ migrating cDCs (Fig. 5A), although these cells did not express detectable Tcf4. Consistent with HBEGF expression in migrating cDC subsets, scRNA-seq of DCs isolated from skin DLN after injection of DT in CLEC4C-DTR+ and DTR– mice after FITC priming showed a dramatic reduction in clusters 1 and 8 as well as significant attrition of cluster 3 in CLEC4C-DTR+ mice (Fig. 6 A–C). Thus, the decrease in CD326+CD103+/− cDCs observed in CLEC4C-DTR+ mice treated with DT during the steady state and FITC CHS (Fig. 4) may result from the direct depletion of these cells.
Fig. 6.
DT treatment of CLEC4C-DTR+ mice induces loss of pDCs and a subset of migratory CD326+ cells in the FITC model of CHS. (A) scRNA-seq of DC subsets in CLEC4C-DTR+ and DTR– mice after DT administration and FITC sensitization showing clusters identified in Fig. 5A. (B) Quantification of the number of cells in each of the subsets shown. (C) Expression of Siglech in DT-treated and untreated mice by scRNA-seq. A circle indicates mainly affected subsets: red, pDCs; blue, CD326+ (CD103+/−) cDCs.
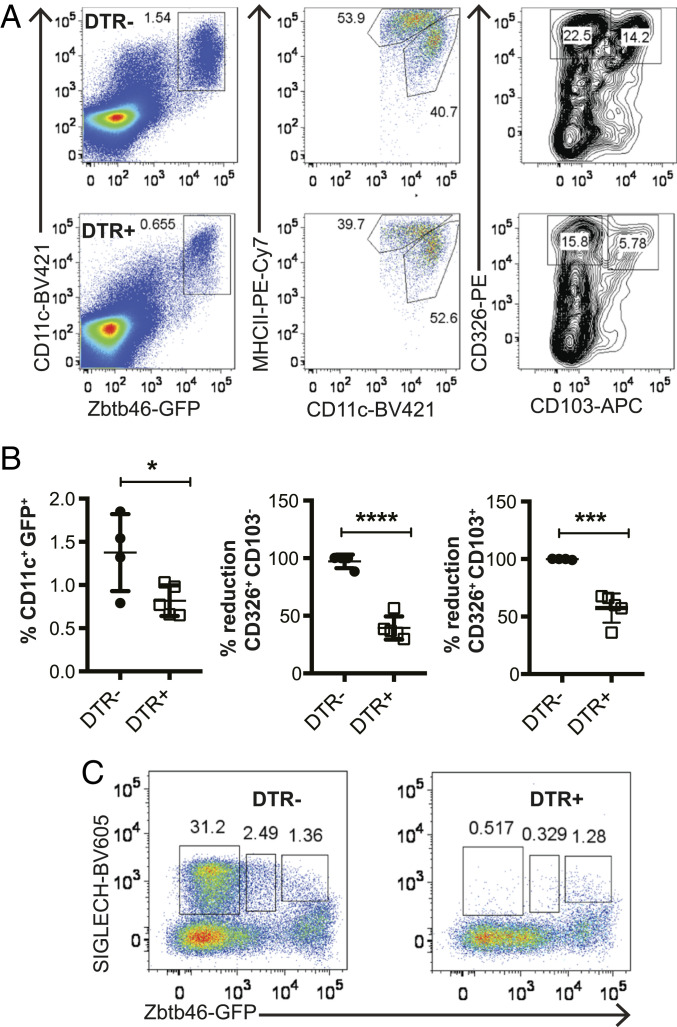
To further corroborate this conclusion, we injected Zbtb46GFP x CLEC4C-DTR+ mice and their DTR– littermates with DT and evaluated the percentages of cDCs and pDCs in skin DLN 24 h later by flow cytometry. Within migratory CD11clowMHChighGFP+ cDCs, both GFP+CD326+CD103– and GFP+CD326+CD103+ cells were reduced by about 50% (Fig. 7 A and B). Within SIGLECH+ pDC subsets, DT treatment caused a reduction of GFP–SIGLECH+ pDCs as expected, as well as GFPlowSIGLECH+ pDCs, which corresponded to activated pDCs. In contrast, DT treatment had no effect on GFPhighSIGLECH+ cells, which corresponded to cluster 7, the CX3CR1+ noncanonical DCs or pDC-like cells (Fig. 7C). These results were consistent with those previously observed in DT-treated CLEC4C-DTR+ and DTR– mice not crossed with Zbtb46GFP at steady state and during CHS (Fig. 4).
Fig. 7.
DT treatment of Zbtb46GFP CLEC4C-DTR+ mice depletes migrating CD103+CD326– and CD103+CD326+ cDCs in skin DLN. (A) Representative dot plots of DC subsets in Zbtb46GFP CLEC4C-DTR– and Zbtb46GFP DTR+ mice 24 h after DT treatment based on expression of GFP, CD11c, MHCII, CD326, and CD103. (B) Quantification of percentages of total cDCs (CD11c+GFPhigh) and percentages of reduction of CD103+CD326+ and CD103+CD326– DCs. Percentages of reduction were calculated because of cage-dependent variations in migrating cDC subsets. CLEC4C-DTR+: n = 4, DTR– n = 5. (C) Depletion of SIGLECH+GFP– and SIGLECH+GFPint cells but not SIGLECH+GPFhigh cells in Zbtb46GFPCLEC4C-DTR+ versus Zbtb46GFPCLEC4C-DTR– mice treated with DT. One experiment representative of two is depicted.
Discussion
This study was prompted by the observation that DT treatment of CLEC4C-DTR+ mice selectively exacerbated Th2-driven CHS induced by FITC but not Th1-driven CHS induced by DNFB. DT treatment acted in the sensitization phase of FITC CHS, causing increased frequency and numbers of GATA3+ CD4+ T cells along with fewer Th1 cells. Since DT treatment of CLEC4C-DTR+ mice depletes pDCs, we initially investigated the role of these cells in FITC CHS. pDCs secrete IFN-I, which promotes Th1 (27), and inhibit ILC2s (28, 29). Accordingly, DT-treated mice harbored fewer IFN-γ–producing CD4+ T cells, as well as increased ILC2s, as did IFNAR-deficient mice. However, global lack of IFN-I signaling did not exacerbate FITC CHS. PDCs have also been reported to attenuate Th2 responses in various disease models by inducing Tregs. Yet, Treg numbers actually increased in DT-treated mice during FITC CHS, suggesting that the exacerbated Th2 responses were not due to defective Treg differentiation.
In searching for a mechanism facilitating Th2 responses beyond pDC depletion, we noted that DT treatment also altered the representation of distinct cDC subsets that migrate from the skin and prime T helper cells. Skin DLN from mice treated with DT during the sensitization phase had markedly reduced numbers of migratory CD103+ cDCs and CD326+ cDCs, which induce Th1 responses (42, 43), along with relatively increased numbers of migratory CD11b+ and TN cDCs that induce Th2 and Th17 responses (19, 44–47). Remarkably, scRNA-seq revealed that CLEC4C-DTR+ mice expressed DTR not only in pDCs, but also in subsets of migrating CD326+ DCs. Thus, in addition to pDCs, DT directly depletes skin DLN CD326+ DCs, which are known to include mainly CD103+CD207+ dermal cDCs and some CD207high CD11bint CD103– LCs (48). CD326+ DCs may impact T cell polarization by providing a source of soluble factors, such as IL-12, that inhibit Th2 polarization, while facilitating Th1 differentiation. Thus, depletion of CD326+ DCs may minimize IL-12 production in the DLN. A Th1 polarizing function of CD103+CD326+ dermal DCs would be consistent with that of CD103+ cDCs recently reported in a tumor model (49). Alternatively, depletion of CD326+ DCs may lead to relative overrepresentation of dermal CD326–/low CD103–/low CD11b–/low TN DCs, which have been shown to be highly responsive to TSLP and to induce Th2 immune responses (19). Whether concomitant depletion of pDCs and CD326+ DCs in skin DLN contributes to the Th2 bias and FITC contact atopy remains to be established.
Our scRNA-seq analysis has revealed an unexpected heterogeneity of pDCs in the skin DLN. We found three subsets of cells expressing the pDC markers Tcf4 and Siglech. The major subset corresponded to prototypic pDCs. A small subset expressed Ccr7, Cd40, and Cd86, which are indicative of activation; interestingly, this subset also expressed Zbtb46 and Id2, two markers of cDCs, suggesting that Zbtb46 and Id2 can be up-regulated by pDCs upon activation, at least in DLN. Accordingly, resting GFP– pDCs sorted from the spleen of Zbtb46GFP reporter mice acquired GFP expression after in vitro activation with imiquimod. It remains to be established whether these pDCs convert into cDCs. A third Siglech+Tcf4+ subset corresponded to the previously described CX3CR1+ noncanonical DCs (38, 39) and the recently appreciated pDC-like cells (40, 41), which may be related populations. These cells reportedly share many features characteristic of pDCs but are also transcriptionally similar to cDC2 (2, 38, 39), which is corroborated by our scRNA-seq data. As suggested by recent studies, canonical pDCs and CX3CR1+ noncanonical DCs may have different progenitors (40, 41), but share similar transcriptional programs.
The expression of DTR on CD326+ cDCs of CLEC4C-DTR+ mice detected by scRNA-seq was surprising. The promoter of the human CLEC4C gene was originally chosen to drive DTR expression in transgenic mice because CLEC4C, although found only in humans, is a highly selective marker for pDCs (16). In contrast, SIGLECH, which identifies pDCs in mouse, is also expressed in some macrophages. Previous studies showed that human CLEC4C promoter is enriched for the DNA motif that binds E2-2, a transcription factor that drives the development of pDCs (15). Moreover, DT treatment of CLEC4C-DTR+ mice effectively depleted pDCs in mouse spleen, suggesting that CLEC4C promoter can also effectively recruit mouse E2-2 (16). However, the current observation that DTR is also expressed in CD326+ cDCs of the DLN which do not express Tcf4 (encoding E2-2) suggests that the CLEC4C promoter contains binding sites for yet-unknown transcription factors that can drive DTR expression in CD326+ cDCs independently of E2-2, at least in skin DLN. Conversely, we noticed that DT treatment did not deplete pDC-like/CX3CR1+ noncanonical DCs despite the fact that these cells express Tcf4, suggesting that other cofactors may control the ability of E2-2 to drive specific pDC expression of DTR.
In conclusion, our data suggest that a derailed balance of cDCs in skin DLN may contribute to subsequent maladaptive Th2 responses to cutaneous antigens, resulting in excessive type 2 inflammation. Thus, preservation of cDC1:cDC2 ratio in the T cell area of the lymph nodes at early stages of sensitization is essential to prevent or attenuate atopic dermatitis.
Materials and Methods
Experimental details on animals, protocols to induce CHS, cell preparations, antibody staining for flow cytometry and sorting, cell cultures and stimulations, enzyme-linked immunosorbent assay (ELISA) and cytometric bead array (CBA) analyses, microarray data analyses, scRNA-seq analyses, and statistical analyses for this study are described in detail in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Patrick Rodrigues and Gary Grajales Reyes for helpful discussions. The Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine conducted microarray experiments. All flow cytometry work was conducted in the Flow Cytometry and Fluorescence Activated Cell Sorting Core in the Department of Pathology and Immunology at Washington University School of Medicine. H.L.M. was supported by Grant F30DK112508 from National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
Competing interest statement: M.K.S., a former post-doc of the Colonna laboratory, is now an employee of Janssen Research & Development.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021364118/-/DCSupplemental.
Data Availability Statement.
RNA seq and scRNA-seq datasets generated during the current studies (GSE160261 and GSE161604) are available in the Gene Expression Omnibus repository (50, 51).
References
- 1.Swiecki M., Colonna M., The multifaceted biology of plasmacytoid dendritic cells. Nat. Rev. Immunol. 15, 471–485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reizis B., Plasmacytoid dendritic cells: Development, regulation, and function. Immunity 50, 37–50 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilliet M., Cao W., Liu Y. J., Plasmacytoid dendritic cells: Sensing nucleic acids in viral infection and autoimmune diseases. Nat. Rev. Immunol. 8, 594–606 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Kawai T., Akira S., Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34, 637–650 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Asselin-Paturel C., et al. , Type I interferon dependence of plasmacytoid dendritic cell activation and migration. J. Exp. Med. 201, 1157–1167 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swiecki M., et al. , Type I interferon negatively controls plasmacytoid dendritic cell numbers in vivo. J. Exp. Med. 208, 2367–2374 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Villadangos J. A., Young L., Antigen-presentation properties of plasmacytoid dendritic cells. Immunity 29, 352–361 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Cella M., Facchetti F., Lanzavecchia A., Colonna M., Plasmacytoid dendritic cells activated by influenza virus and CD40L drive a potent TH1 polarization. Nat. Immunol. 1, 305–310 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Rissoan M. C., et al. , Reciprocal control of T helper cell and dendritic cell differentiation. Science 283, 1183–1186 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Sisirak V., et al. , Plasmacytoid dendritic cells deficient in IFNα production promote the amplification of FOXP3+ regulatory T cells and are associated with poor prognosis in breast cancer patients. OncoImmunology 2, e22338 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vermi W., Soncini M., Melocchi L., Sozzani S., Facchetti F., Plasmacytoid dendritic cells and cancer. J. Leukoc. Biol. 90, 681–690 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Zou W., et al. , Stromal-derived factor-1 in human tumors recruits and alters the function of plasmacytoid precursor dendritic cells. Nat. Med. 7, 1339–1346 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Hadeiba H., et al. , CCR9 expression defines tolerogenic plasmacytoid dendritic cells able to suppress acute graft-versus-host disease. Nat. Immunol. 9, 1253–1260 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Heer H. J., et al. , Essential role of lung plasmacytoid dendritic cells in preventing asthmatic reactions to harmless inhaled antigen. J. Exp. Med. 200, 89–98 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cisse B., et al. , Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell 135, 37–48 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swiecki M., Gilfillan S., Vermi W., Wang Y., Colonna M., Plasmacytoid dendritic cell ablation impacts early interferon responses and antiviral NK and CD8(+) T cell accrual. Immunity 33, 955–966 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaplan D. H., Igyártó B. Z., Gaspari A. A., Early immune events in the induction of allergic contact dermatitis. Nat. Rev. Immunol. 12, 114–124 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honda T., Egawa G., Grabbe S., Kabashima K., Update of immune events in the murine contact hypersensitivity model: Toward the understanding of allergic contact dermatitis. J. Invest. Dermatol. 133, 303–315 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Ochiai S., et al. , CD326(lo)CD103(lo)CD11b(lo) dermal dendritic cells are activated by thymic stromal lymphopoietin during contact sensitization in mice. J. Immunol. 193, 2504–2511 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Li M., et al. , Topical vitamin D3 and low-calcemic analogs induce thymic stromal lymphopoietin in mouse keratinocytes and trigger an atopic dermatitis. Proc. Natl. Acad. Sci. U.S.A. 103, 11736–11741 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munn D. H., et al. , Expression of indoleamine 2,3-dioxygenase by plasmacytoid dendritic cells in tumor-draining lymph nodes. J. Clin. Invest. 114, 280–290 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito T., et al. , Plasmacytoid dendritic cells prime IL-10-producing T regulatory cells by inducible costimulator ligand. J. Exp. Med. 204, 105–115 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanabuchi S., et al. , Thymic stromal lymphopoietin-activated plasmacytoid dendritic cells induce the generation of FOXP3+ regulatory T cells in human thymus. J. Immunol. 184, 2999–3007 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maazi H., Lam J., Lombardi V., Akbari O., Role of plasmacytoid dendritic cell subsets in allergic asthma. Allergy 68, 695–701 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schiering C., et al. , The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature 513, 564–568 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wohlfert E. A., et al. , GATA3 controls Foxp3+ regulatory T cell fate during inflammation in mice. J. Clin. Invest. 121, 4503–4515 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen K. B., et al. , Critical role for STAT4 activation by type 1 interferons in the interferon-γ response to viral infection. Science 297, 2063–2066 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Duerr C. U., et al. , Type I interferon restricts type 2 immunopathology through the regulation of group 2 innate lymphoid cells. Nat. Immunol. 17, 65–75 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moro K., et al. , Interferon and IL-27 antagonize the function of group 2 innate lymphoid cells and type 2 innate immune responses. Nat. Immunol. 17, 76–86 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Thio C. L. P., Lai A. C. Y., Chi P. Y., Webster G., Chang Y. J., Toll-like receptor 9-dependent interferon production prevents group 2 innate lymphoid cell-driven airway hyperreactivity. J. Allergy Clin. Immunol. 144, 682–697.e9 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Coccia E. M., et al. , Viral infection and Toll-like receptor agonists induce a differential expression of type I and lambda interferons in human plasmacytoid and monocyte-derived dendritic cells. Eur. J. Immunol. 34, 796–805 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Merad M., Sathe P., Helft J., Miller J., Mortha A., The dendritic cell lineage: Ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol. 31, 563–604 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Connor L. M., et al. , Th2 responses are primed by skin dendritic cells with distinct transcriptional profiles. J. Exp. Med. 214, 125–142 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kashem S. W., Kaplan D. H., Isolation of murine skin resident and migratory dendritic cells via enzymatic digestion. Curr. Protoc. Immunol. 121, e45 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edelson B. T., et al. , Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha+ conventional dendritic cells. J. Exp. Med. 207, 823–836 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meredith M. M., et al. , Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. J. Exp. Med. 209, 1153–1165 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Satpathy A. T., et al. , Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J. Exp. Med. 209, 1135–1152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bar-On L., et al. , CX3CR1+ CD8α+ dendritic cells are a steady-state population related to plasmacytoid dendritic cells. Proc. Natl. Acad. Sci. U.S.A. 107, 14745–14750 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lau C. M., et al. , Leukemia-associated activating mutation of Flt3 expands dendritic cells and alters T cell responses. J. Exp. Med. 213, 415–431 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dress R. J., et al. , Plasmacytoid dendritic cells develop from Ly6D+ lymphoid progenitors distinct from the myeloid lineage. Nat. Immunol. 20, 852–864 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Rodrigues P. F., et al. , Distinct progenitor lineages contribute to the heterogeneity of plasmacytoid dendritic cells. Nat. Immunol. 19, 711–722 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martínez-López M., Iborra S., Conde-Garrosa R., Sancho D., Batf3-dependent CD103+ dendritic cells are major producers of IL-12 that drive local Th1 immunity against Leishmania major infection in mice. Eur. J. Immunol. 45, 119–129 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mashayekhi M., et al. , CD8α(+) dendritic cells are the critical source of interleukin-12 that controls acute infection by Toxoplasma gondii tachyzoites. Immunity 35, 249–259 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao Y., et al. , Control of T helper 2 responses by transcription factor IRF4-dependent dendritic cells. Immunity 39, 722–732 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Persson E. K., et al. , IRF4 transcription-factor-dependent CD103(+)CD11b(+) dendritic cells drive mucosal T helper 17 cell differentiation. Immunity 38, 958–969 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Schlitzer A., et al. , IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity 38, 970–983 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams J. W., et al. , Transcription factor IRF4 drives dendritic cells to promote Th2 differentiation. Nat. Commun. 4, 2990 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henri S., et al. , CD207+ CD103+ dermal dendritic cells cross-present keratinocyte-derived antigens irrespective of the presence of Langerhans cells. J. Exp. Med. 207, 189–206 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferris S. T., et al. , cDC1 prime and are licensed by CD4+ T cells to induce anti-tumour immunity. Nature 584, 624–629 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller H., et al. , Expression data from FITC challenged skin of CLEC4C-DTR mice vs. littermate controls treated with diptheria toxin. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE160261. Deposited 27 October 2020.
- 51.Miller H., et al. , scRNA-seq analysis of dendritic cells in skin draining lymph nodes of CLEC4C-DTR mice treated with FITC hapten. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE161604. Deposited 17 November 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Miller H., et al. , Expression data from FITC challenged skin of CLEC4C-DTR mice vs. littermate controls treated with diptheria toxin. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE160261. Deposited 27 October 2020.
- Miller H., et al. , scRNA-seq analysis of dendritic cells in skin draining lymph nodes of CLEC4C-DTR mice treated with FITC hapten. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE161604. Deposited 17 November 2020.
Supplementary Materials
Data Availability Statement
RNA seq and scRNA-seq datasets generated during the current studies (GSE160261 and GSE161604) are available in the Gene Expression Omnibus repository (50, 51).