Significance
Platinum resistance remains as a major issue in the therapy for many types of cancer. However, the mechanisms of resistance have not been fully elucidated. ACTL6A gene is frequently amplified in several types of cancer such as lung squamous cell carcinoma, ovarian cancer, and esophageal cancer. ACTL6A is a subunit shared by multiple complexes, including SWI/SNF, INO80, and NuA4/TIP60. We unveil a new role for ACTL6A in repairing cisplatin-induced DNA damage, providing a novel mechanism for cisplatin resistance. We also show that the action of ACTL6A in the repair of cisplatin-induced DNA lesions is through the SWI/SNF remodeling complex. Furthermore, we demonstrate that an HDAC inhibitor can abolish cisplatin resistance caused by ACTL6A overexpression.
Keywords: ACTL6A, DNA repair, cisplatin resistance, SWI/SNF
Abstract
Cisplatin is a mainstay of systemic therapy for a variety of cancers, such as lung cancer, head and neck cancer, and ovarian cancer. However, resistance to cisplatin represents one of the most significant barriers for patient outcome improvement. Actin-like 6A (ACTL6A) is a component of several chromatin remodeling complexes, including SWI/SNF, NuA4/TIP60 histone acetylase, and INO80. Amplification of ACTL6A gene is often seen in lung squamous cell carcinoma, ovarian cancer, and esophageal cancer, but its significance remains to be fully determined. Here we identify ACTL6A overexpression as a novel cause for platinum resistance. High levels of ACTL6A are associated with chemoresistance in several types of human cancer. We show that overexpression of ACTL6A leads to increased repair of cisplatin-DNA adducts and resistance to cisplatin treatment. In contrast, depletion of ACTL6A inhibits the repair of cisplatin-induced DNA lesions, and increases cisplatin sensitivity in cisplatin-resistant ovarian cancer cells. The regulation of repair by ACTL6A is mediated through the SWI/SNF chromatin remodeling complex. Treatment with a histone deacetylase inhibitor can reverse the effect of ACTL6A overexpression on the repair of cisplatin-induced DNA damage and render cancer cells more sensitive to cisplatin treatment in a xenograft mouse model. Taken together, our study uncovers a novel role for ACTL6A in platinum resistance, and provides evidence supporting the feasibility of using HDAC inhibitors for platinum resistant tumors.
Actin-like 6A (ACTL6A), also known as BAF53A, is an accessory subunit of SWI/SNF-like BAF (Brg1/Brm-associated factor) chromatin remodeling complexes and is crucial for the maintenance of stem or progenitor cells during mammalian embryonic development (1–3). ACTL6A is also a subunit of the INO80 chromatin remodeling complex (4) and the NuA4/TIP60 acetyltransferase complex (3). ACTL6A encodes a family member of evolutionally conserved actin-related proteins (Arps) that contain an actin fold for the binding and hydrolysis of adenosine triphosphate (ATP) (5). These Arps are involved in very diverse biological processes, and can be grouped as either cytoskeleton associated (Arp1-3 and 10) or chromatin associated (Arp4-9). Cytoskeleton-associated Arps regulate the assembly and function of the actin and microtubule cytoskeletons. Chromatin-associated Arps (such as Arp4, a yeast homolog of ACTL6A) regulate the structure and function of chromatin. Prior studies have shown that ACTL6A is involved in transcriptional regulation and chromatin remodeling (4, 6, 7). ACTL6A gene is frequently amplified in squamous cell carcinoma of the lung, head, and neck, ovarian and cervical cancers, etc. According to The Cancer Genome Atlas (TCGA) PanCancer Atlas, amplification of ACTL6A gene is found in 37.37% of lung squamous cell carcinoma, 19.52% of ovarian cancer, and 17.58% of esophageal cancer. Recently, the oncogenic role of ACTL6A in tumor progression has been demonstrated by several different groups. ACTL6A can promote epithelial–mesenchymal transition and metastasis or invasion in hepatocellular carcinoma, colon cancer, osteosarcoma, and glioma cells (8–11). The researchers from Dr. Ellisen’s group also found that ACTL6A is coamplified with TP63 in squamous cell carcinoma to drive a YAP-dependent regenerative proliferation, and leads to a poor prognosis in cancer patients (12). However, whether ACTL6A has any effect on the response to chemotherapy has not yet been investigated.
Cisplatin is one of the common chemotherapeutic drugs and is widely used to treat many solid tumors, such as ovarian cancer, testicular cancer, cervical cancer, bladder cancer, lung cancer, head and neck cancer, etc. Despite its effectiveness in a wide spectrum of cancers, many patients either have intrinsic resistance or eventually develop resistance to cisplatin, which presents a major challenge for cisplatin-based anticancer therapy (13). Resistance to cisplatin can be attributed to three molecular mechanisms, including altered accumulation of drug in cells, cytosolic inactivation of drug and altered DNA damage response with enhanced DNA repair process. Among these mechanisms, enhanced DNA repair is the most important cause of cisplatin resistance (14).
Cisplatin causes DNA intrastrand and interstrand cross-links (ICLs) by forming cisplatin-DNA adducts. Intrastrand cross-links are removed by the nucleotide excision repair (NER) mechanism (15). Repair of DNA ICLs involves proteins in NER, Fanconi anemia (FA), homologous recombination (HR), structure-specific endonucleases, and gap-filling translesion DNA synthesis (TLS) (16). Overexpression of some of the NER repair proteins has been linked to cisplatin resistance (14). However, the clinical significance remains unclear since these are not frequent alterations in tumors.
In the present study, we report that ACTL6A regulates the repair of cisplatin-induced DNA damage through the SWI/SNF chromatin complex in cancer cells. Elevated ACTL6A levels in cancer lead to cisplatin resistance in both lung cancer cells and xenograft tumor models, and are also associated with poor prognosis in some cancer patients. Panobinostat is one of the histone deacetylase inhibitors, which is approved for the treatment of multiple myeloma (17). In our study, we found that panobinostat could abolish cisplatin resistance caused by ACTL6A overexpression by inhibiting ACTL6A-mediated DNA repair, which provides a therapeutic strategy to abolish platinum resistance in the cancers with high levels of ACTL6A.
Results
ACTL6A Overexpression Is Associated with Shorter Survival and a Worse Response to Cisplatin-Containing Chemotherapy in Lung Cancer and Ovarian Cancer.
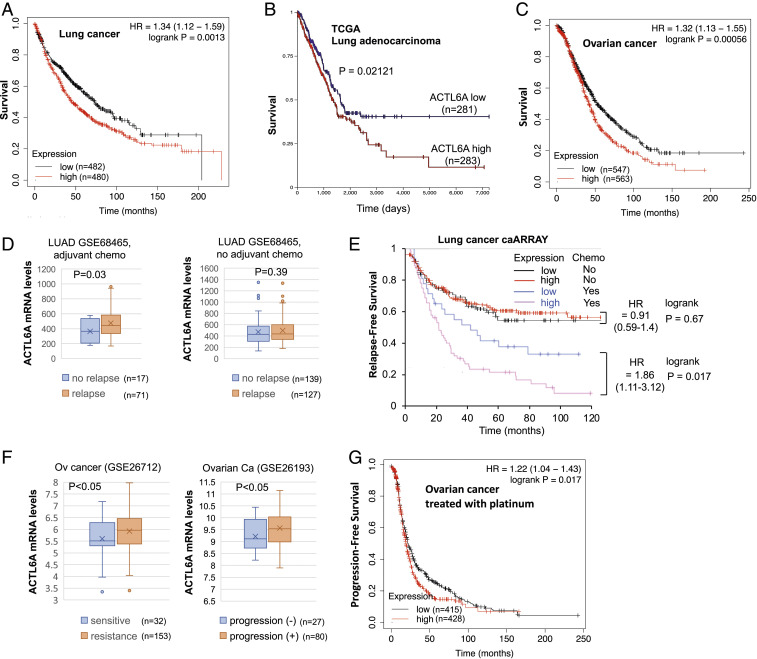
According to Oncomine, ACTL6A mRNA expression is elevated in most cancer types when compared with their normal counterparts, particularly in lung cancer, head and neck, and ovarian cancers which often harbor ACTL6A gene amplification. We therefore used KM Plotter (18), an online tool for metaanalysis of survival from multiple databases including Gene Expression Omnibus, European Genome-Phenome Archive, and TCGA, to assess whether high expression of ACTL6A affects patient outcomes and their response to chemotherapy. Indeed, higher levels of ACTL6A are associated with shorter patient survival in both lung cancer and ovarian cancer (Fig. 1 A–C). ACTL6A overexpression is also associated with a higher relapse rate in lung adenocarcinoma patients who received cisplatin-containing adjuvant chemotherapy, but not in those who did not received adjuvant chemotherapy (Fig. 1 D and E). This result suggests a predictive value for ACTL6A in cisplatin response in lung adenocarcinoma. Likewise, ovarian cancers resistant to cisplatin treatment also express higher levels of ACTL6A than those sensitive to cisplatin (Fig. 1F). Consistently, ACTL6A overexpression is associated with an earlier relapse rate in ovarian cancer patients who received platinum chemotherapy (Fig. 1G). The association between high ACTL6A expression and cisplatin resistance is also observed in cancer cell lines across different types of cancer. We mined Genomics of Drug Sensitivity in Cancer (GDSC) dataset (19). Among GDSC cancer cell lines, the cell lines harboring ACTL6A gene amplification (cnaPANCAN246) are more resistant to cisplatin than those without amplification (SI Appendix, Fig. S1A). Interestingly, the difference of cisplatin IC50 between ACTL6A-amplified and -nonamplified cell lines is bigger than that between TP53 mutations and wild-type cell lines (SI Appendix, Fig. S1 A, Right). GDSC lung cancer cell lines resistant to cisplatin also express higher levels of ACTL6A than those sensitive to cisplatin (SI Appendix, Fig. S1B). Together, these results suggest a role for ACTL6A in determining cisplatin sensitivity in cancer.
Fig. 1.
High ACTL6A levels are associated with shorter survival in lung cancer and ovarian cancer patients. (A) High levels of ACTL6A are associated with shorter overall survival in lung cancer patients. Data were generated using the KM Plotter server (kmplot.com). Patients from all 14 lung cancer datasets in the server were separated into four quartiles based on the expression levels of ACTL6A in tumors. Overall survival is compared between the lowest quartile and the highest quartile. (B) High levels of ACTL6A are associated with shorter survival in lung adenocarcinoma patients. TCGA lung adenocarcinoma data were analyzed in Xena Browser based on ACTL6A expression. (C) High levels of ACTL6A are associated with shorter survival in ovarian cancer patients. Data were generated using the KM Plotter server. Patients from all 15 ovarian cancer datasets in the server were separated into three tertiles based on the expression levels of ACTL6A in tumors. Overall survival is compared between the lowest tertile and the highest tertile. (D and E) High levels of ACTL6A are associated with shorter relapse-free survival in lung cancer patients who received cisplatin-containing adjuvant chemotherapy, but not in patients who did not receive adjuvant chemotherapy. A lung adenocarcinoma (LUAD) dataset GSE68465 (lung cancer caARRAY) (46) was analyzed for ACTL6A expression (D). All adjuvant chemotherapy regimens in this study contain cisplatin. Relapse-free survival from the same dataset was generated using the KM Plotter (E). Since roughly 80% of patients receiving adjuvant chemotherapy and 50% of patients in observation arm had relapsed, lower tertile was chosen as the same cutoff of ACTL6A expression for both groups (i.e., 1/3 of the group are low and 2/3 of the group are high). (F) Two ovarian cancer datasets GSE26712 (47) and GSE26193 (48) were analyzed for ACTL6A expression. All patients in GSE26712 and the majority of patients (93/107) in GSE26193 received cisplatin-containing chemotherapy. (G) High levels of ACTL6A are associated with shorter survival in ovarian cancer patients receiving platinum. Data were generated using the KM Plotter server. Patients were separated into three tertiles based on ACTL6A expression in tumors as described in C except that only patients who received platinum-containing chemotherapy are included for progression-free survival analysis.
Overexpression of ACTL6A Leads to Cisplatin Resistance in Lung Cancer Cells.
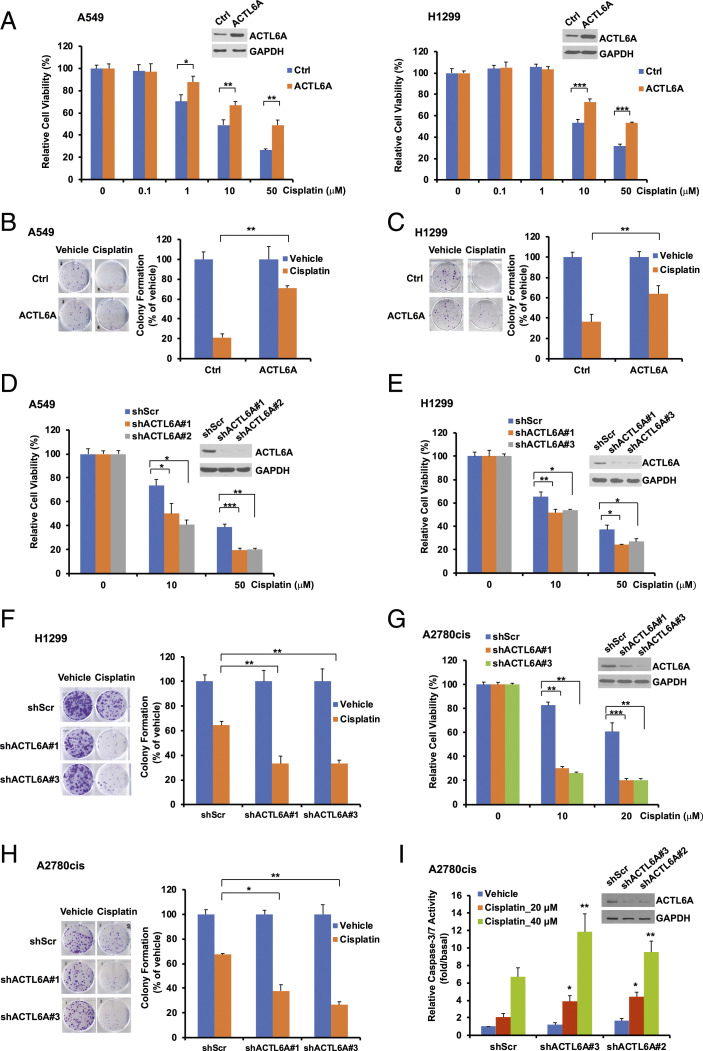
To explore the role of ACTL6A in the regulation of cellular response to cisplatin, we first examined the protein and mRNA levels of ACTL6A in a panel of lung cancer cell lines (SI Appendix, Fig. S2 A and B). As A549 and H1299 express the moderate level of ACTL6A, we decided to either overexpress or knock down ACTL6A in these two cell lines for further studies. We overexpressed ACTL6A in both A549 and H1299 cells by infection of a retrovirus harboring the full-length ACTL6A. After puromycin selection for a week to establish stable cell lines, we performed cell viability and clonogenic cell survival assays. We observed that overexpression of ACTL6A did not affect lung cancer cell growth (SI Appendix, Fig. S2 C and D), but significantly increased cell viability in both lung cancer cell lines following cisplatin treatment (Fig. 2A). The number of colonies after cisplatin treatment was also significantly increased in ACTL6A-overexpressing cells (going up from 21 to 71% in A549 cells, Fig. 2B; and from 36 to 64% in H1299 cells, Fig. 2C). These data indicate that overexpression of ACTL6A enhances cisplatin resistance in lung cancer cells.
Fig. 2.
Overexpression of ACTL6A attenuates sensitivity to cisplatin, whereas depletion of ACTL6A enhances it in lung cancer and ovarian cancer cells. (A) Overexpression of ACTL6A increases cell viability after cisplatin treatment. A549 or H1299 cells that stably harbor an empty vector (Ctrl) or ACTL6A were treated with cisplatin for 48 h. Cell viability was assessed by MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) assay. ACTL6A levels were determined by Western blot analysis. GAPDH (glyceraldehyde 3-phosphate dehydrogenase) serves as a loading control. (B and C) Overexpression of ACTL6A promotes cell survival in both A549 and H1299 lung cancer cells treated with cisplatin. Equal amounts of stable cells as indicated were treated with vehicle (PBS) or 1 μM cisplatin for 24 h. After washing, cells were cultured in fresh medium for 8 to 12 d. After fixation and crystal violet staining, colonies were counted. (D and E) Depletion of ACTL6A increases sensitivity to cisplatin. A549 or H1299 cells stably harboring a scrambled shRNA (shScr) or an ACTL6A shRNA (shACTL6A #1, #2, or #3) were treated with 10 or 50 μM cisplatin for 48 h. Cell viability was determined by MTT assay. Knockdown of ACTL6A was confirmed by Western blot analysis. (F) Depletion of ACTL6A increases cisplatin sensitivity in H1299 lung cancer cells. Clonogenic cell survival assay was performed as described in B. (G and H) Depletion of ACTL6A enhances cisplatin sensitivity in cisplatin-resistant A2780cis ovarian cancer cells. A2780cis stably expressing shScr or shACTL6A were treated with cisplatin and then subjected to MTT assay (G) or clonogenic cell survival assay (H). For each MTT assay, data shown represent the mean ± SD from at least three or four biological replicates. For each clonogenic cell survival assay, data shown represent the mean ± SD from at least three independent experiments, and representative images are shown on the Left of each figure. *P < 0.05, **P < 0.01, and ***P < 0.001. (I) Depletion of ACTL6A enhances cisplatin-induced apoptosis in A2780cis cells. A2780cis cells stably expressing shScr or shACTL6A (#2 or #3) were treated with cisplatin for 20 h. Data shown are the mean ± SEM of four independent experiments. *P < 0.05, **P = 0.01 versus shScr cells treated with 20 μM and 40 μM cisplatin, respectively.
Depletion of ACTL6A Increases Sensitivity to Cisplatin in Cancer Cells.
To further confirm the significant role of ACTL6A in the response to cisplatin, we used lentiviral shRNA constructs (shACTL6A#1, #2, or #3) to knock down endogenous ACTL6A gene expression in several cell lines. Indeed, depletion of ACTL6A significantly decreased cell viability in both A549 and H1299 cells (Fig. 2 D and E). The clonogenic survival assay also showed that the number of colonies was significantly decreased in ACTL6A knockdown H1299 cells after cisplatin treatment (from 64% down to 33%, Fig. 2F), indicating that depletion of ACTL6A increases sensitivity to cisplatin in lung cancer cells.
The effect of ACTL6A in the regulation of cisplatin response is not limited to lung cancer cells. A2780cis is a well-established acquired cisplatin-resistant ovarian cancer cell line, which has been widely used in the study of cisplatin resistance (20). We found that, similar to lung cancer cells, depletion of ACTL6A in A2780cis cells also decreased cell viability (Fig. 2G) and colony formation (from 67% down to 38% or 27%, Fig. 2H) after cisplatin treatment. ACTL6A knockdown also enhances cisplatin-induced apoptosis in A2780cis cells as determined by caspase-3/7 activity assay (Fig. 2I). These findings indicate that depletion of ACTL6A in a cisplatin-resistant ovarian cancer cell line can resensitize these cells to cisplatin.
Taken together, our results strongly suggest that ACTL6A plays an important role in the regulation of cellular response to cisplatin treatment in both lung and ovarian cancer cells.
Overexpression of ACTL6A Causes Cisplatin Resistance in Lung Cancer Xenografts.
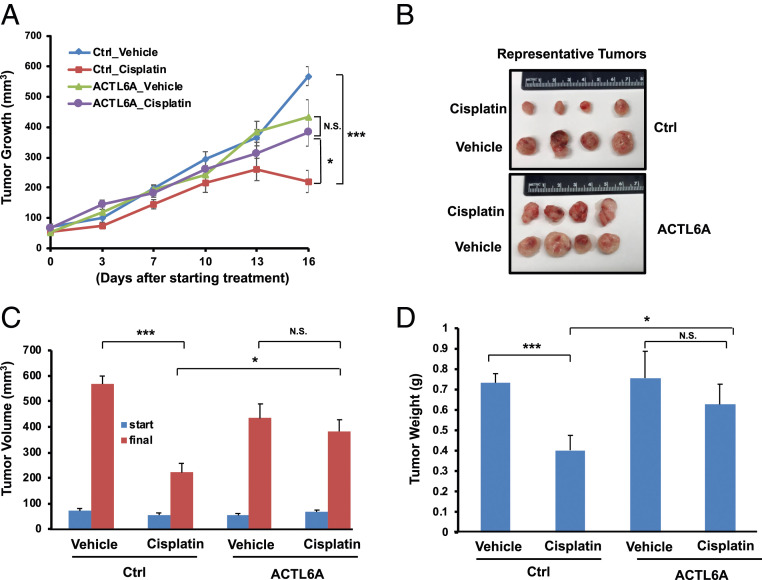
To determine the effect of ACTL6A on the regulation of cisplatin response in vivo, we developed a xenograft lung tumor model in NOD scid IL2 receptor γ chain knockout mice (NSG) mice. Stable H1299 cells harboring a control empty vector (Ctrl) or ACTL6A were injected subcutaneously into both sides of the flank of 6- to 8-wk-old NSG mice. When the tumor volumes reached ∼50 to 100 mm3, mice were randomly separated into two groups, such that each group had about seven to nine mice, with a total of 14 to 18 tumors in each group. Cisplatin or sterile phosphate-buffered saline (PBS) was then given to the mice by intraperitoneal injection twice per week. As shown in Fig. 3, cisplatin treatment effectively blocked the growth of control xenografts. However, ACTL6A-overexpressing xenografts were very resistant to cisplatin treatment, as indicated by the final tumor volumes (Fig. 3 B and C) and weights (Fig. 3D) of the xenografts. Thus, overexpression of ACTL6A causes cisplatin resistance in lung xenografts in vivo.
Fig. 3.
Overexpression of ACTL6A leads to cisplatin resistance in lung cancer xenografts. The H1299 cells stably expressing an empty vector (Ctrl) or ACTL6A were implanted onto both sides of the flanks of NSG mice to establish in vivo xenograft tumors. Mice were randomly divided into two groups and injected with either cisplatin (5 mg/kg) or vehicle (PBS) intraperitoneally twice per week for 2 to 3 wk. (A) Tumor growth curves determined by the tumor volumes measured twice every week. (B) Photographs of representative tumors from each group. (C) Tumor volumes at the start and in the end of cisplatin treatment. (D) Weights of xenograft tumors. The final tumor volumes and tumor weights were calculated by averaging both sides of tumors on each mouse. The number of mice in each group is 7 (Ctrl_Vehicle), 9 (Ctrl_Cisplatin), 8 (ACTL6A_Vehicle), or 9 (ACTL6A_Cisplatin). Data shown represent the mean ± SEM. *P < 0.05, ***P < 0.001, N.S., not significant.
ACTL6A Positively Regulates the Repair of Cisplatin-Induced DNA Damage.
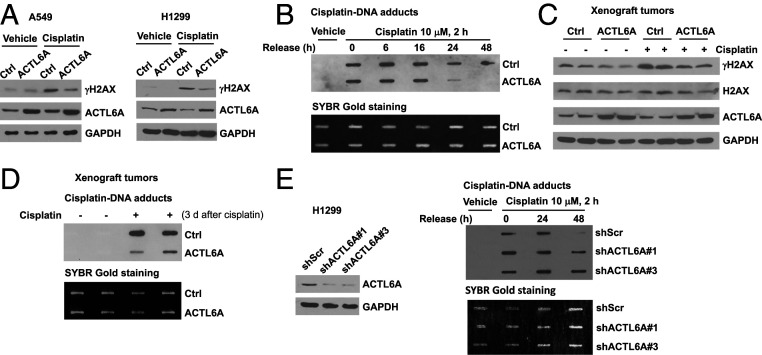
To investigate how ACTL6A promotes cisplatin resistance in cancer cells, we first examined the effect of ACTL6A on DNA damage response and DNA repair process since ACTL6A has been reported to bind most histones as a nuclear actin-related protein, and therefore may be involved in the chromatin remodeling process. We observed that overexpression of ACTL6A blocked cisplatin-induced H2AX activation in both A549 and H1299 cells, as indicated by the inhibition of γH2AX induction (Fig. 4A), which suggests that high levels of ACTL6A may either alter cisplatin efficacy in inflicting DNA damage or enhance DNA repair process after cisplatin treatment. We then performed DNA repair assay with an antibody capable of detecting cisplatin-DNA adducts specifically. Our data showed that there was no difference in the adducts signal between control and ACTL6A-overexpressing cells after cisplatin treatment for 2 h, indicating that ACTL6A overexpression does not prevent cisplatin-induced DNA damage. Nonetheless, after cisplatin was removed, the cisplatin-DNA adducts were cleared more rapidly in ACTL6A-overexpressing cells than in the control cells (Fig. 4B), indicating enhanced DNA repair by ACTL6A. These results highly suggest that overexpression of ACTL6A does not affect cisplatin uptake or its action on DNA, but instead stimulates the DNA repair process.
Fig. 4.
ACTL6A regulates the repair of cisplatin-induced DNA damage in vitro and in vivo. (A) Overexpression of ACTL6A attenuates cisplatin-induced H2AX activation in lung cancer cells. Empty vector control (Ctrl) or ACTL6A-overexpressing A549 or H1299 cells were treated with vehicle or 10 μM cisplatin for 24 h, and the whole cell lysates were subjected to Western blot analysis using an antibody specific to phosphorylated H2AX (γH2AX), ACTL6A, or GAPDH. (B) Overexpression of ACTL6A enhances the repair of cisplatin-DNA adducts in H1299 cells. H1299 stable cells were treated with 10 μM cisplatin for 2 h, and then were released and cultured in fresh medium for various times as indicated. Total genomic DNA was subjected to slot blot analysis using an anti-cisplatin-DNA adducts antibody (Upper) followed by SYBR Gold nucleic acid staining for loading control (Lower). (C) Overexpression of ACTL6A decreases cisplatin-induced H2AX activation in xenograft lung tumors. Figures shown are Western blots of total lysates prepared from two tumors randomly chosen from each group in the experiment described in Fig. 3. (D) Overexpression of ACTL6A enhances the repair of cisplatin-DNA adducts in xenografts. Genomic DNA was extracted from two xenografts randomly selected from each group described in Fig. 3, and then was subjected to slot blot assay using an anti-cisplatin-DNA adducts antibody as described in B. The last dose of cisplatin was given to the mice 3 d before mice were killed and tumors were harvested. (E) Depletion of ACTL6A impairs the efficiency of the repair of cisplatin-DNA adducts. H1299 cells stably expressing shScr or an ACTL6A shRNA (#1 or #3) were treated with 10 μM cisplatin for 2 h, and then released and cultured in fresh medium for various times as indicated, followed by slot blot assay using an anti-cisplatin-DNA adducts antibody as described in B.
To further identify the molecular basis for ACTL6A-mediated cisplatin resistance in vivo, we next measured the cisplatin-DNA adducts in H1299 xenografts from prior experiment shown in Fig. 3. Similar to the results obtained in cell lines, when the mice were killed 3 d after the last injection of cisplatin, cisplatin-induced γH2AX expression (Fig. 4C) and the levels of remaining cisplatin-DNA adducts (Fig. 4D) were much lower in ACTL6A-overexpressing tumors compared to the control tumors, indicating that overexpression of ACTL6A enhanced DNA repair after cisplatin-induced DNA damage in lung cancer xenografts. Conversely, depletion of ACTL6A in H1299 cells delayed the repair of cisplatin-DNA adducts (Fig. 4E).
Taking together, we conclude that ACTL6A positively regulates the DNA repair process following cisplatin-induced DNA damage. The elevation of ACTL6A protein levels promotes resistance to cisplatin in both cultured cancer cells and xenografts by enhancing DNA repair activity.
ACTL6A Regulates the Repair of Cisplatin-Induced DNA Damage through the SWI/SNF Chromatin Remodeling Complex.
Actin-like protein ACTL6A is an essential subunit of SWI/SNF chromatin remodeling complex. The catalytic subunit of the complex, Brahma related gene 1 (Brg1) or Brahma (Brm), has been reported to modulate cisplatin cytotoxicity (21). Down-regulation of Brg1 and Brm results in impaired DNA repair activity and increases cellular sensitivity to cisplatin treatment (21). To test the possibility that ACTL6A regulates cellular response to cisplatin through the SWI/SNF chromatin remodeling complex, we first overexpressed both Brg1 and Brm dominant-negative mutants, which have been used to block the activity of endogenous Brg1 and Brm proteins (22–24), in H1299 cells stably harboring an empty vector or ACTL6A. Our data showed that loss of Brg1 and Brm activity increased the basal levels of γH2AX, and further blocked the inhibitory effect of ACTL6A on H2AX activation (Fig. 5A, comparing lanes 4 and 8). These dominant-negative Brg1/Brm mutants also significantly inhibited ACTL6A-enhanced DNA repair activity (Fig. 5B) after cisplatin treatment. In both cell viability (Fig. 5C) and clonogenic survival assays (Fig. 5D), loss of function of Brg1 and Brm successfully overcame ACTL6A-induced cisplatin resistance in H1299 cells, suggesting that both Brg1 and Brm are required for ACTL6A to enhance DNA repair after cisplatin-induced DNA damage.
Fig. 5.
ACTL6A regulates cellular response to cisplatin and DNA repair process through the SWI/SNF complex. (A) Overexpression of both dominant-negative Brg1 and Brm (dn-Brg1/Brm) increases the basal activity of H2AX, and blocks the effect of ACTL6A on the reduction of cisplatin-induced H2AX activation. H1299 cells stably harboring an empty vector or ACTL6A were transiently transfected with an empty vector or both dn-Brg1 and dn-Brm expression vectors. Cells were treated with 10 μM cisplatin for 24 h before lysis for Western blot analysis. (B) Overexpression of both dn-Brg1 and dn-Brm blocks ACTL6A-enhanced DNA repair after cisplatin treatment. Transfected H1299 cells as described in A were treated with vehicle or 10 μM cisplatin for 2 h, and then released and cultured in fresh medium for indicated times. Genomic DNA was isolated and subjected to slot blot assay as described above. (C and D) Overexpression of both dn-Brg1 and dn-Brm inhibits the effect of ACTL6A on enhancing cell viability and cell survival after cisplatin treatment. Transfected H1299 cells as indicated were treated with 10 or 50 μM cisplatin for 48 h (C), or with 1 μM cisplatin for 24 h (D). Cell viability was determined by MTT assay (C). Clonogenic cell survival assay was performed (D). (E) Depletion of BAF155 enhances the basal activity of H2AX and blocks the inhibitory effect of ACTL6A on cisplatin-induced H2AX activation. H1299 cells stably harboring an empty vector or ACTL6A were infected with a lentivirus expressing either shScr or shBAF155 (#1 or #2). Cells were treated with vehicle or 10 μM cisplatin for 24 h followed by Western blot analysis. (F) Depletion of BAF155 blocks ACTL6A-enhanced DNA repair after cisplatin treatment. Transfected H1299 cells as described in E were treated with vehicle or 10 μM cisplatin for 2 h. After release and culture in fresh medium for indicated times, genomic DNA was isolated and subjected to slot blot analysis. (G and H) Depletion of BAF155 inhibits the effect of ACTL6A on enhancing cell viability and cell survival after cisplatin treatment. Transfected H1299 cells as described in E were treated with 10 or 50 μM cisplatin for 48 h (G) or with 1 μM cisplatin for 24 h (H). Cells were subjected to MTT cell viability assay (G) or clonogenic survival assay (H). For cell viability assay, Data shown represent the mean ± SD from at least three or four biological replicates. For colony formation assay, data shown represent the mean ± SD from at least three independent experiments. Representative images are shown on the Left of each figure. *P < 0.05 and **P < 0.01.
To further confirm that ACTL6A regulates cisplatin-induced DNA repair through the SWI/SNF complex, we knocked down another core protein in this complex, BAF155, which functions as a core subunit to stabilize the whole complex (25, 26). We observed that depletion of BAF155 by its specific shRNA strongly increased cisplatin-induced H2AX activation in ACTL6A-overexpressing cells (Fig. 5E) and completely eliminated ACTL6A-enhanced DNA repair (Fig. 5F). Depletion of BAF155 also completely reversed cisplatin resistance caused by ACTL6A overexpression in H1299 cells as demonstrated in the cell viability (Fig. 5G) and clonogenic survival assays (Fig. 5H), which provides another strong evidence that ACTL6A regulates the repair of cisplatin-induced DNA damage through the SWI/SNF chromatin remodeling complex.
ACTL6A Regulates Cellular Response to Cisplatin by Maintaining the SWI/SNF Complex Integrity.
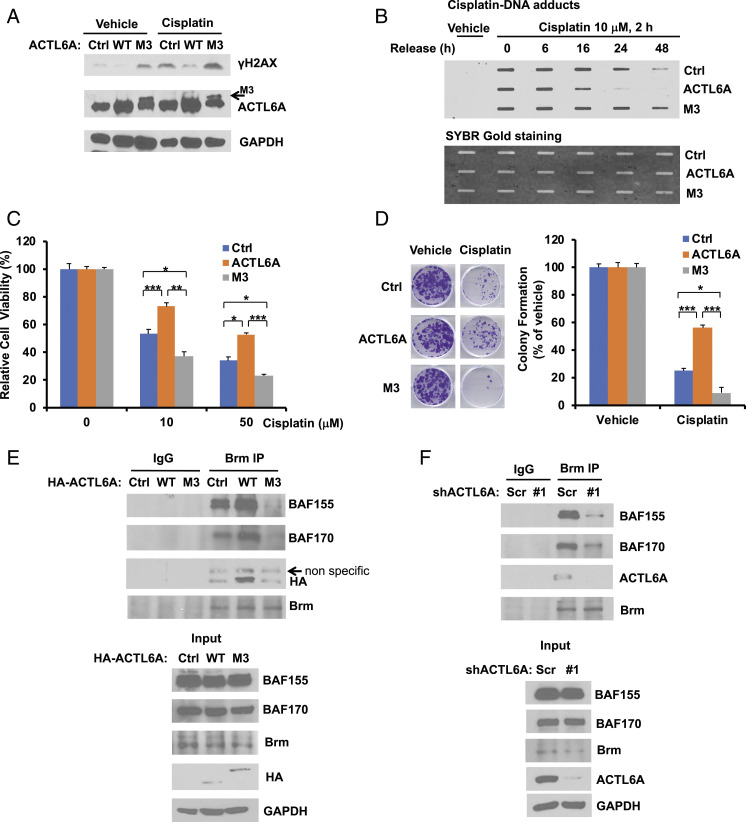
Previous studies have shown that the heterocomplex formation between ACTL6A and β-actin maintains the integrity of Brg1 complex through direct binding to the helicase-SANT-associated (HSA) domain of Brg1 complex (27), whereas an M3 mutant (E388A/R389A/R390A) of ACTL6A, which showed reduced binding to β-actin, is impaired in forming a complex with Brg1 (7). Therefore, we examined the effect of M3 mutant ACTL6A on cisplatin response. Interestingly, contrary to wild-type ACTL6A, overexpression of M3 mutant ACTL6A not only increased the basal activity of H2AX, but also enhanced cisplatin-induced H2AX activation (Fig. 6A); and as a result, it blocked DNA repair activity after cisplatin treatment (Fig. 6B). Indeed, in contrast to the wild-type ACTL6A, overexpression of M3 mutant ACTL6A enhanced cisplatin sensitivity, as demonstrated by cell viability assay (Fig. 6C) and clonogenic cell survival assay (Fig. 6D).
Fig. 6.
The ACTL6A mutant defective in Brg1 binding promotes the H2AX activity and cisplatin sensitivity by blocking DNA repair and the recruitment of the SWI/SNF complex. (A and B) In contrast to wild-type ACTL6A, the M3 mutant of ACTL6A, which is impaired in Brg1 binding, enhances H2AX activation and inhibits the repair of cisplatin-DNA adducts. H1299 cells stably overexpressing an empty vector (Ctrl), wild-type (WT) ACTL6A, or the M3 mutant of ACTL6A were treated with vehicle (PBS) or 10 μM cisplatin for 24 h (A) or 2 h (B). The activation of H2AX was determined by Western blot analysis (A). The slot blot assay was performed to determine the repair of cisplatin-DNA adducts (B). (C and D) Overexpression of the M3 mutant of ACTL6A enhances cisplatin sensitivity. Stable H1299 cells as indicated were treated with cisplatin, followed by MTT cell viability assay (C) or clonogenic cell survival assay (D) as described in Fig. 2 A and B, respectively. For cell viability assay (C), data shown represent the mean ± SD from at least three or four biological replicates. For colony formation assay (D), data shown represent the mean ± SD from at least three independent experiments. Representative images are shown on the Left of each figure. *P < 0.05, **P < 0.01, and ***P < 0.001. (E) Wild-type ACTL6A binds to Brm and promotes the recruitment of both BAF155 and BAF170 to Brm, whereas the M3 mutant of ACTL6A blocks this complex formation. H1299 cells stably overexpressing an empty vector (Ctrl) or HA-tagged ACTL6A (WT or M3 mutant) were harvested for coimmunoprecipitation. Endogenous Brm was immunoprecipitated with an anti-Brm rabbit monoclonal antibody or a control rabbit IgG, followed by immunoblotting. (F) Depletion of ACTL6A impairs the recruitment of both BAF155 and BAF170 to the Brm complex. H1299 cells stably expressing shScr or shACTL6A were harvested, and the endogenous protein coimmunoprecipitation of Brm with BAF155, BAF170, or ACTL6A was performed as described in E.
We next examined the effect of ACTL6A expression on the SWI/SNF family BAF complex formation. Overexpression of wild-type ACTL6A did not affect the protein levels of other components of Brm complex, but increased the complex integrity by enhancing the interaction between Brm and other core subunits, such as BAF155 and BAF170 (Fig. 6E). In contrast, overexpression of the M3 mutant destabilized the complex and blocked Brm binding to both BAF155 and BAF170. As expected, Brm immunoprecipitates pulled down wild-type ACTL6A, but not M3 mutant. Depletion of ACTL6A also impaired the complex integrity by preventing Brm binding to both BAF155 and BAF170 (Fig. 6F), which further confirms the importance of ACTL6A in the maintenance of the SWI/SNF complex integrity, and indicates that ACTL6A regulates cellular response to cisplatin through maintaining the SWI/SNF complex integrity.
Depletion of INO80 or TIP60 and Inactivation of TIP60 Do Not Affect ACTL6A-Mediated Cisplatin Resistance.
ACTL6A is also a component in the INO80 chromatin remodeling complex and the TIP60/p400 acetyltransferase complex. Therefore, we further examined whether INO80 or TIP60/p400 complex also plays a role in the action of ACTL6A in response to cisplatin treatment. We found that depletion of INO80 (SI Appendix, Fig. S3A) slightly delayed the timing of the clearance of cisplatin-DNA adducts, but failed to abolish ACTL6A-enhanced DNA repair (SI Appendix, Fig. S3B). Both cell viability (SI Appendix, Fig. S3C) and clonogenic cell survival assays (SI Appendix, Fig. S3D) showed that overexpression of ACTL6A could still induce cisplatin resistance in INO80-knockdown H1299 cells, indicating that ACTL6A does not regulate cisplatin resistance through the INO80 chromatin remodeling complex. Our data also showed that depletion of TIP60 (SI Appendix, Fig. S4A) did not block ACTL6A-enhanced DNA repair (SI Appendix, Fig. S4B). Inactivation of TIP60 by a chemical inhibitor NU9056, as indicated by the decreased acetylation of Histone H4K8 or H3K18 (SI Appendix, Fig. S4C), also did not affect ACTL6A-mediated repair of cisplatin-DNA adducts (SI Appendix, Fig. S4D). As a result, depletion of TIP60 could not reverse ACTL6A-induced cisplatin resistance (SI Appendix, Fig. S4 E and F). It has been reported that a complete loss of TIP60 or INO80 impairs cell viability (28, 29). Indeed, we observed a decrease in clonogenic viability upon depletion of INO80 or TIP60, particularly when TIP60 expression was almost completely eliminated by shTIP60#1 (SI Appendix, Fig. S4A). Nonetheless, ACTL6A overexpression could still enhance repair and promote cisplatin resistance in these INO80- or TIP60-depleted cells.
Collectively, these results strongly indicate that ACTL6A enhances the repair of cisplatin-induced DNA damage through the SWI/SNF chromatin remodeling complex, but not the INO80 or TIP60/p400 complex.
An HDAC Inhibitor Abolishes ACTL6A-Mediated Cisplatin Resistance.
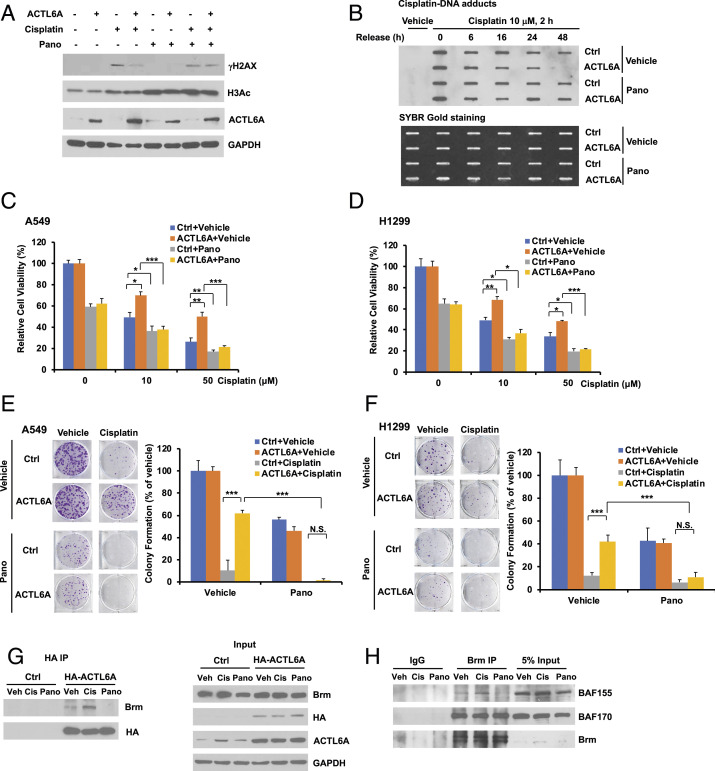
HDAC inhibitors have been used as a promising class of anticancer reagents when combined with common chemotherapeutic drugs, such as cisplatin, doxorubicin, or etoposide, in some cancer patients (30). Panobinostat is one of the pan-HDACs inhibitors with potent inhibitory activity against all class I and II HDAC enzymes at low concentrations, and has been shown to synergize with cisplatin (31, 32). Since ACTL6A causes cisplatin resistance through SWI/SNF, an epigenetic mechanism, we wanted to investigate whether HDAC inhibitors can reverse cisplatin resistance caused by ACTL6A. Indeed, while ACTL6A overexpression could decrease cisplatin-induced H2AX activation, this effect was largely mitigated by cotreatment with panobinostat (Fig. 7A), suggesting that panobinostat may inhibit the effect of ACTL6A on enhancing repair of cisplatin-induced DNA damage. Here the on-target effect of panobinostat was confirmed by the enhanced Histone H3 acetylation (H3Ac). This was further confirmed by slot blotting assay, which showed that with panobinostat treatment, the clearance of cisplatin-DNA adducts was no longer enhanced by ACTL6A overexpression (Fig. 7B), strongly suggesting that panobinostat was able to block the effect of ACTL6A on enhancing DNA repair. We also found that panobinostat treatment resensitized ACTL6A-overexpressing cells to cisplatin as shown by both cell viability assay (Fig. 7 C and D) and clonogenic cell survival assay (Fig. 7 E and F) in two different lung cancer cell lines. These data indicate that panobinostat is able to abolish ACTL6A-induced cisplatin resistance by blocking its enhancing effect on DNA repair.
Fig. 7.
Panobinostat abolishes cisplatin resistance caused by ACTL6A overexpression through the destabilization of the SWI/SNF complex. (A) Panobinostat treatment mitigates the inhibitory effect of ACTL6A on cisplatin-induced H2AX activation. H1299 cells stably overexpressing an empty vector or ACTL6A were treated with 100 nM panobinostat and/or 10 μM cisplatin for 24 h. The whole cell lysates were subjected to Western blot analysis to determine the levels of phosphorylated H2AX (γH2AX) or acetyl Histone H3 (H3Ac). (B) Panobinostat blocks the effect of ACTL6A on enhancing DNA repair after cisplatin treatment. Control or ACTL6A-overexpressing H1299 cells were treated with 10 μM cisplatin and/or 100 nM panobinostat for 2 h. After release and culture in fresh medium for various times as indicated, slot blot analysis was performed using an anti-cisplatin-DNA adducts antibody to determine the DNA repair. (C and D) Panobinostat blocks the effect of ACTL6A on promoting cell viability after cisplatin treatment. A549 (C) or H1299 (D) cells stably overexpressing an empty vector or ACTL6A were treated with vehicle or cisplatin (10 or 50 μM) in the absence or presence of 100 nM panobinostat for 48 h. Cell viability was determined by MTT assay. (E and F) Stable A549 (E) or H1299 (F) cells as described above were treated with 1 μM cisplatin and/or 20 nM panobinostat for 24 h. After washing, cells were cultured in fresh medium until colonies were formed. Representative images are shown on the Left. For cell viability assay, data shown represent the mean ± SD from at least three or four biological replicates. For colony formation assay, data shown represent the mean ± SD from at least three independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001. (G) Panobinostat treatment blocks ACTL6A binding to Brm. H1299 cells stably expressing an empty vector or HA-ACTL6A were treated with vehicle (Veh), 10 μM cisplatin (Cis), or 100 nM panobinostat (Pano) for 24 h. Coimmunoprecipitation was performed by immunoprecipitating HA-ACTL6A with anti-HA monoclonal antibody-conjugated agarose, followed by immunoblotting using an anti-Brm rabbit antibody or an anti-HA antibody. (H) Panobinostat blocks the recruitment of BAF155 but not BAF170, to the Brm complex. H1299 cells were treated with 10 μM cisplatin or 100 nM panobinostat for 24 h. Coimmunoprecipitation of endogenous Brm with BAF155 or BAF170 was performed as described in Fig. 6E.
Since ACTL6A regulates cisplatin response through the SWI/SNF complex, we then performed the coimmunoprecipitation experiment to investigate whether panobinostat had any effect on the integrity of this complex. We observed that cisplatin treatment was able to enhance the interaction between ACTL6A and Brm, but panobinostat completely blocked this binding (Fig. 7G). Furthermore, cisplatin increased the recruitment of BAF155 to Brm, whereas panobinostat impaired this complex formation (Fig. 7H). Neither cisplatin nor panobinostat had an effect on the recruitment of another subunit BAF170 to Brm.
Collectively, our results suggest that panobinostat is able to abolish ACTL6A-induced cisplatin resistance by affecting the integrity of the SWI/SNF complex and inhibiting the repair of cisplatin-DNA adducts.
An HDACs Inhibitor Abolishes ACTL6A-Induced Cisplatin Resistance in a Lung Cancer Xenograft Model.
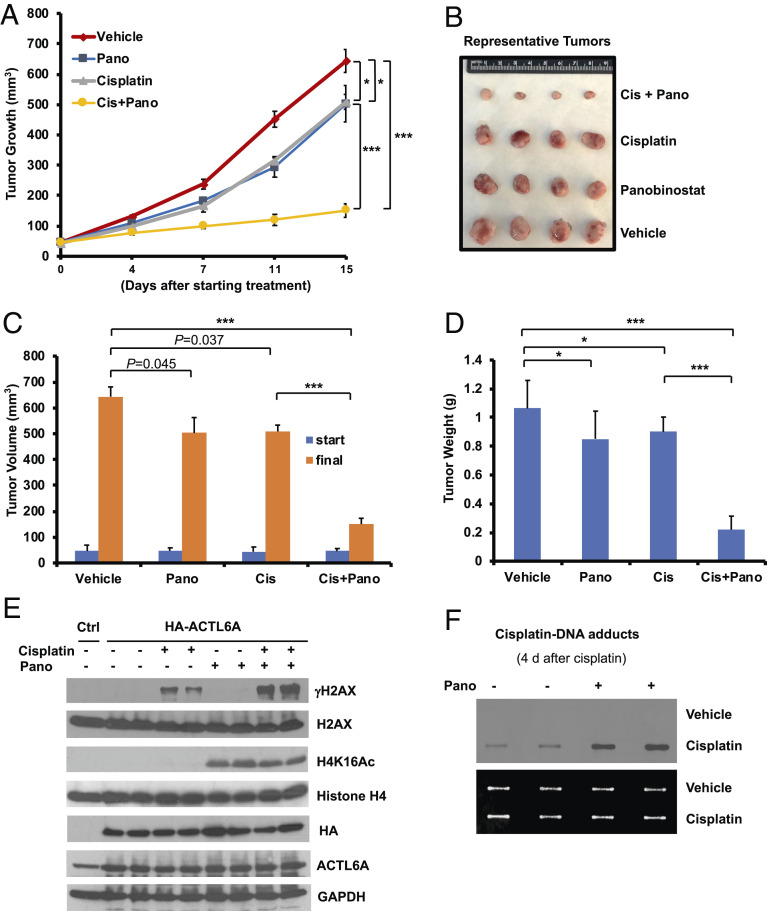
To investigate the therapeutic efficacy of panobinostat in overcoming ACTL6A-induced cisplatin resistance, we next implanted NSG mice with ACTL6A-overexpressing H1299 cancer cells subcutaneously. The mice were randomized to receive the treatment with panobinostat and/or cisplatin, or the vehicle for a total of 15 d. Compared to the vehicle control, cisplatin or panobinostat alone only slightly slowed down the growth of ACTL6A-overexpressing H1299 tumors, whereas the combination of both drugs robustly blocked tumor growth (Fig. 8A). At the end point of the treatment, the size (Fig. 8 B and C) and weights (Fig. 8D) of the harvested xenograft tumors were dramatically reduced by treatment with both cisplatin and panobinostat compared with cisplatin or panobinostat alone. We then randomly selected two tumors from each group for Western blot analysis and cisplatin-DNA adducts assay. Indeed, panobinostat treatment did not alter ACTL6A protein level in vivo, but significantly increased H2AX activation (Fig. 8E) and blocked the repair of cisplatin-DNA adducts (Fig. 8F). The on-target effect of panobinostat was confirmed by strong induction of Histone H4K16 acetylation (H4K16ac) in panobinostat-treated xenografts (Fig. 8E). Taken together, our results demonstrate that panobinostat was able to abolish ACTL6A-induced cisplatin resistance in vivo.
Fig. 8.
Panobinostat abolishes ACTL6A-induced cisplatin resistance in lung cancer xenografts. (A–D) H1299 cells stably overexpressing HA-tagged ACTL6A were used to establish in vivo xenograft tumor models in NSG mice. Mice were randomly divided into four groups and injected with vehicle, cisplatin (Cis, 5 mg/kg), and/or panobinostat (10 mg/kg) intraperitoneally twice weekly for 2 wk. Data shown are the tumor growth curves (A), photographs of representative tumors (B), tumor volumes measured at the start and in the end of the treatment (C), and tumor weights (D). The tumor volumes and weights were calculated by averaging both sides of tumors on each mouse. The number of mice in each group is eight (Veh), nine (Pano), eight (Cis), or nine (Cis+Pano). Data shown represent the mean ± SEM. *P < 0.05, ***P < 0.001. (E and F) Panobinostat enhances cisplatin-induced H2AX activation and blocks DNA repair in xenograft tumors overexpressing ACTL6A. Two tumors randomly selected from each group were processed for Western blot analysis to detect the expression of indicated proteins (E), and cisplatin-DNA adducts slot blot assay was performed 4 d after the last injection of cisplatin (F). The whole cell lysates isolated from control H1299 cells serve as negative control (Ctrl) in E.
Discussion
Despite many advances in cancer therapy, platinum resistance remains a major barrier for patient outcome improvement. In this paper, we identify ACTL6A as a mediator for cisplatin resistance. Amplification of ACTL6A gene is a very common gene alteration event in cancer, which occurs in over one-third of lung squamous cell carcinoma and nearly one-fifth of ovarian cancer. Thus, ACTL6A overexpression could potentially be one of the most common mechanisms for acquired resistance to cisplatin.
To determine its mechanism of action, we performed cisplatin repair assay and demonstrated a role for ACTL6A in the repair of cisplatin-DNA adducts through the SWI/SNF complex, but not INO80 or TIP60 complex. Our results support a role for the SWI/SNF complex in facilitating repair of the cisplatin-DNA lesions as previously proposed (21). ACTL6A is a shared subunit among three remodeling complexes, i.e., SWI/SNF, INO80, and TIP60. All three complexes play a role in DNA repair, particularly during the repair of double strand breaks (DSB) by HR (33). TIP60 acetylates the H4 tail and promotes the shift from repressive to open chromatin for DSB repair (34). INO80 promotes nucleosome eviction and is required for HR (35). However, depletion of INO80 or TIP60 in H1299 cells does not affect ACTL6A-mediated repair of cisplatin-DNA adducts (SI Appendix, Figs. S3B and S4B). In contrast, depletion of BAF155, a core subunit of the SWI/SNF complex, completely blocks the repair of cisplatin-induced DNA damage (Fig. 5F). These results indicate that the SWI/SNF complex, rather than INO80 or TIP60, plays a major role in the repair of cisplatin-induced damage. We also demonstrate the unique role of ACTL6A in regulating the complex formation between Brm and either BAF155 or BAF170. Thus, in cancer cells harboring ACTL6A gene amplification, overexpression of ACTL6A enhances the stability and function of SWI/SNF, and therefore increases the cellular ability in repairing cisplatin-DNA adducts.
The mechanism(s) by which SWI/SNF promotes the repair of cisplatin lesions remain(s) to be fully explored. Efficient DNA repair requires chromatin remodeling activities since nucleosomes inhibit DNA repair as demonstrated in an in vitro system (36). SWI/SNF appears to play an evolutionally conserved role in NER. The yeast SWI/SNF complex can increase the accessibility and incision of damaged DNA by NER proteins on in vitro reconstituted mononucleosomes (37, 38). Alternatively, SWI/SNF may regulate other proteins involved in ICL repair, such as FA, TLS polymerase, HR, etc., to promote repair of cisplatin-DNA lesions. Moreover, SWI/SNF is required for repriming the stalled DNA forks after damage (39, 40). Thus, the severe defect of clonogenic viability found in ACTL6A- or BAF155-depleted cells treated with cisplatin (Figs. 2 F and H and 5H) may be due to not only a defect in repair but also a failure in resuming replication. On the other hand, while our study focuses on the repair of cisplatin-DNA lesions, it is quite likely that ACTL6A plays a similar role in other forms of DNA damage given the fact that SWI/SNF has been implicated in multiple DNA repair pathways, such as DSB repair (41).
Importantly, we identify panobinostat, an HDAC inhibitor, as a potential therapeutic reagent that can reverse cisplatin resistance caused by ACTL6A overexpression. Treatment with panobinostat inhibits the interaction between ACTL6A and Brm, leading to the destabilization of the SWI/SNF complex and inhibition of the repair of cisplatin-DNA lesions. It is worth noting that the biological activity of ACTL6A in promoting repair and the ability of panobinostat in counteracting ACTL6A activity are demonstrated in both cultured cells and in vivo xenograft models. Thus, it is very desirable to investigate if HDAC inhibitors can reverse platinum resistance in future clinical trials. The mechanism by which panobinostat blocks the interaction between ACTL6A and Brm, whereas cisplatin treatment enhances this binding (Fig. 7G) remains to be investigated. We speculate that their interaction might be regulated by posttranslational modifications, such as acetylation, which could be induced by panobinostat. HADC inhibitors not only increase the acetylation of histones, but also prevent the deacetylation of some nonhistone proteins, such as STAT3, E2F, and p53, during gene transcription or DNA damage response (42). Brm and BAF155 contain some potential HDAC deacetylation sites according to the online KAT-specific Acetylation Site Prediction software. Therefore, HDAC inhibitors might directly induce the aceylation of Brm and/or BAF155 and affect the integrity of the SWI/SNF complex. It is intriguing that panobinostat inhibits the repair of cisplatin-induced DNA damage (Figs. 7B and 8F) and only blocks the association of Brm with BAF155, but not with BAF170 (Fig. 7H). Depletion of ACTL6A in H1299 cells also inhibits Brm/BAF155 binding to a greater extent than Brm/BAF170 binding (Fig. 6F). These data suggest that BAF complex that contains BAF155, but not BAF170, is involved in the repair of cisplatin-DNA adducts. It is worthy to investigate this possibility in future experiments.
Many components of SWI/SNF are found to be mutated in certain cancers, e.g., Brg1, Brm, BAF155, and ARID1A/B (41). In contrast, ACTL6A gene is rarely mutated, and often amplified in lung squamous cell carcinoma, ovarian cancer, and esophageal cancer. In addition to gene amplification, ACTL6A can be overexpressed in cancer cells through other mechanisms. ACTL6A is a direct MYC target gene (43, 44), and therefore is also overexpressed in MYC-amplified tumors.
In summary, our study links the overexpression of a subunit of SWI/SNF to the enhanced DNA repair and chemoresistance, and provides evidence for the use of HDAC inhibitors to abolish cisplatin resistance. Overexpression of ACTL6A may serve as a biomarker to predict resistance to cisplatin and identify patients for additional therapies in future clinical trials.
Materials and Methods
DNA Repair Assay.
Cells were treated with cisplatin for indicated hours followed by washing with PBS twice, and were further incubated with fresh cultured medium. Slot blotting assay was performed after cells were harvested at different time points. A total of 0.5 to 1 μg of genomic DNA was denatured as described (45) and vacuum-transferred to nitrocellulose membrane (GE Healthcare), followed by cross-linking with UV light. An anti-cisplatin-DNA adduct monoclonal antibody (Millipore) was used to detect the adducts of cisplatin conjugated to genomic DNA. The SYBR-Gold dye (Life Technologies) was used to detect the total amount of DNA loaded onto the membrane.
Other methods are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Zhou (Sunny) Songyang for the ACTL6A M3 mutant plasmid; Dr. Kwong K. Wong for the A2780cis cell line; and Dr. Ralf Krahe for the RNA samples isolated from SKEMS1, H2170, SW900, or H1395 lung cancer cell lines. This work was supported by NIH R01CA100857, Rivkin Center for Ovarian Cancer Pilot Award (to W.-C.L.), and R01CA203824 and Department of Defense Grants W81XWH-18-1-0329 and W81XWH-19-1-0369 (to W.-C.L and F.-T.L.). Y.X. was supported by T32 Fellowship T32CA174647.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2015808118/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information.
References
- 1.Bao X., et al. , ACTL6a enforces the epidermal progenitor state by suppressing SWI/SNF-dependent induction of KLF4. Cell Stem Cell 12, 193–203 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krasteva V., et al. , The BAF53a subunit of SWI/SNF-like BAF complexes is essential for hemopoietic stem cell function. Blood 120, 4720–4732 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Lu W., et al. , Actl6a protects embryonic stem cells from differentiating into primitive endoderm. Stem Cells 33, 1782–1793 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Brahma S., Ngubo M., Paul S., Udugama M., Bartholomew B., The Arp8 and Arp4 module acts as a DNA sensor controlling INO80 chromatin remodeling. Nat. Commun. 9, 3309 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dion V., Shimada K., Gasser S. M., Actin-related proteins in the nucleus: Life beyond chromatin remodelers. Curr. Opin. Cell Biol. 22, 383–391 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Görzer I., et al. , The nuclear actin-related protein Act3p/Arp4p of Saccharomyces cerevisiae is involved in transcription regulation of stress genes. Mol. Microbiol. 50, 1155–1171 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Nishimoto N., et al. , Heterocomplex formation by Arp4 and β-actin is involved in the integrity of the Brg1 chromatin remodeling complex. J. Cell Sci. 125, 3870–3882 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Xiao S., et al. , Actin-like 6A predicts poor prognosis of hepatocellular carcinoma and promotes metastasis and epithelial-mesenchymal transition. Hepatology 63, 1256–1271 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng Z., Yang H., Xiao S., ACTL6A expression promotes invasion, metastasis and epithelial mesenchymal transition of colon cancer. BMC Cancer 18, 1020 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun W., Wang W., Lei J., Li H., Wu Y., Actin-like protein 6A is a novel prognostic indicator promoting invasion and metastasis in osteosarcoma. Oncol. Rep. 37, 2405–2417 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Meng L., et al. , BAF53a is a potential prognostic biomarker and promotes invasion and epithelial-mesenchymal transition of glioma cells. Oncol. Rep. 38, 3327–3334 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saladi S. V., et al. , ACTL6A is Co-amplified with p63 in squamous cell carcinoma to drive YAP activation, regenerative proliferation, and poor prognosis. Cancer Cell 31, 35–49 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makovec T., Cisplatin and beyond: Molecular mechanisms of action and drug resistance development in cancer chemotherapy. Radiol. Oncol. 53, 148–158 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amable L., Cisplatin resistance and opportunities for precision medicine. Pharmacol. Res. 106, 27–36 (2016). [DOI] [PubMed] [Google Scholar]
- 15.O’Donovan A., Davies A. A., Moggs J. G., West S. C., Wood R. D., XPG endonuclease makes the 3′ incision in human DNA nucleotide excision repair. Nature 371, 432–435 (1994). [DOI] [PubMed] [Google Scholar]
- 16.Hashimoto S., Anai H., Hanada K., Mechanisms of interstrand DNA crosslink repair and human disorders. Genes Environ. 38, 9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cavenagh J. D., Popat R., Optimal management of histone deacetylase inhibitor-related adverse events in patients with multiple myeloma: A focus on panobinostat. Clin. Lymphoma Myeloma Leuk. 18, 501–507 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Nagy Á., Lánczky A., Menyhárt O., Győrffy B., Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci. Rep. 8, 9227 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang W., et al. , Genomics of drug sensitivity in cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 41, D955–D961 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aird R. E., et al. , In vitro and in vivo activity and cross resistance profiles of novel ruthenium (II) organometallic arene complexes in human ovarian cancer. Br. J. Cancer 86, 1652–1657 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kothandapani A., Gopalakrishnan K., Kahali B., Reisman D., Patrick S. M., Downregulation of SWI/SNF chromatin remodeling factor subunits modulates cisplatin cytotoxicity. Exp. Cell Res. 318, 1973–1986 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khavari P. A., Peterson C. L., Tamkun J. W., Mendel D. B., Crabtree G. R., BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 366, 170–174 (1993). [DOI] [PubMed] [Google Scholar]
- 23.Muchardt C., Bourachot B., Reyes J. C., Yaniv M., ras transformation is associated with decreased expression of the brm/SNF2alpha ATPase from the mammalian SWI-SNF complex. EMBO J. 17, 223–231 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu K., Luo Y., Lin F. T., Lin W. C., TopBP1 recruits Brg1/Brm to repress E2F1-induced apoptosis, a novel pRb-independent and E2F1-specific control for cell survival. Genes Dev. 18, 673–686 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang W., et al. , Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 15, 5370–5382 (1996). [PMC free article] [PubMed] [Google Scholar]
- 26.Wang W., et al. , Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 10, 2117–2130 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Szerlong H., et al. , The HSA domain binds nuclear actin-related proteins to regulate chromatin-remodeling ATPases. Nat. Struct. Mol. Biol. 15, 469–476 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Y., et al. , Homozygous disruption of the Tip60 gene causes early embryonic lethality. Dev. Dyn. 238, 2912–2921 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serber D. W., Runge J. S., Menon D. U., Magnuson T., The mouse INO80 chromatin-remodeling complex is an essential meiotic factor for spermatogenesis. Biol. Reprod. 94, 8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suraweera A., O’Byrne K. J., Richard D. J., Combination therapy with histone deacetylase inhibitors (HDACi) for the treatment of cancer: Achieving the full therapeutic potential of HDACi. Front. Oncol. 8, 92 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ha K., et al. , Histone deacetylase inhibitor treatment induces ‘BRCAness’ and synergistic lethality with PARP inhibitor and cisplatin against human triple negative breast cancer cells. Oncotarget 5, 5637–5650 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang G., et al. , Panobinostat synergistically enhances the cytotoxic effects of cisplatin, doxorubicin or etoposide on high-risk neuroblastoma cells. PLoS One 8, e76662 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bennett G., Papamichos-Chronakis M., Peterson C. L., DNA repair choice defines a common pathway for recruitment of chromatin regulators. Nat. Commun. 4, 2084 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gursoy-Yuzugullu O., House N., Price B. D., Patching broken DNA: Nucleosome dynamics and the repair of DNA breaks. J. Mol. Biol. 428, 1846–1860 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morrison A. J., Genome maintenance functions of the INO80 chromatin remodeller. Philos. Trans. R Soc. Lond. B Biol. Sci. 372, 20160289 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hara R., Mo J., Sancar A., DNA damage in the nucleosome core is refractory to repair by human excision nuclease. Mol. Cell. Biol. 20, 9173–9181 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaillard H., et al. , Chromatin remodeling activities act on UV-damaged nucleosomes and modulate DNA damage accessibility to photolyase. J. Biol. Chem. 278, 17655–17663 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Hara R., Sancar A., The SWI/SNF chromatin-remodeling factor stimulates repair by human excision nuclease in the mononucleosome core particle. Mol. Cell. Biol. 22, 6779–6787 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hodges C., Kirkland J. G., Crabtree G. R., The many roles of BAF (mSWI/SNF) and PBAF complexes in cancer. Cold Spring Harb. Perspect. Med. 6, a026930 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niimi A., Chambers A. L., Downs J. A., Lehmann A. R., A role for chromatin remodellers in replication of damaged DNA. Nucleic Acids Res. 40, 7393–7403 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ribeiro-Silva C., Vermeulen W., Lans H., SWI/SNF: Complex complexes in genome stability and cancer. DNA Repair (Amst.) 77, 87–95 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Singh B. N., et al. , Nonhistone protein acetylation as cancer therapy targets. Expert Rev. Anticancer Ther. 10, 935–954 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walz S., et al. , Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature 511, 483–487 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schlosser I., et al. , Dissection of transcriptional programmes in response to serum and c-Myc in a human B-cell line. Oncogene 24, 520–524 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Karbaschi M., Brady N. J., Evans M. D., Cooke M. S., Immuno-slot blot assay for detection of UVR-mediated DNA damage. Methods Mol. Biol. 920, 163–175 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Shedden K. et al.; Director’s Challenge Consortium for the Molecular Classification of Lung Adenocarcinoma , Gene expression-based survival prediction in lung adenocarcinoma: A multi-site, blinded validation study. Nat. Med. 14, 822–827 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bonome T., et al. , A gene signature predicting for survival in suboptimally debulked patients with ovarian cancer. Cancer Res. 68, 5478–5486 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mateescu B., et al. , miR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat. Med. 17, 1627–1635 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.