Significance
Homologous recombination (HR) repairs DNA double-strand breaks by using a homologous template to retrieve sequence information lost at the break site. The broken DNA molecule first engages with the homologous donor molecule and is then separated from it to complete the process. Depending on the HR subpathways used, the separation step can lead to crossovers (COs) between the participating molecules. Such events can cause genomic alterations and eventually cancer if a donor molecule other than the identical sister chromatid is used. Here, we characterize two subpathways of HR with different propensities to form COs. We show the unexpected dominance of the CO-forming subpathway and characterize the processes involved in CO formation and subpathway choice in cancer and normal, untransformed cells.
Keywords: synthesis-dependent strand annealing, Holliday junctions, sister chromatid exchanges, crossovers, DNA repair synthesis
Abstract
Homologous recombination (HR) is an important DNA double-strand break (DSB) repair pathway that copies sequence information lost at the break site from an undamaged homologous template. This involves the formation of a recombination structure that is processed to restore the original sequence but also harbors the potential for crossover (CO) formation between the participating molecules. Synthesis-dependent strand annealing (SDSA) is an HR subpathway that prevents CO formation and is thought to predominate in mammalian cells. The chromatin remodeler ATRX promotes an alternative HR subpathway that has the potential to form COs. Here, we show that ATRX-dependent HR outcompetes RECQ5-dependent SDSA for the repair of most two-ended DSBs in human cells and leads to the frequent formation of COs, assessed by measuring sister chromatid exchanges (SCEs). We provide evidence that subpathway choice is dependent on interaction of both ATRX and RECQ5 with proliferating cell nuclear antigen. We also show that the subpathway usage varies among different cancer cell lines and compare it to untransformed cells. We further observe HR intermediates arising as ionizing radiation (IR)-induced ultra-fine bridges only in cells expressing ATRX and lacking MUS81 and GEN1. Consistently, damage-induced MUS81 recruitment is only observed in ATRX-expressing cells. Cells lacking BLM show similar MUS81 recruitment and IR-induced SCE formation as control cells. Collectively, these results suggest that the ATRX pathway involves the formation of HR intermediates whose processing is entirely dependent on MUS81 and GEN1 and independent of BLM. We propose that the predominant ATRX-dependent HR subpathway forms joint molecules distinct from classical Holliday junctions.
Homologous recombination (HR) is an essential process for the maintenance of genomic integrity and is vital for diverse cellular processes, including replication and double-strand break (DSB) repair during meiosis and in somatic cells. HR relies on the use of a homologous template for repair to ensure the faithful restoration of genetic information. DSB repair is channeled to HR by the 5′ to 3′ resection of break ends to generate 3′ single-stranded DNA (ssDNA) overhangs (1). The recombinase RAD51 is then loaded to ssDNA to direct the homology search for complementary sequences and the formation of a displacement loop (D-loop) to allow DNA repair synthesis and the recovery of the lost sequences (2–4). Repair can subsequently proceed through one of several distinct subpathways. During synthesis-dependent strand annealing (SDSA), the extended break end is displaced from the D-loop and annealed to the complementary sequence at the noninvading break end. Alternatively, the noninvading end anneals to the displaced strand of the D-loop in a second-end capture step, leading to the formation of a structure joining the two recombining molecules, which has been suggested to represent a double Holliday junction (dHJ) (5). This joint molecule (JM) needs to be untangled to allow proper chromosome segregation for which distinct mechanisms are in place. This structure can be resolved by a nuclease complex involving MUS81-EME1 (together with SLX1-SLX4) or by the resolvase GEN1 which induce asymmetrical or symmetrical incisions in the two strands at each HJ, giving rise to both crossover (CO) and non-CO products (6–8). Additionally, dHJs can be dissolved by the BLM/TOPOIIIα/RMI1/2 (BTR) complex, where the two HJs migrate toward each other and merge to form a hemicatenane that is then decatenated by topoisomerases, thus separating the two molecules without exchange of genetic material (9–11). Under specific circumstances, a third subpathway termed break-induced replication (BIR) can mediate conservative DNA synthesis initiated by the D-loop to copy the entire chromosome arm (12, 13).
Pathways leading to COs in somatic cells are often viewed as detrimental, as exchanges between homologous chromosomes can cause loss of heterozygosity (LOH), a contributing factor to carcinogenesis. Therefore, cells harbor multiple mechanisms of CO suppression. Arguably most importantly, somatic cells employ the sister chromatid instead of the homologous chromosome in both yeast and mammalian systems to avoid LOH (14, 15). Additionally, as SDSA is inherently refractory to CO formation, it is believed to be the predominant HR subpathway for the repair of DSBs (16–18). Furthermore, BLM-mediated dissolution of dHJs limits COs, with resolution believed to act as a last resort to handle these intermediates. This is corroborated by the genomic instability of cells from Bloom’s syndrome patients, which harbor mutations in BLM and are characterized by substantially elevated spontaneous levels of sister chromatid exchanges (SCEs) (19, 20), a CO outcome of intersister HR events. Results from HR reporter systems, where COs are rarely observed, and the lower rate of dHJ formation per DSB in somatic versus meiotic cells support a general CO-avoidance tendency (5, 15). This notion is also consistent with historic studies suggesting that DSB-inducing agents, including ionizing radiation (IR), are poor inducers of SCEs (21, 22). However, it is now known that most IR-induced two-ended DSBs are repaired in mammalian cells by nonhomologous end joining (NHEJ) with HR accounting for only a fraction of all repair events even in G2 phase, thereby explaining the relative scarcity of SCEs compared to the number of induced breaks (23–25). Moreover, the sister chromatid represents the natural substrate for HR and COs might not be readily monitored in reporter systems which rely on unequal recombination events between the damaged loci and the provided substrates. Thus, it is unclear how often COs arise during HR in somatic cells.
It has been shown that the chromatin remodeler ATRX promotes extended DNA repair synthesis and SCE formation during HR (26). In this process, ATRX and its partner DAXX deposit the histone variant H3.3 in a step downstream of RAD51 removal (26). Interestingly, the ATRX/DAXX complex interacts with the DNA polymerase processivity factor proliferating cell nuclear antigen (PCNA) after DSB induction, suggesting that histone deposition is tightly coupled to DNA repair synthesis (27). Thus, the model emerged that histone deposition at the D-loop serves to overcome topological constraints arising during the DNA synthesis step which, if not assuaged, preclude extended DNA repair synthesis. The observation that SCEs frequently arise in a manner dependent on ATRX supports previous findings of high IR-induced SCE frequencies (26, 28). It further suggests that extended repair synthesis leads to annealing of the second end to the displaced strand of the D-loop, generating a JM with the potential for CO formation. This notion is consistent with findings showing that gene conversion tracts for CO-forming pathways are substantially larger than for SDSA (29, 30). However, the frequency of the ATRX subpathway versus the use of SDSA has not been investigated and would present an opportunity to quantitatively compare HR subpathway choice, but an SDSA-specific factor is required for this comparison.
Multiple helicases in yeast and mammals have been implicated to facilitate SDSA, including the yeast Srs2 helicase which is known to dissociate RAD51 from ssDNA and to promote non-CO products via SDSA (31–33). Further evidence showed the ability of Srs2 to dismantle extended D-loops post repair synthesis in a manner enhanced by small ubiquitin-like modifier (SUMO)-conjugated PCNA, thereby allowing strand annealing and CO avoidance (34). While Srs2 does not have a known homolog in human cells, multiple helicases have analogous functions. A prominent example is RECQ5, which has multiple roles in the DNA damage response. RECQ5 is recruited to sites of DNA damage in a MRN-dependent manner, resolves replication–transcription conflicts, and promotes processing of stalled forks (35–37). Similar to Srs2, RECQ5 has been shown to interact with and remove RAD51 from ssDNA using an ATPase activity that is stimulated by RPA (38, 39). Further studies suggested that this function serves to promote SDSA by preventing RAD51 filament reformation post strand displacement that could lead to reinvasion cycles and increase the chance of JM formation (40). Additionally, RECQ5 depletion leads to elevated SCE formation, further corroborating its role in promoting SDSA (32, 40).
Here, we show that ATRX and RECQ5 define two distinct subpathways of HR-mediated repair at two-ended DSBs with varying individual contributions to overall HR in different cell lines. We also show that the ATRX-dependent pathway is dominant over SDSA in cells where both factors are present, with PCNA interaction being an important node of pathway choice. A comparison between the number of HR events and the quantity of IR-induced SCEs reveals that COs are a frequent outcome of HR. We propose a model in which ATRX-mediated chromatin remodeling during repair synthesis promotes the formation of JMs that are dependent on MUS81 and GEN1 for resolution and are not subject to BLM-mediated dissolution. This unexpected pathway choice is likely unique to two-ended DSBs and distinct from HR intermediates generated during S phase at stalled or collapsed replication forks.
Results
ATRX and RECQ5 Define Distinct Subpathways of HR.
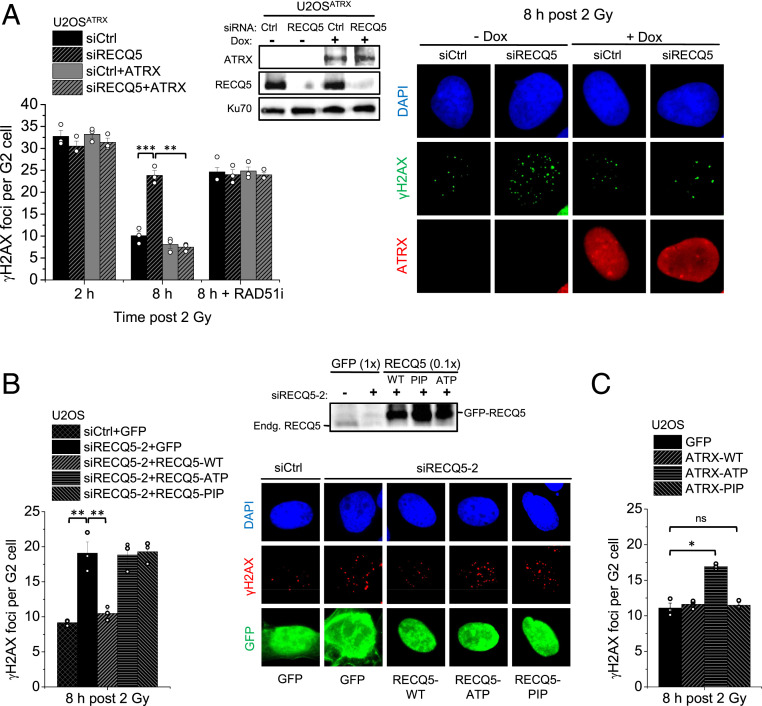
The requirement for ATRX in HR prompted us to investigate DSB repair in cells naturally lacking ATRX. Approximately 10 to 15% of all tumors rely on a telomerase-independent pathway for telomere elongation, called alternative lengthening of telomeres (ALT). This ALT phenotype is strongly associated with inactivating mutations of the ATRX/DAXX/H3.3 complex, with the majority exhibiting loss of expression of the ATRX protein (41, 42). To investigate how these cells perform HR, we focused on the known ALT U2OS cells and used a modified line that expresses ATRX within the range of physiological levels upon induction with doxycycline (43) (SI Appendix, Fig. S1A), from herein referred to as U2OSATRX. To characterize the DSB repair dependency of these cells, we quantified the number of γH2AX foci as a marker of DSBs and followed their resolution over time as a measure for DSB repair, as previously described (44). We specifically analyzed cells irradiated and maintained in G2 phase during repair to avoid any complications from replication-associated processes and investigate HR at two-ended DSBs. Assessment of γH2AX foci in G2-phase U2OSATRX cells revealed a marked decrease in foci numbers between 2 and 8 h post IR, indicating efficient repair (Fig. 1A). Inhibition of RAD51 (RAD51i) resulted in significantly higher levels of γH2AX foci at 8 h post IR compared to control cells, showing that U2OSATRX cells perform HR in an ATRX-independent manner (Fig. 1A). Depletion of the SDSA factor RECQ5 also resulted in elevated γH2AX foci levels similar to those in RAD51i-treated cells and inhibition of RAD51 in U2OSATRX cells lacking RECQ5 did not result in a further increase, indicating that SDSA is used for all HR events (Fig. 1A). Foci levels at 2 h post IR were not altered after RECQ5 depletion, consistent with previous observations that HR defects are only observable at later time points while NHEJ repairs DSBs during the first hours after IR in G2 (23, 24, 26). Expression of ATRX did not significantly affect γH2AX foci levels in control U2OSATRX cells at 2 or 8 h but rescued the elevated foci numbers at 8 h in RECQ5-depeleted cells, showing that ATRX can compensate for the loss of RECQ5-mediated SDSA. However, ATRX expression did not rescue the elevated γH2AX foci level observed in RAD51i-treated cells, consistent with RAD51 being upstream of two subpathways of HR defined by the factors ATRX and RECQ5 (Fig. 1A). A similar result was obtained after transient ATRX expression in unmodified U2OS cells (SI Appendix, Fig. S1 B and C) and another ATRX-deficient ALT cell line, Saos-2 (SI Appendix, Fig. S1D). However, Saos-2 cells showed higher γH2AX foci levels than U2OS cells that were reduced after ATRX expression, consistent with lower RECQ5 levels in Saos-2 compared with U2OS cells that might be insufficient to fully compensate for the lack of ATRX (SI Appendix, Fig. S1E).
Fig. 1.
ATRX and RECQ5 define distinct subpathways of HR. (A) U2OSATRX cells, with and without doxycycline-induced ATRX expression, were transfected with siCtrl or siRECQ5 and treated with RAD51 inhibitor (RAD51i) prior to IR and throughout repair incubation. γH2AX foci were enumerated in EdU-negative G2 cells. Spontaneous foci (four to six) were subtracted. Knockdown and ATRX expression were confirmed by immunoblotting. Representative images of γH2AX foci and ATRX expression are shown. (B) U2OS cells were transfected with siCtrl or siRECQ5-2 and GFP, GFP-RECQ5-WT, GFP-RECQ5-ATP, or GFP-RECQ5-PIP, irradiated and γH2AX foci were enumerated in GFP-positive, EdU-negative G2 cells. Knockdown of the endogenous RECQ5 and GFP-RECQ5 expression levels were confirmed by immunoblotting (SI Appendix, Fig. S2A). Spontaneous foci (four to six) were subtracted. Representative images of γH2AX foci in GFP-positive cells are shown. (C) U2OS cells were transfected with GFP, GFP-ATRX-WT, GFP-ATRX-ATP, or GFP-ATRX-PIP, irradiated and γH2AX foci were enumerated in GFP-positive, EdU-negative G2 cells. Spontaneous foci (four to six) were subtracted. All data show mean ± SEM (n = 3). Results from individual experiments, each derived from 40 cells, are indicated. *P < 0.05; **P < 0.01; ***P < 0.001; ns: not significant (two-tailed t test).
ATRX and RECQ5 Interact with PCNA to Regulate HR Subpathway Choice.
To confirm the specificity of the RECQ5 knockdown and further characterize the role of RECQ5, we used another siRNA (siRECQ5-2) and complemented back with siRNA-resistant GFP-tagged plasmids carrying the wild-type (WT) sequence, mutations in the helicase/ATPase domain (RECQ5-ATP), or mutations in the PCNA-interaction peptide motif (RECQ5-PIP). U2OS cells treated with siRECQ5-2 and GFP showed an elevated level of γH2AX foci 8 h after IR which was rescued by the expression of siRNA-resistant RECQ5-WT, confirming the specificity of the knockdown (Fig. 1B and SI Appendix, Fig. S2A). Interestingly, both RECQ5-ATP and RECQ5-PIP failed to rescue the repair defect, showing that both domains are important for RECQ5 function in HR (Fig. 1B). In contrast to the helicase/ATPase function, the requirement of the PIP domain was unexpected and raised the possibility that both RECQ5 and ATRX are competing for PCNA binding, as ATRX has been already shown to require its PIP domain for HR (26).
To explore this possibility, we overexpressed ATRX defective for either its ATPase function (ATRX-ATP) or its PCNA interaction (ATRX-PIP) in U2OS cells lacking ATRX and asked if they differentially interfere with subpathway choice. We have previously shown that both mutants are defective in the ATRX pathway as they fail to restore repair in HeLa ATRX KO cells, assessed by γH2AX foci analysis and the formation of BrdU repair foci (26). Moreover, the ATRX-PIP but not the ATRX-ATPase mutant fails to interact with PCNA in coimmunoprecipitation (co-IP) studies (26). Of note, the ATRX-PIP mutant did not affect foci levels in U2OS cells (Fig. 1C), indicating that repair could still take place. In contrast, ATRX-ATP caused a repair defect, presumably by outcompeting RECQ5-dependent SDSA through PCNA binding but failing to complete repair due to defective ATPase function (Fig. 1C). Control experiments confirmed that neither ATRX-PIP nor ATRX-ATP is able to rescue the repair defect in siRECQ5-treated U2OS cells (SI Appendix, Fig. S2B). These data suggest that HR subpathway usage is governed by the ability of both ATRX and RECQ5 to interact with PCNA.
ATRX and RECQ5 Differentially Affect Long Tract Gene Conversion and SCE Formation.
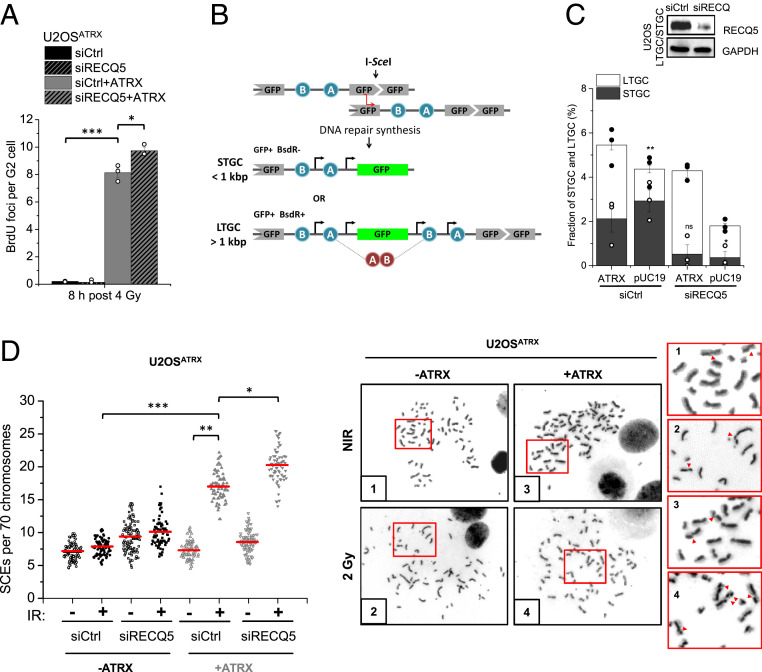
Since extended DNA repair synthesis is a key feature of ATRX-dependent HR, we assessed how RECQ5 affects this step using our previously described BrdU incorporation assay (23, 26). Cells were irradiated and maintained in G2 phase during repair, and the incorporation of the nucleoside analog BrdU at DSB sites was quantified by enumerating visible BrdU foci. Robust BrdU foci formation at 8 h post IR was observed only in ATRX-expressing U2OSATRX cells (Fig. 2A). Depletion of RECQ5, in contrast to the lack of ATRX, slightly increased BrdU foci formation (Fig. 2A). These results show that extended DNA repair synthesis is strictly dependent on ATRX which outcompetes RECQ5-dependent SDSA, as most BrdU foci are not suppressed by RECQ5.
Fig. 2.
ATRX and RECQ5 differentially affect long tract gene conversion and SCE formation. (A) U2OSATRX cells, with and without doxycycline-induced ATRX expression and transfected with siCtrl or siRECQ5, were labeled with EdU, irradiated, and then incubated with BrdU for 8 h. BrdU foci representing DNA repair synthesis were enumerated in EdU-negative G2 cells. Spontaneous foci (fewer than one) were subtracted. (B) Schematic diagram of the LTGC/STGC reporter system. U2OS cells carry a 5′ truncated GFP cassette followed by two incorrectly oriented exons (exon B placed 5′ to exon A) of the Blasticidin resistance (BsdR) gene, and a GFP cassette that is disrupted by the recognition sequence of the I-SceI endonuclease. To obtain a functional GFP protein, the break has to be repaired by HR using the truncated GFP cassette as a template. STGC events of fewer than 1 kbp result in a functional GFP gene but fail to correctly orient the exons of the BsdR gene, and the cells are thus sensitive to Blasticidin. LTGC events of >1 kbp restore the GFP sequence and additionally place exon B of the BsdR gene 3′ to exon A, reinstating correct orientation and conferring Blasticidin resistance. Thus, LTGC events lead to GFP+, BsdR+ cells [image modified from Nagaraju et al. (45)]. For the analysis, GFP+ cells (representing both STGC and LTGC events) and BsdR+ cells (LTGC events) are enumerated. (C) U2OS STGC/LTGC reporter cells were transfected with siCtrl or siRECQ5 and pUC19 or myc-ATRX and 40 h later transfected with I-SceI plasmids. Cells were assessed for GFP expression and Blasticidin resistance and numbers were plotted as a fraction of all cells. Control experiments showed that the fraction of S/G2-phase cells varied by less than 5% between siRECQ5-treated, ATRX-expressing, and control cells, excluding that changes in cell cycle distributions substantially influence the results. Knockdown was confirmed by immunoblotting and myc-ATRX expression efficiency was previously assessed (26). The significance indication above the white bar compares the LTGC fraction of pUC19 to the ATRX-expressing siCtrl-treated cells; indications above the black bars compare STGC fractions upon RECQ5 depletion to the respective siCtrl-treated cells. (D) U2OSATRX cells, with and without doxycycline-induced ATRX expression, were transfected with siCtrl or siRECQ5 and incubated with BrdU for 48 h. Cells were then irradiated with 2 Gy, collected after 16 h, and processed to obtain mitotic spreads. SCEs per spread were quantified and normalized to 70 chromosomes. Representative images of chromosome spreads of nonirradiated and irradiated siCtrl-treated cells with and without ATRX expression are shown; red arrows in the magnifications show individual SCE events. Data in A and C show mean ± SEM (n = 3). Results from individual experiments, each derived from 40 cells in A, are indicated. Individual SCE data are shown and red horizontal lines indicate the mean; 70 to 120 spreads and >4,000 chromosomes per condition were analyzed from three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ns: not significant (two-tailed t test); NIR: nonirradiated.
We additionally investigated the distinct roles of ATRX and RECQ5 during DNA repair synthesis by using a reporter assay that discriminates between short (STGC) and long tract gene conversion (LTGC) events (45). Expression of the I-SceI nuclease leads to a DSB in a GFP gene that can be repaired using a GFP donor sequence positioned 5′ of the acceptor gene as a template for repair (Fig. 2B). The specific feature of this assay is its ability to monitor the extent of DNA repair synthesis that leads to restoration of the GFP gene. During STGC, DSB repair proceeds by limited DNA repair synthesis while LTGC involves copying a cassette that was inserted between the two GFP genes, leading to the duplication of this cassette and expression of an antibiotic resistance gene (Fig. 2B). Hence, LTGC confers antibiotic resistance in addition to GFP expression while STGC involves GFP expression but no antibiotic resistance. To assess the roles of RECQ5 and ATRX in STGC and LTGC, we expressed ATRX in U2OS cells carrying this reporter cassette. Compared to such cells with both factors present, U2OS cells without ATRX showed substantially reduced LTGC (26) (Fig. 2C). In contrast, depletion of RECQ5 in U2OS cells with or without ATRX nearly abolished STGC (Fig. 2C). These results reveal that RECQ5-dependent HR involves short patches of DNA repair synthesis while ATRX-dependent HR proceeds by extended DNA repair synthesis. A comparison with the BrdU incorporation assay suggests that a fraction of the LTGC events assessed in the reporter assay can give rise to microscopically visible BrdU foci while STGC does not. The presence of basal LTGC events in cells lacking ATRX and/or RECQ5 could be the result of an alternative pathway, possibly BIR at one-ended breaks arising when cells replicate.
To further dissect how the subpathways interact, we analyzed SCEs arising in response to G2-induced damage as a known outcome of the ATRX pathway (26). We optimized the time used for SCE measurement for U2OSATRX cells based on their cell cycle progression (SI Appendix, Fig. S2C) and confirmed their DSB repair capacity for the corresponding repair time (SI Appendix, Fig. S2D). We observed IR-induced SCEs only in ATRX-expressing U2OSATRX cells and RECQ5 depletion in those cells caused only modestly higher levels, supporting the notion that the ATRX subpathway dominates over RECQ5-dependent SDSA (Fig. 2D).
HR Subpathway Usage in ATRX-Proficient Cancer and Normal Cells.
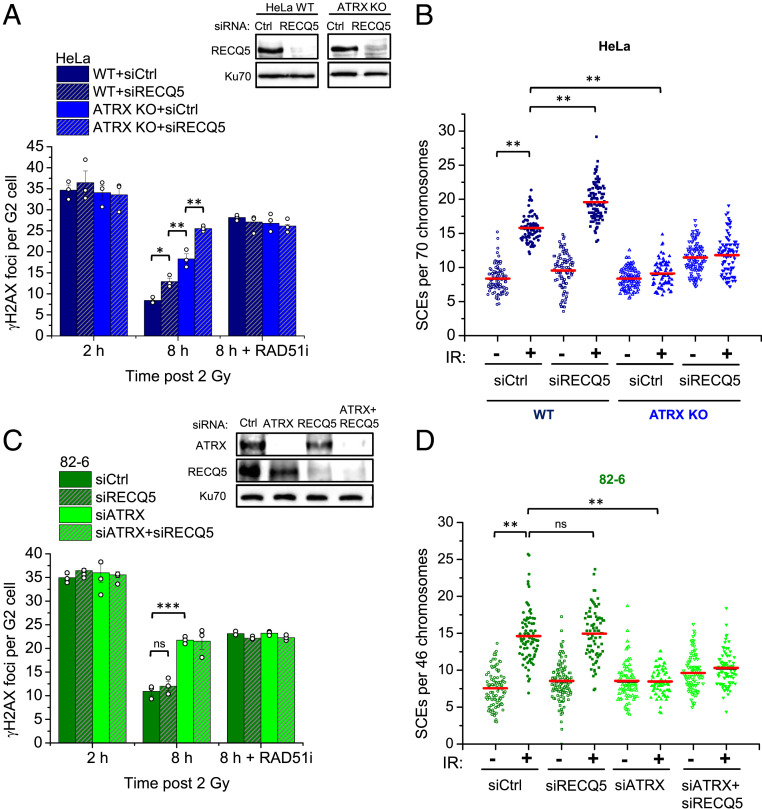
To investigate if similar subpathway usage occurs in other cell types, we then carried out similar repair analyses in ATRX-proficient HeLa WT cells where, unlike U2OS cells, RECQ5 depletion conferred only a mild DSB repair defect at 8 h after IR (Fig. 3A). In contrast, HeLa ATRX knockout (KO) cells showed substantially elevated foci numbers as previously described (26). RECQ5 depletion in ATRX KO cells led to unrepaired DSBs similar to inhibition of RAD51, consistent with ATRX- and RECQ5-dependent HR accounting for the majority of all IR-induced HR events (Fig. 3A). Interestingly, HeLa cells overexpressing RECQ5-ATP but not RECQ5-WT or RECQ5-PIP exhibited partially elevated foci numbers at 8 h post IR (SI Appendix, Fig. S3A), confirming that PCNA interaction is central to HR subpathway choice. We also analyzed SCE formation and repair synthesis and, similar to U2OS cells, observed IR-induced SCEs and BrdU foci only in ATRX-expressing cells where RECQ5 depletion led to only a modest increase in SCE and BrdU foci numbers (Fig. 3B and SI Appendix, Fig. S3B). Taken together, these data show that HeLa cells rely primarily on ATRX for the repair of DSBs in G2 even in the presence of RECQ5.
Fig. 3.
HR subpathway usage in ATRX-proficient cancer and normal cells. (A) HeLa WT and ATRX KO cells were transfected with siCtrl or siRECQ5 and treated with RAD51i prior to IR and throughout repair incubation. γH2AX foci were enumerated in EdU-negative G2 cells. Spontaneous foci (three to four) were subtracted. Knockdown was confirmed by immunoblotting. Data for siCtrl were extracted from supplemental appendix, figure S3A in ref. 26 as they were part of the same experiment with siRECQ5. (B) HeLa WT and ATRX KO cells were transfected with siCtrl or siRECQ5 and incubated with BrdU for 48 h. Cells were then irradiated with 2 Gy, collected after 8 h, and processed to obtain mitotic spreads. SCEs per spread were quantified and normalized to 70 chromosomes. (C) 82-6 cells were transfected with siCtrl, siRECQ5, and/or siATRX and treated with RAD51i prior to IR and throughout repair incubation. γH2AX foci were enumerated in EdU-negative G2 cells. Spontaneous foci (one to two) were subtracted. Knockdown was confirmed by immunoblotting. (D) 82-6 cells were transfected with siCtrl, siRECQ5, and/or siATRX and incubated with BrdU for 48 h. Cells were then irradiated with 2 Gy, collected after 12 h, and processed to obtain mitotic spreads. SCEs per spread were quantified and normalized to 46 chromosomes. Foci data show mean ± SEM (n = 3). Results from individual experiments, each derived from 40 cells, are indicated. Individual SCE data are shown and red horizontal lines indicate the mean; 70 to 120 spreads and >4,000 chromosomes per condition were analyzed from three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ns: not significant (two-tailed t test).
Since cancer cells utilize the RECQ5 pathway to different extents, we next asked how normal, untransformed cells mediate HR subpathway choice. We investigated hTert-immortalized primary human fibroblasts (82-6 cells) and observed elevated γH2AX foci numbers at 8 h after IR following depletion of ATRX while siRECQ5 did not affect foci numbers with or without ATRX depletion (Fig. 3C). RAD51i led to similarly elevated foci numbers as ATRX depletion and siATRX and/or siRECQ5 did not further increase foci numbers in RAD51i-treated cells (Fig. 3C). Additionally, we observed IR-induced SCE formation and BrdU foci whose levels remained unchanged in RECQ5-depleted cells but was abolished by ATRX depletion (Fig. 3D and SI Appendix, Fig. S3 C and D). Thus, unlike U2OS and HeLa cells, normal cells seem to exclusively utilize the ATRX pathway with no apparent contribution of RECQ5-dependent SDSA.
HR and SCE Frequencies at IR-Induced DSBs.
A more quantitative comparison between the number of HR events promoted by the ATRX subpathway and the consequent formation of COs was carried out using RAD51 foci as a specific marker for HR. Total HR events in ATRX-proficient U2OSATRX, HeLa, and 82-6 cells were estimated to be the net difference between the peak number of RAD51 foci formed and the residual foci level at the end of the repair time used for the SCE measurements. All three cell lines showed similar numbers of RAD51 foci being maximally formed (SI Appendix, Fig. S4A) and had a near-identical number of net HR events irrespective of RECQ5 presence (∼20) (SI Appendix, Fig. S4B). These 20 HR events led to about 8 SCEs above background level, which increase in U2OSATRX and HeLa cells upon RECQ5 depletion to about 10 IR-induced SCEs (Figs. 2D and 3 B and D).
We also addressed some of the potential limitations of our SCE and repair assays. First, we confirmed previous data that no substantial RAD51 foci loss occurred until the peak time by depleting RAD54, a factor known to remove RAD51 (3, 26) (SI Appendix, Fig. S5A). We additionally confirmed the total number of HR events by assessing the accumulation of the phosphorylated form of the replication protein A, a marker for resected DSBs (SI Appendix, Fig. S5B). Further, while a dose of 2 Gy can be considered nonphysiological, we show a similar ATRX dependency and SCE frequency at lower doses (SI Appendix, Fig. S5 C and D). It is worth noting in this context that depleting RECQ5 mildly elevates spontaneous SCEs, consistent with RECQ5’s role in resolving replication–transcription conflicts (37), but to a level substantially lower than those induced by 2 Gy. Thus, the analysis of SCEs induced by 2 Gy is less likely to be affected by a small change in spontaneous breaks than analyses at lower doses. Additionally, BrdU-labeled cells used for the SCE assay show similar HR usage as non-BrdU-substituted cells (SI Appendix, Fig. S5E).
ATRX-Dependent HR Leads to the Formation of IR-Induced MUS81-Dependent Intermediates.
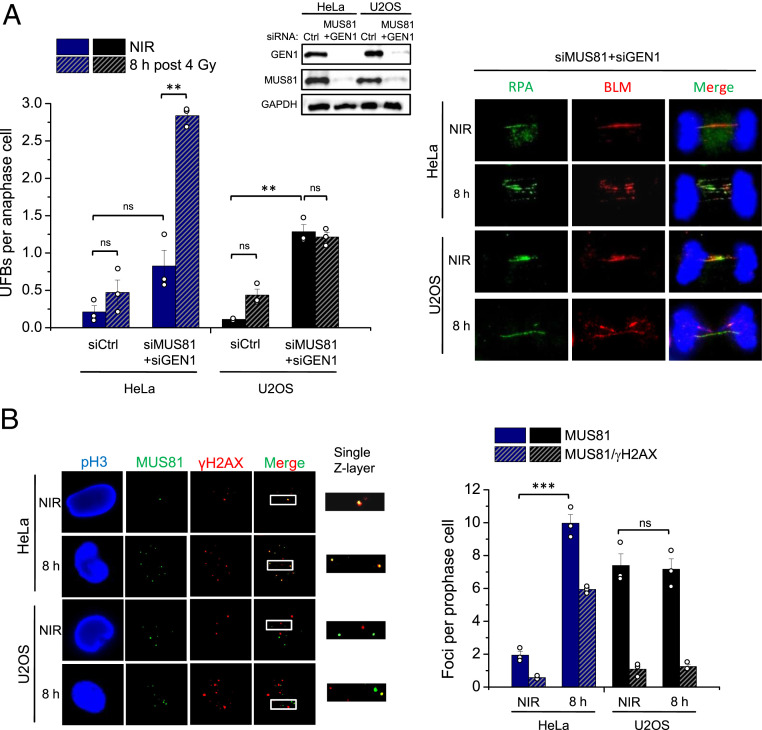
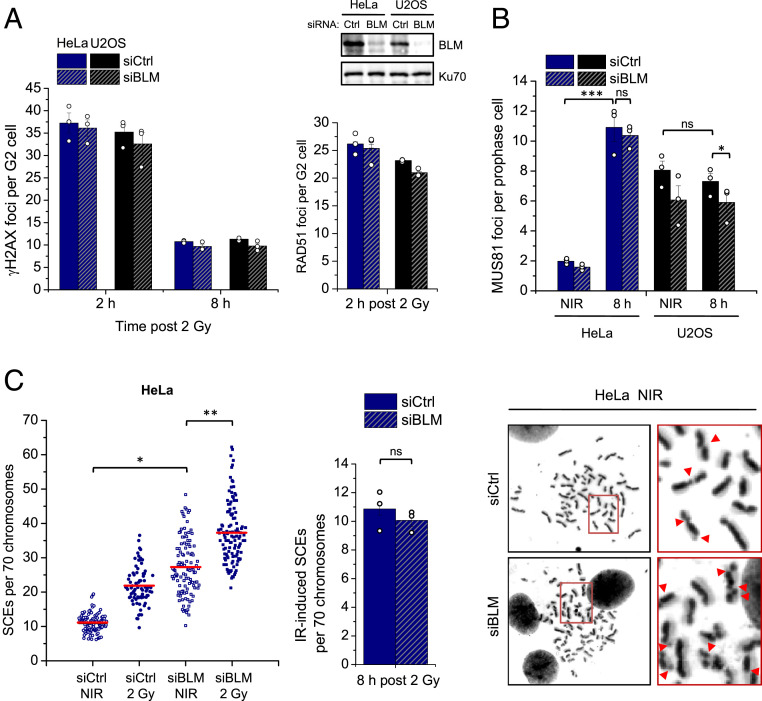
The high SCE frequency observed in the ATRX subpathway suggested a strong reliance on nuclease-dependent resolution of JMs formed after the completion of DNA synthesis. To further characterize this pathway, the formation of HR intermediates in response to IR was analyzed. Several types of JMs can be visualized in anaphase/telophase cells as DAPI-negative ultrafine bridges (UFBs) that immunostain positively for RPA and/or BLM (46, 47). Recently, HR-specific intermediates were shown to arise in cells depleted of MUS81 and GEN1, and increase following DNA damage that is repaired by HR (48). To assess if such intermediates also arise in response to IR, HeLa and U2OS cells were transfected with siMUS81 and siGEN1, irradiated with 4 Gy, and analyzed for UFB formation (Fig. 4A). We chose these two cell systems because their good proliferation allowed the most reliable assessment of mitotic cells in this assay. EdU was added to the cells before IR and only EdU-negative anaphase cells were analyzed to ensure the observed UFBs are specific to HR events in G2. HeLa cells treated with siCtrl exhibited a modest but nonsignificant increase in UFBs 8 h post IR (Fig. 4A). Consistent with published data, unirradiated nuclease-depleted HeLa and U2OS cells showed an increase in UFBs compared to unirradiated siCtrl-treated cells, likely arising from S phase-generated joint structures that were carried over to G2 (48). However, irradiated nuclease-depleted HeLa cells exhibited approximately a fourfold increase in UFBs, demonstrating that IR-induced JMs are processed by nucleases that can give rise to CO products (Fig. 4A). In contrast, U2OS cells failed to show such an IR-induced increase in nuclease-depleted cells, indicating that IR-induced UFBs specifically arise in cells utilizing the ATRX subpathway (Fig. 4A). We also analyzed UFB staining patterns and found that most bridges in nuclease-depleted samples stained for RPA only or RPA plus BLM and only a small fraction was positive for BLM only. No considerable differences in staining patterns were observed between nonirradiated and irradiated as well as between HeLa and U2OS cells (SI Appendix, Fig. S6A).
Fig. 4.
ATRX-dependent HR leads to the formation of IR-induced MUS81-dependent HR intermediates. (A) HeLa and U2OS cells were transfected twice on consecutive days with siCtrl or siMUS81+siGEN1, irradiated, and RPA- and/or BLM-positive UFBs were enumerated in EdU-negative late anaphase cells. Representative images of UFBs in MUS81- and GEN1-depleted HeLa and U2OS cells are shown. Knockdown was confirmed by immunoblotting. (B) HeLa and U2OS cells were irradiated with 2 Gy, prophase cells positive for the mitotic marker pH3 and negative for EdU were selected, and Z-stack images were captured and analyzed for MUS81 foci numbers and colocalization with γH2AX foci. All data show mean ± SEM (n = 3) and results from individual experiments, each derived from 40 (A) or 20 (B) cells, are indicated. **P < 0.01; ***P < 0.001; ns: not significant (two-tailed t test); NIR: nonirradiated.
After establishing a role for MUS81 in resolving these HR structures arising in G2-irradiated cells, MUS81 localization to DSBs in response to IR was also analyzed. HeLa and U2OS cells were assessed for the recruitment of MUS81 to γH2AX foci 8 h after IR. Since discrete MUS81 foci were difficult to visualize in G2, cells in prophase were analyzed, consistent with a peak of MUS81 nuclear localization at G2/M entry (6). HeLa cells exhibited a significant increase in MUS81 foci in response to IR, with ∼60% colocalizing with γH2AX foci (Fig. 4B). In contrast, U2OS cells did not show an IR-induced elevation of MUS81 foci levels, despite having significantly more foci than unirradiated HeLa cells. Furthermore, most MUS81 foci in both irradiated and unirradiated U2OS samples did not colocalize with γH2AX foci, indicating their recruitment to structures other than DSB-derived intermediates, likely 3′ flaps or other structures from replication-associated events (Fig. 4B).
Processing of ATRX-Dependent HR Intermediates Is BLM Independent.
Since HR intermediates can also be targeted by dissolution that prevents COs, the involvement of this pathway was investigated by depleting the BLM helicase. Since BLM has been reported to also be involved in resection (49), the proficiency of HR was first assessed in BLM-depleted cells. HeLa and U2OS cells treated with siBLM showed normal peak formation of RAD51 foci at 2 h post IR and similar γH2AX foci levels at 2 and 8 h compared to control cells, confirming that resection in G2 is unaffected and that repair can take place via HR (Fig. 5A). Since BLM was shown to unwind double-stranded DNA (dsDNA) at UFBs, allowing for RPA binding, BLM deficiency prevents UFB visualization using BLM or RPA immunofluorescence (48). Therefore, this assay could not be used for the analysis of HR intermediate formation. Instead, MUS81 foci formation was assessed in the absence of BLM, with increased levels expected to compensate for loss of the BLM function under investigation. MUS81 foci were enumerated in prophase, prometaphase, and metaphase cells 8 h post 2 Gy in both HeLa and U2OS cells. Consistent with data described above, HeLa cells exhibited a significant IR-induced increase in MUS81 foci in all mitotic stages, with the highest foci numbers in prophase (Fig. 5B) and reduced numbers in prometaphase and metaphase cells (SI Appendix, Fig. S6B). BLM depletion did not impact MUS81 foci levels in any of the mitotic stages analyzed, suggesting that the resolvase already occupies all cleavable structures even in the presence of BLM (Fig. 5B and SI Appendix, Fig. S6B). U2OS cells had similar numbers of MUS81 foci in both unirradiated and irradiated samples in all mitotic stages, which also decreased from prophase to metaphase. Depletion of BLM slightly decreased MUS81 foci in prophase cells, after which they stayed similar to control levels in prometaphase and metaphase cells (Fig. 5B and SI Appendix, Fig. S6B). Therefore, in both cell lines, BLM depletion did not increase MUS81 foci localization, suggesting that it does not enhance the level of resolvase-positive JM structures.
Fig. 5.
Processing of ATRX-dependent HR intermediates is BLM independent. (A) HeLa and U2OS cells were transfected with siCtrl or siBLM, irradiated, and γH2AX (Left) and RAD51 (Right) foci were enumerated in EdU-negative G2 cells. Spontaneous foci (four to six γH2AX and fewer than one RAD51) were subtracted. Knockdown was confirmed by immunoblotting. (B) HeLa and U2OS cells were transfected with siCtrl or siBLM, irradiated with 2 Gy, and MUS81 foci were enumerated in pH3-positive, EdU-negative prophase cells. (C) HeLa cells were transfected with siCtrl or siBLM and incubated with BrdU for 48 h. Cells were then irradiated with 2 Gy, collected after 8 h, and processed to obtain mitotic spreads. SCEs per spread were quantified and normalized to 70 chromosomes. Bar graph shows the IR-induced SCE numbers (total SCEs after background subtraction). Representative images of chromosome spreads from unirradiated siCtrl- and siBLM-treated cells are shown on the right; red arrows in the magnifications show individual SCE events. Individual SCE data are shown and red horizontal lines indicate the mean; 70 to 120 spreads and >4,000 chromosomes per condition were analyzed from three independent experiments. Foci and IR-induced SCE data show mean ± SEM (n = 3) and results from individual experiments, each derived from 40 (A and C) or 20 (B) cells, are indicated. *P < 0.05; **P < 0.01; ***P < 0.001; ns: not significant (two-tailed t test); NIR: nonirradiated.
We also assessed SCE formation to test if the knockdown of BLM would lead to more IR-induced exchanges, as the unprocessed structures would then be resolved by nucleases. Consistent with the literature, BLM depletion significantly increased SCE levels in unirradiated cells, which are known to be caused by the resolution of intermediates arising during S phase that are normally dissolved by the BTR complex (6, 50) (Fig. 5C). BLM-depleted cells also exhibited an increase in SCEs in response to IR, but to an extent similar to BLM-proficient cells (Fig. 5C), indicating that BLM normally does not process HR intermediates arising from two-ended DSBs.
Discussion
HR by ATRX and RECQ5.
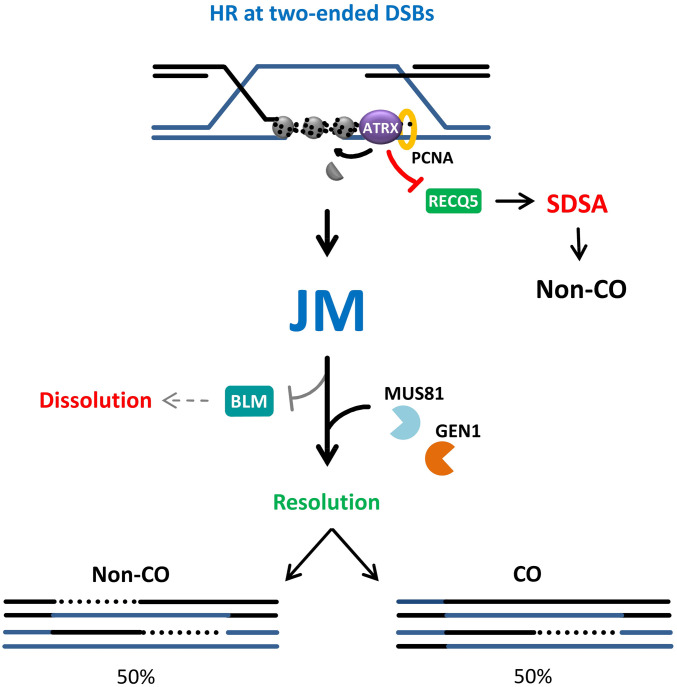
HR of two-ended DSBs can proceed by SDSA or a pathway involving JM formation. Key distinguishing features between the two pathways are the extent of DNA repair synthesis and potential for CO formation. SDSA is believed to involve only limited DNA repair synthesis prior to strand displacement and annealing to the second, noninvasive break end, thus preventing CO formation. In contrast, following extended repair synthesis, the noninvading end can anneal to the D-loop which leads to the formation of a JM with the potential for CO formation. RECQ5 has previously been implicated in SDSA (40) while ATRX has been shown to promote extended DNA repair synthesis and SCE formation (26). Here, we present evidence that RECQ5 and ATRX define distinct RAD51-dependent HR subpathways which together account for the majority, if not all, of HR events at two-ended DSBs. The analysis of RECQ5 mutants revealed that its helicase/ATPase function is essential for SDSA, as previously suggested (40). Interestingly, the ability to interact with PCNA is also important for DSB repair, suggesting that RECQ5 functions during or after DNA synthesis. Interaction with PCNA also likely serves as a node of regulation between the subpathways to define the level at which RECQ5 and ATRX compete for DSB repair (Fig. 6).
Fig. 6.
Model for the interplay between ATRX and RECQ5 to regulate distinct subpathways of HR. Repair of two-ended DSBs by ATRX-mediated HR outcompetes RECQ5, possibly through PCNA binding, thereby suppressing SDSA. Upon the completion of DNA repair synthesis, the resulting JM is not subject to dissolution by BLM and is channelled into the resolution pathway by MUS81 and GEN1, leading to equal probability of CO and non-CO products.
We argue that the ATRX-dependent pathway outcompetes SDSA in cells where both pathways are present based on multiple observations. First, loss of ATRX in HeLa cells causes a greater repair defect than loss of RECQ5 and siATRX in 82-6 cells causes a repair defect similar to RAD51 inhibition while siRECQ5 has no effect. Additionally, HeLa, U2OSATRX and 82-6 cells expressing ATRX show BrdU foci and SCE levels that are only slightly or not at all increased after siRECQ5. Since ATRX-dependent HR has the potential to form COs, these observations challenge the widely held belief that CO formation is suppressed in mammalian cells. Indeed, we discuss below that HR involving ATRX-dependent extended repair synthesis frequently leads to SCEs.
It is important to note that U2OS and to a lesser extent Saos-2 and HeLa cells require SDSA while nontransformed cells do not use RECQ5 for the repair of IR-induced DSBs. This highlights the discrepancy between cell lines in their HR subpathway usage, an aspect that should be taken into consideration when analyzing repair assays and warrants further investigation into how subpathway choice is made. This is also relevant when studying the ALT process in ATRX-deficient cells, which rely on an HR subpathway for telomere maintenance and whose differences in the repair of internal breaks might be paralleled at the telomeres (51).
SCEs Frequently Arise in Irradiated Mammalian Cells.
Here, we quantitatively compare the number of RAD51-dependent HR events in irradiated HeLa, U2OS, and 82-6 cells with the number of induced SCEs, all measured at the same condition. Our γH2AX and RAD51 foci analyses after a dose of 2 Gy reveal ∼20 HR events in HeLa and U2OS cells that give rise to about 10 SCEs. The estimate of 20 HR events is consistent with our previous studies showing that a dose of 2 Gy induces ∼100 DSBs per G2 cell of which 20% undergo repair by HR (23, 24). Also the analysis of 82-6 cells showed robust formation of IR-induced SCEs, although slightly less frequently as in HeLa and U2OS cells. Nevertheless, this demonstrates that the pronounced propensity to employ a CO-forming pathway is not a peculiarity of cancer cells. However, SCE formation is strictly dependent on ATRX as it is not observed in HeLa ATRX KO cells, in U2OS cells without ATRX induction or in 82-6 cells after ATRX depletion. Collectively, these data suggest that approximately every second ATRX-dependent HR event leads to an SCE, a considerably higher frequency than previously thought, while RECQ5-dependent SDSA does not form SCEs, a notion consistent with published data (40). The most likely explanation for this observation is that ATRX promotes extended DNA repair synthesis which leads to second-end capture and the formation of a JM with a 50% likelihood to be resolved into an SCE. Thus, we argue that CO formation is a frequent event in mammalian cells but necessitates the presence of ATRX. It is, therefore, not readily detected in ALT cells (e.g., U2OS cells) which are often used for cellular studies. Additionally, HR reporter assays typically rely on the analysis of unequal recombination events between the damaged loci and the provided substrates which might skew HR subpathway choice. Since the sister chromatid represents the natural substrate for HR events, we argue that SCE measurements more reliably assess CO frequencies.
ATRX-Dependent HR Intermediates Are Exclusively Channeled into the Resolution Pathway.
Our analysis raises the question about the primary pathway used to deal with the ATRX-dependent HR intermediates and suggests that resolution strongly outcompetes dissolution. We explored this idea by analyzing the formation of HR intermediates in the ATRX pathway and found that resolvase-deficient cells accumulate IR-induced UFBs, which are not observed in ATRX-deficient cells. Additionally, the IR-induced recruitment of MUS81 to DSBs was only observed in ATRX-expressing cells and was not suppressed by BLM, further corroborating the notion that the MUS81 and GEN1 resolvases play a major role in processing the HR intermediates formed by the ATRX subpathway (Fig. 6). Moreover, the SCE assay showed no increase in IR-induced SCEs in the absence of BLM, precluding its function in processing these HR intermediates. Of note, in contrast to IR-induced SCEs, the level of spontaneous SCEs increases drastically upon BLM depletion, suggesting that they arise from different substrates, such as dHJs formed at single-stranded gaps or template switches after fork regression, that are specifically handled by the BTR complex (11, 52). The choice between resolution and dissolution pathways could also be influenced by the position in the cell cycle, as the MUS81-EME1 nuclease activity peaks during late G2/M, while the BTR complex is not known to be restricted to a specific cell cycle phase (8). Thus, we cannot rule out that the temporal regulation of the nucleases might affect the differential processing of HR intermediates induced by IR in G2 or arising in S phase at the replication fork.
Notwithstanding such possibilities, our findings suggest the formation of a distinct HR intermediate by the ATRX pathway. One possibility is the formation of a dHJ that is structurally distinct and is resistant to dissolution by BLM, which could also be explained in light of the chromatin-remodeling function of ATRX. Histone deposition within the D-loop could “chromatinize” the dsDNA regions consisting of the two single strands from both sister chromatids and physically impede strand separation and counteract helicase function, suppressing both RECQ5 and BLM (SI Appendix, Fig. S7A). This is reminiscent of the antagonistic effect of the chromatin remodeler CAF-1 on the RecQ-type helicase Rqh1 during repair synthesis observed in fission yeast, where chromatin assembly on the D-loop counteracts strand displacement and favors D-loop stability (53, 54). Therefore, ATRX’s function in D-loop stabilization and repair synthesis promotion through histone deposition and RECQ5 suppression could have the secondary effect of counteracting BLM function, resulting in a high CO frequency. Alternatively, the apparent lack of BLM processing of these intermediates raises the intriguing possibility of the HR intermediates arising during DSB repair being distinct from the classical dHJ structure, such as nicked HJs, D-loops, or other substrates that are more suitable for resolution reactions (SI Appendix, Fig. S7 B and C).
Conclusion
The data presented here show ATRX and RECQ5 define two distinct HR subpathways for repairing DSBs, with ATRX being essential for a CO-forming pathway and RECQ5 promoting SDSA. A quantitative comparison between the relative usages of the two subpathways suggests that ATRX outcompetes RECQ5 for the majority of HR events, likely through PCNA interaction, and leads to the formation of COs in approximately half of these events. The more extended DNA repair synthesis promoted by ATRX likely results in second-end capture and the formation of HR intermediates that are exclusively processed by resolvases and are not subject to dissolution by BLM, therefore constituting distinct substrates from those formed spontaneously during S phase. Our work highlights a potential role for chromatin remodeling in determining HR subpathway choice and the formation of intermediates that are distinct from the classical dHJ. Our model questions the current belief that CO formation is largely suppressed in mammalian cells and warrants reconsideration of studies which addressed the question of HR subpathway usage by investigating ATRX-deficient cell lines or artificial substrates for HR.
Materials and Methods
Cell Lines and Cell Culture.
HeLa-S3 WT (ATCC), HeLa ATRX KO, and U2OS (ATCC) cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM, Sigma) supplemented with 10% fetal calf serum (FCS) and 1% nonessential amino acids (NEAA). Saos-2 (ATCC) cells were cultured in McCoy’s 5A with 15% FCS. U2OS ST/LT GC reporter cells were kindly provided by Ralph Scully, Harvard Medical School, Boston, MA (45) and were cultured in DMEM containing the aforementioned supplements plus 2 mg/mL puromycin. U2OSATRX cells with inducible ATRX expression were a gift from David Clynes, University of Oxford, Oxford, United Kingdom (43) and were grown in DMEM with 10% tetracycline-free fetal bovine serum, 700 µg/mL G418 sulfate, 0.5 µg/mL puromycin, and 1× glutamate. Immortalized human primary fibroblasts (82-6 hTert cells) were a gift from Robert Schiestl, University of California, Los Angeles, CA, and cultured in Minimum Essential Medium supplemented with 20% FCS and 1% NEAA. Cells were routinely tested for mycoplasma contamination by PCR (Minerva Biolabs). All cell lines were maintained at 37 °C in a 5% CO2 incubator.
RNA Interference, Plasmid Transfection, and Inhibitors.
Transfection of cells with siCtrl (5′-AAT TCT CCG AAC GTG TCA CGT-3′), siRECQ5 (Dharmacon smart pool, E-019338-00-0010), siRAD51 (target sequence: 5′-AAG GGA ATT AGT GAA GCC AAA-3′), siRECQ5-2 (5′-GGA GAG TGC GAC CAT GGC T-3′), siATRX (5′-GAG GAA ACC TTC AAT TGT ATT-3′), siMUS81 (5′-AAC AGC CCT GGT GGA TCG ATA-3′), siGEN1 (5′-AAG CGT AAT CTT GGT GGG AAA-3′), siBLM (5′-AAG CTA GGA GTC TGC GTG CGA-3′), siRAD54 (5′-GAA CTC CCA TCC AGA ATG ATT-3′) and siBRCA2 (5′-TTG GAG GAA TAT CGT AGG TAA-3′) was carried out using Lipofectamine RNAiMAX (Thermo Fisher) according to the manufacturer’s instructions. Experiments were performed 48 or 72 h after transfection. For plasmids, GFP-ATRX (Addgene, 45444), myc-ATRX (Origene, RC215176), GFP-ATRX-WT, GFP-ATRX-ATP, GFP-ATRX-PIP, pEGFP (Clontech, 632470), GFP-RECQ5-WT, GFP-RECQ5-ATP, GFP-RECQ5-PIP, pUC19 (New England Biolabs, N3041S) and pBL464_pCBASceI were transfected using Lipofectamine LTX or PEI (Sigma) according to the manufacturer’s instructions. The ATRX mutant constructs were previously described (26) and the RECQ5 mutant constructs were kindly provided by Pavel Janscak, University of Zurich, Zurich, Switzerland (37). For chemical inhibition of RAD51, cells were treated with 60 µM B02 (Calbiochem) 30 min prior to IR and the inhibitor was maintained throughout repair incubation.
Irradiation and Cell Cycle-Specific DSB Repair Analysis.
Irradiation was performed with an X-ray machine operated at 90 kV and 19 mA with an aluminum filter. For cell cycle-specific analysis of DSB repair, the thymidine analog EdU (10 mM, Baseclick, BCN-001) was added to asynchronously growing cells 30 min prior to IR and maintained throughout the repair incubation period. After fixation and immunostaining, DAPI and EdU intensities were measured and plotted in a diagram using a Zeiss microscope and MetaCyte software (Metasystems). EdU-positive cells were identified as S-phase cells, while the EdU-negative cells were categorized as either G1 or G2 based on their DNA content. Cells transitioning from G1 to S and G2 phase incorporated EdU, thus assessed as S-phase cells, and were consequently excluded from analysis.
Immunofluorescence.
Cells grown on glass coverslips were fixed with 3% formaldehyde in phosphate-buffered saline (PBS) for 10 min at room temperature (RT). After washing three times with PBS, cells were permeabilized with 0.5% Triton X-100 in PBS for 10 min at 4 °C, and washed and blocked for 0.5 to 1 h at RT with 1× Rotiblock (Roth). For mitotic cell analyses, cells were fixed with PTEMF buffer (20 mM Pipes pH 6.8, 0.2% Triton X-100, 10 mM ethylene glycol tetraacetic acid, 1 mM MgCl2 and 4% formaldehyde) for 10 min and then permeabilized with 0.2% Triton X-100 in PBS for 5 min. Antibodies against γH2AX (rabbit Abcam, ab81299, mouse Millipore, 05-636, chicken Biozol, BRD-0675MZ), RAD51 (Abcam, ab63801), MUS81 (Santa Cruz, sc-53382), RPA32 (Abcam, ab2175), BLM (Santa Cruz, sc-7790), pH3 (Millipore, 06-570), BrdU (BD Pharmingen, 555627), ATRX (Santa Cruz, sc-15408), GFP (Roche, 11 814 460 001), and RPA32/RPA2 (p-T21) (Abcam, ab109394), were added overnight at 4 °C and EdU was stained using the EdU Click-iT Kit (Roth) according to the manufacturer’s instructions. After secondary antibody incubation, cells were washed and stained with DAPI (0.4 mg/mL) for 10 min at RT and then embedded with Vectashield mounting medium (Vector Laboratories). Cells were examined using a Zeiss microscope and cell cycle-specific analysis was carried out by the manual enumeration of foci in G2 cells (23).
Site-Directed Mutagenesis.
Constructs encoding GFP-RECQ5-WT, GFP-RECQ5-ATP, and GFP-RECQ5-PIP were subjected to site-directed mutagenesis to introduce three silent mutations to confer resistance to siRECQ5-2 using the primers Fwd: 5′-CGCCTTTACAGGAGAGCGCTACGATGGCTGTAGTAAAAGG-3′ and Rev: 5′-CCTTTTACTACAGCCATCGTAGCGCTCTCCTGTAAAGGCG-3′. The mutagenesis reaction was carried out using Q5 high-fidelity polymerase (NEB) and the product was treated with DpnI to degrade the methylated parent plasmid followed by transformation into competent Dh5α Escherichia coli cells. Single colonies were picked after kanamycin selection, expanded, and plasmid DNA was extracted using the ZR plasmid miniprep kit (ZymoResearch) or for higher yield, peqGOLD Xchange plasmid maxi-EF kit (Peqlab). The insertion of the mutations was confirmed by sequencing (Microsynth) using the sequencing primer 5′-GAGCAGCCACCATACCAC CT-3′.
Reporter Assay.
U2OS ST/LT GC reporter cells were transfected with myc-ATRX or pUC19 plasmids and/or siRNA and 40 h later, transfected with I-SceI plasmids. Cells were collected after 48 h and for each sample, a fraction of cells was fixed and another was seeded in defined numbers into Petri dishes and cultured under selection with 2 mg/mL Blasticidin S for 14 d to obtain single colonies. The fixed fraction was stained against GFP and DAPI and the number of GFP-positive cells was determined using a Zeiss microscope and MetaCyte software (MetaSystems). For the cultured fraction, single colonies were fixed with methanol:acetic acid (3:1) and stained with crystal violet. Blasticidin-resistant colonies were counted and represented the LTGC fraction of the GFP-positive HR events.
BrdU Incorporation Assay.
To measure DNA repair synthesis in G2 cells, a specialized BrdU assay was employed. As described previously, incorporation of BrdU following IR can be visualized as foci arising in G2 cells which are dependent on HR factors, but are not impeded by NHEJ factor depletion (23). Briefly, cells were seeded and transfected with siRNA and 48 h later, were labeled with 10 µM EdU for 1 h and irradiated with 4 Gy. Cells were then washed with PBS and incubated with 50 µM BrdU and fixed after 8 h. To allow access of antibodies to the incorporated BrdU, DNA was denatured with 2.5 M HCl for 20 min at RT and the cells were washed thoroughly (10 times) with PBS. Staining was performed as described and BrdU foci were manually enumerated in G2 cells.
Sister Chromatid Exchange Analysis.
HeLa WT, HeLa ATRX KO, U2OSATRX, and 82-6 cells were transfected with siCtrl or siRECQ5 and incubated with 50 µM BrdU for 48 h. Cells were irradiated with 2 Gy and then, 3 h prior to fixation, were treated with 20 mg/mL caffeine and 10 mg/mL colcemid to enrich for mitotic cells. HeLa cells were collected after 8 h post IR and U2OSATRX cells were collected after 16 h due to slower exit from G2 into M phase (SI Appendix, Fig. S2C). The 82-6 cells were treated with 100 nM of the Chk1 inhibitor UCN-01 for 3 h instead of caffeine and collected at 12 h post IR to obtain sufficient mitotic cells. For preparation of chromosome spreads, cells were collected and resuspended in prewarmed 75 mM KCl for 30 min at 37 °C. Cells were then fixed with ice-cold methanol:acetic acid (3:1) three times and chromosomes were spread onto coverslips and air dried overnight. To obtain the differential staining between the two sister chromatids, slides were washed, incubated with 5 µg/mL bisbenzimid (Hoechst 33258) for 1 h, and then covered with 200 mM Na2HPO4 and 4 mM citric acid buffer (pH 7.1 to 7.2) and irradiated with ultraviolet light C at 9 J/cm2. Slides were then washed in 2× saline-sodium citrate buffer at 50 °C for 30 min and stained with Giemsa. Chromosome spreads were captured and analyzed using a Zeiss microscope with Metafer software (MetaSystems).
Immunoblotting.
Cells were lysed in Laemmli buffer [10% sodium dodecyl sulfate (SDS), 300 mM Tris-HCl, 10 mM β-mercaptoethanol, 50% glycine, 0.02% bromophenol blue]. Protein extracts were loaded in equal amounts for SDS-polyacrylamide gel electrophoresis and the separated proteins were transferred to a nitrocellulose membrane, blocked for 1 h at RT in Rotiblock (Roth) and incubated with primary antibodies overnight at 4 °C (anti-RECQ5, Abcam, ab91422 and ab224135 and anti-GEN1, Abgent, AP9493a and the antibodies described above). Membranes were then washed with 0.1% tween-tris-buffered saline and incubated with horseradish peroxidase-conjugated secondary antibodies for 1 h at RT. Immunoblots were developed using WesternBright Quantum or Sirius Kits (Advansta) and signal detection was performed with Fusion FX system (VilberLourmat).
Quantification and Statistical Analysis.
Unless otherwise specified, all data were derived from at least n = 3 replicates and for each experiment at least 40 nuclei or 25 chromosome spreads were analyzed. Background foci were subtracted from the mean values. Column plots show the mean value and the error bars show the SEM between the experiments. P values were obtained by Student’s t test and compare the mean values of independent experiments (*P < 0.05; **P < 0.01; ***P < 0.001).
Supplementary Material
Acknowledgments
We thank Ralph Scully and David Clynes for providing cell lines; Pavel Janscak for providing RECQ5 constructs; Penny Jeggo, Wolf Heyer, Jie Liu, and Hang Phuong Le for comments on the manuscript; and Arthur Mathes, Ratna Weimer, Bettina Basso, Christel Braun, and Cornelia Schmitt for technical assistance. Work in the M.L. laboratory is supported by the Deutsche Forschungsgemeinschaft (GRK1657) and the Bundesministerium für Bildung und Forschung (02NUK054C).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010370118/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information.
References
- 1.Ciccia A., Symington L. S., Stressing out about RAD52. Mol. Cell 64, 1017–1019 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Wright W. D., Shah S. S., Heyer W. D., Homologous recombination and the repair of DNA double-strand breaks. J. Biol. Chem. 293, 10524–10535 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spies J., et al. , Nek1 regulates Rad54 to orchestrate homologous recombination and replication fork stability. Mol. Cell 62, 903–917 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kowalczykowski S. C., An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 7, a016410 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bzymek M., Thayer N. H., Oh S. D., Kleckner N., Hunter N., Double Holliday junctions are intermediates of DNA break repair. Nature 464, 937–941 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wyatt H. D. M., Sarbajna S., Matos J., West S. C., Coordinated actions of SLX1-SLX4 and MUS81-EME1 for Holliday junction resolution in human cells. Mol. Cell 52, 234–247 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Wyatt H. D. M., West S. C., Holliday junction resolvases. Cold Spring Harb. Perspect. Biol. 6, a023192 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfander B., Matos J., Control of Mus81 nuclease during the cell cycle. FEBS Lett. 591, 2048–2056 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Bizard A. H., Hickson I. D., The dissolution of double Holliday junctions. Cold Spring Harb. Perspect. Biol. 6, a016477 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang J., Bachrati C. Z., Ou J., Hickson I. D., Brown G. W., Human topoisomerase IIIalpha is a single-stranded DNA decatenase that is stimulated by BLM and RMI1. J. Biol. Chem. 285, 21426–21436 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ralf C., Hickson I. D., Wu L., The Bloom’s syndrome helicase can promote the regression of a model replication fork. J. Biol. Chem. 281, 22839–22846 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Costantino L., et al. , Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science 343, 88–91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anand R. P., Lovett S. T., Haber J. E., Break-induced DNA replication. Cold Spring Harb. Perspect. Biol. 5, a010397 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kadyk L. C., Hartwell L. H., Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132, 387–402 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson R. D., Jasin M., Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 19, 3398–3407 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zapotoczny G., Sekelsky J., Human cell assays for synthesis-dependent strand annealing and crossing over during double-strand break repair. G3 (Bethesda) 7, 1191–1199 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jasin M., Rothstein R., Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol. 5, a012740 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verma P., Greenberg R. A., Noncanonical views of homology-directed DNA repair. Genes Dev. 30, 1138–1154 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaganti R. S., Schonberg S., German J., A manyfold increase in sister chromatid exchanges in Bloom’s syndrome lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 71, 4508–4512 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarbajna S., West S. C., Holliday junction processing enzymes as guardians of genome stability. Trends Biochem. Sci. 39, 409–419 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Perry P., Evans H. J., Cytological detection of mutagen-carcinogen exposure by sister chromatid exchange. Nature 258, 121–125 (1975). [DOI] [PubMed] [Google Scholar]
- 22.Solomon E., Bobrow M., Sister chromatid exchanges–A sensitive assay of agents damaging human chromosomes. Mutat. Res. 30, 273–278 (1975). [PubMed] [Google Scholar]
- 23.Beucher A., et al. , ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 28, 3413–3427 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeggo P. A., Geuting V., Löbrich M., The role of homologous recombination in radiation-induced double-strand break repair. Radiother. Oncol. 101, 7–12 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Mladenov E., et al. , Strong suppression of gene conversion with increasing DNA double-strand break load delimited by 53BP1 and RAD52. Nucleic Acids Res. 48, 1905–1924 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Juhász S., Elbakry A., Mathes A., Löbrich M., ATRX promotes DNA repair synthesis and sister chromatid exchange during homologous recombination. Mol. Cell 71, 11–24.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Elbakry A., Juhász S., Mathes A., Löbrich M., DNA repair synthesis and histone deposition partner during homologous recombination. Mol. Cell. Oncol. 5, e1511210 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conrad S., Künzel J., Löbrich M., Sister chromatid exchanges occur in G2-irradiated cells. Cell Cycle 10, 222–228 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cole F., et al. , Mouse tetrad analysis provides insights into recombination mechanisms and hotspot evolutionary dynamics. Nat. Genet. 46, 1072–1080 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elliott B., Richardson C., Winderbaum J., Nickoloff J. A., Jasin M., Gene conversion tracts from double-strand break repair in mammalian cells. Mol. Cell. Biol. 18, 93–101 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krejci L., et al. , DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423, 305–309 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Hu Y., et al. , Recql5 and Blm RecQ DNA helicases have nonredundant roles in suppressing crossovers. Mol. Cell. Biol. 25, 3431–3442 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ira G., Malkova A., Liberi G., Foiani M., Haber J. E., Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115, 401–411 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J., et al. , Srs2 promotes synthesis-dependent strand annealing by disrupting DNA polymerase δ-extending D-loops. eLife 6, e22195 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Marco S., et al. , RECQ5 helicase cooperates with MUS81 endonuclease in processing stalled replication forks at common fragile sites during mitosis. Mol. Cell 66, 658–671.e8 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Zheng L., et al. , MRE11 complex links RECQ5 helicase to sites of DNA damage. Nucleic Acids Res. 37, 2645–2657 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urban V., et al. , RECQ5 helicase promotes resolution of conflicts between replication and transcription in human cells. J. Cell Biol. 214, 401–415 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu Y., et al. , RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 21, 3073–3084 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwendener S., et al. , Physical interaction of RECQ5 helicase with RAD51 facilitates its anti-recombinase activity. J. Biol. Chem. 285, 15739–15745 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paliwal S., Kanagaraj R., Sturzenegger A., Burdova K., Janscak P., Human RECQ5 helicase promotes repair of DNA double-strand breaks by synthesis-dependent strand annealing. Nucleic Acids Res. 42, 2380–2390 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lovejoy C. A. et al.; ALT Starr Cancer Consortium , Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 8, e1002772 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heaphy C. M., et al. , Altered telomeres in tumors with ATRX and DAXX mutations. Science 333, 425 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clynes D., et al. , Suppression of the alternative lengthening of telomere pathway by the chromatin remodelling factor ATRX. Nat. Commun. 6, 7538 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Löbrich M., et al. , gammaH2AX foci analysis for monitoring DNA double-strand break repair: strengths, limitations and optimization. Cell Cycle 9, 662–669 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Nagaraju G., Odate S., Xie A., Scully R., Differential regulation of short- and long-tract gene conversion between sister chromatids by Rad51C. Mol. Cell. Biol. 26, 8075–8086 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan Y. W., West S. C., A new class of ultrafine anaphase bridges generated by homologous recombination. Cell Cycle 17, 2101–2109 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan K. L., North P. S., Hickson I. D., BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 26, 3397–3409 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan Y. W., Fugger K., West S. C., Unresolved recombination intermediates lead to ultra-fine anaphase bridges, chromosome breaks and aberrations. Nat. Cell Biol. 20, 92–103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nimonkar A. V., et al. , BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 25, 350–362 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarbajna S., Davies D., West S. C., Roles of SLX1-SLX4, MUS81-EME1, and GEN1 in avoiding genome instability and mitotic catastrophe. Genes Dev. 28, 1124–1136 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lovejoy C. A., Takai K., Huh M. S., Picketts D. J., de Lange T., ATRX affects the repair of telomeric DSBs by promoting cohesion and a DAXX-dependent activity. PLoS Biol. 18, e3000594 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu L., Hickson I. D., The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature 426, 870–874 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Pietrobon V., et al. , The chromatin assembly factor 1 promotes Rad51-dependent template switches at replication forks by counteracting D-loop disassembly by the RecQ-type helicase Rqh1. PLoS Biol. 12, e1001968 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hardy J., et al. , Histone deposition promotes recombination-dependent replication at arrested forks. PLoS Genet. 15, e1008441 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.