Significance
The role of mutations of large effect in adaptive evolution is a question of enduring interest. Large-effect mutations were once seen as unlikely contributors to adaptation, but we now have numerous examples. A major shortcoming of the evidence is the lack of information on fitness effects of mutations. We conducted a quantitative trait locus study that mapped fitness in an experimental field population of stickleback to a large-effect gene, Ectodysplasin (Eda). We compared this result with allele frequency change at the gene in a young lake population, which also revealed strong natural selection and large fitness effects of the Eda gene and/or linked genes. Selection on ancient genetic variants may increase the prevalence of large-effect fitness variants in adaptive evolution.
Keywords: genetics of adaptation, stickleback, natural selection, Ectodysplasin, fitness mapping
Abstract
Mutations of small effect underlie most adaptation to new environments, but beneficial variants with large fitness effects are expected to contribute under certain conditions. Genes and genomic regions having large effects on phenotypic differences between populations are known from numerous taxa, but fitness effect sizes have rarely been estimated. We mapped fitness over a generation in an F2 intercross between a marine and a lake stickleback population introduced to a freshwater pond. A quantitative trait locus map of the number of surviving offspring per F2 female detected a single, large-effect locus near Ectodysplasin (Eda), a gene having an ancient freshwater allele causing reduced bony armor and other changes. F2 females homozygous for the freshwater allele had twice the number of surviving offspring as homozygotes for the marine allele, producing a large selection coefficient, s = 0.50 ± 0.09 SE. Correspondingly, the frequency of the freshwater allele increased from 0.50 in F2 mothers to 0.58 in surviving offspring. We compare these results to allele frequency changes at the Eda gene in an Alaskan lake population colonized by marine stickleback in the 1980s. The frequency of the freshwater Eda allele rose steadily over multiple generations and reached 95% within 20 y, yielding a similar estimate of selection, s = 0.49 ± 0.05, but a different degree of dominance. These findings are consistent with other studies suggesting strong selection on this gene (and/or linked genes) in fresh water. Selection on ancient genetic variants carried by colonizing ancestors is likely to increase the prevalence of large-effect fitness variants in adaptive evolution.
The role of beneficial mutations of large effect during adaptation of wild populations to new and changing environments is a question of enduring interest (1–3). Large-effect mutations were once seen as unlikely to contribute to adaptation, because de novo mutations of small effect are much more likely to be advantageous than mutations of large effect (4). Yet, genetic studies of divergence between natural populations and species frequently detect genomic regions of apparently large phenotypic effect (5–10). Such genes of large effect are easier to detect and validate than genes of small effect, causing an ascertainment bias, but at least they are not rare or peculiar. On the other hand, these loci explaining large phenotypic differences might harbor multiple mutations of individually smaller effect (11–13).
Theory has found a plausible role for large-effect fitness variants during adaptive divergence under certain conditions. New mutations of large effect can be beneficial early in the process of adaptation to a new environment, when the population is still far from the optimum, and when populations perpetually track a distant moving optimum (4, 14). Gene flow between diverging populations can inhibit fixation of small-effect mutations and thereby increase the importance of genes of relatively large effect (15).
Adaptation from standing genetic variation can also favor large-effect variants, especially if these mutations have migrated from populations already adapted to similar selective pressures. Large-effect variants might then fix rapidly in the new environment (16–19). Examples include the repeated fixation of a relatively ancient low-armor Ectodysplasin (Eda) allele in young freshwater populations of threespine stickleback (7), color pattern mutations in Heliconius butterflies (20), opsin variants affecting color vision in Lake Victoria cichlids (21), and mutations adapting Rhagoletis fruit flies to apple (22). Large-effect standing variants that flow in from previously adapted populations might harbor a cluster of multiple beneficial, closely linked mutations constructed over time via a series of mutations and selective sweeps. Even so, in a new population such a group of alleles can behave as a single, large-effect beneficial allele that sweeps to fixation in unison.
A major limitation of the evidence for large-effect variants in divergence of populations is the scarcity of information on fitness effects. Most evidence for large-effect mutations in adaptation is based instead on phenotypic effect sizes. Yet, a gene with a large phenotypic effect need not have a large fitness effect if selection on the trait is not strong. Genetic drift, environmental factors, and natural selection on other traits will also contribute to total variation in fitness. Fitness across a generation, or similarly the change in allele frequency between parent and offspring generations, at loci involved in adaptation has rarely been mapped. Hence, the fitness-effect sizes of variants are almost unknown in wild populations.
We addressed this gap in two ways. First, we carried out a quantitative trait locus (QTL) study to map the number of surviving offspring (hereafter, fitness) in an experimental field population of threespine stickleback (Gasterosteus aculeatus). We crossed an individual from a postglacial freshwater population having reduced armor with an individual from a high-armor marine population representing the ancestral form (Fig. 1). Second-generation (F2) progeny were then introduced to a freshwater pond where fitness was mapped across a generation. Our aim was to test whether genomic regions known to contain genes with large phenotypic effects, such as Eda, also affect fitness in a freshwater environment. The experiment additionally allowed us to test one of the main hypotheses to explain the advantage of the low-armor Eda allele in fresh water, namely that it stems from faster growth associated with the reduced costs of producing armor (23–25). We use QTL mapping to estimate fitness-effect sizes and compare the projected change in allele frequency in the next generation with that observed.
Fig. 1.
Design of the pond experiment. (A) Entrance to the Little Campbell River from the Strait of Georgia, BC. (B) Cranby Lake, BC. (C) A single intercross (F0) was made between a marine (anadromous) stickleback collected in the Little Campbell River and a freshwater-resident stickleback from Cranby Lake. Example specimens are stained with alizarin red to highlight bone. The marine population is fully plated (MM genotype at the Eda locus), whereas the freshwater population has few lateral plates (FF at Eda). First generation (F1) hybrids were crossed in the laboratory to produce second-generation (F2) hybrids that were introduced to a freshwater pond at the University of British Columbia. (D) Author M.E.A. on the experimental pond. (E) Author K.B.M. returning adult F2 hybrids to the pond after measurement.
Second, we compared the fitness effects mapped in a single generation with empirical observations and modeling of multigenerational allele frequency change in an Alaskan lake population recently formed when the lake was recolonized by high-armor marine threespine stickleback (26). A drawback of the QTL and modeling approaches is that they cannot distinguish a single mutation from multiple linked mutations having cumulative effects. Nevertheless, our experimental and observational studies provide an opportunity to rule out the presence of individual regions with large fitness effects and therefore represent a valuable first step in mapping fitness to genes.
Methods
QTL Study Populations.
A single intercross (F0) was made in 2005 between a female marine stickleback from the Little Campbell River (49.01° N, –122.76° W), 45 km south of Vancouver, BC, Canada, and a male threespine stickleback from Cranby Lake, Texada Island, BC (49.70° N, –124.51° W) (Fig. 1). The Little Campbell River marine population represents the fully plated ancestral form having long dorsal and pelvic spines and a fusiform body shape, including a small head (27). The population is anadromous, spending most of the year in the ocean and migrating to the river to reproduce. Anadromous individuals are morphologically similar to fully marine stickleback breeding in shallow coastal areas nearby. Experimentally displaced individuals tend to home to their breeding sites of initial capture, whether salt or fresh water (28), but genetic differences between individuals breeding in the two sites are not known. The Cranby Lake population is a typical “solitary” freshwater population from the region (29), having few lateral plates, reduced dorsal and pelvic spines, a bulkier body shape, and a larger head (30, 31). Its body shape is intermediate between that of the more specialized limnetic and benthic stickleback species pairs that occur in otherwise similar lakes (29, 30). Reduced lateral plates in the Cranby Lake population maps to the location of the Eda gene (7). The F1 offspring from the cross were raised to adulthood in the laboratory using standard laboratory methods (32). We produced the F2 generation between May and July 2006 by crossing six F1 females and four F1 males (each of two males was crossed twice).
Pond Experiment.
Six hundred thirty-six F2 juveniles were introduced to a freshwater pond on 21 August 2006 (mean standard length 22.9 mm ± 3.5 SD), after removing a small tissue sample from the tail fin. The pond was 1 of 13 located on the South Campus of the University of British Columbia (25). The pond was 23 × 23 m in surface area with a bottom that sloped gradually to a depth of 3 m in the center. It was sand-lined and bordered with limestone extracted from surface mines near Cranby Lake on Texada Island, BC. Ponds were constructed in 1991 and seeded with plants and invertebrates from nearby Paxton Lake, Texada Island. Apart from their original construction and use in previous experiments, ponds were unmanipulated environments designed to mimic a natural freshwater environment and having well-developed aquatic communities.
Four hundred eight of the original 636 F2 fish were recovered from the experimental pond using unbaited minnow traps on 3 to 6 March of the following year, prior to the 2007 breeding season. A second round of trapping was conducted on 23 March and 6 April, recovering another 71 individuals not trapped earlier, yielding a total of 479 F2s. The high fraction of previously marked fish among those captured in this second round led to a mark–recapture population size estimate of 547 using the Lincoln–Petersen method (33), indicating that roughly 86% of F2 fish introduced to the ponds the previous year had survived over winter. The opportunity for viability selection was consequently low, and we did not attempt to map survival over the period.
Standard length and lateral plate phenotype were measured on each F2 fish. Standard length was measured in the hand with a ruler as the distance from the tip of the snout to the end of the caudal peduncle. Standard length was strongly correlated with centroid size, a geometric measure of overall body size based on two-dimensional landmark coordinates, in a previous laboratory-raised F2 cross between individuals from these same two populations (r = 0.95, degrees of freedom = 339, P = 2.2 × 10−16) (31). Lateral plate phenotype was scored as low, partial, or complete, according to the number of plates on the left side of the body (34, 35). Low-plated individuals possess a cluster of 1 to 10 anterior plates along the lateral line near the pectoral fin. Partially plated individuals had 11 to 29 plates and were recognized by the presence of a gap of two or more plates between the caudal and midbody regions. Completely plated individuals had 30 or more plates and a gap of at most one plate near the caudal region. A small section of fin was also removed from the tail for genotyping, after which fish were returned to the pond.
Juvenile F3 fish were trapped and removed in October 2007, killed with an overdose of MS-222, and preserved in ethanol. F3 individuals could be distinguished from surviving F2 individuals by their smaller body size. A random sample of 500 F3 individuals was obtained from the collection using computer-generated random numbers and a sample of tail fin was removed for genotyping.
Genotyping.
Using 250 ng of genomic DNA isolated from fin tissues, we genotyped 240 F2 hybrid females, the two wild progenitors (F0), the 10 F1 parents of the F2 generation, and 500 F3 individuals at 1,294 biallelic single-nucleotide polymorphism (SNP) markers of a custom Illumina GoldenGate array (36). Genotyping was carried out in two batches at the HudsonAlpha Institute for Biotechnology (Huntsville, AL) using Illumina’s GoldenGate assay and GenomeStudio software (Illumina Inc.). After quality filtering (SI Appendix, Methods), 458 markers remained for subsequent analyses.
Using four sex-linked markers (SI Appendix, Methods), we determined that seven F2 adults originally scored in the hand as female were actually males and were removed from the analysis; 232 F3 juveniles were female, 249 were male, and the remainder were uncertain. We carried out parentage assignment of F2 females to F1 parents, and of F3 offspring to F2 mothers (SI Appendix, Methods), after removing the four sex-linked markers. Individuals of uncertain parentage were dropped, leaving 224 F2 females and 474 F3s for analysis.
Fitness Measure.
The tally of F3 offspring assigned to each F2 mother provided our estimate of her fitness. Except for sampling error, this measurement incorporates both female reproductive success and survival of her offspring from birth to the sampling date in autumn (October), by which time juveniles had become subadults.
Linkage Mapping and QTL Analysis.
We dropped markers having nonrandom F2 genotype ratios, those with unknown phase, and sex-linked markers (SI Appendix, Methods). Of the 400 remaining markers for mapping, 388 could be assembled into 21 linkage groups corresponding to 21 known chromosomes (SI Appendix, Methods). Twenty autosomal linkage groups spanned an average of 88.8% per chromosome of the corresponding physical map (37).
We used R/qtl to perform QTL mapping on F2 female reproductive success (number of offspring), body size (standard length), and plate phenotype using the F2 intercross setting (38). Traits were mapped using Haley–Knott regression with the scanone function (SI Appendix, Methods). Lateral plate morph was mapped as a quantitative variable, with 0 corresponding to low-plated, 1 to high-plated, and 0.5 to partially plated. F2 female body size (standard length) and number of offspring were analyzed untransformed.
Fraction of Freshwater Alleles.
We determined the fraction of freshwater alleles at markers on each chromosome of all F2 individuals. We used the calc.genoprob function in R/qtl to determine genotype probabilities at every marker. For a given individual, the number of freshwater alleles at a marker was the sum of the probability of genotype FF (the homozygote for freshwater alleles) plus half the probability of genotype MF (the heterozygote). This quantity was summed across all markers to yield the total number for each chromosome. Dividing by the number of markers on the chromosome yielded the fraction of freshwater alleles.
Measuring Selection and Evolution.
Genotypes at the peak marker for fitness were available for 208 (93%) of 224 F2 females. Remaining F2 genotypes were imputed according to the highest probability genotype as determined by calc.genoprob in R/qtl (always >0.80). Genotypes at the same marker were available for 446 (94%) of the 474 offspring assigned to F2 female parents. All but one of the remaining 28 offspring genotypes were determined by the identity of all maximally informative markers (those homozygous for alternate alleles in the F0 progenitors) located within 3 cM of the peak marker. A single F3 genotype could not be called because marker states in the 6-cM interval were not unanimous.
Relative fitness of each F2 genotype at the peak marker was determined by calculating the mean number of F3 offspring for each genotype and dividing by the mean offspring number of the most fit homozygous genotype, in this case the low-armor “freshwater” allele. The selection coefficient s was calculated directly as 1 minus the relative fitness of the least fit homozygote, in this case the high-armor “marine” allele. Finally, the dominance coefficient h was calculated as 1 minus the relative fitness of the heterozygote divided by s. SEs of these estimates were obtained using the bootstrap (39). Values for offspring number (fitness) were randomly resampled with replacement within each F1 × F1 family and genotype and then combined. Mean and relative fitness of each genotype, as well as s and h, were calculated for each bootstrap replicate as described above. This procedure was repeated 1,000 times. The SD of bootstrap replicate values is the bootstrap SE.
Observed change in allele frequency between the F2 and F3 generations at the peak marker for fitness was compared with allele frequency predicted in the F3 generation from the estimated mean fitness of F2 females of each genotype. Observed and predicted frequency changes may differ if mean fitness of genotypes of the unmeasured F2 males is greatly different from that in females. We used equation 1 in Linnen and Hoekstra (40) to predict the change in the frequency of the freshwater allele in the F3 generation:
Here, is the frequency of the low-armor “freshwater” allele in F2 females and is its frequency in the F3 offspring generation. The symbols , , and are the relative fitnesses of the low-armor heterozygote, the heterozygote, and the high-armor “marine” homozygote, here taking the values 1, 1 − hs, and 1 − s. The denominator is mean fitness. Change in allele frequency was calculated as This quantity was also calculated for each bootstrap replicate described above to obtain a SE for the prediction.
Animal Care.
All procedures and protocols were in accordance with the Canadian Council on Animal Care and approved by the University of British Columbia Animal Care Committee.
Longitudinal Study of Loberg Lake Fish.
We examined changes in Eda allele frequency over a larger number of generations in the stickleback population of Loberg Lake, Alaska. Loberg Lake is a natural lake with a surface area of 4.45 ha and a mean depth of 5.4 m. Freshwater fish were exterminated by rotenone poisoning in October of 1982, and no stickleback were detected in the lake in the immediate subsequent years. Adult stickleback with largely marine armor and morphology were subsequently discovered in the lake in 1990, and samples from the newly evolving freshwater population have been collected annually since then (26). Longitudinal population samples from Loberg Lake population were collected in May or June each sampling year as described (26). Fish collected before 1999 were fixed in 10% buffered formalin and preserved in 50% isopropyl alcohol. Fish collected since 1999 were preserved in 95% ethanol.
Eda Genotyping of Loberg Lake Fish.
DNA was extracted and genotyped from caudal fin clips of fish preserved in ethanol or formalin (SI Appendix, Methods). The number of individuals and genotypes from different sampling years are summarized in SI Appendix, Table S1. Genotyping of microsatellite markers Stn60, Stn239, and Stn277 was performed as previously described (41).
Selection at the Eda Locus in Loberg Lake.
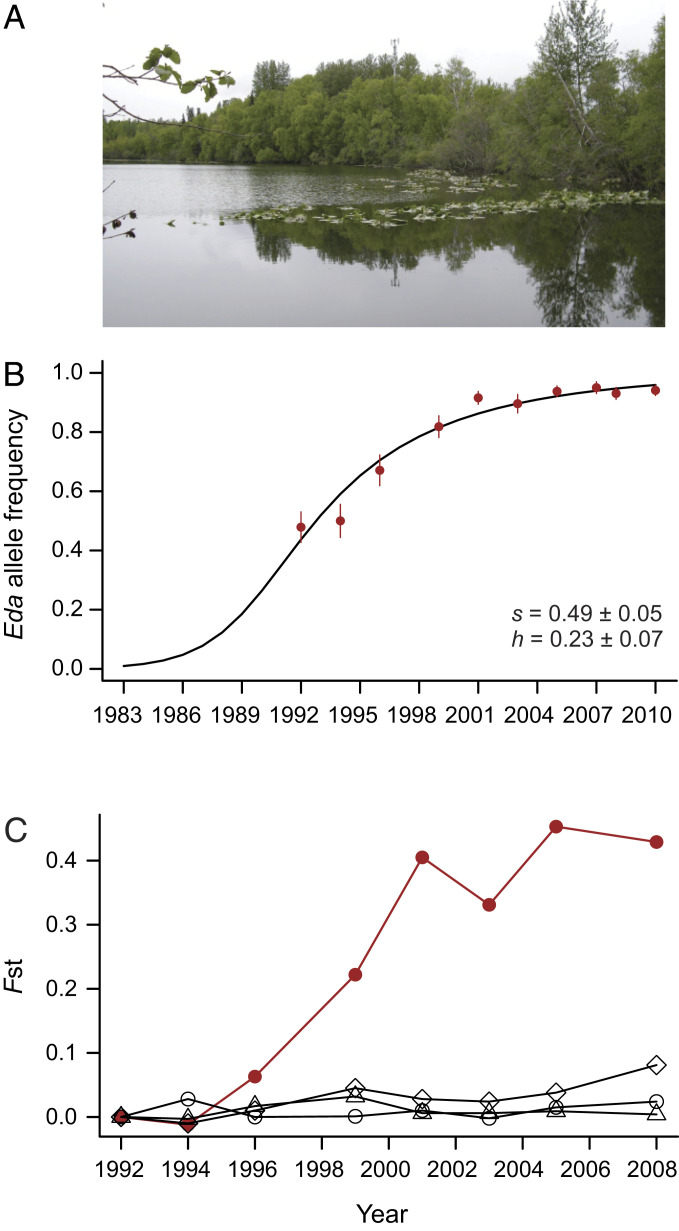
We tracked selection and evolution at the Eda locus in two ways. First, we calculated pairwise Fst between each sample year and the starting 1992 sample using Genepop 4.0 (42). This allowed us to compare the pace of divergence at the Eda locus with several putatively neutral microsatellites. Because of the short time span, divergence at Eda and at microsatellites must both result largely from drift and/or selection on preexisting variation rather than on new mutation. Fst was estimated by a weighted analysis of variance (43). Linear regression was used to test the significance of the difference in slope of the Fst curve over time for Eda and each microsatellite locus.
Second, selection and dominance coefficients for the Eda locus were estimated by modeling the empirically observed allele frequencies in the Loberg Lake time series. This was done using maximum likelihood estimates of the parameters s and h in the standard recursive population genetic formula for biallelic autosomal loci shown above. Modeling of the change in allele frequency over time required a third parameter estimate, , representing the allele frequency at the start of the time series. Estimates for all three parameters were obtained using optimization routines (optim package in R) to find the maximum likelihood estimates of the function. The maximum likelihood function assumed the observed allele frequencies were sampled from a binomial distribution around the fitted frequency. SEs were calculated based on the Fisher information matrix.
Our model assumed a generation time of 1 y, corresponding to the single generation analyzed in the pond experiment. However, lake stickleback in the Cook Inlet region of Alaska often breed at 2 y of age (44, 45). We therefore also analyzed an overlapping generations model, where the allele frequency of fish breeding in a given year t is a mixture, with a proportion Q based on the allele frequency t – 1 y ago and the remaining proportion 1 – Q based on the allele frequency t – 2 y ago:
We then applied the standard population genetic model of selection described above (40). Maximum likelihood estimates of selection in this mixed-generations model were also consistent with strong selection (s = 0.50 ± 0.01; h = 0.22 ± 0.01).
Results
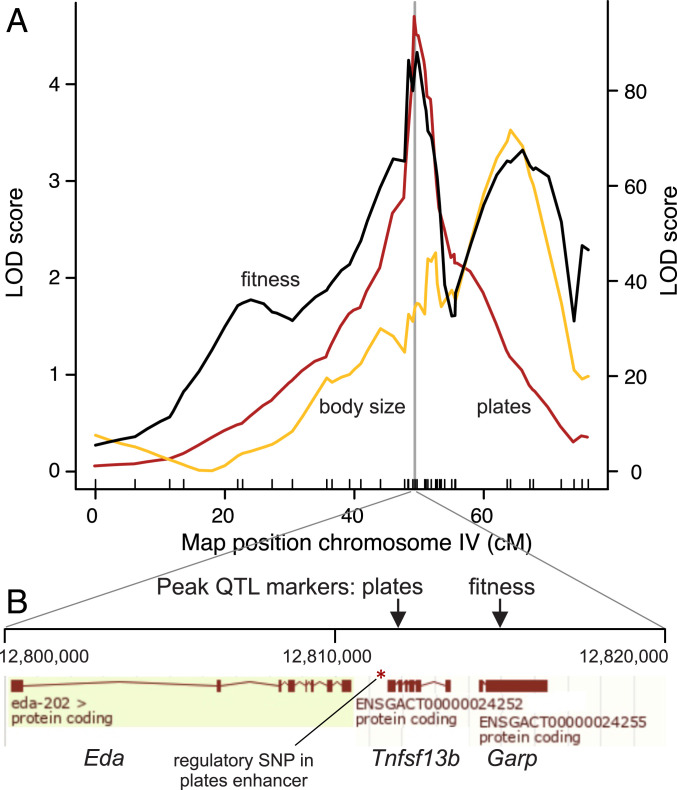
We first confirmed that lateral plate phenotype of F2 females mapped to the region of chromosome IV containing the Eda gene (Fig. 2 and SI Appendix, Fig. S1). The marker corresponding to the peak log odds (LOD) score accounted for 85.8% of the variance in the trait in females of the F2 cross. The peak marker for plates was located at nucleotide position chrIV:12811933 on the reference genome [gasAcu1 assembly (46)], immediately next to the Eda gene (Fig. 2B). The site is close to an enhancer region for Eda, in which a single nucleotide substitution at chrIV:12811481 has been shown experimentally to drive a change in Eda expression in developing lateral plates (47).
Fig. 2.
(A) QTL map of chromosome IV for F2 female fitness measured as the number of surviving offspring. Family identity (unique combination of F1 parents) was included as a covariate. The left vertical axis indicates LOD score for fitness (black line) and body size (standard length; yellow) QTL maps. The right vertical axis indicates LOD score for lateral plates (red). (B) Genomic region surrounding the peak QTL markers for armor and fitness. A small 20-kb region contains the Eda, Tnfsf13b, and Garp genes [ENSEMBL display of gasAcu1 assembly (46)], a previously mapped regulatory SNP located in an enhancer controlling Wnt-responsive Eda expression in developing armor plates [chrIV:12811481 (47)], and the peak QTL markers for armor plate morph (chrIV:12811933) and for fitness (chrIV:12815024) in the pond experiment.
A single QTL for fitness was detected, on chromosome IV (LOD 4.5; Fig. 2), explaining 8.5% of the variance in fitness of F2 females (P = 0.00005; genome scan-adjusted P = 0.008). The peak fitness marker was located at chrIV:12815024, which is again close to the Eda gene and to the causative nucleotide substitution in the enhancer region. However, the 1.5-LOD confidence interval around the peak fitness marker included all markers between chrIV:4034002 and chrIV:32033500, a span of nearly 28 million nucleotides containing many genes including Eda and the peak marker for number of lateral plates. Because the confidence interval for the location of the fitness includes the estimated location of the QTL for plates, the two estimated locations, which are only 3,091 bases apart, are not significantly different from one another (48). No other QTL for fitness was detected (SI Appendix, Fig. S2). Repeating the QTL scan while holding genotype at the peak marker for fitness constant, by including it as a covariate, did not reveal additional markers associated with fitness.
Detection of only a single QTL for fitness might reflect an “omnigenic” basis for fitness (49). Under this view, one or a small number of genes have moderate effect sizes, but most heritable variation results from a multitude of small, difficult-to-detect effects distributed uniformly across the rest of the genome. To investigate this possibility, we used the fraction of freshwater alleles on all chromosomes other than 4 and 19 (the sex chromosome) for each F2 individual. These were used to calculate a rest-of-genome weighted average fraction of freshwater alleles, with chromosomes weighted by their sizes in numbers of nucleotides (37). The expectation from the omnigenic model was that a higher fraction of freshwater alleles across the rest of the genome would result in higher fitness in the freshwater pond. However, we found no such association (F1,217 = 1.19, P = 0.28). We obtained similar results when we tested each Eda genotype separately (FF genotype: F1,55 = 0.20, P = 0.65; MF genotype: F1,94 = 0.87, P = 0.35; MM genotype: F1,54 = 0.92, P = 0.34).
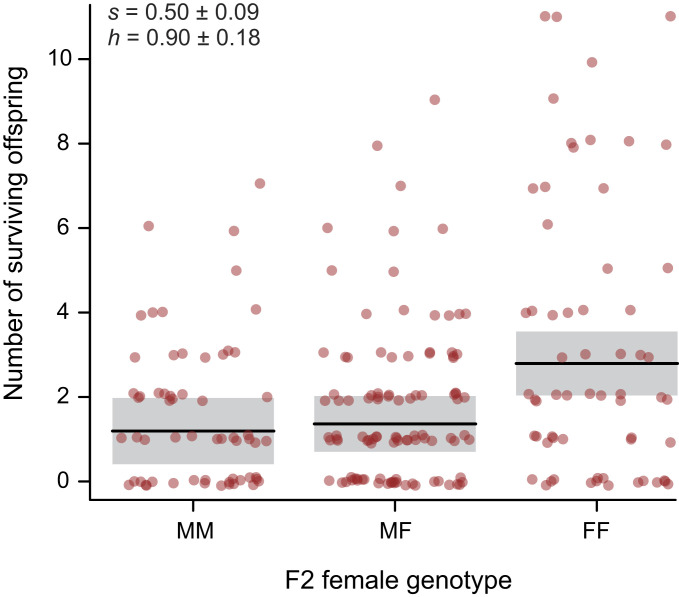
Variation in fitness among F2 females was high, ranging from 0 to 11 F3 offspring (Fig. 3). The mean offspring number per female, 2.12, was set by the number of F3 offspring sampled whose parentage could be assigned to individual F2 females. F2 females having two copies of the marine allele at the peak marker had 1.60 ± 0.30 SE offspring on average, compared with an average of 3.21 ± 0.29 offspring of females having two copies of the freshwater allele instead—a twofold difference. Fitness differences between the two homozygous genotypes at the peak marker corresponded to a selection coefficient of s = 0.50 ± 0.09. Heterozygous females had a mean number of offspring closer to that of the marine genotype at the marker (Fig. 3), corresponding to an estimated dominance coefficient h = 0.90 ± 0.18.
Fig. 3.
Number of surviving offspring (fitness) of individual F2 females differing in genotype at the peak marker in the QTL map for fitness (Fig. 2). MM females are homozygous for the ancestral marine allele, FF females have two copies of the derived freshwater allele, and MF females are heterozygous. Horizontal line segments are means. Vertical span of shaded region is the 95% confidence intervals for the corresponding mean, conditional on F1 × F1 family identity.
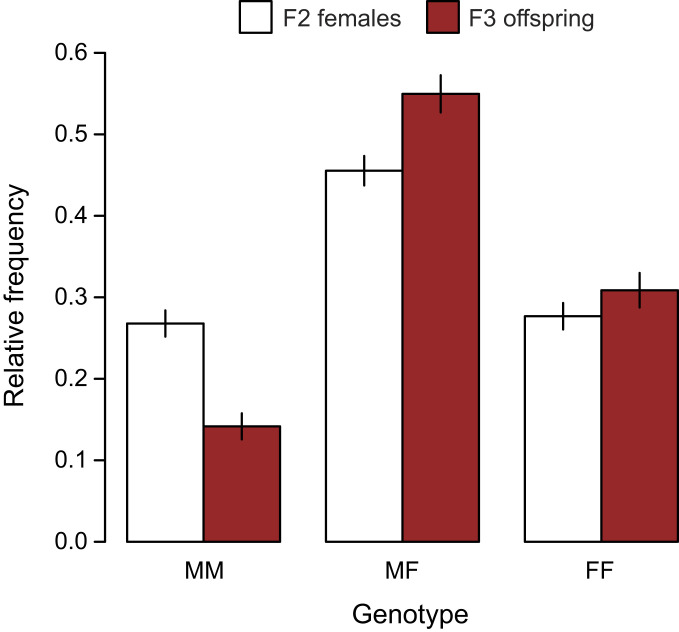
Genotype frequencies at the marker changed significantly between F2 females and their offspring ( = 16.4, P = 0.0003; Fig. 4). Frequency of the low-armor freshwater allele at the peak marker changed from 0.50 ± 0.03 in the F2 females to 0.58 ± 0.02 in the F3 offspring. The frequency of the freshwater allele in the F3 generation is very similar to the frequency predicted from the relative fitness of F2 female genotypes (0.60 ± 0.02) (40). This correspondence implies that the relationship between genotypes and fitness was similar in F2 males, which were not measured, as in F2 females. Possibly, male reproductive performance was affected similarly by genotype at the locus. Alternatively, transgenerational fitness of the F2 males might have been similar to that in females if both were mainly the result of viability selection on their F3 offspring.
Fig. 4.
Relative frequency distribution of genotypes at the peak marker for fitness in the F3 offspring generation (filled bars) compared with that of their F2 mothers (open bars). MM individuals are homozygous for the ancestral marine allele, FF individuals have two copies of the derived freshwater allele, and MF individuals are heterozygous. Vertical lines indicate ±1 SE.
One of the main hypotheses to explain the advantage of the low-armor Eda allele in fresh water is that lateral plates are costly in energy and materials to produce, especially in fresh water (23, 24). Under this hypothesis, fitness of the low-plated genotype is relatively high because individuals experience higher growth rates and thus achieve large body size more rapidly than genotypes bearing the high-armor allele, increasing survival and reproductive success (23, 24). A related hypothesis is that low calcium in fresh water makes plates more costly to produce than in the sea (50). Calcium was unlikely to be limiting in our experiment because the pond (and Cranby Lake, the source of the freshwater male progenitor) contains limestone (calcium carbonate).
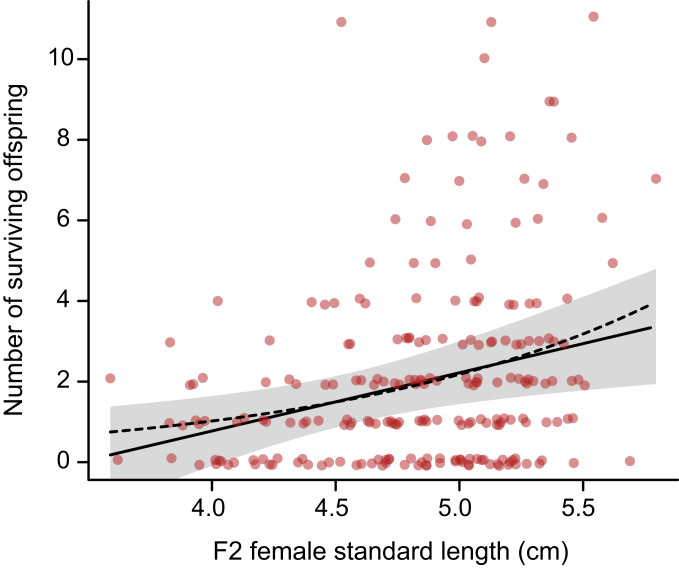
In support of the growth hypothesis, F2 female body size predicted some of the variation in number of surviving F3 offspring (Fig. 5). Also, mean body size was higher in F2 females having freshwater alleles at the peak fitness marker on chromosome IV than females having marine alleles at the peak marker (SI Appendix, Fig. S3). Finally, F2 female body size mapped to chromosome IV (Fig. 2 and SI Appendix, Fig. S2), with the peak size marker near the position chrIV:29763654 (LOD = 4.4, percent variance explained = 8.7%). However, this marker is well downstream of Eda and lies outside the 1.5-LOD interval for the location of the fitness QTL.
Fig. 5.
Relationship between body size (standard length) of F2 females and their numbers of surviving offspring. Regression line and 95% confidence interval for predicted values are conditional upon family identity and F2 sampling date. Points are displaced vertically by a small random amount to reduce overlap. Dashed line indicates the Poisson regression fit to the same data. A single outlier having a standard length of 2.55 cm and 0 offspring was left out of the analysis.
Although size is associated with female fitness and chromosome IV, it is unlikely to have been the main mechanism by which Eda genotype affected fitness in this experiment. First, statistically controlling for F2 female body size did not diminish the evidence for a fitness QTL on chromosome IV (LOD = 4.2, P = 0.00009; genome scan-adjusted P = 0.02). Second, F2 females homozygous for the freshwater genotype at Eda (FF) had higher average numbers of offspring than females of the other two genotypes (MM and MF) across the range of female body sizes (SI Appendix, Fig. S5). Equal slopes of regressions of F2 female fitness on body size among Eda genotypes was not rejected in either linear (F2,212 = 2.41, P = 0.09) or loglinear model fits (F2,212 = 0.48, P = 0.62). Finally, when we statistically compared the mean fitness of the three Eda genotypes at a common body size, the point estimate of selection dropped from s = 0.50 to s = 0.43. These findings suggest that other mechanisms besides F2 female size drove the majority of selection on Eda and the increase in frequency of the freshwater allele in the F3 generation. F2 female fitness was not significantly associated with those marker genotypes on chromosomes IV and VIII most strongly associated with female body size (F2,211 = 1.01, P = 0.37, and F2,211 = 0.30, P = 0.74, respectively) after accounting for Eda genotype.
To examine changes in Eda allele frequency over a greater number of generations in fresh water, we genotyped population samples collected following recolonization of Loberg Lake by marine sticklebacks in Alaska. The earliest year we could successfully genotype Eda in DNA samples from this time series was 1992, when the freshwater Eda allele frequency was already 0.48 (Fig. 6B), similar to the initial frequency in the F2 mapping cross in the experimental pond. Freshwater allele frequency continued to rise in Loberg Lake over subsequent years, reaching a level of 0.96 by 2010. The rate of increase in Fst at the Eda locus between 1992 and later time points to 2008 was significantly higher than that observed at three unlinked microsatellite markers (all F1,7 ≥ 11, all P = 0.01) (Fig. 6C).
Fig. 6.
Allele frequency changes over time at the Eda locus after colonization of a freshwater lake by marine fish in the 1980s. (A) Loberg Lake. (B) Observed frequency change of the low-armor Eda allele (points) compared with the predicted frequency (dotted line) from the fitted model with a selection coefficient estimated as s = 0.49 ± 0.05 SE. (C) Pairwise Fst between the earliest sample in 1992 and subsequent samples to 2008. Significantly faster differentiation over time is observed at Eda (filled circles), than at three putatively neutral microsatellite loci (Stn60, Stn239, and Stn277; open symbols). The two figures use different year spans.
We used the empirical changes in allele frequencies to estimate selection and dominance coefficients for the Eda region using the 1-y generation model (Methods). The maximum likelihood estimates were s = 0.49 ± 0.05 and h = 0.23 ± 0.07 SE across the Loberg Lake time series from 1992 to 2010. Thus, the selection coefficient in Loberg Lake across multiple generations of selection and recombination was almost identical with that in the measurement of selection in a single generation, from the F2 to F3, in the experimental pond.
However, the dominance coefficient differed in the two studies, with the fitness of the heterozygotes resembling the high-armor marine homozygote in the experimental pond and resembling the low-armor Eda homozygote in Loberg Lake. Previous studies show that increasing substitution of freshwater alleles at unlinked armor plate modifier loci can shift the Eda heterozygotes toward low-armor phenotypes (51) and hence modify dominance. The present results are an indication that dominance of fitness effects may be evolvable in stickleback. Although the original marine source population for recolonization of Loberg Lake is still uncertain (26), these results indicate strong selection and intermediate dominance for freshwater alleles of the Eda region over multiple generations in a natural lake system.
Discussion
The role of mutations of large effect during adaptation of wild populations to new environments has been addressed mainly by measuring effect sizes of genes and genomic regions on phenotypic traits. However, a large trait effect need not imply a large fitness effect. We mapped fitness directly across a generation in a recombinant cross between marine and freshwater stickleback transplanted to a freshwater pond. We found evidence consistent with a role for a large-effect variant during stickleback adaptation to fresh water.
A single major QTL for fitness was detected. It mapped to the location of Eda, the major gene controlling lateral plate armor differences between marine and freshwater populations. Eda is a pleiotropic gene that also affects other traits including sensory and behavior phenotypes (7, 47, 52–55). The QTL accounted for 8.5% of the variance in F2 female fitness in our experiment. Mean number of surviving F3 offspring varied twofold between genotypes homozygous for marine and freshwater alleles at the peak fitness marker, a difference that led to an increase in allele frequency from 0.50 to 0.58 in the next (F3) generation. This increase was close to the change in allele frequency predicted from variation in female fitness alone, implying that the genotype–fitness relationship was similar in males, which were not measured. Eda accounted for less F2 variance in fitness (8.5%) than F2 variance in lateral plate phenotype (85.8%), probably because more sources of variation affect fitness than plates, including random and environmental sources. The fitness-effect size of Eda was nevertheless strong, as indicated by a large selection coefficient (s = 0.50).
Our direct measurements of Eda allele frequencies in Loberg Lake allowed a separate estimate of selection on the Eda region, combining both differential survival and differential reproduction in both sexes over multiple generations of selection and recombination. The observed allele frequency patterns were again consistent with a large-effect locus under strong selection (s = 0.49 ± 0.05 SE), producing a freshwater allele frequency of nearly 0.95 within two decades after a new freshwater population was founded by anadromous stickleback.
These results on the fitness effects of the Eda region are consistent with other evidence for strong selection on the Eda gene in freshwater threespine stickleback populations. First, genome scans comparing marine and freshwater threespine stickleback populations consistently report a tall divergence peak at the Eda locus in established extant populations (46, 56–58). Second, lateral plate reduction has been shown to evolve rapidly following human-caused and natural colonization of freshwater lakes and ponds in the recent past (26, 58–60). Third, replicated freshwater pond populations of stickleback established using Eda heterozygotes from a marine population recorded strong selection at the Eda locus within the first offspring generation (25). Selection on Eda was evident even before lateral plates had completed development, and statistically controlling for lateral plate phenotype still recovered variation in fitness associated with Eda genotype (61), implying an influence of the gene on other traits affecting fitness. Evidence that trophic characters (54, 62), and number of neuromasts and schooling behavior (52, 53, 55), map to Eda, and studies showing that Eda alleles are associated with expression differences in multiple tissues (47), indicate that allelic variation at Eda is indeed pleiotropic.
We found no evidence for additional fitness QTL in the experimental cross, despite good marker coverage and differentiation between marine and freshwater populations across multiple regions of the genome (46, 56). The reasons might be several, but low power to detect smaller effects is likely to play a major role (63). Numerous regions of chromosome IV are differentiated between marine and freshwater populations, and linkage between them in the F2 cross might have made it difficult to discern their separate effects on fitness. It is also likely that the contribution of individual genomic regions to fitness varies temporally, changing with environmental conditions among years or with population size and age distribution. A longer-running experiment with additional generations might have detected other fitness loci over time. The freshwater environment used in the QTL experiment was relatively benign in at least one important respect. Vertebrate predators were absent from the pond, whereas diving loons (Gavia immer) and cutthroat trout predators (Oncorhynchus clarkii) are a consistent feature of natural lakes. We note that Loberg Lake is stocked annually with rainbow trout (Oncorhynchus mykiss) and coho salmon (Oncorhynchus kisutch) for recreational fishing (64), and red necked grebes (Podiceps grisegena) and belted kingfishers (Megaceryle alcyon) were always present during early June sampling periods. Thus, the empirical time series data in this lake likely includes multiple sources of predation not present in the QTL mapping experiment.
Limited power in a genome-wide scan might also cause effect sizes to be overestimated for discovered QTL whose true effects are near the detection threshold (63, 65). However, the fitness-effect size of the Eda region estimated here is unlikely to have been greatly biased by this so-called Beavis effect. This is because the Ectodysplasin gene represented a prior fitness candidate, rather than merely a genomic region emerging from a blind scan. The large effect of the Eda region in this experiment is supported by the estimated change of allele frequency in the next generation based on a large sample of F3 offspring, and by the independent estimates of high fitness effects in the multigeneration Loberg Lake times series.
Theories of adaptation that predict the distribution of fitness-effect sizes assume that evolution occurs by the sequential fixation of new beneficial mutations (4). However, although cases of adaptation from new mutation are known in stickleback (6, 66), evidence indicates that the majority of evolution in postglacial stickleback populations occurred via natural selection on old standing genetic variation, which was obtained from other freshwater source populations and was brought to new bodies of fresh water by colonizing marine populations (46, 67). Eda itself represents a case in which natural selection has repeatedly fixed a low-armor allele from standing genetic variation that is still found at low frequency in marine populations (7, 46). The low-armor allele is between 2 and 10 My old and is thought to be maintained in the sea by gene flow from coastal freshwater source populations (7, 16). The strength of negative selection on the low-armor allele in the sea is unknown, but it need not be as strong as positive selection in fresh water. In general, persistent influx of alleles from freshwater source populations is expected to maintain a pool of standing variation in marine sticklebacks at a frequency higher than mutation alone would predict. This pool includes mutations at many loci that have already proved advantageous elsewhere in fresh water, and many are likely to be so again when brought to newly formed freshwater environments by marine colonists. It is not far-fetched to suppose that at the end of the ice age, when new freshwater bodies were forming and being colonized by marine stickleback, and river flows were higher than today (68), that freshwater-adapted alleles were present as standing variation in the sea at much elevated frequencies.
This process of migration–colonization–adaptation likely increases the probability that genes of large fitness effect will contribute to adaptation in a new environment, compared with adaptation by new mutations, for three main reasons. First, large-effect standing genetic variants are likely to be advantageous if the new environment is similar to that of the variant’s source population, in contrast to de novo large-effect mutations. Second, large-effect standing variants should fix rapidly and with high probability, especially if present in multiple individual colonists. Third, some variant alleles present as standing variation might contain several tightly linked beneficial mutations. Such linkage would have built up over a lengthier history of mutation, selection, recombination, and translocation in source populations, but in the new environment they would behave as a single large-effect allele if generations of recombination in the colonizing ancestral population had not yet broken them up into their individual mutations. In this way, multiple mutations can fix in a single selective sweep. This process might help to explain why genetic differences between marine and freshwater stickleback populations tend to be concentrated in regions of low recombination (69), which increases the chances that multiple advantageous mutations remain linked over multiple generations when present as standing variation in the ancestral population.
We also detected a difference in dominance for fitness of Eda alleles in the single-generation pond experiment and the multigenerational Loberg Lake study. The contrast might be caused by a difference between the studies in the frequency of marine alleles at modifier loci. The marine (high-armor) Eda allele is largely dominant for lateral plates if marine alleles are fixed at several modifier loci of lateral plate number. Increasing numbers of freshwater alleles at these modifiers cause Eda heterozygotes to resemble more the low-armor freshwater phenotype instead of the high-armor marine form (51). If freshwater modifier alleles increased in frequency over time in Loberg lake, they might have contributed to different dominance estimates there compared with the single-generation pond study. Freshwater alleles might be favored at the modifier loci in part because of their indirect positive effects on fitness of Eda heterozygotes in freshwater. However, freshwater alleles at modifier loci might also be directly beneficial because like Eda they reduce number of lateral plates. Thus, dominance might also evolve in fresh water as an incidental by-product of selection for reduced armor. Evolution of dominance is known from other natural populations, such as during the spread of industrial melanism in peppered moth (70–72). When Eda modifier loci are further characterized in sticklebacks, it will be interesting to study their frequencies and interactions in marine and freshwater populations, and how these may change during rapid adaptation to new environments.
Nevertheless, the present results do not allow us to conclude that the large effects detected are solely or even predominantly caused by allelic variation at Eda. Our QTL study was carried out on an F2 cross, which maintains high linkage disequilibrium between Eda and many other genes on chromosome IV. For example, Wnt7b and Sult4A are two other loci on chromosome IV for which marine–freshwater divergence peaks are consistently detected (46). Both genes fall within the 1.5-LOD interval for the location of the fitness QTL and could be contributing to the magnitude of its fitness effects. In a linear model, Eda genotype remains significantly associated with F2 female fitness when genotype at the marker nearest Wnt7b (which is closer to Eda than is Sult4A) is included as a covariate (F2,214 = 5.99, P = 0.003). Nevertheless, numerous other genes in this interval could contribute to the apparent fitness effect of the QTL detected near Eda. More advanced generations would be needed for finer scale mapping of fitness. The region of high linkage disequilibrium around Eda is likely to be smaller in Loberg Lake than in the QTL study because many more generations took place. Nevertheless, our result is highly informative despite uncertainty over linked genes. Failing to detect a major QTL in the pond experiment or strong selection on Eda in the observational study would have led us to reject hypotheses predicting that large fitness-effect variants contributed to adaptation in fresh water (with the caveat that power might be limited, and that the strength of selection likely varies over time).
Such large fitness effects at genes and genomic regions differentiating locally adapted natural populations are probably not rare. Few studies of genetic differences between natural populations have measured fitness consequences in a subsequent generation. Nevertheless, within-generation studies mapping QTL for fitness components and lifetime fitness measures (73–76), those measuring genome-wide changes in allele frequencies (77–79), and those measuring selection at locally adapted genes (80, 81) (reviewed in ref. 82) frequently detect selection coefficients as large or larger than those reported here. Similarly large selection coefficients on genes and genomic regions have been documented in studies of selection on polymorphisms within populations (83–86). Selection on genes or genomic regions might fluctuate temporally and spatially, and even oscillate (25), and so large coefficients detected within generations or between a small number of generations might not often be sustained over longer periods (87). Nevertheless, rapid directional evolution at the Eda gene in stickleback, and in lateral plates controlled by Eda, is a repeatable feature of stickleback evolution in freshwater populations (26, 46, 59).
The mechanism producing the large fitness effect associated with Eda remains unclear. The hypothesis that low-armor plating is favored in fresh water because it is especially costly to produce there successfully predicted that high-armored F2 females were smaller in size and had lower fecundity than low-armored females. However, controlling for body size still revealed a major QTL for fitness on chromosome IV near Eda. This suggests that the fitness effects of the QTL on chromosome IV were mainly caused by selection on other phenotypes. It is conceivable that the cost of growing plates was mainly paid by the reduced survival of F3 juveniles, a possibility that we are unable to test with our data. Nevertheless, the Eda gene is highly pleiotropic, affecting multiple phenotypic traits other than armor and body size, including neuromast density and schooling behavior (47, 52–55). A great advantage of the stickleback natural system is that it is feasible to carry out field experiments to test hypotheses about the causes of fitness differences between alternative alleles (81). The magnitude and rapidity of selection on the Eda region, together with the ability to make transgenic sticklebacks carrying modifications of the Eda locus itself (7, 52, 54), or of other genes in the surrounding chromosome region (88), should make it possible to further resolve both the molecular and ecological aspects of strong selection on this major fitness locus in sticklebacks.
Supplementary Material
Acknowledgments
We thank Peter Grant, Hopi Hoekstra, and the D.S. and D.M.K. laboratories for helpful comments. We are grateful for the efforts of Patrick Tamkee, Tim Vines, Arianne Albert, Deb Bryant, Brian Marchinko, and Deanna Yim in the laboratory and field. Funding was provided by Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery grants (D.S.), an NIH Center of Excellence in Genomic Science grant (NHGRI 3P50 HG002568, D.M.K.), NSF grants (DEB-0322818 and DEB-0919184, M.A.B.), a European Research Council Consolidator Grant, Deutsche Forschungsgemeinschaft and the Max Planck Society (F.C.J.), NSERC and Killam Predoctoral fellowships (K.B.M.), and an NSF Postdoctoral Fellowship (M.E.A.). D.M.K. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no competing interest.
See Profiles, e2025633118 and e2025630118, in vol. 118, issue 3.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914889118/-/DCSupplemental.
Data Availability.
Data have been deposited in Dryad (doi: 10.5061/dryad.np5hqbzrc).
References
- 1.Orr H. A., Coyne J. A., The genetics of adaptation: A reassessment. Am. Nat. 140, 725–742 (1992). [DOI] [PubMed] [Google Scholar]
- 2.Rockman M. V., The QTN program and the alleles that matter for evolution: All that’s gold does not glitter. Evolution 66, 1–17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dittmar E. L., Oakley C. G., Conner J. K., Gould B. A., Schemske D. W., Factors influencing the effect size distribution of adaptive substitutions. Proc. R. Soc. 283, 20153065 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orr H. A., The genetic theory of adaptation: A brief history. Nat. Rev. Genet. 6, 119–127 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Shapiro M. D., et al. , Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature 428, 717–723 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Chan Y. F., et al. , Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327, 302–305 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colosimo P. F., et al. , Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science 307, 1928–1933 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Bradshaw H. D. Jr, Otto K. G., Frewen B. E., McKay J. K., Schemske D. W., Quantitative trait loci affecting differences in floral morphology between two species of monkeyflower (Mimulus). Genetics 149, 367–382 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steiner C. C., Weber J. N., Hoekstra H. E., Adaptive variation in beach mice produced by two interacting pigmentation genes. PLoS Biol. 5, e219 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weber J. N., Peterson B. K., Hoekstra H. E., Discrete genetic modules are responsible for complex burrow evolution in Peromyscus mice. Nature 493, 402–405 (2013). [DOI] [PubMed] [Google Scholar]
- 11.McGregor A. P., et al. , Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature 448, 587–590 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Linnen C. R., et al. , Adaptive evolution of multiple traits through multiple mutations at a single gene. Science 339, 1312–1316 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prud’homme B., et al. , Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature 440, 1050–1053 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Kopp M., Hermisson J., The genetic basis of phenotypic adaptation I: Fixation of beneficial mutations in the moving optimum model. Genetics 182, 233–249 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griswold C. K., Whitlock M. C., The genetics of adaptation: The roles of pleiotropy, stabilizing selection and drift in shaping the distribution of bidirectional fixed mutational effects. Genetics 165, 2181–2192 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schluter D., Conte G. L., Genetics and ecological speciation. Proc. Natl. Acad. Sci. U.S.A. 106 (suppl. 1), 9955–9962 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrett R. D. H., Schluter D., Adaptation from standing genetic variation. Trends Ecol. Evol. 23, 38–44 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Hedrick P. W., Adaptive introgression in animals: Examples and comparison to new mutation and standing variation as sources of adaptive variation. Mol. Ecol. 22, 4606–4618 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Seehausen O., Hybridization and adaptive radiation. Trends Ecol. Evol. 19, 198–207 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Dasmahapatra K. K. et al.; Heliconius Genome Consortium , Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487, 94–98 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meier J. I., et al. , Ancient hybridization fuels rapid cichlid fish adaptive radiations. Nat. Commun. 8, 14363 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feder J. L., et al. , Allopatric genetic origins for sympatric host-plant shifts and race formation in Rhagoletis. Proc. Natl. Acad. Sci. U.S.A. 100, 10314–10319 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchinko K. B., Schluter D., Parallel evolution by correlated response: lateral plate reduction in threespine stickleback. Evolution 61, 1084–1090 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Barrett R. D. H., Rogers S. M., Schluter D., Environment specific pleiotropy facilitates divergence at the Ectodysplasin locus in threespine stickleback. Evolution 63, 2831–2837 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Barrett R. D. H., Rogers S. M., Schluter D., Natural selection on a major armor gene in threespine stickleback. Science 322, 255–257 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Bell M. A., Aguirre W. E., Buck N. J., Twelve years of contemporary armor evolution in a threespine stickleback population. Evolution 58, 814–824 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Hagen D. W., Isolating mechanisms in threespine sticklebacks (Gasterosteus). J. Fish. Res. Board Can. 24, 1637–1692 (1967). [Google Scholar]
- 28.Saimoto R. S., “Reproductive and natal homing of marine threespine sticklebacks (Gasterosteus aculeatus),” MSc thesis, University of British Columbia, Vancouver, BC, Canada (1993).
- 29.Schluter D., McPhail J. D., Ecological character displacement and speciation in sticklebacks. Am. Nat. 140, 85–108 (1992). [DOI] [PubMed] [Google Scholar]
- 30.Ingram T., et al. , Intraguild predation drives evolutionary niche shift in threespine stickleback. Evolution 66, 1819–1832 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Rogers S. M., et al. , Genetic signature of adaptive peak shift in threespine stickleback. Evolution 66, 2439–2450 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marchinko K. B., Predation’s role in repeated phenotypic and genetic divergence of armor in threespine stickleback. Evolution 63, 127–138 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Krebs C. J., Ecological Methodology (Addison-Welsey, Menlo Park, CA, 1999). [Google Scholar]
- 34.Marchinko K. B., Matthews B., Arnegard M. E., Rogers S. M., Schluter D., Maintenance of a genetic polymorphism with disruptive natural selection in stickleback. Curr. Biol. 24, 1289–1292 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Hagen D. W., Gilbertson L. G., Geographic variation and environmental selection in Gasterosteus aculeatus L. in the Pacific Northwest, America. Evolution 26, 32–51 (1972). [DOI] [PubMed] [Google Scholar]
- 36.Jones F. C., et al. , A genome-wide SNP genotyping array reveals patterns of global and repeated species-pair divergence in sticklebacks. Curr. Biol. 22, 83–90 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glazer A. M., Killingbeck E. E., Mitros T., Rokhsar D. S., Miller C. T., Genome assembly improvement and mapping convergently evolved skeletal traits in sticklebacks with Genotyping-by-Sequencing. G3 5, 1463–1472 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Broman K. W., Sen S., A Guide to QTL Mapping with R/qtl (Springer, 2009). [Google Scholar]
- 39.Efron B., The Jackknife, the Bootstrap, and Other Resampling Plans (SIAM, 1982). [Google Scholar]
- 40.Linnen C. R., Hoekstra H. E., Measuring natural selection on genotypes and phenotypes in the wild. Cold Spring Harb. Symp. Quant. Biol. 74, 155–168 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peichel C. L., et al. , The genetic architecture of divergence between threespine stickleback species. Nature 414, 901–905 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Rousset F., genepop’007: A complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 8, 103–106 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Weir B. S., Cockerham C. C., Estimating F‐statistics for the analysis of population structure. Evolution 38, 1358–1370 (1984). [DOI] [PubMed] [Google Scholar]
- 44.Havens A. C., Sweet D. E., Baer C. L., Bradley T. J., “Investigation of threespine stickleback abundance in landlocked Matanuska-Susitna Valley Lakes” (Alaska Department of Fish and Game, Juneau, AK, 1984). [Google Scholar]
- 45.Baker J. A., “Life history variation in female threespine stickleback” in The Evolutionary Biology of the Threespine Stickleback, Bell M. A., Foster S. A., Eds. (Oxford University Press, 1994), pp. 144–187. [Google Scholar]
- 46.Jones F. C. et al.; Broad Institute Genome Sequencing Platform & Whole Genome Assembly Team , The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484, 55–61 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O’Brown N. M., Summers B. R., Jones F. C., Brady S. D., Kingsley D. M., A recurrent regulatory change underlying altered expression and Wnt response of the stickleback armor plates gene EDA. eLife 4, e05290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whitlock M., Schluter D., The Analysis of Biological Data (Macmillan Learning, New York, ed. 3, 2020). [Google Scholar]
- 49.Boyle E. A., Li Y. I., Pritchard J. K., An expanded view of complex traits: From polygenic to omnigenic. Cell 169, 1177–1186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giles N., The possible role of environmental calcum levels during the evolution of phenotypic diversity in Outer Hebridean populations of the three‐spined stickleback, Gasterosteus aculeatus. J. Zool. 199, 535–544 (1983). [Google Scholar]
- 51.Colosimo P. F., et al. , The genetic architecture of parallel armor plate reduction in threespine sticklebacks. PLoS Biol. 2, E109 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Greenwood A. K., Mills M. G., Wark A. R., Archambeault S. L., Peichel C. L., Evolution of schooling behavior in threespine sticklebacks is shaped by the Eda gene. Genetics 203, 677–681 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wark A. R., et al. , Genetic architecture of variation in the lateral line sensory system of threespine sticklebacks. G3 2, 1047–1056 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wucherpfennig J. I., Miller C. T., Kingsley D. M., Efficient CRISPR-Cas9 editing of major evolutionary loci in sticklebacks. Evol. Ecol. Res. 20, 107–132 (2019). [PMC free article] [PubMed] [Google Scholar]
- 55.Archambeault S. L., Bärtschi L. R., Merminod A. D., Peichel C. L., Adaptation via pleiotropy and linkage: Association mapping reveals a complex genetic architecture within the stickleback Eda locus. Evol. Lett. 4, 282–301 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hohenlohe P. A., et al. , Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genet. 6, e1000862 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roesti M., Gavrilets S., Hendry A. P., Salzburger W., Berner D., The genomic signature of parallel adaptation from shared genetic variation. Mol. Ecol. 23, 3944–3956 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Terekhanova N. V., et al. , Fast evolution from precast bricks: Genomics of young freshwater populations of threespine stickleback Gasterosteus aculeatus. PLoS Genet. 10, e1004696 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lescak E. A., et al. , Evolution of stickleback in 50 years on earthquake-uplifted islands. Proc. Natl. Acad. Sci. U.S.A. 112, E7204–E7212 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barrett R. D. H., Adaptive evolution of lateral plates in three-spined stickleback Gasterosteus aculeatus: A case study in functional analysis of natural variation. J. Fish. Biol. 77, 311–328 (2010). [DOI] [PubMed] [Google Scholar]
- 61.Rennison D. J., Heilbron K., Barrett R. D. H., Schluter D., Discriminating selection on lateral plate phenotype and its underlying gene, Ectodysplasin, in threespine stickleback. Am. Nat. 185, 150–156 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Miller C. T., et al. , Modular skeletal evolution in sticklebacks is controlled by additive and clustered quantitative trait loci. Genetics 197, 405–420 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Erickson D. L., Fenster C. B., Stenøien H. K., Price D., Quantitative trait locus analyses and the study of evolutionary process. Mol. Ecol. 13, 2505–2522 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Alaska Department of Fish and Game , Top 20 records of fish stocked in Loberg Lake in the MAT-SU area (2019). http://www.adfg.alaska.gov/index.cfm?adfg=SportStockingHatcheriesSearch.locationSearchResults&StockingAreaID=351&AreaID=7&SpeciesID=all&TopRecords=20. Accessed 25 August 2019.
- 65.Albert A. Y. K., et al. , The genetics of adaptive shape shift in stickleback: Pleiotropy and effect size. Evolution 62, 76–85 (2008). [DOI] [PubMed] [Google Scholar]
- 66.Xie K. T., et al. , DNA fragility in the parallel evolution of pelvic reduction in stickleback fish. Science 363, 81–84 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nelson T. C., Cresko W. A., Ancient genomic variation underlies repeated ecological adaptation in young stickleback populations. Evol. Lett. 2, 9–21 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Clague J. J., Luternauer J. L., Hebda R. J., Sedimentary environments and postglacial history of the Fraser Delta and lower Fraser valley, British Columbia. Can. J. Earth Sci. 20, 1314–1326 (1983). [Google Scholar]
- 69.Samuk K., et al. , Gene flow and selection interact to promote adaptive divergence in regions of low recombination. Mol. Ecol. 26, 4378–4390 (2017). [DOI] [PubMed] [Google Scholar]
- 70.Haldane J. B. S., The theory of selection for melanism in Lepidoptera. Proc. R. Soc. 145, 303–306 (1956). [Google Scholar]
- 71.Kettlewell H. B. D., Insect survival and selection for pattern: Most camouflage and survival mechanisms, though highly perfected, can be adapted to changing environments. Science 148, 1290–1296 (1965). [DOI] [PubMed] [Google Scholar]
- 72.Sheppard P. M., The Evolution of Dominance. Natural Selection and Heredity (Hutchinson University Library, London, ed. 4, 1975), pp. 154–171. [Google Scholar]
- 73.Ågrena J., Oakley C. G., McKay J. K., Lovell J. T., Schemske D. W., Genetic mapping of adaptation reveals fitness tradeoffs in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 110, 21077–21082 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Anderson J. T., Lee C. R., Mitchell-Olds T., Strong selection genome-wide enhances fitness trade-offs across environments and episodes of selection. Evolution 68, 16–31 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang X., et al. , The earliest stages of adaptation in an experimental plant population: Strong selection on QTLS for seed dormancy. Mol. Ecol. 19, 1335–1351 (2010). [DOI] [PubMed] [Google Scholar]
- 76.Gardner K. M., Latta R. G., Identifying loci under selection across contrasting environments in Avena barbata using quantitative trait locus mapping. Mol. Ecol. 15, 1321–1333 (2006). [DOI] [PubMed] [Google Scholar]
- 77.Lexer C., Welch M. E., Durphy J. L., Rieseberg L. H., Natural selection for salt tolerance quantitative trait loci (QTLs) in wild sunflower hybrids: Implications for the origin of Helianthus paradoxus, a diploid hybrid species. Mol. Ecol. 12, 1225–1235 (2003). [DOI] [PubMed] [Google Scholar]
- 78.Gompert Z., et al. , Experimental evidence for ecological selection on genome variation in the wild. Ecol. Lett. 17, 369–379 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Laurentino T. G., et al. , Genomic release-recapture experiment in the wild reveals within-generation polygenic selection in stickleback fish. Nat. Commun. 11, 1928 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Barrett R. D. H., et al. , Linking a mutation to survival in wild mice. Science 363, 499–504 (2019). [DOI] [PubMed] [Google Scholar]
- 81.Rennison D. J., Rudman S. M., Schluter D., Genetics of adaptation: Experimental test of a biotic mechanism driving divergence in traits and genes. Evol. Lett. 3, 513–520 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thurman T. J., Barrett R. D. H., The genetic consequences of selection in natural populations. Mol. Ecol. 25, 1429–1448 (2016). [DOI] [PubMed] [Google Scholar]
- 83.Nosil P., et al. , Natural selection and the predictability of evolution in Timema stick insects. Science 359, 765–770 (2018). [DOI] [PubMed] [Google Scholar]
- 84.Johnston S. E., et al. , Life history trade-offs at a single locus maintain sexually selected genetic variation. Nature 502, 93–95 (2013). [DOI] [PubMed] [Google Scholar]
- 85.Mostafavi H., et al. , Identifying genetic variants that affect viability in large cohorts. PLoS Biol. 15, e2002458 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lamichhaney S., et al. , A beak size locus in Darwin’s finches facilitated character displacement during a drought. Science 352, 470–474 (2016). [DOI] [PubMed] [Google Scholar]
- 87.Hoekstra H. E., et al. , Strength and tempo of directional selection in the wild. Proc. Natl. Acad. Sci. U.S.A. 98, 9157–9160 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Howes T. R., Summers B. R., Kingsley D. M., Dorsal spine evolution in threespine sticklebacks via a splicing change in MSX2A. BMC Biol. 15, 115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data have been deposited in Dryad (doi: 10.5061/dryad.np5hqbzrc).