Significance
This paper describes a high-speed fluorescence lifetime imaging method, compressed-sensing fluorescence lifetime imaging microscopy (compressed FLIM), which can produce high-resolution two-dimensional (2D) lifetime images at an unprecedented frame rate. Compared to other state-of-the-art FLIM imagers, compressed FLIM has a striking advantage in acquiring a widefield lifetime image within a single camera snapshot, thereby eliminating motion artifacts and enabling fast recording of biological events.
Keywords: FLIM, high-speed, compressed imaging
Abstract
We present high-resolution, high-speed fluorescence lifetime imaging microscopy (FLIM) of live cells based on a compressed sensing scheme. By leveraging the compressibility of biological scenes in a specific domain, we simultaneously record the time-lapse fluorescence decay upon pulsed laser excitation within a large field of view. The resultant system, referred to as compressed FLIM, can acquire a widefield fluorescence lifetime image within a single camera exposure, eliminating the motion artifact and minimizing the photobleaching and phototoxicity. The imaging speed, limited only by the readout speed of the camera, is up to 100 Hz. We demonstrated the utility of compressed FLIM in imaging various transient dynamics at the microscopic scale.
Ultrafast biological dynamics occur at atomic or molecular scales. Resolving these transient phenomena requires a typical frame rate at gigahertz, which is beyond the bandwidth of most electronic sensors, posing a significant technical challenge on the detection system (1, 2). Ultrafast optical imaging techniques (3–8) can provide a high temporal resolution down to a femtosecond and have established themselves as indispensable tools in blur-free observation of fast biological events (7–12).
Fluorescence lifetime imaging microscopy (FLIM) (13, 14)—a powerful ultrafast imaging tool for fingerprinting molecules—has been extensively employed in a wide spectrum of biomedical applications, ranging from single molecule analysis to medical diagnosis (15, 16). Rather than imaging the time-integrated fluorescent signals, FLIM measures the time-lapse fluorescent decay after pulsed or time-modulated laser excitation. Because the lifetime of a fluorophore is dependent on its molecular environment but not on its concentration, FLIM enables a more quantitative study of molecular effects inside living organisms compared with conventional intensity-based approaches (16–21).
To detect the fast fluorescence decay in FLIM, there are generally two strategies. Time-domain FLIM illuminates the sample with pulsed laser, followed by using time-correlated single-photon counting (TCSPC) or time-gated sensors to directly measure the fluorescence decay (14, 22, 23). By contrast, frequency-domain FLIM excites the sample with high-frequency modulated light and infers the fluorescence lifetime through measuring the relative phase shift in the modulated fluorescence (24, 25). Despite being quantitative, the common drawback of these approaches is their dependence on scanning and/or repetitive measurements. For example, to acquire a two-dimensional (2D) image, a confocal FLIM imager must raster scan the entire field of view (FOV), resulting in a trade-off between the FOV and frame rate. To avoid motion artifacts, the sample must remain static during data acquisition. Alternatively, widefield FLIM systems acquire spatial data in parallel. Nonetheless, to achieve high temporal resolution, they still need to temporally scan either a gated window (26, 27) or detection phase (28), or they must use TCSPC, which requires a large number of repetitive measurements to construct a temporal histogram (29, 30). Limited by the scanning requirement, current FLIM systems operate at only a few frames per second when acquiring high-resolution images (22, 31, 32). The slow frame rate thus prevents these imagers from capturing transient biological events, such as neural spiking (33) and calcium oscillation (34). Therefore, there is an unmet need to develop new and efficient imaging strategies for high-speed, high-resolution FLIM.
To overcome the above limitations, herein we introduced the paradigm of compressed sensing into FLIM and developed a snapshot widefield FLIM system, termed compressed FLIM, which can image fluorescence lifetime at an unprecedented speed. Through compressed sensing (CS), temporal information is encoded in each measured frame from which temporal dynamics can be estimated using CS reconstruction algorithms. Our method is made possible by a unique integration of 1) compressed ultrafast photography (CUP) (6) for data acquisition, 2) a dual-camera detection scheme for high-resolution image reconstruction (35), and 3) computer cluster hardware and graphic processing unit (GPU) parallel computing technologies for real-time data processing. The synergistic effort enables high-resolution (500 × 400 pixels) widefield lifetime imaging at 100 frames per second (fps). To demonstrate compressed FLIM, we performed experiments sequentially on fluorescent beads and live neurons, measuring the dynamics of bead diffusion and firing of action potential, respectively.
Results
Compressed FLIM.
Compressed FLIM operates in two steps: data acquisition and image reconstruction (both further described in Methods). Briefly, the sample is first imaged by a high-resolution fluorescence microscope. The output image is then passed to the CUP camera for time-resolved measurement. Finally, we use a GPU-accelerated CS reconstruction algorithm—two-step iterative shrinkage/thresholding (TwIST) (36)—to process the image in real time.
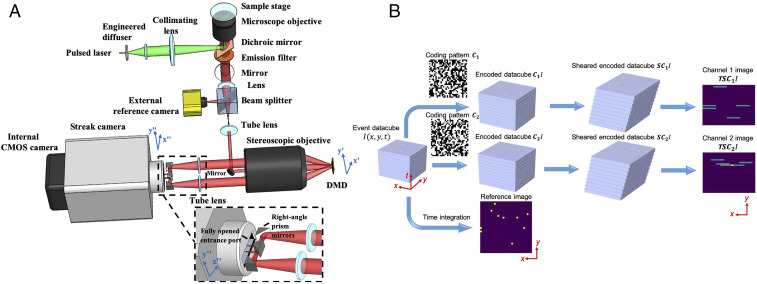
A compressed FLIM system (Fig. 1A) consists of an epi-fluorescence microscope and a CUP camera. An animated video (Movie S1) shows the system in action. Fig. 1B shows the data acquisition pipeline. Upon excitation by a single laser pulse, the fluorescence is collected by an objective lens with a high numerical aperture (NA) and forms an intermediate image at the microscope’s side image port. A beam splitter then divides the fluorescence into two beams. The reflected light is directly captured by a reference complementary metal–oxide–semiconductor (CMOS) camera, generating a time-integrated image:
| [1] |
where denotes the time-lapse fluorescence decay.
Fig. 1.
(A) Schematic of compressed FLIM. (Lower Right Inset) A close-up of the configuration at the streak camera’s entrance port. Light beams in two complementary encoding channels are folded by two right angle prisms before entering the fully opened entrance port of the streak camera. (B) Data acquisition pipeline. For simple illustration, we assume the object consists of individual static points sparsely distributed in space and constantly illuminated in time.
The transmitted light is relayed to a digital micromirror device (DMD) through a 4f imaging system consisting of a tube lens, a mirror, and a stereoscope objective. A static, pseudorandom, binary pattern is displayed on the DMD to encode the image. Each encoding pixel is turned either on (tilted −12° with respect to the DMD surface norm) or off (tilted +12° with respect to the DMD surface norm) and reflects the incident fluorescence in one of the two directions. Two reflected fluorescence beams, masked with the complementary patterns, are both collected by the same stereoscope objective and enter corresponding subpupils at the objective’s back focal plane. The fluorescence from these two subpupils are then imaged by two tube lenses, folded by right-angle prism mirrors (the lower right inset in Fig. 1A), and form two complementary channel images at the entrance port of a streak camera. To accept the encoded 2D image, this entrance port is fully opened (∼5 mm width) to its maximum, exposing the entire photocathode to the incident light. Inside the streak camera, the encoded fluorescence signals are temporally deflected along the vertical axis according to the time of arrival of incident photons. The final image is acquired by a CMOS camera—the photons are temporally integrated within the camera’s exposure time and spatially integrated within the camera’s pixel. The formation of complementary channel images can be written as:
| [2] |
Here, is a temporal shearing operator, and T is a spatiotemporal integration operator. They describe the functions of the streak camera and CMOS camera, respectively. C1 and C2 are spatial encoding operators, depicting the complementary masks applied to two channel images, and , in which I is a matrix of ones. This complementary-encoding setup minimizes the light throughput loss, a fact that is critical for photon-starved imaging. Also, because there is no information loss, our encoding scheme enriches the observation and favors the compressed-sensing-based image reconstruction.
During data acquisition, we synchronize the streak camera with the reference camera in the transmission optical path. Therefore, each excitation event yields three images: one time-integrated fluorescence image, , and two spatially encoded, temporally sheared channel images, and .
The image reconstruction of compressed FLIM is the solution of the inverse problem of the above image formation process (Eqs. 1 and 2). Because the two complementary channel images , are essentially associated with the same scene, the original fluorescence decay event can be reasonably estimated by applying a CS algorithm TwIST (35) to the concatenated data . Additionally, to further improve the resolution, we apply the time-integrated image recorded by the reference camera, , as the spatial constraint. Finally, we fit a nonlinear least-squares exponential curve to the reconstructed fluorescence decay at each spatial sampling location and produce the high-resolution fluorescence lifetime map. To reconstruct the image in real time, we implement our algorithm on GPU and computer cluster hardware.
The synergistic integration of hardware and algorithm innovations enables acquisition of high-quality microscopic fluorescence lifetime images. Operating in a snapshot format, the frame rate of compressed FLIM is limited by only the readout speed of the streak camera and up to 100 fps. The spatial resolution, determined by the NA of the objective lens, is in a submicron range, providing a resolving power to uncover the transient events inside a cell.
Imaging Fluorescent Beads in Flow.
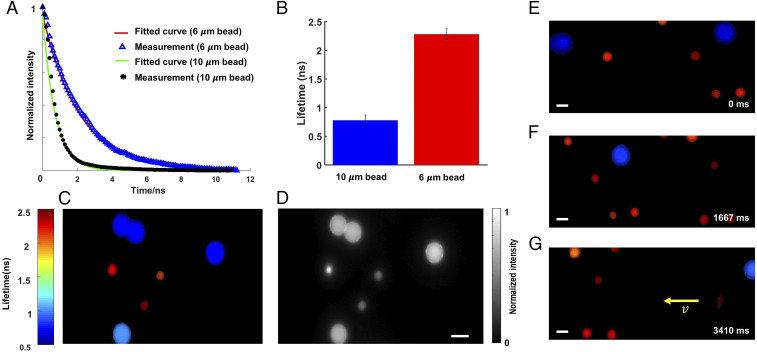
We demonstrated compressed FLIM by imaging fluorescent beads in flow. We mixed two types of fluorescent beads (diameters 6 and 10 μm) in phosphate buffer solutions (PBS) and flowed them in a microtubing at a constant speed using a syringe pump. We excited the beads at 532 nm and continuously imaged the fluorescence using compressed FLIM at 75 fps. As an example, the reconstructed time-lapse fluorescence decays at two beads’ locations are shown in Fig. 2A, and the mean lifetimes of two types of the beads are shown in Fig. 2B. We pseudocolored the bead image based on the fitted lifetimes (Fig. 2C). The result indicates that compressed FLIM can differentiate these two types of beads with close lifetimes. For comparison, the corresponding time-integrated image captured by the reference camera is shown in Fig. 2D.
Fig. 2.
Lifetime imaging of fluorescent beads in flow. (A) Reconstructed fluorescence decays of two types of fluorescent beads. (B) Mean lifetimes. The SEMs are shown as error bars. (C) Reconstructed snapshot lifetime image. (D) Reference intensity image. (E–G) Lifetime images of fluorescent beads in flow at representative temporal frames 0 ms, 1667 ms, and 3410 ms, respectively. (Scale bar, 10 μm.) Camera frame time: 13 ms. Average laser power at the sample: 0.45 mW.
We further reconstructed the entire flow dynamics and show the movie and representative temporal frames in Movie S2 and Fig. 2E, respectively. Because the lifetime image was acquired in a snapshot format, no motion blur is observed.
Lifetime Unmixing of Neural Cytoskeletal Proteins.
Next, we applied compressed FLIM to cell imaging and demonstrated fluorescence lifetime unmixing of multiple fluorophores with highly overlapped emission spectra. Multitarget fluorescence labeling is commonly used to differentiate intracellular structures. Separation of multiple fluorophores can be accomplished by spectrally resolved detection and multicolor analysis (37–39) or time-resolved detection by FLIM (40–42). The spectral method fails when the emission spectra of the fluorophores strongly overlap. By contrast, FLIM has a unique edge in this case provided a difference in fluorophores’ lifetimes.
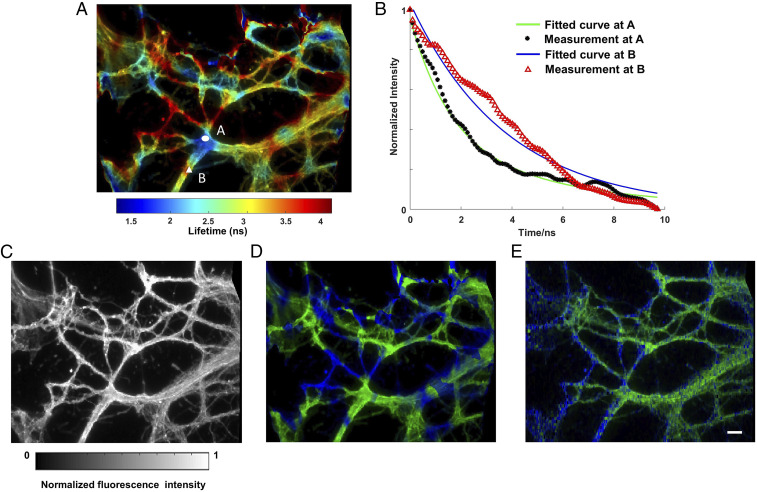
We imaged two protein structures in the cytoskeleton of neurons and unmixed them based on the lifetime. We immunolabeled Vimentin and Tubulin in the cytoskeleton with Alexa Fluor 555 and Alexa Fluor 546, respectively. The emission spectra of two fluorophores highly overlap, but their fluorescence lifetimes differ (1.3 ns versus 4.1 ns). Within a single snapshot, we captured a high-resolution lifetime image of immunofluorescently stained neurons (Fig. 3A). Movie S3 records the time-lapse fluorescence decay process after a single pulse excitation, and two representative decay curves associated with Alexa Fluor 555 and 546 are shown in Fig. 3B. The reference fluorescence intensity image is shown in Fig. 3C. Next, we applied a regularized unmixing algorithm (Methods) to the lifetime data and separated Vimentin and Tubulin into two channels in Fig. 3D (green channel, Vimentin; blue channel, Tubulin). To acquire the ground-truth unmixing result, we operated our system in a slit-scanning mode and imaged the same FOV (Methods). The resultant unmixed result (Fig. 3E) matches qualitatively well with compressed FLIM measurement. To further validate the distribution pattern of Vimentin and Tubulin in neuron cytoskeleton structures, we imaged the sample using a benchmark confocal FLIM system (ISS Alba FCS), and SI Appendix, Fig. S1 presents the two proteins distribution. The neurons exhibit a similar protein distribution pattern as that inferred by compressed FLIM.
Fig. 3.
Lifetime imaging of neuronal cytoskeleton immunolabeled with two fluorophores. Alexa Fluor 555 immunolabels Vimentin. Alexa Fluor 546 immunolabels Tubulin. (A) Reconstructed lifetime image of immunofluorescently stained neurons. (B) Reconstructed fluorescence decays at two fluorophore locations. (C) Reference intensity image. (D) Lifetime unmixed image. Green channel, Vimentin; Blue channel, Tubulin. (E) Ground-truth lifetime unmixed image captured by line-scanning streak imaging. (Scale bar, 10 µm.) Camera frame time: 10 ms. Average laser power at the sample: 0.60 mW.
Imaging Neural Spikes in Live Cells.
The complex functions of the brain depend on coordinated activity of multiple neurons and neuronal circuits. Therefore, visualizing the spatial and temporal patterns of neuronal activity in single neurons is essential to understand the operating principles of neural circuits. Recording neuronal activity using optical methods has been a long-standing quest for neuroscientists as it promises a noninvasive means to probe real-time dynamic neuronal function. Imaging neuronal calcium transients (somatic calcium concentration changes) with genetically encoded calcium indicators (43, 44), fluorescent calcium indicator stains (45), and two-photon excitation methods using galvanometric (46) and target-path scanners (47) have been used to resolve suprathreshold spiking (electrical) activity.
However, calcium imaging is an indirect method of assessing neuronal activity, and the spike number and firing rates using fluorescence recording are error prone, especially when being used in cell populations that contain heterogeneous spike-evoked calcium signals. Therefore, the capability of directly assessing neuronal function is invaluable for advancing our understanding of neurons and the nervous system.
Genetically encoded voltage indicators (GEVIs) offer great promise for directly visualizing neural spike dynamics (48, 49). Compared with calcium imaging, GEVIs provide much faster kinetics that faithfully capture individual action potentials and subthreshold voltage dynamics. Förster resonance energy transfer (FRET)-opsin fluorescent voltage sensors report neural spikes in brain tissue with superior detection fidelity compared with other GEVIs (50).
As a molecular ruler, FRET involves the nonradiative transfer of excited state energy from a fluorophore, the donor, to another nearby absorbing but not necessarily fluorescent molecule, the acceptor. When FRET occurs, both the fluorescence intensity and lifetime of the donor decrease. So far, most fluorescence voltage measurements using FRET-opsin–based GEVIs report relative fluorescence intensity changes and fail to reveal the absolute membrane voltage because of illumination intensity variations, photobleaching, and background autofluorescence. By contrast, because FLIM is based on absolute lifetime measurement, it is insensitive to the environmental factors. Therefore, FLIM enables a more quantitative study with FRET-opsin–based GEVIs and provides a readout of the absolute voltage (51).
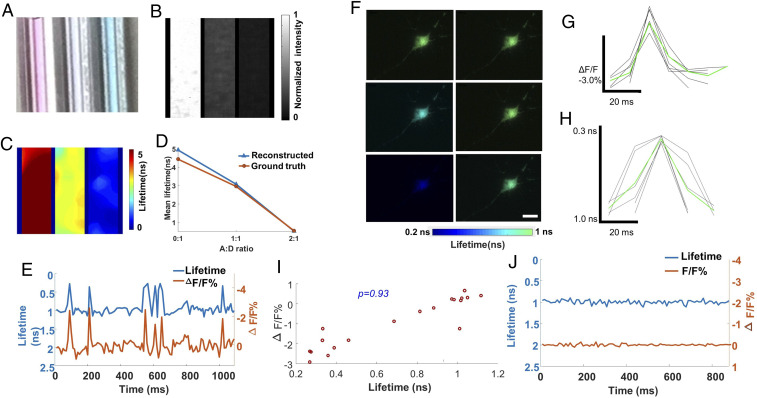
To demonstrate how compressed FLIM can be used to detect FRET, we first imaged two fluorescence dyes (the donor with Alexa Fluor 546 and the acceptor with Alexa Fluor 647 in PBS) with varied mixed concentration ratios. The emission spectrum of the donor overlaps considerably with the absorption spectrum of the acceptor, meeting the requirement for FRET. Acceptor bleed-through contamination, that is, the direct excitation and emission of the acceptor, is minimized by properly choosing the excitation wavelength and emission filter (Methods). We prepared three samples (Fig. 4A) with different concentration ratios (1:0, 1:1, and 1:2) of donor and acceptor and imaged the fluorescence intensities and lifetimes using the reference time-integrated camera and compressed FLIM, respectively (Fig. 4 B and C). As expected, fluorescence emission intensity of the donor gradually diminishes with more acceptor presence and stronger fluorescence quenching. Also, as revealed by compressed FLIM, there is a decrease in the donor’s fluorescence lifetime along with an increase in the acceptor’s concentration. Furthermore, we performed the ground-truth measurement by switching the system to the line-scanning mode (Methods). The lifetime results acquired by compressed FLIM match well with the ground truth (Fig. 4D).
Fig. 4.
High-speed lifetime imaging of neural spiking in live neurons expressing MacQ-mOrange2. (A–D) Lifetime imaging of FRET in phantoms. Acceptor Alexa 647 and donor Alexa 546 were mixed with varied concentration ratios (A:D ratio). (A) Photograph of three mixed solutions with different A:D ratios (0:1, 1:1, and 2:1). (B) Reference intensity image. (C) Reconstructed lifetime image. (D) Comparison between measurement and ground truth. (E–J) Lifetime imaging of FRET in live neurons. The FRET sensor MacQ-mOrange2 was expressed in a live neuron. (E) Reconstructed time-lapse lifetime and intensity recording of neural spiking. The signals were averaged inside a cell. (F) Reconstructed lifetime images of the neuron at representative temporal frames upon potassium stimulation. (G and H) Intensity and lifetime waveforms of neural spikes (black lines) and their mean (green line). (I) Scatter plot between lifetime and intensity of MacQ-mOrange2 measured at different times. The Pearson correlation coefficient is 0.93. (J) Negative control. (Scale bar, 10 µm.) Camera frame time: 10 ms. Average laser power at the sample: 0.60 mW.
Next, we evaluated compressed FLIM in imaging a FRET-opsin–based GEVI, MacQ-mOrange2 (50), to detect spiking in cultured neurons. During voltage depolarization, the optical readout is fluorescence quenching of the FRET donor mOrange2. We transfected neurons with plasmid DNA MacQ-mOrange2 and stimulated with high potassium treatment. We then used compressed FLIM to image the neural spikes. To determine fluorescence lifetime and intensity traces for individual cells, we extracted the pixels that rank in the top 50% of the signal-to-noise ratio values, defined as , where is the voltage-dependent change in fluorescence intensity (50), and is a pixel’s mean baseline fluorescence intensity (50). Average fluorescence lifetime and fluorescence intensity were calculated from these pixels in each frame. Fig. 4E shows the fluorescence intensity and lifetime traces of MacQ-mOrange2 sensor expressed in a cultured hippocampal neuron within 50 mM potassium environment imaged at 100 Hz. Movie S4 records the lifetime dynamics due to neural pulsing. Representative snapshots at 50 ms, 60 ms, 70 ms, 80 ms, 90 ms, and 100 ms shown in Fig. 4F indicate occurrence of lifetime oscillations. The average relative fluorescence intensity change and absolute lifetime change in response to one spiking event are -2.9% and -0.7 ns, respectively. Fig. 4 G and H present the experimentally determined fluorescence intensity and lifetime waveforms of single action potentials from MacQ-mOrange2 (black trace) and their mean (green trace, average over n = 6 spikes). To further study the correlation between fluorescence intensities and lifetimes, we scatter plotted their relationship in Fig. 4I. The calculated Pearson coefficient is 0.93, indicating a high correlation between measured fluorescence intensities and lifetimes. Finally, to provide a negative control, we imaged MacQ-mOrange2 within a subthreshold nonactivated 20 μM potassium stimulation (Fig. 4J). Both the fluorescence intensities and lifetimes were stable during the entire time trace, and no oscillations were observed.
Noteworthily, herein we measured lifetimes rather than absolute voltages. Because previous studies (51, 52) have established the relation between the lifetime and absolute membrane voltage for the FRET-based GEVI sensors, the conversion of lifetime to absolute voltage can be done through the conventional electrophysiological approach. Also, the full recording of the neuron spiking typically requires a kilohertz frame rate. The lifetime imaging rate of our current system is limited by the sweeping rate of the streak camera, and it is up to 100 Hz. However, this speed can be readily improved by replacing it with a high repetition streak camera (53), which allows a megahertz sweeping rate at an expense of a reduced signal to noise ratio.
Discussion
Compared with conventional scanning-based FLIM imagers, compressed FLIM features three predominant advantages. First, based on a compressed-sensing architecture, compressed FLIM can produce high-resolution 2D widefield fluorescence lifetime maps of optically-thin specimens at 100 fps, allowing quantitative capture of transient biological dynamics. The gain in the imaging speed is attributed to the compressibility of a fluorescence scene in a specific domain. To show the dependence of reconstructed image quality on the compression ratio (CR) of a fluorescence event, we calculated the CR when imaging a biological cell stained with a typical fluorophore with a lifetime of 4 ns. The observation time window (i.e., the temporal sweeping range of the streak camera) was set to be 20 ns. Here, we define CR as the ratio of the total number of voxels ( and , samplings along spatial axes x, y and temporal axis t, respectively) in the reconstructed event datacube to the total number of pixels ( and , samplings along spatial axes , in the camera coordinate, respectively) in the raw image:
| [3] |
In compressed FLIM, the spatial information y and temporal information t occupy the same axis in the raw image. Therefore, their sum, , cannot exceed the total number of camera pixels along axis. Here, we consider the equality case . Also, for simplicity, we assume the two complementary image channels fully occupy the entire axis on the camera, that is, . We then rewrite CR as follows:
| [4] |
To evaluate the relation between CR and the reconstructed image quality, we used the peak signal-to-noise ratio (PSNR) as the metric. For a given output image format (Ny = 200 pixels), we varied the CR by increasing and calculated the corresponding PSNRs (SI Appendix, Fig. S3A). The corresponding for data points presented are 10, 20, 50, 100, 200, and 500, respectively. The figure implies that the larger the temporal sequence depth of the event datacube, the higher the CR, and the worse the image quality. For high-quality imaging (PSNR ≥ 20 dB), the CR must be no greater than 70 (i.e., the temporal sequence depth ≤ 200). Therefore, in compressed FLIM, the sequence depth of the reconstructed event datacube must be balanced for image quality.
Second, because of using a complementary encoding scheme, compressed FLIM minimizes the light throughput loss. The light throughput advantage can be characterized by the snapshot advantage factor, which is defined as the portion of datacube voxels that are continuously visible to the instrument (54). When measuring a high-resolution image in the presented format (500 × 400 pixels), we gain a factor of 2 × 105 and 400 in light throughput compared with those in the point-scanning and line-scanning–based counterparts. Such a throughput advantage makes compressed FLIM particularly suitable for low light imaging applications. To study the dependence of the reconstructed image quality on the number of photons received at a pixel, we simulated the imaging performance of compressed FLIM for a shot-noise–limited system. Provided that the pixel with the maximum count in the image collects M photons, the corresponding shot noise is photons. We added photon noises to all pixels both in the temporally sheared and integrated images and performed the image reconstruction with various Ms (SI Appendix, Fig. S3B). We kept CR the same for all the data points in the plot. SI Appendix, Fig. S3B shows that a larger M (i.e., more photons) leads to a higher PSNR. For high-quality image reconstruction (PSNR ≥ 20 dB), M must be greater than 80 photons.
Lastly, operating in a snapshot format, compressed FLIM eliminates the motion artifacts and enables fast recording of biological events. We quantitatively computed the maximum blur-free motion allowed by our system. Assuming we image a typical fluorophore using a 1.4 NA objective, the maximum blur-free movement during a single-shot acquisition equals the system’s spatial resolution (∼0.2 μm). To estimate the correspondent blur-free speed limit, we use the streak camera’s observation time window (20 ns) as the camera’s effective exposure time. The blur-free speed limit is thus calculated as 0.2 μm/20 ns = 10 μm/us. A lower NA objective lens or a shorter observation window will increase this threshold, however, at the expense of a reduced resolution and temporal sequence depth. The ability to capture rapid motion at the microscopic scale will be valuable to studying fast cellular events, such as protein folding (55) or ligand binding (56). Moreover, compressed FLIM employs widefield illumination to excite the sample, presenting a condition that is favorable for live cell imaging. Cells exposed to radiant energy may experience physiological damages because of heating and/or the generation of reactive oxygen species during extended fluorescence microscopy (57). Because phototoxicity has a nonlinear relation with illumination radiance (58), widefield compressed FLIM prevails in preserving cellular viability compared with its scanning-based counterpart.
In compressed FLIM, the spatial and temporal information are multiplexed and measured by the same detector array. The system therefore faces two constrains. First, there is a trade-off between lifetime estimation accuracy and illumination intensity. The observation time window T (i.e., the temporal sweeping range) of the steak camera can be computed as , where L is the height of the phosphor screen of the streak tube, and v is the temporal shearing velocity. For the streak camera used, T is tunable from 0.5 ns to 1 ms. Given 1,016 detector rows sampling this sweeping range, the temporal resolution of the streak camera is . Provided that the fluorescence decay has a duration of , we can calculate the number of temporal samplings as . Therefore, using a faster temporal shearing velocity in the streak camera will increase . Given ample photons, this will improve the lifetime estimation accuracy approximately in the manner of. However, a larger will reduce the signal-to-noise ratio because the fluorescence signals are sampled by more temporal bins. To maintain the photon budget at each temporal bin, we must accordingly increase the illumination intensity at the sample, a fact which may introduce photobleaching and shorten the overall observation time. The second trade-off exists between y-axis spatial sampling and t-axis temporal sampling. For a given format detector array, without introducing temporal shearing, the final raw image occupies pixel rows. With temporal shearing, the image is sheared to pixel rows with the constraint . Increasing the image size will decrease the and thereby the lifetime estimation accuracy. Therefore, the image FOV and lifetime estimation accuracy must be balanced for a given application.
As a widefield imager, compressed FLIM does not have the sectioning capability. The crosstalk from out-of-focus depth layers may affect the accuracy of fluorescence lifetime measured. However, operating in the passive imaging mode, compressed FLIM can be readily combined with structured or light-sheet illumination, thereby enabling depth-resolved lifetime imaging.
In summary, we have developed a high-speed, high-resolution fluorescent lifetime imaging method, compressed FLIM, and demonstrated its utility in imaging dynamics. Capable of capturing a 2D lifetime image within a snapshot, we expect compressed FLIM would have broad applications in blur-free observation of transient biological events, enabling new avenues of both basic and translational biomedical research.
Methods
Forward Model.
We describe compressed FLIM’s image formation process using the forward model (6). Compressed FLIM generates three projection channels: a time-unsheared channel and two time-sheared channels with complementary encoding. Upon laser illumination, the fluorescence decay scene is first imaged by the microscope to the conjugated plane, and denotes the intensity distribution. A beam splitter then divides the conjugated dynamic scene into two beams. The reflected beam is directly captured by a reference CMOS camera, generating a time-integrated image. The optical energy, , measured at pixel m, n, on the reference CMOS camera is as follows:
| [5] |
where d is the pixel size of the reference camera.
The transmitted beam is then relayed to an intermediate plane (DMD plane) by an optical imaging system. Assuming unit magnification and ideal optical imaging, the intensity distribution of the resultant intermediate image is identical to that of the original scene. The intermediate image is then spatially encoded by a pair of complementary pseudorandom binary patterns displayed at the DMD plane. The two reflected spatially encoded images have the following intensity distribution:
| [6] |
Here, C1,i,j and C2,i,j are elements of the matrix representing the complementary patterns with C1,i,j + C2,i,j = 1, i, j are matrix element indices, and d' is the binned DMD pixel size. For each dimension, the rectangular function (rect) is defined as:
The two reflected light beams masked with complimentary patterns are then passed to the entrance port of a streak camera. By applying a voltage ramp, the streak camera acts as a shearing operator along the vertical axis (y'' axis in Fig. 1) on the input image. Assuming ideal optics with unit magnification again, the sheared images can be expressed as
| [7] |
where v is the shearing velocity of the streak camera.
is then spatially integrated over each camera pixel and temporally integrated over the exposure time. The optical energy, , measured at pixel m, n, is as follows:
| [8] |
Here, d'' is the camera pixel size. Accordingly, we can voxelize the input scene, , into Ii,j,k as follows:
| [9] |
where Δt = d''/v. If the pattern elements are mapped 1:1 to the camera pixels (that is, d'=d'') and perfectly registered, and the reference CMOS camera and the internal CMOS camera of the streak camera have the same pixel size (that is, d =d''), combining Eqs. 5–9 yields
| [10] |
Here Ci,m,n-k Im,n-k,k (i = 1, 2) represents the complimentary-coded, sheared scene, and the inverse problem of Eq. 10 can be solved using existing compressed-sensing algorithms (36, 59, 60).
Compressed FLIM Image Reconstruction Algorithm.
Given prior knowledge of the binary pattern, to estimate the original scene from the compressed FLIM measurement, we need to solve the inverse problem of Eq. 10. Because of the sparsity in the original scene, the image reconstruction can be realized by solving the following optimization problem
| [11] |
where is the regularization function and β is the weighing factor between the fidelity and sparsity. To further impose space and intensity constraint, we construct the new constrained solver:
| [12] |
Here, M is a set of possible solutions confined by a spatial mask extracted from the reference intensity image and defines the zone of action in reconstruction (61). To obtain M, we first compute a locally adaptive threshold for the grayscale reference image (62) and generate an initial mask. Next, we use a median filter to remove the salt-and-pepper noise in the binary image. The resultant M is then used as the spatial constraint in optimization: the pixels outside the mask remain zero and are not updated. This spatial constraint improves the image resolution and accelerates the reconstruction (61). s is the low intensity threshold constraint to reduce the low-intensity artifacts in the data cube. In compressed FLIM image reconstruction, we adopt an algorithm called TwIST) (36), with in the form of total variation:
| [13] |
Here, we assume that the discretized form of I has dimensions (, and are respectively the numbers of voxels along x, y, and t), and m, n, k are three indices. Im, In, Ik denote the 2D lattices along the dimensions m, n, k, respectively. and are horizontal and vertical first-order local difference operators on a 2D lattice. After the reconstruction, nonlinear least squares exponential fitting is applied to data cube along the temporal dimension at each spatial sampling point to extract the lifemap. As shown in Fig. 2A, compressed FLIM reconstructs a fluorescence decay process at one sampling point which matches well with the ground truth.
Lifetime-Based Fluorophore Unmixing Algorithm.
For simplicity, here we consider only fluorophores with single-exponential decays. Provided that the sample consists of n mixed fluorophores with lifetimes τ = (τ1, …, τn) and concentration x = (x1, …, xn), upon a delta pulse excitation, the discretized time-lapse fluorescence decay is as follows:
| [14] |
Here, k is the fluorophore index, is an element of the fluorescence decay component matrix A, and . The inverse problem of Eq. 14 is a least squares problem with constraints. We choose a two-norm penalty and form the solvent for l2-regularized least squares problem:
| [15] |
where the first term represents the measurement fidelity, and the regularization term penalizes large norm of x. The regularization parameter λ adjusts the weight ratio between fidelity and two-norm penalty.
In our experiment, to construct fluorescence decay component matrix A, we first directly imaged Alexa Fluorophore 555 and Alexa Fluorophore 546 in solution and captured their time-lapse fluorescence decay. Then we computed their lifetimes by fitting the asymptotic portion of the decay data with single exponential curves. Finally, we applied the regularized unmixing algorithm to the lifetime data and separated the fluorophores into two channels.
GPU Assisted Real-Time Reconstruction Using Computer Cluster.
Based on the iterative construction, compressed FLIM is computationally extensive. For example, to reconstruct a 500 × 400 × 617 (x, y, t) event datacube and compute a single lifetime image, it takes tens of minutes on a single personal computer (PC). The time of constructing a dynamic lifetime movie is prohibitive. To accelerate this process, we 1) implemented the reconstruction algorithm using a parallel programming framework on two NVIDIA Tesla K40 GPUs and 2) performed all reconstructions simultaneously on a computer cluster (Illinois Campus Cluster). The synergistic effort significantly improved the reconstruction speed and reduced the movie reconstruction time to seconds. Table 1 illustrates the improvement in reconstruction time when the computation is performed on a single PC versus the GPU-assisted computer cluster.
Table 1.
Reconstruction time comparison when the computation is performed on a single PC versus a GPU-accelerated cluster
| Single PC | GPU-assisted computer cluster | |
| Single lifetime image | 155 s | 2.7 s |
| Lifetime movie (100 frames) | 4.3 h | 2.7 s |
Compressed FLIM: Hardware.
In the compressed FLIM system, we used an epi-fluorescence microscope (Olympus IX83) as the front-end optics (SI Appendix, Fig. S4A). We excited the sample using a 515 nm picosecond pulse laser (Genki XPC, NKT Photonics) and separated the fluorescence from excitation using a combination of a 532 nm dichroic mirror (ZT532rdc, Chroma) and a 590/50 nm band-pass emission filter (ET590/50m, Chroma). The laser fluence at the sample is ∼0.5 J/cm2, which is far below the cell damage threshold 4 J/cm2 (57, 63). Upon excitation, an intermediate fluorescence image was formed at the side image port of the microscope. A beam splitter (BSX16, Thorlabs) transmitted 10% of light to a temporal-integration camera (CS2100M-USB, Thorlabs) and reflected 90% of light to the temporal-shearing channels. The reflected image was then relayed to a DMD (DLP LightCrafter 6500, Texas Instruments) through a 4f system consisting of a tube lens (AC508-100-A, Thorlabs) and a stereoscopic objective (MV PLAPO 2XC, Olympus; NA, 0.50). At the DMD, we displayed a random, binary pattern to encode the image. The reflected light from both the “on” mirrors (tilted +12° with respect to the norm) and “off” mirrors (tilted -12° with respect to the norm) were collected by the same stereoscopic objective, forming two complementary channel images at the entrance port of a streak camera (C13410-01A, Hamamatsu) (SI Appendix, Fig. S4B). The streak camera deflected the image along the vertical axis depending on the time of arrival of incident photons. The resultant spatially encoded, temporally sheared images were acquired by an internal CMOS camera (ORCA-Flash 4.0 V3, C13440-20CU, Hamamatsu) with a sensor size of 1344 (H) × 1016 (V) pixels (1 × 1 binning; pixel size d = 6.5 µm). We synchronized the data acquisition of cameras using a digital delay generator (DG645, Stanford Research Systems).
Compressed FLIM is based on the CUP technology (6) that was previously developed for macroscopic imaging. However, it differs from the original CUP system in two aspects. First, our system is built on a high-resolution microscope, allowing transient imaging at a micrometer scale. Second, we minimize the light throughput loss by using a complementary dual-channel encoding scheme. This is particularly important for fluorescence microscopy where the photon budget is limited.
Filter Selection for FRET-FLIM Imaging.
To assure only fluorescence emission from the donor was collected during FRET-FLIM imaging, we chose a filter set (a 515 nm excitation filter and a 590/50 nm emission filter) to separate excitation and fluorescence emission. This filter combination suppressed the direct excitation of the acceptor to <3% and minimized the collection of the acceptor’s fluorescence (SI Appendix, Fig. S5), thereby eliminating the acceptor bleed-through contamination.
Spatial Registration among Three Imaging Channels.
Because compressed FLIM images a scene in three channels (one temporally integrated channel and two complementary temporally sheared channels), the resultant images must be spatially registered. We calibrated the system using a point-scanning–based method. We placed an illuminated pinhole at the microscope’s sample stage and scanned it across the FOV. At each point-scanning position, we operated the streak camera in the “focus” mode (i.e., without temporal shearing) and captured two impulse response images with all DMD’s pixels turned “on” and “off,” respectively. Meanwhile, the reference CMOS camera captured another impulse response image in the temporally integrated channel. We then extracted the pinhole locations in these three impulse response channel images and calculated the homography matrix by which a pixel-to-pixel correspondence was established among the three channels. The reprojection error using the homography is no greater than one pixel. This homography matrix is later used to register the three channel images for concatenated image reconstruction. In addition, in the two complimentary channels, we made the optical path lengths the same within the accuracy of the depth of focus of the relay lens. Therefore, the resultant channel images are both in focus at the detector plane.
Acquisition of Encoding Matrices C1 and C2.
To acquire the encoding matrices C1 and C2, we imaged a uniform scene and operated the streak camera in the “focus” mode. The streak camera directly captured the encoding patterns without temporal shearing. Additionally, we captured two background images with all DMD’s pixels turned “on” and “off,” respectively. To correct for the nonuniformity of illumination, we then divided the coded pattern images by the corresponding background images pixelwise.
Slit-Scanning Streak Camera Imaging.
To form a ground-truth lifetime image, we employed the DMD as a line scanner and scanned the sample along the direction perpendicular to the streak camera entrance slit. We turned on the DMD’s (binned) mirror rows sequentially and imaged the temporally sheared line image in the correspondent imaging channel. Given no spatiotemporal mixing along the vertical axis, the fluorescence decay data along this line direction could be directly extracted from the streak image. Next, we computed the fluorescence lifetimes by fitting the decay data to single exponential curves. The resultant line lifetime images were stacked to form a 2D representation. For slit-scanning streak imaging, the frame time is equal to the product of streak camera’s frame readout time (10 ms) and the number of lines in the images. Therefore, the larger the image format, the longer the acquisition time. For example, in the lifetime unmixing of neural cytoskeletal proteins experiment, the image contains 46 lines, leading to a frame acquisition time of 460 ms.
Confocal FLIM Imaging.
For confocal FLIM imaging, we used a benchmark commercial system (ISS Alba FCS). The sample was excited by a Ti:Sapphire laser, and fluorescence was collected by a Nikon Eclipse Ti inverted microscope. The time-lapse fluorescence decay was measured by a TCSPC unit. To form a 2D lifetime image, the system raster scanned the sample. The typical time to capture a 2D lifetime image (256 × 256 pixels) was ∼60 s.
Fluorescence Beads.
We used a mixture of 6- and 10-μm diameter fluorescence beads (C16509, Thermo Fisher; F8831, Thermo Fisher) in our experiment. To prepare the mixed beads solution, we first diluted the 6- and 10-μm diameter bead suspensions. After sonicating the two fluorescence bead suspensions, we pipetted 10 μL of the 6-μm bead suspension (∼1.7 × 107 beads/mL) and 10-μm bead suspension (∼3.6 × 106 beads/mL) into 1 mL and 1 mL PBS, respectively. Next, we mixed 100 μL diluted 10-μm bead solution with 100 µL diluted 6-μm bead solution. The final mixed beads solution contained approximately 8.5 × 104 6-μm beads/mL and 1.8 × 104 10-μm beads/mL
FRET Phantom.
We used Alexa Fluor 546 (A-11003, Thermo Fisher) as the donor and Alexa Fluor 647 (A-21235, Thermo Fisher) as the acceptor. We prepared the acceptor solutions with three different concentrations (0 mg/mL, 1 mg/mL, and 2 mg/mL) and mixed them with the same donor solution (1 mg/mL). We then injected them into three glass tubes (14705-010, VWR) for imaging.
Primary Cell Culture.
Primary hippocampal neurons were cultured from dissected hippocampi of Sprague Dawley rat embryos. Hippocampal neurons were then plated on 29 mm glass bottom Petri dishes that are precoated with poly-d-lysine (0.1 mg/mL; Sigma-Aldrich). To help the attachment of neurons (300 cells/mm2) on to the glass bottom dish, neurons were initially incubated with a plating medium containing 86.55% Minimum Essential Medium Eagle’s with Earle’s Balanced Salt Solution (Lonza), 10% fetal bovine serum (refiltered and heat inactivated; ThermoFisher), 0.45% of 20% (wt/vol) glucose, 1× 100 mM sodium pyruvate (100×; Sigma-Aldrich), 1× 200 mM glutamine (100×; Sigma-Aldrich), and 1× penicillin/streptomycin (100×; Sigma-Aldrich). After 3 h of incubation in the incubator (37 °C and 5% CO2), the plating media was aspirated and replaced with maintenance media containing Neurobasal growth medium supplemented with B-27 (Invitrogen), 1% 200 mM glutamine (Invitrogen), and 1% penicillin/streptomycin (Invitrogen) at 37 °C, in the presence of 5% CO2. Half the media was aspirated once a week and replaced with fresh maintenance media. The hippocampal neurons were grown for 10 d before imaging.
Immunofluorescently Stained Neurons.
We immunolabeled the Vimentin (MA5-11883, Thermo Fisher) with Alexa Fluor 555 (A-21422, Thermo Fisher) and Tubulin (PA5-16891, Thermo Fisher) with Alexa Fluor 546 (A-11010, Thermo Fisher) in the neurons.
Transfect the Neurons to Express MacQ-mOrange2.
We used Mac mutant plasmid DNA Mac-mOrange2 (48761, Addgene) to transfect hippocampal neurons. In DNA collection, as soon as we received the agar stab, we grew bacteria in Luria–Bertani broth with ampicillin in 1:1,000 dilution for overnight in 37 °C. Standard Miniprep (Qiagen) protocol was performed in order to collect DNA. DNA concentration was measured by Nanodrop 2000c (Thermo Fisher). In neuron transfection, lipofectamine 2000 (Invitrogen) was used as transfecting reagent. In an Eppendorf tube, we stored 1 mL the conditioned culture media from 29 mm Petri plates neuron culture with 1 mL fresh media. We prepared two separate Eppendorf tubes and added 100 μL Neurobasal medium to each tube. For one tube, 3 μg DNA was added, while 4 uL lipofectamine 2000 was added to the other tube. After 5 min, two tubes were mixed together and incubated in room temperature for 20 min. This mixture was added to neuron culture in dish for 4 h in the incubator (37 °C and 5% CO2). We took out the media containing lipofectamine 2000 reagent and added the stored, conditioned, and fresh culture media to neuron culture in dish. Hippocampal neurons were imaged 40 h after transfection.
Potassium Stimulation and Imaging.
We used high potassium (50 mM) treatment to stimulate neuron spike. The extracellular solution for cultured neurons (150 mM NaCl, 4 mM KCl, 10 mM glucose, 10 mM HEPES, 2 mM CaCl2, and 2 mM MgCl2) was adjusted to reach the desired final K+ concentration (50 mM). To maintain physiological osmolality, we increased the concentration of KCl in the solution to 50 mM with an equimolar decrease in NaCl concentration. We started imaging approximately 30 s before the high potassium treatment. At each stimulation, we removed the media in the plate followed by adding high potassium extracellular solution with a pipette.
Supplementary Material
Acknowledgments
This work was supported partially by NIH (R01EY029397, R35GM128761, and R21EB028375) and NSF (1652150).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2004176118/-/DCSupplemental.
Data Availability.
Image and code data have been deposited in GitHub (10.5281/zenodo.4408547).
References
- 1.Gu M., Reid D., Ben-Yakar A., Ultrafast biophotonics. J. Optics 12, 080301 (2010). [Google Scholar]
- 2.Mikami H., Gao L., Goda K., Ultrafast optical imaging technology: Principles and applications of emerging methods. Nanophotonics 5, 497–509 (2016). [Google Scholar]
- 3.Llull P., et al. , Coded aperture compressive temporal imaging. Opt. Express 21, 10526–10545 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Goda K., Tsia K. K., Jalali B., Serial time-encoded amplified imaging for real-time observation of fast dynamic phenomena. Nature 458, 1145–1149 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Nakagawa K., et al. , Sequentially timed all-optical mapping photography (STAMP). Nat. Photonics 8, 695–700 (2014). [Google Scholar]
- 6.Gao L., et al. , Single-shot compressed ultrafast photography at one hundred billion frames per second. Nature 516, 74–77 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matlis N. H., Axley A., Leemans W. P., Single-shot ultrafast tomographic imaging by spectral multiplexing. Nat. Commun. 3, 1111 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Diebold E. D., Buckley B. W., Gossett D. R., Jalali B., Digitally synthesized beat frequency multiplexing for sub-millisecond fluorescence microscopy. Nat. Photonics 7, 806–810 (2013). [Google Scholar]
- 9.Goda K., Jalali B., Dispersive Fourier transformation for fast continuous single-shot measurements. Nat. Photonics 7, 102–112 (2013). [Google Scholar]
- 10.Goda K., et al. , High-throughput single-microparticle imaging flow analyzer. Proc. Natl. Acad. Sci. U.S.A. 109, 11630–11635 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Z., Zgadzaj R., Wang X., Chang Y.-Y., Downer M. C., Single-shot tomographic movies of evolving light-velocity objects. Nat. Commun. 5, 3085 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang J., et al. , Single-shot real-time video recording of a photonic Mach cone induced by a scattered light pulse. Sci. Adv. 3, e1601814 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borst J. W., Visser A. J. W. G., Fluorescence lifetime imaging microscopy in life sciences. Meas. Sci. Technol. 21, 102002 (2010). [Google Scholar]
- 14.Becker W., Fluorescence lifetime imaging–Techniques and applications. J. Microsc. 247, 119–136 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Marcu L., Fluorescence lifetime techniques in medical applications. Ann. Biomed. Eng. 40, 304–331 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Manen H.-J., et al. , Refractive index sensing of green fluorescent proteins in living cells using fluorescence lifetime imaging microscopy. Biophys. J. 94, L67–L69 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wallrabe H., Periasamy A., Imaging protein molecules using FRET and FLIM microscopy. Curr. Opin. Biotechnol. 16, 19–27 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Sun Y., Day R. N., Periasamy A., Investigating protein-protein interactions in living cells using fluorescence lifetime imaging microscopy. Nat. Protoc. 6, 1324–1340 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun Y., et al. , Endoscopic fluorescence lifetime imaging for in vivo intraoperative diagnosis of oral carcinoma. Microsc. Microanal. 19, 791–798 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bower A. J., et al. , High-speed imaging of transient metabolic dynamics using two-photon fluorescence lifetime imaging microscopy. Optica 5, 1290–1296 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Becker W., Frere S., Slutsky I., “Recording Ca++ transients in neurons by TCSPC FLIM” in Advanced Optical Methods for Brain Imaging, Kao F. J., Keiser G., Gogoi A., Eds. (Springer, Singapore, 2019), pp. 103–110. [Google Scholar]
- 22.Becker W., The bh TCSPC Handbook (Becker & Hickl GmbH, Berlin, 6th ed., 2014). [Google Scholar]
- 23.Krishnan R. V., Masuda A., Centonze V. E., Herman B., Quantitative imaging of protein-protein interactions by multiphoton fluorescence lifetime imaging microscopy using a streak camera. J. Biomed. Opt. 8, 362–367 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Redford G. I., Clegg R. M., Polar plot representation for frequency-domain analysis of fluorescence lifetimes. J. Fluoresc. 15, 805–815 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Digman M. A., Caiolfa V. R., Zamai M., Gratton E., The phasor approach to fluorescence lifetime imaging analysis. Biophys. J. 94, L14–L16 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dowling K., Hyde S. C. W., Dainty J. C., French P. M. W., Hares J. D., 2-D fluorescence lifetime imaging using a time-gated image intensifier. Opt. Commun. 135, 27–31 (1997). [Google Scholar]
- 27.Cole M. J., et al. , Time-domain whole-field fluorescence lifetime imaging with optical sectioning. J. Microsc. 203, 246–257 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Eichorst J. P., Teng K. W., Clegg R. M., “Fluorescence lifetime imaging techniques: Frequency-domain FLIM” in Fluorescence Lifetime Spectroscopy and Imaging: Principles and Applications in Biomedical Diagnostics, Marcu P. F. L., Elson D. S., Eds. (CRC Press, 2014), pp. 165–186. [Google Scholar]
- 29.Gersbach M., et al. , High frame-rate TCSPC-FLIM using a novel SPAD-based image sensor. Proc. SPIE 7780, 77801H–77801H (2010). [Google Scholar]
- 30.Parmesan L., et al. , A 256 x 256 SPAD array with in-pixel time to amplitude conversion for fluorescence lifetime imaging microscopy. Memory 900, M4–M5 (2015). [Google Scholar]
- 31.van der Oord C. J., Stoop K. W., van Geest L. K., “Fluorescence lifetime attachment LIFA” in Advances in Fluorescence Sensing Technology V. (International Society for Optics and Photonics, 2001), 4252, pp. 115–118. [Google Scholar]
- 32.Krämer B., et al. , Compact FLIM and FCS Upgrade kit for laser scanning microscopes (LSMs) (PicoQuant GmbH, Berlin, 2009). [Google Scholar]
- 33.Grinvald A., Frostig R. D., Lieke E., Hildesheim R., Optical imaging of neuronal activity. Physiol. Rev. 68, 1285–1366 (1988). [DOI] [PubMed] [Google Scholar]
- 34.Herron T. J., Lee P., Jalife J., Optical imaging of voltage and calcium in cardiac cells & tissues. Circ. Res. 110, 609–623 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L., Xiong Z., Gao D., Shi G., Wu F., Dual-camera design for coded aperture snapshot spectral imaging. Appl. Opt. 54, 848–858 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Bioucas-Dias J. M., Figueiredo M. A. T., A new twIst: Two-step iterative shrinkage/thresholding algorithms for image restoration. IEEE Trans. Image Process. 16, 2992–3004 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Liyanage M., et al. , Multicolour spectral karyotyping of mouse chromosomes. Nat. Genet. 14, 312–315 (1996). [DOI] [PubMed] [Google Scholar]
- 38.Garini Y., Gil A., Bar-Am I., Cabib D., Katzir N., Signal to noise analysis of multiple color fluorescence imaging microscopy. Cytometry 35, 214–226 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Tsurui H., et al. , Seven-color fluorescence imaging of tissue samples based on Fourier spectroscopy and singular value decomposition. J. Histochem. Cytochem. 48, 653–662 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Bastiaens P. I., Squire A., Fluorescence lifetime imaging microscopy: Spatial resolution of biochemical processes in the cell. Trends Cell Biol. 9, 48–52 (1999). [DOI] [PubMed] [Google Scholar]
- 41.Chang C. W., Sud D., Mycek M. A., Fluorescence lifetime imaging microscopy. Methods Cell Biol. 81, 495–524 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Wahl M., Koberling F., Patting M., Rahn H., Erdmann R., Time-resolved confocal fluorescence imaging and spectrocopy system with single molecule sensitivity and sub-micrometer resolution. Curr. Pharm. Biotechnol. 5, 299–308 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Wallace D. J., et al. , Single-spike detection in vitro and in vivo with a genetic Ca2+ sensor. Nat. Methods 5, 797–804 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Stosiek C., Garaschuk O., Holthoff K., Konnerth A., In vivo two-photon calcium imaging of neuronal networks. Proc. Natl. Acad. Sci. U.S.A. 100, 7319–7324 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greenberg D. S., Houweling A. R., Kerr J. N. D., Population imaging of ongoing neuronal activity in the visual cortex of awake rats. Nat. Neurosci. 11, 749–751 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Lillis K. P., Eng A., White J. A., Mertz J., Two-photon imaging of spatially extended neuronal network dynamics with high temporal resolution. J. Neurosci. Methods 172, 178–184 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zanin N., et al. “STAM Interaction with Hrs Controls JAK/STAT Activation by Interferon-α at the Early Endosome.” 10.1101/509968 (2 January 2019). [DOI]
- 48.Gong Y., The evolving capabilities of rhodopsin-based genetically encoded voltage indicators. Curr. Opin. Chem. Biol. 27, 84–89 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mollinedo-Gajate I., Song C., Knöpfel T., Genetically encoded fluorescent calcium and voltage indicators. 260, 209–229 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Gong Y., Wagner M. J., Li J. Z., Schnitzer M. J., Imaging neural spike in brain tissue using FRET-opsin protein voltage sensors. Nat. Commun. 5, 3674 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brinks D., Klein A. J., Cohen A. E., Two-photon lifetime imaging of voltage indicating proteins as a probe of absolute membrane voltage. Biophys. J. 109, 914–921 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hou J. H., Venkatachalam V., Cohen A. E., Temporal dynamics of microbial rhodopsin fluorescence reports absolute membrane voltage. Biophys. J. 106, 639–648 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hamamatsu, High repetition streak camera C14831-130. https://www.hamamatsu.com/us/en/product/type/C14831-130/index.html. Accessed November 2020.
- 54.Hagen N., Kester R. T., Gao L., Tkaczyk T. S., Snapshot advantage: A review of the light collection improvement for parallel high-dimensional measurement systems. Opt. Eng. 51, 111702 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feng R., Gruebele M., Davis C. M., Quantifying protein dynamics and stability in a living organism. Nat. Commun. 10, 1179 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nitsche C., Otting G., NMR studies of ligand binding. Curr. Opin. Struct. Biol. 48, 16–22 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Dixit R., Cyr R., Cell damage and reactive oxygen species production induced by fluorescence microscopy: Effect on mitosis and guidelines for non-invasive fluorescence microscopy. Plant J. 36, 280–290 (2003). [DOI] [PubMed] [Google Scholar]
- 58.Hopt A., Neher E., Highly nonlinear photodamage in two-photon fluorescence microscopy. Biophys. J. 80, 2029–2036 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Y., et al. , Rank minimization for snapshot compressive imaging. IEEE Trans. Pattern Anal. Mach. Intell. 41, 2990–3006 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Liao X., Li H., Carin L., Generalized alternating projection for weighted minimization with applications to model-based compressive sensing. SIAM J. Imaging Sci. 7, 797–823 (2014). [Google Scholar]
- 61.Zhu L., et al. , Space- and intensity-constrained reconstruction for compressed ultrafast photography. Optica 3, 694–697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bradley D., Roth G., Adaptive thresholding using the integral image. J. Graphics Tools 12, 13–21 (2007). [Google Scholar]
- 63.Yuan X., et al. , Effect of laser irradiation on cell function and its implications in Raman spectroscopy. Appl. Environ. Microbiol. 84, 8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Image and code data have been deposited in GitHub (10.5281/zenodo.4408547).