Significance
Satellites are virus-like agents which require both a host and a virus to complete their life cycle. The only human-infecting satellite is hepatitis delta virus (HDV), which exacerbates liver disease in patients co-infected with hepatitis B virus (HBV). How HDV originated is a long-standing evolutionary puzzle. Using terabase-scale data mining, coevolutionary analyses, and field studies in bats, we show that deltaviruses can jump between highly divergent host species. Our results further suggest that the contemporary association between HDV and HBV likely arose following zoonotic transmission from a yet-undiscovered animal reservoir in the Americas. Plastic host and virus associations open prospects that deltaviruses might alter the virulence of multiple viruses in multiple host species.
Keywords: satellite virus, hepatitis delta virus, zoonosis, host shifting
Abstract
Hepatitis delta virus (HDV) is an unusual RNA agent that replicates using host machinery but exploits hepatitis B virus (HBV) to mobilize its spread within and between hosts. In doing so, HDV enhances the virulence of HBV. How this seemingly improbable hyperparasitic lifestyle emerged is unknown, but it underpins the likelihood that HDV and related deltaviruses may alter other host–virus interactions. Here, we show that deltaviruses diversify by transmitting between mammalian species. Among 96,695 RNA sequence datasets, deltaviruses infected bats, rodents, and an artiodactyl from the Americas but were absent from geographically overrepresented Old World representatives of each mammalian order, suggesting a relatively recent diversification within the Americas. Consistent with diversification by host shifting, both bat and rodent-infecting deltaviruses were paraphyletic, and coevolutionary modeling rejected cospeciation with mammalian hosts. In addition, a 2-y field study showed common vampire bats in Peru were infected by two divergent deltaviruses, indicating multiple introductions to a single host species. One vampire bat-associated deltavirus was detected in the saliva of up to 35% of individuals, formed phylogeographically compartmentalized clades, and infected a sympatric bat, illustrating horizontal transmission within and between species on ecological timescales. Consistent absence of HBV-like viruses in two deltavirus-infected bat species indicated acquisitions of novel viral associations during the divergence of bat and human-infecting deltaviruses. Our analyses support an American zoonotic origin of HDV and reveal prospects for future cross-species emergence of deltaviruses. Given their peculiar life history, deltavirus host shifts will have different constraints and disease outcomes compared to ordinary animal pathogens.
Hepatitis delta virus (HDV), the only member of the only species (Hepatitis delta virus) in the genus Deltavirus, is a globally distributed human pathogen which causes the most severe form of viral hepatitis in an estimated 20 million people (1). Unlike typical viruses, HDV is an obligate “satellite” virus that is replicated by diverse host cells but requires the envelope of an unrelated “helper” virus (classically hepatitis B virus, HBV, family Hepadnaviridae) for cellular entry, egress, and transmission (1). The peculiar life history of HDV, together with its lack of sequence homology to known viral groups, have made the evolutionary origins of HDV a long-standing puzzle. Geographic associations of most HDV genotypes point to an Old World origin. Yet historical explanations of the mechanistic origin of HDV spanned from emergence from the messenger RNA of an HBV-infected human (2) to ancient evolution from viroids (circular, single-stranded RNA pathogens of plants) (3). More recently, discoveries of HDV-like genomes in vertebrates and invertebrates (4–7) overturned the decades-long belief that deltaviruses exclusively infect humans. These discoveries also suggested new models of deltavirus evolution in which these satellites either cospeciated with their hosts over ancient timescales or possess an unrecognized capacity for host shifting, which would imply their potential to emerge in novel species. The latter scenario has been presumed unlikely since either both satellite and helper would need to be compatible with the novel host or deltaviruses would need to simultaneously switch host species and helper virus, possibly altering the virulence of newly acquired helpers as a result.
Efforts to distinguish competing evolutionary hypotheses for deltaviruses have been precluded by the remarkably sparse distribution of currently known HDV-like agents across the animal tree of life. Single representatives are reported from arthropods (subterranean termite, Schedorhinotermes intermedius), fish (a pooled sample from multiple species), birds (a pooled sample from three duck species, Anas gracilis, A. castanea, and A. superciliosa), reptiles (common boa, Boa constrictor), and mammals (Tome’s spiny rat, Proechimys semispinosus), and only two are known from amphibians (Asiatic toad, Bufo gargarizans and the Chinese fire belly newt, Cynops orientalis) (4–7). Most share minimal homology with HDV, even at the protein level (<25%), frustrating robust phylogenetic reconstructions of evolutionary histories (SI Appendix, Fig. S1). On the one hand, the distribution of deltaviruses may reflect rare host shifting events among divergent taxa. Alternatively, reliance on untargeted metagenomic sequencing (a relatively new and selectively applied tool) to find novel species may mean that the distribution of deltaviruses in nature is largely incomplete (8, 9). Additional taxa could reveal ancient cospeciation of HDV-like agents with their hosts or evidence for host shifting.
Results
We sought to fill gaps in the evolutionary history of mammalian deltaviruses, the group most likely to clarify the origins of HDV. We used a two-pronged approach (Materials and Methods). First, we used data from Serratus, a newly developed bioinformatic platform which screens RNA sequences from the NCBI SRA (National Center for Biotechnology Information Short Read Archive) for similarity to known viruses, and which is described by Edgar et al. (10). We focused on search results from 96,695 transcriptomic and metagenomic datasets, comprising 348 terabases of RNA sequences from 403 species across 24 mammalian orders (22 terrestrial, two aquatic; see Dataset S1). Although domesticated animals comprised the largest single fraction of the dataset (67.2%), remaining data were from a variety of globally distributed species (Fig. 1 A and B). Our second search was prompted by our earlier detection of uncharacterized deltavirus-like sequences in a Neotropical bat (11) and evidence of underrepresentation in the volume of Neotropical bat data in the SRA (Fig. 1A). We therefore carried out metagenomic sequencing of 23 frugivorous, insectivorous, nectarivorous, and sanguivorous bat species from Peru, using 59 samples available within our laboratory (SI Appendix, Table S1). All datasets containing sequences with significant protein homology to deltaviruses were subjected to de novo genome assembly.
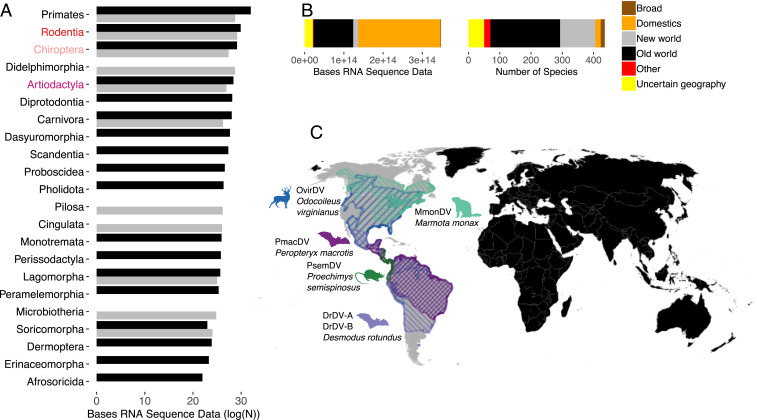
Fig. 1.
The geographic and taxonomic distribution of mammalian datasets and novel deltaviruses. (A) The host and geographic distribution of metagenomic and transcriptomic datasets searched for novel deltaviruses; note the log scale. Text colors indicate orders (red = Rodentia; pink = Chiroptera; purple = Artiodactyla); bar colors indicate geography (black = Old World; gray = New World). (B) Stacked bar charts show the volume of mammalian datasets in units of RNA bases and the number of species searched, separated by species geography. Additional segments describe widely distributed domesticated animals (Domestics), datasets with genus-level metadata from broadly distributed genera (Broad), datasets from cell lines or with taxonomic information only at the class level (Other), and those which had no geographic range data available (Uncertain geography, mostly aquatic mammals). (C) Host distributions of newly discovered and recently reported deltaviruses, color coded by mammalian species (data from The International Union for Conservation of Nature). All except PsemDV were discovered through our search.
Searches revealed five deltaviruses spanning three mammalian orders: Artiodactyla (n = 1), Chiroptera (n = 3), and Rodentia (n = 1; Fig. 1C). No deltaviruses were detected in nonhuman primates, indicating HDV as the sole known representative infecting the order Primates. Strikingly, despite overrepresentation of Old World–derived data by factors of 4.3 (Artiodactyla), 5.8 (Chiroptera), and 2.1 (Rodentia), all new mammalian deltaviruses originated from North and South American species (Fig. 1 A and C and SI Appendix, Supplementary Results, section 1). Chiropteran deltaviruses included two genotypes from common vampire bats (Desmodus rotundus), which shared only 48.4 to 48.6% genome-wide nucleotide (nt) identity (hereafter, DrDV-A and DrDV-B; SI Appendix, Fig. S1). A third deltavirus was identified in a liver transcriptome (accession SRR7910143 (12)) from a lesser dog-like bat (Peropteryx macrotis) from Mexico (PmacDV) but was more closely related to recently described deltaviruses from Tome’s spiny rat from Panama (PsemDV (7)), sharing 95.9 to 97.4% amino acid (aa) and 93.0 to 95.7% nt identity. Additional genomes were recovered from transcriptomes derived from the pedicle tissue of white-tailed deer [Odocoileus virginianus; OvirDV; accession SRR4256033 (13)] and from a captive-born Eastern woodchuck [Marmota monax; MmonDV; accession SRR2136906 (14)]. Bioinformatic screens recovered additional reads matching each genome in related datasets (either different individuals from the same study or different tissues from the same individuals), suggesting active infections (SI Appendix, Table S2). All genomes had lengths of 1,669 to 1,771 nt, high intramolecular base pairing, and contained genomic and antigenomic ribozymes characteristic of deltaviruses. The DrDV-A and DrDV-B genomes are more fully characterized in SI Appendix (SI Appendix, Figs. S2 and S3 and Table S3 and Supplementary Results, section 2). The other genomes and a case study on MmonDV infections in animals inoculated with woodchuck hepatitis virus are described by Edgar et al. (10).
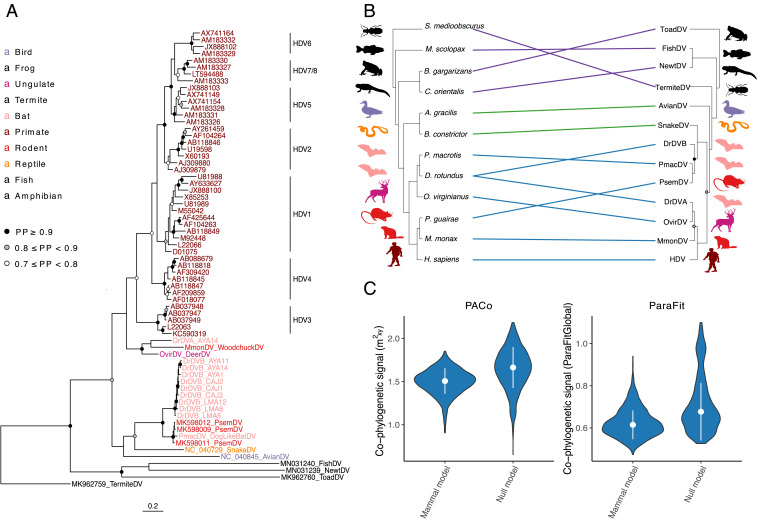
Phylogenetic analysis of the small delta antigen (DAg) protein sequences using MrBayes (Fig. 2A) and a multispecies coalescent model in StarBeast (Fig. 2B) revealed multiple putative host shifts within the evolutionary history of mammalian deltaviruses. For instance, vampire bat deltaviruses were paraphyletic, suggesting at least two independent incursions into this species. Specifically, DrDV-A formed a clade with OvirDV and MmonDV (posterior probability, PP = 0.99 and 0.80 in MrBayes and StarBeast, respectively), which was basal to HDV (PP = 0.65 and 0.81), while DrDV-B shared a most recent common ancestor with PmacDV and PsemDV (PP = 1 and 1). Rodent-associated deltaviruses (MmonDV and PsemDV) were also highly divergent and paraphyletic. Consequently, cophylogenetic analyses using 1,000 randomly sampled topologies from StarBeast failed to reject independence of mammal and deltavirus phylogenies, consistent with a model of diversification by host shifting (Fig. 2 B and C). Analyses of all deltavirus host pairs (including highly divergent HDV-like agents) and an “ingroup” clade containing mammalian, along with avian and snake, deltaviruses revealed somewhat greater dependence of the deltavirus phylogeny on the host phylogeny (SI Appendix, Fig. S4). However, statistical significance varied across cophylogenetic approaches and topological incongruences were evident among nonmammals, excluding cospeciation as the sole diversification process, even in deeper parts of the coevolutionary history (SI Appendix, Figs. S4 and S5 and Supplementary Results, section 3).
Fig. 2.
The evolutionary history of deltaviruses reveals host shifts among mammals. (A) Bayesian phylogeny of a 192-aa alignment of the DAg. Ingroup taxa, including mammal, snake, and avian deltaviruses, are colored by order; other HDV-like taxa are shown in black. (B) Cophylogeny depicting connections between the consensus deltavirus phylogeny from StarBeast and the host tree (56). Links are colored according to subsets of data used in cophylogenetic analyses; all taxa (purple + green + blue), ingroup (green + blue), or mammal (blue). Host taxa are Schedorhinotermes medioobscurus, Macroramphosus scolopax, Bufo gargarizans, Cynops orientalis, Anas gracilis, Boa constrictor, Peropteryx macrotis, Desmodus rotundus, Odocoileus virginianus, Proechimys guirae, Marmota monax, and Homo sapiens. (C) An absence of phylogenetic dependence of the mammalian deltavirus phylogeny on the host phylogeny. Violin plots show distributions of test statistics from two cophylogenetic approaches across 1,000 posterior trees relative to null models, along with medians and SDs. For PACo, higher values would indicate greater phylogenetic dependence; for ParaFit, lower values would indicate greater phylogenetic dependence. Both approaches rejected a global model of cospeciation (P > 0.05).
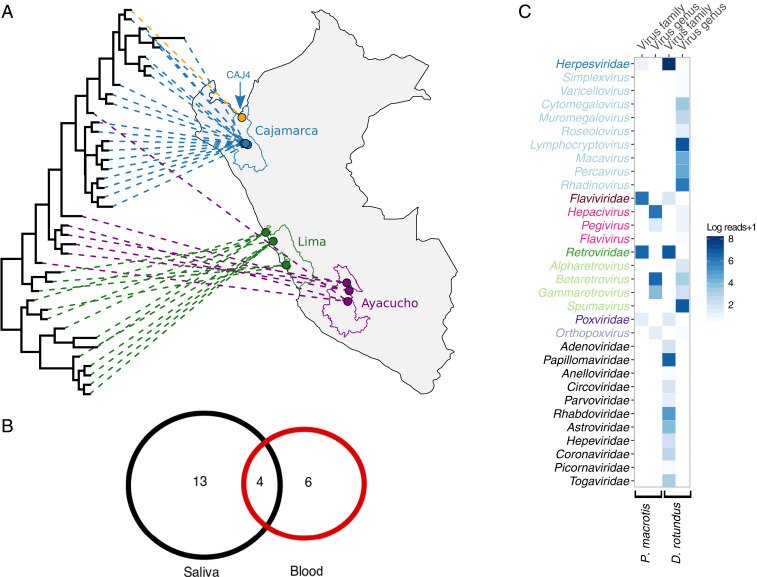
Having extended the mammalian host range of deltaviruses to Neotropical bats, we subsequently explored the transmission dynamics, host range, and candidate helper virus associations within this group through a field study in three regions of Peru (Fig. 3A). Out of 240 D. rotundus saliva samples from 12 bat colonies collected in 2016 and 2017, RT-PCR targeting the DAg detected DrDV-A in single adult females from one of the two metagenomic pools that contained this genotype (bat identification 8299, site AYA14, see SI Appendix, Tables S4 and S5). In contrast, DrDV-B was detected in 17.1% of D. rotundus saliva samples (colony level prevalence: 0 to 35%). Prevalence varied neither by region of Peru (likelihood-ratio test; χ2 = 3.21; degree of freedom = 2; P = 0.2) nor by bat age or sex (binomial generalized linear mixed model, age: P = 0.38; sex: P = 0.87), suggesting geographic and demographic ubiquity of infections. Given that vampire bats subsist on blood, deltavirus sequences encountered in bat saliva might represent contamination from infected prey. A small set of blood samples screened for DrDV-A (n = 60, including bat 8299) were negative. However, 6 out of 41 bats that were DrDV-B negative and 4 out of 18 bats with DrDV-B in saliva also contained DrDV-B in blood samples (Fig. 3B). In the four individuals with paired saliva and blood samples, DAg sequences were identical, supporting systemic infections. Significant spatial clustering of DrDV-B sequences at both regional and bat colony levels further supported local infection cycles driven by horizontal transmission among vampire bats (Fig. 3A and SI Appendix, Table S6).
Fig. 3.
The transmission biology and candidate helper viruses for bat deltaviruses. (A) Bayesian phylogeny of a 214-nt alignment of DrDV-B DAg projected onto vampire bat capture locations in Peru. Lines and points are colored by administrative regions. Site CAJ4, where the C. perspicillata sequence was detected, is depicted in orange. (B) DrDV-B detections in saliva and blood. The numbers represent individual bats; the four bats in the center had genetically identical DrDV-B sequences in saliva and blood. (C) Mammal-infecting viral communities are shown for the P. macrotis liver transcriptome, which contained PmacDV and combined D. rotundus saliva metagenomes. Viral taxa are colored by family, with lighter shades indicating genera within families for overlapping viral families in both bat species. Viral families only present in one bat species are shown in black. Candidate helpers for OvirDV and MonDV are shown in SI Appendix, Fig. S9.
Given the evolutionary evidence for deltavirus host shifts, we hypothesized that spillover infections might also occur at detectable frequencies in sympatric Neotropical bats. Among 87 non-D. rotundus bats captured in or outside of D. rotundus-occupied roosts, RT-PCR detected deltavirus RNA in the saliva of a single Seba’s short-tailed bat (Carollia perspicillata; n = 31 individuals; SI Appendix, Fig. S6). This result was unlikely attributable to erroneous bat species assignment or laboratory contamination (SI Appendix, Supplementary Results, section 4). The partial DAg recovered from the C. perspicillata was identical to a DrDV-B strain collected from a vampire bat in the same roost (CAJ4; Fig. 3A). Given the rapid evolution expected in deltaviruses (ca. 10−3 substitutions/site/year), genetic identity is most parsimoniously explained as horizontal transmission from D. rotundus to C. perspicillata, which was followed by an absence of (or short-lived) transmission among C. perspicillata at the time of sampling (15). This finding therefore demonstrates cross-species transmission on ecological timescales, a defining prerequisite for evolutionary diversification of deltaviruses through host shifting.
Finally, we evaluated whether bat deltaviruses use hepadnavirus helpers akin to HDV (1). Consistent with a previous study, PCR screens of DrDV-positive and -negative saliva (n = 54) and blood samples (n = 119) found no evidence of hepadnaviruses in vampire bats (16). To rule out divergent hepadnaviruses missed by PCR, we next used a bioinformatic pipeline to characterize viral communities in the metagenomic and transcriptomic datasets from deltavirus-infected bat species (Materials and Methods). Hepadnaviruses were again absent from all datasets (Fig. 3C). Together with the finding that all three bat-infecting deltavirus genomes lacked the farnesylation site thought to facilitate acquisition of the hepadnaviral envelope (SI Appendix, Supplementary Results, section 2), use of hepadnavirus helpers by bat deltaviruses seems unlikely. To identify alternative plausible candidates, we quantified the abundance (approximated by sequence reads) of viral taxa that overlapped between the two deltavirus-infected bat species, P. macrotus and D. rotundus. In the P. macrotus liver, which contained PMacDV, reads from hepaciviruses (Flaviviridae) spanned a complete viral genome (GenBank third party annotation [TPA]: BK013349) and outnumbered all other viral genera with the exception of Betaretroviruses, whose abundance may reflect endogenization in the host genome (Fig. 3C). Lower, but detectable, hepacivirus abundance in vampire bats may reflect the tissue tropism of these viruses or pooling of samples from multiple individuals. Intriguingly, hepaciviruses experimentally mobilize HDV in vitro and were found in 26 out of 30 PsemDV-infected rats (7, 17). Phylogenetic analysis of hepaciviruses associated with the deltavirus-positive host species (P. macrotus, D. rotundus, and P. semispinosus) revealed the candidate helpers to be highly divergent (SI Appendix, Fig. S7) despite the apparently close relationships between the deltaviruses infecting these hosts. Reads matching Poxviridae formed small contigs in both libraries (P. macrotus: 229 to 1,386 nt and D. rotundus: 358 nt) and could not be excluded as false positives, particularly in D. rotundus. Although nonopportunistic sampling is required to decisively identify the helpers of bat deltaviruses, existing evidence points to hepaciviruses as top contenders, perhaps using alternative enveloping mechanisms to HDV.
Discussion
Unlike conventional pathogens (e.g., viruses, bacteria, and protozoans), the obligatory dependence of deltaviruses on evolutionarily independent helpers creates a barrier to cross-species transmission that might be expected to promote host specificity. Data to test this hypothesis have been unavailable until now. Our study demonstrates transmission of deltaviruses among divergent mammals on both ecological and evolutionary timescales.
Deltavirus host shifts could conceivably arise through several processes. Mobilization by nonviral microorganisms (e.g., intracellular bacteria) is conceivable but has never been observed. Unaided spread through a yet-undefined mechanism is also possible. However, given that the best-studied deltavirus (HDV) depends on viral envelopes to complete its life cycle and that conserved genomic features in related deltaviruses suggest a similar life history strategy, helper virus–mediated host shifting is the most reasonable expectation. We and others have excluded hepadnavirus helpers for PmacDV, DrDVs, and PsemDV, yet natural HDV infections consistently involve HBV (1, 7). In light of this and the evidence presented here for host shifts among mammals, the contemporary HDV–HBV association must have arisen through acquisition of the hepadnaviral helper somewhere along the evolutionary divergence separating human- and other mammal-infecting deltaviruses. Evidence that deltaviruses can exploit diverse enveloped viruses experimentally adds further weight to this conclusion (17, 18). As several new mammalian deltaviruses were detected with hepacivirus and poxvirus coinfections, either simultaneous host shifts of deltaviruses and helpers or preferential deltavirus shifting among host species that are already infected with compatible helpers are plausible. If hepaciviruses are functioning as helpers, divergent phylogenetic relationships between species detected in bat and rodent hosts (SI Appendix, Fig. S7) suggest they have been acquired independently of deltaviruses rather than representing simultaneous host shifts or cospeciation of deltaviruses and helpers. Conclusively identifying the helper associations of novel mammalian deltaviruses and their evolutionary relationships will be crucial to disentangling these possibilities.
A limitation of our study was that the species in which novel deltaviruses were discovered were presumed to be definitive hosts (i.e., capable of sustained horizontal transmission). Consequently, some putative host shifts detected in our cophylogenetic analysis may represent short-lived transmission chains in novel hosts or singleton infections, analogous to our observation of DrDV-B in C. perspicillata. For example, PmacDV from the lesser dog-like bat clustered within the genetic diversity of PsemDV from rats but infected two out of three individual bats analyzed, suggesting a recent cross-species transmission event followed by some currently unknown amount of onward transmission. Irrespective of the long-term outcomes of index infections, our results unequivocally support the conclusion that deltaviruses can transmit between divergent mammalian orders. The global distribution of deltavirus positive and negative datasets provides additional, independent evidence for host shifts. Even allowing for subdetection due to variation in infection prevalence and dataset quality, deltaviruses should have been more widespread across the mammalian phylogeny than we observed (ca. 1% of species analyzed) if they cospeciated with their hosts. Given the presence of HDV in humans, nonhuman primates in particular would have been expected to host HDV-like deltaviruses in a cospeciation scenario, which was not observed. Moreover, the three deltavirus-infected mammalian orders occur in both the New and Old Worlds, yet nonhuman deltaviruses occurred exclusively in the Americas. Sampling biases cannot readily explain this pattern. By most metrics of sequencing effort, Old World mammals were overrepresented in each deltavirus-infected mammalian order, including at finer continental scales (SI Appendix, Supplementary Results, section 1). Despite the large scale of our search, we evaluated <10% of mammalian species. We therefore anticipate further discoveries of mammalian deltaviruses. Crucially, however, new viruses could not reunite the paraphyletic rodent and bat deltaviruses or resolve widespread incongruence between mammal and deltavirus phylogenies, making our conclusions on host shifting robust.
The origin of HDV has been a long-standing mystery thwarted by the absence of closely related deltaviruses. The addition of six new mammalian deltaviruses by ourselves and others allowed us to reevaluate this question (7, 10). The pervasiveness of host shifting among deltaviruses and our discovery of a clade of mammalian deltaviruses basal to HDV (albeit with variable support depending on phylogeny, PP = 0.64 to 0.81) strongly points to a zoonotic origin. Although the exact progenitor virus remains undiscovered, the exclusive detection of mammalian deltaviruses in New World species supports an “out of the Americas” explanation for the origin and global spread of HDV (Fig. 1). The basal placement of the highly divergent Amazonian HDV genotype (HDV-3) within the phylogeny lends further credence to this scenario. It is therefore conceivable that mammal-infecting deltaviruses evolved in the Americas and the hypothesized arrival of HBV via human dispersal along the Bering land bridge facilitated the zoonotic emergence of HDV (19). Though circumstantial, the earliest records of HDV are from the Amazon basin in the 1930s (20). The greater diversity of HDV genotypes outside of the Americas—long argued to support an Old World origin—may instead reflect diversification arising from geographic vicariance within human populations. We suggest that American mammals should be the focus of future efforts to discover the direct ancestor and zoonotic reservoir of HDV.
Our results show that deltaviruses jump between mammalian host species through an unusual process that most likely requires parasitizing evolutionarily independent viruses. Since satellite viruses in general and HDV in particular tend to alter the pathogenesis and transmissibility of their helpers (21), our findings imply the potential for deltaviruses to act as host-switching virulence factors that could alter the progression of viral infections in multiple host species. The presence of deltaviruses in several mammalian orders, including in the saliva of sanguivorous bats, which feed on humans, wildlife, and domestic animals, provides ecological opportunities for cross-species transmission. Constraints on future host shifts are likely to differ from those of conventional animal pathogens. Specifically, given the broad cellular tropism of deltaviruses, interactions with viral helpers would likely be more important determinants of cross-species transmission than interactions with their hosts (17, 18). Consequently, anticipating future host shifts requires understanding the determinants and plasticity of deltavirus and helper compatibility along with the ecological factors that enable cross-species exposure.
Materials and Methods
Virus Discovery.
Serratus.
The Serratus platform was used to search published mammalian metagenomic and transcriptomic sequence datasets available in the NCBI SRA. Briefly, Serratus uses a cloud computing infrastructure to perform ultra-high-throughput alignment of publicly available SRA short read datasets to viral genomes of interest (10). Due to the exceptional computational demands of this search and mutual interest between ourselves and another research team, Serratus searches were designed by both teams and carried out at the nucleotide and amino acid levels by Edgar et al. (10). Query sequences for Serratus searches included all HDV genotypes and all deltavirus-like genomes which were publicly available at the time of the search (July 2020), along with representative genomes from DrDV-A and DrDV-B, which our team had already discovered. Results were shared among the two teams who subsequently pursued complementary lines of investigation. Mammalian deltaviruses were discovered using nucleotide (OvirDV) and amino acid level (Mmon DV and PmacDV) searches of the mammalian SRA search space.
Neotropical bat metagenomic sequencing.
Total nucleic acid was extracted from archived saliva swabs from Neotropical bats on a Kingfisher Flex 96 automated extraction machine (Thermo Fisher Scientific) with the Biosprint One-For-All Vet Kit (Qiagen) using a modified version of the manufacturer’s protocol as described previously (11). Ten pools of nucleic acids from vampire bats and other bat species were created for shotgun metagenomic sequencing (SI Appendix, Table S1). Eight pools comprised samples from bats in the same genus (two to 10 individuals per pool depending on availability of samples, 30 μL total nucleic acid per individual). The CAJ1_SV vampire bat pool from (22) which contained deltavirus reads was included as a sequencing control. The final pool (“rare species”) comprised eight other bat species that had only one individual sampled each. Pools were treated with DNase I (Ambion) and purified using RNAClean XP beads (Agencourt) following (11). Libraries were prepared using the SMARTer Stranded Total RNA-Seq Kit v2–Pico Input Mammalian (Clontech) and sequenced on an Illumina NextSeq 500 at The University of Glasgow Polyomics Facility. Samples were bioinformatically processed for viral discovery as described previously (11), with a slight modification to the read trimming step to account for shorter reads and a different library preparation kit.
DrDV genome assembly and annotation.
DrDV genomes were assembled using SPAdes (23) and refined by mapping cleaned reads back to SPAdes-generated contigs within Geneious v 7.1.7 (24). Regions of overlapping sequence at the ends of genomes due to linear de novo assemblies of circular genomes were resolved manually. Genome circularity was confirmed based on the presence of overlapping reads across the entire circular genome of both DrDVs. The amino acid sequence of the small DAg was extracted from sequences using getorf (25). Other smaller identified open reading frames did not exhibit significant homology when evaluated by protein blast against GenBank. Nucleotide sequences of full deltavirus genomes and amino acid sequences of DAg were aligned along with representative sequences from other deltaviruses using the E-INS-i algorithm in MAFFT v 7.017 (26). Genetic distances as percent identities were calculated based on an untrimmed full genome alignment of 2,321 nt and an untrimmed delta antigen alignment of 281 aa. Protein domain homology of the DAg was analyzed using HHpred (27). Ribozymes were identified manually by examining the region upstream of the delta antigen open reading frame where ribozymes are located in other deltavirus genomes (4, 5). RNA secondary structure and self-complementarity were determined using the web servers for Mfold (28) and RNAstructure (29). We found no evidence of recombination in nucleotide alignments of DrDV DAg according to the program GARD (Genetic Algorithm for Recombination Detection) (30) on the Datamonkey web server (31). Genome assembly and annotation of PmacDV, OvirDV, and MmonDV are described in ref. 10. We used blast searches of novel mammalian deltavirus genomes (blastn) and deltavirus antigen protein sequences (tblastn) against published host genomes on GenBank to evaluate the possible presence of endogenous deltavirus-like elements. DrDV-A and DrDV-B sequences were searched against the Desmodus rotundus genome assembly (GCA_002940915.2), MmonDV sequences were searched against the Marmota monax genome assembly (GCA_901343595.1), and OvirDV sequences were searched against the Odocoileus virginianus Reference Sequence (RefSeq) genome (NC_015247.1). There was no published genome of Peropteryx macrotis available on GenBank, so we used blastn and tblastn to compare the genome and delta antigen protein sequence against all of GenBank, restricting results by organism P. macrotis (taxid:249015). None of these searches yielded any hits, suggesting there is currently no evidence these deltaviruses have endogenized in their hosts.
Evaluation of deltavirus positive cohorts.
To establish that deltaviruses were likely to be actively infecting hosts in which they were detected, and not laboratory contamination or incidental detection of environmentally derived RNA, we searched for evidence of deltavirus infections in additional samples from the various studies that detected full genomes. Samples included sequencing libraries derived from both different individuals in the same study and different tissues from the same individuals. Searches used the program bwa (Burrows–Wheeler Alignment) (32) to map raw reads from deltavirus positive cohorts to the corresponding novel deltavirus genomes which had been detected in those same cohorts. Genome remapping was performed for all vampire bat libraries, two other Peropteryx macrotis libraries, and all other Neotropical bat species sequenced in the same study (12) and other pooled tissue samples sequenced by RNA-sequencing from Odocoileus virginianus in the same study (13). Results are shown in SI Appendix, Table S2. Deltavirus reads from additional individuals and timepoints from the Marmota monax study (14) are described in (10).
Global biogeographic analysis of deltavirus presence and absence.
To characterize the global distribution of mammal-infecting deltaviruses, we used the metadata of each SRA accession queried in the Serratus search to identify the associated host. We focused primarily on the mammalian dataset, which was generated by the SRA search query (“Mammalia”[Organism] NOT “Homo sapiens”[Organism] NOT “Mus musculus”[Organism]) AND (“type_rnaseq”[Filter] OR “metagenomic”[Filter] OR “metatranscriptomic”[Filter]) AND “platform illumina”[Properties]. All analyses were performed in R version 3.5.1 (33). We removed libraries with persistent errors which had not completed in the Serratus search. For remaining libraries, when host identification information was available to the species level, we matched Latin binomial species names to PanTHERIA, a dataset which contains the centroids of mammalian geographic distributions (see Dataset S1), and used an R script to assign species to continents using these geographic data (34). Subspecies present in scientific names of SRA metadata were reassigned to species level and recently updated binomial names were changed to match the PanTHERIA dataset. Due to the fact that mammalian taxonomic data in PanTHERIA date to 2005, some former orders which are no longer in use (e.g., Soricomorpha) appear in our data but are not expected to affect the results of analyses. A total of 48 species whose centroids occurred in water bodies were assigned to continents by manually inspecting species ranges. Widely distributed domesticated animals, datasets with genus-level metadata from broadly distributed genera, datasets from cell lines or with taxonomic information only at the class level, and those which had no geographic range data available (mostly aquatic mammals) were searched by Serratus but excluded from geographic comparisons. We quantified geographic and taxonomic biases in our dataset both in units of bases of RNA sequenced and number of species investigated.
Although most mammalian metagenomic and transcriptomic libraries were captured in the mammalian search, we also examined datasets from SRA search queries for vertebrates, metagenomes, and viromes to ensure that all relevant libraries were captured in our measures of search effort. For these datasets, we removed libraries with persistent errors and calculated search effort as bases of RNA sequenced across the three orders in which deltaviruses were discovered, removing libraries named for specific viral taxa which may have been enriched for these taxa and therefore do not represent a likely source of novel viruses. As these libraries lacked species-level metadata (hence their exclusion from the mammalian search above), we could not systematically calculate number of species in these datasets. Additional search queries are available at https://github.com/ababaian/serratus/wiki/SRA-queries.
Phylogenetic Analyses.
Bayesian phylogeny using MrBayes.
Phylogenetic analysis was performed on complete DAg amino acid sequences to place mammalian deltaviruses relative to HDV and other described deltaviruses. Representative sequences from each clade of HDV and other previously published deltaviruses were aligned with DrDVs (sequences generated by RT-PCR, described in RT-PCR and sequencing of blood and saliva samples), PmacDV, OvirDV, and MmonDV using the E-INS-i algorithm and JTT2000 scoring matrix of MAFFT within Geneious. Large delta antigen sequences from HDV were trimmed manually to small delta antigen length, and the alignment was further trimmed in trimAl using the automated1 setting to a final length of 192 aa (see Dataset S2). The best substitution model (JTTDCMut + F + G4) was determined using ModelFinder (35) within IQ-TREE 2 (36). Bayesian phylogenetic analysis was performed using the most similar model available within MrBayes (JTT + F + G4). The analysis was run for 5,000,000 generations and sampled every 2,500 generations, with the first 500 trees discarded as burn-in to generate the consensus tree.
Bayesian phylogeny using StarBeast.
We used StarBeast to generate a species-level phylogeny for the cophylogenetic analysis, using the same amino acid alignment of complete DAg which was used in the MrBayes analysis. StarBeast is typically used with multilocus sequence data from multiple individuals per species but can also be applied to single gene alignments (37). Notably, a preliminary analysis suggested this approach was more conservative than a constant effective population size coalescent model in BEAST which substantially inflated posterior probabilities on nodes across the tree relative to the MrBayes analysis (Fig. 2A). The StarBeast multispecies coalescent analysis was carried out as two duplicate runs of 50 million generations (sampling every 5000 generations) in BEAST2, using the JTT+G substitution model, the linear with constant root model for the species tree population size, and a Yule speciation model. Combined log files were assessed for convergence and effective sample size values >200 using Tracer. Twenty percent of trees were discarded as burn-in prior to generating the consensus tree.
Cophylogeny.
Cophylogenetic analyses were performed in R using PACo (38, 39) and ParaFit (40, 41). Analyses were performed on three subsets of matched host-deltavirus data: all taxa, ingroup taxa (mammals, bird, and snake), and mammals only. Host datasets consisted of distance matrices derived from a TimeTree phylogeny (http://timetree.org). For metagenomic libraries which contained individuals of multiple species (AvianDV and FishDV), one host was selected for inclusion (Anas gracilis and Macroramphosus scolopax, respectively). Host data were not available in TimeTree for two species in which deltaviruses were discovered (Proechimys semispinosus and Schedorhinotermes intermedius), so available congeners were substituted (Proechimys guairae and Schedorhinotermes medioobscurus, respectively). Virus datasets consisted of distances matrices from posterior species trees generated in StarBeast. Cophylogeny analyses performed using virus distance matrices derived from posterior MrBayes trees, pruned to contain only relevant taxa, yielded qualitatively similar results. For both analyses, the principal coordinates analysis of distance matrices was performed with the “Cailliez” correction. Since units of branch length differed between host and virus trees, all distance matrices were normalized prior to coevolutionary analyses. To account for phylogenetic uncertainty in the evolutionary history of deltaviruses, analyses were carried out using 1,000 trees randomly selected from the posterior distribution of the Bayesian phylogenetic analyses (separately for StarBeast and MrBayes). Due to uncertain placement of HDV3 in both phylogenies, one representative of HDV was randomly selected for each iteration. For each tree, we calculated summary statistics (see below) describing the dependence of the deltavirus phylogeny on the host phylogeny. P values were estimated using 1,000 permutations of host–virus associations.
For PACo analyses, the null model selected was r0, which assumes that virus phylogeny tracks the host phylogeny. Levels of cophylogenetic signal were evaluated as the median global sum of squared residuals (m2xy) and mean significance (P values), averaged over the 1,000 posterior trees. Empirical distributions were compared to null distributions for each dataset. For ParaFit analyses, the levels of cophylogenetic signal in each dataset were evaluated as the median sum of squares of the fourth corner matrix (ParaFitGlobal) and mean significance (P values), averaged over 1,000 posterior trees. ParaFit calculates the significance of the global host–virus association statistic by randomly permuting hosts in the host–virus association matrix to create a null distribution. Since ParaFit does not provide this distribution to users, we approximated it for visualization by manually reestimating the global host–virus association statistic for 1,000 random permutations of hosts in the host–virus association matrix. Phylogenies and cophylogenies were visualized in R using the packages “ape” (41), “phangorn” (42), “phytools” (43), and “ggtree” (44).
Deltaviruses in Neotropical Bats.
Capture and sampling of wild bats.
To examine DrDV prevalence in vampire bats, we studied 12 sites in three departments of Peru between 2016 and 2017 (Fig. 3A). Age and sex of bats were determined as described previously (11). Saliva samples were collected by allowing bats to chew on sterile cotton-tipped wooden swabs (Fisherbrand). Blood was collected from vampire bats only by lancing the propatagial vein and saturating a sterile cotton-tipped wooden swab with blood. Swabs were stored in 1 mL of RNALater (Ambion) overnight at 4 °C before being transferred to dry ice and stored in −70 °C freezers.
Bat sampling protocols were approved by the Research Ethics Committee of the University of Glasgow School of Medical, Veterinary and Life Sciences (Ref081/15), the University of Georgia Animal Care and Use Committee (A2014 04-016-Y3-A5), and the Peruvian Government (RD-009-2015-SERFOR-DGGSPFFS, RD-264-2015-SERFOR-DGGSPFFS, RD-142-2015-SERFOR-DGGSPFFS, and RD-054-2016-SERFOR-DGGSPFFS).
RT-PCR and sequencing of blood and saliva samples.
Primers were designed to screen bat saliva and blood samples for a conserved region of the DAg protein of DrDV-A (236 nt) and DrDV-B (231 nt) by heminested and nested RT-PCR, respectively (SI Appendix, Table S4). Alternative primers were designed to amplify the complete DAg for DrDV-A (707 nt) and DrDV-B (948 nt) using a one-step RT-PCR (SI Appendix, Table S4). We used RT-PCR to screen vampire bat saliva samples (described in Capture and sampling of wild bats) as well as saliva samples from additional Neotropical bat species included in metagenomic sequencing pools (described in Neotropical bat metagenomic sequencing) and further archived samples from nonvampire bat species which were withheld from metagenomic pools in order to balance pool sizes (SI Appendix, Fig. S6). A subset of vampire bat blood samples were also screened by RT-PCR; blood samples were unavailable from nonvampire bat species. Complementary DNA (cDNA) was generated from total nucleic acid extracts using the ProtoScript II First Strand cDNA Synthesis Kit with random hexamers; RNA and random hexamers were heated for 5 min at 65 °C then placed on ice. ProtoScript II reaction mix and ProtoScript II enzyme mix were added to a final concentration of 1×, and the reaction was incubated at 25 °C for 5 min, 42 °C for 15 min, and 80 °C for 5 min. PCR was performed using Q5 High-Fidelity DNA Polymerase (NEB). Each reaction contained 1× Q5 reaction buffer, 200 μM of dNTPs, 0.5 μM of each primer, 0.02 U/μL of Q5 High-Fidelity DNA polymerase, and either 2.5 μL of cDNA or 1 μL of Round 1 PCR product. Reactions were incubated at 98 °C for 30 s, followed by 40 cycles of 98 °C for 10 s, 61 to 65 °C for 30 s (or 58 to 60 °C for 30 s for the complete DAg), 72 °C for 40 s, and a final elongation step of 72 °C for 2 min. PCR products of the correct size were confirmed by reamplification from cDNA or total nucleic acid extracts and/or Sanger sequencing (Eurofins Genomics).
Bat species confirmation.
We confirmed the morphological species assignment of the C. perspicillata individual in which DrDV-B was detected by sequencing cytochrome B. Cytochrome B was amplified from the same saliva sample in which DrDV-B was detected using primers Bat 05A and Bat 04A (45) and GoTaq Green Master Mix (Promega) according to the manufacturer’s instructions, and the resulting product was Sanger sequenced (Eurofins Genomics) then evaluated by nucleotide blast against GenBank.
Genetic diversity and distribution of DrDV-B.
To examine relationships among DrDV-B sequences, Bayesian phylogenetic analysis was performed on a 214-nt fragment of the DAg. Sequences from saliva and blood of 41 D. rotundus and saliva from one C. perspicillata were aligned using MAFFT within Geneious (see Dataset S3). Duplicate sequences originating from the blood and saliva of the same individuals were removed. Alignments were trimmed using trimAl (46) with automated1 settings, and the best model of sequence evolution was determined using jModelTest2 (47). Phylogenetic analysis was performed using MrBayes 3.6.2 (48) with the GTR+I model. The analysis was run for 4,000,000 generations and sampled every 2,000 generations, with the first 1,000 trees removed as burn-in. The association between phylogenetic relationships and location at both the regional and colony level was tested using BaTS (49) with 1,000 posterior trees and 1,000 replicates to generate the null distribution.
Statistical analyses of DrDV-B.
We modeled the effects of age and sex on DrDV-B presence in saliva using a binomial generalized linear mixed model in the package lme4 (50) in R (see Dataset S4). Sex (female/male) and age (adult/subadult) were modeled as categorical variables with site included as a random effect. We also evaluated differences in DrDV-B prevalence between regions of Peru using a binomial generalized linear model and used the Anova function of the car package (51) to calculate the likelihood ratio χ2 test statistic.
Identifying Candidate Helper Viruses for Mammalian Deltaviruses.
Hepadnavirus screening in vampire bats.
We tested samples for the presence of bat hepadnavirus as a candidate helper virus to DrDV. DNA from saliva and blood samples was screened for HBV-like viruses using pan-Hepadnaviridae primers (HBV-F248, HBV-R397, HBV-R450a, and HBV-R450b; SI Appendix, Table S4) and PCR protocols (16). We used a plasmid carrying a 1.3-mer genome of human HBV that is particle assembly defective but replication competent as a positive control.
Bioinformatic screening of published metagenomic datasets.
We performed comprehensive virus discovery using an in-house bioinformatic pipeline (11) on sequence datasets containing deltaviruses to identify candidate helper viruses. Datasets analyzed included all vampire bat datasets [22 from (11) and 46 from (22)], P. macrotis datasets (SRR7910142, SRR7910143, and SRR7910144), O. virginianus datasets (SRR4256025 to SRR4256034), and M. monax datasets (SRR2136906 and SRR2136907). Briefly, after quality trimming and filtering, reads were analyzed by BLASTX using DIAMOND (52) against a RefSeq database to remove bacterial and eukaryotic reads. Remaining reads were then de novo assembled using SPAdes (23) and resulting contigs were analyzed by BLASTX using DIAMOND against a nonredundant protein database (53). KronaTools (54) and MEGAN (55) were used to visualize and report taxonomic assignments.
Supplementary Material
Acknowledgments
We thank Jaime Pacheco, Luigi Carrasco, Yosselym Luzon, Saori Grillo, and Megan Griffiths for field and laboratory assistance; Megan Griffiths, Joseph Hughes, and Matt Hutchinson for analysis advice; and Ana da Silva Filipe, Felix Drexler, Pablo Murcia, and Mafalda Viana for comments on earlier versions of the manuscript. We thank the Serratus team, particularly Artem Babaian and Robert Edgar, for assistance with Serratus. Funding was provided by the Wellcome Trust (Institutional Strategic Support Fund Early Career Researcher Catalyst Grant; Wellcome-Beit Prize:102507/Z/13/A; Senior Research Fellowship: 102507/Z/13/Z), the Human Frontier Science Program (RGP0013/2018), and the Medical Research Council (MC_UU_12014/12). Additional support was provided by the NSF (Graduate Research Fellowship and DEB‐1601052), Achievement Rewards for College Scientists Foundation, Sigma Xi, Animal Behavior Society, Bat Conservation International, American Society of Mammalogists, Odum School of Ecology, University of Georgia (UGA) Graduate School, UGA Latin American and Caribbean Studies Institute, UGA Biomedical and Health Sciences Institute, and the Explorers Club.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2019907118/-/DCSupplemental.
Data Availability.
DrDV genome sequences are available on GenBank (accession numbers MT649206–MT649209). Serratus data and a description of the platform are available at https://serratus.io/access and the deltavirus genome sequences analyzed here are available on a GitHub (PmacDV: https://raw.githubusercontent.com/wiki/ababaian/serratus/assets/lassie.fa, OvirDV: https://raw.githubusercontent.com/wiki/ababaian/serratus/assets/bambi.fa, and MmonDV: https://raw.githubusercontent.com/wiki/ababaian/serratus/assets/murray.fa). The Peropteryx macrotis hepacivirus genome sequence is available in the TPA section of the DDBJ (DNA Data Bank of Japan)/ENA (European Nucleotide Archive)/GenBank databases under the accession number TPA: BK013349. Vampire bat hepacivirus contigs are available on GenBank (accession numbers MW249008 and MW249009). Peruvian bat metagenomes are available in ENA project PRJEB35111. Scripts used for bioinformatic analyses are available on GitHub (https://github.com/rjorton/Allmond). Datasets S1–S4 are provided as supplementary information.
References
- 1.Magnius L. et al.; ICTV Report Consortium , ICTV virus taxonomy profile: Deltavirus. J. Gen. Virol. 99, 1565–1566 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Salehi-Ashtiani K., Lupták A., Litovchick A., Szostak J. W., A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science 313, 1788–1792 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Elena S. F., Dopazo J., Flores R., Diener T. O., Moya A., Phylogeny of viroids, viroidlike satellite RNAs, and the viroidlike domain of hepatitis delta virus RNA. Proc. Natl. Acad. Sci. U.S.A. 88, 5631–5634 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wille M., et al. , A divergent hepatitis D-like agent in birds. Viruses 10, 720–729 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hetzel U., et al. , Identification of a novel deltavirus in Boa constrictors. MBio 10, 1447–1448 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang W.-S., et al. , Novel hepatitis D-like agents in vertebrates and invertebrates. Virus Evol. 5, vez021 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paraskevopoulou S., et al. , Mammalian deltavirus without hepadnavirus coinfection in the neotropical rodent Proechimys semispinosus. Proc. Natl. Acad. Sci. U.S.A. 117, 17977–17983 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi M., et al. , Redefining the invertebrate RNA virosphere. Nature 540, 539–543 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Shi M., et al. , The evolutionary history of vertebrate RNA viruses. Nature 556, 197–202 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Edgar R. C., et al. , Petabase-scale sequence alignment catalyses viral discovery. bioRxiv: 10.1101/2020.08.07.241729 (10 August 2020). [DOI] [PubMed]
- 11.Bergner L. M., et al. , Using noninvasive metagenomics to characterize viral communities from wildlife. Mol. Ecol. Resour. 19, 128–143 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moreno-Santillán D. D., Machain-Williams C., Hernández-Montes G., Ortega J., De novo transcriptome assembly and functional annotation in five species of bats. Sci. Rep. 9, 6222 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seabury C. M., et al. , Genome-wide polymorphism and comparative analyses in the white-tailed deer (Odocoileus virginianus): A model for conservation genomics. PLoS One 6, e15811–e15819 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fletcher S. P., et al. , Intrahepatic transcriptional signature associated with response to Interferon-α treatment in the woodchuck model of chronic hepatitis B. PLoS Pathog. 11, e1005103 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krushkal J., Li W. H., Substitution rates in hepatitis delta virus. J. Mol. Evol. 41, 721–726 (1995). [DOI] [PubMed] [Google Scholar]
- 16.Drexler J. F., et al. , Bats carry pathogenic hepadnaviruses antigenically related to hepatitis B virus and capable of infecting human hepatocytes. Proc. Natl. Acad. Sci. U.S.A. 110, 16151–16156 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez-Vargas J., et al. , Enveloped viruses distinct from HBV induce dissemination of hepatitis D virus in vivo. Nat. Commun. 10, 2098 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szirovicza L., et al. , Snake deltavirus utilizes envelope proteins of different viruses to generate infectious particles. MBio 11, e03250-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasche A., Sander A.-L., Corman V. M., Drexler J. F., Evolutionary biology of human hepatitis viruses. J. Hepatol. 70, 501–520 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alvarado-Mora M. V., et al. , Dynamics of hepatitis D (delta) virus genotype 3 in the Amazon region of South America. Infect. Genet. Evol. 11, 1462–1468 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Gnanasekaran P., Chakraborty S., Biology of viral satellites and their role in pathogenesis. Curr. Opin. Virol. 33, 96–105 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Bergner L. M., et al. , Demographic and environmental drivers of metagenomic viral diversity in vampire bats. Mol. Ecol. 29, 26–39 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bankevich A., et al. , SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kearse M., et al. , Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rice P., Longden I., Bleasby A., EMBOSS: The European molecular biology open software suite. Trends Genet. 16, 276–277 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Katoh K., Misawa K., Kuma K., Miyata T., MAFFT: A novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Söding J., Biegert A., Lupas A. N., The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuker M., Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellaousov S., Reuter J. S., Seetin M. G., Mathews D. H., RNAstructure: Web servers for RNA secondary structure prediction and analysis. Nucleic Acids Res. 41, W471–W474 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kosakovsky Pond S. L., Posada D., Gravenor M. B., Woelk C. H., Frost S. D. W., GARD: A genetic algorithm for recombination detection. Bioinformatics 22, 3096–3098 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Weaver S., et al. , Datamonkey 2.0: A modern web application for characterizing selective and other evolutionary processes. Mol. Biol. Evol. 35, 773–777 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.R Core Team , R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2019). https://www.r-project.org/.
- 34.Jones K. E., et al. , PanTHERIA: A species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecology 90, 2648 (2009). [Google Scholar]
- 35.Kalyaanamoorthy S., Minh B. Q., Wong T. K. F., von Haeseler A., Jermiin L. S., ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minh B. Q., et al. , IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drummond A. J., Suchard M. A., Xie D., Rambaut A., Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balbuena J. A., Míguez-Lozano R., Blasco-Costa I., PACo: A novel procrustes application to cophylogenetic analysis. PLoS One 8, e61048 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hutchinson M. C., Cagua E. F., Balbuena J. A., Stouffer D. B., Poisot T., Paco: Implementing procrustean approach to Cophylogeny in R. Methods Ecol. Evol. 8, 932–940 (2017). [Google Scholar]
- 40.Legendre P., Desdevises Y., Bazin E., A statistical test for host-parasite coevolution. Syst. Biol. 51, 217–234 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Paradis E., Schliep K., Ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Schliep K. P., phangorn: Phylogenetic analysis in R. Bioinformatics 27, 592–593 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Revell L. J., phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223 (2011). [Google Scholar]
- 44.Yu G., Smith D. K., Zhu H., Guan Y., Lam T. T.-Y., ggtree: An rpackage for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36 (2016). [Google Scholar]
- 45.Martins F. M., Ditchfield A. D., Meyer D., Morgante J. S., Mitochondrial DNA phylogeography reveals marked population structure in the common vampire bat, Desmodus rotundus (Phyllostomidae). J Zoological System 45, 372–378 (2007). [Google Scholar]
- 46.Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T., trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Darriba D., Taboada G. L., Doallo R., Posada D., jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 9, 772 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ronquist F., et al. , MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parker J., Rambaut A., Pybus O. G., Correlating viral phenotypes with phylogeny: Accounting for phylogenetic uncertainty. Infect. Genet. Evol. 8, 239–246 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Bates D., Mächler M., Bolker B., Walker S., Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48 (2015). [Google Scholar]
- 51.Fox J., Weisberg S., An R Companion to Applied Regression (Sage, Thousand Oaks, CA: ), ed. 2, 2011). [Google Scholar]
- 52.Buchfink B., Xie C., Huson D. H., Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Pruitt K. D., Tatusova T., Maglott D. R., NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35, D61–D65 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ondov B. D., Bergman N. H., Phillippy A. M., Interactive metagenomic visualization in a Web browser. BMC Bioinformatics 12, 385 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huson D. H., et al. , MEGAN community edition–Interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput. Biol. 12, e1004957 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar S., Stecher G., Suleski M., Hedges S. B., TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 34, 1812–1819 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DrDV genome sequences are available on GenBank (accession numbers MT649206–MT649209). Serratus data and a description of the platform are available at https://serratus.io/access and the deltavirus genome sequences analyzed here are available on a GitHub (PmacDV: https://raw.githubusercontent.com/wiki/ababaian/serratus/assets/lassie.fa, OvirDV: https://raw.githubusercontent.com/wiki/ababaian/serratus/assets/bambi.fa, and MmonDV: https://raw.githubusercontent.com/wiki/ababaian/serratus/assets/murray.fa). The Peropteryx macrotis hepacivirus genome sequence is available in the TPA section of the DDBJ (DNA Data Bank of Japan)/ENA (European Nucleotide Archive)/GenBank databases under the accession number TPA: BK013349. Vampire bat hepacivirus contigs are available on GenBank (accession numbers MW249008 and MW249009). Peruvian bat metagenomes are available in ENA project PRJEB35111. Scripts used for bioinformatic analyses are available on GitHub (https://github.com/rjorton/Allmond). Datasets S1–S4 are provided as supplementary information.