Significance
N6-methyladenosine (m6A) modification occurs in cellular RNAs and viral transcripts and regulates the fate of cellular and viral RNAs. Hepatitis B virus (HBV) transcripts are m6A-methylated in the consensus DRACH motif in the epsilon stem–loop, and this modification differentially regulates the viral life cycle depending on the m6A position at the 5′- or 3′-epsilon stem–loop. Here, we report that HBV X (HBx) protein recruits cellular m6A machinery onto HBV minichromosome and host PTEN chromosomal locus to add m6A modification cotranscriptionally. Induced m6A modification of HBV RNAs and PTEN mRNA by HBx decreases their stability. This study identifies the mechanism by which a viral protein regulates m6A modification of RNA.
Keywords: N6-methyladenosine modification, METTL3/14, hepatitis B virus, hepatitis B virus X protein, cotranscriptional modification
Abstract
Chronic hepatitis B virus (HBV) infections are one of the leading causes of cirrhosis and hepatocellular carcinoma. N6-methyladenosine (m6A) modification of cellular and viral RNAs is the most prevalent internal modification that occurs cotranscriptionally. Previously, we reported the dual functional role of m6A modification of HBV transcripts in the viral life cycle. Here, we show that viral HBV X (HBx) protein is responsible for the m6A modifications of viral transcripts. HBV genomes defective in HBx failed to induce m6A modifications of HBV RNAs during infection/transfection, while ectopic expression of HBx restores m6A modifications of the viral RNAs but not the mutant HBx carrying the nuclear export signal. Using chromatin immunoprecipitation assays, we provide evidence that HBx and m6A methyltransferase complexes are localized on the HBV minichromosome to achieve cotranscriptional m6A modification of viral RNAs. HBx interacts with METTL3 and 14 to carry out methylation activity and also modestly stimulates their nuclear import. This role of HBx in mediating m6A modification also extends to host phosphatase and tensin homolog (PTEN) mRNA. This study provides insight into how a viral protein recruits RNA methylation machinery to m6A-modify RNAs.
Hepatitis B virus (HBV), a member of the Hepadnaviridae family, causes chronic hepatitis and liver cirrhosis, leading to the development of hepatocellular carcinoma (1). HBV genome consists of 3.2-kb partially double-stranded DNA, which encodes surface (HBs), precore or “e” (HBe), and core (HBc) antigen proteins, polymerase (pol, a reverse transcriptase), and X (HBx) proteins. Although HBV is a DNA virus, it replicates via an RNA intermediate termed pregenomic RNA (pgRNA) to produce a relaxed circular DNA, which ultimately transforms into a covalently closed DNA (cccDNA) in the nucleus (1). Despite an effective vaccine, there are an estimated 350 million people infected with HBV worldwide, with 600,000 deaths reported annually (2). Current HBV therapies have limited efficacy and do not eliminate the cccDNA, which maintains its presence and resumes the infectious process upon withdrawal of antivirals. The inability to completely eliminate HBV infection remains a challenge today.
HBx, a regulatory protein, has been intensely studied and shown to play an important role in viral transcription and replication (3–6). HBV transcription occurs from the cccDNA that is organized as an episomal minichromosome in association with histones and nonhistone proteins (7). HBx has been previously shown to be associated with cccDNA in concert with host protein complexes (7). A large body of literature describes the various functional activities of HBx in the viral life cycle from its cytoplasmic as well as nuclear locations (4, 5). HBx is traditionally considered as a transactivator of both viral and host gene expression and a factor required for the viral life cycle shown convincingly in in vivo model systems (5, 8). HBx does not bind DNA, but interacts with both basal transcription factors and coactivators (4, 5, 8–11). These activities place HBx at the sites of transcription initiation.
The mammalian mRNAs are epigenetically modified by diverse chemical modifications to regulate RNA stability and turnover (12). Among the RNA chemical modifications, m6A methylation is the most prevalent internal RNA modification of eukaryotic cells (13). Over 25% of mammalian RNAs are m6A-modified, and these modifications have been linked to various biological processes, which include innate immune response, sex determination, stem cell differentiation, circadian clock, meiosis, stress response, and cancer (13). m6A methylation is installed cotranscriptionally by m6A methyltransferases (“writers”) in a sequence motif RRACH/DRACH context (14). The cotranscriptional activity of these enzymes, however, requires their presence at the sites of transcription initiation on the open chromatin of the chromosomes interacting with a large body of assembled transcription factors and coactivators (15). The enzymes that catalyze the m6A internal modifications of the RNA are a complex of methyltransferase-like 3 (METTL3) catalytic subunit, a functional subunit of METTL14, and additional adapter proteins, including WTAP (13). m6A modification is reversibly catalyzed by Fat mass and obesity-associated protein (FTO) and ALKBH5 (“erasers”) and is typically enriched near the stop codons and the 3′-untranslated region (UTR) of cellular mRNA (13). Generally, the YTH domain family (YTHDF) proteins (“readers”) regulate m6A-modified RNA stability, translation, and localization by direct interaction (16, 17). Recently, the presence of m6A modification has been shown to affect various aspects of the viral life cycle and associated pathogenesis (18–22). m6A methyltransferases (METTL3/14) and reader proteins (YTHDF) regulate the life cycle of both DNA and RNA viruses. Surprisingly, m6A methylation is more frequent in viruses than cellular mRNAs, and their effects on viral replication and translation are being characterized (18–22).
We have previously reported that HBV transcripts are m6A-methylated during infection and identified a single m6A site within the HBV genome (SI Appendix, Fig. S1 A and B) (21). The m6A modification of the 5′-epsilon stem–loop structure induces reverse transcription activity, while m6A modification of the 3′-epsilon stem–loop structure reduces translation activity due to their interactions with YTHDF2 proteins leading to their degradation (21). HBV infection also affects m6A profiling of host RNAs including PTEN mRNA in chronic HBV patients (SI Appendix, Fig. S1C) (23). The PTEN 3′-UTR region is m6A-modified, and the increased m6A modification of PTEN by HBV infection promotes destabilization of PTEN RNA via its interaction with YTHDF2 protein (23). In this study, we show that HBx interacts with METTL3/14 proteins and recruits them at transcription initiation sites to affect internal m6A modification of RNAs while they are being nascently transcribed from the cccDNA template. Evidence is presented by chromatin immunoprecipitation (ChIP) analysis of the wild type HBV and HBx-null virus-infected primary human hepatocytes (PHHs) and HepG2-NTCP cells. Further, using mutant 1.3-mer HBV genomes defective in HBx and rescue efforts with ectopic expression of wild type HBx and HBx plasmids carrying nuclear localization (NLS) and nuclear export (NES) signals of HepG2-NTCP–infected cells, we confirm that nuclear HBx is necessary for m6A modification of viral RNAs.
Results
HBx Regulates m6A Modification of HBV and Host PTEN RNAs.
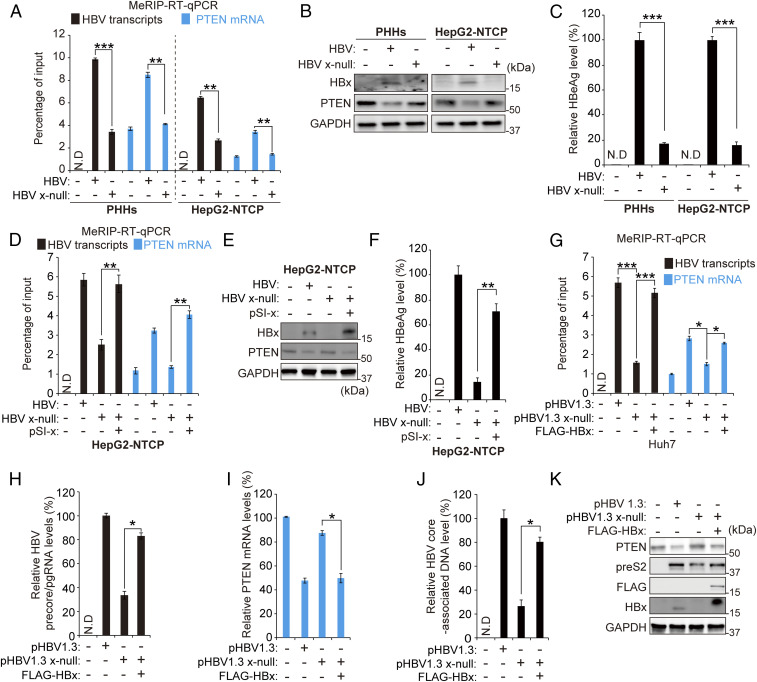
To determine whether HBx protein affects m6A modification of HBV RNAs or a representative host PTEN mRNA, we conducted a methylated RNA immunoprecipitation (MeRIP) qRT-PCR assay using the m6A-specific antibody in HBV-infected cells (Fig. 1A). PHHs or HepG2-NTCP cells were infected with HBV or HBx-defective (x-null) HBV particles. Total RNA was extracted and subjected to MeRIP qRT-PCR using primers that recognize a shared 3′-UTR sequence presented in all HBV transcripts, including pgRNA. Interestingly, we found that the HBx-defective HBV infection drastically reduced m6A modification of HBV transcripts and PTEN mRNA. In the MeRIP qRT-PCR assay, CREBBP and HPRT1 were used as positive and negative controls, respectively (SI Appendix, Fig. S1 E and D) (24). Similar results were also observed in HBV-transfected Huh7 cells (SI Appendix, Fig. S2A). HBx-defective HBV genome transfection reduced m6A modification of viral transcripts and PTEN mRNA compared to WT HBV transfection. Viral protein, precore/pgRNA, and HBV core-associated DNA levels were reduced in the absence of HBx (Fig. 1 B and C, SI Appendix, Fig. S2 B–D). These results are consistent with previous studies showing a generalized decrease in viral RNA and proteins in the absence of HBx, and HBx plays a critical role in viral replication (5, 6). HBx expression alone did not affect m6A in PTEN mRNA, indicating that the viral life cycle is required for modulating the m6A process by HBx (SI Appendix, Fig. S2 G–I). In addition, HBV infection reduced PTEN expression, while the HBx-defective HBV infection did not affect PTEN protein level (Fig. 1B). Because it was reported that HBV enhances m6A modification of PTEN mRNA, leading to reduced PTEN protein expression (23), these results suggest that HBx protein decreases PTEN expression by the up-regulation of m6A modification.
Fig. 1.
HBx protein regulates m6A modification of HBV RNAs and host PTEN mRNA during HBV infection of PHHs and HepG2 NTCP cells and HBV 1.3 transfection. (A–C) PHHs and HepG2-NTCP cells were infected with 2.5 × 103 genome equivalents per cell of HBV WT or x-null particles. After 10 d, total RNA and protein were extracted from HBV-infected PHHs. m6A-modified HBV transcripts and PTEN mRNA levels were quantified by MeRIP qRT-PCR (A). The indicated protein expression levels were analyzed by immunoblotting (B). HBeAg levels in media were analyzed using culture media by ELISA (C). (D–F) HepG2-NTCP cells were infected with HBV WT or x-null particles. After 7 d, pSI-x plasmids were transfected into HBV x-null–infected HepG2-NTCP cells. After 3 d, cells were harvested to assess m6A modification levels of HBV transcripts and PTEN mRNA (D) or to perform immunoblotting analysis for the indicated proteins (E) or HBeAg levels (F). (G–K) Huh7 cells were transfected with pHBV 1.3 or pHBV 1.3 x-null or cotransfected with pHBV 1.3 x-null and pcDNA3.1 FLAG-HBx. After 72 h, cells were harvested to assess m6A-methylated HBV transcripts and PTEN mRNA (G), HBV precore/pgRNA (H), PTEN mRNA (I), HBV core-associated DNA (J), or the indicated protein expressions (K). In all panels, data are mean ± SD (*P < 0.05; **P < 0.01; ***P < 0.001; unpaired one-tailed Student’s t test).
It has been reported that, during HBV infection, the absence of HBx dramatically decreases viral transcription but does not affect HBV cccDNA stability (25). Thus, decreased m6A modification of viral RNAs in HBV x-null–infected PHHs or HepG2-NTCP cells may be due to insufficient viral RNA levels. To confirm this, we investigated whether an ectopic expression of HBx by transfection of HBV x-null–infected cells can restore m6A modification of viral transcripts and PTEN RNA. Indeed, we observed that, in the HepG2-NTCP–infected cells with HBx-null virus particles, ectopic HBx transfection restored m6A methylation of viral transcripts and PTEN mRNA (Fig. 1 D and E). Similarly, the viral protein (HBeAg) expression was also restored (Fig. 1F). Similar results were obtained using HBV DNA transfection of Huh7 or HepG2 cells (Fig. 1 G–K and SI Appendix, Fig. S3). These results collectively suggest that HBx protein plays a pivotal role in regulating m6A modification of viral transcripts and host PTEN mRNA.
HBx Protein Promotes the Recruitment of m6A Methyltransferases onto the HBV cccDNA and PTEN Chromosomal Locus.
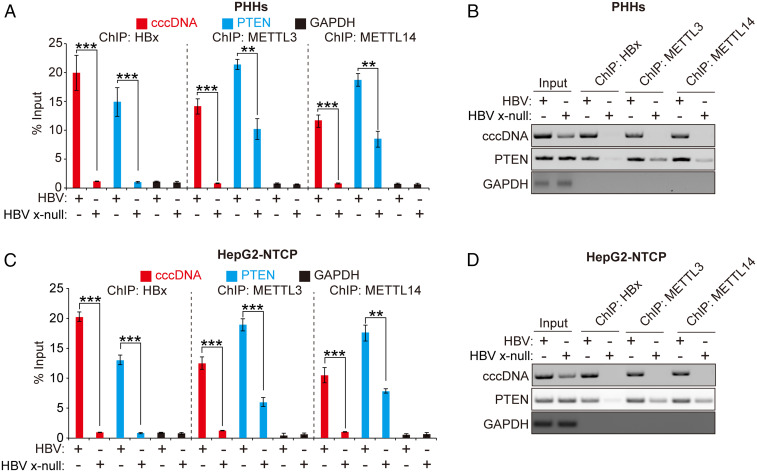
Next, we investigated whether HBx affects the recruitment of m6A methyltransferases onto the HBV cccDNA or PTEN chromosomal locus because m6A methyltransferases are guided to the chromosome for adding m6A to RNAs by histone H3 trimethylation at Lys36 (H3K36me3) (15). ChIP assays allow identification of in vivo DNA binding sites of chromosomal components (7). We performed ChIP assays using antibodies specific to HBx, METTL3, METTL14, or IgG in HBV-infected PHHs and HepG2-NTCP nuclear lysates to determine if HBx directly recruits METTL enzymes to transcription initiation sites (Fig. 2 and SI Appendix, Fig. S4). These assays revealed that HBx binds cccDNA, as shown previously (7), and, most importantly, that, in the absence of HBx, there was a dramatic reduction of interaction between m6A methyltransferases and cccDNA during HBV infection (Fig. 2 A–D). Similarly, the binding affinity between m6A methyltransferases and PTEN 3′-UTR chromosomal region was decreased in HBx-defective HBV-infected PHHs and HepG2-NTCP cells (Fig. 2 A–D). We further confirmed these results using HBV-transfected Huh7 cells and found that the absence of HBx reduced the recruitment of METTL3/14 to PTEN chromosomal locus (SI Appendix, Fig. S4 E–I). We also show that the interactions of m6A methyltransferases with PTEN mRNA and HBV transcript were reduced in HBx-null–transfected cells (SI Appendix, Fig. S4 J and K). This result shows that HBx induced interaction between METTL3/14 and HBV RNAs or PTEN mRNA. Altogether, these data suggested that HBx expression induced recruitment of m6A writer METTL enzymes onto the HBV cccDNA as well as PTEN 3′-UTR chromosomal locus.
Fig. 2.
ChIP assay to determine the HBx protein-mediated recruitment of m6A methyltransferases onto HBV cccDNA and PTEN locus on the chromosome. (A and B) PHHs were infected with HBV or HBV x-null particles. Cross-linked chromatins prepared 10 d after infection were immunoprecipitated with anti-HBx or anti-METTL3 or anti-METTL14 antibodies and analyzed by qRT-PCR (A) or semiquantitative PCR (B). (C and D) Chromatins extracted from HBV- or HBV Δx-infected HepG2-NTCP cells were immunoprecipitated with anti-HBx or anti-METTL3 or anti-METTL14 antibodies. Immunoprecipitated chromatins were analyzed by qRT-PCR (C) or semiquantitative PCR (D). In all panels, data are mean ± SD (**P < 0.01; ***P < 0.001; unpaired one-tailed Student’s t test).
HBx-Mediated m6A Modification Affects the Stability of HBV and PTEN RNAs.
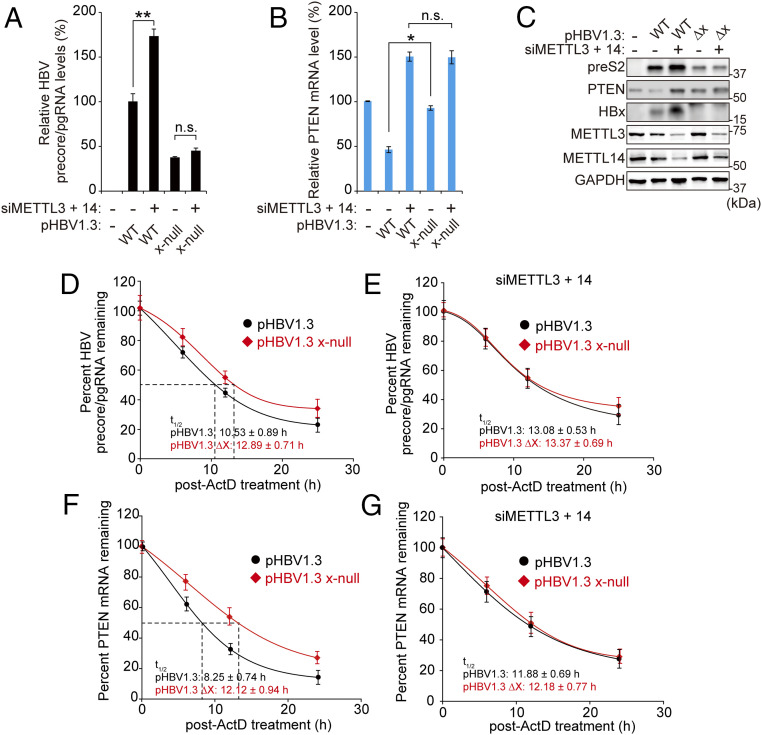
Since increased m6A modified RNAs are recognized by the YTH domain family of proteins, which affects RNA stability (16, 26), we investigated the stability of both HBV and PTEN RNAs in this context (Fig. 3). The silencing of m6A methyltransferases increased HBV precore/pgRNA and viral protein expression, but, in the absence of HBx, HBV replication and translation activities were not changed by knockdown of METTL3/14 (Fig. 3 A and C). HBV transfection reduced PTEN mRNA and protein levels in an m6A modification-dependent manner, while HBx-defective HBV genome transfection did not affect PTEN mRNA and protein levels (Fig. 3 B and C). We then determined the half-life of precore/pgRNA and PTEN mRNA in HBV WT or defective HBx HBV-transfected cells following actinomycin D treatment. The stabilities of HBV pgRNA and PTEN were induced in HBx-defective HBV-transfected cells compared to HBV WT transfection (Fig. 3 D and F), whereas the effect of HBx on HBV RNAs and PTEN mRNA stabilities were unaffected in the METTL3/14-silenced cells (Fig. 3 E and G). These results demonstrated that HBx reduces viral transcripts and PTEN mRNA stabilities via induction of m6A methylation.
Fig. 3.
HBx protein-mediated m6A modification affects the stability of viral RNAs and PTEN RNA. (A–C) Huh7 cells, which were transfected with pHBV 1.3 or pHBV 1.3 x-null, were treated with control or METTL3/14 siRNAs. After 72 h, total RNA and lysates were extracted to assess HBV precore/pgRNA (A), PTEN mRNA (B), or the indicated protein expressions (C). (D–G) Analysis of HBV precore/pgRNA (D and E) and PTEN mRNA (F and G) stabilities in Huh7 cells transfected with pHBV 1.3 or pHBV 1.3 x-null. After 24 h, the transfected cells were treated with control or METTL3/14 siRNAs by incubation for 2 d, and cells were incubated with actinomycin D. The cells were harvested at 0, 6, 12, or 24 h following actinomycin D treatment, and relative levels of remaining HBV pgRNA (D and E) and PTEN mRNA (F and G) were analyzed. In all panels, data are mean ± SD (*P < 0.05 by unpaired one-tailed Student’s t test). n.s., not significant.
HBx Protein Binds to m6A Methyltransferases and Stimulates Their Nuclear Import.
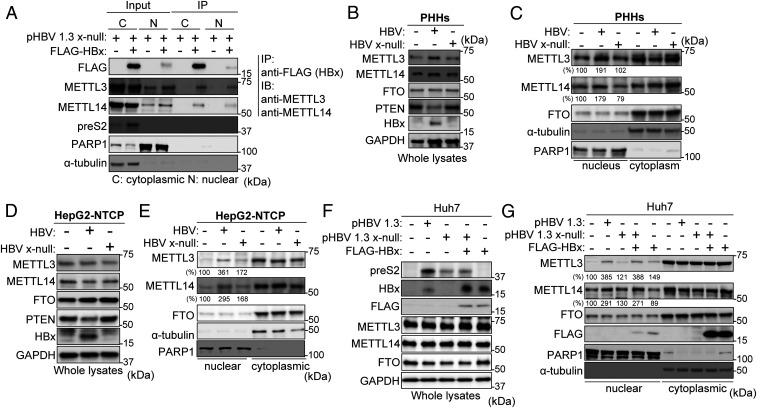
Because HBx protein did not affect METTL3/14 expression levels in HBV DNA-transfected cells or ectopic HBx expression plasmid-transfected cells (SI Appendix, Fig. S2 C and I), we investigated the interaction between HBx and METTL3/14 to determine how HBx protein affects m6A methyltransferase activity to induce m6A modification of viral transcripts and PTEN mRNA. Using the coimmunoprecipitation strategy, we show that FLAG-tagged HBx specifically interacted with the endogenous METTL3/14 (SI Appendix, Fig. S5 A–C). We further analyzed HBx interactions with m6A methyltransferases in both the nucleus and cytoplasm lysates because METTL enzymes are expressed both in the nucleus and cytoplasm and the nuclear localization of METTL3/14 is important in m6A modification processing. We conducted the coimmunoprecipitation experiments using both nuclear and cytoplasmic fractions from the HBV x-null–transfected Huh7 cells in which FLAG-tagged HBx was ectopically expressed (Fig. 4A and SI Appendix, Fig. S5D). We observed that HBx protein interacts with endogenous METTL3/14 in both nuclear and cytoplasmic fractions during HBV replication. Interestingly, HBx protein expression in HBV x-null DNA-transfected cells modestly induced METTL3 and METTL14 expression levels in the nuclear fraction (input samples; Fig. 4A and SI Appendix, Fig. S5D). We confirmed these results using HBV infection systems. Increased levels of nuclear METTL3/14 were observed in HBV WT-infected PHHs and HepG2-NTCP cells compared to HBx-defective HBV infection (Fig. 4 B–E). Notably, ectopic HBx expression restored induced METTL3/14 levels in the nucleus during HBV x-null replication, while HBx expression in non-HBV replicating cells did not induce nuclear localization of METTL3/14 (Fig. 4 F and G and SI Appendix, Fig. S5E), suggesting that HBx protein expression induces nuclear import of METTL3/14 only during HBV replication. Taken together, these results demonstrate that HBx protein interacts with METTL3/14, which promotes recruitment onto the HBV cccDNA and PTEN chromosomal locus during HBV infection.
Fig. 4.
HBx protein interacts with METTL3 and 14 and stimulates their nuclear import. (A) Huh7 cells, which were transfected with pHBV 1.3 x-null, were treated with control or pcDNA 3.1 FLAG-HBx. Cytoplasm and nuclear lysates were immunoprecipitated with anti-FLAG, followed by immunoblotting for the indicated proteins. (B–E) PHHs were infected with HBV WT or x-null particles for 10 d (B and C). HepG2-NTCP cells were infected with the same amount of virus particles of HBV WT or x-null for 24 h and then further incubated for 9 d (D and F). The HBV-infected PHHs and HepG2-NTCP cells were harvested to assess immunoblotting from whole lysates (B and D) or isolated nuclear and cytoplasmic biochemical fractions (C and E). (F and G) Huh7 cells were transfected with pHBV 1.3 or HBV 1.3 x-null together with pcDNA3.1 FLAG-HBx or pcDNA3.1 (control). The indicated proteins were analyzed by immunoblotting (F). Immunoblot analysis of isolated nuclear and cytoplasmic biochemical fraction from indicated cells (G). In C, E, and G, the METTL3 and 14 expression levels relative to FTO from three independent experiments were quantified using ImageJ.
Nuclear HBx Is Required for m6A Modification.
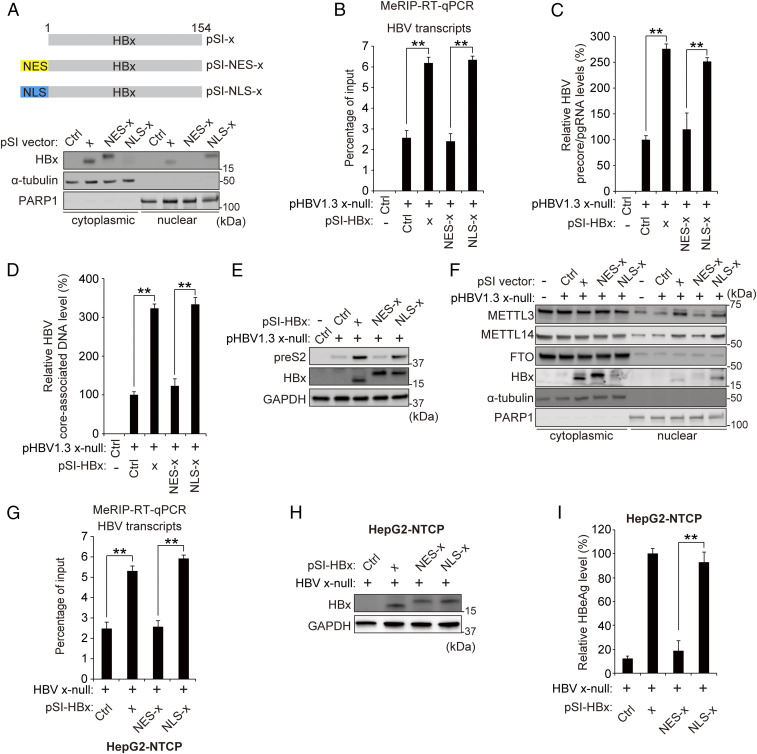
Slagle and colleagues reported that nuclear HBx was necessary for its functional role in viral replication and gene expression (5, 27). Using the HBx expression plasmids carrying an NES or NLS (Fig. 5A), we investigated whether HBx nuclear import affects m6A methylation of HBV RNAs and PTEN mRNA. NLS-tagged HBx expression increased m6A modification of the viral transcripts and PTEN mRNA, but NES-tagged HBx expression failed to induce m6A modification (Fig. 5B and SI Appendix, Fig. S6). HBV precore/pgRNA and HBV core-associated DNA levels were rescued by NLS-tagged HBx expression, but not NES-HBx, as shown previously (Fig. 5 C–E). We further confirmed these results in HBV-infected HepG2-NTCP cells and observed similar results (Fig. 5 G–I). These results suggest that the nuclear localization of HBx induces the m6A modifications of viral transcripts and host mRNA, which in turn has a bearing on RNA stability. Furthermore, we analyzed whether nuclear localization of HBx affects the subcellular localization of m6A methyltransferases. We found that the nuclear imports of METTL3/14 were induced by NLS-tagged HBx expression during HBV replication, but not NES-tagged HBx expression (Fig. 5F). These data also indicate that nuclear HBx induces METTL3/14 nuclear import.
Fig. 5.
Nuclear localization of HBx is required for inducing m6A modification of viral transcripts. (A) Subcellular localization of HBx, NES-fused HBx, or NLS-fused HBx protein transiently expressed in Huh7 cells for 48 h was analyzed by immunoblotting. (B–E) Huh7 cells were transfected with HBV 1.3 x-null together with pSI-x, pSI-NES-x, or pSI-NLS-x for 48 h prior to quantification of MeRIP qRT-PCR (B), precore/pgRNA (C), core-associated DNA (D), or immunoblotting analysis for the indicated proteins (E). (F) Analysis of m6A methyltransferase levels in the cytoplasmic or nuclear fraction of HBV 1.3 x-null–expressed Huh7 cells, which were cotransfected with pSI-x, pSI-NES-x, or pSI-NLS-x. The indicated proteins were analyzed from cytoplasmic and nuclear lysates by immunoblotting. (G–I) HepG2-NTCP cells were infected with HBV WT or x-null particles. After 7 d, cells were transfected with pSI-x, pSI-NLS-x, or pSI-NES-x plasmid for 3 d. Cells were harvested to assess m6A methylation levels of HBV RNAs and PTEN mRNA (G) or for immunoblotting analysis for the indicated proteins (H) or HBeAg levels (I). In all panels, data are mean ± SD (**P < 0.01 by unpaired one-tailed Student’s t test).
Generally, m6A RNA methylation enzymes are localized both in the cytoplasm and nucleus (14). In cancer cells and primary cells, these have been found in both locations (Fig. 4). However, the functional significance of their cytoplasmic location remains to be characterized. Herein, we investigated whether cytoplasmically localized m6A methyltransferases may modify HBV transcripts and host RNA. MeRIP qRT-PCR analysis using nuclear or cytoplasmic RNA from HBV DNA-transfected cells was carried out (SI Appendix, Fig. S7). We observed that the absence of HBx could not induce m6A modification of viral RNAs and PTEN mRNA in either nuclear or cytoplasmic location (SI Appendix, Fig. S7 A and B). Notably, the NLS-tagged HBx expression increased m6A-methylated HBV transcripts and PTEN mRNA in both nuclear and cytoplasmic fractions, but NES-tagged HBx did not further stimulate m6A modification viral transcripts (SI Appendix, Fig. S7C). These results suggest that HBV transcripts are m6A-modified in the nucleus and that the cytoplasmic HBx (pSI-NES transfections) did not additionally induce m6A modification of HBV transcripts. Similarly, m6A-methylated PTEN RNA is increased by only NLS-tagged HBx expression (SI Appendix, Fig. S7C), suggesting that nuclear-localized HBx induces m6A modification of PTEN mRNA.
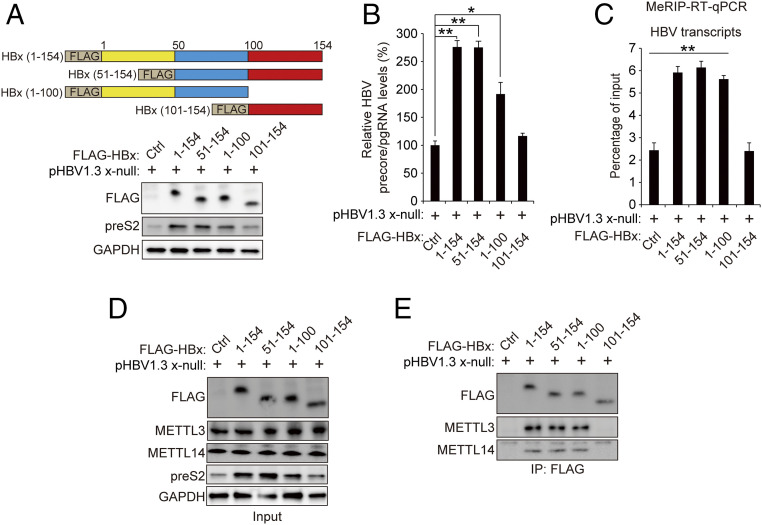
The HBx aa 50 to 100, Containing Signal Transduction and Transactivation Domains, Are Required for the Up-regulation of m6A Modification via Interaction with METTL3/14.
HBx protein consists of 154 amino acids (aa). HBx protein has been characterized to contain four functional domains, including the transrepression domain (aa 1 to 20), the signal transduction domain (aa 58 to 119), the transactivation domain (aa 58 to 140), and the nuclear transactivation domain (aa 120 to 140) (28). HBx C-terminal domain (aa 51 to 154) is required for transactivation of viral and cellular promoters, and the regions spanning aa 52 to 65 and aa 88 to 154 play important roles in stimulatory function in HBV transcription (28). Here, we tested which domain of HBx affects m6A modification of HBV RNAs using FLAG-tagged HBx truncation expression plasmids (Fig. 6 A, Top). We found that the full-length HBx protein (1 to 154) and its truncation mutants (51 to 154) and (1 to 100) increased HBV RNAs and preS2 expression levels (Fig. 6 A and B). In addition, HBx (51 to 154) and HBx (1 to 100) expressions induced the m6A-methylated HBV transcripts to levels similar to the full-length HBx-expressing cells (Fig. 6C). In contrast, the other truncation HBx mutant (101 to 154) failed to increase HBV replication and m6A modification of HBV RNAs. These results identify that aa 51 to 100 of HBx are essential for the role of HBx in modulating m6A modification. We further investigated whether aa 51 to 100 of HBx are required for interaction with METTL3/14 (Fig. 6 D and E). Coimmunoprecipitation results showed that FLAG-HBx protein (1 to 154) and its truncation mutants HBx (51 to 154) and HBx (1 to 100) interact with METTL3/14 complex, while HBx (101 to 154) did not interact with METTL3/14. These results suggest that aa 51 to 100 of HBx are necessary for interaction with METTL3/14 to induce the m6A modification of HBV transcripts.
Fig. 6.
HBx amino acids 51 to 100 are essential for interaction with METTL3/14 and regulating m6A modification of HBV RNAs. (A–C) Huh7 cells, which expressed pHBV 1.3 x-null, were transfected with control, pcDNA 3.1 FLAG-HBx, or HBx truncated mutant plasmids. Cells were harvested to assess immunoblotting analysis for the indicated proteins (A), HBV precore/pgRNA levels (B), or m6A methylation levels of HBV RNAs (C). (D and E) Huh7 cells, which were transfected with pHBV 1.3 x-null, were treated with control; truncated HBx 50 to 154, 1 to 100, or 100 to 154; or HBx plasmid. The indicated proteins were analyzed by immunoblotting (D). Lysates were immunoprecipitated with anti-FLAG, followed by immunoblotting for the indicated proteins (E). In B and C, data are mean ± SD (*P < 0.05; **P < 0.01; ***P < 0.001; unpaired one-tailed Student’s t test).
Discussion
The cotranscriptional deposition of m6A on the transcripts has been highlighted via the H3K36me3 histone modification, a marker of transcription elongation globally guiding m6A modification (15). This study implicates HBx protein in the control of m6A modification of RNAs and its direct role in binding to methyltransferases and bringing them to the sites of transcription initiation (SI Appendix, Fig. S8). Our results clearly establish that HBx, a regulatory protein encoded by the HBV genome, binds and recruits METTL3/14 enzymes onto the viral cccDNA template organized as HBV minichromosome as well as host chromosome loci (Fig. 2). In the absence of HBx, RNA methylation of HBV transcripts did not occur (Fig. 1). This lack of methylation enhances their stability and led to an increase in viral proteins (Fig. 3). However, the pgRNA that is encapsidated within the core particle to program RT of RNA into DNA was severely affected (Fig. 1J). m6A DRACH motif located in the lower stem of the 5′ epsilon structure containing the RT priming site upstream in the bulge structure aids in the RT activity (21). Mutation of the DRACH motif, which locally distorts the helical structure, also affects RT activity of the core-associated pgRNA. A compensatory mutation that restores the helical structure of the mutant genome did not reverse the RT defect, clearly suggesting the impact of the DRACH motif on the priming activity of the RT enzyme (21). How m6A methylation affects RT priming activity remains to be characterized. The role of HBx protein in affecting m6A methylation occurring by recruiting the METTL3/14 complex onto the HBV episomal minichromosome supports the view of its function in regulating the viral life cycle in addition to its transactivating function and those affecting Smc5/6 complex and HBx-DDB–mediated degradation activities (29, 30). Of relevance in this context are previous major findings that HBx directly binds to transcription factors and was found associated with the viral cccDNA, respectively; these form the basis of the various ways HBx may achieve these transactivating functions of gene expression (4, 7, 9, 10). Thus, these studies provide a platform for the various ways HBx may achieve these transactivating functions of gene expression. But the unique property of HBx reported here in inducing RNA methylation is distinguished from its widely known conventional function of being a transcriptional transactivator. The pivotal role of HBx in maintaining the regulation of the viral life cycle has prompted consideration as a therapeutic target in the quest for schemes to eliminate cccDNA and/or its transcription function. This report further reinforces the role of HBx in the viral life cycle as a central player. As previously noted, the overall role of m6A methylation in the HBV life cycle, seemingly to regulate the HBV gene expression, in which the site of this modification, which exists only once in the genome identified by the currently available methods used, renders the viral transcripts less stable but positively regulates RT activity of the pgRNA (21). This additional layer of regulation may dictate events of chronic hepatitis B pathogenesis.
In addition to its role as a viral protein to activate the HBV life cycle, HBx protein is also known to be involved in immune evasion and the development of hepatocellular carcinoma (HCC) (31). The tumor-suppressor activity of PTEN is well characterized in PTEN-knockout mice (32). Thus, our results imply that the HBx protein may contribute to HCC development by increasing m6A modification of PTEN mRNA to destabilize RNA stability. Decreased PTEN expression resulting from HBx expression during HBV infection may in part explain the oncogenic phenotype previously reported in transgenic mice expressing the HBx protein. We recently reported that modification of HBV pgRNA inhibits RIG-I signaling (33). m6A-modified motif of HBV RNA was also recently shown to be the target of interferon-mediated decay of viral RNA (34). In light of these results, HBx protein may contribute to HCC development by increasing m6A modification of PTEN mRNA to destabilize RNA stability and may affect the RIG-I signaling pathway via regulating m6A modification of pgRNA.
Despite an effective HBV vaccine and partially effective antivirals, elimination of cccDNA and its functions remains a challenge in curing chronic hepatitis B infection worldwide (35). The work presented here highlights an additional role of HBx in directly affecting the schemes of cotranscriptional synthesis/modification of viral RNA via its interaction with METTL3/14 enzymes and recruitment to the HBV cccDNA template and chromosomal loci. These studies offer new avenues for possible therapeutic intervention of the activities of cccDNA aimed at its clearance from infected cells.
Materials and Methods
Plasmids, Antibodies, and Reagents.
HBV 1.3-mer (no. 65462), HBV 1.3-mer x-null (no. 65461), and pcDNA3.1-FLAG-HBx (no. 42596) were obtained from Addgene. pcDNA3.1-FLAG-HBx (51 to 154), (1 to 100), and (101 to 154) were constructed by recombinational cloning from pcDNA3.1-FLAG-HBx. pSI-X, pSI-NES-X, and pSI-NLS-X were a gift from Betty L. Slagle (Baylor College of Medicine, Houston, TX). Antibodies were obtained as follows: anti-m6A antibody from Synaptic Systems, anti-HBx antibody from BioVendor, anti-preS2 and anti-GAPDH antibodies from Santa Cruz Biotechnology, anti-METTL3 antibody from Proteintech Group, anti-METTL14 antibody from Sigma-Aldrich, anti-FTO antibody from Abcam, and anti-FLAG, anti–α-tubulin, anti-PARP1, and anti-PTEN antibodies from Cell Signaling Technology. Anti-HBx and preS2 antibodies were diluted at a 1:200 ratio in 5% BSA buffer for immunoblotting. The other antibodies were used at a 1:1,000 ratio in 5% BSA buffer for immunoblotting. The ON-TARGET plus siRNAs of METTL3 (L-005170-02-0005) and METTL14 (L-014169-02-0005) were obtained from Dharmacon.
Cell Culture and Transfection.
HepG2 and Huh7 cells were obtained from American Type Culture Collection. The HepG2-NTCP cells were provided by Wenhui Li (National Institute of Biological Sciences, Beijing, China). Huh7 cells were cultured with Roswell Park Memorial Institute medium (Gibco). The HepG2-NTCP and HepG2 cells were grown in Dulbecco’s modified Eagle’s medium (Gibco). Both media were supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin, and 0.1 mM nonessential amino acid under standard culture conditions (5% CO2, 37 °C). The HepG2-NTCP cells were treated with G418 (geneticin 400 µg/mL; Sigma-Aldrich) during maintenance. PHHs were purchased from Gibco and cultured according to the manufacturer’s protocol. HepG2 and Huh7 cells were transfected with plasmids using Mirus TransIT-LT1 according to the manufacturer’s protocol. siRNAs were formulated with Lipofectamine RNA iMAX (Invitrogen) for transfecting into cells according to the manufacturer’s instructions.
Virus Production and Cell Infection.
HBV wild type and HBV HBx-deficient particles were harvested from the supernatants of Huh7 cells transfected with HBV 1.3-mer plasmid or HBV 1.3-mer x-null plasmid. The culture medium was centrifuged at 4 °C, 10,000 × g for 15 min. The clarified supernatants were incubated with 5% polyethylene glycol (PEG) 8000 overnight at 4 °C and then centrifuged at 4,000 rpm for 30 min at 4 °C. Pellet was redissolved in a serum-free culture medium at 1% volume of the original supernatant. For infection, the HepG2-NTCP and PHHs were split in collagen-coated plates and incubated for 24 h with HBV particles, which are diluted in a serum-free culture medium with 4% PEG 8000 and 2% dimethyl sulfoxide (DMSO). After incubation with HBV particles, the cells were washed with culture medium. Cells were incubated for 10 d, with medium changed every 2 d, in medium containing 2% DMSO.
Real-Time qRT-PCR.
Total RNA was isolated using TRIzol reagent (Invitrogen). The cDNAs were synthesized from extracted total RNA using iScript Reverse Transcription Supermix (Bio-Rad). qPCR was assessed with Ssoadvanced Universal SYBR Green supermix (Bio-Rad). Each viral RNA and mRNA expression level, normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH), was analyzed using the ΔΔCt method. The primers for qRT-PCR are shown in SI Appendix, Table S1.
MeRIP Sequencing.
For MeRIP sequencing, three independent RNA samples were prepared. PolyA RNA was isolated from total RNA by using a polyA Spin mRNA isolation kit (New England Biolabs). The isolated polyA RNA was fragmented using Ambion RNA fragmentation reagent and then heated to 75 °C for 5 min and placed on ice for 3 min. The fragmented RNA was incubated with anti-m6A antibody (Synaptic Systems) conjugated to Protein G Dynabeads (Thermo Fisher Scientific) in MeRIP buffer (50 mM Tris⋅HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, and 0.1% Nonidet P-40) overnight at 4 °C. RNA–bead complexes were washed with MeRIP buffer five times, and bound RNA was eluted in MeRIP buffer containing 6.7 mM m6A 5′-monophosphate sodium salt (Sigma). Eluted RNA was isolated using the RNeasy mini kit (Qiagen). The cDNA libraries were synthesized from this RNA, as well as input RNA, for Illumina sequencing using a TruSeq RNA sequencing kit (Illumina). The deep-sequencing libraries were sequenced to 1 × 50 base-pair reads on an Illumina HiSeq2500. The sequencing data were aligned to a combined human (hg38) using Spliced Transcripts Alignment to a Reference. Mean coverage was plotted for all three replicates using CovFuzze (https://github.com/al-mcintyre/covfuzze). The raw data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database and are accessible through GEO Series accession number GSE114486. MeRIP qRT-PCR followed the same protocol, except that total RNA was not fragmented. Eluted RNA was reverse-transcribed into cDNA and subjected to qRT-PCR.
Western Blotting and Immunoprecipitation.
Cell lysates were prepared in a Nonidet P-40 lysis buffer (1% Nonidet P-40, 50 mM Tris⋅HCl, pH 8.0, 150 mM NaCl) supplemented with a protease inhibitor (Thermo Fisher Scientific). FLAG-tagged HBx protein was immunoprecipitated using anti–FLAG-M2 Magnetic Beads (Sigma-Aldrich) from cell lysates following incubation for 2 h on a rotator at 4 °C. Immunoprecipitates or lysates were resolved by sodium dodecyl sulfate/polyacrylamide gel electrophoresis and blotted onto a nitrocellulose membrane (Bio-Rad). Membranes were blocked with 5% BSA for 1 h 30 min and then with primary antibodies diluted in 5% BSA overnight at 4 °C. After washing, the membranes were incubated with secondary antibody-conjugated horseradish peroxidase (Cell Signaling Technology) for 1 h. Antibody complexes were detected using a chemiluminescence substrate (Millipore). Chemiluminescence signals were detected using the ChemiDoc MP Imaging System (Bio-Rad).
Subcellular Fractionation.
Cell pellets were harvested with trypsin/EDTA solution (Gibco) and washed twice with ice-cold PBS. Cytoplasmic and nuclear protein fractions were isolated using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific) according to the manufacturer’s protocol.
ChIP.
Before harvesting, cells were treated with 1% formaldehyde for 10 min at room temperature and then added to 125 mM glycine solution for 5 min. Cells were washed twice with ice-cold PBS. Chromatin fractions were prepared using a Pierce Chromatin Prep Module (Thermo Fisher Scientific) according to the manufacturer’s protocol. Cross-linked chromatin was incubated with antibodies specific to HBx, METTL3, and METLL14 for 16 h at 4 °C. Relative IgG was used in each experiment as negative control. Immunoprecipitated chromatin was analyzed by semiquantitative PCR and real-time PCR.
Isolation of Core Particles.
Cells were treated with 1 mL of transfection lysis buffer (50 mM Tris⋅HCl, pH 8.0, 1 mM EDTA, and 1% Nonidet P-40) supplemented with a protease inhibitor (Thermo Fisher Scientific) at 37 °C for 10 min. The lysates were centrifuged for 1 min at 14,000 rpm, and then the supernatants were transferred to a new centrifuge tube. The supernatants were incubated with 5 mM CaCl2 and 75 U micrococcal nuclease (New England Biolabs) for 45 min at 37 °C. After centrifugation, 75 U micrococcal nuclease was added to the supernatant again and incubated for 45 min at 37 °C. Then, the supernatants were centrifuged for 1 min at 14,000 rpm and transferred to a new centrifuge tube, and 32 µL of 0.5M EDTA and 260 µL of 35% PEG in 1.75 M NaCl was added and kept at 4 °C for 1 h. After centrifugation at 13,000 rpm for 5 min at 4 °C, the pellet was resuspended in 300 µL TNE buffer.
HBeAg ELISA.
HBeAg levels in HBV-infected PHHs and HepG2-NTCP cells were determined using a commercial ELISA kit (Cusabio) according to the manufacturer’s protocol.
Statistical Analysis.
Statistical analyses were performed using GraphPad Prism 6.01. All results are representative of three independent experiments. For each result, error bars represent the SD from at least three independent experiments. The P value was calculated using a one-tailed unpaired Student’s t test.
Supplementary Material
Acknowledgments
We thank Dr. Betty L. Slagle (Baylor College of Medicine, Houston, TX) for the generous gift of NES and NLS HBx plasmids. This work was supported by NIH Grants AI125350 and AI139234 (to A.S.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2019455118/-/DCSupplemental.
Data Availability.
All data that support the findings of this study are available in the manuscript and supporting information. Source data for Figs. 1–6 and SI Appendix, Table S1 and Figs. S1–S8 are provided with the paper.
References
- 1.Seeger C., Zoulim F., Mason W., Hepadnaviruses, in Fields Virology, Knipe D. M., Howley P.M., Eds. (Lippincott Williams & Wilkins, Philadelphia,, ed. 6, 2013), pp. 2185–2221. [Google Scholar]
- 2.de Martel C., Maucort-Boulch D., Plummer M., Franceschi S., World-wide relative contribution of hepatitis B and C viruses in hepatocellular carcinoma. Hepatology 62, 1190–1200 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aufiero B., Schneider R. J., The hepatitis B virus X-gene product trans-activates both RNA polymerase II and III promoters. EMBO J. 9, 497–504 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouchard M. J., Schneider R. J., The enigmatic X gene of hepatitis B virus. J. Virol. 78, 12725–12734 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slagle B. L., Bouchard M. J., Hepatitis B virus X and regulation of viral gene expression. Cold Spring Harb. Perspect. Med. 6, a021402 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slagle B. L., et al. , Technical standards for hepatitis B virus X protein (HBx) research. Hepatology 61, 1416–1424 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belloni L., et al. , Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc. Natl. Acad. Sci. U.S.A. 106, 19975–19979 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balsano C., et al. , Hepatitis B virus X gene product acts as a transactivator in vivo. J. Hepatol. 21, 103–109 (1994). [DOI] [PubMed] [Google Scholar]
- 9.Maguire H. F., Hoeffler J. P., Siddiqui A., HBV X protein alters the DNA binding specificity of CREB and ATF-2 by protein-protein interactions. Science 252, 842–844 (1991). [DOI] [PubMed] [Google Scholar]
- 10.Oropeza C. E., et al. , The regulation of HBV transcription and replication. Adv. Exp. Med. Biol. 1179, 39–69 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Qadri I., Conaway J. W., Conaway R. C., Schaack J., Siddiqui A., Hepatitis B virus transactivator protein, HBx, associates with the components of TFIIH and stimulates the DNA helicase activity of TFIIH. Proc. Natl. Acad. Sci. U.S.A. 93, 10578–10583 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roundtree I. A., Evans M. E., Pan T., He C., Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yue Y., Liu J., He C., RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 29, 1343–1355 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi H., Wei J., He C., Where, when, and how: Context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell 74, 640–650 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang H., et al. , Histone H3 trimethylation at lysine 36 guides m6A RNA modification co-transcriptionally. Nature 567, 414–419 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du H., et al. , YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 7, 12626 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer K. D., Jaffrey S. R., Rethinking m6A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 33, 319–342 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gokhale N. S., et al. , N6-Methyladenosine in flaviviridae viral RNA genomes regulates infection. Cell Host Microbe 20, 654–665 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzales-van Horn S. R., Sarnow P., Making the mark: The role of adenosine modifications in the life cycle of RNA viruses. Cell Host Microbe 21, 661–669 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lichinchi G., et al. , Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat. Microbiol. 1, 16011 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imam H., et al. , N6-methyladenosine modification of hepatitis B virus RNA differentially regulates the viral life cycle. Proc. Natl. Acad. Sci. U.S.A. 115, 8829–8834 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu M., et al. , N6-methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG-I. Nat. Microbiol. 5, 584–598 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim G. W., et al. , HBV-induced increased N6 methyladenosine modification of PTEN RNA affects innate immunity and contributes to HCC. Hepatology, 10.1002/hep.31313 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X., et al. , N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lucifora J., et al. , Hepatitis B virus X protein is essential to initiate and maintain virus replication after infection. J. Hepatol. 55, 996–1003 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Shi H., et al. , YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 27, 315–328 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keasler V. V., Hodgson A. J., Madden C. R., Slagle B. L., Hepatitis B virus HBx protein localized to the nucleus restores HBx-deficient virus replication in HepG2 cells and in vivo in hydrodynamically-injected mice. Virology 390, 122–129 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murakami S., Cheong J. H., Kaneko S., Human hepatitis virus X gene encodes a regulatory domain that represses transactivation of X protein. J. Biol. Chem. 269, 15118–15123 (1994). [PubMed] [Google Scholar]
- 29.Decorsière A., et al. , Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor. Nature 531, 386–389 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Minor M. M., et al. , Hepatitis B virus HBx protein mediates the degradation of host restriction factors through the Cullin 4 DDB1 E3 ubiquitin ligase complex. Cells 9, 834 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang C., et al. , Hepatitis B virus X (HBx) induces tumorigenicity of hepatic progenitor cells in 3,5-diethoxycarbonyl-1,4-dihydrocollidine-treated HBx transgenic mice. Hepatology 55, 108–120 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Carnero A., Paramio J. M., The PTEN/PI3K/AKT pathway in vivo, cancer mouse models. Front. Oncol. 4, 252 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim G. W., Imam H., Khan M., Siddiqui A., N6-Methyladenosine modification of hepatitis B and C viral RNAs attenuates host innate immunity via RIG-I signaling. J. Biol. Chem. 295, 13123–13133 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Imam H., Kim G. W., Mir S. A., Khan M., Siddiqui A., Interferon-stimulated gene 20 (ISG20) selectively degrades N6-methyladenosine modified Hepatitis B Virus transcripts. PLoS Pathog. 16, e1008338 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu J., Protzer U., Siddiqui A., Revisiting Hepatitis B., Revisiting hepatitis B virus: Challenges of curative therapies. J. Virol. 93, e01032-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data that support the findings of this study are available in the manuscript and supporting information. Source data for Figs. 1–6 and SI Appendix, Table S1 and Figs. S1–S8 are provided with the paper.