Abstract
The COVID-19 pandemic is a shocking reminder of how our world would look in the absence of vaccination. Fortunately, new technologies, the pace of understanding new and existing pathogens, and the increased knowledge of the immune system allow us today to develop vaccines at an unprecedented speed. Some of the vaccine technologies that are fast-tracked by the urgency of COVID-19 may also be the answer for other health priorities, such as antimicrobial resistance, chronic infections, and cancer, that the post-COVID-19 world will urgently need to face. This perspective analyzes the way COVID-19 is transforming vaccinology and the opportunities for vaccines to have an increasingly important role in health and well-being.
Keywords: COVID-19, vaccinology, vaccines
The path toward fully synthetic vaccines made using genomic information started in 2013. Sunday, March 31, 2013, was a nice Easter festivity when the World Health Organization (WHO) was notified about a new H7N9 avian influenza virus that had infected three people in China and killed two of them (1). It was a new, potentially pandemic, virus for which the world was not prepared. The experience of the H2N2 in 1957, the H3N2 in 1968, and even of the H1N1 pandemic in 2009 had shown that vaccines had become available only after the pandemic peak, and therefore they were too late to be useful. On Monday, April 1, 2013, scientists at the J. Craig Venter Institute in San Diego, CA, accessed the sequence of the hemagglutinin and neuraminidase genes posted by the Chinese Center for Disease Control and Prevention on the Global Initiative for Sharing All Influenza Data system and used the enzymatic isothermal assembly method with self-error correction for the cell-free synthesis of the two genes (2). The synthetic genes were then shipped overnight from California to Massachusetts. There, scientists from Novartis Vaccines used the synthetic genes to generate, in only 5 d, a synthetic influenza virus seed ready for vaccine manufacturing. In addition, they produced an RNA vaccine ready for animal immunization in the record time of 1 wk (3). Fortunately, the H7N9 influenza virus did not transmit efficiently between humans, and, although it caused a few hundred cases during the next few years, it did not cause a pandemic, limiting the use of these vaccines only to clinical trials.
Anticipated by the work of Craig Venter (4), teleportation of DNA code through great distances was not Star Trek’s fiction anymore. For the first time, a fully synthetic viral vaccine was developed by in vitro cell-free synthesis of genes using the genomic sequence that had been teleported across the planet at the speed of light via the Internet. The process of teleporting the genomic sequence has the ambition to change forever the old—and dangerous—way we used to make viral vaccines by shipping viruses across the world. We use the term “Internet-based vaccines” to describe this new way of making vaccines using the Internet to share the genomic information, without the need to transport, access, and grow the real virus.
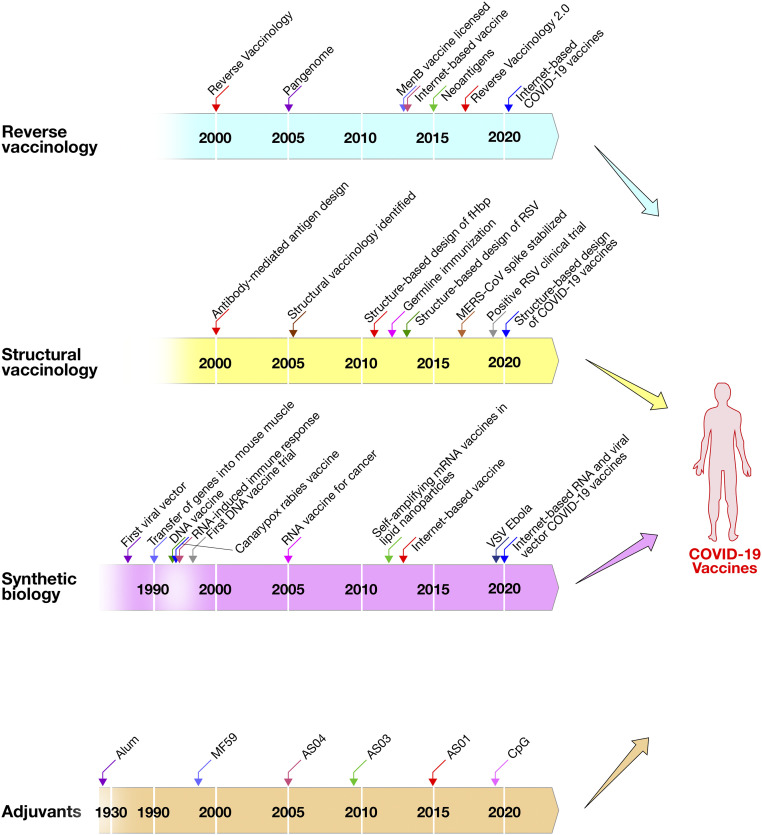
When, in January 2020, scientists from Fudan University and their collaborators posted on the Internet the genomic sequence of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), responsible for the current COVID-19 pandemic, most of the laboratories across the world were ready for the challenge. They not only had the technology to make vaccines starting from synthetic genes, but some of them could also use computer modeling of the atomic structure of the spike protein of similar coronaviruses to design, up front, an antigen stabilized in the prefusion conformation (5, 6). Synthetic genes were used to rapidly start the development of more than 200 different vaccines. The remarkable quality and speed used for COVID-19 vaccine development was possible because the scientists combined, for the first time, three decades of scientific progress in independent fields: reverse vaccinology, structural vaccinology, synthetic biology, and vaccine adjuvants (Fig. 1). The advances in antigen selection and design (reverse and structural vaccinology) together with the use of innovative synthetic platforms such as nucleic acid vaccines (RNA and DNA based), viral vectors, and the availability of licensed adjuvants allowed for an unprecedented speed in the discovery of several COVID-19 vaccine candidates, many of which were already in clinical development stage.
Fig. 1.
Technological advances that merged to develop a COVID-19 vaccine.
Technologies Used for COVID-19 Vaccine Development
Reverse vaccinology, structural vaccinology, synthetic biology, and vaccine adjuvants, that so far had been used independently to develop vaccines, were combined in an unprecedented worldwide effort to design and develop COVID-19 vaccines.
Reverse vaccinology, the science that identifies vaccine antigens from the genome of pathogens, was used for the first time in 2000 to identify novel antigens for vaccine against meningococcus B, which, up to that moment, had been an impossible task for conventional technologies (7, 8). The vaccine was licensed by the European Medicines Agency in 2013 and by Food and Drug Administration in 2015 and was recently shown to reduce by 74% the incidence of disease in United Kingdom and by 91% in Italy (9, 10). During the last two decades, genomics has been used in the development of most vaccines, exploiting the pangenome of bacterial and viral species. Remarkable progress in genome-based vaccines was made in 2013 when an RNA vaccine against a potentially pandemic H7N9 influenza virus was produced in 1 wk without culturing the virus but using the genome sequence available in public databases (3). During the last few years, tumor immunologists used the genome of cancer cells to identify mutations coding for neoantigens to be incorporated in cancer vaccines (11).
Structural vaccinology, or structure-based antigen design, was predicted as an emerging field in 2007 when it became clear that high-throughput structure determination was going to be possible in the near future (12). However, it had already been anticipated in 2002 that the study of antibodies recognizing protective epitopes was going to inform vaccine design (13). The first example, published in 2011, was the design of a single meningococcal antigen containing the epitopes of three antigenic variants of the same molecule (14). In 2013, structure-based vaccine design was used for the first time to develop a vaccine that had been impossible for other technologies, when McLellan et al. (15, 16) described the stabilization of the Respiratory Syncytial virus (RSV) Fusion (F) protein in the prefusion conformation. In 2019, the prefusion stabilized F protein was shown to induce unprecedented levels of neutralizing antibodies and to be ready for phase III clinical trials (17). In 2013, structure-based design was also used for germline immunization to generate broadly neutralizing antibodies against HIV (18). Finally, in 2015, structural vaccinology was used to stabilize the spike protein of the Middle East respiratory syndrome-related coronavirus (MERS-CoV) in the prefusion conformation (5). In 2017, a perspective in the Journal of Experimental Medicine predicted the merging of reverse and structural vaccinology and named it reverse vaccinology 2.0 (19).
Synthetic biology is the ability to use synthetic genes for vaccination or cancer therapy. It was pioneered in 1986 by the use of a cloned gene into a viral vector for gene therapy (20), and, in 1992, by the cloning of the glycoprotein gene of rabies virus into a canarypox viral vector for the development of a rabies vaccine (21). In parallel, it was shown that protein expression could be achieved by the direct transfer of genes into mouse muscle cells (22). This observation suggested the use of naked DNA (23, 24) and of RNA (25) for vaccination. DNA vaccination became very popular during the following decade, until it was realized that, while successful in most animal models, DNA vaccination has not been, until to date, successful in humans. The decline of DNA popularity led to the rediscovery of viral vectors and RNA at the end of the first decade of the 21st century. In this period, viral vectors became very popular and were extensively used for the rapid generation of vaccines to fight the Ebola epidemic of 2014, which led to the licensure of the first viral vector vaccine in 2019. In the meantime, the technology to make, stabilize, and deliver RNA matured in the pharmaceutical industry for the development of antisense RNA therapeutics. This technology, which employed delivery of RNA using lipid nanoparticles, was transferred to vaccines and allowed the efficient delivery of RNA vaccines (26) and the rapid development of fully synthetic RNA vaccines in 1 wk against an emerging pathogen (3). During the last few years, the production and clinical testing of RNA vaccines and viral vectors increased exponentially so that both technologies were ready to tackle the SARS-CoV-2 pandemic.
Adjuvants are substances added to vaccines to increase their potency. Aluminum phosphate or aluminum hydroxide has been used since the 1920s (27). MF59, the first modern adjuvant, was licensed in 1997 to improve an influenza vaccine (28). Since then, several novel adjuvants have been licensed and used in millions of people. The other adjuvants licensed today are AS03, AS04, AS01, and CpG oligonucleotides which are used for pandemic influenza, papillomavirus, herpes zoster, and hepatitis B, respectively (29). In the clinical evaluation setting, alum, AS03, MF59, CpG, and Matrix-M are being used for COVID-19 vaccines.
In January 2020, these four technologies were used together for the development of a number of COVID-19 vaccines. A SARS-CoV-2 nucleotide sequence coding for the spike protein was derived from the genome sequence uploaded on public databases (reverse vaccinology); the synthetic gene was modified upfront to introduce the mutations previously identified to stabilize the coronavirus antigen in the prefusion form (structural vaccinology), and used for RNA and viral vector vaccines (synthetic biology). Finally, the protein-based vaccines (as stabilized recombinant trimers, viral-like particles, and nanoparticles) were combined with adjuvants. Although several SARS-CoV-2 vaccines in the clinic use other approaches such as inactivation or attenuation of the virus, the combination of these technologies and the coordinated global effort allowed for an unprecedented speed in the discovery of several COVID-19 vaccine candidates.
Vaccines for COVID-19
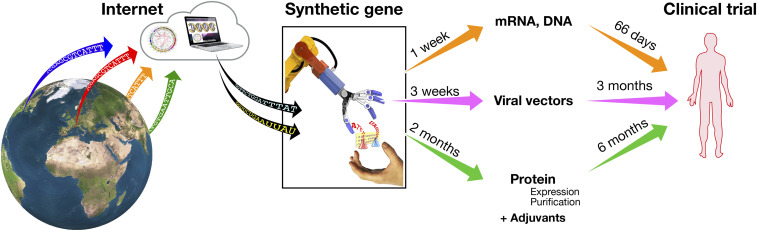
Several approaches are used to make COVID-19 vaccines, including nucleic acid-based vectors, inactivated or live attenuated viruses, recombinant proteins, and virus-like particles (30). In this manuscript, we focus on the three main categories for which Internet-based vaccines are demonstrating massive developmental acceleration: synthetic RNA vaccines, viral vectors, and adjuvanted protein-based vaccines (Fig. 2).
Fig. 2.
COVID-19 vaccines in development and their timeline to clinical testing in humans.
Synthetic RNA vaccines are the fastest to develop. A fully synthetic gene is cloned in a plasmid vector, which is then used as template for the in vitro synthesis of the RNA vaccine (31). There are two types of RNA vaccines: those encoding only the antigen and those encoding for both the antigen and the enzymatic machinery for RNA template replication following vaccination [self-amplifying RNA (26)]. Given that RNA vaccines are fully synthetic and do not need a biological phase, they were able to reach clinical trials in the record time of 66 d, to move from phase I to phase II clinical trials in less than 5 mo (32), to produce promising immunogenicity and efficacy data in humans in 10 mo (33, 34). Today, RNA vaccines are among the most promising vaccine technologies, and they will very likely be one of the most important platforms of the future. However, we need to be aware that, today, we do not have a licensed RNA vaccine yet; therefore, this type of vaccine still needs to go through the challenges of demonstrating safety, immunogenicity, and efficacy in a large human population. In addition, the manufacturing of RNA vaccines, despite being much simpler than conventional vaccines, has never been scaled up beyond the need of clinical trials, so that we have not yet developed the industrial capacity to make tens or hundreds of millions of doses. The urgency to cope with COVID-19 is providing an unprecedented opportunity to fast-track this technology and accelerate its maturation by several years.
In the case of viral vectors, the synthetic gene coding for the spike protein is inserted into one of many viruses that usually have been engineered so that they cannot replicate in the human host. The virus is then grown in culture and used to deliver the synthetic gene during vaccination. There are many viruses that can be used for this purpose. The most popular ones are adenoviruses (chimpanzee adenovirus, human adenoviruses 5 and 26), measles virus, modified vaccinia Ankara, vesicular stomatitis virus (VSV), cytomegalovirus (CMV), and others (35, 36). So far, the only licensed vaccine based on viral vectors are the Ebola vaccines based on VSV and human adenovirus 26. Although we have a long experience of clinical trials with viral vector vaccines, these vaccines have never been used in millions of people, and therefore we still need to move forward cautiously. Large-scale manufacturing capacity to produce hundreds of millions of doses is not yet present in the industry, and it is being accelerated with unprecedented public and private investments to fast-track COVID-19 vaccines. It is also important to point out that vector immunity is a concern with viral and bacterial vectors. Boosting with the same vector has limitations, and this could impact using the same vector for a different pathogen, an issue that should be carefully considered.
Protein-based vaccines are the only ones for which we have large experience. In this case, the synthetic gene coding for the spike protein, prefusion stabilized or also receptor binding domain only, is used to engineer mammalian cells, baculovirus, or plant cells to produce the recombinant protein that then is purified, combined with adjuvants, and used as vaccine. The initial phase of these vaccines involving the generation of the cell line and the purification of the protein requires more time compared to RNA or viral vector vaccines, and therefore at least 6 mo were needed before the first protein-based COVID-19 vaccine started clinical trials (37). Preliminary data on immunogenicity in humans show that these vaccines induce very high neutralizing titers which exceed those found in convalescent people. However, given the industrial and clinical experience accumulated with protein-based vaccines combined with licensed adjuvants, there is confidence that these vaccines will be well tolerated, effective, and available in large quantities.
Post−COVID-19 Health Priorities
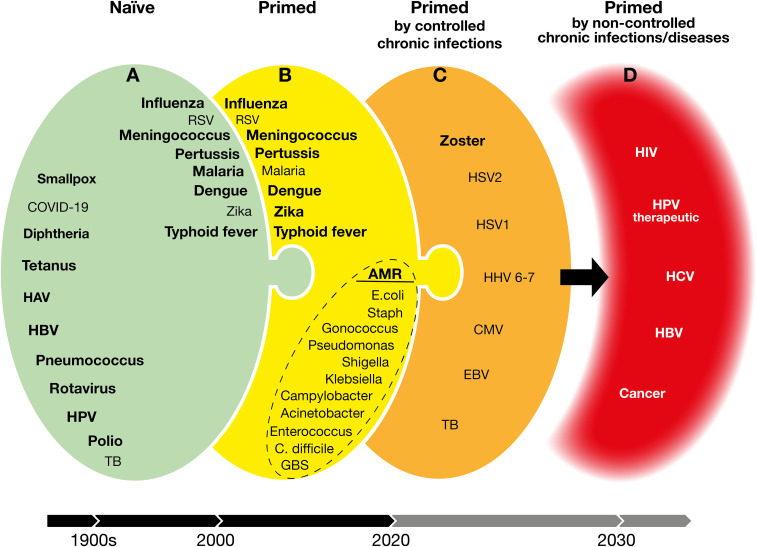
Reverse vaccinology, structure-based design, synthetic biology, and adjuvants are the tools that we have today to design vaccines that can be delivered as purified antigens, or by RNA and viral vectors. The COVID-19 pandemic has accelerated the maturation of RNA and viral vectors by at least a decade and made these new platforms available not only for emerging infections but also for the other health priorities such as antimicrobial resistance (AMR), chronic infections, and cancer that our world will need to face with urgency as soon as the COVID-19 emergency is over. To analyze the new challenges for vaccines, in Fig. 3, we divided vaccines into four groups. On the opposite sides, there are vaccines that we already have or that can be made with existing technologies (group A; Fig. 3A) and vaccines that we cannot yet approach with today’s knowledge (group D; Fig. 3D). Vaccines in groups B and C (Fig. 3 B and C) are intermediate. A closer look at these groups shows that we can divide vaccination into two big categories, depending on whether we vaccinate a naïve immune system or vaccinate an immune system that has already encountered the antigen (primed immune system).
Fig. 3.
Vaccines developed addressing naïve, previously exposed, and chronic infections. Green (A) are vaccines available or doable with existing technologies. Bold, available vaccines. Yellow (B) and orange (C) are doable vaccines with increasing challenges for today’s technologies. Red (D) are targets for which we do not yet have the scientific knowledge and technologies. HAV, hepatitis A virus; HBV, hepatitis B virus; HPV, human papillomavirus; TB, tuberculosis; RSV, respiratory syncytial virus; AMR, antimicrobial resistance; E. coli, Escherichia coli; Staph, Staphylococcus aureus; C. difficile, Clostridium difficile; GBS, group B Streptococcus; HSV1, herpes simplex virus 1; HSV2, herpes simplex virus 2; HHV, 6-7 human herpes viruses 6 and 7; CMV, cytomegalovirus; EBV, Epstein-Barr virus; HIV, human immunodeficiency virus; HCV, hepatitis C virus.
Vaccines for a Naïve Immune System.
The vaccine against smallpox developed more than two centuries ago and the vaccines in development today against COVID-19 are based on a similar principle. They both introduce, into the body, antigens that had never been seen before by the immune system, aiming at stimulating a long-term protection for a future encounter with the virus. The large majority of the vaccines in use today are also based on antigens that had never been seen before by the naïve immune system (diphtheria toxin, tetanus toxin, measles, mumps, rubella, poliomyelitis, hepatitis B, papillomavirus, and infant vaccination against influenza, pneumococcus, and meningococcus) (Fig. 3A). When these vaccines are used, the antigens are taken up by professional antigen-presenting cells and presented to naïve B and T cells which mount an adaptive immune response. An important step in this process is the formation of germinal centers where follicular T helper cells and B cells cooperate to increase the potency of the B cells specific for the new antigen, via affinity maturation of antigen-reactive antibodies. This is the textbook vaccination for which we have both mechanistic and animal models, and is the vaccinology that we study when we inject animals (mostly mice) with a variety of antigens that are new for their immune system. In most cases, we have sufficient technologies and knowledge to develop vaccines against pathogens for which the immune system is naïve. There are cases, however, where we are not yet able to make vaccines. Examples are HIV, where the virus changes so rapidly that vaccines are not effective, or malaria, where the antigenic profile is very complex, and we struggle to make effective vaccines.
Vaccines for a Primed Immune System.
Some of the vaccines described above, when delivered to adolescents, adults, or the elderly, may find an immune system that has already been exposed to the antigen, following natural infection or by other microorganisms carrying cross-reacting antigens (Fig. 3B). In this case, the immune system is not naïve any longer, and the vaccines are required to modify the preexisting immunity of antigen-experienced people. Seasonal influenza is probably the best example. In this case, we deliver a vaccine specific for a new influenza virus strain to an immune system that has already gone through the process of developing the response to the same antigen and has already generated specific memory B and T cells. The new vaccine quickly expands the preexisting memory B cells and, at the same time, triggers the expansion and affinity maturation of naïve B cells (38). However, it is clear that the first exposure to the antigen has already shaped forever the way the immune system reacts to subsequent encounters with the same antigen. This phenomenon is known as “antigenic sin” (39). Another recent example is vaccination against dengue virus. In this case, a vector-based vaccine was effective in boosting a preexisting immunity in seropositive people, while it was unable to effectively prime the naïve immune system of naïve children where it induced antibody-dependent disease enhancement, which increased the risk of hospitalization (40). Meningococcal and pneumococcal conjugate vaccines are another example (41). When they are given to naïve infants, they prime the immune system to the new antigen, and it takes at least two immunizations to have a good immune response. However, when the same vaccine is given to adolescents or the elderly, who have already been exposed to these pathogens, one dose of vaccine is sufficient to get an excellent immune response. Although there are no definitive studies in humans describing the germinal center response in this context, it is likely that the single vaccination elicits an immediate antibody response—probably by an extrafollicular transformation of memory B cells into plasma cells—and then the immune system becomes refractory to any booster immunization for a long period (as long as 2 y). In this period, more affinity maturation happens, and new memory B cells are generated. Only after that, the immune system is ready to respond to a booster immunization with a massive level of antibodies which can be as high as 10 times the response to the first immunization (41). Unfortunately, we do not have animal models able to reproduce what is described in the examples above, and we do not have a mechanistic understanding of what it takes to vaccinate an “experienced” immune system. The absence of animal models and the lack of knowledge are serious limitations for the development of new vaccines that target pathogens to which most people have already been exposed by natural infection.
A big and urgent example in this category is bacteria resistant to antibiotics and responsible for recurrent infections. AMR is a slowly evolving pandemic, with predicted catastrophic consequences for health and economy during the next 10 to 20 y (42). Vaccines can help to tackle AMR (43). We urgently need vaccines for pathogenic Escherichia coli, Staphylococcus aureus, Clostridium difficile, Klebsiella pneumoniae, Pseudomonas aeruginosa, Neisseria gonorrhoeae, Salmonella typhi, Shigella, Acinetobacter baumannii, Enterococcus faecium, and Campylobacter (Fig. 3B). Experimental vaccines against some of these pathogens are based on proteins or polysaccharides which induce normal or low response to the first vaccination when tested in naïve mice, followed by a better response to the second and third vaccinations. However, when adult volunteers were immunized with the same vaccines, a strong response was observed already after the first immunization, with no increased response to the second vaccination (at least in the short term). The main reason for this is that adult volunteers have already been colonized by these bacteria or by their relatives, and they already have memory B and T cells that recognize them and respond to vaccination. In this setting, adjuvants failed to increase the antibody response. The consequence is that, during vaccine development, in most cases, we make the choice to make a one-dose vaccine without adjuvant (44). However, we are not sure whether this is the right choice for long-term protection, and some of the vaccines failed even the primary efficacy endpoint (45). While we do not yet fully understand the mechanistics of immunizing a primed immune system, or the lack of a protective immune response that allows reinfection, we have enough technologies and empirical knowledge to develop new vaccines for AMR. Similarly, we have enough knowledge to develop vaccines for some viral diseases such as respiratory syncytial virus, dengue, and Zika viruses even in adults and the elderly, where the immune system has been usually primed by natural infection.
Vaccines for an Immune System Primed by Controlled Chronic Infections.
The difficulty of making vaccines increases when the immune system not only has already been primed by the exposure to the pathogen but somehow has already been defeated by it. The immune system has not been able to clear the pathogen, which has established a lifelong chronic infection. In some cases, once chronic infections are established, the immune system is still able to keep at bay the pathogen for most of the time. This is the case for herpes viruses (zoster, HSV1 and HSV2, EBV, and CMV) and for bacteria such as Mycobacterium tuberculosis (Fig. 3C). The pathogen establishes a latent infection and persists quietly in the body without causing disease. However, due to concomitant infections, immunosuppressive pharmacological treatments, or aging, the immune system becomes weak, and the pathogen takes over, causing disease.
Up to a few years ago, we had not a single example of a successful vaccine against chronic infections. It took us 20 y of research to start conquering some of them. The first step in this direction was the licensure of the live attenuated vaccine against herpes zoster in 2006 (46). Although this vaccine was not able to eliminate the chronic infection, it was able to keep the chronic virus silent and avoid reactivation in 60% of the cases. Recently, a new vaccine composed of a protein antigen and the potent AS01 adjuvant (a liposome containing a TLR4 agonist and a saponin) showed an efficacy of 97% against herpes zoster (47). This was followed by encouraging results against tuberculosis, where the combination of a protein antigen and the AS01 adjuvant was able to prevent reactivation and disease in 50% of the chronically infected people (48). The successful vaccines against herpes zoster and the encouraging results against tuberculosis represent an incredible milestone in the history of vaccination, because, for the first time, we have been able to make effective vaccines against chronic infections.
Vaccines for a Primed and Failed Immune System.
There are cases in which the immune system has been exposed to pathogens and has been completely defeated. Examples are chronic infections, such as HIV, papillomavirus, hepatitis C virus (HCV), hepatitis B virus (HBV), and cancer, where the immune system is not able to control the pathogen or the cancer cells, which continue to replicate forever (Fig. 3D). So far, we have not been able to make successful vaccines against these diseases, and we do not have the scientific knowledge to make them. However, even this area is not without hope, because the progress made by immunotherapy in the area of cancer has shown that the defeated immune system is characterized by dormant regulatory T cells that can be activated using antibodies against the checkpoint inhibitors, removing the constrains imposed on the immune system (49). The success of immunotherapy in the field of cancer and the increased understanding of mechanistic features of the defeated immune system suggest that, in the near future, vaccination may also be able to conquer cancer and chronic diseases.
Conclusions
The urgent need for COVID-19 vaccines has accelerated the time required to develop vaccines and the availability of powerful technologies. It is possible that evolution of the new technologies fast-tracked for COVID-19 (RNA vaccines, viral vectors, and protein-based vaccines with potent adjuvants) combined with the learning coming from immunotherapy will be the answer for some of the new challenges of modern society such as emerging infections, AMR, chronic infections, and cancer. For instance, RNA vaccines and viral vectors may be designed to encode not only antigens but also molecules able to reactivate the dormant immune system.
Acknowledgments
We thank Giorgio Corsi for the artwork and Catherine Mallia for the editorial assistance.
Footnotes
Competing interest statement: All authors are full-time employees of the GlaxoSmithKline group of companies. This work was sponsored by GlaxoSmithKline Biologicals SA.
This article is a PNAS Direct Submission.
Data Availability.
All study data are included in the article.
References
- 1.Gao R., et al. , Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J. Med. 368, 1888–1897 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Dormitzer P. R., et al. , Synthetic generation of influenza vaccine viruses for rapid response to pandemics. Sci. Transl. Med. 5, 185ra68 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Hekele A., et al. , Rapidly produced SAM(®) vaccine against H7N9 influenza is immunogenic in mice. Emerg. Microbes Infect. 2, e52 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craig Venter J., Life at the Speed of Light: From the Double Helix to the Dawn of Digital Life (Little, Brown, 2013). [Google Scholar]
- 5.Pallesen J., et al. , Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. U.S.A. 114, E7348–E7357 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wrapp D., et al. , Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pizza M., et al. , Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 287, 1816–1820 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Rappuoli R., Reverse vaccinology. Curr. Opin. Microbiol. 3, 445–450 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Ladhani S. N., et al. , Vaccination of infants with meningococcal group B vaccine (4CMenB) in England. N. Engl. J. Med. 382, 309–317 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Azzari C., et al. , Effectiveness and impact of the 4CMenB vaccine against group B meningococcal disease in two Italian regions using different vaccination schedules: A five-year retrospective observational study (2014–2018). Vaccines (Basel) 8, 469 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schumacher T. N., Schreiber R. D., Neoantigens in cancer immunotherapy. Science 348, 69–74 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Serruto D., Rappuoli R., Post-genomic vaccine development. FEBS Lett. 580, 2985–2992 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Burton D. R., Antibodies, viruses and vaccines. Nat. Rev. Immunol. 2, 706–713 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Scarselli M., et al. , Rational design of a meningococcal antigen inducing broad protective immunity. Sci. Transl. Med. 3, 91ra62 (2011). [DOI] [PubMed] [Google Scholar]
- 15.McLellan J. S., et al. , Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science 340, 1113–1117 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McLellan J. S., et al. , Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science 342, 592–598 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crank M. C. et al.; VRC 317 Study Team , A proof of concept for structure-based vaccine design targeting RSV in humans. Science 365, 505–509 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Jardine J., et al. , Rational HIV immunogen design to target specific germline B cell receptors. Science 340, 711–716 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rappuoli R., Bottomley M. J., D’Oro U., Finco O., De Gregorio E., Reverse vaccinology 2.0: Human immunology instructs vaccine antigen design. J. Exp. Med. 213, 469–481 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilboa E., Eglitis M. A., Kantoff P. W., Anderson W. F., Transfer and expression of cloned genes using retroviral vectors. Biotechniques 4, 504–512 (1986). [Google Scholar]
- 21.Cadoz M., et al. , Immunisation with canarypox virus expressing rabies glycoprotein. Lancet 339, 1429–1432 (1992). [DOI] [PubMed] [Google Scholar]
- 22.Wolff J. A., et al. , Direct gene transfer into mouse muscle in vivo. Science 247, 1465–1468 (1990). [DOI] [PubMed] [Google Scholar]
- 23.Tang D. C., DeVit M., Johnston S. A., Genetic immunization is a simple method for eliciting an immune response. Nature 356, 152–154 (1992). [DOI] [PubMed] [Google Scholar]
- 24.Ulmer J. B., et al. , Heterologous protection against influenza by injection of DNA encoding a viral protein. Science 259, 1745–1749 (1993). [DOI] [PubMed] [Google Scholar]
- 25.Martinon F., et al. , Induction of virus-specific cytotoxic T lymphocytes in vivo by liposome-entrapped mRNA. Eur. J. Immunol. 23, 1719–1722 (1993). [DOI] [PubMed] [Google Scholar]
- 26.Geall A. J., et al. , Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. U.S.A. 109, 14604–14609 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.HogenEsch H., O’Hagan D. T., Fox C. B., Optimizing the utilization of aluminum adjuvants in vaccines: You might just get what you want. NPJ Vaccines 3, 51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Podda A., The adjuvanted influenza vaccines with novel adjuvants: Experience with the MF59-adjuvanted vaccine. Vaccine 19, 2673–2680 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Del Giudice G., Rappuoli R., Didierlaurent A. M., Correlates of adjuvanticity: A review on adjuvants in licensed vaccines. Semin. Immunol. 39, 14–21 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Krammer F., SARS-CoV-2 vaccines in development. Nature 586, 516–527 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Maruggi G., Zhang C., Li J., Ulmer J. B., Yu D., mRNA as a transformative technology for vaccine development to control infectious diseases. Mol. Ther. 27, 757–772 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jackson L. A. et al.; mRNA-1273 Study Group , An mRNA vaccine against SARS-CoV-2—Preliminary report. N. Engl. J. Med. 383, 1920–1931 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sahin U., et al. , COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature 586, 594–599 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Anderson E. J. et al.; mRNA-1273 Study Group , Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N. Engl. J. Med. 383, 2427–2438 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bloom D. E., Black S., Rappuoli R., Emerging infectious diseases: A proactive approach. Proc. Natl. Acad. Sci. U.S.A. 114, 4055–4059 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Folegatti P. M. et al.; Oxford COVID Vaccine Trial Group , Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: A preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet 396, 467–478 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keech C., et al. , First-in-human trial of a SARS CoV 2 recombinant spike protein nanoparticle vaccine. medRxiv [Preprint] (2020). https://www.medrxiv.org/content/10.1101/2020.08.05.20168435v1 (accessed 6 August 2020). [DOI] [PMC free article] [PubMed]
- 38.Turner J. S., et al. , Human germinal centres engage memory and naive B cells after influenza vaccination. Nature 586, 127–132 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Francis T., On the doctrine of original antigenic sin. Proc. Am. Philos. Soc. 104, 572–578 (1960). [Google Scholar]
- 40.Sridhar S., et al. , Effect of dengue serostatus on dengue vaccine safety and efficacy. N. Engl. J. Med. 379, 327–340 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Rappuoli R., Glycoconjugate vaccines: Principles and mechanisms. Sci. Transl. Med. 10, eaat4615 (2018). [DOI] [PubMed] [Google Scholar]
- 42.World Health Organization , Antimicrobial resistance. https://www.who.int/antimicrobial-resistance/interagency-coordination-group/final-report/en/. Accessed 4 October 2020.
- 43.Bloom D. E., Black S., Salisbury D., Rappuoli R., Antimicrobial resistance and the role of vaccines. Proc. Natl. Acad. Sci. U.S.A. 115, 12868–12871 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harro C. D., et al. , The immunogenicity and safety of different formulations of a novel Staphylococcus aureus vaccine (V710): Results of two phase I studies. Vaccine 30, 1729–1736 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Fowler V. G., et al. , Effect of an investigational vaccine for preventing Staphylococcus aureus infections after cardiothoracic surgery: A randomized trial. JAMA 309, 1368–1378 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Oxman M. N. et al.; Shingles Prevention Study Group , A vaccine to prevent herpes zoster and postherpetic neuralgia in older adults. N. Engl. J. Med. 352, 2271–2284 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Lal H. et al.; ZOE-50 Study Group , Efficacy of an adjuvanted herpes zoster subunit vaccine in older adults. N. Engl. J. Med. 372, 2087–2096 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Tait D. R., et al. , Final analysis of a trial of M72/AS01E vaccine to prevent tuberculosis. N. Engl. J. Med. 381, 2429–2439 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Sharma P., Allison J. P., The future of immune checkpoint therapy. Science 348, 56–61 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All study data are included in the article.