Abstract
The production of tree seedlings in forest nurseries and their use in the replanting of clear-cut forest sites is a common practice in the temperate and boreal forests of Europe. Although conifers dominate on replanted sites, in recent years, deciduous tree species have received more attention due to their often-higher resilience to abiotic and biotic stress factors. The aim of the present study was to assess the belowground fungal communities of bare-root cultivated seedlings of Alnus glutinosa, Betula pendula, Pinus sylvestris, Picea abies and Quercus robur in order to gain a better understanding of the associated fungi and oomycetes, and their potential effects on the seedling performance in forest nurseries and after outplanting. The study sites were at the seven largest bare-root forest nurseries in Lithuania. The sampling included the roots and adjacent soil of 2–3 year old healthy-looking seedlings. Following the isolation of the DNA from the individual root and soil samples, these were amplified using ITS rRNA as a marker, and subjected to high-throughput PacBio sequencing. The results showed the presence of 161,302 high-quality sequences, representing 2003 fungal and oomycete taxa. The most common fungi were Malassezia restricta (6.7% of all of the high-quality sequences), Wilcoxina mikolae (5.0%), Pustularia sp. 3993_4 (4.6%), and Fusarium oxysporum (3.5%). The most common oomycetes were Pythium ultimum var. ultimum (0.6%), Pythium heterothallicum (0.3%), Pythium spiculum (0.3%), and Pythium sylvaticum (0.2%). The coniferous tree species (P. abies and P. sylvestris) generally showed a higher richness of fungal taxa and a rather distinct fungal community composition compared to the deciduous tree species (A. glutinosa, B. pendula, and Q. robur). The results demonstrated that the seedling roots and the rhizosphere soil in forest nurseries support a high richness of fungal taxa. The seedling roots were primarily inhabited by saprotrophic and mycorrhizal fungi, while fungal pathogens and oomycetes were less abundant, showing that the cultivation practices used in forest nurseries secured both the production of high-quality planting stock and disease control.
Keywords: fungal diversity, community composition, ectomycorrhiza, pathogens, oomycetes, tree seedlings
1. Introduction
In Europe, forest tree planting has increased considerably in recent decades [1,2], thereby increasing the demand for planting stock. The increased planting is primarily due to the commitment in many countries to increase the forest area and/or to rehabilitate degraded forest ecosystems, and thus to reclaim disturbed sites for forestry. This also allows us to maintain and increase biodiversity, and to mitigate the negative effects of global climate change [2,3]. In European forests, tree planting is a principal reforestation practice that also contributes to the sustainability and productivity of forests. Nowadays, most of the planting stock, which is used in forestry, is produced in forest nurseries [4]. The quality of the seedlings produced is one of the critical factors that contributes to successful forest restoration programs [5]. Although the seedling quality may depend on different factors, the associated microbial communities can be of key importance, and may determine the success of the seedlings’ survival, establishment and growth after outplanting [6,7,8].
In nature, healthy and asymptomatic plants cohabit with diverse microbes that form complex microbial consortia and impact plant growth and productivity [9,10]. Several studies have reported a wide range of beneficial effects of microbiota members on plant health, including disease suppression [11,12], the priming of the plants’ immune systems [13], the induction of systemic resistance [14], increased nutrient acquisition [15], increased tolerance to abiotic stresses [16], better adaptation to different environmental conditions [17], and the promotion of root mycorrhization [18]. Among the beneficial microbes, ectomycorrhizal (ECM) fungi are known to provide nutritional benefits to host trees, and may also mitigate negative effects of different abiotic and biotic factors [19]. For example, Zak [20] has postulated several mechanisms by which ECM fungi may provide disease protection to the feeder roots of plants. However, the conditions that often prevail in forest nurseries—namely confined space, high soil fertility, the use of fungicides, and abundant watering—may discourage the ECM colonization of seedling roots [21,22,23]. Such nursery conditions may select for opportunistic ECM fungal species, while limiting root colonisation by ECM fungi present in forest ecosystems [23]. In contrast to ECM fungi, pathogenic fungi or bacteria and plant nematodes may have a negative effect on seedling health, thereby causing abnormal growth or even mortality [24]. Infestations caused by oomycetes, including Phytophthora species, can be another threat to seedling production in forest nurseries. Jung et al. [25] demonstrated that nursery seedlings across Europe are commonly infested with a large array of Phytophthora spp. Nursery diseases may also have a negative effect on the field performance of outplanted seedlings. Besides this, nursery diseases may also be a threat to local forests when infected seedlings are outplanted, especially in areas where the disease was not present [26]. For example, nursery stock has been shown to be the most common means for the introduction of new Phytophthora species into landscapes and habitats worldwide [27,28]. Although many previous studies have linked Phytophthora diseases to ornamental plants [25,29,30,31], recent studies suggest that these are also found on native plants produced in forest nurseries [32,33,34].
Interactions between plants and soil microbes are highly dynamic in nature [35,36,37,38], and rhizosphere microbial communities may differ between different plant species [39,40,41], different genotypes within the species [42,43], and between the different developmental stages of a given plant [44,45]. For example, the assessment of fungal communities in the roots of healthy-looking P. sylvestris and P. abies seedlings in Swedish forest nurseries has showed the dominance of ECM and/or endophytic fungi, but has also revealed some differences in the fungal community composition between the two tree species [46]. Bzdyk et al. [47] demonstrated that the roots of nursery-grown Fagus sylvatica and Q. robur were inhabited by rather different communities of fungi. By contrast, Beyer-Ericson et al. [48] studied the root-associated fungi of diseased seedlings in Swedish forest nurseries, showing that the commonly-detected fungi were fungal pathogens, including species from the genera Cylindrocarpon, Fusarium, Pythium, Botrytis, Alternaria and Ulocladium. Similarly, in Norwegian forest nurseries, the roots of diseased seedlings were associated with pathogens from the genera Pythium and Rhizoctonia [49,50,51], while in Finnish forest nurseries, there were pathogens from the genera Fusarium, Rhizoctonia, Pythium and Phytophthora [52,53]. The above studies demonstrate that the root-associated fungal communities in forest nurseries may depend not only on the health status of the tree seedlings, but also on the tree species. Although fungal communities in the healthy and decaying roots of nursery-grown tree seedlings are relatively well understood, studies comparing the belowground fungal communities associated with different tree species are still scarce.
Incidences of locally-occurring, and especially of invasive, forest pathogens have increased exponentially in the last two centuries, causing extensive economic and ecological damage [54]. As nursery seedlings may also serve as a probable source for forest infestation [55,56,57], a better understanding of the fungal and oomycete communities associated with different tree species could be of considerable practical importance. The aim of the present study was to assess the belowground fungal communities of five principle bare-root cultivated tree species in the seven largest forest nurseries in Lithuania using DNA-metabarcoding in order to gain a better understanding of the beneficial and pathogenic fungi and oomycetes, and their potential effects on seedling performance in forest nurseries and after outplanting.
2. Materials and Methods
2.1. Study Site and Sampling
The study sites were at the seven largest bare-root forest nurseries in Lithuania, namely: Alytus (N 54°24′21.91″, E 24°2′33.27″), Anykščiai (N 55°34′15.07″, E 25°7′3.73″), Dubrava (N 54°50′11.42″, E 24°1′59.23″), Kaišiadorys (54°48′41.97″, 24°33′15.98″), Kretinga (N 56°1′29.82″, E 21°14′2.53″), Panevėžys (N 55°45′32.72″, E 24°30′28.33″) and Trakai (N 54°30′13.05″, E 24°49′46.97″). All of these forest nurseries were situated within a radius of 300 km. Information on each forest nursery, the climate within the area and the soil type is in Table 1. The sampling was conducted between April and May 2018, i.e., at the end of dormancy, but before the time of the seedlings’ outplanting in the forest, by sampling the roots and adjacent soil of 2–3 year old healthy-looking seedlings of A. glutinosa, B. pendula, P. sylvestris, P. abies and Q. robur. The latter seedlings were selected as they represent the principal tree species in the given geographical area. The approximate seedling height was 20 cm for P. sylvestris and P. abies, 30 cm for Q. robur, and 50 cm for B. pendula and A. glutinosa. During the seedling cultivation in the forest nurseries, fungicides are not used, but fertilizers are applied according to the established routines.
Table 1.
Characteristics of the seven largest bare-root forest nurseries in Lithuania that were sampled in the present study.
| Forest Nursery | Area of the Nursery, ha | Total No. of Seedlings Produced, Millions | No. of Seedlings Produced by Tree Species, Millions | Mean Monthly Temperature, °C * | Mean Monthly Precipitation, mm * | Soil Type | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Alnus glutinosa | Betula pendula | Picea abies | Pinus sylvestris | Quercus robur | ||||||
| Alytus | 23.1 | 4.7 | 0.2 | 0.4 | 2.6 | 1.1 | 0.3 | 10.5 | 49.6 | Nc |
| Anykščiai | 34.0 | 5.6 | 0.3 | 0.3 | 4.6 | 0.2 | 0.2 | 10.1 | 38.2 | Nc |
| Dubrava | 56.3 | 7.1 | 0.3 | 0.2 | 5.2 | 1.2 | 0.2 | 10.2 | 64.8 | Nc |
| Kaišiado-rys | 27.0 | 4.6 | 0.2 | 0.2 | 3.6 | 0.2 | 0.3 | 9.9 | 52.1 | Nc |
| Kretinga | 66.0 | 5.5 | 0.1 | 0.1 | 4.4 | 0.4 | 0.2 | 8.8 | 61.6 | Nc |
| Panevėžys | 66.0 | 8.8 | 0.4 | 0.3 | 7.4 | 0.3 | 0.5 | 9.8 | 46.0 | Lc |
| Trakai | 31.4 | 5.1 | 0.1 | 0.2 | 3.2 | 1.3 | 0.1 | 10.3 | 41.0 | Nb |
* The climate data is for the period of sampling, i.e., April–May 2018. the soil type: Nb—oligotrophic mineral soils of normal humidity; Nc—mesotrophic mineral soils of normal humidity; Lc—eutrophic gleyic sandy loam [58].
The sampling in each forest nursery included 10 seedling root systems of each tree species, and 10 soil samples that were collected in the vicinity of each collected root system. In total, 350 root samples (7 nurseries × 5 tree species × 10 root samples) and 350 soil samples (7 nurseries × 5 tree species × 10 soil samples) were collected. For the collection of the roots, seedlings were randomly selected, their roots were excavated, they were gently shaken to remove the larger particles of soil, and they were cut off from the shoots. The soil samples (ca. 100 g) were taken using a 2 cm diameter soil core down to a 20 cm depth. The soil core was carefully cleaned between taking different samples. The collected root and soil samples were individually placed into plastic bags, labelled, transported to the laboratory and stored at −20 °C before being further processed.
In the laboratory, the roots were carefully washed in tap water in order to remove any of the remaining soil, and the fine roots with root tips were separated from the coarse roots (which were discarded). The fine roots were cut into ca. 1 cm-long segments, and each forest nursery and tree species was pooled together, resulting in a total of 35 root samples (7 nurseries × 5 tree species). The soil samples were sieved (mesh size 2 × 2 mm) in order to remove larger particles and roots, and each forest nursery and tree species was pooled together, resulting in a total of 35 samples (7 nurseries × 5 tree species).
2.2. DNA Isolation, Amplification and Sequencing
The principles of the DNA work followed the study by Marčiulynas et al. [59]. No surface sterilization of the roots was carried out. Prior to the isolation of the DNA, each sample (soil or roots) was freeze-dried (Labconco FreeZone Benchtop Freeze Dryer, Cole-Parmer, Vernon Hills, IL, USA) at −60 °C for two days. After the freeze-drying, ca. 0.03 g dry weight of each root sample was placed into a 2-mL screw-cap centrifugation tube together with glass beads. All of the samples were homogenized using a Fast prep shaker (Montigny-le-Bretonneux, France). The DNA from the roots was isolated using CTAB extraction buffer (0.5 M EDTA pH 8.0, 1 M Tris-HCL pH 8.0, 5 M NaCl, 3% CTAB) followed by incubation at 65 °C for 1 h. After the centrifugation, the supernatant was transferred to a new 1.5-mL Eppendorf tube, mixed with an equal volume of chloroform, centrifuged at 13,000 rpm for 8 min, and the upper phase was transferred to new 1.5-mL Eppendorf tubes. Then, an equal volume of 2-propanol was used to precipitate the DNA into a pellet by centrifugation at 13,000 rpm for 20 min. The pellet was washed in 500 μL 70% ethanol, dried, and dissolved in 50 μL sterile milli-Q water. Unlike the roots, ca. 1 g of freeze-dried soil was used for the isolation of the DNA from each sample using a NucleoSpin®Soil kit (Macherey-Nagel GmbH & Co. Duren, Germany) according to the manufacturer’s recommendations. Following the isolation of the DNA, the DNA concentration in the individual samples was determined using a NanoDrop™ One spectrophotometer (Thermo Scientific, Rodchester, NY, USA). The amplification of the ITS rRNA region was achieved using a forward primer ITS6 [60] and barcoded universal primer ITS4 [61]. This primer pair was shown to amplify both fungi and oomycetes [60]. The polymerase chain reaction (PCR) was performed in 50 μL reactions, and consisted of the following final concentrations: 0.02 ng/μL template DNA, 200 μM dNTPs, 750 μM MgCl2, 0.025 μM DreamTaq Green polymerase (5 U/μL) (Thermo Scientific, Waltham, MA, USA), and 200 nM of each primer; sterile milli-Q water was added in order to make up the final volume (50.0 μL) of the reaction. The amplifications were performed using a Applied Biosystems 2720 thermal cycler (Applied Biosystems, Foster City, CA, USA). The PCR program started with an initial denaturation step at 95 °C for 2 min, followed by 35 cycles of 95 °C for 30 s, and annealing at 55 °C for 30 s and 72 °C for 1 min, followed by a final extension step at 72 °C for 7 min. The PCR products were assessed using gel electrophoresis on 1% agarose gels stained with GelRed (Biotium, Fremont, CA, USA). The PCR products were purified using 3 M sodium acetate (pH 5.2) (Applichem GmbH, Darmstadt, Germany) and 96% ethanol mixture (1:25). After the quantification of all of the PCR products using a Qubit fluorometer 4.0 (Life Technologies, Stockholm, Sweden), they were pooled in an equimolar mix and sequenced using a PacBio platform and one Sequel SMRT cell at a SciLifeLab facility in Uppsala, Sweden.
2.3. Bioinformatics
The sequences were filtered and clustered using the Sequence Clustering and Analysis of Tagged Amplicons (SCATA) next-generation sequencing (NGS) pipeline (http://scata.mykopat.slu.se). The sequences were filtered for quality, removing short sequences (<200 bp), sequences with low mean read quality (Q < 20), primer dimers, and homopolymers, which were collapsed to three base pairs (bp) before clustering. The sequences were screened for primers and sample-identifying barcodes, and those sequences that were missing a barcode or primer were removed. The sequences were clustered into different taxa by single linkage clustering, with a 2.0% maximum distance allowed for the sequences to enter the clusters. The fungal taxa were taxonomically classified using a Ribosomal Database Project (RDP) pipeline classifier (https://pyro.cme.msu.edu/index.jsp), and sequences showing <80% similarity to the phylum level were considered to be of non-fungal or oomycete origin, and were excluded from further analyses. All of the fungal and oomycete sequences were taxonomically identified using the GenBank (NCBI) database and the Blastn algorithm (Table S1). The following criteria were used for the identification: sequence coverage > 80%, similarity to taxon level 98–100%, and similarity to genus level 94–97%. Sequences not matching these criteria were considered unidentified, and were given unique names. Representative sequences of the fungal and oomycete non-singletons are available from GenBank under accession numbers MW214720–MW216333.
2.4. Statistical Analysis
The rarefaction analysis was performed using Analytical Rarefaction v.1.3, which is available at http://www.uga.edu/strata/software/index.html. The differences in the richness of the fungal and oomycete operational taxonomic units (OTUs) in the roots and soil among the different tree species were compared using the nonparametric chi-square test [62]. As each of the datasets was subjected to multiple comparisons, the confidence limits for the p-values of the chi-square tests were reduced the corresponding number of times required by the Bonferroni correction [63]. The Shannon diversity index, the qualitative Sørensen similarity index, and non-metric multidimensional scaling (NMDS) in Canoco v.5.02 (Microcomputer Power, Ithaca, NY, USA) [62,64] were used in order to characterise the diversity and composition of the fungal communities. A MANOVA in Minitab v. 18.1 (Pennsylvania State University, State College, Pennsylvania, PA, USA) was used in order to evaluate the degree of separation (along NMDS axes 1 and 2) between the fungal communities in the different types of samples (roots and soil) and among the different tree species. For each type of sample (roots or soil), the nonparametric Mann-Whitney test in Minitab was used to test whether the Shannon diversity index differed among the different tree species. The assignment of ecological roles was based on FUNGuild [65].
3. Results
A total of 305,139 sequences was generated by the PacBio sequencing. The quality filtering showed the presence of 161,302 (52.9%) high-quality sequences, while the remaining 143,837 (47.1%) low quality sequences were excluded from further analyses. The clustering of the high-quality sequences showed the presence of 3564 non-singleton contigs representing different OTUs. Singletons were excluded. The taxonomic classification showed that 2003 (56.2%) of the OTUs represented fungi (Table S1 and Table 2), while 1561 (43.8%) non-fungal OTUs were excluded. Among all of the fungal OTUs, 390 (19.5%) were identified to the species level, 345 (17.2%) were identified to the genus level, and 1268 (63.3%) were identified only to a higher taxonomic level.
Table 2.
Generated high-quality fungal sequences and the detected diversity of the fungal taxa in the roots and soil of the five different tree species from the seven forest nurseries in Lithuania. The data from the different forest nurseries are combined.
| Samples | Tree Species | No. of Fungal Sequences/Taxa | Shannon Diversity Index H a |
|---|---|---|---|
| Roots | Alnus glutinosa | 6979/691 | 2.16–4.33 |
| Betula pendula | 2620/256 | 1.93–4.09 | |
| Picea abies | 12,532/591 | 1.75–4.28 | |
| Pinus sylvestris | 11,910/718 | 2.62–4.32 | |
| Quercus robur | 5270/453 | 2.14–4.23 | |
| Total Roots | 39,311/1414 | ||
| Soil | Alnus glutinosa | 4292/664 | 3.28–5.03 |
| Betula pendula | 7525/836 | 2.57–5.20 | |
| Picea abies | 7961/873 | 3.28–4.89 | |
| Pinus sylvestris | 8745/881 | 3.56–4.69 | |
| Quercus robur | 3862/646 | 3.83–5.17 | |
| Total Soil | 32,385/1737 | ||
| All | 71,696/2003 |
a The Shannon diversity index H column shows a variation among the different forest nurseries.
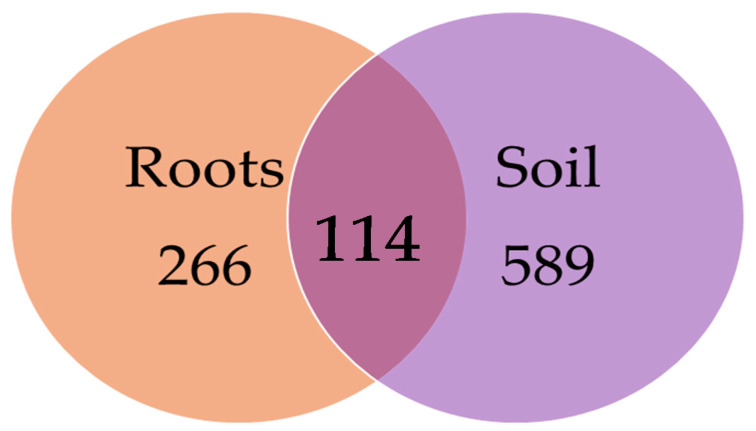
Of the 2003 fungal OTUs (all of which identified at least to the phylum level, Table S1) across all of the soil and root samples, Ascomycota was the most dominant phylum, which accounted for 50.4% of all of the fungal OTUs, followed by Basidiomycota (31.4%), Mucoromycota (6.2%), Chytridiomycota (6.0%), Oomycota (3.4%) and Zoopagomycota (2.0%). Entorrhizomycota, Blastocladiomycota, Cryptomycota, Zygomycota, Olpidiomycota and Blastoclamidiomycota were also detected, but in a very low proportions (each < 0.2%). Among all of the fungal OTUs (different tree species and nurseries combined), 266 (13.3%) were exclusively found in roots, 589 (29.4%) were exclusively found in the soil, and 1148 (57.3%) were shared between both types of samples (Figure 1).
Figure 1.
Venn diagram showing the diversity of the fungal OTUs found in the seedling roots and soil, and the number of fungal OTUs shared between both types of samples. The samples from different tree species and forest nurseries are combined.
Information on the 30 most common fungal OTUs—representing 62.03% of all of the high-quality fungal sequences in the roots and 38.95% of all of the high-quality fungal sequences in the soil of the different tree species—is in Table 3. The most common fungi (all samples combined) were Malassezia restricta (6.7% of all of the high-quality sequences), Wilcoxina mikolae (5.0%), Pustularia sp. 3993_4 (4.6%), Fusarium oxysporum (3.5%), Tomentella sp. 3993_7 (2.6%) and Suillus luteus (1.3%) (Table 3). Among the 30 most common fungal OTUs, five OTUs represented plant pathogens, including F. oxysporum, Dactylonectria macrodidyma (1.1%), Fusarium solani (0.9%), Pestalotiopsis sp. 3993_40 (0.8%), and Fusarium sp. 3993_41 (0.8%) (Table 3). The phylum Oomycota was represented by 68 OTUs, which comprised 1.5% of all of the high-quality sequences in the roots, and 3.2% of all of the high-quality sequences in the soil. Among these, there were 35 (51.5%) OTUs of Pythium spp., 4 (5.9%) of Phytophthora spp., 3 (4.4%) of Phytopythium spp., 2 (2.9%) of Hyaloperonospora spp., 2 (2.9%) of Peronospora spp., and 1 (1.5%) of Pythiogeton sp., while 21 (30.9%) could not be identified to the species or genus level (Table S1). Information on the 30 most common oomycete OTUs is in Table 4. The most common oomycetes were Pythium ultimum var. ultimum (0.6% of all of the high-quality sequences), Pythium heterothallicum (0.3%), Pythium spiculum (0.3%), Pythium sylvaticum (0.2%), Pythium irregulare (0.2%) and Peronospora sp. 3993_148 (0.1%) (Table 4).
Table 3.
Occurrence and relative abundance of the 30 most common fungal OTUs (show as a proportion of all of the high-quality fungal sequences) in the roots and soil of the five tree species that were bare-root cultivated in the forest nurseries. The data from the different forest nurseries are combined.
| OTU | Phylum | Reference | Similarity,% a | Alnus glutinosa | Betula pendula | Picea abies | Pinus sylvestris | Quercus robur | All | All | Total% | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Roots% | Soil% | Roots% | Soil% | Roots% | Soil% | Roots% | Soil% | Roots% | Soil% | Roots% | Soil% | |||||
| Malassezia restricta | Basidiomycota | CP030254 | 727/728 (99) | 5.79 | 3.59 | 8.02 | 4.96 | 6.57 | 2.85 | 10.43 | 6.00 | 20.75 | 0.85 | 9.01 | 4.05 | 6.70 |
| Wilcoxina mikolae | Ascomycota | KU061020 | 592/593 (99) | 0.11 | 0.09 | 0.38 | 0.07 | 22.28 | 0.95 | 3.82 | 1.46 | 0.88 | 0.08 | 8.82 | 0.67 | 5.03 |
| Pustularia sp. 3993_4 | Ascomycota | MF352743 | 513/518 (99) | - | 0.26 | 0.15 | 4.52 | 7.50 | 0.04 | 15.53 | 0.03 | 0.82 | - | 7.56 | 1.10 | 4.56 |
| Fusarium oxysporum | Ascomycota | GU136492 | 547/547 (100) | 2.41 | 5.24 | 0.73 | 4.44 | 3.28 | 4.81 | 2.99 | 4.63 | 1.91 | 2.54 | 2.72 | 4.46 | 3.53 |
| Tomentella sp. 3993_7 | Basidiomycota | KX095160 | 648/663 (98) | 0.01 | 0.05 | - | 0.07 | 14.20 | 0.15 | 0.07 | 0.01 | 0.15 | 0.00 | 4.80 | 0.06 | 2.60 |
| Suillus luteus | Basidiomycota | KU721223 | 688/688 (100) | - | 0.02 | 0.04 | 0.00 | 0.06 | 0.01 | 6.90 | 0.77 | 0.12 | 0.08 | 2.23 | 0.22 | 1.30 |
| Mycoarthris corallina | Ascomycota | AH009124 | 467/473 (99) | 6.53 | 0.44 | 0.57 | 9.13 | 0.02 | 0.82 | 0.06 | 0.11 | 0.09 | 0.16 | 1.30 | 2.43 | 1.82 |
| Paraphaeosphaeria sporulosa | Ascomycota | KY977581 | 594/594 (100) | 1.83 | 1.49 | 0.53 | 1.62 | 1.29 | 2.42 | 1.22 | 3.90 | 0.52 | 1.11 | 1.25 | 2.36 | 1.76 |
| Chaetomium cochliodes | Ascomycota | KT895345 | 570/570 (100) | 0.42 | 2.10 | 0.27 | 1.73 | 1.51 | 3.13 | 1.20 | 3.56 | 0.40 | 1.32 | 1.02 | 2.57 | 1.74 |
| Pleotrichocladium opacum | Ascomycota | NR155696 | 545/549 (99) | 0.44 | 0.40 | 28.66 | 0.54 | 0.80 | 0.78 | 0.42 | 0.29 | 0.79 | 0.54 | 2.57 | 0.51 | 1.61 |
| Halenospora varia | Ascomycota | AJ608987 | 538/538 (100) | 1.09 | 0.91 | 4.77 | 0.43 | 1.39 | 1.88 | 1.39 | 1.44 | 3.71 | 1.58 | 1.78 | 1.26 | 1.54 |
| Tomentella sublilacina | Basidiomycota | HM189981 | 648/662 (98) | 12.94 | 1.28 | 0.04 | 0.00 | 0.06 | - | 0.03 | 0.01 | 0.18 | 0.05 | 2.47 | 0.18 | 1.41 |
| Umbelopsis vinacea | Mucoromycota | KC489498 | 628/633 (99) | 0.46 | 0.91 | 0.42 | 0.33 | 1.39 | 3.49 | 1.11 | 2.53 | 0.27 | 0.13 | 0.96 | 1.75 | 1.33 |
| Cladosporium cladosporioides | Ascomycota | MG664765 | 547/547 (100) | 4.28 | 0.70 | 1.22 | 4.16 | 0.22 | 0.14 | 0.49 | 0.27 | 1.12 | 0.54 | 1.22 | 1.23 | 1.22 |
| Suillus granulatus | Basidiomycota | AJ272409 | 677/678 (99) | - | - | - | - | 0.01 | - | 6.83 | 0.01 | 0.03 | - | 2.18 | 0.00 | 1.17 |
| Solicoccozyma terricola | Basidiomycota | KY558367 | 641/641 (100) | 0.50 | 2.89 | 0.15 | 1.10 | 0.26 | 3.52 | 0.29 | 1.53 | 0.12 | 1.84 | 0.29 | 2.14 | 1.15 |
| Phialocephala fortinii | Ascomycota | AY078131 | 560/561 (99) | 0.96 | 0.19 | 0.27 | 0.09 | 1.61 | 1.42 | 2.34 | 0.35 | 1.22 | 0.05 | 1.59 | 0.50 | 1.08 |
| Tuber sp. 3993_24 | Ascomycota | KT215193 | 646/646 (100) | 0.03 | 0.02 | 0.04 | 0.00 | 2.35 | 0.06 | 2.64 | 0.00 | 5.20 | 0.03 | 2.10 | 0.02 | 1.13 |
| Dactylonectria macrodidyma | Ascomycota | JN859422 | 541/541 (100) | 0.63 | 2.10 | 0.38 | 0.69 | 0.46 | 0.90 | 2.17 | 0.89 | 1.22 | 1.04 | 1.10 | 1.03 | 1.06 |
| Mortierella sp. 3993_26 | Mucoromycota | KP311420 | 641/641 (100) | 0.99 | 1.10 | 0.31 | 0.98 | 0.78 | 1.53 | 0.97 | 1.02 | 0.30 | 1.97 | 0.80 | 1.26 | 1.02 |
| Saitozyma podzolica | Basidiomycota | KY320605 | 511/511 (100) | 1.00 | 1.56 | 0.11 | 1.48 | 0.34 | 1.75 | 0.26 | 1.97 | 0.21 | 1.14 | 0.41 | 1.65 | 0.99 |
| Unidentified sp. 3993_29 | Ascomycota | FN393100 | 546/550 (99) | 0.34 | 0.12 | 0.34 | 0.56 | 1.33 | 0.40 | 1.90 | 1.01 | 0.91 | 0.41 | 1.22 | 0.57 | 0.92 |
| Pseudogymnoascus sp. 3993_33 | Ascomycota | KY977601 | 562/562 (100) | 0.19 | 3.84 | 3.21 | 1.01 | 0.33 | 1.39 | 0.22 | 0.59 | 0.46 | 0.67 | 0.48 | 1.33 | 0.87 |
| Penicillium sp. 3993_47 | Ascomycota | MK226541 | 579/579 (100) | 0.29 | 2.54 | 0.04 | 1.05 | 0.29 | 1.04 | 0.43 | 2.24 | 0.12 | 0.80 | 0.30 | 1.54 | 0.87 |
| Mortierella sp. 3993_38 | Mucoromycota | HG935763 | 640/640 (100) | 1.43 | 1.77 | - | 0.74 | 0.34 | 1.42 | 0.39 | 1.36 | 0.15 | 1.29 | 0.52 | 1.28 | 0.87 |
| Trichoderma crassum | Ascomycota | NR134370 | 610/610 (100) | 0.97 | 0.98 | 0.15 | 0.74 | 0.51 | 0.88 | 0.79 | 1.22 | 1.64 | 0.70 | 0.76 | 0.93 | 0.84 |
| Fusarium solani | Ascomycota | MF782768 | 562/562 (100) | 0.14 | 3.87 | 0.08 | 1.09 | 0.31 | 1.65 | 0.31 | 0.82 | 0.12 | 1.45 | 0.25 | 1.57 | 0.86 |
| Pestalotiopsis sp. 3993_40 | Ascomycota | KT963804 | 588/588 (100) | 1.65 | 0.70 | 0.08 | 0.52 | 0.26 | 0.92 | 0.26 | 1.77 | 0.49 | 1.68 | 0.53 | 1.12 | 0.80 |
| Fusarium sp. 3993_41 | Ascomycota | MH550484 | 559/559 (100) | 1.03 | 0.70 | 1.11 | 1.24 | 0.41 | 1.19 | 0.29 | 1.18 | 0.18 | 0.44 | 0.51 | 1.04 | 0.76 |
| Sebacina sp. 3993_35 | Basidiomycota | JX844771 | 641/643 (99) | 0.03 | 0.00 | 0.27 | 0.03 | 3.67 | 0.00 | 0.06 | 0.48 | 0.06 | - | 1.28 | 0.14 | 0.75 |
| Total of 30 OTUs | 46.51 | 39.84 | 52.33 | 43.31 | 73.83 | 39.56 | 65.78 | 41.48 | 44.15 | 22.50 | 62.03 | 38.95 | 51.3 | |||
a Sequence similarity column shows base pairs compared between the query sequence and the reference sequence in the NCBI databases, with the percentage of the sequence similarity in parentheses.
Table 4.
Occurrence and relative abundance of the 30 most common oomycete OTUs (shown as a proportion of all of the high-quality fungal sequences) in the roots and soil of the five tree species that were bare-root cultivated in the forest nurseries. The data from the different forest nurseries are combined.
| OTU | Reference | Similarity % a | Alnus glutinosa | Betula pendula | Picea abies | Pinus sylvestris | Quercus robur | All | All | Total % | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Roots % | Soil % | Roots % | Soil % | Roots % | Soil % | Roots % | Soil % | Roots % | Soil % | Roots % | Soil % | ||||
| Pythium ultimum var. ultimum | AY598657 | 917/917 (100) | 0.258 | 0.559 | 0.076 | 1.435 | 0.048 | 0.829 | 0.067 | 1.349 | 0.030 | 1.554 | 0.094 | 1.161 | 0.59 |
| Pythium heterothallicum | AY598654 | 882/889 (99) | 0.143 | 0.210 | 0.076 | 0.545 | - | 0.251 | 0.008 | 0.812 | 0.030 | 0.829 | 0.038 | 0.534 | 0.27 |
| Pythium spiculum | HQ643790 | 972/978 (99) | 0.072 | 0.047 | 0.038 | 0.040 | 0.032 | 0.088 | 1.243 | 0.114 | - | - | 0.423 | 0.068 | 0.26 |
| Pythium sylvaticum | AY598645 | 997/999 (99) | 0.172 | 0.326 | 0.496 | 0.385 | 0.088 | 0.138 | 0.243 | 0.057 | 0.030 | 0.466 | 0.177 | 0.238 | 0.21 |
| Pythium irregulare | AY598702 | 1026/1029 (99) | 0.043 | 0.280 | 0.076 | 0.133 | 0.128 | 0.301 | 0.151 | 0.114 | 0.091 | 0.207 | 0.113 | 0.198 | 0.15 |
| Peronospora sp. 3993_148 | MF372507 | 803/852 (94) | - | - | - | - | - | - | - | - | 2.765 | - | 0.244 | - | 0.13 |
| Pythium intermedium | KU211482 | 957/959 (99) | - | 0.489 | 0.038 | 0.133 | - | 0.038 | 0.059 | 0.057 | - | 0.026 | 0.021 | 0.124 | 0.07 |
| Pythium amasculinum | AY598671 | 856/857 (99) | 0.029 | - | - | 0.159 | 0.008 | 0.025 | 0.008 | 0.023 | - | 0.363 | 0.011 | 0.093 | 0.05 |
| Pythium sp. 3993_349 | KU211471 | 894/907 (99) | 0.014 | 0.047 | - | 0.053 | 0.024 | - | 0.008 | 0.057 | - | 0.363 | 0.013 | 0.077 | 0.04 |
| Unidentified sp. 3993_508 | MF570293 | 101/119 (85) | 0.029 | 0.047 | 0.038 | 0.040 | 0.008 | 0.050 | 0.025 | 0.069 | 0.030 | 0.104 | 0.021 | 0.059 | 0.04 |
| Pythium acanthicum | AY598617 | 858/859 (99) | 0.014 | - | - | 0.013 | 0.040 | 0.050 | 0.017 | 0.057 | 0.030 | 0.104 | 0.024 | 0.043 | 0.03 |
| Pythium apiculatum | HQ643443 | 948/954 (99) | 0.072 | 0.023 | - | 0.120 | - | 0.063 | - | 0.011 | 0.061 | - | 0.019 | 0.049 | 0.03 |
| Pythium rostratifingens | KU211363 | 1053/1064 (99) | 0.115 | 0.023 | 0.038 | 0.066 | - | - | - | 0.034 | 0.030 | 0.104 | 0.027 | 0.040 | 0.03 |
| Phytopythium citrinum | HM061322 | 852/857 (99) | - | - | 0.496 | - | - | - | 0.008 | - | 0.030 | - | 0.040 | - | 0.02 |
| Unidentified sp. 3993_709 | KJ716873 | 724/865 (84) | - | - | - | - | 0.016 | - | 0.109 | 0.011 | - | - | 0.040 | 0.003 | 0.02 |
| Pythium pleroticum | AY598642 | 958/959 (99) | - | 0.093 | - | - | - | 0.063 | - | - | - | 0.078 | - | 0.037 | 0.02 |
| Pythium sp. 3993_1159 | AY598639 | 940/966 (97) | 0.086 | - | - | - | - | 0.075 | - | - | - | - | 0.016 | 0.019 | 0.02 |
| Unidentified sp. 3993_729 | HQ643756 | 226/240 (94) | - | - | - | - | 0.064 | 0.013 | 0.008 | 0.023 | - | - | 0.024 | 0.009 | 0.02 |
| Pythium rostratifingens | KU209835 | 962/969 (99) | - | 0.047 | - | 0.053 | - | 0.050 | - | - | - | - | - | 0.031 | 0.01 |
| Pythium sp. 3993_1163 | AY598696 | 993/1036 (96) | 0.043 | - | - | - | - | 0.013 | - | 0.034 | - | 0.078 | 0.008 | 0.022 | 0.01 |
| Unidentified sp. 3993_943 | MH671329 | 870/883 (99) | - | - | - | - | 0.048 | 0.025 | - | 0.011 | - | 0.026 | 0.016 | 0.012 | 0.01 |
| Unidentified sp. 3993_1191 | KF318041 | 606/754 (80) | - | 0.023 | - | - | - | - | - | 0.046 | - | 0.104 | - | 0.028 | 0.01 |
| Phytophthora fragariae | KJ755093 | 896/905 (99) | 0.029 | 0.116 | 0.038 | - | - | - | - | - | - | - | 0.008 | 0.015 | 0.01 |
| Phytophthora pseudosyringae | EU074793 | 838/848 (99) | - | - | - | - | - | - | - | 0.011 | - | 0.181 | - | 0.025 | 0.01 |
| Pythium violae | AY598717 | 1001/1006 (99) | 0.043 | 0.023 | - | 0.013 | - | 0.025 | - | 0.011 | - | - | 0.008 | 0.015 | 0.01 |
| Unidentified sp. 3993_1954 | LC176476 | 143/157 (91) | 0.057 | - | - | 0.053 | - | - | - | - | - | - | 0.011 | 0.012 | 0.01 |
| Unidentified sp. 3993_981 | KF318041 | 602/754 (80) | - | 0.023 | - | 0.040 | - | - | - | - | - | 0.104 | - | 0.025 | 0.01 |
| Pythium sp. 3993_1117 | JF431913 | 804/842 (95) | - | - | - | 0.013 | - | 0.050 | - | - | - | 0.052 | - | 0.022 | 0.01 |
| Unidentified sp. 3993_1171 | KJ716873 | 797/854 (93) | 0.014 | - | - | 0.040 | - | - | - | - | - | 0.078 | 0.003 | 0.019 | 0.01 |
| Hyaloperonospora brassicae | MG757782 | 940/943 (99) | - | - | - | - | - | - | 0.050 | - | - | - | 0.016 | - | 0.01 |
| Total of 30 OTUs | 1.232 | 2.377 | 1.412 | 3.336 | 0.503 | 2.148 | 2.007 | 2.905 | 3.130 | 4.816 | 1.414 | 2.977 | 2.14 | ||
a Sequence similarity column shows base pairs compared between the query sequence and the reference sequence in the NCBI databases, with the percentage of the sequence similarity in parentheses.
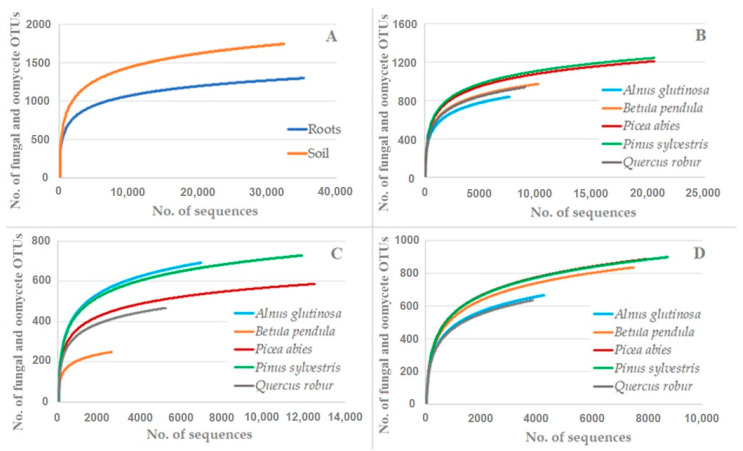
The rarefaction analysis showed that the number of fungal and oomycete OTUs did not reach species saturation (Figure 2). When the same number of fungal and oomycete sequences had been taken, the richness of the fungal and oomycete OTUs was significantly lower in the roots than in the soil (tree species and nurseries combined) (p < 0.0001) (Figure 2A). A similar comparison among the different tree species (root and soil data combined) showed that the deciduous tree species (A. glutinosa, B. pendula and Q. robur) had a significantly lower richness of fungal and oomycete OTUs taxa compared to the coniferous tree species (P. abies and P. sylvestris) (p < 0.0001) (Figure 2B). A higher variation in the richness of the fungal and oomycete OTUs was observed when the root and soil samples were analysed separately (nurseries combined) (Figure 2C,D). For example, in the roots, the richness of the fungal and oomycete OTUs was significantly lower compared B. pendula vs. other tree species (p < 0.0001), Q. robur vs. A. glutinosa, P. abies or P. sylvestris (p < 0.0001), and P. abies vs. A. glutinosa or P. sylvestris (p < 0.02) (Figure 2C). In a similar comparison, P. sylvestris and A. glutinosa did not differ significantly from each other (p > 0.05) (Figure 2C). In the soil, the richness of the fungal and oomycete OTUs was significantly lower compared to A. glutinosa vs. other tree species (p < 0.0001) (except A. glutinosa vs. Q. robur, p > 0.05) and Q. robur vs. B. pendula, P. abies or P. sylvestris (p < 0.0001), while B. pendula, P. abies and P. sylvestris did not differ significantly from each other (p > 0.05) (Figure 2D).
Figure 2.
Rarefaction curves showing the relationship between the cumulative number of fungal and oomycete OTUs and the number of ITS rRNA sequences compared: (A) roots vs. soil; (B) the different tree species (roots and soil samples combined); (C) root samples of the different tree species; and (D) soil samples of the different tree species.
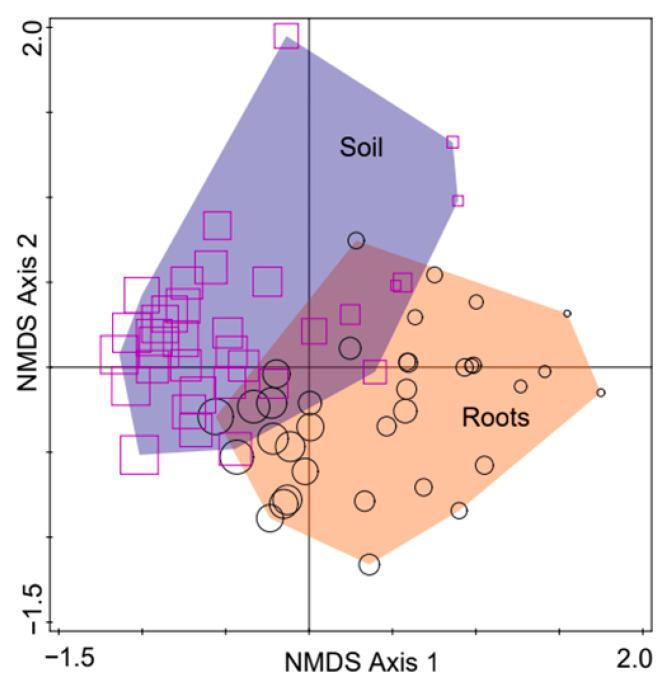
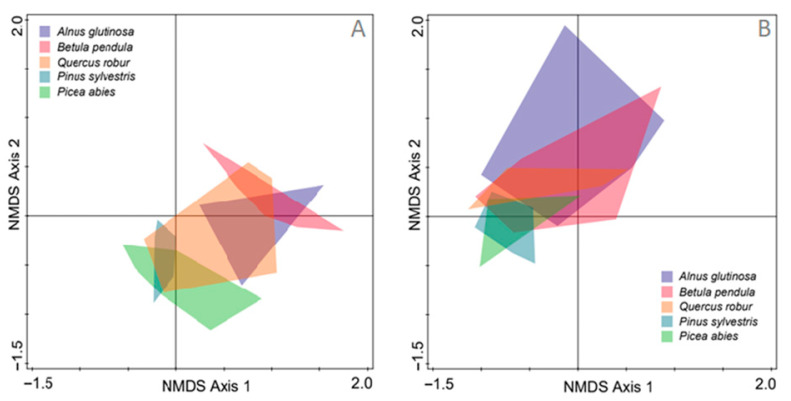
The non-metric multidimensional scaling of the fungal and oomycete communities showed a partial separation of the root and soil samples (tree species and nurseries combined) (Figure 3). The MANOVA showed that this separation was statistically significant (p < 0.0001) (Figure 3). For both the root and soil samples, a higher degree of separation of the fungal communities was between the coniferous tree species (P. abies and P. sylvestris) and the deciduous tree species (A. glutinosa, B. pendula and Q. robur) (Figure 4A,B). The MANOVA showed that the fungal and oomycete communities in the roots of P. abies and P. sylvestris differed significantly from those in A. glutinosa and B. pendula (p < 0.0001), while the fungal community in the roots of Q. robur were similar to all of the other tree species (Figure 4A). A similar comparison of fungal communities between P. abies and P. sylvestris, and between A. glutinosa and B. pendula showed that these did not differ significantly from each other (p > 0.05), respectively. In the soil, the fungal communities did not differ significantly among the different tree species (p > 0.05) (Figure 4B).
Figure 3.
Nonmetric multidimensional scaling (NMDS) of the fungal and oomycete communities in the roots and soil of the five tree species grown in the forest nurseries. Each point (circles for the roots and squares for the soil) represents a separate sample of different tree species, and the size of each point reflects the relative richness of the fungal and oomycete OTUs. The NMDS of the fungal and oomycete communities explained 52.8% of the variation on Axis 1 and 26.8% of the variation on Axis 2.
Figure 4.
NMDS of the fungal and oomycete communities in the roots (A) and soil (B) of the five tree species grown in the forest nurseries. The data from the different forest nurseries are combined.
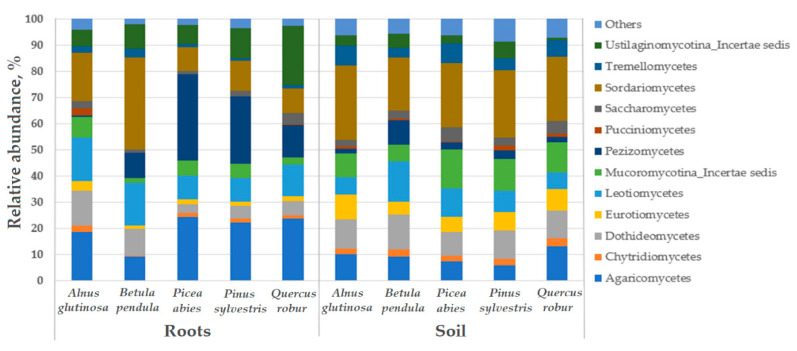
A total of 31 fungal classes was detected. A comparison among the different tree species showed that the relative abundance of fungal classes was more uniform among the soil samples than among the root samples (Figure 5). Nevertheless, in both the root and soil samples, Sordariomycetes, Leotiomycetes, Dothideomycetes and Agaricomycetes were the most common (Figure 5). An exception was the class Pezizomycetes, which showed a high relative abundance in the roots of P. abies and P. sylvestris, while Ustilaginomycotina_Incertae sedis showed a high relative abundance in the roots of Q. robur (Figure 5).
Figure 5.
Relative abundance of the fungal classes in the roots and soil of the different tree species, and the other fungal classes that presented with a relative abundance of <1%. The data from the different forest nurseries are combined.
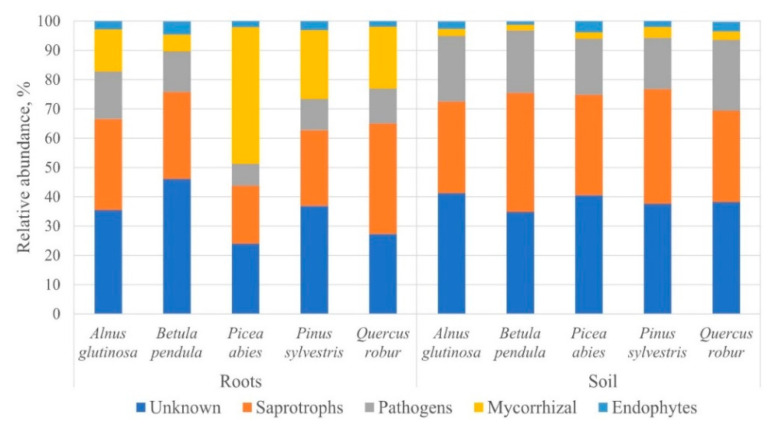
In the different tree species, the Sørensen similarity index of the fungal and oomycete communities was moderate, and ranged between 0.35 and 0.55 among the root samples, and between 0.43 and 0.56 among the soil samples. The Shannon diversity (H) index of the fungal and oomycete communities was high in both the root and soil samples (Table 2). The Mann-Whitney test showed that the Shannon diversity index did not differ significantly among the root or soil samples of the different tree species (p > 0.05). The assignment of fungal and oomycete ecological roles (nurseries combined) revealed a higher variation in the relative abundance among the root samples than among the soil samples of the different tree species (Figure 6). In the roots, the most common fungal and oomycete OTUs were of unknown ecological roles (23.9–46.1%, a range represents different tree species), followed by saprotrophs (19.6–37.7%), mycorrhizal fungi (5.7–46.6%), and pathogens (7.5–16.3%), and the least common were endophytes (1.9–4.5%) (Figure 6). Similarly, in the soil, the most common fungal and oomycete OTUs were unknown (34.7–41.2%), followed by saprotrophs (31.2–40.7%), pathogens (17.6–24.2%), and endophytes (1.8–3.6%), and the least common were mycorrhizal fungi (1.2–3.8%) (Figure 6).
Figure 6.
Ecological roles (shown as a proportion of the high-quality sequences) of the fungal and oomycete OTUs detected in the roots and soil of the different tree species. The data from the different forest nurseries are combined.
4. Discussion
In the present study, a comparison of the five economically-important tree species cultivated under similar conditions in bare-root forest nurseries provided valuable insights into the specificity of the associated fungal and oomycete OTUs. Firstly, the results demonstrated that the seedling roots and the rhizosphere soil were inhabited by a high diversity of fungal and oomycete OTUs (Figure 1 and Figure 2, Table 2), thereby corroborating previous observations that the belowground habitat in forest nurseries supports species-rich communities of fungi [66]. Interestingly, the detected richness of the fungal OTUs can be comparable to those present in the forest stands of the same geographical area [59], even though the rarefaction analysis showed that the observed richness of fungal OTUs can potentially be higher with increased sequencing effort (Figure 2). Secondly, our results revealed that the diversity and composition of the fungal and oomycete communities were partly dependant on the substrate (roots or soil) and/or on the host tree species (Figure 1, Figure 2, Figure 3, Figure 4 and Figure 5). As a result, there was generally a higher richness of fungal and oomycete OTUs in the soil than in the seedling roots, which was probably due to a higher heterogeneity of the soil environment compared to the roots [67], even though the intensive soil preparations (e.g. plowing and harrowing) in bare-root forest nurseries may have a homogenising effect on the soil’s fungal communities (Figure 4, Figure 5 and Figure 6). The segregation of the fungal and oomycete communities between the root and soil samples (Figure 3) was likely influenced by the host tree species, owing to a higher degree of modification of the associated fungal and oomycete communities in the roots than in the soil. In agreement with this, several studies have shown that plants may modify the structure of the associated microbial communities in their roots [68,69]. By contrast, the fungal communities in the soil can only be indirectly controlled by plants through the release of organic compounds that may contribute to the unique rhizosphere nutrient pool which is accessible to the soil microorganisms [70,71,72].
The coniferous tree species (P. abies and P. sylvestris) generally showed a higher richness of fungal and oomycete OTUs, and a rather distinct community composition compared to the deciduous tree species (A. glutinosa, B. pendula and partially Q. robur) (Figure 2 and Figure 4). These distinctions were more apparent among the fungal and oomycete communities in the roots, but were expressed less in the soil (Figure 4, Figure 5 and Figure 6). As certain root-associated fungi can be host-dependent, and some can even be host-specific [73,74,75,76,77], this demonstrates the relative importance of the host [78]. For example, Ishida et al. [79] showed that taxonomically-close host species harbour similar communities of mycorrhizal fungi. In agreement with this, it appears that mycorrhizal fungi play a key role in shaping the fungal communities in the roots of different tree species, as the abundance of fungi of other ecological roles was rather similar among the different tree species and substrates (roots or soil) (Figure 6). The latter may suggest that fungi of unknown ecological roles, saprotrophs, pathogens and endophytes generally possess a lower host or substrate specificity than mycorrhizal fungi; thus, the former were likely often represented by fungal generalists. Furthermore, in agreement with the previous studies on fungal communities in forest nurseries [7,46,80], the results have shown the dominance of fungal OTUs belonging to Ascomycota (50.4%) and Basidiomycota (31.4%). A higher relative abundance of ascomycetes in the soil and roots from all of the forest nurseries may reflect their better adaptation to a highly-transformed forest nursery environment compared to basidiomycetes. Indeed, members of Ascomycota have been shown to dominate on sites following site disturbances [81].
Among the dominant fungi, Malassezia restricta was shown to be exceedingly widespread and ecologically diverse in the environmental samples [82]. It was found in deep-sea sediments [83], hydrothermal vents [84], stony corals [85], Antarctic soils [86,87], on the exoskeleton of soil nematodes [88], and on various plant roots [89]. A recent study has also indicated that M. restricta is one of the most frequently-occurring species in the irrigation water of forest nurseries [59]. Despite many investigations, remarkably little is known about the impact of M. restricta on plant health. Wilcoxina mikolae, which was the second most commonly-detected fungus, was found in different forest nurseries, tree species and substrates (roots and soil) (Table 3), thereby showing a broad ecological niche. In agreement with this, this mycorrhizal symbiont was commonly reported in association with the roots of forest nursery seedlings [90,91]. Besides this, it was shown that Wilcoxina fungi can reduce the negative effect of salt stress on the plants [92] and support tree growth in high-altitude marginal habitats [93]. Its common occurrence in forest nurseries and on different hosts raises the question of its potential effect on seedling performance in forest nurseries, but such information is scarce. For example, Smaill and Walbert [94] showed that, on the roots of Pinus radiata seedlings, the abundance of Wilcoxina increases with increased applications of fertilizers and fungicides. Jones et al. [95] found that seedlings colonised by Wilcoxina showed an increased accumulation of 15N. Suillus luteus was another mycorrhizal fungus commonly detected in forest nurseries (Table 3). Suillus spp. are known as pioneer fungi, occurring in association with Pinus spp. in forest nurseries and in newly established forest plantations [6,96]. Pinus sylvestris seedlings inoculated with S. luteus were shown to have significantly better survival and growth rates after outplanting compared to controls [97], thereby demonstrating that this fungus can benefit the host trees.
Although the fungal communities were dominated by saprotrophs (Figure 6, Table S1), pathogens were also detected, indicating their potential threat to the plants. Fusarium oxysporum was the most commonly detected pathogen (Table 3), and it is known as one of the most destructive soil-borne pathogens, causing seedling diseases in forest nurseries worldwide [98]. Fusarium solani was also detected, but at lower proportions (Table 3). It is often found on dead organic matter, but under certain conditions it can cause disease in various hosts [99]. Dactylonectria macrodidyma (previously Neonectria macrodidyma) was also commonly recorded both in the root and soil samples of different tree seedlings (Table 3). Dactylonectria macrodidyma was shown to be an economically-important pathogen in forest nurseries [48,52,100,101,102]. Interestingly, the above-mentioned fungal pathogens showed generally low host or habitat specificity, but their relative abundance was often higher in the soil than in the roots of the different tree species (Table 3).
Oomycetes represent one of the most problematic groups of disease-causing microorganisms in different growing environments, including forest nurseries [25]. They can also cause diseases in different hosts, including trees, ornamental plants, and crops [103]. In the present study, oomycetes were often more abundant in the soil than in the seedlings’ roots (Table 4), suggesting that, in healthy roots, their development was largely restricted. The oomycetes that were most common in this study (Table 4) are also known to cause seedling diseases in forest nurseries, including taxa such as P. ultimum var. ultimum, P. heterothallicum [104,105] and P. spiculum [106,107,108]. Pythium spiculum was previously detected in the feeder roots and in the rhizosphere soil of declining oaks [109], but information on its pathogenicity to oaks is limited [106]. Among other oomycetes, P. irregulare is known to be associated with more than 200 host plants, and can cause root rot in deciduous and coniferous trees [110]. Interestingly, Peronospora sp. 3993_148 was exclusively detected in oak roots (Table 4), which may be due to the fact that representatives of this genus are known to cause downy mildew disease [111]. Among the 30 most common oomycetes, only two Phytophthora species were detected, i.e., Phytophthora pseudosyringae and Phytophthora fragariae (Table 4). Phytophthora pseudosyringae is known to cause root and collar rot in deciduous trees [112]. Phytophthora fragariae is a root pathogen that causes red stele disease in strawberries [113,114], but in the present study, it was associated with the soil and roots of A. glutinosa, and with the roots of B. pendula (Table 4).
5. Conclusions
The results demonstrated that the seedling roots and the rhizosphere soil in bare-root forest nurseries support a high richness of fungal taxa, which can be comparable to those present in forest stands.
Although the fungal communities in the roots were generally different between coniferous and deciduous tree species, the corresponding fungal communities in the soil were similar, thereby showing the relative importance of fungal generalists. The seedling roots were primarily inhabited by saprotrophic and mycorrhizal fungi, while fungal pathogens and oomycetes were less abundant, showing that the cultivation practices used in forest nurseries secured both the production of high-quality planting stock and disease control.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/9/1/150/s1. Table S1: Relative abundance of fungal and oomycete OTUs sequenced from the root and soil samples of the five tree species bare-root cultivated in forest nurseries in Lithuania.
Author Contributions
Conceptualization, D.M., A.M. (Audrius Menkis); methodology, D.M., A.M. (Adas Marčiulynas), A.M. (Audrius Menkis); validation, D.M., A.M. (Adas Marčiulynas), J.L., A.G., M.V.; formal analysis, D.M., A.M. (Audrius Menkis); investigation, D.M., A.M. (Adas Marčiulynas), J.L., A.G., M.V.; resources, D.M., A.G.; data curation, D.M., A.M. (Audrius Menkis); writing—original draft preparation, D.M.; writing—review and editing, D.M., A.M. (Adas Marčiulynas), J.L., A.G., M.V., A.M. (Audrius Menkis); visualization, D.M., A.M. (Audrius Menkis); supervision, A.M. (Audrius Menkis); project administration, D.M., A.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Research Council of Lithuania, grant no. S-MIP-17-6.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in Supplementary Material, Table S1.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Karjalainen T., Liski J., Pussinen A., Lapvetela Èinen T. Sinks in the Kyoto Protocol and Considerations for Ongoing Work in the UNFCCC Work. 2000. The Nordic Ministry Council. in press. [Google Scholar]
- 2.APAT—Auditory Processing Abilities Test . Nursery Production and Stand Establishment of Broad-Leaves to Promote Sustainable Forest Management. APAT—Auditory Processing Abilities Test; Roma, Italy: 2003. [Google Scholar]
- 3.Bastin J.F., Finegold Y., Garcia C., Mollicone D., Rezende M., Routh D., Zohner C.M., Crowther T.W. The global tree restoration potential. Science. 2019;365:76–79. doi: 10.1126/science.aax0848. [DOI] [PubMed] [Google Scholar]
- 4.Krasowski M.J. Forests and Forest Plants—Volume III: Producing Planting Stocks in Forest Nurseries. Eolss Publishers Co.; Oxford, UK: 2012. [Google Scholar]
- 5.Grossnickle S.C., MacDonald J.E. Seedling Quality: History, Application and Plant Attributes. Forests. 2018;9:283. doi: 10.3390/f9050283. [DOI] [Google Scholar]
- 6.Menkis A., Vasiliauskas R., Taylor A.F.S., Stenlid J., Finlay R. Fungal communities in mycorrhizal roots of conifer seedlings in forest nurseries under different cultivation systems, assessed by morphotyping, direct sequencing and mycelial isolation. Mycorrhiza. 2005;16:33–41. doi: 10.1007/s00572-005-0011-z. [DOI] [PubMed] [Google Scholar]
- 7.Menkis A., Vasiliauskas R., Taylor A.F.S., Stenström E., Stenlid J., Finlay R. Fungi in decayed roots of conifer seedlings in forest nurseries, afforested clear-cuts and abandoned farmland. Plant. Pathol. 2006;55:117–129. doi: 10.1111/j.1365-3059.2005.01295.x. [DOI] [Google Scholar]
- 8.Menkis A., Vasiliauskas R., Taylor A.F.S., Stenlid J., Finlay R. Afforestation of abandoned farmland with conifer seedlings inoculated with three ectomycorrhizal fungi—Impact on plant performance and ectomycorrhizal community. Mycorrhiza. 2007;17:337–348. doi: 10.1007/s00572-007-0110-0. [DOI] [PubMed] [Google Scholar]
- 9.Berendsen R.L., Pieterse C.M.J., Bakker P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant. Sci. 2012;17:478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Vorholt J.A. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 2012;10:828–840. doi: 10.1038/nrmicro2910. [DOI] [PubMed] [Google Scholar]
- 11.Mendes R., Kruijt M., de Bruijn I., Dekkers E., van der Voort M., Schneider J.H.M., Piceno Y.M., DeSantis T.Z., Andersen G.L., Bakker P.A., et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- 12.Ritpitakphong U., Falquet L., Vimoltust A., Berger A., Metraux J.P., L’Haridon F. The microbiome of the leaf surface of Arabidopsis protects against a fungal pathogen. New Phytol. 2016;210:1033–1043. doi: 10.1111/nph.13808. [DOI] [PubMed] [Google Scholar]
- 13.Van der Ent S., Van Hulten M., Pozo M.J., Czechowski T., Udvardi M.K., Pieterse C.M.J., Ton J. Priming of plant innate immunity by rhizobacteria and β-aminobutyric acid: Differences and similarities in regulation. New Phytol. 2009;183:419–431. doi: 10.1111/j.1469-8137.2009.02851.x. [DOI] [PubMed] [Google Scholar]
- 14.Zamioudis C., Korteland J., Van Pelt J.A., van Hamersveld M., Dombrowski N., Bai Y., Hanson J., Van Verk M.C., Ling H.Q., Schulze-Lefert P., et al. Rhizobacterial volatiles and photosynthesis-related signals coordinate MYB72 expression in Arabidopsis roots during onset of induced systemic resistance and iron-deficiency responses. Plant. J. 2015;84:309–322. doi: 10.1111/tpj.12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van der Heijden M., Bruin S., Luckerhoff L., van Logtestijn R.S., Schlaeppi K. A widespread plant-fungal-bacterial symbiosis promotes plant biodiversity, plant nutrition and seedling recruitment. ISME J. 2016;10:389–399. doi: 10.1038/ismej.2015.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rolli E., Marasco R., Vigani G., Ettoumi B., Mapelli F., Deangelis M.L., Gandolfi C., Casati E., Previtali F., Gerbino R., et al. Improved plant resistance to drought is promoted by the root-associated microbiome as a water stress-dependent trait. Environ. Microbiol. 2015;17:316–331. doi: 10.1111/1462-2920.12439. [DOI] [PubMed] [Google Scholar]
- 17.Haney C.H., Samuel B.S., Bush J., Ausubel F.M. Associations with rhizosphere bacteria can confer an adaptive advantage to plants. Nat. Plants. 2015;1:15051. doi: 10.1038/nplants.2015.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garbaye J. Helper bacteria—A new dimension to the mycorrhizal symbiosis. New Phytol. 1994;128:197–210. doi: 10.1111/j.1469-8137.1994.tb04003.x. [DOI] [PubMed] [Google Scholar]
- 19.Smith S.E., Read D.J. Mycorrhizal Symbiosis. 2nd ed. Academic Press; London, UK: 1997. p. 605. [Google Scholar]
- 20.Zak B. The role of mycorrhizae in root disease. Ann. Rev. Phytopathol. 1964;2:377–392. doi: 10.1146/annurev.py.02.090164.002113. [DOI] [Google Scholar]
- 21.Vaario L.M., Tervonen A., Haukioja K., Haukioja M., Pennanen T., Timonen S. The effect of nursery substrate and fertilization on the growth and ectomycorrhizal status of containerized and out planted seedlings of Picea abies. Can. J. For. Res. 2009;39:64–75. doi: 10.1139/X08-156. [DOI] [Google Scholar]
- 22.Kazantseva O., Bingham M., Simard S.W., Berch S.M. Effects of growth medium, nutrients, water, and aeration on mycorrhization and biomass allocation of greenhouse-grown interior Douglas-fir seedlings. Mycorrhiza. 2009;20:51–66. doi: 10.1007/s00572-009-0263-0. [DOI] [PubMed] [Google Scholar]
- 23.Henry C., Raivoarisoa J., Razafimamonjy A., Ramanankierana H., Andrianaivomahefa P., Ducousso M., Selosse M.A. Transfer to forest nurseries significantly affects mycorrhizal community composition of Asteropeia mcphersonii wildings. Mycorrhiza. 2017;27:321–330. doi: 10.1007/s00572-016-0750-z. [DOI] [PubMed] [Google Scholar]
- 24.Raj A.J., Lal S.B. Forestry Principles and Applications. Scientific Publishers; Rajasthan, India: 2013. [Google Scholar]
- 25.Jung T., Orlikowski L., Henricot B., Abad-Campos P., Aday A.G., Aguín Casal O., Bakonyi J., Cacciola S.O., Cech T., Chavarriaga D., et al. Widespread Phytophthora infestations in European nurseries put forest, semi-natural and horticultural ecosystems at high risk of Phytophthora diseases. For. Path. 2016;46:134–163. doi: 10.1111/efp.12239. [DOI] [Google Scholar]
- 26.Peterson G.W., Smith R.S., Jr. Forest Nursery Diseases in the United States. U.S. Department of Agriculture; Washington, DC, USA: 1975. p. 125. Agriculture Hand-Book No 470. Forest Service 1975. Library of Congress Catalog No. 74-600103. [Google Scholar]
- 27.Brasier C.M. The biosecurity threat to the UK and global environment from international trade in plants. Plant. Pathol. 2008;57:792–808. doi: 10.1111/j.1365-3059.2008.01886.x. [DOI] [Google Scholar]
- 28.Liebhold A.M., Brockerhoff E.G., Garrett L.J., Parke J.L., Britton K.O. Live plant imports: The major pathway for forest insect and pathogen invasions of the US. Front. Ecol. Environ. 2012;10:135–143. doi: 10.1890/110198. [DOI] [Google Scholar]
- 29.Yakabe L.E., Blomquist C.L., Thomas S.L., MacDonald J.D. Identification and frequency of Phytophthora species associated with foliar diseases in California ornamental nurseries. Plant. Dis. 2009;93:883–890. doi: 10.1094/PDIS-93-9-0883. [DOI] [PubMed] [Google Scholar]
- 30.Bienapfl J.C., Balci Y. Movement of Phytophthora spp. in Maryland’s nursery trade. Plant. Dis. 2014;98:134–144. doi: 10.1094/PDIS-06-13-0662-RE. [DOI] [PubMed] [Google Scholar]
- 31.Parke J.L., Knaus B.J., Fieland V.J., Lewis C., Grünwald N.J. Phytophthora community structure analyses in Oregon nurseries inform systems approaches to disease management. Phytopathology. 2014;104:1052–1062. doi: 10.1094/PHYTO-01-14-0014-R. [DOI] [PubMed] [Google Scholar]
- 32.Reeser P.W., Sutton W., Hansen E.M., Goheen E.M., Fieland V.J., Grunwald N.J. First report of Phytophthora occultans causing root and collar rot on Ceanothus, boxwood, rhododendron, and other hosts in horticultural nurseries in Oregon, USA. Plant. Dis. 2015;99:1282. doi: 10.1094/PDIS-02-15-0156-PDN. [DOI] [Google Scholar]
- 33.Rooney-Latham S., Blomquist C., Swiecki T., Bernhardt E., Frankel S.J. First detection in the USA: New plant pathogen, Phytophthora tentaculata, in native plant nurseries and restoration sites in California. Native Plants J. 2015;16:23–25. doi: 10.3368/npj.16.1.23. [DOI] [Google Scholar]
- 34.Rooney-Latham S., Blomquist C.L., Kosta K.L., Gou Y.Y., Woods P.W. Phytophthora species are common on nursery stock grown for restoration and revegetation purposes in California. Plant. Dis. 2019;103:448–455. doi: 10.1094/PDIS-01-18-0167-RE. [DOI] [PubMed] [Google Scholar]
- 35.Dobbelaere S., Vanderleyden J., Okon Y. Plant growth-promoting effects of diazotrophs in the rhizosphere. Crit. Rev. Plant. Sci. 2003;22:107–149. doi: 10.1080/713610853. [DOI] [Google Scholar]
- 36.Duffy B., Keel C., Defago G. Potential role of pathogen signaling in multitrophic plant-microbe interactions involved in disease protection. Appl. Environ. Microbiol. 2004;70:1836–1842. doi: 10.1128/AEM.70.3.1836-1842.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morgan J.A.W., Bending G.D., White P.J. Biological costs and benefits to plant-microbe interactions in the rhizosphere. J. Exp. Bot. 2005;56:1729–1739. doi: 10.1093/jxb/eri205. [DOI] [PubMed] [Google Scholar]
- 38.Reinhart K.O., Callaway R.M. Soil biota and invasive plants. New Phytol. 2006;170:445–457. doi: 10.1111/j.1469-8137.2006.01715.x. [DOI] [PubMed] [Google Scholar]
- 39.Batten K.M., Scow K.M., Davies K.F., Harrison S.P. Two invasive plants alter soil microbial community composition in serpentine grasslands. Biol. Invasions. 2006;8:217–230. doi: 10.1007/s10530-004-3856-8. [DOI] [Google Scholar]
- 40.Innes L., Hobbs P.J., Bardgett R.D. The impacts of individual plant species on rhizosphere microbial communities in soils of different fertility. Biol. Fertil. Soils. 2004;40:7–13. doi: 10.1007/s00374-004-0748-0. [DOI] [Google Scholar]
- 41.Priha O., Grayston S.J., Pennanen T., Smolander A. Microbial activities related to C and N cycling and microbial community structure in the rhizospheres of Pinus sylvestris, Picea abies and Betula pendula seedlings in an organic and mineral soil. FEMS Microbiol. Ecol. 1999;30:187–199. doi: 10.1111/j.1574-6941.1999.tb00647.x. [DOI] [PubMed] [Google Scholar]
- 42.Kowalchuk G.A., Hol W.H.G., Van Veen J.A. Rhizosphere fungal communities are influenced by Senecio jacobaea pyrrolizidine alkaloid content and composition. Soil Biol. Biochem. 2006;38:2852–2859. doi: 10.1016/j.soilbio.2006.04.043. [DOI] [Google Scholar]
- 43.Klavina D., Zaluma A., Pennanen T., Velmala S., Gaitnieks T., Gailis A., Menkis A. Seed provenance impacts growth and ectomycorrhizal colonisation of Picea abies seedlings. Balt. For. 2015;21:184–191. [Google Scholar]
- 44.Mougel C., Offre P., Ranjard L., Corberand T., Gamalero E., Robin C., Lemanceau P. Dynamic of the genetic structure of bacterial and fungal communities at different developmental stages of Medicago truncatula Gaertn. cv. Jemalong line J5. New Phytol. 2006;170:165–175. doi: 10.1111/j.1469-8137.2006.01650.x. [DOI] [PubMed] [Google Scholar]
- 45.Weisskopf L., Tomasi N., Santelia D., Martinoia E., Langlade N.B., Tabacchi R., Abou-Mansour E. Isoflavonoid exudation from white lupin roots is influenced by phosphate supply, root type and cluster-root stage. New Phytol. 2006;171:657–668. doi: 10.1111/j.1469-8137.2006.01776.x. [DOI] [PubMed] [Google Scholar]
- 46.Stenström E., Ndobe N.E., Jonsson M., Stenlid J., Menkis A. Root associated fungi of healthy-looking Pinus sylvestris and Picea abies seedlings in Swedish forest nurseries. Scand. J. For. Res. 2014;29:12–21. doi: 10.1080/02827581.2013.844850. [DOI] [Google Scholar]
- 47.Bzdyk R.M., Olchowik J., Studnicki M., Oszako T., Sikora K., Szmidla H., Hilszczańska D. The impact of effective microorganisms (em) and organic and mineral fertilizers on the growth and mycorrhizal colonization of Fagus sylvatica and Quercus robur seedlings in a bare-root nursery experiment. Forests. 2018;9:597. doi: 10.3390/f9100597. [DOI] [Google Scholar]
- 48.Beyer-Ericson L., Damm E., Unestam T. An overview of root dieback and its causes in Swedish forest nurseries. Eur. J. For. Pathol. 1991;21:439. doi: 10.1111/j.1439-0329.1991.tb00781.x. [DOI] [Google Scholar]
- 49.Galaaen R., Venn K. Pythium sylvaticum Campbell & Hendrix and other fungi associated with root dieback of 2-0 seedlings of Picea abies (L.) Karst. in Norway. Meddelerser Norsk Inst. Skogforsk. 1979;34:221–228. [Google Scholar]
- 50.Venn K. Norwegian, English Summary. Volume 11 Norwegian Forest Research Institute; Ås, Norway: 1985. Rotavdoing hos bartreplanter i skogplanteskoler (Root dieback of coniferous seedlings in forest nurseries) Research Paper 3/85. [Google Scholar]
- 51.Venn K., Sandvik M., Langerud B. Nursery routines, growth media and pathogens affect growth and root dieback in Norway spruce seedlings. Meddelelser Norsk Inst. Skogforsk. 1986;34:314–328. [Google Scholar]
- 52.Lilja A., Lilja S., Poteri M., Ziren L. Conifer seedling root fungi and root dieback in Finnish nurseries. Scand. J. For. Res. 1992;7:547–556. doi: 10.1080/02827589209382746. [DOI] [Google Scholar]
- 53.Lilja A. The occurrence and pathogenicity of uni- and binucleate Rhizoctonia and Pythiaceae fungi among conifer seedlings in Finnish forest nurseries. Eur. J. Plant. Pathol. 1994;24:181–192. doi: 10.1111/j.1439-0329.1994.tb00984.x. [DOI] [Google Scholar]
- 54.Santini A., Ghelardini L., De Pace C., Desprez-Loustau M.L., Capretti P., Chandelier A., Cech T., Chira D., Diamandis S., Gaitniekis T., et al. Biogeographical patterns and determinants of invasion by forest pathogens in Europe. New Phytol. 2013;197:238–250. doi: 10.1111/j.1469-8137.2012.04364.x. [DOI] [PubMed] [Google Scholar]
- 55.Cram M.M., Hansen E.M. Phytophthora Root Rot. In: Cram M.M., Frank M.S., Mallams K.M., editors. Forest Nursery Pests. Department of Agriculture, Forest Service; Washington, DC, USA: 2012. pp. 126–128. Agriculture Handbook 680 Rev. [Google Scholar]
- 56.Goheen E.M., Kanaskie A., Navarro S., Hansen E. Sudden oak death management in Oregon tanoak forests. For. Phytophthoras. 2017;7:45–53. doi: 10.5399/osu/fp.7.1.4030. [DOI] [Google Scholar]
- 57.Stenlid J., Oliva J., Boberg J.B., Hopkins A.J.M. Emerging diseases in European forest ecosystems and responses in society. Forests. 2011;2:486–504. doi: 10.3390/f2020486. [DOI] [Google Scholar]
- 58.Vaičys M. Lietuvos Dirvožemiai [Forest Site Types. Lithuanian Soils] Mokslas Publishers; Vilnius, Lithuania: 2001. Miško dirvožemių klasifikacija; pp. 1040–1043. (In Lithuanian) [Google Scholar]
- 59.Marčiulynas A., Marčiulynienė D., Lynikienė J., Gedminas A., Vaičiukynė M., Menkis A. Fungi and oomycetes in the irrigation water of forest nurseries. Forests. 2020;11:459. doi: 10.3390/f11040459. [DOI] [Google Scholar]
- 60.Cooke D.E.L., Drenth A., Duncan J.M., Wagels G., Brasier C.M. A molecular phylogeny of Phytophthora and related oomycetes. Fungal Genet. Biol. 2000;30:17–32. doi: 10.1006/fgbi.2000.1202. [DOI] [PubMed] [Google Scholar]
- 61.White T.J., Bruns T., Lee S., Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. PCR Protocols: A Guide to Methods and Applications. Academic Press Inc.; San Diego, CA, USA: 1990. pp. 315–322. [Google Scholar]
- 62.Magurran A.E. Ecological Diversity and Its Measurement. Princeton University Press; Princeton, NJ, USA: 1988. p. 192. [Google Scholar]
- 63.Sokal R.R., Rohlf F.J. Biometry: The Principles and Practice of Statistics in Biological Research. 3rd ed. W.H. Freeman and Company; New York, NY, USA: 1995. [Google Scholar]
- 64.Shannon C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948;27:379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x. [DOI] [Google Scholar]
- 65.Nguyen D., Boberg J., Ihrmark K., Stenstrom E., Stenlid J. Do foliar fungal communities of Norway spruce shift along a tree species diversity gradient in mature European forests? Fungal Ecol. 2016;23:97–108. doi: 10.1016/j.funeco.2016.07.003. [DOI] [Google Scholar]
- 66.Menkis A., Burokienė D., Stenlid J., Stenström E. High-throughput sequencing shows high fungal diversity and community segregation in the rhizospheres of container-grown conifer seedlings. Forests. 2016;7:44. doi: 10.3390/f7020044. [DOI] [Google Scholar]
- 67.Mickan B.S., Hart M.M., Solaiman Z.M., Jenkins S., Siddique K.H.M., Abbott L.K. Molecular divergence of fungal communities in soil, roots and hyphae highlight the importance of sampling strategies. Rhizosphere. 2017;4:104–111. doi: 10.1016/j.rhisph.2017.09.003. [DOI] [Google Scholar]
- 68.Pascale A., Proietti S., Pantelides I.S., Stringlis I.A. Modulation of the root microbiome by plant molecules: The basis for targeted disease suppression and plant growth promotion. Front. Plant. Sci. 2020;10:1741. doi: 10.3389/fpls.2019.01741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aponte C., García L.V., Marañón T. Tree species effects on nutrient cycling and soil biota: A feedback mechanism favouring species coexistence. For. Ecol. Manage. 2013;309:36–46. doi: 10.1016/j.foreco.2013.05.035. [DOI] [Google Scholar]
- 70.Klaubauf S., Inselsbacher E., Zechmeister-Boltenstern S., Wanek W., Gottsberger R., Strauss J., Gorfer M. Molecular diversity of fungal communities in agricultural soils from Lower Austria. Fungal Divers. 2010;44:65–75. doi: 10.1007/s13225-010-0053-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jiang Y.M., Chen C.R., Xu Z.H., Liu Y.Q. Effects of single and mixed species forest ecosystems on diversity and function of soil microbial community in subtropical China. J. Soils Sediments. 2012;12:228–240. doi: 10.1007/s11368-011-0442-4. [DOI] [Google Scholar]
- 72.Han L.L., Wang J.T., Yang S.H., Chen W.F., Zhang L.M., He J.Z. Temporal dynamics of fungal communities in soybean rhizosphere. J. Soils Sediments. 2016;17:491–498. doi: 10.1007/s11368-016-1534-y. [DOI] [Google Scholar]
- 73.Petrini O. Fungal Endophytes of Tree Leaves. In: Andrews J.H., Hirano S.S., editors. Microbial Ecology of Leaves. Springer; New York, NY, USA: 1991. pp. 179–197. [Google Scholar]
- 74.King B.C., Waxman K.D., Nenni N.V., Walker L.P., Bergstrom G.C., Gibson D.M. Arsenal of plant cell wall degrading enzymes reflects host preference among plant pathogenic fungi. Biotechnol. Biofuels. 2011;4:1–14. doi: 10.1186/1754-6834-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lang C., Seven J., Polle A. Host preferences and differential contributions of deciduous tree species shape mycorrhizal species richness in a mixed Central European forest. Mycorrhiza. 2011;21:297–308. doi: 10.1007/s00572-010-0338-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mayerhofer M.S., Kernaghan G., Harper K.A. The effects of fungal root endophytes on plant growth: A meta-analysis. Mycorrhiza. 2013;23:119–128. doi: 10.1007/s00572-012-0456-9. [DOI] [PubMed] [Google Scholar]
- 77.Austin A.T., Vivanco L., Gonzalez-Arzac A., Perez L.I. There’s no place like home? An exploration of the mechanisms behind plant litter-decomposer affinity in terrestrial ecosystems. New Phytol. 2014;204:307–314. doi: 10.1111/nph.12959. [DOI] [PubMed] [Google Scholar]
- 78.Jumpponen A., Egerton-Warburton L.M. Mycorrhizal fungi in successional environments—A community assembly model incorporating host plant, environmental and biotic filters. In: Dighton J., White J.F., Oudemans P., editors. The Fungal Community. CRC Press; New York, NY, USA: 2005. pp. 139–180. [Google Scholar]
- 79.Ishida T.A., Nara K., Hogetsu T. Host effects on ectomycorrhizal fungal communities: Insight from eight host species in mixed conifer–broadleaf forests. New Phytol. 2007;174:430–440. doi: 10.1111/j.1469-8137.2007.02016.x. [DOI] [PubMed] [Google Scholar]
- 80.Menkis A., Vasaitis R. Fungi in roots of nursery grown Pinus sylvestris: Ectomycorrhizal colonisation, genetic diversity and spatial distribution. Microb Ecol. 2011;61:52–63. doi: 10.1007/s00248-010-9676-8. [DOI] [PubMed] [Google Scholar]
- 81.Vrålstad T., Myhre E., Schumacher T. Molecular diversity and phylogenetic affinities of symbiotic root-associated ascomycetes of the Helotiales in burnt and metal polluted habitats. New Phytol. 2002;155:131–148. doi: 10.1046/j.1469-8137.2002.00444.x. [DOI] [PubMed] [Google Scholar]
- 82.Sugita T., Boekhout T., Velegraki A., Guillot J., Hađina S., Cabañes F.J. Epidemiology of Malassezia-related skin diseases. In: Boekhout T., Guého-Kellermann E., Mayser P., Velegraki A., editors. Malassezia and the Skin: Science and Clinical Practice. 1st ed. Springer; Berlin/Heidelberg, Germany: 2010. pp. 65–120. [Google Scholar]
- 83.Lai X., Cao L., Tan H., Fang S., Huang Y., Zhou S. Fungal communities from methane hydrate-bearing deep-sea marine sediments in South China Sea. ISME J. 2007;1:756–762. doi: 10.1038/ismej.2007.51. [DOI] [PubMed] [Google Scholar]
- 84.Le Calvez T., Burgaud G., Mahe S., Barbier G., Vandenkoornhuyse P. Fungal diversity in deep-sea hydrothermal ecosystems. Appl. Environ. Microbiol. 2009;75:6415–6421. doi: 10.1128/AEM.00653-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Amend A.S., Barshis D.J., Oliver T.A. Coral-associated marine fungi form novel lineages and heterogeneous assemblages. ISME J. 2012;6:1291–1301. doi: 10.1038/ismej.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Arenz B.E., Held B.W., Jurgens J.A., Farrell R.L., Blanchette R.A. Fungal diversity in soils and historic wood from the Ross Sea Region of Antarctica. Soil Biol. Biochem. 2006;38:3057–3064. doi: 10.1016/j.soilbio.2006.01.016. [DOI] [Google Scholar]
- 87.Fell J.W., Scorzetti G., Connell L., Craig S. Biodiversity of micro-eukaryotes in Antarctic Dry Valley soils with. Soil Biol. Biochem. 2006;38:3107–3119. doi: 10.1016/j.soilbio.2006.01.014. [DOI] [Google Scholar]
- 88.Renker C., Alphei J., Buscot F. Soil nematodes associated with the mammal pathogenic fungal genus Malassezia (Basidiomycota: Ustilaginomycetes) in Central European forests. Biol. Fertil Soils. 2003;37:70–72. doi: 10.1007/s00374-002-0556-3. [DOI] [Google Scholar]
- 89.Roy M., Watthana S., Stier A., Richard F., Vessabutr S., Selosse M.A. Two mycoheterotrophic orchids from Thailand tropical dipterocarpacean forests associate with a broad diversity of ectomycorrhizal fungi. BMC Biol. 2009;7:51. doi: 10.1186/1741-7007-7-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Klavina D., Menkis A., Gaitnieks T., Pennanen T., Lazdiņš A., Velmala S., Vasaitis R. Low impact of stump removal on mycorrhization and growth of replanted Picea abies: Data from three types of hemiboreal forest. Balt For. 2016;22:16–24. [Google Scholar]
- 91.Renseigné N., Rudawska M., Leski T. Mycorrhizal associations of nursery grown Scots pine (Pinus sylvestris L.) seedlings in Poland. Ann. For. Sci. 2006;63:715–723. [Google Scholar]
- 92.Zwiazek J.J., Equiza M.A., Karst J., Senorans J., Wartenbe M., Calvo-Polanco M. Role of urban ectomycorrhizal fungi in improving the tolerance of lodgepole pine (Pinus contorta) seedlings to salt stress. Mycorrhiza. 2019;29:303–312. doi: 10.1007/s00572-019-00893-3. [DOI] [PubMed] [Google Scholar]
- 93.Lazarevic J., Menkis A. Fungi inhabiting fine roots of Pinus heldreichii in the Montenegrin montane forests. Symbiosis. 2018;74:189–197. doi: 10.1007/s13199-017-0504-5. [DOI] [Google Scholar]
- 94.Smaill S.J., Walbert K. Fertilizer and fungicide use increases the abundance of less beneficial ectomycorrhizal species in a seedling nursery. Appl. Soil Ecol. 2013;65:60–64. doi: 10.1016/j.apsoil.2013.01.007. [DOI] [Google Scholar]
- 95.Jones M.D., Grenon F., Peat H., Fitzgerald M., Holt L., Philip L.J., Bradley R. Differences in N-15 uptake amongst spruce seedlings colonized by three pioneer ectomycorrhizal fungi in the field. Fungal Ecol. 2009;2:110–120. doi: 10.1016/j.funeco.2009.02.002. [DOI] [Google Scholar]
- 96.Chu-Chou M., Grace L.J. Mycorrhizal fungi of radiata pine in different forests of the North and South Islands in New Zealand. Soil Biol. Biochem. 1988;20:883–886. doi: 10.1016/0038-0717(88)90098-3. [DOI] [Google Scholar]
- 97.Menkis A., Lygis V., Burokienė D., Vasaitis R. Establishment of ectomycorrhiza-inoculated Pinus sylvestris seedlings on coastal dunes following a forest fire. Balt For. 2012;18:33–40. [Google Scholar]
- 98.James R.L., Dumroese R.K. Investigations of Fusarium diseases within Inland Pacific Northwest forest nurseries. In: Guyon J.C., editor. Proceedings of the 53rd Western International Forest Disease Work Conference, Jackson, WY, USA, 26–29 August 2005. USDA Forest Service, Intermountain Region; Ogden, UT, USA: 2007. pp. 3–11. [Google Scholar]
- 99.Adesemoye A., Eskalen A., Faber B., O’Connell N. Current nowledge on Fusarium dry root rot of citrus. Citrograph. 2011;2:29–33. [Google Scholar]
- 100.Adesemoye A.O., Mayorquin J.S., Peacock B.B., Moreno K., Hajeri S., Yokomi R., Eskalen A. Association of Neonectria macrodidyma with dry root rot of citrus in California. J. Plant. Pathol. Microbiol. 2016;8:391. [Google Scholar]
- 101.Hamelin R.C., Berube P., Gignac M., Bourassa M. Identification of root rot fungi in nursery seedlings by nested multiplex PCR. Appl. Environ. Microbiol. 1996;62:4026–4031. doi: 10.1128/AEM.62.11.4026-4031.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Menkis A., Burokienė D. Distribution and genetic diversity of the root-rot pathogen Neonectria macrodidyma in a forest nursery. For. Pathol. 2012;42:79–83. doi: 10.1111/j.1439-0329.2011.00712.x. [DOI] [Google Scholar]
- 103.Derevnina L., Petre B., Kellner R., Dagdas Y.F., Sarowar M.N., Giannakopoulou A., De la Concepcion J.C., Chaparro-Garcia A., Pennington H.G., van West P., et al. Emerging oomycete threats to plants and animals. Philosophical transactions of the Royal Society of London. Proc. R. Soc. B. 2016;371:20150459. doi: 10.1098/rstb.2015.0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lazerg F., Belabid L., Sanchez J., Gallego E. Root rot and damping-off of Aleppo pine seedlings caused by Pythium spp. in Algerian forest nurseries. For. Sci. 2016;62:322–328. doi: 10.17221/66/2014-JFS. [DOI] [Google Scholar]
- 105.Paulitz T.C., Adams K. Composition and distribution of Pythium communities in wheat fields in eastern washington state. Ecol. Popul. Biol. 2003;93:867. doi: 10.1094/PHYTO.2003.93.7.867. [DOI] [PubMed] [Google Scholar]
- 106.Jiménez J.J., Sánchez J.E., Romero M.A., Belbahri L., Trapero A., Lefort F., Sánchez M.E. Pathogenicity of Pythium spiculum and P. sterilum on feeder roots of Quercus rotundifolia. Plant. Pathol. 2008;57:369. doi: 10.1111/j.1365-3059.2007.01627.x. [DOI] [Google Scholar]
- 107.Serrano M.D., de Vita P., Fernández-Rebollo P., Coelho A.C., Belbahri L., Sánchez M.E. Phytophthora cinnamomi and Pythium spiculum as main agents of Quercus decline in southern Spain and Portugal. IOBC/WPRS Bull. 2012;76:97–100. [Google Scholar]
- 108.de Vita P., Serrano M.S., Belbahri L., García L.V., Ramo C., Sánchez M.E. Germination of hyphal bodies of Pythium spiculum isolated from declining cork oaks at Doñana National Park (Spain) Phytopathol. Mediterr. 2011;50:478–481. [Google Scholar]
- 109.Paul B., Bala K., Belbahri L., Calmin G., Sánchez E., Lefort F. A new species of Pythium with ornamented oogonia: Morphology, taxonomy, ITS region of its rDNA, and its comparison with related species. FEMS Microbiol. Lett. 2006;254:317–323. doi: 10.1111/j.1574-6968.2005.00048.x. [DOI] [PubMed] [Google Scholar]
- 110.Matsumoto C., Kageyama K., Suga H., Hyakumachi M. Intraspecific DNA polymorphisms of Pythium irregulare. Mycol. Res. 2000;104:1333–1341. doi: 10.1017/S0953756200002744. [DOI] [Google Scholar]
- 111.Thines M., Choi Y.J. Evolution, diversity, and taxonomy of the Peronosporaceae, with focus on the genus Peronospora. Phytopathology. 2016;106:6–18. doi: 10.1094/PHYTO-05-15-0127-RVW. [DOI] [PubMed] [Google Scholar]
- 112.Scanu B., Jones B., Webber J.F. A new disease of Nothofagus in Britain caused by Phytophthora pseudosyringae. New Dis. Rep. 2012;25:27. doi: 10.5197/j.2044-0588.2012.025.027. [DOI] [Google Scholar]
- 113.Scheewe P. Identification of pathogenic races of Phytophthora fragariae Hickman in Germany. In: Schmidt H., Kellerhals M., editors. Progress in Temperate Fruit Breeding, Developments in Plant Breeding. Volume 1. Springer; Dordrecht, Germany: 1994. pp. 67–71. [Google Scholar]
- 114.Jung T., Nechwatal J., Cooke D.E., Hartmann G., Blaschke M., Oßwald W.F., Duncan J.M., Delatour C. Phytophthora pseudosyringae sp. nov., a new species causing root and collar rot of deciduous tree species in Europe. Mycol Res. 2003;107:772–789. doi: 10.1017/S0953756203008074. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available in Supplementary Material, Table S1.