Abstract
Curculigo orchioides and C. latifolia have been used as traditional medicines such as antidiabetic and anticancer. This study measured the total phenolics and flavonoid contents as well as analyzed the functional groups and chemical compounds using Fourier-transform infrared (FTIR) spectra and UHPLC-Q-Orbitrap-HRMS profiling for the discrimination of plant parts, geographical origin, and compounds that presumably have a significant contribution as antioxidant and α-glucosidase inhibitors on both plants. The total phenolics and flavonoids contents in Curculigo species varied from 142.09 to 452.47 mg gallic acid equivalent (GAE/g) and from 0.82 to 5.44 mg quercetin equivalent (QE/g), respectively. The lowest IC50 for antioxidant and α-glucosidase inhibitory activities is presented by C. latifolia from a higher altitude region. Principal component analysis (PCA) from FTIR and UHPLC-Q-Orbitrap-HRMS data could discriminate the plant parts and geographical origin. Partial least squares (PLS) analysis has identified several functional groups, such as O–H, C–H, C=O, C–C, C–O, and chemical compounds, unknown-185 and unknown-85, that are most likely to contribute to the antioxidant and α-glucosidase inhibitory activities.
Keywords: antioxidant activity, α-glucosidase inhibitor, Curculigo spp., FTIR, metabolomics, UHPLC-Q-Orbitrap HRMS
1. Introduction
Curculigo orchioides Gaertn. and Curculigo latifolia Dryand. ex W.T.Aiton are widely distributed in Asia’s tropical and subtropical regions, mainly India, China, Indonesia, Malaysia, and Singapore. In Indonesia, the two Curculigo species are found from Sumatera to Papua. Curculigo spp. have several therapeutic properties because of their bioactive metabolites content. The rhizome of Curculigo species is traditionally used as estrogenic, neuroprotective, antibacterial, diuretics, aphrodisiac, antiapoptosis, anticancer, antidiabetic, and it neutralizes free radicals that likely could trigger various diseases. The leaves and flowers of C. latifolia are used to treat high fever, stomach-ache, and frequent urination [1,2]. The pharmacological effects of this species are attributed to the variety of their bioactive compounds, such as triterpene glycosides, phenolics, phenolic glycosides, and alkaloids.
A previous study [3] stated that the chemical components in Curculigo spp. possess some biological activities, such as cycloartane glycosides, which are used for leukemia treatment. Curculigosides F was found to inhibit hepatitis B virus (HBV) and antigen (HBeAg) in the HepG2.2.15 cell line, while chlorophenolic glycoside groups, such as curculigine M, curculigine N, and curculigine O, were reported to induce the proliferation of osteoblasts, orcinol glycosides, and 5,7,4′-trihydroxy-3′-methoxy flavone, which have antioxidant activity [4,5,6]. The phenolic group of Curculigo spp. is known as an antioxidant and has α-glucosidase inhibition activity (hydrolysis of α-1,6-glycosidic bonds) [7,8]. The metabolites in complex biological samples of Curculigo species could be identified rapidly and accurately using a metabolomics analysis to figure out their bioactive compounds. In the metabolomics analysis, there are two approaches to identifying the metabolites: the targeted and non-targeted analysis [9,10]. In the non-targeted analysis, metabolite fingerprinting and profiling could provide the complete identification of metabolites from a biological sample of Curculigo species. The analytical instrumentations used to study the metabolites of Curculigo species were Fourier-transform infrared (FTIR) spectrophotometer and UHPLC-Q-Orbitrap HRMS to obtain qualitative and quantitative data. Ultra-high performance liquid chromatography-mass spectrometry (UHPLC-MS) platforms have been widely used in plant science for metabolomics applications to identify and quantify compounds [10], and this approach is more sensitive and accurate for detecting metabolites [11].
It is not easy to find the correlation between FTIR spectra and UHPLC-Q-Orbitrap HRMS chromatograms with the contribution of biological activities related to its functional groups and metabolites due to the complexity of metabolomics data. Therefore, chemometrics analysis is needed in order to resolve this issue. The chemometric technique enables measurements of chemical systems through applications between mathematics and statistics, and it is an important method of analyzing much information from chromatographic data and can be used to evaluate the chemical similarities and dissimilarity of fingerprints between chromatographs of the sample [12]. Principal component analysis (PCA) and partial least squares (PLS) are the most frequent chemometrics analyses. PCA techniques make it possible to extract analytical information from chromatograms or spectra of samples to identify similarities and differences among highly complex datasets. PLS is one of the most popular calibration methods, which is based on determining the linear relationship between the independent dataset of measurement X (such as peak, absorbance values, fingerprint, spectra, or peak area from the detected metabolites) and the dependent set of variable Y (biological activity) in order to predict the bioactive compounds [13,14,15]. For the first time, we have performed metabolomics analysis using FTIR and UHPLC-Q-Orbitrap HRMS to identify major functional groups and metabolites in the two Curculigo spp. with a focus on substances having antioxidant and α-glucosidase inhibitory activities. We also differentiate the two Curculigo species based on their plant parts and their origin of geographical location.
2. Results
We designed the experiment based on the code samples, designating the species, the plant parts, and geographical location origin, which are summarized in Table 1. The highest total phenolics content was found in the rhizome of C. latifolia originated from Sinjai-Puncak (RLSK) (452.47 ± 0.12 mg GAE/g). In contrast, the lowest content of total phenolic contents was found in the leaves of C. latifolia from Sinjai-Biji Nangka (LLSB) (142.09 ± 0.63 mg GAE/g). The rhizome of C. latifolia harvested from different regions (RLSB and RLSP) also presented high total phenolics content, while C. orchioides rhizomes, regardless of their region, contained low phenolic content. In both species, phenolic compounds are accumulated more in the rhizomes than in the leaves and petiole of plant parts (Table 2).
Table 1.
Sources of plant samples in South Sulawesi.
| Sample Species | Regency (District) |
Coordinate | Organ | Sample Code | Collection Time | Season | Altitude (MASL) |
|---|---|---|---|---|---|---|---|
| C. orchioides | Barru (Mallawa) |
4°12′00′′ S 19°38′31′′E |
Rhizome Leaves |
ROBM LOBM |
July 2018 | Dry | 14 |
| Maros (Bengo-bengo) |
5°00′09′′ S 19°45′56′′ E |
Rhizome Leaves |
ROMB LOMB |
July 2018 | Dry | 462 | |
| Gowa (Malakaji) |
5°26′06′′ S 19°50′12′′ E |
Rhizome Leaves |
ROGM LOGM |
July 2018 | Dry | 750 | |
| C. latifolia | Sinjai (Palangka) |
5°18′04′′ S 20°06′33′′ E |
Rhizome Leaves Petiole |
RLSP LLSP PLSP |
July 2018 | Dry | 483 |
| Sinjai (Biji Nangka) |
5°18′43′′ S 120°05′25′′ E |
Rhizome Leaves Petiole |
RLSB LLSB PLSB |
February 2019 | Rainy | 640 | |
| Sinjai (Puncak) |
5°13′25′′ S 120°02′46′′ E |
Rhizome Leaves Petiole |
RLSK LLSK PLSK |
February 2019 | Rainy | 959 |
MASL: meters above sea level.
Table 2.
Total phenolics, total flavonoids, antioxidant, and α-glucosidase inhibitory activities in 70% ethanol extract of C. orchioides and C. latifolia.
| Sample Code | Total Phenolics (mgGAE/g) | Total Flavonoids (mgQE/g) | IC50 (mg/mL Extract) | |
|---|---|---|---|---|
| Antioxidant (DPPH) | α-Glucosidase Inhibitory | |||
| ROBM | 242.58 ± 0.49 e | 0.85 ± 0.01 a | 71.09 ± 2.25 b | 537.82 ± 6.87 h |
| ROMB | 288.20 ± 0.90 h | 0.89 ± 0.01 a | 102.93 ± 0.53 c | 522.00 ± 5.32 h |
| ROGM | 254.66 ± 0.44 f | 0.82 ± 0.01 a | 165.54 ± 1.26 e | 434.65 ± 5.55 fg |
| RLSP | 429.63 ± 0.42 l | 0.88 ± 0.02 a | 52.30 ± 0.07 a | 378.39 ± 8.17 cde |
| RLSB | 435.98 ± 0.32 m | 1.37 ± 0.01 b | 50.68 ± 0.41 a | 182.73 ± 3.87 a |
| RLSK | 452.47 ± 0.12 n | 1.57 ± 0.01 c | 47.08 ± 1.35 a | 155.25 ± 0.68 a |
| LOBM | 303.38 ± 0.95 j | 5.10 ± 0.01 h | 165.84 ± 0.57 e | 553.38 ± 12.22 h |
| LOMB | 170.90 ± 0.95 c | 2.72 ± 0.01 d | 161.44 ± 4.82 e | 349.14 ± 4.09 bd |
| LOGM | 194.22 ± 0.80 d | 5.44 ± 0.01 i | 205.89 ± 4.31 f | 349.93 ± 4.17 bd |
| LLSP | 168.95 ± 0.44 c | 4.21 ± 0.03 e | 363.12 ± 4.00 h | 325.45 ± 2.89 b |
| LLSB | 142.09 ± 0.63 a | 4.85 ± 0.01 g | 473.04 ± 0.12 j | 328.82 ± 21.48 bc |
| LLSK | 151,86 ± 1.16 b | 4.41 ± 0.01 f | 447.70 ± 0.76 i | 300.02 ± 0.97 b |
| PLSP | 278.23 ± 0.24 g | 4.39 ± 0.04 f | 140.45 ± 0.79 d | 462.72 ± 23.22 g |
| PLSB | 331.83 ± 0.42 k | 4.85 ± 0.01 g | 223.88 ± 0.91 g | 397.56 ± 11.19 df |
| PLSK | 229.35 ± 0.53 i | 4.18 ± 0.02 e | 213.23 ± 1.24 fg | 404.43 ± 2.53 ef |
The reported value is the mean ± SD of a triplicate assay for each sample. The values in the same column, followed with different superscript letters, indicate a significant difference at the level of p < 0.05 (Welch’s test).
The concentrations of total flavonoid in C. latifolia and C. orchioides are shown in Table 2. According to the plant parts and their origin location, the flavonoid concentrations in these two species are also varied. We found that the highest total flavonoid content was obtained in C. orchioides leaves (LOGM) as 5.44 ± 0.01 mg QE/g, while the lowest content was from C. orchioides rhizome (ROGM) as 0.82 ± 0.01 mg QE/g. Both samples were derived from the Gowa-Malakaji area in the southern part of Sulawesi island. The abundance of flavonoid compounds is more accumulated in the leaves than in the other plant parts (rhizome and/or petiole) of the two species.
The rhizome has a relatively higher antioxidant activity than the leaves and petiole of both species (Table 2), with the IC50 values varying between 47.08 and 473.04 mg/mL. C. latifolia rhizome has demonstrated the highest antioxidant activity, whereas the leaves of C. orchioides shows the second-highest antioxidant activity. The inhibitory concentration of α-glucosidase inhibitor from separated parts of the plant shows a different pattern with the values of IC50 varying from 155.25 to 553.38 mg/mL. The C. latifolia extract from the rhizome and leaves parts showed the highest α-glucosidase inhibitory activity among the other extracts from Sinjai-Puncak (RLSK and LLSK). Meanwhile, the C. orchioides extract from leaves and rhizome parts showed a weak effect as an inhibitor for α-glucosidase activity.
All samples of Curculigo species used in this study showed a specific FTIR spectrum profile, especially in the fingerprint region. The FTIR spectra derived from the ethanol extract of plant parts with the highest phenolic content are shown in Figure S1. At 3300 cm–1, the broad absorption bands indicated a stretching vibration from the O–H group. All samples of Curculigo species show the broad absorption band at a similar wavenumber. A stretching vibration at 3000 cm−1 was identified as C–H (sp2), which corresponds to the benzene ring, and a similar band was detected in all samples. However, the intensity of the absorption peak at 3000 cm−1 from the leaves of C. orchioides (LOBM), leaves of C. latifolia (LLSP), and petiole of C. latifolia (PLSP) samples was higher than that of the rhizome of C. orchioides (ROBM) and the rhizome of C. latifolia (RLSP) samples. The absorption band at 2800 cm−1 was related to the stretching vibration of C–H (alkane), and it was only observed in LOBM, LLSP, and PLSP samples. In the area of fingerprint wavenumber (1800–400 cm−1), only the absorption band in the range of 1152–1000 cm−1 was considered as C–O bending vibration. The absorption band in this range (1152–1000 cm−1) was detected in all samples, but the rhizomes of both Curculigo species manifested the highest intensity.
A typical chromatogram of UHPLC-Q-Orbitrap HRMS from both Curculigo species is shown in Figure S2, and it indicates abundant chemical compounds in their respective plant parts. This chromatogram profile shows a distinct variation in the retention times of 0.50–3.50, 14.00–16.00, and 23.00–27.00 min among parts of samples, i.e., leaves, petiole, and rhizome. The variation profile from sample ion chromatogram at these retention times can be distinguished in the number of peaks detected, peak height, and area.
We identified alkaloids, norlignan, phenolics, steroids, and terpenoids in C. orchioides and C. latifolia (Table S1). The compound identified from the triterpenoid (cycloartane) class in the Curculigo sample was curculigosaponin G, H, and I. Curculigosaponin G (m/z 783.488, ion mode [M + H]+) is fragmented into m/z 621.867 [M + H-C6H12O5]+, which indicates the release of C6H12O5 molecules followed by m/z 459.726 [M + H-C6H12O5]+ due to the release of C6H12O5 (Figure S3A). Curculigosaponin H (m/z 915.529, ion mode [M + H]+) is fragmented into m/z 751.350 [M + H-C6H12O5]+ and indicates the loss of C6H12O5 molecules followed by m/z 589.137 [M + H-C6H12O5]+ and 458.650 [M + H-C5H10O4]+, which indicates the release of C6H12O5 and C5H10O4 molecules (Figure S3B). Curculigosaponin I with m/z 945.150 in ion mode [M + H]+ fragmented into m/z 783.009, 621.867, and 458.726, which indicates the release of three C6H12O5 molecules (Figure S3C). The occurrence of fragmentations at m/z 459.726, 458.650, and 458.726 indicates the basic framework of triterpenoid saponins.
Curculigoside B (m/z 453. 136, ion mode [M + H]+) belongs to the phenolic compound class. It is fragmented into m/z 290.271 [M + H-C6H12O5]+ after releasing the C6H12O5 molecule and then becomes 276.244 [M + H-CH2]+ due to the release of CH2 (Figure S3D). Orchioside B (m/z 463.149, ion mode [M + H]+) is fragmented into m/z 350.143 [M + H-C6H8O2]+, which indicates the release of C6H8O2 molecules followed by m/z 213.143 [M + H-C8H9O2]+ due to the release of C8H9O2 molecules (Figure S3E).
One of the compounds identified from the norlignan class is (1S,2R)-O-Methylnyasicoside (m/z 493.167, ion mode [M + H]+). This compound is fragmented into m/z 475.159 and becomes 313.117 [M + H-C6H12O5]+ with the loss of C6H12O5 molecules (Figure S3F). The heatmap model (Figure S4A,B) depicts the variation of compounds found in the ion [M + H]+ and [M – H]−, which corresponds to its intensity from each plant part of both Curculigo species.
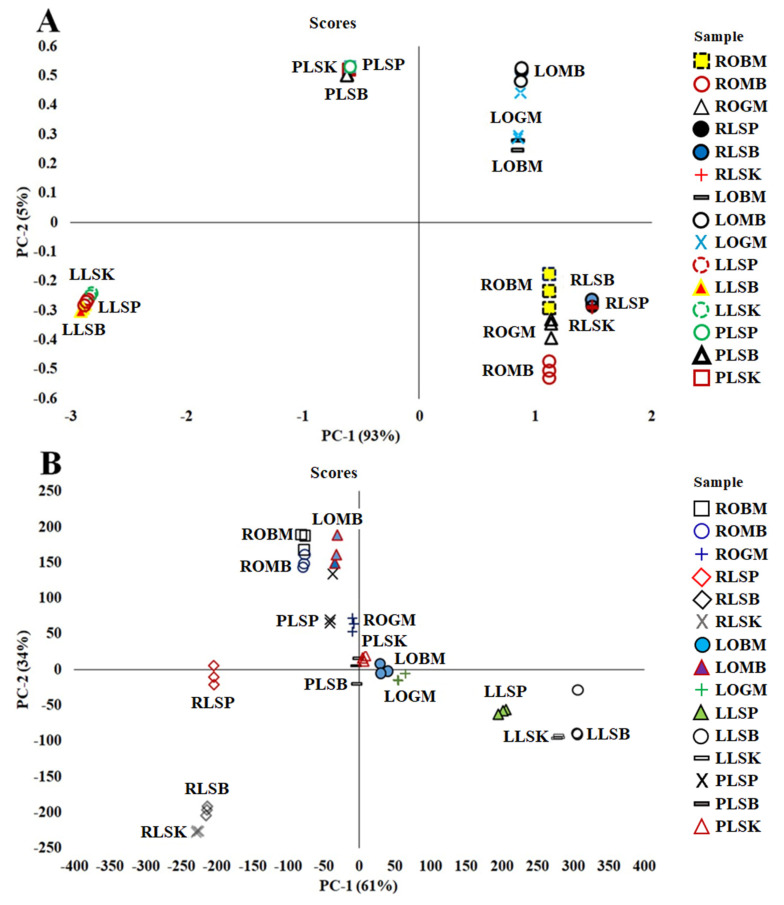
Based on the PCA analysis shown (Figure 1A,B), each sample from the plant parts of the Curculigo species was divided into their respective group, namely, leaves, petiole, and rhizome, with a total variance value of the first two main components of 98% (PC-1, 93% and PC-2, 5%) and 95% (PC-1, 61% and PC-2, 34%).
Figure 1.
Principal component analysis (PCA) score plot for principal component (PC–1) and (PC–2) in 70% ethanol extract of C. orchioides and C. latifolia using Fourier-transform infrared (FTIR) spectra (A) and UHPLC-Q-Orbitrap HRMS (B).
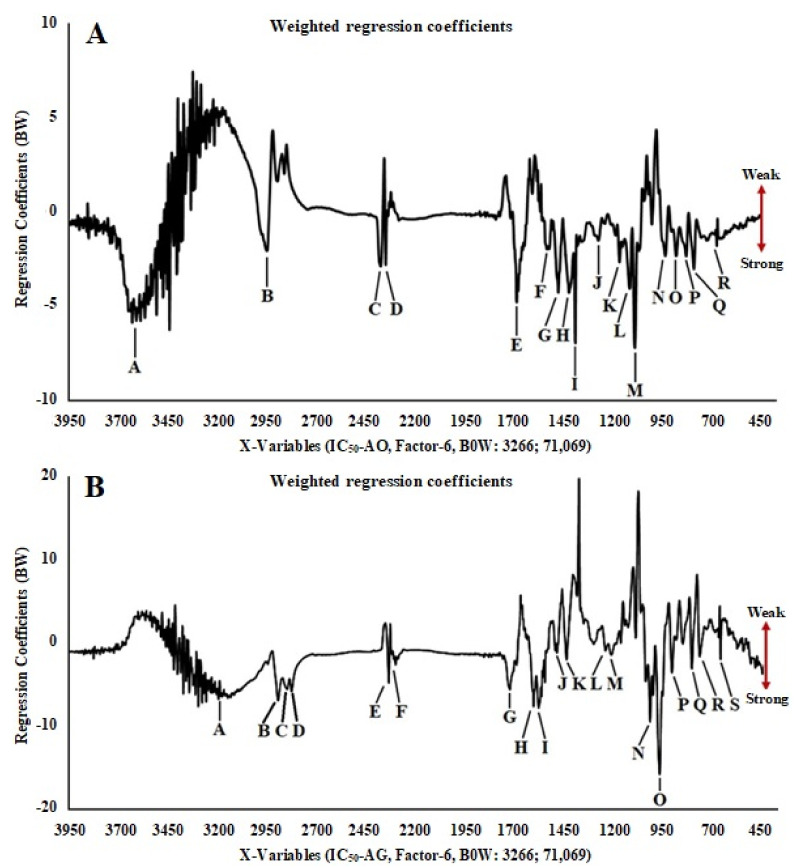
The PLS model was made from each spectral/chromatogram data variable (FTIR and UHPLC-Q-Orbitrap HRMS) and the biological activity assay. LV-1 and LV-2 values in the PLS model for antioxidant activity are 92% and 3%, and for α-glucosidase inhibition, they are 90% and 6%, respectively. The negative value of the PLS U score (factor-1) in the x-y relation outliers plot shows a strong contribution of each sample to the antioxidant (A) and α-glucosidase inhibition (B) activities (Figure S5). Line loading plots (Figure 2A,B) provided information on the functional groups and metabolites that contribute a major part to antioxidant activity and the inhibition of the α-glucosidase. Table 3 shows the wavenumbers, functional groups, and vibration modes present in all samples. The arrow mark at the right position of the line loading plots in Figure 2 indicates the level of bioactivity of Curculigo samples. The peaks at M (1083.920 cm−1) and I (1384.795 cm−1) with negative values and regression coefficients (BW) are −7.23 × 104 and −7.90 × 104, respectively. These peaks indicating the stretching vibrations of the C–O group (OH, COOH) and the bending vibrations of the O–H (phenol) group could have a major contribution to antioxidant activity. Meanwhile, the peaks in the area of O (975.914 cm–1) and N (1024.131 cm–1) with the smallest regression coefficients (BW) −5.91 × 104 and −3.52 × 104, respectively, indicate the stretching vibrations of the C–O group (trans disubstituent) and the bending vibrations of the C–H (alkenes), as well as the bending vibrations of the C–C group (cyclohexane). The three latter groups might have a major contribution to the inhibition of the α-glucosidase.
Figure 2.
Partial least squares (PLS) plot weighted regression coefficients (important variables) of antioxidant (A) and α-glucosidase inhibitory (B) activities in 70% ethanol extract of C. orchioides and C. latifolia using FTIR spectra.
Table 3.
The results of partial least squares (PLS) analysis on the wavenumbers, functional group, and vibration mode contribute to the antioxidant and α-glucosidase inhibitory activities.
| In Vitro Assay | Peak Code | Wavenumber (cm−1) | Functional Group | Vibration Mode |
|---|---|---|---|---|
| Antioxidant activity | A | 3606.638 | O–H | Stretching |
| B | 2945.009 | C–H | Stretching | |
| C | 2370.352 | O=C=O, N–H | Stretching | |
| D | 2345.2 | O=C=O | Stretching | |
| E | 1681.812 | C=O | Stretching | |
| F | 1527.517 | C=C (aromatic) | Stretching | |
| G | 1471.584 | O–H | Bending | |
| H | 1417.582 | –C–H (CH3) | Bending | |
| I | 1384.795 | C–O | Bending | |
| J | 1267.145 | C–O (OH, COOH) | Bending | |
| K | 1162.996 | C–C | Bending | |
| L | 1108.993 | C–O | Stretching (aliphatic of ether) | |
| M | 1083.92 | C–O | Stretching (secondary alcohol) | |
| N | 933.483 | –C–H (alkene) | Bending (trans disubtituen) | |
| O | 875.622 | –C–H (aromatic) | Bending | |
| P | 821.619 | –C–H (aromatic) | Bending (trisubtituen) | |
| Q | 784.974 | C–Cl | Bending | |
| R | 669.253 | C–X (halogen) | Halogen and aromatic disubtituen | |
| α-glucosidase inhibitory activity | A | 3170.755 | O–H | Stretching |
| B | 2916.169 | C–H | Stretching | |
| C | 2869.881 | C–H | Stretching | |
| D | 2842.879 | C–H | Stretching | |
| E | 2352.994 | O=C=O | Stretching | |
| F | 2318.277 | N–H, O–H, S–H | Streching | |
| G | 1737.744 | C=O | Stretching | |
| H | 1616.237 | N–H (NH2) | Bending | |
| C=C (alkene) | Stretching | |||
| I | 1591.164 | C=C (aromatic) | Streching | |
| J | 1496.658 | C=N, N=O | Bending | |
| C=C (alkene) | Stretching | |||
| K | 1448.441 | –C–H (–CH3) | Bending | |
| L | 1251.715 | C–O | Stretching | |
| M | 1220.857 | C–O | Stretching | |
| N | 1024.131 | C–N | Stretching | |
| O | 975.914 | C–H | Bending | |
| P | 914.196 | C–N | Bending | |
| Q | 811.976 | C–H | Bending | |
| R | 775.437 | C–Cl | Stretching | |
| S | 665.396 | C–X (halogen) | Halogen and aromatic disubtituen |
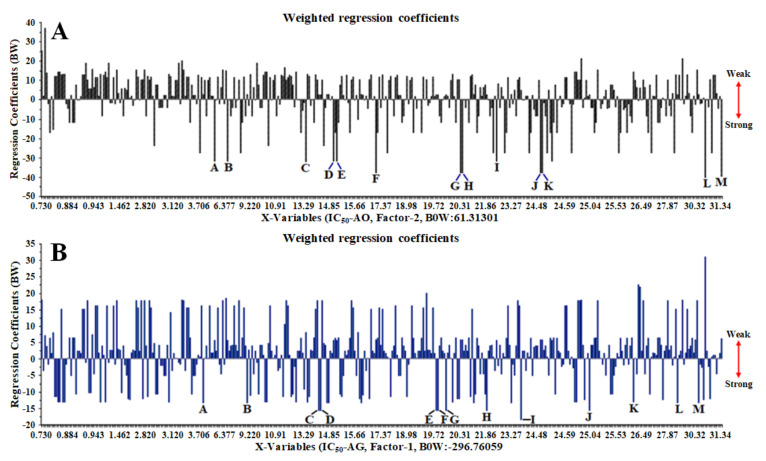
The chemical compounds, such as unknown-185 at a retention time of 30.920 min (peak L) and unknown-85 at 23.900 min (peak I), represent a strong (negative) signal to the antioxidant activity and α-glucosidase inhibition (Figure 3A,B). These chemical compounds predicted having a significant contribution to antioxidant activity and the inhibition of α-glucosidase, which are accumulated the highest in LLSB and LOBM samples and are all found in the leaves of C. latifolia and C. orchioides, respectively.
Figure 3.
Partial least squares (PLS) plot weighted regression coefficients (important variables) of antioxidant (A) and α-glucosidase inhibitory (B) activities in 70% ethanol extract of C. orchioides and C. latifolia using UHPLC-Q-Orbitrap HRMS.
The FTIR and PLS analysis from C. latifolia rhizome can be obtained qualitatively, the functional groups of –OH, C=O, C–O, –C–H, C–C, and C–OH belong to these two triterpene compounds. These triterpene compounds are the major groups that might contribute the most to the antioxidant activity and α-glucosidase inhibition. Table 4 gives information about the retention times and BW values of these triterpene compounds. Their functional groups from the identified compounds may contribute to the antioxidant activity and the inhibition of α-glucosidase, which are summarized in Figure S6.
Table 4.
The compounds identified in the 70% ethanol extracts of C. orchioides and C. latifolia using UHPLC-Q-Orbitrap HRMS that contributed to the antioxidant and α-glucosidase inhibitory activity.
| Peak Code | RT [min] | BW | Type | Metabolite Name | Formula | Chemical Type |
|---|---|---|---|---|---|---|
| Antioxidant Activity | ||||||
| A | 5.945 | −31.8958 | [M + H]+ | Curculigoside B | C21H24O11 | Phenolic glycosides |
| B | 6.680 | −31.8958 | [M + H]+ | Unknown-173 | C6H10O13 | - |
| C | 13.320 | −32.1579 | [M − H]− | Unknown-99 | C37H39N14O20 | - |
| D | 14.930 | −31.8958 | [M + H]+ | Unknown-176 | C15H13O8 | - |
| E | 15.050 | −31.8958 | [M − H]− | Unknown-175 | C26H35O16 | - |
| F | 16.848 | −37.8736 | [M + H]+ | (1S,2R)-O-Methylnyasicoside | C24H28O11 | Norlignan |
| G | 20.320 | −37.8736 | [M − H]− | 1,1-Bis(3,4-dihydroxyphenyl-1-(2-furan)-methane | C17H14O5 | Norlignan |
| H | 20.320 | −37.8736 | [M − H]− | Curculigosaponin G | C18H18O6 | Cycloartane (Triterpene) |
| I | 22.410 | −31.8958 | [M + H]+ | Unknown-179 | C47H49O | - |
| J | 24.506 | −37.8736 | [M − H]− | Curculigoside B | C21H24O11 | Phenolic glycosides |
| K | 24.506 | −37.8736 | [M − H]− | Orchioside B | C23H26O10 | Phenolic glycosides |
| L | 30.920 | −40.3125 | [M + H]+ | Unknown-185 | C47H59O7 | - |
| M | 31.435 | −39.7257 | [M − H]− | 2,4-Dichloro-5-methoxy-3-methylphenol | C8H8Cl2O2 | Phenolic |
| α-Glucosidase inhibitory | ||||||
| A | 4.060 | −13.3962 | [M + H]+ | Unknown-76 | C13H9O11 | - |
| B | 8.770 | −13.3962 | [M − H]− | Unknown-77 | C13H30N5O11 | - |
| C | 14.272 | −15.7379 | [M + H]+ | Orcinol glucoside | C13H18O7 | Phenolic glycosides |
| D | 14.272 | −15.7379 | [M + H]+ | 1,1-Bis(3,4-dihydroxyphenyl-1-(2-furan)-methane | C17H14O5 | Phenolic |
| E | 19.801 | −15.7379 | [M + H]+ | 5-Hydroxymethylfurfural | C6H6O3 | Aldehyde |
| F | 19.801 | −15.7379 | [M + H]+ | Curculigosaponin G | C42H70O13 | Cycloartane (Triterpene) |
| G | 20.049 | −15.7379 | [M + H]+ | Curculigosaponin I | C48H80O18 | Cycloartane (Triterpene) |
| H | 22.200 | −15.7379 | [M − H]− | Unknown-84 | C30H61O19 | - |
| I | 23.900 | −18.5262 | [M + H]+ | Unknown-85 | C42H51O6 | - |
| J | 25.040 | −15.7379 | [M + H]+ | Unknown-87 | C57H86N5O5 | - |
| K | 26.030 | −13.1626 | [M − H]− | Unknown-10 | C7H12NO12 | - |
| L | 29.010 | −13.3962 | [M + H]+ | Curculigosaponin H | C47H78O17 | Cycloartane (Triterpene) |
| M | 30.901 | −13.3962 | [M − H]− | Unknown-88 | C54H68O2 | - |
3. Discussion
The total phenolic and flavonoid contents in C. latifolia and C. orchioides varied depend on their harvest time, the origin of growth location, and the plant parts. The total phenolics compounds are all accumulated in the rhizomes part of plants. In this study, the total phenolic and flavonoid contents in RLSP, RLSB, and RLSK of C. latifolia samples are affected by its altitude. Its soil nutrient condition might influence the phenolic content in all C. latifolia samples, the soil moisture (microclimate), and the harvest time. RLSB and RLSK samples were harvested during the monsoon season and contained higher phenolic compound contents than the RLSP sample, which was harvested during the drought season. Oxidative stress caused by excessive or low soil water availability and unfavorable temperature can induce phenolic compounds’ synthesis for the plant survival [16,17].
The highest total flavonoid compounds are found in C. orchioides’ leaves and the leaves and petiole of C. latifolia. Flavonoids are accumulated in the aerial part of plants, especially leaves, and serve as phytoalexin to protect against pathogenic attacks and solar radiation, particularly ultraviolet radiation [18].
Some compounds are present in higher concentrations when certain exogenous effects are present. At higher altitudes, soil pH and soil micronutrient content decrease, which can be attributed to lower mineralization processes at lower pH. Some compounds (phenols, flavonoids, and terpenoids) increase in response to different altitudes. The accumulation of secondary metabolites is reported to have a vital role in tolerance to environmental biotic and abiotic stresses. These secondary metabolite help in the adaptation of stressed plants to different environmental conditions, for example, the accumulation of phenols and flavonoids as antioxidants [19].
Phenolic compounds prevent the enzymatic hydrolysis of carbohydrates by inhibiting α-amylase and α-glucosidase, with a mixed mechanism (competitive and uncompetitive) [20]. Previous studies using in vitro and in vivo testing [21,22,23] demonstrated an antidiabetic activity from the rhizome extracts of C. orchioides and C. latifolia. However, in this work, we found that only the rhizome of C. latifolia showed high antioxidant activity and potent inhibition of α-glucosidase enzyme associated with the high phenolic compounds in plants. In general, high phenolic content, along with high flavonoids in the samples, strongly correlates with antioxidants and α-glucosidase inhibition activities [24,25]. In our studies, the result is not in agreement with previous reports due to antioxidant and α-glucosidase inhibitory activity related to the other compounds, such as terpenoids, steroids, or alkaloids [26,27].
The FTIR spectra derived from Curculigo species show a distinct difference even though both plants are from the same genus. This is associated with both species having differences in the compound composition and content level of metabolites. The functional groups obtained from both extracts of Curculigo species are C–O, C=C, –CH, –COOH, and –OH, which are likely to be associated with antioxidant activity and α-glucosidase inhibitor. This assumption has been reported and described by Saleh et al. [28].
UHPLC-Q-Orbitrap HRMS can be operated with complete automatization with a short analysis time, and the extracted sample does not need further derivatization. The heatmap model was used to analyze each metabolite’s area from the Curculigo species’ extract and compare it for a similar compound. The heatmap model’s color intensity was used to depict the relative abundance in the sample extract of Curculigo species, and its variation can tell us about the compounds diversity in the plant [29]. We used the peak area from the detected metabolites because it is widely accepted that the peak area of an analyte in a chromatogram is directly proportional to its concentration [30].
C. orchioides has been reported to have an antioxidant activity associated with its compounds such as orcinol glycosides, curculigoside, curculigoside B, curculigoside C, curculigosaponin, 2,6-dimethoxyl benzoic acid, vanillin, and syringic acid [31,32]. A similar compound is also found in C. latifolia. Vanillin, which is phenolic acids, is used as a standard in several antioxidant studies. Both types of phenolic acids have more significant antioxidant activity than gallic acid [33]. At the same time, lycorine has also been determined in the rhizome and leaves of both Curculigo species. This compound, a type of alkaloid widely found in the genus Amaryllidaceae, has been reported before having an antioxidant and α-glucosidase inhibition activity [34].
In this study, PCA, as an unsupervised pattern recognition technique, has been used for obtaining the information of patterns of sample grouping, similarities, and differences among species and plant parts [35,36]. A PCA plot will consist of two principal components (PCs) because the first two PCs capture most of the variant of the data the closer the PC values of the samples. The cumulative percentages of total variance (PC-1 and PC-2) obtained from FTIR spectra and UHPLC-Q-Orbitrap HRMS data were 98% and 95%, respectively. It can be concluded that the PCA analysis used for sample clustering shows good accuracy in both terms of species and plant parts.
PLS, as a supervised pattern recognition method, is often used to correlate medicinal plants’ biological activities with chromatographic or spectroscopic data (peak area or peak and absorbance value) [15,30,37]. The parameters used to create the PLS model are latent variables (LV), and its full cross-validation was applied for data validation and avoiding overfitting [38,39]. As suggested by Easmin et al. [40], line loading plots (weighted regression coefficients) in the PLS model were used to evaluate functional groups and chemical compounds, and also for samples clustering having low or high activity (x-y relation outliers) from the FTIR spectrum and UHPLC-Q-Orbitrap HRMS data. Based on the results from PLS analysis, O–H, C–H, C=O, C–O, and C–C functional groups in the chemical structure of unknown-185 and unknown-85 are highly observed in the LLSB of C. latifolia and LOBM samples of C. orchioides. This is predicted to significantly contribute to the antioxidant activity and α-glucosidase inhibition, whereas the phenolics (curculigoside B, 2,4-dichloro-5-methoxy-3-methylphenol, orcinol glucoside, and orchioside B), cycloartane triterpene (curculigosaponin G, H, and I), and norlignan compounds (1,1-Bis(3,4-dihydroxyphenyl)-1-(2-furan)-methane and (1S,2R)-O-Methylnyasicoside) have an only minor contribution to the biological activities.
Interestingly, from this study, the active compounds are not always derived from the most abundant compounds found in the extract of Curculigo species. Other functional groups are also present in the chemical compounds from cycloartane triterpene (curculigosaponin G, H, and I), i.e., hydroxyl groups at the positions C–3 and C–11, and a double bond at C23=C24. It has been pointed out that the double bonds at C24=C25 of the tetracyclic triterpenoids have an essential role as antioxidant and α-glucosidase inhibitors [26,41].
4. Materials and Methods
4.1. Chemicals and Reagents
Alpha-glucosidase enzymes (≥98%), gallic acid (≥98%), quercetin (≥98%), p-nitrophenyl-α-D-glucopyranoside (≥99%), and 1,1-diphenyl-2-picrylhydrazyl (DPPH) (≥98%) were purchased from Sigma Aldrich (St Louis, USA). Water (LC-MS grade), acetonitrile (LC-MS grade ≥ 99.9%), methanol (LC-MS grade ≥ 99.9%), formic acid (≥98%), potassium bromide (KBr, FTIR grade), aluminum chloride (≥98%), sodium carbonate (≥98%), and DMSO (≥99.9%) were purchased from Merck (Darmstadt, Germany).
4.2. Plant Material
The plant samples from Curculigo species were collected during 2018–2019 from four regencies in Sulawesi Island, Indonesia: Barru, Maros, Gowa, and Sinjai. The coordinates, plant parts, sample code, collection time (season), and altitude are summarized in Table 1, and the geographical locations are shown in Figure S7. All samples were identified based on local knowledge and by comparing them to the published flora and voucher specimens stored at the Tropical Biopharmaca Research Center, IPB University, Indonesia.
4.3. Sample Preparation
The plant samples were air-dried for three days in a drying cabinet at 40 °C. Each sample (0.5 kg) was pulverized and sieved with a 40-mesh sieve. The samples were extracted by the maceration method using 5 L of ethanol (70%) for 24 h at room temperature. A similar procedure was repeated twice. Then, the sample was filtrated, which was followed by filtrate evaporation using a rotary evaporator (Rotavapor R-210, BÜCHII Labortechnik, Switzerland) below 50 °C until obtaining a dry condition.
4.4. Determination of Total Phenolics
The total phenolics content was determined using the method described by Margraf et al. [42] with slight modifications. Approximately a 20 µL sample was transferred to a 96-well microplate (Costar®, Washington, DC, USA), which was followed by the addition of 110 µL of Folin–Ciocalteu and 70 µL sodium carbonate (0.01 M). After vortexing, the sample mixture was incubated for 30 min at 37 °C. The absorbance was measured at 750 nm. Phenolics content was expressed as gallic acid equivalent (mg GAE/g) by using a calibration curve of gallic acid (y = 0.0091x − 0.0123, with R2 = 0.9982).
4.5. Determination of Total Flavonoids
A procedure from Zhu et al. [43] was used for the determination of total flavonoids with slight modifications. Briefly, 10 µL of the sample extract was mixed with 60 µL ethanol, 10 µL AlCl3 (10% b/v), 10 µL potassium acetate (1 M), and 120 µL distilled water. The mixture was added to a 96-well plate, incubated for 30 min at 37 °C, and the absorbance was read at 450 nm. The total flavonoid content was expressed as quercetin equivalent (mg QE/g) by using the calibration curve of quercetin (y = 0.003x − 0.0157, with R2 = 0.9935).
4.6. Antioxidant Activity by DPPH Method
We used the DPPH method for the measurement of radical scavenging activity, according to Ananthi et al. [44], with slight modifications. About 10 mg of each sample extract was dissolved in 1 mL of ethanol 70%. We added 100 µL of sample solution into a 96-well plate and then mixed it with 100 µL of DPPH 125 µM (in ethanol). The mixture was incubated at 37 °C for 30 min, and the absorbance was measured at 517 nm. The percentage of scavenging activity of the extract was calculated by using the following equation:
| (1) |
where A = absorbance of sample, B = absorbance of sample correction, C = absorbance of blank, and D = absorbance correction of the blank. Antioxidant activity was determined using the following equation: y = 0.06665x − 0.0333, with R2 = 0.9984. Quercetin was used as the standard.
4.7. α-Glucosidase Enzyme Inhibition Assay
The method for determining α-glucosidase enzyme inhibitory activity has been previously described by Hamid et al. [45] with slight modifications. The reaction mixture consisted of a 10 µL sample extract, 50 µL phosphate buffer (0.1 M; pH 7), 25 µL 4-nitrophenyl α-d-glucopyranoside, 0.01 M substrate in phosphate buffer (pH 7), and 25 µL α-glucosidase enzyme (0.1 U/mL α-glucosidase solution in 0.01 M phosphate buffer pH 6). The mixture was added to a 96-well plate, and the plate was agitated slowly for 5 min. After being homogenized, the mixture was incubated at 37 °C for 20 min. The reaction process was stopped by adding 100 µL of sodium carbonate. The α-glucosidase inhibitory activity was determined by measuring the absorbance at 410 nm in a microplate reader. The following equation was used for the determination of α-glucosidase inhibitory activity according to antioxidant activity.
The inhibitory activity was determined using the following equation: y = 13.516 ln(x) + 68.941, with R2 = 0.9272. Acarbose was used as the standard.
4.8. Measurement of FTIR Spectrum
Dried extracts (5 mg) of each sample were mixed with 95 mg of KBr and then compressed to form a tablet (3 mm) before the analysis by FTIR spectrophotometry. The FITR spectrophotometer (Tensor 37, Bruker Optik GmbH, Germany) was equipped with a DTGS (deuterated detectors triglycine sulfate). FTIR spectra were recorded in the region of 4000–400 cm–1, in absorbance mode, 32 scans/min, and resolution of 4 cm–1. The spectra were processed by OPUS ver. 4.2 software (Bruker, Germany) and Spectragryph v1.2.11. This analysis was used to obtain the spectrum pattern from the sample of plant parts, i.e., leaves, petiole, and rhizome. Then, the obtained spectrum was analyzed by chemometrics for the identification, discrimination, groupings, and detecting any functional groups that might contribute to the antioxidant activity and α-glucosidase inhibition.
4.9. Identification of Metabolites by UHPLC-Q-Orbitrap HRMS
This qualitative analysis referred to the method developed by He et al. [31], with various modifications. Approximately 50 mg of dried extract was ultrasonically extracted (Branson Ultrasonic Corporation, Danbury, CT, USA) with 1.5 mL of methanol at 30 °C for 15 min. Then, the mixture was filtered through a 0.2 µm syringe filter membrane (SY25TF PTFE mdi), and the filtrate was collected in a vial. Metabolite profiling was performed in a Vanquish Flex UHPLC-Q Exactive Plus Orbitrap High-Resolution Mass Spectrometer using AccucoreTM Phenyl Hexyl (100 × 2.1 mm, 2.6 µm) as the separation column and UV detector at 254 nm. The source of MS ionization used was electrospray ionization (ESI) and Q-Orbitrap was used as the mass analyzer. The collision energy used for fragmentation was 18, 35, and 53 eV. Other conditions were as follows: spray voltage 3.8 kV, a capillary temperature of about 320 °C, sheath gas and auxiliary gas flow rates of 15 and 3 mL/min, respectively.
The flow rate from the delivery system was adjusted at 0.3 mL/min, the autosampler temperature was maintained at 10 °C, and the sample injection volume was 0.5 µL. The mobile phase consisted of (A) 0.1% formic acid in water and (B) acetonitrile. A linear gradient elution program was applied as follows: 0–1.5 min (5% B), 1.5–9 min (5–10% B), 9–13 min (10–20% B), 13–17 min (20–28% B), 17–23 min (28–78% B), 23–26 min (70–95% B), 26–29 min (95% B), and 29–32 min (5% B). The total run time was 32 min, with relative abundance 0–100 and MS full-scan type (100–1500) in positive and negative mode.
4.10. Data Analysis
The experimental results of total phenolics and flavonoid contents and the determination of antioxidant and α-glucosidase inhibitory are expressed as means ± SD of three replicates for each sample. One-way ANOVA used R ver. I386 3.6.0 and significance levels were tested at p < 0.05 (Welch’ s test).
The processed data from FTIR (spectra data) [15] and UHPLC-Q-Orbitrap HRMS (peak area value) [46] were analyzed using the UnscramblerX ver. 10.4. (CAMO, Norwegia) for multivariate data analysis (MVDA). In order to access the information of patterns of sample grouping, similarities, and differences among species and plant parts, principal component analysis (PCA) was used. In this PCA method, data preprocessing uses a spectroscopic model, validation with full cross-validation, and an algorithm using singular value decomposition (SVD). The partial least squares (PLS) method was conducted to analyze further the correlation of antioxidant and α-glucosidase inhibitory activity with functional groups and the chemical compounds in the Curculigo spp. samples. In the PLS method, data preprocessing uses a spectroscopic model, validation with full cross-validation, and an algorithm using the Kernel PLS.
Qualitative analysis of the compounds was analyzed using the MS-DIAL ver. 3.82 (peak discrimination, filtering, and alignment). MS raw data (*.raw format) were imported to Abf Converter.4.0.0 (*.abf format) and MS FileReader 2.2.62. The peak detection parameters used are minimum peak height 10,000 (Orbitrap), smoothing method (linear weighted moving average), smoothing level (2), and minimum peak width (3). Identification used an in-house database with a score cut off (75%), adduct type [M + H]+ and [M − H]−, and alignment using reference blanks. Then, for the compounds identified, we calculated the mass error value (10 ppm) as the filtering criteria to scan experiments. Fragmentation patterns (MS and MS/MS) were analyzed using Compound Discoverer™ Software (Thermo Scientific™, Waltham, MA, USA), and there are freely available software fragmentation algorithms such as MS-FINDER, CFM-ID (https://cfmid.wishartlab.com/), MetFrag, and CSI:FingerID (https://www.csi-fingerid.uni-jena.de/).
In addition, a heatmap of hierarchical cluster analysis was conducted to present the results of putatively identified compounds in plant parts extracts of Curculigo spp. and can be used to discover clustering patterns in the datasets. Heatmap was constructed with MetaboAnalyst 4.0. (https://www.metaboanalyst.ca) using peak areas data (*.csv format) from all compounds detected in the adduct type ([M + H]+ and [M − H]−) of each organ of Curculigo species. For hierarchical clustering analysis, distance measures using Euclidean, clustering algorithms using complete, and standardization using autoscale were chosen [30,47].
5. Conclusions
C. latifolia’s rhizome contains a more significant amount of total phenolics than all parts of C. orchioides, and the phenolic compounds are more active as an antioxidant and α-glucosidase inhibitor than flavonoids. FTIR, UHPLC-Q-Orbitrap HRMS, and assays for biological activities, combined with PCA, can be used as a comprehensive method to distinguish the two species, C. orchioides and C. latifolia, and also their plant parts. Using PLS analysis, we can predict the functional groups and the chemical compounds contributing to antioxidant and α-glucosidase inhibitory activities. The chemical compounds that contribute to the antioxidant activity and the inhibition of α-glucosidase, unknown-185 and unknown-85, are accumulated the highest in LLSB and LOBM samples, and all are founded in leaves of Curculigo species. Based on FTIR and PLS analysis, –OH, C=O, C–O, –C–H, C–C, and C–OH groups from these compounds are the major groups that might contribute the most to the biological activities in Curculigo plant species.
Acknowledgments
The authors gratefully acknowledged The Directorate of Higher Education, The Ministry of Research and Higher Education the Republic of Indonesia, for funding this research, which is part of the scholarship (BPP-DN Scheme 2017–2020) granted to Abdul Halim Umar.
Supplementary Materials
The following are available online at https://www.mdpi.com/2218-1989/11/1/42/s1. Figure S1: FTIR spectra of a 70% ethanol extract of C. orchioides and C. latifolia plant parts, Figure S2: Typical total ion chromatograms profiles of 70% ethanol extracts of C. orchioides and C. latifolia plant parts, Figure S3: The proposed fragmentation patterns of representative curculigosaponin G (triterpenoid) (A), curculigosaponin H (triterpenoid) (B), curculigosaponin I (triterpenoid) (C), curculigoside B (phenolic) (D), orchioiside B (phenolic) (E), and (1S,2R)-O-Methylnyasicoside (norlignan) (F) at ion mode [M + H]+) and [M − H]−) in C. orchioides and C. latifolia, Figure S4: Heatmap correlation between chemical compounds in 70% ethanol extract of C. orchioides and C. latifolia and their relative abundance. Positive mode (A) and negative mode (B). The color scale differentiates values: high (red), moderate (black), and low (green), Figure S5: Partial least squares (PLS) plots x-y relation outliers antioxidant (A) and α-glucosidase inhibitory (B) activity of a 70% ethanol extract of C. orchioides and C. latifolia. Axis and ordinate plots show PLS scores for each organ sample, Figure S6: The chemical structure of curculigosaponin G (A), curculigosaponin H (B), curculigosaponin I (C), curculigoside B (D), orchioiside B (E), and (1S,2R)-O-Methylnyasicoside (F) and their functional groups might have a major contribution to the antioxidant activity and the inhibition of α-glucosidase, Figure S7: Sampling location of Curculigo spp. in South Sulawesi, Sulawesi Island, Indonesia. Curculigo orchioides Gaertn. (A) and Curculigo latifolia Dryand. ex W.T.Aiton (B), Table S1: The chemical compounds identified in all sample extracts of C. orchioides and C. Latifolia.
Author Contributions
Conceptualization and methodology, A.H.U., D.R. and M.R.; software, validation, data curation, and formal analysis, A.H.U. and M.R.; investigation, D.R., M.R. and Y.C.S.; writing-original draft preparation, A.H.U.; writing—review and editing, A.H.U., D.R., M.R. and Y.C.S.; supervision, D.R. All authors have read and agreed to the published the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in https://www.mdpi.com/2218-1989/11/1/42/s1.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ishak N.A., Ismail M., Hamid M., Ahmad Z., Abd Ghafar S.A. Antidiabetic and hypolipidemic activities of Curculigo latifolia fruit: Root extract in high fat fed diet and low dose STZ induced diabetic rats. J. Evid. Based Complement. Altern. Med. 2013;2013:601838. doi: 10.1155/2013/601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farzinebrahimi R., Mat Taha R., Rashid K.A., Ali Ahmed B., Danaee M., Rozali S.E. Preliminary screening of antioxidant and antibacterial activities and establishment of an efficient callus induction in Curculigo latifolia Dryand (Lemba) J. Evid. Based Complement. Altern. Med. 2016;2016:6429652. doi: 10.1155/2016/6429652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yokosuka A., Sato K., Yamori T., Mimaki Y. Triterpene glycosides from Curculigo orchioides and their cytotoxic activity. J. Nat. Prod. 2010;73:1102–1106. doi: 10.1021/np100111s. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z.H., Gong X.Y., Zhou D.J., Xu P.F., Huang M., Zhang Q.L., Meng Y.L., Niu C., Zhang Y.R. Three new chlorophenolic glucosides from Curculigo orchioides Gaertn. Phytochem. Lett. 2018;26:9–11. doi: 10.1016/j.phytol.2018.05.005. [DOI] [Google Scholar]
- 5.Zuo A.X., Shen Y., Jiang Z.Y., Zhang X.M., Zhou J., Lü J., Chen J.J. Three new phenolic glycosides from Curculigo orchioides G. Fitoterapia. 2010;81:910–913. doi: 10.1016/j.fitote.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Zabidi N.A., Ishak N.A., Hamid M., Efliza Ashari S. Subcritical water extraction of antioxidants from Curculigo latifolia root. J. Chem. 2019;2019:8047191. doi: 10.1155/2019/8047191. [DOI] [Google Scholar]
- 7.Sales P.M., Souza P.M., Simeoni L.A., Silveira D. α-Amylase inhibitors: A review of raw material and isolated compounds from plant source. J. Pharm. Pharm. Sci. 2012;15:141–183. doi: 10.18433/J35S3K. [DOI] [PubMed] [Google Scholar]
- 8.Nie Y., Dong X., He Y., Yuan T., Han T., Rahman K., Qin L., Zhang Q. Medicinal plants of genus Curculigo: Traditional uses and a phytochemical and ethnopharmacological review. J. Ethnopharmacol. 2013;147:547–563. doi: 10.1016/j.jep.2013.03.066. [DOI] [PubMed] [Google Scholar]
- 9.Kramberger K., Barlič-Maganja D., Bandelj D., Baruca Arbeiter A., Peeters K., Miklavčič Višnjevec A., Jenko Pražnikar Z. HPLC-DAD-ESI-QTOF-MS determination of bioactive compounds and antioxidant activity comparison of the hydroalcoholic and water extracts from two Helichrysum italicum species. Metabolites. 2020;10:403. doi: 10.3390/metabo10100403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim V., Gorji S.G., Daygon V.D., Fitzgerald M. Untargeted and targeted metabolomic profiling of Australian indigenous fruits. Metabolites. 2020;10:114. doi: 10.3390/metabo10030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong X., Guo Y., Xiong C., Sun L. Evaluation of two major Rhodiola species and the systemic changing characteristics of metabolites of Rhodiola crenulata in different altitudes by chemical methods combined with UPLC-QqQ-MS-based metabolomics. Molecules. 2020;25:4062. doi: 10.3390/molecules25184062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bystrzanowska M., Tobiszewski M. Chemometrics for selection, prediction, and classification of sustainable solutions for green chemistry—A review. Symmetry. 2020;12:2055. doi: 10.3390/sym12122055. [DOI] [Google Scholar]
- 13.Stocchero M., Locci E., d’Aloja E., Nioi M., Baraldi E., Giordano G. PLS2 in metabolomics. Metabolites. 2019;9:51. doi: 10.3390/metabo9030051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim T.J., Park J.G., Ahn S.K., Kim K.W., Choi J., Kim H.Y., Ha S.H., Seo W.D., Kim J.K. Discrimination of Adzuki bean (Vigna angularis) geographical origin by targeted and non-targeted metabolite profiling with gas chromatography time-of-flight mass spectrometry. Metabolites. 2020;10:112. doi: 10.3390/metabo10030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hawrył A., Hawrył M., Hajnos-Stolarz A., Abramek J., Bogucka-Kocka A., Komsta Ł. HPLC fingerprint analysis with the antioxidant and cytotoxic activities of selected lichens combined with the chemometric calculations. Molecules. 2020;25:4301. doi: 10.3390/molecules25184301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Albergaria E.T., Oliveira A.F.M., Albuquerque U.P. The effect of water deficit stress on the composition of phenolic compounds in medicinal plants. S. Afr. J. Bot. 2020;131:12–17. doi: 10.1016/j.sajb.2020.02.002. [DOI] [Google Scholar]
- 17.Sirooeinejad B., Zamani Z., Fatahi M.R. Study of physiological and biochemical responses to freezing stress in pomegranate (Punica granatum L.) trees during acclimation and deaclimation cycle. J. Hortic. Sci. Biotechnol. 2019;95:341–355. doi: 10.1080/14620316.2019.1674699. [DOI] [Google Scholar]
- 18.Groenbaek M., Tybirk E., Neugart S., Sundekilde U.K., Schreiner M., Kristensen H.L. Flavonoid glycosides and hydroxycinnamic acid derivatives in baby leaf rapeseed from white and yellow flowering cultivars with repeated harvest in a 2-years field study. Front. Plant Sci. 2019;10:355. doi: 10.3389/fpls.2019.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hashim A.M., Alharbi B.M., Abdulmajeed A.M., Elkelish A., Hozzein W.N., Hassan H.M. Oxidative stress responses of some endemic plants to high altitudes by intensifying antioxidants and secondary metabolites content. Plants. 2020;9:869. doi: 10.3390/plants9070869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costa T.M., Mayer D.A., Siebert D.A., Micke G.A., Alberton M.D., Tavares L.B.B., de Oliveira D. Kinetics analysis of the inhibitory effects of alpha-glucosidase and identification of compounds from Ganoderma lipsiense Mycelium. Appl. Biochem. Biotechnol. 2020;10:1007. doi: 10.1007/s12010-020-03239-4. [DOI] [PubMed] [Google Scholar]
- 21.Tacchini M., Spagnoletti A., Marieschi M., Caligiani A., Bruni R., Efferth T., Sacchetti G., Guerrini A. Phytochemical profile and bioactivity of traditional ayurvedic decoctions and hydro-alcoholic macerations of Boerhaavia diffusa L. and Curculigo orchioides Gaertn. Nat. Prod. Res. 2015;29:2071–2079. doi: 10.1080/14786419.2014.1003299. [DOI] [PubMed] [Google Scholar]
- 22.Alqahtani A.S., Hidayathulla S., Rehman M.T., ElGamal A.A., Al-Massarani S., Razmovski-Naumovski V., Alqahtani M.S., El Dib R.A., AlAjmi M.F. Alpha-amylase and alpha-glucosidase enzyme inhibition and antioxidant potential of 3-Oxolupenal and katononic acid isolated from Nuxia oppositifolia. Biomolecules. 2020;10:61. doi: 10.3390/biom10010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anacleto-Santos J., López-Camacho P., Mondragón-Flores R., Vega-Ávila E., Islas G.B., Mondragón-Castelán M., Carrasco-Ramírez E., Rivera-Fernández N. Anti-toxoplasma, antioxidant and cytotoxic activities of Pleopeltis crassinervata (Fée) T. Moore hexane fraction. Saudi J. Biol. Sci. 2020;27:812–819. doi: 10.1016/j.sjbs.2019.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Craft B.D., Kerrihard A.L., Amarowicz R., Pegg R.B. Phenol-based antioxidants and the in vitro methods used for their assessment. Compr. Rev. Food Sci. Food Saf. 2012;11:148–173. doi: 10.1111/j.1541-4337.2011.00173.x. [DOI] [Google Scholar]
- 25.Shu P., Li J., Fei Y., Zhu H., Yu M., Liu A., Niu H., Zou S., Wei X., Ju Z., et al. Isolation, structure elucidation, tyrosinase inhibitory, and antioxidant evaluation of the constituents from Angelica dahurica roots. J. Nat. Med. 2020;74:456–462. doi: 10.1007/s11418-019-01375-8. [DOI] [PubMed] [Google Scholar]
- 26.Teng H., Yuan B., Gothai S., Arulselvan P., Song X., Chen L. Dietary triterpenes in the treatment of type 2 diabetes: To date. Trends Food Sci. Technol. 2018;72:34–44. doi: 10.1016/j.tifs.2017.11.012. [DOI] [Google Scholar]
- 27.Kakar M., Amin M.U., Alghamdi S., Sahibzada M.U.K., Ahmad N., Ullah N. Antimicrobial, cytotoxic, and antioxidant potential of a novel flavone “6,7,4′-Trimethyl Flavone” isolated from Wulfenia amherstiana. J. Evid. Based Complement. Altern. Med. 2020;2020:3903682. doi: 10.1155/2020/3903682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saleh M.S.M., Siddiqui M.J., Mat So’ad S.Z., Roheem F.O., Saidi-Besbes S., Khatib A. Correlation of FT-IR fingerprint and α-glucosidase inhibitory activity of salak (Salacca zalacca) fruit extracts utilizing orthogonal partial least square. Molecules. 2018;23:1434. doi: 10.3390/molecules23061434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Z., Meenu M., Chen P., Xu B. Comparative study on phytochemical profiles and antioxidant capacities of chestnuts produced in different geographic area in China. Antioxidants. 2020;9:190. doi: 10.3390/antiox9030190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daliri E.B.-M., Ofosu F.K., Chelliah R., Kim J.H., Kim J.R., Yoo D., Oh D.H. Untargeted metabolomics of fermented rice using UHPLC Q-TOF MS/MS reveals an abundance of potential antihypertensive compounds. Foods. 2020;9:1007. doi: 10.3390/foods9081007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He Y., Dong X., Jia X., Li M., Yuan T., Xu H., Qin L., Han T., Zhang Q. Qualitative and quantitative analysis on chemical constituents from Curculigo orchioides using ultra high performance liquid chromatography coupled with electrospray ionization quadrupole time-of-flight tandem mass spectrometry. J. Pharm. Biomed. Anal. 2015;102:236–245. doi: 10.1016/j.jpba.2014.09.024. [DOI] [PubMed] [Google Scholar]
- 32.Saha N., Trivedi P., Dutta Gupta S. Surface plasmon resonance (SPR) based optimization of biosynthesis of silver nanoparticles from rhizome extract of Curculigo orchioides Gaertn. and its antioxidant potential. J. Clust. Sci. 2016;27:1893–1912. doi: 10.1007/s10876-016-1050-7. [DOI] [Google Scholar]
- 33.Xu M., Shen C., Zheng H., Xu Y., Xue C., Zhu B., Hu J. Metabolomic analysis of acerola cherry (Malpighia emarginata) fruit during ripening development via UPLC-Q-TOF and contribution to the antioxidant activity. Food Res. Int. 2020;130:108915. doi: 10.1016/j.foodres.2019.108915. [DOI] [PubMed] [Google Scholar]
- 34.Ghane S., Attar U., Yadav P., Lekhak M. Antioxidant, anti-diabetic, acetylcholinesterase inhibitory potential and estimation of alkaloids (lycorine and galanthamine) from Crinum species: An important source of anticancer and anti-alzheimer drug. Ind. Crops. Prod. 2018;125:168–177. doi: 10.1016/j.indcrop.2018.08.087. [DOI] [Google Scholar]
- 35.Batsukh Z., Toume K., Javzan B., Kazuma K., Cai S.Q., Hayashi S., Kawahara N., Maruyama T., Komatsu K. Metabolomic profiling of Saposhnikoviae radix from Mongolia by LC-IT-TOF-MS/MS and multivariate statistical analysis. J. Nat. Med. 2020;74:170–188. doi: 10.1007/s11418-019-01361-0. [DOI] [PubMed] [Google Scholar]
- 36.Rohaeti E., Rafi M., Syafitri U.D., Heryanto R. Fourier transform infrared spectroscopy combined with chemometrics for discrimination of Curcuma longa, Curcuma xanthorrhiza and Zingiber cassumunar. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2015;137:1244–1249. doi: 10.1016/j.saa.2014.08.139. [DOI] [PubMed] [Google Scholar]
- 37.Tsiokanos E., Tsafantakis N., Termentzi A., Aligiannis N., Skaltsounis L.A., Fokialakis N. Phytochemical characteristics of bergamot oranges from the Ionian Islands of Greece: A multi-analytical approach with emphasis in the distribution of neohesperidose flavanones. Food Chem. 2020:128400. doi: 10.1016/j.foodchem.2020.128400. [DOI] [PubMed] [Google Scholar]
- 38.Garza-Juárez A., de la Luz Salazar-Cavazos M., Salazar-Aranda R., Pérez-Meseguer J., de Torres N.W. Correlation between chromatographic fingerprint and antioxidant activity of Turnera diffusa (Damiana) Planta Med. 2011;77:958–963. doi: 10.1055/s-0030-1250684. [DOI] [PubMed] [Google Scholar]
- 39.Innamorato V., Longobardi F., Lippolis V., Cortese M., Logrieco A.F., Catucci L., Agostiano A., De Girolamo A. Tracing the geographical origin of lentils (Lens culinaris Medik.) by infrared spectroscopy and chemometrics. Food Anal. Methods. 2019;12:773–779. doi: 10.1007/s12161-018-1406-8. [DOI] [Google Scholar]
- 40.Easmin S., Sarker M.Z.I., Ghafoor K., Ferdosh S., Jaffri J., Ali M.E., Mirhosseini H., Al-Juhaimi F.Y., Perumal V., Khatib A. Rapid investigation of α-glucosidase inhibitory activity of Phaleria macrocarpa extracts using FTIR-ATR based fingerprinting. J. Food Drug Anal. 2017;25:306–315. doi: 10.1016/j.jfda.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sánchez-Marzo N., Lozano-Sánchez J., de la Luz Cádiz-Gurrea M., Herranz-López M., Micol V., Segura-Carretero A. Relationships between chemical structure and antioxidant activity of isolated phytocompounds from lemon verbena. Antioxidants. 2019;8:324. doi: 10.3390/antiox8080324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Margraf T., Karnopp A.R., Rosso N.D., Granato D. Comparison between Folin-Ciocalteu and Prussian Blue Assays to estimate the total phenolic content of juices and teas using 96-well microplates. J. Food Sci. 2015;80:C2397–C2403. doi: 10.1111/1750-3841.13077. [DOI] [PubMed] [Google Scholar]
- 43.Zhu M.Z., Wu W., Jiao L.L., Yang P.F., Guo M.Q. Analysis of flavonoids in lotus (Nelumbo nucifera) leaves and their antioxidant activity using macroporous resin chromatography coupled with LC-MS/MS and antioxidant biochemical assays. Molecules. 2015;20:10553–10565. doi: 10.3390/molecules200610553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ananthi S., Raghavendran H.R.B., Sunil A.G., Gayathri V., Ramakrishnan G., Vasanthi H.R. In vitro antioxidant and in vivo anti-inflammatory potential of crude polysaccharide from Turbinaria ornata (marine brown alga) Food Chem. Toxicol. 2010;48:187–192. doi: 10.1016/j.fct.2009.09.036. [DOI] [PubMed] [Google Scholar]
- 45.Hamid H.A., Yusoff M.M., Liu M., Karim M.R. α-Glucosidase and α-amylase inhibitory constituents of Tinospora crispa: Isolation and chemical profile confirmation by ultra-high performance liquid chromatography-quadrupole time-of-flight/mass spectrometry. J. Funct. Foods. 2015;16:74–80. doi: 10.1016/j.jff.2015.04.011. [DOI] [Google Scholar]
- 46.Li C., Tu C., Che Y., Zhang M., Dong B., Zhou X., Shi Y., Li G., Wang J. Bioassay based screening for the antiplatelet aggregation quality markers of Polygonum multiflorum with UPLC and chemometrics. J. Pharm. Biomed. Anal. 2019;166:264–272. doi: 10.1016/j.jpba.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 47.Hemmer S., Manier S.K., Fischmann S., Westphal F., Wagmann L., Meyer M.R. Comparison of three untargeted data processing workflows for evaluating LC-HRMS metabolomics data. Metabolites. 2020;10:378. doi: 10.3390/metabo10090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available in https://www.mdpi.com/2218-1989/11/1/42/s1.