Abstract
The cell wall plays an important role in responses to various stresses. The cellulose synthase-like gene (Csl) family has been reported to be involved in the biosynthesis of the hemicellulose backbone. However, little information is available on their involvement in plant tolerance to low-temperature (LT) stress. In this study, a total of 42 Csls were identified in Musa acuminata and clustered into six subfamilies (CslA, CslC, CslD, CslE, CslG, and CslH) according to phylogenetic relationships. The genomic features of MaCsl genes were characterized to identify gene structures, conserved motifs and the distribution among chromosomes. A phylogenetic tree was constructed to show the diversity in these genes. Different changes in hemicellulose content between chilling-tolerant and chilling-sensitive banana cultivars under LT were observed, suggesting that certain types of hemicellulose are involved in LT stress tolerance in banana. Thus, the expression patterns of MaCsl genes in both cultivars after LT treatment were investigated by RNA sequencing (RNA-Seq) technique followed by quantitative real-time PCR (qPCR) validation. The results indicated that MaCslA4/12, MaCslD4 and MaCslE2 are promising candidates determining the chilling tolerance of banana. Our results provide the first genome-wide characterization of the MaCsls in banana, and open the door for further functional studies.
Keywords: banana (Musa spp.), cellulose synthase-like genes, genome-wide identification, hemicellulose, low temperature stress
1. Introduction
The plant cell wall consists of polysaccharides (cellulose, hemicellulose and pectin), proteins and other compounds. It plays critical roles in the maintenance of cell integrity, and the regulation of many developmental processes in plants [1,2,3,4,5,6]. The cell wall represents not only a mechanical barrier, but also a signaling component during plant responses to various biotic [7,8] and abiotic stresses [9,10].
Hemicelluloses are a diverse group of complex, non-cellulosic polysaccharides, which constitute approximately one-third of the plant cell wall. The backbones of hemicellulosic polysaccharides in plants are made of the cellulose synthase-like (Csl), which are members of a much larger superfamily of genes referred to as glycosyltransferase 2 [11,12]. Early studies of cellulose synthase (CesA) homologs in model plant organisms established that there are nine Csl families: CslA, CslB, CslC, CslD, CslE, CslF, CslG, CslH and CslJ [11,13,14]. Recent research in other flowering plants has identified a new CslM family [15].
It has been reported that CslAs are involved in the biosynthesis of mannan and glucomannan backbones [5,16,17,18,19], while CslCs are related to the synthesis of xyloglucan backbone [20,21,22]. CslD genes may participate in either cellulose or mannan synthesis in tip-growing cells [23,24,25,26,27], as well as xylan and homogalacturonan [28]. CslF, CslH and CslJ subfamilies are responsible for the synthesis of (1,3; 1,4)-β-glucan, also known as mixed-linkage glucan (MLG) synthases [29,30,31,32,33]. However, the functions of CslB/E/G/M remain poorly characterized [34].
Plant Csl genes play substantial roles in developmental processes, such as root hair formation [6], the control of organ size [4], tiller number [3] and the maintenance of adherent mucilage structure [1,2,5]. Csl genes were also reported to be involved in plant resistance/tolerance to biotic or abiotic stresses, such as salt [35,36], boron (B) [37,38] or heavy metal [36,39] stress, as well as pathogen infection [32,40,41]. The responses of Csl genes or hemicellulose components to low temperatures (LTs) have been reported in many plant species, including banana [42,43,44,45,46]. Recently, some hemicellulose metabolism-related genes were proven to play important roles in plant tolerance to LT stress [47,48]. However, the role of Csl genes in plant tolerance to LT stress has not been reported.
A detailed characterization of the plant Csl genes will be helpful for better understanding their functional and biochemical properties. To date, the whole Csl gene repertoire has been cataloged in rice (Oryza sativa) [13], poplar (Populus trichocarpa) [49,50], moss (Physcomitrella patens) [51], maize (Zea mays) [52,53], barley (Hordeum vulgare) [54], pine (Pinus taed) [55], tomato (Solanum lycopersicum) [56], pineapple (Ananas comosus) [57], bread wheat (Triticum aestivum L.) [58] and white pears (Pyrus bretschneideri) [59]. However, these genes have not been extensively studied in banana (Musa spp.), one of the most important fruit and food crops in the tropical and subtropical regions [60,61].
To screen potential Csl candidates that determine the chilling tolerance of banana, we provide a genome-wide characterization of Csls in banana (Musa acuminata). Moreover, changes in the hemicellulose contents in chilling-tolerant (CT) and chilling-sensitive (CS) banana cultivars after exposure to LT stress were investigated, and the expression of 42 MaCsl members was examined by RNA sequencing (RNA-Seq) techniques followed by validation with quantitative real-time PCR (qPCR) in the present study. These results will significantly facilitate studies focused on the functions of MaCsls in plant growth and development, as well as tolerance/resistance to biotic and abiotic stresses. Furthermore, we present the potential MaCsls candidates determining banana chilling tolerance, which might be used for the development of new banana genotypes tolerant to chilling conditions.
2. Results
2.1. The Response of Hemicellulose in Banana to LT Stress
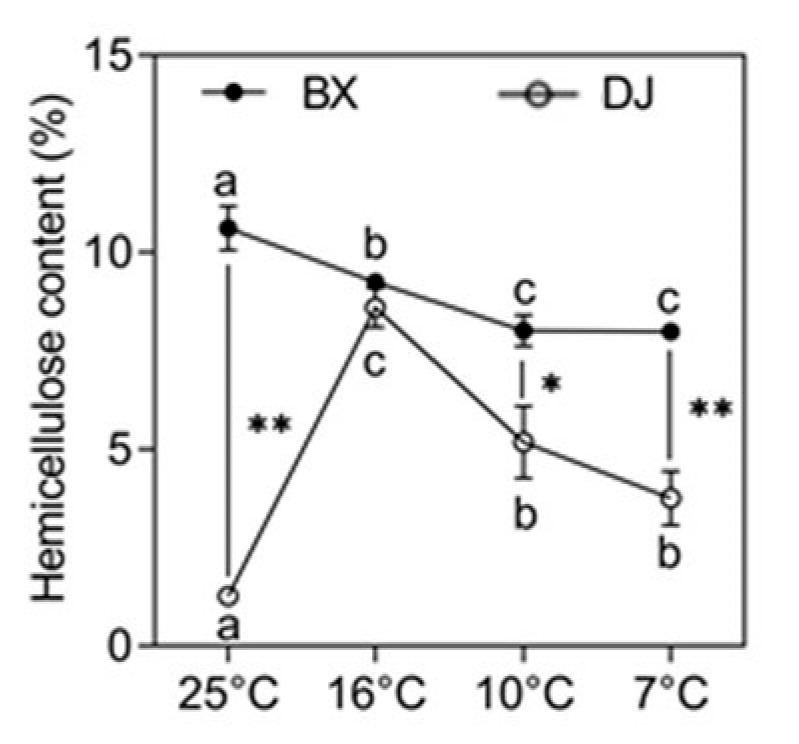
The CS cultivar ‘Baxijiao’ (BX) exhibited an eight-times higher content of hemicellulose compared to the CT cultivar ‘Dongguandajiao’ (DJ) in the control condition (25 °C; Figure 1). However, the hemicellulose content dramatically increased in the CT cultivar grown at 16 °C, followed by a significant decrease with the further decline in temperature, but it still maintained significantly high levels. Contrary, the hemicellulose content in the CS cultivar continuously decreased with the drop in temperature. The hemicellulose content in the CS cultivar was significantly higher than that in the CT plants grown at 10 °C and 7 °C.
Figure 1.
Changes in the hemicellulose content of banana (Musa spp.) under low temperature stress. Hemicellulose content is expressed as a percentage per gram of fresh leaves. The data represent an average of three replicates ± SE. Values followed by the same letter are not significantly different using a Duncan’s multiple range test at p < 0.05 after angular transformation of the data for each cultivar. Values marked with a star were considered significant at p < 0.05, while values marked with two stars were considered significant at p < 0.01 when evaluated using Student’s t-test. BX ‘Baxijiao’, chilling-sensitive (CS); DJ ‘Dongguandajiao’, chilling-tolerant (CT).
2.2. MaCsl and Their Molecular Structural Features
2.2.1. Phylogenetic Analysis of the MaCsls
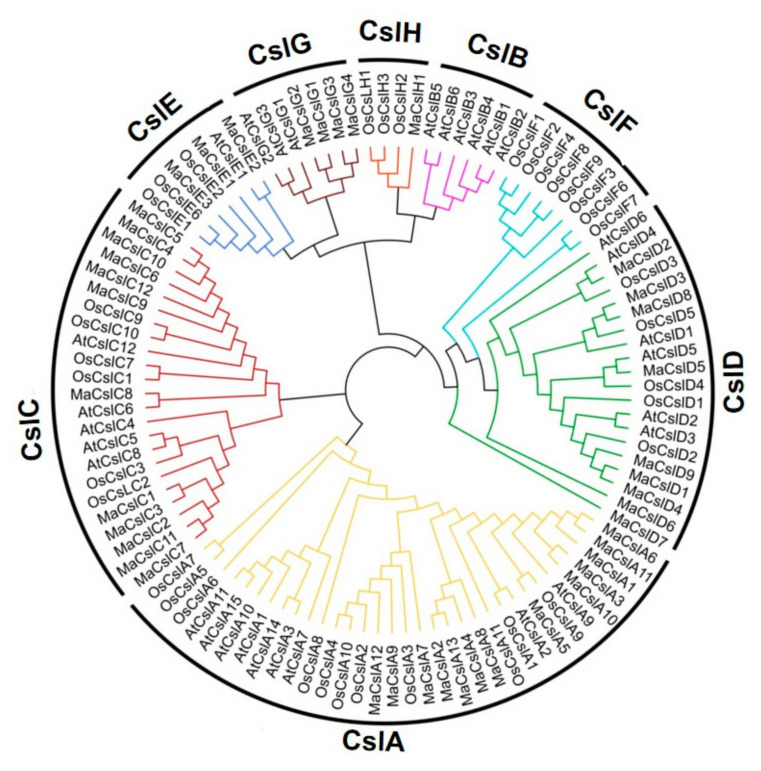
In this study, we constructed a multi-species phylogenetic tree of Arabidopsis (Arabidopsis thaliana, model species for dicots), rice (monocots), and banana (monocots) Csl genes based on full-length protein sequences using MEGA software. MaCsl subfamilies clustered together with similar Csl subfamilies from Arabidopsis and rice, indicating a shared evolutionary history (Figure 2). The most abundant MaCsl subfamilies are CslA and CslC, with 13 and 12 members, respectively. Remarkably, the CslA family is abundant also in Arabidopsis (9 genes) and rice (11 genes). The MaCslD and MaCslE subfamilies contain 9 and 3 genes, respectively. The CslG subfamily, previously reported to be specific to dicots, harbors four Csl members in monocotyledonous banana. Only one CslH member was identified in Musa acuminata, while the rice genome possesses three CslHs. In contrast to rice, which contains eight CslFs, this subfamily is missing in banana (as well as Arabidopsis). Both banana and rice do not have the CslB subfamily, the specific subfamily for dicots.
Figure 2.
Phylogenetic tree of Csl proteins in banana (Musa acuminata), rice (Oryza sativa), and Arabidopsis (Arabidopsis thaliana). Totals 42 Csl proteins in banana, 36 in rice, and 30 in Arabidopsis were analyzed using Clustal W. Neighbor-joining trees were constructed using MEGA7.0. The bootstrap value was 1000 replicates.
2.2.2. Identification of Csl Genes in Musa acuminata
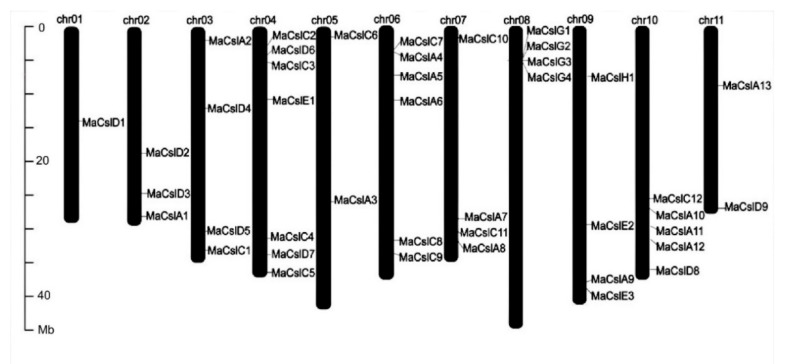
In total, we have identified 42 candidate Csl genes in the banana (Musa acuminata) genome. Based on phylogenetic relationships with Arabidopsis and rice, these 42 MaCsls are grouped into six subfamilies: MaCslA, MaCslC, MaCslD, MaCslE, MaCslG, and MaCslH. The 42 MaCsl genes are distributed over all 11 banana chromosomes. Interestingly, the MaCslG subfamily members are located solely on Chr8, and no other subfamily members are present on this chromosome (Figure 3, Table 1). The basic characterization of the MaCsl gene family, including the corresponding proteins, is shown in Table 1. The length of their open reading frames ranged from 1245 bp (MaCslD7) to 3657 bp (MaCslD5), encoding polypeptides with 415 to 1219 amino acids. The molecular weight (MW) of the polypeptides varied from 46.1 to 134.9 kD, with isoelectric points (pI) ranging from 6.4 (MaCslD2) to 9.5 (MaCslD7). The diversity in the amino acid length, MW and pI of MaCsls may indicate functional differences among the members.
Figure 3.
Chromosomal localization of the MaCsl genes in the banana (Musa acuminata) genome.
Table 1.
42 MaCsl genes identified in Musa acuminata and their sequence characteristics.
| Gene Name | Gene ID | Chr | Start | End | Length (bp) | Strand | Size (aa) | pI | MW (kD) |
|---|---|---|---|---|---|---|---|---|---|
| MaCslA1 | Ma02_t22150 | chr02 | 27,565.079 | 27,570.275 | 1602 | − | 534 | 8.9 | 60,834.3 |
| MaCslA2 | Ma03_t01730 | chr03 | 1207.899 | 1213.633 | 1629 | + | 543 | 9.0 | 61,799.5 |
| MaCslA3 | Ma05_t18900 | chr05 | 25,534.488 | 25,538.961 | 1602 | − | 534 | 9.0 | 60,875.2 |
| MaCslA4 | Ma06_t04300 | chr06 | 3100.141 | 3102.901 | 1635 | − | 545 | 8.7 | 62,234.1 |
| MaCslA5 | Ma06_t09180 | chr06 | 6479.551 | 6484.223 | 1668 | + | 556 | 9.0 | 63,946.1 |
| MaCslA6 | Ma06_t14920 | chr06 | 10,164.997 | 10,168.70 | 1602 | + | 534 | 8.9 | 60,781.3 |
| MaCslA7 | Ma07_t19410 | chr07 | 27,422.775 | 27,428.514 | 1719 | + | 573 | 9.1 | 65,060.1 |
| MaCslA8 | Ma07_t22600 | chr07 | 30,481.370 | 30,484.454 | 1677 | + | 559 | 9.0 | 64,175.8 |
| MaCslA9 | Ma09_t27610 | chr09 | 38,562.778 | 38,574.772 | 1626 | + | 542 | 8.8 | 61,639.2 |
| MaCslA10 | Ma10_t11450 | chr10 | 24,986.818 | 24,991.666 | 1602 | + | 534 | 8.9 | 61,025.5 |
| MaCslA11 | Ma10_t15510 | chr10 | 27,582.419 | 27,587.095 | 1602 | − | 534 | 9.1 | 60,994.7 |
| MaCslA12 | Ma10_t18740 | chr10 | 29,549.996 | 29,554.980 | 1755 | − | 585 | 8.9 | 65,936.2 |
| MaCslA13 | Ma11_t08690 | chr11 | 6915.612 | 6920.893 | 1626 | + | 542 | 9.3 | 62,165.2 |
| MaCslC1 | Ma03_t29290 | chr03 | 32,234.549 | 32,238.695 | 2106 | + | 702 | 8.9 | 79,991.8 |
| MaCslC2 | Ma04_t02130 | chr04 | 1873.146 | 1876.963 | 2094 | − | 698 | 8.7 | 79,922.4 |
| MaCslC3 | Ma04_t05930 | chr04 | 4437.079 | 4440.382 | 2085 | + | 695 | 9.1 | 79,398.2 |
| MaCslC4 | Ma04_t29650 | chr04 | 30,511.868 | 30,515.518 | 2115 | + | 705 | 8.5 | 79,553.3 |
| MaCslC5 | Ma04_t38760 | chr04 | 36,164.056 | 36,168.517 | 2106 | + | 702 | 7.8 | 79,537.2 |
| MaCslC6 | Ma05_t01870 | chr05 | 1142.347 | 1146.294 | 2112 | − | 704 | 8.5 | 79,580.2 |
| MaCslC7 | Ma06_t03600 | chr06 | 2622.347 | 2625.771 | 2067 | + | 689 | 8.8 | 79,142.5 |
| MaCslC8 | Ma06_t29550 | chr06 | 30,901.418 | 30,906.541 | 2091 | + | 697 | 9.1 | 78,791.9 |
| MaCslC9 | Ma06_t31890 | chr06 | 32,901.867 | 32,906.161 | 2115 | + | 705 | 8.1 | 79,750.6 |
| MaCslC10 | Ma07_t00740 | chr07 | 619,565 | 624,194 | 2121 | − | 707 | 8.0 | 79,935.6 |
| MaCslC11 | Ma07_t20970 | chr07 | 28,958.080 | 28,961.838 | 2106 | − | 702 | 8.8 | 80,597.2 |
| MaCslC12 | Ma10_t09350 | chr10 | 23,545.265 | 23,549.303 | 2124 | − | 708 | 7.5 | 79,820.2 |
| MaCslD1 | Ma01_t18500 | chr01 | 13,756.260 | 13,761.905 | 3450 | + | 1150 | 6.9 | 128,482.2 |
| MaCslD2 | Ma02_t07580 | chr02 | 18,211.558 | 18,215.380 | 3447 | − | 1149 | 6.4 | 128,432.9 |
| MaCslD3 | Ma02_t17080 | chr02 | 24,143.640 | 24,146.984 | 2709 | − | 903 | 8.9 | 100,568.6 |
| MaCslD4 | Ma03_t14070 | chr03 | 11,229.456 | 11,235.431 | 3453 | + | 1151 | 7.5 | 128,361.1 |
| MaCslD5 | Ma03_t25420 | chr03 | 29,461.721 | 29,465.754 | 3657 | − | 1219 | 8.1 | 134,887.3 |
| MaCslD6 | Ma04_t04560 | chr04 | 3486.824 | 3489.813 | 2757 | + | 919 | 8.8 | 103,194.6 |
| MaCslD7 | Ma04_t33100 | chr04 | 32,908.316 | 32,909.842 | 1245 | − | 415 | 9.5 | 46,143.7 |
| MaCslD8 | Ma10_t26210 | chr10 | 34,009.560 | 34,013.193 | 2721 | − | 907 | 8.9 | 100,850.9 |
| MaCslD9 | Ma11_t21750 | chr11 | 25,761.358 | 25,765.90 | 3480 | + | 1160 | 6.8 | 128,916.7 |
| MaCslE1 | Ma04_t13090 | chr04 | 9902.007 | 9907.918 | 2187 | − | 729 | 8.3 | 83,277.4 |
| MaCslE2 | Ma09_t20060 | chr09 | 27,488.282 | 27,493.065 | 2241 | + | 747 | 8.2 | 84,590.1 |
| MaCslE3 | Ma09_t28670 | chr09 | 39,320.301 | 39,325.310 | 2193 | + | 731 | 8.5 | 82,679.5 |
| MaCslG1 | Ma08_t05160 | chr08 | 3537.407 | 3540.538 | 2193 | − | 731 | 8.2 | 81,197.7 |
| MaCslG2 | Ma08_t05170 | chr08 | 3544.521 | 3548.562 | 2169 | − | 723 | 6.9 | 80,645.9 |
| MaCslG3 | Ma08_t05180 | chr08 | 3548.894 | 3551.285 | 1638 | − | 546 | 6.7 | 60,810.7 |
| MaCslG4 | Ma08_t05190 | chr08 | 3561.425 | 3563.941 | 1602 | − | 534 | 6.5 | 59,472.2 |
| MaCslH1 | Ma09_t08420 | chr09 | 5566.954 | 5569.888 | 2223 | + | 741 | 7.2 | 83,277.7 |
aa: amino acids; bp: base pair; kD: kilodaltons; MW: molecular weight; pI: isoelectric point.
2.2.3. Phylogenetic Evolutionary Tree, Gene Structure, and Conserved Motifs
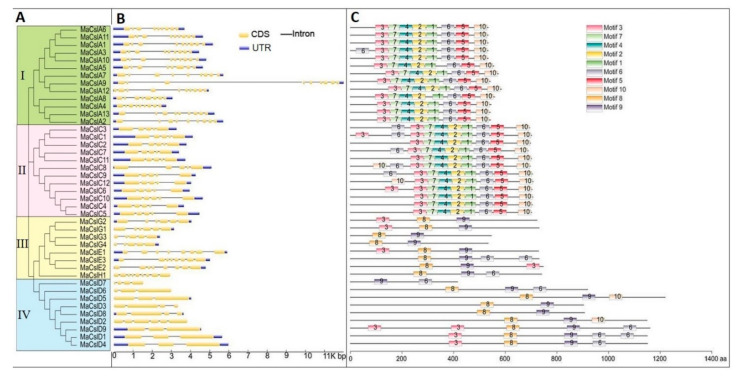
As shown in Figure 4A, the MaCsl proteins could be divided into four subgroups according to the phylogenetic distribution. MaCslA proteins are grouped into subgroup I, while subgroup II consists of MaCslC proteins. The MaCslD proteins belong to subgroup III, while the MaCslE, MaCslG and MaCslH proteins are present in subgroup IV.
Figure 4.
Phylogenetic relationships, gene structure, and motif distribution of MaCsls. (A) Unrooted phylogenetic tree of 42 MaCsls, generated with the MEGA7.0 software by the neighbor-joining method with 1000 bootstrap replicates after alignment of the full-length protein sequence by Clustal W. (B) Exon/intron structures of MaCsl genes. (C) Conserved motifs of the MaCsl gene family, analyzed by Multiple Em for Motif Elucidation (MEME). Different motifs are represented by different colored boxes with numbers 1–10 (color figure online).
The analysis of the exon–intron structure of the gene family can help to better understand its evolutionary trajectory. The intron/exon arrangement of 42 MaCsls was constructed based on the phylogenetic tree. The results showed that the exon–intron structures of the MaCsl genes are similar within subgroups I and II. The genes of subgroup I (MaCslAs) possess 7–9 introns and 8–10 exons, while only 4 introns and 5 exons are present in the genes of subgroup II (MaCslCs). In addition, most MaCslA and MaCslC genes possess both an upstream 5′ untranslated region (5′UTR) and a downstream 3′UTR. These results suggest a conserved evolutionary pattern of MaCsl genes in these two subgroups. However, a different pattern was observed in subgroups III and IV, which have a higher variation in the gene structure. For example, there are five introns in MaCslD2, but only two to three are found in other MaCslDs. Moreover, MaCslD5 has no 5′UTR, while the other four MaCslD genes (MaCslD2/3/6/7) have neither 5′UTR nor 3′UTR (Figure 4B).
Next, the Multiple Em for Motif Elucidation (MEME) web-based application was utilized to further analyze the putative motifs of MaCsl proteins. A total of 10 conserved motifs were identified, and the relative positions of these motifs in the amino acid sequences are shown in Figure 4C. The MaCslA and MaCslC members are more closely related and contain motifs 1–7 and motif 10, while the rest, with the exception of MaCslH, contain motifs 8 and 9. Members with similar motif compositions can be clustered together, indicating functional similarity among the MaCsl proteins of the same subfamily.
2.3. Differences in the Responses of MaCsls to LT Stress between CS and CT Cultivars
In order to know the response of MaCsls to LT and find the MaCsls potentially determining banana LT stress tolerance, their expressions in CT and CS banana cultivars under different LTs were investigated using RNA-Seq techniques. The number of differentially expressed genes (DEGs) in each comparison group is shown in Table S1. Cold responsive genes are listed in Table S2. The different responses of the MaCsls to LT between the CT and CS banana cultivars are shown in Table 2. Most MaCsls were downregulated by LT (s). Nine MaCsls (MaCslA2/6, MaCslC1/4, MaCslD3/6/7/8, MaCslG4) did not respond to LTs in both cultivars. Four (MaCslC3 and MaCslD1/5/9) were upregulated in both cultivars. Some other MaCsls were only regulated in the CT (MaCslA4/12, MaCslD4) or CS (MaCslA8, MaCslC5, MaCslD2) cultivar. When compared to the CS cultivar, MaCslE2 showed a significantly higher expression level in the CT cultivar at all four tested temperature points, MaCslC7 showed higher expression at 25 °C, 10 °C and 7 °C while MaCslG1 at 16 °C, 10 °C, and 7 °C. Relative to the CS cultivar, the expression levels of MaCslA10/11, MaCslC11 and MaCslD4 in the CT cultivar were significantly higher at two temperature points, and the expression levels of MaCslA13 and MaCslG2 were higher only at 7 °C. Table S3 lists the p-values of differentially expressed MaCsls, while the fragments per kilobase of exon per million reads mapped values of 42 MaCsls in the two cultivars before and after LT treatments are shown in Table S4.
Table 2.
Analysis of differentially expressed MaCsl genes in banana (Musa spp.) leaves under low temperatures.
| Gene Name | log2 Fold Change | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CKDJ vs. CKBX | LT16DJ vs. LT16BX | LT10DJ vs. LT10BX | LT7DJ vs. LT7BX | LT16BX vs. CKBX | LT10BX vs. CKBX | LT7BX vs. CKBX. | LT16DJ vs. CKDJ | LT10DJ vs. CKDJ | LT7DJ vs. CKDJ | |
| MaCslA1 | −2.52 | |||||||||
| MaCslA2 | ||||||||||
| MaCslA3 | −1.73 | −2.44 | ||||||||
| MaCslA4 | −2.10 | 1.51 | 2.00 | 2.31 | ||||||
| MaCslA5 | −1.70 | −1.99 | −2.20 | −2.46 | −2.88 | −2.81 | ||||
| MaCslA6 | ||||||||||
| MaCslA7 | −2.63 | −1.18 | −2.20 | |||||||
| MaCslA8 | 2.31 | −1.82 | 3.98 | |||||||
| MaCslA9 | −1.35 | −1.42 | −1.41 | −1.44 | ||||||
| MaCslA10 | 3.11 | 3.94 | −3.60 | −4.16 | −3.94 | |||||
| MaCslA11 | 1.96 | 2.12 | −1.76 | −2.17 | −2.20 | |||||
| MaCslA12 | −1.59 | −1.73 | 1.14 | |||||||
| MaCslA13 | 1.85 | −3.81 | −1.71 | |||||||
| MaCslC1 | ||||||||||
| MaCslC2 | −3.54 | −6.28 | −4.44 | −5.34 | −5.66 | |||||
| MaCslC3 | 4.90 | 3.93 | ||||||||
| MaCslC4 | ||||||||||
| MaCslC5 | 2.96 | −3.23 | 3.17 | 3.75 | −2.28 | −3.07 | −1.95 | |||
| MaCslC6 | −1.90 | −1.65 | ||||||||
| MaCslC7 | 3.86 | 2.59 | 3.37 | −2.97 | −3.22 | −3.20 | ||||
| MaCslC8 | −1.05 | −1.22 | −1.10 | |||||||
| MaCslC9 | −1.61 | |||||||||
| MaCslC10 | −3.45 | −3.96 | −4.22 | −3.94 | −3.53 | |||||
| MaCslC11 | 1.68 | 1.94 | −3.00 | −3.78 | −2.85 | −3.27 | −4.51 | |||
| MaCslC12 | −1.26 | |||||||||
| MaCslD1 | 1.58 | 1.34 | ||||||||
| MaCslD2 | 4.04 | |||||||||
| MaCslD3 | ||||||||||
| MaCslD4 | 1.27 | 1.34 | 1.52 | 2.16 | ||||||
| MaCslD5 | 2.46 | 2.70 | ||||||||
| MaCslD6 | ||||||||||
| MaCslD7 | ||||||||||
| MaCslD8 | ||||||||||
| MaCslD9 | −1.28 | 3.71 | 1.89 | 4.41 | ||||||
| MaCslE1 | −1.27 | |||||||||
| MaCslE2 | 1.50 | 1.98 | 1.37 | 2.58 | −1.55 | −1.70 | −2.11 | −1.16 | −1.89 | |
| MaCslE3 | −1.01 | −1.97 | −1.16 | |||||||
| MaCslG1 | 1.33 | 1.30 | 2.74 | −2.31 | ||||||
| MaCslG2 | −1.17 | 1.79 | −1.03 | −4.03 | ||||||
| MaCslG3 | −1.54 | −1.41 | −3.18 | |||||||
| MaCslG4 | ||||||||||
| MaCslH1 | −3.09 | −1.38 | −2.41 | −4.62 | −3.46 | |||||
BX: ‘Baxijiao’, chilling-sensitive; DJ: ‘Dongguandajiao’, chilling-tolerant; CK: 25 °C control; LT16: low temperature of 16 °C; LT10: low temperature of 10 °C; LT7: low temperature of 7 °C.
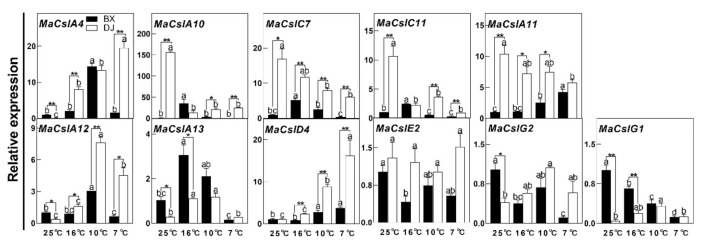
To validate the results of the RNA-Seq analysis, the expressions of genes with higher expression levels under LT(s) in the CT cultivar when compared with the CS one (MaCslA10/11/13, MaCslC7/11, MaCslD4, MaCslE2, MaCslG1/2), and the ones induced by LTs only in the CT cultivar (MaCslA4/12, and overlapped MaCslD4), were analyzed by qPCR. As shown in Figure 5, MaCslA4 was induced by all tested LT points in the CT cultivar, while it was upregulated only at 10 °C in the CS one. As a result, the CT cultivar showed significantly higher expression levels at 10 °C and 7 °C, though the result was opposite at 25 °C. The relative expression level of MaCslA10 in the control CT cultivar was 151 times higher than in the CS one, and decreased dramatically under LTs. Though there was a small peak in MaCslA10 expression in the CS cultivar at 16 °C, it was still lower than in the CT one at 10 °C and 7 °C. Similar trends were observed for MaCslC7 and MaCslC11. The LT treatment resulted in an increase in MaCslA11 expression in the CS banana, while the opposite was found in the CT cultivar, but the latter still showed a significantly higher expression level at all tested temperatures except 7 °C. MaCslA12 was upregulated by LTs of 10 °C and 7 °C in the CT cultivar, but only 10 °C in the CS one. Furthermore, the CT banana showed higher MaCslA12 expression at all tested LTs. MaCslA13 was upregulated by LTs of 16 °C and 10 °C in both cultivars. MaCslD4 was induced by LTs of 10 °C and 7 °C in both cultivars, and showed significantly higher expression levels in the CT cultivar under LTs. A decrease in expression of MaCslE2 and MaCslG2 was observed in LT-treated CS banana, but this was not the case for the CT one. Opposite to MaCslC7, the expression level of MaCslG1 in the CS cultivar was much higher than in the CT one, and it was downregulated by LTs. Though the expression level in the CT cultivar increased at 10 °C, it was lower than that in the CS cultivar at 25 °C and 16 °C. In most cases, the qPCR confirmed the results from the RNA-Seq analysis, and contradictory results were observed only for MaCslA13 and MaCslG1. In conclusion, our results suggest MaCslD4, MaCslA4/12 and MaCslE4 as genes with highest potential for the determination of banana chilling tolerance.
Figure 5.
Quantitative real-time PCR (qPCR) analysis of 11 MaCsl gene expressions in banana (Musa spp.) leaves under low temperature stress. The data represent an average of three replicates ± SE. Values followed by the same letter are not significantly different using Duncan’s multiple range test at p < 0.05 after the angular transformation of the data for each cultivar (comparison among different low-temperature points). Values marked with a star were considered significant at p < 0.05, while values marked with two stars were considered significant at p < 0.01 when evaluated using Student’s t-test (comparison between the two cultivars at the same low-temperature point). BX ‘Baxijiao’, chilling-sensitive; DJ ‘Dongguandajiao’, chilling-tolerant.
3. Discussion
3.1. The Features of MaCsls
Hemicelluloses encompass heteromannans, xyloglucan, heteroxylans, and MLG, and constitute roughly one-third of the cell wall biomass [62], and their backbones are considered to be synthesized by Csl proteins. The first report on the function of Csl-encoded proteins demonstrated their mannan-synthase activity in soybean (Glycine max) [16]. Later on, numerous evidences were reported on their capability to synthesize hemicellulose backbones [5,17,18,19,21,22,23,27,29,30,31,33,63]. However, little is known about their function in plant growth, development and stress tolerance.
A genome-wide analysis of gene family is an efficient approach for understanding gene structure, function, and evolution. To date, detailed genome-wide explorations of Csls have been limited to less than 20 plant species, such as Arabidopsis, poplar, rice, sorghum (Sorghum bicolor), maize, and various grasses [50,53,56,57,59,64,65]. In this study, we conducted a comprehensive analysis of the MaCsl gene family, including the identification of members, phylogenetic relationships, chromosomal distribution, and expression profiles in two LT-treated banana cultivars with different tolerances to LT. A total of 42 putative Csl genes were identified in the Musa acuminata genome. The number of Csl genes varies among the plant species, ranging from 21 in Dendrobium catenatum [66] to 108 in bread wheat [58]. Further, the number of Csl genes in these species is not proportional with the genome size, likely because the genomes of some species have undergone significant gene losses [53].
Plant Csl gene family could be classified into ten subfamilies (CslA–CslH, CslJ and CslM) [11,13,14,15]. Among them, CslA, CslC, and CslD are conserved in all land plants. The CslB and CslG families have been known to be specific for dicots [24], whereas CslF and CslH are restricted to grasses [14,67]. We have found that the genome of banana does not have the CslF family, but it contains CslGs. Similarly, CslJ was originally believed to be specific to grasses, but it was recently identified in some dicots [14]. On the other hand, a recent report established the presence of the CslB subfamily in monocots as well [68]. These results suggest that the knowledge about plant Csl gene family needs further examination. Banana contains only one CslH gene, and lacks CslF and CslJ genes, suggesting that the abundance/level of MLG in banana is much lower than in the other monocotyledonous crops, such as rice, wheat and maize, because they possess much more CslF/H/J genes responsible for the biosynthesis of MLG [13,53,58,69].
In bread wheat, more than half the TaCsl genes are concentrated on two chromosomes (chr2 and chr3 of each sub-genome) [58]. However, in banana, most chromosomes contain 4–5 Csl genes. This suggests a relatively even distribution of the MaCsl genes on these chromosomes.
3.2. The Involvement of MaCsls in Tolerance to LT Stress
Banana production is seriously threated by various biotic and abiotic stresses, such as Fusarium wilt and chilling stress. In the present study, the changes in hemicellulose content and the expressions of genes related to the biosynthesis of hemicellulose backbone were compared between CS and CT cultivars.
It was proposed that increased amounts of hemicelluloses were connected to enhanced cell wall stiffening, and prevented cell collapse caused by dehydration, thus contributing to plant tolerance to LTs [70,71]. In agreement, increase of hemicellulose content was observed only in the LT-treated CT cultivar in the present study, though the hemicellulose content in the CS cultivar was always higher than in the CT one, except at 16 °C. This suggests that the amount of specific types of hemicellulose, and not the total hemicellulose content, that was affecting the chilling tolerance of banana. Similarly, XXXG-rich xyloglucan, arabinoxylan and acetylated galactomannan were reported to be involved in plant desiccation tolerance [72]. The CT cultivar showed a striking increase in hemicellulose content when the temperature dropped from 25 °C to 16 °C, indicating that this increase is very important for the acclimation of banana to LT stress.
In the present study, RNA-Seq techniques were employed to compare the changes in the expression of 42 MaCsls between the CT and CS banana cultivars subjected to LT conditions. The results revealed that these genes were differentially regulated by LT stress. For example, some genes (e.g., MaCslC3 and MaCslD1) were upregulated by LTs in both cultivars, while others (e.g., MaCslA5 and MaCslC10) were downregulated. Some genes showed different expression patterns under LT stress in the CS and CT cultivars. For example, MaCslC5 in the CT cultivar showed lower expression levels under LT stress in comparison to 25 °C, but it showed opposite trend in the CS cultivar. On the other hand, all LTs induced the expression of MaCslD4 in the CT cultivar, but this was not the case for the CS one. Similarly, the CslD1 and CslD4 levels of chilling-tolerant indica rice were upregulated by LTs, while the result was the opposite for CslA1 and CslF6 [46]. The phenomenon that genes from the same family differently respond to the same stress is frequently observed in the plant kingdom, such as MaFLAs (fasciclin-like AGP) in banana under LT stress [10] and barley HvCslFs upon the infection of cereal cyst nematodes [40]. These results suggested that members from certain gene families play diverse roles in plant tolerance/resistance to biotic and abiotic stresses.
In the present study, most MaCsls were suppressed by LTs in both CS and CT banana cultivars. Similarly, boron deficiency resulted in the downregulation of CslB5 and several xyloglucan endotransglucosylase/hydrolase proteins (XTHs) in Arabidopsis roots [37,38]. In spinach (Spinacia oleracea L.), CslE1 was found to be inhibited by salinity stress and a combination of salinity and cadmium stress [36]. Some Csl and CesA genes in Arabidopsis were also found to negatively modulate salt tolerance [73]. Besides abiotic stress, plant Csl genes were also reported to be suppressed by pathogens. For instance, the expression of several CesA genes (homologs of Csl genes) in rice was downregulated by rice tungro spherical virus at the early stage of infection [74], while the expression level of HvCslF6 in barley decreased immediately after pathogen infection [40].
On the other hand, MaCslD4 was induced by LT in the CT cultivar, and showed significantly higher expression levels in the CT cultivar under LTs when compared to the CS one, suggesting this gene plays an important role in the chilling tolerance of banana. Besides MaCslD4, MaCslA4/12 and MaCslE2 are also likely related to banana chilling tolerance, because they showed higher expression levels in the CT cultivar when compared to the CS, and were induced by LT or remained stable under LT stress in the CT cultivar. Similarly, the AtCslG3 in Arabidopsis showed a higher expression level in the Cd-tolerant ecotype after exposure to Cd stress, likely being involved in enhanced Cd retention in the cell wall and reduction of Cd toxicity [39]. Another Arabidopsis Csl gene, AtCslD5, was suggested to play a critical role in osmotic stress, and is required for osmotic tolerance, because hypersensitive sos6-1 (encodes AtCslD5) mutant plants accumulate high levels of reactive oxygen species under osmotic stress [35]. The silencing of HvCslD2 resulted in the increased susceptibility of barley to powdery mildew, suggesting that HvCslD2-mediated cell wall changes represented an important defense reaction [41,75]. Thus, Csl genes are involved in plant tolerance/resistance to biotic and abiotic stresses [76]. The MaCsl genes found by this study could help the banana cell wall withstand the LT conditions. As proposed earlier, they might affect the cell turgor pressure [76], or increase cell wall flexibility/extensibility and reconstruction under LT-induced dehydration [72,75,76,77,78].
4. Materials and Methods
4.1. Plant Materials and Natural LT Conditions
The plant material for this study included two banana genotypes, Musa spp. AAA cv. Baxijiao and Musa spp. ABB cv. Dongguandajiao, which are CS and CT, respectively [9,79]. Three biological replicates of each genotype were subjected to low temperature treatments, following the method described by Yan et al. [9]. The leaves of plants growing for 3 days at 25 °C, 16 °C, 10 °C and 7 °C were used for analyses.
4.2. Measurement of Hemicellulose Content of Banana Leaves
The leaf samples were treated according to the analytical procedure recommended by the national renewable energy laboratory (NREL) [80] to obtain the filtrate. The content of hemicellulose in the filtrate was determined by the orcinol colorimetric method [81].
4.3. RNA-Seq Analysis
The RNA preparation, and the library preparation for RNA-Seq and data analysis, were carried out as described by Klepikova et al. [82]. False discovery rate was used to determine the threshold of the p-value in multiple tests and analyses. In the present study, |log2 (fold change)| > 1 and a threshold of false discovery rate values <0.05 were used as the threshold to evaluate the significance of differentially expressed genes.
4.4. Identification of Csls in Banana
To study the Csl gene family in banana, all Musa acuminata protein sequences were obtained from Banana-Genome-Hub (https://banana-genome-hub.southgreen.fr/download) Musa acuminata DH Pahang v2 (updated in January 2016). The Csl amino acid sequences of Arabidopsis and Oryza sativa were downloaded from the Arabidopsis Information Resource (TAIR) (http://www.Arabidopsis.org/download) and the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/downloads_gad.shtml).
Double-directional BLAST was employed to obtain potential Csl members; the BLAST function of TBTools [83] was used to retrieve potential MaCsl sequences referring to the amino acid sequence of Csl proteins of Arabidopsis. The obtained potential MaCsl sequences were thereafter compared with Csls in the Swissprot database, and those without typical characteristics of Csl proteins were removed. Candidate genes were obtained by analyzing the obtained genes using the Search pfam (http://pfam.xfam.org/search) online tool and eliminating sequences that lack the typical functional domain of Csls. All candidate MaCsls should contain one of the two PFAM domain models, namely PF00535 or PF03552.
4.5. Physicochemical Properties and Phylogenetic Analysis
Expert protein analysis system (ExPASy, http://web.expasy.org/compute_pi/) were employed to predict the pI and MW of MaCsl amino acid sequences.
Amino acid sequence alignments of all Csl members from the Arabidopsis, rice, or banana were performed using Clustal W2 [84] under default settings, while Molecular Evolutionary Genetics Analysis (MEGA) 7.0 [85] software was used to construct the phylogenetic trees, followed by visualization with FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/). The default parameters were manually adjusted using the neighbor-joining method (the JTT+I+G substitution model and 1000 bootstrap replicates).
4.6. Conserved Motif and Gene Structure Analysis
The online software MEME (http://meme-suite.org/tools/meme) was used to identify the conserved motifs of MaCsl from the deduced protein sequence. The number of conserved motifs selected was 10, while the other default parameters were set automatically by the software. The results were visualized with TBtools. The structural analysis of the introns, exons and non-coding regions of all MaCsls was performed by Gene Structure Display Server 2.0 (GSDS, https://gsds.cbi.pku.edu.cn/) [86] using corresponding CDS sequences and genomic sequences of MaCsls retrieved from Banana-Genome-Hub (https://banana-genome-hub.southgreen.fr/download).
4.7. Quantification of the Expression Level of MaCsls Using qPCR
The experiment was carried out according to the method described by Meng et al. [10] and the primers used are listed in Table S5.
4.8. Statistical Analysis
One-way analysis of variance (one-way ANOVA) was done using IBM SPSS Statistics software for Windows, Version 26.0 (IBM Corporation, Armonk, NY, USA). The results of hemicellulose content and qPCR were expressed as mean ± SE. Statistical differences between the two species at each temperature point were determined using the Student’s t-test.
5. Conclusions
This study provides a comprehensive analysis of the MaCsl gene family in banana. In total, 42 members of the MaCsl gene family were identified in the Musa acuminata genome, and were classified into six subfamilies: CslA, CslC, CslD, CslE, CslG, and CslH. This information provides an important basis for studying the physiological role played by the MaCsl genes in response to biotic and abiotic stresses. In addition, the different expression patterns of MaCsl genes in CT and CS banana cultivars under LT stress indicate their involvement in plant chilling tolerance, suggesting their potential utilization in the breeding of CT banana. Further research should focus on the function of specific MaCsl genes in chilling tolerance and the underlying mechanisms.
Abbreviations
| bp | Base pair |
| BX | Baxijiao |
| CK | The control |
| CS | Chilling-sensitive |
| Csl | Cellulose synthase-like gene |
| CT | Chilling-tolerant |
| DEG | Differentially expressed gene |
| DJ | Dongguandajiao |
| kD | Kilodalton |
| LT | Low temperature |
| LT16 | LT of 16 °C |
| LT10 | LT of 10 °C |
| LT7 | LT of 7 °C |
| MEME | Multiple Em for Motif Elucidation |
| MLG | Mixed-linkage glucan |
| MW | Molecular weight |
| pI | Isoelectric point |
| qPCR | Quantitative real-time PCR |
| RNA-Seq | RNA sequencing |
| UTR | Untranslated region |
Supplementary Materials
The following are available online at https://www.mdpi.com/2223-7747/10/1/122/s1, Table S1: The number of DEGs in banana (Musa spp.) under low temperature stress, Table S2: Cold response genes present in banana (Musa spp.) under low temperature stress, Table S3: The p-value of differentially expressed MsCsl genes in banana (Musa spp.) under low temperature stress, Table S4: The fragments per kilobase of exon per million reads mapped values of MaCsl genes in banana (Musa spp.) under low temperature stress, Table S5: Primer sequences used for qPCR.
Author Contributions
Conceptualization, W.Y. and C.X.; data curation, W.Y., J.L., J.M., Y.T., T.N., Z.H. and C.X.; formal analysis, W.Y., J.L., and C.X.; funding acquisition, C.X., G.Y., H.C., X.L. and T.T.; investigation, W.Y.; methodology, W.Y., J.L., J.M. and C.X.; project administration, C.X.; resources, W.Y., J.L., J.M., Y.T., T.N. and Z.H.; supervision, C.X.; validation, W.Y. and C.X.; visualization, W.Y.; writing—original draft, W.Y. and C.X.; writing—review and editing, W.Y., T.T. and C.X. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key Research and Development Program of China (2018YFD1000300), the Key Research and Development Program of Guangdong Province for Modern Plant Breeding (2018B020202005), the Guangdong Province Special Fund for Modern Agriculture Industry Technology Innovation Teams (2021KJ109), the Earmarked Fund for Modern Agro-industry Technology Research System (CARS-31-04), and by the European Regional Development Fund (ERDF) for the project “Plants as a tool for sustainable development”, number CZ.02.1.01/0.0/16_019/0000827.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hu H., Zhang R., Dong S., Li Y., Fan C., Wang Y., Xia T., Chen P., Wang L., Feng S., et al. AtCslD3 and GtCslD3 mediate root growth and cell elongation downstream of the ethylene response pathway in Arabidopsis. J. Exp. Bot. 2018;69:1065–1080. doi: 10.1093/jxb/erx470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunter C.T., Kirienko D.H., Sylvester A.W., Peter G.F., McCarty D.R., Koch K.E. Cellulose synthase-like D1 is integral to normal cell division, expansion, and leaf development in maize. Plant Physiol. 2012;158:708–724. doi: 10.1104/pp.111.188466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hyles J., Vautrin S., Pettolino F., MacMillan C., Stachurski Z., Breen J., Berges H., Wicker T., Spielmeyer W. Repeat-length variation in a wheat cellulose synthase-like gene is associated with altered tiller number and stem cell wall composition. J. Exp. Bot. 2017;68:1519–1529. doi: 10.1093/jxb/erx051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li N., Han X., Xu S., Li C., Wei X., Liu Y., Zhang R., Tang X., Zhou J., Huang Z. Glycoside hydrolase family 39 #-xylosidase of sphingomonas showing salt/ethanol/trypsin tolerance, low-pH/low-temperature activity, and transxylosylation activity. J. Agric. Food Chem. 2018;66:9465–9472. doi: 10.1021/acs.jafc.8b03327. [DOI] [PubMed] [Google Scholar]
- 5.Yu L., Shi D., Li J., Kong Y., Yu Y., Chai G., Hu R., Wang J., Hahn M.G., Zhou G. Cellulose synthase-like A2, a glucomannan synthase, is involved in maintaining adherent mucilage structure in Arabidopsis seed. Plant Physiol. 2014;164:1842–1856. doi: 10.1104/pp.114.236596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng X., Pang H., Abbas M., Yan X., Dai X., Li Y., Li Q. Characterization of cellulose synthase-like D (CslD) family revealed the involvement of PtrCslD5 in root hair formation in Populus trichocarpa. Sci. Rep. 2019;9 doi: 10.1038/s41598-018-36529-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma L., Jiang S., Lin G., Cai J., Ye X., Chen H., Li M., Li H., Takáč T., Šamaj J., et al. Wound-induced pectin methylesterases enhance banana (Musa spp. AAA) susceptibility to Fusarium oxysporum f. sp. cubense. J. Exp. Bot. 2013;64:2219–2229. doi: 10.1093/jxb/ert088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niu Y., Hu B., Li X., Chen H., Takáč T., Šamaj J., Xu C. Comparative digital gene expression analysis of tissue-cultured plantlets of highly resistant and susceptible banana cultivars in response to Fusarium oxysporum. Int. J. Mol. Sci. 2018;19:350. doi: 10.3390/ijms19020350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan Y., Takáč T., Li X., Chen H., Wang Y., Xu E., Xie L., Su Z., Šamaj J., Xu C. Variable content and distribution of arabinogalactan proteins in banana (Musa spp.) under low temperature stress. Front. Plant Sci. 2015;6 doi: 10.3389/fpls.2015.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng J., Hu B., Yi G., Li X., Chen H., Wang Y., Yuan W., Xing Y., Sheng Q., Su Z., et al. Genome-wide analyses of banana fasciclin-like AGP genes and their differential expression under low-temperature stress in chilling sensitive and tolerant cultivars. Plant Cell Rep. 2020;39:693–708. doi: 10.1007/s00299-020-02524-0. [DOI] [PubMed] [Google Scholar]
- 11.Richmond T.A., Somerville C.R. The cellulose synthase superfamily. Plant Physiol. 2000;124:495–498. doi: 10.1104/pp.124.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lombard V., Golaconda Ramulu H., Drula E., Coutinho P.M., Henrissat B. The carbohydrate-active enzymes database (CAZy) Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hazen S.P., Scott-Craig J.S., Walton J.D. Cellulose synthase-like genes of rice. Plant Physiol. 2002;128:336–340. doi: 10.1104/pp.010875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farrokhi N., Burton R.A., Brownfield L., Hrmova M., Wilson S.M., Bacic A., Fincher G.B. Plant cell wall biosynthesis: Genetic, biochemical and functional genomics approaches to the identification of key genes. Plant Biotechnol. J. 2006;4:145–167. doi: 10.1111/j.1467-7652.2005.00169.x. [DOI] [PubMed] [Google Scholar]
- 15.Little A., Schwerdt J.G., Shirley N.J., Khor S.F., Neumann K., O’ Donovan L.A., Lahnstein J., Collins H.M., Henderson M., Fincher G.B., et al. Revised phylogeny of the cellulose synthase gene superfamily: Insights into cell wall evolution. Plant Physiol. 2018;177:1124–1141. doi: 10.1104/pp.17.01718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhugga K.S., Barreiro R., Whitten B., Stecca K., Hazebroek J., Randhawa G.S., Dolan M., Kinney A.J., Tomes D., Nichols S., et al. Guar seed β-mannan synthase is a member of the cellulose synthase super gene family. Science. 2004;303:363–366. doi: 10.1126/science.1090908. [DOI] [PubMed] [Google Scholar]
- 17.Goubet F., Barton C.J., Mortimer J.C., Yu X., Zhang Z., Miles G.P., Richens J., Liepman A.H., Seffen K., Dupree P. Cell wall glucomannan in Arabidopsis is synthesised by CslA glycosyltransferases, and influences the progression of embryogenesis. Plant J. 2009;60:527–538. doi: 10.1111/j.1365-313X.2009.03977.x. [DOI] [PubMed] [Google Scholar]
- 18.Liepman A.H., Wilkerson C.G., Keegstra K. Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc. Natl. Acad. Sci. USA. 2005;102:2221–2226. doi: 10.1073/pnas.0409179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He C., Zhang J., Liu X., Zeng S., Wu K., Yu Z., Wang X., Teixeira da Silva J.A., Lin Z., Duan J. Identification of genes involved in biosynthesis of mannan polysaccharides in Dendrobium officinale by RNA-seq analysis. Plant Mol. Biol. 2015;88:219–231. doi: 10.1007/s11103-015-0316-z. [DOI] [PubMed] [Google Scholar]
- 20.Dwivany F.M., Yulia D., Burton R.A., Shirley N.J., Wilson S.M., Fincher G.B., Bacic A., Newbigin E., Doblin M.S. The Cellulose-synthase like C (CslC) family of barley includes members that are integral membrane proteins targeted to the plasma membrane. Mol. Plant. 2009;2:1025–1039. doi: 10.1093/mp/ssp064. [DOI] [PubMed] [Google Scholar]
- 21.Cocuron J.C., Lerouxel O., Drakakaki G., Alonso A.P., Liepman A.H., Keegstra K., Raikhel N., Wilkerson C.G. A gene from the cellulose synthase-like C family encodes a beta-1,4 glucan synthase. Proc. Natl. Acad. Sci. USA. 2007;104:8550–8555. doi: 10.1073/pnas.0703133104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim S., Chandrasekar B., Rea A.C., Danhof L., Zemelis-Durfee S., Thrower N., Shepard Z.S., Pauly M., Brandizzi F., Keegstra K. The synthesis of xyloglucan, an abundant plant cell wall polysaccharide, requires CslC function. Proc. Natl. Acad. Sci. USA. 2020;117:20316–20324. doi: 10.1073/pnas.2007245117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verhertbruggen Y., Yin L., Oikawa A., Scheller H.V. Mannan synthase activity in the CslD family. Plant Signal. Behav. 2015;6:1620–1623. doi: 10.4161/psb.6.10.17989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dhugga K.S. Biosynthesis of non-cellulosic polysaccharides of plant cell walls. Phytochemistry. 2012;74:8–19. doi: 10.1016/j.phytochem.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 25.Park S., Szumlanski A.L., Gu F., Guo F., Nielsen E. A role for CslD3 during cell-wall synthesis in apical plasma membranes of tip-growing root-hair cells. Nat. Cell Biol. 2011;13:973–980. doi: 10.1038/ncb2294. [DOI] [PubMed] [Google Scholar]
- 26.Yin L., Verhertbruggen Y., Oikawa A., Manisseri C., Knierim B., Prak L., Jensen J.K., Knox J.P., Auer M., Willats W.G.T., et al. The cooperative activities of CslD2, CslD3, and CslD5 are required for normal Arabidopsis development. Mol. Plant. 2011;4:1024–1037. doi: 10.1093/mp/ssr026. [DOI] [PubMed] [Google Scholar]
- 27.Yang J., Bak G., Burgin T., Barnes W.J., Mayes H.B., Peña M.J., Urbanowicz B.R., Nielsen E. Biochemical and genetic analysis identify CslD3 as a beta-1,4-glucan synthase that functions during plant cell wall synthesis. Plant Cell. 2020;32:1749–1767. doi: 10.1105/tpc.19.00637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernal A.J., Jensen J.K., Harholt J., Sørensen S., Moller I., Blaukopf C., Johansen B., De Lotto R., Pauly M., Scheller H.V., et al. Disruption of AtCslD5 results in reduced growth, reduced xylan and homogalacturonan synthase activity and altered xylan occurrence in Arabidopsis. Plant J. 2007;52:791–802. doi: 10.1111/j.1365-313X.2007.03281.x. [DOI] [PubMed] [Google Scholar]
- 29.Danilova T.V., Friebe B., Gill B.S., Poland J., Jackson E. Development of a complete set of wheat–barley group-7 robertsonian translocation chromosomes conferring an increased content of β-glucan. Theor. Appl. Genet. 2017;131:377–388. doi: 10.1007/s00122-017-3008-z. [DOI] [PubMed] [Google Scholar]
- 30.Burton R.A., Wilson S.M., Hrmova M., Harvey A.J., Shirley N.J., Medhurst A., Stone B.A., Newbigin E.J., Bacic A., Fincher G.B. Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-β-d-glucans. Science. 2006;311:1940–1942. doi: 10.1126/science.1122975. [DOI] [PubMed] [Google Scholar]
- 31.Doblin M.S., Pettolino F.A., Wilson S.M., Campbell R., Burton R.A., Fincher G.B., Newbigin E., Bacic A. A barley cellulose synthase-like CslH gene mediates (1,3;1,4)-beta-D-glucan synthesis in transgenic Arabidopsis. Proc. Natl. Acad. Sci. USA. 2009;106:5996–6001. doi: 10.1073/pnas.0902019106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vega-Sánchez M.E., Verhertbruggen Y., Christensen U., Chen X., Sharma V., Varanasi P., Jobling S.A., Talbot M., White R.G., Joo M., et al. Loss of cellulose synthase-like F6 function affects mixed-linkage glucan deposition, cell wall mechanical properties, and defense responses in vegetative tissues of rice. Plant Physiol. 2012;159:56–69. doi: 10.1104/pp.112.195495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Little A., Lahnstein J., Jeffery D.W., Khor S.F., Schwerdt J.G., Shirley N.J., Hooi M., Xing X., Burton R.A., Bulone V. A novel (1,4)-β-linked glucoxylan is synthesized by members of the cellulose synthase-like F gene family in land plants. ACS Cent. Sci. 2019;5:73–84. doi: 10.1021/acscentsci.8b00568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scheible W.R., Pauly M. Glycosyltransferases and cell wall biosynthesis: Novel players and insights. Curr. Opin. Plant Biol. 2004;7:285–295. doi: 10.1016/j.pbi.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 35.Zhu J., Lee B., Dellinger M., Cui X., Zhang C., Wu S., Nothnagel E.A., Zhu J. A cellulose synthase-like protein is required for osmotic stress tolerance in Arabidopsis. Plant J. 2010;63:128–140. doi: 10.1111/j.1365-313X.2010.04227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bagheri R., Bashir H., Ahmad J., Iqbal M., Qureshi M.I. Spinach (Spinacia oleracea L.) modulates its proteome differentially in response to salinity, cadmium and their combination stress. Plant Physiol. Bioch. 2015;97:235–245. doi: 10.1016/j.plaphy.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 37.Camacho-Cristóbal J.J., Herrera-Rodríguez M.B., Beato V.M., Rexach J., Navarro-Gochicoa M.T., Maldonado J.M., González-Fontes A. The expression of several cell wall-related genes in Arabidopsis roots is down-regulated under boron deficiency. Environ. Exp. Bot. 2008;63:351–358. doi: 10.1016/j.envexpbot.2007.12.004. [DOI] [Google Scholar]
- 38.İşkil R., Surgun-Acar Y. Expression analysis of cell wall assembly and remodelling-related genes in Arabidopsis roots subjected to boron stress and brassinosteroid at different developmental stages. Acta Bot. Bras. 2018;32:546–554. doi: 10.1590/0102-33062018abb0023. [DOI] [Google Scholar]
- 39.Xiao Y., Wu X., Liu D., Yao J., Liang G., Song H., Ismail A.M., Luo J., Zhang Z. Cell wall polysaccharide-mediated cadmium tolerance between two Arabidopsis thaliana ecotypes. Front. Plant Sci. 2020;11:473. doi: 10.3389/fpls.2020.00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aditya J., Lewis J., Shirley N.J., Tan H., Fincher M.H.B., Burton R.A., Mather D.E., Tucker M.R. The dynamics of cereal cyst nematode infection differ between susceptible and resistant barley cultivars and lead to changes in (1,3;1,4)-β-glucan levels and HvCslF gene transcript abundance. New Phytol. 2015;207:135–147. doi: 10.1111/nph.13349. [DOI] [PubMed] [Google Scholar]
- 41.Douchkov D., Lueck S., Hensel G., Kumlehn J., Rajaraman J., Johrde A., Doblin M.S., Beahan C.T., Kopischke M., Fuchs R., et al. The barley (Hordeum vulgare) cellulose synthase-like D2 gene (HvCslD2) mediates penetration resistance to host-adapted and nonhost isolates of the powdery mildew fungus. New Phytol. 2016;212:421–433. doi: 10.1111/nph.14065. [DOI] [PubMed] [Google Scholar]
- 42.Domon J., Baldwin L., Acket S., Caudeville E., Arnoult S., Zub H., Gillet F., Lejeune-Hénaut I., Brancourt-Hulmel M., Pelloux J., et al. Cell wall compositional modifications of Miscanthus ecotypes in response to cold acclimation. Phytochemistry. 2013;85:51–61. doi: 10.1016/j.phytochem.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 43.Le Gall H., Philippe F., Domon J., Gillet F., Pelloux J., Rayon C. Cell wall metabolism in response to abiotic stress. Plants. 2015;4:112–166. doi: 10.3390/plants4010112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le M.Q., Pagter M., Hincha D.K. Global changes in gene expression, assayed by microarray hybridization and quantitative RT-PCR, during acclimation of three Arabidopsis thaliana accessions to sub-zero temperatures after cold acclimation. Plant Mol. Biol. 2015;87:1–15. doi: 10.1007/s11103-014-0256-z. [DOI] [PubMed] [Google Scholar]
- 45.Yang Q., Gao J., He W., Dou T., Ding L., Wu J., Li C., Peng X., Zhang S., Yi G. Comparative transcriptomics analysis reveals difference of key gene expression between banana and plantain in response to cold stress. BMC Genom. 2015;16 doi: 10.1186/s12864-015-1551-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pradhan S.K., Pandit E., Nayak D.K., Behera L., Mohapatra T. Genes, pathways and transcription factors involved in seedling stage chilling stress tolerance in indica rice through RNA-Seq analysis. BMC Plant Biol. 2019;19 doi: 10.1186/s12870-019-1922-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Panter P.E., Kent O., Dale M., Smith S.J., Skipsey M., Thorlby G., Cummins I., Ramsay N., Begum R.A., Sanhueza D., et al. MUR1-mediated cell-wall fucosylation is required for freezing tolerance in Arabidopsis thaliana. New Phytol. 2019;224:1518–1531. doi: 10.1111/nph.16209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takahashi D., Johnson K.L., Hao P., Tuong T., Erban A., Sampathkumar A., Bacic A., Livingston D.P., Kopka J., Kuroha T., et al. Cell wall modification by the xyloglucan endotransglucosylase/hydrolase XTH19 influences freezing tolerance after cold and sub-zero acclimation. Plant Cell Environ. 2020 doi: 10.1111/pce.13953. [DOI] [PubMed] [Google Scholar]
- 49.Djerbi S., Lindskog M., Arvestad L., Sterky F., Teeri T.T. The genome sequence of black cottonwood (Populus trichocarpa) reveals 18 conserved cellulose synthase (CesA) genes. Planta. 2005;221:739–746. doi: 10.1007/s00425-005-1498-4. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki S., Li L., Sun Y., Chiang V.L. The cellulose synthase gene superfamily and biochemical functions of xylem-specific cellulose synthase-like genes in Populus trichocarpa. Plant Physiol. 2006;142:1233–1245. doi: 10.1104/pp.106.086678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roberts A.W., Bushoven J.T. The cellulose synthase (CesA) gene superfamily of the moss Physcomitrella patens. Plant Mol. Biol. 2007;63:207–219. doi: 10.1007/s11103-006-9083-1. [DOI] [PubMed] [Google Scholar]
- 52.Appenzeller L., Doblin M., Barreiro R., Wang H., Niu X., Kollipara K., Carrigan L., Tomes D., Chapman M., Dhugga K.S. Cellulose synthesis in maize: Isolation and expression analysis of the cellulose synthase (CesA) gene family. Cellulose. 2004;11:287–299. doi: 10.1023/B:CELL.0000046417.84715.27. [DOI] [Google Scholar]
- 53.Li Y., Cheng X., Fu Y., Wu Q., Guo Y., Peng J., Zhang W., He B. A genome-wide analysis of the cellulose synthase-like (Csl) gene family in maize. Biol. Plantarum. 2019;63:721–732. doi: 10.32615/bp.2019.081. [DOI] [Google Scholar]
- 54.Burton R.A., Shirley N.J., King B.J., Harvey A.J., Fincher G.B. The CesA gene family of barley quantitative analysis of transcripts reveals two groups of co-expressed genes. Plant Physiol. 2004;134:224–236. doi: 10.1104/pp.103.032904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nairn C.J., Haselkorn T. Three loblolly pine CesA genes expressed in developing xylem are orthologous to secondary cell wall CesA genes of angiosperms. New Phytol. 2005;166:907–915. doi: 10.1111/j.1469-8137.2005.01372.x. [DOI] [PubMed] [Google Scholar]
- 56.Song X., Xu L., Yu J., Tian P., Hu X. Genome-wide characterization of the cellulose synthase gene superfamily in Solanum lycopersicum. Gene. 2018;688:71–83. doi: 10.1016/j.gene.2018.11.039. [DOI] [PubMed] [Google Scholar]
- 57.Cao S., Cheng H., Zhang J., Aslam M., Yan M., Hu A., Lin L., Ojolo S.P., Zhao H., Priyadarshani S., et al. Genome-wide identification, expression pattern analysis and evolution of the Ces/Csl gene superfamily in pineapple (Ananas comosus) Plants. 2019;8:275. doi: 10.3390/plants8080275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaur S., Dhugga K.S., Beech R., Singh J. Genome-wide analysis of the cellulose synthase-like (Csl) gene family in bread wheat (Triticum aestivum L.) BMC Plant Biol. 2017;17:193. doi: 10.1186/s12870-017-1142-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li G., Liu X., Liang Y., Zhang Y., Cheng X., Cai Y. Genome-wide characterization of the cellulose synthase gene superfamily in Pyrus bretschneideri and reveal its potential role in stone cell formation. Funct. Integr. Genomic. 2020;20:723–738. doi: 10.1007/s10142-020-00747-8. [DOI] [PubMed] [Google Scholar]
- 60.Perrier X., De Langhe E., Donohue M., Lentfer C., Vrydaghs L., Bakry F., Carreel F., Hippolyte I., Horry J.P., Jenny C., et al. Multidisciplinary perspectives on banana (Musa spp.) domestication. Proc. Natl. Acad. Sci. USA. 2011;108:11311–11318. doi: 10.1073/pnas.1102001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.FAOSTAT. [(accessed on 6 March 2020)];2020 Available online: https://www.fao.org/faostat/en/#data/QC/visualize.
- 62.Pauly M., Gille S., Liu L., Mansoori N., de Souza A., Schultink A., Xiong G. Hemicellulose biosynthesis. Planta. 2013;238:627–642. doi: 10.1007/s00425-013-1921-1. [DOI] [PubMed] [Google Scholar]
- 63.Yin Y., Huang J., Xu Y. The cellulose synthase superfamily in fully sequenced plants and algae. BMC Plant Biol. 2009;9:99. doi: 10.1186/1471-2229-9-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Holland N., Holland D., Helentjaris T., Dhugga K.S., Xoconostle-Cazares B., Delmer D.P. A comparative analysis of the plant cellulose synthase (CesA) gene family. Plant Physiol. 2000;123:1313–1324. doi: 10.1104/pp.123.4.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang L., Guo K., Li Y., Tu Y., Hu H., Wang B., Cui X., Peng L. Expression profiling and integrative analysis of the CesA/Csl superfamily in rice. BMC Plant Biol. 2010;10:282. doi: 10.1186/1471-2229-10-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cai C., Li Q., Duan C., Chen D., Liu J. Bioinformatics and expression analysis on Csl gene family in Dendrobium catenatum [in Chinese with English abstract] GAB. 2019;38:2159–2166. doi: 10.13417/j.gab.038.002159. [DOI] [Google Scholar]
- 67.Burton R.A., Collins H.M., Kibble N.A., Smith J.A., Shirley N.J., Jobling S.A., Henderson M., Singh R.R., Pettolino F., Wilson S.M., et al. Over-expression of specific HvCslF cellulose synthase-like genes in transgenic barley increases the levels of cell wall (1,3;1,4)-beta-D-glucans and alters their fine structure. Plant Biotechnol. J. 2011;9:117–135. doi: 10.1111/j.1467-7652.2010.00532.x. [DOI] [PubMed] [Google Scholar]
- 68.Yin Y., Johns M.A., Cao H., Rupani M. A survey of plant and algal genomes and transcriptomes reveals new insights into the evolution and function of the cellulose synthase superfamily. BMC Genom. 2014;15:260. doi: 10.1186/1471-2164-15-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carpita N.C., McCann M.C. The maize mixed-linkage (1→3),(1→4)-β-D-glucan polysaccharide is synthesized at the golgi membrane. Plant Physiol. 2010;153:1362–1371. doi: 10.1104/pp.110.156158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kubacka-Zębalska M., Kacperska A. Low temperature-induced modifications of cell wall content and polysaccharide composition in leaves of winter oilseed rape (Brassica napus L. Var. Oleifera L.) Plant Sci. 1999;148:59–67. doi: 10.1016/s0168-9452(99)00122-3. [DOI] [Google Scholar]
- 71.Weiser R.L., Wallner S.J., Waddell J.W. Cell wall and extensin mRNA changes during cold acclimation of pea seedlings. Plant Physiol. 1990;93:1021–1026. doi: 10.1104/pp.93.3.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Plancot B., Gügi B., Mollet J., Loutelier-Bourhis C., Govind S.R., Lerouge P., Follet-Gueye M., Vicré M., Alfonso C., Nguema-Ona E., et al. Desiccation tolerance in plants: Structural characterization of the cell wall hemicellulosic polysaccharides in three Selaginella species. Carbohyd. Polym. 2019;208:180–190. doi: 10.1016/j.carbpol.2018.12.051. [DOI] [PubMed] [Google Scholar]
- 73.Zang D., Wang J., Zhang X., Liu Z., Wang Y. Arabidopsis heat shock transcription factor HSFA7b positively mediates salt stress tolerance by binding to an E-box-like motif to regulate gene expression. J. Exp. Bot. 2019;70:5355–5374. doi: 10.1093/jxb/erz261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Budot B.O., Encabo J.R., Ambita I.D.V., Atienza-Grande G.A., Satoh K., Kondoh H., Ulat V.J., Mauleon R., Kikuchi S., Choi I. Suppression of cell wall-related genes associated with stunting of Oryza glaberrima infected with rice tungro spherical virus. Front. Microbiol. 2014;5 doi: 10.3389/fmicb.2014.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chowdhury J., Schober M.S., Shirley N.J., Singh R.R., Jacobs A.K., Douchkov D., Schweizer P., Fincher G.B., Burton R.A., Little A. Down-regulation of the glucan synthase-like 6 gene (HvGsl6) in barley leads to decreased callose accumulation and increased cell wall penetration by Blumeria graminis f. sp. hordei. New Phytol. 2016;212:434–443. doi: 10.1111/nph.14086. [DOI] [PubMed] [Google Scholar]
- 76.Kesten C., Menna A., Sanchez-Rodriguez C. Regulation of cellulose synthesis in response to stress. Curr. Opin. Plant Biol. 2017;40:106–113. doi: 10.1016/j.pbi.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 77.Wang T., McFarlane H.E., Persson S. The impact of abiotic factors on cellulose synthesis. J. Exp. Bot. 2016;67:543–552. doi: 10.1093/jxb/erv488. [DOI] [PubMed] [Google Scholar]
- 78.Moore J.P., Nguema-Ona E.E., Vicré-Gibouin M., Sørensen I., Willats W.G.T., Driouich A., Farrant J.M. Arabinose-rich polymers as an evolutionary strategy to plasticize resurrection plant cell walls against desiccation. Planta. 2013;237:739–754. doi: 10.1007/s00425-012-1785-9. [DOI] [PubMed] [Google Scholar]
- 79.Huang X., Ji Z., Li P. A study on the injury symptoms and physiological quota of banana and an effective measure for cold injury protection [in Chinese with English abstract] J. South China Agri. Univ. 1982;3:1–12. [Google Scholar]
- 80.Sluiter A.D., Hames B., Ruiz R., Scarlata C., Sluiter J., Templeton D.W., Crocker D. Determination of Structural Carbohydrates and Lignin in Biomass. NREL; Golden, CO, USA: 2012. [(accessed on 2 April 2020)]. Available online: http://www.nrel.gov/biomass/analytical_procedures.html. [Google Scholar]
- 81.Hu G., Ellberg S., Burton C., Evans C., Satterfield K., Bockelman H. Application of an orcinol-ferric chloride colorimetric assay in barley and wheat accessions for water-extractable and total arabinoxylan. J. Cereal Sci. 2020;93:102962. doi: 10.1016/j.jcs.2020.102962. [DOI] [Google Scholar]
- 82.Klepikova A.V., Logacheva M.D., Dmitriev S.E., Penin A.A. RNA-seq analysis of an apical meristem time series reveals a critical point in Arabidopsis thaliana flower initiation. BMC Genom. 2015;16 doi: 10.1186/s12864-015-1688-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen C., Xia R., Chen H., He Y. TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv. 2018;13:1194–1202. doi: 10.1101/289660. [DOI] [Google Scholar]
- 84.Thompson J.D., Higgins D.G., Gibson T.J. Clustal W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kumar S., Stecher G., Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hu B., Jin J., Guo A., Zhang H., Luo J., Gao G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.