Abstract
The aim of this study is to describe the drug resistance and virulence of enterococci in river water sampled downstream (DRW) and upstream (URW) from the wastewater discharge point, to determine the pool of virulent and drug-resistant enterococci in untreated wastewater (UWW) and the extent to which these bacteria are eliminated from hospital wastewater (HWW) and municipal wastewater treated (TWW) by biological and mechanical methods in a wastewater treatment plant (WWTP). A total of 283 strains were identified with the use of culture-dependent methods and PCR, including seven different species including E. faecalis and E. faecium which were predominant in all analyzed samples. Majority of the strains were classified as multidrug resistant (MDR), mostly on streptomycin and trimethoprim. Strains isolated from wastewater and DRW harbored van genes conditioning phenotypic resistance to vancomycin, the highest percentage of vancomycin-resistant strains (57.0%), mostly strains harboring vanC1 genes (27.6%), was noted in TWW. More than 65.0% of the isolated strains had different virulence genes, the highest number of isolates were positive for cell wall adhesin efaA and sex pheromones cob, cpd, and ccf which participate in the induction of virulence. Many of the strains isolated from TWW were resistant to a higher number of drugs and were more virulent than those isolated from UWW and HWW. The enterococci isolated from DRW and wastewater were characterized by similar multidrug resistance and virulence profiles, and significant correlations were observed between these groups of isolates. These findings suggest that pathogenic enterococci are released with TWW and can spread in the river, pose a serious epidemiological threat and a risk to public health.
Keywords: enterococci, multidrug-resistant (MDR), virulence, vancomycin, river water, wastewater
1. Introduction
Wastewater is treated to reduce pollutant concentrations to environmentally safe levels. However, various compounds and pathogenic fecal bacteria, including antibiotic-resistant bacteria (ARB) and antibiotic-resistance genes (ARGs), are evacuated to surface water bodies with treated wastewater [1,2,3]. These micropollutants influence the trophic status, microbiological quality, and epidemiological safety of natural ecosystems. Bacteria and their metabolites are evacuated with wastewater, and they disrupt the natural balance of microbiota which colonize aquatic systems, actively participate in the biogeochemical cycle, restore and maintain ecological balance in water ecosystems. The inflow of allochthonous microorganisms as well as other organic and inorganic pollutants to aquatic ecosystems disrupts the ecological balance and exerts a negative impact on a given biotope [4,5,6]. Gram positive enterococci rapidly evolve and spread in the environment. These oval-shaped bacteria often occur in pairs or short chains, and they are able to grow in a wide range of temperatures (10 °C to 45 °C), pH of 9.6, and NaCl concentration of up to 6.5% [7,8]. Enterococcus strains can be both pathogenic and commensal. Commensal bacteria colonize the digestive tract of humans and animals, and they are not highly virulent. Enterococci are fecal indicator bacteria that are used to assess fecal pollution of the natural environment. Enterococci are also indirect indicators of the risks associated with the spread of other pathogens in aquatic ecosystems and the transmission of waterborne diseases [9,10,11,12]. Therefore, these bacteria are used to monitor water quality and detect sanitary and epidemiological threats worldwide. In non-contaminated waters, enterococcus counts generally do not exceed several to less than 20 colony-forming units in 100 mL of water. However, the inflow of treated wastewater, surface runoffs from agricultural land, and pollutants from recreational areas considerably increases enterococcus counts in aquatic systems [4]. Enterococcus counts ranged from 4.5 × 101 to 1.2 × 104 cfu 100 mL–1 in the rivers of the Seine watershed [13]. The abundance of enterococci in less polluted rivers was similar to that in nonpoint source pollution loads in forests, and their concentrations in the most heavily contaminated rivers were similar to those in treated wastewater. In untreated municipal wastewater, the abundance of fecal enterococci and bacteria of the family Enterobacteriaceae was determined in the range of 4 to 8 log cfu mL−1 [6,13,14]. Wastewater treatment reduces the counts of indicator microorganisms by even several orders of magnitude, but large amounts of these bacteria reach water receptacles with the evacuated effluents and compromise the microbiological safety of surface water bodies.
Selected species of the family Enterobacteriaceae are used in the production of probiotics and starter cultures in the food-processing industry and veterinary medicine [15,16]. However, because of the growing levels of drug resistance, enterococci are increasingly often classified as alert pathogens in the hospital environment. These strains are particularly dangerous for patients with respiratory diseases and patients receiving broad-spectrum antimicrobial drugs [17,18,19]. Enterococcus faecalis and E. faecium pose the greatest epidemiological risk, whereas E. avium, E. casseliflavus, E. durans, E. gallinarum, and E. raffinosus are less likely to cause infections in hospitals [20,21]. Recent years have witnessed a steady increase in the abundance of vancomycin-resistant E. faecium belonging to the ESKAPE group of bacteria (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter spp.) responsible for hospital infections [18,22]. Enterococci cause various diseases, including urinary tract infections, infectious diseases in infants, postoperative infections, infections of the central nervous system, and endocarditis [23,24,25]. Virulent and vancomycin-resistant enterococci (VRE) are particularly important from the medical point of view. Enterococcus strains identified both in and outside of hospitals are increasingly often resistant to vancomycin. Until recently, vancomycin was regarded as the most potent antimicrobial drug, but it is no longer effective against the VRE strains [18,26]. Several resistance phenotypes have been identified in enterococci depending on their resistance to vancomycin and teicoplanin (vanA B, C, D, E, G, L, M, and van N). The most prevalent phenotypes, vanA and vanB, have been described mainly in E. faecalis and E. faecium, and phenotype vanC1—in E. casseliflavus and E. gallinarum [27,28,29]. In 2018, the highest number of clinical VRE isolates of E. faecium were determined in Romania (40.3%), Ireland (40.2%), and Poland (35.8%), (ECDC 2020). Vancomycin-resistant enterococci are isolated in various countries (Ethiopia—4.8%, North America—21%, Asia—24%, Europe—20%), and their prevalence is influenced by the applied isolation method, time of analysis, and the size of the studied population [30,31]. Some species harbor virulence factors, including cytolysin which facilitates cell lysis (cylA), aggregation substance (AS), gelatinase which hydrolyzes gelatin, casein, and hemoglobin (gel E), hyaluronidase which increases the invasiveness of bacteria (hyl), endocarditis antigen (efaA), factor-encoding surface protein that participates in biofilm formation (esp), and sex pheromones that carry ARGs (cob, cpd, and ccf) [32,33,34]. According to the Polish National Program for the Protection of Antibiotics (NPOA) and the European Center for Disease Prevention and Control (ECDC), the prevalence of antibiotic-resistant E. faecalis and E. faecium strains continues to increase around the world. In recent years, the spread of virulent and multidrug-resistant (MDR) enterococci was also observed outside healthcare facilities in various environments [13,18,35,36]. The scale and sources of bacterial dissemination can differ, and they have not been fully elucidated. Human activities, in particular the overuse of antibiotics in veterinary medicine, livestock production (mostly avoparcin which is used as a feed additive), agriculture, and human medicine have contributed to the expansion of pathogenic strains [37]. The plastic genome enables enterococci to exchange genes on mobile genetic elements such as transposons and plasmids. These microorganisms can accumulate and transfer ARGs to other bacteria. As a result, contact with infected livestock, handling of food products of animal origin, and runoffs from agricultural areas fertilized with manure or slurry can promote the transfer of MDR strains, including VRE [38,39,40]. Hospitals are also hotspots of ARB and ARGs because of a large number of antibiotic treatments and the presence of significant clinically resistant pathogenic bacteria. Municipal WWTPs that process municipal and hospital wastewater can also act as critical points in the transfer of resistant strains to the environment, and transfer of antibiotic resistance. Multidrug-resistant strains, including VRE, are increasingly often identified in wastewater and wastewater receptacles [14,36,37,41]. Research into the prevalence of ARB and ARGs in rivers and streams plays a particularly important role. Microbiological pollutants are carried along the river continuum, and resistance genes can be transmitted to native bacteria colonizing the aquatic environment. As a result, rivers become potential reservoirs of ARB and ARGs, and they pose a significant sanitary and epidemiological threat for users of water.
In the present study, samples of river water were collected 500 m downstream and upstream from the wastewater discharge point to determine the sources of contamination with virulent and MDR enterococci. The prevalence of bacteria was also determined in hospital wastewater and in untreated and treated wastewater. The extent to which virulent and MDR enterococci were eliminated by biological and mechanical treatment methods in the WWTP was analyzed. Sampling sites were compared against markers of bacterial virulence and multidrug resistance. The main sources of contamination with enterococci should be identified in lotic ecosystems to support their protection and to prevent the spread of virulent and MDR bacteria in the environment.
In the light of Polish regulations, treated wastewater does not have to be disinfected before it is evacuated to surface water bodies, which promotes the transfer of pathogenic enterococci and their antibiotic resistance genes (ARGs) to the environment and poses health risks for humans and animals. In this context, the results of this study contribute important knowledge and can be used to plan effective measures to minimize bacterial emissions to the environment and reduce epidemiological risks.
2. Materials and Methods
2.1. Sampling Sites and Methods
The study was conducted in the Łyna Municipal Wastewater Treatment Plant in Olsztyn, the capital of the Region of Warmia and Mazury in north-eastern Poland. The plant is equipped with biological and mechanical wastewater treatment systems and has a daily processing capacity of around 32,000 m3. Effluents from three hospitals in Olsztyn account for around 2% of the treated wastewater. Technological scheme of the wastewater treatment plant is presented in Figure S1 (Supplementary Materials). These hospitals do not operate on their own WWTPs or wastewater disinfection systems. Each year, they admit around 46,000 patients on average. Wastewater is produced by hospital laboratories, departments, and wards, including surgery, oncology, cardiology, urology, gynecology, ophthalmology, anesthesiology, dermatology and otolaryngology wards, and intensive care units [42].
In the studied WWTP, wastewater is treated in mechanical and biological systems with the involvement of the activated sludge technology and chemical processes. Treated wastewater is not disinfected through chlorination or exposure to UV light before it is discharged to the river. Mechanical treatment involves preliminary purification (screening, sedimentation of mineral suspensions in grit chambers, and organic suspensions in preliminary sedimentation tanks). Pretreated wastewater is pumped to the biological treatment station where nitrogen, phosphorus, and carbon are removed in a series of nitrification, denitrification, and dephosphatation processes. The sediment is separated in sedimentation tanks, and treated wastewater is evacuated directly to the Łyna River which is one of the main lowland watercourses in the southern watershed of the Baltic Sea (northern Poland). The Łyna River has been characterized in detail by Gotkowska-Płachta et al. [6].
Samples of river water were collected 500 m downstream (DRW) and upstream (URW) from the wastewater discharge point. Hospital wastewater (HWW) was sampled at the place of generation, directly from hospital sewer sumps (before reaching the main sewer pipe). Samples of untreated wastewater (UWW) were collected directly behind the grating screen, and treated wastewater (TWW) was sampled from the outflow pipe before it was evacuated to the Łyna River. River water and wastewater samples were collected at approximately eight-week intervals over a period of 12 months, taking into account the research seasons. Municipal wastewater was sampled 22 h after the collection of HWW samples to account for the time needed to reach the WWTP. Water and wastewater samples were collected into sterile glass containers (1000 mL). River water was sampled at a depth of 0.3–0.5 m. A total of 16 river water samples and 40 wastewater samples (8 UWW samples, 8 TWW samples, and 24 HWW samples) were collected in the analyzed period. The samples were transported to the laboratory in insulated containers at a temperature of 4 °C. They were analyzed in the laboratory within 24 h after transport.
2.2. Determination of Physicochemical Parameters
The following physicochemical parameters of river water and wastewater samples were determined: temperature (°C) and pH. The measurements were conducted with the YSI 556 Multiprobe System (MPS) with an accuracy of ±0.1 °C and ±0.01 pH.
2.3. Isolation of Enterococci from Samples of River Water and Wastewater
Enterococci were enumerated by membrane filtration on Slanetz and Bartley (SB) medium after 48 h of incubation at a temperature of 36 ± 2 °C. The results were validated on Bile Aesculin Azide Agar (Marck) after 2 h of incubation at a temperature of 44 ± 0.5 °C according to Polish Standard [43]. The emerged colonies were counted and expressed in colony-forming units (cfu) in 100 mL of water or wastewater. Ten representative colonies were selected from each sample, plated on brain–heart infusion (BHI) broth and incubated at 37 °C for 24–48 h. Pure strains were initially identified to the genus Enterococcus based on the results of Gram stains, the catalase test, bacterial proliferation in BHI broth with the addition of 6.5% NaCl, and esculin hydrolysis into esculentin on Bile Aesculin Azide Agar (Merck) [44]. A total of 430 tested strains were preliminarily classified as Enterococcus spp. The isolates were stored on LB Miller medium (Merck) with 10% glycerol at a temperature of −80 °C until further analysis.
2.4. Identification of Enterococci to Genus and Species Level
The isolated strains were identified to be the genus Enterococcus by identifying the tuf gene encoding elongation factor EF–Tu, which has a strongly conserved sequence in the analyzed bacteria. An internal primer targeting a fragment (sodA int) of the sodA gene which encodes manganese dismutase was designed. Bacterial species were identified by amplifying ddl genes that encode D–alanyl–D–alanine ligase (D–Ala–D–Ala). Primers specific for this fragment of the gene supported the identification of Enterococcus faecalis and Enterococcus faecium which play a key role in human and animal infections. Strains belonging to E. durans, E. hirae, E. avium, and E. gallinarum/E. casseliflavus were identified. Primer sequences (synthesized by Genomed), product size, and PCR conditions are presented in Table S1—Supplementary Materials. The reactions were controlled with the use of the following reference strains: E. faecalis ATCC 29212, E. faecium ATCC 19434, E. casseliflavus ATCC 49605; E. gallinarum ATCC700425, E. durans ATCC 6056, and E. hirae ATCC 8043.
2.5. Determination of Enterococcus Susceptibility to Antimicrobial Drugs
All enterococcus isolates were analyzed for susceptibility to 13 antimicrobial drugs by the disc diffusion method with the use of antibiotics produced by Oxoid (Baningstoke, Hampshire, England). The selected antimicrobials belonged to 10 groups of antibiotics that are widely used in medicine and agriculture: (1) penicillins: ampicillin (AMP—2 μg); (2) carbapenems: imipenem (IPM—10 μg), (3) aminoglycosides: gentamicin (GEN—30 μg); streptomycin (S—300 μg); (4) glycopeptides: teicoplanin (TEC—30 μg), vancomycin (VAN—5 μg); (5) streptogramins: quinupristin/dalfopristin (QD—15 μg); (6) glycylcyclines: tigecycline (TGC—15 μg); (7) oxazolidinones: linezolid (LZD—10 μg); (8) tetracyclines: doxycycline (DO—30 μg); (9) fluoroquinolones: ciprofloxacin (CIP 5—5 μg), (10) chemotherapeutics: nitrofurantoin (NIT—100 μg), trimethoprim (W5—5 μg). The analyzed strains were classified as multidrug resistant (MDR) and extensively drug resistant (XDR). Multidrug-resistant bacteria were defined as microorganisms that were resistant to at least one antibiotic from at least three groups of antimicrobial drugs targeting a given species. Extensively, drug-resistant bacteria are defined as microorganisms resistant to at least one antibiotic in all but two antibiotic groups targeting a given species [45]. The sensitivity of the tested strains was determined on Mueller–Hinton agar (bioMerieux). Bacteria were incubated at 37 °C for 16–18 h, and resistance to vancomycin was determined after 24 h of incubation. The diameter of the inhibition zone was expressed in millimeters, and strains were classified as susceptible or resistant. The reference strain Enterococcus faecalis ATCC 29212 was used as the control strain. Susceptibility analyses were based on the guidelines of the European Committee on Antimicrobial Susceptibility Testing (EUCAST 2012) [46] and Clinical and Laboratory Standards Institute (CLSI 2012) [47].
2.6. Isolation of Genomic DNA and PCR Conditions
To isolate genomic DNA, a single bacterial colony was collected from the agar plate (BHI agar) after 24 h of incubation, suspended in Tris–EDTA buffer, digested with lysozyme (0.6 mg L−1) and heated at 95 °C for 10 min (QBD2 block heater, Grant). The colony was centrifuged at 5000 rpm for 5 min at 4 °C, and the concentration and quality of the isolated DNA was determined using a spectrophotometer (Eppendorf BioSpectrometer® kinetic, Eppendorf, Hamburg, Germany). If DNA was of poor quality, it was isolated again with the Genomic Mini AX Bacteria Mini Kit (SPIN) (A&A Biotechnology, Gdynia, Poland) according to the manufacturer’s instructions. Genomic DNA was isolated in triplicate and stored at a temperature of −20 °C until further analysis. Single and multiplex polymerase chain reactions (PCR) were carried out in a thermal cycler (Eppendorf, Mastercycler Family, Hamburg, Germany). The total volume of the reaction mix was 20–50 μL for different reactions and the evaluated bacterial properties. The phases of the amplification process (denaturation, hybridization, and elongation) were adapted to a given reaction type (Tables S1–S3, Supplementary Materials). Amplified PCR products were separated and visualized on 1.5% agarose gel stained with ethidium bromide (1 mg L−1) (Sub–Cell® GT system, Bio–Rad, CA, USA). 100–1000 bp and 142–3794 bp DNA ladders (A&A Biotechnology, Gdynia, Poland) were used as molecular size markers. DNA bands were visualized under UV light with a gel documentation kit (DGelScan, Kucharczyk, Warsaw, Poland). Randomly selected amplicons were sequenced and identified in the BLAST program available on the website of the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/BLAST) to confirm the identity of the isolated Enterococcus strains.
2.7. Identification of Enterococci and Detection of Vancomycin Resistance Genes and Virulence Factors by PCR
The isolated strains were identified to species level, and vancomycin resistance genes and virulence factors were detected by single and multiplex PCR. The most prevalent vancomycin resistance genes—vanA, vanB, vanC1, and vanC2/C3—were identified. The ten most important enterococcus virulence factors were identified: cylA—cytolysin toxin also known as hemolysin; hyl—hyaluronidase enzyme; ace—collagen–binding surface protein; EfaA—cell wall adhesins; gelE—gelatinase; as—aggregation substance; esp—extracellular surface protein; cpd, cob, ccf—sex pheromones that participate in the exchange of genetic material between strains. Primer sequences (synthesized by Genomed), product size, and PCR conditions are presented in Tables S1–S3—Supplementary Materials.
2.8. Statistical Analysis
Statistical analyses were performed in the Statistica 13.2 program software package (StatSoft Inc., 1984–2019, Tulsa, OK, USA) at a significance level of 0.05. Spearman’s rank correlation coefficient was calculated to determine the correlations between the analyzed physicochemical parameters, total enterococcus counts, and the number of MDR and virulent strains. Because of abnormally distributed data, the Kruskal–Wallis (KW) test, a non-parametric version of classical one-way analysis of variance ANOVA, was used to determine the differences in enterococcus counts in wastewater and river water depending on the time of the sample collection and the type of investigated wastewater. A heatmap was generated, and a cluster analysis was performed with the use of Ward’s hierarchical cluster method in RStudio (Version 1.2.5033) using “heatmap.2” and “gplots” packages.
3. Results
3.1. Physicochemical Parameters
The physicochemical parameters of river water and wastewater samples are presented in Table 1. The average pH of wastewater samples was similar during the entire period of the study in the range of 8.35 (HWW) to 8.40 (TWW). The average temperature of wastewater samples did not exceed 16 °C. The temperature in most URW sampling sites was above 20 °C in summer and below 10 °C in winter.
Table 1.
Physicochemical properties of wastewater and river water.
| Sampling Sites | Parameters | |
|---|---|---|
| pH | Temperature (°C) | |
| HWW | 8.35 ± 0.62 a | 14.37 ± 6.96 |
| (7.60−9.20) b | (7.58−25.89) | |
| UWW | 8.39 ± 0.42 | 14.74 ± 6.60 |
| (8.02−9.22) | (7.58−25.85) | |
| TWW | 8.40 ± 0.35 | 15.55 ± 8.67 |
| (8.02−9.01) | (7.47−26.80) | |
| URW | 7.16 ± 0.41 | 11.76 ± 4.87 |
| (6.40−7.50) | (0.0−21.10) | |
| DRW | 7.21 ± 0.65 | 14.31 ± 6.0 |
| (6.50−8.40) | (9.30−24.70) | |
| p value c | 0.0011 * | 0.8452 |
a mean and standard deviation (±SD); b range c Kruskal–Wallis test, statistically significant differences [* p ≤ 0.001] between sampling stations. HWW—hospital waste water; UWW and TWW—untreated and treated wastewater; URW and DRW—upstream and downstream river water.
3.2. Microbial Counts in Wastewater and River Water Determined by the Culture-Dependent Method
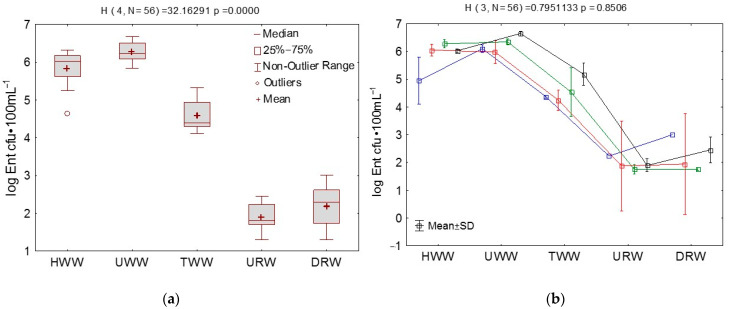
Enterococcus counts ranged from 1.30 log cfu·100 mL−1 in URW and DRW to 6.68 log cfu·100 mL−1 in UWW. The highest average bacterial counts were noted in UWW and HWW (6.27 and 5.83 log cfu·100 mL−1, respectively). In URW and DRW samples, the average enterococcus counts did not exceed 2.2 log cfu·100 mL−1 and were several orders of magnitude lower than in wastewater (Figure 1). Bacterial counts were highest in HWW in summer (6.28 log cfu·100 mL−1) and in TWW and UWW in fall (5.18 and 6.65 log cfu·100 mL−1, respectively). In URW and DRW samples, enterococcus counts were higher in winter (2.23 and 3.0 log cfu·100 mL−1, respectively) and in fall (1.89 and 2.45 log cfu·100 mL−1, respectively). Significant differences (p < 0.05) in bacterial counts were observed between the sampling sites. Enterococcus counts did not differ significantly across seasons (Figure 1).
Figure 1.
Enterococcus counts [log Ent cfu·100 mL−1] determined by the culture-dependent method in river water and wastewater sampled from different (a) sites and (b) seasons during the study. Kruskal–Wallis test (ANOVA), SD—standard deviation; N—number of samples; p—significance level.
3.3. Analysis of Correlations between Enterococci, Vancomycin Resistance Genes, Virulence Factors, Physicochemical Parameters, Sampling Site, and Season
The correlation analysis did not reveal significant differences (p < 0.05) between any of the examined parameters (excluding temperature) and season. Therefore, microbial parameters determined in different seasons are not presented. A negative correlation (r = −0.886) was noted between enterococcus counts and sampling site, whereas significant positive (r = 0.792) and negative (r = −0.606) correlations were determined between bacterial counts vs. water pH. Sampling sites were also significantly correlated with selected enterococcus species (E. faecalis, E. faecium, E. avium) (r = −0.599–0.620), resistance to the analyzed antibiotic groups (excluding TEC, VAN and DO), VC2/VC3 genes (r = −0.497), virulence factors (as, cpd, ccf), and water pH (r = −0.703), (Table S4A, Supplementary Materials). Enterococcus species were correlated with resistance to antibiotics, selected resistance genes (mainly vanB and vanC1) and virulence factors. Enterococcus faecalis and E. faecium were correlated with all examined virulence factors (r = 0.482–0.906) (excluding E. faecium and COB). Significant correlations were also noted between drug-resistant strains, virulent strains, and strains harboring van genes, subject to the analyzed factor (Table S4B,D, Supplementary Materials).
3.4. Species Diversity, Percentage Composition and Multidrug Resistance of Enterococci in Samples of River Water and Wastewater
Seven Enterococcus species were identified in the total number of 283 strains isolated from water and wastewater samples: E. faecalis, E. faecium, E. durans, E. avium, E. hirae, E. gallinarum, and E. casseliflavus/flavescens. Strains not identified to species level were labeled as other Enterococcus spp. Enterococcus faecium was the dominant species in wastewater samples regardless of the sampling site, and its percentage share ranged from 38.8% to 42.9%. The second most abundant species was E. faecalis which represented 29.4% to 31.0% of the species identified in wastewater samples. The abundance of the remaining species in wastewater was much lower and did not exceed several percent. Enterococcus gallinarum (31.3%) and E. faecium (25%) were dominant in DRW samples, and E. faecalis (29.4%) was most abundant in URW samples. Enterococci spp. accounted for several to more than 10% of all bacterial species in wastewater samples and more than 70% in DRW samples (Table 2).
Table 2.
Number and percentage of multidrug-resistant (MDR) and extensively drug-resistant (XDR) enterococci in wastewater and river water.
| Sewage | River | ||||
|---|---|---|---|---|---|
| Species | HWW | UWW | TWW | URW | DRW |
| Strains: number (%) | |||||
| MDR (%) | |||||
| XDR (%) | |||||
| E. faecium | 36 (42.9) | 28 (41.2) | 38 (38.8) | 0 | 4 (25.0) |
| 34 (94.4) | 27 (96.4) | 33 (86.8) | 0 | 4 (100) | |
| 2 (5.6) | 1 (3.57) | 1 (2.6) | 0 | 0 | |
| E. faecalis | 26 (31.0) | 20 (29.4) | 29 (29.6) | 5 (29.4) | 1 (6.3) |
| 23 (88.5) | 19 (95.0) | 28 (96.6) | 3 (60) | 1 (100) | |
| 1(3.8) | 0 | 0 | 0 | 0 | |
| E. durans | 1 (1.2) | 0 | 3 (3.1) | 0 | 0 |
| 1 (100) | 0 | 3 (100) | 0 | 0 | |
| E. avium | 7 (8.3) | 4 (5.9) | 2 (2.0) | 0 | 0 |
| 7 (100) | 4 (100) | 2 100) | 0 | 0 | |
| E. hirae | 2 (2.4) | 1 (1.5) | 4 (4.1) | 0 | 0 |
| 2 (100) | 1 (100) | 4 (100) | 0 | 0 | |
| E. gallinarum | 1 (1.2) | 3 (4.4) | 1 (1.0) | 0 | 5 (31.3) |
| 1 (100) | 3 (100) | 1 (100) | 0 | 4 (80) | |
| E. casseliflavus/ | 1 (1.2) | 0 | 0 | 0 | 0 |
| flawescens | 1 (100) | 0 | 0 | 0 | 0 |
| Other | 10 (11.9) | 12 (17.6) | 21 (21.4) | 12 (70.6) | 6 (37.5) |
| Enterococcus spp. | 10 (100) | 13 (100) | 20 (95.2) | 5 (41.6) | 6 (100) |
| 1 (10.0) | 0 | 0 | 0 | 0 | |
| Number of strains | 84 | 68 | 98 | 17 | 16 |
| MDR (%) | 79 (94.0) | 67 (98.5) | 91 (92.9) | 8 (47.0) | 15 (93.8) |
| XDR (%) | 4 (4.8) | 1(1.5) | 1(1.0) | 0 | 0 |
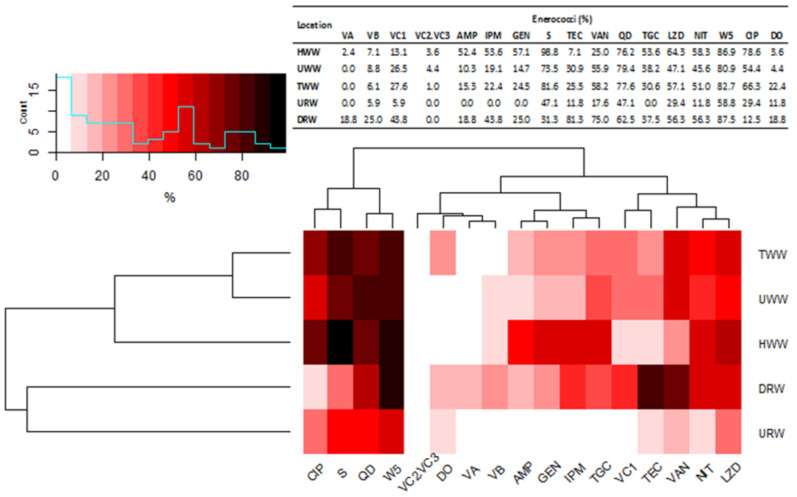
The isolated strains were tested for resistance to 13 antimicrobials belonging to 10 drug groups. The results were used to classify the bacterial strains as MDR and XDR. Two hundred and thirty-five isolates representing 94% of all enterococci identified in wastewater and 23 isolates representing 69.7% of all strains identified in river water were classified as MDR. Samples of HWW contained 79 MDR strains (94.0%) and 4 XDR strains (4.8%). In UWW and TWW samples, 67 (98.5%) and 91 (92.9%) strains were classified as MDR, respectively, and 1 strain in each sample (1.5% and 1.0%) was classified as XDR. Multidrug-resistant E. faecium was predominant in HWW (34 strains, 94.4%) and UWW (27 strains, 96.4%) samples, and E. faecalis was most abundant in TWW samples (28 strains, 96.6%). The remaining species isolated from wastewater samples were classified as MDR (Table 2).
Three E. faecalis strains in URW samples (60.0%) and four E. faecium strains in DRW samples (100%) were classified as MDR. Most of the strains (50–83.0%) isolated from wastewater samples were resistant to streptomycin (S) and trimethoprim (W5). More than 78% of E. faecium and E. faecalis isolates were resistant to these antibiotics. The percentage of isolates resistant to low concentrations of vancomycin ranged from 22% (E. faecium) in HWW to 70% (E. faecalis) in UWW samples. Most of the identified isolates were sensitive to doxycycline (DO). Bacterial species in DRW and URW samples had similar antibiotic resistance profiles to the strains isolated from wastewater. Most strains identified in URW samples were resistant to one, three, or six antibiotics, whereas most strains isolated from DRW samples were resistant to ten antimicrobials (Figure 2). Enterococcus faecium and E. faecalis, which cause most human and animal infections, were resistant mainly to teicoplanin (TEC), trimethoprim (W5), and streptomycin (S). None of the strains isolated from URW samples were resistant to ampicillin (AMP), imipenem (IMP), gentamicin (GEN), or tigecycline (TGC) (Table S5 in Supplementary Materials).
Figure 2.
Heatmap with percentage content of studied multidrug resistant enterococcus among wastewater and river water (HWW—hospital wastewater; UWW—untreated wastewater; TWW—treated wastewater URW—upstream river water; DRW—downstream river).
3.5. Identification of Vancomycin Resistance Genes (Van) in Enterococci Isolated from Wastewater and River Water
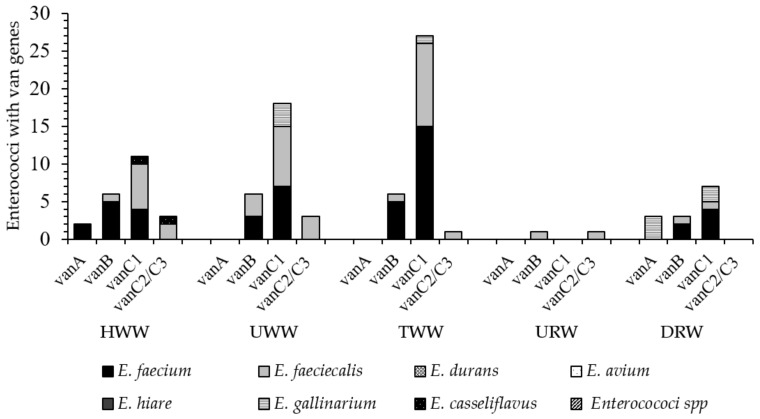
Out of the 250 strains isolated from wastewater, only the strains identified in HWW samples harbored all vancomycin resistance genes. The number of vancomycin-resistant enterococci in HWW samples ranged from 2 strains (2.4%) with vanA genes to 15 strains (39.5%) with vanC1 genes. Six strains harboring vanB genes were identified in UWW and TWW samples each (8.8% and 6.1%, respectively). The presence of the vanC1 genes was noted in 18 (26.5%) and 27 (27.6%) of UWW and TWW isolates, respectively. One strain (1.02%) in TWW samples and three strains (4.41%) in UWW samples, mostly E. faecium and E. faecalis, harbored vanC2/C3 genes. The predominant VRE species were E. faecium (van B genes, 5 strains, 13.8%) in HWW and E. faecalis (van C genes, 8 strains, 40%) in UWW samples. Five strains (13.1%) with vanB genes and 15 strains (39.5%) with vanC1 were isolated from TWW samples. All E. gallinarum and E. casseliflavus strains identified in wastewater harbored vanC1 and vanC2/C3 genes that confer natural resistance to vancomycin. Vancomycin-resistance genes were not identified in the remaining species isolated from the wastewater (Figure 3).
Figure 3.
The number of enterococci with van genes in wastewater and river water (HWW—hospital wastewater; UWW—untreated wastewater; TWW—treated wastewater URW—upstream river water; DRW—downstream river).
In URW samples, only one E. faecalis (5.88%) strain with phenotypic resistance to vancomycin harbored van B and van C2/C3 genes. The percentage of VRE strains was much higher in DRW samples, and it ranged from 18.7% (3 E. galinarium strains with van A genes) to 43.75% (1 E. faecalis strains, 4 E. faecium strains and 2 E. gallinarum strains with van C1 genes) Figure 2.
3.6. Identification of Genes Encoding Virulence Factors in Enterococci Isolated from Wastewater and River Water
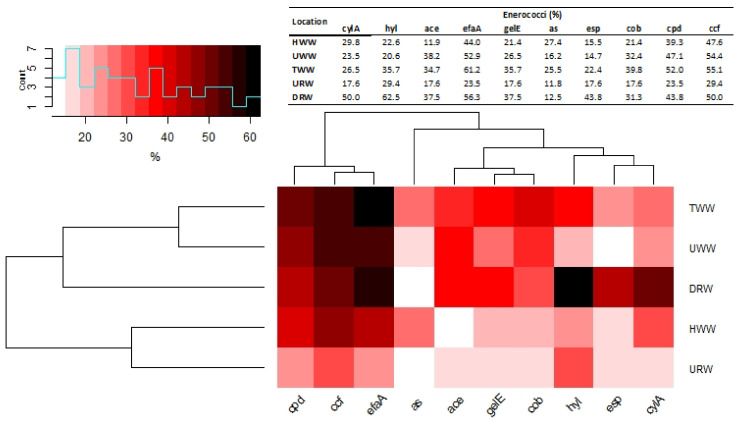
Genes encoding virulence factors were detected in 188 strains (75.2%) in wastewater and in 22 strains (66.6%) in river water samples. Strains with the highest number of virulence factors were detected mainly in TWW samples, and strains with the smallest number of virulence factors were determined in URW samples. Most virulence factors were associated with efaA and ccf genes. These genes were most abundant in the strains isolated from TWW samples (61.2% and 55.1%, respectively), followed by the strains isolated from UWW (52.9% and 54.4%) and HWW (44.0% and 47.6%) samples. The parentage of selected virulence genes (hyl, ace, efaA, gelE, as, esp) was higher in TWW (several to around 10%) than in HWW and UWW. In URW samples, 23.5% and 29.4% of the strains harbored efaA and ccf genes, respectively, but their percentage was lower than in wastewater isolates. The smallest number of strains in HWW (12.0%) harbored ace genes, and the smallest number of strains in UWW (14.7%) and TWW (22.4%) samples harbored esp genes. Strains with as genes were least frequently isolated from URW (11.8%) and DRW (12.5%) samples. (Figure 4). In TWW samples, the percentage of E. faecalis harboring various virulence genes ranged from 26.9% to 100% (Table S6, Supplementary Materials). Virulent E. faecium strains accounted for 0% (HWW) to 55.3% (TWW) of the isolates. More than 70% of E. faecalis strains and 40–60% of E. faecium strains isolated from UWW and TWW samples harbored efaA, cpd, and ccf genes. Enterococcus faecium strains with cylA, hyl, and ace genes were predominant (more than 20%) only in HWW samples on average, 10 virulence genes were identified in E. faecium and E. faecalis. In the remaining enterococci, the number of virulence genes was around 50% lower on average (Table S6, Supplementary Materials).
Figure 4.
Heatmap with percentage content of studied virulence enterococcus among wastewater and river water (HWW—hospital wastewater; UWW—untreated wastewater; TWW—treated wastewater URW—upstream river water; DRW—downstream river).
The presence of correlations between the abundance of vancomycin resistance genes (van) and phenotypic resistance to antibiotics in strains isolated from wastewater and river water samples was determined in a correlation analysis using the Ward’s method, and it revealed three main clusters (Figure 3). The first cluster was composed of strains resistant to CIP, S, QD, and W5. The second cluster comprised strains harboring vanC2/C3, vanA, and vanB genes, as well as strains resistant to AMP, GEN, IPM, and TGC. The third cluster grouped strains with vanC1 genes which were resistant to TEC, VAN, NIT, and LZD. The strains harboring van genes and MDR strains were divided into two main clusters based on their source. The first cluster was composed of strains isolated from TWW, UWW, and HWW samples, and the second cluster comprised strains isolated from DRW and URW samples.
Two clusters were identified based on the results of the virulence analysis. The first cluster grouped strains with cpd, ccf, and efaA genes that were most abundant in TWW, UWW, and DRW samples. The second cluster was subdivided into two groups composed of strains with ace, gelE, and cob genes (that were also most abundant in TWW, UWW, and DRW samples) and strains harboring hyl, esp, and cylA, which were most prevalent in DRW samples. Strains harboring as genes were isolated from both subgroups, and they were detected infrequently, mostly in HWW and TWW samples (Figure 4).
4. Discussion
4.1. Enterococcus Counts in Wastewater and River Water Samples
Rapid economic growth and industrialization contribute to growing levels of anthropogenic pressure on water resources around the world. Biological micropollutants that reach water bodies with treated wastewater are a serious and often marginalized problem. These pathogens are often correlated with the incidence of gastrointestinal diseases, the presence of ARGs and ARB, and they pose a significant epidemiological risk. Despite the above, there are no detailed guidelines for disinfecting treated wastewater or controlling microbiological pollution in water bodies (Council Directive 91/271/EEC; OJ, item 1800), [48]. This study analyzed the pool of virulent and antibiotic-resistant enterococci in municipal wastewater (including hospital wastewater) before and after mechanical and biological treatment in WWTP and in river water sampled downstream and upstream from the discharge point. Significant differences (p < 0.05) in enterococcus counts were noted between sampling sites, but not between seasons. The average enterococcus counts in UWW and HWW samples were determined at 6 log cfu·100 mL−1. Wastewater treatment reduced the abundance of enterococci by around 97%, but TWW samples contained more than several dozen thousand bacterial cells per 100 mL. The analyzed WWTP has a daily processing capacity of around 32,000 m3, which indicates that millions of bacterial cells reach the river each day. Enterococcus counts were more than two-fold lower in the samples collected upstream from the effluent discharge point, which suggests that the evacuation of treated wastewater compromises the microbiological quality of river water. The abundance of indicator bacteria, including enterococci, as well as ARGs and ARB is higher in water bodies that act as receptacles of municipal wastewater in urban areas [5,6,49,50,51]. An increase in enterococcus counts points to the contamination of river water with human and animal feces. Fecal contamination is also associated with higher abundance of potentially pathogenic bacteria such as Aeromonas hydrophila, Listeria monocytogenes, Salmonella spp., Pseudomonas aeruginosa, Campylobacter spp., Vibrio spp., and Yersinia spp. [52,53,54,55,56]. Human activities (wastewater treatment, agriculture, tourism) also contribute to the spread of MDR enterococci and VRE both in and outside the hospital environment around the world [14,35,36,57].
4.2. Species Composition, Multidrug Resistance and the Presence of Van Genes in Enterococci Isolated from Water and Wastewater Samples
In the group of seven identified enterococcus species, E. faecium (102 strains, 40.8%), E. faecalis (75 strains, 30.0%) were most abundant in all wastewater and river water samples, excluding DRW samples where E. gallinarium was most prevalent (29.4%). The percentage of the remaining enterococcus species (E. durans, E. avium, E. hirae, E. casseliflavus/flavescens) did not exceed 8% in the analyzed samples. Higher counts and percentage of E. faecium and E. faecalis in samples of untreated and treated wastewater and in samples of river water collected in the vicinity of the discharge point were also reported in other studies [49,58,59,60]. Enterococcus faecalis and E. faecium are the most abundant species in human and animal feces, wastewater and water contaminated with fecal matter [15,36,61,62]. In turn, E. hirae was most prevalent in wastewater in Portugal and the USA [63,64], whereas E. durans (24%) was the second most prevalent species after E. faecalis in treated and untreated hospital waste in Eastern Cape in the Republic of South Africa [65]. The composition of wastewater microbiota and their antibiotic resistance profiles can differ depending on the type of wastewater and the applied treatment methods [50,66]. A pilot study investigating the effectiveness of UV radiation in wastewater treatment demonstrated that UV light decreased enterococcus counts in treated wastewater by 3.1–3.3 log CFU relative to untreated wastewater. Ultraviolet radiation decreased the abundance of enterococci in treated wastewater to a level that is generally noted in unpolluted waters [8,12]. Despite the fact that treated wastewater contributes to environmental pollution, it does not have to be disinfected before it is evacuated to surface water bodies in Poland [67].
Multidrug-resistant enterococci accounted for 94% and 69.7% of the strains isolated from wastewater and river water samples, respectively. More than 87% of E. faecium and E faecalis strains isolated from wastewater, and all E. faecium and E faecalis strains isolated from DRW were classified as MDR. Multidrug-resistant strains represented 60% of the isolates from URW samples. Individual strains of the remaining enterococcus species (E. durans, E. avium, E. hirae, E. gallinarum, and E casseliflavus/flavescens) were also resistant to multiple antimicrobials. In wastewater samples, up to 5.6% of E. faecium and E. faecalis were XDR. These strains are particularly dangerous because they are resistant to at least one antibiotic in all but two antimicrobial groups [45]. Multidrug-resistant enterococci are responsible for a high percentage of hospital infections that are difficult to diagnose and treat [8,63]. Most of the isolated MDR strains were resistant to 7–8 of the tested antibiotics. More than 80% of those strains were resistant to streptomycin (aminoglycosides) and trimethoprim (chemotherapeutics), whereas the smallest number of strains (up to 22%) were resistant to doxycycline (tetracyclines). Enterococcus faecalis and E. faecium are highly resistant to aminoglycosides around the world [12,68], including in Poland [14]. More than 50% of E. faecalis and E. faecium strains identified in Ethiopia were resistant to vancomycin, penicillin, amoxicillin, doxycycline, and tetracycline, and 60.0% of all identified enterococci were classified as MDR [31]. In the present study, enterococcus strains isolated 500 m downstream from the wastewater discharge point had similar antibiotic resistance profiles, and most of them were resistant to trimethoprim (87.5%), vancomycin, and teicoplanin (more than 75% of the strains). These results clearly indicate that antibiotic-resistant enterococci are transferred to rivers with TWW. Numerous studies have demonstrated that the percentage of MDR enterococci and other bacterial groups continues to increase in TWW evacuated to water bodies [33,36,37,41,57,58].
Various van genes were identified in enterococcus strains characterized by phenotypic resistance to vancomycin and isolated from river water and wastewater. The cluster analysis involving Ward’s method revealed two groups of enterococcus strains whose antibiotic resistance profiles were most highly correlated with van genes. The first group comprised the strains isolated from UWW, TWW, and HWW samples, and the second group consisted of strains isolated from DRW and URW samples. Strains harboring all of the examined resistance genes (vanA, vanB, vanC1, and vanC2/C3) were identified mainly in HWW samples, but their percentage was generally low in the range of 2.4% (vanA) to 13.1% (vanC1). The above genes were detected in 1.02% (vanC2/C3) to 27.6% (vanC1) of the strains isolated from UWW and TWW samples. vanC1 and vanC2/3 genes conditioning natural resistance to vancomycin were most abundant in E. gallinarum and E. casseliflavus. The abundance of van genes was highest in E. faecalis and E. faecium isolates. These strains (20–70%) were also resistant to vancomycin in the disc diffusion test, and they included isolates resistant to high concentrations of vancomycin (MIC ≥ 32 to ≥ 256 mg·L–1; data not shown), which can be classified as VRE. The proportion of VRE strains exceeded 10%, and it was somewhat higher in TWW than UWW samples. Individual VRE strains were also noted in DRW samples. A similar percentage of VRE strains (27.0%) was reported in a WWTP in the United States [69]. In other studies, the proportions of VRE strains in wastewater ranged from 2% to 52%, depending on the treatment method, type of wastewater, and treatment stage [69,70,71]. Enterococci resistant to high concentrations of vancomycin (20 mg·L–1) were isolated from surface water (7%) in many European countries (Sweden, Spain, United Kingdom). These bacteria were also identified in UWW (71.0%) and TWW (36.0%). Resistance to vancomycin was determined mainly in E. faecalis strains harboring vanA genes [60]. In the current study, vanB, vanC1, and vanC2/C3 were the main genetic determinants of resistance to vancomycin. The presence of vanC1 genes in E. faecium and E. faecalis was surprising. Until recently, it was believed that vanC1 is encoded chromosomally and is not horizontally transferred. However, this gene is increasingly often identified in E. faecium and E. faecalis, which suggests that it can be acquired from E. gallinarum and E. casseliflavus by horizontal transfer [72]. Multidrug-resistant enterococci and VRE have been spreading at an alarming rate in both hospitals [31,73] and the natural environment [41,65]. These pathogens could originate from various sources which have not been fully elucidated to date. Enterococci harboring virulence factors that cause human infections and spread in the environment also pose a considerable problem [35,74].
4.3. Characteristics of Virulent Enterococci in Wastewater and River Water Samples
In wastewater samples, 75.2% of the isolated enterococcus strains harbored virulence factors (cylA, hyl, ace, efaA, as, gelE, esp, cob, cpd, ccf). The cluster analysis involving Ward’s method revealed the strongest correlations between sampling sites and the prevalence of strains with virulence genes in untreated (UWW) and treated (TWW) municipal wastewater as well as in river water sampled downstream from the wastewater discharge point (DRW). These samples were characterized by the highest percentage of strains containing sex pheromones cpd and ccf (21.4–55.1%) and cell wall adhesin efaA (44.1–61.2%). Sex pheromones participate in the exchange of genetic material and, consequently, induction of virulence. They also colonize hosts and cause inflammations [75,76]. Enterococcus strains with dominant sex pheromone genes were also identified in the South Nation River watershed in Ontario, Canada [65]. In the present study, 11.9% to 38.2% of the strains isolated from wastewater also harbored genes encoding cytolysin (cylA), collagen-binding surface protein (ace) (UWW) and gelatinase (gelE). Virulence genes are often associated with hospital infections caused by pathogenic enterococci [35,77]. In this study, E. faecalis (5.0–100%) and E. faecium (0–57.1%) were characterized by the highest number and diversity of virulence genes, with a predominance of efaA, cpd, and ccf. Similar observations were made in hospital strains in Bulgaria [36]. Virulence genes asa (aggregation substance) and cylA (cytolytic toxin) are often predominant in enterococci isolated from hospitals, the natural environment, animals, and wastewater [35,78]. Enterococci isolated from the Ganges River in India were characterized by a similar virulence profile [12]. In the Łyna River, 87% of the strains isolated downstream from the effluent discharge point harbored different virulence genes, but only 47.1% of such strains were isolated upstream from the discharge point. The isolated strains had similar virulence profiles to the strains identified in UWW and TWW samples. However, the highest percentage of these strains (62.5%) harbored genes encoding hyaluronidase (hyl) which contributes to connective tissue damage and facilitates the spread of bacteria in the host organism [18]. These genes were most abundant in vancomycin-resistant strains that harbored van genes and were isolated from URW samples. Hyl genes are frequently identified in clinical VRE isolates, but they are also present in vancomycin-susceptible enterococci (VSE) [79]. Most E. faecalis and E. faecium strains containing virulence genes, including esp, gelE, cylA and hyl, are associated with human infections [35,77]. The spread of MDR enterococci and VRE in the environment is particularly dangerous because these pathogens are not easy to detect or treat. These strains cause life-threatening infections and pose a significant epidemiological risk.
5. Conclusions
The results of this study indicate that WWTPs are significant sources of MDR and virulent enterococci in the environment. The strains isolated from TWW samples were characterized by higher levels of antibiotic resistance and virulence than those isolated from UWW and, in many cases, HWW samples. This observation indicates that bacteria undergo selective pressure during wastewater treatment and are transformed into MDR strains by accumulating genes that encode resistance to various antimicrobials. The enterococci isolated from DRW and wastewater were characterized by similar multidrug resistance and virulence profiles, and significant correlations were observed between these groups of isolates. The counts of these bacteria were two-fold lower in river water sampled upstream from the effluent discharge point. These findings suggest that large numbers of VRE and MDR enterococci are released to water bodies even from WWTPs equipped with highly effective treatment systems. The spread of these bacteria along the river continuum poses a significant epidemiological risk and a threat to public health. Therefore, enterococcus emissions should be monitored, in the hospital environment, treated wastewater should be disinfected before it is evacuated to surface water bodies and that new wastewater treatment technologies should be developed to minimize the risk of exposure to these pathogens.
Supplementary Materials
The following are available online at https://www.mdpi.com/1660-4601/18/2/563/s1. Figure S1: Technological scheme of wastewater treatment plant. Table S1: Oligonucleotide sequences and product size of primers for identifying enterococci. Table S2: Sequences of oligonucleotides and primers for identifying vancomycin resistance genes in enterococci. Table S3: Sequences of oligonucleotides and primers for identifying virulence factors in enterococci. Table S4: (Part A–B) Correlations between the analyzed physicochemical parameters, total enterococcus counts, different species, and the number of MDR and virulent strains (Spearman’s rank correlation coefficient; marked in red are values with p < 0.05). Table S5: Number and percentage of antibiotic resistant enterococci in wastewater and river water. Table S6: Number and percentage of virulent enterococci in wastewater and river water.
Funding
This study was supported by Ministry of Science and Higher Education (Poland), grant number 29.610.021-110. This research was also funded by the National Science Center, grant number 2017/27/B/NZ9/00267. Project financially co-supported by Minister of Science and Higher Education in the range of the program entitled “Regional Initiative of Excellence” for the years 2019–2022, Project No. 010/RID/2018/19, amount of funding 12,000,000 PLN.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article or Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Harnisz M., Kiedrzyńska E., Kiedrzyński M., Korzeniewska E., Czatzkowska M., Koniuszewska I., Joźwik A., Szklarek S., Niestępski S., Zalewski M. The impact of WWTP size and sampling season on the prevalence of antibiotic resistance genes in wastewater and the river system. Sci. Total Environ. 2020;741:140466. doi: 10.1016/j.scitotenv.2020.140466. [DOI] [PubMed] [Google Scholar]
- 2.Lekunberri I., Villagrasa M., Balcázar J.L., Borrego C.M. Contribution of bacteriophage and plasmid DNA to the mobilization of antibiotic resistance genes in a river receiving treated wastewater discharges. Sci. Total Environ. 2017;601–602:206–209. doi: 10.1016/j.scitotenv.2017.05.174. [DOI] [PubMed] [Google Scholar]
- 3.Niestępski S., Harnisz M., Ciesielski S., Korzeniewska E., Osińska A. Environmental fate of Bacteroidetes, with particular emphasis on Bacteroides fragilis group bacteria and their specific antibiotic resistance genes, in activated sludge wastewater treatment plants. J. Hazard. Mater. 2020;394:122544. doi: 10.1016/j.jhazmat.2020.122544. [DOI] [PubMed] [Google Scholar]
- 4.Bojarczuk A., Jelonkiewicz Ł., Lenart-Boroń A. The effect of anthropogenic and natural factors on the prevalence of physicochemical parameters of water and bacterial water quality indicators along the river Białka, southern Poland. Environ. Sci. Pollut. Res. 2018;25:10102–10114. doi: 10.1007/s11356-018-1212-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glińska-Lewczuk K., Gołaś I., Koc J., Gotkowska-Płachta A., Harnisz M., Rochwerger A. The impact of urban areas on the water quality gradient along a lowland river. Environ. Monit. Assess. 2016;188:624–638. doi: 10.1007/s10661-016-5638-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gotkowska-Płachta A., Gołaś I., Korzeniewska E., Koc J., Rochwerger A., Solarski K. Evaluation of the distribution of fecal indicator bacteria in a river system depending on different types of land use in the southern watershed of the Baltic Sea. Environ. Sci. Pollut. Res. 2016;23:4073–4085. doi: 10.1007/s11356-015-4442-6. [DOI] [PubMed] [Google Scholar]
- 7.Byappanahalli M.N., Nevers M.B., Korajkic A., Staley Z.R., Harwood V.J. Enterococci in the environment. Microbiol. Mol. Biol. Rev. 2012;76:685–706. doi: 10.1128/MMBR.00023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Staley C., Dunny G.M., Sadowsky M.J. Environmental and Animal-associated enterococci. Adv. Appl. Microbiol. 2014:147–186. doi: 10.1016/B978-0-12-800261-2.00004-9. [DOI] [PubMed] [Google Scholar]
- 9.Kampmeier S., Kossow A., Clausen L.M., Knaack D., Ertmer C., Gottschalk A., Freise H., Mellmann A. Hospital acquired vancomycin resistant enterococci in surgical intensive care patients—A prospective longitudinal study. Antimicrob. Resist. Infect. Control. 2018;7:103. doi: 10.1186/s13756-018-0394-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Recreational Water Quality Criteria. Office of Water, United States Environmental Protection Agency; Washington, DC, USA: 2012. [Google Scholar]
- 11.Zielińska D., Kolożyn-Krajewska D. Food-origin lactic acid bacteria may exhibit probiotic properties: Review. Biomed Res. Int. 2018:1–15. doi: 10.1155/2018/5063185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lata P., Ram S., Agrawal M., Shanker R. Enterococci in river Ganga surface waters: Propensity of species distribution; dissemination of antimicrobial–resistance and virulence–markers among species along landscape. BMC Microbiol. 2009;9:140. doi: 10.1186/1471-2180-9-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Servais P., Passerat J. Antimicrobial resistance of fecal bacteria in waters of the Seine river watershed (France) Sci. Total Environ. 2009;408:365–372. doi: 10.1016/j.scitotenv.2009.09.042. [DOI] [PubMed] [Google Scholar]
- 14.Sadowy E., Łuczkiewicz A. Drug–resistant and hospital–associated Enterococcus faecium from wastewater, riverine estuary and anthropogenically impacted marine catchment basin. BMC Microbiol. 2014;14:66. doi: 10.1186/1471-2180-14-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boehm A.B., Sassoubre L.M. Enterococci as indicators of environmental fecal contamination. In: Gilmore M.S., Clewell D.B., Ike Y., Shankar N., editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infections. Massachusetts Eye and Ear Infirmary; Boston, MA, USA: 2014. pp. 1–31. [PubMed] [Google Scholar]
- 16.Franz C.M.A.P., Huch M., Abriouel H., Holzapfel W., Galvez A. Enterococci as probiotics and their implications in food safety. Int. J. Food Microbiol. 2011;151:125–140. doi: 10.1016/j.ijfoodmicro.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 17.Gao W., Howden B.P., Stinear T.P. Evolution of virulence in Enterococcus faecium, a hospital–adapted opportunistic pathogen. Curr. Opin. Microbiol. 2018;41:76–82. doi: 10.1016/j.mib.2017.11.030. [DOI] [PubMed] [Google Scholar]
- 18.Talaga-Ćwiertnia K., Bulanda M. Drug resistance in the genus enterococcus-current problem in humans and animals. Post. Mikrob. Adv. Microbiol. 2018;57:244–250. doi: 10.21307/PM-2018.57.3.244. [DOI] [Google Scholar]
- 19.O’Driscoll T., Crank C.W. Vancomycin–resistant enterococcal infections: Epidemiology, clinical manifestations, and optimal management. Infect. Drug Resist. 2015;8:217–230. doi: 10.2147/IDR.S54125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghoshal U., Garg A., Tiwari D., Ayyagiri A. Emerging vancomycin resistance in enterococci in India. Indian. J. Pathol. Microbiol. 2006;49:620–622. [PubMed] [Google Scholar]
- 21.Gordon S., Swenson J., Hill B., Pigott N., Facklam R., Cooksey R., Thornsberry C., Jarvis W., Tenover F. Antimicrobial susceptibility patterns of common and unusual species of enterococci causing infections in the United States. J. Clin. Microbiol. 1992;30:2373–2378. doi: 10.1128/JCM.30.9.2373-2378.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Oliveira D.M.P., Forde B.M., Kidd T.J., Harris P.N.A., Schembri M.A., Beatson S.A., Paterson D.L., Walker M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020;33:e00181-19. doi: 10.1128/CMR.00181-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel T., Lewis M.E., Niesley M.L., Chowdhury M. Postneurosurgical Central Nervous System Infection Due to Enterococcus faecalis Successfully Treated With Intraventricular Vancomycin. Infect. Dis. Clin. Pract. 2016;24:174–176. doi: 10.1097/IPC.0000000000000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sievert D.M., Ricks P., Edwards J.R., Schneider A., Patel J., Srinivasan A., Kallen A., Limbago B., Fridkin S., National Healthcare Safety Network (NHSN) Team and Participating NHSN Facilities Antimicrobial-resistant pathogens associated with healthcare-associated infections: Summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect. Control Hosp. Epidemiol. 2013;34:1–14. doi: 10.1086/668770. [DOI] [PubMed] [Google Scholar]
- 25.Yuen G.J., Ausubel F.M. Enterococcus infection biology: Lessons from invertebrate host models. J. Microbiol. 2014;52:200–210. doi: 10.1007/s12275-014-4011-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levitus M., Perera T.B. StatPearls. StatPearls Publishing LLC; Treasure Island, FL, USA: 2019. [(accessed on 21 July 2020)]. Vancomycin-Resistant Enterococci (VRE) Available online: https://www.ncbi.nlm.nih.gov/books/NBK513233/ [PubMed] [Google Scholar]
- 27.Monticelli J., Knezevich A., Luzzati R., Di Bella S. Clinical management of non-faecium non-faecalis vancomycin-resistant enterococci infection. Focus on Enterococcus gallinarum and Enterococcus casseliflavus/flavescens. J. Infect. Chemother. 2018;24:237–246. doi: 10.1016/j.jiac.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 28.Protonotariou E., Dimitroulia E., Pournaras S., Pitiriga V., Sofianou D., Tsakris A. Trends in antimicrobial resistance of clinical isolates of Enterococcus faecalis and Enterococcus faecium in Greece between 2002 and 2007. J. Hosp. Infect. 2010;75:225–227. doi: 10.1016/j.jhin.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 29.Yang J.X., Li T., Ning Y.Z., Shao D.H., Liu J., Wang S.Q., Liang G.W. Molecular characterization of resistance, virulence and clonality in vancomycin-resistant Enterococcus faecium and Enterococcus faecalis: A hospital-based study in Beijing, China. Infect. Genet. Evol. 2015;33:253–260. doi: 10.1016/j.meegid.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 30.Alevizakos M., Gaitanidis A., Nasioudis D., Tori K., Flokas M.E., Mylonakis E. Colonization With Vancomycin–Resistant Enterococci and Risk for Bloodstream Infection Among Patients With Malignancy: A Systematic Review and Meta–Analysis. Open Forum Infect. Dis. 2017;4:ofw246. doi: 10.1093/ofid/ofw246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melese A., Genet C., Andualem T. Prevalence of Vancomycin resistant enterococci (VRE) in Ethiopia: A systematic review and meta–analysis. BMC Infect. Dis. 2020;20:124. doi: 10.1186/s12879-020-4833-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barbosa-Ribeiro M., De Jesus-Soares A., Zaia A.A., Ferraz C.C., Almeida J.F., Gomes B.P. Antimicrobial susceptibility and characterization of virulence genes of Enterococcus faecalis isolates from teeth with failure of the endodontic treatment. J. Endod. 2016;42:1022–1028. doi: 10.1016/j.joen.2016.03.015. [DOI] [PubMed] [Google Scholar]
- 33.Comerlato C.B., Resende M.C., Caierão J., d’Azevedo P.A. Presence of virulence factors in Enterococcus faecalis and Enterococcus faecium susceptible and resistant to vancomycin. Mem. Inst. Oswaldo Cruz. 2013;108:590–595. doi: 10.1590/S0074-02762013000500009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strateva T., Atanasova D., Savov E., Petrova G., Mitov I. Incidence of virulence determinants in clinical Enterococcus faecalis and Enterococcus faecium isolates collected in Bulgaria. Braz. J. Infect. Dis. 2016;20:127–133. doi: 10.1016/j.bjid.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferguson D.M., Talavera G.N., Hernandez L.A., Weisberg S.B., Ambrose R.F., Jay J.A. Virulence genes among Enterococcus faecalis and Enterococcus faecium isolated from coastal beaches and human and nonhuman sources in Southern California and Puerto Rico. J. Pathog. 2016 doi: 10.1155/2016/3437214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lata P., Ram S., Shanker R. Multiplex PCR based genotypic characterization of pathogenic vancomycin resistant Enterococcus faecalis recovered from an Indian river along a city landscape. Springer Plus. 2016;5:1199. doi: 10.1186/s40064-016-2870-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Varela A.R., Ferro G., Vredenburg J., Yanık M., Vieira L., Rizzo L., Lameiras C., Manaia C.M. Vancomycin resistant enterococci: From the hospital effluent to the urban wastewater treatment plant. Sci. Total Environ. 2013;450–451:155–161. doi: 10.1016/j.scitotenv.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 38.Bartley P.B., Schooneveldt J.M., Looke D.F.M., Morton A., Johnson D.W., Nimmo G.R. The relationship of a clonal outbreak of Enterococcus faecium vanA to methicillin-resistant Staphylococcus aureus incidence in a Australian hospital. J. Hosp. Infect. 2001;48:43–54. doi: 10.1053/jhin.2000.0915. [DOI] [PubMed] [Google Scholar]
- 39.Talebi M., Sadeghi J., Rahimi F., Pourshafie M.R. Isolation and Biochemical Fingerprinting of Vancomycin–Resistant Enterococcus faecium From Meat, Chicken and Cheese. Jundishapur J. Microbiol. 2015;8:e15815. doi: 10.5812/jjm.8(4)2015.15815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Torres C., Alonso C.A., Ruiz-Ripa L., León-Sampedro R., Del Campo R., Coque T.M. Antimicrobial resistance in Enterococcus spp. of animal origin. Microbiol. Spectr. 2018;6:ARBA-0032-2018. doi: 10.1128/microbiolspec.ARBA-0032-2018. [DOI] [PubMed] [Google Scholar]
- 41.Nishiyama M., Iguchi A., Suzuki Y. Identification of Enterococcus faecium and Enterococcus faecalis as vanC–type vancomycin–resistant enterococci (VRE) from sewage and river water in the provincial city of Miyazaki, Japan. J. Environ. Sci. Health. 2015;50:16–25. doi: 10.1080/10934529.2015.964599. [DOI] [PubMed] [Google Scholar]
- 42.Korzeniewska E., Korzeniewska A., Harnisz M. Antibiotic resistant Escherichia coli in hospital and municipal sewage and their emission to the environment. Ecotoxicol. Environ. Saf. 2013;91:96–102. doi: 10.1016/j.ecoenv.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 43.Polish Standard PN–EN ISO 7899–2:2004 . Water Quality—Detection and Enumeration of Intestinal Enterococci—Part 2: Membrane Filtration Method. PKN Warsaw; Płock, Poland: 2004. [Google Scholar]
- 44.Facklam R.R., Collins M.D. Identification of enterococcus species isolated from human infections by a conventional test scheme. J. Clin. Microbiol. 1989;27:731–734. doi: 10.1128/JCM.27.4.731-734.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Magiorakos A.P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G., Harbart S., Hindler J.F., Kahlmeter G., Olsson-Liljequist B., et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 46.European Committee on Antimicrobial Susceptibility Testing (EUCAST) Breakpoint Tables for Interpretation of MICs and Zone Diameters. [(accessed on 20 May 2020)];2012 Available online: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Disk_test_documents/EUCAST_breakpoints_v_2.0_120101.pdf.
- 47.CLSI, Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Disk Susceptibility Tests, Approved Standard. 11th ed. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2012. CLSI document M02-A11. [Google Scholar]
- 48.Council of the European Communities Council Directive 91/271/ECC of Concerning Urban Waste Water Treatment. Off. J. Eur. Communities. 1991:40–52. [Google Scholar]
- 49.Giebułtowicz J., Tyski S., Wolinowska R., Grzybowska W., Zaręba T., Drobniewska A., Wroczyński P., Nałęcz-Jawecki G. Occurrence of antimicrobial agents, drug–resistant bacteria, and genes in the sewage–impacted Vistula River (Poland) Environ. Sci. Pollut. Res. 2018;25:5788–5807. doi: 10.1007/s11356-017-0861-x. [DOI] [PubMed] [Google Scholar]
- 50.Korzeniewska E., Harnisz M. Relationship between modification of activated sludge wastewater treatment and changes in antibiotic resistance of bacteria. Sci. Total Environ. 2018;639:304–315. doi: 10.1016/j.scitotenv.2018.05.165. [DOI] [PubMed] [Google Scholar]
- 51.Osińska A., Korzeniewska E., Harnisz M., Felis E., Bajkacz S., Jachimowicz P., Niestępski S., Konopka I. Small–scale wastewater treatment plants as a source of the dissemination of antibiotic resistance genes in the aquatic environment. J. Hazard. Mater. 2020;381:121221. doi: 10.1016/j.jhazmat.2019.121221. [DOI] [PubMed] [Google Scholar]
- 52.Lipp E.K., Farrah S.A., Rose J.B. Assessment and impact of microbial fecal pollution and human enteric pathogens in a coastal community. Mar. Pollut. Bull. 2001;42:286–293. doi: 10.1016/S0025-326X(00)00152-1. [DOI] [PubMed] [Google Scholar]
- 53.Hörman A., Rimhanen-Finne R., Maunula L., von Bonsdorff C.-H., Torvela N., Heikinheimo A., Hanninen M.L. Campylobacter spp., Giardia spp., Cryptosporidium spp., Noroviruses, and indicator organisms in surface water in southwestern Finland, 2000–2001. Appl. Environ. Microbiol. 2004;70:87–95. doi: 10.1128/AEM.70.1.87-95.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fries J.S., Characklis G.W., Noble R.T. Sediment–water exchange of Vibrio sp. and fecal indicator bacteria: Implications for persistence and transport in the Neuse River Estuary, North Carolina. USA. Water Res. 2008;42:941–950. doi: 10.1016/j.watres.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 55.Wilkes G., Edge T., Gannon V., Jokinen C., Lyautey E., Medeiros D., Neumann N., Ruecker N., Topp E., Lapen D.R. Seasonal relationships among indicator bacteria, pathogenic bacteria, Cryptosporidium oocysts, Giardia cysts, and hydrological indices for surface waters within an agricultural landscape. Water Res. 2009;43:2209–2223. doi: 10.1016/j.watres.2009.01.033. [DOI] [PubMed] [Google Scholar]
- 56.Rodrigues V., Ramaiah N., Kakti S., Samant D. Long–term variations in abundance and distribution of sewage pollution indicator and human pathogenic bacteria along the central west coast of India. Ecol. Indic. 2011;11:318–327. doi: 10.1016/j.ecolind.2010.05.010. [DOI] [Google Scholar]
- 57.Giebułtowicz J., Nałęcz-Jawecki G., Harnisz M., Kucharski D., Korzeniewska E., Płaza G. Environmental Risk and Risk of Resistance Selection Due to Antimicrobials’ Occurrence in Two Polish Wastewater Treatment Plants and Receiving Surface Water. Molecules. 2020;25:1470. doi: 10.3390/molecules25061470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lanthier M., Scott A., Zhang Y., Cloutier M., Durie D., Henderson V.C., Wilkes G., Lapen D.R., Topp E. Distribution of selected virulence genes and antibiotic resistance in Enterococcus species isolated from the South Nation River drainage basin; Ontario; Canada. J. Appl. Microbiol. 2011;110:407–421. doi: 10.1111/j.1365-2672.2010.04893.x. [DOI] [PubMed] [Google Scholar]
- 59.Leclercq R., Oberlé K., Galopin S., Cattoir V., Budzinski H., Petit F. Changes in enterococcal populations and related antibiotic resistance along a medical center–wastewater treatment plant–river continuum. Appl. Environ. Microbiol. 2013;79:2428–2434. doi: 10.1128/AEM.03586-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kühn I., Iversen A., Burman L.G., Olsson-Liljequist B., Franklin A., Finn M., Aarestrup F., Seyfarth A.M., Blanch A.R., Vilanova X., et al. Comparison of enterococcal populations in animals; humans; and the environment—A European study. Int. J. Food Microbiol. 2003;88:133–145. doi: 10.1016/S0168-1605(03)00176-4. [DOI] [PubMed] [Google Scholar]
- 61.Lebreton F., Willems R.J.L., Gilmore M.S. Enterococcus Diversity, Origins in Nature, and Gut Colonization. In: Gilmore M.S., Clewell D.B., Ike Y., Shankar N., editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Massachusetts Eye and Ear Infirmary; Boston, MA, USA: 2014. pp. 5–64. [PubMed] [Google Scholar]
- 62.Layton B.A., Walters S.P., Lam L.H., Boehm A.B. Enterococcus species distribution among human and animal hosts using multiplex PCR. J. Appl. Microbiol. 2010;109:539–547. doi: 10.1111/j.1365-2672.2010.04675.x. [DOI] [PubMed] [Google Scholar]
- 63.Bonilla T.D., Nowosielski K., Esiobu N., McCorquodale D.S., Rogerson A. Species assemblages of Enterococcus indicate potential sources of fecal bacteria at a south Florida recreational beach. Mar. Pollut. Bull. 2006;52:807–810. doi: 10.1016/j.marpolbul.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 64.Da Silva M.F., Tiago I., Veríssimo A., Boaventura R.A.R., Nunes O.C., Manaia C.M. Antimicrobial resistance patterns in Enterobacteriaceae isolated from an urban wastewater treatment plant. FEMS Microbiol. Ecol. 2007;60:166–176. doi: 10.1111/j.1574-6941.2006.00268.x. [DOI] [PubMed] [Google Scholar]
- 65.Iweriebor B.C., Gaqavu S., Obi L.C., Nwodo U.U., Okoh A.I. Antibiotic susceptibilities of enterococcus species isolated from hospital and domestic wastewater effluents in alice, eastern cape province of South Africa. Int. J. Environ. Res. Public Health. 2015;12:4231–4246. doi: 10.3390/ijerph120404231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gotkowska-Płachta A., Filipkowska Z., Korzeniewska Z., Janczukowicz W., Dixon B., Gołaś I., Szwalgin D. Airborne microorganisms emitted from wastewater treatment plant treating domestic wastewater and meat processing industry wastes. Clean Soil Air Water. 2013;41:429–436. doi: 10.1002/clen.201100466. [DOI] [Google Scholar]
- 67.Aarestrup F.M., Butaye P., Witte W. Nonhuman reservoirs of enterococci. In: Gilmore M.S., Clewell D.B., Courvalin P., Dunny G.M., Murray B.E., Rice L.B., editors. The Enterococci: Pathogenesis. Molecular Biology and Antibiotic Resistance. ASM Press; Washington, DC, USA: 2002. pp. 55–99. [Google Scholar]
- 68.Osińska A., Korzeniewska E., Harnisz M., Niestępski S. The prevalence and characterization of antibiotic–resistant and virulent Escherichia coli strains in the municipal wastewater system and their environmental fate. Sci. Total Environ. 2017;577:367–375. doi: 10.1016/j.scitotenv.2016.10.203. [DOI] [PubMed] [Google Scholar]
- 69.Rosenberg Goldstein R.E., Micallef S.A., Gibbs S.G., George A., Claye E., Sapkota A., Joseph S.W., Sapkota A.R. Detection of vancomycin–resistant enterococci (VRE) at four U.S. wastewater treatment plants that provide effluent for reuse. Sci. Total Environ. 2014;1:404–411. doi: 10.1016/j.scitotenv.2013.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kotzamanidis C., Zdragas A., Kourelis A., Moraitou E., Papa A., Yiantzi V., Pantelidou C., Yiangou M. Characterization of vanA–type Enterococcus faecium isolates from urban and hospital wastewater and pigs. J. Appl. Microbiol. 2009;107:997–1005. doi: 10.1111/j.1365-2672.2009.04274.x. [DOI] [PubMed] [Google Scholar]
- 71.Morris D., Galvin S., Boyle F., Hickey P., Mulligan M., Cormican M. Enterococcus faecium of the vanA genotype in rural drinking water, effluent, and the aqueous environment. Appl. Environ. Microbiol. 2012;78:596–598. doi: 10.1128/AEM.06636-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moura T.M., Cassenego A.P.V., Campos F.S., Ribeiro A.M.L., Franco A.C., d’Azevedo P.A., Frazzon J., Frazzon A.P.G. Detection of vanC1 gene transcription in vancomycin–susceptible Enterococcus faecalis. Mem. Inst. Oswaldo Cruz. 2013;108:453–456. doi: 10.1590/S0074-0276108042013009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Toner L., Papa N., Aliyu S.H., Dev H., Lawrentschuk N., Al-Hayek S. Vancomycin resistant enterococci in urine cultures: Antibiotic susceptibility trends over a decade at a tertiary hospital in the United Kingdom. Investig. Clin. Urol. 2016;57:129–134. doi: 10.4111/icu.2016.57.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hancock L.E., Murray B.E., Sillanpää J. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Massachusetts Eye and Ear Infirmary; Boston, MA, USA: 2014. Enterococcal Cell Wall Components and Structures; pp. 503–546. [PubMed] [Google Scholar]
- 75.Kayaoglu G., Orstavik D. Virulence factors of Enterococcus faecalis: Relationship to endodontic disease. Crit. Rev. Oral Biol. Med. 2004;15:308–320. doi: 10.1177/154411130401500506. [DOI] [PubMed] [Google Scholar]
- 76.Valenzuela A.S., Omar N.B., Abriouel H., López R.L., Ortega E., Cañamero M.M., Gálvez A. Risk factors in enterococci isolated from foods in Morocco: Determination of antimicrobial resistance and incidence of virulence traits. Food Chem. Toxicol. 2008;46:2648–2652. doi: 10.1016/j.fct.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 77.Shokoohizadeh L., Ekrami A., Labibzadeh M., Ali L., Alavi S.M. Antimicrobial resistance patterns and virulence factors of enterococci isolates in hospitalized burn patients. BMC Res. Notes. 2018;11:1. doi: 10.1186/s13104-017-3088-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Creti R., Imperi M., Bertuccini L., Fabretti F., Orefici G., Di Rosa R., Baldassarri L. Survey for virulence determinants among Enterococcus faecalis isolated from different sources. J. Med. Microbiol. 2004;53:13–20. doi: 10.1099/jmm.0.05353-0. [DOI] [PubMed] [Google Scholar]
- 79.Biswas P.P., Dey S., Sen A., Adhikari L. Molecular Characterization of Virulence Genes in Vancomycin–Resistant and Vancomycin–Sensitive Enterococci. J. Glob. Infect. Dis. 2016;8:16–24. doi: 10.4103/0974-777X.176141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is contained within the article or Supplementary Materials.