Abstract
Simple Summary
The Lachninae is a unique aphid lineage with some particular characteristics. The first complete mitochondrial genome from this subfamily is reported here, with Stomaphis sinisalicis as a representative. The gene arrangement of the new mitogenome differs greatly from the normal gene order of other aphids and insects. The existence of a tandem repeat region in the Lachninae mitogenome may suggest it is probably an ancient feature of aphid mitogenomes. Phylogenetic analysis indicates a basal position of Lachninae within Aphididae.
Abstract
Complete mitochondrial genomes are valuable resources for different research fields such as genomics, molecular evolution and phylogenetics. The subfamily Lachninae represents one of the most ancient evolutionary lineages of aphids. To date, however, no complete Lachninae mitogenome is available in public databases. Here we report the Stomaphis sinisalicis mitogenome, representing the first complete mitogenome of Lachninae. The S. sinisalicis mitogenome is consist of 13 protein-coding genes (PCGs), two rRNA genes (rRNAs), 22 tRNA genes (tRNAs), a control region and a large tandem repeat region. Strikingly, the mitogenome exhibits a novel, highly rearranged gene order between trnE and nad1 compared with that of other aphids. The presence of repeat region in the basal Lachninae may further indicate it is probably an ancestral feature of aphid mitogenomes. Collectively, this study provides new insights on mitogenome evolution and valuable data for future comparative studies across different insect lineages.
Keywords: mitogenome, aphid, repeat region, gene rearrangement
1. Introduction
Complete mitochondrial genomes of insects are highly conserved circular molecules, which are valuable resources for investigating the molecular evolution, phylogenetic relationships and gene arrangement among different lineages, due to their rare recombination, rapid evolutionary rate and evolutionary conservation in organization [1,2,3]. The double-stranded molecules are generally composed of 13 protein coding genes (PCGs), two ribosomal genes (rRNAs), 22 tRNA genes (tRNAs) and a large non-coding control region (also called AT-rich region). In some taxa, such as some hemipteran aphid species, however, there is an additional unique non-coding tandem repeat region comprising different repeat units and copy numbers [4,5,6,7]. The gene order within insect mitogenomes is basically pretty conservative and identical to that of proposed Insecta ancestral gene arrangement pattern, with some exceptions of gene rearrangements involving tRNAs, rRNAs or PCGs [1,8,9,10,11,12]. Hemipteroids are one of the most distinct examples having a highly rearrangement frequency among insects, including species from orders Phthiraptera, Psocoptera, and Thysanoptera [1,13]. High rearrangement rates are also found in some Hemipterans including planthoppers (Delphacida), bugs (Aradidae, Enicocephalidae, Largidae, Pyrrhocoridae), whiteflies (Aleyrodidae), and more recently, scale insects (Coccoidea) [1,14,15,16,17,18]. While very few gene rearrangements are found in other hemipteran taxa such as aphids, those gene arrangement patterns remain to be confirmed by investigating more taxa. Mitogenome arrangement data have been supposed to provide useful information for investigating evolutionary relationships and insect evolutionary history because gene rearrangement of typical 37 genes in mitogenome is extremely uncommon and unlikely to replicate through convergent evolution [2,3]. With the growing availability of complete mitogenomes of insects, comprehensive comparative studies across different lineages should promote our understanding for their molecular evolution and phylogenetic relationships. However, mitogenomes of some insect taxa are still underrepresented.
Aphids are an important and diverse group within Hemiptera that includes many plant pests with complex life cycles and a large number of host plants. In the Aphididae, more than 5000 aphid species from 24 subfamilies have been described [19]. Of the thousands of extant aphid species, complete mitochondrial genomes are known for only 43 species from six subfamilies (Table S1). For some major aphid lineages, not even a single complete mitochondrial genome has been reported. The limited mitogenomic resources and small sampling has hampered widely comparative studies for the mitogenome sequences, gene arrangements and molecular evolution among aphids. Moreover, an extensive comparative analysis among aphids will favor gaining insights into the evolution of some unique characteristics for aphid mitogenomes, such as the presence of a large tandem repeat region. The unique tandem repeat region in the aphid mitogenomes is generally locating between the trnE and trnF but differ largely in length and copy number of repeat units [5,7]. This repeat region was first reported in several species from the subfamily Aphidinae and was once considered to be a lineage-specific feature which occurred from independent evolutionary events [20,21], whereas several species from other subfamilies have subsequently been found to contain a repeat region as well. So far, 18 aphid species from four subfamilies (Aphidinae, Eriosomatinae, Greenideinae and Hormaphidinae) have been reported to contain this tandem repeat region [4,22,23,24]. It was proposed that the repeat region in aphid mitogenomes either originated from the recent common ancestor of aphids followed by subsequent loss events throughout aphid diversification or independently evolved among different aphid lineages [5,6,25]. The former hypothesis has been increasingly accepted recently as more species from other subfamilies have been found to contain this region, and high sequence similarity of this region among species has been discovered [25]. However, there has been no data from a more ancient aphid lineage with repeat region to support this hypothesis, which will help to better understand the origin and evolution of repeat region in aphid mitogenomes.
The Lachninae represent an important and unusual aphid lineage with complicated host associations, encompassing distinct species feeding on both angiosperm and gymnosperm hosts [26]. This subfamily has been supposed to be one of the most ancient clades in the Aphididae according to several recent phylogenetic studies [27,28]. Up to now, however, no complete mitochondrial genome from Lachninae has been deposited in the GenBank (http://www.ncbi.nlm.nih.gov; accessed on October 19, 2020). Stomaphis sinisalicis is one representative species of the aphid genus Stomaphis, which represent a unique evolutionary lineage in the aphid subfamily Lachninae and are known for their large body size and particularly long rostrum (see the aphid in Figure 1) [29,30]. S. sinisalicis is mainly distributed in northern China with temperate climate, including Beijing, Hebei, Liaoning, Ningxia and Shandong. This species inhabits and feeds on the base of tree trunk or crevices of thick bark of Salix spp. and Populus sp., and generally establishes close mutualistic relationship with ants [29].
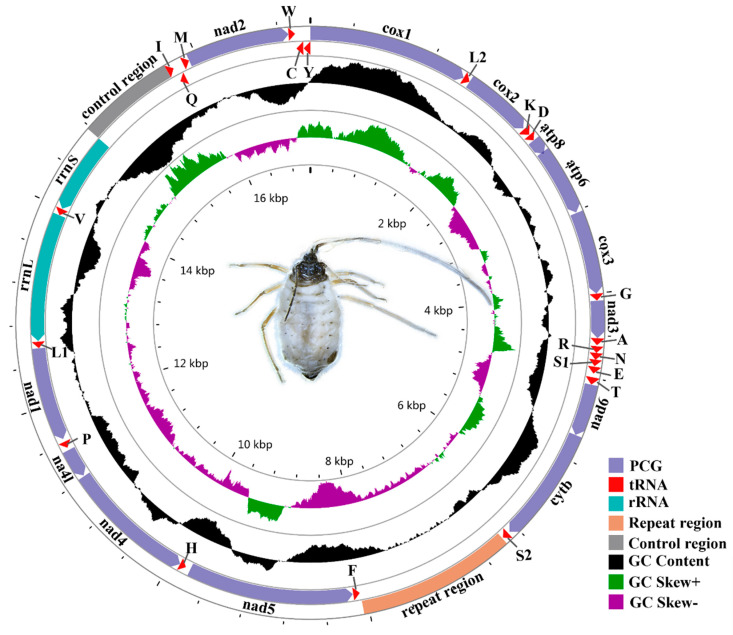
Figure 1.
Circular map of the complete mitogenome of Stomaphis sinisalicis. Different colors indicate different types of genes and regions. Genes shown at the outer circle are located on the J strand, and those at the inner circle are located at the N-strand.
In this study, we generated and analyzed the first complete mitochondrial genome for S. sinisalicis. The structural organization, nucleotide composition, codon usage and gene order arrangement were analyzed and compared against that of well characterized complete mitogenomes of other aphids to investigate the unique features and mitogenome evolution of aphids. This complete mitogenome reported here contributes to aphid higher phylogeny reconstruction based on mitogenome sequences and promotes future comparative mitogenome studies, which should help understand the mitogenome evolution and gene arrangement pattern across different aphid lineages.
2. Materials and Methods
2.1. Sample Preparation and DNA Extraction
The S. sinisalicis samples used in this study were collected on Salix babylonica from Beijing Olympic Forest Park, China (40°1′30.8″ N, 116°23′48.4″ E), in June 2018. All collected samples and voucher specimen (20180615-12) were preserved in 95% ethanol under −80 °C and deposited in the Insect Systematics and Diversity Lab at Fujian Agriculture and Forestry University, Fuzhou, China. Samples of S. sinisalicis were morphologically identified based on previous morphological descriptions of this species by the corresponding author.
2.2. Mitogenome Sequencing and Assembly
The next-generation sequencing was performed at the Biomarker Technologies Co., Ltd. (Beijing, China). For library construction, genomic DNA concentration was detected using ExKubit dsDNA HS test kit (ExCell Biotech Co., Ltd., Shanghai, China) with a Qubit fluorimeter (Invitrogen, Carlsbad, CA, USA). The qualified high-quality genomic DNA was used for next-generation sequencing subsequently. For library construction, TrueLib DNA Library Rapid Prep Kit for Illumina (ExCell Biotech Co., Ltd.) was used with 250 ng DNA as template, then sequencing was conducted using the NovaSeq 6000 System (Illumina, San Diego, CA, USA) (2 × 150 bp paired-end reads) with an insert size of 350 bp. Finally, approximately 10.4 Gb raw data was obtained and further filtered to remove adapters and low-quality reads, including reads with an N base content reached 10% of the read length and those with low quality bases (quality value ≤ 10) over 50% of the read length. Mitogenome was assembled using a total of 7.1 Gb clean data (clean reads: 23,773,262) by NovoPlasty v. 2.7.1 [31] with default parameters, using a COI sequence from S. sinisalicis as the seed sequence. The final genome assembly showed a high coverage and depth (Figure S1).
2.3. Annotation and Analysis of S. sinisalicis Mitogenome
The assembled mitogenome was first annotated using MITOS2 web server (http://mitos2.bioinf.uni-leipzig.de/index.py) [32] with the invertebrate mitochondrial genetic code. The boundaries of 13 PCGs and two rRNA genes were further determined by alignment with the known corresponding sequences of closely related aphid species. All tRNA genes in S. sinisalicis were identified using the MITOS2 web server and program ARWEN v. 1.2 [33], and their clover-leaf secondary structure were further drawn by RNAplot integrated in the ViennaRNA Package [34]. Tandem repeat region was detected by using the Tandem Repeats Finder Web server (http://tandem.bu.edu/trf/trf.html) [35]. The AT-rich control region was identified via boundaries of adjacent genes. Circular maps of the assembled mitogenome of S. sinisalicis was finally drawn with CGView Server [36]. MEGA7 software [37] was used to analyze the nucleotide composition and relative synonymous codon usage (RSCU). To better understand the base usage bias of the mitogenome, we also calculated the nucleotide skew values using the formulae AT-skew = (A − T)/(A + T) and GC-skew = (G − C)/(G + C) [38].
2.4. Phylogenetic Analysis
For phylogenetic analysis, all 13 PCG sequences from S. sinisalicis were collated to that of 24 available mitogenomes of representative species from different subfamilies of Aphididae and that of Adelges tsugae (Adelgidae) as ourgroup (Table 1). All PCG sequences (excluding the stop codons) of these 26 aphid species was aligned individually using codon-based multiple alignments by MAFFT v7.149 software [39] with default settings. After removing ambiguously aligned sites, alignments of 13 aligned PCGs were concatenated into a single alignment dataset using PhyloSuite v1.1.15 [40]. The best-fit substitution model was determined using the ModelFinder [41] integrated in IQ-TREE [42], and GTR+F+R4 was inferred as the optimal model according to the Bayesian Information Criterion (BIC) and was then used for subsequent phylogenetic analysis. Maximum-likelihood (ML) phylogenetic tree was reconstructed with the A. tsugae as the outgroup using IQ-TREE with 1000 ultrafast bootstrapping replicates. We also repeated the phylogenetic reconstruction with the dataset partitioned by each PCG, which yielded phylogenetic tree with identical topological relationships (Figure S2).
Table 1.
Information for representative complete mitochondrial genomes of aphids used in this study.
| Family | Subfamily | Species | Length (bp) | Accession Number | References |
|---|---|---|---|---|---|
| Adelgidae | Adelges tsugae | 16,056 | MT263947 | [43] | |
| Aphididae | Lachninae | Stomaphis sinisalicis | 17,109 | MW006542 | This study |
| Hormaphidinae | Hamamelistes spinosus | 15,089 | MT010853 | [44] | |
| Hormaphis betulae | 15,088 | NC_029495 | [45] | ||
| Pseudoregma bambucicola | 16,632 | NC_044640 | [24] | ||
| Mindarinae | Mindarus keteleerifoliae | 15,199 | NC_033410 | [20] | |
| Eriosomatinae | Eriosoma lanigerum | 15,640 | NC_033352 | [21] | |
| Paracolopha morrisoni | 16,330 | NC_045103 | [46] | ||
| Baizongia pistaciae | 15,602 | NC_035314 | [47] | ||
| Floraphis choui | 15,308 | NC_035310 | [48] | ||
| Floraphis meitanensis | 15,301 | NC_035316 | [48] | ||
| Kaburagia rhusicola ensigallis | 16,164 | MF043984 | [47] | ||
| Kaburagia rhusicola ovatirhusicola | 16,184 | MF043985 | [47] | ||
| Kaburagia rhusicola ovogallis | 16,164 | MF043986 | [47] | ||
| Kaburagia rhusicola rhusicola | 16,159 | MF043987 | [47] | ||
| Meitanaphis elongallis | 16,191 | NC_035315 | [48] | ||
| Meitanaphis flavogallis | 16,150 | NC_035312 | [48] | ||
| Meitanaphis microgallis | 16,191 | NC_047419 | [49] | ||
| Melaphis rhois | 15,436 | NC_036065 | [50] | ||
| Nurudea ibofushi | 16,054 | NC_035311 | [47] | ||
| Nurudea shiraii | 15,389 | NC_035301 | [47] | ||
| Nurudea yanoniella | 15,858 | NC_035313 | [48] | ||
| Schlechtendalia chinensis | 16,047 | NC_032386 | [51] | ||
| Schlechtendalia peitan | 15,609 | NC_035302 | [47] | ||
| Greenideinae | Cervaphis quercus | 15,272 | NC_024926 | [52] | |
| Greenidea ficicola | 17,361 | NC_048525 | [22] | ||
| Greenidea psidii | 16,202 | NC_041198 | [6] | ||
| Schoutedenia ralumensis | 16,051 | MT381994 | [53] | ||
| Calaphidinae | Appendiseta robiniae | 15,049 | NC_042165 | [23] | |
| Therioaphis trifolii | 16,068 | MK766411 | [54] | ||
| Aphidinae | Acyrthosiphon pisum | 16,971 | NC_011594 | NA | |
| Aphis aurantii | 15,469 | MN397939 | [55] | ||
| Aphis citricidus | 16,763 | NC_043903 | [5] | ||
| Aphis craccivora | 15,308 | NC_031387 | [56] | ||
| Aphis fabae mordvilkoi | 15,346 | NC_039988 | [23] | ||
| Aphis glycines | 17,954 | NC_045236 | [25] | ||
| Aphis gossypii | 15,869 | NC_024581 | [57] | ||
| Cavariella salicicola | 16,317 | NC_022682 | [4] | ||
| Diuraphis noxia | 15,784 | NC_022727 | [7] | ||
| Hyalopterus pruni | 15,410 | MT898422 | NA | ||
| Indomegoura indica | 15,220 | NC_045897 | [58] | ||
| Myzus persicae | 17,382 | NC_029727 | [23] | ||
| Rhopalosiphum nymphaeae | 15,594 | NC_046740 | [59] | ||
| Schizaphis graminum | 15,721 | NC_006158 | [14] | ||
| Sitobion avenae | 15,180 | NC_024683 | [60] |
Note: Only aphid species with complete mitogenomes and annotation information are listed; “NA” indicates sequences that are direct submission to GenBank.
2.5. Comparative Mitogenomic Analysis
The base composition, gene length, and genomic organization of all available aphid complete mitogenomes were collected and calculated for comparative genomic analysis. Furthermore, the online Interactive Tree of Life (iTOL) v 4 tool [61] was used for gene order visualization of known aphid mitogenomes, with the gene arrangement pattern mapping onto an inferred phylogenetic tree. We also detected the putative gene rearrangement events occurred in mitogenomes of each aphid species using the web-based program Common interval Rearrangement Explorer (CREx) [62] with arrangement of Drosophila yakuba as the inferred ancestral gene order [12,63]. And the genome rearrangement scenarios including transpositions, reverse transpositions, reversals, and tandem duplication random loss (TDRL) were reconstructed between D. yakuba and each aphid.
3. Results and Discussion
3.1. Mitogenome Features of S. sinisalicis and Other Aphids
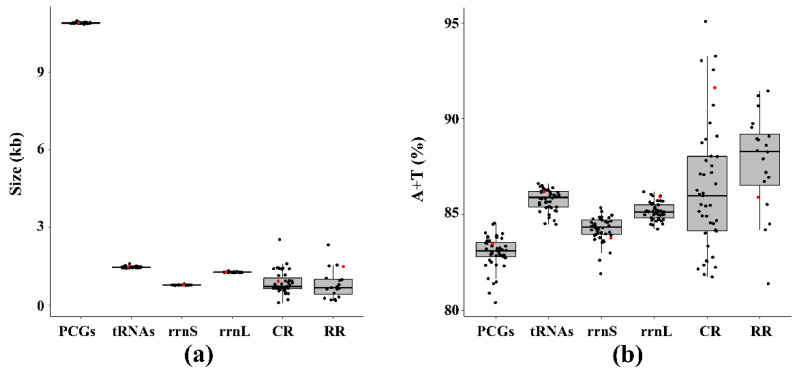
Known complete mitogenomes of aphids range from 15,049 (Appendiseta robiniae) to 17,954 bp (Aphis glycines) (Table 1). The mitochondrial genome of S. sinisalicis was 17,109 bp in length (the circular map is shown in Figure 1), longer than most other known aphid mitogenomes (Table 1). As in the case in other insects, the S. sinisalicis mitogenome contained the typical 13 PCGs, two rRNAs and 22 tRNAs, in which 14 tRNAs and 9 PCGs were transcribed on the majority strand (J-strand), while the others (eight tRNAs, two rRNAs and four PCGs) were located on the minority strand (N-strand) (Table 2). Besides, the S. sinisalicis mitogenome also contained a typical AT-rich control region locating between rrnS and trnI, and a large non-coding tandem repeat region. For aphid mitogenomes, the sizes of control region and repeat region vary greatly among different species, whereas the PCGs, tRNAs and two rRNAs show little variation in length (Figure 2a), suggesting that the mitogenome size of different aphids is largely determined by the size of non-coding regions.
Table 2.
Features of the mitochondrial genome of Stomaphis sinisalicis.
| Name | Strand | Location | Length (bp) | Start Codons | Stop Codons | Intergenic Sequence (bp) |
|---|---|---|---|---|---|---|
| cox1 | J | 1–1531 | 1531 | ATA | T | −1 |
| trnL2 (taa) | J | 1531–1599 | 67 | −1 | ||
| cox2 | J | 1599–2273 | 675 | ATC | TAA | 9 |
| trnK (ctt) | J | 2283–2354 | 72 | 3 | ||
| trnD (gtc) | J | 2358–2421 | 64 | 0 | ||
| atp8 | J | 2422–2580 | 159 | ATT | TAA | −20 |
| atp6 | J | 2561–3214 | 654 | ATT | 4 | |
| cox3 | J | 3219–4004 | 786 | ATG | TAA | −1 |
| trnG (tcc) | J | 4004–4070 | 67 | 0 | ||
| nad3 | J | 4071–4424 | 354 | ATT | TAA | −1 |
| trnA (tgc) | J | 4424–4490 | 67 | 8 | ||
| trnR (tcg) | J | 4499–4564 | 64 | −3 | ||
| trnN (gtt) | J | 4562–4629 | 66 | −2 | ||
| trnS1 (gct) | J | 4628–4687 | 60 | 5 | ||
| trnE (ttc) | J | 4693–4760 | 66 | 34 | ||
| trnT (tgt) | J | 4795–4863 | 69 | 0 | ||
| nad6 | J | 4864–5361 | 498 | ATT | −1 | |
| cytb | J | 5361–6473 | 1113 | ATG | TAA | 17 |
| trnS2 (tga) | J | 6491–6557 | 65 | 7 | ||
| repeat region | J | 6565–8053 | 1489 | 6 | ||
| trnF (gaa) | N | 8060–8125 | 66 | 3 | ||
| nad5 | N | 8129–9796 | 1668 | ATA | TAA | 50 |
| trnH (gtg) | N | 9847–9910 | 64 | 0 | ||
| nad4 | N | 9911–11,219 | 1309 | ATA | T | 8 |
| nad4l | N | 11,228–11,521 | 294 | ATG | TAA | 24 |
| trnP (tgg) | N | 11,546–11,614 | 67 | 28 | ||
| nad1 | N | 11,643–12,578 | 936 | ATT | TAA | 0 |
| trnL1 (tag) | N | 12,579–12,643 | 65 | 6 | ||
| rrnL | N | 12,650–13,927 | 1278 | 0 | ||
| trnV (tac) | N | 13,928–13,994 | 67 | 3 | ||
| rrnS | N | 13,998–14,774 | 777 | 0 | ||
| control region | J | 14,775–15,693 | 919 | 0 | ||
| trnI (gat) | J | 15,694–15,759 | 66 | −3 | ||
| trnQ (ttg) | N | 15,757–15,822 | 66 | 35 | ||
| trnM (cat) | J | 15,858–15,922 | 65 | 0 | ||
| nad2 | J | 15,923–16,903 | 981 | ATA | TAA | −1 |
| trnW (tca) | J | 16,903–16,977 | 60 | −9 | ||
| trnC (gca) | N | 16,969–17,039 | 69 | 2 | ||
| trnY (gta) | N | 17,042–17,108 | 67 | 1 |
Note: Intergenic sequences refer to intergenic regions between each gene and the gene after it.
Figure 2.
The size (a) and AT content (b) of PCGs, rrnL, rrnS, CR and RR of 44 aphid species with the red dots representing the Stomaphis sinisalicis. Lower edge of the gray rectangle, 25 percentile; central black bar within the rectangle, median; upper edge of the rectangle, 75 percentile.
Eleven gene overlaps with a total of 43 bp were present in S. sinisalicis mitogenome, ranging from 1 to 20 bp, with the longest overlap (20 bp) occurring between atp6 and atp8 (Table 2). The overlap between atp8-atp6 is also found in mitogenomes of other arthropods [7,64]. Moreover, there were also 19 intergenic spacers between genes, with the longest intergenic spacer (50 bp) existing between nad5 and trnH. Longer intergenic regions were also found in the junctions between trnE–trnT (34 bp) and trnQ–trnM (35 bp) (Table 2).
The S. sinisalicis mitogenome contained an obviously higher A+T content (84.8%) and a lower G+C content (15.2%). These values are comparable to that of other aphid species, which have an average A+T content of 84.0%, varying from 81.7% (A. robiniae) to 85.7% (Greenidea ficicola) (Table S1). This A+T bias in S. sinisalicis mitogenome was reflected in all components of its genome. The A+T content of control region and repeat region were 91.5% and 85.9%, and the PCGs, tRNAs and rRNAs had an average A+T content of 83.5%, 86.4% and 85.1%, respectively (Table 3). In aphids, the A+T content varies greatly for the repeat region and control region, but a small variation for PCGs, tRNAs and two rRNAs (Figure 2b), indicating that higher A+T contents in the control region and repeat region probably raise the total A+T content of aphid mitogenomes.
Table 3.
Nucleotide composition of the Stomaphis sinisalicis mitogenome.
| Regions | Size (bp) | T(U) | C | A | G | A+T (%) | G+C (%) | AT-Skew | GC-Skew |
|---|---|---|---|---|---|---|---|---|---|
| Whole genome | 17,109 | 40.5 | 9.9 | 44.3 | 5.3 | 84.8 | 15.2 | 0.044 | −0.305 |
| PCGs * | 10,923 | 48.3 | 8.8 | 35.2 | 7.7 | 83.5 | 16.5 | −0.156 | −0.067 |
| 1st codon | 3641 | 40.3 | 8.7 | 40.4 | 10.6 | 80.7 | 19.3 | 0.001 | 0.098 |
| 2nd codon | 3641 | 53.5 | 13.1 | 23.1 | 10.3 | 76.6 | 23.4 | −0.397 | −0.120 |
| 3rd codon | 3641 | 51.1 | 4.6 | 42.3 | 2.1 | 93.4 | 6.7 | −0.094 | −0.373 |
| tRNAs | 1478 | 41.5 | 5.7 | 44.9 | 7.9 | 86.4 | 13.6 | 0.038 | 0.164 |
| rRNAs | 2055 | 42.9 | 4.9 | 42.2 | 10.0 | 85.1 | 14.9 | −0.007 | 0.340 |
| Repeat region | 1489 | 41.5 | 9.7 | 44.4 | 4.3 | 85.9 | 14.0 | 0.034 | −0.385 |
| Control region | 919 | 46.8 | 4.6 | 44.7 | 3.5 | 91.5 | 8.1 | −0.023 | −0.135 |
* Stop codons are excluded from the statistics of protein-coding genes.
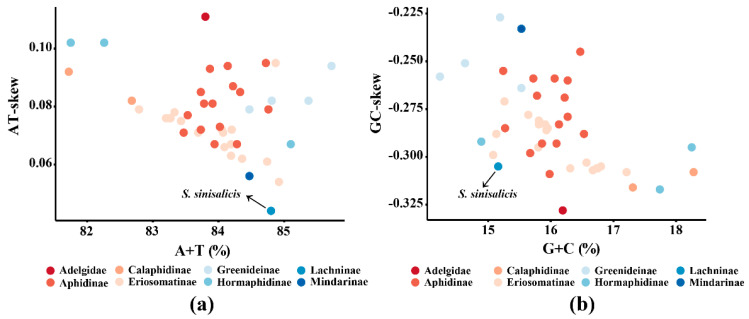
The nucleotide skew analysis showed that the whole S. sinisalicis mitogenome exhibit a positive AT-skew (0.044) and a negative GC-skew (−0.305). A similar pattern has been found in other aphid mitogenomes [4], with the AT-skew ranging from 0.054 in Paracolopha morrisoni to 0.102 in Hamamelistes spinosus and a GC-skew varying from −0.317 in Hormaphis betulae to −0.227 in Cervaphis quercus. These results indicate that S. sinisalicis mitogenome has a weakest A skew and a stronger C skew in compared to other known aphid mitogenomes (Figure 3a,b). This base composition bias has been suggested to be associated with replication and transcription of mitochondrial genome [65].
Figure 3.
Nucleotide composition of Stomaphis sinisalicis and 44 others complete aphid mitogenomes (including the Adelges tsugae). (a) The A+T content and AT skew. (b) The G+C content and GC-skew. See Table S1 for details of aphid species.
3.2. Gene Arrangement Patterns
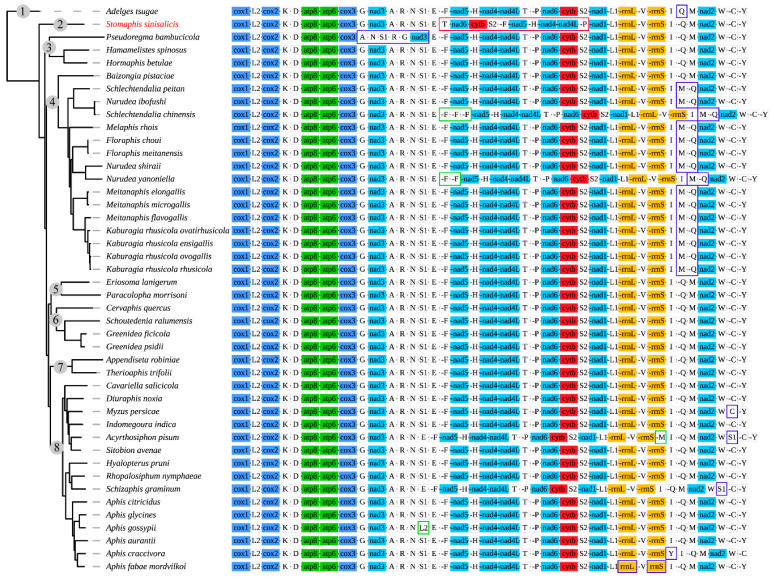
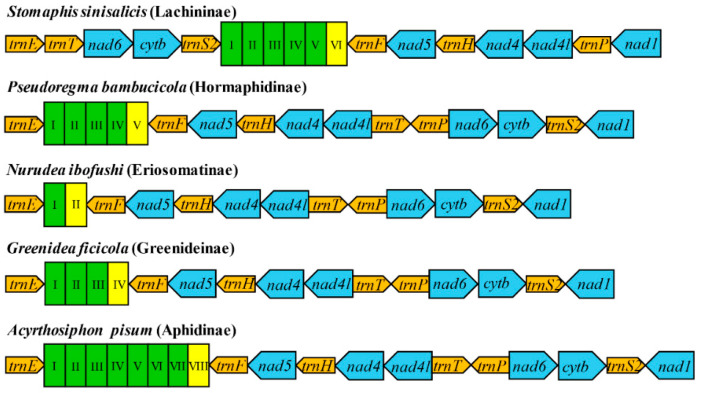
Gene arrangement of mitogenomes has been supposed to be quite conservative among insects and identical to that of the ancestral pancrustacean mitogenomes [1,12,66]. Various gene rearrangements have been observed in some insect taxa, including major rearrangement involving the PCGs and rRNAs, as well as minor rearrangement involving only tRNAs [1]. Mitochondrial genome rearrangement is rarely reported among aphids, and most of them have a gene order resembling the ancestral insect arrangement. However, in S. sinisalicis mitogenome, the gene order between trnE and nad1 was highly rearranged, with the order of gene cluster trnF-nad5-trnH-nad4-nad4l-trnT-trnP-nad6-cob-trnS2 shifting into the trnT-nad6-cob-trnS2-trnF-nad5-trnH-nad4-nad4l-trnP (Figure 4). This gene arrangement pattern differed greatly from that of other aphid species, which has been thought highly conserved within Hemipteran insects and similar to the ancestral gene order [4,15,63].
Figure 4.
Gene arrangement in mitochondrial genomes of aphids (including Adelges tsugae). Numbers labeled on clades of the left phylogenetic tree indicate different family, subfamily or tribe of aphids, which correspond to clades in the phylogenetic tree below, 1: Adelgidae; 2: Lachninae; 3: Hormaphidinae; 4: Fordini (Eriosomatinae); 5: Eriosomatini (Eriosomatinae); 6: Greenideinae; 7: Calaphidinae; 8: Aphidinae. Within the right gene arrangement image, the color blocks represent different categories of genes, dark blue: cytochrome oxidase complex; turquoise: NADH dehydrogenase complex; red: cytochrome b; ginger: ribosomal RNA genes; grey white: transfer RNA genes. The transfer RNA genes are indicated by the corresponding capital letters. Putative gene rearrangement events identified by CREx analysis (with the Drosophila yakuba as referenced ancestral gene arrangement) are marked by different color boxes. The predicted tandem-duplication-random-loss events (TDRL) occurred in Stomaphis sinisalicis are indicated with red box; the other gene rearrangement events (including the transposition, inversion, inverse transposition) are indicated by blue box; gene duplications are labeled with green boxes. Non-coding region was excluded from the analysis.
Rearrangement of mitogenomes can generally be classified into several gene movements: transposition, inversion, and inverse transposition [66]. Several models have been proposed to explain the rearrangement of mitogenome genes, such as the tandem duplication random loss (TDRL) model [67,68], tandem duplication/non-random loss (TDNL) model [69], recombination model [70] and tRNA mispriming model [71]. According to the results of CREx analysis, the rearrangement event occurred in S. sinisalicis mitogenome can be better explained by the widely accepted tandem duplication random loss (TDRL) model [67,68]. Based on this model, the gene cluster locating trnE and nad1 in S. sinisalicis mitogenome can be separated into four gene blocks: trnF-nad5-trnH-nad4-nad4l, trnT, trnP and nad6-cob-trnS2, and the present gene arrangement pattern may be derived from tandem duplications and random losses of these gene blocks. The gene order observed in S. sinisalicis mitogenome represents a new arrangement pattern that has never been discovered in any other aphid species, which may have important implications for understanding the evolution of aphid mitogenomes. It’s unclear whether this rearrangement is a mitochondrial signature for species of the subfamily Lachninae or an independent evolutionary event only for S. sinisalicis. A broader sampling of this subfamily will be needed to further investigate whether this new rearrangement pattern also occurs in other species.
Most aphids of other subfamilies possess a conserved gene order identical to that of ancestral insects. There are some exceptions with mostly involving the duplications, transpositions, reverse transpositions or reversals of some tRNA genes, such as several species from Fordini (Eriosomatinae) and Aphidinae (Figure 4). Notably, all species within the Fordini exhibit a transposition of trnQ and trnM except the Baizongia pistaciae, a basal species within Fordini. Whereas gene order of two Eriosomatini (Eriosomatinae) species is arranged in the same order as the ancestral insect. These indicate that the most recent common ancestor of Fordini aphids may have a typical arrangement of trnQ-trnM, which may be rearranged through an independent evolutionary event that occurred during subsequent species diversification of this tribe. In addition to the cases involving tRNAs, two rRNA gene reversals are found in Aphis fabae mordvilkoi from Aphidinae. Besides, a more prominent rearrangement including five tRNAs and one PCG is observed in Pseudoregma bambucicola (Hormaphidinae), with transpositions occurred within the gene cluster trnG-nad3-trnA-trnR-trnN-trnS1 resulting in a novel arrangement pattern of trnA-trnN-trnS1-trnR-trnG-nad3, while the other two Hormaphidinae species have an ancestral gene order as most other aphids, suggesting that gene rearrangement in P. bambucicola seems to occur independently.
3.3. Protein Coding Genes and Codon Usage Patterns
The overall size (excluding stop codons) of 13 PCGs of S. sinisalicis was 10,923 bp, accounting for 63.8% of the total genome (Table 3). All PCGs showed a high A+T bias, ranging from 76.3% (cox1) to 91.2% (atp8). The AT-skew (−0.156) and GC-skew (−0.067) were both negative for the whole PCGs, reflecting a bias towards nucleotides T and C than their counterparts. The third codon position presented a much higher A+T content than that of the first and second codon positions, and tended to use T and C bases (Table 3). All protein-coding genes were initiated with the typical ATN codons, and terminated with TAA, except the cox1 and nad4 terminating with a truncated termination codon T, which is common in other metazoan mitogenomes [4,72]. These incomplete stop codons have been supposed to be completed through post-transcriptional polyadenylation [73].
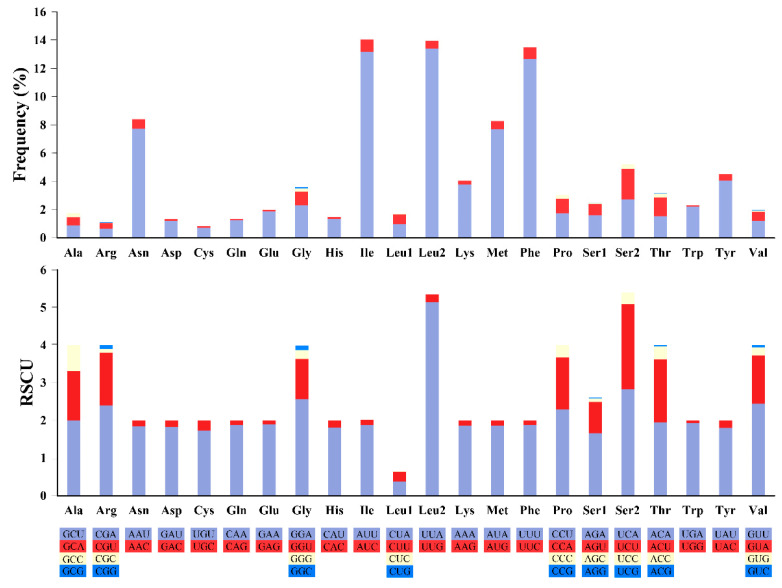
The codon usage analysis showed that the most frequently used codons were UUA-Leu2 (13.4%), AUU-Ile (13.2%) and UUU-Phe (12.7%), while the codons CUG-Leu1, UCG-Ser2, CCG-Pro and GCG-Ala were not used in the mitogenome (Figure 5, Table S2). The UUA (Leu2) also had the highest RSCU values, further indicating that UUA was the most preferred codon. The RSCU values of the PCGs revealed that there was a higher frequency in the usage of AT than that of GC in the third codon positions (Table S2).
Figure 5.
Codon usage in the mitochondrial genome of Stomaphis sinisalicis. The bar chart above indicates codon usage frequency of amino acids used for construction of 13 protein coding genes; and the bar chart below represents the relative synonymous codon usage (RSCU) of mitochondrial protein-coding genes. Codon families are labeled below the X-axis.
3.4. Transfer and Ribosomal RNA Genes
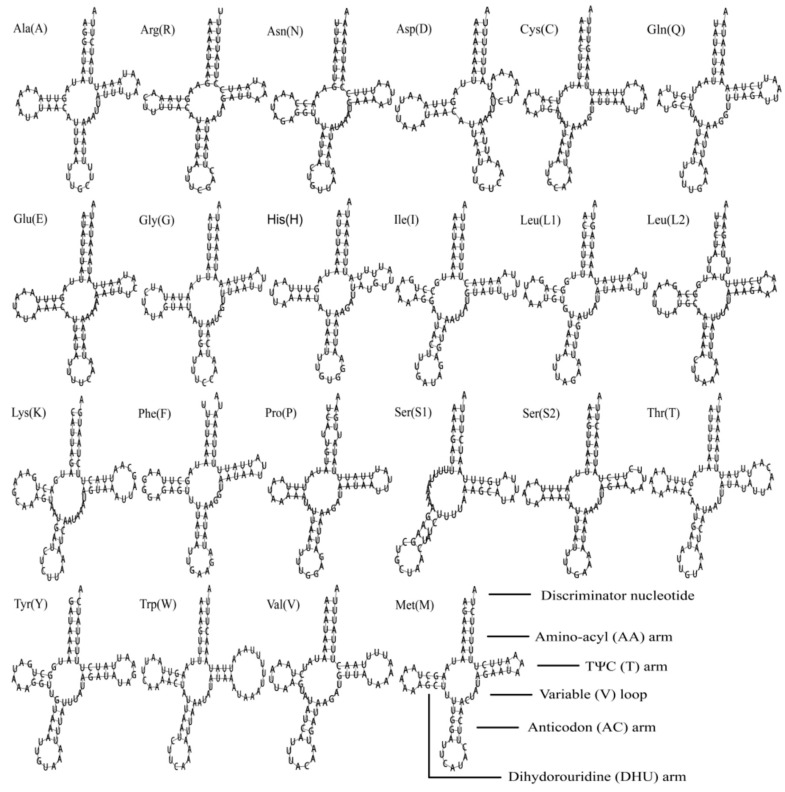
The complete set of 22 typical tRNAs were all found in S. sinisalicis mitogenome, their secondary structures were shown in the Figure 6. The total length of tRNAs was 1478 bp, ranging from 60 bp to 75 bp in size. Both the AT-skew value (0.025) and GC-skew (0.170) were positively biased in the tRNAs, which showed a slight bias toward using A and an obvious bias toward G (Table 3). Most tRNAs exhibited the typical clover-leaf structures with two exceptions: trnS1 missing the DHU arm and trnD without the TΨC stem. Lacking of DHU arm in trnS1 is a common feature for most metazoan mitogenomes [1]. This aberrant tRNAs are supposed to sustain their function via a posttranscriptional RNA editing mechanism [74,75].
Figure 6.
Secondary structures of the transfer RNA genes (tRNAs) in Stomaphis sinisalicis mitogenome.
The rrnL (1278 bp) and rrnS (777 bp) genes were located between trnL1 and trnV, trnV and the control region, respectively. There was a little variation in the size of both rRNAs among different aphid species (Figure 2a). The overall rRNAs of S. sinisalicis mitogenome showed a negative AT-skew (−0.007) and a positive GC-skew (0.340), indicating a preference for using T base over A, and G over C (Table 3). The overall length and A+T content were rather conservative and identical to that of other aphid species.
3.5. Non-Protein Coding Regions
The AT-rich control region of S. sinisalicis mitogenome located between rrnS and trnI, spanning 919 bp in length. This is comparable to that of most other aphids, which have an average length of 867 bp (Table S1). Generally, the length of control region varies greatly with the size of embedded tandem repeats, which was not detected in the S. sinisalicis mitogenome. The control region is supposed to be involved in the initiation of replication and transcription of mitogenomes [76]. The control region in S. sinisalicis mitogenome had a higher A+T content (95.07%) compared to most aphid mitogenomes, which is varying from 81.75% (Kaburagia rhusicola rhusicola) to 95.07% (P. bambucicola). Both the AT-skew (−0.023) and GC-skew (−0.135) were negative, showing a bias towards using T and C (Table 3).
In addition to the control region, a large non-coding repeat region (1489 bp) comprising five tandem repeat units ranging from 262 bp to 264 bp and a 171 bp partial repeat unit was also present in the S. sinisalicis mitogenome. Such special tandem repeats have been found in some aphid species from different subfamilies, and vary widely in size and number of repeat units, leading to varied lengths of repeat regions, ranging from 452 bp in Nurudea yanoniella to 2322 bp in A. glycines. However, in our analysis, obvious tandem repeats were not found in some aphid mitogenomes previously reported with repeat regions (A. robiniae, A. fabae mordvilkoi, Myzus persicae, Rhopalosiphum nymphaeae and Sitobion avenae; Table S1), indicating the validity of the annotation of repeat regions in these species need to be further verified. Figure 7 presents the repeat regions of S. sinisalicis and other representative species from four aphid subfamilies. Typically, the unique non-coding tandem repeat region found in known aphid mitogenomes from other subfamilies (Hormaphidinae, Eriosomatinae, Greenideinae and Aphidinae) is located between trnE and trnF [4,22,23,24], which has been thought an alternative origin of the mitogenome replication as in other insects [77]. However, the repeat region in S. sinisalicis mitogenome was found located between trnS2 and trnF. According to the gene rearrangement analysis above, the repeat region along with the gene cluster trnF-nad5-trnH-nad4-nad4l may constitute a gene block, and go through the TRDL event together. The function of this unique tandem repeat region is currently unknown. However, the existence of repeat region in mitogenome of the basal species S. sinisalicis but not in A. tsugae may suggest that it is probably an ancient feature acquired by the common ancestor of Aphididae, which along with the scatter distribution within different subfamilies further favors the hypothesis about a single origin and subsequent diversification of the repeat region in aphid mitogenomes [5,6,25].
Figure 7.
Repeat regions of the mitochondrial genome from Stomaphis sinisalicis and selected representative species from other aphid subfamilies. Green bars indicate tandem repeats, and the yellow bars refer to partial repeat unit; PCGs and tRNAs are indicated by the turquoise and orange squares, respectively.
3.6. Phylogenetic Analysis
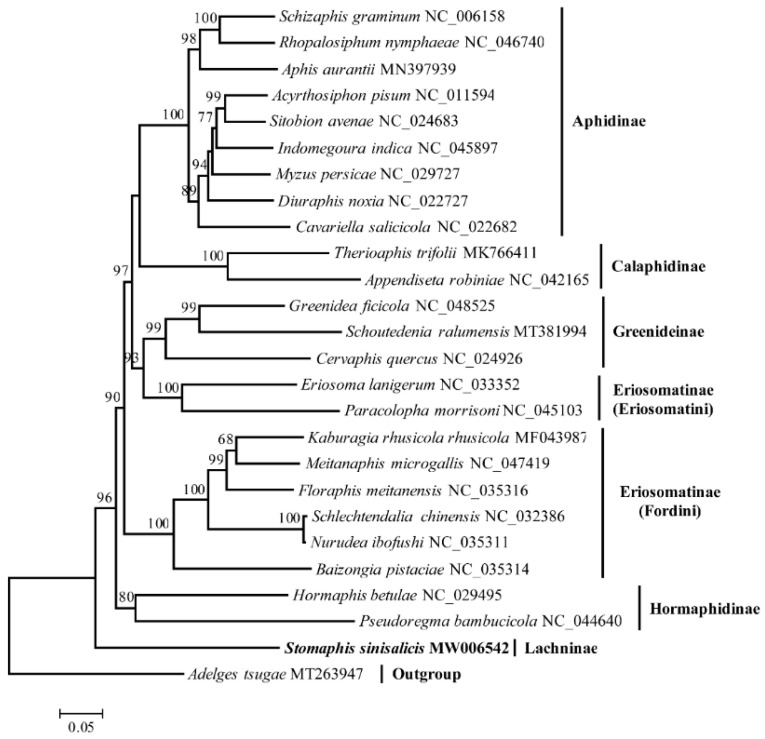
The phylogenetic analysis using mitogenome data supported the monophyly of most subfamilies with high support values except the Eriosomatinae, which was found to be paraphyletic consistent with previous phylogenetic studies, with the Eriosoamtini clade clustering with the Greenideinae (Figure 8) [22,78]. The S. sinisalicis formed a separate clade locating at the basal position of Aphididae, which further confirms the previous proposed phylogenetic hypothesis [28]. This primitive taxonomic status makes this mitogenome an important data resource for future comparative studies to investigate the mitogenome evolution across different aphid lineages.
Figure 8.
The maximum-likelihood phylogenetic tree based on 13 protein-coding genes of Stomaphis sinisalicis and 24 other aphids with the Adelges tsugae as outgroup. Bootstrap support values >50% are indicated above the branches.
4. Conclusions
In this study, we reported the S. sinisalicis mitogenome, the first complete mitogenome from the subfamily Lachninae. The gene content, base composition and overall organization of mitogenome was analyzed in detail and compared to other aphid species. This mitogenome is similar to that of ancestral insect mitogenomes in terms of the gene content, base composition and overall organization; however, the gene order between the trnE and nad1 is highly rearranged and differs greatly with other aphids. This gene rearrangement involving tRNAs, PCGs and the repeat region occurred in S. sinisalicis mitogenome has probably evolved from the ancestral gene order due to the tandem duplication and random loss events. Comparative analysis also indicates that the presence of repeat region may be an ancestral feature of aphid mitogenomes. Besides, phylogenetic analysis supports the basal position of Lachninae within Aphididae. Overall, our study provides valuable information for investigating the molecular evolution, mitogenome arrangement patterns and phylogenetic relationships across different aphid lineages.
Acknowledgments
We would like to thank Dan Wang for help in sample collection and Zhentao Cheng for help in illustration.
Supplementary Materials
The following are available online at https://www.mdpi.com/2075-4450/12/1/55/s1, Table S1: Information of aphid species with their representative complete mitochondrial genome sequences. Table S2: Codon usage of 13 PCGs in the mitogenome of Stomaphis sinisalicis.
Author Contributions
Conceptualization, X.H. and H.Z.; methodology, H.Z., Q.L., C.L.; investigation, Q.L.; data curation, formal analysis and visualization, H.Z., Q.L., C.L. and J.D.; writing—original draft preparation, H.Z.; writing—review and editing, X.H.; project administration and funding acquisition, X.H. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by National Natural Science Foundation of China (Grant number: 31970446) and National Key R&D Program of China (Grant number: 2016YFE0203100).
Data Availability Statement
The complete mitochondrial genome of S. sinisalicis is available in the GenBank under accession number MW006542. The raw sequencing data are deposited in the NCBI Sequence Read Archive (SRA) database with accession number PRJNA668672.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cameron S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014;59:95–117. doi: 10.1146/annurev-ento-011613-162007. [DOI] [PubMed] [Google Scholar]
- 2.Boore J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boore J.L., Collins T.M., Stanton D., Daehler L.L., Brown W.M. Deducing the pattern of arthropod phylogeny from mitochondrial DNA rearrangements. Nature. 1995;376:163–165. doi: 10.1038/376163a0. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y., Huang X.-L., Qiao G.-X. Comparative analysis of mitochondrial genomes of five aphid species (Hemiptera: Aphididae) and phylogenetic implications. PLoS ONE. 2013;8:e77511. doi: 10.1371/journal.pone.0077511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei D.D., Lang N., Tao Y., He W., Wang J.J. The mitochondrial genome of the brown citrus aphid Aphis (Toxoptera) citricidus: Insights into the repeat regions in aphids and phylogenetic implications. Int. J. Biol. Macromol. 2019;136:531–539. doi: 10.1016/j.ijbiomac.2019.06.101. [DOI] [PubMed] [Google Scholar]
- 6.Chen J., Wang Y., Qin M., Jiang L.-Y., Qiao G.-X. The mitochondrial genome of Greenidea psidii van der Goot (Hemiptera: Aphididae: Greenideinae) and comparisons with other Aphididae aphids. Int. J. Biol. Macromol. 2019;122:824–832. doi: 10.1016/j.ijbiomac.2018.10.209. [DOI] [PubMed] [Google Scholar]
- 7.Zhang B., Ma C., Edwards O., Fuller S., Kang L. The mitochondrial genome of the Russian wheat aphid Diuraphis noxia: Large repetitive sequences between trnE and trnF in aphids. Gene. 2014;533:253–260. doi: 10.1016/j.gene.2013.09.064. [DOI] [PubMed] [Google Scholar]
- 8.Chen L., Chen P.-Y., Xue X.-F., Hua H.-Q., Li Y.-X., Zhang F., Wei S.-J. Extensive gene rearrangements in the mitochondrial genomes of two egg parasitoids, Trichogramma japonicum and Trichogramma ostriniae (Hymenoptera: Chalcidoidea: Trichogrammatidae) Sci. Rep. 2018;8:7034. doi: 10.1038/s41598-018-25338-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshizawa K., Johnsonb K.P., Sweet A.D., Yao I., Ferreira R.L., Cameron S.L. Mitochondrial phylogenomics and genome rearrangements in the barklice (Insecta: Psocodea) Mol. Phylogenet. Evol. 2018;119:118–127. doi: 10.1016/j.ympev.2017.10.014. [DOI] [PubMed] [Google Scholar]
- 10.Shao R., Barker S.C. The highly rearranged mitochondrial genome of the plague thrips, Thrips imaginis (Insecta : Thysanoptera): Convergence of two novel gene boundaries and an extraordinary arrangement of rRNA genes. Mol. Biol. Evol. 2003;20:362–370. doi: 10.1093/molbev/msg045. [DOI] [PubMed] [Google Scholar]
- 11.Shao R., Campbell N.J.H., Barker S.C. Numerous gene rearrangements in the mitochondrial genome of the wallaby louse, Heterodoxus macropus (Phthiraptera) Mol. Biol. Evol. 2001;18:858–865. doi: 10.1093/oxfordjournals.molbev.a003867. [DOI] [PubMed] [Google Scholar]
- 12.Boore J.L., Lavrov D.V., Brown W.M. Gene translocation links insects and crustaceans. Nature. 1998;392:667–668. doi: 10.1038/33577. [DOI] [PubMed] [Google Scholar]
- 13.Shao R., Campbell N.J.H., Schmidt E.R., Barker S.C. Increased rate of gene rearrangement in the mitochondrial genomes of three orders of hemipteroid insects. Mol. Biol. Evol. 2001;18:1828–1832. doi: 10.1093/oxfordjournals.molbev.a003970. [DOI] [PubMed] [Google Scholar]
- 14.Thao M.L., Baumann L., Baumann P. Organization of the mitochondrial genomes of whiteflies, aphids, and psyllids (Hemiptera, Sternorrhyncha) BMC Evol. Biol. 2004;4:25. doi: 10.1186/1471-2148-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y., Chen J., Jiang L.-Y., Qiao G.-X. Hemipteran mitochondrial genomes: Features, structures and implications for phylogeny. Int. J. Mol. Sci. 2015;16:12382–12404. doi: 10.3390/ijms160612382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng J., Lu C., Huang X. The first mitochondrial genome of scale insects (Hemiptera: Coccoidea) Mitochondrial DNA B. 2019;4:2094–2095. doi: 10.1080/23802359.2019.1622464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu H.-L., Chen Q.-D., Chen S., Pu D.-Q., Chen Z.-T., Liu Y.-Y., Liu X. The highly rearranged mitochondrial genomes of three economically important scale insects and the mitochondrial phylogeny of Coccoidea (Hemiptera: Sternorrhyncha) PeerJ. 2020;8:e9932. doi: 10.7717/peerj.9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu C., Huang X., Deng J. The challenge of Coccidae (Hemiptera: Coccoidea) mitochondrial genomes: The case of Saissetia coffeae with novel truncated tRNAs and gene rearrangements. Int. J. Biol. Macromol. 2020;158:854–864. doi: 10.1016/j.ijbiomac.2020.04.257. [DOI] [PubMed] [Google Scholar]
- 19.Favret C. Aphid Species File. Version 5.0/5.0. [(accessed on 7 November 2020)]; Available online: http://Aphid.SpeciesFile.org.
- 20.Wang Y., Chen J., Jiang L.-Y., Qiao G.-X. The complete mitochondrial genome of Mindarus keteleerifoliae (Insecta: Hemiptera: Aphididae) and comparison with other Aphididae insects. Int. J. Mol. Sci. 2015;16:30091–30102. doi: 10.3390/ijms161226219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y., Jiang L., Liu Y., Chen J., Qiao G. General methods to obtain and analyze the complete mitochondrial genome of aphid species: Eriosoma lanigerum (Hemiptera: Aphididae) as an example. Zool. Syst. 2016;41:123–132. doi: 10.11865/zs.201610. [DOI] [Google Scholar]
- 22.Liu Q., Zhang H., Deng J., Lin X., Huang X. The complete mitochondrial genome of Greenidea ficicola (Hemiptera: Aphididae: Greenideinae), a pest of Ficus. Mitochondrial DNA B. 2020;5:254–256. doi: 10.1080/23802359.2019.1699472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voronova N.V., Levykina S., Warner D., Shulinski R., Bandarenka Y., Zhorov D. Characteristic and variability of five complete aphid mitochondrial genomes: Aphis fabae mordvilkoi, Aphis craccivora, Myzus persicae, Therioaphis tenera and Appendiseta robiniae (Hemiptera; Sternorrhyncha; Aphididae) Int. J. Biol. Macromol. 2020;149:187–206. doi: 10.1016/j.ijbiomac.2019.12.276. [DOI] [PubMed] [Google Scholar]
- 24.Zhang H., Deng J., Liu Q., Huang X. The mitochondrial genome of a social aphid, Pseudoregma bambucicola (Hemiptera: Aphididae: Hormaphidinae) Mitochondrial DNA B. 2019;4:2100–2101. doi: 10.1080/23802359.2019.1622470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song H., Donthu R.K., Hall R., Hon L., Weber E., Badger J.H., Giordano R. Description of soybean aphid (Aphis glycines Matsumura) mitochondrial genome and comparative mitogenomics of Aphididae (Hemiptera: Sternorrhyncha) Insect Biochem. Mol. Biol. 2019;113:103208. doi: 10.1016/j.ibmb.2019.103208. [DOI] [PubMed] [Google Scholar]
- 26.Chen R., Favret C., Jiang L., Wang Z., Qiao G. An aphid lineage maintains a bark-feeding niche while switching to and diversifying on conifers. Cladistics. 2016;32:555–572. doi: 10.1111/cla.12141. [DOI] [PubMed] [Google Scholar]
- 27.Ortiz-Rivas B., Moya A., Martínez-Torres D. Molecular systematics of aphids (Homoptera: Aphididae): New insights from the long-wavelength opsin gene. Mol. Phylogenet. Evol. 2004;30:24–37. doi: 10.1016/S1055-7903(03)00155-6. [DOI] [PubMed] [Google Scholar]
- 28.Ortiz-Rivas B., Martínez-Torres D. Combination of molecular data support the existence of three main lineages in the phylogeny of aphids (Hemiptera: Aphididae) and the basal position of the subfamily Lachninae. Mol. Phylogenet. Evol. 2010;55:305–317. doi: 10.1016/j.ympev.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 29.Qiao G., Zhang G. A revision of Stomaphis Walker from China with descriptions of three new species (Homoptera: Lachnidae) Insect Sci. 1999;6:289–298. doi: 10.1111/j.1744-7917.1999.tb00125.x. [DOI] [Google Scholar]
- 30.Depa Ł., Mróz E. Central European Acer- and salicaceae-feeding aphids of the genus Stomaphis (Insecta: Aphidoidea: Lachnidae) —Separate species or populations? Zool. Sci. 2013;30:509–518. doi: 10.2108/zsj.30.509. [DOI] [PubMed] [Google Scholar]
- 31.Dierckxsens N., Mardulyn P., Smits G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2016;45:e18. doi: 10.1093/nar/gkw955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernt M., Donath A., Jühling F., Externbrink F., Florentz C., Fritzsch G., Pütz J., Middendorf M., Stadler P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013;69:313–319. doi: 10.1016/j.ympev.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 33.Laslett D., Canbäck B. ARWEN: A program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 2008;24:172–175. doi: 10.1093/bioinformatics/btm573. [DOI] [PubMed] [Google Scholar]
- 34.Lorenz R., Bernhart S.H., zu Siederdissen C.H., Tafer H., Flamm C., Stadler P.F., Hofacker I.L. ViennaRNA Package 2.0. Algorithms. Mol. Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benson G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999;37:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grant J.R., Stothard P. The CGView server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36:W181–W184. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar S., Stecher G., Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perna N.T., Kocher T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995;41:353–358. doi: 10.1007/BF01215182. [DOI] [PubMed] [Google Scholar]
- 39.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang D., Gao F., Jakovlić I., Zou H., Zhang J., Li W.X., Wang G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020;20:348–355. doi: 10.1111/1755-0998.13096. [DOI] [PubMed] [Google Scholar]
- 41.Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., Jermiin L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen L.-T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2014;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yeh H.-T., Ko C.-C., Wu L.-W. The first complete mitochondrial genome of Adelges tsugae Annand (Hemiptera: Adelgidae) Mitochondrial DNA B. 2020;5:2288–2290. doi: 10.1080/23802359.2020.1772682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu Z.-H., Liang Y.-K., Wen J., Ren Z.-M. Complete mitochondrial genome of the witch-hazel leaf gall aphid Hamamelistes spinosus (Hemiptera: Aphididae: Hormaphidinae) Mitochondrial DNA B. 2020;5:1388–1389. doi: 10.1080/23802359.2020.1735965. [DOI] [Google Scholar]
- 45.Nong X., Liu Y., Wang L., Zhong S., Yu X., Xie Y. Mitochondrial genome of Hormaphis betulae and its comparative analysis with Pseudoregma bambucicola (Hemiptera: Hormaphidinae) Mitochondrial DNA B. 2020;5:906–907. doi: 10.1080/23802359.2020.1715878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J., Park J., Lee H., Park J., Lee W. The complete mitochondrial genome of Paracolopha morrisoni (Baker, 1919) (Hemiptera: Aphididae) Mitochondrial DNA B. 2019;4:3037–3039. doi: 10.1080/23802359.2019.1666046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ren Z., Harris A.J., Dikow R.B., Ma E., Zhong Y., Wen J. Another look at the phylogenetic relationships and intercontinental biogeography of eastern Asian - North American Rhus gall aphids (Hemiptera: Aphididae: Eriosomatinae): Evidence from mitogenome sequences via genome skimming. Mol. Phylogenet. Evol. 2017;117:102–110. doi: 10.1016/j.ympev.2017.05.017. [DOI] [PubMed] [Google Scholar]
- 48.Ren Z., von Dohlen C.D., Harris A.J., Dikow R.B., Su X., Wen J. Congruent phylogenetic relationships of Melaphidina aphids (Aphididae: Eriosomatinae: Fordini) according to nuclear and mitochondrial DNA data with taxonomic implications on generic limits. PLoS ONE. 2019;14:e0213181. doi: 10.1371/journal.pone.0213181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liang Y.-K., Wen J., Ren Z.-M. Complete mitochondrial genome of Rhus gall aphid Meitanaphis microgallis (Hemiptera: Aphididae: Eriosomatinae) Mitochondrial DNA B. 2019;4:2363–2364. doi: 10.1080/23802359.2019.1629846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ren Z.-M., Wen J. Complete mitochondrial genome of the North American Rhus gall aphid Melaphis rhois (Hemiptera: Aphididae: Eriosomatinae) Mitochondrial DNA B. 2017;2:169–170. doi: 10.1080/23802359.2017.1303345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ren Z.-M., Bai X., Harris A.J., Wen J. Complete mitochondrial genome of the Rhus gall aphid Schlechtendalia chinensis (Hemiptera: Aphididae: Eriosomatinae) Mitochondrial DNA B. 2016;1:849–850. doi: 10.1080/23802359.2016.1241678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y., Huang X.-L., Qiao G.-X. The complete mitochondrial genome of Cervaphis quercus (Insecta: Hemiptera: Aphididae: Greenideinae) Insect Sci. 2014;21:278–290. doi: 10.1111/1744-7917.12112. [DOI] [PubMed] [Google Scholar]
- 53.Jing C., Jiang L., Zhang X., Qiao G. The complete mitochondrial genome of Schoutedenia ralumensis Rübsaamen, 1905 (Hemiptera: Aphididae: Greenideinae) Mitochondrial DNA B. 2020;5:2217–2218. doi: 10.1080/23802359.2020.1768938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu X., Wei S., He J., Song F., Cai W. Complete mitochondrial genome of the spotted alfalfa aphid, Therioaphis trifolii (Hemipera: Aphididae) Mitochondrial DNA B. 2019;4:3260–3261. doi: 10.1080/23802359.2019.1644232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pu D., Liu C., Liu H., Chen Z.-T., Wu X., Xiao K., Luo X., Mao J., Huang Q. Complete mitochondrial genome of Sichuan’s population of Aphis aurantii (Hemiptera: Aphididae) Mitochondrial DNA B. 2020;5:2119–2120. doi: 10.1080/23802359.2020.1715303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song N., Zhang H., Li H., Cai W. All 37 mitochondrial genes of aphid Aphis craccivora obtained from transcriptome sequencing: Implications for the evolution of aphids. PLoS ONE. 2016;11:e0157857. doi: 10.1371/journal.pone.0157857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang S., Luo J., Wang C., Lv L., Li C., Jiang W., Cui J., Rajput L.B. Complete mitochondrial genome of Aphis gossypii Glover (Hemiptera: Aphididae) Mitochondrial DNA A. 2016;27:854–855. doi: 10.3109/19401736.2014.919474. [DOI] [PubMed] [Google Scholar]
- 58.Hong B., Zhang F., Hu Z., Zhao H. The complete mitochondrial genome of Indomegoura indica (Hemiptera: Aphididae) Mitochondrial DNA B. 2019;4:882–883. doi: 10.1080/23802359.2019.1574674. [DOI] [Google Scholar]
- 59.Park J., Kim Y., Xi H., Park J., Lee W. The complete mitochondrial genome of Rhopalosiphum nymphaeae (Linnaeus, 1761) (Hemiptera: Aphididae) Mitochondrial DNA B. 2020;5:1613–1615. doi: 10.1080/23802359.2020.1745711. [DOI] [Google Scholar]
- 60.Zhang B., Zheng J., Liang L., Fuller S., Ma C.-S. The complete mitochondrial genome of Sitobion avenae (Hemiptera: Aphididae) Mitochondrial DNA A. 2016;27:945–946. doi: 10.3109/19401736.2014.926498. [DOI] [PubMed] [Google Scholar]
- 61.Letunic I., Bork P. Interactive tree of life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bernt M., Merkle D., Ramsch K., Fritzsch G., Perseke M., Bernhard D., Schlegel M., Stadler P.F., Middendorf M. CREx: Inferring genomic rearrangements based on common intervals. Bioinformatics. 2007;23:2957–2958. doi: 10.1093/bioinformatics/btm468. [DOI] [PubMed] [Google Scholar]
- 63.Clary D.O., Wolstenholme D.R. The mitochondrial DNA molecule of Drosophila yakuba: Nucleotide sequence, gene organization, and genetic code. J. Mol. Evol. 1985;22:252–271. doi: 10.1007/BF02099755. [DOI] [PubMed] [Google Scholar]
- 64.Lavrov D.V., Boore J.L., Brown W.M. The complete mitochondrial DNA sequence of the horseshoe crab Limulus polyphemus. Mol. Biol. Evol. 2000;17:813–824. doi: 10.1093/oxfordjournals.molbev.a026360. [DOI] [PubMed] [Google Scholar]
- 65.Wei S.-J., Shi M., Chen X.-X., Sharkey M.J., van Achterberg C., Ye G.-Y., He J.-H. New views on strand asymmetry in insect mitochondrial genomes. PLoS ONE. 2010;5:e12708. doi: 10.1371/journal.pone.0012708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dowton M., Castro L.R., Austin A.D. Mitochondrial gene rearrangements as phylogenetic characters in the invertebrates: The examination of genome ‘morphology’. Invertebr. Syst. 2002;16:345–356. doi: 10.1071/IS02003. [DOI] [Google Scholar]
- 67.Boore J.L. Comparative Genomics. Springer; Dordrecht, The Netherlands: 2000. The duplication/random loss model for gene rearrangement exemplified by mitochondrial genomes of deuterostome animals; pp. 133–147. [DOI] [Google Scholar]
- 68.Chaudhuri K., Chen K., Mihaescu R., Rao S. On the tandem duplication-random loss model of genome rearrangement. Proc. Seventeenth Annu. ACM-SIAM Symp. 2006:564–570. doi: 10.1145/1109557.1109619. [DOI] [Google Scholar]
- 69.Lavrov D.V., Boore J.L., Brown W.M. Complete mtDNA sequences of two millipedes suggest a new model for mitochondrial gene rearrangements: Duplication and nonrandom loss. Mol. Biol. Evol. 2002;19:163–169. doi: 10.1093/oxfordjournals.molbev.a004068. [DOI] [PubMed] [Google Scholar]
- 70.Lunt D.H., Hyman B.C. Animal mitochondrial DNA recombination. Nature. 1997;387:247. doi: 10.1038/387247a0. [DOI] [PubMed] [Google Scholar]
- 71.Cantatore P., Gadaleta M.N., Roberti M., Saccone C., Wilson A.C. Duplication and remoulding of tRNA genes during the evolutionary rearrangement of mitochondrial genomes. Nature. 1987;329:853–855. doi: 10.1038/329853a0. [DOI] [PubMed] [Google Scholar]
- 72.Xu S.-Y., Long J.-K., Chen X.-S. Comparative analysis of the complete mitochondrial genomes of five Achilidae species (Hemiptera: Fulgoroidea) and other Fulgoroidea reveals conserved mitochondrial genome organization. PeerJ. 2019;7:e6659. doi: 10.7717/peerj.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ojala D., Montoya J., Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 74.Lavrov D.V., Brown W.M., Boore J.L. A novel type of RNA editing occurs in the mitochondrial tRNAs of the centipede Lithobius forficatus. Proc. Natl. Acad. Sci. USA. 2000;97:13738–13742. doi: 10.1073/pnas.250402997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Masta S.E., Boore J.L. The complete mitochondrial genome sequence of the spider Habronattus oregonensis reveals rearranged and extremely truncated tRNAs. Mol. Biol. Evol. 2004;21:893–902. doi: 10.1093/molbev/msh096. [DOI] [PubMed] [Google Scholar]
- 76.Zhang D.-X., Hewitt G.M. Insect mitochondrial control region: A review of its structure, evolution and usefulness in evolutionary studies. Biochem. Syst. Ecol. 1997;25:99–120. doi: 10.1016/S0305-1978(96)00042-7. [DOI] [Google Scholar]
- 77.Crozier R.H., Crozier Y.C. The mitochondrial genome of the honeybee Apis mellifera: Complete sequence and genome organization. Genetics. 1993;133:97–117. doi: 10.1093/genetics/133.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nováková E., Hypša V., Klein J., Foottit R.G., von Dohlen C.D., Moran N.A. Reconstructing the phylogeny of aphids (Hemiptera: Aphididae) using DNA of the obligate symbiont Buchnera aphidicola. Mol. Phylogenet. Evol. 2013;68:42–54. doi: 10.1016/j.ympev.2013.03.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete mitochondrial genome of S. sinisalicis is available in the GenBank under accession number MW006542. The raw sequencing data are deposited in the NCBI Sequence Read Archive (SRA) database with accession number PRJNA668672.