Abstract
Colletotrichum spp. are important pathogens of citrus that cause dieback of branches and postharvest disease. Globally, several species of Colletotrichum have been identified as causing anthracnose of citrus. One hundred and sixty-eight Colletotrichum isolates were collected from anthracnose symptoms on citrus stems, leaves, and fruit from Victoria, New South Wales, and Queensland, and from State herbaria in Australia. Colletotrichum australianum sp. nov., C. fructicola, C. gloeosporioides, C. karstii, C. siamense, and C. theobromicola were identified using multi-gene phylogenetic analyses based on seven genomic loci (ITS, gapdh, act, tub2, ApMat, gs, and chs-1) in the gloeosporioides complex and five genomic loci (ITS, tub2, act, chs-1, and his3) in the boninense complex, as well as morphological characters. Several isolates pathogenic to chili (Capsicum annuum), previously identified as C. queenslandicum, formed a clade with the citrus isolates described here as C. australianum sp. nov. The spore shape and culture characteristics of the chili and citrus isolates of C. australianum were similar and differed from those of C. queenslandicum. This is the first report of C. theobromicola isolated from citrus and the first detection of C. karstii and C. siamense associated with citrus anthracnose in Australia.
Keywords: anthracnose, citrus, Colletotrichum australianum, phylogenetic analysis, taxonomy
1. Introduction
Edible citrus (Citrus spp.) are important fruit crops globally, produced in temperate and tropical climates [1]. Cumquat (Citrus japonica), grapefruit (Citrus × paradisi), lemon (Citrus limon), lime (Citrus aurantifolia), mandarin (Citrus reticulata), and orange (Citrus × sinensis) are all commercially important citrus species [1,2]. Australia is a major citrus producer with citrus grown in every mainland state [3,4]. In 2019, there was approximately 25,500 ha of citrus production in Australia [5]. Citrus is one of the largest fresh fruit exports from Australia. Australia exported 251,594 tonnes of citrus in 2018, with a total value of $A452.9 million [6].
In citrus, anthracnose caused by Colletotrichum spp. is a serious disease limiting production globally. Preharvest anthracnose reduces yield, while postharvest anthracnose affects fruit quality, negatively impacting fruit export and marketability [7]. Colletotrichum species are difficult to identify based on morphological characters. Molecular phylogeny has reinvigorated Colletotrichum taxonomy [8], with over 220 Colletotrichum species in 14 species complexes now recognised [9,10].
Globally, multiple Colletotrichum species within several species complexes have been identified as causing citrus anthracnose. Colletotrichum gloeosporioides was reported to be associated with anthracnose in Australia [8], Vietnam [11], China [12], Italy [8,13], Morocco [14], Mexico [15,16], Pakistan [17], Ghana [18,19], Brazil [11,20], Algeria [21], Greece [8], Malta [8], New Zealand [8], Portugal [8,22], South Africa [11], Spain [8], Tunisia [23,24], United States [8] and Zimbabwe [11]. Colletotrichum karstii was reported in Southern Italy [13], China [25,26,27], Portugal [23], South Africa [11], Europe [8], United States [28], Tunisia [16], Turkey [29], and New Zealand [25]; C. fructicola was reported in China [26,27,30]; and C. siamense was reported in Vietnam [11], Bangladesh [11], Egypt [11], China [31], Mexico [22], and Pakistan [17,32]. Additionally, C. abscissum, C. acutatum, C. boninense, C. brevisporum, C. catinaense, C. citri, C. citricola, C. citri-maximae, C. constrictum, C. godetiae, C. helleniense, C. hystricis, C. johnstonii, C. cigarro, C. limetticola, C. limonicola, C. novae-zelandiae, C. queenslandicum, C. simmondsii, C. tropicicola, and C. truncatum have all been associated with citrus anthracnose [8,11,25,27,33,34,35,36].
Colletotrichum acutatum, C. fructicola, C. gloeosporioides, and C. nymphaeae have been reported as pathogens associated with citrus anthracnose in Australia. However, C. acutatum was identified based on morphology, and C. nymphaeae was verified by a single tub2 sequence [37,38]. Citrus fruits and plants with anthracnose symptoms are very common both in home gardens and in commercial orchards in Australia. Hence, it is necessary to accurately characterize the Colletotrichum species causing anthracnose diseases of citrus in Australia to help develop appropriate disease management strategies and provide a baseline for plant biosecurity, trade, and market access.
In this study, a representative collection of Colletotrichum isolates from eastern Australian citrus was established from symptomatic leaves, twigs, and fruit, and from culture collections. Colletotrichum species were determined by utilising a polyphasic approach, in which informative gene loci were sequenced. Multigene phylogenetic analyses, morphological characters, and pathogenicity bioassays were used to confirm the taxonomy and phylogenetic relationships of Colletotrichum spp. pathogens causing citrus anthracnose in Australia.
2. Materials and Methods
2.1. Sample Collection
A total of 147 Colletotrichum isolates were collected from anthracnose lesions on citrus stems and leaves of trees growing in Victoria and New South Wales and from citrus fruits with anthracnose disease symptoms from supermarkets in Melbourne, Victoria. In addition, 21 isolates originating from citrus plants were obtained from State fungaria (the Victorian Plant Pathology Herbarium (VPRI), the Queensland Plant Pathology Herbarium (BRIP), and the NSW Plant Pathology Collection (DAR)).
2.2. Isolate Preparation
Infected fruits, stems, and leaves were surface sterilized by dipping in 2.3% (active ingredient) sodium hypochlorite (NaOCl) for 2 min and rinsed five times with sterile distilled water (SDW). Tissue pieces (2 mm2) were excised from the margins of infected lesions and plated onto potato dextrose agar (PDA). The plates were incubated at 25 °C in continuous dark for 7 d as described by Guarnaccia et al. [8]. Subcultures of mycelia on PDA plates were maintained under the same growing conditions for a further 7 d. All isolates were established as single spore cultures, as described in De Silva et al. [39].
2.3. Morphological and Cultural Analyses
Plugs (2 mm2) of actively growing mycelia were taken from the edge of 7-d-old cultures and transferred onto PDA and synthetic nutrient-poor agar (SNA), as described by Guarnaccia et al. [8]. After 7 d of incubation at 25 °C under continuous near-ultraviolet light, colony growth was determined by measuring two diameters perpendicular to each other per plate and determining the average of six plates. At 10 d, colony colour was determined using colour charts [40]. Acervuli were induced by inoculating pieces of sterilized mandarin rind with mycelia and incubating on water agar (WA) and SNA, at 25 °C for 10 d.
Appressoria were induced using the slide culture technique described by Johnston and Jones [41]. The length and width of 30 appressoria/slide were measured using X1000 magnification with a Leica DM6000 LED compound microscope, Leica DMC2900 camera, and Leica LAS v. 4.5.0 software.
Slide preparations of morphological structures were prepared in lactic acid, and at least 30 observations were recorded for conidia, conidiophores, and conidiogenous cells per isolate, as well as presence or absence of setae. The range, mean, and standard error (SE) were calculated for each isolate.
2.4. Multigene Phylogenetic Analysis
2.4.1. DNA Extraction, PCR Amplification, and Sequencing
1. DNA extraction
Genomic DNA was extracted from pure (single-spored) mycelia of Colletotrichum isolates grown on PDA at 25 °C for 7 d using DNeasy Plant Mini kits (Qiagen, Australia), following the manufacturer’s instructions. DNA concentration was determined using NanoDrop, then diluted to 2 ng∙µL−1 and stored at −20 °C until further use [39].
2. PCR amplification and sequencing
Isolates were assigned to a species complex based on morphology and internal transcribed spacer and intervening 5.8S nrDNA gene (ITS) and β-tubulin (tub2) gene sequences data. Isolates in the gloeosporioides species complex were further characterised using seven gene loci: ITS, glyceraldehyde-3-phosphate dehydrogenase (gapdh), actin (act), tub2, the Apn2–Mat1–2 intergenic spacer and partial mating type (Mat1–2) (ApMat), glutamine synthetase (gs), and chitin synthase 1 (chs-1) genes. Isolates in the boninense species complex were further characterised using five gene loci: ITS, tub2, act, chs-1, and histone (his3). These gene sequences were amplified and sequenced by using primer pairs: ITS-1F (ITS; [42]) and ITS4 (ITS; [43]), GDF1 and GDR1 (gapdh; [44]), ACT-512F + ACT-783R (act; [45]), Btub2Fd and Btub4Rd (tub2; [46]), AMF1 and AMR1 (ApMat; [47]), GSF1 and GSR1 (gs; [48]), CHS-79F and CHS-354R (chs-1; [45]), and CYLH3F and CYLH3R (his3; [49]).
PCR was performed in a 2720 Thermal Cycler (Applied Biosystems, Australia). The total volume of PCR mixture was 25 µL. The PCR of the ITS, gapdh, act, tub2, gs, chs-1, and his3 genes followed the protocol described by De Silva et al. [39] and contained 1× PCR buffer, 2 mM MgCl2, 0.2 mM dNTP, 1 U Taq DNA polymerase (MangoTaq DNA polymerase; Bioline, Australia), 0.4 µM of each primer, and 6 ng template DNA. The PCR annealing temperatures were adjusted to 55 °C for ITS, gapdh, and his3; 58 °C for act, tub2, and gs; and 66 °C for chs-1.
For ApMat, in the 25 µL PCR mixture, the concentration of each primer was adjusted to 0.5 µM, and the template DNA was adjusted to 10 ng. The PCR amplification protocols were performed according to Silva et al. [47], except the annealing temperature of ApMat was adjusted to 62 °C.
All PCR products were purified using QIA-quick PCR Purification Kit (Qiagen, Australia) following the manufacturer’s instructions. Purified PCR products were sequenced in both the forward and reverse sense at the Australian Genome Research Facility (AGRF, Melbourne), then aligned to produce a consensus sequence for each isolate using ClustalW in MEGA 6.06 [50]. The consensus sequences were deposited in GenBank.
2.4.2. Phylogenetic Analyses
The sequences of reference isolates were retrieved from GenBank for use in phylogenetic analyses (Table 1). All the sequences were aligned by using ClustalW in MEGA 6.06 and manually edited when necessary. The ITS and tub2 sequences of morphologically different isolates were compared to determine which species complex each isolate belonged based on maximum likelihood analysis (ML) by using MEGA 6.06 [10]. For isolates from the gloeosporioides species complex, phylogenetic analyses of combined seven gene sequences (ITS, gapdh, act, tub2, ApMat, gs, and chs-1) and combined two gene sequences (ApMat and gs) were carried out with selected reference sequences [39,51]. For isolates from the boninense species complex, phylogenetic analysis of combined five gene sequences (ITS, tub2, act, chs-1, and his3) was constructed [8].
Table 1.
Strains of Colletotrichum species used in the phylogenetic analyses with details of host and location, and GenBank accession numbers of the sequences.
| Species | Accession Number | Host | Location | GenBank Accession Numbers | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ITS | GAPDH | ACT | TUB2 | gs | ApMat | CHS-1 | HIS3 | ||||
| Gloeosporioides complex | |||||||||||
| C. aenigma | ICMP 18608 * | Persea americana | Israel | JX010244 | JX010044 | JX009443 | JX010389 | JX010078 | KM360143 | JX009774 | – |
| C. aeschynomenes | ICMP 17673; ATCC 201874 * | Aeschynomene virginica | USA | JX010176 | JX009930 | JX009483 | JX010392 | JX010081 | KM360145 | JX009799 | – |
| C. alatae | ICMP 17919 * | Dioscorea alata | India | JX010190 | JX009990 | JX009471 | JX010383 | JX010065 | KC888932 | JX009837 | – |
| C. alienum | ICMP 12071 * | Malus domestica | New Zealand | JX010251 | JX010028 | JX009572 | JX010411 | JX010101 | KM360144 | JX009882 | – |
| C. asianum | ICMP 18580; CBS 130418 * | Coffea arabica | Thailand | FJ972612 | JX010053 | JX009584 | JX010406 | JX010096 | FR718814 | JX009867 | – |
| C. aotearoa | ICMP 18537 * | Coprosma sp. | New Zealand | JX010205 | JX010005 | JX009564 | JX010420 | JX010113 | KC888930 | JX009853 | – |
| C. artocarpicola | MFLUCC 18–1167 * | Artocarpus heterophyllus | Thailand | MN415991 | MN435568 | MN435570 | MN435567 | – | – | MN435569 | – |
| C. australianum | VPRI 43074; UMC001 | Citrus reticulata | Australia, Vic | MG572137 | MG572126 | MK473452 | MG572148 | MG572159 | MG572170 | MW091986 | – |
| VPRI 43075; UMC002 * | Citrus sinensis | Australia, Vic | MG572138 | MG572127 | MN442109 | MG572149 | MG572160 | MG572171 | MW091987 | – | |
| BRIP 63695 | Capsicum annuum | Australia | KU923677 | MN442115 | MN442105 | KU923693 | KU923737 | KU923727 | MW092000 | – | |
| BRIP 63696 | Capsicum annuum | Australia | KU923678 | – | – | KU923694 | KU923738 | KU923728 | – | – | |
| BRIP 63697 | Capsicum annuum | Australia | KU923679 | – | – | KU923695 | KU923739 | KU923729 | – | – | |
| BRIP 63698 | Capsicum annuum | Australia | KU923680 | MN442116 | MN442106 | KU923696 | KU923740 | KU923730 | MW092001 | – | |
| BRIP 63699 | Capsicum annuum | Australia | KU923681 | MN442117 | MN442107 | KU923697 | KU923741 | KU923731 | MW092002 | – | |
| BRIP 63700 | Capsicum annuum | Australia | KU923682 | MN442118 | MN442108 | KU923698 | KU923742 | KU923732 | MW092003 | – | |
| C. camelliae | CGMCC 3.14925 * | Camellia sinensis | China | KJ955081 | KJ954782 | KJ954363 | KJ955230 | KJ954932 | KJ954497 | – | – |
| Glomella cingulate f. sp. camelliae | ICMP 10643 * | Camellia × williamsii | UK | JX010224 | JX009908 | JX009540 | JX010436 | JX010119 | KJ954625 | JX009891 | – |
| C. changpingense | MFLUCC 15-0022 | Fragaria ×ananassa | China | KP683152 | KP852469 | KP683093 | KP852490 | – | – | KP852449 | – |
| C. chrysophilum | CMM4268 * | Musa sp. | Brazil | KX094252 | KX094183 | KX093982 | KX094285 | KX094204 | – | KX094083 | – |
| C. conoides | CAUG17 * | Capsicum annuum | China | KP890168 | KP890162 | KP890144 | KP890174 | – | – | KP890156 | – |
| C. cordylinicola | MFLUCC 090551; ICMP 18579 * | Cordyline fruticosa | Thailand | JX010226 | JX009975 | HM470235 | JX010440 | JX010122 | JQ899274 | JX009864 | – |
| C. clidemiae | ICMP 18658 * | Clidemia hirta | USA, Hawaii | JX010265 | JX009989 | JX009537 | JX010438 | JX010129 | KC888929 | JX009877 | – |
| C. endophytica | CAUG28 | Capsicum annuum | China | KP145441 | KP145413 | KP145329 | KP145469 | – | – | KP145385 | – |
| C. fructicola | ICMP 18581; CBS 130416 * | Coffea arabica | Thailand | JX010165 | JX010033 | FJ907426 | JX010405 | JX010095 | JQ807838 | JX009866 | – |
| LC2923; LF130 | Camellia sinensis | China | KJ955083 | KJ954784 | KJ954365 | KJ955232 | KJ954934 | KJ954499 | – | – | |
| VPRI 43079; UMC006 | Citrus reticulata | Australia, Qld | MG572142 | MG572131 | MK473454 | MG572153 | MG572164 | MG572175 | MW091991 | – | |
| BRIP 65028a; VPRI 43034; B03-43034 | Citrus latifolia | Australia, Qld | MK470007 | MK470025 | MK470097 | MK470061 | MK470043 | MK470079 | MW091983 | – | |
| C. fructicola (syn. C. ignotum) | ICMP 18646 | Tetragastris panamensis | Panama | JX010173 | JX010032 | JX009581 | JX010409 | JX010099 | JQ807839 | JX009874 | – |
| C. fructivorum | CBS 133125 * | Vaccinium macrocarpon | USA | JX145145 | – | – | JX145196 | – | – | – | – |
| C. gloeosporioides | IMI 356878; ICMP 17821; CBS 112999 * | Citrus sinensis | Italy | JX010152 | JX010056 | JX009531 | JX010445 | JX010085 | JQ807843 | JX009818 | – |
| LC3110; LF318 | Camellia sinensis | China | KJ955127 | KJ954828 | KJ954407 | KJ955275 | KJ954978 | KJ954541 | – | – | |
| LC3312; LF534 | Camellia sinensis | China | KJ955158 | KJ954859 | KJ954434 | KJ955305 | KJ955009 | KJ954569 | – | – | |
| LC3382; LF604 | Camellia sinensis | China | KJ955176 | KJ954877 | KJ954450 | KJ955323 | KJ955026 | KJ954584 | – | – | |
| LC3686; LF916 | Camellia sinensis | China | KJ955226 | KJ954927 | KJ954493 | KJ955371 | KJ955076 | KJ954629 | – | – | |
| VPRI 43076; UMC003 | Citrus sinensis | Australia, Vic | MG572139 | MG572128 | MN442110 | MG572150 | MG572161 | MG572172 | MW091988 | – | |
| VPRI 43078; UMC005 | Citrus aurantifolia | Australia, Qld | MG572141 | MG572130 | MN442111 | MG572152 | MG572163 | MG572174 | MW091990 | – | |
| VPRI 43080; UMC007 | Citrus reticulata | Australia, Qld | MG572143 | MG572132 | MK473455 | MG572154 | MG572165 | MG572176 | MW091992 | – | |
| VPRI 43081; UMC008 | Citrus reticulata | Australia, Qld | MG572144 | MG572133 | MN442112 | MG572155 | MG572166 | MG572177 | MW091993 | – | |
| VPRI 43082; UMC009 | Citrus reticulata | Australia, Qld | MG572145 | MG572134 | MN442113 | MG572156 | MG572167 | MG572178 | MW091994 | – | |
| VPRI 43084; UMC011 | Citrus japonica | Australia, Vic | MG572147 | MG572136 | MN442114 | MG572158 | MG572169 | MG572180 | MW091996 | – | |
| VPRI 43648; UMC012 | Citrus sinensis | Australia, Vic | MW081160 | MW081163 | MW081166 | MW081169 | MW081175 | MW081172 | MW091997 | – | |
| VPRI 43649; UMC013 | Citrus limon | Australia, Vic | MW081161 | MW081164 | MW081167 | MW081170 | MW081176 | MW081173 | MW091998 | – | |
| VPRI 43650; UMC014 | Citrus japonica | Australia, Vic | MW081162 | MW081165 | MW081168 | MW081171 | MW081177 | MW081174 | MW091999 | – | |
| VPRI 10312; A01-10312 | Citrus sinensis | Australia, Vic | MK469996 | MK470014 | MK470086 | MK470050 | MK470032 | MK470068 | MW091972 | – | |
| VPRI 10347; A02-10347; BRIP 54771 | Citrus limon | Australia, Vic | MK469997 | MK470015 | MK470087 | MK470051; KU221374 | MK470033 | MK470069 | MW091973 | – | |
| WAC 12803; BRIP 63680a; VPRI 43024; A05-43024 | Citrus sinensis | Australia, WA | MK469998 | MK470016 | MK470088 | MK470052 | MK470034 | MK470070 | MW091974 | – | |
| BRIP 66210a; VPRI 43026; A07-43026 | Citrus reticulata | Australia, SA | MK470000 | MK470018 | MK470090 | MK470054 | MK470036 | MK470072 | MW091976 | – | |
| BRIP 66210b; VPRI 43027; A08-43027 | Citrus reticulata | Australia, SA | MK470001 | MK470019 | MK470091 | MK470055 | MK470037 | MK470073 | MW091977 | – | |
| BRIP 28546a; VPRI 43028; A09-43028 | Citrus sinensis Navel | Australia, Qld | MK470002 | MK470020 | MK470092 | MK470056 | MK470038 | MK470074 | MW091978 | – | |
| BRIP 28754a; VPRI 43030; A11-43030 | Citrus reticulata | Australia, Qld | MK470003 | MK470021 | MK470093 | MK470057 | MK470039 | MK470075 | MW091979 | – | |
| BRIP 53157d; VPRI 43031; A12-43031 | Citrus aurantifolia Tahiti | Australia, Qld | MK470004 | MK470022 | MK470094 | MK470058 | MK470040 | MK470076 | MW091980 | – | |
| BRIP 66135a; VPRI 43032; B01-43032 | Citrus reticulata Imperial Blanco | Australia, Qld | MK470005 | MK470023 | MK470095 | MK470059 | MK470041 | MK470077 | MW091981 | – | |
| BRIP 28831a; VPRI 43033; B02-43033 | Citrus sinensis | Australia, Qld | MK470006 | MK470024 | MK470096 | MK470060 | MK470042 | MK470078 | MW091982 | – | |
| VPRI 42955; G01-42955 | Citrus limon | Australia, NSW | MK470008 | MK470026 | MK470098 | MK470062 | MK470044 | MK470080 | MW091984 | – | |
| VPRI 42956; H01-42956 | Citrus sinensis | Australia, NSW | MK470009 | MK470027 | MK470099 | MK470063 | MK470045 | MK470081 | MW091985 | – | |
| C. grevilleae | CBS 132879 * | Grevillea sp. | Italy | KC297078 | KC297010 | KC296941 | KC297102 | KC297033 | – | KC296987 | – |
| C. grossum | CGMCC3.17614T; CAUG7 * | Capsicum sp. | China | KP890165 | KP890159 | KP890141 | KP890171 | – | – | KP890153 | – |
| CAU31 | Capsicum sp. | China | KP890166 | KP890160 | KP890142 | KP890172 | – | – | KP890154 | – | |
| CAUG32 | Capsicum sp. | China | KP890167 | KP890161 | KP890143 | KP890173 | – | – | KP890155 | – | |
| C. hebeiense | MFLUCC13-0726 * | Vitis vinifera cv. Cabernet Sauvignon | China | KF156863 | KF377495 | KF377532 | KF288975 | – | – | KF289008 | – |
| C. helleniense | CPC 26844; CBS 142418 * | Poncirus trifoliata | Greece | KY856446 | KY856270 | KY856019 | KY856528 | – | – | KY856186 | – |
| C. henanense | LC3030; CGMCC 3.17354; LF238 * | Camellia sinensis | China | KJ955109 | KJ954810 | KM023257 | KJ955257 | KJ954960 | KJ954524 | – | – |
| C. horii | ICMP 10492 * | Diospyros kaki | Japan | GQ329690 | GQ329681 | JX009438 | JX010450 | JX010137 | JQ807840 | JX009752 | – |
| C. hystricis | CPC 28153; CBS 142411 * | Citrus hystrix | Italy | KY856450 | KY856274 | KY856023 | KY856532 | – | – | KY856190 | – |
| C. jiangxiense | LF687 * | Camellia sinensis | China | KJ955201 | KJ954902 | KJ954471 | KJ955348 | KJ955051 | KJ954607 | – | – |
| C. cigarro | ICMP 18534 | Kunzea ericoides | New Zealand | JX010227 | JX009904 | JX009473 | JX010427 | JX010116 | HE655657 | JX009765 | – |
| C. kahawae | IMI 319418; ICMP 17816 * | Coffea arabica | Kenya | JX010231 | JX010012 | JX009452 | JX010444 | JX010130 | JQ894579 | JX009813 | – |
| C. musae | ICMP 19119; CBS 116870 * | Musa sp. | USA | JX010146 | JX010050 | JX009433 | HQ596280 | JX010103 | KC888926 | JX009896 | – |
| ICMP 17817 | Musa sapientum | Kenya | JX010142 | JX010015 | JX009432 | JX010395 | JX010084 | – | JX009815 | – | |
| C. nupharicola | ICMP 18187 * | Nuphar lutea subsp. polysepala | USA | JX010187 | JX009972 | JX009437 | JX010398 | JX010088 | JX145319 | JX009835 | – |
| C. pandanicola | MFLUCC 17-0571 | Pandanaceae | Thailand | MG646967 | MG646934 | MG646938 | MG646926 | – | – | MG646931 | – |
| C. proteae | CBS 132882 * | Protea sp. | South Africa | KC297079 | KC297009 | KC296940 | KC297101 | KC297032 | – | KC296986 | – |
| C. psidii | ICMP 19120 * | Psidium sp. | Italy | JX010219 | JX009967 | JX009515 | JX010443 | JX010133 | KC888931 | JX009901 | – |
| C. queenslandicum | ICMP 1778 * | Carica papaya | Australia | JX010276 | JX009934 | JX009447 | JX010414 | JX010104 | KC888928 | JX009899 | – |
| CPC 17123 | Syzygium australa | Australia | KP703357 | KP703282 | – | KP703439 | KP703693 | KP703778 | – | – | |
| ICMP 18705 | Coffea sp. | Fiji | JX010185 | JX010036 | JX009490 | JX010412 | JX010102 | – | JX009890 | – | |
| CMM3233 | Anacardium occidentale | Brazil, Pernambuco state | – | MF110849 | – | MF111058 | MF110996 | MF110639 | – | – | |
| CMM3241 | Anacardium occidentale | Brazil, Pernambuco state | – | MF110848 | – | MF111059 | MF111000 | MF110642 | – | – | |
| CMM3236 | Anacardium occidentale | Brazil, Pernambuco state | – | MF110850 | – | MF111060 | MF110997 | MF110640 | – | – | |
| CMM3240 | Anacardium occidentale | Brazil, Pernambuco state | – | MF110852 | – | MF111061 | MF110999 | MF110644 | – | – | |
| CMM3237 | Anacardium occidentale | Brazil, Pernambuco state | – | MF110853 | – | MF111062 | MF110998 | MF110641 | – | – | |
| CMM3242 | Anacardium occidentale | Brazil, Pernambuco state | – | – | – | MF111063 | MF111001 | MF110643 | – | – | |
| C. rhexiae | CBS 133134 * | Rhexia virginica | USA | JX145128 | – | – | JX145179 | – | – | – | – |
| C. salsolae | ICMP 19051 * | Salsola tragus | Hungary | JX010242 | JX009916 | JX009562 | JX010403 | JX010093 | KC888925 | JX009863 | – |
| C. siamense | ICMP 18578 CBS 130417 * | Citrus arabica | Thailand | JX010171 | JX009924 | FJ907423 | JX010404 | JX010094 | JQ899289 | JX009865 | – |
| VPRI 43077; UMC004 | Citrus limon | Australia, NSW | MG572140 | MG572129 | MK473453 | MG572151 | MG572162 | MG572173 | MW091989 | – | |
| BRIP 54270b; VPRI 43029; A10-43029 | Citrus australasica | Australia, Qld | MK469995 | MK470013 | MK470085 | MK470049 | MK470031 | MK470067 | MW091971 | – | |
| C. siamense (syn. C. jasmini-sambac) | CBS 130420; ICMP 19118 | Jasminum sambac | Vietnam | HM131511 | HM131497 | HM131507 | JX010415 | JX010105 | JQ807841 | JX009895 | – |
| C. siamense (syn. C. hymenocallidis) | CBS 125378; ICMP 18642; LC0043 | Hymenocallis americana | China | JX010278 | JX010019 | GQ856775 | JX010410 | JX010100 | JQ899283 | GQ856730 | – |
| C. siamense (syn. C. murrayae) | GZAAS 5.09506 | Murraya sp. | China | JQ247633 | JQ247609 | JQ247657 | JQ247644 | JQ247621 | – | – | – |
| C. syzygicola | DNCL021; MFLUCC 10-0624 * | Syzygium samarangense | Thailand | KF242094 | KF242156 | KF157801 | KF254880 | KF242125 | – | – | – |
| C. temperatum | CBS 133122 * | Vaccinium macrocarpon | USA | JX145159 | – | – | JX145211 | – | – | – | – |
| C. theobromicola | ICMP 18649; CBS 124945 * | Theobroma cacao | Panama | JX010294 | JX010006 | JX009444 | JX010447 | JX010139 | KC790726 | JX009869 | – |
| C. theobromicola (syn. C. fragariae) | ICMP 17927; CBS 142.31; MTCC 10325T | Fragaria × ananassa | USA | JX010286 | JX010024 | JX009516 | JX010373 | JX010064 | JQ807844 | JX009830 | – |
| VPRI 43083; UMC010 | Citrus aurantifolia | Australia, Qld | MG572146 | MG572135 | MK473456 | MG572157 | MG572168 | MG572179 | MW091995 | – | |
| C. ti | ICMP 4832 * | Cordyline sp. | New Zealand | JX010269 | JX009952 | JX009520 | JX010442 | JX010123 | KM360146 | JX009898 | – |
| C. tropicale | ICMP 18653; CBS 124949 * | Theobroma cacao | Panama | JX010264 | JX010007 | JX009489 | JX010407 | JX010097 | KC790728 | JX009870 | – |
| C. viniferum | GZAAS 5.08601 * | Vitis vinifera, cv. ‘Shuijing’ | China | JN412804 | JN412798 | JN412795 | JN412813 | JN412787 | – | – | – |
| C. wuxiense | CGMCC 3.17894 * | Camellia sinensis | China | KU251591 | KU252045 | KU251672 | KU252200 | KU252101 | KU251722 | KU251939 | – |
| C. xanthorrhoeae | BRIP 45094; ICMP 17903; CBS 127831 * | Xanthorrhoea preissii | Australia | JX010261 | JX009927 | JX009478 | JX010448 | JX010138 | KC790689 | JX009823 | – |
| Colletotrichum sp. | BRIP 58074a; VPRI 43025; A06-43025 | Citrus australasica | Australia, Qld | MK469999 | MK470017 | MK470089 | MK470053 | MK470035 | MK470071 | MW091975 | – |
| Boninense complex | |||||||||||
| C. annellatum | CBS 129826 * | Hevea brasiliensis | Colombia | JQ005222 | – | JQ005570 | JQ005656 | – | – | JQ005396 | JQ005483 |
| C. beeveri | CBS 128527 * | Brachyglottis repanda | New Zealand | JQ005171 | – | JQ005519 | JQ005605 | – | – | JQ005345 | JQ005432 |
| C. boninense | ICMP 17904; CBS 123755 * | Crinum asiaticum ‘Sinicum’ | Japan | JQ005153 | – | JQ005501 | JQ005588 | – | – | JQ005327 | JQ005414 |
| C. brassicicola | CBS 101059 | Brassica oleracea var. gemmifera | New Zealand | JQ005172 | – | JQ005520 | JQ005606 | – | – | JQ005346 | JQ005433 |
| C. brasiliense | CBS 128501* | Passiflora edulis | Brazil | JQ005235 | – | JQ005583 | JQ005669 | – | – | JQ005409 | JQ005496 |
| C. catinaense | CBS 142417; CPC 27978 * | Citrus reticulata | Italy, Catania | KY856400 | – | KY855971 | KY856482 | – | – | KY856136 | KY856307 |
| C. citricola | CBS 134228 * | Citrus unchiu | China | KC293576 | – | KC293616 | KC293656 | – | – | KY856140 | KY856311 |
| C. constrictum | CBS 128504 | Citrus limon | New Zealand | JQ005238 | – | JQ005586 | JQ005672 | – | – | JQ005412 | KY856313 |
| C. colombiense | CBS 129818 * | Passiflora edulis | Colombia | JQ005174 | – | JQ005522 | JQ005608 | – | – | JQ005348 | JQ005435 |
| C. cymbidiicola | IMI 347923 * | Cymbidium sp. | Australia | JQ005166 | – | JQ005514 | JQ005600 | – | – | JQ005340 | JQ005427 |
| C. dacrycarpi | CBS 130241 * | Dacrycarpus dacrydioides | New Zealand | JQ005236 | – | JQ005584 | JQ005670 | – | – | JQ005410 | JQ005497 |
| C. hippeastri | CBS 125376 * | Hippeastrum vittatum | China | JQ005231 | – | JQ005579 | JQ005665 | – | – | JQ005405 | JQ005492 |
| C. karstii | CBS 126532 | Citrus sp. | South Africa | JQ005209 | – | JQ005557 | JQ005643 | – | – | JQ005383 | JQ005470 |
| CBS 128551 | Citrus sp. | New Zealand | JQ005208 | – | JQ005556 | JQ005642 | – | – | JQ005382 | JQ005469 | |
| CBS 129829 | Gossypium hirsutum | Germany | JQ005189 | – | JQ005537 | JQ005623 | – | – | JQ005363 | JQ005450 | |
| CPC 27853 | Citrus sinensis | Italy, Catania | KY856461 | – | KY856034 | KY856543 | – | – | KY856202 | KY856377 | |
| CPC 31139 | Citrus sinensis | Italy, Catania | KY856467 | – | KY856040 | KY856549 | – | – | KY856208 | KY856383 | |
| CBS 129833 | Musa sp. | Mexico | JQ005175 | – | JQ005523 | JQ005609 | – | – | JQ005349 | JQ005436 | |
| CBS 861.72 | Bombax aquaticum | Brazil | JQ005184 | – | JQ005532 | JQ005618 | – | – | JQ005358 | JQ005445 | |
| DAR 25017a; VPRI 42941; D02-42941 | Citrus sinensis Valencia | Australia, NSW | MK470103 | – | MK470109 | MK470106 | – | – | MK470115 | MK470112 | |
| DAR 29821a; VPRI 42943; F02-42943 | Citrus sinensis Valencia | Australia, NSW | MK470104 | – | MK470110 | MK470107 | – | – | MK470116 | MK470113 | |
| DAR 29826a; VPRI 42944; G02-42944 | Citrus sinensis Valencia | Australia, NSW | MK470105 | – | MK470111 | MK470108 | – | – | MK470117 | MK470114 | |
| VPRI 43651; UMC015 | Citrus limon | Australia, Vic | MW081178 | – | MW081186 | MW081182 | – | – | MW081190 | MW081194 | |
| VPRI 43652; UMC016 | Citrus sinensis | Australia, Vic | MW081179 | – | MW081187 | MW081183 | – | – | MW081191 | MW081195 | |
| VPRI 43653; UMC017 | Citrus sinensis | Australia, Vic | MW081180 | – | MW081188 | MW081184 | – | – | MW081192 | MW081196 | |
| VPRI 43654; UMC018 | Citrus sinensis | Australia, Vic | MW081181 | – | MW081189 | MW081185 | – | – | MW081193 | MW081197 | |
| C. limonicola | CBS 142410; CPC 31141 * | Citrus limon | Malta, Gozo | KY856472 | – | KY856045 | KY856554 | – | – | KY856213 | KY856388 |
| C. novae-zelandiae | CBS 128505 * | Capsicum annuum | New Zealand | JQ005228 | – | JQ005576 | JQ005662 | – | – | JQ005402 | JQ005489 |
| C. oncidii | CBS 129828 * | Oncidium sp. | Germany | JQ005169 | – | JQ005517 | JQ005603 | – | – | JQ005343 | JQ005430 |
| C. parsonsiae | CBS 128525 * | Parsonsia capsularis | New Zealand | JQ005233 | – | JQ005581 | JQ005667 | – | – | JQ005407 | JQ005494 |
| C. petchii | CBS 378.94 * | Dracaena marginata | Italy | JQ005223 | – | JQ005571 | JQ005657 | – | – | JQ005397 | JQ005484 |
| C. phyllanthi | CBS 175.67 * | Phyllanthus acidus | India | JQ005221 | – | JQ005569 | JQ005655 | – | – | JQ005395 | JQ005482 |
| C. torulosum | CBS 128544 * | Solanum melongena | New Zealand | JQ005164 | – | JQ005512 | JQ005598 | – | – | JQ005338 | JQ005425 |
| Truncatum complex | |||||||||||
| C. truncatum | CBS 151.35 * | Phaseolus lunatus | USA | GU227862 | – | GU227960 | GU228156 | – | – | GU228352 | GU228058 |
Vic: Victoria, NSW: New South Wales, Qld: Queensland, WA: Western Australia, SA: South Australia. * Ex-holotype or ex-epitype cultures.
Further phylogenetic analyses were based on Bayesian Inference analyses (BI) by using MrBayes v. 3.1.2 and ML analysis by using MEGA 6.06 [39]. For BI analyses, MrModeltest2.3 was used to determine the best-fit model for each locus [52] (Table 2). MrBayes v. 3.2.6 was used to generate phylogenetic trees. Four chains were used in the Markov Chain Monte Carlo (MCMC) analysis and were run for 1,000,000,000 generations. The trees were sampled every 100 generations and the heating parameter was set to 0.2. Analyses stopped once the average standard deviation of split frequencies was below 0.01. For ML analysis, analyses were done by using MEGA 6.06. The phylogeny test was the Bootstrap method with 1000 replicates. The substitution model was the Tamura–Nei model based on nucleotide type. The tree inference option was Nearest-Neighbor-Interchange (NNI) ML heuristic method.
Table 2.
Best-fit model for each gene locus selected by MrModeltest.
| Dataset | Substitution Models | ||||||
|---|---|---|---|---|---|---|---|
| ITS | tub2 | act | chs-1 | his3 | |||
| boninense complex | SYM + I+G | HKY + I | HKY + G | GTR + G | HKY + I | ||
| ITS | gapdh | tub2 | act | ApMat | gs | chs-1 | |
| gloeosporioides complex | SYM + I | HKY + I | SYM + I | HKY + I | HKY + G | GTR + G | K80 + G |
2.5. Pathogenicity Testing
One isolate of each Colletotrichum species (except for C. siamense, which did not sporulate in culture) was used in the pathogenicity tests to inoculate orange (Washington Navel) fruits, orange leaves, lemon (Myer) leaves, and orange flower petals according to the method of Guarnaccia et al. [8].
2.5.1. Fruit Bioassay
Conidial suspensions of each isolate were prepared by adding 10 mL of SDW to 7-d-old cultures, scraping the mycelia then filtering through muslin cloth. The concentration of spore suspension was adjusted to 106 conidia mL−1. Organically grown orange fruits (Citrus sinensis) purchased from a market (Queen Victoria Market in Melbourne) were washed with tap water and then submerged in 70% ethanol for 10 min, and finally rinsed in SDW twice. The orange fruits were marked in the middle to divide into two parts and inoculated with both wound (W) and non-wound (NW) methods. For the wound method, the orange skin was pricked with a sterilized pipette tip to about 1 mm depth. Six wound points were made, and each inoculated with 6 µL spore suspension. In the non-wound method, six drops of 6 µL spore suspension were placed directly on the orange skin. For the control group, 6 µL of SDW was used to treat orange fruit in both wound and non-wound methods. There were three replicates per treatment per isolate and the experiments replicated twice. The fruit was transferred to a plastic box and incubated at 25 °C with 100% humidity in dark. After 10 d, fruits were examined for symptom development, and the percentage of infection was calculated ().
2.5.2. Leaf Bioassay
Young, healthy, fully expanded orange and lemon leaves were collected from trees growing in pots. The leaves were washed with tap water, then submerged in 70% ethanol for 2 min, and finally rinsed in SDW twice. The petioles of leaves were wrapped with damp cotton wool and the leaves were placed into petri dishes, three leaves per dish. Three drops of 6 µL spore suspension (106 conidia/mL) were individually placed directly onto the leaf upper surfaces. For the control group, 6 µL of SDW was used. Each set of three leaves per petri dish was inoculated with a different isolate. The petri dishes were placed inside a plastic box and the leaves incubated at 25 °C with 100% humidity and 12/12 h fluorescent light/dark cycle. After 10 d, the leaves were examined for symptom development, and the percentage of infection was calculated ().
2.5.3. Petal Bioassay
Healthy orange flower petals were collected from the same trees. Petals were washed in tap water, then submerged in 70% ethanol for 30 s, and finally rinsed in SDW twice. One drop of 6 µL spore suspension (103 conidia/mL) was carefully placed on the middle of each petal without wounding. For the control group, 6 µL of SDW was used. Seven flower petals were used per isolate. The inoculated petals were put in a plastic box and incubated at 25 °C with 100% humidity and 12/12 h fluorescent light/dark cycle. After 3 d, the petals were examined for symptom development, and the percentage of infection was calculated ().
3. Results
3.1. Phylogenetic Analyses
The 147 isolates were separated into 18 morphological groups based on culture characteristics. One isolate from each morphological group and 18 isolates from State fungaria from different hosts and location were selected for phylogenetic analyses. Among the 36 Colletotrichum isolates, 29 were identified to be in the gloeosporioides complex and seven were identified to be in the boninense complex based on analysis of combined ITS and tub2 gene sequences. All the isolates in the gloeosporioides complex were isolated from stems, leaves, or fruit, while six of the seven isolates in the boninense complex were isolated from infected orange leaf, while another one was from infected lemon leaf (Table S1).
3.1.1. Gloeosporioides Species Complex
1. Seven-gene tree of citrus isolates in gloeosporioides species complex
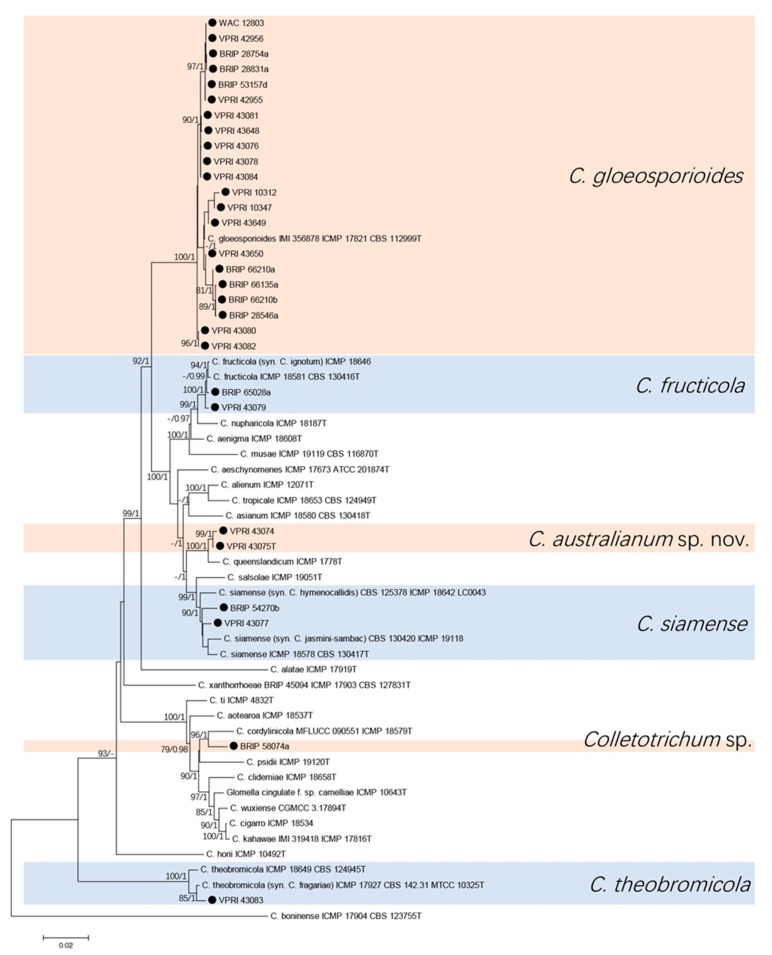
The seven-gene phylogenetic analysis consisted of 29 citrus isolates and 29 reference sequences from the gloeosporioides species complex. Colletotrichum boninense (ICMP 17904T) was used as the out-group. A total of 3703 characters (ITS: 504, gapdh: 271, act: 271, tub2: 510, ApMat: 898, gs: 914, chs-1: 275 and 10 N to separate each two sequences) were analysed. The Bayesian analysis lasted 825,000 generations, resulting in 11,995 total trees, of which 8997 trees were used to calculate the posterior probabilities. The BI posterior probabilities were plotted on the ML tree (Figure 1).
Figure 1.
Phylogenetic analysis of the combined ITS, gapdh, act, tub2, ApMat, GS, and chs-1 sequence alignment of Colletotrichum isolates in the gloeosporioides complex. The bootstrap support values (ML > 75%) of maximum likelihood analysis and Bayesian posterior probabilities (PP > 0.90) are displayed at the nodes (ML/PP). Black circle denotes isolates from Citrus spp.
2. Two-gene tree of citrus isolates in gloeosporioides species complex
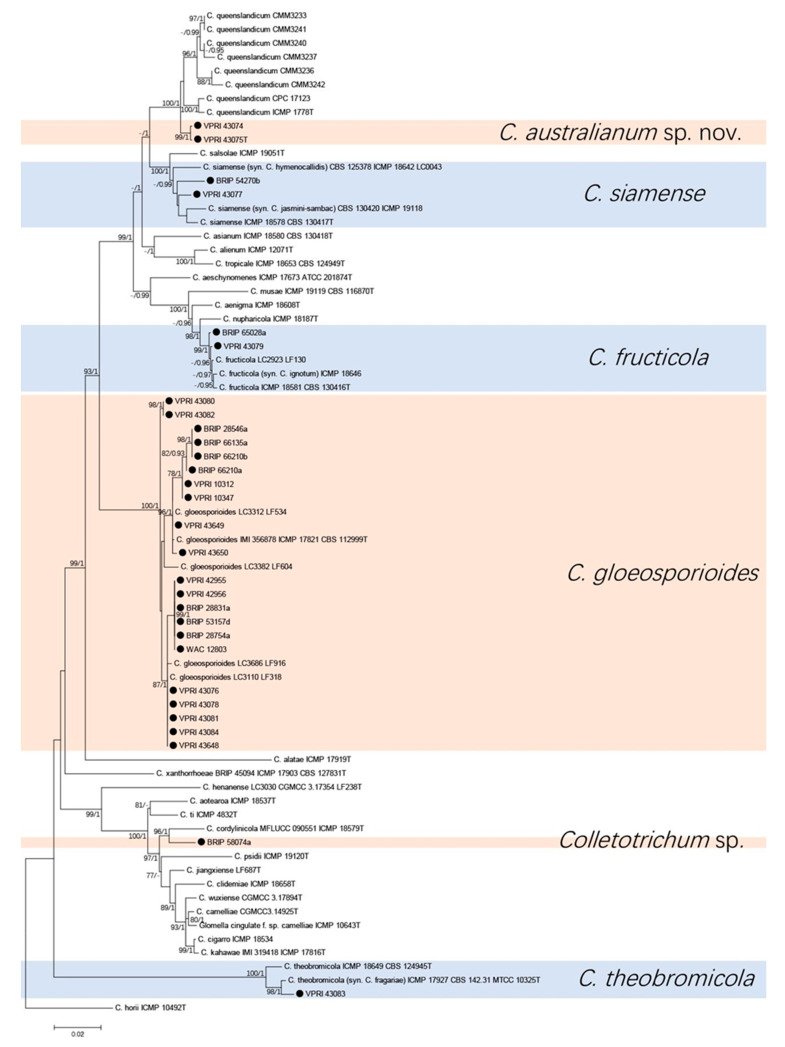
Analysis using the ApMat and gs sequence alignment consisted of 29 citrus isolates and 44 reference sequences from the gloeosporioides species complex. Colletotrichum horii (ICMP 10492T) was used as the out-group. A total of 1832 characters (ApMat: 903, gs: 919 and 10 N to separate two sequences) were analysed. The Bayesian analysis lasted 240,000 generations, resulting in 3601 total trees of which 2701 trees were used to calculate the posterior probabilities. The BI posterior probabilities were plotted on the ML tree (Figure 2).
Figure 2.
Phylogenetic analysis of the combined ApMat and GS sequence alignment of Colletotrichum isolates in the gloeosporioides complex. The bootstrap support values (ML > 75%) of maximum likelihood analysis and Bayesian posterior probabilities (PP > 0.90) are displayed at the nodes (ML/PP). Black circle denotes isolates from Citrus spp.
Five species and one unknown Colletotrichum sp. were identified from the two trees (Figure 1 and Figure 2). Twenty-one (72%) of citrus isolates were identified as C. gloeosporioides, two isolates clustered with three reference isolates of C. siamense, two isolates clustered with three reference isolates of C. fructicola, and one isolate was identified to be C. theobromicola. Two isolates were identified and described as a new species, which was phylogenetically close but significantly different to C. queenslandicum with high support (100/1 in both trees). Isolate BRIP 58074a formed a significantly separate clade (96/1 in both trees) close to C. cordylinicola.
3.1.2. Boninense Species Complex
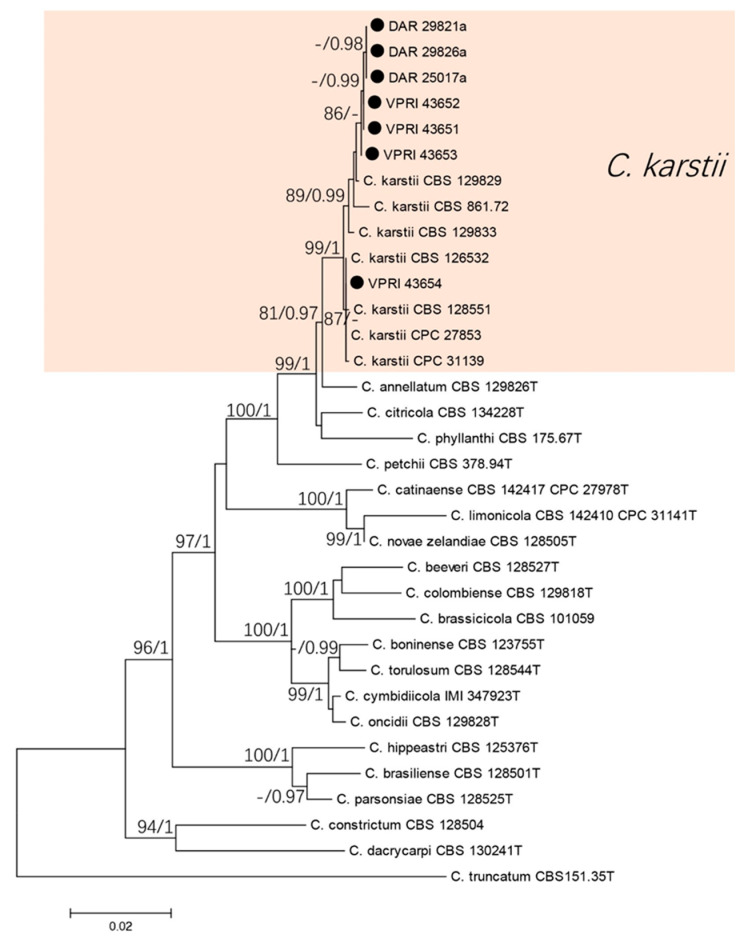
The five gene phylogenetic analysis consisted of seven citrus isolates and 26 reference sequences from the boninense complex. Colletotrichum truncatum (CBS 151.35T) was used as the out-group. A total of 2048 characters (ITS: 559, tub2: 503, act: 280, chs-1: 282, his3: 395) were analysed. The Bayesian analysis lasted 135,000 generations, resulting in 1994 total trees, of which 1496 trees were used to calculate the posterior probabilities. The BI posterior probabilities were plotted on the ML tree. The phylogenetic analysis of the boninense species complex identified the seven citrus isolates as C. karstii (Figure 3).
Figure 3.
Phylogenetic analysis of the combined ITS, tub2, act, chs-1, and his3 sequence alignment of Colletotrichum isolates in the boninense complex. The bootstrap support values (ML > 75%) of maximum likelihood analysis and Bayesian posterior probabilities (PP > 0.90) are displayed at the nodes (ML/PP). Black circle denotes isolates from Citrus spp.
3.2. Morphological Analysis
Morphological characters including conidial size, conidial shape, and growth rate of seven Colletotrichum species were recorded (Table 3). Their conidial size, conidial shape, and growth rate overlapped.
Table 3.
Morphological characters of Colletotrichum species.
| Taxon | Conidial Length (μm) | Conidial Width (μm) | Conidial Shape | Growth Rate (mm/day) 1 |
|---|---|---|---|---|
| C. gloeosporioides | (10.2–) 13.8–14.3 (–16.1) | (4.2–) 5.3–5.5 (–7.3) | Subcylindrical | 10.4–10.8 |
| C. siamense | (12.0–) 13.1–13.4 (–15.8) | (4.8–) 5.4–5.5 (–5.9) | Fusoid | 10.9–11.5 |
| C. fructicola | (12.7–) 14.2–14.6 (–17.1) | (4.6–) 5.1–5.2 (–5.7) | Cylindrical | 10.5–11.1 |
| C. theobromicola | (10.8–) 15.2–16 (–21.2) | (4.0–) 4.8–5 (–5.8) | Cylindrical | 10.5–10.7 |
| Colletotrichum sp. | (13.1–) 15.6–16 (–18.0) | (4.6–) 6.1–6.3 (–7.7) | Cylindrical | 8.9–9.7 |
| C. karstii | (11.3–) 13.2–13.6 (–14.8) | (6.4–) 7.1–7.3 (–8.3) | Cylindrical | 9.4–9.6 |
| New species | (12.7–) 14.1–14.5 (–17.2) | (3.9–) 4.5–4.7 (–5.5) | Cylindrical with one end acute | 9.7–10.3 |
| C. queenslandicum 2 | (12–) 14.5–16.5 (–21.5) | (3.5–) 4.5–5 (–6) | Cylindric, straight, sometimes slightly constricted near center, ends broadly rounded | / |
1 Seven Colletotrichum species incubated at 25 °C for 7 d. Colony growth was determined by measuring two diameters perpendicular to each other per plate and determining the average of six plates. 2 C. queenslandicum ICMP 1778, MycoBank MB563593 [36].
Morphological characters of the type specimen of C. queenslandicum (ICMP 1778) were according to Weir et al. [36] (Table 3). The new species varied morphologically from the type specimen of C. queenslandicum (ICMP 1778) by having different spore shape. Although the range of spore size overlapped between the new species and C. queenslandicum, the average conidial length of the new species was smaller than that of C. queenslandicum [36].
3.3. New Colletotrichum Species
3.3.1. Two-Gene Tree of New Colletotrichum Species
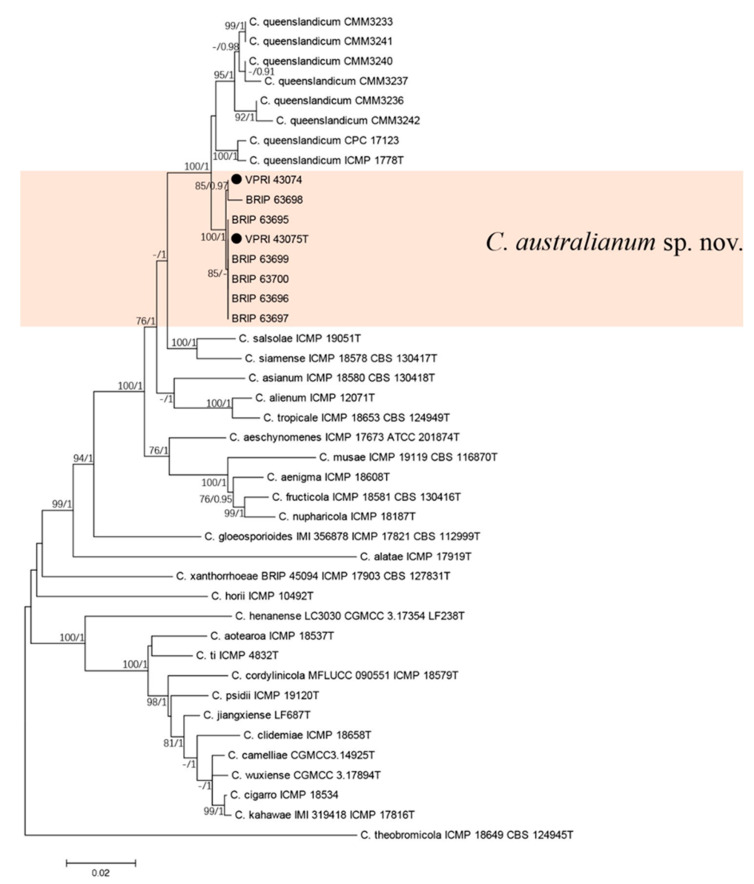
The two gene phylogenetic analysis consisted of six chili (Capsicum annuum) and two citrus isolates of the new Colletotrichum species, 34 reference sequences from the C. gloeosporioides species complex, including eight isolates of C. queenslandicum. Colletotrichum theobromicola (ICMP 18649T) was used as the out-group. A total of 1820 characters (ApMat: 900, gs: 910 and 10 N to separate two sequences) were analysed. The Bayesian analysis lasted 115,000 generations, resulting in 1709 total trees, of which 1282 trees were used to calculate the posterior probabilities. The BI posterior probabilities were plotted on the ML tree (Figure 4).
Figure 4.
Phylogenetic analysis of the combined ApMat and GS sequence alignment of Colletotrichum australianum sp. nov. The bootstrap support values (ML > 75%) of maximum likelihood analysis and Bayesian posterior probabilities (PP > 0.90) are displayed at the nodes (ML/PP). Black circle denotes isolates from Citrus spp.
The six isolates from chili [39] clustered with the two citrus isolates of the new Colletotrichum species in the two-gene tree, which were significantly different from C. queenslandicum (Figure 4).
Taxonomy
Morphological characters and phylogenetic analyses indicated that the Colletotrichum species isolated from infected mandarin and orange fruits collected from Melbourne and Dunkeld, Victoria, respectively, and isolated from infected chili fruit collected from Brisbane, Queensland, Australia, was a new species, for which the name Colletotrichum australianum is proposed.
Colletotrichum australianum W. Wang, D. D. De Silva, and P. W. J. Taylor, sp. nov. (Figure 5).
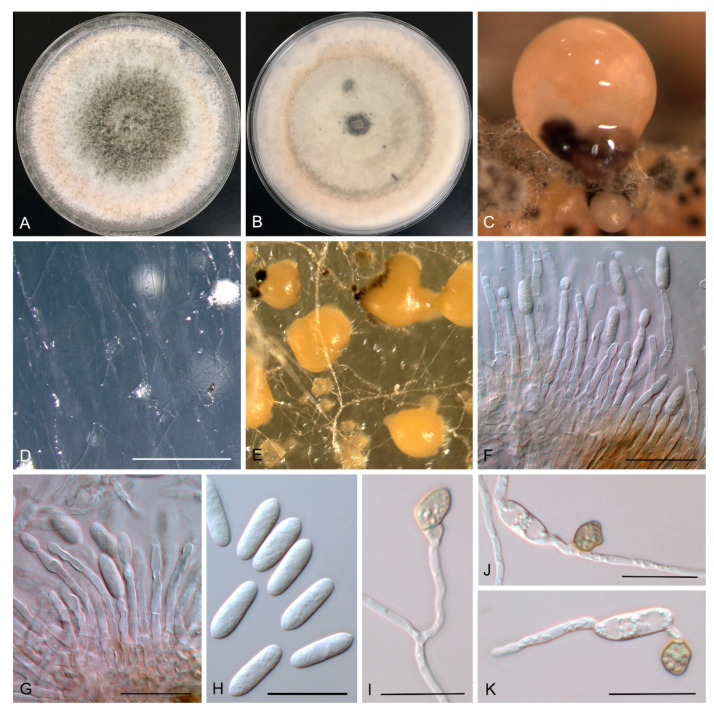
Figure 5.
Morphological characteristics of Colletotrichum australianum sp. nov.: One-week-old culture on PDA (A,B), conidiomata on mandarin rind (C), conidiomata on SNA (D), conidiomata on PDA (E), conidiophores (F,G), conidia (H) and appressoria (I–K). Scale bars: D, 500 µm; F, G, H, I, J, K, 20 µm.
MycoBank Number: MB830323.
Etymology: Named after the country where the pathogen was first isolated, Australia.
Holotype: Australia, Victoria, Dunkeld, on fruit of Citrus sinensis, May 2016, J. Kennedy (VPRI 43075–holotype; UMC002–ex-type culture).
Asexual morph on SNA. Conidiomata on SNA inconspicuous or absent, 41–140 µm diam, formed from hyphae, lacking setae. Conidia hyaline, smooth, aseptate, straight, cylindrical with one end slightly acute, granular, and guttulate, (13.2–) 14.4–14.6 (–15.9) × (4.8–) 5.6–5.7 (–6.1) µm. Appressoria single, medium to dark brown, ovoid with an undulate margin, (6.1–) 8.5–8.9 (–12.2) × (4.6–) 6.7–7.1 (–9.3) µm.
Asexual morph on PDA. Conidiomata on PDA formed on hyphae or on a brown central stroma, lacking setae. Conidiophores hyaline, smooth-walled, septate, branched, 28–58 × 2–3 μm. Conidiogenous cells hyaline, smooth-walled, subcylindrical, straight to curved, phialidic with visible periclinal thickening at the apex, 14–30 × 2–3 µm. Conidia hyaline, smooth, aseptate, straight, cylindrical with one end acute, granular and guttulate, (12.7–) 14.1–14.5 (–17.2) × (3.9–) 4.5–4.7 (–5.5) µm. Appressoria single, medium to dark brown, ovoid with an undulate margin, (7.2–) 8.1–8.3 (–9.5) × (5.4–) 6.5–6.7 (–7.6) µm.
Mycelia on mandarin rind were colourless to white. Conidiomata salmon, smooth. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical with one end acute, granular and guttulate, (12.9–) 14.7–15.1 (–16.1) × (4.3–) 4.8–5 (–5.4) µm.
Culture characteristics: Colonies on SNA flat, entire margin, hyaline, 45–55 mm diam in 7 d. Colonies on PDA 65–75 mm in 7 d; pale yellow to white aerial mycelia, changing to grey in the centre, reverse have a uniform concentric ring with pinkish outside and inside pale grey to grey in the centre. Colonies on MEA flat, entire margin, white aerial mycelia, 52–78 mm in 7 d.
Notes: Colletotrichum australianum is phylogenetically close to C. queenslandicum but are separable using ApMat and gs sequences. The closest match in a Blastn search with the gs sequence was GenBank KP703693, C. queenslandicum strain CPC 17123, with 98 % identity.
3.4. Pathogenicity Assay
For the fruit bioassay, C. australianum, C. fructicola, C. theobromicola, Colletotrichum sp., and C. karstii developed brown lesions on wounded orange fruits. Colletotrichum karstii had the highest infection incidence at 100%, while the C. gloeosporioides isolate did not cause obvious symptoms (Table 4). None of the Colletotrichum species were able to infect non-wounded orange fruit.
Table 4.
Incidence of infection on Washington Navel orange fruit and leaves and Meyer lemon leaves by Colletotrichum species.
| Culture | Fungus Species | Infection Incidence % | |||
|---|---|---|---|---|---|
| Fruit Bioassay (Wound) | Leaf Bioassay | Petal Bioassay | |||
| Orange Leaf | Lemon Leaf | ||||
| VPRI 43075 | C. australianum sp. nov. | 95.8 | 0 | 0 | 100 |
| VPRI 43076 | C. gloeosporioides | 0 | 0 | 0 | 100 |
| VPRI 43079 | C. fructicola | 75 | 0 | 0 | 100 |
| VPRI 43083 | C. theobromicola | 95.8 | 0 | 83.3 | 100 |
| BRIP 58074a | Colletotrichum sp. | 95.8 | 0 | 0 | 100 |
| VPRI 43654 | C. karstii | 100 | 100 | 100 | 100 |
For the leaf bioassay, C. karstii developed lesions on both orange and lemon leaves, while C. theobromicola only developed lesions on lemon leaves (Table 4). Other Colletotrichum isolates did not cause obvious symptoms on both orange and lemon leaves.
In the petal bioassay, all isolates infected orange petals.
4. Discussion
Six Colletotrichum species were identified from citrus stems, leaves, and fruits with anthracnose symptoms in Australia. Colletotrichum australianum was isolated from orange and mandarin fruit in Victoria, Australia, and identified and described as a new species causing anthracnose of citrus in Australia. Isolates from chili (Capsicum annuum) from Queensland and previously identified as C. queenslandicum [39] were also reidentified as C. australianum. Phylogenetic analyses clearly showed C. australianum to be a new species closely related to C. queenslandicum. There were also differences in morphological characters between these two species. The ApMat and gs sequences clearly distinguished C. australianum. These genes are considered as informative markers to identify species within the C. gloeosporioides species complex [10,36,51,53].
Colletotrichum gloeosporioides sensu lato was the most frequently isolated in diseased citrus. There was no preference for a particular Citrus sp. or infected organ tissue. Colletotrichum gloeosporioides was isolated from various citrus species, including cumquat, finger lime, grapefruit, lemon, lime, mandarin, orange, Persian lime, and Tahitian lime. Colletotrichum gloeosporioides was previously cultured from lemon (Citrus limon) and orange (Citrus sinensis) in Australia [37]. The isolate VPRI 10347 from lemon from Victoria and previously identified as C. nymphaeae [37] was also reidentified as C. gloeosporioides. The prevalence of Colletotrichum species that cause anthracnose of citrus in Australia, is in accordance with recent global studies on the major cause of anthracnose of citrus [8,11,12,13,14,19,20,21,23,24,27].
This is the first report in Australia of Colletotrichum siamense being associated with citrus anthracnose. Colletotrichum siamense was isolated from lemon fruit and finger lime fruit and has been recorded as a pathogen of a broad range of plants in Australia [37,39]. Colletotrichum siamense was previously reported to be isolated from catmon (Citrus pennivesiculata) in Bangladesh and Egypt, mandarin (C. reticulata Blanco cv. Shiyue Ju) in China, and mandarin (C. reticulata cv. Kinnow) in Pakistan [11,31,32,54]. Colletotrichum siamense isolate BRIP 54270b was collected in 2011 in Queensland, suggesting C. siamense has been a citrus pathogen for at least 10 years in Australia. However, both C. siamense isolates were collected from citrus fruits, and no C. siamense isolate was found on citrus leaves or stems, suggesting C. siamense is more likely to be a postharvest pathogen of citrus in Australia.
Colletotrichum theobromicola is for the first time reported as a pathogen of citrus. Colletotrichum theobromicola was isolated from lime fruit from Queensland but was recently neotypified from cacao tree (Theobroma cacao) in Panama [36]. Colletotrichum theobromicola has been recorded as a pathogen of a broad range of plants in Australia including jointvetch (Aeschynomene falcata), arabica coffee (Coffea arabica), olive (Olea europaea), pomegranate (Punica granatum), stylo (Stylosanthes guianensis), and sticky stylo (Stylosanthes viscosa) [37].
Colletotrichum fructicola was reported for the first time, associated with anthracnose symptoms from mandarin fruit in Australia. Isolate BRIP 65028 from Tahitian lime growing in Queensland was previously identified as C. fructicola in 2018 [38]. Colletotrichum fructicola was also isolated from avocado (Persea americana) in Australia [37]. In China, C. fructicola was reported to be associated with bergamot orange (Citrus bergamia), pomelo (C. grandis), mandarin (C. reticulata cv. nanfengmiju), oranges (C. sinensis), and kumquat (Fortunella margarita) [26,27,30]. Colletotrichum fructicola was found to cause both preharvest and postharvest citrus disease in Australia.
Colletotrichum karstii was the second dominant pathogen and was isolated from infected orange and lemon leaves in both New South Wales and Victoria. Colletotrichum karstii is the only species in the boninense species complex found to be associated with citrus anthracnose in Australia. Three C. karstii isolates were collected from orange leaves in the 1970s and were maintained in State fungaria, suggesting C. karstii has been a citrus pathogen for over 50 years in Australia but was misidentified as C. gloeosporioides. Colletotrichum karstii was reported to infect citrus and to have a wide global distribution [8,11,13,16,23,25,26,27,28,29]. Previously, C. karstii was reported from other hosts such as black plum (Diospyros australis), strawberry (Fragaria x ananassa), and banana (Musa banksia) in Australia [37].
Six Colletotrichum isolates from chili (Capsicum annuum) that had been previously identified as causing anthracnose fruit rot of chili in Brisbane, Queensland, Australia [39], were also identified as C. australianum. These six Colletotrichum isolates were morphologically similar to C. australianum from citrus rather than the type specimen of C. queenslandicum (ICMP 1778), which was originally isolated from infected papaya. The identification of C. australianum from diverse hosts such as orange, mandarin, and chili, suggests that C. australianum may have a broad host range. Further studies are required on the host range of this pathogen, which may have biosecurity implication for the export of Australian fruit. The occurrence of C. australianum in both Victoria and Queensland indicates the wide geographic spread across different climatic zones in Australia.
The species identification of Colletotrichum isolates based on ApMat and gs gene sequences were as similar as the results from phylogenetic analysis of seven-gene combination, proving that the locus ApMat was effective in identifying Colletotrichum species within the gloeosporioides species complex. The phylogenetic analysis of combined ApMat and gs sequences can identify species within the gloeosporioides species complex [10,47,51,53]. The efficiency of the ApMat gene to identify species was also supported by Sharma et al. [55] and Sharma, Pinnaka, and Shenoy [56], who differentiated Colletotrichum isolates in India. The isolate VPRI 10347 was identified to be Colletotrichum nymphaeae in Shivas et al. [37] based on single tub2 sequence. However, in this study, ApMat and gs gene sequences identified isolate VPRI 10347 as C. gloeosporioides, same as the result from phylogenetic analysis of the seven-gene combination. However, the limitation of using the ApMat gene in constructing phylogenetic trees is that several reference Colletotrichum species in the gloeosporioides species complex in GenBank have not been sequenced for ApMat. For example, the isolate VPRI 43083 was phylogenetically close to C. grevilleae and C. grossum based on analysis of combined ITS and tub2 gene sequences (Supplementary Figure S1) but due to a lack of ApMat sequence of C. grevilleae and C. grossum, these species were not included in either the seven-gene nor the two-gene trees, whereas VPRI 43083 was identified as C. theobromicola based on seven gene combination and two gene combination analyses with high bootstrap value. Due to a lack of replicate isolates, as well as a lack of reference sequences, especially ApMat gene data of Colletotrichum species close to BRIP 58074a, the unknown Colletotrichum sp. (BRIP 58074a) isolate cannot be further described taxonomically or phylogenetically at this stage.
Colletotrichum acutatum has been reported from lemon (DAR 80516, from Tasmania in 2009, and DAR 72160, from NSW in 1998) previously [38]. However, C. acutatum was not found in this study. The two C. acutatum isolates were identified based on morphology but have not been confirmed by molecular analysis. Gene sequences of isolates DAR 80516 and DAR 72160 should be analysed to accurately identify these two isolates.
Pathogenicity tests of five Colletotrichum species from citrus showed that all species except for C. gloeosporioides were capable of infecting wounded fruit. In contrast, none of the five Colletotrichum species caused disease on the non-wounded fruit. These results are consistent with previous reports where wound inoculated citrus fruits were used in postharvest pathogenicity testing of Colletotrichum species [8,27]. Variable maturity of the fruit may also be a reason for lack of infection. Mature fruits are reported to be more sensitive to Colletotrichum species [57]. The fruit used for inoculation in this study may not have been fully mature, although they were selected based on the colour of mature fruit; thus, they were not conducive for Colletotrichum spores to attach to the cuticle, germinate, and form appressoria prior to infection.
Different Colletotrichum species had various degrees of aggressiveness on wounded orange fruit and non-wounded orange and lemon leaves. Colletotrichum karstii was the most aggressive species when infecting orange fruit and orange and lemon leaves. The variable aggressiveness of different Colletotrichum species has been reported by Guarnaccia et al. [8]. Colletotrichum gloeosporioides isolate VPRI 43076 was non-pathogenic on fruit and leaves but was pathogenic on orange petals. Conversely, Guarnaccia et al. [8] reported C. gloeosporioides to be the most aggressive species when infecting orange fruit. Pathogenic variation has been reported within populations of a Colletotrichum species [10,58,59]. Hence, VPRI 43076 was likely to have been an isolate of Colletotrichum gloeosporioides, which had weak aggressiveness on citrus fruit. Further assessment of pathogenicity of isolates from each species needs to be undertaken to determine the variability of aggressiveness.
5. Conclusions
Six Colletotrichum spp. were identified to cause anthracnose of citrus in Australia that included one novel species C. australianum, and one undetermined species. In addition, this was the first report of C. theobromicola as a pathogen of citrus globally, and the first report of C. karstii and C. siamense to be associated with citrus anthracnose in Australia.
Supplementary Materials
The following are available online at https://www.mdpi.com/2309-608X/7/1/47/s1: Figure S1: Phylogram generated from maximum likelihood analysis of all available Colletotrichum species in the gloeosporioides species complex and the boninense species complex based on combined ITS and tub2 sequences data, Table S1: Information of the 36 Colletotrichum isolates selected for phylogenetic analyses.
Author Contributions
Conceptualization: W.W., J.E., P.K.A. and P.W.J.T.; data curation: W.W. and J.E.; formal analysis: W.W., D.D.d.S., A.M., and J.E.; funding acquisition: P.W.J.T. and J.E.; methodology: W.W., D.D.d.S., P.K.A., and P.W.J.T.; project administration: P.W.J.T.; resources: W.W. and P.W.J.T.; supervision: P.K.A., P.W.C., and P.W.J.T.; writing—original draft: W.W.; writing—review and editing: W.W., D.D.d.S., A.M., J.E., P.K.A., P.W.C., and P.W.J.T. All authors have read and agreed to the published version of the manuscript.
Funding
W.W. was supported by a University of Melbourne postgraduate scholarship. Financial support was also received from the Innovation Seed Fund for Horticulture Development between Agriculture Victoria Research and The University of Melbourne.
Institutional Review Board Statement
Not applicable
Informed Consent Statement
Not applicable
Data Availability Statement
Alignments generated during the current study are available in TreeBASE (accession http://purl.org/phylo/treebase/phylows/study/TB2:S27542). All sequence data are available in NCBI GenBank following the accession numbers in the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Spiegel-Roy P., Goldschmidt E.E. The Biology of Citrus. Cambridge University Press; Cambridge, UK: 2008. [Google Scholar]
- 2.Ladaniya M. Citrus Fruit. Academic Press; London, UK: 2008. [Google Scholar]
- 3.Fruit and Vegetables. [(accessed on 4 March 2020)]; Available online: https://www.austrade.gov.au/search.aspx?ModuleID=8367&keywords=citrus&multiSite=False.
- 4.Cooke T., Persley D., House S. Diseases of Fruit Crops in Australia. CSIRO Publishing; Collingwood, ON, Canada: 2009. [Google Scholar]
- 5.Australian Citrus Production Regions. [(accessed on 4 October 2020)]; Available online: https://citrusaustralia.com.au/growers-industry.
- 6.The 2018 Annual Report. [(accessed on 4 October 2020)]; Available online: https://citrusaustralia.com.au/wp-content/uploads/CAL-Annual-Report-2018-FINAL.pdf.
- 7.Phoulivong S., McKenzie E.H.C., Hyde K.D. Cross Infection of Colletotrichum Species; A Case Study with Tropical Fruits. Curr. Res. Environ. Appl. Mycol. 2012;2:99–111. doi: 10.5943/cream/2/2/2. [DOI] [Google Scholar]
- 8.Guarnaccia V., Groenewald J.Z., Polizzi G., Crous P.W. High Species Diversity in Colletotrichum Associated with Citrus Diseases in Europe. Pers. Mol. Phylogeny Evol. Fungi. 2017;39:32–50. doi: 10.3767/persoonia.2017.39.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marin-Felix Y., Groenewald J.Z., Cai L., Chen Q., Marincowitz S., Barnes I., Bensch K., Braun U., Camporesi E., Damm U., et al. Genera of Phytopathogenic Fungi: GOPHY 1. Stud. Mycol. 2017;86:99–216. doi: 10.1016/j.simyco.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Silva D.D., Groenewald J.Z., Crous P.W., Ades P.K., Nasruddin A., Mongkolporn O., Taylor P.W.J. Identification, Prevalence and Pathogenicity of Colletotrichum Species Causing Anthracnose of Capsicum Annuum in Asia. IMA Fungus. 2019;10:1–32. doi: 10.1186/s43008-019-0001-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Douanla-Meli C., Unger J. Phylogenetic Study of The Colletotrichum Species on Imported Citrus Fruits Uncovers A Low Diversity and A New Species in The Colletotrichum gigasporum Complex. Fungal Biol. 2017;121:858–868. doi: 10.1016/j.funbio.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Yin L., Du S., Cai M., Luo C. Identification of Anthracnose of Wild Citrus. Southwest China J. Agric. Sci. 2017;30:590–594. [Google Scholar]
- 13.Aiello D., Carrieri R., Guarnaccia V., Vitale A., Lahoz E., Polizzi G. Characterization and Pathogenicity of Colletotrichum gloeosporioides and C. karstii causing Preharvest Disease on Citrus Sinensis in Italy. J. Phytopathol. 2014;163:168–177. doi: 10.1111/jph.12299. [DOI] [Google Scholar]
- 14.Benyahia H., Jrifia A., Smaili C., Afellah M., Lamsetef Y., Timmer L.W. First Report of Colletotrichum gloeosporioides Causing Withertip on Twigs and Tear Stain on Fruit of Citrus in Morocco. Plant Pathol. 2003;52:798. doi: 10.1111/j.1365-3059.2003.00907.x. [DOI] [Google Scholar]
- 15.Cruz-Lagunas B., Ortega-Acosta S.A., Reyes-García G., Toribio-Jiménez J., Juárez-López P., Guillén-Sánchez D., Damián-Nava A., Romero-Ramírez Y., Palemón-Alberto F. Colletotrichum gloeosporioides Causes Anthracnose on Grapefruit (Citrus paradisi) In Mexico. Australas. Plant Dis. Notes. 2020;15:1–4. doi: 10.1007/s13314-020-00401-z. [DOI] [Google Scholar]
- 16.Pérez-Mora J.L., Mora-Romero G.A., Beltrán-Peña H., García-León E., Lima N.B., Camacho-Tapia M., Tovar-Pedraza J.M. First Report of Colletotrichum siamense and C. gloeosporioides Causing Anthracnose of Citrus Spp. In Mexico. [(accessed on 21 December 2020)];Plant Dis. 2020 Available online: https://pubmed.ncbi.nlm.nih.gov/32910725/ [Google Scholar]
- 17.Fayyaz A., Sahi S.T., Nawaz-ul-Rehman M.S., Amrao L. Molecular Characterization of Fungal Pathogens Associated with Citrus Withertip/Dieback from Major Citrus Growing Areas of Punjab, Pakistan. Int. J. Agric. Biol. 2020;23:1165–1170. [Google Scholar]
- 18.Honger J.O., Offei S.K., Oduro K.A., Odamtten G.T., Nyaku S.T. Identification and Species Status of The Mango Biotype of Colletotrichum gloeosporioides In Ghana. Eur. J. Plant Pathol. 2014;140:455–467. doi: 10.1007/s10658-014-0480-z. [DOI] [Google Scholar]
- 19.Honger J.O., Offei S.K., Oduro K.A., Odamtten G.T., Nyaku S.T. Identification and Molecular Characterisation of Colletotrichum species From Avocado, Citrus and Pawpaw in Ghana. S. Afr. J. Plant Soil. 2016;33:177–185. doi: 10.1080/02571862.2015.1125958. [DOI] [Google Scholar]
- 20.Lima W.G., Spósito M.B., Amorim L., Gonçalves F.P., de Filho P.A.M. Colletotrichum gloeosporioides, A New Causal Agent of Citrus Post-Bloom Fruit Drop. Eur. J. Plant Pathol. 2011;131:157–165. doi: 10.1007/s10658-011-9795-1. [DOI] [Google Scholar]
- 21.Mahiout D., Bendahmane B.S., Benkada M.Y., Mekouar H., Berrahal N., Rickauer M. First Report of Colletotrichum gloeosporioides on Citrus in Algeria. Phytopathol. Mediterr. 2018;57:355–359. [Google Scholar]
- 22.Ramos A.P., Talhinhas P., Sreenivasaprasad S., Oliveira H. Characterization of Colletotrichum gloeosporioides, As the Main Causal Agent of Citrus Anthracnose, and C. karstii As Species Preferentially Associated with Lemon Twig Dieback in Portugal. Phytoparasitica. 2016;44:549–561. doi: 10.1007/s12600-016-0537-y. [DOI] [Google Scholar]
- 23.Daoud H.B.H., Baraldi E., Iotti M., Leonardi P., Boughalleb-M’hamdi N. Characterization and Pathogenicity of Colletotrichum Spp. Causing Citrus Anthracnose in Tunisia. Phytopathol. Mediterr. 2019;58:175–185. [Google Scholar]
- 24.Rhaiem A., Taylor P.W.J. Colletotrichum gloeosporioides Associated with Anthracnose Symptoms on Citrus, A New Report for Tunisia. Eur. J. Plant Pathol. 2016;146:219–224. doi: 10.1007/s10658-016-0907-9. [DOI] [Google Scholar]
- 25.Damm U., Cannon P.F., Woudenberg J.H.C., Johnston P.R., Weir B.S., Tan Y.P., Shivas R.G., Crous P.W. The Colletotrichum boninense Species Complex. Stud. Mycol. 2012;73:1–36. doi: 10.3114/sim0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng L., Yang Y., Hyde K.D., Bahkali A.H., Liu Z. Colletotrichum Species on Citrus Leaves in Guizhou And Yunnan Provinces, China. Cryptogam. Mycol. 2012;33:267–283. [Google Scholar]
- 27.Huang F., Chen G.Q., Hou X., Fu Y.S., Cai L., Hyde K.D., Li H.Y. Colletotrichum Species Associated with Cultivated Citrus in China. Fungal Divers. 2013;61:61–74. doi: 10.1007/s13225-013-0232-y. [DOI] [Google Scholar]
- 28.Mayorquin J.S., Nouri M.T., Peacock B.B., Trouillas F.P., Douhan G.W., Kallsen C., Eskalen A. Identification, Pathogenicity, And Spore Trapping of Colletotrichum karstii Associated with Twig and Shoot Dieback in California. Plant Dis. 2019;103:1464–1473. doi: 10.1094/PDIS-08-18-1425-RE. [DOI] [PubMed] [Google Scholar]
- 29.Uysal A., Kurt Ş. First Report of Colletotrichum karstii Causing Anthracnose on Citrus in The Mediterranean Region of Turkey. J. Plant Pathol. 2019;101:753. doi: 10.1007/s42161-018-00215-0. [DOI] [Google Scholar]
- 30.Hu W.L., Ma Y.Z., Chen J.Z. First Report of Citrus sinensis Anthracnose Caused by Colletotrichum fructicola in China. Plant Dis. 2019;103:1018. doi: 10.1094/PDIS-08-18-1466-PDN. [DOI] [Google Scholar]
- 31.Cheng B.P., Huang Y.H., Song X.B., Peng A.T., Ling J.F., Chen X. First Report of Colletotrichum siamense Causing Leaf Drop and Fruit Spot of Citrus reticulata Blanco cv. Shiyue Ju in China. Plant Dis. 2013;97:1508. doi: 10.1094/PDIS-04-13-0352-PDN. [DOI] [PubMed] [Google Scholar]
- 32.Fayyaz A., Bonello P., Tufail M.R., Amrao L., Habib A., Gai Y., Sahi S.T. First Report of Citrus Withertip (Tip Dieback), A Disease Complex Caused by Colletotrichum siamense And Lasiodiplodia iraniensis, On Citrus reticulata cv. Kinnow in Punjab, Pakistan. Plant Dis. 2018;102:2659. doi: 10.1094/PDIS-04-18-0576-PDN. [DOI] [Google Scholar]
- 33.Damm U., Cannon P.F., Woudenberg J.H.C., Crous P.W. The Colletotrichum acutatum Species Complex. Stud. Mycol. 2012;73:37–113. doi: 10.3114/sim0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kunta M., Park J.W., Vedasharan P., da Graça J.V., Terry M.D. First Report of Colletotrichum queenslandicum on Persian Lime Causing Leaf Anthracnose in The United States. Plant Dis. 2018;102:677. doi: 10.1094/PDIS-09-17-1382-PDN. [DOI] [Google Scholar]
- 35.Perrone G., Magistà D., Ismail A.M. First report of Colletotrichum kahawae subsp. Ciggaro on mandarin in italy. J. Plant Pathol. 2016;98:682. [Google Scholar]
- 36.Weir B.S., Johnston P.R., Damm U. The Colletotrichum gloeosporioides Species Complex. Stud. Mycol. 2012;73:115–180. doi: 10.3114/sim0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shivas R.G., Tan Y.P., Edwards J., Dinh Q., Maxwell A., Andjic V., Liberato J.R., Anderson C., Beasley D.R., Bransgrove K., et al. Colletotrichum Species in Australia. Australas. Plant Pathol. 2016;45:447–464. doi: 10.1007/s13313-016-0443-2. [DOI] [Google Scholar]
- 38.Australian Plant Pest Database|Plant Health Australia. [(accessed on 22 November 2020)]; Available online: https://www.planthealthaustralia.com.au/resources/australian-plant-pest-database/
- 39.De Silva D.D., Ades P.K., Crous P.W., Taylor P.W.J. Colletotrichum species Associated with Chili Anthracnose in Australia. Plant Pathol. 2016;66:254–267. doi: 10.1111/ppa.12572. [DOI] [Google Scholar]
- 40.Rayner R.W. A Mycological Colour Chart. Taylor & Francis, Ltd.; Abingdon, UK: 1970. [Google Scholar]
- 41.Johnston P.R., Jones D. Relationships Among Colletotrichum Isolates from Fruit-Rots Assessed Using Rdna Sequences. Mycologia. 1997;89:420–430. doi: 10.1080/00275514.1997.12026801. [DOI] [Google Scholar]
- 42.Gardes M., Bruns T.D. ITS Primers with Enhanced Specificity for Basidiomycetes—Application to the Identification of Mycorrhizae and Rusts. Mol. Ecol. 1993;2:113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 43.White T.J., Bruns T.D., Lee S.B., Taylor J.W. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. PCR Protoc. Guide Methods Appl. 1990;18:315–322. [Google Scholar]
- 44.Guerber J.C., Liu B., Correll J.C., Johnston P.R. Characterization of Diversity in Colletotrichum acutatum sensu lato By Sequence Analysis of Two Gene Introns, Mtdna And Intron Rflps, And Mating Compatibility. Mycologia. 2003;95:872. doi: 10.1080/15572536.2004.11833047. [DOI] [PubMed] [Google Scholar]
- 45.Carbone I., Kohn L.M. A Method for Designing Primer Sets for Speciation Studies in Filamentous Ascomycetes. Mycologia. 1999;91:553–556. doi: 10.1080/00275514.1999.12061051. [DOI] [Google Scholar]
- 46.Woudenberg J.H.C., Aveskamp M.M., de Gruyter J., Spiers A.G., Crous P.W. Multiple Didymella Teleomorphs Are Linked to The Phoma Clematidina Morphotype. Pers. Mol. Phylogeny Evol. Fungi. 2009;22:56–62. doi: 10.3767/003158509X427808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silva D.N., Talhinhas P., Várzea V., Cai L., Paulo O.S., Batista D. Application of The Apn2/MAT Locus to Improve the Systematics of The Colletotrichum gloeosporioides Complex: An Example from Coffee (Coffea spp.) Hosts. Mycologia. 2012;104:396–409. doi: 10.3852/11-145. [DOI] [PubMed] [Google Scholar]
- 48.Stephenson S.A., Green J.R., Manners J.M., Maclean D.J. Cloning and Characterisation of Glutamine Synthetase From Colletotrichum gloeosporioides And Demonstration of Elevated Expression During Pathogenesis on Stylosanthes guianensis. Curr. Genet. 1997;31:447–454. doi: 10.1007/s002940050228. [DOI] [PubMed] [Google Scholar]
- 49.Crous P.W., Groenewald J.Z., Risède J.M., Simoneau P., Hywel-Jones N.L. Calonectria Species and Their Cylindrocladium Anamorphs: Species with Sphaeropedunculate Vesicles. Stud. Mycol. 2004;50:415–430. doi: 10.3114/sim.55.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu F., Weir B.S., Damm U., Crous P.W., Wang Y., Liu B., Wang M., Zhang M., Cai L. Unravelling Colletotrichum Species Associated with Camellia: Employing Apmat And GS Loci to Resolve Species in The C. gloeosporioides Complex. Pers. Mol. Phylogeny Evol. Fungi. 2015;35:63–86. doi: 10.3767/003158515X687597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nylander J.A.A. MrModeltest v. 2. Program Distributed by the Author. Evolutionary Biology Centre, Uppsala University; Uppsala, Sweden: 2004. [Google Scholar]
- 53.Liu F., Wang M., Damm U., Crous P.W., Cai L. Species Boundaries in Plant Pathogenic Fungi: A Colletotrichum Case Study. BMC Evol. Biol. 2016;16:81. doi: 10.1186/s12862-016-0649-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.James R.S., Ray J., Tan Y.P., Shivas R.G. Colletotrichum siamense, C. theobromicola and C. queenslandicum from Several Plant Species and The Identification of C. asianum in The Northern Territory, Australia. Australas. Plant Dis. Notes. 2014;9:1–6. doi: 10.1007/s13314-014-0138-x. [DOI] [Google Scholar]
- 55.Sharma G., Kumar N., Weir B.S., Hyde K.D., Shenoy B.D. The Apmat Marker Can Resolve Colletotrichum Species: A Case Study with Mangifera Indica. Fungal Divers. 2013;61:117–138. doi: 10.1007/s13225-013-0247-4. [DOI] [Google Scholar]
- 56.Sharma G., Pinnaka A.K., Shenoy B.D. Resolving the Colletotrichum siamense Species Complex Using Apmat Marker. Fungal Divers. 2014;71:247–264. doi: 10.1007/s13225-014-0312-7. [DOI] [Google Scholar]
- 57.Pitt J.I., Hocking A.D. Fungi and Food Spoilage. 3rd ed. Springer-Verlag; New York, NY, USA: 2009. [Google Scholar]
- 58.Montri P., Taylor P.W.J., Mongkolporn O. Pathotypes of Colletotrichum capsici, The Causal Agent of Chili Anthracnose, in Thailand. Plant Dis. 2009;93:17–20. doi: 10.1094/PDIS-93-1-0017. [DOI] [PubMed] [Google Scholar]
- 59.Talhinhas P., Gonçalves E., Sreenivasaprasad S., Oliveira H. Virulence Diversity of Anthracnose Pathogens (Colletotrichum acutatum and C. gloeosporioides Species Complexes) On Eight Olive Cultivars Commonly Grown in Portugal. Eur. J. Plant Pathol. 2014;142:73–83. doi: 10.1007/s10658-014-0590-7. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Alignments generated during the current study are available in TreeBASE (accession http://purl.org/phylo/treebase/phylows/study/TB2:S27542). All sequence data are available in NCBI GenBank following the accession numbers in the manuscript.