Abstract
Rice grain yield is a complex trait determined by three components: panicle number, grain number per panicle (GNPP) and grain weight. GNPP is the major contributor to grain yield and is crucial for its improvement. GNPP is determined by a series of physiological and biochemical steps, including inflorescence development, formation of rachis branches such as primary rachis branches and secondary rachis branches, and spikelet specialisation (lateral and terminal spikelets). The molecular genetic basis of GNPP determination is complex, and it is regulated by numerous interlinked genes. In this review, panicle development and the determination of GNPP is described briefly, and GNPP-related genes that influence its determination are categorised according to their regulatory mechanisms. We introduce genes related to rachis branch development and their regulation of GNPP, genes related to phase transition (from rachis branch meristem to spikelet meristem) and their regulation of GNPP, and genes related to spikelet specialisation and their regulation of GNPP. In addition, we describe other GNPP-related genes and their regulation of GNPP. Research on GNPP determination suggests that it is possible to cultivate rice varieties with higher grain yield by modifying GNPP-related genes.
Keywords: grain number per panicle, grain yield, phase transition, rachis branch, rice panicle, spikelet specialisation
1. Introduction
Rice is one of the most important food crops and feeds half of the world’s population [1,2]. Because of reductions in available land and the increasing global population, increasing the rice grain yield per unit area is crucial for food security, particularly in developing countries in Asia, such as in China and India [3].
Rice grain yield is primarily determined by three traits—grain number per panicle (GNPP), grain weight and number of panicles [4]. Because the rice grain yield per unit area is high, increasing the GNPP could further improve the grain yield [4,5,6]. Much research has been focused on GNPP determination, and considerable progress has been made in understanding the underlying regulatory mechanism. Here, we review progress in the molecular and genetic aspects of GNPP determination in rice.
2. Panicle Development and GNPP Determination in Rice
2.1. Panical Development
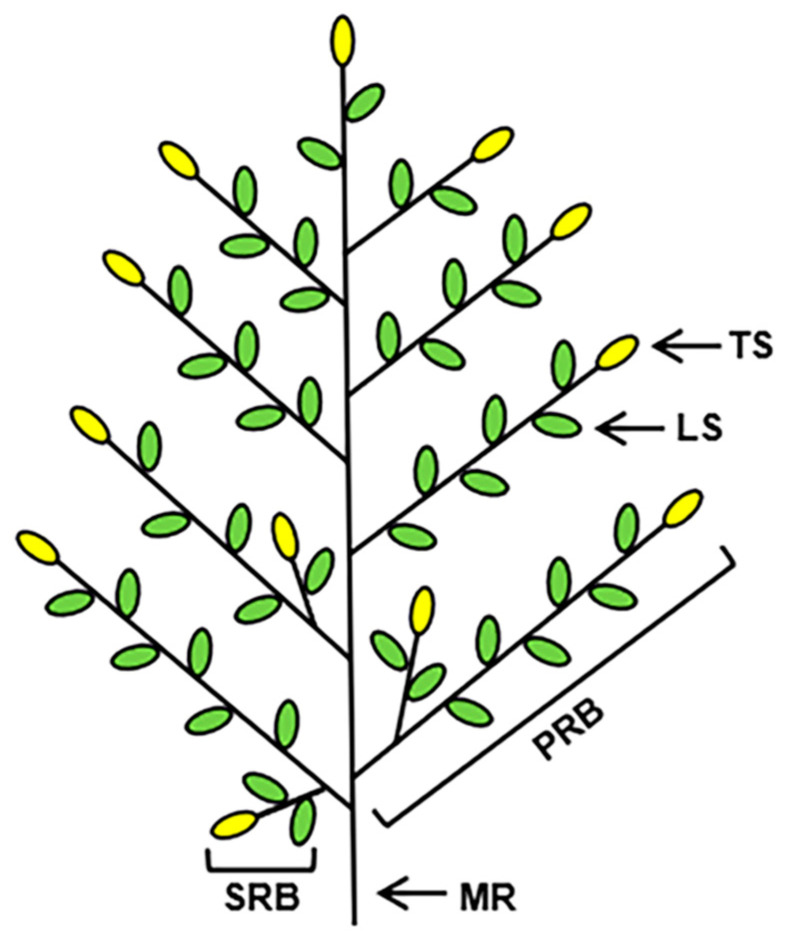
The rice panicle is composed of the main rachis, rachis branches (primary rachis branches (PRBs) and secondary rachis branches (SRBs)) and spikelets (lateral and terminal spikelets) (Figure 1) [3,7]. In a few rice varieties, there are tertiary rachis branches in the panicle. GNPP determination involves the development of the inflorescence, formation of rachis branches and spikelet specialisation. During rice panicle development, the inflorescence meristem (IM) is an important regulator of GNPP formation [8]. In rice, transition to the reproductive phase involves the transformation of the shoot apical meristem (SAM) into the IM, initiating the growth of several lateral meristems as PRBs. Next, the IM loses its activity, leaving a vestige at the base of the uppermost PRB. The PRB meristem produces next-order branches as lateral meristems. The few initially formed lateral meristems grow as SRBs and later meristems directly from spikelet meristems. Lateral spikelets differentiate directly from newly formed lateral meristems, and terminal spikelets are converted from rachis branch meristems. Therefore, three factors—rachis branch formation, the transition from rachis branch meristem to spikelet meristem and spikelet specialisation—determine the overall architecture of the panicle and the GNPP in rice [9].
Figure 1.
Panicle architecture of rice. The green ellipses show the lateral spikelets and the yellow ellipses show the terminal spikelets. LS, lateral spikelet; MR, main rachis; PRB, primary rachis branch; SRB, secondary rachis branch; TS, terminal spikelet.
2.2. GNPP Determination in Rice
Plant hormones, such as auxin, gibberellin (GA), cytokinin (CK), abscisic acid (ABA) and ethylene, are involved in regulating panicle development and GNPP in rice [10,11]. Auxin has a pivotal role in panicle development, as it is required for the initiation and maintenance of axillary meristems. Auxin is produced mainly in growing shoot apices and is transported basipetally down the site along specific transport routes through polar transport machinery. Consequently, disruption in auxin synthesis or auxin transport results in fewer rachis branches and reduced GNPP in rice [12,13,14]. GA can affect panicle-associated traits including panicle length, rachis branch number and GNPP in rice [15]. A previous study demonstrated that OsCYP71D8L controls panicle-related traits by regulating GA homeostasis. Gain-of-function of OsCYP71D8L leads to shorter panicles, fewer rachis branches, and reduced GNPP in rice [16]. It has been reported that the fine-tuning of bioactive CK level in the IM is a critical trait for controlling the number of rachis branches and GNPP in rice. The decreased level of bioactive CK in rice IM is usually accompanied by fewer rachis branches and reduced GNPP [17,18], and the weakened CK signalling in rice IM also results in fewer rachis branches and reduced GNPP [19,20]. This evidence suggests that CK positively regulates GNPP in rice. On the other hand, the stress hormones such as ABA and ethylene negatively regulate the GNPP in rice [11,21]. In addition, signalling cascades and responses of several plant hormones overlap, and the molecular components are often shared among them. A complex network of effectors of multiple hormonal pathways collide and communicate to regulate critical agronomic traits including GNPP in rice [10].
Increasing evidence indicates that plant hormones mediate GNPP determination mainly through the transcriptional or post-transcriptional regulation of GNPP-related genes in rice [10,22,23]. Additionally, GNPP-related genes control panicle development mainly by regulating three factors, including rachis branch formation, the transition from rachis branch meristem to spikelet meristem and spikelet specialisation [9,17,18].
3. Functional Classification of GNPP-Related Genes and Their Regulation of GNPP
GNPP-related genes control the GNPP mainly by regulating rachis branch formation, the transition from the rachis branch meristem to the spikelet meristem and spikelet specialisation. To date, numerous genes that control GNPP by regulating rachis branch formation [24,25,26], and several that regulate the transition from the rachis branch meristem to the spikelet meristem [9,27,28,29,30], have been investigated. However, few genes involved in regulating spikelet specialisation have been reported [31,32,33]. Typically, inhibition of rachis branch meristem formation and acceleration of the conversion from the rachis branch meristem to the spikelet meristem reduce the GNPP by decreasing the number of rachis branches and vice versa [30,32,34]. In addition, the inhibition of spikelet specialisation may reduce the GNPP by inhibiting spikelet formation and vice versa [32,35].
3.1. Rachis Branch Development-Related Genes and Their Regulation of GNPP
3.1.1. Positive Regulation of GNPP by Rachis Branch Development-Related Genes
LAX PANICLE (LAX) positively regulates the number of rachis branches and GNPP (Table 1). LAX encodes a plant-specific bHLH transcription factor. During rice panicle development, LAX is mainly expressed at the boundary region between the apical meristem and the newly formed lateral meristem, and it plays an important role in lateral meristem formation. Loss of function of LAX leads to a decreased number of rachis branches and a reduced GNPP. LAX cDNA is 1080 bp in length and contains one exon, encoding a protein of 215 amino acids. In the lax-1 mutant, retrotransposon insertion causes a premature termination codon, and translation is terminated prematurely. In the lax-2 mutant, there is a 36-kb deletion encompassing LAX. In the lax-3 mutant, there is a 59-bp deletion in the bHLH domain of LAX. In the lax-4 mutant, alanine 49 in the bHLH domain of LAX is changed to threonine. In the lax-5 mutant, there is an arginine at position 50 of the bHLH domain of LAX [32]. Additionally, lax-1, lax-4 and lax-5 are lax mutants with mild changes in phenotype, with a reduced number of rachis branches, inhibited lateral spikelet development, normal terminal spikelet development and a reduced GNPP. By contrast, the panicles of severe lax mutants (lax-2 and lax-3) have the main rachis but no rachis branches, and lateral spikelet development is completely blocked, whereas terminal spikelet development is inhibited [32]. Thus, the GNPP is reduced significantly in severe lax mutants. In a stronger allelic variant of the lax mutant, lax1-2, the initiation and maintenance of the rachis branch meristem, lateral spikelet meristem and terminal spikelet meristem are severely impaired. The transition from the partial rachis branch meristem to the spikelet meristem is delayed in the lax1-2 mutant. The mild mutant lax1-1 has no lateral spikelets but terminal spikelets and a small GNPP instead, whereas the severe mutant lax1-2 has no spikelets and a few PRBs and SRBs [26].
Table 1.
GNPP-related genes and their regulation of GNPP. GNPP, grain number per panicle; Phase transition, from rachis branch meristem to spikelet meristem; PRB, primary rachis branch; SRB, secondary rachis branch; −, negative regulation; +, positive regulation.
| Gene | RGAP Locus ID | Protein Products | PRB | SRB | Phase Transition | Spikelet Specialisation | Lateral Spikelet | Terminal Spikelet | GNPP | References |
|---|---|---|---|---|---|---|---|---|---|---|
| LAX1 | LOC_Os01g61480 | bHLH transcription factor | + | + | + | + | + | + | [26,32] | |
| IPA1/WFP/OsSPL14 | LOC_Os08g39890 | SOUAMOSA PROMOTER BINDING PROTEIN-LIKE protein | + | + | + | [36,37] | ||||
| MOC1 | LOC_Os06g40780 | GRAS-family nuclear protein | + | + | + | [24,38] | ||||
| OSH1 | LOC_Os03g51690 | A protein product of 364 amino acids | + | + | + | [22,39,40,41] | ||||
| SPA | Not reported | Small panicle | + | + | + | + | [32] | |||
| SP1 | LOC_Os11g12740 | A putative transporter of the peptide transporter (PTR) family | + | + | + | [25] | ||||
| DEP1 | LOC_Os09g26999 | G protein gamma subunit | + | + | + | [42] | ||||
| OsNAC2/OMTN2/Ostil1 | LOC_Os04g38720 | NAC transcription factor | + | + | + | [43,44,45,46] | ||||
| GN4-1 | Not reported | Grain number4-1 | + | + | + | [4,47] | ||||
| GNP1 | LOC_Os03g63970 | GA20-oxidase 1 | + | + | [48] | |||||
| Gnp4/LAX2 | LOC_Os04g32510 | A nuclear protein with a plant-specific conserved domain | + | + | [49,50] | |||||
| PAY1 | LOC_Os08g31470 | Peptidase | + | + | [51] | |||||
| LP/EP3 | LOC_Os02g15950 | F-box protein | − | − | − | [52,53,54] | ||||
| DEP3/OspPLAIIIδ | LOC_Os06g46350 | Patatin-related phospholipase A | − | − | − | [55,56] | ||||
| PAP2/OsMADS34 | LOC_Os03g54170 | SEP-like MADS box transcription factor | − | − | + | − | [34,57,58,59,60] | |||
| An-1 | LOC_Os04g28280 | bHLH protein | − | − | − | [61] | ||||
| PROG1 | LOC_Os07g05900 | A 161-amino-acid Cys2-His2 zinc-finger protein | − | − | − | [62,63,64] | ||||
| DST | LOC_Os03g57240 | Zinc-finger transcription factor | − | − | − | [8,65] | ||||
| GN2 | Not reported | Gain number2 | − | − | [5] | |||||
| APO1/SCM2 | LOC_Os06g45460 | F-box protein of 429 amino acids | + | + | − | + | [9,27,28] | |||
| RCN1 | LOC_Os11g05470 | ATP-binding cassette transporter | + | − | + | [29] | ||||
| RCN2 | LOC_Os02g32950 | ATP-binding cassette transporter | + | − | + | [29] | ||||
| TAW1 | LOC_Os10g33780 | Nuclear protein | + | + | − | − | + | [30] | ||
| FZP/BFL1 | LOC_Os07g47330 | ERF transcription factor | − | + | + | + | [33,35,66] | |||
| FZP2 | Not reported | Frizzy panicle 2 | − | + | + | + | [31] | |||
| GN1a/OsCKX2 | LOC_Os01g10110 | Cytokinin oxidase/dehydrogenase | − | − | − | [17] | ||||
| LOG | LOC_Os01g40630 | Cytokinin riboside 50-monophosphate phosphoribohydro-lase | + | + | + | [18] | ||||
|
PYL1
PYL4 PYL6 |
LOC_Os01g61210 LOC_Os03g18600 LOC_Os05g39580 |
ABA receptor protein | − | − | − | [11] | ||||
| Ghd7 | LOC_Os07g15770 | CCT(CO, CO-LIKE and TIMING OF CAB1) | + | + | + | [67,68,69] | ||||
| Ghd8/DTH8/OsHAP3H/LHD1 | LOC_Os08g07740 | HAP3 subunit of the HAP (heterotrimeric haem activator) complex | + | + | + | [70,71,72,73] | ||||
| DTH7/Ghd7.1/OsPRR37 | LOC_Os07g49460 | A pseudo-response regulator protein | + | + | + | [74,75,76,77] | ||||
| GAD1 | LOC_Os08g37890 | A cysteine-rich secretory peptide | + | [78] | ||||||
| NOG1 | LOC_Os01g54860 | Enoyl-CoA hydratase/isomerase | + | [79] |
Ideal Plant Architecture 1 (IPA1)/WEALTHY FARMER’S PANICLE (WFP)/SOUAMOSA PROMOTER BINDING PROTEIN-LIKE 14 (OsSPL14) positively regulates the number of rachis branches and the GNPP (Table 1). IPA1/WFP/OsSPL14 encodes a SOUAMOSA PROMOTER BINDING PROTEIN-LIKE protein. Rice varieties with IPA1/WFP/OsSPL14 have an ideal plant architecture, including more rachis branches and a greater GNPP. Overexpression of OsSPL14 increases the number of rachis branches and the GNPP [36]. Moreover, OsmiR156 interacts with and lyses OsSPL14. As a result of a point mutation, OsSPL14ipa1 cannot be lysed by OsmiR156, resulting in a significant increase in the number of PRBs and SRBs and in the GNPP in panicles of OsSPL14ipa1 plants [37]. Therefore, IPA1/WFP/OsSPL14 positively regulates the number of rachis branches and the GNPP, and upregulating IPA1/WFP/OsSPL14 or enhancing the IPA1/WFP/OsSPL14 protein level increases the number of rachis branches and the GNPP, resulting in an increased grain yield [36,37].
MONOCULM1(MOC1) positively regulates tiller number, rachis branch number and GNPP in rice (Table 1). The MOC1 cDNA is 1666 bp in length and contains four exons. MOC1 encodes a GRAS-family nuclear protein consisting of 441 amino acids with a VHIID motif and an SH2-like domain. MOC1 is expressed mainly in the axillary buds, initiating the buds and promoting their outgrowth. In moc1, a 1.9-kb retrotransposon is inserted at position 948, causing a premature translation stop, resulting in a truncated 338-amino-acid fusion protein with the last 22 residues encoded by the retrotransposon sequence. The moc1 mutant has only a main culm without tillers as a result of a defect in tiller bud formation [24]. Moreover, MOC1 regulates the GNPP mainly by controlling IM activity and bract primordium initiation in rice. Therefore, in moc1 mutant plants, the number of PRBs and SRBs and the GNPP are decreased by the reduced IM activity and inhibited rachis primordium initiation [24,38]. These findings indicate that MOC1 positively regulates the tiller number, number of rachis branches and GNPP in rice.
Oryza sativa Homeobox1 (OSH1) positively regulates the number of rachis branches and the GNPP in rice (Table 1). OSH1 cDNA is 1086 bp in length and contains five exons, encoding a protein product of 364 amino acids. OSH1, a rice homolog of Knotted1-like Homeobox (KNOX), a KNOX family class 1 homeobox gene, participates in SAM initiation and maintenance [39,40]. OSH1 is self-induced and binds to five KNOX loci (OSH1, OSH6, OSH15, OSH43 and OSH71), inducing their expression [22]. KNOX inhibits the expression of the GA biosynthesis gene OsGA20ox to reduce the GA content and promotes the expression of CK biosynthesis genes (including OsIPT2 and OsIPT3) to increase the CK content [23]. KNOX induces SAM formation and maintains SAM activity by maintaining a high CK content and low GA content [22]. The self-induced expression of OSH1 is necessary for SAM self-maintenance. Compared with the wild type, osh1 mutant plants cannot maintain SAM activity, resulting in reduced numbers of PRBs and SRBs and a reduced GNPP [41].
Small Panicle (SPA) positively regulates the number of rachis branches and the GNPP (Table 1). SPA is functionally redundant with LAX1 and regulates the rice axillary meristem. In spa, a SPA loss-of-function mutant, the rachis branches are short and abnormal, most base PRBs are missing and the numbers of SRBs and lateral spikelets are decreased significantly, resulting in a reduced number of rachis branches and a smaller GNPP. In a spa and lax1-1 (a weak allelic mutant of lax1) double mutant, the panicle becomes a linear structure lacking all rachis branches. In addition, spa or lax1 has little effect on tillering, which is almost completely inhibited in the spa lax1 double mutant [32]. Therefore, SPA positively regulates the GNPP by promoting PRB, SRB and lateral spikelet formation.
Short Panicle 1 (SP1) positively regulates panicle size, the length of the main rachis and rachis branches, the number of rachis branches and GNPP (Table 1). SP1 encodes a putative transporter of the peptide transporter (PTR) family. SP1 contains a conserved PTR2 domain consisting of 12 transmembrane domains, and the SP1-GFP fusion protein was found to be localised in the plasma membrane. SP1 is highly expressed in the phloem of the rachis branches of young panicles and may control panicle size by regulating rachis growth. SP1 has been suggested to be a nitrate transporter. However, transport of neither nitrate nor any other compound transported by PTR proteins has been detected, suggesting that SP1 requires other component(s) to function as a transporter, or that it transports unknown substrates [25]. Compared with the wild type, panicle elongation in sp1 plants (loss of function of SP1) is defective, resulting in a shorter panicle. Moreover, the number of SRBs and the GNPP are significantly reduced as a result of the decreased number of PRBs [25]. Therefore, SP1 positively regulates the GNPP by regulating the growth of the main rachis and rachis branches in rice.
DENSE AND ERECT PANICLE1 (DEP1) positively regulates the number of rachis branches and the GNPP (Table 1). DEP1 is located at a major quantitative trait locus (QTL) that controls rice grain yield. DEP1, a key gene with multiple functions including the control of rice grain yield, was isolated from Shennong 265, a super rice variety in northeast China. The dominant allele at the DEP1 locus is a gain-of-function mutation causing truncation of a phosphatidylethanolamine-binding protein-like domain protein [42]. This allele increases meristem activity, increasing the number of rachis branches (PRBs and SRBs) and the GNPP, consequently increasing grain yield by 15–20% [42]. This allele is common to many high-yield rice varieties planted in a large area of China, suggesting that DEP1 has played an important role in increasing rice grain yield in China.
OsNAC2/OMTN2/Ostil1 positively regulates the panicle length, rachis branch number and GNPP (Table 1). OsNAC2/OMTN2/Ostil1 encodes a NAC transcription factor. OsNAC2 is located at the same locus as OMTN2 and Ostil1 [43,44,45]. The microRNA miR164b interacts with and lyses OsNAC2. Overexpressing OErN (mutation of OsNAC2), which is not lysed by miR164b, leads to increased panicle length, stem thickness and number of vascular bundles in stem and leaves, resulting in increased numbers of PRBs and SRBs and an increased GNPP and grain yield [46]. IPA1 and DEP1 are significantly upregulated in OErN plants, and regulation by OsNAC2 of the number of rachis branches and the GNPP may be related to CK signalling. OsNAC2-RNAi plants, transgenic rice plants with low expression of OsNAC2, have a shorter panicle, thinner stem, decreased number of vascular bundles in the stem and leaf, decreased numbers of PRBs and SRBs, and a smaller GNPP and thus have a lower grain yield. In addition, overexpression of miR164b in the transgenic rice OE164 resulted in the fragmentation of OsNAC2. Compared with wild-type plants, OE164 had shorter panicles, lower GNPP and a lower grain yield [46].
Grain Number4-1 (GN4-1) positively regulates the number of rachis branches, and GN4-1 has a marked effect on the GNPP (Table 1) [47]. Insertion of a near-isogenic line (NIL; GN4-1) from Wuyunjing 8 into Zhonghui 8006 increases the number of PRBs and SRBs, the GNPP and the grain yield. Compared with Zhonghui 8006, the expression levels of OsCKX2 (cytokinin oxidase) and other cytokinin oxidase genes (OsCKX1, OsCKX4, OsCKX7, OsCKX8, OsCKX9, OsCKX10 and OsCKX11) were decreased significantly in NIL (GN4-1), elevating the contents of CKs (zeatin, zeatin riboside) and IM activity and increasing the number of PRBs and SRBs and the GNPP in NIL (GN4-1) [4]. GN4-1 from Wuyunjing 8 promotes CK accumulation in rice inflorescences and increases the GNPP by 17% [4]. Therefore, GN4-1 regulates the GNPP by controlling the CK content in rice inflorescences.
Grain Number per Panicle1 (GNP1) positively regulates the number of SRBs and the GNPP (Table 1). A T-DNA insertion mutant (gnp1-D) with enhanced GNP1 expression exhibits increased plant height, more rachis branches and a greater GNPP. Compared with the NIL-GNP1LT (isogenic control) line, the total and solid grain numbers in the NIL-GNP1TQ line increased by 56% and 28%, respectively. In the NIL-GNP1TQ line, although there was no obvious increase in PRB number, the SRB number increased significantly, significantly increasing grain yield at multiple experimental sites in China [48]. GNP1 may upregulate KNOX expression, inducing the expression of the CK biosynthesis gene OsIPT, and enhance IM activity by increasing the CK content and enhancing CK signalling. Moreover, GNP1 reduces the contents of active GAs (GA1 and GA3) by promoting their inactivation via the upregulation of GA2ox expression, which may positively regulate SRB number and GNPP [48]. Therefore, GNP1 may enhance IM activity by enhancing CK signalling and suppressing GA signalling, thus increasing the SRB number and GNPP [48].
Grain Number per Panicle Gene4 (Gnp4)/LAX PANICLE2 (LAX2) encodes a nuclear protein that regulates the formation of the axillary meristem and positively regulates the number of SRBs and the GNPP (Table 1). Gnp4 is located within a 10.7-kb region in the long arm of rice chromosome 4. There is no sequence difference between the mutant and the wild type in this region, but there are differences in cytosine methylation levels in the CpG island region of the candidate gene promoter [49]. LAX2 is a nuclear protein with a plant-specific conserved domain that interacts with LAX1 and regulates axillary meristem formation in rice [50]. Gnp4 is located at the same locus as LAX2 [49,50]. The SRB of the gnp4 mutant does not produce spikelets, and the SRB number and GNPP of the gnp4 lax1-1 double mutant are significantly reduced [49]. The spikelets in the panicles of the lax2 mutant are sparse, and the SRB number and GNPP of the lax2-1 mutant are significantly reduced, although there is no difference in PRB number between lax2-1 and wild-type plants. The decreases in rachis branch number and the GNPP are greater in the lax1 lax2 double mutant than in either single mutant [50]. Therefore, Gnp4/LAX2 positively regulates the number of SRBs and the GNPP in rice.
PLANT ARCHITECTURE AND YIELD1 (PAY1) positively regulates the SRB number and the GNPP (Table 1). Zhao et al. constructed a mutant library by mutating YIL55 (infiltration system of wild-type rice) via EMS mutagenesis and investigated a mutant (PAY1) with an upright and compact architecture. YIL55 had a shorter plant height, more tillers, a larger tiller angle, a lower GNPP and lower grain yield [51]. By contrast, PAY1 mutants had a taller plant height, fewer tillers, a smaller tiller angle, thicker culm, more SRBs, a larger GNPP and lower grain yield but exhibited no significant changes in PRB number. Auxin polar transport activity is weakened in PAY1, and so PAY1 may regulate rice phenotype by influencing auxin polar transport and altering the distribution of endogenous indole-3-acetic acid (the most important auxin in higher plants) [51]. Compared with wild-type plants, PAY1-OE plants had a thicker culm, fewer tillers, taller plant height, no significant change in PRB number, more SRBs, a larger GNPP and a higher grain yield. By contrast, PAY1-RNAi plants had a thinner culm, more tillers, shorter plant height, fewer SRBs, a lower GNPP and a lower grain yield [51]. Therefore, PAY1 positively regulates GNPP by increasing the number of SRBs in rice.
LAX1, IPA1/WFP/OsSPL14, MOC1, OSH1, SPA, SP1, DEP1, OsNAC2 and GN4-1 positively regulate the number of rachis branches and the GNPP. Loss of function of these genes reduces the number of PRBs and SRBs and the grain yield, whereas their upregulation or gain of function increases PRB and SRB numbers and the grain yield. By contrast, although GNP1, GNP4/LAX2 and PAY1 positively regulate the number of SRBs and the GNPP, they do not regulate the number of PRBs. However, the mechanisms by which these genes regulate the number of rachis branches and the GNPP are unclear, and some have not been cloned yet. Therefore, the positive regulation by these genes of the number of rachis branches and the GNPP warrants further investigation. It may be possible to increase the number of rachis branches and the GNPP using GNPP-related genes, thus improving grain yield.
3.1.2. Negative Regulation of GNPP by Rachis Branch Development-Related Genes
LARGER PANICLE (LP)/ERECT PANICLE3 (EP3) negatively regulates the number of rachis branches and the GNPP (Table 1). LP encodes an F-box protein rich in Kelch, and in situ hybridisation showed that LP is mainly expressed in the rachis primordium [52]. The F-box protein encoded by EP3 may function as a subunit of E3 ubiquitin ligase in the recognition and degradation of specific substrates [53]. EP3 and LP are located at the same locus [52,53,54]. LP interacts with SKP1-like protein to upregulate the expression of OsCKX2 and decrease the CK level in rice inflorescence, leading to more PRBs and SRBs and a higher grain yield [52]. Compared to wild-type plants, the number of rachis branches, particularly of PRBs, increased significantly in an lp (loss of function of LP) mutant, increasing the GNPP and grain yield in lp mutant plants [52]. In addition, lp mutant plants are more lodging resistant compared to wild-type plants [52].
DENSE AND ERECT PANICLE3 (DEP3)/OspPLAIIIδ negatively regulates the number of rachis branches and the GNPP (Table 1). DEP3 and OspPLAIIIδ are located at the same locus [55,56]. Compared with wild-type DEP3 gene, the dep3 mutant allele loses 408 bp at LOC_Os06g46350, including the back 47 bp of the region encoding the third exon and the front 361 bp of the untranslated region (UTR) of the 3’-end [55]. The panicles of wild-type plants typically begin to droop after the flowering stage, but the panicles of dep3 mutants remain upright from the flowering stage to the fully mature stage. Moreover, there are differences in the number of vascular bundles and the vascular bundle size, and in other phenotypic characteristics (including panicle length, rachis branch length and culm thickness) between dep3 and wild-type plants. The dep3 mutant plants have more vascular bundles in the upmost internode, a smaller vascular bundle, shorter panicle, shorter rachis branch and thicker culm [55]. In addition, compared with wild-type plants, dep3 mutant plants have more PRBs and SRBs, a larger GNPP and a higher grain yield [55]. These results show that DEP3 negatively regulates the number of rachis branches and the GNPP, thus increasing the grain yield.
PANICLE PHYTOMER 2 (PAP2)/OsMADS34 negatively regulates the number of rachis branches and the GNPP (Table 1). PAP2 encodes an SEP-like MADS box transcription factor. PAP2 is expressed only in the inflorescence meristem, PRB meristem, lateral spikelet meristem, floret meristem, glumes and degenerated glumes during the panicle development stage [34]. The PRB number, SRB number and GNPP of a pap2-1 (PAP2 loss of function) mutant increased significantly compared with those of wild-type plants. The panicle size of pap2-1 is slightly smaller than that of wild-type plants because panicle elongation in the mutant is inhibited, but plant height, tillering number and leaf number do not differ significantly between pap2-1 and wild-type plants [34]. OsMADS34, also known as PAP2, is a specific SEP-like MADS box gene in gramineous plants that regulates panicle morphology by controlling the rachis number and GNPP [34,57,58]. OsMADS34 is expressed in the root, stem, leaf, leaf sheath, panicle, glume and degenerated glume and strongly expressed in developing organs such as young panicles. OsMADS34 determines the grain size and rice grain yield. In osmads34-t mutants, the numbers of PRBs and SRBs are increased but the panicle length is reduced, and the GNPP is increased but the grain size and seed-setting rate are reduced [59]. Six MADS-box genes—OsMADS50, OsMADS56, OsMADS22, OSMADS47, OsMADS55 and OsMADS34—have been detected in rice [60]. These MADS-box genes significantly increase the number of rachis branches, including PRBs and SRBs, by inhibiting the expression of REDUCED CULM NUMBER 4. In addition, knockout of OsMADS50, OsMADS56, OsMADS22, OsMADS47 and OsMADS55 in an osmads34 mutant significantly increased the number of PRBs and SRBs and the GNPP [60]. These findings indicate that PAP2/OsMADS34 negatively regulates the number of rachis branches and the GNPP in rice.
Awn-1 (An-1) encodes a bHLH protein that positively regulates awn length and negatively regulates rachis branch number and GNPP in rice (Table 1). During spikelet development, An-1 is first expressed in two degenerate glumes and two empty glume primordia, next in lemma primordia and palea primordia, and finally in stamens and carpel primordia. An-1 expression gradually increases at the tip of the lemma primordium and is significantly enhanced in the awn primordium from the sixth to the eighth stage, gradually decreasing thereafter [61]. The An-1 allele was introduced into Guanglu-Ai-4, an indica rice without awns, yielding the near-isogenic line NIL-An-1. The awn length and grain length increased, but the number of rachis branches and the GNPP decreased significantly in NIL-An-1 compared with wild-type plants. Upregulation of An-1 expression in inflorescences decreases IM activity, thus reducing the number of rachis branches and the GNPP. An-1-OX, a transgenic plant with high expression of An-1, has fewer PRBs and SRBs and a lower GNPP. By contrast, the number of rachis branches and the GNPP are increased in An-1-RNAi plants, in which An-1 expression is knocked down [61].
PROSTRATE GROWTH1 (PROG1) positively regulates prostate growth and negatively regulates the number of PRBs and SRBs and the GNPP in rice (Table 1). The semi-dominant gene PROG1 is located between RM298 and RM481, the short-arm simple sequence repeat markers of chromosome 7 [62]. PROG1 has been isolated and cloned by two research teams [62,63]. The PROG1 cDNA is 833 bp long and encompasses a 486-bp open reading frame, a 147-bp 5’ UTR and a 200-bp 3’ UTR. PROG1 encodes a 161-amino-acid Cys2-His2 zinc-finger protein, which is mainly expressed in the axillary meristem [62,64]. There is a base mutation in the coding region of the gene in cultivated rice that causes an amino acid substitution, which may be selected during artificial domestication [63]. During the evolution of rice, PROG1 of wild rice evolved into prog1 of cultivated rice, resulting in the loss of function of PROG1, which mediates not only the transition from prostrate growth to erect growth but also changes in panicle architecture—e.g., increasing the numbers of PRBs and SRBs, the GNPP and the grain yield of cultivated rice. One hundred eighty-two varieties of cultivated rice, including indica and japonica cultivars from 17 countries, carry identical mutations in the prog1 coding region, suggesting that prog1 has become fixed during artificial domestication in rice [62]. Therefore, PROG1 negatively regulates the number of rachis branches and the GNPP in rice.
DROUGHT AND SALT TOLERANCE (DST) negatively regulates the CK content, reducing the number of rachis branches and the GNPP in rice (Table 1). DST is a zinc-finger transcription factor in rice that directly regulates the expression of the cytokinin oxidase-encoding gene OsCKX2, thus increasing the number of rachis branches, GNPP and grain yield [8,65]. DST is mainly expressed in the SAM, PRB, SRB and young spikelets of the developing panicle [8]. DST promotes the expression of OsCKX2, thus reducing the CK content, SAM activity, number of rachis branches and GNPP. DST is a transcription factor with a C2H2 zinc-finger domain, by which DST proteins bind to DST-binding sequence (DBS) elements [42]. DBS elements are present in the promoter region of OsCKX2 and other OsCKX genes [8]. In the semi-dominant mutant reg1, a single base insertion in DST led to premature termination of protein translation, resulting in the loss of the transcriptional activation ability of DST, accompanied by decreased expression of OsCKX2 and other OsCKX genes, an increased CK content in the IM, increased numbers of PRBs and SRBs, and an enhanced GNPP in rice [8].
Grain Number2 (GN2) negatively regulates the number of rachis branches and the GNPP in rice (Table 1). Chen et al. inserted GN2, a gene from the wild rice Yuanjiang (O. rufipogon Griff.), into indica Teqing and obtained an introgression line (YIL9). Compared with Teqing, the GNPP, panicle length, PRB length and SRB number, but not the PRB number, of YIL9 were reduced [5]. Compared with wild-type plants, GN2-OE, a transgenic plant overexpressing GN2, had a reduced GNPP, shorter panicles and fewer SRBs [5]. Therefore, GN2 negatively regulates grain yield mainly by reducing the number of SRBs and the GNPP in rice.
The above findings indicate that LP/EP3, DEP3, PAP2/OsMADS34, An-1, PROG1 and DST negatively regulate the number of PRBs and SRBs and the GNPP in rice. Mutants with loss of function of these genes have more PRBs and SRBs and a larger GNPP compared to wild-type plants. GN2 does not regulate the number of PRBs, but it negatively regulates the number of SRBs and the GNPP. However, the molecular mechanisms by which these genes regulate the formation of rachis branches and spikelets warrant further study. Elucidation of the molecular regulatory mechanisms of these genes will enable the breeding of rice varieties with more rachis branches, a larger GNPP and higher grain yield using molecular markers and gene editing.
3.2. Phase Transition (Rachis Branch Meristem to Spikelet Meristem)-Related Genes and Their Regulation of GNPP
ABERRANT PANICLE ORGANIZATION 1 (APO1)/SCM2 plays an important role during the phase transition from rachis branch meristem to spikelet meristem and positively regulates the number of rachis branches and the GNPP in rice (Table 1). APO1/SCM2 contains two exons and encodes an F-box protein of 429 amino acids. APO1 is mainly expressed in the SAM and lateral organ primordia. APO1 regulates the timing of meristem transition, and loss of function of APO1 resulted in premature spikelet formation and an extended period of lodicule and carpel formation in an apo1 mutant [27]. In plants with apo1-D, a gain-of-function mutation of APO1, IM activity is prolonged, and conversion from the rachis branch meristem to the lateral spikelet meristem and from the rachis branch meristem to the terminal spikelet meristem is delayed, resulting in a larger IM, more PRBs and SRBs, and higher grain yield than in wild-type plants [9]. By contrast, in plants with apo1, a loss-of-function mutation of APO1, IM activity is shortened, resulting in a smaller IM, fewer PRBs and SRBs, and a decreased GNPP compared to wild-type plants [27]. In addition, the QTL containing SCM2 controls culm thickness. Mapping cloning results showed that SCM2 is equivalent to APO1, and that NILs carrying SCM2 (NIL-SCM2) exhibited increased stem strength, more tillers and a larger GNPP, indicating that SCM2 is pleiotropic. Although SCM2 is a gain-of-function mutant of APO1, there are differences in panicle architecture, including the GNPP and spikelet shape, between SCM2 and APO1 plants [28]. Compared with wild-type plants, the number of rachis branches and the GNPP in NIL-SCM2 plants are obviously increased. However, the GNPP of APO1-OE plants is not larger than that of wild-type plants, and the reason why overexpression of APO1 does not increase the GNPP is unclear. Therefore, APO1/SCM2 plays an important role in controlling the phase transition from rachis branch meristem to spikelet meristem as well as regulating the number of rachis branches and the GNPP in rice [28].
RCN1 and RCN2 increase the number of rachis branches and the GNPP by delaying the phase transition from rachis branch meristem to spikelet meristem (Table 1). RCN1 and RCN2 are rice TERMINAL FLOWER 1 /CENTRORADIALIS-like homologs, which regulate rice plant architecture (tiller number, plant height, panicle architecture) mainly by regulating meristem phase transition [29]. In 35S::RCN1 and 35S::RCN2 transgenic rice plants, phase transition to the reproductive stage is delayed, and transgenic rice plants have more tillers and denser panicles. Observation of the panicle structure revealed that the transition from the rachis branch meristem to the spikelet meristem is delayed, leading to the generation of higher-order rachis branches. Although there is no significant difference in PRB number between transgenic rice (35S::RCN1 and 35S::RCN2) plants and wild-type plants, the number of higher-order rachis branches, including SRBs and tertiary rachis branches, increases significantly, increasing the GNPP in 35S::RCN1 and 35S::RCN2 plants [29]. These findings indicate that RCN1 and RCN2 increase the number of rachis branches and the GNPP by delaying the phase transition to the spikelet meristem.
TAWAWA1 (TAW1) positively regulates the number of rachis branches and the GNPP by regulating the phase transition of the meristem (Table 1). TAW1 encodes a nuclear protein of unknown function and is highly expressed in the SAM, IM and rachis branch meristem. TAW1 regulates inflorescence development by extending IM activity and delaying the phase transition from rachis branch meristem to spikelet meristem [30]. In the dominant gain-of-function mutant tawawa1-D, IM activity is extended and spikelet specialisation is delayed, resulting in delayed IM abortion and prolonged rachis branch formation, thus increasing the number of rachis branches, including SRBs and terminal rachis branches, and the GNPP. By contrast, in TAW1-RNAi transgenic plants, the decreased TAW1 activity causes precocious IM abortion and spikelet formation, resulting in the generation of small inflorescences with reduced numbers of PRBs and SRBs and a decreased GNPP [30].
Therefore, RCN1, RCN2 and TAW1 positively regulate the number of rachis branches and the GNPP by regulating the phase transition from rachis branch meristem to spikelet meristem. Overexpression of these genes and their dominant gain-of-function mutation may delay the phase transition from rachis branch meristem to spikelet meristem and prolong the formation of higher-order rachis branches, including SRBs and terminal rachis branches, thus increasing the GNPP. In addition, compared with wild-type plants, the dominant gain-of-function mutant apo1-D and NIL-SCM2 have more rachis branches and a larger GNPP. Therefore, rational use of these genes can enable the breeding of rice varieties with a large number of rachis branches, greater GNPP and high grain yield.
3.3. Spikelet-Specialisation-Related Genes and Their Regulation of GNPP
FRIZZY PANICLE (FZP)/BFL1 positively regulates spikelet specialisation and GNPP (Table 1). FZP is a single-copy gene in rice, located in chromosome 7, that encodes an ERF transcription factor and is the rice ortholog of maize BD1. FZP is required to prevent the formation of an axillary meristem instead of a spikelet meristem and for the subsequent establishment of spikelet meristem identity [33]. In a fzp mutant, instead of proceeding to spikelet formation, axillary meristems are formed in the axils of rudimentary glumes and are either arrested or develop into higher-order rachis branches, such as SRBs and terminal branches. Therefore, although the fzp mutant has more SRBs and terminal rachis branches, it has fewer spikelets and a reduced GNPP. In addition, there is no significant difference in PRB number between fzp-11 mutant and wild-type plants, but fzp-11 mutants have more SRBs and a lower GNPP compared with wild-type plants [35]. Therefore, FZP may positively regulate the GNPP by promoting spikelet specialisation in rice. Additionally, FZP and BFL1 are located at the same locus [66]. BFL1 encodes a transcription factor with an EREBP/AP2 domain, and BFL1 is involved in mediating spikelet specialisation in rice. The bfl1 mutant harbours a single Ds insertion in the upstream region of BFL1, and Ds insertion drastically reduces the BFL1 transcript level in the bfl1 mutant. Compared to the wild type, the bfl1 mutant has a similar PRB number and more higher-order rachis branches but fewer spikelets and a reduced GNPP as a result of defective spikelet formation [66].
FRIZZY PANICLE 2 (FZP2) positively regulates spikelet specialisation and GNPP (Table 1). FZP2 plays an important role in spikelet specialisation. In fzp2, a loss-of-function mutant, rachis branch meristem activity is prolonged, and the phase transition from rachis branch meristem to spikelet meristem is delayed, significantly increasing the number of rachis branches and inhibiting lateral and terminal spikelet formation, resulting in a decreased GNPP in rice [31].
The above results indicate that FZP/BFL1 and FZP2 regulate GNPP by regulating spikelet specialisation. Compared with wild-type plants, although the number of higher-order rachis branches (PRBs and terminal rachis branches) increases, the number of spikelets and GNPP decrease in fzp and fzp2. Therefore, FZP/BFL1 and FZP2 play a key role in balancing the numbers of rachis branches and spikelets, and the regulation of these genes’ functions increases rice grain yield by balancing the number of rachis branches and spikelets.
4. Other GNPP-Related Genes and Their Regulation of GNPP
Grain Number1a (GN1a)/Cytokinin Oxidase2 (OsCKX2) negatively regulate GNPP by reducing the CK content. The Gn1a locus is the main QTL affecting GNPP in rice. A QTL-Gn1 was identified in the short arm of chromosome 1 using an indica × japonica cross (Habataki/Koshihikari). Further, 96 F2 plants produced by heterozygous NIL-Gn1 (Gn1/gn1) plants were used to divide Gn1 into two loci—Gn1a and Gn1b—with similar functions. Gn1a is located in the region (less than 2 cM) between R3192 and C12072S, and Gn1b is located upstream of Gn1a [17]. Additionally, using 13,000 F2 plants generated from hybrid plants of NIL-Gn1a (Gn1a/gn1a), Gn1a was confirmed to be located in the 6.3-kb region between 3A28 and 3A20, where there is only one open reading frame, namely OsCKX2. Complementary transformation experiments showed that Gn1a is OsCKX2 [17]. Decreased expression or loss of function of OsCKX2 increases the GNPP and grain yield of rice. OsCKX2 is highly expressed in the leaf, stem, IM and spikelet, weakly expressed in the SAM and not expressed in the root or embryo. The SAM is responsible for the development of the aboveground organs, such as leaves, stems and flowers, after embryo transfer. CK plays a key role in maintaining SAM activity. Decreased OsCKX2 expression results in the accumulation of CK in the IM, increasing the number of rachis branches and GNPP (Table 1), and ultimately increasing the grain yield [17].
LONELY GUY (LOG) positively regulates GNPP by promoting CK biosynthesis. LOG encodes a CK-activating enzyme that mediates the final step of CK biosynthesis. LOG is expressed at the apex of the SAM, indicating that CK activation occurs in a specific developmental region. CK promotes the development of the SAM. Loss of function of LOG results in premature termination of SAM activity, decreasing the number of rachis branches and GNPP. LOG may regulate SAM activity by controlling the concentration and spatial distribution of CK [18].
Pyrabactin Resistance-Like (PYL) positively regulates ABA signalling and negatively regulates GNPP in rice. PYLs are ABA receptors implicated in ABA signal transduction [11]. Mutations in ABA receptor genes can promote rice growth and increase grain yield [11]. Rice PYL genes are divided into two groups. Group I includes PYL1–PYL6 and PYL12, and Group II includes PYL7–PYL11 and PYL13. CRISPR/Cas9 has been used to edit PYL genes. Polygenic mutations in Group I can promote the growth of rice, but no such effect has been found for Group II. Compared with the wild type, a pyL1/4/6 mutant exhibited enhanced growth and a higher grain yield in paddy fields with significantly increased panicle length, number of PRBs and SRBs, and GNPP [11]. Therefore, PYLs regulate rice panicle development, possibly in a manner dependent on the ABA signal transduction pathway.
Grain Number, Plant Height, and Heading Date7 (Ghd7) is a major QTL that simultaneously controls the GNPP, plant height and heading stage of rice [67]. Ghd7 encodes a nuclear protein with a CTT (CO-like and TIMING OF CAB1) domain [67]. Proteins containing this structural domain, such as Arabidopsis CONSTANS (CO) and rice Hd1, are implicated in the regulation of flowering time [68,69]. In the photoperiod-regulated flowering pathway, Ghd7 is located upstream of Ehd1 and Hd3a and acts via the Ghd7-Ehd1-Hd3A pathway. Ghd7 does not affect the expression of Hd1, but it does affect the expression of Ehd1 and Hd3a. Under long-day conditions, Ghd7 expression is upregulated, Hd3a expression is inhibited, and the heading date is delayed, enabling rice to make full use of light and temperature, thus increasing panicle length, plant height, GNPP and grain yield. In temperate regions, the short growth period of rice weakens or eliminates the function of Ghd7 in rice varieties in these regions, reducing or avoiding the effect of delayed heading date on rice grain yield. Therefore, the main function of Ghd7 is to prolong the differentiation period of rice panicles and increase panicle length, thus enhancing the production of PRBs and SRBs and the GNPP as well as increasing the grain yield [67].
Grain Number, Plant Height, and Heading Date8 (Ghd8)/DTH8/OsHAP3H/LHD1 regulates GNPP by adjusting the photoperiodic pathway in rice. Ghd8, DTH8, LHD1 and OsHAP3H are located at the same locus [70,71,72,73]. Ghd8 encodes the HAP3 subunit of the HAP (heterotrimeric haem activator) complex [71]. Ghd8 expression is maintained at a high level in meristems at all developmental stages under long-day conditions but is low under short-day conditions. Ghd8 has bidirectional regulatory effects on the Ehd1-Hd3a pathway. Under short-day conditions, Ghd8 upregulates Ehd1, Hd3a and RFT1, resulting in premature heading and flowering in rice. Under long-day conditions, Ghd8 inhibits the expression of these three genes and delays the heading and flowering dates of rice, thus increasing the number of rachis branches and GNPP. Ghd8 may increase the number of tillers, PRBs and SRB as well as the GNPP of rice by upregulating the expression of MOC1 [71].
DTH7/Ghd7.1/OsPRR37 is a pleiotropic gene that controls heading date, plant height and GNPP in rice (Table 1). DTH7/Ghd7.1/OsPRR37 encodes a pseudo-response regulator protein, and its expression is regulated by photoperiod. OsPRR37, Ghd7.1 and DTH7 are located at the same locus [74,75,76,77]. Under long-day conditions, DTH7 acts downstream of photosensitive pigment B and inhibits the expression of the anthocyanin genes Hd3a and Ehd1 in rice, thus delaying flowering [76] and inducing an increase in the number of rachis branches, GNPP and grain yield per plant. Ghd7.1 encodes pseudo-response REGULATOR37 (OsPRR37), which has a CCT domain [77]. OsPRR37 is strongly expressed in the leaf and panicle, especially in the meristems of young panicles. Similar to Ghd7, under long-day conditions, Ghd7.1 does not affect the expression of Hd1, but it does affect the expression of Ehd1 and Hd3a [77]. In indica rice Zhenshan 97B, an eight-base deletion in OsPRR37 leads to premature heading, shorter plant height, fewer rachis branches and a smaller GNPP [77]. Therefore, DTH7/Ghd7.1/OsPRR37 positively regulates GNPP via the photoperiodic pathway in rice.
GRAIN NUMBER, GRAIN LENGTH AND AWN DEVELOPMENT1 (GAD1) negatively regulates GNPP (Table 1). GAD1, located in the long arm of chromosome 8 of rice, encodes a cysteine-rich secretory peptide and has greater homology with the EPIDERMAL PATTERNING FACTOR-LIKE family of Arabidopsis thaliana [78]. The GAD1 protein has a signal peptide site at the N terminus, and the mature peptide has conserved cysteine residues at the C terminus. Common wild rice (W2014) has GAD1, but cultivated rice 93-11 harbours gad1. Wild rice typically has a lower GNPP, longer grains and long awns atop the grains. GAD1 may reduce the CK content by activating the expression of DST and OsCKX2, thus decreasing the GNPP of wild rice [78]. By contrast, a code-shifting mutation of GAD1 in cultivated rice destroys the conserved cysteine structure, leading to loss of function of GAD1, thereby increasing the GNPP, shortening grain length and inhibiting awn development [78].
NUMBER OF GRAINS 1 (NOG1) positively regulates the GNPP in rice (Table 1). In 2017, Chinese scientists cloned the NOG1 gene, which is involved in the regulation of GNPP in rice. The introduction of NOG1 increased grain yield by 25.8% in the NOG1-deficient rice cultivar Zhonghua 17, and overexpression of NOG1 further increased grain yield by 19.5% in the NOG1-containing variety Teqing. NOG1, which encodes an enoyl-CoA hydratase/isomerase, increases the grain yield of rice by enhancing GNPP without reducing the number of panicles per plant or grain weight. Furthermore, NOG1 plays important roles in regulating jasmonic acid homeostasis and the β-oxidation of fatty acids, which may be associated with its regulation of GNPP [79]. The mechanism by which NOG1 regulates GNPP warrants further investigation.
5. Conclusions
GNPP, a grain-yield component in rice, is a hot research topic for breeders and molecular biologists. Several rice GNPP-related genes have been cloned, and their regulation of GNPP has been investigated. LAX1, IPA1/WFP/OsSPL14, MOC1, OSH1, SPA, SP1, DEP1, OsNAC2 and GN4-1 positively regulate the number of rachis branches and GNPP, whereas GNP1, GNP4/LAX2 and PAY1 positively regulate the number of SRBs and GNPP but not the number of PRBs (Table 1). By contrast, LP/EP3, DEP3, PAP2/OsMADS34, An-1, PROG1 and DST negatively regulate the number of PRBs and SRBs and the GNPP in rice. GN2 does not regulate the number of PRBs, but it negatively regulates the number of SRBs and the GNPP. Moreover, RCN1, RCN2, TAW1 and APO1/SCM2 positively regulate the number of rachis branches and the GNPP by regulating the phase transition from rachis branch meristem to spikelet meristem. In addition, FZP/BFL1 and FZP2 regulate the GNPP by regulating spikelet specialisation via balancing the numbers of rachis branches and spikelets (Table 1). Further, other GNPP-related genes and their regulation of GNPP have been reported (Table 1). However, the mechanisms by which some GNPP-related genes regulate GNPP determination are unclear in rice, and some have not been cloned yet. Therefore, the molecular regulatory networks of GNPP determination in rice need to be investigated. These molecular regulatory networks can be uncovered by constructing mutants and using molecular, genetic, physiological and -omics techniques, as well as bioinformatics. Furthermore, molecular marker-assisted selection, transgene techniques, gene editing and genome selection enable directional modification of GNPP-related genes and the polymerisation of favourable GNPP-related genes, allowing the cultivation of rice varieties with higher GNPP and grain yields.
Abbreviations
| ABA | Abscisic acid |
| An-1 | Awn-1 |
| APO1 | ABERRANT PANICLE ORGANIZATION 1 |
| CK | Cytokinin |
| DEP1 | DENSE AND ERECT PANICLE1 |
| DEP3 | DENSE AND ERECT PANICLE3 |
| DST | DROUGHT AND SALT TOLERANCE |
| EP3 | ERECT PANICLE3 |
| FZP | FRIZZY PANICLE |
| FZP2 | FRIZZY PANICLE 2 |
| GA | Gibberellin |
| GAD1 | GRAIN NUMBER, GRAIN LENGTH AND AWN DEVELOPMENT1 |
| Ghd7 | Grain Number, Plant Height, and Heading Date7 |
| Ghd8 | Grain Number, Plant Height, and Heading Date8 |
| GN1a | Grain Number1a |
| GN2 | Grain Number2 |
| GN4-1 | Grain Number4-1 |
| GNP1 | Grain Number per Panicle1 |
| Gnp4 | Grain Number per Panicle Gene4 |
| GNPP | Grain number per panicle |
| HAP | Heterotrimeric haem activator |
| LOG | LONELY GUY |
| IM | Inflorescence meristem |
| IPA1 | Ideal Plant Architecture 1 |
| KNOX | Knotted1-like Homeobox |
| LAX | LAX PANICLE |
| LAX2 | LAX PANICLE2 |
| LP | LARGER PANICLE |
| MOC1 | MONOCULM1 |
| NOG1 | NUMBER OF GRAINS 1 |
| OsCKX2 | Cytokinin oxidase2 |
| OSH1 | Oryza sativa Homeobox1 |
| PAP2 | PANICLE PHYTOMER 2 |
| PAY1 | PLANT ARCHITECTURE AND YIELD1 |
| PROG1 | PROSTRATE GROWTH1 |
| PRR37 | Pseudo-response REGULATOR37 |
| PTR | Peptide transporter |
| PYL | Pyrabactin Resistance-Like |
| QTL | Quantitative trait locus |
| SAM | Shoot apical meristem |
| SRBs | Secondary rachis branches |
| SP1 | Short Panicle 1 |
| SPA | Small Panicle |
| SPL14 | SOUAMOSA PROMOTER BINDING PROTEIN-LIKE 14 |
| TAW1 | TAWAWA1 |
| UTR | Untranslated region |
| WFP | WEALTHY FARMER’S PANICLE |
Author Contributions
Writing—original draft preparation, C.Y.; writing—review and editing, C.Y., Y.Z. and X.L.; supervision, Y.L.; funding acquisition, C.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Fundamental Research Funds for the Central Universities (No. 2662018PY076), the National Key Research and Development Program of China (No. 2016YFD0300102) and the Science and Technology Major Projects of Guangxi Province of China (GuiKeChuang 010106).
Conflicts of Interest
The authors declare that they have no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Demont M., Stein A.J. Global value of GM rice: A review of expected agronomic and consumer benefits. New Biotechnol. 2013;30:426–436. doi: 10.1016/j.nbt.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 2.Xing Y., Zhang Q. Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 2010;61:421–442. doi: 10.1146/annurev-arplant-042809-112209. [DOI] [PubMed] [Google Scholar]
- 3.Lu H., Shi Z. Molecular research progress of rice panicle development. Plant Physiol. J. 2013;49:111–121. [Google Scholar]
- 4.Zhou Y., Tao Y., Yuan Y., Zhang Y., Miao J., Zhang R., Yi C., Gong Z., Yang Z., Liang G. Characterisation of a novel quantitative trait locus, GN4-1, for grain number and yield in rice (Oryza sativa L.) Theor. Appl. Genet. 2018;131:637–648. doi: 10.1007/s00122-017-3025-y. [DOI] [PubMed] [Google Scholar]
- 5.Chen H., Tang Y., Liu J., Tan L., Jiang J., Wang M., Zhu Z., Sun X., Sun C. Emergence of a novel chimeric gene underlying grain number in rice. Genetics. 2017;205:993–1002. doi: 10.1534/genetics.116.188201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L., Bian J., Shi S., Yu J., Khanzada H., Wassan G.M., Zhu C., Luo X., Tong S., Yang X., et al. Genetic analysis for the grain number heterosis of a super-hybrid rice WFYT025 combination using RNA-Seq. Rice. 2018;11:37. doi: 10.1186/s12284-018-0229-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu K., Tao H., Min C., Yang Y. Research progress on rice panicle development. Mol. Plant Breed. 2015;13:2109–2117. doi: 10.13271/j.mpb.013.002109. [DOI] [Google Scholar]
- 8.Li S., Zhao B., Yuan D., Duan M., Qian Q., Tang L., Wang B., Liu X., Zhang J., Wang J., et al. Rice zinc finger protein DST enhances grain production through controlling Gn1a/OsCKX2 expression. Proc. Natl. Acad. Sci. USA. 2013;110:3167–3172. doi: 10.1073/pnas.1300359110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ikeda-Kawakatsu K., Yasuno N., Oikawa T., Iida S., Nagato Y., Maekawa M., Kyozuka J. Expression level of ABERRANT PANICLE ORGANIZATION1 determines rice inflorescence form through control of cell proliferation in the meristem. Plant Physiol. 2009;150:736–747. doi: 10.1104/pp.109.136739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deveshwar P., Prusty A., Sharma S., Tyagi A.K. Phytohormone-Mediated molecular mechanisms involving multiple genes and QTL govern grain number in rice. Front. Genet. 2020;11:586462. doi: 10.3389/fgene.2020.586462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miao C., Xiao L., Hua K., Zou C., Zhao Y., Bressan R.A., Zhu J.-K. Mutations in a subfamily of abscisic acid receptor genes promote rice growth and productivity. Proc. Natl. Acad. Sci. USA. 2018;115:6058–6063. doi: 10.1073/pnas.1804774115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshida A., Ohmori Y., Kitano H., Taguchi-Shiobara F., Hirano H.Y. Aberrant spikelet and panicle1, encoding a TOPLESS-related transcriptional co-repressor, is involved in the regulation of meristem fate in rice. Plant J. 2012;70:327–339. doi: 10.1111/j.1365-313X.2011.04872.x. [DOI] [PubMed] [Google Scholar]
- 13.Li Y., Zhu J., Wu L., Shao Y., Wu Y., Mao C. Functional divergence of PIN1 paralogous genes in rice. Plant Cell Physiol. 2019;60:2720–2732. doi: 10.1093/pcp/pcz159. [DOI] [PubMed] [Google Scholar]
- 14.Malik N., Ranjan R., Parida S.K., Agarwal P., Tyagi A.K. Mediator subunit OsMED14_1 plays an important role in rice development. Plant J. 2020;101:1411–1429. doi: 10.1111/tpj.14605. [DOI] [PubMed] [Google Scholar]
- 15.Gao S., Chu C. Gibberellin metabolism and signaling: Targets for improving agronomic performance of crops. Plant Cell Physiol. 2020;61:1902–1911. doi: 10.1093/pcp/pcaa104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou J., Li Z., Xiao G., Zhai M., Pan X., Huang R., Zhang H. CYP71D8L is a key regulator involved in growth and stress responses by mediating gibberellin homeostasis in rice. Science. 2020;71:1160–1170. doi: 10.1093/jxb/erz491. [DOI] [PubMed] [Google Scholar]
- 17.Ashikari M., Sakakibara H., Lin S., Yamamoto T., Takashi T., Nishimura A., Angeles E.R., Qian Q., Kitano H., Matsuoka M. Cytokinin oxidase regulates rice grain production. Science. 2005;309:741–745. doi: 10.1126/science.1113373. [DOI] [PubMed] [Google Scholar]
- 18.Kurakawa T., Ueda N., Maekawa M., Kobayashi K., Kojima M., Nagato Y., Sakakibara H., Kyozuka J. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature. 2007;445:652–655. doi: 10.1038/nature05504. [DOI] [PubMed] [Google Scholar]
- 19.Sun L., Zhang Q., Wu J., Zhang L., Jiao X., Zhang S., Zhang Z., Sun D., Lu T., Sun Y. Two rice authentic histidine phosphotransfer proteins, OsAHP1 and OsAHP2, mediate cytokinin signaling and stress responses in rice. Plant Physiol. 2014;165:335–345. doi: 10.1104/pp.113.232629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirose N., Makita N., Kojima M., Kamada-Nobusada T., Sakakibara H. Overexpression of a type-A response regulator alters rice morphology and cytokinin metabolism. Plant Cell Physiol. 2007;48:523–539. doi: 10.1093/pcp/pcm022. [DOI] [PubMed] [Google Scholar]
- 21.Wuriyanghan H., Zhang B., Cao W.-H., Ma B., Lei G., Liu Y.-F., Wei W., Wu H.-J., Chen L.-J., Chen H.W., et al. The ethylene receptor ETR2 delays floral transition and affects starch accumulation in rice. Plant Cell. 2009;21:1473–1494. doi: 10.1105/tpc.108.065391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sentoku N., Sato Y., Kurata N., Ito Y., Kitano H., Matsuoka M. Regional expression of the Rice KN1-Type homeobox gene family during embryo, shoot, and flower development. Plant Cell. 1999;11:1651–1664. doi: 10.1105/tpc.11.9.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakamoto T., Sakakibara H., Kojima M., Yamamoto Y., Nagasaki H., Inukai Y., Sato Y., Matsuoka M. Ectopic expression of KNOTTED1-like homeobox protein induces expression of cytokinin biosynthesis genes in rice. Plant Physiol. 2006;142:54–62. doi: 10.1104/pp.106.085811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X., Qian Q., Fu Z., Wang Y., Xiong G., Zeng D., Wang X., Liu X., Teng S., Hiroshi F., et al. Control of tillering in rice. Nature. 2003;422:618–621. doi: 10.1038/nature01518. [DOI] [PubMed] [Google Scholar]
- 25.Li S., Qian Q., Fu Z., Zeng D., Meng X., Kyozuka J., Maekawa M., Zhu X., Zhang J., Li J., et al. Short panicle1 encodes a putative PTR family transporter and determines rice panicle size. Plant J. 2009;58:592–605. doi: 10.1111/j.1365-313X.2009.03799.x. [DOI] [PubMed] [Google Scholar]
- 26.Oikawa T., Kyozuka J. Two-step regulation of LAX PANICLE1 protein accumulation in axillary meristem formation in rice. Plant Cell. 2009;21:1095–1108. doi: 10.1105/tpc.108.065425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikeda K., Ito M., Nagasawa N., Kyozuka J., Nagato Y. Rice ABERRANT PANICLE ORGANIZATION 1, encoding an F-box protein, regulates meristem fate. Plant J. 2007;51:1030–1040. doi: 10.1111/j.1365-313X.2007.03200.x. [DOI] [PubMed] [Google Scholar]
- 28.Ookawa T., Hobo T., Yano M., Murata K., Ando T., Miura H., Asano K., Ochiai Y., Ikeda M., Nishitani R., et al. New approach for rice improvement using a pleiotropic QTL gene for lodging resistance and yield. Nat. Commun. 2010;1:132. doi: 10.1038/ncomms1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakagawa M., Shimamoto K., Kyozuka J. Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J. 2002;29:743–750. doi: 10.1046/j.1365-313X.2002.01255.x. [DOI] [PubMed] [Google Scholar]
- 30.Yoshida A., Sasao M., Yasuno N., Takagi K., Daimon Y., Chen R., Yamazaki R., Tokunaga H., Kitaguchi Y., Sato Y., et al. TAWAWA1, a regulator of rice inflorescence architecture, functions through the suppression of meristem phase transition. Proc. Natl. Acad. Sci. USA. 2013;110:767–772. doi: 10.1073/pnas.1216151110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Komatsu M., Maekawa M., Shimamoto K., Kyozuka J. The LAX1 and FRIZZY PANICLE 2 genes determine the inflorescence architecture of rice by controlling rachis-branch and spikelet development. Dev. Biol. 2001;231:364–373. doi: 10.1006/dbio.2000.9988. [DOI] [PubMed] [Google Scholar]
- 32.Komatsu K., Maekawa M., Ujiie S., Satake Y., Furutani I., Okamoto H., Shimamoto K., Kyozuka J. LAX and SPA: Major regulators of shoot branching in rice. Proc. Natl. Acad. Sci. USA. 2003;100:11765–11770. doi: 10.1073/pnas.1932414100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komatsu M., Chujo A., Nagato Y., Shimamoto K., Kyozuka J. FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development. 2003;130:3841–3850. doi: 10.1242/dev.00564. [DOI] [PubMed] [Google Scholar]
- 34.Kobayashi K., Maekawa M., Miyao A., Hirochika H., Kyozuka J. PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiol. 2010;51:47–57. doi: 10.1093/pcp/pcp166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bai X., Huang Y., Mao D., Wen M., Zhang L., Xing Y. Regulatory role of FZP in the determination of panicle branching and spikelet formation in rice. Sci. Rep. 2016;6:19022. doi: 10.1038/srep19022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miura K., Ikeda M., Matsubara A., Song X.-J., Ito M., Asano K., Matsuoka M., Kitano H., Ashikari M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 2010;42:545–549. doi: 10.1038/ng.592. [DOI] [PubMed] [Google Scholar]
- 37.Jiao Y., Wang Y., Xue D., Wang J., Yan M., Liu G., Dong G., Zeng D., Lu Z., Zhu X., et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010;42:541–544. doi: 10.1038/ng.591. [DOI] [PubMed] [Google Scholar]
- 38.Shao G., Lu Z., Xiong J., Wang B., Jing Y., Meng X., Liu G., Ma H., Liang Y., Chen F., et al. Tiller bud formation regulators MOC1 and MOC3 cooperatively promote tiller bud outgrowth by activating FON1 expression in rice. Mol. Plant. 2019;12:1090–1102. doi: 10.1016/j.molp.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 39.Postma-Haarsma A.D., Verwoert I.I.G.S., Stronk O.P., Koster J., Lamers G.E.M., Hoge J.H.C., Meijer A.H. Characterization of the KNOX class homeobox genes Oskn2 and Oskn3 identified in a collection of cDNA libraries covering the early stages of rice embryogenesis. Plant Mol. Biol. 1999;39:257–271. doi: 10.1023/A:1006153506868. [DOI] [PubMed] [Google Scholar]
- 40.Sato Y., Hong S.K., Tagiri A., Kitano H., Yamamoto N., Nagato Y., Matsuoka M. A rice homeobox gene, OSH1, is expressed before organ differentiation in a specific region during early embryogenesis. Proc. Natl. Acad. Sci. USA. 1996;93:8117–8122. doi: 10.1073/pnas.93.15.8117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsuda K., Ito Y., Sato Y., Kurata N. Positive autoregulation of a KNOX gene is essential for shoot apical meristem maintenance in rice. Plant Cell. 2011;23:4368–4381. doi: 10.1105/tpc.111.090050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang X., Qian Q., Liu Z., Sun H., He S., Luo D., Xia G., Chu C., Li J., Fu X. Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 2009;41:494–497. doi: 10.1038/ng.352. [DOI] [PubMed] [Google Scholar]
- 43.Chen X., Lu S., Wang Y., Zhang X., Lv B., Luo L., Xi D., Shen J., Ma H., Ming F. OsNAC2 encoding a NAC transcription factor that affects plant height through mediating the gibberellic acid pathway in rice. Plant J. 2015;82:302–314. doi: 10.1111/tpj.12819. [DOI] [PubMed] [Google Scholar]
- 44.Fang Y., Xie K., Xiong L. Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice. J. Exp. Bot. 2014;65:2119–2135. doi: 10.1093/jxb/eru072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mao C., Ding W., Wu Y., Yu J., He X., Shou H., Wu P. Overexpression of a NAC-domain protein promotes shoot branching in rice. New Phytol. 2007;176:288–298. doi: 10.1111/j.1469-8137.2007.02177.x. [DOI] [PubMed] [Google Scholar]
- 46.Jiang D., Chen W., Dong J., Li J., Yang F., Wu Z., Zhou H., Wang W., Zhuang C. Overexpression of miR164b-resistant OsNAC2 improves plant architecture and grain yield in rice. J. Exp. Bot. 2018;69:1533–1543. doi: 10.1093/jxb/ery017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deshmukh R., Singh A., Jain N., Anand S., Gacche R., Singh A., Gaikwad K., Sharma T., Mohapatra T., Singh N. Identification of candidate genes for grain number in rice (Oryza sativa L.) Funct. Integr. Genom. 2010;10:339–347. doi: 10.1007/s10142-010-0167-2. [DOI] [PubMed] [Google Scholar]
- 48.Wu Y., Wang Y., Mi X.-F., Shan J.-X., Li X.-M., Xu J.-L., Lin H.-X. The QTL GNP1 encodes GA20ox1, which increases grain number and yield by increasing cytokinin activity in rice panicle meristems. PLoS Genet. 2016;12:e1006386. doi: 10.1371/journal.pgen.1006386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Z., Li J., Yao G., Zhang H., Dou H., Shi H., Sun X., Li Z. Fine mapping and cloning of the grain number per-panicle gene (Gnp4) on chromosome 4 in rice (Oryza sativa L.) Agric. Sci. China. 2011;10:1825–1833. doi: 10.1016/S1671-2927(11)60182-X. [DOI] [Google Scholar]
- 50.Tabuchi H., Zhang Y., Hattori S., Omae M., Shimizu-Sato S., Oikawa T., Qian Q., Nishimura M., Kitano H., Xie H., et al. LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell. 2011;23:3276–3287. doi: 10.1105/tpc.111.088765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao L., Tan L., Zhu Z., Xiao L., Xie D., Sun C. PAY1 improves plant architecture and enhances grain yield in rice. Plant J. 2015;83:528–536. doi: 10.1111/tpj.12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li M., Tang D., Wang K., Wu X., Lu L., Yu H., Gu M., Yan C., Cheng Z. Mutations in the F-box gene LARGER PANICLE improve the panicle architecture and enhance the grain yield in rice. Plant Biotechnol. J. 2011;9:1002–1013. doi: 10.1111/j.1467-7652.2011.00610.x. [DOI] [PubMed] [Google Scholar]
- 53.Yu H., Murchie E.H., González-Carranza Z.H., Pyke K.A., Roberts J.A. Decreased photosynthesis in the erect panicle 3 (ep3) mutant of rice is associated with reduced stomatal conductance and attenuated guard cell development. J. Exp. Bot. 2015;66:1543–1552. doi: 10.1093/jxb/eru525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Piao R., Jiang W., Ham T.-H., Choi M.-S., Qiao Y., Chu S.-H., Park J.-H., Woo M.-O., Jin Z., An G., et al. Map-based cloning of the ERECT PANICLE 3 gene in rice. Theor. Appl. Genet. 2009;119:1497–1506. doi: 10.1007/s00122-009-1151-x. [DOI] [PubMed] [Google Scholar]
- 55.Qiao Y., Piao R., Shi J., Lee S.-I., Jiang W., Kim B.-K., Lee J., Han L., Ma W., Koh H.-J. Fine mapping and candidate gene analysis of dense and erect panicle 3, DEP3, which confers high grain yield in rice (Oryza sativa L.) Theor. Appl. Genet. 2011;122:1439–1449. doi: 10.1007/s00122-011-1543-6. [DOI] [PubMed] [Google Scholar]
- 56.Liu G., Zhang K., Ai J., Deng X., Hong Y., Wang X. Patatin-related phospholipase A, pPLAIIIα, modulates the longitudinal growth of vegetative tissues and seeds in rice. J. Exp. Bot. 2015;66:6945–6955. doi: 10.1093/jxb/erv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zahn L.M., Kong H., Leebens-Mack J.H., Kim S., Soltis P.S., Landherr L.L., Soltis D.E., de Pamphilis C.W., Ma H. The evolution of the SEPALLATA subfamily of MADS-box genes: A preangiosperm origin with multiple duplications throughout angiosperm history. Genetics. 2005;169:2209–2223. doi: 10.1534/genetics.104.037770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gao X., Liang W., Yin C., Ji S., Wang H., Su X., Guo C., Kong H., Xue H., Zhang D. The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiol. 2010;153:728–740. doi: 10.1104/pp.110.156711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y., Yu H., Liu J., Wang W., Sun J., Gao Q., Zhang Y., Ma D., Wang J., Xu Z., et al. Loss of function of OsMADS34 leads to large sterile lemma and low grain yield in rice (Oryza sativa L.) Mol. Breed. 2016;36:147. doi: 10.1007/s11032-016-0578-4. [DOI] [Google Scholar]
- 60.Liu C., Teo Z.W.N., Bi Y., Song S., Xi W., Yang X., Yin Z., Yu H. A conserved genetic pathway determines inflorescence architecture in Arabidopsis and rice. Dev. Cell. 2013;24:612–622. doi: 10.1016/j.devcel.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 61.Luo J., Liu H., Zhou T., Gu B., Huang X., Shangguan Y., Zhu J., Li Y., Zhao Y., Wang Y., et al. An-1 encodes a basic helix-loop-helix protein that regulates awn development, grain size, and grain number in rice. Plant Cell. 2013;25:3360–3376. doi: 10.1105/tpc.113.113589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tan L., Li X., Liu F., Sun X., Li C., Zhu Z., Fu Y., Cai H., Wang X., Xie D., et al. Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 2008;40:1360–1364. doi: 10.1038/ng.197. [DOI] [PubMed] [Google Scholar]
- 63.Jin J., Huang W., Gao J.-P., Yang J., Shi M., Zhu M.-Z., Luo D., Lin H.-X. Genetic control of rice plant architecture under domestication. Nat. Genet. 2008;40:1365–1369. doi: 10.1038/ng.247. [DOI] [PubMed] [Google Scholar]
- 64.Wang Y., Li J. Rice, rising. Nat. Genet. 2008;40:1273–1275. doi: 10.1038/ng1108-1273. [DOI] [PubMed] [Google Scholar]
- 65.Guo T., Lu Z.-Q., Shan J.-X., Ye W.-W., Dong N.-Q., Lin H.-X. ERECTA1 acts upstream of the OsMKKK10-OsMKK4-OsMPK6 cascade to control spikelet number by regulating cytokinin metabolism in rice. Plant Cell. 2020;32:2763–2779. doi: 10.1105/tpc.20.00351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu Q.-H., Hoque M.S., Dennis E.S., Upadhyaya N.M. Ds tagging of BRANCHED FLORETLESS 1 (BFL1) that mediates the transition from spikelet to floret meristem in rice (Oryza sativa L) BMC Plant Biol. 2003;3:6. doi: 10.1186/1471-2229-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xue W., Xing Y., Weng X., Zhao Y., Tang W., Wang L., Zhou H., Yu S., Xu C., Li X., et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 2008;40:761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- 68.Putterill J., Robson F., Lee K., Simon R., Coupland G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell. 1995;80:847–857. doi: 10.1016/0092-8674(95)90288-0. [DOI] [PubMed] [Google Scholar]
- 69.Yano M., Katayose Y., Ashikari M., Yamanouchi U., Monna L., Fuse T., Baba T., Yamamoto K., Umehara Y., Nagamura Y., et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell. 2000;12:2473–2484. doi: 10.2307/3871242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wei X., Xu J., Guo H., Jiang L., Chen S., Yu C., Zhou Z., Hu P., Zhai H., Wan J. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 2010;153:1747–1758. doi: 10.1104/pp.110.156943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yan W.-H., Wang P., Chen H.-X., Zhou H.-J., Li Q.-P., Wang C.-R., Ding Z.-H., Zhang Y.-S., Yu S.-B., Xing Y.-Z., et al. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol. Plant. 2011;4:319–330. doi: 10.1093/mp/ssq070. [DOI] [PubMed] [Google Scholar]
- 72.Dai X., Ding Y., Tan L., Fu Y., Liu F., Zhu Z., Sun X., Sun X., Gu P., Cai H., et al. LHD1, an allele of DTH8/Ghd8, controls late heading date in common wild rice (Oryza rufipogon) J. Integr. Plant Biol. 2012;54:790–799. doi: 10.1111/j.1744-7909.2012.01166.x. [DOI] [PubMed] [Google Scholar]
- 73.Thirumurugan T., Ito Y., Kubo T., Serizawa A., Kurata N. Identification, characterization and interaction of HAP family genes in rice. Mol. Genet. Genom. 2008;279:279–289. doi: 10.1007/s00438-007-0312-3. [DOI] [PubMed] [Google Scholar]
- 74.Murakami M., Matsushika A., Ashikari M., Yamashino T., Mizuno T. Circadian-associated rice pseudo response regulators (OsPRRs): Insight into the control of flowering time. Biosci. Biotech. Biochem. 2005;69:410–414. doi: 10.1271/bbb.69.410. [DOI] [PubMed] [Google Scholar]
- 75.Koo B.-H., Yoo S.-C., Park J.-W., Kwon C.-T., Lee B.-D., An G., Zhang Z., Li J., Li Z., Paek N.-C. Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol. Plant. 2013;6:1877–1888. doi: 10.1093/mp/sst088. [DOI] [PubMed] [Google Scholar]
- 76.Gao H., Jin M., Zheng X.-M., Chen J., Yuan D., Xin Y., Wang M., Huang D., Zhang Z., Zhou K., et al. Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc. Natl. Acad. Sci. USA. 2014;111:16337–16342. doi: 10.1073/pnas.1418204111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yan W., Liu H., Zhou X., Li Q., Zhang J., Lu L., Liu T., Liu H., Zhang C., Zhang Z., et al. Natural variation in Ghd7.1 plays an important role in grain yield and adaptation in rice. Cell Res. 2013;23:969–971. doi: 10.1038/cr.2013.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jin J., Hua L., Zhu Z., Tan L., Zhao X., Zhang W., Liu F., Fu Y., Cai H., Sun X., et al. GAD1 encodes a secreted peptide that regulates grain number, grain length, and awn development in rice domestication. Plant Cell. 2016;28:2453–2463. doi: 10.1105/tpc.16.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huo X., Wu S., Zhu Z., Liu F., Fu Y., Cai H., Sun X., Gu P., Xie D., Tan L., et al. NOG1 increases grain production in rice. Nat. Commun. 2017;8:1497. doi: 10.1038/s41467-017-01501-8. [DOI] [PMC free article] [PubMed] [Google Scholar]