Abstract
STUDY QUESTION
What are the genetic factors that increase the risk of aneuploid egg production?
SUMMARY ANSWER
A non-synonymous variant rs2303720 within centrosomal protein 120 (CEP120) disrupts female meiosis in vitro in mouse.
WHAT IS KNOWN ALREADY
The production of aneuploid eggs, with an advanced maternal age as an established contributing factor, is the major cause of IVF failure, early miscarriage and developmental anomalies. The identity of maternal genetic variants contributing to egg aneuploidy irrespective of age is missing.
STUDY DESIGN, SIZE, DURATION
Patients undergoing fertility treatment (n = 166) were deidentified and selected for whole-exome sequencing.
PARTICIPANTS/MATERIALS, SETTING, METHODS
Patients self-identified their ethnic groups and their ages ranged from 22 to 49 years old. The study was performed using genomes from White, non-Hispanic patients divided into controls (97) and cases (69) according to the number of aneuploid blastocysts derived during each IVF procedure. Following a gene prioritization strategy, a mouse oocyte system was used to validate the functional significance of the discovered associated genetic variants.
MAIN RESULTS AND THE ROLE OF CHANCE
Patients producing a high proportion of aneuploid blastocysts (considered aneuploid if they missed any of the 40 chromatids or had extra copies) were found to carry a higher mutational burden in genes functioning in cytoskeleton and microtubule pathways. Validation of the functional significance of a non-synonymous variant rs2303720 within Cep120 on mouse oocyte meiotic maturation revealed that ectopic expression of CEP120:p.Arg947His caused decreased spindle microtubule nucleation efficiency and increased incidence of aneuploidy.
LIMITATIONS, REASONS FOR CAUTION
Functional validation was performed using the mouse oocyte system. Because spindle building pathways differ between mouse and human oocytes, the defects we observed upon ectopic expression of the Cep120 variant may alter mouse oocyte meiosis differently than human oocyte meiosis. Further studies using knock-in ‘humanized’ mouse models and in human oocytes will be needed to translate our findings to human system. Possible functional differences of the variant between ethnic groups also need to be investigated.
WIDER IMPLICATIONS OF THE FINDINGS
Variants in centrosomal genes appear to be important contributors to the risk of maternal aneuploidy. Functional validation of these variants will eventually allow prescreening to select patients that have better chances to benefit from preimplantation genetic testing.
STUDY FUNDING/COMPETING INTEREST(S)
This study was funded through R01-HD091331 to K.S. and J.X. and EMD Serono Grant for Fertility Innovation to N.R.T. N.R.T. is a shareholder and an employee of Genomic Prediction.
TRIAL REGISTRATION NUMBER
N/A.
Keywords: fertility, IVF, aneuploidy, exome sequencing, centrosomal protein 120, oocyte maturation, chromosome segregation, microtubule-organizing centers, spindle
Introduction
Aneuploidy is the most common genetic abnormality in human embryos leading to early miscarriage and failure in ART (Nagaoka et al., 2012; Franasiak et al., 2014). Maternal age is associated with increased risk of embryo aneuploidy, where the prevalence of aneuploid egg formation or aneuploid pregnancy rapidly increases after the age of 35 years (Hassold and Chiu, 1985; Carp et al., 2001; Hassold and Hunt, 2001; Hogge et al., 2003; Kuliev et al., 2011; Franasiak et al., 2014; Kubicek et al., 2019). However, reports that documented aneuploidy frequency of eggs or embryos also showed a large variation in the prevalence of aneuploidy from women of the same age (Franasiak et al., 2014; McCoy et al., 2015b). Therefore, age alone is not sufficient to predict the chances to achieve a healthy pregnancy for every woman, and other risk factors must be evaluated.
One risk factor to consider is that maternal genetic variants affect chromosome segregation fidelity. In fact, recent studies indicated that variants in several genes can predispose women to a higher incidence of embryonic aneuploidy at younger than average ages. For example, variants in tubulin β class VIII (TUBB8) (Feng et al., 2016a,b), PAT1 homolog 2 (PATL2) (Chen et al., 2017) and WEE2 oocyte meiosis inhibiting kinase (WEE2) (Sang et al., 2018; Yang et al., 2019; Zhang et al., 2019) genes were associated with oocyte meiosis defects. Variants in polo-like kinase 4 (PLK4) (McCoy et al., 2015a, 2018; Zhang et al., 2017) and transducin-like enhancer (TLE) family member 6, subcortical maternal complex member (TLE6) (Alazami et al., 2015) in maternal genomes were associated with embryonic mitosis defects. However, a comprehensive understanding of all genes contributing to embryonic aneuploidy and the intricate relationship between variants in these genes and aneuploidy is still lacking.
At least three hurdles impeded the progress in this area of research: the inability to define cases and controls with respect to a woman’s risk of producing aneuploid eggs, the lack of DNA from women who have had their eggs evaluated for aneuploidy and the inability to systematically characterize DNA variations which may be associated with the risk of an aneuploid conception. This study capitalizes on using a unique DNA bank collection of deidentified patients who had their blastocysts evaluated for aneuploidy during their fertility procedure. Because of these samples and their associated aneuploidy phenotype, we overcame these hurdles and identified gene variants that increase the risk of producing aneuploid eggs. Specifically, we performed exome sequencing of women that produced high and low proportions of aneuploid blastocysts and identified an enrichment of coding variants in genes important for spindle building in patients that produced a high proportion of aneuploid blastocysts. As proof of principle, we then used mouse oocytes as a meiotic bioassay to evaluate the functional consequences of a centrosomal protein 120 (CEP120) variant, a gene required for centrosome function and microtubule stability in mitosis (Mahjoub et al., 2010; Comartin et al., 2013; Lin et al., 2013). In particular, we report a non-synonymous single-nucleotide variant (SNV), rs2303720, within CEP120 as a potential novel variant that reduces female fertility. Importantly, we found that expression of CEP120:p.R947H in otherwise wild-type (WT) mouse oocytes decreased spindle quality and increased the incidence of aneuploidy at metaphase of meiosis II (MII), suggesting a negative dominant effect.
Materials and methods
Ethical Approval
Analysis of DNA samples from the Reproductive Medicine Associates of New Jersey (RMANJ) DNA Bank was approved by the Institutional Review Board (IRB) #RMA1-09-165 at Copernicus Group IRB and IRB #Pro2018000106 at Rutgers. All animals were maintained following the Rutgers Institutional Animal Use and Care Committee (#11-032) and the National Institutes of Health guidelines.
Aneuploidy rate calculation
Information on patients that had their embryos evaluated for chromosomal number prior to selecting embryos for uterine transfer was obtained from RMANJ. The fertility treatment was performed by ICSI. Standard quality assessments were performed for the semen samples (volume, concentration, percentage normal, total motile, percentage motile, etc.). Because the fertility treatment was performed by ICSI, the overall sperm sample quality is less crucial than in the traditional IVF procedures. Therefore, the sperm samples are not required to be normazoospermic, and a sperm with the best morphology was selected for each egg fertilization. Patients self-identified their ethnic groups and their ages ranged from 22 to 49 years old. Patients self-reported as ‘White, non-Hispanic’ and a minimum of four embryos tested were used for aneuploidy rate calculation as follows (Supplementary Table SI):
According to this formula, a person was said to have an aneuploidy rate of 50% if half of the blastocysts were aneuploid.
Sequencing and variant call set generation
Patient genomic DNA was isolated from blood using QIAamp DNA Blood Maxi Kit (Qiagen, Hilden, Germany). Whole-exome sequencing experiments were carried out on an Ion Proton instrument (Thermo Fisher Scientific, Waltham, MA, USA) using an Ion AmpliSeq Exome kit (Thermo Fisher Scientific, Waltham, MA, USA). The sequenced reads were aligned to the human reference genome (version hg19) with the Torrent Mapping Alignment Program as part of the Ion Torrent Suite software v4.4 (Thermo Fisher Scientific, Waltham, MA, USA). Samples with at least 20× coverage at more than 60% of the exome target regions were included in the variant calling step. For an individual sequenced more than once, sequencing runs that passed the quality threshold were merged prior to variant calling on that individual. To generate individual variant call format (VCF) files, the Torrent Variant Caller (TVC) plugin was applied with default parameters. Two rounds of variant calling were carried out. The first round of variant calling identified variant site candidates. Biallelic SNV candidates from all samples were then pooled and a second round of variant calling was carried out for these variants only, resulting in genotype calls for all SNV candidates.
Variant filtering
Individual VCFs were merged with vcf-merge from the VCFtools (version 0.1.12b) (Danecek et al., 2011). Variants within exome target regions were determined by the ‘intersect’ function from bedtools (version 2.25.0) (Quinlan and Hall, 2010). Sites with a per-SNV failure rate >20%, samples with a per-individual genotype call failure rate >10% and SNVs that deviated from the Hardy–Weinberg equilibrium (threshold P < 1 × 10−6) were removed with VCFtools.
Principal component analysis
The principal component analysis (PCA) space of worldwide populations was generated using the genotype file from the 1000 Genomes Project: ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/release/20130502/supporting/hd_genotype_chip/ALL.chip.omni_broad_sanger_combined.20140818.snps.genotypes.vcf.gz.
Variants with allele frequency (AF) >2% in our variant set were used in PCA. Variants present in both our and the 1000 Genomes Project dataset were extracted with function ‘isec’ in bcftools (version 1.6-36-gd945e75; htslib 1.6-32-g3357f86) (Li et al., 2009). PCA was performed using R package SNPRelate (Bioconductor 3.3 (BiocInstaller 1.22.3), R 3.3.0 (2016-05-03); SNPRelate_1.6.6) (Zheng et al., 2012). Individuals on principal components 1 and 2 that were more than 4 SD away from the mean of our samples were removed.
Gene prioritization with TRAPD
SNVs were annotated with Variant Effect Predictor (VEP) (McLaren et al., 2016). Variants were considered rare if the AF was smaller than 0.05 among the gnomAD non-Finnish European (NFE) samples (r2.0.1, using GRCh37 coordinates) and common if AF ≥ 0.05. Variants not observed in gnomAD (unknown/novel to our study) were counted as both rare and common, irrespective of their allele count in the data. Testing Rare vAriants using Public Data (TRAPD) strategy (Guo et al., 2018) was applied with the following modifications. Control and case VCF files were both processed with make_snp_file.py (where ‘file.vcf’ stands for either ‘control.vcf’ or ‘case.vcf’):
$ python make_snp_file.py --vcffile file.vcf --outfile file.out --genecolname SYMBOL --excludevep gnomAD_NFE_AF[>=]0.05 --snpformat "CHRPOSREFALT" --bedfile regions.bed --vep –snponly
The ‘[>=]’ in the above command indicates that only rare and novel variants are counted. Similarly, ‘[<]’ in the command will instruct to count only common and novel variants.
The resulting control and case VCF files were both processed with count_cases.py:
$ python count_cases.py -v file.vcf -s file.out -o file.Counts.txt --snpformat “CHRPOSREFALT” --bedfile regions.bed
The significance test of mutational load was performed using burden.R script with four lines modified:
#Dominant model
case_count<-dat[i,]$CASE_COUNT_HET.x+dat[i,]$CASE_COUNT_CH.x+dat[i,]$CASE_COUNT_HOM.x
control_count<-
dat[i,]$CASE_COUNT_HET.y+dat[i,]$CASE_COUNT_CH.y+dat[i,]$CASE_COUNT_HOM.y
#Recessive model
case_count_rec<-dat[i,]$CASE_COUNT_CH.x+dat[i,]$CASE_COUNT_HOM.x
control_count_rec<-dat[i,]$CASE_COUNT_CH.y+dat[i,]$CASE_COUNT_HOM.y
$Rscript burden.R --casefile cases.Counts.txt --casesize 68 --controlfile controls.Counts.txt --controlsize 92 --outfile case_vs_control_testing.out
Gene prioritization with VAAST
VAAST (Variant Annotation, Analysis and Search Tool) (Hu et al., 2013) was run following user guidelines in four steps:
- vaast_converter --build hg19 --path pathto/gvf_files/--format VCF file.vcf
- VAT --features ref_gff --fasta ref_fasta file.gvf > file.vat.gvf
- VST -o "U(0.$indx)" -b hg19 --input_files controlsIDs.txt > controls.cdr
- VST -o "U(0.$indx)" -b hg19 --input_files casesIDs.txt > cases.cdr
- VAAST --lh y -r 0.05 -d 1e5 -o outputfile -m lrt -k ref_gff controls.cdr cases.cdr
Gene ontology enrichment analysis
The tool DAVID (v6.8) (Huang da et al., 2009a,b) was used to perform gene ontology (GO) enrichment analysis.
Validation of CEP120 variant
Sanger sequencing validation was performed using a Sanger sequencing primer (hs00790274_ce) and 3730 DNA analyzer (Thermo Fisher Scientific, Waltham, MA, USA) at the Foundation for Embryonic Competence. Four probands (three predicted to be heterozygotes, and one homozygous for the variant) were selected for validation of the CEP120 variant. All samples were confirmed.
Mouse oocyte isolation
Oocytes were harvested from sexually mature CF-1 female mice (6–10 weeks of age) (Envigo, Indianapolis, IN, USA). Mice were first sacrificed by cervical dislocation and the dissected ovaries were transferred to minimal essential medium (MEM) supplemented with 2.5 μM milrinone (#M4659, Sigma-Aldrich, St. Louis, MO, USA). For the microtubule nucleation assay, the oocytes were collected from the ovaries of female mice 48 h after i.p. injection of 5 IU of pregnant mare’s serum gonadotrophin (#367222, Calbiochem, St. Louis, MO, USA). Oocytes were isolated from follicles by pipetting using a Pasteur pipette not exceeding 80 μm. After isolation, the oocytes were transferred to Chatot, Ziomek and Bavister (CZB) media containing 2.5 μM milrinone (Chatot et al., 1989). The CZB medium was also supplemented with 100 mM glutamine (#25030-08, Gibco, Waltham, MA, USA). Oocytes were then incubated at 37°C in an atmosphere of 5% CO2 in air. To induce meiotic resumption, oocytes were cultured in milrinone-free CZB medium and those that had not undergone nuclear envelope breakdown within 1 h 30 min of removing milrinone were removed from the experiment. To assess chromosome number, oocytes were matured for 13 h after nuclear envelope breakdown and incubated with monastrol (#M8515, Sigma-Aldrich, St. Louis, MO, USA) for 2 h 15 min prior to fixation. Monastrol was dissolved in dimethyl sulfoxide (#472301, Sigma-Aldrich, St. Louis, MO, USA) and added to the CZB culture media at a final concentration of 100 μM. IVM of monastrol-treated oocytes was performed in organ culture dishes (#353037, Becton Dickinson, Franklin Lakes, NJ, USA) under humidified conditions. The microtubule nucleation assay was performed as previously described (Ma and Viveiros, 2014). Briefly, oocytes that reached metaphase of meiosis I (MI) were incubated for 1 h in organ culture dishes containing pre-chilled MEM on ice. After cold treatment, the oocytes were incubated in pre-warmed CZB at 37°C for 30 min and fixed for immunofluorescence analyses.
Constructs and cRNA generation
Conservation of the variant site between mouse and human CEP120 was assessed using MacVector v12.7.5 (Apex, NC, USA) with mouse transcript Cep120 NM_178686.4 and human transcript CEP120 NM_153223.3. To generate enhanced green fluorescent protein (eGFP)-CEP120 cRNA for overexpression experiments, the full-length open reading frame of the gene encoding mouse Cep120 was amplified from a mouse Cep120 cDNA clone (#MC203364, Origene, Rockville, MD, USA) and cloned into the plasmid in vitro transcription (pIVT) vector (Igarashi et al., 2007) containing an N-terminal GFP fusion (forward primer: 5′-ATATCCCGGGCATGGTCCCCAAG-3′, reverse primer: 5′-CGGGAGCTCTTAACTGGCACTGTTTT-3′). Mutagenesis was performed by amplifying the pIVT-Cep120 plasmid (forward primer: 5′-GACTACCTGACACACCTGATAGAGGAAA-3′, reverse primer: 5′-TTTCCTCTATCAGGTGTGTCAGGTAGTC-3′). The PCR product was digested with DpnI restriction enzyme (#R0176S, New England Biolabs, Ipswich, MA, USA) to remove the parental plasmid. The above plasmids were sequenced and linearized with NdeI (#R0111S, New England Biolabs, Ipswich, MA, USA). pGEMHE-Cherry-TRIM21 (#105522, Addgene, Watertown, MA, USA) was linearized with AscI (#R0558S, New England Biolabs, Ipswich, MA, USA). All linearized plasmids were purified using a QIAquick PCR Purification kit (#28104, Qiagen, Hilden, Germany) and in vitro transcribed using a T7 mMessage mMachine Kit (#AM1340, Ambion, Waltham, MA, USA). The cRNAs were purified using an RNAeasy kit (#74104, Qiagen, Hilden, Germany) and stored at −80°C.
Microinjection
Germinal vesicle-intact oocytes were microinjected with eGfp-Cep120 or eGfp-Cep120-R947H cRNA. mCherry-Trim21 was co-microinjected with the rabbit anti-Cep120 antibody that was purified using an Amicon Ultra 0.5-ml Centrifugal Filter (#UFC500308, Merk Millipore, St. Louis, MO, USA). All cRNAs were injected at 700 ng/μl. The anti-Cep120 antibody was injected at 0.025 mg/ml. Injections were performed in MEM plus 2.5 μM milrinone using a Xenoworks digital microinjector (Sutter Instruments, Novato, CA, USA). Following microinjection, the oocytes were maintained at prophase I of MI in CZB supplemented with milrinone and glutamine with a mineral oil overlay (#M5310, Sigma-Aldrich, St. Louis, MO, USA) to prevent evaporation. Microinjected oocytes were incubated at 37°C for at least 3 h or overnight according to the experiment.
Antibodies and immunofluorescence
The following antibodies were used: rabbit anti-Cep120 antibody (1:50, #PA5-55985, Invitrogen, Waltham, MA, USA), rabbit anti-pAurkA/B/C (1:100, #D13A11, Cell Signaling Technology, Danvers, MA, USA), mouse anti-pericentrin antibody (1:100, #611814, BD Biosciences, San Jose, CA, USA), mouse anti-acetylated α-tubulin (1:100, #T7451, Sigma-Aldrich, St. Louis, MO, USA), mouse anti-α-tubulin FITC (1:100, #322588, Invitrogen, Waltham, MA, USA) and human anti-centromeric antigen (1:30, #15-234, Antibodies Incorporated, Davis, CA, USA). Secondary antibodies (1:200) were conjugated with Alexa Fluor 488 (goat anti-mouse, #A11029) or Alexa Fluor 568 (goat anti-mouse, #A10037, goat anti-rabbit, #A10042, Invitrogen, Waltham, MA, USA).
Immunofluorescence was performed as follows: oocytes were fixed at metaphase I (germinal vesicle breakdown (GVBD) + 7 h) or metaphase II (GVBD + 15 h) in 2% paraformaldehyde in PBS for 20 min. For pericentrin staining, oocytes were fixed in 4% paraformaldehyde for 20 min. Oocytes were washed twice with blocking buffer (1% PBS, 0.3% bovine serum albumin). Prior to immunostaining, the oocytes were permeabilized with 1% PBS containing 0.1% Triton X-100 and 0.3% bovine serum albumin for 20 min followed by 10 min incubation in blocking buffer. The oocytes were then incubated in primary antibodies for 3 h at room temperature. After washing three times for 10 min in blocking buffer, the oocytes were incubated in secondary antibodies for 1 h 30 min at room temperature. The oocytes were washed again three times for 10 min in blocking buffer. The oocytes were mounted on slides in vectashield (#H-1000, Vector Laboratories, Burlingame, CA, USA) containing 4(6-diamidino-2-phenylindole), dihydrochloride (1:170; #D1306, Thermo Fisher Scientific, Waltham, MA, USA).
Microscopy and image analysis
Images were captured using a Leica (Wetzlar, Germany) SP8 confocal microscope with a 63×/1.40 objective. For each image, optical z-slices were obtained using 0.5- to 1-μm steps with a zoom setting of 3. For comparison of pixel intensities, the laser power was kept constant for each oocyte in an experiment. Images of oocytes at prophase of MI were acquired using an EVOS FL Auto Imaging System (Life Technologies, Waltham, MA, USA) with a 20× objective.
ImageJ software (NIH, Bethesda, MD, USA) was used to process all images. For analysis, z-slices for each image were merged into a max projection. For α-tubulin and pericentrin, pixel intensity analyses of the average pixel intensity were recorded using the measurement tool. For chromosome alignment analysis, the distance between the two most extreme chromosome homologs from either side of the spindle midzone was measured and normalized to the total area of the spindle using freehand line and selection tools. For in situ chromosome spreads, the numbers of kinetochores were counted in metaphase MII eggs using Crest immunostaining as a marker. Eggs were considered euploid if they contained 40 chromatids and aneuploid if they missed any or had extra copies.
Immunoblotting
Following microinjection at prophase of MI, 20 oocytes expressing eGFP-CEP120 or eGFP-CEP120R947H were mixed with Laemli buffer (#1610737, BioRad, Hercules, CA, USA) containing β-mercaptoethanol and denatured at 95°C for 5 min. Western blotting was performed by separating proteins on a 7.5% sodium dodecyl sulfate–polyacrylamide gel, as previously described (Fellmeth et al., 2016), followed by incubation of the membrane with blocking solution overnight at 4°C. The membrane was then incubated for 1 h with an anti-GFP primary antibody (1:500, G1544, Sigma-Aldrich, St. Louis, MO, USA) followed by washing three times for 15 min each and incubated again for 1 h with a peroxidase-conjugated secondary antibody from the KwikQuant Western Blot Kit (#R1006, Kindle Biosciences, Greenwich, CT, USA). After detection, the membrane was treated for 1 h with Restore Plus Western blot Stripping Buffer (#46430, Thermo Scientific, Waltham, MA, USA) to remove the antibodies prior to probing with anti-α-tubulin (1:1000, 11H10, Cell Signaling Technology, Danvers, MA, USA) for normalization. Protein bands were detected using the ECL detection reagents from the KwikQuant Western Blot Kit following the manufacturer’s protocol.
Data analysis
All experiments were performed at least in triplicate unless otherwise noted. Averaged results from all experiments performed were reported. Scatter plots were performed with PRISM6 (Graphpad, San Diego, CA, USA). Student’s t-test was performed with Microsoft Excel (Redmond, WA, USA) and a value of P < 0.05 was considered significant.
Results
Defining low and high aneuploidy groups
An overview of the strategy and results of this study is outlined in Fig. 1. To identify genes associated with increased risk of aneuploid conception, we first selected patients producing low and high proportions of aneuploid blastocysts using phenotype data obtained from RMANJ (Materials and methods). Based on their proportions of aneuploid blastocysts, we defined the individuals with <30% aneuploid blastocysts as Low Aneuploidy Rate Group (LRG), and individuals with 50% or more aneuploid blastocysts as High Aneuploidy Rate Group (HRG). To control for genetic difference among populations, we limited our study to individuals that self-reported as ‘White, non-Hispanic’. In total, we selected 166 individuals for downstream analysis: 97 in LRG and 69 in HRG. The median ages were similar between the LRG individuals [36 years, interquartile range (IQR) 4 years] and HRG individuals [34 years, IQR 5 years]. The paternal ages of the embryos were also similar between the two groups with median paternal age of 37 years [IQR 5 years] for LRG and 36 years [IQR 9 years] for HRG.
Figure 1.
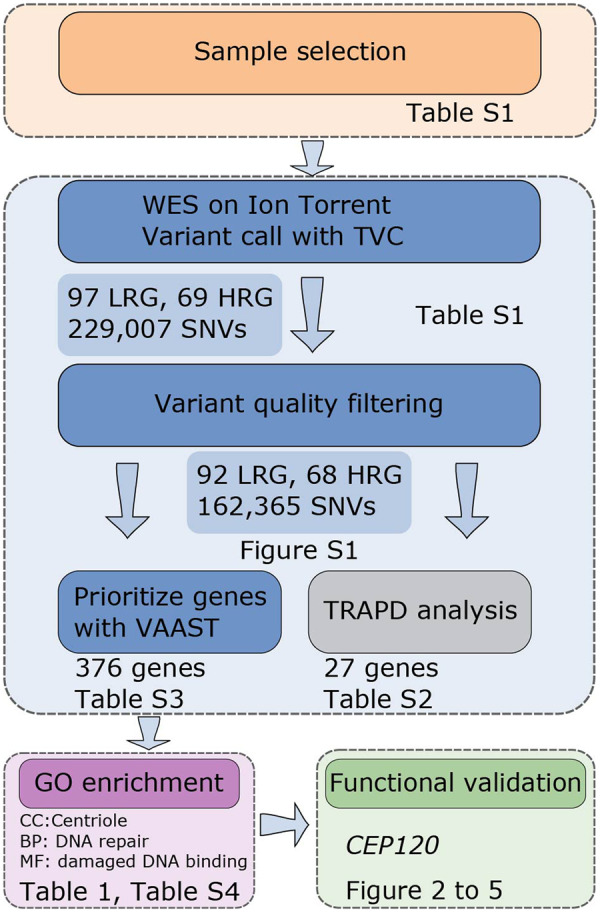
Overall strategy to identify candidate aneuploidy genes in human. Selected patients were subjected to exome sequencing using the Ion Torrent platform. Variants were identified using Torrent Variant Caller (TVC) software and high-quality single-nucleotide variant (SNV) were used for association analysis. Mutation burden analysis was performed with TRAPD (Testing Rare vAriants using Public Data) and VAAST (Variant Annotation, Analysis and Search Tool). Genes identified by VAAST were used for gene ontology (GO) enrichment and pathway analysis. Centrosomal protein 120 (CEP120) was selected for functional studies in a mouse oocyte model.
Sequencing and variant identification
To identify coding variants in the patient genomes, we performed exome sequencing. The sequencing achieved good coverage among individuals: on average, we obtained 46 069 739 mapped reads per individual (minimum 20 716 571; maximum 88 661 679) with a minimum base coverage of 20× for 84.53% of the target regions (minimum 63.16%, maximum 96.66% of the target region). Per-individual mapping statistics are detailed in Supplementary Table SI.
Using the mapped reads, we identified a total of 229 007 SNVs in the patients. We excluded two individuals with apparent admixed ancestry, as determined by the population structure inspection via PCA (Supplementary Fig. S1A). We filtered low-quality samples and SNVs, which reduced the dataset to 92 LRG and 68 HRG individuals, and 162 365 SNVs. We observed a similar overall accumulation of rare synonymous variants among LRG and HRG individuals (Supplementary Fig. S1B). This similarity indicates a good calibration of the rare variants (Guo et al., 2018). This set of 162 365 SNVs was used in the following variant association tests and gene discovery analyses.
Burden analysis implicates several genes in aneuploid conception risk
To identify candidate genes associated with increased risk of aneuploid egg production, we performed a gene-level burden test with TRAPD (Guo et al., 2018). By counting the mutational load within each gene in HRG (i.e. our cases) compared to LRG (i.e. controls), TRAPD identified 8 and 19 genes that reached genome-wide significance level for the rare and common variant analysis, respectively (Supplementary Table SII). Several are likely to have functions in meiosis. For example, from the rare variant analysis we identified FAM184A (family with sequence similarity 184 member A), a gene that functions in a protein complex with CENPJ (centromere protein J). CENPJ is a centrosome protein implicated in centriole biogenesis (http://hu.proteincomplexes.org/displayComplexes?complex_key=2611, 27 June 2020, date last accessed). Among significant genes obtained from common variant analysis, Mcm5 (minichromosome maintenance complex component 5) was implicated in embryonic lethality in mouse (http://www.mousemine.org/mousemine/template.do?name=HGene_MPhenotype&scope=all, 27 June 2020, date last accessed). Because meiosis is a highly conserved biological process, lower eukaryotes are a valuable resource to further our understanding of molecular mechanisms associated with aneuploidy. Indeed, RNAi depletion of mcm-5 in Caenorhabditis elegans causes sister chromatid segregation defects (https://wormbase.org/species/c_elegans/gene/WBGene00003157#0-b-3, 27 June 2020, date last accessed). Similarly, in budding yeast Saccharomyces cerevisiae, MCM5 is implicated in chromosome segregation defects (https://www.yeastgenome.org/locus/S000004264, 27 June 2020, date last accessed). The above examples from model organisms suggest that TRAPD gene burden analysis effectively identified candidate genes important for meiosis.
Cohesin and centrosome components are associated with increased risk of aneuploid conception
The TRAPD analysis treats all variants as having similar damaging effects. To account for differential functional impact of variants in the association testing, we performed a second gene-level variant analysis by VAAST (Hu et al., 2013). Importantly, VAAST scores account for the population AF, functional impact and evolutionary conservation of variants within a gene. Over 5000 genes received positive scores in the VAAST analysis and 376 genes had a P-value smaller than 0.05 (Supplementary Table SIII). Because of the sample size, no single gene reached a genome-wide significance level after multiple testing correction. Nevertheless, among the top-scoring candidate genes, several function in either meiotic or mitotic processes and were reportedly linked to fertility in mouse (e.g. NAT1 (N-acetyltransferase 1), HINFP (histone H4 transcription factor), Supplementary Table SIII). For example, we identified a deleterious (as predicted by SIFT) non-synonymous SNV rs61735519 within the gene SMC1B (structural maintenance of chromosomes protein 1B), causing a p.Phe1055Leu amino acid change. SMC1B is a documented fertility gene and encodes a meiosis-specific cohesin subunit required for maintaining meiotic cohesion of sister chromatids (Revenkova et al., 2004). Female mice that harbor this allele are fertile. However, a close analysis of the published data (Singh and Schimenti, 2015) suggests subfertility in the mice which in turn suggests a potential mild but discernable effect of this variant. The variant showed a strong enrichment among HRG individuals in our dataset: its AF is more than 2-fold higher in the HRG (13.43%) than in the LRG (4.35%) and European populations (gnomAD NFE AF = 5.64%).
After individual gene analysis, we investigated whether the top genes are enriched for specific molecular functions, cellular components or biological processes through the GO enrichment analysis. For the analysis, we focused on 376 genes with VAAST P-value below 0.05 before multiple testing correction (Supplementary Table SIII). Among the enriched GO Cellular Compartment terms, ‘centriole’ was the third most significant term (P = 0.016; HERC2 (HECT and RLD domain-containing e3 ubiquitin protein ligase 2), CEP120, LRRCC1 (leucine-rich repeat and coiled-coil centrosomal protein 1), NIN (ninein), PCNT (pericentrin), PCM1 (pericentriolar material 1), TUBD1 (tubulin delta 1); Supplementary Table SIV). Five of the genes, CEP120, LRRCC1, NIN, PCNT and PCM1, are centrosomal proteins, and the term ‘centrosome’ was also enriched (P = 0.022; 15 genes in total; Supplementary Table SIV). Even though mammalian oocytes lack centrioles, some of these genes were previously linked to fertility. For instance, Pcnt is a known mouse fertility gene and its depletion by small interfering RNA results in acentriolar microtubule-organizing center (aMTOC) defects and meiotic errors (Ma and Viveiros, 2014; Baumann et al., 2017). In Table I, we provided the VAAST rankings of the above ‘centriole’ genes, variants scored and brief description of each gene. Taken together, our results showed that our candidate gene discovery approach was effective because known fertility genes ranked high in the prioritized list.
Table I.
Genes associated with gene ontology term ‘centriole’ together with identified mutations.
| VAAST rank | Gene | Variant (hg19) | dbSNP ID | Amino acid change | Allele frequency | Functional consequence |
|---|---|---|---|---|---|---|
| 25 | CEP120 | NM_153223.3:c.2840G>A | rs2303720 | p.Arg947His | 3.2 x 10−2 | D/PD |
| 37 | LRRCC1 | NM_033402.4:c.207T>A | rs16913589 | p.His69Gln | 4.7 x 10−2 | D/PSD |
| NM_033402.4:c.2339T>C | rs147274148 | p.Leu780Pro | 6.3 x 10−3 | D/PSD | ||
| NM_033402.4:c.2405G>A | rs779167117 | p.Arg802Gln | 9.8 x 10−6 | D/PSD | ||
| 262 | NIN | NM_182944.2:c.5996G>A | rs141405524 | p.Arg1999His | 1 x 10−3 | T/B |
| NM_182944.2:c.2987C>T | rs41313507 | p.Ala996Val | 3.8 x 10−3 | T/B | ||
| 282 | PCNT | NM_006031.5:c.2545G>T | rs776223654 | p.Gly849Trp | 0 | n.a./PD |
| NM_006031.5:c.6563T>G | rs1044998 | p.Met2188Arg | 7.6 x 10−2 | n.a./B | ||
| NM_006031.5:c.7130C>T | rs61735814 | p.Pro2377Leu | 6.2 x 10−2 | n.a./PD | ||
| NM_006031.5:c.8924T>C | rs35881595 | p.Leu2975Pro | 1.4 x 10-3 | n.a./PSD | ||
| NM_006031.5:c.7118G>A | rs147670568 | p.Gly2373Glu | 2.3 x 10-4 | n.a./PD | ||
| NM_006031.5:c.5771C>T | rs184420466 | p.Ala1924Val | 8.6 x 10-3 | n.a./PSD | ||
| NM_006031.5:c.4109G>A | rs145055342 | p.Arg1370Gln | 4.5 x 10-4 | n.a./B | ||
| 309 | TUBD1 | NM_001193609.1:c.775G>T | chr17:57944105 | p.Asp314Tyr | Novel | D/B |
| 349 | PCM1 | NM_006197.3:c.808A>G | rs201598166 | p.Met270Val | 2.7 x 10−3 | T/B |
| NM_006197.3:c.2168A>T | rs150814716 | p.Asp723Val | 6 x 10−5 | D/PD | ||
| NM_006197.3:c.2612G>T | rs7009117 | p.Gly871Val | 6 x 10−4 | D/PD | ||
| NM_006197.3:c.3980C>T | rs1254061869 | p.Pro1327Leu | 6.5 x 10−5 | T/PD | ||
| 352 | HERC2 | NM_004667.5:c.13919A>G | rs138725743 | p.His4640Arg | 5 x 10−3 | n.a./B |
| NM_004667.5:c.9110G>A | rs369787802 | p.Arg3037Gln | 7 x 10−5 | n.a./PD | ||
| NM_004667.5:c.5040C>G | chr15:28474686 | p.Ser1680Arg | 1.2 x 10−2 | n.a./B |
CEP120, centrosomal protein 120; LRRCC1, leucine-rich repeat and coiled-coil centrosomal protein 1; NIN, ninein; PCNT, pericentrin; TUBD1, tubulin delta 1; PCM1, pericentriolar material 1; HERC2, HECT and RLD domain-containing E3 ubiquitin protein ligase 2.
For known variants, allele frequencies and predicted functional consequences were extracted from gnomAD NFE. For the novel variant in TUBD1, amino acid change was annotated using ANNOVAR and functional impact was predicted using SIFT and PolyPhen2 (D, deleterious; T, tolerated; PD, probably damaging; PSD, possibly damaging; B, benign; n.a., information not available).
CEP120 localizes to MTOCs independent of Arg947
The goal of this study was to identify and validate novel fertility genes associated with aneuploidy. As such, an intriguing candidate among the top-scoring genes was CEP120. In our dataset CEP120 contains a non-synonymous variant (rs2303720) that replaces a conserved amino acid residue in CEP120, p.Arg947His. The variant is predicted to be deleterious by SIFT and probably damaging by Polyphen 2. Variant rs2303720 is common in NFE populations (gnomAD NFE AF = 3.16%). In our dataset, the AF in the HRG (6.62%) is 2-fold higher than in NFE populations (3.16%) and 12-fold higher than in the LRG (0.56%). CEP120 is also a strong candidate because it is among the centriole components enriched in the GO analysis. CEP120 was implicated to regulate spindle assembly and chromosome segregation (Hutchins et al., 2010), as well as to control centriole duplication and elongation during mitosis (Comartin et al., 2013; Lin et al., 2013). Although CEP120 was recently described to localize to aMTOCs in mouse oocytes (So et al., 2019), its role during mammalian oocyte meiotic maturation remains unknown.
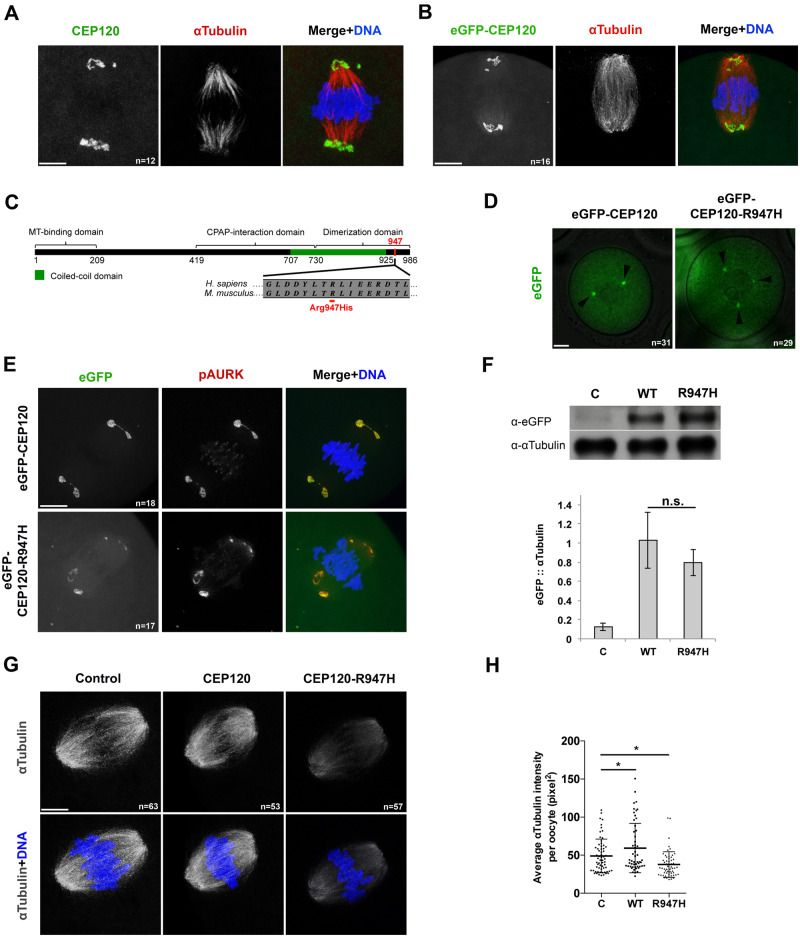
To assess the role of CEP120 during meiotic maturation, we exploited the mouse oocyte system which is suitable for studying meiosis and can be manipulated in vitro. Notably, mouse and human oocytes both lack centrioles. Mouse oocytes possess aMTOCs, which are the main organizing center for microtubule nucleation; the presence of aMTOCs in human oocytes is still under investigation (George et al., 1996; Schuh and Ellenberg, 2007; Holubcová et al., 2015; Virant-Klun et al., 2016; Roeles and Tsiavaliaris, 2019). Therefore, a requirement for CEP120 in spindles that form without centrioles is unprecedented. We first investigated the subcellular localization of endogenous CEP120 at metaphase of MI in mouse oocytes. By performing immunocytochemistry, we found that CEP120 localized to the polar ends of metaphase MI spindles (Fig. 2A). Furthermore, exogenous eGFP-tagged CEP120 also localized to metaphase MI spindle poles and to aMTOCs in prophase I-arrested oocytes, thereby confirming that eGFP-tagged CEP120 has the same localization as endogenous CEP120 in MI (Fig. 2B and D).
Figure 2.
CEP120:p.R947H negatively regulates meiotic spindle microtubules in mouse oocytes. (A) Localization of endogenous CEP120 at metaphase of meiosis I (MI). In overlay: CEP120 (green), α-tubulin (red) and DNA (blue). (B) Metaphase MI oocytes expressing exogenous enhanced green fluorescent protein (eGFP)-CEP120. In overlay: eGFP-CEP120 (green), α-tubulin (red) and DNA (blue). (C) Schematic of secondary structure of CEP120 protein (adapted from Lin et al. (2013)) with the amino acid change indicated in red. MT, microtubule. Protein alignment indicates Arg947 amino acid conservation between human and mouse. (D) Localization of the wild type (WT) and mutant eGFP-CEP120 constructs at prophase of MI. Arrowheads indicate GFP-positive foci. (E) Localization of exogenous CEP120 at metaphase of MI. In the overlay: eGFP-CEP120 (green), phosphorylated Aurora kinase (pAURK; red) and DNA (blue). (F) Western blot detecting eGFP-CEP120 and eGFP-CEP120:p.R947H exogenous proteins. α-Tubulin served as loading control. The graph below the western blot image is the quantification of eGFP/α-tubulin ratio. Bars and error bars indicate mean ± SEM. (G) Oocytes injected with the indicated constructs were fixed at metaphase of MI and stained to detect the spindle (α-tubulin; gray) and DNA (blue). (H) Quantification of the relative pixel intensity a-tubulin in (G) (n = 65, 53, 57 for C, WT and R947H, respectively). Bars and error bars indicate mean ± SD. In panel A, B, D, E, G: a representative oocyte is shown for each group. n, total number of oocytes analyzed in each group. In panel F, H: C, control; WT, eGFP-CEP120; R947H, eGFP-CEP120:p.R947H. Student’s t-test was used to determine significance. n.s., not significant; *P < 0.05. Scale bar: 5 μm.
Next, we asked whether the variant identified in this study, p.Arg947His (rs2303720), could affect the localization of CEP120. The variant p.Arg947His resides at the C-terminus of the protein within the CEP120 dimerization domain (Fig. 2C). The amino acid 700-988 region of CEP120 protein is both necessary and sufficient for centriolar localization of CEP120 in somatic cells (Mahjoub et al., 2010). To assess whether p.Arg947His affects the localization of CEP120, we overexpressed either GFP-tagged WT or the CEP120:p.R947H mutant in otherwise WT mouse oocytes, and compared their localization at prophase of MI and at metaphase of MI. Localization of the eGFP-CEP120:p.R947H at aMTOCs at prophase I (Fig. 2D) and at spindle poles (Fig. 2E) at metaphase of MI was indistinguishable from WT CEP120, indicating that the variant does not impact protein localization in mouse oocytes.
Arg947 is important for microtubule nucleation during meiosis
To understand how the p.R947H change could affect CEP120 function in acentriolar spindle formation, we performed gain-of-function experiments through overexpression of WT and mutant cRNAs. First, we confirmed that WT and mutant constructs were expressed to similar levels by western blotting (Fig. 2F). Then, we measured the intensity of the spindle by assessing α-tubulin intensity at metaphase of MI. Overexpression of eGFP-CEP120 in mouse oocytes caused a modest increase in intensity of α-tubulin (Fig. 2G and H). In contrast, overexpression of the eGfp-Cep120R947H construct caused a reduction in α-tubulin intensity compared to control oocytes (Fig. 2G and H), suggesting that the p.R947H residue plays a regulatory role in microtubule nucleation or stability.
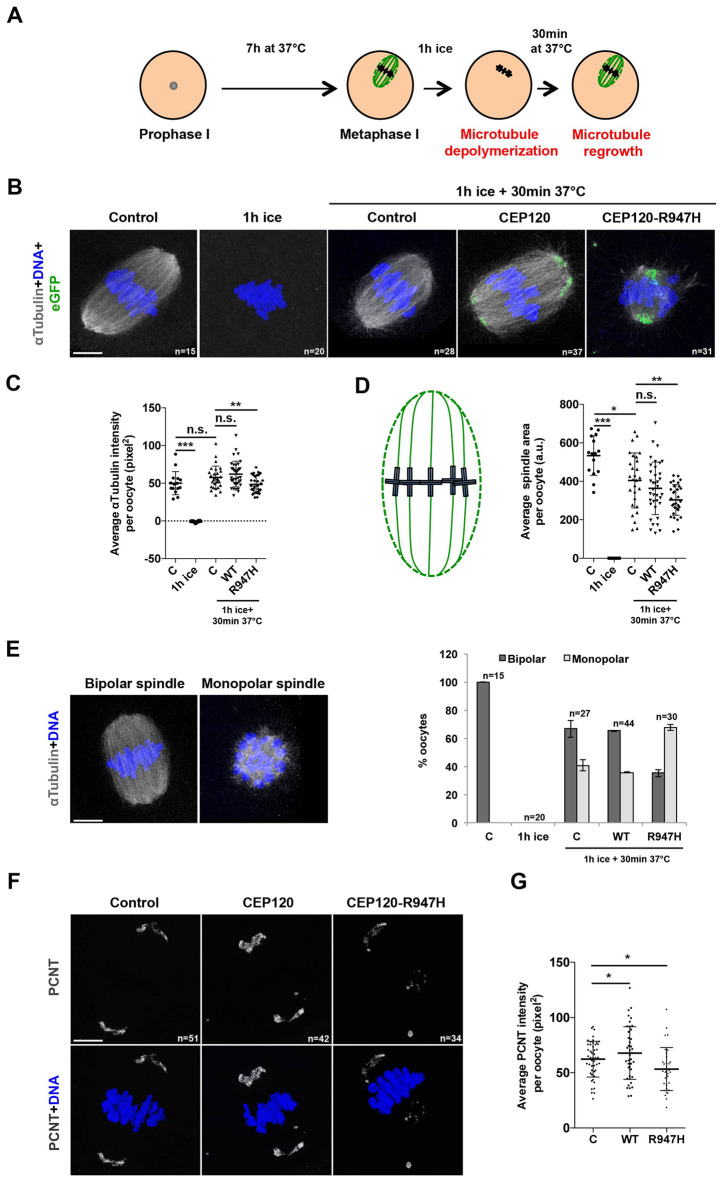
To test CEP120’s involvement in microtubule nucleation, we performed a microtubule nucleation assay whereby microtubules were depolymerized by cold treatment and their regrowth was monitored upon warming (Fig. 3A). At metaphase of MI, control oocytes contained bipolar spindles and chromosomes aligned along the metaphase plate (Fig. 3B, first panel). As expected, cold treatment of the oocytes resulted in complete microtubule depolymerization (Fig. 3B, second panel). Warming the oocytes was sufficient to restore the bipolar spindles in control oocytes (Fig. 3B, third panel; Fig. 3C). Importantly, the average intensity of microtubule regrowth in oocytes expressing eGFP-CEP120:p.R947H was reduced compared to control and to eGFP-CEP120-expressing oocytes (Fig. 3B and C). Consistent with reduced microtubules, overexpression of the mutant construct significantly reduced the spindle area compared to control and eGFP-CEP120-injected oocytes (Fig. 3D). Taken together, these results suggest that p.R947H delays microtubule regrowth.
Figure 3.
CEP120:p.R947H delays meiotic microtubule nucleation by interfering with acentriolar microtubule-organizing centers (aMTOCs) in mouse oocytes. (A) Experimental scheme: prophase MI oocytes injected with the indicated construct were matured in vitro until metaphase of MI. The oocytes were then cold treated for 1 h to depolymerize all microtubules followed by recovery by incubation in warm culture media for 30 min for microtubule regrowth assessment. (B–E) Control oocytes were fixed at metaphase of MI before cold treatment, 1 h after cold treatment, or 30 min after recovery; whereas, eGFP-CEP120 and eGFP-CEP120:p.R947H overexpressing oocytes were fixed 30 min after recovery. All fixed oocytes were then stained to detect the spindle with α-tubulin (gray) and DNA (blue). (B) Representative figures showing the shape of the spindles in each condition. (C) Quantification of the pixel intensity of α-tubulin in (B). (D) Quantification of the spindle area (green dashed line) in oocytes according to the schematic shown (a.u., arbitrary unit) in (B). (E) Left: Representative images of bipolar and monopolar spindles. Right: Quantifications of bipolar and monopolar spindles in each condition. (F) Oocytes injected with the indicated constructs were fixed at metaphase of MI and stained to detect aMTOCs with PCNT (gray) and DNA (blue). (G) Quantification of the pixel intensity PCNT in (F). Student’s t-test was used to determine significance. n, total number of oocytes examined. Indicated are mean ± SD (C, D) or mean ± SEM (E). **P < 0.001, ***P < 0.0001. Scale bar: 5 μm.
CEP120:p.Arg947His abrogates aMTOC formation
While performing the microtubule nucleation assay, we noticed a higher number of eGFP-CEP120:p.R947H-expressing oocytes with a monopolar spindle compared to control (by 1.6-fold) and eGFP-CEP120-expressing (by 1.8-fold) oocytes (Fig. 3E). We speculated that these oocytes were arrested at a stage resembling early prometaphase, most likely due to defective formation of aMTOCs. To test this hypothesis, we examined the appearance of PCNT upon overexpression of WT and CEP120:p.R947H proteins. PCNT is an integral component of mouse aMTOCs and plays an important role in regulating meiotic spindle formation (Ma and Viveiros, 2014; Baumann et al., 2017). PCNT immunostaining of metaphase MI oocytes significantly increased when oocytes overexpressed WT CEP120 compared to control-injected oocytes. In contrast, PCNT-foci intensities significantly decreased following eGfp-Cep120R947H overexpression (Fig. 3F and G). Taken together, these results suggest that CEP120:p.R947H plays a role in the organization of the microtubule arrays in meiosis, possibly via controlling aMTOC formation and/or structure. The presented data are consistent with R947 being important for CEP120 function and that the variant identified has a negative effect.
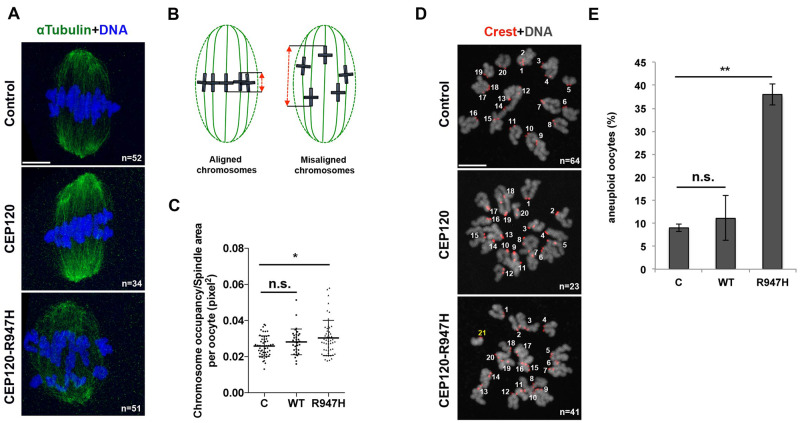
CEP120:p.R947H perturbs chromosome alignment and increases aneuploidy frequency
While evaluating the effects of the mutant overexpression on the spindle and aMTOC formation, we noticed that some oocytes showed aberrant chromosome alignment along the metaphase plate. Consistent with microtubule defects triggered by p.R947H, oocytes expressing the p.R947H mutant had significantly more mis-aligned chromosomes along the metaphase plate compared to oocytes overexpressing WT CEP120 or to controls (Fig. 4A–C).
Figure 4.
CEP120 p.R947 is important to maintain euploid mouse eggs. (A) Oocytes injected with the indicated construct were fixed at metaphase of MI and stained to detect spindles with α-tubulin (green) and DNA (blue). (B) Quality of chromosome alignment was quantified according to the schematic. The distance between the two extreme chromosome bivalents (red dashed line) was normalized to the total spindle area (green dashed line). (C) Quantification of the chromosome alignment in oocytes from (A). Student’s t-test was used to determine significance. (D) Oocytes injected with the indicated construct were matured to metaphase of meiosis II (MII) to perform an in situ chromosome counting assay. Centromeres were detected with anti-centromeric antigen (Crest; red) and DNA was detected with 4′,6-diamidino-2-phenylindole staining (gray). Shown is an eGFP-CEP120:p.R947H-expressing egg with an abnormal number of chromosomes. The extra chromosome is indicated in yellow. (E) Aneuploid eggs at metaphase of MII were counted and expressed as a proportion. Student’s t-test was used to determine significant differences in aneuploid egg frequency between conditions. n, total number of analyzed oocytes. Indicated are mean ± SD (C) or mean ± SEM (E). Student’s t-test was used to determine significance. n.s., not significant; **P < 0.001. Scale bar: 5 μm.
Finally, to assess the consequences of these chromosome alignment and spindle formation defects on chromosome segregation errors, we matured oocytes to metaphase of MII and performed in situ chromosome spreads to assess chromosome number. As anticipated, overexpression of eGfp-Cep120R947H caused an increase in the proportion of aneuploid MII eggs (∼40%) compared to control MI oocytes or MI oocytes microinjected with WT CEP120 (Fig. 4D and E). In our experiment, the injected mutant competes with the endogenous WT CEP120, which follows a definition of a dominant negative mutation. Of note, we only observe about 40% of the oocytes to be affected, which could be explained by either of the following. One, there is not a complete penetrance of the injected mutant to the oocytes. Two, the level to which this construct is expressed in a given oocyte is crucial in order to observe the phenotype. This could in turn suggest a dosage effect acting upon the mutation we describe. Together, these data suggest that the R947 residue in CEP120 is critical for chromosome alignment, a fundamental prerequisite for faithful chromosome segregation in MI.
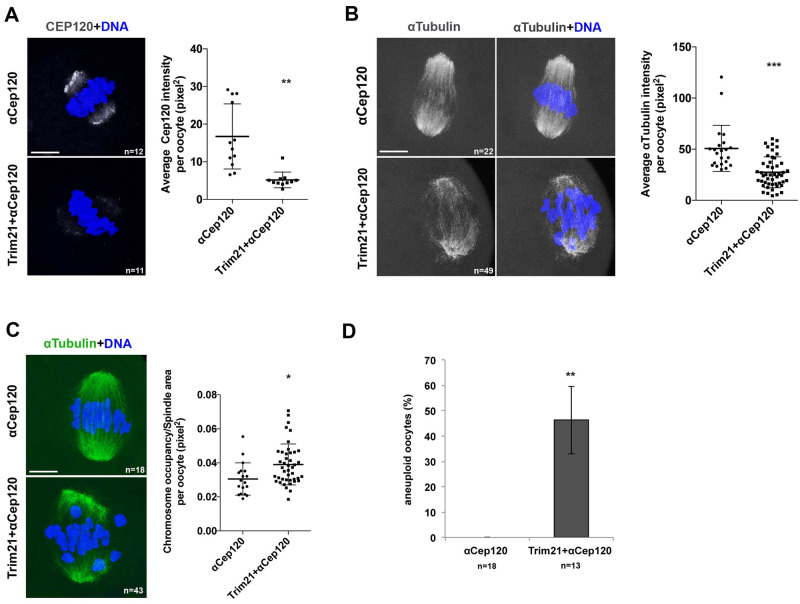
Depletion of endogenous CEP120 phenocopies eGFP-CEP120:p.R947H overexpression
Seven of eight HRG patients that harbor the rs2303720 variant were heterozygous. To explore the mechanism by which Cep120:pR947 could be acting, we compared the effects of eGFP-CEP120:p.R947H overexpression to those where CEP120 has been depleted. To deplete CEP120, we used the Trim-Away strategy (Clift et al., 2017). By immunostaining oocytes co-injected with Trim21-mCherry cRNA and anti-CEP120 antibody, we found that the levels of endogenous CEP120 protein were significantly reduced, by ∼60%, compared to those microinjected with anti-CEP120 alone (Fig. 5A). Similar to overexpression of eGFP-CEP120:p.R947H, we observed that CEP120 depletion resulted in a significant decrease of α-tubulin intensity (Fig. 5B), severe MI chromosome alignment defects (Fig. 5C) and a significant increase in aneuploid egg frequency (Fig. 5D). In summary, these data suggest that CEP120 participates in microtubule nucleation during meiotic spindle formation, which in turn directs chromosome alignment and fidelity of chromosome segregation during MI. Also, we infer that R947 is critical for CEP120 function in meiosis, and it is possible either that its mutation could transform CEP120 into a non-functional dominant negative protein or that this mutant may have a dosage effect when present in the heterozygous state.
Figure 5.
In mouse oocytes, CEP120 depletion phenocopies WT oocytes overexpressing CEP120:p.R947H. (A–C) Prophase MI oocytes were microinjected with α-Cep120 or co-microinjected with mCherry-Trim21 and α-Cep120 and matured to metaphase of MI prior to detecting CEP120 (A) and α-tubulin (B, C). Chromosomes are shown in blue. On the right of each panel are corresponding quantifications of CEP120 intensity (A), α-tubulin intensity (B) or chromosome alignment (C). (D) Quantification of the number of the aneuploid eggs in control (α-Cep120) and CEP120-depleted oocytes (mCherry-Trim21 + α-Cep120). n, number of oocytes (A–C) or eggs (D) analyzed. Indicated are mean ± SD (A–C) or mean ± SEM (D). Student’s t-test was used to determine significance. *P < 0.05, **P < 0.001, ***P < 0.0001. Scale bar: 5 μm.
Discussion
According to the latest Centers for Disease Control statistics, ∼12% of women in the USA of reproductive age are subfertile, taking longer than average to achieve a pregnancy. When pregnancies are not achieved or maintained, some of these women may seek fertility treatments and require multiple cycles before success. An estimated 50% of these idiopathic sub- or infertility cases have genetic causes, with aneuploidy accounting for a significant proportion of IVF failures and early miscarriages. Yet despite advances in understanding genetic basis of many diseases, we know little about human gene variants causative of female subfertility. As a consequence, advanced maternal age, prior miscarriage or failed IVF cycles are the most common referral reasons for a preimplantation genetic testing for aneuploidy (PGT-A)—a diagnostic test to select euploid embryos for transfer to improve implantation chances and achieve a healthy pregnancy. Such predictors, however, do not fully rule out an unhealthy pregnancy and giving birth to aneuploid offspring. For genomic studies, it would be ideal to have egg aneuploidy data from a large collection of patients, but this would require polar body biopsies. To our knowledge, a DNA bank of samples associated with this phenotype does not exist. For this reason and because maternal meiosis is the primary source of aneuploid embryos, we compared patients with high and low frequencies of aneuploid blastocysts to identify genetic factors responsible for chromosome mis-segregation errors. This sample selection approach was designed to identify genes and variants that show clear discrepancy between the two groups with opposite phenotypes and have been previously used to study genetic factors of complex diseases, including aneuploidy (Bruse et al., 2016; Nguyen et al., 2017).
Following exome sequencing and gene burden analysis, we identified several candidate genes with roles in meiosis or mitosis associated with increased risk of aneuploidy. Notably, we identified an enrichment of human candidate genes that likely perturb spindle building in the female germline thereby increasing the incidence of aneuploid egg production. For functional analysis we selected CEP120 with an identified non-synonymous variant, p.Arg947His, located in the C-terminus of the protein. We showed that the p.Arg947His change does not affect CEP120 localization at metaphase of MI in oocyte meiosis. However, it impairs microtubules during meiotic spindle formation and decreases meiotic chromosome segregation fidelity. We also showed that the p.R947H substitution impairs PCNT recruitment to aMTOCs, which together suggests that p.Arg947His may be reducing affinity for CEP120-binding partners, interfering with their localization and/or proper functioning, such as aMTOC clustering. If this CEP120 function is specific to aMTOC-containing cell types, this defect would not be detrimental for a carrier’s health, consistent with the otherwise healthy presentation of these patients.
Oocytes from most organisms lack centrioles, the organelle that nucleates microtubules and establishes spindle bipolarity in somatic cells (Radford et al., 2017). Instead, mouse oocytes use aMTOCs to build a meiotic, bipolar spindle. These aMTOCs contain many of the proteins found in mitotic spindles such as pericentriolar material, regulatory proteins, motor proteins and proteins that make up a newly described liquid-like spindle domain (So et al., 2019). For instance, CEP120 localizes with proteins that constitute pericentriolar material such as CDK5 regulatory subunit-associated protein 2 (CDK5RAP2), PCNT and γ-tubulin. Intriguingly, an analysis of human oocytes that were donated as discarded material from IVF cycles suggested that meiotic spindles lack aMTOCs and that a RanGTP chromatin-driven pathway is instead responsible for spindle building in humans (Holubcová et al., 2015). The spindle gene enrichment in our dataset raises the possibility that there are some aMTOC components in human oocytes that evaded detection limits or are defective in low-quality donated oocyte samples. There are at least two reports documenting evidence that human oocytes do contain aMTOCs (George et al., 1996; Roeles and Tsiavaliaris, 2019). Therefore, evaluation of the candidate genes we provide and their mechanism of action in human oocytes will be an important step forward to understand the biological significance of the identified genetic variants during human female meiosis.
It is noteworthy that mutations in CEP120 can cause different types of diseases, such as ciliopathies (Shaheen et al., 2015; Roosing et al., 2016; Joseph et al., 2018). For instance, another nearby variant (p.I975S) disrupts CEP120 function by decreasing its affinity toward C2 domain-containing 3 centriole elongation regulator (C2CD3) binding and impairing cilia formation (Tsai et al., 2019). Given the broad spectrum of clinical manifestation related to CEP120 mutagenesis, a comprehensive understanding of CEP120 binding partners is necessary to elucidate the molecular mechanisms underlying the breadth of observed clinical features. Nevertheless, we identified a common variant with potential severe consequences on embryo chromosome status, which makes it a compelling subfertility variant for prescreening of mothers. Also, the identified heterozygous mutation p.Arg947His has an apparent dominant negative effect exerted upon CEP120 function. This is in stark contrast to all nine previously reported pathogenic CEP120 variants which were observed in biallelic state and were either rare or novel (Roosing et al., 2016). Interestingly, mice heterozygous for a mutant BUB1 mitotic checkpoint serine/threonine kinase (BUB1) protein also show rapid fertility decline, suggesting either a dominant negative or dosage-dependent mechanism (Leland et al., 2009). As such, these effects of gene variants causing subfertility are mechanisms that need further exploration.
The strength of our study is the use of exome sequencing on a unique collection of subfertile patients. To control for genetic difference among populations, we limited our study to the largest self-reported ethnic group in the collection: White, non-Hispanic. Even with a limited sample size, we saw an enrichment for the centriole-related genes in our top candidate gene list, many of which are likely to be associated with aneuploidy. Because of the limitation of the Ion Torrent technology, we only focused on SNVs. With a larger sample size and a more comprehensive set of genetic variants, we expect to uncover additional genes that contribute to female aneuploidy. Another limitation of our study is the use of mouse oocytes to study the function of identified variants. Human oocytes appear to use a chromatin-driven spindle building pathway in contrast to mouse oocytes which use an MTOC-driven pathway (Holubcová et al., 2015). Thus, the defects we observed upon ectopic expression of the Cep120 variant may alter mouse oocyte meiosis differently than human oocyte meiosis. These differences could further extend into the mitotic divisions of the preimplantation embryo, where human embryos inherit centrioles from the sperm (Sathananthan et al., 1991), and mouse embryos do not (Schatten et al., 1986). We therefore must evaluate the effects of this variant in a human meiotic spindle building system that appears to lack MTOCs to further implicate this variant in causing human egg aneuploidy.
One important finding of this study is that many candidate variants are common in the general population. An extensive list of infertility variants opens possibilities for developing diagnostic tests that would allow screening of subfertile patients to identify those that have a better prognosis of IVF treatment success, those that would require multiple IVF cycles or those that could be counseled regarding alternative family planning options. Once a list is examined, we envision the next step involving gene editing to create ‘humanized’ animal models to assess any biological consequences of expressing the gene variants on the organisms (Singh and Schimenti, 2015). We anticipate that genes we continue to identify will extend the list of key mutations contributing to the risk of aneuploidy, with a scope of allowing prescreening patients that have better chances to benefit from assisted reproduction practice.
Data availability
The SNVs identified in this study, along with their allele frequencies in sequenced samples, are available at Rutgers University Community Repository (RUcore): https://rucore.libraries.rutgers.edu/rutgers-lib/61172/.
Authors’ roles
N.R.T., R.T.S., K.S. and J.X. conceived the project and designed the overall strategy. R.T.S. recruited the samples. K.M.T., W.E., A.B., J.L., Y.Z. and X.T. performed the data analysis. W.E. performed the functional validation. X.T. performed the Sanger validation. K.M.T., W.E., K.S. and J.X. drafted the manuscript. All authors reviewed and edited the manuscript.
Funding
National Institutes of Health/National Institute for Child Health and Development (R01-HD091331 to K.S., J.X. and X.T.); an EMD Serono Grant for Fertility Innovation (to N.R.T.) for sequencing.
Conflict of interest
N.R.T. is a shareholder and an employee of Genomic Prediction.
Supplementary Material
References
- Alazami AM, Awad SM, Coskun S, Al-Hassan S, Hijazi H, Abdulwahab FM, Poizat C, Alkuraya FS. TLE6 mutation causes the earliest known human embryonic lethality. Genome Biol 2015;16: 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann C, Wang X, Yang L, Viveiros MM. Error-prone meiotic division and subfertility in mice with oocyte-conditional knockdown of pericentrin. J Cell Sci 2017;130:1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruse S, Moreau M, Bromberg Y, Jang JH, Wang N, Ha H, Picchi M, Lin Y, Langley RJ, Qualls C et al. Whole exome sequencing identifies novel candidate genes that modify chronic obstructive pulmonary disease susceptibility. Hum Genomics 2016;10:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carp H, Toder V, Aviram A, Daniely M, Mashiach S, Barkai G. Karyotype of the abortus in recurrent miscarriage. Fertil Steril 2001;75:678–682. [DOI] [PubMed] [Google Scholar]
- Chatot CL, Ziomek CA, Bavister BD, Lewis JL, Torres I. An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. J Reprod Fertil 1989;86:679–688. [DOI] [PubMed] [Google Scholar]
- Chen B, Zhang Z, Sun X, Kuang Y, Mao X, Wang X, Yan Z, Li B, Xu Y, Yu M et al. Biallelic mutations in PATL2 cause female infertility characterized by oocyte maturation arrest. Am J Hum Genet 2017;101:609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clift D, McEwan WA, Labzin LI, Konieczny V, Mogessie B, James LC, Schuh M. A method for the acute and rapid degradation of endogenous proteins. Cell 2017;171:1692–1706.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comartin D, Gupta GD, Fussner E, Coyaud E, Hasegan M, Archinti M, Cheung SW, Pinchev D, Lawo S, Raught B et al. CEP120 and SPICE1 cooperate with CPAP in centriole elongation. Curr Biol 2013;23:1360–1366. [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST et al. The variant call format and VCFtools. Bioinformatics 2011;27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellmeth JE, Ghanaim EM, Schindler K. Characterization of macrozoospermia-associated AURKC mutations in a mammalian meiotic system. Hum Mol Genet 2016;25:2698–2711. [DOI] [PubMed] [Google Scholar]
- Feng R, Sang Q, Kuang Y, Sun X, Yan Z, Zhang S, Shi J, Tian G, Luchniak A, Fukuda Y et al. Mutations in TUBB8 and human oocyte meiotic arrest. N Engl J Med 2016. a;374:223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng R, Yan Z, Li B, Yu M, Sang Q, Tian G, Xu Y, Chen B, Qu R, Sun Z et al. Mutations in TUBB8 cause a multiplicity of phenotypes in human oocytes and early embryos. J Med Genet 2016. b;53:662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franasiak JM, Forman EJ, Hong KH, Werner MD, Upham KM, Treff NR, Scott RT. Aneuploidy across individual chromosomes at the embryonic level in trophectoderm biopsies: changes with patient age and chromosome structure. J Assist Reprod Genet 2014;31:1501–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George MA, Pickering SJ, Braude PR, Johnson MH. The distribution of alpha- and gamma-tubulin in fresh and aged human and mouse oocytes exposed to cryoprotectant. Mol Hum Reprod 1996;2:445–456. [DOI] [PubMed] [Google Scholar]
- Guo MH, Plummer L, Chan YM, Hirschhorn JN, Lippincott MF. Burden testing of rare variants identified through exome sequencing via publicly available control data. Am J Hum Genet 2018;103:522–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold T, Chiu D. Maternal age-specific rates of numerical chromosome abnormalities with special reference to trisomy. Hum Genet 1985;70:11–17. [DOI] [PubMed] [Google Scholar]
- Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet 2001;2:280–291. [DOI] [PubMed] [Google Scholar]
- Hogge WA, Byrnes AL, Lanasa MC, Surti U. The clinical use of karyotyping spontaneous abortions. Am J Obstet Gynecol 2003;189:397–400. [DOI] [PubMed] [Google Scholar]
- Holubcová Z, Blayney M, Elder K, Schuh M. Human oocytes. Error-prone chromosome-mediated spindle assembly favors chromosome segregation defects in human oocytes. Science 2015;348:1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Huff CD, Moore B, Flygare S, Reese MG, Yandell M. VAAST 2.0: improved variant classification and disease-gene identification using a conservation-controlled amino acid substitution matrix. Genet Epidemiol 2013;37:622–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009. a;37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009. b;4:44–57. [DOI] [PubMed] [Google Scholar]
- Hutchins JR, Toyoda Y, Hegemann B, Poser I, Heriche JK, Sykora MM, Augsburg M, Hudecz O, Buschhorn BA, Bulkescher J et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 2010;328:593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi H, Knott JG, Schultz RM, Williams CJ. Alterations of PLCbeta1 in mouse eggs change calcium oscillatory behavior following fertilization. Dev Biol 2007;312:321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph N, Al-Jassar C, Johnson CM, Andreeva A, Barnabas DD, Freund SMV, Gergely F, van Breugel M. Disease-associated mutations in CEP120 destabilize the protein and impair ciliogenesis. Cell Rep 2018;23:2805–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek D, Hornak M, Horak J, Navratil R, Tauwinklova G, Rubes J, Vesela K. Incidence and origin of meiotic whole and segmental chromosomal aneuploidies detected by karyomapping. Reprod Biomed Online 2019;38:330–339. [DOI] [PubMed] [Google Scholar]
- Kuliev A, Zlatopolsky Z, Kirillova I, Spivakova J, Cieslak Janzen J. Meiosis errors in over 20,000 oocytes studied in the practice of preimplantation aneuploidy testing. Reprod Biomed Online 2011;22:2–8. [DOI] [PubMed] [Google Scholar]
- Leland S, Nagarajan P, Polyzos A, Thomas S, Samaan G, Donnell R, Marchetti F, Venkatachalam S. Heterozygosity for a Bub1 mutation causes female-specific germ cell aneuploidy in mice. Proc Natl Acad Sci U S A 2009;106:12776–12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-N, Wu C-T, Lin Y-C, Hsu W-B, Tang C-JC, Chang C-W, Tang TK. CEP120 interacts with CPAP and positively regulates centriole elongation. J Cell Biol 2013;202:211–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Viveiros MM. Depletion of pericentrin in mouse oocytes disrupts microtubule organizing center function and meiotic spindle organization. Mol Reprod Dev 2014;81:1019–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahjoub MR, Xie Z, Stearns T. Cep120 is asymmetrically localized to the daughter centriole and is essential for centriole assembly. J Cell Biol 2010;191:331–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy RC, Demko Z, Ryan A, Banjevic M, Hill M, Sigurjonsson S, Rabinowitz M, Fraser HB, Petrov DA. Common variants spanning PLK4 are associated with mitotic-origin aneuploidy in human embryos. Science 2015. a;348:235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy RC, Demko ZP, Ryan A, Banjevic M, Hill M, Sigurjonsson S, Rabinowitz M, Petrov DA. Evidence of selection against complex mitotic-origin aneuploidy during preimplantation development. PLoS Genet 2015. b;11:e1005601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy RC, Newnham LJ, Ottolini CS, Hoffmann ER,, Chatzimeletiou K, Cornejo OE, Zhan Q, Zaninovic N, Rosenwaks Z, Petrov DA et al. Tripolar chromosome segregation drives the association between maternal genotype at variants spanning PLK4 and aneuploidy in human preimplantation embryos. Hum Mol Genet 2018;27:2573–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol 2016;17:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka SI, Hassold TJ, Hunt PA. Human aneuploidy: mechanisms and new insights into an age-old problem. Nat Rev Genet 2012;13:493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AL, Marin D, Zhou A, Gentilello AS, Smoak EM, Cao Z, Fedick A, Wang Y, Taylor D, Scott RT Jr et al. Identification and characterization of Aurora kinase B and C variants associated with maternal aneuploidy. Mol Hum Reprod 2017;23:406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010;26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford SJ, Nguyen AL, Schindler K, McKim KS. The chromosomal basis of meiotic acentrosomal spindle assembly and function in oocytes. Chromosoma 2017;126:351–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenkova E, Eijpe M, Heyting C, Hodges CA, Hunt PA, Liebe B, Scherthan H, Jessberger R. Cohesin SMC1 beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat Cell Biol 2004;6:555–562. [DOI] [PubMed] [Google Scholar]
- Roeles J, Tsiavaliaris G. Actin-microtubule interplay coordinates spindle assembly in human oocytes. Nat Commun 2019;10:4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosing S, Romani M, Isrie M, Rosti RO, Micalizzi A, Musaev D, Mazza T, Al-Gazali L, Altunoglu U, Boltshauser E et al. Mutations in CEP120 cause Joubert syndrome as well as complex ciliopathy phenotypes. J Med Genet 2016;53:608–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Q, Li B, Kuang Y, Wang X, Zhang Z, Chen B, Wu L, Lyu Q, Fu Y, Yan Z et al. Homozygous mutations in WEE2 cause fertilization failure and female infertility. Am J Hum Genet 2018;102:649–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathananthan AH, Kola I, Osborne J, Trounson A, Ng SC, Bongso A, Ratnam SS. Centrioles in the beginning of human development. Proc Natl Acad Sci U S A 1991;88:4806–4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatten H, Schatten G, Mazia D, Balczon R, Simerly C. Behavior of centrosomes during fertilization and cell division in mouse oocytes and in sea urchin eggs. Proc Natl Acad Sci U S A 1986;83:105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M, Ellenberg J. Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell 2007;130:484–498. [DOI] [PubMed] [Google Scholar]
- Shaheen R, Schmidts M, Faqeih E, Hashem A, Lausch E, Holder I, Superti-Furga A, Mitchison HM, Almoisheer A, Alamro R et al. A founder CEP120 mutation in Jeune asphyxiating thoracic dystrophy expands the role of centriolar proteins in skeletal ciliopathies. Hum Mol Genet 2015;24:1410–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Schimenti JC. The genetics of human infertility by functional interrogation of SNPs in mice. Proc Natl Acad Sci U S A 2015;112:10431–10436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So C, Seres KB, Steyer AM, Monnich E, Clift D, Pejkovska A,, Mobius W, Schuh M. A liquid-like spindle domain promotes acentrosomal spindle assembly in mammalian oocytes. Science 2019;364:eaat9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J-J, Hsu W-B, Liu J-H, Chang C-W, Tang TK. CEP120 interacts with C2CD3 and Talpid3 and is required for centriole appendage assembly and ciliogenesis. Sci Rep 2019;9: 6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virant-Klun I, Leicht S, Hughes C, Krijgsveld J. Identification of maturation-specific proteins by single-cell proteomics of human oocytes. Mol Cell Proteomics 2016;15:2616–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Shu L, Cai L, Sun X, Cui Y, Liu J. Homozygous missense mutation Arg207Cys in the WEE2 gene causes female infertility and fertilization failure. J Assist Reprod Genet 2019;36:965–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Li G, Zhang L, Sun X, Zhang D, Lu J, Ma J, Yan J, Chen ZJ. Maternal common variant rs2305957 spanning PLK4 is associated with blastocyst formation and early recurrent miscarriage. Fertil Steril 2017;107:1034–1040.e5. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Mu J, Zhao J, Zhou Z, Chen B, Wu L, Yan Z, Wang W, Zhao L, Dong J et al. Novel mutations in WEE2: expanding the spectrum of mutations responsible for human fertilization failure. Clin Genet 2019;95:520–524. [DOI] [PubMed] [Google Scholar]
- Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 2012;28:3326–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The SNVs identified in this study, along with their allele frequencies in sequenced samples, are available at Rutgers University Community Repository (RUcore): https://rucore.libraries.rutgers.edu/rutgers-lib/61172/.