Abstract
Alternative translation initiation (ATLI) refers to the existence of multiple translation initiation sites per gene and is a widespread phenomenon in eukaryotes. ATLI is commonly assumed to be advantageous through creating proteome diversity or regulating protein synthesis. We here propose an alternative hypothesis that ATLI arises primarily from nonadaptive initiation errors presumably due to the limited ability of ribosomes to distinguish sequence motifs truly signaling translation initiation from similar sequences. Our hypothesis, but not the adaptive hypothesis, predicts a series of global patterns of ATLI, all of which are confirmed at the genomic scale by quantitative translation initiation sequencing in multiple human and mouse cell lines and tissues. Similarly, although many codons differing from AUG by one nucleotide can serve as start codons, our analysis suggests that using non-AUG start codons is mostly disadvantageous. These and other findings strongly suggest that ATLI predominantly results from molecular error, requiring a major revision of our understanding of the precision and regulation of translation initiation.
Keywords: evolution, molecular error, natural selection, start codon, translational amount
Introduction
The translation of mRNA in eukaryotes is a complex multistep process consisting of initiation, elongation, and termination, among which initiation is considered the most important step because it largely determines the amount of translation (Jackson et al. 2010; Aitken and Lorsch 2012). During the initiation, the initiator tRNA and 40S and 60S ribosomal subunits are assembled by initiation factors into an 80S ribosome at a start codon in an mRNA molecule to initiate the translation (Pestova et al. 2001). Most initiation events occur via a cap-dependent mechanism requiring the recognition of the m7G(5′)ppp(5′)N structure termed cap that is located at the 5′ end of most mRNA molecules, whereas a minority of initiation events occur by a cap-independent mechanism that relies on internal ribosome entry site (IRES) elements in the 5′ untranslated region (UTR) of mRNAs (Sonenberg and Hinnebusch 2009; Martinez-Salas et al. 2012). Both mechanisms primarily abide by a scanning model in which the 40S ribosome subunit does not bind to a start codon directly but first binds to the cap or IRES and then scans the mRNA until meeting a start codon (Sonenberg and Hinnebusch 2009; Jackson et al. 2010). Needless to say, translation initiation regulation is a crucial step in the control of protein synthesis (Pestova et al. 2001; Sonenberg and Hinnebusch 2009; Jackson et al. 2010; Martinez-Salas et al. 2012).
For a given gene, translation may start from one of several translation initiation sites (TISs), a phenomenon known as alternative translation initiation (ATLI) (Kochetov 2008). ATLI may occur in the same transcript by leaky scanning, reinitiation, or IRES-dependent initiation (Kochetov 2008). It may also occur because of the presence of multiple transcript isoforms resulting from alternative transcriptional start or alternative splicing (de Klerk and 't Hoen 2015). ATLI may lead to the formation of upstream open reading frames (uORFs), whose translation tends to repress the translation of the main ORF (Morris and Geballe 2000; Johnstone et al. 2016). For example, a mutation in the 5′ UTR of human CDKN2A creates a uORF that suppresses the translation of CDKN2A, which predisposes the affected individuals to melanoma (Liu et al. 1999). ATLI may cause the production, from the same gene, of multiple proteins with different sequences due to in-frame extensions, truncations, or frameshifts (Kochetov 2008) and these different protein isoforms may possess different functions. For instance, the human la antigen-associated invariant chain (ln) exists in two major forms, p33 and p35, as a result of ATLI (Strubin et al. 1986). In another example, human MAVS produces a full-length MAVS and a truncated mini-MAVS that are functionally different from each other; although both proteins positively regulate cell death, mini-MAVS interferes with interferon production induced by full-length MAVS (Brubaker et al. 2014).
ATLI may also arise from the use of non-AUG start codons; in most of such cases, a near-cognate codon, which differs from AUG by one nucleotide (e.g., CUG, AGG, and AUA), serves as the start codon (Peabody 1989; Kearse and Wilusz 2017). Despite translation initiation from non-AUG codons, proteins synthesized may still be functional. For example, DAP5, a translation initiation factor, plays a critical role in IRES-mediated translation initiation; its translation is initiated from a GUG start codon in human and mouse (Imataka et al. 1997).
Thanks to recent developments of high-throughput technologies for genome-wide studies of translation, ATLI and non-AUG initiation are now known to be widespread in eukaryotes (Ingolia et al. 2011; Fritsch et al. 2012; Lee et al. 2012). Even within one cell line, on average 2.5 and 2 TISs per gene have been reported in human and mouse, respectively (Lee et al. 2012; Wan and Qian 2014). Along with the previously known cases, the high prevalence of ATLI recently revealed from genomic studies led to the prevailing view that ATLI is a widely used, regulated, and beneficial mechanism to increase proteome diversity and regulate protein translation (Kochetov et al. 2005; Kochetov 2008; Bazykin and Kochetov 2011; Ivanov et al. 2011, 2017; Lee et al. 2012; Ingolia 2014, 2016; de Klerk and 't Hoen 2015; Kearse and Wilusz 2017). We will refer to this hypothesis as the adaptive hypothesis. It is important to stress that the adaptive hypothesis asserts that the vast majority if not all instances of ATLI are beneficial. For instance, Lee et al. (2012) discussed extensively the potential benefits of ATLI without mentioning the possibility that ATLI could be deleterious or neutral. Similarly, Kearse and Wilusz (2017) rejected the notion that non-AUG translation initiation is an error and wrote that “non-AUG initiation events are not simply errors but instead are used to generate or regulate proteins with key cellular functions.”
Nevertheless, only a tiny fraction of ATLI has verified functions, whereas most ATLI has no known functions. Furthermore, the number of AUGs in 5′ UTR is significantly lower than the random expectation (Iacono et al. 2005; Lynch et al. 2005; Zur and Tuller 2013), suggesting that upstream alternative TISs are selected against rather than selected for. These observations question whether ATLI is generally adaptive as many have assumed, because ATLI could also arise from imprecise translation initiation. We thus propose and test the hypothesis that most ATLI reflects nonadaptive molecular errors. Such errors could occur because of the chance occurrence of sequence motifs that resemble translation initiation signals. The error hypothesis makes a series of distinct predictions about genomic patterns of ATLI that are not expected a priori under the adaptive hypothesis. By analyzing high-throughput translation initiation data from multiple cell lines and tissues of humans and mice, we provide unequivocal evidence for the error hypothesis.
Results
TIS Diversity Decreases with the Amount of Translation
If ATLI is mostly caused by molecular errors, it can be deleterious for a number of reasons. First, it may lower the proportion of protein molecules with normal functions. Second, it could waste cellular resource and energy in protein synthesis and degradation. Third, some proteins produced via ATLI may even be toxic. Given the rate of translation initiation error, the detriment from the above second and third reasons should rise with the number of protein molecules synthesized. Consequently, the overall harm of imprecise translation initiation of a gene is expected to increase with its amount of translation. Hence, natural selection against translation initiation error of a gene intensifies with its amount of translation. As a result, the error rate and the extent of ATLI should decline with translational amount. By contrast, the adaptive hypothesis of ATLI does not predict this negative correlation a priori, because, under this hypothesis, the extent of ATLI of a gene depends on the specific function and regulation of the gene.
To distinguish between the error hypothesis and the adaptive hypothesis, we analyzed the TISs identified by quantitative translation initiation sequencing (QTI-seq) in human and mouse cells. QTI-seq enriches mRNA fragments protected by initiating ribosomes, enabling the capture of real-time translation initiation events in a quantitative manner (Gao et al. 2015). The TIS data sets used here are from five human samples (HeLa and its YTHDF1 mutant, HEK293 and its eIF2α mutant, and HEK293 under amino acid starvation) and four mouse samples (MEF cell line, MFF under amino acid starvation, normal mouse liver, and fasting mouse liver) (supplementary table S1, Supplementary Material online). Because only AUG and its nine near-cognate codons are thought to be able to serve as start codons (Peabody 1989; Kearse and Wilusz 2017), TISs in this study were predicted using these ten codons unless otherwise noted. To measure the extent of ATLI of a gene in a sample, we quantified its TIS diversity using the Simpson index (Simpson 1949) and Shannon index (Shannon 1948), following the recent studies of alternative transcriptional start and alternative polyadenylation (Xu and Zhang 2018; Xu et al. 2019). Briefly, both Simpson and Shannon indices consider the number of TISs and fractional use of each TIS, but the Simpson index gives more weights to the frequently used TISs than does the Shannon index. The higher the Simpson and Shannon indices, the greater the extent of ATLI. Because TIS diversity is poorly estimated when the number of QTI-seq reads mapped to a gene is too small, only genes with at least ten reads in a sample were analyzed. By counting TISs as well as QTI-seq reads mapped to each TIS in a gene, we calculated the gene’s TIS diversity. We measured the translational amount of the gene by the total number of QTI-seq reads mapped to the gene per million reads mapped in the entire sample (RPM; see Materials and Methods).
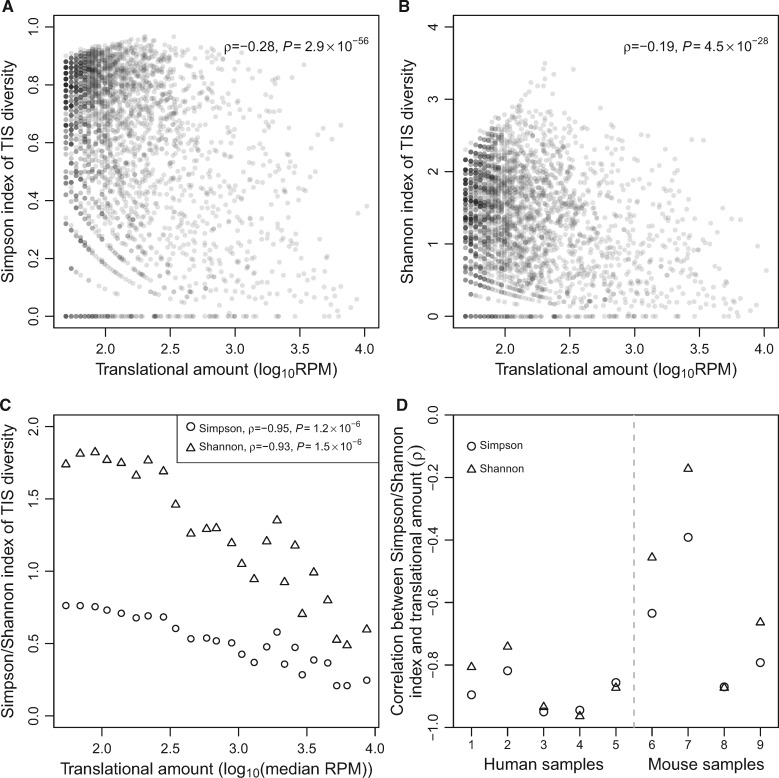
We started by analyzing the QTI-seq data from human HEK293 cells. Consistent with the prediction of the error hypothesis, the rank correlation (ρ) between the translational amount of a gene and its Simpson index of TIS diversity is significantly negative (ρ = −0.28, P = 2.9 × 10−56; fig. 1A). Using the Shannon index to measure TIS diversity similarly yielded a negative correlation (ρ = −0.19, P = 4.5 × 10−28; fig. 1B).
Fig. 1.
TIS diversity of a gene generally decreases with the translational amount of the gene. (A) The Simpson index of TIS diversity in human HEK293 cells declines with the translational amount. (B) The Shannon index of TIS diversity in human HEK293 cells declines with translational amount. In (A) and (B), each dot represents a gene. Spearman’s rank correlation coefficient (ρ) and associated P value are presented. RPM, number of QTI-seq reads mapped to a given gene per million reads mapped to all genes in the sample. (C) Simpson and Shannon indices of TIS diversity in HEK293 decline with translational amount. Each triangle/circle represents a supergene that is composed of many individual genes. All supergenes have the same total translational amount. X-axis shows the median translational amount of all genes belonging to the supergene. (D) Spearman’s correlation between median translational amount and Simpson or Shannon index of TIS diversity among supergenes in each sample examined. Sample IDs listed in the X-axis refer to those in supplementary table S1, Supplementary Material online. P < 0.05 for all correlations except for the mouse liver under fasting (sample #7).
Because sequencing depth and the precision of TIS survey for a gene rise with its translational amount, it is possible that the correlations in figure 1A and B originate from unequal TIS surveys among genes. To eliminate this potential bias, we constructed “supergenes” with equal sequencing depths. Specifically, we ranked genes by their QTI-seq read number and then grouped all genes into 25 bins such that each bin contained the same total read number. We then considered all genes in a bin together as a supergene. Briefly, reads from the most commonly used TIS of each gene were combined to represent the reads of the most commonly used TIS of the supergene, reads from the second most commonly used TIS of each gene were combined to represent the reads of the second most commonly used TIS of the supergene, and so on (see Materials and Methods). The number of TISs of the supergene is the maximum number of TISs of any gene belonging to the supergene. We then computed the TIS diversity of the supergene. The uniformity of read number among supergenes eliminates the potential bias aforementioned. Yet, the TIS diversity of a supergene decreases with the median read number (i.e., translational amount) of all genes belonging to the supergene, regardless of whether Simpson (ρ = −0.95, P = 1.2 × 10−6; fig. 1C) or Shannon index (ρ = −0.93, P = 1.5 × 10−6, fig. 1C) is used in measuring TIS diversity. Importantly, this negative correlation is apparent across the entire range of translational amount (fig. 1C). Other human and mouse samples similarly exhibit a negative correlation (fig. 1D).
To exclude the possibility that the above results are statistical artifacts of our analyses, we performed a computer simulation where we randomly generated genes whose read numbers and relative TIS usages respectively follow the actual distributions of the data from HEK293. We analyzed the simulated genes as if they were real genes but found a positive correlation between translational amount and TIS diversity among genes (supplementary fig. S1A and B, Supplementary Material online); this correlation was even stronger in the supergene analysis (supplementary fig. S1C, Supplementary Material online). These simulation results suggest that, if anything, our analyses are conservative in detecting the negative correlation between translational amount and TIS diversity among actual genes.
To investigate the robustness of our findings, we performed several additional analyses using the supergene approach. First, the TISs above considered were identified using the cutoff of P < 0.05 in Ribo-TISH (Zhang et al. 2017) (see Materials and Methods), so may contain some false positives. Using only the TISs under the cutoff of Q < 0.05 (i.e., after correcting for multiple testing) should reduce false positives. Nevertheless, we found the negative correlation between translational amount and TIS diversity among supergenes qualitatively unchanged (supplementary fig. S2A, Supplementary Material online). Second, even when only AUG was considered to be the start codon, the obtained result was qualitatively unchanged (supplementary fig. S2B, Supplementary Material online). Third, despite the validity of our analysis demonstrated by the simulation (supplementary fig. S1, Supplementary Material online), one might argue that it is better to measure translational amount and TIS diversity independently in order to study their biological relationship. Because the translational amount of a gene should rise with its mRNA concentration, we predict a negative correlation between mRNA concentration and TIS diversity. Indeed, mRNA concentration quantified by RNA sequencing (RNA-seq) and TIS diversity measured using QTI-seq are negatively correlated (supplementary fig. S2C, Supplementary Material online).
Because the likelihood of ATLI increases with the transcript length, we further computed the partial correlation between the translational amount of a gene and its TIS diversity after controlling for the transcript length of the gene, which is defined by the total length of its annotated UTRs and exons. However, because the transcript length cannot be controlled in the supergene approach, we used another statistical approach referred to as downsampling. Specifically, we downsampled our data by randomly picking ten QTI-seq reads per gene for all genes with at least ten reads to equalize the sequencing depths of different genes before estimating the Simpson or Shannon index (see Materials and Methods). We observed a negative correlation between the translational amount and Simpson (supplementary fig. S3A, Supplementary Material online) or Shannon (supplementary fig. S3B, Supplementary Material online) index of TIS diversity among genes before and after controlling for the transcript length.
Because different genes differ in multiple aspects in addition to translational amount, we further minimized the influences of potential confounding factors by comparing between human paralogous genes, because paralogous genes are similar in gene structure, DNA sequence, regulation, and function (Zhang 2013). Because the supergene approach cannot pair paralogous genes, we used the downsampling approach before estimating the Simpson or Shannon index. Consistent with the error hypothesis, Simpson and Shannon indices tend to be higher for the relatively lowly translated paralog than the relatively highly translated one in a pair of paralogous genes (supplementary fig. S4, Supplementary Material online). For example, in human HEK293, ∼62% of gene pairs show such a trend when the Simpson index is considered, significantly more than the random expectation of 50% (P = 6.4 × 10−4, binomial test; supplementary fig. S4A, Supplementary Material online). This pattern holds in most samples analyzed, with two exceptions (supplementary fig. S4B, Supplementary Material online). Nevertheless, in each of these exceptions, although the corresponding fraction is below 50%, it is not significantly below 50%. Similar patterns were observed when we downsampled 20 instead of 10 reads per gene (supplementary fig. S4C, Supplementary Material online).
Usages of All but the Major TISs Decline with Translational Amount
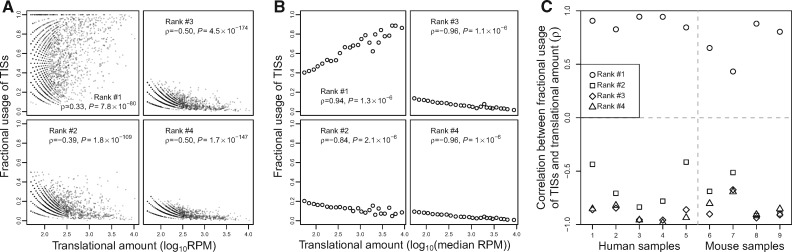
Although the above analyses suggest that using many TISs of a gene is generally deleterious such that the overall TIS diversity declines with the translational amount as a result of natural selection against detrimental ATLI, it does not tell us using which TISs is harmful. To address this question, we ranked all TISs of a gene by their fractional usages. The fractional usage of a TIS is measured by the number of QTI-seq reads mapped to the TIS divided by the total number of reads mapped to all TISs of the gene. For a given gene, the TIS with the highest fractional usage (i.e., ranked #1) is referred to as the major TIS, whereas all others are referred to as minor TISs. Intuitively, the major TIS should be the TIS preferred by natural selection. Because natural selection against translation initiation error intensifies with the translational amount, the fractional usage of each preferred TIS should increase, whereas that of each unpreferred TIS should reduce as the translational amount increases. We first examined human HEK293 cells. Again, we considered only genes with at least ten QTI-seq reads to ensure a certain level of accuracy in TIS usage estimation. Indeed, the fractional usage of the major TIS in a gene tends to increase with its translational amount (upper-left plot in fig. 2A). By contrast, each minor TIS examined shows the opposite trend, suggesting that none of them is preferred. For example, among all genes with at least two TISs, the fractional usage of the second most frequently used TIS in a gene decreases with the translational amount of the gene (lower-left plot in fig. 2A). A similar negative correlation is observed for the third most frequently used TIS among genes with at least three TISs (upper-right plot in fig. 2A) and for the fourth most frequently used TIS among genes with at least four TISs (lower-right plot in fig. 2A). We verified the above results by the supergene approach that equalizes the translational amount among supergenes (fig. 2B). Further, the trends remain unchanged when only AUG TISs are considered (supplementary fig. S5A, Supplementary Material online) and when the analysis is extended to the fifth, sixth, seventh, and eighth most frequently used TISs among genes with at least five, six, seven, and eight TISs, respectively (supplementary fig. S6A and B, Supplementary Material online). Other human and mouse samples show similar patterns (fig. 2C and supplementary figs. S5B and S6C, Supplementary Material online).
Fig. 2.
Increased fractional use of the most frequently used TIS and decreased fractional use of each other TIS when translational amount rises. (A) Spearman’s correlation between the translational amount of a gene and the fractional uses of ranked TISs of the gene in human HEK293 cells. Each dot represents a gene. (B) Spearman’s correlation between the median translational amount of a supergene and the fractional uses of its ranked TISs in HEK293. Each dot represents a supergene. (C) Spearman’s correlation between the median expression level of a supergene and the fractional uses of its ranked TISs in each human and mouse sample examined. Sample IDs listed in the X-axis of (C) refer to those in supplementary table S1, Supplementary Material online. P < 0.05 in all cases.
Translation of uORFs of a gene has been proposed as an adaptive strategy to negatively regulate the translation of the main ORF. To evaluate whether translating uORFs is adaptive, we considered all upstream minor TISs of a gene together but found that the total fractional usage of these upstream TISs declines with the translational amount of the gene (supplementary fig. S7A, Supplementary Material online), and this trend remains even when only AUG TISs are considered (supplementary fig. S7B, Supplementary Material online) or when the annotated TIS of a gene is considered the main TIS in defining uORFs (supplementary fig. S7C, Supplementary Material online). The same trend is observed when translational amount is replaced with mRNA concentration in these correlations (supplementary fig. S7A–C, Supplementary Material online). Together, these findings strongly suggest that, for most genes, only the major TIS is selectively preferred, whereas all other TISs are unpreferred, arguing against the adaptive hypothesis of translating uORFs.
We also validated the above human results using paralogous genes and again used the downsampling approach aforementioned. For HEK293, the major TIS is used more often in the relatively highly translated paralog than in the relatively lowly translated one in 67% of the 189 pairs of paralogous genes analyzed, significantly more than the random expectation of 50% (supplementary fig. S8A, Supplementary Material online). By contrast, for the second, third, and fourth most frequently used TISs respectively, significantly smaller than 50% of gene pairs show this pattern (supplementary fig. S8A, Supplementary Material online). Similar results were observed across all human and mouse samples (supplementary fig. S8B, Supplementary Material online).
Out-of-frame translation from minor TISs is likely to be more deleterious than in-frame translation from minor TISs. Consistently, the fraction of minor TISs that are out-of-frame is only 3.9% (supplementary fig. S9A, Supplementary Material online) and the fraction of translation initiations from minor TISs that are out-of-frame is only 6.3% (supplementary fig. S9B, Supplementary Material online).
Variations in TIS Usage between Cell Types Support the Error Hypothesis
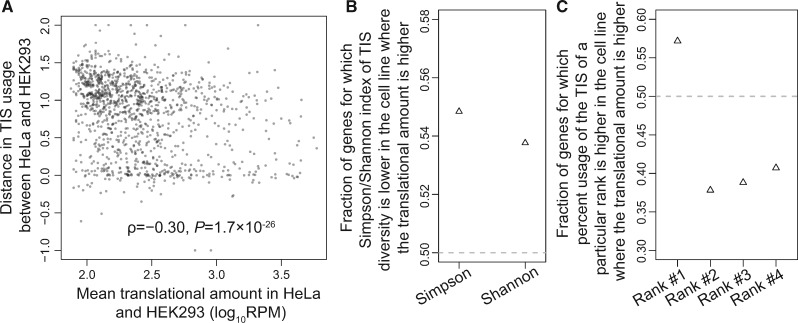
Under the error hypothesis of ATLI, differences in TIS usage of the same gene between cell types are due to the stochastic nature of translation initiation error. Hence, upon natural selection against error, the differences should decrease with the translational amount, because the sensitivity of fitness to a change in relative TIS usage increases with the translational amount. By contrast, no such prediction is made a priori by the adaptive hypothesis, because the difference in ATLI between cell types would depend on the specific cell types and genes. To investigate whether the error hypothesis is supported by between cell type comparisons, we compared the QTI-seq data of HeLa and HEK293, the only different cell types of the same species in our data (other comparisons would involve the environment, mutations, and/or a tissue that is a mixture of multiple cell types). We considered only genes with at least ten QTI-seq reads in both cell lines to ensure a certain level of accuracy in TIS usage estimation. We used the downsampling approach instead of the supergene approach, because the latter could not pair the same gene from different cell types. For each gene, we randomly picked ten reads from each cell line to measure the distance in fractional uses of TISs between the two cell lines (see Materials and Methods) and then correlated this distance with the average translational amount of the gene in these cell lines. We found that between HeLa and HEK293, the correlation is significantly negative (ρ = −0.30, P = 1.7 × 10−26; fig. 3A), as predicted by the error hypothesis.
Fig. 3.
Comparison of TIS usage between human HeLa and HEK293 cells. (A) Distance in TIS usage between the two cell lines for a gene decreases with the mean translational amount of the gene in these cell lines. (B) Fraction of genes for which the Simpson/Shannon index of TIS diversity is lower in the cell line where the translational amount of the gene is higher. Both fractions significantly exceed the random expectation of 50% (P < 0.05, binomial test). (C) Fraction of genes for which the percent usage of the TIS of a particular rank is higher in the cell line where the translational amount of the gene is higher. All fractions deviate significantly from the random expectation of 50% (P < 0.05, binomial test). Downsampled data are used in all panels.
Because a gene may have different translational amounts in different cell types, the error hypothesis predicts that its TIS diversity should be lower in the cell type where its translational amount is higher. To verify this prediction, we compared the TIS diversity of each gene between HeLa and HEK293. We used downsampled data to guard against the influence of unequal sequencing depths between cell lines. Indeed, significantly more than 50% of genes exhibit lower TIS diversities in the cell line where their translational amounts are higher, regardless of whether the Simpson or Shannon index of TIS diversity is used (fig. 3B).
Furthermore, the error hypothesis predicts that, for each gene, the fractional use of the major TIS tends to be higher and that of each minor TIS tends to be lower in the cell type where the translational amount of the gene is higher. To verify this prediction, for each gene, we defined the major and minor TISs in each cell line separately and then compared the fractional usage of each TIS between HeLa and HEK293 based on the downsampled data. Indeed, 57% of genes show higher fractional use of the major TIS in the cell line where the translational amount is higher (fig. 3C), significantly more than the random expectation of 50%, whereas only 38–41% of genes show the similar pattern for each minor TIS examined, significantly lower than the random expectation (fig. 3C).
Although our evidence so far suggests that, for most genes, each cell type has only one selectively preferred TIS, it remains possible that the selectively preferred TIS varies among cell types such that the variation in translation initiation among cell types is adaptive. To assess this possibility, the simplest approach is to count genes that have different major TISs in different cell types, but this assessment would be inaccurate due to sampling error caused by limited sequencing depths. To rectify this problem, for each gene with different major TISs in the two cell lines compared, we randomly shuffled its QTI-seq reads from HeLa and HEK293 without altering the number of reads in each cell line. We repeated this process 10,000 times and estimated the fraction of times (f) when the number of major TISs observed in the shuffled data is 2. Here, f is an estimate of the one-tailed P value in testing the null hypothesis that the two cell lines share the same major TIS. We converted the P values to Q values to guard against false positives due to multiple testing and used Q < 0.05 to call significance. Of 1,198 genes examined, only 212 genes have significantly more than one major TIS in the two cell lines. Thus, selection appears to have favored different TISs in the two cell lines for only a minority (24.3%) of genes, consistent with the hypothesis that among cell-type variations in translation initiation are largely nonadaptive. It would be interesting and important to revisit this issue when comparable data from more cell types become available. In terms of the molecular mechanism, the variation of TIS usage between cell types for the same gene can be achieved by differences in the concentrations of trans-regulatory factors of translation initiation in different cell types.
Patterns of Natural Selection on Kozak Regions of Various TISs
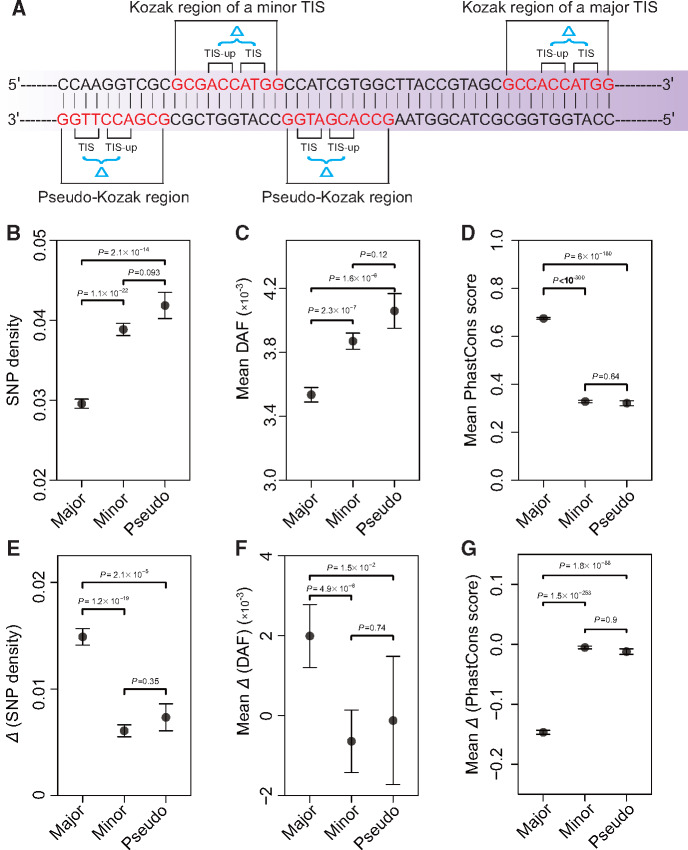
The Kozak region is a ten-nucleotide DNA segment including the translation start codon, its upstream six nucleotides, and its downstream one nucleotide (Kozak 1984). This region plays a major role in translation initiation because its sequence affects the initiation efficiency (Kozak 1986). Our finding of the relative uses of various TISs predicts that, as the translational amount of a gene rises, the mean strength of Kozak regions of all minor TISs in initiating translation relative to the strength of the Kozak region of the major TIS should decline. To verify this prediction, we used translation initiation strengths of various Kozak sequences measured in a reporter assay (Noderer et al. 2014; Diaz de Arce et al. 2018). Indeed, a significant, negative correlation was observed in the vast majority of human and mouse samples (supplementary fig. S10, Supplementary Material online). This is true regardless of whether we considered all TISs observed or only those that appeared in the downsampled data (supplementary fig. S10, Supplementary Material online).
Our finding that, for most human and mouse genes, the major TIS is likely selectively preferred, whereas all minor TISs are likely unpreferred predicts that Kozak regions corresponding to major TISs are evolutionarily constrained, whereas those corresponding to minor TISs are unconstrained. To verify this prediction, we merged all QTI-seq reads of five independent human samples to determine the global major and minor TISs of each gene. For each observed TIS, its Kozak region is −6 to +3 nucleotides, where the coordinates of the start codon are 0 to +2 (fig. 4A). For comparison, we define pseudo-Kozak regions, which inmmediately flank but are on the oppsoite strand of observed Kozak regions and contain a potential start codon (AUG or one of its nine near-cognate codons) at appropriate positions (fig. 4A). Pseudo-Kozak regions presumbly do not initiate translation so are comparable with Kozak regions in all aspects except natural selection in relation to the function of translation initiation (fig. 4A; see Materials and Methods). Because a Kozak region in the 5′ UTR of a transcript may be located in the coding or intron region of another transcript of the same gene, we considered only Kozak regions that have never been annotated as coding sequence, intron, or 3′ UTR in any transcript; the same criterion was used for pseudo-Kozak regions. The error hypothesis predicts purifying selection acting on the Kozak regions of major TISs but not on the Kozak regions of minor TISs nor pseudo-Kozak regions. Purifying selection reduces the single-nucleotide polymophism (SNP) density and derived allele frequencies (DAFs) but increases the PhastCons conservation score from among-mammal comparions (Siepel et al. 2005). Note that we examine both intraspecific polymorphsims and interspecific divergences, because the former is potentially affected by selection of neighboring sites whereas the latter is not. Indeed, on average, Kozak regions of major TISs have a significantly lower SNP density (fig. 4B), a significantly lower DAF (fig. 4C), and a significantly higher PhastCons score (fig. 4D) than Kozak regions of minor TISs and pseudo-Kozak regions, whereas no significant difference exists between the latter two regions in any of the three measures (fig. 4B–D).
Fig. 4.
Purifying selection on human Kozak regions of various TISs. (A) A schematic showing the Kozak region of a major TIS, Kozak region of a minor TIS, and pseudo-Kozak regions. A Kozak region contains ten nucleotides, including a TIS, its six upstream nucleotides, and one downstream nucleotide. Pseudo-Kozak regions inmmediately flank but are on the oppsoite strand of observed Kozak regions and contain one of the ten potential start codons at appropriate positions. TIS-up is the immediately upstream codon of the TIS codon. (B–D) SNP density (B), mean DAF (C), and mean PhastCons conservation score (D) for Kozak regions of major TISs, Kozak regions of minor TISs, and pseudo-Kozak regions, respectively. (E–G) Mean difference between TIS-up and TIS in SNP density (E), DAF (F), and PhastCons conservation score (G) for Kozak regions of major TISs, Kozak regions of minor TISs, and pseudo-Kozak regions, respectively. Fisher’s exact test was used to compute the P values for (B) and (E), whereas Mann–Whitney U test was used to compute the P values for (C), (D), (F), and (G). Error bars show standard errors.
Although the above comparison takes the entire Kozak region into consideration, variation in purifying selection among sites within a Kozak region can also be informative. Specifically, a functional Kozak region spans from the relatively unconserved UTR (i.e., nucleotide positions −6 to −1) to the relatively conserved protein-coding region (i.e., positions 0 to +3) (fig. 4A). By contrast, a nonfunctional Kozak region should not show this feature. To this end, we compared SNP density, DAF, and PhastCons score between the TIS codon and its immediate upstream (TIS-up) codon (fig. 4A). As predicted by the error hypothesis, the difference in SNP density between the TIS-up codon and the TIS codon is significantly greater for Kozak regions of major TISs than for Kozak regions of minor TISs and pseudo-Kozak regions, whereas no significant difference is observed between the latter two regions (fig. 4E). The same is true in terms of the difference in DAF between TIS-up and TIS (fig. 4F). Furthermore, the difference in PhastCons between TIS-up and TIS is significantly more negative for Kozak regions of major TISs than for Kozak regions of minor TISs and pseudo-Kozak regions, whereas no significant difference is observed between the latter two regions (fig. 4G), again consistent with the error hypothesis of ATLI.
Non-AUG Initiation Is Generally Nonadaptive
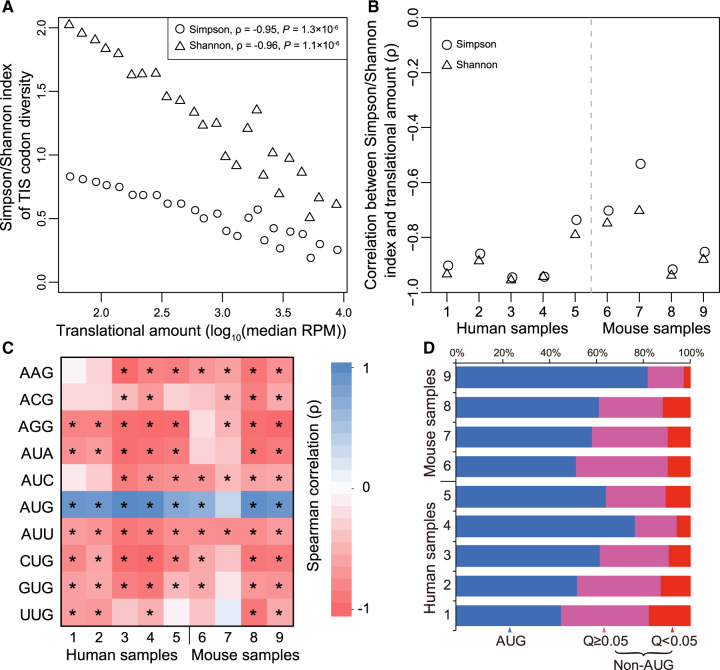
As mentioned, in addition to AUG, its nine near-cognate codons often serve as start codons (Peabody 1989; Ingolia et al. 2011). Because some non-AUG initiations appear to be beneficial (Gerashchenko et al. 2010; Starck et al. 2012, 2016), it has been proposed that non-AUG initiations generally increase the proteome diversity and are adaptive (Kearse and Wilusz 2017). However, whether non-AUG initiations generally reflect initiation errors or adaptations is unclear. Although our above analyses suggest that most ATLI is nonadaptive, they do not directly answer the question about non-AUG initiations because our analyses were based on TISs, not start codon identities. To this end, we followed the analysis in figure 1 but lumped all TISs with the same initiation codon identity (see Materials and Methods). Using the supergene approach, we estimated start codon diversity by the Simpson or Shannon index and the fractional use of each of the ten possible start codons. Among 25 supergenes, we observed a negative correlation between start codon diversity and median translational amount in HEK293 (fig. 5A) as well as other human and mouse samples (fig. 5B). Additionally, among the supergenes, only the fractional use of AUG increases with the median translational amount, whereas the fractional use of each of the nine near-cognate codons decreases with the median translational amount (fig. 5C). Thus, it appears that only AUG is generally selectively preferred as the start codon, whereas all nine near-cognate codons are generally unpreferred.
Fig. 5.
Non-AUG initiation is generally nonadaptive. (A) Simpson or Shannon index of TIS codon diversity of a supergene in human HEK293 cells declines with the median translational amount of genes belonging to the supergene. Each circle or triangle represents a supergene. (B) Spearman’s correlation between translational amount and Simpson/Shannon index of TIS codon diversity among supergenes in each sample examined. P < 0.01 for all correlations. (C) Spearman’s correlation between the fractional use of each of the ten possible start codons in a supergene and the median translational amount of genes belonging to the supergene, shown by different colors. * indicates a statistically significant correlation (P < 0.05). (D) Fraction of genes in each sample for which the most frequently used translation start codon is AUG (blue), is not AUG but the AUG usage is not significantly lower than that of the most commonly used start codon (Q ≥ 0.05; purple), and is not AUG and the AUG usage is significantly lower than that of the most frequently used start codon (Q < 0.05; red), respectively. In (B) and (C), sample IDs listed in the X-axis refer to those in supplementary table S1, Supplementary Material online.
We further examined individual genes to estimate the prevalence of using a non-AUG codon as the major start codon of a gene. In the nine samples examined, AUG is the major start codon in 61.1% of genes on average (blue bars in fig. 5D). In some genes, although AUG is not the major start codon, the major start codon is not used significantly more often than AUG (purple bars in fig. 5D). In only a small fraction (on average ∼10%) of genes did we observe a significantly higher use of a non-AUG than AUG start codon (red bars in fig. 5D).
Discussion
Genome-scale discoveries of abundant ATLI in mammals (Ingolia et al. 2011; Fritsch et al. 2012; Lee et al. 2012) prompted the prevailing view that ATLI generates proteome diversity and regulate protein synthesis so is generally adaptive (Kochetov 2008; Bazykin and Kochetov 2011; Ivanov et al. 2011, 2017; Lee et al. 2012; Ingolia 2014, 2016; de Klerk and 't Hoen 2015; Kearse and Wilusz 2017). In this study, we proposed and tested an alternative hypothesis that ATLI arises largely from nonadaptive, translation initiation errors. Analyzing genome-wide data of translation initiation in several human and mouse cell line and tissue samples, we observed that 1) the TIS diversity of a gene declines with its translational amount, 2) the fractional use of the major TIS increases, but that of each minor TIS decreases with the translational amount, and 3) Kozak regions of major TISs but not those of minor TISs are selectively constrained. The first two observations also apply to the diversity of start codon identity. Together, these findings strongly support the error hypothesis and refute the common view that ATLI is generally adaptive. That ATLI is mostly deleterious does not preclude its occasional adaptive use, as has been found in a small number of cases (Strubin et al. 1986; Liu et al. 1999; Brubaker et al. 2014; Zhang et al. 2018). But the general pattern revealed in this study argues that ATLI should be considered nonadaptive unless proven otherwise.
It is reasonable to assume that the fractions of beneficial, neutral, and deleterious ATLI for a gene before the action of natural selection are independent of the gene’s translational amount. Deleterious ATLI is expected to be purged by natural selection, especially for genes with large amounts of translation. Assuming that deleterious ATLI has not been purged in genes with the lowest translational amounts but has been completely removed in those with the highest translational amounts, we can treat the extent of ATLI of the most weakly translated genes as the total ATLI (T) and that of the most strongly translated genes as nondeleterious ATLI (ND). Thus, the fraction of ATLI that is deleterious is (T − ND)/T = 1 − ND/T. We used the Simpson index of TIS diversity to measure the amount of ATLI because both the number of TISs and their relative usages are considered. In figure 1C, the most weakly translated supergene has a Simpson index of 0.76 (T), whereas the most strongly translated supergene has a Simpson index of 0.25 (ND). Hence, the fraction of deleterious ATLI is 1 − ND/T = 1 − 0.25/0.76 = 67%. A similar fraction of deleterious ATLI (1 − 0.60/1.74 = 66%) is estimated using Shannon index of TIS diversity. If we compare 20 most weakly translated genes with 20 most strongly translated genes (Xu et al. 2019), the fraction of deleterious ATLI is ∼75%. Thus, two-thirds to three quarters of ATLI are deleterious. Note that the above estimate is conservative, because slightly deleterious ATLI may not have been fully removed by selection in the most strongly translated genes and because some strongly deleterious ATLIs may have been removed by selection even in the most weakly translated genes. The above finding is broadly consistent with our estimate that only 24.3% of genes have evidence for different selectively favored TISs in different cell types and explains why Kozak regions of minor TISs are generally unconserved.
The negative correlation between the translational amount of a gene and its ATLI diversity is a primary line of evidence supporting the error hypothesis. One might argue that this negative correlation can be explained if ATLI is largely neutral or deleterious in genes with large amounts of translation but adaptive in genes with low amounts of translation. This hypothesis is at odds with multiple observations. For example, it could not explain why the average selective constraint on the Kozak regions of minor TISs is not greater than that on pseudo-Kozak regions. Previous studies reported that the total number of upstream TISs of all genes tends to be lower than expected by chance (Iacono et al. 2005; Lynch et al. 2005; Zur and Tuller 2013). We found that this trend is also true for every bin after we stratified genes by their translational amounts (supplementary fig. S11, Supplementary Material online), supporting the lack of positive selection for ATLI even in genes with low translational amounts.
Minor TISs in this study include both upstream and downstream TISs relative to the major TIS. Use of upstream TISs leads to the translation of uORFs that are thought to suppress the translation of the main ORF (Wethmar et al. 2014; Johnstone et al. 2016). Our results suggest that this suppression is unlikely to be an adaptive strategy of translation regulation, because we found that the fractional use of upstream TISs decreases with the translational amount of the gene (supplementary fig. S7A–C, Supplementary Material online). One might contend that this negative correlation could be a result of uORFs’ repression of translation instead of a result of natural selection against initiation error. This hypothesis is untenable for two reasons. First, translation of uORFs suppresses translation but has only modest impacts on mRNA concentrations (Calvo et al. 2009), so this hypothesis cannot explain the strong negative correlation between upstream TIS usage and mRNA concentration across genes (supplementary fig. S7A–C, Supplementary Material online). Second, under this hypothesis, an increase in the use of upstream TISs and uORFs from 5% to 15% on average suppresses translation by ∼100-fold (supplementary fig. S7D, Supplementary Material online), but experimentally confirmed effect of uORF translation on the translational amount of the main ORF, based on the comparison between constructs with and without the uORF, is typically no more than 5-fold (Calvo et al. 2009). Some authors observed the use of uORFs as a cellular response to stress (Chen et al. 2010; Barbosa and Romao 2014; Gao et al. 2015), but this stress response may not be adaptive; it could be a passive consequence of the disruption of cellular homeostasis under stress (Ho and Zhang 2018). Although several uORFs of mammals are evolutionarily conserved (Churbanov et al. 2005; Chew et al. 2016; Spealman et al. 2018), using uORFs as an adaptive strategy is unlikely to be general (Churbanov et al. 2005; Neafsey and Galagan 2007), because the number of upstream TISs and 5′UTR length tend to be lower than expected by chance (Iacono et al. 2005; Lynch et al. 2005; Zur and Tuller 2013). Apart from upstream TISs, some downstream TISs were reported to be evolutionarily conserved, but such cases are rare (e.g., in ∼10.6% of human genes) (Bazykin and Kochetov 2011). Thus, the previous studies are not inconsistent with our finding that ATLI is largely nonadaptive.
Recent ribosome footprint mapping revealed that non-AUG start codons are widespread (Ingolia et al. 2011; Lee et al. 2012; Gao et al. 2015; Kearse and Wilusz 2017) and these non-AUG initiations are thought to play special functional/regulatory roles (Kearse and Wilusz 2017). Our results do not support this adaptive view; instead, they suggest that most non-AUG initiations are errors (fig. 5), consistent with the fact that near-cognate non-AUG start codons have much lower initiation efficiencies than AUG (Diaz de Arce et al. 2018). Our conclusion, of course, does not preclude the existence of a small number of cases where non-AUG initiations are beneficial (Gerashchenko et al. 2010; Starck et al. 2012, 2016) or non-AUG start codons are conserved (Ivanov et al. 2011; Spealman et al. 2018).
It should be noted that we did not classify ATLI by how it is generated. Leaky scanning, reinitiation, and IRES-dependent initiation are well-known mechanisms of ATLI (Kochetov 2008). In addition, alternative transcriptional start and alternative splicing can provide different transcripts to the translation machinery to create additional ATLI (de Klerk and 't Hoen 2015). QTI-seq data do not allow differentiating among these mechanisms. Hence, relative contributions of the various mechanisms to translation initiation error remain unknown.
Our results on ATLI echo recent findings about variations in multiple steps of transcription and translation that generate transcriptome and/or proteome diversities, including, for example, alternative transcriptional start (Xu et al. 2019), alternative splicing (Saudemont et al. 2017), RNA editing (Xu and Zhang 2014; Liu and Zhang 2018a, 2018b; Jiang and Zhang 2019), alternative polyadenylation (Xu and Zhang 2018, 2020), translational stop-codon read-through (Li and Zhang 2019), and posttranslational modification (Landry et al. 2009; Park and Zhang 2011). They have all been shown to be largely the results of molecular errors instead of adaptive regulations. Thus, despite the central importance of the precision of transcription and translation in cellular life, errors are present and abundantly observed via modern high-throughput sequencing technologies. Although the most harmful errors have likely been suppressed by natural selection, many slightly deleterious errors still exist presumably because selection against them is too weak and/or the cost of removing these errors exceeds the benefit. The astonishing imprecision of key molecular processes in the cell, revealed here and elsewhere, contrasts the common view of an exquisitely perfected cellular life and has fundamental implications for our understanding of biology (Lynch 2007; Warnecke and Hurst 2011; Lynch et al. 2014; Zhang and Yang 2015).
Materials and Methods
Translation Initiation Sites
In this study, we used QTI-seq data to identify TISs in human and mouse. Unlike ribo-seq that captures all ribosome-protected mRNA fragments, QTI-seq captures and sequences only the mRNA fragments protected by initiating ribosomes; QTI-seq outperforms other methods that identify TISs (Gao et al. 2015).
We analyzed nine QTI-seq samples, including five human samples and four mouse samples (supplementary table S1, Supplementary Material online). QTI-seq reads were downloaded from the SRA database or provided by the original authors. We used FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/; last accessed March 13, 2020) to control the data quality and used Trimmomatic (Bolger et al. 2014) to filter out low-quality reads when necessary. The filtered reads with lengths between 25 and 35 nucleotides were mapped to the human genome (hg19) or mouse genome (mm9) with Ensembl gene annotation release 75 using TopHat2 (Kim et al. 2013) with default parameters. Nonuniquely mapped reads were discarded. Ribo-TISH, which outperforms several established methods in both efficiency and accuracy (Zhang et al. 2017), was employed to identify TISs. Briefly, Ribo-TISH first evaluates data quality and estimates the P-site position in a read, then utilizes data-driven methods to model the background distribution of QTI-seq data, and finally determines a TIS according to the P or Q value (statistical significance before and after the control for multiple testing) calculated for each TIS from the distribution (Zhang et al. 2017).
Translation can start from AUG or its near-cognate codons that differ from AUG by one nucleotide (Peabody 1989; Kearse and Wilusz 2017). Thus, only AUG and its near-cognate codons are considered as true translation start codons in the TIS prediction by Ribo-TISH. The coordinate of the 5′ most nucleotide of a translation start codon was used to represent the TIS. In total, 114,655 and 69,517 TISs found in at least one sample were identified for human and mouse, respectively.
Translational Amount of a Gene
The number of QTI-seq reads of a gene is proportional to the number of translational initiation events per unit time. Because the number of initiation events should be proportional to the number of protein molecules synthesized under the assumption of equal probabilities of complete protein synthesis among genes, the number of QTI-seq reads of a gene is proportional to the number of protein molecules synthesized per unit time (i.e., translational amount) for the gene. To make the number of QTI-seq reads of a gene comparable among samples, we report the number of reads per million total reads in the sample (RPM).
Because the number of translation initiation events per unit time for a gene equals its mRNA concentration multiplied by the translation initiation rate per mRNA molecule, mRNA concentration is strongly positively correlated with the number of translation initiation events per unit time. We used RNA-seq to measure mRNA concentrations and downloaded RNA-seq data from SRA (supplementary table S1, Supplementary Material online). Upon the quality control, reads were mapped to human (hg19) or mouse (mm9) genome using TopHat2 (Kim et al. 2013). Fragment per kilobase of transcripts per million mapped reads (FPKM) of a gene was first calculated by cufflinks (Trapnell et al. 2012) and then converted to TPM using the formula of TPM = (FPKM × 106)/(sum of FPKM) (Li and Dewey 2011).
Statistical Methods for Correcting Unequal Surveys of TISs among Genes
Due to different translational amounts among genes, their TISs are not surveyed to the same depth by QTI-seq. To remove the potential influence of this unequal survey, we used two different approaches. The first is the supergene approach. Briefly, we first ranked all genes by their translational amounts. We then grouped the genes into 25 bins representing 25 supergenes, requiring the total translational amount per bin to be the same for all bins. Within a bin, reads from the most commonly used TIS of each gene were combined to represent reads of the most commonly used TIS of the supergene. Similarly, reads from the second most commonly used TIS of each gene were combined to represent reads of the second most commonly used TIS of the supergene, and so on. The supergene approach may not be usable under certain circumstances (e.g., for a comparison between two paralogous genes). Under these circumstances, we used the second approach—downsampling. Briefly, we randomly picked ten QTI-seq reads per gene from all genes with at least ten reads unless otherwise mentioned to calculate TIS diversities and fractional usages. The supergene approach is preferred over downsampling when both are usable, because the former uses all data, whereas the latter uses only part of the data.
Measures of TIS Diversity
Following our recent studies of alternative polyadenylation and alternative transcriptional initiation (Xu and Zhang 2018; Xu et al. 2019), we used the Simpson index (Simpson 1949) and Shannon index (Shannon 1948) to quantify TIS diversity of each gene in a sample. Both indices are commonly used in biodiversity researches and tend to rise with the number of TISs as well as the evenness of the relative uses of these TISs, but the Simpson index gives more weights to the frequently used TISs than does the Shannon index. For a single gene, Simpson and Shannon indices of TIS diversity are respectively defined by 1 − and −, where S is the number of TISs in a gene and pi is the fractional use of the ith TIS. When the above formulas are used to calculate the TIS diversity of a supergene, S is the maximum number of TISs of any gene belonging to the supergene, whereas pi is the sum of the number of reads mapped to the TIS of rank #i of each gene belonging to the supergene, divided by the total number of reads mapped to all TISs of all genes belonging to the supergene.
Computer Simulation
To confirm that the patterns observed in figure 1A–C are not statistical artifacts, we performed computer simulations as follows. We randomly generated each gene whose translational amount (measured by the total number of reads mapped to all TISs of the gene) and relative TIS usages (measured by the numbers of reads mapped to various TISs of the gene), respectively, follow the gene translational amount distribution and relative TIS usage distribution of the data from HEK293. Specifically, the total read number of a simulated gene was randomly sampled from the collection of actual read numbers of all genes with replacement, whereas the numbers of reads mapped to the TISs of the gene were multinomial random variables drawn according to the TIS usages of a randomly picked real gene. We then analyzed the read data from the simulated genes as if they were the actual data.
Distance in TIS Usage between Cell Lines
To measure the difference in TIS usage for a gene between samples A and B, we used a net correlational distance defined by dAB − 0.5dA − 0.5dB. Here, dAB equals 1 minus Pearson’s correlation coefficient between samples A and B in the fractional uses of all TISs of the gene, dA is the same as dAB except that the two samples used are two QTI-seq bootstrap samples derived from sample A, and dB is the same as dAB except that the two samples used are two QTI-seq bootstrap samples derived from sample B (Xu and Zhang 2018; Xu et al. 2019). We set zero usage in sample A for TISs found in sample B only, and vice versa. The appropriateness of the distance measure used here was previously confirmed (Xu and Zhang 2018).
Paralogs
Human paralogous genes were downloaded from Ensembl (release 89; May 2017). We obtained 3,678 human gene families, including 51,657 pairs of human paralogous protein-coding genes. We randomly selected from each gene family only one paralogous pair that exhibits a 2-fold or greater difference in translational amount to allow a sufficient statistical power.
Kozak Strength
The Kozak strengths used were from studies reporting the translation initiation efficiencies of all potential Kozak sequences (Noderer et al. 2014; Diaz de Arce et al. 2018). These studies combined fluorescence-activated cell sorting and high-throughput DNA sequencing to measure the translation initiation efficiency associated with every start codon (AUG and its nine neighbors) in combination with every possible nucleotide sequence at the adjacent −4 to −1 and +3 positions. The value of translation initiation efficiency was relative to that of CACCAUGG, then multiplied by 100. Each TIS identified by QTI-seq was assigned a Kozak strength value according to its eight-nucleotide Kozak sequence. Here, we considered eight nucleotides instead of the typical ten-nucleotide Kozak region, because the above assay considered only −4 to +3 positions.
Conservation Score and Polymorphisms
To assess purifying selection acting on TISs and Kozak regions, we downloaded from UCSC (http://hgdownload.cse.ucsc.edu/goldenpath/hg19/phastCons46way/placentalMammals/; last accessed March 13, 2020) PhastCons scores (Siepel et al. 2005) computed from genome alignments of 46 placental mammals including the human (hg19). Human polymorphism data, including allele frequencies, from Interim Phase 3 of the 1000 Genomes project (Sudmant et al. 2015), were downloaded from ftp://ftp.1000genomes.ebi.ac.ukVol03707ftp/release/20130502/ on June 29, 2018. This data set comprises the genotypes of 2,504 individuals from 26 populations and includes a total of 78,136,341 autosomal SNPs. Only SNPs were included in the analysis. The nucleotide observed at a SNP was categorized as ancestral if it is the same as the nucleotide of the “AA” field in the polymorphism VCF file; other nucleotides at the SNP are derived. The DAF at a SNP is the frequency of the derived allele at the SNP.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors thank Dr Chuan He and Dr Xiao Wang for sharing the QTI-seq data of HeLa cells and Daohan Jiang, Zhengting Zou, and two anonymous reviewers for valuable comments. This work was supported by the research grant GM120093 from U.S. National Institutes of Health to J.Z.
References
- Aitken CE, Lorsch JR.. 2012. A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol. 19(6):568–576. [DOI] [PubMed] [Google Scholar]
- Barbosa C, Romao L.. 2014. Translation of the human erythropoietin transcript is regulated by an upstream open reading frame in response to hypoxia. RNA 20(5):594–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazykin GA, Kochetov AV.. 2011. Alternative translation start sites are conserved in eukaryotic genomes. Nucleic Acids Res. 39(2):567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brubaker SW, Gauthier AE, Mills EW, Ingolia NT, Kagan JC.. 2014. A bicistronic MAVS transcript highlights a class of truncated variants in antiviral immunity. Cell 156(4):800–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Pagliarini DJ, Mootha VK.. 2009. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci U S A. 106(18):7507–7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YJ, Tan BC, Cheng YY, Chen JS, Lee SC.. 2010. Differential regulation of CHOP translation by phosphorylated eIF4E under stress conditions. Nucleic Acids Res. 38(3):764–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew GL, Pauli A, Schier AF.. 2016. Conservation of uORF repressiveness and sequence features in mouse, human and zebrafish. Nat Commun. 7:11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churbanov A, Rogozin IB, Babenko VN, Ali H, Koonin EV.. 2005. Evolutionary conservation suggests a regulatory function of AUG triplets in 5′-UTRs of eukaryotic genes. Nucleic Acids Res. 33(17):5512–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Klerk E, 't Hoen PA.. 2015. Alternative mRNA transcription, processing, and translation: insights from RNA sequencing. Trends Genet. 31(3):128–139. [DOI] [PubMed] [Google Scholar]
- Diaz de Arce AJ, Noderer WL, Wang CL.. 2018. Complete motif analysis of sequence requirements for translation initiation at non-AUG start codons. Nucleic Acids Res. 46:985–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch C, Herrmann A, Nothnagel M, Szafranski K, Huse K, Schumann F, Schreiber S, Platzer M, Krawczak M, Hampe J, et al. 2012. Genome-wide search for novel human uORFs and N-terminal protein extensions using ribosomal footprinting. Genome Res. 22(11):2208–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Wan J, Liu B, Ma M, Shen B, Qian SB.. 2015. Quantitative profiling of initiating ribosomes in vivo. Nat Methods. 12(2):147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerashchenko MV, Su D, Gladyshev VN.. 2010. CUG start codon generates thioredoxin/glutathione reductase isoforms in mouse testes. J Biol Chem. 285(7):4595–4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho W-C, Zhang J.. 2018. Evolutionary adaptations to new environments generally reverse plastic phenotypic changes. Nat Commun. 9(1):350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono M, Mignone F, Pesole G.. 2005. uAUG and uORFs in human and rodent 5′untranslated mRNAs. Gene 349:97–105. [DOI] [PubMed] [Google Scholar]
- Imataka H, Olsen HS, Sonenberg N.. 1997. A new translational regulator with homology to eukaryotic translation initiation factor 4G. EMBO J. 16(4):817–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT. 2014. Ribosome profiling: new views of translation, from single codons to genome scale. Nat Rev Genet. 15(3):205–213. [DOI] [PubMed] [Google Scholar]
- Ingolia NT. 2016. Ribosome footprint profiling of translation throughout the genome. Cell 165(1):22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS.. 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147(4):789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Firth AE, Michel AM, Atkins JF, Baranov PV.. 2011. Identification of evolutionarily conserved non-AUG-initiated N-terminal extensions in human coding sequences. Nucleic Acids Res. 39(10):4220–4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Wei J, Caster SZ, Smith KM, Michel AM, Zhang Y, Firth AE, Freitag M, Dunlap JC, Bell-Pedersen D, et al. 2017. Translation initiation from conserved non-AUG codons provides additional layers of regulation and coding capacity. MBio 8(3):e00844–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV.. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 11(2):113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Zhang J.. 2019. The preponderance of nonsynonymous A-to-I RNA editing in coleoids is nonadaptive. Nat Commun. 10(1):5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone TG, Bazzini AA, Giraldez AJ.. 2016. Upstream ORFs are prevalent translational repressors in vertebrates. EMBO J. 35(7):706–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse MG, Wilusz JE.. 2017. Non-AUG translation: a new start for protein synthesis in eukaryotes. Genes Dev. 31(17):1717–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL.. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14(4):R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochetov AV. 2008. Alternative translation start sites and hidden coding potential of eukaryotic mRNAs. BioEssays 30(7):683–691. [DOI] [PubMed] [Google Scholar]
- Kochetov AV, Sarai A, Rogozin IB, Shumny VK, Kolchanov NA.. 2005. The role of alternative translation start sites in the generation of human protein diversity. Mol Genet Genomics. 273(6):491–496. [DOI] [PubMed] [Google Scholar]
- Kozak M. 1984. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucl Acids Res. 12(2):857–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 1986. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44(2):283–292. [DOI] [PubMed] [Google Scholar]
- Landry CR, Levy ED, Michnick SW.. 2009. Weak functional constraints on phosphoproteomes. Trends Genet. 25(5):193–197. [DOI] [PubMed] [Google Scholar]
- Lee S, Liu B, Lee S, Huang SX, Shen B, Qian SB.. 2012. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci U S A. 109(37):E2424–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN.. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12(1):323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Zhang J.. 2019. Stop-codon read-through arises largely from molecular errors and is generally nonadaptive. PLoS Genet. 15(5):e1008141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Dilworth D, Gao L, Monzon J, Summers A, Lassam N, Hogg D.. 1999. Mutation of the CDKN2A 5′ UTR creates an aberrant initiation codon and predisposes to melanoma. Nat Genet. 21(1):128–132. [DOI] [PubMed] [Google Scholar]
- Liu Z, Zhang J.. 2018a. Human C-to-U coding RNA editing is largely nonadaptive. Mol Biol Evol. 35(4):963–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Zhang J.. 2018b. Most m6A RNA modifications in protein-coding regions are evolutionarily unconserved and likely nonfunctional. Mol Biol Evol. 35(3):666–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. 2007. The frailty of adaptive hypotheses for the origins of organismal complexity. Proc Natl Acad Sci U S A. 104(Suppl 1):8597–8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Field MC, Goodson HV, Malik HS, Pereira-Leal JB, Roos DS, Turkewitz AP, Sazer S.. 2014. Evolutionary cell biology: two origins, one objective. Proc Natl Acad Sci U S A. 111(48):16990–16994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Scofield DG, Hong X.. 2005. The evolution of transcription-initiation sites. Mol Biol Evol. 22(4):1137–1146. [DOI] [PubMed] [Google Scholar]
- Martinez-Salas E, Pineiro D, Fernandez N.. 2012. Alternative mechanisms to initiate translation in eukaryotic mRNAs. Comp Funct Genomics. 2012:391546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DR, Geballe AP.. 2000. Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol. 20(23):8635–8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neafsey DE, Galagan JE.. 2007. Dual modes of natural selection on upstream open reading frames. Mol Biol Evol. 24(8):1744–1751. [DOI] [PubMed] [Google Scholar]
- Noderer WL, Flockhart RJ, Bhaduri A, Diaz de Arce AJ, Zhang J, Khavari PA, Wang CL.. 2014. Quantitative analysis of mammalian translation initiation sites by FACS-seq. Mol Syst Biol. 10(8):748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C, Zhang J.. 2011. Genome-wide evolutionary conservation of N-glycosylation sites. Mol Biol Evol. 28(8):2351–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody DS. 1989. Translation initiation at non-AUG triplets in mammalian cells. J Biol Chem. 264(9):5031–5035. [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG, Lomakin IB, Pilipenko EV, Shatsky IN, Agol VI, Hellen CU.. 2001. Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci U S A. 98(13):7029–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saudemont B, Popa A, Parmley JL, Rocher V, Blugeon C, Necsulea A, Meyer E, Duret L.. 2017. The fitness cost of mis-splicing is the main determinant of alternative splicing patterns. Genome Biol. 18(1):208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon CE. 1948. A mathematical theory of communication. Bell Syst Tech J. 27(3):379–423. and 623–656. [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15(8):1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson EH. 1949. Measurement of diversity. Nature 163(4148):688–688. [Google Scholar]
- Sonenberg N, Hinnebusch AG.. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136(4):731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spealman P, Naik AW, May GE, Kuersten S, Freeberg L, Murphy RF, McManus J.. 2018. Conserved non-AUG uORFs revealed by a novel regression analysis of ribosome profiling data. Genome Res. 28(2):214–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck SR, Jiang V, Pavon-Eternod M, Prasad S, McCarthy B, Pan T, Shastri N.. 2012. Leucine-tRNA initiates at CUG start codons for protein synthesis and presentation by MHC class I. Science 336(6089):1719–1723. [DOI] [PubMed] [Google Scholar]
- Starck SR, Tsai JC, Chen K, Shodiya M, Wang L, Yahiro K, Martins-Green M, Shastri N, Walter P.. 2016. Translation from the 5′ untranslated region shapes the integrated stress response. Science 351(6272):aad3867–aad3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strubin M, Long EO, Mach B.. 1986. Two forms of the Ia antigen-associated invariant chain result from alternative initiations at two in-phase AUGs. Cell 47(4):619–625. [DOI] [PubMed] [Google Scholar]
- Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Fritz MH, et al. 2015. An integrated map of structural variation in 2,504 human genomes. Nature 526(7571):75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L.. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 7(3):562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Qian SB.. 2014. TISdb: a database for alternative translation initiation in mammalian cells. Nucleic Acids Res. 42(D1):D845–D850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnecke T, Hurst LD.. 2011. Error prevention and mitigation as forces in the evolution of genes and genomes. Nat Rev Genet. 12(12):875–881. [DOI] [PubMed] [Google Scholar]
- Wethmar K, Barbosa-Silva A, Andrade-Navarro MA, Leutz A.. 2014. uORFdb—a comprehensive literature database on eukaryotic uORF biology. Nucleic Acids Res. 42(D1):D60–D67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Park JK, Zhang J.. 2019. Evidence that alternative transcriptional initiation is largely nonadaptive. PLoS Biol. 17(3):e3000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Zhang J.. 2018. Alternative polyadenylation of mammalian transcripts is generally deleterious, not adaptive. Cell Syst. 6(6):734–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Zhang J.. 2020. A different perspective on alternative cleavage and polyadenylation. Nat Rev Genet. 21(1):63–63. [DOI] [PubMed] [Google Scholar]
- Xu G, Zhang J.. 2014. Human coding RNA editing is generally nonadaptive. Proc Natl Acad Sci U S A. 111(10):3769–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Dou S, He F, Luo J, Wei L, Lu J.. 2018. Genome-wide maps of ribosomal occupancy provide insights into adaptive evolution and regulatory roles of uORFs during Drosophila development. PLoS Biol. 16(7):e2003903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. 2013. Gene duplication In: Losos J, editor. The princeton guide to evolution. Princeton (NJ: ): Princeton University Press; p. 397–405. [Google Scholar]
- Zhang J, Yang JR.. 2015. Determinants of the rate of protein sequence evolution. Nat Rev Genet. 16(7):409–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, He D, Xu Y, Hou J, Pan BF, Wang Y, Liu T, Davis CM, Ehli EA, Tan L, et al. 2017. Genome-wide identification and differential analysis of translational initiation. Nat Commun. 8(1):1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zur H, Tuller T.. 2013. New universal rules of eukaryotic translation initiation fidelity. PLoS Comput Biol. 9(7):e1003136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.