Abstract
Background
Epigenetic variation in the serotonin transporter gene (SLC6A4) has been shown to modulate the functioning of brain circuitry associated with the salience network and may heighten the risk for mental illness. This study is, to our knowledge, the first to test this epigenome–brain–behaviour pathway in patients with anorexia nervosa.
Methods
We obtained resting-state functional connectivity (rsFC) data and blood samples from 55 acutely underweight female patients with anorexia nervosa and 55 age-matched female healthy controls. We decomposed imaging data using independent component analysis. We used bisulfite pyrosequencing to analyze blood DNA methylation within the promoter region of SLC6A4. We then explored salience network rsFC patterns in the group × methylation interaction.
Results
We identified a positive relationship between SLC6A4 methylation levels and rsFC between the dorsolateral prefrontal cortex and the salience network in patients with anorexia nervosa compared to healthy controls. Increased rsFC in the salience network mediated the link between SLC6A4 methylation and eating disorder symptoms in patients with anorexia nervosa. We confirmed findings of rsFC alterations for CpG-specific methylation at a locus with evidence of methylation correspondence between brain and blood tissue.
Limitations: This study was cross-sectional in nature, the sample size was modest for the method and methylation levels were measured peripherally, so findings cannot be fully generalized to brain tissue.
Conclusion
This study sheds light on the neurobiological process of how epigenetic variation in the SLC6A4 gene may relate to rsFC in the salience network that is linked to psychopathology in anorexia nervosa.
Introduction
The serotonin system has been associated with mood regulation, anxiety and the modulation of appetite,1 all of which nominate this neurotransmitter system as a potential candidate for involvement in the pathogenesis of anorexia nervosa.2 Anorexia nervosa is characterized by severe food restriction and affective symptoms (such as depressed mood and difficulties in emotion regulation),3 as well as a heightened prevalence of anxious and obsessive personality traits.4 A number of studies have suggested reduced serotonergic tone during the acute undernourished state of the disorder: for example the serotonin metabolite 5-hydroxyindolacetic acid in cerebrospinal fluid5 was decreased, plasma prolactin response to serotonin agonists were blunted,6 and whole blood serotonin content and tryptophan levels7 were reduced.
However, patients with anorexia nervosa in the weight-recovered state may exhibit hyperserotonergic dysfunction.8 The assumption that increased serotonin transmission is a trait marker of the disorder is supported by findings of decreased platelet monoamine oxidase B activity9 and altered cerebral serotonin 1A and 2A receptor binding.10,11 It has been hypothesized that serotonergic dysfunctions may drive body-image distortions and self-starvation in anorexia nervosa.12 However, associations between genetic polymorphisms of the serotonergic system (in particular the serotonin transporter SLC6A4) and anorexia nervosa could not be replicated.13 Therefore, an interest in the epigenome has gained momentum. Epigenetic processes such as DNA methylation may mediate environmental effects on gene expression without changing the DNA sequence. During DNA methylation, a methyl group is added to a cytosine residue, often adjacent to a guanine nucleotide. These so-called CpG sites tend to cluster together in CpG islands. In the promoter region of genes, CpG islands are often demethylated, whereas the methylated state is commonly associated with reduced gene expression.14 Within the SLC6A4 promoter region, DNA methylation appears to be of functional relevance, because it has been found to predict lower SLC6A4 mRNA expression.15 Furthermore, we previously reported that DNA methylation at CpG site 13 (cg14692377) showed a significant correlation between blood and brain tissue obtained simultaneously from paired blood and temporal lobe biopsy samples.16 This CpG was also shown to be correlated in paired premortem blood and postmortem brain samples.17 However, although SLC6A4 methylation has been reported to play a role in depressive disorders and schizophrenia,18 the only existing candidate-gene study in anorexia nervosa found no group differences compared to healthy controls.19 As well, a study using a methylome-wide approach did not provide support for a link between SLC6A4 DNA methylation and a diagnosis of anorexia nervosa.20 However, related phenotypes such as cortisol stress reactivity have been repeatedly associated with altered SLC6A4 methylation.21 Expanding the scope by integrating epigenetic and neuroimaging data may allow us to test the complex interplay between genes, brain function and abnormal behaviour related to eating disorders.22,23
One promising neuroimaging technique is resting-state functional connectivity (rsFC), which is defined as the temporally correlated, low-frequency, blood oxygen level–dependent (BOLD) signal fluctuation of distinct brain regions while the participant is at rest. This method is sensitive to changes in communication between spatially separated brain regions and is thought to be more sensitive to the effects of genetic and environmental variation in heterogeneous phenotypes such as psychiatric disorders.24,25
Recent evidence from rsFC studies points to a dysfunction in the salience network as a prominent feature of several mental disorders,24 including anorexia nervosa. The salience network — consisting of the dorsal anterior cingulate cortex, the insular cortices and additional limbic structures such as the amygdala24 — is associated with detecting, filtering and integrating biologically relevant sources of salience, such as emotional information, homeostatic regulation and reward value.26 Some studies have demonstrated aberrant functional within- and between-network connectivity of the salience network in anorexia nervosa, but others found only indirect evidence for salience network abnormalities in patients in the acutely underweight state.27,28 Importantly, studies in healthy participants have observed that activity in key nodes of the salience network — including the amygdala29 and insula,30 as well as amygdala connectivity31 — might depend on SLC6A4 methylation. Using a monozygotic twin design, a recent study demonstrated an association between SLC6A4 methylation and brain responses to negative stimuli in frontal-limbic regions overlapping the salience network that was independent of DNA sequence variations.32
Given that previous studies in anorexia nervosa have identified neural alterations in the salience network, and that this network has also been identified as a neural correlate of SLC6A4 methylation, we assumed that a serotonin-related epigenome–brain–behaviour process, discussed in detail in a study by Palma-Gaudiel and Fañanás,18 could also be highly relevant for anorexia nervosa. These authors describe a neurobiological pathway that is rooted in a reduction of serotonin transporter expression mediated by SLC6A4 methylation. This reduction may lead to changes in serotonergic tone, associated with altered reactivity and connectivity of brain regions and mental health problems relevant to the salience network.
In light of the proposed epigenome–brain–behaviour pathway for anorexia nervosa, the aim of this study was to examine the associations between SLC6A4 methylation, salience network rsFC and eating disorder symptoms in acutely ill patients with anorexia nervosa compared to matched healthy controls.
Methods
Participants
Our sample consisted of 110 female volunteers: 55 participants with acute anorexia nervosa according to the Diagnostic and Statistical Manual of Mental Disorders, 5th edition (DSM-5; age 12–28 yr) and 55 pair-wise age-matched female healthy controls (age 12–28 yr). All patients with anorexia nervosa were assessed within 96 hours after the beginning of a behaviourally oriented nutritional rehabilitation program. In the anorexia nervosa group, 53 of the patients were of the restrictive and 2 of the binge/purge subtype; 7 had comorbid psychiatric disorders (4 patients with depressive disorders including dysthymia, 3 with anxiety disorder, and 1 with obsessive–compulsive disorder). Six patients with anorexia nervosa and 10 healthy controls reported that they smoked cigarettes currently or had smoked in the past.
We applied several exclusion criteria for each group (Appendix 1, Supplemental Materials 1.1, available at jpn.ca/190015-a1), most importantly psychotropic medication other than selective serotonin reuptake inhibitors (SSRIs) within 4 weeks before the study (n = 1), binge eating or diagnosis of bulimia nervosa, substance abuse, or neurologic or medical conditions. This study was approved by the review board of the Technische Universität Dresden, and all participants (and, if underage, their guardians) gave written informed consent.
Clinical measures
To ascertain the absence or presence of a current eating disorder, we administered the expert form of the Structured Interview for Anorexia and Bulimia Nervosa for DSM-IV (SIAB-EX33) in all participants. Interviews were adapted to DSM-5 criteria (no amenorrhea criteria) and carried out by clinically experienced and trained research assistants under the supervision of the attending child and adolescent psychiatrist. We estimated intelligence quotient using a short version of the German adaptation of the Wechsler Adult Intelligence Scale,34 or a short version of the German adaptation of the Wechsler Intelligence Scale for Children35 for participants aged 15 years or younger.
We assessed eating disorder–specific psychopathology using the short version of the Eating Disorders Inventory (EDI-2).36 We examined expressive symptoms using the German version of the Beck Depression Inventory (BDI-II).37 We investigated symptoms of anxiety using the anxiety subscale of the Symptom Checklist 90 Revised (SCL-90-R).38
Blood DNA methylation, sample processing and quality control
Quantitative peripheral DNA methylation at 15 CpG sites in the promoter-associated CpG island of SLC6A4 initially described by Philibert and colleagues39 was performed by Varionostic GmbH (www.varionostic.de). We focused on 15 CpG sites within amplicon 3 of a 799 bp region (originally CpG 43–57, referred to in the current study as CpG 1–15), because a previous study observed significant interindividual variation in methylation in this subregion (but not in others) and found a significant correlation with rsFC in the salience network.31 As well, DNA methylation in this subregion has been shown to be correlated between blood and brain tissue.16 Moreover, previous studies have argued that CpG-specific methylation (rather than average methylation) corresponds with mRNA expression.15,40 To further investigate the correlation between peripheral, blood-based measures of SLC6A4 DNA methylation with those in postmortem brain tissue, we queried BECon, an online database of cross-tissue DNA methylation.41 Based on whole-blood and postmortem brain tissue from 3 regions (frontal, temporal and parietal) obtained from 16 individuals, cg14692377 showed moderate-rank correlations between tissues (ρ BA10 = 0.29 [frontal]; ρ BA20 = 0.36 [temporal]; ρBA10 = 0.48 [parietal]), indicating that in this specific case peripheral DNA methylation might indeed index brain-based methylation patterns. For analysis, genomic DNA was extracted from EDTA whole-blood samples and bisulfite-treated using the EZ DNA Methylation Gold Kit (Zymo Research). Subsequent pyrosequencing was performed on the Q24/ID System, and percent DNA methylation at each CpG site was quantified using the PyroMark Q24 software (Qiagen). For a detailed protocol containing amplicon, sequencing, and cross-CpG correlations and transcription factor binding site within the region, see Appendix 1, Supplemental Materials 1.2. We visually inspected pyrograms to individually verify DNA methylation estimates if they were at least 3 standard deviations away from the mean, or if internal quality scores indicated a deviation from the expected pattern. Based on these criteria, we excluded data from 1 CpG (CpG 15). The mean missingness after exclusion was 0.58% across CpGs (range: 0%–1.82%) and participants (range: 0%–35.71%). Missing values were imputed using predictive mean matching with default settings as implemented in the R mice package.42
MRI data acquisition and preprocessing
Data were acquired with a 3T Siemens Trio. We acquired the T1-weighted structural brain scans with a rapid acquisition gradient-echo (MP-RAGE) sequence (parameters are described in Appendix 1, Supplemental Materials 1.3). We acquired an 8-minute resting functional MRI (fMRI) scan using gradient-echo T2*-weighted echo planar imaging with standard parameters (Appendix 1, Supplemental Materials 1.3). During fMRI, participants were instructed to lie still with closed eyes and not fall asleep. Functional and structural images were processed using SPM8 (www.fil.ion.ucl.ac.uk/spm/) within the Nipype framework (http://nipy.sourceforge.net/nipype) following standard procedures (Appendix 1, Supplemental Materials 1.4).
We evaluated the quality of the fMRI data using manual inspection and artifact detection tools (Appendix 1, Supplemental Materials 1.4).
Independent component analysis and identification of component of interest
To extract temporally coherent networks that reflected well-defined circumscribed functional properties, we conducted a spatial group independent component analysis (ICA)43 for all participants using the Group ICA fMRI Toolbox (GIFT) implemented in Matlab (http://mialab.mrn.org/software/gift). Unlike seed-based approaches, this technique allows for identification of a resting-state network that is independent of neuroanatomical variability. After initial data reduction using principal component analysis, we decomposed the fMRI data into 24 maximally independent components by applying the infomax algorithm (Appendix 1, Supplemental Materials 1.5). For each participant, we back-reconstructed component spatial maps using GICA and converted to z values.44 As in our previous reports,45,46 we identified components of interest by spatial correlation with the relevant templates (salience, frontoparietal and visual network) from a study by Yeo and colleagues.47 We mapped independent component numbers 9 (IC9) and 13 (IC13) onto the salience network template defined by Yeo and colleagues47 (Appendix 1, Supplemental Materials 1.6).
Statistical models
We averaged DNA methylation levels across the 14 individual CpG sites after quality control and imputation to calculate a mean SLC6A4 methylation score (methylationmean) for each participant.31 Although SLC6A4 methylationmean was our main measure of interest, we also investigated the effects of CpG site 13 (methylationCpG13), because there is evidence16 for small to moderate blood–brain correspondence of methylationCpG13. To identify the effects of potentially confounding variables on DNA methylation, we investigated associations with age (using Pearson r).
For rsFC, we entered spatial maps of the components of each participant into SPM8 to conduct the following analyses. First, to test for basic group differences, we performed a 2-sample t test for the 2 components identified as salience network (step 1).47 Second, we examined components of the main model for which we had found a basic group difference at step 1. The rsFC in components that showed a group difference in step 1 were regressed using group, SLC6A4 methylationmean, their interaction, and age as covariates; the interaction term was our main effect of interest (step 2). Third, based on previous findings as outlined earlier15,16 we reran the main model described in step 2 using methylationCpG13 instead of methylationmean (step 3). Fourth, to investigate the specificity of the effect of DNA methylation on the rsFC of the salience network, we examined the group × methylationmean interaction win a control analysis for the visual network (also identified by spatial correlation with a template47), a brain network that is thought to be somewhat less influenced by the serotonergic system48 (step 4). The results of all models were masked using the aforementioned RSN templates47 and had to exceed p < 0.05 (family-wise error [FWE] correction) to guard against type I errors. To test whether our main analysis (step 2) was influenced by smoking, we ran additional subanalyses excluding participants who smoked currently or had smoked in the past (anorexia nervosa = 6; healthy controls = 10; including occasional smoking) or were taking SSRIs (anorexia nervosa = 1).
To develop a better understanding of the group × methylationmean interaction, we extracted β-values using the MarsBaR toolbox (http://marsbar.sourceforge.net/) at a threshold of p < 0.001 (uncorrected to account for greater anatomic variability) and subjected those to further testing using SPSS version 21.0 (SPSS Inc.). We examined associations with the physiologic and psychometric parameters body mass index standard deviation score (BMI-SDS),49 EDI-2, BDI-II and SCL-90-R anxiety using Pearson r for each diagnostic group separately.
To probe the proposed serotonin-related epigenome–brain–behaviour pathway, we conducted a mediation analysis using the PROCESS package implemented in SPSS. To this end, we tested whether the link between alterations in SLC6A4 methylationmean and elevated psychopathology (EDI-2 total, BDI-II and SCL-90-R anxiety) was mediated by altered rsFC in the salience network (indirect effect) in patients with anorexia nervosa using the bootstrapping procedure on 20 000 samples to compute the lower and upper levels of the 95% confidence interval.
Results
Participants
As shown in Table 1, we found no significant group differences for age. As expected, patients with anorexia nervosa had lower BMI-SDS and a higher EDI-2 total, BDI-II and SCL-90-R anxiety scores than healthy controls. Patients with anorexia nervosa did not differ from healthy controls with respect to SLC6A4 methylationmean or methylationCpG13. We did find that SLC6A4 methylationmean was significantly associated with age (rp = 0.22; p = 0.021), so we included age as a covariate in our main statistical models.
Table 1.
Demographic and clinical characteristics*
| Characteristic | Anorexia nervosa (n = 55) | Healthy controls (n = 55) | Statistics | |
|---|---|---|---|---|
|
| ||||
| t | p value | |||
| Demographic | ||||
| Age, yr | 16.15 ± 3.07 | 16.17 ± 3.07 | −0.37 | 0.97 |
| BMI-SDS | −3.21 ± 1.13 | 0.09 ± 1.04 | −15.91 | < 0.001 |
| BMI | 14.55 ±1.35 | 20.68 ± 2.37 | −16.44 | < 0.001 |
| Clinical | ||||
| Age at onset, yr | 13.82 ± 2.63 | NA | ||
| EDI-2 total | 202.27 ± 47.28 | 142.85 ± 26.82 | −7.99 | < 0.001 |
| BDI-II | 21.49 ± 10.92 | 5.78 ± 6.27 | 9.24 | < 0.001 |
| Anxiety (SCL-90-R) | 0.81 ± 0.80 | 0.31 ± 0.41 | 4.04 | < 0.001 |
| SLC6A4 DNA methylation | ||||
| Mean SLC6A4 methylation | 5.45 ± 1.51 | 5.48 ± 2.28 | −0.08 | 0.94 |
| DNA methylation at CpG 13 | 8.05 ± 2.14 | 8.50 ± 7.11 | −0.44 | 0.66 |
BDI-II = Beck Depression Inventory II; BMI = body mass index; BMI-SDS = body mass index standard deviation score; EDI-2 = Eating Disorder Inventory 2; NA = not applicable; SCL-90-R = Symptom Checklist 90 Revised.
Data are mean ± standard deviation unless otherwise indicated.
We investigated whether SLC6A4 methylationCpG13 was dependent on genetic variation, but based on publicly available data, we could not identify underlying methylation quantitative trait loci in blood (http://mqtldb.org) or human brain tissue (http://epigenetics.essex.ac.uk.mQTL; Appendix 1, Supplemental Materials 2.6).
Group comparison of independent components and association with methylation
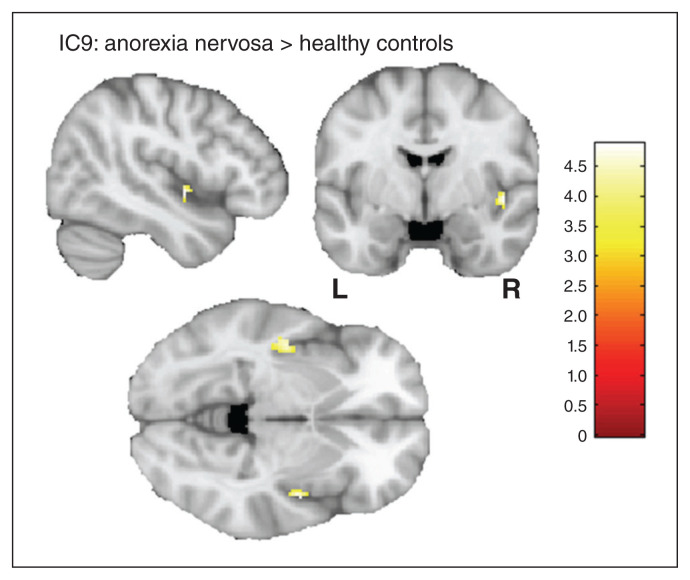
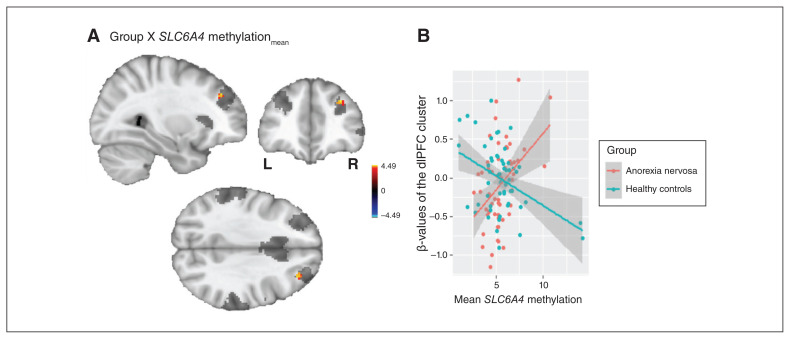
A 2-sample t test of the 2 salience network components (IC13 and IC9) revealed increased rsFC in the left (t = 4.68; pFWE = 0.006; k = 15) and right (t = 4.88; pFWE = 0.014; k = 7) posterior insula and IC9 for patients with anorexia nervosa compared to healthy controls (step 1; Fig. 1). We identified no group difference for IC13. In step 2, we identified a group × methylationmean interaction in the right dorsolateral prefrontal cortex (dlPFC) for IC9, controlling for age (t = 4.49; pFWE = 0.016; k = 6; x = 32, y = 40, z = 34; Fig. 2, panel A). A detailed analysis of this interaction revealed that SLC6A4 methylationmean was correlated positively with rsFC between this region and other areas of IC9 in patients with anorexia nervosa (r = 0.425; p = 0.002), but negatively in healthy controls (r = −0.408; p = 0.002; Fig. 2, panel B). In step 3, we specifically tested the group × methylationCpG13 interaction and observed an identical neural pattern in the right dlPFC (t = 4.73; pFWE = 0.018; k = 5; x = 34, y = 38, z = 30; Appendix 1, Supplemental Materials 2.1).
Fig. 1.
A 2-sample t test of the salience network component IC9 revealed increased functional connectivity between the left (t = 4.68; pFWE = 0.006) and right (t = 4.88; pFWE = 0.014) posterior insula and the rest of the component for patients with anorexia compared to healthy controls. FWE = family-wise error; IC9 = independent component 9.
Fig. 2.
(A) Results of the group × SLC6A4 methylationmean interaction analysis in the salience network at the dorsolateral prefrontal cortex (dlPFC; for illustrative purposes shown at p < 0.001, uncorrected). The applied salience network mask45 is depicted in transparent grey. The colour bar represents t values. (B) Plot of the association of the extracted β-values of the dlPFC cluster and mean SLC6A4 methylation (in %) separately for patients with anorexia nervosa and healthy controls. methylationmean = mean methylation.
Control analysis in the visual network revealed no group × methylationmean interaction, suggesting that the finding described above was specific to the salience network (step 4). Exclusion of current or previous smokers confirmed our initial results in the salience network (t = 5.09; pFWE = 0.007; k = 13; x = 30, y = 40, z = 32). Exclusion of the participant who took SSRIs also had no influence on the results (t = 4.46; pFWE = 0.018; k = 5; x = 32, y = 40, z = 34).
Increased rsFC between the significant cluster in the dlPFC (for which we reported the group × methylationmean interaction) and other regions of the salience network was associated with eating disorder psychopathology, indicated by a correlation with the EDI-2 total score, but not with the BMI-SDS, BDI-II or SCL-90-R anxiety scores, in patients with anorexia nervosa (rp = 0.349; p = 0.010; withstood Bonferroni correction for 4 tests). Mediation analysis showed that the effect of SLC6A4 methylationmean on EDI-2 total (but not on BDI-II or on SCL-90-R anxiety) was mediated by alterations in the rsFC of the salience network in patients with anorexia nervosa (Table 2 and Appendix 1, Supplemental Materials 2.4).
Table 2.
Mediation analysis between mean SCL6A4 methylation score and outcome variables with rsFC of the salience network as the mediator
| Outcome | Total effect, 95% CI | Direct effect, 95% CI | Indirect effect, 95% CI | |||
|---|---|---|---|---|---|---|
|
|
|
|
||||
| Lower limit | Upper limit | Lower limit | Upper limit | Lower limit | Upper limit | |
| EDI-2 total | −2.642 | 14.385 | −7.435 | 10.654 | 0.019 | 9.296 |
| BDI-II | 0.087 | 3.953 | −0.561 | 3.706 | −0.524 | 1.474 |
BDI-II = Beck Depression Inventory II; CI = confidence interval; EDI-2 = Eating Disorder Inventory 2; rsFC = resting-state functional connectivity.
Discussion
The present study is, to our knowledge, the first to investigate a potential epigenome–brain–behaviour pathway in anorexia nervosa by examining the association between SLC6A4 methylation and rsFC of the salience network in patients acutely ill with anorexia nervosa and healthy controls. At a group level, we observed increased rsFC in the salience network at the posterior insula in patients compared with controls, but (in agreement with previous reports19,50) no difference in SLC6A4 methylation levels. However, we did identify a positive association between methylation levels and rsFC in the dlPFC and other regions of the salience network in patients with anorexia nervosa that was also associated with elevated eating disorder symptoms. In fact, alterations in salience network rsFC mediated the link between SLC6A4 methylation changes and general eating disorder psychopathology, but not depressive or anxiety-related psychopathology. These results may suggest that this methylation-related rsFC pattern was specific to eating disorder symptoms. However, to confirm this notion, further research that includes other clinical groups is needed. In healthy controls, increased SLC6A4 methylation levels were associated with reduced rsFC in this brain region. Importantly, these associations could be demonstrated for mean methylation as well as for CpG-specific methylation (SLC6A4 methylationCpG13) at a locus with previous evidence for the correspondence of methylation levels across blood and brain tissue.16 We detected no association between methylation and rsFC in a negative control network chosen a priori (the visual network), indicating that the association we found may have been specific for the salience network.
Alterations in the salience network have been described to underlie abnormal sensitivity to salient external or internal events, with severe consequences for cognition and behaviour across several mental disorders,51 including anorexia nervosa.27 The posterior insula, the location of the general group difference in the salience network, has been associated with pain and sensorimotor representation and is thought to function as a major interoceptive region.52 In line with this, we have demonstrated altered functional network characteristics in the posterior insula in anorexia nervosa,53,54 which may reflect inefficient information transfer to and from this brain region. Functional alterations in the insula have also repeatedly been shown in anorexia nervosa using task-based fMRI (for an overview of selected studies, see Appendix 1, Supplemental Materials 3). These dysfunctions in the insular cortices relate to impaired integration of visuospatial, homeostatic and interoceptive signals, which may account for some of symptoms in anorexia nervosa, such as body image distortions, elevated pain threshold and severe self-starvation.55
Our finding of methylation-related alterations in rsFC of the salience network, which were also associated with eating disorder severity in anorexia nervosa, fit well with a previously reported notion of a clinically relevant brain-mediated association between SLC6A4 methylation and illness-related behaviour.29,31,56,57 Previous neuroimaging studies in health and disease (for a review, see Palma-Gudiel and Fañanás18) — as well as our present findings in anorexia nervosa — suggest that increased SLC6A4 methylation may be linked to heightened activity and connectivity in brain regions related to the salience network in people with severe mental health conditions. In contrast to our observations for anorexia nervosa, we found a negative (rather than a positive) association between SLC6A4 methylation and rsFC in the salience network in healthy controls. Similarly, Frodl and colleagues30 demonstrated a negative association between methylation and neural reactivity in the posterior insula, pons and temporal pole when shifting attention away from emotional stimuli in a sample that included healthy controls and patients with major depressive disorder. The direction and exact localization of the association between SLC6A4 methylation and brain functioning is still somewhat unclear and difficult to compare because of different fMRI methods used (resting state [seed-based31 versus ICA] or task fMRI) and the contrast conditions in task fMRI.
Interestingly, although previous studies investigating SLC6A4 methylation-related brain regions focused primarily on ventral-limbic parts of the brain such as the amygdala, we found a methylation-related increase in rsFC between the dlPFC and other regions of the salience network in anorexia nervosa. To further probe the specificity of this finding regarding the salience network, we also tested the frontoparietal network (network with the dlPFC as a central hub) as part of an additional follow-up analysis (Appendix 1, Supplemental Materials 2.3). Similar to the visual network (negative control network), we could not detect a significant association between methylation and rsFC in the frontoparietal network. Given that the methylation-related increase of rsFC was present in a section of the dlPFC belonging to the salience network but not for sub-regions of the dlPFC, which are part of the frontoparietal network, we suggest that the associations with SLC6A4 methylation may be specific to the frontolimbic circuit. This circuit is a component of the salience network and part of the serotonin pathway (i.e., it is strongly innervated by serotonergic neurons and rich in various serotonin receptors).58 With regard to processing emotions and saliency — a major function of the salience network — the dlPFC (and its coupling with limbic regions) is also engaged in bottom–up appraisal of emotions59 and in emotion regulation via cognitive strategies.60 Modulation of frontolimbic circuits via the serotonergic system has previously been shown by studies focusing on the serotonin-transporter-linked polymorphic region. Based on this, a speculative interpretation of our findings could be that methylation-related increased involvement of the dlPFC in the salience network is related to elevated cognitive or self-control — a much debated characteristic of anorexia nervosa.2 This tentative interpretation of the findings dovetails with the notion of anorexia nervosa as a model-disorder of increased cognitive control, an interpretation that may be particularly reflected in the tendency of patients to respond in a strategic rather than hedonic manner to salient stimuli.61,62 Interestingly, in a recent study we discovered a sustained lateral prefrontal cortex responsivity in patients with anorexia nervosa when food-related social but also neutral stimuli were presented.62 We speculated that this may reflect an inability of patients with anorexia nervosa to disengage cognitive control. In light of this, the current findings of increased rsFC between dlPFC and the rest of the salience network associated with SLC6A4 — although correlational in nature — may be interpreted as a tendency of anorexia nervosa patients to “stay in control,” which could be a neural downstream effect of elevated SLC6A4 methylation, leading to unwanted effects such as increased eating disorder symptoms.
Limitations
Interpretation of the present study rests on the following limitations. First, the cross-sectional nature of the study did not allow us to determine a causal link between epigenetic variation, brain network characteristics and the emergence of eating disorder symptoms, or whether the observed alterations were an effect of undernutrition. To shed light on temporal relationships, longitudinal studies are needed. Second, we assessed methylation measures peripherally, so our findings cannot be fully generalized to neural tissue. However, previous work could demonstrate a substantial association between methylation levels in blood and brain for SLC6A4 methylationCpG13,16,17 for which we could confirm our initial findings. Furthermore, although methylation at this site was somewhat low, which could add noise due to technical variation, the methylation levels in our study were in line with those of previous reports.63 Third, the present study focused only on epigenetic variation and failed to incorporate genetic variability and other influencing factors such as stress and childhood adversity. Although the contribution of both epigenetic and genetic factors might be relevant to predicting the expression of SLC6A4,18 SLC6A4 methylationCpG13 does not seem to be under strong genetic control. Fourth, rates of comorbidity were lower than previously reported,64 which could have been due to the fact that no structured interview other than the SIAB-EX was used (Appendix 1, Supplemental Materials 1.1).
Conclusion
If replicated, our findings suggest that a serotonin-related epigenome–brain–behaviour pathway is also relevant for anorexia nervosa. They shed light on the neurobiological process of how epigenetic variation of the SLC6A4 gene may be associated with rsFC in the salience network that is specifically linked to eating-disorder symptoms. Such knowledge may spur future research into the potential of epigenetic markers to assess risk for disordered eating and pave the way for potential novel pharmacological and behavioural treatment strategies that modulate the methylation levels of specific genes.65,66
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (DFG EH 367/5-1, EH 367/7-1 & SFB 940) and the Technische Universität Dresden. The authors thank the Center for Information Services and High Performance Computing (ZIH) at Technische Universität Dresden for generous allocations of computer time.
Footnotes
Competing interests: V. Roessner reports lecture fees from Eli Lilly, Janssen-Cilag, Medice and Novartis and was a member of advisory boards for Eli Lilly and Novartis in the past 2 years. No other authors reported competing interests.
Contributors: E. Walton, D. Geisler, V. Roessner and S. Ehrlich designed the study. I. Boehm, M. Seidel, J. King and K. Weidener acquired the data, which I. Boehm, E. Walton, N. Alexander, V.-L. Batury and S. Ehrlich analyzed. I. Boehm, J. King and S. Ehrlich wrote the article, which all authors reviewed. All authors approved the final version to be published and can certify that no other individuals not listed as authors have made substantial contributions to the paper.
References
- 1.Halford JCG, Blundell JE. Separate systems for serotonin and leptin in appetite control. Ann Med. 2000;32:222–32. doi: 10.3109/07853890008998829. [DOI] [PubMed] [Google Scholar]
- 2.Kaye W, Fudge J, Paulus M. New insights into symptoms and neurocircuit function of anorexia nervosa. Nat Rev Neurosci. 2009;10:573–84. doi: 10.1038/nrn2682. [DOI] [PubMed] [Google Scholar]
- 3.Haynos AF, Fruzzetti AE. Anorexia nervosa as a disorder of emotion dysregulation: evidence and treatment implications. Clin Psychol Sci Pract. 2011;18:183–202. [Google Scholar]
- 4.Marzola E, Fassino S, Amianto F, et al. Affective temperaments in anorexia nervosa: the relevance of depressive and anxious traits. J Affect Disord. 2017;218:23–9. doi: 10.1016/j.jad.2017.04.054. [DOI] [PubMed] [Google Scholar]
- 5.Demitrack MA, Heyes MP, Altemus M, et al. Cerebrospinal fluid levels of kynurenine pathway metabolites in patients with eating disorders: relation to clinical and biochemical variable. Biol Psychiatry. 1995;37:512–20. doi: 10.1016/0006-3223(94)00173-Z. [DOI] [PubMed] [Google Scholar]
- 6.Monteleone P, Brambilla F, Bortolotti F, et al. Prolactin response to d-fenfluramine is blunted in people with anorexia nervosa. Br J Psychiatry. 1998;172:439–42. doi: 10.1192/bjp.172.5.439. [DOI] [PubMed] [Google Scholar]
- 7.Gauthier C, Hassler C, Mattar L, et al. Symptoms of depression and anxiety in anorexia nervosa: links with plasma tryptophan and serotonin metabolism. Psychoneuroendocrinology. 2014;39:170–8. doi: 10.1016/j.psyneuen.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 8.Kaye W, Gwirtsman HE, George DT, et al. Altered serotonin activity in anorexia nervosa after long-term weight restoration: does elevated cerebrospinal fluid 5-hydroxyindoleacetic acid level correlate with rigid and obsessive behavior? Arch Gen Psychiatry. 1991;48:556–62. doi: 10.1001/archpsyc.1991.01810300068010. [DOI] [PubMed] [Google Scholar]
- 9.Ehrlich S, Franke L, Schott R, et al. Platelet monoamine oxidase activity in underweight and weight-recovered females with anorexia nervosa. Pharmacopsychiatry. 2008;41:226–31. doi: 10.1055/s-2008-1078749. [DOI] [PubMed] [Google Scholar]
- 10.Bailer U, Frank G, Henry S, et al. Altered brain serotonin 5-HT1A receptor binding after recovery from anorexia nervosa measured by positron emission tomography and [Carbonyl11C]WAY-100635. Arch Gen Psychiatry. 2005;62:1032–41. doi: 10.1001/archpsyc.62.9.1032. [DOI] [PubMed] [Google Scholar]
- 11.Frank G, Kaye W, Meltzer CC, et al. Reduced 5-HT2A receptor binding after recovery from anorexia nervosa. Biol Psychiatry. 2002;52:896–906. doi: 10.1016/s0006-3223(02)01378-1. [DOI] [PubMed] [Google Scholar]
- 12.Riva G. Neurobiology of anorexia nervosa: serotonin dysfunctions link self-starvation with body image disturbances through an impaired body memory. Front Hum Neurosci. 2016;10:600. doi: 10.3389/fnhum.2016.00600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Solmi M, Gallicchio D, Collantoni E, et al. Serotonin transporter gene polymorphism in eating disorders: data from a new biobank and META-analysis of previous studies. World J Biol Psychiatry. 2016;17:244–57. doi: 10.3109/15622975.2015.1126675. [DOI] [PubMed] [Google Scholar]
- 14.Abdolmaleky HM, Smith CL, Faraone SV, et al. Methylomics in psychiatry: modulation of gene–environment interactions may be through DNA methylation. Am J Med Genet B Neuropsychiatr Genet. 2004;127B:51–9. doi: 10.1002/ajmg.b.20142. [DOI] [PubMed] [Google Scholar]
- 15.Wankerl M, Miller R, Kirschbaum C, et al. Effects of genetic and early environmental risk factors for depression on serotonin transporter expression and methylation profiles. Transl Psychiatry. 2014;4:e402. doi: 10.1038/tp.2014.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walton E, Hass J, Liu J, et al. Correspondence of DNA methylation between blood and brain tissue and its application to schizophrenia research. Schizophr Bull. 2016;42:406–14. doi: 10.1093/schbul/sbv074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hannon E, Lunnon K, Schalkwyk L, et al. Interindividual methylomic variation across blood, cortex, and cerebellum: implications for epigenetic studies of neurological and neuropsychiatric phenotypes. Epigenetics. 2015;10:1024–32. doi: 10.1080/15592294.2015.1100786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palma-Gudiel H, Fañanás L. An integrative review of methylation at the serotonin transporter gene and its dialogue with environmental risk factors, psychopathology and 5-HTTLPR. Neurosci Biobehav Rev. 2017;72:190–209. doi: 10.1016/j.neubiorev.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 19.Pjetri E, Dempster E, Collier DA, et al. Quantitative promoter DNA methylation analysis of four candidate genes in anorexia nervosa: a pilot study. J Psychiatr Res. 2013;47:280–2. doi: 10.1016/j.jpsychires.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 20.Steiger H, Booij L, ‘Esther Kahan, et al. A longitudinal, epigenome-wide study of DNA methylation in anorexia nervosa: results in actively ill, partially weight-restored, long-term remitted and non-eating-disordered women. J Psychiatry Neurosci. 2019;44:205–13. doi: 10.1503/jpn.170242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller R, Wankerl M, Stalder T, et al. The serotonin transporter gene-linked polymorphic region (5-HTTLPR) and cortisol stress reactivity: a meta-analysis. Mol Psychiatry. 2013;18:1018–24. doi: 10.1038/mp.2012.124. [DOI] [PubMed] [Google Scholar]
- 22.Bogdan R, Salmeron BJ, Carey CE, et al. Imaging genetics and genomics in psychiatry: a critical review of progress and potential. Biol Psychiatry. 2017;82:165–75. doi: 10.1016/j.biopsych.2016.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Booij L, Tremblay RE, Szyf M, et al. Genetic and early environmental influences on the serotonin system: consequences for brain development and risk for psychopathology. J Psychiatry Neurosci. 2015;40:5–18. doi: 10.1503/jpn.140099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menon V. Large-scale brain networks and psychopathology: a unifying triple network model. Trends Cogn Sci. 2011;15:483–506. doi: 10.1016/j.tics.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 25.Meyer-Lindenberg A. Neural connectivity as an intermediate phenotype: brain networks under genetic control. Hum Brain Mapp. 2009;30:1938–46. doi: 10.1002/hbm.20639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seeley WW, Menon V, Schatzberg AF, et al. Dissociable intrinsic connectivity networks for salience processing and executive control. J Neurosci. 2007;27:2349–56. doi: 10.1523/JNEUROSCI.5587-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaudio S, Wiemerslage L, Brooks SJ, et al. A systematic review of resting-state functional-MRI studies in anorexia nervosa: evidence for functional connectivity impairment in cognitive control and visuospatial and body-signal integration. Neurosci Biobehav Rev. 2016;71:578–89. doi: 10.1016/j.neubiorev.2016.09.032. [DOI] [PubMed] [Google Scholar]
- 28.Uniacke B, Wang Y, Biezonski D, et al. Resting-state connectivity within and across neural circuits in anorexia nervosa. Brain Behav. 2019;9:e01205. doi: 10.1002/brb3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikolova YS, Koenen KC, Galea S, et al. Beyond genotype: serotonin transporter epigenetic modification predicts human brain function. Nat Neurosci. 2014;17:1153–5. doi: 10.1038/nn.3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frodl T, Szyf M, Carballedo A, et al. DNA methylation of the serotonin transporter gene (SLC6A4) is associated with brain function involved in processing emotional stimuli. J Psychiatry Neurosci. 2015;40:296–305. doi: 10.1503/jpn.140180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muehlhan M, Kirschbaum C, Wittchen H-U, et al. Epigenetic variation in the serotonin transporter gene predicts resting state functional connectivity strength within the salience-network. Hum Brain Mapp. 2015;36:4361–71. doi: 10.1002/hbm.22923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ismaylova E, Lévesque ML, Pomares FB, et al. Serotonin transporter promoter methylation in peripheral cells and neural responses to negative stimuli: a study of adolescent monozygotic twins. Transl Psychiatry. 2018;8:147. doi: 10.1038/s41398-018-0195-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fichter M, Quadflieg N. SIAB. Struckturiertes Inventar fuer anorektische und Bulimische Essstoerungen nach DSM-IV und ICD-10. Bern, Switzerland: Huber; 1999. [Google Scholar]
- 34.Von Aster M, Neubauer A, Horn R. Wechsler Intelligenztest für Erwachsene (WIE) Deutschsprachige Bearbeitung und Adaptation des WAIS-III von David Wechsler. Frankfurt am Main, Germany: Harcourt Test Services; 2006. [Google Scholar]
- 35.Petermann F, Petermann U. HAWIK-IV: Hamburg-Wechsler Intelligenztest für Kinder-IV. New York: Huber; 2008. [Google Scholar]
- 36.Paul T, Thiel A. Eating Disorder Inventory-2 (EDI-2): Deutsche Version. Goettingen, Germany: Hogrefe; 2005. [Google Scholar]
- 37.Hautzinger M, Beck AT, Keller F, et al. Beck Depressions-Inventar: BDI II; Manual. Frankfurt am Main: Pearson Assessment; 2009. [Google Scholar]
- 38.Franke G. SCL-90-R Symptom-Checklist von L.R. Derogatis—Deutsche Version. Goettingen, Germany: Beltz Test GmbH; 2002. [Google Scholar]
- 39.Philibert R, Madan A, Andersen A, et al. Serotonin transporter mRNA levels are associated with the methylation of an upstream CpG island. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:101–5. doi: 10.1002/ajmg.b.30414. [DOI] [PubMed] [Google Scholar]
- 40.Iga J-I, Watanabe S-Y, Numata S, et al. Association study of polymorphism in the serotonin transporter gene promoter, methylation profiles, and expression in patients with major depressive disorder. Hum Psychopharmacol. 2016;31:193–9. doi: 10.1002/hup.2527. [DOI] [PubMed] [Google Scholar]
- 41.Edgar RD, Jones MJ, Meaney MJ, et al. BECon: a tool for interpreting DNA methylation findings from blood in the context of brain. Transl Psychiatry. 2017;7:e1187. doi: 10.1038/tp.2017.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Buuren S, Groothuis-Oudshoorn K. mice: Multivariate Imputation by Chained Equations in R. J Stat Softw. 2011;45:1–67. [Google Scholar]
- 43.Calhoun VD. Group ICA of fMRI toolbox (GIFT) Atlanta (GA): TReNDS Center; 2004. [accessed 2019 Dec 2]. Available: http://icatb.sourceforge.net. [Google Scholar]
- 44.Erhardt EB, Rachakonda S, Bedrick EJ, et al. Comparison of multi-subject ICA methods for analysis of fMRI data. Hum Brain Mapp. 2011;32:2075–95. doi: 10.1002/hbm.21170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boehm I, Geisler D, Tam F, et al. Partially restored resting-state functional connectivity in women recovered from anorexia nervosa. J Psychiatry Neurosci. 2016;41:150259. doi: 10.1503/jpn.150259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boehm I, Geisler D, King J, et al. Increased resting state functional connectivity in the fronto-parietal and default mode network in anorexia nervosa. Front Behav Neurosci. 2014;8:346. doi: 10.3389/fnbeh.2014.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yeo BT, Krienen FM, Sepulcre J, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106:1125–65. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reader TA, Grondin L. Distribution of catecholamines, serotonin, and their major metabolites in the rat cingulate, piriform-entorhinal, somatosensory, and visual cortex: a biochemical survey using high-performance liquid chromatography. Neurochem Res. 1987;12:1087–97. doi: 10.1007/BF00971709. [DOI] [PubMed] [Google Scholar]
- 49.Kromeyer-Hauschild K, Wabitsch M, Kunze D, et al. Percentiles of body mass index in children and adolescents evaluated from different regional German studies. Monatsschr Kinderheilkd. 2001;149:807–18. [Google Scholar]
- 50.Booij L, Casey KF, Antunes JM, et al. DNA methylation in individuals with anorexia nervosa and in matched normal-eater controls: a genome-wide study. Int J Eat Disord. 2015;48:874–82. doi: 10.1002/eat.22374. [DOI] [PubMed] [Google Scholar]
- 51.Uddin LQ. Salience processing and insular cortical function and dysfunction. Nat Rev Neurosci. 2015;16:55–61. doi: 10.1038/nrn3857. [DOI] [PubMed] [Google Scholar]
- 52.Craig A. Interoception: the sense of the physiological condition of the body. Curr Opin Neurobiol. 2003;13:500–5. doi: 10.1016/s0959-4388(03)00090-4. [DOI] [PubMed] [Google Scholar]
- 53.Ehrlich S, Lord A, Geisler D, et al. Reduced functional connectivity in the thalamo-insular subnetwork in patients with acute anorexia nervosa. Hum Brain Mapp. 2015;36:1772–81. doi: 10.1002/hbm.22736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Geisler D, Borchardt V, Lord AR, et al. Abnormal functional global and local brain connectivity in female patients with anorexia nervosa. J Psychiatry Neurosci. 2016;41:6–15. doi: 10.1503/jpn.140310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nunn K, Frampton I, Gordon I, et al. The fault is not in her parents but in her insula—a neurobiological hypothesis of anorexia nervosa. Eur Eat Disord Rev. 2008;16:355–60. doi: 10.1002/erv.890. [DOI] [PubMed] [Google Scholar]
- 56.Ismaylova E, Di Sante J, Szyf M, et al. Serotonin transporter gene promoter methylation in peripheral cells in healthy adults: neural correlates and tissue specificity. Eur Neuropsychopharmacol. 2017;27:1032–41. doi: 10.1016/j.euroneuro.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 57.Swartz JR, Hariri AR, Williamson DE. An epigenetic mechanism links socioeconomic status to changes in depression-related brain function in high-risk adolescents. Mol Psychiatry. 2017;22:209–14. doi: 10.1038/mp.2016.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dayer A. Serotonin-related pathways and developmental plasticity: relevance for psychiatric disorders. Dialogues Clin Neurosci. 2014;16:29–41. doi: 10.31887/DCNS.2014.16.1/adayer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Comte M, Schön D, Coull JT, et al. Dissociating bottom-up and top-down mechanisms in the cortico-limbic system during emotion processing. Cereb Cortex. 2016;26:144–55. doi: 10.1093/cercor/bhu185. [DOI] [PubMed] [Google Scholar]
- 60.Kanske P, Heissler J, Schönfelder S, et al. How to regulate emotion? Neural networks for reappraisal and distraction. Cereb Cortex. 2011;21:1379–88. doi: 10.1093/cercor/bhq216. [DOI] [PubMed] [Google Scholar]
- 61.Wagner A, Aizenstein H, Venkatraman V, et al. Altered reward processing in women recovered from anorexia nervosa. Am J Psychiatry. 2007;164:1842–9. doi: 10.1176/appi.ajp.2007.07040575. [DOI] [PubMed] [Google Scholar]
- 62.Boehm I, King J, Bernardoni F, et al. Subliminal and supraliminal processing of reward-related stimuli in anorexia nervosa. Psychol Med. 2017;48:790–800. doi: 10.1017/S0033291717002161. [DOI] [PubMed] [Google Scholar]
- 63.Alexander N, Illius S, Stalder T, et al. Serotonin transporter gene methylation predicts long-term cortisol concentrations in hair. Psychoneuroendocrinology. 2019;106:179–82. doi: 10.1016/j.psyneuen.2019.03.033. [DOI] [PubMed] [Google Scholar]
- 64.Salbach-Andrae H, Lenz K, Simmendinger N, et al. Psychiatric comorbidities among female adolescents with anorexia nervosa. Child Psychiatry Hum Dev. 2008;39:261–72. doi: 10.1007/s10578-007-0086-1. [DOI] [PubMed] [Google Scholar]
- 65.Roberts S, Lester KJ, Hudson JL, et al. Serotonin transporter [corrected] methylation and response to cognitive behaviour therapy in children with anxiety disorders. Transl Psychiatry. 2014;4:e444. doi: 10.1038/tp.2014.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Asai T, Bundo M, Sugawara H, et al. Effect of mood stabilizers on DNA methylation in human neuroblastoma cells. Int J Neuropsychopharmacol. 2013;16:2285–94. doi: 10.1017/S1461145713000710. [DOI] [PubMed] [Google Scholar]