Abstract
Approximately 15% of patients with multiple myeloma (MM) harbour the t(4;14) chromosomal translocation, leading to the overexpression of the histone methyltransferase NSD2. Patients with this translocation display increased tumour dissemination, accelerated disease progression and rapid relapse. Using publicly available gene expression profile data from NSD2high (n=135) and NSD2low (n=878) MM patients, we identified 39 epithelial-mesenchymal transition (EMT)-associated genes which are overexpressed in NSD2high MM plasma cells. In addition, our analyses identified Twist-1 as a key transcription factor upregulated in NSD2high MM patients and t(4;14)-positive cell lines. Overexpression and knockdown studies confirmed that Twist-1 is involved in driving the expression of EMT-associated genes in the human MM cell line KMS11 and promoted the migration of myeloma cell lines in vitro. Notably, Twist-1 overexpression in the mouse MM cell line 5TGM1 significantly increased tumour dissemination in an intratibial tumour model. These findings demonstrate that Twist-1, downstream of NSD2, contributes to the induction of an EMT-like signature in t(4;14)-positive MM and enhances the dissemination of MM plasma cells in vivo, which may, in part, explain the aggressive disease features associated with t(4;14)-positive MM.
Keywords: multiple myeloma, epithelial-mesenchymal transition, NSD2, Twist-1, dissemination
1. Introduction
Multiple myeloma (MM) is a haematological malignancy characterised by proliferation of clonal plasma cells (PCs) within the bone marrow (BM) [1]. While PC malignancies may present as a solitary plasmacytoma, without evidence of dissemination, symptomatic MM is characterised by multiple tumours throughout the skeleton, suggesting that the dissemination of MM PCs is a key component of myeloma disease progression. Similar to the metastatic process which results in the dissemination of epithelial cancer cells, it is thought that MM PCs exit the BM, circulate via the peripheral blood and home to and establish at distant BM sites [2–4].
The reciprocal chromosomal translocation t(4;14) is present in approximately 15% of MM patients [5–7] and is associated with poor prognosis and high risk of relapse [8, 9]. The t(4;14) translocation occurs between the immunoglobulin heavy chain locus at 14q32 and breakpoints between the FGFR3 and NSD2 genes at 4p16, leading to the simultaneous upregulation of FGFR3 and NSD2. NSD2 (also known as WHSC1 or MMSET), is a histone methyltransferase that is specific for dimethylation of histone H3 lysine tail residue 36 (H3K36me2) [10, 11]. Overexpression of NSD2 results in an increase in the gene activation mark H3K36me2 and a decrease in the repressive mark H3K27me3 across the genome, leading to changes in the chromatin structure allowing it to be more accessible to active transcription [12, 13]. Notably, while FGFR3 expression is lost in approximately 25% of t(4;14)-positive patients, the poor prognosis associated with the t(4;14) translocation is independent of FGFR3 expression [8], suggesting that NSD2 is the key oncogenic factor in this subgroup of MM patients. t(4;14)-positive MM is associated with acquisition of a migratory phenotype, as evidenced by an increased incidence of elevated circulating tumour cells and extramedullary dissemination [14–19]. Elevated NSD2 expression is also observed in aggressive, metastatic solid tumours such as neuroblastoma, hepatocellular carcinoma and ovarian carcinoma [20]. Furthermore, NSD2 has been identified as a novel driver of epithelial to mesenchymal transition (EMT) in prostate cancer, at least in part via upregulation of the transcription factor Twist-1 [21]. Interestingly, elevated expression of mesenchymal markers N-cadherin and vimentin have been reported in t(4;14)-positive MM patients [22, 23]. However, whether NSD2 activates an more extensive EMT-like programme in t(4;14)-positive MM, leading to increased tumour cell dissemination, remains to be determined.
In this study, we utilised previously published microarray datasets from newly diagnosed MM patients and human MM cell lines to investigate whether NSD2 promotes the acquisition of an EMT-like gene signature in t(4;14)-positive MM, which may facilitate increased MM PC dissemination in this patient cohort. These analyses identified Twist-1 as a key transcription factor that was upregulated by NSD2 and was associated with an EMT-like gene expression signature in t(4;14)-positive MM PCs. Moreover, we investigated the functional role of Twist-1 in MM pathogenesis in vitro and in the 5TGM1/KaLwRij model of myeloma [24–26] in vivo.
2. Materials and Methods
2.1. Publicly available microarray data
For analysis of gene expression in CD138-purified BM PCs from newly-diagnosed MM patients, CEL files from 4 independent Affymetrix Human Genome U133 Plus 2.0 cDNA microarray datasets [GSE19784 (n = 328 patients) [27], GSE26863 (n = 304) [28], E-MTAB-363 (n = 155) [29], E-MTAB-317 (n = 226) [30] and GSE16558 (n = 60) [31]] were downloaded from NCBI Gene Expression Omnibus (GEO) and ArrayExpress. CEL files were normalised using RMA, as previously described [32, 33]. As shown in Supp. Fig. 1, the distribution of NSD2 expression and FGFR3 expression was used to divide patients in each dataset into NSD2low and NSD2high subgroups. An additional dataset (GSE16558), which included both gene expression and FISH analysis from an independent cohort of MM patients [31], was used to confirm that the NSD2high cohort included all t(4;14) patients, as identified by FISH (Supp. Fig. 1E). For GSE19784, GSE26863, E-MTAB-363 and E-MTAB-317, linear models for microarray data analysis (LIMMA; [34]) was used to compare gene expression in the NSD2high and NSD2low groups and the adjusted p-values from the four individual microarrays were combined using Fisher’s method [35]. Gene set enrichment analysis (GSEA) was performed [36] transcriptional regulators were identified by cross-referencing with the databases of Messina, et al (2004) and Moreland, et al (2009). Gene expression levels (counts) in a panel of 66 human MM cell lines was assessed using publicly available RNA-Seq data downloaded from www.keatslab.org [39].
2.2. Stable overexpression and transient knock down of NSD2 or Twist-1 in MM cell lines
KMS11 cells in which the translocated NSD2 allele has been knocked out (KMS11-TKO cells [40]), or RPMI-8226 cells, were retrovirally transduced with RetroX-IRES-NSD2-DsRedExpress or the empty vector (EV) [21]. 5TGM1 cells overexpressing murine Twist-1 were generated by lentiviral transduction of 5TGM1-luc [26, 33] cells with pLEGO-iT2-mTWIST1 [41], generated as described in Supplementary Methods. The WL2 cell line overexpressing Twist-1 was generated by retroviral transduction with pRUF-IRES-GFP harbouring full-length cDNA encoding human TWIST1 [42].
For Twist-1 knockdown, KMS11 cells were transfected with 50 nM Silencer Select siRNA targeting human TWIST1 (s14523, Thermo Fisher Scientific, Waltham, MA, USA) or Silencer Select Negative Control siRNA (4390863, Thermo Fisher Scientific) using Lipofectamine RNAiMAX (Thermo Fisher Scientific).
2.3. Microarray analysis
Gene expression in KMS11-TKO-EV and KMS11-TKO-NSD2 cells, by Illumina Sentrix Human-6 v3 Expression BeadChip microarrays, has previously been described (NCBI GEO accession GSE24726) [12].
2.4. cDNA preparation and real-time PCR
Total RNA was isolated using TRIzol (Life Technologies, Mulgrave, Australia) and cDNA was synthesized using Superscript III (Life Technologies). Quantitative real-time PCR (qRT-PCR) were performed using RT2 SYBR Green qPCR Mastermix (Qiagen, Chadstone, Australia) using a CFX Connect real-time thermal cycler (Bio-Rad, Hercules, USA). Primers used are detailed in the Supplementary Methods.
2.5. Western blotting
Nuclear lysates were isolated using a Nuclear Complex Co-IP kit (Active Motif, Carlsbad, USA). Whole cell lysates were prepared in reducing lysis buffer, as described previously [43]. Protein lysates (30 μg/lane) were resolved using 10% or 12% SDS-polyacrylamide gels, transferred to PVDF and Western blotting was performed as previously described previously [43], using antibodies detailed in the Supplementary Methods.
2.6. Migration assays
Trans-endothelial or trans-well migration assays were performed using 8-μm polycarbonate membrane trans-wells (Corning, Corning, USA) in 24-well plates, towards 20% FCS (5TGM1) or 10% FCS (KMS11 or WL2) as previously described [44].
2.7. Cell proliferation assays
For BrdU assays, cells were seeded at 1×106 cells/mL and cell proliferation was assessed by using a BrdU Cell Proliferation ELISA kit (Merck) as previously described [32]. For WST-1 assays, cells were `ded at 1×105 cells/mL and relative cell number/well was quantitated using WST-1 reagent (Merck). Where indicated, cells were treated with bortezomib (in 0.01% DMSO [final concentration]; Selleck Chemicals, Houston, USA) or dexamethasone (in 0.13% saline [final concentration]; Mayne Pharma, Mulgrave, Australia), or vehicle alone, for 72 hrs, prior to conducting the WST-1 assay.
2.8. C57Bl/KaLwRij murine model of MM
All procedures were performed with the approval of the SAHMRI and University of Adelaide (Adelaide, Australia) animal ethics committees. For the systemic MM model, 6–8-week-old C57BL/KalwRij mice were injected via the tail vein with 5×105 5TGM1-EV or 5TGM1-TWIST1 cells and tumour development was monitored using bioluminescence imaging, as previously described [44]. For the intratibial MM model, 5–6-week-old mice were injected intratibially with 1×105 5TGM1-EV or 5TGM1-TWIST1 cells. After 3.5 weeks, BM was collected from the tibia of the injected leg, and from the tibia and femur of the contralateral leg, by flushing with PBS containing 2% FCS and 2 mM EDTA (PFE). Red blood cells were lysed by incubation for 10 minutes at room temperature with red blood cell lysis solution (155 mM NH4Cl, 10 mM KHCO3, and 1 mM EDTA). GFP-positive tumour cells were analysed on a FACSCanto II flow cytometer (BD, Franklin Lakes, USA), and assessed as a percentage of the total number of viable mononuclear cells.
3. Results
3.1. Identification of an epithelial to mesenchymal-like gene expression signature in NSD2high MM patients
To identify genes that were differentially expressed in t(4;14)-positive MM compared with non-t(4;14) MM, we interrogated four independent cDNA microarray datasets examining the gene expression in CD138-selected BM PCs from newly diagnosed MM patients. MM patients in each dataset were classified as NSD2high (135 of 1013 patients; 13.3%) or NSD2low (878 of 1013 patients; 86.7%), using specific cut-offs for NSD2, as shown in Supp. Fig. 1. Genes that were consistently and significantly up or down regulated in NSD2high patients compared with NSD2low patients across all 4 datasets were included in subsequent analyses. A total of 1958 protein coding genes were significantly upregulated (Fisher’s combined p-value < 0.05) and 1840 genes were significantly downregulated (Fisher’s combined p-value < 0.05) in the NSD2high patient group, when compared with the NSD2low patient group across the four datasets (Supp. Table I). Consistent with previous studies [22, 45, 46], GSEA revealed that upregulated genes in NSD2high patients were predominantly associated with proliferation-related genesets (Figure 1A). Notably, GSEA also identified an enrichment for genes in the Epithelial Mesenchymal Transition (EMT) gene set (39 genes; FDR q-value: 3.47 × 10−15) (Figure 1A–B).
Figure 1. Identification of an EMT-like gene expression signature in t(4;14)-positive MM.
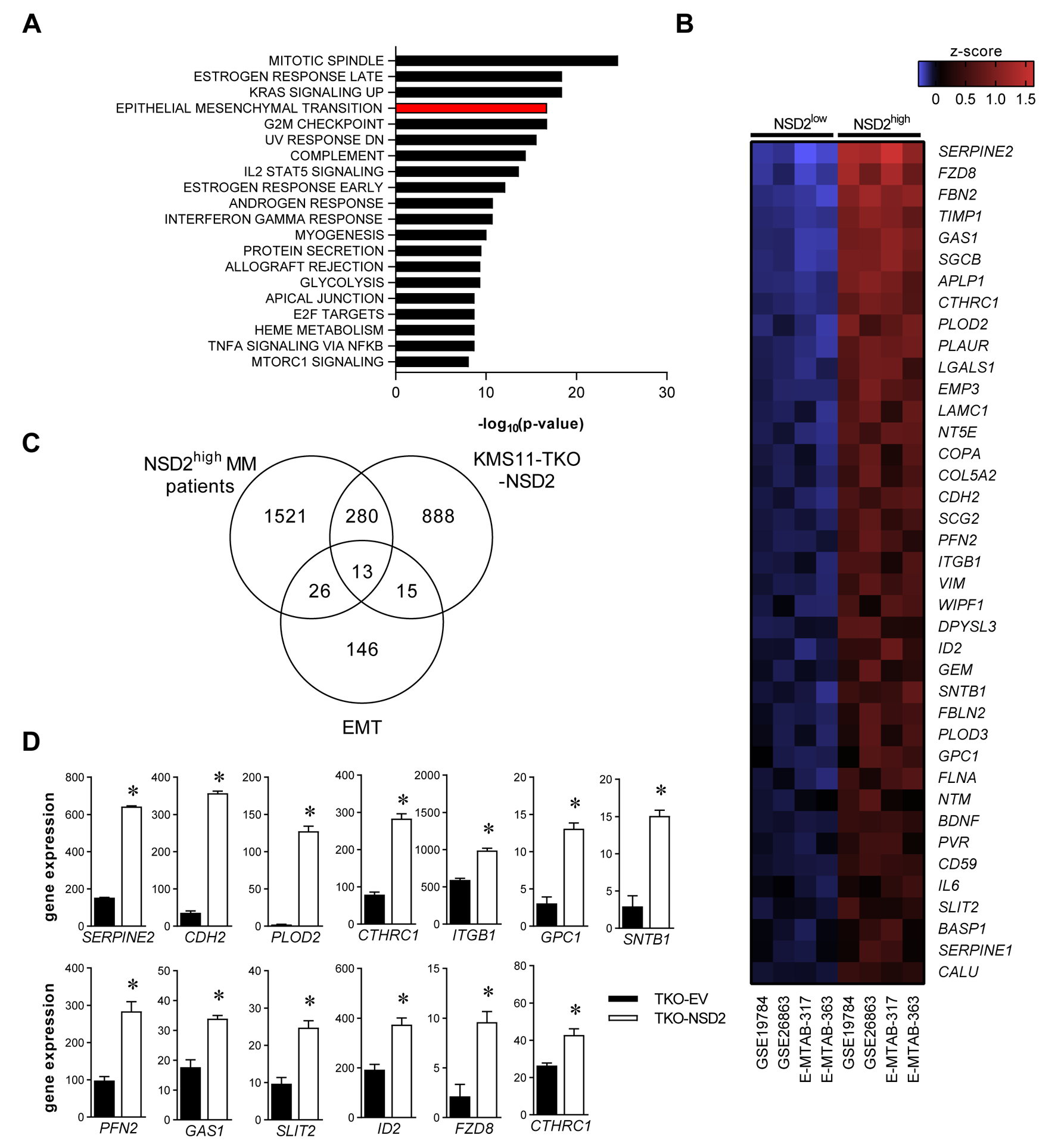
In silico analyses of gene expression in CD138+ BM MM PCs from NSD2high and NSD2low MM patients in publicly available microarray datasets (GSE19784 [NSD2low, n = 288; NSD2high, n = 40], GSE26863 [NSD2low, n = 270; NSD2high, n = 34], E-MTAB-317 [NSD2low, n = 192; NSD2high, n = 34], E-MTAB-363 [NSD2low, n = 128; NSD2high, n = 27]). Gene set enrichment analysis (GSEA) for MSigDB Hallmark gene sets was performed on genes that were significantly upregulated (Fishers’ combined p-value < 0.05) across the four datasets demonstrated enrichment for EMT-associated genes. The 20 most significantly enriched gene sets are shown (A). Heat map shows the average z-score for NSD2high and NSD2low patients for the 39 significantly upregulated genes in the MSigDB Hallmark EMT gene set (B). Venn diagram shows the overlap between the upregulated genes in MM PC from NSD2high MM patients, genes that were significantly upregulated by NSD2 overexpression in KMS11-TKO cells and the 200 genes in the MSigDB Hallmark EMT gene set (C). Expression of the 13 EMT-associated genes that were significantly upregulated by NSD2 overexpression in KMS11-TKO cells (TKO-NSD2) and in NSD2high MM patients is shown for control (TKO-EV) and TKO-NSD2 cells. Graphs depict mean + SEM; * p < 0.05 (LIMMA) (D).
Canonical EMT markers that were upregulated in the NSD2high group included the mesenchymal markers CDH2 (N-cadherin; mean log2 fold-change: 1.05-fold; range: 0.59–1.54; Fisher’s combined p-value: 3×10−6) and VIM (vimentin; mean log2 fold-change: 0.68-fold; range: 0.54–0.87; Fisher’s combined p-value: 2×10−5) (Supp. Table I). Other EMT-associated genes upregulated in NSD2high patients include those associated with cell adhesion (APLP1, ITGB1, NTM, PVR, SGCB), extracellular matrix (CTHRC1, COL5A2, FBN2, FBLN2, LGALS1, LAMC1, PLOD2), migration (SERPINE2, FLNA, GPC1, PLAUR, SCG2, SLIT2) and cytoskeletal organisation (WIPF1, FLNA, PFN2) (Figure 1B; Supp. Table I). While EMT in epithelial cancers is generally associated with gain of mesenchymal markers accompanied with loss of expression of epithelial markers, epithelial genes were generally not expressed in CD138-positive BM PCs from MM patients, regardless of NSD2 expression, consistent with the haematopoietic origin of these cells. For example, the mean relative expression of the canonical epithelial marker CDH1 (E-cadherin) was generally low across all four datasets (CDH1 expression: 4.61, 95% CI: 4.56–4.66) and was not significantly different between NSD2high and NSD2low MM patients (Supp. Table I).
In order to further investigate the EMT-like gene expression signature associated with the t(4;14) translocation, we assessed the gene expression profile of the t(4;14)-positive human MM cell line KMS11 following NSD2 overexpression. Microarray analysis was conducted on KMS11 cells in which the translocated NSD2 gene has been knocked out (KMS11-TKO-EV control cell line) and KMS11-TKO cells in which NSD2 was overexpressed (KMS11-TKO-NSD2 cell line), as described previously [12]. This analysis identified 28 genes in the hallmark EMT geneset [36] that were significantly upregulated in KMS11-TKO-NSD2 cells (Figure 1C; Supp. Table II), including 13 genes that were also upregulated in NSD2high MM patients (Figure 1D). Taken together, these data suggest that NSD2 upregulation in MM PCs is associated with a gene expression signature that is similar to that of cells that have undergone EMT.
3.2. The transcription factor Twist-1 is upregulated by NSD2 in MM
In order to investigate the potential mechanism whereby NSD2 promotes the expression of an EMT-like gene expression signature in t(4;14)-positive MM, we initially identified transcription factors that were commonly upregulated in NSD2high MM patients and following NSD2 overexpression in KMS11-TKO cells. This identified 28 transcription factors that are commonly associated with NSD2 expression in MM (Supp. Table III). The most significantly upregulated transcription factor in NSD2high MM patients was TWIST1 (Fisher’s combined p-value: 2.5×10−56; mean log2 fold-change: 0.79; Supp. Table III; Fig. 2A–B) which has previously been implicated in driving EMT in epithelial cancers and has been shown to be regulated by NSD2 in prostate cancer [21]. In line with the MM patient microarray data, TWIST1 expression was expressed in over 80% of t(4;14)-positive cell lines, as assessed by RNA-Seq and qRT-PCR, while TWIST1 expression was undetectable in the majority of non-t(4;14) cell lines (Fig. 2C–D). To validate the NSD2-mediated TWIST1 upregulation observed in KMS11-TKO cells, we overexpressed NSD2 in the non-t(4;14) myeloma cell line RPMI-8226 (Fig. 2E–F). Overexpression of NSD2 in RPMI-8226 cells resulted in a 1.7-fold increase in TWIST1 mRNA levels (Fig. 2G), supporting the role for NSD2 in regulating Twist-1 expression in MM.
Figure 2. TWIST1 expression is upregulated by NSD2 in MM.
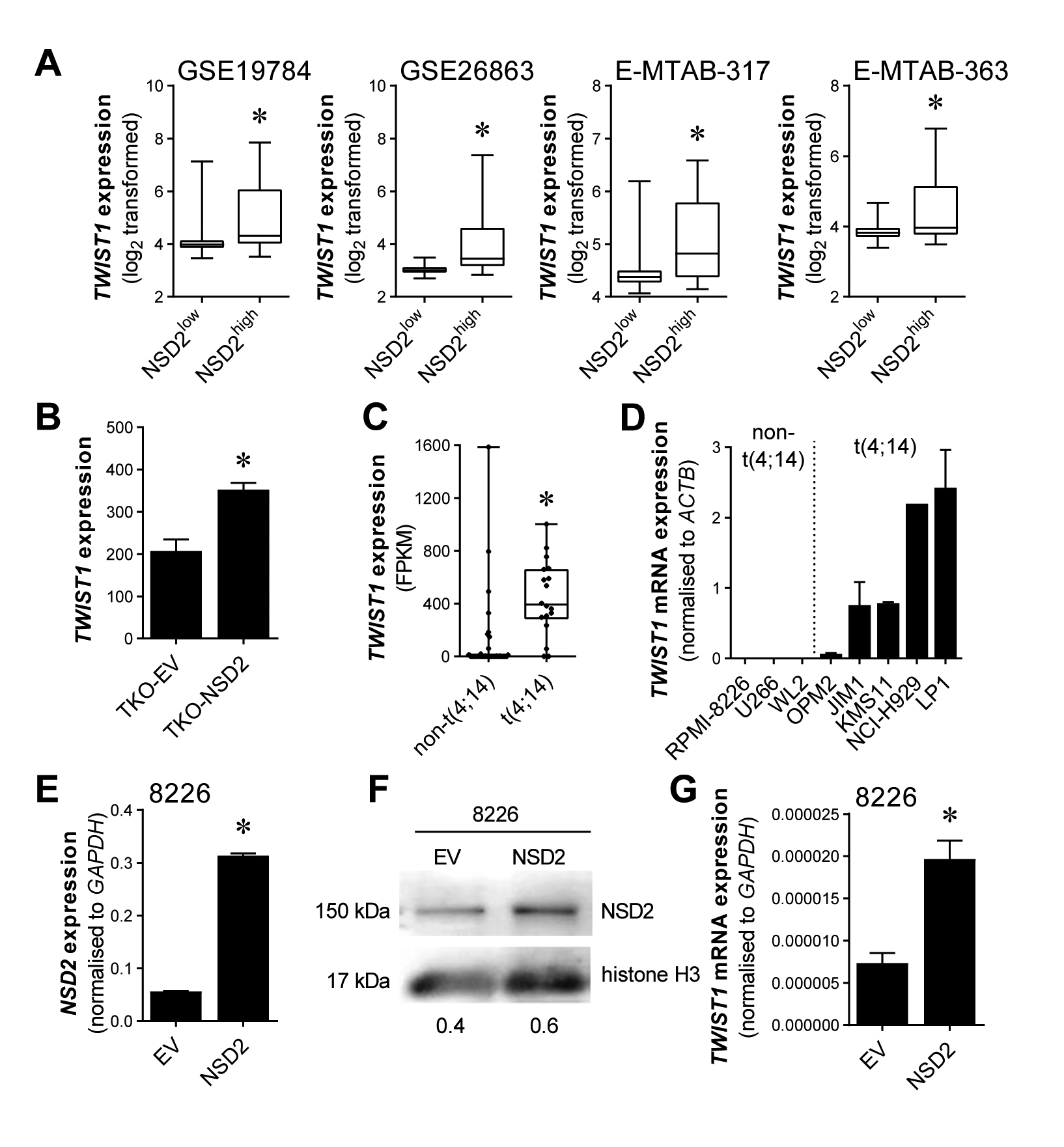
TWIST1 expression, as determined by microarray, was assessed in CD138-selected BM PCs from NSD2low and NSD2high MM patients (A). Upregulation of TWIST1 expression with NSD2 overexpression is shown in KMS11-TKO-NSD2 cells compared with control cells (KMS11-TKO-EV), as assessed by microarray (B). Expression of TWIST1 in t(4;14)-positive and non-t(4;14) MM cell lines was assessed in a publicly available RNAseq dataset (C) and by qRT-PCR in a panel of human B-cell and MM cell lines (D). The non-t(4;14) MM cell line RPMI-8226 was retrovirally transduced to constitutively overexpress NSD2 and NSD2 expression, normalised to GAPDH, was assessed by qRT-PCR (E). Upregulation of NSD2 protein in RPMI-8226-NSD2 cells, relative to RPMI-8226-EV cells, was confirmed by Western blotting on nuclear lysates, run on 10% SDS-PAGE gels, using histone H3 as a loading control. Numerical values indicate NSD2 protein levels relative to histone H3 protein levels (F). Relative TWIST1 mRNA expression, normalised to GAPDH, was measured by qRT-PCR in RPMI-8226-EV and RPMI-8226-NSD2 cell lines (G). Graphs depict the median and interquartile ranges (GSE19784 [NSD2low, n = 288; NSD2high, n = 40], GSE26863 [NSD2low, n = 270; NSD2high, n = 34], E-MTAB-317 [NSD2low, n = 192; NSD2high, n = 34], E-MTAB-363 [NSD2low, n = 128; NSD2high, n = 27], A; t(4;14), n = 18; non-t(4;14), n = 48; C) or mean + SEM of three independent biological replicates (B, D, E, G). * p < 0.05 (LIMMA [A, C] or unpaired t-test [B, E, G]).
3.3. Twist-1 knockdown downregulates the expression of EMT-associated genes and inhibits migration in vitro
In order to investigate the potential role of Twist-1 in the regulation of EMT-associated genes in t(4;14)-positive MM PCs, we used siRNA to transiently knockdown Twist-1 in KMS11 cells (Fig. 3A–B) and assessed the expression of 4 EMT-associated genes by qRT-PCR. Knockdown of Twist-1 significantly downregulated the expression of EMT-associated genes CDH2, SERPINE2 and PLOD2 (Fig. 3C), consistent with a potential role for Twist-1 in driving the expression of an EMT-like gene expression signature in these cells. In contrast, expression of VIM, which was not upregulated by NSD2 overexpression in KMS11-TKO cells (data not shown), was not modulated by Twist-1 knockdown in KMS11 cells (Fig. 3C). As Twist-1 has been implicated in mediating NSD2-driven invasiveness in epithelial cancers [21], we also investigated the effect of Twist-1 knockdown on the migratory capacity of KMS11 cells in vitro. Knockdown of Twist-1 significantly reduced trans-well migration of KMS11 cells by 30% (Fig. 3D), while having no effect on cell proliferation (Fig. 3E), supporting the potential role for Twist-1 regulation of genes involved in migration and invasion.
Figure 3. Twist-1 knockdown downregulates EMT-associated gene expression and inhibits cell migration in KMS11 cells.
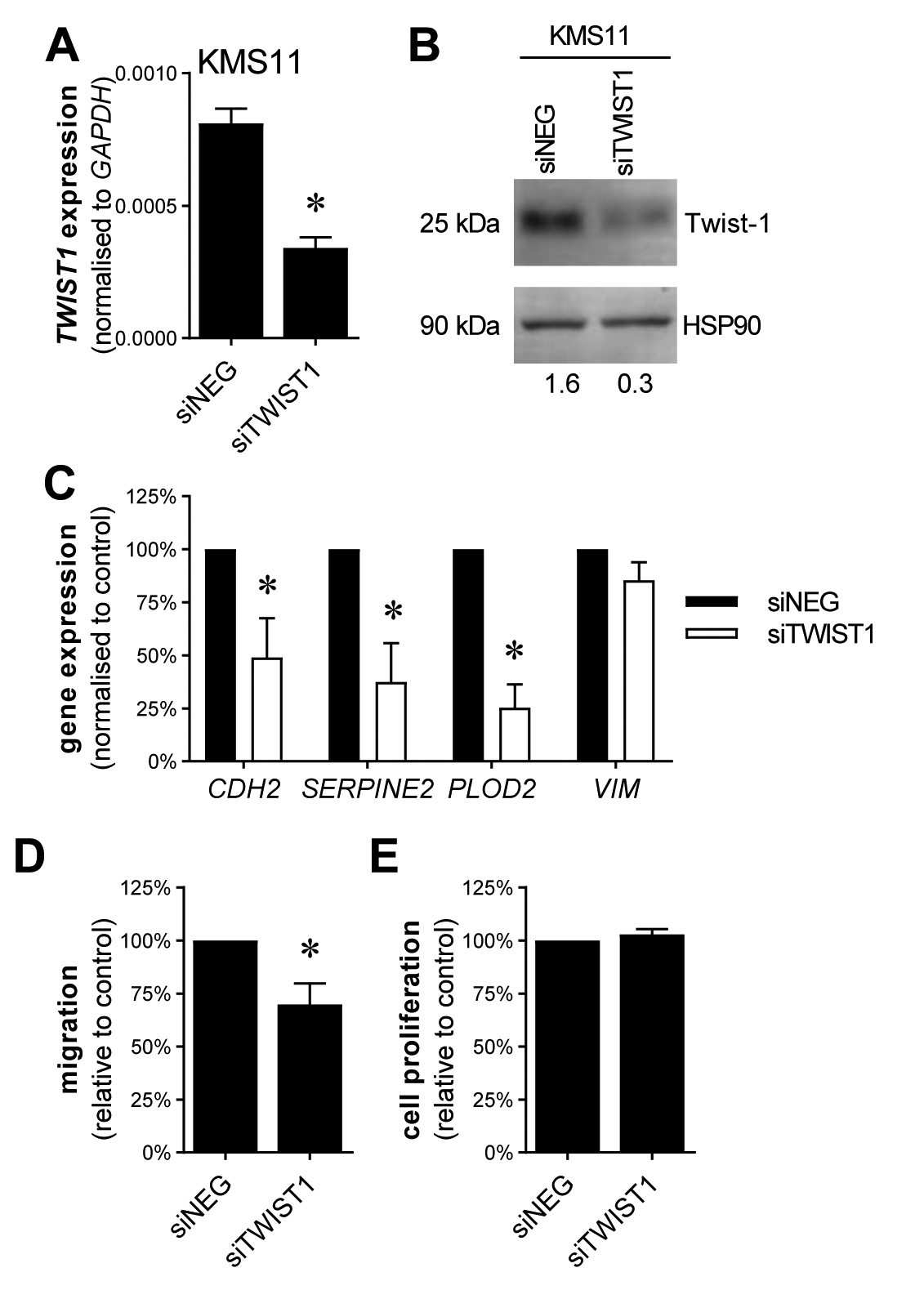
TWIST1 expression was knocked down in t(4;14)-positive KMS11 cells by transfection with TWIST1 siRNAs (siTWIST1) or negative control siRNA (siNEG). Knockdown of Twist-1 was confirmed by qRT-PCR, normalised to GAPDH (A) and by Western blot on whole cell lysates, resolved on 12% SDS-PAGE gels, using anti-Twist-1 and HSP90 antibodies (B). Numerical values indicate Twist-1 protein levels relative to HSP90 protein levels. Expression of EMT-associated genes CDH2, SERPINE2, PLOD2 and VIM was assessed in siTWIST1 or siNEG KMS11 cells by qRT-PCR, normalised to GAPDH (C). Trans-well migration of KMS11 cells following Twist-1 siRNA knockdown was assessed in a trans-well assay in response to an FCS gradient, expressed relative to siNEG controls (D). Proliferation of KMS11 cells following Twist-1 siRNA knockdown was assessed using the BrdU assay, expressed relative to the siNEG control (E). Graphs depict mean + SEM of three (A, C, E) or four (D) independent experiments. *P < 0.05, unpaired t-test.
3.4. Twist-1 overexpression increases MM tumour dissemination in vivo
To examine the consequences of increased Twist-1 expression in MM pathogenesis, we overexpressed mouse Twist-1 in a luciferase-tagged 5TGM1 mouse MM cell line. While endogenous Twist-1 expression was undetectable in 5TGM1-EV control cells, the level of Twist-1 expression in 5TGM1-TWIST1 cells was comparable with that observed in murine fibroblastic NIH-3T3 cells (Fig. 4A–B).
Figure 4. Twist-1 overexpression in the murine MM cell line 5TGM1 increases cell migration in vitro and tumour dissemination in vivo.
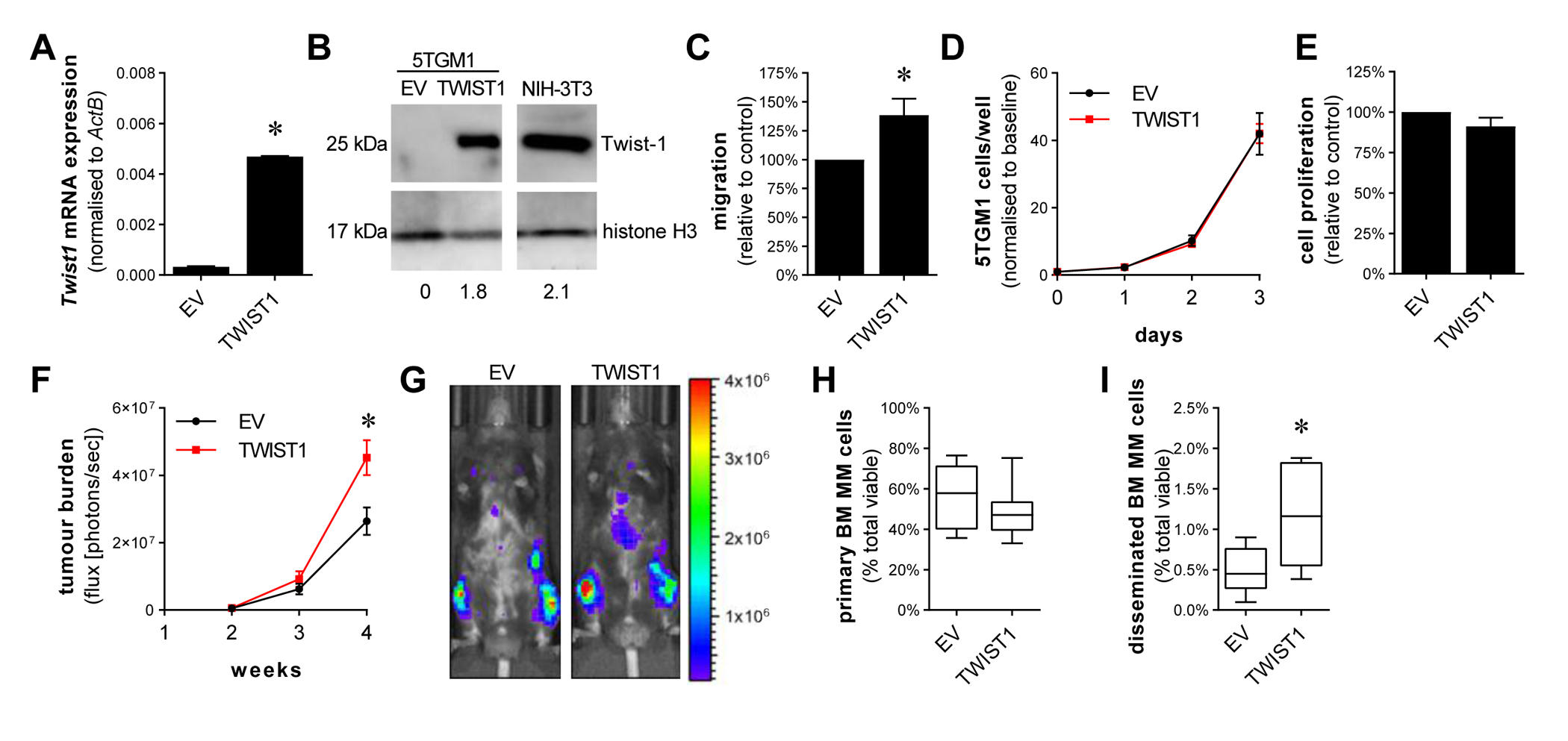
5TGM1 cells were lentivirally transduced with a Twist1 expression vector or empty vector (EV) control and Twist1 expression, normalised to ActB was assessed by qRT-PCR (A). Overexpression of Twist-1 protein in 5TGM1-TWIST1 cells was validated by Western blot on nuclear protein extracts of 5TGM1-EV, 5TGM1-TWIST1 and positive control NIH-3T3 cells, resolved on 12% SDS-PAGE gels and immunoblotted with anti-Twist-1 and anti-histone H3 antibodies. Numerical values indicate Twist-1 protein levels relative to histone H3 protein levels (B). Trans-endothelial migration of 5TGM1-Twist1 and 5TGM1-EV controls cells (1×105 cells/well) in response to FCS gradient was assessed in a trans-well assay (C). Proliferation of 5TGM1-Twist1 and 5TGM1-EV controls cells was assessed by WST-1, normalised to day 0 (D), or by BrdU assay, normalised to EV controls (E). C57BL/KalwRij mice were intravenously injected with 5×105 5TGM1-EV or 5TGM1-TWIST1 cells. Total body tumour burden was assessed using bioluminescence imaging at 2, 3 and 4 weeks (F). Bioluminescence images of a representative mouse from each group at week 4 are shown (G). The left tibiae of C57BL/KalwRij mice were injected with either 1×105 5TGM1-EV or 1×105 5TGM1-TWIST1 cells. After 3.5 weeks, tumour burden in the injected tibia (H) and in the contralateral tibia and femur (I) was assessed by detection of GFP+ tumour cells by flow cytometry, shown as a percentage of total mononuclear cells. Graphs depict mean ± SEM of three (A, D-E) or five (C) independent experiments, mean ± SEM of 8–10 mice/group (F) or median and interquartile range of 7–11 mice/group (H, I). * p < 0.05, unpaired t-test (A, C), two-way ANOVA with Sidak’s multiple comparison test (F) or Mann-Whitney test (H, I).
Consistent with the inhibition of migration observed with Twist-1 siRNA knockdown in KMS11 cells, Twist-1 overexpression significantly increased the trans-endothelial migration of 5TGM1 cells by 39% (Fig. 4C), while having no effect on their proliferation, as assessed by WST-1 assay (Fig. 4D) or BrdU incorporation (Fig. 4E). Similarly, Twist-1 overexpression increased trans-endothelial migration, but not proliferation, of the non-t(4;14) human transformed B-cell line WL2 (Supp. Fig. 3).
Twist-1 expression has been previously implicated in drug resistance in human epithelial cancer cell lines [47–49]. In order to investigate whether Twist-1 may play a role in resistance to anti-myeloma therapies in MM, we treated 5TGM1-TWIST1 and WL2-TWIST1 cells, or empty vector controls, with bortezomib or dexamethasone and assessed cell numbers using WST-1 assay. As shown in Supp. Fig. 4, Twist-1 overexpression had no effect on response to bortezomib or dexamethasone in these lines.
To investigate the role of Twist-1 expression in MM cell disease progression in vivo, the 5TGM1-EV or 5TGM1-TWIST1 cells were injected intravenously into C57BL/KaLwRij mice (n = 8–10 mice/group) and tumour development was monitored over 4 weeks by in vivo bioluminescence imaging. Tumour burden in 5TGM1-TWIST1-bearing mice was 73% higher than that of 5TGM1-EV-bearing mice after 4 weeks (P = 0.042, two-way ANOVA with Sidak’s multiple comparisons test; Fig. 4F–G). In order to determine whether this increase in tumour burden was due to an effect of Twist-1 on tumour growth or resulted from increased tumour cell BM homing and/or dissemination, we then injected 5TGM1-EV or 5TGM1-TWIST1 cells directly into the tibia and, after 3.5 weeks, analysed 5TGM1 cell numbers in the injected tibia and contralateral tibia and femur, using flow cytometry (Fig. 4H–I). Primary tumour burden in the injected tibia was not significantly different between mice bearing 5TGM1-TWIST1 and 5TGM1-EV tumours, suggesting that Twist-1 overexpression does not affect tumour growth in vivo (Fig. 4H). However, 5TGM1-TWIST1-bearing mice exhibited a significant increase in the number of cells that had spontaneously disseminated to the contralateral limb (Fig. 4I), when compared with that observed in 5TGM1-EV-bearing mice. These data demonstrate that Twist-1 overexpression increases the dissemination of 5TGM1 cells in vivo, suggesting that Twist-1 may be responsible, at least in part, for increased tumour cell dissemination in t(4;14)-positive MM.
4. Discussion
One of the defining features of t(4;14)-positive MM is an increased propensity for dissemination, with an increased incidence of high numbers of tumour cells in the peripheral circulation and of dissemination to soft tissues [14–19]. The process of tumour cell dissemination requires migration from the BM into the vasculature and subsequent trans-endothelial migration and homing to a secondary site. These steps require adhesive interactions with other non-tumour cells and extracellular matrix protein, increased migratory capacity and an ability to degrade extracellular matrix and other proteins. In order to investigate the potential mechanisms whereby NSD2 may increase migration and dissemination in t(4;14)-positive MM, we conducted in silico analysis of transcriptomic data from primary MM cells and the human MM cell line KMS11 in which NDS2 was overexpressed. These studies revealed that NSD2 expression is associated with the upregulation of genes that are commonly elevated in cells that have undergone EMT. These included genes that are involved in key processes required for dissemination, such as cell adhesion (CDH2, ITGB1), extracellular matrix remodelling (CTHRC1, PLOD2), migration (SERPINE2, GPC1, SLIT2) and cytoskeletal organisation (PFN2), suggesting potential candidates that may mediate the enhanced migration associated with NSD2 upregulation in t(4;14)-positive MM.
While an extensive EMT-like gene expression signature has not previously been described in MM, expression of some EMT-associated genes have been found to be induced under certain conditions in MM cells. For example, interleukin-17 [50] and heparanase [51] have been suggested to increase vimentin expression in human MM cell lines in vitro. In addition, hypoxia and interleukin-17 have both been suggested to decrease E-cadherin expression in human MM cells lines MM.1S and NCI-H929 [50, 52]. A key EMT-associated gene that was upregulated in NSD2high patients is CDH2 (N-cadherin), a critical driver of dissemination in solid tumours [53]. This is consistent with previous studies which have demonstrated that CDH2 is regulated by NSD2 in prostate cancer [21] and in human MM cell lines [40]. Notably, studies from our group [44] and others [23] have demonstrated that N-cadherin plays an important role in the adhesion of MM PC to endothelial cells, thereby increasing homing of MM PCs to the BM. These studies suggest a direct role for N-cadherin in the increased dissemination of NSD2-positive MM PCs. Other genes that are upregulated in t(4;14)-positive MM have also been shown to increase migration and invasion of tumour cells in vitro and in vivo. Expression of the serine protease inhibitor SerpinE2 has been associated with the incidence of metastases in patients with gastric cancer [54] and endometrial cancer [55]. Furthermore, knockdown of SERPINE2 expression has been shown to decrease migration and invasion in endometrial, gastric cancer or melanoma cells lines in vitro [54–56] and to decrease invasion in a pancreatic tumour cell model in vivo [57]. Procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 (PLOD2) is an enzyme that is involved in collagen matrix synthesis. Overexpression and knockdown studies suggest that PLOD2 plays a role in cancer cell migration and invasion in solid cancer cell lines in vitro [58–60]. In addition, knockdown of PLOD2 decreased local invasion and lung and lymph node metastasis in mouse models of breast cancer [61] and sarcoma [59]. Further investigation is required to confirm whether upregulation of these, and other, EMT-associated genes could contribute to the increased dissemination associated with t(4;14)-positive MM.
In order to investigate how NSD2 may regulate an EMT-like signature in MM patients, we examined the differential expression of transcriptional regulators in NSD2high MM patients and in KMS11-TKO-NSD2 cells. Notably, the most significantly differentially expressed transcription factor in NSD2high MM patients that was also modulated by NSD2 in KMS11-TKO cells was Twist-1, a basic helix-loop-helix transcription factor which is a well-established driver of EMT in epithelial cancers [48, 62]. Previous gene expression profiling studies have suggested that t(4;14)-positive human MM cell lines have increased expression of TWIST1 [21, 63, 64]. Furthermore, Twist-1 has been shown to regulate the expression of occludin, E-cadherin and vimentin downstream of NSD2 in immortalised prostatic epithelial cell lines [21]. Consistent with the known role of Twist-1 as an inducer of EMT in epithelial cancer cells [65], we found that knockdown of Twist-1 in KMS11 cells decreased the expression of EMT-associated genes CDH2, SERPINE2 and PLOD2, suggesting that Twist-1 contributes to driving an EMT-like gene expression signature in t(4;14)-positive MM.
Given the association between NSD2 and Twist-1 expression, we postulated that upregulation of Twist-1 by NSD2 in t(4;14)-positive MM may, in part, contribute to the poor prognosis of this MM sub-group. While Twist-1 has previously been shown to play a role in self-renewal in haematopoietic and leukaemic stem cells [66–68], we found Twist-1 overexpression or knockdown in MM cell lines had no effect on cell proliferation in vitro. Furthermore, Twist-1 overexpression in the murine MM cell line 5TGM1 did not affect primary tumour burden following direct intratibial tumour injection in C57BL/KaLwRij mice, suggesting that Twist-1 does not play a role in 5TGM1 tumour growth in vivo. In contrast, the spontaneous dissemination of the 5TGM1 cells from the injected tibia to the BM of the contralateral tibia was significantly increased by Twist-1 overexpression. Furthermore, when 5TGM1 cells were injected via the tail vein, Twist-1 overexpression was associated with a significant increase in subsequent BM tumour burden, suggesting that Twist-1 overexpression may increase BM homing of circulating MM PCs. In support of this, we found that knockdown of Twist-1 in KMS11 cells decreased cell migration, while overexpression in WL2 or 5TGM1 cell lines increased cell migration in vitro. The role of Twist-1 in MM invasion and dissemination shown here is in agreement with a previous study that showed nuclear-localised Twist-1 expression was higher in paraskeletal MM tumours when compared with BM-localised MM PCs in newly diagnosed MM patients, suggesting a potential role for Twist-1 in local invasion [69]. Furthermore, our results are consistent with previous studies which showed that overexpression of Twist-1 promotes metastasis in breast cancer [62, 70, 71], gastric carcinoma [72], glioma [73, 74], oral squamous cell carcinoma [75] and pancreatic cancer [76]. Additionally, knockdown of Twist-1 has previously been shown to counteract the pro-migration and invasion effects of NSD2 expression in prostatic epithelial cells [21], supporting the role of Twist-1 as a mediator of the promigratory effects of NSD2 in cancer. Twist-1 expression has also been associated with migration and dissemination in haematological malignancies. Twist-1 expression has been suggested to play a role in the pathogenesis of haematological cancers including lymphoma [77–80], chronic myeloid leukaemia (CML) [66] and acute myeloid leukaemia (AML) [81]. Notably, shRNA-mediated Twist-1 knockdown has been shown to decrease the homing, establishment and/or growth of AML cells in the brain and BM following intravenous injection in a mouse model of AML [81]. Twist-1 knockdown has also been shown to decrease the invasion of ALK-positive paediatric anaplastic large cell lymphoma (ALCL) cells in vitro [78].
Here, we describe, for the first time, a critical role for NSD2 in driving an EMT-like gene expression signature in t(4;14)-positive MM. In conjunction with the previously described association between NSD2 and EMT in prostate cancer [21], these studies suggest that an EMT-like gene expression signature may be a common feature of cancers characterised by high expression of NSD2. The identification of an EMT-like gene expression signature in the t(4;14)-positive subset of MM patients increases our understanding of how this translocation leads to a poor prognostic outcome. Furthermore, our studies have identified a role for Twist-1 in promoting MM tumour dissemination downstream of NSD2, by increasing MM PC migration. Thus, Twist-1-mediated MM PC dissemination may, in part, explain the aggressive disease progression of t(4;14)-positive MM, and adds to the current understanding of the role of Twist-1 in haematological malignancies.
Supplementary Material
5. Acknowledgements
This research was supported by a Priority-driven Collaborative Cancer Research Scheme (PdCCRS) grant, co-funded by Cancer Australia and the Leukaemia Foundation, awarded to A.Z., a Young Investigator Project Grant from the Cancer Australia PdCCRS, funded by Cure Cancer Australia, awarded to K.V., and US National Institutes of Health grant CA195732 and a Leukemia & Lymphoma Society Specialized Center of Research grant, awarded to J.D.L. C.M.C. received an Adelaide Graduate Research Scholarship from the University of Adelaide. B.P. was supported by a Hospital Research Foundation Early Career Research Fellowship and a National Health and Medical Research Council Early Career Fellowship. J.E.N. was supported by a Veronika Sacco Clinical Cancer Research Fellowship from the Florey Medical Research Foundation. K.V. was supported by a Mary Overton Early Career Research Fellowship from the Royal Adelaide Hospital Research Foundation and an Early Career Cancer Research Fellowship from the Cancer Council SA Beat Cancer Project on behalf of its donors and the State Government of South Australia through the Department of Health.
6. References
- [1].Dutta AK, Hewett DR, Fink JL, Grady JP, Zannettino ACW, Cutting edge genomics reveal new insights into tumour development, disease progression and therapeutic impacts in multiple myeloma, Br. J. Haematol, (2017). [DOI] [PubMed]
- [2].Nowakowski GS, Witzig TE, Dingli D, Tracz MJ, Gertz MA, Lacy MQ, Lust JA, Dispenzieri A, Greipp PR, Kyle RA, Rajkumar SV, Circulating plasma cells detected by flow cytometry as a predictor of survival in 302 patients with newly diagnosed multiple myeloma, Blood, 106 (2005) 2276–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rawstron AC, Owen RG, Davies FE, Johnson RJ, Jones RA, Richards SJ, Evans PA, Child JA, Smith GM, Jack AS, Morgan GJ, Circulating plasma cells in multiple myeloma: characterization and correlation with disease stage, Br J Haematol, 97 (1997) 46–55. [DOI] [PubMed] [Google Scholar]
- [4].Ghobrial IM, Myeloma as a model for the process of metastasis: implications for therapy, Blood, 120 (2012) 20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bergsagel PL, Kuehl WM, Chromosome translocations in multiple myeloma, Oncogene, 20 (2001) 5611–5622. [DOI] [PubMed] [Google Scholar]
- [6].Chng WJ, Kumar S, Vanwier S, Ahmann G, Price-Troska T, Henderson K, Chung TH, Kim S, Mulligan G, Bryant B, Carpten J, Gertz M, Rajkumar SV, Lacy M, Dispenzieri A, Kyle R, Greipp P, Bergsagel PL, Fonseca R, Molecular dissection of hyperdiploid multiple myeloma by gene expression profiling, Cancer Res, 67 (2007) 2982–2989. [DOI] [PubMed] [Google Scholar]
- [7].Fonseca R, Bergsagel PL, Drach J, Shaughnessy J, Gutierrez N, Stewart AK, Morgan G, Van Ness B, Chesi M, Minvielle S, Neri A, Barlogie B, Kuehl WM, Liebisch P, Davies F, Chen-Kiang S, Durie BG, Carrasco R, Sezer O, Reiman T, Pilarski L, Avet-Loiseau H, International Myeloma Working Group molecular classification of multiple myeloma: spotlight review, Leukemia, 23 (2009) 2210–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Keats JJ, Reiman T, Maxwell CA, Taylor BJ, Larratt LM, Mant MJ, Belch AR, Pilarski LM, In multiple myeloma, t(4;14)(p16;q32) is an adverse prognostic factor irrespective of FGFR3 expression, Blood, 101 (2003) 1520–1529. [DOI] [PubMed] [Google Scholar]
- [9].Chang H, Sloan S, Li D, Zhuang L, Yi QL, Chen CI, Reece D, Chun K, Keith Stewart A, The t(4;14) is associated with poor prognosis in myeloma patients undergoing autologous stem cell transplant, Br J Haematol, 125 (2004) 64–68. [DOI] [PubMed] [Google Scholar]
- [10].Li Y, Trojer P, Xu CF, Cheung P, Kuo A, Drury WJ 3rd, Qiao Q, Neubert TA, Xu RM, Gozani O, Reinberg D, The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate, The Journal of biological chemistry, 284 (2009) 34283–34295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kuo AJ, Cheung P, Chen K, Zee BM, Kioi M, Lauring J, Xi Y, Park BH, Shi X, Garcia BA, Li W, Gozani O, NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming, Mol. Cell, 44 (2011) 609–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Martinez-Garcia E, Popovic R, Min DJ, Sweet SM, Thomas PM, Zamdborg L, Heffner A, Will C, Lamy L, Staudt LM, Levens DL, Kelleher NL, Licht JD, The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells, Blood, 117 (2011) 211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Popovic R, Martinez-Garcia E, Giannopoulou EG, Zhang Q, Ezponda T, Shah MY, Zheng Y, Will CM, Small EC, Hua Y, Bulic M, Jiang Y, Carrara M, Calogero RA, Kath WL, Kelleher NL, Wang JP, Elemento O, Licht JD, Histone methyltransferase MMSET/NSD2 alters EZH2 binding and reprograms the myeloma epigenome through global and focal changes in H3K36 and H3K27 methylation, PLoS Genet, 10 (2014) e1004566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chandesris MO, Soulier J, Labaume S, Crinquette A, Repellini L, Chemin K, Malphettes M, Fieschi C, Asli B, Uzunhan Y, Fermand JP, Bories JC, Arnulf B, Detection and follow-up of fibroblast growth factor receptor 3 expression on bone marrow and circulating plasma cells by flow cytometry in patients with t(4;14) multiple myeloma, Br J Haematol, 136 (2007) 609–614. [DOI] [PubMed] [Google Scholar]
- [15].Bink K, Haralambieva E, Kremer M, Ott G, Beham-Schmid C, de Leval L, Peh SC, Laeng HR, Jutting U, Hutzler P, Quintanilla-Martinez L, Fend F, Primary extramedullary plasmacytoma: similarities with and differences from multiple myeloma revealed by interphase cytogenetics, Haematologica, 93 (2008) 623–626. [DOI] [PubMed] [Google Scholar]
- [16].Gonsalves WI, Rajkumar SV, Gupta V, Morice WG, Timm MM, Singh PP, Dispenzieri A, Buadi FK, Lacy MQ, Kapoor P, Gertz MA, Kumar SK, Quantification of clonal circulating plasma cells in newly diagnosed multiple myeloma: implications for redefining high-risk myeloma, Leukemia, 28 (2014) 2060–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Qu X, Chen L, Qiu H, Lu H, Wu H, Liu P, Guo R, Li J, Extramedullary manifestation in multiple myeloma bears high incidence of poor cytogenetic aberration and novel agents resistance, Biomed Res Int, 2015 (2015) 787809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Besse L, Sedlarikova L, Greslikova H, Kupska R, Almasi M, Penka M, Jelinek T, Pour L, Adam Z, Kuglik P, Krejci M, Hajek R, Sevcikova S, Cytogenetics in multiple myeloma patients progressing into extramedullary disease, Eur. J. Haematol, 97 (2016) 93–100. [DOI] [PubMed] [Google Scholar]
- [19].Granell M, Calvo X, Garcia-Guinon A, Escoda L, Abella E, Martinez CM, Teixido M, Gimenez MT, Senin A, Sanz P, Campoy D, Vicent A, Arenillas L, Rosinol L, Sierra J, Blade J, de Larrea CF, Prognostic impact of circulating plasma cells in patients with multiple myeloma: implications for plasma cell leukemia definition, Haematologica, 102 (2017) 1099–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hudlebusch HR, Santoni-Rugiu E, Simon R, Ralfkiaer E, Rossing HH, Johansen JV, Jorgensen M, Sauter G, Helin K, The histone methyltransferase and putative oncoprotein MMSET is overexpressed in a large variety of human tumors, Clinical cancer research : an official journal of the American Association for Cancer Research, 17 (2011) 2919–2933. [DOI] [PubMed] [Google Scholar]
- [21].Ezponda T, Popovic R, Shah MY, Martinez-Garcia E, Zheng Y, Min DJ, Will C, Neri A, Kelleher NL, Yu J, Licht JD, The histone methyltransferase MMSET/WHSC1 activates TWIST1 to promote an epithelial-mesenchymal transition and invasive properties of prostate cancer, Oncogene, 32 (2013) 2882–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Dring AM, Davies FE, Fenton JA, Roddam PL, Scott K, Gonzalez D, Rollinson S, Rawstron AC, Rees-Unwin KS, Li C, Munshi NC, Anderson KC, Morgan GJ, A global expression-based analysis of the consequences of the t(4;14) translocation in myeloma, Clin. Cancer Res, 10 (2004) 5692–5701. [DOI] [PubMed] [Google Scholar]
- [23].Groen RW, de Rooij MF, Kocemba KA, Reijmers RM, de Haan-Kramer A, Overdijk MB, Aalders L, Rozemuller H, Martens AC, Bergsagel PL, Kersten MJ, Pals ST, Spaargaren M, N-cadherin-mediated adhesion of multiple myeloma cells inhibits osteoblast differentiation, Haematologica, 96 (2011) 1653–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Paton-Hough J, Chantry AD, Lawson MA, A review of current murine models of multiple myeloma used to assess the efficacy of therapeutic agents on tumour growth and bone disease, Bone, 77 (2015) 57–68. [DOI] [PubMed] [Google Scholar]
- [25].Asosingh K, Radl J, Van Riet I, Van Camp B, Vanderkerken K, The 5TMM series: a useful in vivo mouse model of human multiple myeloma, Hematol J, 1 (2000) 351–356. [DOI] [PubMed] [Google Scholar]
- [26].Hewett DR, Vandyke K, Lawrence DM, Friend N, Noll JE, Geoghegan JM, Croucher PI, Zannettino ACW, DNA barcoding reveals habitual clonal dominance of myeloma plasma cells in the bone marrow microenvironment, Neoplasia, 19 (2017) 972–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Broyl A, Hose D, Lokhorst H, de Knegt Y, Peeters J, Jauch A, Bertsch U, Buijs A, Stevens-Kroef M, Beverloo HB, Vellenga E, Zweegman S, Kersten MJ, van der Holt B, el Jarari L, Mulligan G, Goldschmidt H, van Duin M, Sonneveld P, Gene expression profiling for molecular classification of multiple myeloma in newly diagnosed patients, Blood, 116 (2010) 2543–2553. [DOI] [PubMed] [Google Scholar]
- [28].Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet JP, Ahmann GJ, Adli M, Anderson KC, Ardlie KG, Auclair D, Baker A, Bergsagel PL, Bernstein BE, Drier Y, Fonseca R, Gabriel SB, Hofmeister CC, Jagannath S, Jakubowiak AJ, Krishnan A, Levy J, Liefeld T, Lonial S, Mahan S, Mfuko B, Monti S, Perkins LM, Onofrio R, Pugh TJ, Rajkumar SV, Ramos AH, Siegel DS, Sivachenko A, Stewart AK, Trudel S, Vij R, Voet D, Winckler W, Zimmerman T, Carpten J, Trent J, Hahn WC, Garraway LA, Meyerson M, Lander ES, Getz G, Golub TR, Initial genome sequencing and analysis of multiple myeloma, Nature, 471 (2011) 467–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Reme T, Hose D, Theillet C, Klein B, Modeling risk stratification in human cancer, Bioinformatics, 29 (2013) 1149–1157. [DOI] [PubMed] [Google Scholar]
- [30].Hose D, Reme T, Hielscher T, Moreaux J, Messner T, Seckinger A, Benner A, Shaughnessy JD Jr., Barlogie B, Zhou Y, Hillengass J, Bertsch U, Neben K, Mohler T, Rossi JF, Jauch A, Klein B, Goldschmidt H, Proliferation is a central independent prognostic factor and target for personalized and risk-adapted treatment in multiple myeloma, Haematologica, 96 (2011) 87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gutierrez NC, Sarasquete ME, Misiewicz-Krzeminska I, Delgado M, De Las Rivas J, Ticona FV, Ferminan E, Martin-Jimenez P, Chillon C, Risueno A, Hernandez JM, Garcia-Sanz R, Gonzalez M, San Miguel JF, Deregulation of microRNA expression in the different genetic subtypes of multiple myeloma and correlation with gene expression profiling, Leukemia, 24 (2010) 629–637. [DOI] [PubMed] [Google Scholar]
- [32].Noll JE, Vandyke K, Hewett DR, Mrozik KM, Bala RJ, Williams SA, Kok CH, Zannettino AC, PTTG1 expression is associated with hyperproliferative disease and poor prognosis in multiple myeloma, J Hematol Oncol, 8 (2015) 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Cheong CM, Chow AW, Fitter S, Hewett DR, Martin SK, Williams SA, To LB, Zannettino AC, Vandyke K, Tetraspanin 7 (TSPAN7) expression is upregulated in multiple myeloma patients and inhibits myeloma tumour development in vivo, Exp. Cell Res, 332 (2015) 24–38. [DOI] [PubMed] [Google Scholar]
- [34].Smyth GK, Linear models and empirical bayes methods for assessing differential expression in microarray experiments, Stat Appl Genet Mol Biol, 3 (2004) Article3. [DOI] [PubMed] [Google Scholar]
- [35].Campain A, Yang YH, Comparison study of microarray meta-analysis methods, BMC Bioinformatics, 11 (2010) 408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P, The Molecular Signatures Database (MSigDB) hallmark gene set collection, Cell Syst, 1 (2015) 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Moreland RT, Ryan JF, Pan C, Baxevanis AD, The Homeodomain Resource: a comprehensive collection of sequence, structure, interaction, genomic and functional information on the homeodomain protein family, Database (Oxford), 2009 (2009) bap004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Messina DN, Glasscock J, Gish W, Lovett M, An ORFeome-based analysis of human transcription factor genes and the construction of a microarray to interrogate their expression, Genome Res, 14 (2004) 2041–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Yellapantula V, Allen K, Jelinek DF, Bergsagel L, Keats JJ, The comprehensive genomic characterisation of all commercially and non-commercially available multiple myeloma cell lines, Blood, 122 (2013) 1914–1914.23900238 [Google Scholar]
- [40].Lauring J, Abukhdeir AM, Konishi H, Garay JP, Gustin JP, Wang Q, Arceci RJ, Matsui W, Park BH, The multiple myeloma associated MMSET gene contributes to cellular adhesion, clonogenic growth, and tumorigenicity, Blood, 111 (2008) 856–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Weber K, Bartsch U, Stocking C, Fehse B, A multicolor panel of novel lentiviral “gene ontology” (LeGO) vectors for functional gene analysis, Mol Ther, 16 (2008) 698–706. [DOI] [PubMed] [Google Scholar]
- [42].Isenmann S, Arthur A, Zannettino ACW, Turner JL, Shi S, Glackin CA, Gronthos S, TWIST family of basic helix-loop-helix transcription factors mediate human mesenchymal stem cell growth and commitment, Stem Cells, 27 (2009) 2457–2468. [DOI] [PubMed] [Google Scholar]
- [43].Vandyke K, Dewar AL, Diamond P, Fitter S, Schultz CG, Sims NA, Zannettino ACW, The tyrosine kinase inhibitor dasatinib dysregulates bone remodeling through inhibition of osteoclasts in vivo, J. Bone Miner. Res, 25 (2010) 1759–1770. [DOI] [PubMed] [Google Scholar]
- [44].Mrozik KM, Cheong CM, Hewett D, Chow AW, Blaschuk OW, Zannettino AC, Vandyke K, Therapeutic targeting of N-cadherin is an effective treatment for multiple myeloma, Br. J. Haematol, 171 (2015) 387–399. [DOI] [PubMed] [Google Scholar]
- [45].Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S, Epstein J, Yaccoby S, Sawyer J, Burington B, Anaissie E, Hollmig K, Pineda-Roman M, Tricot G, van Rhee F, Walker R, Zangari M, Crowley J, Barlogie B, Shaughnessy JD Jr., The molecular classification of multiple myeloma, Blood, 108 (2006) 2020–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Brito JL, Walker B, Jenner M, Dickens NJ, Brown NJ, Ross FM, Avramidou A, Irving JA, Gonzalez D, Davies FE, Morgan GJ, MMSET deregulation affects cell cycle progression and adhesion regulons in t(4;14) myeloma plasma cells, Haematologica, 94 (2009) 78–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wang X, Ling MT, Guan XY, Tsao SW, Cheung HW, Lee DT, Wong YC, Identification of a novel function of TWIST, a bHLH protein, in the development of acquired taxol resistance in human cancer cells, Oncogene, 23 (2004) 474–482. [DOI] [PubMed] [Google Scholar]
- [48].Kwok WK, Ling MT, Lee TW, Lau TC, Zhou C, Zhang X, Chua CW, Chan KW, Chan FL, Glackin C, Wong YC, Wang X, Up-regulation of TWIST in prostate cancer and its implication as a therapeutic target, Cancer Res, 65 (2005) 5153–5162. [DOI] [PubMed] [Google Scholar]
- [49].Cheng GZ, Chan J, Wang Q, Zhang W, Sun CD, Wang LH, Twist transcriptionally up-regulates AKT2 in breast cancer cells leading to increased migration, invasion, and resistance to paclitaxel, Cancer Res, 67 (2007) 1979–1987. [DOI] [PubMed] [Google Scholar]
- [50].Sun Y, Pan J, Mao S, Jin J, IL-17/miR-192/IL-17Rs regulatory feedback loop facilitates multiple myeloma progression, PLoS One, 9 (2014) e114647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Li J, Pan Q, Rowan PD, Trotter TN, Peker D, Regal KM, Javed A, Suva LJ, Yang Y, Heparanase promotes myeloma progression by inducing mesenchymal features and motility of myeloma cells, Oncotarget, 7 (2016) 11299–11309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Azab AK, Hu J, Quang P, Azab F, Pitsillides C, Awwad R, Thompson B, Maiso P, Sun JD, Hart CP, Roccaro AM, Sacco A, Ngo HT, Lin CP, Kung AL, Carrasco RD, Vanderkerken K, Ghobrial IM, Hypoxia promotes dissemination of multiple myeloma through acquisition of epithelial to mesenchymal transition-like features, Blood, 119 (2012) 5782–5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Mrozik KM, Blaschuk OW, Cheong CM, Zannettino ACW, Vandyke K, N-cadherin in cancer metastasis, its emerging role in haematological malignancies and potential as a therapeutic target in cancer, BMC Cancer, 18 (2018) 939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Wang K, Wang B, Xing AY, Xu KS, Li GX, Yu ZH, Prognostic significance of SERPINE2 in gastric cancer and its biological function in SGC7901 cells, J. Cancer Res. Clin. Oncol, 141 (2015) 805–812. [DOI] [PubMed] [Google Scholar]
- [55].Shen Y, Wang X, Xu J, Lu L, SerpinE2, a poor biomarker of endometrial cancer, promotes the proliferation and mobility of EC cells, Cancer Biomark, 19 (2017) 271–278. [DOI] [PubMed] [Google Scholar]
- [56].Wu QW, Serpine2, a potential novel target for combating melanoma metastasis, Am J Transl Res, 8 (2016) 1985–1997. [PMC free article] [PubMed] [Google Scholar]
- [57].Buchholz M, Biebl A, Neesse A, Wagner M, Iwamura T, Leder G, Adler G, Gress TM, SERPINE2 (protease nexin I) promotes extracellular matrix production and local invasion of pancreatic tumors in vivo, Cancer Res, 63 (2003) 4945–4951. [PubMed] [Google Scholar]
- [58].Xu F, Zhang J, Hu G, Liu L, Liang W, Hypoxia and TGF-beta1 induced PLOD2 expression improve the migration and invasion of cervical cancer cells by promoting epithelial-to-mesenchymal transition (EMT) and focal adhesion formation, Cancer Cell Int, 17 (2017) 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Eisinger-Mathason TS, Zhang M, Qiu Q, Skuli N, Nakazawa MS, Karakasheva T, Mucaj V, Shay JE, Stangenberg L, Sadri N, Pure E, Yoon SS, Kirsch DG, Simon MC, Hypoxia-dependent modification of collagen networks promotes sarcoma metastasis, Cancer Discov, 3 (2013) 1190–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Song Y, Zheng S, Wang J, Long H, Fang L, Wang G, Li Z, Que T, Liu Y, Li Y, Zhang X, Fang W, Qi S, Hypoxia-induced PLOD2 promotes proliferation, migration and invasion via PI3K/Akt signaling in glioma, Oncotarget, 8 (2017) 41947–41962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Gilkes DM, Bajpai S, Wong CC, Chaturvedi P, Hubbi ME, Wirtz D, Semenza GL, Procollagen lysyl hydroxylase 2 is essential for hypoxia-induced breast cancer metastasis, Mol Cancer Res, 11 (2013) 456–466. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [62].Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA, Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis, Cell, 117 (2004) 927–939. [DOI] [PubMed] [Google Scholar]
- [63].Lombardi L, Poretti G, Mattioli M, Fabris S, Agnelli L, Bicciato S, Kwee I, Rinaldi A, Ronchetti D, Verdelli D, Lambertenghi-Deliliers G, Bertoni F, Neri A, Molecular characterization of human multiple myeloma cell lines by integrative genomics: insights into the biology of the disease, Genes. Chromosomes Cancer, 46 (2007) 226–238. [DOI] [PubMed] [Google Scholar]
- [64].Moreaux J, Klein B, Bataille R, Descamps G, Maiga S, Hose D, Goldschmidt H, Jauch A, Reme T, Jourdan M, Amiot M, Pellat-Deceunynck C, A high-risk signature for patients with multiple myeloma established from the molecular classification of human myeloma cell lines, Haematologica, 96 (2011) 574–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Zhu QQ, Ma C, Wang Q, Song Y, Lv T, The role of TWIST1 in epithelial-mesenchymal transition and cancers, Tumour Biol, 37 (2016) 185–197. [DOI] [PubMed] [Google Scholar]
- [66].Cosset E, Hamdan G, Jeanpierre S, Voeltzel T, Sagorny K, Hayette S, Mahon FX, Dumontet C, Puisieux A, Nicolini FE, Maguer-Satta V, Deregulation of TWIST-1 in the CD34+ compartment represents a novel prognostic factor in chronic myeloid leukemia, Blood, 117 (2011) 1673–1676. [DOI] [PubMed] [Google Scholar]
- [67].Dong CY, Liu XY, Wang N, Wang LN, Yang BX, Ren Q, Liang HY, Ma XT, Twist-1, a novel regulator of hematopoietic stem cell self-renewal and myeloid lineage development, Stem Cells, 32 (2014) 3173–3182. [DOI] [PubMed] [Google Scholar]
- [68].Wang N, Guo D, Zhao YY, Dong CY, Liu XY, Yang BX, Wang SW, Wang L, Liu QG, Ren Q, Lin YM, Ma XT, TWIST-1 promotes cell growth, drug resistance and progenitor clonogenic capacities in myeloid leukemia and is a novel poor prognostic factor in acute myeloid leukemia, Oncotarget, 6 (2015) 20977–20992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Yang JZ, Lian WG, Sun LX, Qi DW, Ding Y, Zhang XH, High nuclear expression of Twist1 in the skeletal extramedullary disease of myeloma patients predicts inferior survival, Pathol. Res. Pract, 212 (2016) 210–216. [DOI] [PubMed] [Google Scholar]
- [70].Mironchik Y, Winnard PT Jr., Vesuna F, Kato Y, Wildes F, Pathak AP, Kominsky S, Artemov D, Bhujwalla Z, Van Diest P, Burger H, Glackin C, Raman V, Twist overexpression induces in vivo angiogenesis and correlates with chromosomal instability in breast cancer, Cancer Res, 65 (2005) 10801–10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Croset M, Goehrig D, Frackowiak A, Bonnelye E, Ansieau S, Puisieux A, Clezardin P, TWIST1 expression in breast cancer cells facilitates bone metastasis formation, Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research, 29 (2014) 1886–1899. [DOI] [PubMed] [Google Scholar]
- [72].Bexell D, Gunnarsson S, Tormin A, Darabi A, Gisselsson D, Roybon L, Scheding S, Bengzon J, Bone marrow multipotent mesenchymal stroma cells act as pericyte-like migratory vehicles in experimental gliomas, Mol Ther, 17 (2009) 183–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Elias MC, Tozer KR, Silber JR, Mikheeva S, Deng M, Morrison RS, Manning TC, Silbergeld DL, Glackin CA, Reh TA, Rostomily RC, TWIST is expressed in human gliomas and promotes invasion, Neoplasia, 7 (2005) 824–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Mikheeva SA, Mikheev AM, Petit A, Beyer R, Oxford RG, Khorasani L, Maxwell JP, Glackin CA, Wakimoto H, Gonzalez-Herrero I, Sanchez-Garcia I, Silber JR, Horner PJ, Rostomily RC, TWIST1 promotes invasion through mesenchymal change in human glioblastoma, Mol Cancer, 9 (2010) 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].da Silva SD, Alaoui-Jamali MA, Soares FA, Carraro DM, Brentani HP, Hier M, Rogatto SR, Kowalski LP, TWIST1 is a molecular marker for a poor prognosis in oral cancer and represents a potential therapeutic target, Cancer, 120 (2014) 352–362. [DOI] [PubMed] [Google Scholar]
- [76].Chen S, Chen JZ, Zhang JQ, Chen HX, Yan ML, Huang L, Tian YF, Chen YL, Wang YD, Hypoxia induces TWIST-activated epithelial-mesenchymal transition and proliferation of pancreatic cancer cells in vitro and in nude mice, Cancer Lett, 383 (2016) 73–84. [DOI] [PubMed] [Google Scholar]
- [77].van Doorn R, Dijkman R, Vermeer MH, Out-Luiting JJ, van der Raaij-Helmer EM, Willemze R, Tensen CP, Aberrant expression of the tyrosine kinase receptor EphA4 and the transcription factor twist in Sezary syndrome identified by gene expression analysis, Cancer Res, 64 (2004) 5578–5586. [DOI] [PubMed] [Google Scholar]
- [78].Zhang J, Wang P, Wu F, Li M, Sharon D, Ingham RJ, Hitt M, McMullen TP, Lai R, Aberrant expression of the transcriptional factor Twist1 promotes invasiveness in ALK-positive anaplastic large cell lymphoma, Cell. Signal, 24 (2012) 852–858. [DOI] [PubMed] [Google Scholar]
- [79].Lemma S, Karihtala P, Haapasaari KM, Jantunen E, Soini Y, Bloigu R, Pasanen AK, Turpeenniemi-Hujanen T, Kuittinen O, Biological roles and prognostic values of the epithelial-mesenchymal transition-mediating transcription factors Twist, ZEB1 and Slug in diffuse large B-cell lymphoma, Histopathology, 62 (2013) 326–333. [DOI] [PubMed] [Google Scholar]
- [80].Wong HK, Gibson H, Hake T, Geyer S, Frederickson J, Marcucci G, Caligiuri MA, Porcu P, Mishra A, Promoter-Specific Hypomethylation Is Associated with Overexpression of PLS3, GATA6, and TWIST1 in the Sezary Syndrome, The Journal of investigative dermatology, 135 (2015) 2084–2092. [DOI] [PubMed] [Google Scholar]
- [81].Xu J, Zhang W, Yan XJ, Lin XQ, Li W, Mi JQ, Li JM, Zhu J, Chen Z, Chen SJ, DNMT3A mutation leads to leukemic extramedullary infiltration mediated by TWIST1, J Hematol Oncol, 9 (2016) 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Livak KJ, Schmittgen TD, Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method, Methods, 25 (2001) 402–408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


