Abstract
Gastrointestinal (GI) tissue biopsies provide critical diagnostic information for a wide variety of conditions such as neoplastic diseases (colorectal, small bowel and stomach cancers) and non-neoplastic diseases (inflammatory disorders, infection, celiac disease). Endoscopic biopsies collect small tissue samples that require resource intensive processing to permit histopathological analysis. Unfortunately, the sparsely collected biopsy samples may fail to capture the pathologic condition because selection of biopsy sites relies on macroscopic superficial tissue features and clinician judgement. Here, we present the first all-optical non-contact label-free non-interferometric photoacoustic microscopy system capable of performing “virtual biopsies”. A modular photoacoustic remote sensing (PARS™) architecture is used facilitating imaging of unprocessed tissues providing information similar to conventional histopathological staining techniques. Prospectively this would allow gastroenterologists to assess subcellular tissue morphology in situ when selecting biopsy location. Tested on preserved unstained human and freshly resected murine tissues, the presented PARS microscope rapidly retrieves images of similar area to current biopsies, while maintaining comparable quality to the current standard for histopathological analysis. Additionally, results show the first label free assessment of subsurface cellular morphology in FFPE GI tissue blocks. Clinically relevant features are recovered including cellular details such as lamina propria within colon tissue and cell nuclear structure in resected smooth muscle. Constructed with a modular architecture, this system facilitates the future development of compact imaging heads. The modular PARS system overcomes many of the challenges with imaging unstained thick tissue in situ, representing a significant milestone in the development of a clinical microscope providing virtual biopsy capabilities.
Keywords: Photoacoustic remote sensing microscopy (PARS), photoacoustics, histology, pathology, gastrointestinal (GI)
Introduction
Each year gastrointestinal (GI) diseases impact 60 to 70 million people in the United States alone, more than any other common disease group (1,2). GI diseases represent a spectrum of conditions including common benign afflictions such as gastroesophageal reflux disease (GERD) with a prevalence of 20% of North Americans, and life-threatening problems such as colorectal cancer (3). In GI diseases, endoscopy is the most common diagnostic maneuver, with 15 million procedures annually for colonoscopies alone (2,4). Endoscopy provides a minimally invasive visual assessment of otherwise inaccessible tissues, identifying polyps, macroscopic lesions, bleeding and other tissue abnormalities. However, current endoscopy techniques are limited by the discordance between macroscopic superficial features and microscopic tissue morphology, and the pragmatic limits on the number of biopsies that can be safely obtained. The current workflow requires clinicians to perform endoscopic biopsies on areas of macroscopic interest, and the tissue fragments then undergo histopathological assessment. In the case of colorectal cancer diagnosis, a limited number of biopsies are taken for histological analysis (5).
Indeed, the number of samples collected is usually restricted to reduce the risks of bleeding, infection, and perforation. Additionally, collection of multiple samples can be complicated by bleeding from ulcerated tumors. Furthermore, sample analysis is limited as histopathological processing is a resource intensive method requiring tissue fixation, substrate embedding, thin sectioning, and sample staining. While there are standard guidelines for biopsy site selection, the lack of real-time microscopic morphological feedback risks the underdiagnosis of malignant diseases (6,7). Ultimately, biopsy location is decided by the judgement of the clinician and the collected specimens may not be fully representative of the disease, leading to delays in definitive diagnosis and associated poor patient outcomes. These problems could be mitigated by gastroenterologists receiving real-time histological information through an endoscopic tool, allowing live in situ “virtual biopsies”. Such a technique would improve the diagnostic accuracy of endoscopy biopsies by providing rapid and in situ morphological tissue assessment to ensure that adequate and diagnostic tissue samples were obtained for definitive histopathological analysis.
An ideal virtual biopsy device would provide in situ contrast akin to standard staining processes such as hematoxylin and eosin (H&E); this would permit rapid adoption without extensive user re-training. Contrary to current histopathological standards which depend on exogenous dyes, providing H&E like contrast label-free in situ requires imaging of endogenous nuclear contrast. Furthermore, imaging thick tissues in situ requires a reflection mode architecture as current tissue fixation and thin sectioning techniques cannot be used. While meeting these design criteria, the virtual biopsy system should conform to the stringent restrictions (<15 mm diameter) on device construction for viable endoscopy-based tools (8). The requirements for a label-free reflection-mode architecture rule out current histopathological imaging methods, along with many emerging alternative techniques. While Stimulated Raman Spectroscopy has presented H&E like images without requiring exogenous labeling, this system typically operates in transmission mode requiring thin sectioned tissue (9). Microscopy with ultraviolet surface excitation (MUSE) is a reflection mode system showing promising H&E-like images, but requires exogenous dyes to achieve contrast (10). Optical coherence tomography (OCT) meets most of the desired criteria and has been implemented in endoscopes. However, while OCT can image cellular scale structures in human tissues, the scattering contrast mechanism does not permit the biomolecule-specific contrast required to generate H&E-like images (11,12). To simulate H&E, OCT systems must use external processing techniques, such as artificial intelligence-based image processing (12).
Photoacoustic remote sensing microscopy (PARS™) is a recently reported imaging technique that provides virtual biopsy capabilities (13,14). PARS captures the endogenous optical absorption contrast of biomolecules in an all optical, label-free non-interferometric reflection-mode architecture (13,14). A picosecond to nanosecond scale pulsed excitation laser is used to deposit optical energy into the sample, inducing local refractive index modulations proportional to the absorbed optical energy. These modulations are observed by a secondary co-focused continuous wave detection laser. The use of optical absorption contrast provides high imaging specificity compared to alternative techniques. By using an ultraviolet (UV) excitation source, PARS has previously recovered nuclear morphology to provide absorption contrast reminiscent of H&E staining (15,16). Furthermore, the reflection-mode architecture of PARS readily enables imaging of thick tissues. Here, we present an all-optical non-interferometric non-contact photoacoustic technique optimized to image unstained GI tissue specimens. We show the first PARS histology-like images recovered from thick FFPE human GI tissues, including volumetric images visualizing subsurface nuclear morphology. Additionally, we validate the proposed technique in unprocessed freshly resected murine GI tissues. While the current PARS system is a non-compact table-top embodiment, a modular architecture is utilized, facilitating future development of compact imaging heads.
Methods
PARS system architecture
The proposed microscope is depicted in Figure 1. In this system, the optical processing for the excitation and detection pathways (Figure 1 red outline) is segmented from the remote imaging head (Figure 1 blue outline) to allow testing of differing imaging heads. To achieve nuclear contrast, DNA excitation is provided by a picosecond-pulsed 266 nm source (WEDGE XF 266, Bright Solutions). The detection is provided by a 1,310-nm continuous-wave superluminescent diode (S5FC1018P, ThorLabs Inc.). Vertically polarized light from the detection source is passed through a polarizing beam splitter (CCM1-PBS254, Thorlabs Inc.) and becomes circularly polarized by a quarter wave plate (WPQ10M-1310, Thorlabs Inc.). The excitation and detection are then combined via a dichroic mirror (HBSY234, ThorLabs Inc.) and passed to the imaging head. Within the imaging head, the beams are co-aligned through a set of galvo-mirrors and co-focused onto the sample using a 15×, 0.3 NA reflective objective lens (LMM-15X-UVV, Thorlabs Inc.). The polarization of back reflected detection light is shifted to a horizontal state by the quarter wave plate such that it is directed towards the photodetector through a long pass filter (FELH1000, ThorLabs Inc.) and an aspheric condenser lens (ACL25416U, Thorlabs Inc.).
Figure 1.
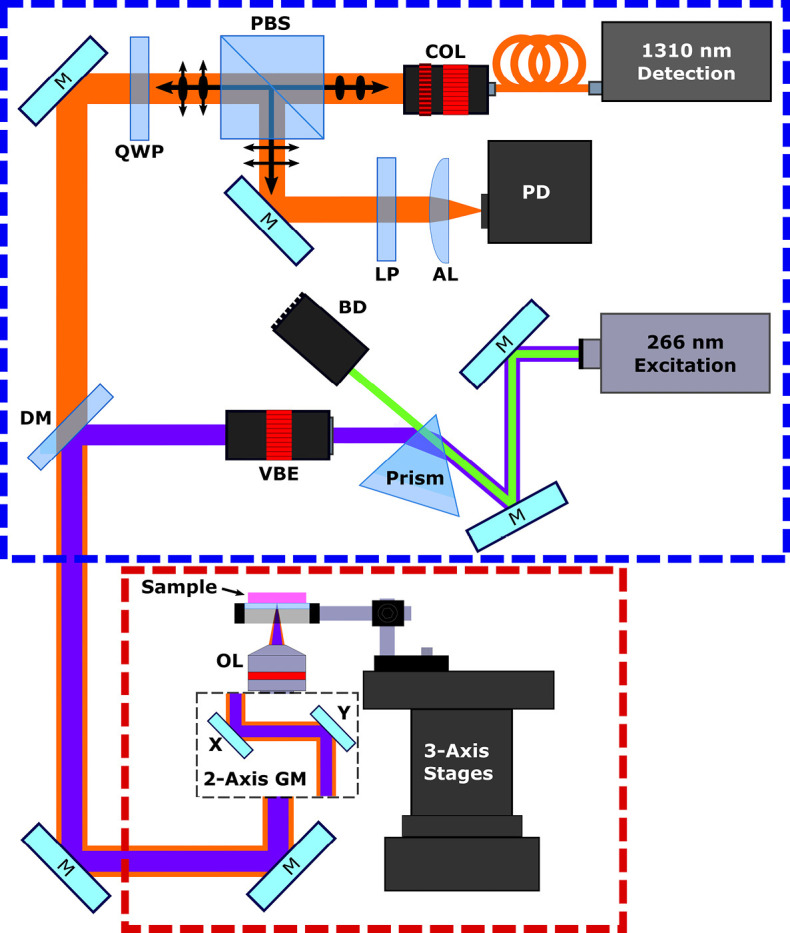
PARS system experimental setup. The system imaging head is enclosed in the red box, while the system back-end optical processing section is enclosed in the blue box. Components are labeled as follows: quarter wave plate (QWP), polarizing beam splitter (PBS), collimator (COL), long pass filter (LP), aspheric lens (AL), dichroic mirror (DM), variable beam expander (VBE), objective lens (OL), beam dump (BD), galvo-mirrors (GM).
Image acquisition
Wide area images were formed by gathering a sequence of optically scanned subframes in a two-dimensional grid, then registering the subframes to form a complete wide field of view (FOV) image. The mechanical stage system was used to move the sample in the two-dimensional grid pattern, while the galvo-mirrors were used to optically scan each subframe. The galvo-mirrors were driven by a positional waveform from a function generator, where each the x and y axis were driven separately to ensure consistent coverage of the scanning area. With each excitation pulse, the position signal from each galvo-mirror, and short sample of photodiode output was captured, using a 16-bit digitizer (CSE161G4, DynamicSignals LLC, Lockport, IL, USA). From the average scanning mirror voltages an x and y coordinate was extracted relative to the center of the scanned area. In conjunction, the upper and lower envelopes of the photodiode signal were extracted and used to calculate the absolute magnitude of the PARS signal for the corresponding scatter location. Nearest neighbor search was then used to fit the scattered PARS data to a cartesian grid. With the data fitted to a grid, linear interpolation was then used to determine the signal at each cartesian location. Based on the x and y location of the scanning stages at the beginning of each subframe acquisition, the independent subframes were registered together to form a completed image.
Results and discussion
Recovering histological information from a single standard 2.8 mm biopsy requires time consuming and resource intensive processing which usually takes 2 days (17,18). The presented system can recover equivalent information in seconds from unstained unprocessed tissues. To perform rapid wide area imaging, an optical and mechanical scanning method is used here. Optical scanning is performed with galvo-mirrors allowing for real-time capture of small fields, while the mechanical stage is shifted to capture different areas. By tracking the location of the imaging head relative to the sample, the multitude of small area optical scans are co-registered to rapidly reconstruct a large FOV image. One such PARS image, of unstained normal colonic mucosal tissue is presented in Figure 2A. This 100 subframe 1.21 mm2 scan captures over one third the area of a standard GI biopsy channel. For each subframe, 10 seconds was spent collecting 200,000 interrogation points, while 0.1 seconds was spent shifting the stages between images. In total, capturing the entire collection of subframes required 16.8 minutes.
Figure 2.
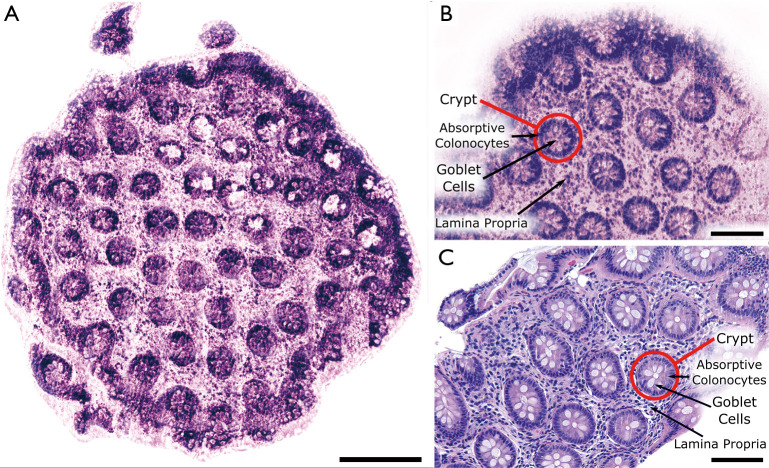
Comparison of bright field H&E stained colonic mucosa and PARS images of unstained colonic mucosa. (A) Bright field H&E image of a normal colonic mucosa in sectioned formalin fixed paraffin embedded (FFPE) form. Scale bar: 100 µm; (B) wide field of view PARS image of unstained sectioned FFPE normal colonic mucosa biopsy. Scale bar: 200 µm; (C) small field PARS image of unstained sectioned FFPE normal colonic mucosa biopsy. Scale bar: 100 µm. Artificial H&E-like coloring has been applied to the PARS images.
Considering the proposed systems low imaging rate, recovering such scans in vivo presents several hurdles, as the GI tract is prone to movement. Since PARS is non-interferometric, and therefore can be designed for low susceptibility to axial movement artifacts, the main challenges arise from lateral movements. To reduce lateral motion artifacts and significantly improve clinical compatibility, the excitation rate could be increased, thus reducing imaging time accordingly. While previous reports have explored excitation rates of up to 10 MHz, even a 1 MHz excitation, would reduce imaging time to 0.2 seconds per frame, or about 30 seconds total for an image equivalent to Figure 2A (19). Additionally, imaging acuity could be further improved by implementing motion tracking techniques akin to those used in OCT retinal imaging (20). Employing these strategies, PARS could be used in an endoscopic embodiment allowing for rapid wide area scans of entire biopsy locations prior to collection.
An enhanced smaller section of the normal colonic mucosa sample is shown in Figure 2B, while a similar section of tissue which has undergone standard H&E processing is shown in Figure 2C. While these three images are not all from the same piece of tissue, in each case characteristic crypt structures are clearly visible, as outlined in Figure 2C and C. Furthermore, individual cells and their organization are easily differentiable with high acuity, including the absorptive colonocytes and goblet cells within the crypt structures and the surrounding lamina propria.
The different FOV images recovered by the PARS system in Figure 1, exemplify the versatility of the proposed scanning mechanism. By recovering a larger number of optical scans with lower point densities, large FOV images can be recovered as in Figure 2A. Concurrently, by capturing a smaller number of optical scans with an increased point density, higher resolution images can be recovered as in Figure 2B. Used in conjunction, the large FOV scans may provide rapid imaging of broad areas maintaining a minimum diagnostic quality, while the small FOV scans may provide finer details within areas of specific interest. Moving forward, the quality and efficiency of these optical scans could be enhanced with improved spatial sampling methods. Currently, optical scanning is performed with scanning mirrors, where each axis is driven independently by a positional wave form. Scanning frequencies are then selected to generate approximately equidistant interrogation points. Unfortunately, this method results in substantial variability across each frame. Utilizing a more structured scanning mechanism could therefore reduce the points required to form an image, while improving the visual acuity of the results.
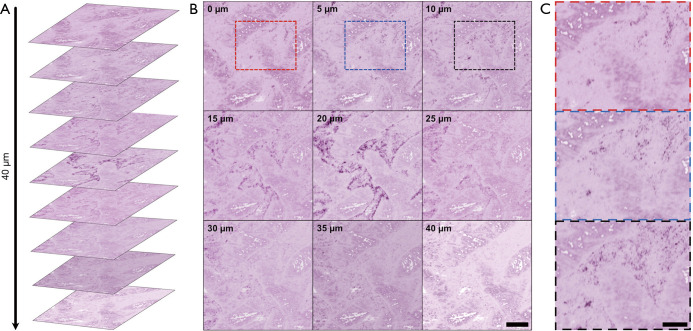
In addition to capturing large FOV surface scans, PARS has the ability to visualize subsurface structures in thick tissue. Standard H&E tissue preparation methods generate 4 µm sections, therefore, recovering structures from differing depths requires multiple thin sections of tissue to be cut and stained. Using optical sectioning PARS achieves depth discrimination within tissues without increasing the complexity of acquisition. Shown in Figure 3A is a multi-layer volumetric scan penetrating 40 µm into a paraffin embedded GI tissue sample. Each of the individual volumetric images is shown in Figure 3B, labelled by the corresponding depth. The nuclear morphology of this sample can be discerned, where an area of significant variation in nuclear structure with axial focus adjustment can be seen enclosed in the red blue and black boxes (Figure 3B). These sections of morphological variation are shown in greater detail in Figure 3C. Applied to endoscopy, this technique would enable investigation of subsurface cellular morphology without resection. Notably, these results are the first label-free histology-like images recovered from unprocessed FFPE human GI tissues, without prior modification to the sample or the paraffin substrate. Previous reports using PARS have depended on deparaffinizing the tissue samples (21).
Figure 3.
Optically sectioned PARS volumetric images within formalin fixed paraffin embedded (FFPE) human gastrointestinal tissues. (A) Optical sections of differing depths in FFPE human gastrointestinal tissues arranged in the same orientation as imaging; (B) optical sections of differing depths in FFPE human gastrointestinal tissues. Scale bar: 200 µm; (C) notable areas of structural changes between the 0 to 10 micron depth are enclosed in the red blue and black boxes. Scale bar: 100 µm. Artificial H&E-like coloring has been applied to the PARS images.
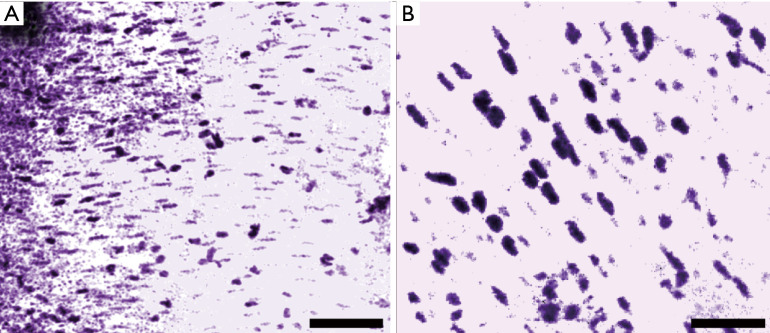
In addition to FFPE preparations, the system was characterized on freshly resected murine small intestine samples. These tissues were retrieved, placed in phosphate buffer solution (PBS), then transported to the microscope for imaging. Imaging was performed within 20 minutes of sample acquisition. Figure 4A and B shows images obtained from the freshly resected unprocessed tissues. These images were acquired from a transverse cross section of the small intestine and show smooth muscle structure, identifiable by the parallel stripes of elongated nuclei. Here, the shape, separation and cytological detail of the individual cell nuclei are easily recovered with quality comparable to H&E preparations of similar tissue sections (22). Visualizing the UV absorption of DNA, cell nuclei contrast is isolated from the surrounding tissues without the use of exogenous dyes.
Figure 4.
PARS images of freshly resected mouse small intestine, highlighting smooth muscle structure. (A) Large field of view image. Scale bar: 200 µm; (B) small field of view image. Scale bar: 75 µm. Artificial H&E-like coloring has been applied to the PARS images.
The proposed PARS microscope overcomes many of the pragmatic hurdles that have delayed the clinical development of a system capable of performing virtual biopsies. We have for the first time demonstrated the retrieval of H&E-like images from unprocessed GI tissues using an all-optical non-interferometric reflection-mode photoacoustic microscopy system. Presented here, clinically relevant cellular and subcellular structures were acquired from large areas of tissues using an optical and mechanical scanning technique. Furthermore, for the first time subsurface nuclear morphology was imaged within FFPE human GI tissue blocks using an all-optical reflection mode system. The presented results exemplify PARS suitability for imaging unprocessed thick tissue samples. These imaging capabilities could significantly increase the morphological information available to clinicians to optimize biopsy site selection and improve patient outcomes. To improve imaging speeds with larger imaging fields, while providing additional selective contrast for biomolecules such as lipids and hemoglobin, future efforts will focus on the incorporation of faster repetition rate lasers and additional excitation wavelengths. While the present feasibility results use a non-compact table-top PARS microscopy system, moving forward focus will be placed on incorporating a fiber based optical path from the optical processing unit to the imaging head, potentially utilizing photonic crystal fiber, or UV compatible fibers. Furthermore, future work will focus on miniaturizing the footprint of the imaging head to meet endoscope design requirements.
Acknowledgments
Funding: Natural Sciences and Engineering Research Council of Canada (DGECR-2019-00143, RGPIN2019-06134); Canada Foundation for Innovation (JELF #38000); Mitacs Accelerate (IT13594); University of Waterloo Startup funds; Centre for Bioengineering and Biotechnology (CBB Seed fund); illumiSonics Inc. (SRA #083181); New frontiers in research fund—exploration.
Ethical Statement: Formalin fixed paraffin embedded gastrointestinal tissue samples were obtained under protocols approved by Research Ethics Board of Alberta (Protocol ID: HREBA.CC-18-0277) and University of Waterloo Health Research Ethics Committee (Humans: #40275). The ethics committees waived the requirement for patient consent as the selected samples were archival tissue no longer required for diagnostic purposes, and no patient identifiers were provided to the researchers. All experiments were performed in accordance with the requirements of the Government of Canada, Panel on Research Ethics Guidelines. All procedures were regarding the acquisition and use of resected murine tissues were approved by the University of Waterloo Health Research Ethics Committee under the protocol Animals: #41543.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Footnotes
Provenance and Peer Review: With the arrangement by the Guest Editors and the editorial office, this article has been reviewed by external peers.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/qims-20-722). The special issue “Advanced Optical Imaging in Biomedicine” was commissioned by the editorial office without any funding or sponsorship. KB, DD, JRM and PHR have financial interests in illumiSonics Inc. IllumiSonics partially supported this work. The authors have no other conflicts of interest to declare.
References
- 1.Everhart JE, Ruhl CE. Burden of Digestive Diseases in the United States Part I: Overall and Upper Gastrointestinal Diseases. Gastroenterology 2009;136:376-86. 10.1053/j.gastro.2008.12.015 [DOI] [PubMed] [Google Scholar]
- 2.Peery AF, Crockett SD, Murphy CC, Lund JL, Dellon ES, Williams JL, Jensen ET, Shaheen NJ, Barritt AS, Lieber SR, Kochar B, Barnes EL, Fan YC, Pate V, Galanko J, Baron TH, Sandler RS. Burden and Cost of Gastrointestinal, Liver, and Pancreatic Diseases in the United States: Update 2018. Gastroenterology 2019;156:254-272.e11. 10.1053/j.gastro.2018.08.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El-Serag HB, Sweet S, Winchester CC, Dent J. Update on the epidemiology of gastro-oesophageal reflux disease: a systematic review. Gut 2014;63:871-80. 10.1136/gutjnl-2012-304269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seeff LC, Richards TB, Shapiro JA, Nadel MR, Manninen DL, Given LS, Dong FB, Winges LD, McKenna MT. How many endoscopies are performed for colorectal cancer screening? Results from CDC’s survey of endoscopic capacity. Gastroenterology 2004;127:1670-7. 10.1053/j.gastro.2004.09.051 [DOI] [PubMed] [Google Scholar]
- 5.Choi Y, Choi HS, Jeon WK, Kim BI, Park DI, Cho YK, Kim HJ, Park JH, Sohn CI. Optimal Number of Endoscopic Biopsies in Diagnosis of Advanced Gastric and Colorectal Cancer. J Korean Med Sci 2012;27:36-9. 10.3346/jkms.2012.27.1.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peixoto A, Silva M, Pereira P, Macedo G. Biopsies in Gastrointestinal Endoscopy: When and How. GE Port J Gastroenterol 2015;23:19-27. 10.1016/j.jpge.2015.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonsalves N, Policarpio-Nicolas M, Zhang Q, Rao S, Hirano I. Histopathologic variability and endoscopic correlates in adults with eosinophilic esophagitis. Gastrointest Endosc 2006;64:313-9. 10.1016/j.gie.2006.04.037 [DOI] [PubMed] [Google Scholar]
- 8.Varadarajulu S, Banerjee S, Barth BA, Desilets DJ, Kaul V, Kethu SR, Pedrosa MC, Pfau PR, Tokar JL, Wang A, Song LMWK, Rodriguez SA. GI endoscopes. Gastrointest Endosc 2011;74:1-6. 10.1016/j.gie.2011.01.061 [DOI] [PubMed] [Google Scholar]
- 9.Orringer DA, Pandian B, Niknafs YS, Hollon TC, Boyle J, Lewis S, Garrard M, Hervey-Jumper SL, Garton HJL, Maher CO, Heth JA, Sagher O, Wilkinson DA, Snuderl M, Venneti S, Ramkissoon SH, McFadden KA, Fisher-Hubbard A, Lieberman AP, Johnson TD, Xie S, Trautman JK, Freudiger CW, Camelo-Piragua S. Rapid intraoperative histology of unprocessed surgical specimens via fibre-laser-based stimulated Raman scattering microscopy. Nat Biomed Eng 2017;1:0027. [DOI] [PMC free article] [PubMed]
- 10.Fereidouni F, Harmany ZT, Tian M, Todd A, Kintner JA, McPherson JD, Borowsky AD, Lechpammer M, Bishop J, Demos SG, Levenson R. Microscopy with ultraviolet surface excitation for rapid slide-free histology. Nat Biomed Eng 2017;1:957-66. 10.1038/s41551-017-0165-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sivak MV, Kobayashi K, Izatt JA, Rollins AM, Ung-runyawee R, Chak A, Wong RCK, Isenberg GA, Willis J. High-resolution imaging of the GI tract using optical coherence tomography. Gastrointest Endosc 2000;51:474-9. 10.1016/S0016-5107(00)70450-0 [DOI] [PubMed] [Google Scholar]
- 12.Tsai S, Chan C, Chen HH, Tiju J, Huang S. Segmentation based OCT Image to H&E-like Image Conversion. J Biomed Opt 2020. Available online: https://doi.org/ 10.1364/MICROSCOPY.2020.MM3A.5 [DOI]
- 13.Hajireza P, Shi W, Bell K, Paproski RJ, Zemp RJ. Non-interferometric photoacoustic remote sensing microscopy. Light-Sci Appl 2017;6:e16278. 10.1038/lsa.2016.278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haji Reza P, Bell K, Shi W, Shapiro J, Zemp RJ. Deep non-contact photoacoustic initial pressure imaging. Optica 2018;5:814-20. 10.1364/OPTICA.5.000814 [DOI] [Google Scholar]
- 15.Abbasi S, Le M, Sonier B, Dinakaran D, Bigras G, Bell K, Mackey JR, Haji Reza P. All-optical Reflection-mode Microscopic Histology of Unstained Human Tissues. Sci. Rep 2019;9:13392. 10.1038/s41598-019-49849-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abbasi S, Le M, Sonier B, Bell K, Dinakaran D, Bigras G, Mackey JR, Haji Reza P. Chromophore selective multi-wavelength photoacoustic remote sensing of unstained human tissues. Biomed Opt Express 2019;10:5461-9. 10.1364/BOE.10.005461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Odze R, Goldblum J. Surgical Pathology of the GI Tract, Liver, Biliary Tract, and Pancreas, 2nd ed. Philadelphia: Saunders Elsevier, 2008. [Google Scholar]
- 18.Patel S, Smith JB, Kurbatova E, Guarner J. Factors that impact turnaround time of surgical pathology specimens in an academic institution. Hum Pathol 2012;43:1501-5. 10.1016/j.humpath.2011.11.010 [DOI] [PubMed] [Google Scholar]
- 19.Snider L, Bell K, Haji Reza P, Zemp RJ. Toward wide-field high-speed photoacoustic remote sensing microscopy. Proc. SPIE 10494, Photons Plus Ultrasound: Imaging and Sensing 2018. Available online: https://doi.org/ 10.1117/12.2291036 [DOI]
- 20.Vienola KV, Braaf B, Sheehy CK, Yang Q, Tiruveedhula P, Arathorn DW, de Boer JF, Roorda A. Real-time eye motion compensation for OCT imaging with tracking SLO. Biomed Opt Express 2012;3:2950-63. 10.1364/BOE.3.002950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haven NJM, Kedarisetti P, Restall BS, Zemp RJ. Reflective objective-based ultraviolet photoacoustic remote sensing virtual histopathology. Opt Lett 2020;45:535-8. 10.1364/OL.382415 [DOI] [Google Scholar]
- 22.Chapter 10: Muscle Tissue. In: Mescher AL. editor. Junqueira’s Basic Histology: Text and Atlas, 15th ed. Lange McGraw Hill Education, New York, New York, 2018. [Google Scholar]