Abstract
This study discussed the use of antimicrobials in the commercial chicken production system and the possible factors influencing the presence of Extended-spectrum β-lactamase (ESBL)/AmpC producers strains in the broiler production chain. The aim of this study was to perform longitudinal monitoring of ESBL-producing and fosfomycin-resistant Escherichia coli from poultry farms in southern Brazil (Paraná and Rio Grande do Sul states) and determine the possible critical points that may be reservoirs for these strains. Samples of poultry litter, cloacal swabs, poultry feed, water, and beetles (Alphitobius sp.) were collected during three distinct samplings. Phenotypic and genotypic tests were performed for characterization of antimicrobial resistant strains. A total of 117 strains were isolated and 78 (66%) were positive for ESBL production. The poultry litter presented ESBL positive strains in all three sampled periods, whereas the cloacal swab presented positive strains only from the second period. The poultry litter represents a significant risk factor mainly at the beginning poultry production (odds ratio 6.43, 95% confidence interval 1–41.21, p < 0.05). All beetles presented ESBL positive strains. The predominant gene was blaCTX–M group 2, which occurred in approximately 55% of the ESBL-producing E. coli. The cit gene was found in approximately 13% of the ESBL-producing E. coli as AmpC type determinants. A total of 19 out of 26 fosfomycin-resistant strains showed the fosA3 gene, all of which produced ESBL. The correlation between fosA3 and blaCTX–M group 1 (blaCTX–M55) genes was significant among ESBL-producing E. coli isolated from Paraná (OR 3.66, 95% CI 1.9–9.68) and these genetic determinants can be transmitted by conjugation to broiler chicken microbiota strains. Our data revealed that poultry litter and beetles were critical points during poultry production and the presence of fosfomycin-resistant strains indicate the possibility of risks associated with the use of this antimicrobial during production. Furthermore, the genetic determinants encoding CTX-M and fosA3 enzymes can be transferred to E. coli strains from broiler chicken microbiota, thereby creating a risk to public health.
Keywords: Avian, multidrug resistance (MDR), Enterobacteriaceae, fosfomycin, poultry litter, public health
Introduction
Antimicrobial resistance is one of the most alarming public health problems in recent years. According to O’Neill (2014), by 2050, bacterial resistance could cause the deaths of approximately 10 million people each year. The widespread use of antimicrobial drugs, both in humans and animals (including livestock animals), has favored the selection and dissemination of bacterial resistance worldwide (World Health Organization, 2014).
Extended-spectrum β-lactamase (ESBL) and AmpC-like enzymes are among the best-known mechanisms of bacterial resistance, which are both mediated by plasmid genes (Ceccarelli et al., 2019) and can be achieved through the horizontal transfer of mobile genetic elements, in both intestinal and extra-intestinal environments (Lazarus et al., 2014). ESBL and AmpC enzymes are capable of hydrolyzing various β-lactam antimicrobials such as cephalosporins and monobactams, increasing the difficulty of treating these infections (Paterson and Bonomo, 2005; Bush and Fisher, 2011).
Initially, the detection of ESBL/AmpC-producing bacteria was related to cases of infection in humans (Smet et al., 2009; Ewers et al., 2012). However, several studies have reported the presence of these resistant strains in animals, whether domestic (Wieler et al., 2011; Bortolami et al., 2019) or livestock (Pitout and Laupland, 2008; Smet et al., 2009; Dierikx et al., 2013; Laube et al., 2013; Dominguez et al., 2018). ESBL/AmpC enzymes are found in Enterobacteriaceae family members, such as Escherichia coli, which have often been isolated in livestock, especially during poultry production (Blanc et al., 2006; Carattoli, 2008; Li et al., 2015). Among the most relevant β-lactamases, CTX-M is one of the main enzymes present in E. coli that colonize and infect poultry (Olsen et al., 2014).
Worldwide, ESBL-producing Enterobacteriaceae have emerged in farm animals in recent decades (Carattoli, 2008). The pressure exerted by antimicrobial use, particularly in broiler chickens, led to sensitive strains elimination and selection of resistant ones (Saliu et al., 2017). Although antimicrobial resistance is a natural phenomenon, the prevalence of ESBL-producing strains in broilers has increased due to the antimicrobial use in production (Dierikx et al., 2013). Another relevant finding regarding the use of antimicrobials in production is the presence of fosfomycin-resistant strains in poultry carcasses (Cyoia et al., 2019). Fosfomycin is approved in several countries for urinary tract infections treatment in humans (Keating, 2013; Falagas et al., 2019). Thus, the presence of fosfomycin-resistant strains in poultry raises public health safety concerns.
The possibility that bacterial strains, especially ESBL/AmpC-producing and fosfomycin-resistant E. coli, may reach the human population via chicken meat consumption is a public health concern because, compared to other types of meat (e.g., pork and beef), chicken meat has been found to become highly contaminated with ESBL-producing bacteria (Friese et al., 2013). Koga et al. (2015) reported the presence of ESBL/AmpC-producing E. coli strains isolated from poultry carcasses in southern Brazil. Cyoia et al. (2019) demonstrated that ESBL-producing E. coli strains were capable of transferring genes encoding CTX-M enzymes to a human E. coli strain. Plasmid incompatibility groups (Inc groups) are categorized by the propagation inability, in the same cell, of two plasmids belonging to the same group. These plasmids may carry resistance genes and can be found in E. coli (Datta and Hedges, 1971; Carattoli, 2011). The blaCTX-M (e.g., CTX-M15 and CTX-M55) and blaTEM genes are associated with plasmids belonging to the IncF and IncI groups (Cantón and Coque, 2006; Carattoli, 2013).
The presence of ESBL/AmpC-producing and fosfomycin-resistant E. coli in poultry carcasses reveals that there are possibly critical points during industrial broiler production where these strains can be found and selected by antimicrobials. Thus, the present study aimed to monitor ESBL/AmpC-producing and fosfomycin-resistant E. coli in poultry production and determine the possible critical points that may be reservoirs of these strains.
Materials and Methods
Farm Characterization
Monitoring was undertaken in the biggest broiler producers states from Brazil. Five farms in the Rio Grande do Sul (RS) state were sampled between February and May 2016, and three farms in the Paraná (PR) state were sampled between January and March 2018. All farms sampled employed an all-in all-out system. In this system, the barns are emptied for slaughter; the poultry litter is turned and covered with a plastic canvas, remaining inside the barn to be reused; the feeders and drinking fountains are disinfected; and the place remains closed for 15 days (depopulation period), until the arrival of new chicks. Sampling was performed in one barn per farm and monitored at three different times: (1) first day (one-day-old restocking chicks), (2) between 20th and 25th days, and (3) between 36th and 38th days of fattening period (Supplementary Table 1). It is important to note that in the first sampling time, chicks samples were collected, but they did not come into contact with water, feed or poultry litter of barn, so that this would not interfere with the analysis of the results. Besides, a questionnaire was given to the producers to obtain information regarding property characteristics, management, and biosecurity.
In the RS state, the production system was characterized as manual. On average, the farmers restocked approximately 15,000 one-day-old chicks in the poultry houses, one or two poultry houses per farm, and the slaughtering period ranged from 38 to 45 days. The water came from artesian wells and was chlorinated in a reservoir present in each poultry house. In general, the poultry litter was composed of rice husk and reused in up to five subsequent flocks without undergoing treatment. The antimicrobials used in the RS farms were enrofloxacin (ENR), halquinol, and virginiamycin.
In the PR state, the production system used was automated (dark house system). The average restocking was approximately 25,000 one-day-old chicks, one or two poultry houses per farm, and the slaughtering period ranged from 38 to 42 days. Similar to the RS farms, the water used in the bird drinkers was chlorinated. The poultry litter was made of wood shavings and reused without undergoing treatment. The antimicrobials used for treatment in the PR farms were norfloxacin (NOR), ciprofloxacin (CIP), and fosfomycin. All barns investigated were disinfected between a previous and a subsequent flock, followed by a depopulation period to approximately 15 days.
Ethics Statement
The present study was approved by the Animal Ethics Committee of State University of Londrina (CEUA/UEL) (processing number – 22867.2015.23).
Sampling
The sampling methodology was based on the procedure described by Laube et al. (2013), with some modifications. During each fattening period, samples of poultry litter (boot swab), cloacal swab (20 randomly selected broiler), poultry feed (500 g), and water (500 mL) were collected. In the PR state, samples of Alphitobius sp. (approximately 100 beetles), popularly known as “darkling beetle,” were also collected from the farms. The poultry litter was collected using a sterile boot swab by walking the entire house length in a “zigzag” pattern. The poultry feed was obtained directly from the reservoir and water was collected from the farm reservoir taps. Therefore, a total of 45 samples from PR, and 60 samples in RS were collected.
The collected samples were refrigerated (4°C) and sent to the laboratory for processing on the same day. A total of 25 g of feed was weighed and diluted in 225 mL of buffered peptone water, followed by manual homogenization (approximately 10 min). The boot swab was soaked with buffered peptone water, followed by manual homogenization. The water was processed using the multi-tube method (most probable number method), using Lauryl Triptose broth, Brilliant Green broth, and E. coli broth (Himedia Laboratories Pvt. Ltd., Mumbai, India). The beetles were processed following the protocol described by Segabinazi et al. (2005). After preprocessing, all samples were plated on MacConkey agar (MC) (Neogen Corporation, Lansing, Michigan, United States) and cefotaxime-supplemented MacConkey agar (MC/CTX) at a concentration of 8 μg/mL to select positive ESBL/AmpC strains (Jarlier et al., 1988).
E. coli Isolation
MacConkey agar and MC/CTX plates were analyzed for the growth of characteristic colonies of E. coli. The grew colonies in MC/CTX (possible ESBL/AmpC-producing E. coli) were prioritized, with a collection of one to five colonies. In MC/CTX plates that presented no growth, one to five colonies from the MC plates were collected. The colonies were submitted for biochemical identification using media Escola Paulista de Medicina (Probac, Brazil) (Toledo et al., 1982a; Edwards and Ewing, 1986), Motility, Indole and Lysine (Probac, Brazil) (Toledo et al., 1982b; Edwards and Ewing, 1986), and Simmons citrate (Merck, Darmstadt, Germany). E. coli positive colonies were stored in Brain Heart Infusion broth (Himedia Laboratories Pvt. Ltd., Mumbai, India) supplemented with 30% glycerol at −20 and −80°C for subsequent phenotypic and genotypic characterization.
Antimicrobial Susceptibility Test
The antimicrobial susceptibility test was performed using the standard disk diffusion method recommended by the Clinical and Laboratory Standards Institute (CLSI, 2018). The antimicrobials used included several classes, such as β-lactams: cefotaxime (CTX, 30 μg), ceftazidime (CAZ, 30 μg), cefepime (FEP, 30 μg), aztreonam (ATM, 30 μg), cefoxitin (FOX, 30 μg), imipenem (IPM, 30 μg) and amoxicillin-clavulanic acid (AMC, 20/10 μg); quinolones: CIP (5 μg), NOR (10 μg), enrofloxacin (ENR, 10 μg), and nalidixic acid (NAL, 30 μg); sulfonamides: trimethoprim-sulfamethoxazole (SXT, 1.25/23.75 μg); tetracycline (TET, 30 μg); aminoglycosides: gentamicin (GEN, 10 μg), phenicols: chloramphenicol (CHL, 30 μg); nitrofurans: nitrofurantoin (NIT, 300 μg); and fosfomycins: fosfomycin-trometamol (FOT, 200 μg) (Oxoid Ltd., Basingstoke, Hants, United Kingdom). All strains were confirmed for ESBL production using the double-disk approximation test, described by Jarlier et al. (1988). E. coli strain ATCC 25922 was used as quality control and results were interpreted based on the CLSI (2018) criteria.
DNA Template
DNA samples used for polymerase chain reaction (PCR) assays and sequencing, were extracted using Pure Link® Genomic DNA Mini Kit (Invitrogen®).
Detection of ESBL/AmpC Genes in E. coli Strains
A previously described PCR method was used for the detection of the following antimicrobial resistance genes: ESBL producer (blaCTX–M groups – 1, 2, 8, 9, and 25) (Arlet and Philippon, 1991; Woodford et al., 2005); and AmpC-type producer (mox, fox, ebc, acc, dha, and cit) (Pérez-Pérez and Hanson, 2002). The PCR products positive to blaCTX–M group 1 were characterized for bidirectional Sanger sequencing on ABI-PRISM 3500 XL (Applied Biosystems), following the manufacturer’s recommendations.
Detection of Other Antimicrobial Resistance Genes
The presence of fosfomycin (fosA3) and colistin (mcr-1) resistance was examined as described previously by Sato et al. (2013) and Liu et al. (2016), respectively.
Phylogenetic Analysis
All E. coli isolates were assigned to phylogenetic groups A, B1, B2, or D using PCR according to the methodology described by Clermont et al. (2000). Isolates were grouped into the following groups: group A (chuA–, yjaA–, and TspE4.C2–); group B1 (chuA–, yjaA+, TspE4.C2–); group B2 (chuA+, yjaA+, TspE4.C2- or chuA+, yjaA+, TspE4.C2+); and group D (chuA+, yjaA–, TspE4.C2+).
Conjugation Experiments
Horizontal transmission of blaCTX–M and fosA3 genes was investigated using conjugation assays. ESBL-producing E. coli from poultry litter, harboring the resistance genes, were chosen as donor strains. Non-ESBL-producing E. coli from chicks microbiota were selected as possible recipients, based on the antibiogram test and phylogenetic profile. The selected colonies were strains were grown overnight in Luria Bertani (LB) broth (Difco, Sparks, MD, United States) under agitation at 36°C. Then, 1 mL of each strain was centrifuged at 12,000 × g for 2 min at 25°C, the supernatants were discarded and resuspended in new LB broth. A total of 100 μL of a recipient strain and 50 μL of a donor strain were added in 3 mL of new LB broth and incubated overnight without shaking at 36°C. Then, 100 μL of the conjugated samples were seeded on LB agar supplemented with GEN (10 μg/mL) and CTX (4 μg/mL), and the plates were incubated at 36°C for growth. The transconjugants selected were used for phylogenetic analysis and tested for the presence of blaCTX–M and fosA3 genes.
Plasmid-Based Replicon Typing
All isolates were characterized by the Inc group using plasmid-based replicon typing (PBRT; Carattoli et al., 2005). Simplex-PCR was used to recognize the eight incompatibility plasmids: FIA, FIB, FIC, FII, I1, HI1, HI2, and N (Carattoli et al., 2005).
Enterobacterial Repetitive Intergenic Consensus Sequence–PCR Analysis
A template DNA was used to amplify the repetitive elements from bacterial isolates using the PCR technique to generate DNA fingerprint patterns. The isolates’ clonality was determined by homology among fragments amplified using enterobacterial repetitive intergenic consensus sequence (ERIC)–PCR (Versalovic et al., 1991). Gel analysis was performed using BioNumerics software, version 7.6 (Applied Maths, Sint-Martens-Laten, Belgium). Similarities in the amplicon profile were compared using a DICE coefficient at 1% tolerance and 0.5% optimization. A dendrogram was constructed with the unweighted-pair group method using the arithmetic mean clustering method with a cut-off of 80% similarity (McLellan et al., 2003).
Statistical Analysis
The data obtained were analyzed using a logistic regression model to calculate the odds ratio (OR) and 95% confidence interval (CI), with a significance level set at p < 0.05, and the statistical software R version 3.5.1. In general, the analysis was performed to verify whether there was any significant trend between the isolates producing ESBL and their origin.
Results
Poultry Farm Samples
A total of 117 E. coli strains were isolated from eight poultry farms in two states, 58 strains from three PR farms, and 59 strains from five RS farms. E. coli samples grown in MC/CTX were obtained from poultry litter in all periods in both states, whereas it was only present in cloacal samples isolated from the second period (Table 1). ESBL-producing strains were detected in poultry feed and beetle samples from PR state. In both states, the water samples showed no E. coli growth in MC/CTX (Table 1).
TABLE 1.
Detection of ESBL-producing E. coli isolated in MC/CTX agar from farms in PR and RS states.
| E. coli source | PR farms |
RS farms |
||||||||||||||||||||||
| Farm 1 |
Farm 2 |
Farm 3 |
Farm 4 |
Farm 5 |
Farm 6 |
Farm 7 |
Farm 8 |
|||||||||||||||||
| 1° | 2° | 3° | 1° | 2° | 3° | 1° | 2° | 3° | 1° | 2° | 3° | 1° | 2° | 3° | 1° | 2° | 3° | 1° | 2° | 3° | 1° | 2° | 3° | |
| Poultry Litter | + | + | + | + | + | + | − | + | + | − | − | + | + | − | + | + | + | − | + | + | + | − | − | + |
| Poultry | − | + | + | − | + | + | − | + | + | − | + | + | − | + | + | − | + | − | − | + | + | − | + | + |
| Poultry Feed | − | + | − | − | + | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + |
| Water | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Beetle | + | − | + | − | + | + | − | + | + | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS |
(+) Presence; (−) Absence; (NS) Non Sampling.
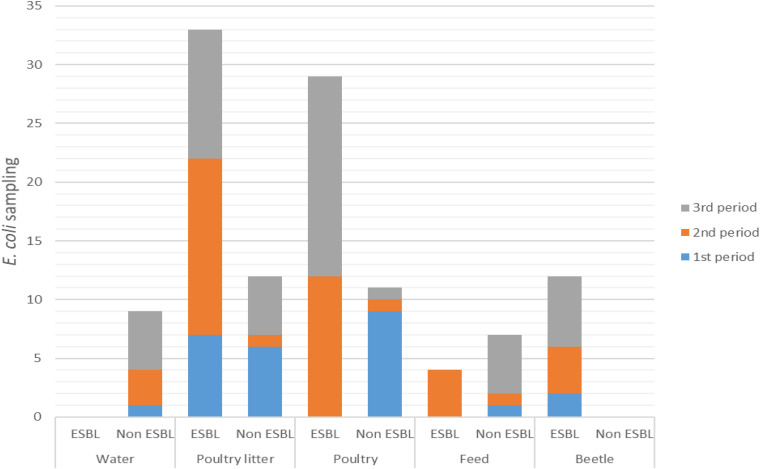
ESBL-producing E. coli were confirmed, respectively, in 49/58 (84%) and 29/59 (49%) isolates from PR and RS. The distribution of ESBL-producing and non-ESBL-producing E. coli strains over the sampling periods are shown in Figure 1. Among the 117 samples, ESBL-producing E. coli strains were detected in poultry litter in all sampling periods: first period (OR 6.42, 95% CI 1–41.21, p < 0.05), second period (OR 3.75, 95% CI 0.4–35.54, p < 0.05), and third period (OR 1.05, 95% CI 0.28–3.78, p < 0.05). The profile of cloacal poultry strains revealed that ESBL-producing E. coli was also present in the second sampling period (OR 2.61, 95% CI 0.27–24.94, p < 0.05) and third sampling period (OR 15, 95% CI 1.78–126.59, p < 0.05). All the strains isolated from beetles were characterized as ESBL producers from all poultry farms in the PR state (OR > 100, 95% CI 0–inf., p < 0.05). No ESBL-producing strains were found in water samples. Moreover, low frequency was found in poultry feed only in the second sampling (OR 0.65, 95% CI 0.06–7.01 p < 0.05).
FIGURE 1.
Distribution of ESBL-producing and non-ESBL-producing E. coli during the sampling periods.
E. coli Antimicrobial Resistance
The antimicrobial susceptibility test indicated that the strains isolated from the poultry farms presented a high frequency of antimicrobial resistance, with 90 and 73% of strains considered as multidrug-resistant (MDR), from PR and RS, respectively (Supplementary Tables 2,3).
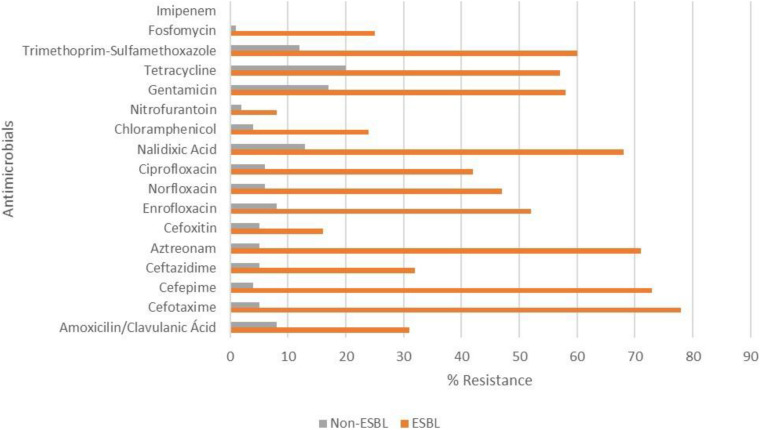
In the PR state, most isolates were resistant to CTX (85%), NAL (85%), FEP (81%), TET (79%), SXT (78%), ATM (74%), ENR (66%), CIP, and NOR (55%). Most isolates from the RS state were resistant to GEN (70%), TET (63%), CTX (58%), ATM (56%), NAL (54%), and FEP (51%), presenting a different resistance profile from PR state.
The frequency of resistance observed for fosfomycin in PR and RS strains was 40 and 5%, respectively. In both states, no IPM-resistant E. coli strain was found. ESBL-producing E. coli were resistant to a higher number of antimicrobials (except FOX, NIT, and IPM) compared to non-ESBL-producing E. coli (p < 0.05) (Figure 2).
FIGURE 2.
Resistance of ESBL-producing and non-ESBL-producing E. coli strains isolated from poultry farms in southern Brazil. There were significant differences between ESBL-producing and non-producing strains (p < 0.05), except for cefoxitin, nitrofurantoin and imipenem (all isolates were sensitive).
Detection of ESBL/AmpC, fosA3, and mcr-1 Genes
Polymerase chain reaction analysis identified ESBL genes in 92/117 (77%) strains isolated from poultry farms (Supplementary Tables 2,3). Approximately 77/78 (99%) of the ESBL-producing E. coli strains were positive for the ESBL genes investigated. In these strains, the genes blaCTX–M group 2 (55%), blaCTX–M group 1 (38%), and blaCTX–M group 8 (10%) were detected. The only AmpC gene detected was cit, found in 14/117 isolates, which was associated with blaCTX–M genes in nine strains. The blaCTX–M group 9 and blaCTX–M group 25 genes were not detected and one ESBL-producing strain carried none of the investigated genes. By sequencing, we found that 100% of blaCTX–M group 1 positive isolates were identified as CTX-M55.
Among the 26 fosfomycin-resistant E. coli strains, 19 (73%) harbored the fosA3 gene, all had ESBL phenotypes features and 17 of these also harbored the blaCTX–M55 gene (OR 3.66, 95% CI 1.39–9.68, p < 0.05). All of these fosA3 positive strains were from PR farms that used fosfomycin in broiler production. Three fosfomycin-resistant strains were isolated from the RS state and none of them presented the fosA3 gene. The mcr-1 gene was detected in only one isolate (EcRS60) from RS. Among the ESBL-producing strains, 14 combinations between the resistance genes detected were observed and most of them are distributed among poultry and poultry litter isolates (Table 2).
TABLE 2.
Distribution of ESBL-producing E. coli strains based on a combination of genes (or CTX-M group genes) and sources in PR and RS states.
| Poultry litter |
Poultry |
Feed |
Beetle |
|||||
| PR | RS | PR | RS | PR | RS | PR | RS | |
| CTX-M1 | 1 | – | 3 | 1 | 1 | – | 5 | NS |
| CTX-M1 + fosA3 | 6 | – | 4 | – | 2 | – | 3 | NS |
| CTX-M1 + cit | – | – | – | 1 | – | – | – | NS |
| CTX-M1 + CTX-M2 + CTX-M8 | 1 | – | – | – | – | – | – | NS |
| CTX-M1 + CTX-M8 + fosA3 | 1 | – | – | – | – | – | – | NS |
| CTX-M1 + cit + fosA3 | – | – | 1 | – | – | – | – | NS |
| CTX-M2 | 7 | 9 | 6 | 10 | – | – | 4 | NS |
| CTX-M2 + mcr-1 | – | 1 | – | – | – | – | – | NS |
| CTX-M2 + cit | – | 1 | – | – | – | – | – | NS |
| CTX-M2 + CTX-M8 | – | – | – | 1 | – | – | – | NS |
| CTX-M2 + CTX-M8 + cit | – | 2 | – | 1 | – | – | – | NS |
| CTX-M8 + cit | – | – | 2 | – | – | – | – | NS |
| fosA3 | – | – | 1 | – | 1 | – | – | NS |
| cit | – | – | – | 2– | – | – | – | NS |
(–) Absence; (NS) Non Sampling; Water not presented ESBL-producing E. coli positive.
Inc Group Plasmid
Tables 2, 3 show the results obtained from the strains submitted to the PBRT technique. Replicons were detected in 97/117 (83%) isolates and the Inc typing showed the presence of FIB (n = 89; 76%), I1 (n = 37; 32%), HI2 (n = 7; 6%), FIA (n = 5; 4%), FIC (n = 5; 4%), and N (n = 4; 3%). The presence of HI1 and FII groups was not detected and 19 strains were negative for all tested replicons. The most frequent replicons among the ESBL-producing samples were FIB (71%) and I1 (36%).
TABLE 3.
Conjugation experiment between ESBL-producing E. coli from poultry litter and non-ESBL-producing E. coli from the microbiota of chicks.
| Strain | Phylogenetic group | blaCTX–M gene | fosA3 gene | |
| Donors | EcPR1 | B1 | blaCTX–M55 | + |
| EcPR2 | B1 | blaCTX–M55 | + | |
| Recipients | EcRS1 | D | blaCTX–M8 | − |
| EcRS2 | D | blaCTX–M8 | − | |
| Transconjugants | T1 (EcPR1+EcRS1) | D | blaCTX–M8 / blaCTX–M55 | + |
| T4 (EcPR1+EcRS2) | D | blaCTX–M8 / blaCTX–M55 | + |
(+) Presence; (−) Absence.
Conjugation Experiments
Two transconjugant strains were obtained in the conjugation assay (T1 and T4), which presented blaCTX-M1 and fosA3 genes and ESBL phenotype (Table 3).
Phylogenetic Group and ERIC-PCR Analysis
All strains were assigned to four main phylogenetic groups (Clermont et al., 2000). Most strains belonged to the phylogenetic group D (n = 54; 46%), followed by group B1 (n = 28; 24%), group A (n = 27; 23%), and group B2 (n = 8; 7%) (Table 4). The ERIC-PCR analysis showed 16 sub-clusters in the PR state and 15 clusters in the RS state samples, with 19 and 17 singletons remaining, respectively (Supplementary Figures 1,2).
TABLE 4.
Phylogenetic distribution of 78 ESBL-producing E. coli strains and 39 non-ESBL-producing E. coli strains based on the isolate source.
| ESBL producer – number of strains (%) |
Non-ESBL producer – number of strains (%) |
||||||||||
| A | B1 | B2 | D | Total | A | B1 | B2 | D | Total | ||
| Poultry Litter | 12 (15.4%) | 9 (11.5%) | 0 (0%) | 8 (10.3%) | 29 | 2 (5.1%) | 2 (5.1%) | 3 (7.7%) | 5 (12.8%) | 12 | |
| Poultry | 6 (7.7%) | 3 (3.8%) | 1 (1.3%) | 23 (29.5%) | 33 | 1 (2.6%) | 5 (12.8%) | 1 (2.6%) | 4 (10.3%) | 11 | |
| Poultry Feed | 0 (0%) | 1 (1.3%) | 0 (0%) | 3 (3.8%) | 4 | 4 (10.3%) | 0 (0%) | 2 (5.1%) | 1 (2.6%) | 7 | |
| Water | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 | 2 (5.1%) | 4 (10.3%) | 0 (0%) | 3 (7.7%) | 9 | |
| Beetle | 0 (0%) | 4 (5.1%) | 1 (1.3%) | 7 (9%) | 12 | NS (0%) | NS (0%) | NS (0%) | NS (0%) | NS | |
(NS) Non Sampling.
Discussion
The present study revealed a high occurrence of ESBL/AmpC-producing and fosfomycin-resistant E. coli in poultry farms from southern Brazil. ESBL phenotype positive strains were found in poultry litter in all sampled periods, and similar was observed by Laube et al. (2013) in Germany. We showed in our study that the chance of ESBL-producing E. coli occurrence in poultry litter was significant in all sampling periods, mainly at the beginning of poultry production (OR 6.42). Furthermore, ESBL-producing E. coli strains were detected in poultry since the second period, with an increased occurrence in the third period. These data indicate that poultry litter could be a risk factor for ESBL-producing E. coli dissemination in poultry houses, including the colonization of one-day-old chicks at the beginning of production. Besides, poultry litter may present a potential risk in the formation of bioaerosols containing antimicrobial-resistant bacteria. Brooks et al. (2010) observed a high concentration of these aerosolized bacteria in poultry houses. The results showed an increase in bacterial concentrations between pre-flock (29%) and late-flock (66%). Studies must be carried out to verify the potential risk of bioaerosols on the health of producers that handle poultry daily. Our results proved that water (OR 0) and poultry feed (OR 0.65) were not sources of ESBL-producing E. coli strains.
Besides poultry litter, all E. coli strains isolated from beetles were positive for ESBL production (OR > 100). These beetles were collected only in the PR state due their presence and abundance in the local farms. These insects are omnivorous scavengers that feed on fecal matter and debris; therefore, they are commonly found in poultry litter (Axtell, 1999). Many insects that inhabit poultry houses can be carriers or reservoirs of MDR bacteria. Studies performed by Blaak et al. (2014) and Solà-Ginés et al. (2015) showed the presence of ESBL-producing E. coli in flies isolated from poultry farms.
Daehre et al. (2018) demonstrated the influence of previous fattening flocks on ESBL-producing strains of the following broiler flock. Through the whole-genome analysis, the authors showed the transmission of ESBL-producing strains between the contaminated environment and poultry, suggesting that cleaning and disinfection practices should be applied. These practices are essential for decreasing the risk of ESBL-producing E. coli spreading to the next flocks, according to Mo et al. (2016). The authors analyzed the risk factors for the presence of cephalosporin-resistant E. coli during broiler production, which is 0.1 times after a rigorous disinfection process and 9 times after people entering the sheds. These authors suggest that good cleaning practices and the control of people’s access to the barn can reduce the occurrence of these resistant strains in the barn environment. In line with these studies, our work reaffirms the importance of applying disinfection techniques to reduce MDR strains in the breeding broiler environment.
Another biosecurity measure in poultry production is the poultry litter processing by composting, a widely used method to ensure that organic waste is safe before use (Wilkinson et al., 2011). Composting implies a reduction in organic waste volume and considerably decreases pathogenic microorganisms (Bernal et al., 2009). Gazal et al. (2015) analyzed poultry litter after the composting process, observing that few isolates (6.3%) contained virulence genes from extra-intestinal pathogenic E. coli and were susceptible to a large number of antimicrobials. Siller et al. (2020) demonstrated that short-term storage reduces the amount of ESBL-producing E. coli in poultry litter. Therefore, the practice of composting or storage is a good alternative for eliminating possible pathogens and multiresistant strains.
Escherichia coli strains found in water samples were negative for ESBL production. All water from the poultry houses was chlorinated, which excludes the possibility of being a source of ESBL-producing E. coli.
The MDR strain frequency in the present study is concerning, and many strains were detected as ESBL-producing. A similar resistance profile has been detected in E. coli strains from poultry farms in Italy (Ghodousi et al., 2015) and Germany (Laube et al., 2013). Cyoia et al. (2019) detected MDR in 80% of commercialized chicken carcasses in both PR and RS states. Moreover, 30% of these strains were characterized as ESBL producers. Poultry-derived products are considered the main ESBL-producing bacteria sources among all animal products (Saliu et al., 2017). In Brazil, many antimicrobial agents that have been previously used as growth promoters are now prohibited (MAPA, 2003, 2009); however, some antimicrobials are still used for treatment or as a prophylactic measure. In both states, the investigated farms reported the use of quinolones during poultry production. Moreover, we found high resistance to quinolones, especially in ESBL strains (Figure 2), which also showed high resistance to other antimicrobials, whose use in poultry production was not mentioned, such as GEN, SXT, and TET. Cyoia et al. (2019) also found that ESBL-producing E. coli were more resistant to a higher number of antimicrobials than non-ESBL strains. Besides, Zeng and Lin (2017) reported reported the association between the resistance of ESBL and other antimicrobial classes, such as aminoglycosides and fluoroquinolones.
Currently, resistance to third-generation cephalosporins induced by ESBL production represents a major public health problem (Blaak et al., 2015; Centers for Disease Control and Prevention, 2015). These enzymes are very relevant as they confer resistance to antimicrobials used in veterinary medicine, such as penicillin, aminopenicillin, and cephalosporins (e.g., ceftiofur) (Poirel et al., 2018). In the present study, 92/117 (77%) E. coli isolates showed bla genes (CTX-M1, 2 and 8 groups). Approximately 66% were confirmed as ESBL-producing, and the majority presented the blaCTX–M group 2 (55%) gene, followed by blaCTX–M group 1 (38%) and blaCTX–M group 8 (10%). These data corroborate the description made by Silva and Lincopan (2012) regarding the epidemiology of ESBL genes in Brazilian territory, especially in animal production. Four strains were negative for ESBL phenotype, but harbored both ESBL and AmpC genes (Supplementary Table 2). According to Nishimura et al. (2018), the presence of AmpC determinants interfere with ESBL phenotype and increases false negative detection.
The selective pressure of antimicrobials in poultry farming leads to the death of sensitive strains and also selects the resistant ones (Poole, 2012; Saliu et al., 2017). Moreover, resistance to determined antimicrobials can lead to cross-resistance and co-selection, with stronger promoters increasing ESBL genes’ expression (Gniadkowski, 2008).
Approximately 73% of strains harbored the fosA3 gene among the 26 phenotypical fosfomycin-resistant strains. Fosfomycin is a bactericidal antimicrobial that inhibits peptidoglycan synthesis and is widely used in human medicine to treat urinary tract infections (Falagas et al., 2016, 2019). Notably, all the fosA3 positive strains were also characterized as ESBL-producing. The relation between fosA3 and blaCTX–M55 (blaCTX–M group 1) genes was significant among ESBL-producing E. coli isolated from PR (OR 3.66) and is more evident on strains isolated from RS (OR 6.44, CI 95% 2.89–14.34, p < 0.05) since only two strains presented blaCTX–M55 and none strains harboring the fosA3 gene were isolated. The fosA3 gene was found in isolates from properties that used fosfomycin during poultry production in the PR state. Sato et al. (2013) suggested that ESBL-producing strains that harbor the blaCTX–M gene (especially the CTX-M55 enzyme) associated with the fosA3 gene naturally reside in the intestinal microbiota of individuals in clinical and veterinary environments. Thus, the use of fosfomycin in Brazil’s poultry production may lead to the co-selection of ESBL-producing strains. This is the first study reporting positive strains for the fosA3 gene isolated from poultry production in Brazil.
Another important finding was the high heterogeneity among E. coli strains analyzed by ERIC-PCR between PR and RS strains that showed several sub-clusters in both states. These results demonstrated that ESBL-producing E. coli strains isolated from poultry production in southern Brazil were not caused by clonal spreading. Ghodousi et al. (2015) presented similar data regarding the analysis of ESBL-producing E. coli strains isolated from poultry farms in Italy. The spread of ESBL genes among strains isolated from animals occurs by horizontal gene transference (Poirel et al., 2018). Thus, the large number of clusters associated with the high genetic diversity showed in our study suggests that the dissemination of resistance genes is due to horizontal transfer by conjugative plasmids.
IncFIB (n = 89; 76%) followed by IncI1 (N = 37, 32%) were the dominant replicon types in our samples. Several plasmid families carry ESBL/AmpC genes in ESBL/AmpC-producing Enterobacteriaceae (Carattoli, 2009), of which IncF, IncI, and IncK types are frequently found (Liebana et al., 2012). Our results corroborate with other studies that investigated the presence of ESBL/AmpC-producing E. coli strains in poultry (Leverstein-van Hall et al., 2011; Mnif et al., 2012).
The present study also demonstrated the conjugation between ESBL-producing E. coli from poultry litter and non-ESBL-producing E. coli from chicken microbiota, in which the recipient strains became positive to blaCTX–M55 and fosA3 genes (T1 and T4). Cyoia et al. (2019) showed that the genetic determinants encoding CTX-M enzymes were transferred to the J53 strain from a human source. Thus, the conjugation results indicated two relevant points. First, it reinforces the association between blaCTX–M55 and fosA3 genes because both were found in the transconjugants. Second, poultry litter could be a starting point for the spreading of resistance genes to poultry and, consequently, to humans via the food chain. Future research intends to clarify the relation between ESBL-producing strains and the production chain in Brazil. In the Netherlands, Apostolakos et al. (2019) showed that ESBL/AmpC producing E. coli was prevalent in the broiler production chain, with significant transference to subsequent production levels, indicating that all production levels need to be investigated.
These results show the need for monitoring systems to investigate and understand antimicrobial resistance spreading in animal production, and thus encourage the promotion of antimicrobial rational use. Brazil is the second-largest producer of poultry meat and the leader of exports worldwide (Associação Brasileira de Proteína Animal, 2018). Therefore, Brazil’s production has great relevance in the world poultry scenario and, consequently, influences public health through the food chain.
Currently, the One Health concept has been presented as a way to raise awareness regarding the relation among human, animal, and environmental health (Kahn, 2011). This approach is considered by international organizations, such as the World Health Organization, as an important element in disease control and prevention (Lerner and Berg, 2015). However, this awareness should not only be limited to the control and prevention of pathogens, but also address aspects of antimicrobial resistance. Lammie and Hughes (2016) suggest that due to the challenge presented by antimicrobial resistance and its relation to human, animal, and environmental health, it is essential to include this issue in the One Health approach.
Conclusion
The antimicrobial use in poultry production has a great influence on MDR bacteria selection and severely influences public health, as well as the national productive sector. A longitudinal monitoring program in poultry production should be implemented to provide data regarding antimicrobial use. The improvement in management techniques, such as poultry litter treatment and good clean practices, may decrease MDR strain frequency in poultry farms. These measures can optimize poultry production and increase animal and human health preservation.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by Animal Ethics Committee of State University of Londrina (CEUA/UEL).
Author Contributions
LG contributed to the development of experimental research, data analysis, and wrote the manuscript. LM, MD, VK, BG, TG, TC, and JP contributed to the development of experimental research. EN contributed to the statistical analysis. RK, GN, KB, BB, and EV contributed to and assisted in the design of the work and in preparation of the article, and critically reviewed the manuscript. All authors have participated in this study and commented on the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the National Council for Scientific and Technological Development and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior who enabled the execution of this study. We would also like to thank Benito Guimarães de Brito Ph.D. and Kelly Cristina Tagliari de Brito Ph.D. from the Avian Health Laboratory, Veterinary Research Institute Desidério Finamor for their contributions to this study. We would like to thank Mariangela Hungria da Cunha Ph.D. and Renan Augusto Ribeiro Ph.D. from the Soil Biotechnology Laboratory at EMBRAPA for helping with the analysis using BioNumerics. We would also like to thank Silvia Helena Sofia Ph.D. and Caroline Apolinário da Silva M.Sc. from the Genetics and Animal Ecology Laboratory at the State University of Londrina for their help in sequencing.
Footnotes
Funding. This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brazil – Finance Code 001 (CAPES). This work was also supported by the National Council for Scientific and Technological Development – Process 433656/2018-2 Call MCTIC/CNPq 28/2018. These funding agencies paid scholarships to students and the consumption materials used in this study. Additional support was provided by the Bill and Melinda Gates Foundation’s Grand Challenges Explorations Brazil – New Approaches to Characterize the Global Burden of Antimicrobial Resistance (number OPP1193112). None of these funding agencies had any role in this study’s design, collection, analysis, data interpretation and manuscript writing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.604544/full#supplementary-material
References
- Apostolakos I., Mughini-Gras L., Fasolato L., Piccirillo A. (2019). Assessing the occurrence and transfer dynamics of ESBL/pAmpC-producing Escherichia coli across the broiler production pyramid. PLoS One 14:e0217174. 10.1159/000500311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlet G., Philippon A. (1991). Construction by polymerase chain reaction and intragenic DNA probes for three main types of transferable β-lactamases (TEM, SHV, CARB). FEMS Microbiol. Lett. 82 19–25. 10.1111/j.1574-6968.1991.tb04833.x [DOI] [PubMed] [Google Scholar]
- Associação Brasileira de Proteína Animal (2018). Annual Report of Brazilian Poultry Association. São Paulo: Brazilian Poultry Union. [Google Scholar]
- Axtell R. C. (1999). Poultry integrated pest management: status and future. Integr. Pest Manag. Rev. 4 53–73. 10.1023/A:1009637116897 [DOI] [Google Scholar]
- Bernal M. P., Alburquerque J. A., Moral R. (2009). Composting of animal manures and chemical criteria for compost maturity assessment. A review. Bioresour. Technol. 100 5444–5453. 10.1016/j.biortech.2008.11.027 [DOI] [PubMed] [Google Scholar]
- Blaak H., Hamidjaja R. A., van Hoek A. H., de Heer L., de Roda Husman A. M., Schets F. M. (2014). Detection of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli on flies at poultry farms. Appl. Environ. Microbiol. 80 239–246. 10.1128/AEM.02616-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaak H., van Hoek A. H., Hamidjaja R. A., van der Plaats R. Q., Kerkhof-de Heer L., de Roda Husman A. M., et al. (2015). Distribution, numbers, and diversity of ESBL-producing E. coli in the poultry farm environment. PLoS One 10:e0135402. 10.1371/journal.pone.0135402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc V., Mesa R., Saco M., Lavilla S., Prats G., Miró E., et al. (2006). ESBL-and plasmidic class C β-lactamase-producing E. coli strains isolated from poultry, pig and rabbit farms. Vet. Microbiol. 118 299–304. 10.1016/j.vetmic.2006.08.002 [DOI] [PubMed] [Google Scholar]
- Bortolami A., Zendri F., Maciuca E., I, Wattret A., Ellis C., Schmidt V., et al. (2019). Diversity, virulence, and clinical significance of extended-spectrum β-Lactamase-and pAmpC-producing Escherichia coli from companion animals. Front. Microbiol. 10:1260. 10.3389/fmicb.2019.01260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks J. P., McLaughlin M. R., Scheffler B., Miles D. M. (2010). Microbial and antibiotic resistant constituents associated with biological aerosols and poultry litter within a commercial poultry house. Sci. Total Env. 408 4770–4777. 10.1016/j.scitotenv.2010.06.038 [DOI] [PubMed] [Google Scholar]
- Bush K., Fisher J. F. (2011). Epidemiological expansion, structural studies, and clinical challenges of new β-lactamases from gram-negative bacteria. Annu. Rev. Microbiol. 65 455–478. 10.1146/annurev-micro-090110-102911 [DOI] [PubMed] [Google Scholar]
- Cantón R., Coque T. M. (2006). The CTX-M β-lactamase pandemic. Curr. Opin. Microbiol. 9 466–475. 10.1016/j.mib.2006.08.011 [DOI] [PubMed] [Google Scholar]
- Carattoli A. (2008). Animal reservoirs for extended spectrum β-lactamase producers. Clin. Microbiol. Infect. 14 117–123. 10.1111/j.1469-0691.2007.01851.x [DOI] [PubMed] [Google Scholar]
- Carattoli A. (2009). Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53 2227–2238. 10.1128/AAC.01707-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A. (2011). Plasmids in Gram negatives: molecular typing of resistance plasmids. Int. J. Med. Microbiol. 301 654–658. 10.1016/j.ijmm.2011.09.003 [DOI] [PubMed] [Google Scholar]
- Carattoli A. (2013). Plasmids and the spread of resistance. Int. J. Med. Microbiol. 303 298–304. 10.1016/j.ijmm.2013.02.001 [DOI] [PubMed] [Google Scholar]
- Carattoli A., Bertini A., Villa L., Falbo V., Hopkins K. L., Threlfall E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods. 63 219–228. 10.1016/j.mimet.2005.03.018 [DOI] [PubMed] [Google Scholar]
- Ceccarelli D., Kant A., van Essen-Zandbergen A., Dierikx C., Hordijk J., Wit B., et al. (2019). Diversity of plasmids and genes encoding resistance to extended spectrum cephalosporin’s in commensal Escherichia coli from Dutch livestock in 2007-2017. Front. Microbiol. 10:76. 10.3389/fmicb.2019.00076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (2015). Antibiotic Resistance Threats in the United States, 2013. Atlanta: CDC. [Google Scholar]
- Clermont O., Bonacorsi S., Bingen E. (2000). Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66 4555–4558. 10.1128/AEM.66.10.4555-4558.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2018). Performance Standards for Antimicrobial Susceptibility Testing. Twenty-Fifth Informational Supplement CLSI Document M100. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Cyoia P. S., Koga V. L., Nishio E. K., Houle S., Dozois C. M., de Brito K. C. T., et al. (2019). Distribution of ExPEC virulence factors, blaCTX-M, fosA3, and mcr-1 in Escherichia coli isolated from commercialized chicken carcasses. Front. Microbiol. 9:3254. 10.3389/fmicb.2018.03254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daehre K., Projahn M., Semmler T., Roesler U., Friese A. (2018). Extended-spectrum beta-lactamase-/AmpC beta-lactamase-producing enterobacteriaceae in broiler farms: transmission dynamics at farm level. Microb. Drug Resist. 24 511–518. 10.1089/mdr.2017.0150 [DOI] [PubMed] [Google Scholar]
- Datta N., Hedges R. W. (1971). Compatibility groups among fi–R factors. Nature 234 222. 10.1038/234222a0 [DOI] [PubMed] [Google Scholar]
- Dierikx C. M., van der Goot J. A., Smith H. E., Kant A., Mevius D. J. (2013). Presence of ESBL/AmpC-producing Escherichia coli in the broiler production pyramid: a descriptive study. PLoS One 8:e79005. 10.1371/journal.pone.0079005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez J. E., Redondo L. M., Figueroa Espinosa R. A., Cejas D., Gutkind G. O., Chacana P. A., et al. (2018). Simultaneous carriage of mcr-1 and other antimicrobial resistance determinants in Escherichia coli from poultry. Front. Microbiol. 9:1679. 10.3389/fmicb.2018.01679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards P. R., Ewing W. H. (1986). Edwards and Ewing’s Identification of Enterobacteriaceae. Amsterdam: Elsevier Publishing Company. [Google Scholar]
- Ewers C., Bethe A., Semmler T., Guenther S., Wieler L. H. (2012). Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clin. Microbiol. Infect. 18 646–655. 10.1111/j.1469-0691.2012.03850.x [DOI] [PubMed] [Google Scholar]
- Falagas M. E., Athanasaki F., Voulgaris G. L., Triarides N. A., Vardakas K. Z. (2019). Resistance to fosfomycin: mechanisms, frequency and clinical consequences. Int. J. Antimicrob. Agents. 53 22–28. 10.1016/j.ijantimicag.2018.09.013 [DOI] [PubMed] [Google Scholar]
- Falagas M. E., Vouloumanou E. K., Samonis G., Vardakas K. Z. (2016). Fosfomycin. Clin. Microbiol. Rev. 29 321–347. 10.1128/CMR.00068-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friese A., Schulz J., Laube H., Hartung J., Roesler U. (2013). Faecal occurrence and emissions of livestock-associated methicillin-resistant Staphylococcus aureus (laMRSA) and ESBL/AmpC-producing E. coli from animal farms in Germany. Berl. Munch. Tierarztl. Wochenschr. 126 175–180. 10.1037/e411862008-001 [DOI] [PubMed] [Google Scholar]
- Gazal L. E. S., Puño-Sarmiento J. J., Medeiros L. P., Cyoia P. S., da Silveira W. D., Kobayashi R. K., et al. (2015). Presence of pathogenicity islands and virulence genes of extraintestinal pathogenic Escherichia coli (ExPEC) in isolates from avian organic fertilizer. Poult. Sci. 94 3025–3033. 10.3382/ps/pev278 [DOI] [PubMed] [Google Scholar]
- Ghodousi A., Bonura C., Di Noto A. M., Mammina C. (2015). Extended-spectrum ß-lactamase, AmpC-producing, and fluoroquinolone-resistant Escherichia coli in retail broiler chicken meat, Italy. Foodborne Pathog. Dis. 12 619–625. 10.1089/fpd.2015.1936 [DOI] [PubMed] [Google Scholar]
- Gniadkowski M. (2008). Evolution of extended-spectrum β-lactamases by mutation. Clin. Microbiol. Infect. 14 11–32. 10.1111/j.1469-0691.2007.01854.x [DOI] [PubMed] [Google Scholar]
- Jarlier V., Nicolas M. H., Fournier G., Philippon A. (1988). Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Clin. Infect. Dis. 10 867–878. 10.1093/clinids/10.4.867 [DOI] [PubMed] [Google Scholar]
- Kahn L. H. (2011). The need for one health degree programs. Infect. Ecol. Epidemiol. 1 7919. 10.3402/iee.v1i0.7919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating G. M. (2013). Fosfomycin trometamol: a review of its use as a single-dose oral treatment for patients with acute lower urinary tract infections and pregnant women with asymptomatic bacteriuria. Drugs 73 1951–1966. 10.1007/s40265-013-0143-y [DOI] [PubMed] [Google Scholar]
- Koga V. L., Rodrigues G. R., Scandorieiro S., Vespero E. C., Oba A., de Brito B. G., et al. (2015). Evaluation of the antibiotic resistance and virulence of Escherichia coli strains isolated from chicken carcasses in 2007 and 2013 from Paraná, Brazil. Foodborne Pathog. Dis. 12 479–485. 10.1089/fpd.2014.1888 [DOI] [PubMed] [Google Scholar]
- Lammie S. L., Hughes J. M. (2016). Antimicrobial resistance, food safety, and one health: the need for convergence. Annu. Ver. Food Sci. T. 7 287–312. 10.1146/annurev-food-041715-033251 [DOI] [PubMed] [Google Scholar]
- Laube H., Friese A., Von Salviati C., Guerra B., Käsbohrer A., Kreienbrock L., et al. (2013). Longitudinal monitoring of extended-spectrum-beta-lactamase/AmpC-producing Escherichia coli at German broiler chicken fattening farms. Appl. Environ. Microbiol. 79 4815–4820. 10.1128/AEM.00856-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus B., Paterson D. L., Mollinger J. L., Rogers B. A. (2014). Do human extraintestinal Escherichia coli infections resistant to expanded-spectrum cephalosporins originate from food-producing animals? A systematic review. Clin. Infect. Dis. 60 439–452. 10.1093/cid/ciu785 [DOI] [PubMed] [Google Scholar]
- Lerner H., Berg C. (2015). The concept of health in One Health and some practical implications for research and education: what is One Health? Infect. Ecol. Epidemiol. 5 25300 10.3402/iee.v5.2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leverstein-van Hall M. A., Dierikx C. M., Cohen Stuart J., Voets G. M., Van Den Munckhof M. P., van Essen-Zandbergen A., et al. (2011). Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin. Microbiol. Infect. 17 873–880. 10.1111/j.1469-0691.2011.03497.x [DOI] [PubMed] [Google Scholar]
- Li Y., Chen L., Wu X., Huo S. (2015). Molecular characterization of multidrug-resistant avian pathogenic Escherichia coli isolated from septicemic broilers. Poult. Sci. 94 601–611. 10.3382/ps/pev008 [DOI] [PubMed] [Google Scholar]
- Liebana E., Carattoli A., Coque T. M., Hasman H., Magiorakos A. P., Mevius D., et al. (2012). Public health risks of enterobacterial isolates producing extended-spectrum β-lactamases or AmpC β-lactamases in food and food-producing animals: an EU perspective of epidemiology, analytical methods, risk factors, and control options. Clin. Infect. Dis. 56 1030–1037. 10.1093/cid/cis1043 [DOI] [PubMed] [Google Scholar]
- Liu Y. Y., Wang Y., Walsh T. R., Yi L. X., Zhang R., Spencer J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16 161–168. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- MAPA (2003). “Pecuária e Abastecimento,” in 27 de junho de 2003. Proíbe a Fabricação, a Manipulação, o Fracionamento, a Comercialização, a Importação e o Uso dos Princípios Ativos Cloranfenicol e Nitrofuranos e os Produtos que contenham estes Princípios Ativos, para uso Veterinário e Susceptível de Emprego na Alimentação de Todos os Animais e Insetos. Instrução Normativa (Portuguese). Available online at: https://www.gov.br/agricultura/pt-br/assuntos/insumos-agropecuarios/insumos-pecuarios/alimentacao-animal/arquivos-alimentacao-animal/legislacao/instrucao-normativa-no-9-de-27-de-junho-de-2003.pdf [Google Scholar]
- MAPA (2009). “Pecuária e Abastecimento,” in Regulamento Técnico Para a Fabricação, o Controle de Qualidade, a Comercialização e o Emprego de Produtos Antimicrobianos de Uso Veterinário. Instrução Normativa 26, Ministério da Agricultura, Pecuária e Abastecimento Brasília (Portuguese). Available online at: https://www.gov.br/agricultura/pt-br/assuntos/insumos-agropecuarios/insumos-pecuarios/alimentacao-animal/arquivos-alimentacao-animal/legislacao/instrucao-normativa-no-26-de-9-de-julho-de-2009.pdf [Google Scholar]
- McLellan S. L., Daniels A. D., Salmore A. K. (2003). Genetic characterization of Escherichia coli populations from host sources of fecal pollution by using DNA fingerprinting. Appl. Environ. Microbiol. 69 2587–2594. 10.1128/AEM.69.5.2587-2594.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mnif B., Ktari S., Rhimi F. M., Hammami A. (2012). Extensive dissemination of CTX-M-1-and CMY-2-producing Escherichia coli in poultry farms in Tunisia. Lett. Appl. Microbiol. 55 407–413. 10.1111/j.1472-765X.2012.03309.x [DOI] [PubMed] [Google Scholar]
- Mo S. S., Kristoffersen A. B., Sunde M., Nødtvedt A., Norström M. (2016). Risk factors for occurrence of cephalosporin-resistant Escherichia coli in Norwegian broiler flocks. Prev. Vet. Med. 130 112–118. 10.1016/j.prevetmed.2016.06.011 [DOI] [PubMed] [Google Scholar]
- Nishimura F., Morinaga Y., Akamatsu N., Matsuda J., Kaku N., Takeda K., et al. (2018). Plasmid-mediated AmpC β-lactamase and underestimation of extended-spectrum β-lactamase in cefepime-susceptible, elevated-ceftazidime-MIC Enterobacteriaceae isolates. Jpn. J. Infect. Dis. 71 281–285. 10.7883/yoken.jjid.2017.469 [DOI] [PubMed] [Google Scholar]
- Olsen R. H., Bisgaard M., Löhren U., Robineau B., Christensen H. (2014). Extended-spectrum β-lactamase-producing Escherichia coli isolated from poultry: a review of current problems, illustrated with some laboratory findings. Avian Pathol. 43 199–208. 10.1080/03079457.2014.907866 [DOI] [PubMed] [Google Scholar]
- O’Neill J. (2014). The Review on Antimicrobial Resistance. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. Available online at: https://amr-review.org/sites/default/files/AMR%20Review%20Pa-per%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20-wealth%20of%20nations_1.pdf (accessed June 20, 2019). [Google Scholar]
- Paterson D. L., Bonomo R. A. (2005). Extended-spectrum β-lactamases: a clinical update. Clin. Microbiol. Rev. 18 657–686. 10.1128/CMR.18.4.657-686.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez F. J., Hanson N. D. (2002). Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40 2153–2162. 10.1128/JCM.40.6.2153-2162.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitout J. D., Laupland K. B. (2008). Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect. Dis. 8 159–166. 10.1016/S1473-3099(08)70041-0 [DOI] [PubMed] [Google Scholar]
- Poirel L., Madec J. Y., Lupo A., Schink A. K., Kieffer N., Nordmann P., et al. (2018). Antimicrobial resistance in Escherichia coli. Microbiol. Spectr. 6 289–316. 10.1128/9781555819804.ch13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K. (2012). Bacterial stress responses as determinants of antimicrobial resistance. J. Antimicrob. Chemother. 67 2069–2089. 10.1093/jac/dks196 [DOI] [PubMed] [Google Scholar]
- Saliu E. M., Vahjen W., Zentek J. (2017). Types and prevalence of extended–spectrum beta–lactamase producing Enterobacteriaceae in poultry. Anim. Health Res. Rev. 18 46–57. 10.1017/S1466252317000020 [DOI] [PubMed] [Google Scholar]
- Sato N., Kawamura K., Nakane K., Wachino J. I., Arakawa Y. (2013). First detection of fosfomycin resistance gene fosA3 in CTX-M-producing Escherichia coli isolates from healthy individuals in Japan. Microb. Drug Resis. 19 477–482. 10.1089/mdr.2013.0061 [DOI] [PubMed] [Google Scholar]
- Segabinazi S. D., Flôres M. L., da Silva Barcelos A., Jacobsen G., Eltz R. D. (2005). Bactérias da família Enterobacteriaceae em Alphitobius diaperinus oriundos de granjas avícolas dos Estados do Rio Grande do Sul e Santa Catarina, Brasil. Acta Sci. Vet. 33 51–55. 10.22456/1679-9216.14449 [DOI] [Google Scholar]
- Siller P., Daehre K., Thiel N., Nṻbel U., Roesler U. (2020). Impact of short-term storage on the quantity of extended-spectrum beta-lactamase–producing Escherichia coli in broiler litter under practical conditions. Poultry Sci. 99 2125–2135. 10.1016/j.psj.2019.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva K. C. D., Lincopan N. (2012). Epidemiologia das betalactamases de espectro estendido no Brasil: impacto clínico e implicações para o agronegócio. J. Bras. Patol. Med. Lab. 48 91–99. 10.1590/S1676-24442012000200004 [DOI] [Google Scholar]
- Smet A., Martel A., Persoons D., Dewulf J., Heyndrickx M., Herman L., et al. (2009). Broad-spectrum β-lactamases among Enterobacteriaceae of animal origin: molecular aspects, mobility and impact on public health. FEMS Microbiol. Rev. 34 295–316. 10.1111/j.1574-6976.2009.00198.x [DOI] [PubMed] [Google Scholar]
- Solà-Ginés M., González-López J. J., Cameron-Veas K., Piedra-Carrasco N., Cerdà-Cuéllar M., Migura-Garcia L. (2015). Houseflies (Musca domestica) as vectors for extended-spectrum β-lactamase-producing Escherichia coli on Spanish broiler farms. Appl. Environ. Microbiol. 81 3604–3611. 10.1128/AEM.04252-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo M. R. F., Fontes C. F., Trabulsi L. R. (1982a). EPM-A modification of Rugai and Araujo medium for simultaneous test of gas production from glucose, H2S, urease and tryptophan deaminase. Rev. Microbiol. 13 230–235. [Google Scholar]
- Toledo M. R. F., Fontes C. F., Trabulsi L. R. (1982b). MILi-a medium for detection of motility, indole and lysine decarboxylase. Rev. Microbiol. 13 230–235. [Google Scholar]
- Versalovic J., Koeuth T., Lupski R. (1991). Distribution of repetitive DNA sequences in eubacteria and application to finerpriting of bacterial enomes. Nucleic Acids Res. 19 6823–6831. 10.1093/nar/19.24.6823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieler L. H., Ewers C., Guenther S., Walther B., Lübke-Becker A. (2011). Methicillin-resistant staphylococci (MRS) and extended-spectrum beta-lactamases (ESBL)-producing Enterobacteriaceae in companion animals: nosocomial infections as one reason for the rising prevalence of these potential zoonotic pathogens in clinical samples. Int. J. Med. Microbiol. 301 635–641. 10.1016/j.ijmm.2011.09.009 [DOI] [PubMed] [Google Scholar]
- Wilkinson K. G., Tee E., Tomkins R. B., Hepworth G., Premier R. (2011). Effect of heating and aging of poultry litter on the persistence of enteric bacteria. Poul. Sci. 90 10–18. 10.3382/ps.2010-01023 [DOI] [PubMed] [Google Scholar]
- Woodford N., Fagan E. J., Ellington M. J. (2005). Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum β-lactamases. J. Antimicrob. Chemother. 57 154–155. 10.1093/jac/dki412 [DOI] [PubMed] [Google Scholar]
- World Health Organization (2014). Antimicrobial resistance: global report on surveillance. Avaliable online at: https://apps.who.int/iris/bitstream/handle/10665/112642/9789241564748_eng.pdf;jsessionid=9BD9CFC92083023155E1028C22D10F1F?sequence=1 (accessed June 20, 2019). [Google Scholar]
- Zeng X., Lin J. (2017). Factors influencing horizontal gene transfer in the intestine. Anim. Health Res. Rev. 18 153–159. 10.1017/S1466252317000159 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.